Abstract
Broadly neutralizing antibodies are critical for protection against both drifted and shifted influenza viruses. Here, we reveal first exposure to the 2009 pandemic H1N1 influenza virus recalls memory B cells that are specific to the conserved receptor-binding site (RBS) or lateral patch epitopes of the hemagglutinin (HA) head domain. Monoclonal antibodies (mAbs) generated against these epitopes are broadly neutralizing against H1N1 viruses spanning 40 years of viral evolution and provide potent protection in vivo. Lateral patch-targeting antibodies demonstrated near universal binding to H1 viruses and RBS-binding antibodies commonly cross-reacted with H3N2 viruses and influenza B viruses. Lateral patch-targeting mAbs were restricted to expressing the variable heavy chain gene VH3–23 with or without the variable kappa chain gene VK1–33 and often possessed a Y-x-R motif within the heavy chain complementarity determining region 3 to make key contacts with HA. Moreover, lateral patch antibodies that utilized both VH3–23 and VK1–33 maintained neutralizing capability with recent pH1N1 strains that acquired mutations near the lateral patch. RBS-binding mAbs used a diverse repertoire, but targeted the RBS epitope similarly and made extensive contacts with the major antigenic site Sb. Together, our data indicate RBS- and lateral patch-targeting clones are abundant within the human memory B cell pool and universal vaccine strategies should aim to drive antibodies against both conserved head and stalk epitopes.
One Sentence Summary:
Structural and experimental determination reveals broadly neutralizing antibodies targeting conserved epitopes on the HA head of H1N1 viruses.
Introduction:
Hemagglutinin (HA) is the major surface antigen of influenza viruses and is composed of two major domains: the head and the stalk. Seasonal influenza virus vaccines largely induce potently neutralizing antibodies against the immunodominant variable epitopes of the head domain, which provide narrow protection against only a few strains. Influenza viruses rapidly mutate to evade antibodies targeting the major antigenic sites of the HA head, necessitating constant reformulation of the seasonal influenza virus vaccine. Therefore, new vaccine strategies that elicit potent broadly neutralizing antibodies are desperately needed to provide universal protection against influenza viruses.
Broadly neutralizing antibodies against conserved epitopes on the HA stalk have been well characterized, but little is known about the structure, repertoire, and viral binding features of broadly neutralizing antibodies against the HA head. Two conserved HA head epitopes have been identified on H1 influenza viruses: the receptor-binding site (RBS (1, 2)) and the lateral patch (3). RBS-binding antibodies exhibit broad cross-reactivity amongst H1N1 viruses and can occasionally cross-react with H3N2 viruses (1, 2). Although only one definitive lateral patch-binding antibody has been identified (3), antibodies binding within this region are susceptible to recent mutations in the major antigenic site, Sa (3–5). However, it is not known which factors limit or promote the recall of memory B cells (MBCs) targeting the lateral patch and RBS. Moreover, very little is known about lateral patch-binding antibodies, including their relative abundance, repertoire, and binding features, and whether all lateral patch-binding antibodies have lost binding to recent antigenically drifted variants of the 2009 pandemic H1N1 virus (pH1N1).
By comprehensively analyzing the specificities of B cells targeting the HA head after vaccination, we determined first exposure to pHN1 preferentially recalls MBCs targeting the conserved RBS and lateral patch epitopes, suggesting pre-existing immunity against the variable epitopes of the HA head limits the recall of MBCs against conserved head epitopes. Antibodies elicited against these epitopes are broadly neutralizing against antigenically drifted and shifted H1N1 viruses, and even against a recent H1N1 strain that has mutated near the lateral patch. This study highlights the structural and repertoire features of RBS- and lateral patch-binding monoclonal antibodies (mAbs) and reveals a public binding motif shared across lateral patch-binding mAbs.
Results:
First exposure to pH1N1 virus exposure robustly induced antibodies against conserved head epitopes.
The head domain of HA is more variable and mutates more frequently than the stalk domain (Fig. 1A, fig. S1A) (6, 7). Five major antigenic sites of the HA head have been observed (Ca1, Ca2, Cb, Sa, Sb) and exhibit strong overlap with the variable regions of the HA head (Fig. 1A). However, two epitopes of the HA head, the RBS and the lateral patch, encompass more conserved regions of the HA head, although mAbs binding the RBS (CH65 (2)) and the lateral patch (Fab6649 (3)) exhibit some overlap with the major antigenic sites (Fig. 1A, fig. S1B).
Fig. 1: First exposure to pH1N1 induces antibodies against the RBS and lateral patch epitopes of the HA head.
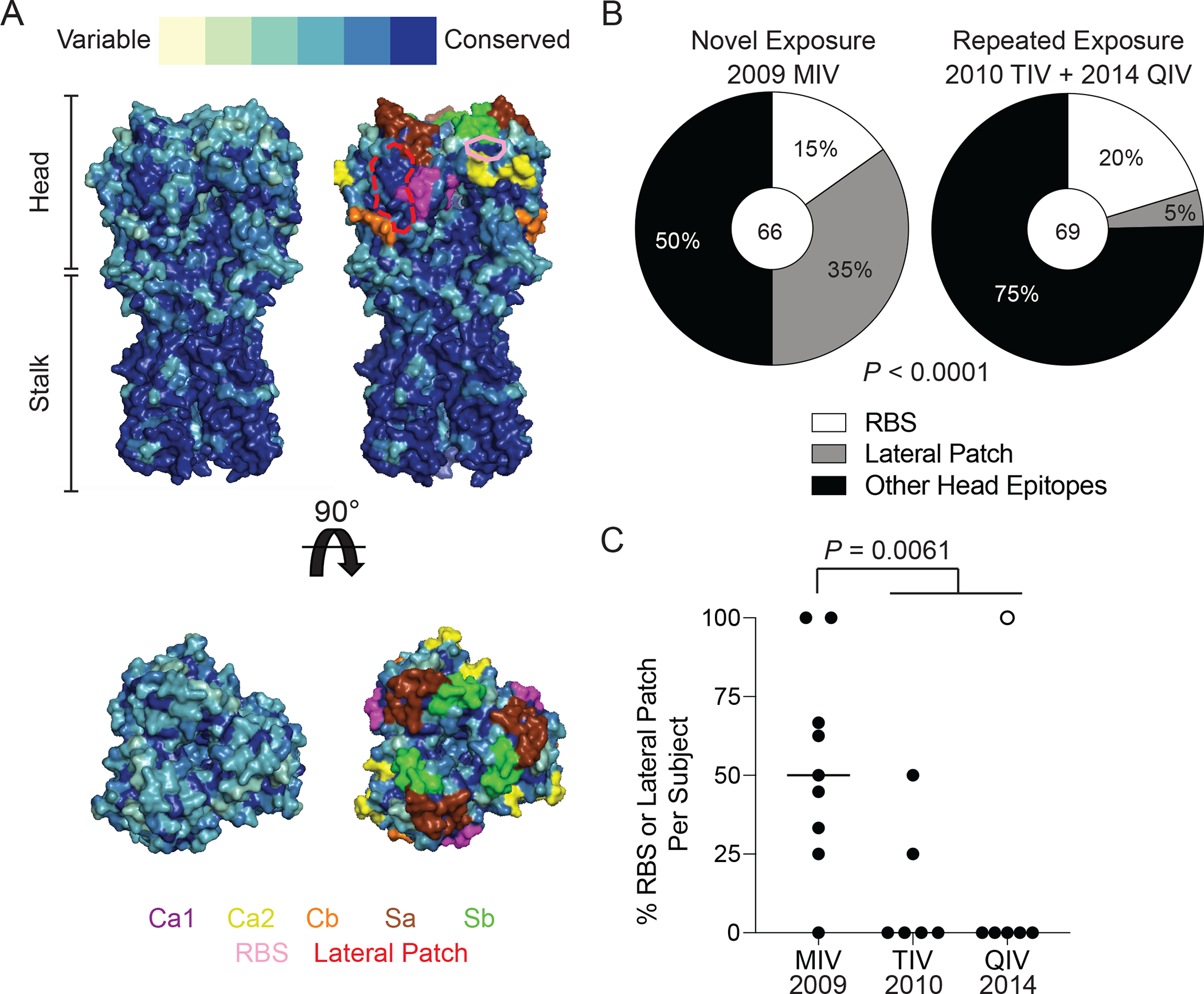
(A) The conservation of H1 over 102 years of H1N1 evolution with the major antigenic sites highlighted and the RBS epitope outlined in pink and lateral patch epitopes outlined in dashed red line projected onto the structure of A/California/04/2009 (side and top views; PDB:4jtv). (B) The proportion of HAI+ mAbs induced by the 2009 MIV or seasonal influenza virus vaccines (2010 TIV and 2014 QIV) binding the RBS, lateral patch, or other head epitopes. (C) The proportion of RBS- and lateral patch-binding mAbs out of all HAI+ mAbs per individual isolated from the 2009 MIV, 2010 TIV, and 2014 QIV vaccine cohorts. Each symbol represents one participant. The hollow symbol represents participant 240. Statistical significance was determined using Chi-square test (B) and an unpaired two-sided non-parametric Mann-Whitney Test (C). Bar in (C) represents the median. Numbers in center of pie graphs represent number of individual mAbs analyzed.
Immunization with the 2009 pH1N1 monovalent virus vaccine (MIV) robustly induced plasmablasts (PBs) targeting conserved epitopes of the HA stalk (8). However, we identified that head-binding mAbs were the dominant product of PBs induced by the 2009 MIV and seasonal influenza virus vaccination, with over 50% of mAbs generated from PBs binding the HA head, as determined by their ability to inhibit hemagglutination (HAI+; fig. S1C and D). Strikingly, 50% of head binding mAbs induced by the MIV targeted either the RBS or lateral patch in comparison to 25% of head binding mAbs induced by seasonal vaccination (Fig. 1B). Lateral patch-targeting PBs were particularly boosted, comprising 35% of all HAI+ mAbs isolated from the 2009 MIV relative to only 5% of all HAI+ mAbs isolated from the 2010 trivalent influenza virus vaccine (TIV) and 2014 quadrivalent influenza virus vaccine (QIV) cohorts (Fig. 1B).
On average, individuals who received the MIV had a greater proportion of HAI+ mAbs binding either the RBS or lateral patch compared to individuals who received either the 2010 TIV or the 2014 QIV (P = 0.0061; Fig. 1C). Notably, only 2 individuals in the 2010 TIV cohort (33.3%) and 1 individual in the 2014 QIV cohort (16.7%) induced mAbs against the RBS and the lateral patch, relative to 8 out of 9 individuals in the 2009 MIV cohort (88.9%, Fig. 1C). Moreover, all head antibodies isolated from participant 240 in the 2014 QIV cohort targeted either the RBS or lateral patch (hollow circle; Fig. 1C) and accounted for all the RBS-binding mAbs recorded from individuals that received a seasonal vaccine (table S1). The 2014 QIV may have been the first time that participant 240 had been exposed to the newly circulating pH1N1 virus, as this individual had no record of prior influenza virus vaccinations (table S1). We cannot rule out that this individual had been naturally infected with pH1N1. However, all isolated HAI+ PBs cross-reacted with pre-pH1N1 viruses and targeted the RBS and lateral patch, similar to PBs induced by 2009 MIV (fig. S1E). Therefore, participant 240 likely had low pre-existing immunity against the pH1N1 virus, which led to the recall of MBCs targeting conserved epitopes of the HA head. The only participant in the 2009 MIV cohort that did not mount an antibody response against the RBS or lateral patch was SFV020, who was born in 1945 and 64 years old at the time of vaccination (table S1). Drifted viral variants of the 1918 H1N1 were the only circulating influenza A viruses during SFV020’s childhood, and therefore, vaccination of participant SFV020 recalled MBCs targeting variable epitopes of the H1 head that were likely similar to those during this individual’s childhood, as the 2009 pH1N1 virus and 1918 H1N1 were antigenically similar (9, 10).
Using Δ-1 mutants, in which an individual major antigenic site was replaced with the corresponding epitope from H5 or H13 (11), we identified mAbs binding each major antigenic site, with most antibodies targeting Ca1, Cb, and Sb (Fig. S1F and G). Notably, the 64-year-old SFV020 was the only participant to mount a response against Ca2 (fig. S1F and G), further suggesting this individual could recall MBCs targeting variable epitopes of the HA head. Moreover, 8 mAbs had ablated binding to multiple Δ-1 mutants, with 50% susceptible to mutations in the Ca1 and Sb (fig. S1H). Together, these data reveal that first exposure to pH1N1 preferentially induces antibodies against conserved epitopes of the HA head. These data also indicate that, in the absence of pre-existing immunity against variable epitopes of the HA head of pH1N1, individuals can recall MBCs targeting conserved epitopes of the HA head, as was the case for those in the 2009 MIV cohort and participant 240.
Structural basis for lateral patch-binding mAbs
We identified 10 distinct antibody clones targeting the lateral patch from 7 individuals using negative stain electron microscopy (EM; fig. S2A and table S2). Of the individual clones targeting the lateral patch, all mAbs utilized variable heavy (VH) and kappa (VK) genes VH3–23 or VK1–33, with four clones utilizing both VH3–23 and VK1–33 (Fig. 2A to C; table S2). However, lateral patch-binding mAbs used a variety of heavy chain diversity (DH), heavy chain joining (JH), and kappa chain joining (JK) genes, with a preference for JH4 and JK4 (fig. S2B to D). These data indicate that B cells targeting the lateral patch utilize a highly-restricted VH and VK gene repertoire. To understand the nature of lateral patch-binding antibodies that utilized both VH3–23 and VK1–33, we determined the structure of 045–09 2B05 binding to the pH1N1 A/California/04/2009 HA by cryo-EM (table S3). 2D class averages of the complex exhibited diverse views with visible secondary structure and 3 fragment antigen binding regions (Fabs) bound per HA trimer (fig. S2E). Iterative classification and refinement in 3D resulted in a final map at 3.2 Å resolution (Fig. 2D), with the highest resolution features corresponding to the paratope:epitope region and the interior of the HA head (fig. S2F to G, table S2). The dominant contacts of 045–09 2B05 occurred in the heavy chain complementarity determining region 3 (H-CDR3), where residue Y100b interacted with the hydrophobic groove between the HA β-sheets and Q100a and R100d formed salt bridges with K174 and E122 on the HA head, respectively (Fig. 2E). We also observed a salt bridge formed between R30 of K-CDR1 and D171 on the HA head (Fig. 2E). Crucially, 045–09 2B05 binds the HA head through multiple hydrophobic and electrostatic interactions spanning the lateral patch epitope.
Fig. 2: Lateral patch-binding antibodies utilize a restricted repertoire.
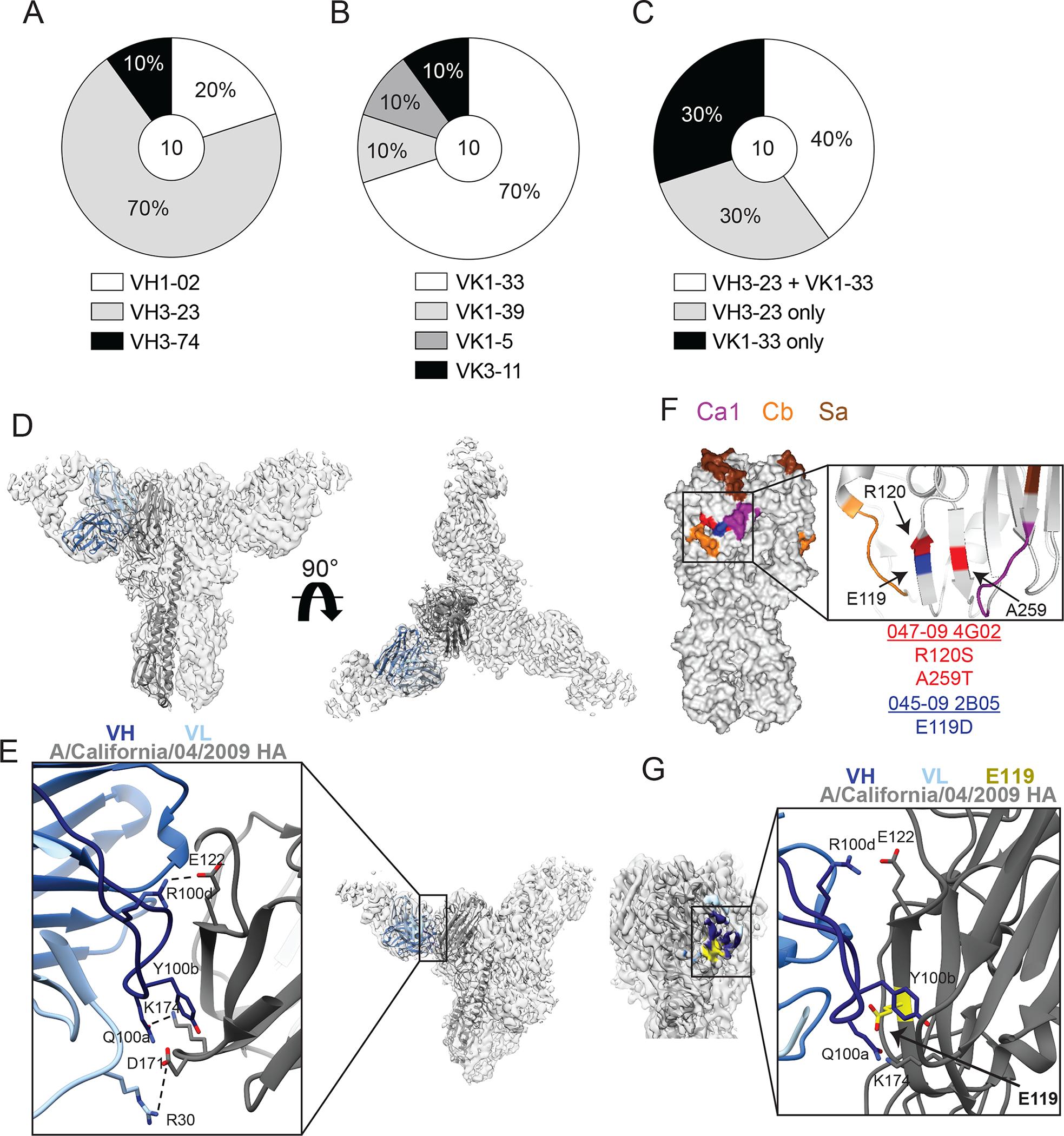
(A to C) The repertoire of unique B cell clones targeting the lateral patch including VH gene usage (A), VK gene usage (B), and proportion of clones utilizing VH3–23 or VK1–33 (C). Numbers in center of pie graphs represents number of distinct clones analyzed. (D and E) Cryo-EM structure of 045–09 2B05 in complex with A/California/7/2009 HA. Side and top views of 045–09 2B05 binding HA (D) and zoomed in view of paratope:epitope interactions (E). Dashed lines represent contacts between 045–09 2B05 and HA. (F and G) Virus escape mutants for 045–09 2B05 and 047–09 4G02 were generated using A/Netherlands/602/2009 H1N1. (F) Location of mutations (arrows) driven by 045–09 2B05 (blue; E119D) and 047–09 4G02 (red; R120S and A259T) shown on A/California/04/2009 HA (PDB: 4jtv). (G) The location of E119 (arrow) within the 045–09 2B05 and HA paratope:epitope interactions.
Because of the critical role of Y100b and R100d within the H-CDR3 of 045–09 2B05, we assessed whether these two residues were common among additional lateral patch-targeting mAbs. Of the 10 clones evaluated, we identified 5 distinct clones possessed a Y-x-R/H motif and another 3 clones possessed a Y-R motif within the H-CDR3 (fig. S2H). Notably, the Y-x-R/H and Y-R motifs were generated as a result of junctional diversity via non-templated DNA additions, suggesting B cells with these specific H-CDR3 motifs were preferentially selected to bind to the lateral patch. Additionally, a recent report also identified a broadly neutralizing mAb targeting the side of the HA head that utilized VH3–23/VK1–33 and possessed a Y-x-R motif within the H-CDR3 (12), suggesting the Y-x-R motif is a common feature of lateral patch-binding antibodies across individuals and is a public binding motif.
To understand how influenza viruses can escape lateral patch mAbs that utilize the VH3–23/VK1–33 pairing, we generated virus escape mutants using two lateral patch mAbs, 047–09 4G02 and 045–09 2B05. 047–09 4G02 resulted in mutations R120S and A259T, which are parallel on the β-sheets of the lateral patch epitope and lie next to the major antigenic sites Ca1 and Cb (Fig. 2F). Notably, only the A259T mutation is found in recent circulating pH1N1 viruses, and is suggested to improve HA stability, rather than be an antibody escape mutant (13). However, the specific isolation of A259T from virus grown in the presence of 047–09 4G02, but not parallel mock-treated virus, suggests that mutant selection driven lateral patch-binding antibodies could have led to this mutation being fixed into circulating pH1N1 viruses. 045–09 2B05 resulted in the mutation E119D, which sits between Ca1 and Cb (Fig. 2F). Although residue E119 does not directly contact 045–09 2B05, it resides within the H-CDR3 footprint, and mutation to Asp could potentially destabilize the epitope:paratope interaction (Fig. 2G). Together, these data indicate that mutations in residues next to antigenic sites Ca1 and Cb escape restriction from VH3–23/VK1–33 lateral patch-binding mAbs.
RBS-binding mAbs derive from a diverse repertoire but possess a similar binding footprint
We identified 10 distinct RBS-targeting clones from 6 individuals using negative stain EM (fig. S3A and table S4). Unlike mAbs targeting the lateral patch, RBS-binding mAbs used a diverse range of V(D)J genes across both the heavy and light chains (Fig. 3A and B; fig. S3B and C). However, we observed three repertoire trends: RBS-binding mAbs frequently utilized VH4–59 (Fig. 3A), a lambda light chain (Fig. 3B), and 5 out of 10 clones used JH6 (Fig. 3C), which often recombines to encode for the critical dipeptide residues responsible for sialic acid-mimicking contacts of RBS-binding mAbs (2, 14). However, not all RBS-binding mAbs utilized JH6; mAbs utilizing JH4 also demonstrated H-CDR3 dipeptides similar to those in JH6-utilizing mAbs (fig. S3D and E). Our data show mAbs against the RBS utilize H-CDR3s with similar lengths as mAbs targeting the lateral patch or other head epitopes (fig. S3F). Furthermore, no apparent differences were observed in the light chain CDR3 (L-CDR3) length and the H-CDR3 and L-CDR3 isoelectric points of mAbs targeting distinct head epitopes (fig. S3G to I).
Fig. 3: RBS-binding antibodies utilize a diverse repertoire and are susceptible to mutations in Sb.
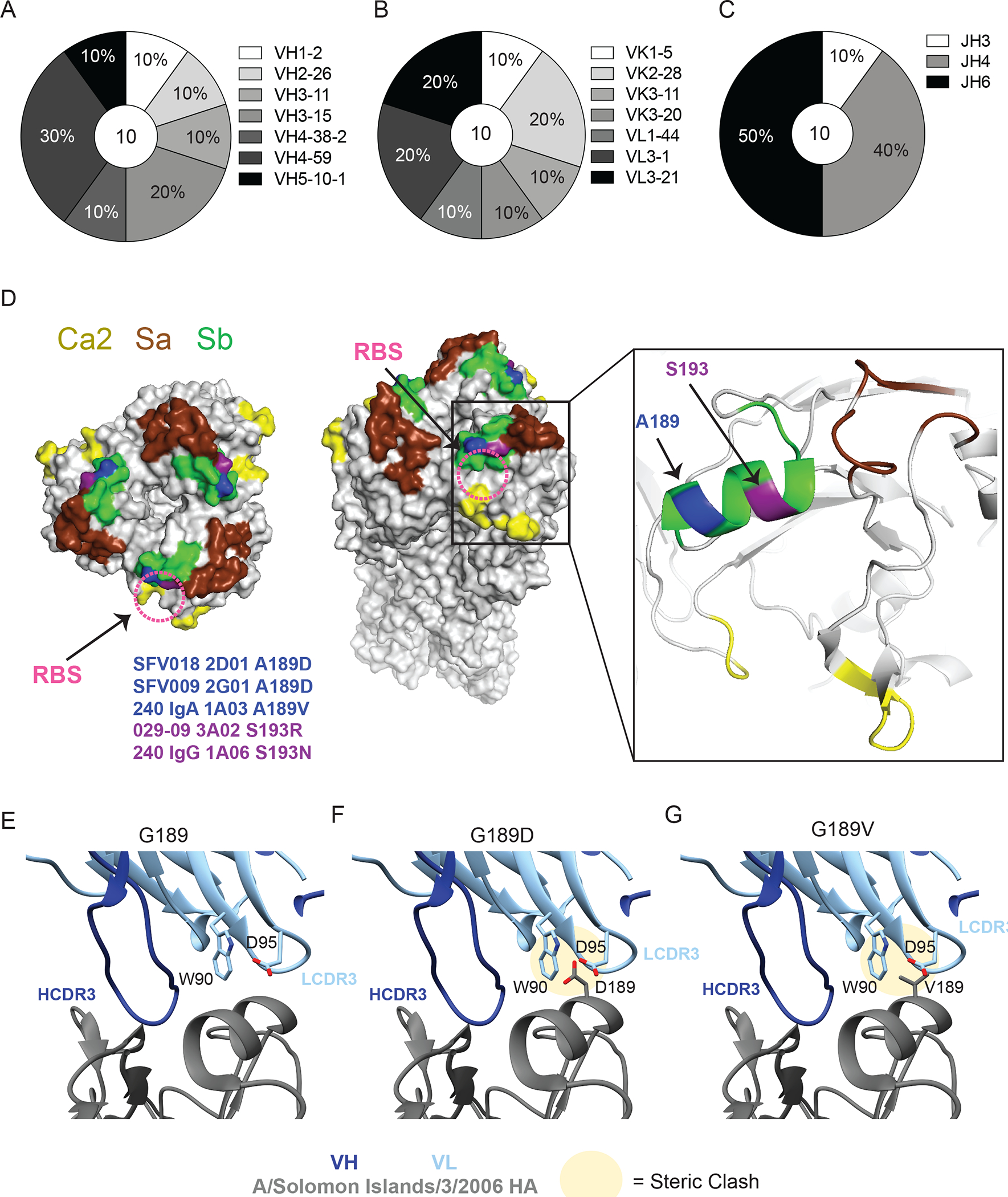
(A-C) Repertoire of unique B cell clones targeting the RBS including VH gene usage (A), VK or VL gene usage (B), and JH gene usage (C). Numbers in center of pie graphs represents number of distinct clones analyzed. (D) Location of virus escape mutations at A189 (blue; A189D or A189V) and S193 (purple; S193R or S193N) shown on A/California/04/2009 HA (PDB: 4jtv) for SFV018 2D01, SFV009 2G01, 240 IgA 1A03, 029–09 3A02, and 240 IgG 1A06, generated using A/Netherlands/602/2009 H1N1. The RBS is shown with a pink dashed circle. (E to G) CH65 binding to WT G189 (E), and steric clashes between L-CDR3 and HA resulting from G189D (F) and G189V (G) mutations in A/Solomon Islands/3/2006 HA (PDB:5ugy).
To better understand how influenza viruses can evade RBS-targeting mAbs, we generated viral escape mutants for 5 distinct non-clonal RBS-binding mAbs. All escaped viruses mutated at residues A189 and S193 in the major antigenic site Sb (Fig. 3D). Residues A189 and S193 sit adjacent on the 190-helix of the Sb epitope and protrude out from HA (Fig. 3D). Notably, structures of RBS-binding mAbs, particularly the light chain, have extensive interactions with the 190-helix of the Sb epitope (2, 14, 15), and mAb CH65 selects for a G189D mutation with the pre-pH1N1 virus A/Solomon Islands/03/2006 (14). Through molecular analysis, we found G189D and G189V escape mutations would result in steric clashes with W90 and D95 of the L-CDR3 of CH65 (Fig. 3E to G), likely destabilizing mAb binding. Together, these data indicate H1N1 viruses evade RBS-targeting mAbs by mutating in the antigenic site Sb.
Binding breadth and affinity of mAbs targeting the RBS and lateral patch
We next evaluated the viral binding breadth of HAI+ mAbs induced by the 2009 MIV and mAbs targeting the lateral patch, RBS, or other head epitopes. HAI+ mAbs induced by the MIV bound more H1N1 strains than mAbs induced by seasonal influenza virus vaccines (P = 0.0003, Fig. 4A, fig. S4A) due to RBS- and lateral patch-binding mAbs exhibiting broad viral binding breadth with pre- and post-pH1N1 seasonal H1 viruses (Fig. 4B; fig. S4B). Lateral patch-binding mAbs exhibited the broadest viral binding breath with nearly 100% of mAbs binding the 1918 H1 and 1977–2009 H1N1 viruses (P < 0.0001, fig. S4B). Viral binding breadth of RBS- and lateral patch-binding mAbs varied by clone, but lateral patch mAbs were consistently broadly reactive across H1N1 subtypes (fig. S4C). In contrast, RBS-binding clones varied greatly by their ability to bind to distinct H1N1 viruses (fig. S4C). Consistent with this notion, the lateral patch viral escape mutant residues were highly conserved whereas the RBS generated viral escape mutant residues were poorly conserved (fig. S4D). Together, these observations indicate that the lateral patch is generally more conserved than the RBS across H1N1 viruses or, alternatively, that the lateral patch has less antibody mediated pressure than the RBS epitope and mutates less frequently as a result. Nearly all lateral patch- and RBS-binding mAbs cross-reacted with a swine H1N2 virus (A/swine/Mexico/AVX8/2011 (16); P < 0.0001, Fig. 4C), suggesting MBCs against the lateral patch and RBS could be robustly recalled upon swine H1 virus spillovers into humans. Despite having less binding breadth amongst H1N1 viruses than lateral patch-binding mAbs, RBS-binding mAbs were more likely to cross-react with H3N2 and influenza B viruses than mAbs targeting the lateral patch (P = 0.0078) or other head epitopes (P = 0.0002, Fig. 4D and E). Moreover, 4 distinct RBS-binding clones were isolated from participant 240, with 3 clones demonstrating broad breadth to H1N1 and H3N2 (fig. S4E). These data indicate that lateral patch-binding mAbs demonstrate the greatest viral binding breadth amongst H1 viruses, whereas RBS mAbs have greater potential for cross-reactivity with other influenza A virus subtypes and influenza B viruses.
Fig. 4: Recalled RBS- and lateral patch-binding B cells are broadly reactive and were likely initially induced during childhood.
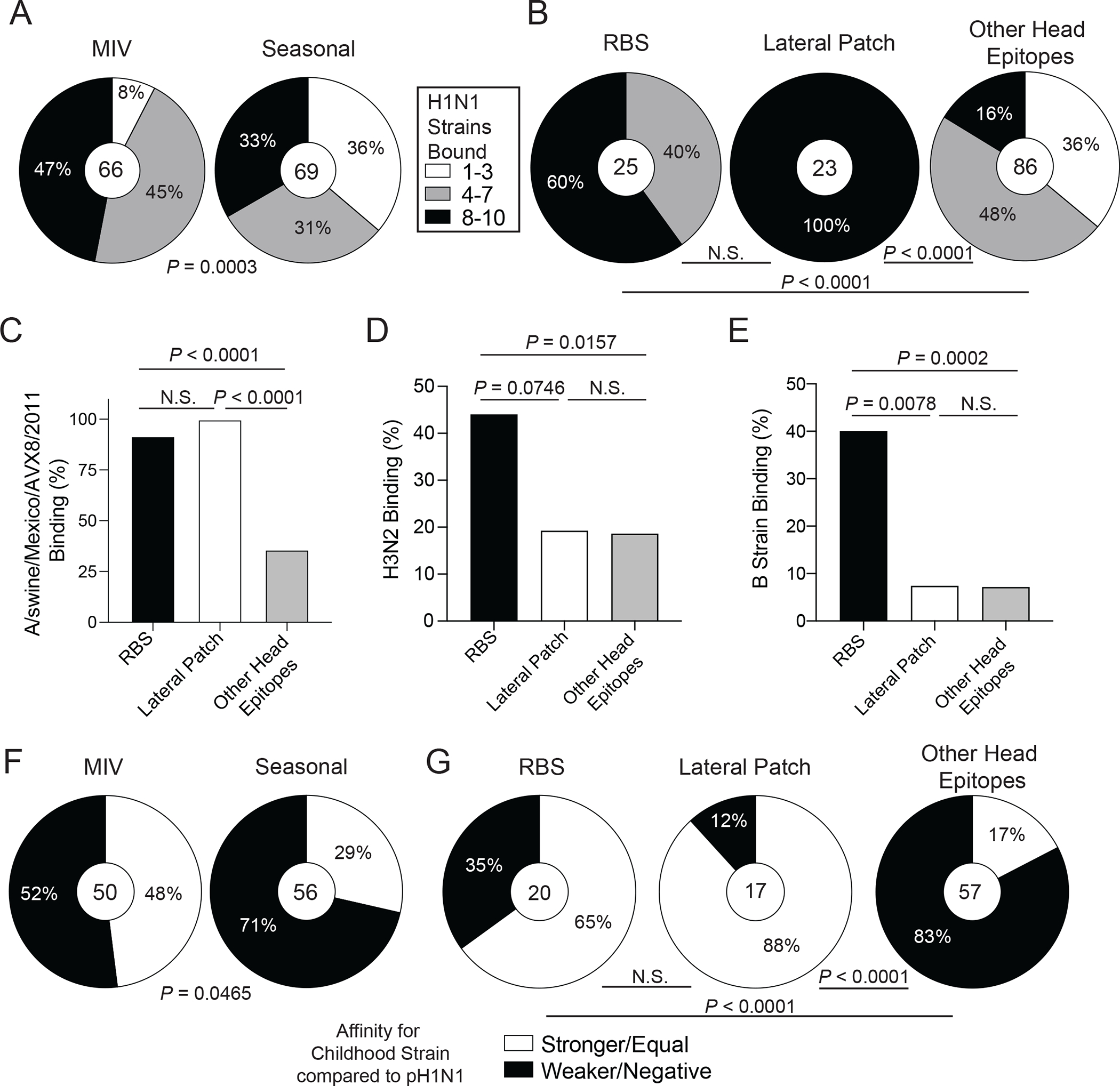
(A-B) HAI+ mAbs induced by the 2009 MIV or seasonal vaccines (2010 TIV and 2014 QIV; A) or binding distinct head epitopes (B) were tested for binding to seasonal and pandemic H1N1 strains. Data are shown as percent of mAbs binding to different numbers of strains. (C to E) Proportion of mAbs targeting the RBS, lateral patch, or other head epitopes binding a swine H1N2 virus (C), H3N2 viruses (D), or influenza B viruses (E). (F and G) Percent of MIV- or seasonal vaccine-induced HAI+ mAbs (F) and RBS, lateral patch, and other head epitopes binding mAbs (G) with equal or greater binding affinity to childhood strains relative to A/California/7/2009. Statistical significance was determined using Chi-square tests (A and B) and Fisher’s exact tests (C-G). Numbers in center of pie graphs (A, B, F, and G) represent number of individual mAbs analyzed. All mAbs were tested in duplicate and each ELISA was performed twice.
Head binding mAbs induced by the 2009 MIV had weaker affinity for A/California/7/2009 virus than head mAbs induced by seasonal vaccination (P = 0.0032, fig. S4F). Because mAbs were generated from acutely activated PBs, MBCs recalled from prior exposures constitute the PB response to vaccination and represent the immune history of B cells against particular epitopes. MIV-induced head mAbs had nearly twice as many mutations as mAbs induced by seasonal vaccination (P < 0.0001, fig. S4G), suggesting the PBs isolated after seasonal vaccination are derived from MBCs that were more recently generated in response to pH1N1. Moreover, nearly 50% of MIV-induced mAbs had higher or equal affinity for an H1N1 strain circulating in childhood (table S5) relative to pH1N1 compared to only 29% of seasonal vaccine-induced mAbs (P = 0.0465, Fig. 4F). We did not observe a statistical difference in the relative affinity of mAbs targeting the RBS, lateral patch, or other head epitopes (fig. S4H). MAbs targeting the lateral patch tended to have a median of 27 mutations, compared to only 17 and 16 mutations for mAbs binding the RBS and other head epitopes, respectively (P = 0.0008 and P < 0.0001, fig. S4I), indicating extensive affinity maturation of lateral patch-binding B cells. The number of mutations varied by distinct RBS clones (fig. S4J), suggesting that these clones undergo differential affinity maturation pathways, which could lead to distinct viral binding patterns. Furthermore, 88% of mAbs against the lateral patch and 65% of mAbs against the RBS had higher or equal affinity for a viral strain circulating during the individuals’ childhoods, compared to only 17% of mAbs targeting other head epitopes that reacted with the childhood strain (P < 0.0001, Fig. 4G; fig. S4K), demonstrating that RBS- and lateral patch-binding antibodies likely derive from B cells primed early in life. However, affinity for pH1N1 and a childhood strain varied across and within clonal expansion (fig. S4K), further indicating B cells targeting distinct HA head epitopes undergo differential affinity maturation paths. Together, our data show that first exposure to pH1N1 harnessed immune history to recall MBCs from childhood that demonstrate broad viral binding against the HA head.
RBS- and lateral patch-targeting mAbs are broadly neutralizing
Head binding mAbs induced by the MIV neutralized more H1N1 strains than head binding mAbs induced by seasonal vaccination (P < 0.0001, Fig. 5A), which was likely the result of increased neutralizing breadth of mAbs targeting the RBS or lateral patch, including mAbs within and across clonal expansions (P < 0.0001, Fig. 5B to D and fig. S5A). MAbs targeting distinct immunodominant epitopes exhibited differential neutralizing breadth, with mAbs against Ca1 being more strain-specific and mAbs against Sb having greater neutralizing breadth (P = 0.0003, fig. S5B and C). To assess neutralization potency, we performed HAI assays to specifically measure inhibition of HA binding to sialic acid, the HA receptor, and microneutralization (MN) assays that measure the ability of an antibody to inhibit infection of a cell, which includes inhibition of receptor binding, membrane fusion, and viral egress. MIV-induced and seasonal vaccine-induced mAbs generally had similar neutralization potency against the pH1N1 virus strain (fig. S6A and B), although seasonal vaccine-induced mAbs tended to be more potent by HAI. MAbs targeting the RBS or lateral patch were less potent against A/California/7/2009 by HAI relative to mAbs targeting other head epitopes (RBS, P = 0.0032; lateral patch, P < 0.0001, Fig. 5E). Only mAbs targeting the lateral patch were less potently neutralizing by MN assays relative to mAbs targeting other head epitopes (P = 0.0038, Fig. 5F). Moreover, mAbs binding any head epitope were 10-fold more potent than mAbs targeting the HA stalk (P < 0.0001, fig. S6C), indicating the potent neutralizing potential of all head binding mAbs, independent of precise epitope targeting. MN potency across lateral patch-binding clones was consistent, whereas RBS-binding clones varied dramatically in their neutralization potency, suggesting each RBS-binding clone and individual clonal members target the epitope slightly different (fig. S6D). MAbs targeting distinct immunodominant epitopes demonstrated different neutralization potency by HAI assay, with mAbs targeting Ca1 and Sb demonstrating the highest potency by HAI, likely because these epitopes are close to the RBS (fig. S6E). However, no difference in MN potency was observed for mAbs targeting distinct immunodominant epitopes (fig. S6F). We found that the affinity of mAbs against A/California/7/2009 strongly correlated with MN potency (P < 0.0001, fig. S6G). However, mAb HAI breadth inversely correlated with MN potency (P = 0.0066, fig. S6H), suggesting that as mAbs increase neutralization breadth, they reduce neutralization potency. Together, these data indicate mAbs targeting the RBS or lateral patch are broadly neutralizing against H1N1 viruses.
Fig. 5: Recalled RBS- and lateral patch-binding B cells are broadly neutralizing against H1N1 strains.
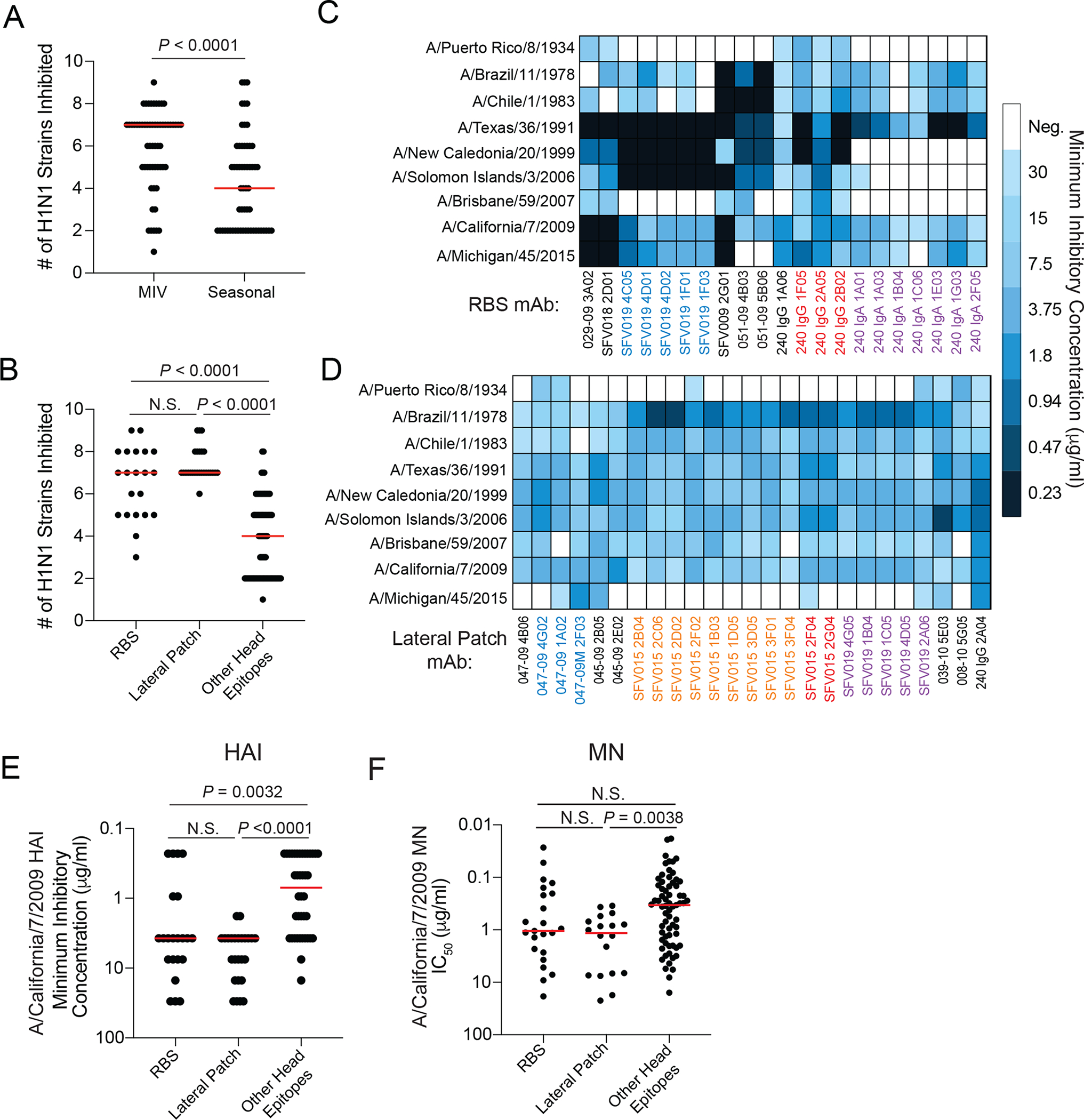
(A) Number of H1N1 strains inhibited by HAI+ mAbs induced by the 2009 MIV or seasonal vaccines (2010 TIV and 2014 QIV). (B) Number of H1N1 strains inhibited in an HAI assay by mAbs targeting the RBS, lateral patch, or other head epitopes. (C and D) Heat maps of HAI activity of RBS (C) and lateral patch (D) binding antibodies. MAb names of non-black colors represent mAbs within a clone. (E) HAI potency of mAbs targeting the RBS, lateral patch, or other head epitopes against A/California/7/2009 strain. (F) MN potency of mAbs targeting the RBS, lateral patch, and other head antibodies against A/California/7/2009. Statistical significance was determined by an unpaired two-sided non-parametric Mann-Whitney test (A) and unpaired two-sided non-parametric Kruskal-Wallis test (B, E, and F). Bars (A, B, E, and F) represent the median. Each symbol (A, B, E, and F) represents a distinct mAb. All mAbs were tested in duplicate and each HAI and MN assay was performed twice.
RBS- and lateral patch-binding antibodies are potently protective in vivo
We next determined whether mAbs targeting distinct head epitopes provided differential protection in vivo using a lethal influenza virus infection mouse model. We prophylactically treated mice intraperitoneally with mAb cocktails (5 mAbs per cocktail) 2 hours prior to intranasal infection with a lethal dose of the mouse-adapted A/Netherlands/602/2009 H1N1 virus. The cocktail included mAbs targeting either the RBS, lateral patch, or other head epitopes (table S6; fig. S7A), which are more representative of the total antibody response against these epitopes. Notably, mAbs included within each cocktail demonstrated similar neutralization potency to the mouse-adapted A/Netherlands/602/2009 and A/California/7/2009 (fig. S7B). Mice receiving the 5 mg/kg dose were 100% protected from weight loss and death, regardless of mAb cocktail (fig. S7C and D). Furthermore, mice at the 1 mg/kg dose exhibited some weight loss (Fig. 6A), with mice receiving the other head epitope cocktail exhibiting the most weight loss (fig. S7E), although no mice succumbed to infection (Fig. 6B). All mice in the 0.2 mg/kg treatment groups lost weight, and 50–70% of mice succumbed to the infection, regardless of mAb cocktail specificity (Fig. 6C and D). As antibodies against the RBS and lateral patch do not compete for binding on the HA trimer by negative stain EM (Fig. 6E), these data suggest that RBS- and lateral patch-binding antibodies could synergize with each other to provide potent protection against distinct H1-expressing viruses.
Fig. 6: MAbs targeting the lateral patch and RBS are potently protective in vivo.
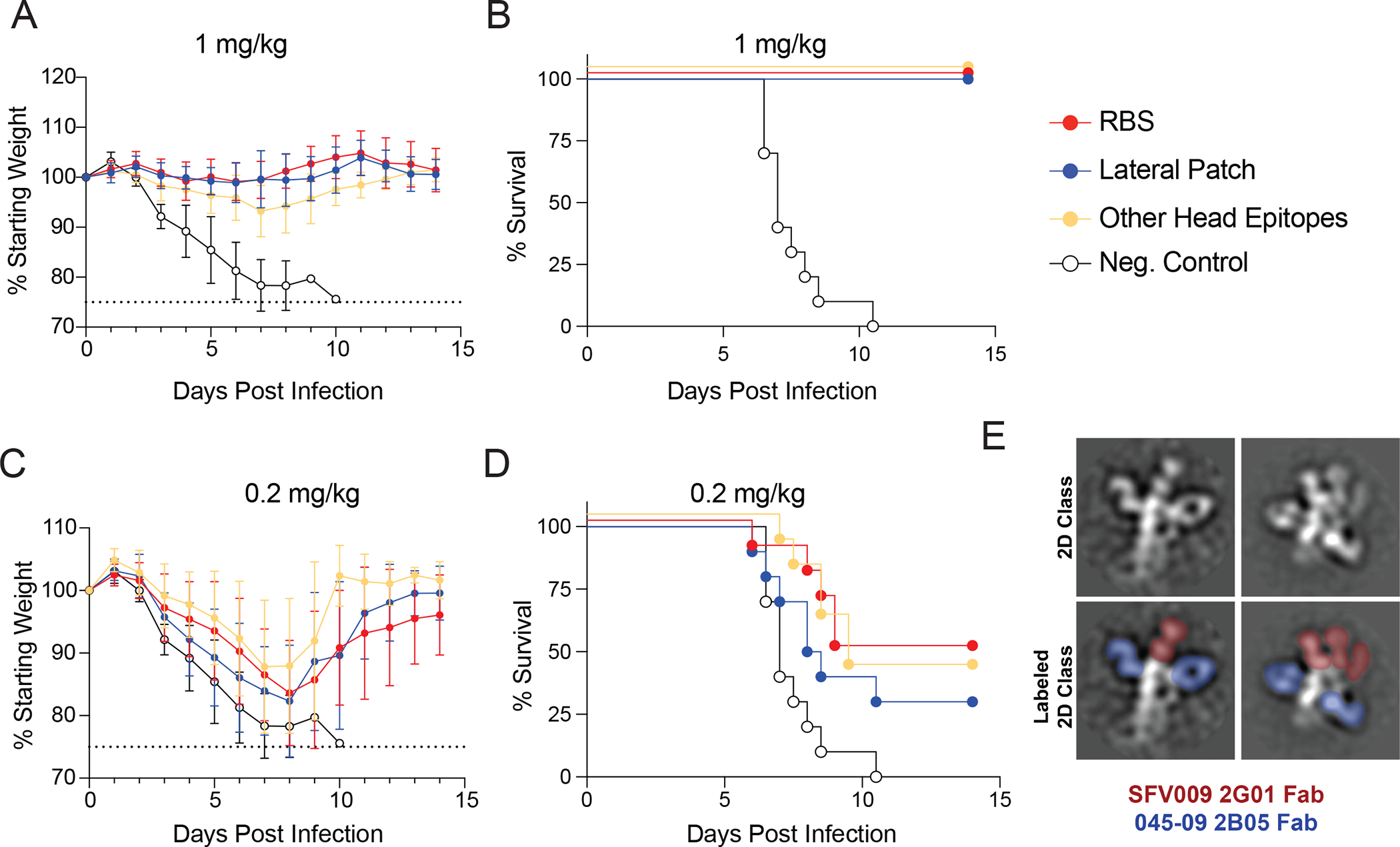
(A to D) Epitope mAb cocktails (5 mAbs per cocktail) were administered prophylactically by intraperitoneal injection, and mice were infected 2 hours later with 10 LD50 of A/Netherlands/602/2009 H1N1 virus intranasally. Weight loss (A) and survival (B) were measured for mice treated with 1 mg/kg of mAb cocktails. Weight loss (C) and survival (D) were measured for mice treated with 0.2 mg/kg of mAb cocktails (E) 2D class averages of a RBS clone (SFV009 2G01; red) and lateral patch clone (045–09 2B05; blue) binding the same HA. Data in A and C are the mean ± standard deviation (n=10 mice per group), and data from A-D are pooled from two independent experiments. Dashed lines in A and C represent the humane end point of 25% weight loss.
Certain lateral patch-targeting mAbs can still bind a natural escape mutant
In 2013, pH1N1 viruses acquired S165N and K166Q mutations in the Sa major antigenic site (4). The seasonal influenza virus vaccine was updated in 2017 to include A/Michigan/45/2015, an H1N1 strain that possesses N165 and Q166. Notably, the lateral patch-binding mAb, Fab6649, was unable to bind and neutralize A/Michigan/45/2015 (3), suggesting this mutation arose due to the selective pressure by antibodies against this epitope (4). From our cohorts, only 39% of lateral patch-targeting mAbs bound A/Michigan/45/2015 (Fig. 7A). Although A/Michigan/45/2015 binding ability varied greatly between and within clones, the only clonal expansion with all members binding A/Michigan/45/2015 expressed VH3–23 and VK1–33 (clone 1; fig. S8A). Moreover, A/Michigan/45/2015 acquired the same A259T mutation that arose for the 047–09 4G02 escape mutant virus. However, A259T alone likely does not explain the loss of lateral patch-binding mAbs, as 047–09 4G02 (clone 1) can still bind and neutralize A/Michigan/45/2015 (fig. S8A). MAbs that could still bind A/Michigan/45/2015 had reduced binding affinity (P = 0.0137) and MN potency (P = 0.0420) for A/Michigan/45/2015 relative to A/California/7/2009 (Fig. 7B and C). Fab6649 directly bound the mutated residues of A/Michigan/45/2015 (Fig. 7D), limiting its ability to bind to post-2013 pH1N1 viruses (3). However, 045–09 2B05 bound residues below the S165N and K166Q mutations (Fig. 7D) and still bound and neutralized A/Michigan/45/2015 to a similar degree as A/California/7/2009 (Fig. 7E). Moreover, the HA binding contacts of 045–09 2B05 were generally more conserved than the binding contacts of Fab6649, as two out of three contacts made by 045–09 2B05 were 100% conserved, in contrast to only one out of seven contacts of Fab6649 (fig. S8B). In addition to 045–09 2B05, all mAbs utilizing VH3–23 and VK1–33 were still capable of binding and neutralizing A/Michigan/45/2015 (fig. S8C). Alignment of the VH3–23/VK1–33 expressing antibody sequences revealed that all antibodies possessed the Y-x-R motif within the H-CDR3 (fig. S8D and E), indicating lateral patch-binding antibodies with this motif are capable of binding recent H1N1 viruses. We next generated chimeric mAbs that utilized the heavy or light chain of 045–09 2B05 and the heavy or light chain of mAbs that did not utilize both VH3–23 and VK1–33 to investigate whether the heavy and light chains of 045–09 2B05 could complement mAbs with other repertoire features and allow for binding to this drifted variant. Only natively paired heavy and light chains could bind to A/California/7/2009 (fig. S8F), suggesting that the identified lateral patch-binding clones were specifically selected against this epitope and these antibody chains are not interchangeable for epitope binding. However, as the heavy and light chains of these chimeric mAbs affinity matured independently, they may not bind HA as well as heavy and light chains that have naturally affinity matured together.
Fig. 7: VH3–23/VK1–33 lateral patch-binding antibodies can bind and neutralize a natural escape mutant.
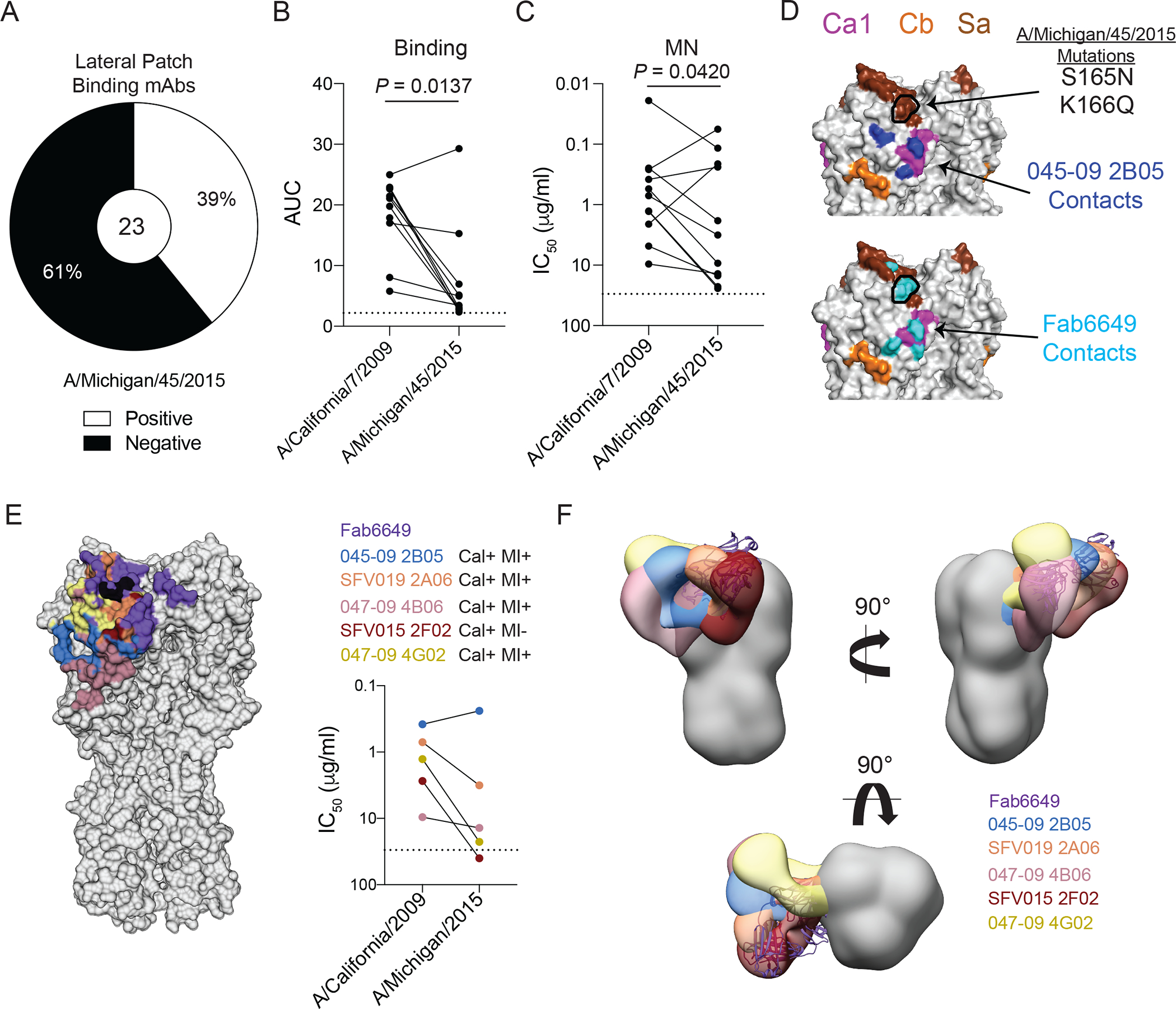
(A) Proportion of lateral patch-targeting mAbs binding A/Michigan/45/2015. (B and C) Binding strength as represented as area under the curve (AUC) from virus-specific ELISAs (B) and MN potency (C) of A/Michigan/45/2015 binding lateral patch-targeting mAbs against A/California/7/2009 and A/Michigan/45/2015 viruses. (D) Binding contacts of 045–09 2B05 (blue) and Fab6649 (cyan) relative to the S165N and K166Q mutations (outlined in black) found in A/Michigan/45/2015 projected on A/California/04/2009 (PDB: 4jtv). (E) Footprints of 6 lateral patch-binding antibodies in reference to the S165N and K166Q mutations (black) and their respective MN potencies against A/California/7/2009 and A/Michigan/45/2015 viruses. (F) Side and top view of negative stain EM reconstructions of lateral patch-targeting mAbs binding HA relative to Fab6649 (based on PDB: 5w6g). Statistical significance was determined by a paired two-sided non-parametric Wilcoxon matched-pairs signed rank tests (B-C). Dashed lines in B, C, and E represent the limit of detection (LOD). Each line in B, C, and E connects a single mAb (paired symbols) binding to each virus. All mAbs were tested in duplicate and each ELISA and MN was performed twice.
To understand the structural basis of lateral patch-targeting mAbs binding to A/Michigan/45/2015, we evaluated the antibody footprint and angle of approach of 6 lateral patch-binding mAbs. Notably, the footprints of all mAbs assayed were largely overlapping (Fig. 7E). Similar to Fab6649, S165N and K166Q mutations fell directly within the footprint of SFV015 2F02, which was incapable of binding and neutralizing A/Michigan/45/2015 (Fig. 7E and fig. S8G). Additionally, Fab6649 and SFV015 2F02 largely overlapped in the angle of approach (Fig. 7F), suggesting these two antibodies are targeting the lateral patch in a similar manner. The S165N and K166Q mutations were located on the edge of the binding footprint of mAbs capable of binding and neutralizing A/Michigan/45/2015 (Fig. 7E and fig. S8G). Furthermore, both the binding footprints and the angles of approach of 045–09 2B05, 047–09 4B06, and 047–09 4G02, which all utilize VH3–23/VK1–33, were more proximal to the RBS of the corresponding HA protomer relative to Fab6649, limiting the overlap with S165 and K166 (Fig. 7E and F and S8G. These data suggest the binding footprint and angle of approach may allow some lateral patch-binding antibodies to overcome mutations within the Sa epitope.
Discussion
In this study, we report that first exposure to pH1N1 recalls MBCs against conserved epitopes of the HA head, whereas repeated exposure to pH1N1 elicits MBCs against the variable major antigenic sites of the HA head. As individuals in 2009 MIV cohort had little to no pre-existing immunity against pH1N1, MBCs targeting conserved epitopes of both the HA head and stalk were preferentially recalled in response to this threat to provide robust and broad protection. Prior studies have shown first exposure to pH1N1 virus (8, 17–19), H1N1 infection (20), and avian H5N1 and H7N9 virus vaccination (21–24) robustly recruited MBCs targeting conserved epitopes of the HA stalk. As the stalk is capable of escaping broadly neutralizing antibodies (25, 26), further investigation into whether vaccination with other viruses induces cross-reactive antibodies targeting conserved epitopes of the HA head is warranted. A recent study demonstrated that H5 vaccination could induce a polyclonal antibody response against RBS and lateral patch epitopes of the H5 head (27), although whether these antibodies can cross-react with other influenza virus subtypes has not been determined.
Despite the robust recall of MBCs targeting the lateral patch and RBS, naïve B cells against variable epitopes were likely recruited into germinal centers following first exposure to the pH1N1 to diversify the humoral immune response, as observed with seasonal influenza virus vaccinations (28). Consistent with this, our studies indicated an increase in pH1N1-specific mAbs in the seasonal vaccine cohorts and the identification of mAbs binding specific variable epitopes of the HA head. These data also indicate that pre-existing immunity against the variable major antigenic sites of the HA head may limit the activation and recruitment of B cells targeting conserved epitopes of the HA head and stalk domains. The preferential recruitment of B cells against variable epitopes may be due to serum antibodies masking conserved HA head epitopes (29, 30) or reduced epitope immunogenicity of conserved HA head epitopes, as previously observed for the HA stalk (31, 32). Moreover, influenza virus vaccine-induced plasma cells within the bone marrow are short-lived (33), consistent with continued susceptibility to influenza virus infections, despite the generation of antibodies against conserved epitopes. Therefore, new vaccine formulations that robustly induce B cells, both into the memory B cell pool and bone marrow resident plasma cells, against the conserved epitopes of the HA head and stalk are urgently needed to provide broad and durable humoral immunity.
Antibodies binding the RBS and lateral patch were likely induced in response to the antigenically distinct pH1N1 exposure due to their broad viral binding and neutralization breadth. The repertoire features of RBS-binding mAbs identified in our study are similar to those described previously (14), suggesting a similar vaccine strategy could be utilized to induce antibodies against the RBS in most people with pre-existing immunity against H1N1 viruses. Although RBS-binding mAbs were inconsistently neutralizing against drifted seasonal H1N1 strains, similar to other RBS directed mAbs CH65 and C05 (1, 2), a previous report found RBS-binding B cell clones expand their breadth as they are exposed to more influenza viruses (34, 35), which could explain the differences in RBS antibody neutralizing potential. Mutations in the RBS region, including the K133a insertion and G158N/N159G mutations within the 150-loop in 1918- and 2009-pandemic H1N1 strains, may also contribute to the binding and neutralizing differences to seasonal H1N1 strains observed with RBS antibodies (2, 35–37). Moreover, reports have indicated that head-binding antibodies induced by egg-grown vaccines exhibit better binding to avian-adapted H1 viruses than human-adapted H1 viruses (38, 39). Depending on an individual’s natural exposure history to human H1 viruses and H1 viruses grown in eggs for vaccination, it is of interest to understand whether these differences in viral exposure account for differences in viral binding strength and HAI potency of mAbs to historical and recent H1N1 viruses.
We also observed many RBS-binding mAbs cross-reacted with H3 and influenza B viruses, suggesting the key residues involved in mAb binding to the H1 RBS were similar to those residues within the H3 and influenza B virus RBS epitopes. These data suggest RBS-binding B cells could affinity mature against not only H1N1 but also H3N2 and influenza B viruses and could provide near universal protection against influenza viruses. However, whether RBS antibodies are induced to high enough concentrations to protect against other influenza virus subtypes and influenza B virus is yet to be determined. Additionally, the need for extensive affinity-maturation against many influenza viruses may limit the induction of RBS antibodies with potent and broadly neutralizing potential.
Lateral patch-targeting mAbs are mostly restricted to VH3–23 or VK1–33 and feature a crucial, conserved Y-x-R or Y-R motif within the H-CDR3. KPF1, a potent H1 head-binding mAb binding the lateral patch, also utilizes VH3–23/VK1–33 and similarly possesses a Y-x-R motif within the H-CDR3 (12). It is unclear as to why there is an enrichment for antibodies utilizing VH3–23 and VK1–33, as the main contacts of 045–09 2B05 were within the junctional diversity of the H-CDR3 and a mutation within the K-CDR1. The selection of VH3–23 and VK1–33 may be associated with CDR compatibilities, as heavy and light chains are frequently co-selected to create a functional binding site (40). A recent study identified that VK gene usage altered H-CDR3 loop confirmations for a public clonotype against the SARS-CoV-2 receptor-binding domain (41). A similar complementation between heavy chain and light chains may be necessary for lateral patch epitope binding. However, further structural analysis of lateral patch-binding mAbs are needed to understand why antibodies with these two variable gene are preferentially selected against the lateral patch. Despite this, our data identify B cells utilizing VH3–23/VK1–33 and possessing a Y-x-R motif as public antibody clonotypes, as mAbs with these features were identified in 3 individuals from our study and 1 individual in Nogales et al. (12). The lateral patch-binding mAbs identified within our study do not share the key features of Fab6649 (3), as Fab6649 does not use VH3–23 or VK1–33 and does not possess the Y-x-R or Y-R motifs within the H-CDR3. Moreover, all the mAbs utilizing VH3–23/VK1–33 could still bind and neutralize A/Michigan/45/2015. These data indicate that the lateral patch is an important vaccine target to drive potent broadly neutralizing antibodies against, and that VH3–23/VK1–33 utilizing antibodies are a good indicator of B cells targeting this epitope.
There are several limitations to our study. Although our study identified that first exposure to pH1N1 was able to robustly induce antibodies against the lateral patch and RBS in the context of monoclonal B cells, it is unknown whether the plasmablast response accurately represents the long-term antibody response following vaccination. As influenza virus vaccination recruits both MBCs and naïve B cells into the germinal center response (28), the antibody landscape likely shifts overtime to include antibodies against the new variable epitopes of the H1 head. Consistent with this, participants in the repeated exposure cohorts preferentially induced a plasmablast response against the variable epitopes of pH1N1 head domain. We identified multiple antibody classes targeting the lateral patch with variations in antibody footprints and angles of approach, binding motif properties, and neutralization properties, several of which still targeted recent pH1N1 viruses. However, our study only generated the structure and viral escape mutants for one class of antibodies targeting the lateral patch. It is of continued interest to understand the structural similarities of distinct antibody classes binding the lateral patch, including Fab6649 (3), and how these have potentially driven pH1N1 viral evolution.
Although conserved stalk epitopes are the primary targets of many candidate universal influenza vaccines, a concentrated antibody response against stalk epitopes could lead to viral escape at these sites (25, 26). A combined epitope approach is necessary to generate a more robust vaccine that can safeguard against viral resistance. Several investigative vaccine platforms have the potential to robustly induce RBS- and lateral patch-targeting antibodies. Platforms that are engineered to remove or mask the major antigenic sites while preserving conserved epitopes have the potential to divert the antibody response against conserved epitopes of the HA stalk and head domain, including the newly identified head-interface binding antibodies (42–44), as opposed to variable epitopes of the HA head. Notably, a phase 1 clinical trial of a chimeric HA vaccine that swapped the head domain of H1 for that of exotic avian influenza virus subtypes (H5 or H8), was able to overcome pre-existing immunity against the HA head to induce antibodies against conserved epitopes of the HA stalk and the head-interface epitope (45). A variation of the chimeric HA approach is the development of mosaic HA antigens that replace the major antigenic sites with those from avian influenza viruses. Pre-clinical studies have shown mosaic HAs are capable of inducing antibodies that provide broad protection against divergent influenza viruses (46, 47). Additionally, several platforms have “masked” major antigenic sites using glycan shields (42) or PEGylation (48) to drive antibody responses against conserved epitopes of HA, and these approaches could be modified to drive antibodies specifically to the RBS and lateral patch epitopes. Furthermore, heterotypic nanoparticles that display HA head domains from multiple H1 strains were shown to induce broadly neutralizing HAI+ antibodies (49). These vaccination platforms should be investigated to determine if pre-existing immunity in humans can be harnessed to robustly recall MBCs targeting the RBS and lateral patch.
In summary, our study identifies that, in the absence of pre-existing immunity against the variable epitopes of the HA head domain, individuals can preferentially recall MBCs against broadly neutralizing epitopes of the HA head including the lateral patch and RBS. To date, our study provides the most comprehensive analysis of antibodies targeting the lateral patch, revealing that lateral patch antibodies utilize a restricted repertoire and are broadly neutralizing across H1-expressing viruses. Furthermore, our study shows the VH3–23/VK1–33 class of lateral patch-binding antibodies can still neutralize a recent pH1N1 virus, which was believed to have mutated to circumvent antibodies targeting this epitope. Altogether, our study provides a framework for next-generation vaccines to circumvent antibodies against variable epitopes of the HA head and robustly induce potently protective broadly neutralizing antibodies against the lateral patch and RBS
Materials and Methods
Study design
We initiated this study to compare the specificities of vaccine-induced plasmablast responses upon first and repeated vaccination with the pH1N1 virus. For the first pH1N1 exposure cohort, participants were recruited to receive the 2009 MIV. For the repeated pH1N1 cohort, participants that had previously received the 2009 MIV were recruited in 2010 to receive the 2010 TIV. An additional repeated exposure cohort was recruited in 2014 to receive the 2014 QIV. Plasmablasts were single-cell sorted from peripheral blood mononuclear cells (PBMCs) between days 5 and 7 post-vaccination and mAbs were generated to determine specificity. Several participants in the 2009 MIV cohort were recruited back 14 days and 6 months following vaccination to determine pH1N1 specific MBC responses, as HA+ B cells were single-cell sorted and mAbs were generated to determine epitope specificity. Because these three groups were vaccinated in different years with the vaccine available each year, this study was unblinded and not randomized. We enrolled any healthy adult 18 years or older that had not received an influenza virus vaccine that year. We generated and characterized mAbs from 9 participants in the 2009 MIV cohort, 6 participants in the 2010 TIV cohort, and 7 participants in the 2014 QIV cohort. The number of participants recruited each year was limited by the number of participants we could vaccinate before the vaccine expired. We characterized all HA head-specific mAbs that could be isolated from participants within the limits of the labor-intensive process of making mAbs.
All studies were performed with the approval of the University of Chicago and Emory University institutional review boards. The 2009 MIV vaccine participants were recruited at the University of Chicago and Emory University (8); the 2010 TIV and 2014 QIV vaccine participants were recruited at the University of Chicago (8, 50). Informed consent was obtained after the research applications and possible consequences of the studies were disclosed to study participants.
All experiments were done in accordance with the University of Chicago Institutional Animal Care and Use Committee and in adherence to the NIH Guide for the Care and Use of Laboratory Animals. Six- to eight-week-old female BALB/c mice were used for infection studies, as the virus was titrated in mice of the same strain, age, and sex. Ten mice were used per group, and a power analysis was used to determine the number of mice needed per experiment. All mice from two independent were included in the data analysis until the point of euthanasia, which resulted from a 25% weight loss from initial starting weight or upon competition of the experiment (14 days). Mice were provided a standard diet chow and water and were housed in the ABSL-2 (animal biosafety level-2) facility within the Carlson Animal Research Facility at the University of Chicago.
Cell Culture
Human embryo kidney (HEK) 293T cells (Thermo Fisher Scientific) were maintained at 37°C with 5% CO2 in Advanced Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 2% ultralow IgG fetal bovine serum (FBS; Gibco), 1% L-glutamine (Gibco), 1% antibiotic-antimycotic (Gibco). Madin-Darby Canine Kidney (MDCK) cells (American Type Culture Collection) were maintained in culture at at 37°C with 5% CO2 in DMEM (Gibco) supplemented with 10% FBS (Gibco), 1% L-glutamine (Gibco), 1% penicillin/streptomycin (Gibco).
Viruses and recombinant proteins
Influenza viruses used in all assays were grown in-house in specific pathogen free (SPF) eggs, harvested, purified, and titered (51). The A/swine/Mexico/AVX8/2011 H1N2 virus (16) was provided by Ignacio Mena and Adolfo García-Sastre at Icahn School of Medicine at Mount Sinai. Recombinant HA used for enzyme-linked immunosorbent assays (ELISAs) were obtained from BEI Resources or provided by the Krammer laboratory at Icahn School of Medicine at Mount Sinai. All HA residue numbering is based on FluDB H3 numbering and annotated HA figures were produced using Pymol (Schrödinger). Recombinant HA (A/California/04/2009, with stabilizing mutations E47K or E47G in HA2 (15)) used for negative stain and cryo-EM were produced in-house or kindly provided by Nicholas Wu and Ian Wilson at The Scripps Research Institute.
Monoclonal antibodies
Monoclonal antibodies were generated as previously described (52–54). Peripheral blood was obtained from each participant approximately 7 or 28+ after vaccination. For the 2014 QIV cohort, IgG+ and IgA+ PBs were sorted as part of another study (50). Lymphocytes were isolated and enriched for B cells using RosetteSep (StemCell Technologies). Total PBs (CD3−CD19+CD27hiCD38hi; 2009 MIV and 2010 TIV; fig. S9A), IgG+ PBs (CD3−CD19+IgM−CD27hiCD38hiIgG+IgA−; 2014 QIV; fig. S9B), or IgA+ PBs (CD3−CD19+IgM−CD27hiCD38hiIgG−IgA+; 2014 QIV cohort; fig. S9B), or HA+ bait-sorted MBCs (CD3−CD19+CD27+CD38lo/+HA+; fig. S9C) were single-cell sorted into 96-well plates. Immunoglobulin heavy and light chain genes were amplified by reverse transcriptase polymerase chain reaction (RT-PCR), sequenced, cloned into human IgG1, human kappa chain, or human lambda expression vectors. Primer sequences used for initial amplification of antibody sequences and for cloning PCR steps are listed in table 1 of a previously published protocol from our group (52, 54). Plasmids for the heavy and light chains of a corresponding mAb, 9 μg each, were mixed with 100 μg of polyethyleneimine (PEI; Sigma-Aldrich) and were co-transfected into confluent HEK 293T cells for 12 to 18 hours, after which media was changed to Protein-Free Hybridoma Medium (PFMH-II; Gibco). Secreted mAbs were purified from the supernatant using protein A agarose beads (Pierce). For chimeric mAb generation, the heavy and light chains for various lateral patch-binding mAbs were co-transfected and purified as described above. Expression of chimeric mAbs was comparable to natively paired mAbs. For Fab generation, heavy chain sequences were cloned into a Fab vector and co-transfected with the corresponding light chain. Fabs were purified using Ni-NTA beads (Qiagen). For mAbs generated from the 2014 QIV cohort, mAb names include the original isotype of the sorted PB, and all mAbs were expressed as human IgG1. B cell clones were determined by aligning all the V(D)J sequences sharing identical progenitor sequences, as predicted by IgBLAST using Vgenes. Repertoire analysis was performed using mini-prepped sequences that were analyzed using IgBLAST and Clustal Omega (European Bioinformatics Institute).
Antigen-Specific ELISA
High protein-binding microtiter plates (Costar) were coated with 8 hemagglutination units (HAU) of virus in carbonate buffer or with recombinant HA at 1 μg/ml in phosphate-buffered saline (PBS) overnight at 4°C. Plates were washed with PBS/0.05% Tween and blocked with PBS containing 20% fetal bovine serum (FBS) for 1 hour at 37°C. Antibodies were then serially diluted 1:3 starting at 10 μg/ml and incubated for 1.5 hour at 37°C. Horseradish peroxidase (HRP)-conjugated goat anti-human IgG antibody diluted 1:1000 (Jackson ImmunoResearch) was used to detect binding of mAbs, and plates were subsequently developed with Super Aquablue ELISA substrate (eBiosciences). Absorbance was measured at 405 nm on a microplate spectrophotometer (BioRad). To standardize the assays, control antibodies with known binding characteristics were included on each plate, and the plates were developed when the absorbance of the control reached 3.0 optical density (OD) units. All experiments were performed in duplicate and replicated 2–3 times. Affinity measurements, as represented as Kd at a molar concentration (M), were calculated using Prism 9 (Graphpad) by performing a non-linear regression. Area under the curve (AUC) values were calculated using Prism 9 (Graphpad).
Negative stain EM
Immune complexes were prepared by incubating Fab with HA (A/California/04/2009 with E47K or E47G stabilizing mutations) at greater than 3:1 molar ratio for 2 hours at room temperature (RT). Samples were deposited at ~10μg/ml on glow-discharged, carbon-coated 400 mesh copper grids (Electron Microscopy Sciences) and stained with 2% w/v uranyl formate. Samples were imaged at 73,000x magnification, 200kV, on a Talos 200C microscope equipped with a Falcon II direct electron detector and a CETA 4k camera (FEI). Micrographs were collected with Leginon, single particles were processed with Appion and Relion, footprints were mapped with University of California, San Francisco (UCSF) Chimera, and figures were made with UCSF Chimera (55–58).
Cryo-EM
Immune complexes were prepared as described in “Negative stain EM.” Samples diluted to 750μg/ml with 5μM final concentration of Lauryl Maltose Neopentyl Glycol (Anatrace) were deposited on glow-discharged 1.2/1.3 quantifoil 400 grids (Electron Microscopy Sciences) and plunge frozen in liquid ethane using a Vitrobot (FEI). Samples were imaged at 29,000x magnification, 300kV, on a Titan Krios microscope (FEI) in counting mode equipped with a Gatan K2 summit detector. 1298 micrographs were collected at a defocus range of −0.8 to −2 μm using Leginon (55), with a total exposure time of 9.25s and a total dose of 49.27e/Å2. Micrograph frames were aligned using MotionCor2 (59). Using CryoSPARC2 (60), data were subject to CTF Estimation (Gctf BETA), a subset of single particles were manually picked and used as templates, particles were picked using the template picker, particles were cleaned using 2D classification, and particles were refined using heterogenous, homogenous, and non-uniform refinements. The final map consisted of 27,888 particles and refined to 3.22Å with 3-fold imposed symmetry. Figures were made with UCSF Chimera (58).
Model building and refinement
ROSIE (The Rosetta Online Server that Includes Everyone (61)) was used to generate a predicted model of the variable regions of 045–09 2B05 mAb. This predicted model was docked together with the model of A/California/04/2009 HA with E47G HA2 (Protein Database (PDB): 4m4y) into the cryo-EM map. These combined models were used as an initial model for iterative manual adjustment using COOT and refinement using Rosetta. The model was numbered based on the H3 numbering scheme for HA and the Kabat numbering scheme for Fab. The final model and map were evaluated using MolProbity, EMRinger, and the PDB validation server (62–65).
Structural analysis
Atomic distance and steric clash analyses were performed using tools embedded in UCSF Chimera (58).
Microneutralization and Hemagglutination Inhibition Assays (HAI)
Microneutralization assay for mAb characterization was carried out as previously described (66, 67). The day before the experiment, 25,000 MDCK cells were added to each well of a 96-well plate. Serial two-fold dilutions of mAb were mixed with an equal volume of 100 50% tissue culture infectious doses (TCID50) of virus for 1 hour and added to MDCK cells for 1 hour at 37°C. The mixture was removed, and cells were cultured for 20 hours at 37°C with 1X minimum essential medium (MEM) supplemented with 1 μg/ml tosyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin and appropriate mAb concentration. Cells were washed twice with PBS, fixed with 80% ice cold acetone at −20°C for at least 1 hour, washed 3 times with PBS, blocked for 30 minutes with 3% BSA-PBS, and then treated for 30 minutes with 2% H2O2. Cells were incubated with a mouse anti-NP antibody (1:1000; Millipore) in 3% BSA-PBS for 1 hour at RT, followed by goat anti-mouse IgG-HRP (1:1000; Southern Biotech) in 3% BSA-PBS for 1 hour at RT. The plates were developed with Super Aquablue ELISA substrate, and absorbance was measured at 405 nm until virus only controls reached an OD of 1. The signal from uninfected wells was averaged to represent 100% inhibition. The signal from infected wells without mAb was averaged to represent 0% inhibition. Duplication wells were used to calculate the mean and SD of neutralization, and inhibitory concentration 50 (IC50) was determined by a sigmoidal dose response curve. The inhibition ratio (%) was calculated as below: ((OD Pos. Control – OD Sample) / (OD Pos. Control – OD Neg. Control)) * 100. The final IC50 was determined using Prism 9 (GraphPad). All experiments were performed in duplicate twice.
For HAI assays, viruses were diluted to 8 HAU/50 μl. 25 μl was combined with an equal volume of mAb (starting concentration 30 μg/ml), serially diluted 1:2 in PBS in duplicate, and subsequently incubated at RT for 1 hour. 50 μl of 0.5% turkey red blood cells (Lampire Biological) were added to each well and incubated for 45 minutes at RT. Minimum inhibitory concentrations were determined based on the final dilution of mAb for which hemagglutination inhibition was observed. All experiments were performed in duplicate twice.
Virus Escape Mutant Generation
Virus escape mutants were generated as previously described (68). Ten TCID50 of A/Netherlands/602/2009 virus was incubated with 1:5 IC50 of mAb for 1 hour. Virus/mAb mixtures were added to 500,000 MDCK cells for 1 hour. Cells were washed with PBS and plated with 2ml Ultra MDCK media (Lonza) supplemented with 1 μg/ml TPCK-treated trypsin and corresponding mAb concentrations. After two days, supernatants were collected, and HA assays were performed to determine viral growth. Wells with virus were incubated with 2x increase of mAb for one hour before addition to MDCK cells. Virus grown in the absence of mAb was included for all mAbs to control for passage-associated mutations. The virus was grown with increasing concentration of mAb until 64–128x IC50 of mAb was achieved, typically 10–16 passages. After an escape mutant was generated, supernatants were collected, viral RNA was collected and purified using QIAamp Viral RNA Mini Kit (Qiagen), cDNA was generated using SuperScript III (Invitrogen), HA sequences were amplified using A/Netherlands/602/2009 specific HA primers, and sequenced by Sanger sequencing. Sequences were analyzed using Librator (in-house software) and Clustal Omega (European Bioinformatics Institute). Mutations were visualized on A/California/4/2009 HA (PDB: 4jtv) using Pymol (Schrödinger).
HA conservation modeling
To avoid potential bias from highly imbalanced sample years (more than 90% of publicly available H1 sequences are from the last 10 years), we built a representative dataset of H1N1 viruses (table S5). In this dataset, for each year, only the most representative sequence (most identical to the consensus sequence of the year) was selected and was used to calculate conservation scores for all residues on this dataset. We quantified the peptide variations for residues on HA proteins of H1 sequences by adopting an entropy function (69).
denotes the frequency of the -th amino acid of this residue, N denotes the total number of all possible amino acids (N = 20), and denotes the total number of the -th amino acid of this residue. The theoretical maximum value of is log2(20) ≈ 4.32, which indicates the most variable; and the minimal value of is 0, which indicates the most conserved. We then normalized all entropy scores into [0,1] by dividing all entropy scores by the theoretical maximum value (in practices, we set it to 4.32). All residues were divided into 6 groups with different variation rates: 0, (0,0.2], (0.2,0.4], (0.4,0.6], (0.6,0.8] and (0.8,1]. As shown in Figure 1, we assigned gradient colors for those 6 groups to visualize single residue conservation on the structure of strain A/California/04/2009 HA (PDB ID: 4jtv). Ratio of minor amino acid, which represents variability of a given residue of HA (where 0 indicates 100% conservation and 1 indicates 0% conservation), was calculated based on the sequences of viruses listed in table S7.
Δ-1 Major Antigenic Site Staining
Δ-1 mutant HAs were generated by substituting the major antigenic sites of A/Michigan/45/2015 H1N1 with those of A/Vietnam/1203/2004 H5 or A/black headed gull/Sweden/1/1999 H13 using reverse genetics, as previously described (11). Δ-1 mutant HAs HEK293T cells were plated into a 96-well plate and transfected the following day with 100 ng Δ-1 mutant HA plasmid and TransIT-LT1 (MirusBio) for 24 hours. Cells were fixed with methanol overnight at −20°C. Cells were washed 3x with PBS and blocked with 5% milk powder in PBS. Full-length mAbs were diluted 5–20 μg/ml in blocking buffer and added to PBS-washed cells for 2 hours. Cells were washed 3x with PBS and stained with a goat anti-human IgG AF488 (Invitrogen) for 1 hour. Cells were washed 3x with PBS and imaged with an EVOS inverted fluorescence microscope (AMG).
In vivo challenge infections
MAb cocktails (Table S6) were passively transferred into 6- to 8-week-old female BALB/c mice (Jackson Laboratories) by intraperitoneal injection of 0.2, 1, and 5 mg/kg mAb cocktail. Negative control mice received 5 mg/kg of the anthrax-specific mAb 003-15D03 by intraperitoneal injection as an isotype control. Two hours post-mAb injection, mice were anesthetized with isoflurane and intranasally challenged with 10 LD50 of mouse-adapted A/Netherlands/602/2009 H1N1 virus. As a read out, survival and weight loss were monitored 1–2 times daily for two weeks. Mice were euthanized upon 25% weight loss. All experiments were done in accordance with the University of Chicago Institutional Animal Care and Use Committee.
Statistical analysis
All statistical analyses were performed using Prism software (Graphpad Version 9.0). Sample sizes (n) for the number of mAbs tested are indicated in corresponding figures, and number of biological repeats for experiments and specific tests for statistical significance used are indicated in the corresponding figure legends. P values less than or equal to 0.05 were considered significant. * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001, **** P < 0.0001.
Supplementary Material
Acknowledgments:
We are thankful to all individuals who participated in this study. We thank Sarah Andrews, Rafi Ahmed, Jens Wrammert, and Karlynn Neu for initiating studies on the 2009 MIV, 2010 TIV, and 2014 QIV cohorts and Jiaolong Wang for technical assistance. We thank Ignacio Mena and Adolfo García-Sastre for providing the H1N2 virus used in this study and Nicholas Wu and Ian Wilson for providing recombinant HA and feedback on experimental design.
Funding:
This project was funded in part by the National Institutes of Health grant numbers U19AI109946 (P.C.W.), U19AI057266 (P.C.W.), P01 AI097092 (P.P.), R01AI145870-01 (P.P.), T32HL007605-35 (J.J.G.), and T32AI007244-36 (J.H.). This work was also partially supported by the National Institute of Allergy and Infectious Diseases (NIAID) Centers of Excellence for Influenza Research and Surveillance (CEIRS) grant numbers HHSN272201400005C (P.C.W.) and HHSN272201400008C (F.K., R.N., P.P.) and the NIAID Collaborative Influenza Vaccine Innovation Centers (CIVIC) contract 75N93019C00051 (F.K., P.P., A.B.W., P.C.W.). Icahn School of Medicine at Mount Sinai received funds from GlaxoSmithKline to study universal influenza virus vaccine development. Structural figures and analyses were performed with UCSF Chimera; developers received support from the National Institutes of Health R01-GM129325 and P41-GM103311 and the Office of Cyber Infrastructure and Computational Biology (NIAID).
Footnotes
Competing interests: F.K. reports consulting for Curevac, Merck, Pfizer, Seqirus, and Avimex on topics unrelated to this manuscript, and consulting for Dynavax and GlaxoSmithKline on the development of influenza virus vaccines. A.F., F.K., P.P., R.N. are inventors on a patent application (63002682) submitted by the University of Pennsylvania regarding influenza virus vaccines. F.K., P.P., and R.N. are listed as inventors on a patent (20190125859) submitted by the Icahn School of Medicine at Mount Sinai regarding influenza virus vaccines. A.F. and R.N. are currently employed by Moderna, Inc.
Data and materials availability:
All data to understand the conclusions of this research are available in the main text, supplementary materials, and the following repositories: Heavy and light chain mAb sequences specific to this study are deposited in GenBank (MW857987–MW858070) and all mAb accession numbers are summarized in Data file S2. Electron microscopy maps are deposited on to Electron Microscopy Data Bank (EMD-23792 – EMD23978) and the 045–09 2B05 + HA model to is deposited on Protein Data Bank (PDB: 7MEM).
References:
- 1.Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, Lee JH, Dillon MA, O’Neil RE, Faynboym AM, Horowitz M, Horowitz L, Ward AB, Palese P, Webby R, Lerner RA, Bhatt RR, Wilson IA, Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489, 526–532 (2012); published online EpubSep 27 ( 10.1038/nature11414). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, Kepler TB, Liao HX, Harrison SC, Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A 108, 14216–14221 (2011); published online EpubAug 23 ( 10.1073/pnas.1111497108). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raymond DD, Bajic G, Ferdman J, Suphaphiphat P, Settembre EC, Moody MA, Schmidt AG, Harrison SC, Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody. Proc Natl Acad Sci U S A 115, 168–173 (2018); published online EpubJan 2 ( 10.1073/pnas.1715471115). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Linderman SL, Chambers BS, Zost SJ, Parkhouse K, Li Y, Herrmann C, Ellebedy AH, Carter DM, Andrews SF, Zheng NY, Huang M, Huang Y, Strauss D, Shaz BH, Hodinka RL, Reyes-Teran G, Ross TM, Wilson PC, Ahmed R, Bloom JD, Hensley SE, Potential antigenic explanation for atypical H1N1 infections among middle-aged adults during the 2013–2014 influenza season. Proc Natl Acad Sci U S A 111, 15798–15803 (2014); published online EpubNov 4 ( 10.1073/pnas.1409171111). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang KY, Rijal P, Schimanski L, Powell TJ, Lin TY, McCauley JW, Daniels RS, Townsend AR, Focused antibody response to influenza linked to antigenic drift. J Clin Invest 125, 2631–2645 (2015); published online EpubJul 1 ( 10.1172/JCI81104). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirkpatrick E, Qiu X, Wilson PC, Bahl J, Krammer F, The influenza virus hemagglutinin head evolves faster than the stalk domain. Sci Rep 8, 10432 (2018); published online EpubJul 11 ( 10.1038/s41598-018-28706-1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henry C, Zheng NY, Huang M, Cabanov A, Rojas KT, Kaur K, Andrews SF, Palm AE, Chen YQ, Li Y, Hoskova K, Utset HA, Vieira MC, Wrammert J, Ahmed R, Holden-Wiltse J, Topham DJ, Treanor JJ, Ertl HC, Schmader KE, Cobey S, Krammer F, Hensley SE, Greenberg H, He XS, Wilson PC, Influenza Virus Vaccination Elicits Poorly Adapted B Cell Responses in Elderly Individuals. Cell Host Microbe 25, 357–366 e356 (2019); published online EpubMar 13 ( 10.1016/j.chom.2019.01.002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrews SF, Huang Y, Kaur K, Popova LI, Ho IY, Pauli NT, Henry Dunand CJ, Taylor WM, Lim S, Huang M, Qu X, Lee JH, Salgado-Ferrer M, Krammer F, Palese P, Wrammert J, Ahmed R, Wilson PC, Immune history profoundly affects broadly protective B cell responses to influenza. Sci Transl Med 7, 316ra192 (2015); published online EpubDec 02 ( 10.1126/scitranslmed.aad0522). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manicassamy B, Medina RA, Hai R, Tsibane T, Stertz S, Nistal-Villan E, Palese P, Basler CF, Garcia-Sastre A, Protection of mice against lethal challenge with 2009 H1N1 influenza A virus by 1918-like and classical swine H1N1 based vaccines. PLoS Pathog 6, e1000745 (2010); published online EpubJan 29 ( 10.1371/journal.ppat.1000745). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE Jr., Wilson IA, Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science 328, 357–360 (2010); published online EpubApr 16 ( 10.1126/science.1186430). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu STH, Behzadi MA, Sun W, Freyn AW, Liu WC, Broecker F, Albrecht RA, Bouvier NM, Simon V, Nachbagauer R, Krammer F, Palese P, Antigenic sites in influenza H1 hemagglutinin display species-specific immunodominance. J Clin Invest 128, 4992–4996 (2018); published online EpubNov 1 ( 10.1172/JCI122895). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nogales A, Piepenbrink MS, Wang J, Ortega S, Basu M, Fucile CF, Treanor JJ, Rosenberg AF, Zand MS, Keefer MC, Martinez-Sobrido L, Kobie JJ, A Highly Potent and Broadly Neutralizing H1 Influenza-Specific Human Monoclonal Antibody. Sci Rep 8, 4374 (2018); published online EpubMar 12 ( 10.1038/s41598-018-22307-8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castelan-Vega JA, Magana-Hernandez A, Jimenez-Alberto A, Ribas-Aparicio RM, The hemagglutinin of the influenza A(H1N1)pdm09 is mutating towards stability. Adv Appl Bioinform Chem 7, 37–44 (2014) 10.2147/AABC.S68934). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt AG, Therkelsen MD, Stewart S, Kepler TB, Liao HX, Moody MA, Haynes BF, Harrison SC, Viral receptor-binding site antibodies with diverse germline origins. Cell 161, 1026–1034 (2015); published online EpubMay 21 ( 10.1016/j.cell.2015.04.028). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong M, Lee PS, Hoffman RM, Zhu X, Krause JC, Laursen NS, Yoon SI, Song L, Tussey L, Crowe JE Jr., Ward AB, Wilson IA, Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. J Virol 87, 12471–12480 (2013); published online EpubNov ( 10.1128/JVI.01388-13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mena I, Nelson MI, Quezada-Monroy F, Dutta J, Cortes-Fernandez R, Lara-Puente JH, Castro-Peralta F, Cunha LF, Trovao NS, Lozano-Dubernard B, Rambaut A, van Bakel H, Garcia-Sastre A, Origins of the 2009 H1N1 influenza pandemic in swine in Mexico. Elife 5, (2016); published online EpubJun 28 ( 10.7554/eLife.16777). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, Mehta A, Razavi B, Del Rio C, Zheng NY, Lee JH, Huang M, Ali Z, Kaur K, Andrews S, Amara RR, Wang Y, Das SR, O’Donnell CD, Yewdell JW, Subbarao K, Marasco WA, Mulligan MJ, Compans R, Ahmed R, Wilson PC, Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med 208, 181–193 (2011); published online EpubJan 17 ( 10.1084/jem.20101352). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li GM, Chiu C, Wrammert J, McCausland M, Andrews SF, Zheng NY, Lee JH, Huang M, Qu X, Edupuganti S, Mulligan M, Das SR, Yewdell JW, Mehta AK, Wilson PC, Ahmed R, Pandemic H1N1 influenza vaccine induces a recall response in humans that favors broadly cross-reactive memory B cells. Proc Natl Acad Sci U S A 109, 9047–9052 (2012); published online EpubJun 05 ( 10.1073/pnas.1118979109). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guthmiller JJ, Lan LY, Fernandez-Quintero ML, Han J, Utset HA, Bitar DJ, Hamel NJ, Stovicek O, Li L, Tepora M, Henry C, Neu KE, Dugan HL, Borowska MT, Chen YQ, Liu STH, Stamper CT, Zheng NY, Huang M, Palm AE, Garcia-Sastre A, Nachbagauer R, Palese P, Coughlan L, Krammer F, Ward AB, Liedl KR, Wilson PC, Polyreactive Broadly Neutralizing B cells Are Selected to Provide Defense against Pandemic Threat Influenza Viruses. Immunity 53, 1230–1244 e1235 (2020); published online EpubDec 15 ( 10.1016/j.immuni.2020.10.005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dugan HL, Guthmiller JJ, Arevalo P, Huang M, Chen YQ, Neu KE, Henry C, Zheng NY, Lan LY, Tepora ME, Stovicek O, Bitar D, Palm AE, Stamper CT, Changrob S, Utset HA, Coughlan L, Krammer F, Cobey S, Wilson PC, Preexisting immunity shapes distinct antibody landscapes after influenza virus infection and vaccination in humans. Sci Transl Med 12, (2020); published online EpubDec 9 ( 10.1126/scitranslmed.abd3601). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henry Dunand CJ, Leon PE, Huang M, Choi A, Chromikova V, Ho IY, Tan GS, Cruz J, Hirsh A, Zheng NY, Mullarkey CE, Ennis FA, Terajima M, Treanor JJ, Topham DJ, Subbarao K, Palese P, Krammer F, Wilson PC, Both Neutralizing and Non-Neutralizing Human H7N9 Influenza Vaccine-Induced Monoclonal Antibodies Confer Protection. Cell Host Microbe 19, 800–813 (2016); published online EpubJun 08 ( 10.1016/j.chom.2016.05.014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellebedy AH, Krammer F, Li GM, Miller MS, Chiu C, Wrammert J, Chang CY, Davis CW, McCausland M, Elbein R, Edupuganti S, Spearman P, Andrews SF, Wilson PC, Garcia-Sastre A, Mulligan MJ, Mehta AK, Palese P, Ahmed R, Induction of broadly cross-reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans. Proc Natl Acad Sci U S A 111, 13133–13138 (2014); published online EpubSep 09 ( 10.1073/pnas.1414070111). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andrews SF, Chambers MJ, Schramm CA, Plyler J, Raab JE, Kanekiyo M, Gillespie RA, Ransier A, Darko S, Hu J, Chen X, Yassine HM, Boyington JC, Crank MC, Chen GL, Coates E, Mascola JR, Douek DC, Graham BS, Ledgerwood JE, McDermott AB, Activation Dynamics and Immunoglobulin Evolution of Pre-existing and Newly Generated Human Memory B cell Responses to Influenza Hemagglutinin. Immunity 51, 398–410 e395 (2019); published online EpubAug 20 ( 10.1016/j.immuni.2019.06.024). [DOI] [PubMed] [Google Scholar]
- 24.Andrews SF, Joyce MG, Chambers MJ, Gillespie RA, Kanekiyo M, Leung K, Yang ES, Tsybovsky Y, Wheatley AK, Crank MC, Boyington JC, Prabhakaran MS, Narpala SR, Chen X, Bailer RT, Chen G, Coates E, Kwong PD, Koup RA, Mascola JR, Graham BS, Ledgerwood JE, McDermott AB, Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans. Sci Immunol 2, (2017); published online EpubJul 14 ( 10.1126/sciimmunol.aan2676). [DOI] [PubMed] [Google Scholar]
- 25.Wu NC, Thompson AJ, Lee JM, Su W, Arlian BM, Xie J, Lerner RA, Yen HL, Bloom JD, Wilson IA, Different genetic barriers for resistance to HA stem antibodies in influenza H3 and H1 viruses. Science 368, 1335–1340 (2020); published online EpubJun 19 ( 10.1126/science.aaz5143). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park JK, Xiao Y, Ramuta MD, Rosas LA, Fong S, Matthews AM, Freeman AD, Gouzoulis MA, Batchenkova NA, Yang X, Scherler K, Qi L, Reed S, Athota R, Czajkowski L, Han A, Morens DM, Walters KA, Memoli MJ, Kash JC, Taubenberger JK, Pre-existing immunity to influenza virus hemagglutinin stalk might drive selection for antibody-escape mutant viruses in a human challenge model. Nat Med, (2020); published online EpubJun 29 ( 10.1038/s41591-020-0937-x). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han J, Schmitz AJ, Richey ST, Dai YN, Turner HL, Mohammed BM, Fremont DH, Ellebedy AH, Ward AB, Polyclonal epitope mapping reveals temporal dynamics and diversity of human antibody responses to H5N1 vaccination. Cell Rep 34, 108682 (2021); published online EpubJan 26 ( 10.1016/j.celrep.2020.108682). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turner JS, Zhou JQ, Han J, Schmitz AJ, Rizk AA, Alsoussi WB, Lei T, Amor M, McIntire KM, Meade P, Strohmeier S, Brent RI, Richey ST, Haile A, Yang YR, Klebert MK, Suessen T, Teefey S, Presti RM, Krammer F, Kleinstein SH, Ward AB, Ellebedy AH, Human germinal centres engage memory and naive B cells after influenza vaccination. Nature 586, 127–132 (2020); published online EpubOct ( 10.1038/s41586-020-2711-0). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zarnitsyna VI, Ellebedy AH, Davis C, Jacob J, Ahmed R, Antia R, Masking of antigenic epitopes by antibodies shapes the humoral immune response to influenza. Philos Trans R Soc Lond B Biol Sci 370, (2015); published online EpubSep 5 ( 10.1098/rstb.2014.0248). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zarnitsyna VI, Lavine J, Ellebedy A, Ahmed R, Antia R, Multi-epitope Models Explain How Pre-existing Antibodies Affect the Generation of Broadly Protective Responses to Influenza. PLoS Pathog 12, e1005692 (2016); published online EpubJun ( 10.1371/journal.ppat.1005692). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Angeletti D, Kosik I, Santos JJS, Yewdell WT, Boudreau CM, Mallajosyula VVA, Mankowski MC, Chambers M, Prabhakaran M, Hickman HD, McDermott AB, Alter G, Chaudhuri J, Yewdell JW, Outflanking immunodominance to target subdominant broadly neutralizing epitopes. Proc Natl Acad Sci U S A 116, 13474–13479 (2019); published online EpubJul 2 ( 10.1073/pnas.1816300116). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan HX, Jegaskanda S, Juno JA, Esterbauer R, Wong J, Kelly HG, Liu Y, Tilmanis D, Hurt AC, Yewdell JW, Kent SJ, Wheatley AK, Subdominance and poor intrinsic immunogenicity limit humoral immunity targeting influenza HA stem. J Clin Invest 129, 850–862 (2019); published online EpubFeb 1 ( 10.1172/JCI123366). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis CW, Jackson KJL, McCausland MM, Darce J, Chang C, Linderman SL, Chennareddy C, Gerkin R, Brown SJ, Wrammert J, Mehta AK, Cheung WC, Boyd SD, Waller EK, Ahmed R, Influenza vaccine-induced human bone marrow plasma cells decline within a year after vaccination. Science 370, 237–241 (2020); published online EpubOct 9 ( 10.1126/science.aaz8432). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt AG, Do KT, McCarthy KR, Kepler TB, Liao HX, Moody MA, Haynes BF, Harrison SC, Immunogenic Stimulus for Germline Precursors of Antibodies that Engage the Influenza Hemagglutinin Receptor-Binding Site. Cell Rep 13, 2842–2850 (2015); published online EpubDec 29 ( 10.1016/j.celrep.2015.11.063). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarthy KR, Watanabe A, Kuraoka M, Do KT, McGee CE, Sempowski GD, Kepler TB, Schmidt AG, Kelsoe G, Harrison SC, Memory B Cells that Cross-React with Group 1 and Group 2 Influenza A Viruses Are Abundant in Adult Human Repertoires. Immunity 48, 174–184 e179 (2018); published online EpubJan 16 ( 10.1016/j.immuni.2017.12.009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ni F, Kondrashkina E, Wang Q, Determinant of receptor-preference switch in influenza hemagglutinin. Virology 513, 98–107 (2018); published online EpubJan 1 ( 10.1016/j.virol.2017.10.010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koel BF, Burke DF, Bestebroer TM, van der Vliet S, Zondag GC, Vervaet G, Skepner E, Lewis NS, Spronken MI, Russell CA, Eropkin MY, Hurt AC, Barr IG, de Jong JC, Rimmelzwaan GF, Osterhaus AD, Fouchier RA, Smith DJ, Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 342, 976–979 (2013); published online EpubNov 22 ( 10.1126/science.1244730). [DOI] [PubMed] [Google Scholar]
- 38.Garretson TA, Petrie JG, Martin ET, Monto AS, Hensley SE, Identification of human vaccinees that possess antibodies targeting the egg-adapted hemagglutinin receptor binding site of an H1N1 influenza vaccine strain. Vaccine 36, 4095–4101 (2018); published online EpubJun 27 ( 10.1016/j.vaccine.2018.05.086). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raymond DD, Stewart SM, Lee J, Ferdman J, Bajic G, Do KT, Ernandes MJ, Suphaphiphat P, Settembre EC, Dormitzer PR, Del Giudice G, Finco O, Kang TH, Ippolito GC, Georgiou G, Kepler TB, Haynes BF, Moody MA, Liao HX, Schmidt AG, Harrison SC, Influenza immunization elicits antibodies specific for an egg-adapted vaccine strain. Nat Med 22, 1465–1469 (2016); published online EpubDec ( 10.1038/nm.4223). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Czerwinski M, Siemaszko D, Siegel DL, Spitalnik SL, Only selected light chains combine with a given heavy chain to confer specificity for a model glycopeptide antigen. J Immunol 160, 4406–4417 (1998); published online EpubMay 1( [PubMed] [Google Scholar]
- 41.Tan TJC, Yuan M, Kuzelka K, Padron GC, Beal JR, Chen X, Wang Y, Rivera-Cardona J, Zhu X, Stadtmueller BM, Brooke CB, Wilson IA, Wu NC, Sequence signatures of two IGHV3-53/3-66 public clonotypes to SARS-CoV-2 receptor binding domain. bioRxiv, (2021); published online EpubJan 27 ( 10.1101/2021.01.26.428356). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bajic G, Maron MJ, Adachi Y, Onodera T, McCarthy KR, McGee CE, Sempowski GD, Takahashi Y, Kelsoe G, Kuraoka M, Schmidt AG, Influenza Antigen Engineering Focuses Immune Responses to a Subdominant but Broadly Protective Viral Epitope. Cell Host Microbe 25, 827–835 e826 (2019); published online EpubJun 12 ( 10.1016/j.chom.2019.04.003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bangaru S, Lang S, Schotsaert M, Vanderven HA, Zhu X, Kose N, Bombardi R, Finn JA, Kent SJ, Gilchuk P, Gilchuk I, Turner HL, Garcia-Sastre A, Li S, Ward AB, Wilson IA, Crowe JE Jr., A Site of Vulnerability on the Influenza Virus Hemagglutinin Head Domain Trimer Interface. Cell 177, 1136–1152 e1118 (2019); published online EpubMay 16 ( 10.1016/j.cell.2019.04.011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watanabe A, McCarthy KR, Kuraoka M, Schmidt AG, Adachi Y, Onodera T, Tonouchi K, Caradonna TM, Bajic G, Song S, McGee CE, Sempowski GD, Feng F, Urick P, Kepler TB, Takahashi Y, Harrison SC, Kelsoe G, Antibodies to a Conserved Influenza Head Interface Epitope Protect by an IgG Subtype-Dependent Mechanism. Cell 177, 1124–1135 e1116 (2019); published online EpubMay 16 ( 10.1016/j.cell.2019.03.048). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nachbagauer R, Feser J, Naficy A, Bernstein DI, Guptill J, Walter EB, Berlanda-Scorza F, Stadlbauer D, Wilson PC, Aydillo T, Behzadi MA, Bhavsar D, Bliss C, Capuano C, Carreno JM, Chromikova V, Claeys C, Coughlan L, Freyn AW, Gast C, Javier A, Jiang K, Mariottini C, McMahon M, McNeal M, Solorzano A, Strohmeier S, Sun W, Van der Wielen M, Innis BL, Garcia-Sastre A, Palese P, Krammer F, A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nat Med, (2020); published online EpubDec 7 ( 10.1038/s41591-020-1118-7). [DOI] [PubMed] [Google Scholar]
- 46.Broecker F, Liu STH, Suntronwong N, Sun W, Bailey MJ, Nachbagauer R, Krammer F, Palese P, A mosaic hemagglutinin-based influenza virus vaccine candidate protects mice from challenge with divergent H3N2 strains. NPJ Vaccines 4, 31 (2019) 10.1038/s41541-019-0126-4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun W, Kirkpatrick E, Ermler M, Nachbagauer R, Broecker F, Krammer F, Palese P, Development of Influenza B Universal Vaccine Candidates Using the “Mosaic” Hemagglutinin Approach. J Virol 93, (2019); published online EpubJun 15 ( 10.1128/JVI.00333-19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weidenbacher PA, Kim PS, Protect, modify, deprotect (PMD): A strategy for creating vaccines to elicit antibodies targeting a specific epitope. Proc Natl Acad Sci U S A 116, 9947–9952 (2019); published online EpubMay 14 ( 10.1073/pnas.1822062116). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanekiyo M, Joyce MG, Gillespie RA, Gallagher JR, Andrews SF, Yassine HM, Wheatley AK, Fisher BE, Ambrozak DR, Creanga A, Leung K, Yang ES, Boyoglu-Barnum S, Georgiev IS, Tsybovsky Y, Prabhakaran MS, Andersen H, Kong WP, Baxa U, Zephir KL, Ledgerwood JE, Koup RA, Kwong PD, Harris AK, McDermott AB, Mascola JR, Graham BS, Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat Immunol 20, 362–372 (2019); published online EpubMar ( 10.1038/s41590-018-0305-x). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neu KE, Guthmiller JJ, Huang M, La J, Vieira MC, Kim K, Zheng NY, Cortese M, Tepora ME, Hamel NJ, Rojas KT, Henry C, Shaw D, Dulberger CL, Pulendran B, Cobey S, Khan AA, Wilson PC, Spec-seq unveils transcriptional subpopulations of antibody-secreting cells following influenza vaccination. J Clin Invest 129, 93–105 (2019); published online EpubJan 2 ( 10.1172/JCI121341). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brauer R, Chen P, Influenza virus propagation in embryonated chicken eggs. J Vis Exp, (2015); published online EpubMar 19 ( 10.3791/52421). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guthmiller JJ, Dugan HL, Neu KE, Lan LY, Wilson PC, An Efficient Method to Generate Monoclonal Antibodies from Human B Cells. Methods Mol Biol 1904, 109–145 (2019) 10.1007/978-1-4939-8958-4_5). [DOI] [PubMed] [Google Scholar]
- 53.Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng NY, Mays I, Garman L, Helms C, James J, Air GM, Capra JD, Ahmed R, Wilson PC, Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature 453, 667–671 (2008); published online EpubMay 29 ( 10.1038/nature06890). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith K, Garman L, Wrammert J, Zheng NY, Capra JD, Ahmed R, Wilson PC, Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat Protoc 4, 372–384 (2009) 10.1038/nprot.2009.3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B, Automated molecular microscopy: the new Leginon system. J Struct Biol 151, 41–60 (2005); published online EpubJul ( 10.1016/j.jsb.2005.03.010). [DOI] [PubMed] [Google Scholar]
- 56.Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, Lyumkis D, Potter CS, Carragher B, Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol 166, 95–102 (2009); published online EpubApr ( 10.1016/j.jsb.2009.01.002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scheres SH, RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530 (2012); published online EpubDec ( 10.1016/j.jsb.2012.09.006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE, UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 (2004); published online EpubOct ( 10.1002/jcc.20084). [DOI] [PubMed] [Google Scholar]
- 59.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA, MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017); published online EpubApr ( 10.1038/nmeth.4193). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296 (2017); published online EpubMar ( 10.1038/nmeth.4169). [DOI] [PubMed] [Google Scholar]
- 61.Lyskov S, Chou FC, Conchuir SO, Der BS, Drew K, Kuroda D, Xu J, Weitzner BD, Renfrew PD, Sripakdeevong P, Borgo B, Havranek JJ, Kuhlman B, Kortemme T, Bonneau R, Gray JJ, Das R, Serverification of molecular modeling applications: the Rosetta Online Server that Includes Everyone (ROSIE). PLoS One 8, e63906 (2013) 10.1371/journal.pone.0063906). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emsley P, Cowtan K, Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132 (2004); published online EpubDec ( 10.1107/S0907444904019158). [DOI] [PubMed] [Google Scholar]
- 63.Weitzner BD, Jeliazkov JR, Lyskov S, Marze N, Kuroda D, Frick R, Adolf-Bryfogle J, Biswas N, Dunbrack RL Jr., Gray JJ, Modeling and docking of antibody structures with Rosetta. Nat Protoc 12, 401–416 (2017); published online EpubFeb ( 10.1038/nprot.2016.180). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC, MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21 (2010); published online EpubJan ( 10.1107/S0907444909042073). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barad BA, Echols N, Wang RY, Cheng Y, DiMaio F, Adams PD, Fraser JS, EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat Methods 12, 943–946 (2015); published online EpubOct ( 10.1038/nmeth.3541). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Henry Dunand CJ, Leon PE, Kaur K, Tan GS, Zheng NY, Andrews S, Huang M, Qu X, Huang Y, Salgado-Ferrer M, Ho IY, Taylor W, Hai R, Wrammert J, Ahmed R, Garcia-Sastre A, Palese P, Krammer F, Wilson PC, Preexisting human antibodies neutralize recently emerged H7N9 influenza strains. J Clin Invest 125, 1255–1268 (2015); published online EpubMar 2 ( 10.1172/JCI74374). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen YQ, Wohlbold TJ, Zheng NY, Huang M, Huang Y, Neu KE, Lee J, Wan H, Rojas KT, Kirkpatrick E, Henry C, Palm AE, Stamper CT, Lan LY, Topham DJ, Treanor J, Wrammert J, Ahmed R, Eichelberger MC, Georgiou G, Krammer F, Wilson PC, Influenza Infection in Humans Induces Broadly Cross-Reactive and Protective Neuraminidase-Reactive Antibodies. Cell 173, 417–429 e410 (2018); published online EpubApr 5 ( 10.1016/j.cell.2018.03.030). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leon PE, Wohlbold TJ, He W, Bailey MJ, Henry CJ, Wilson PC, Krammer F, Tan GS, Generation of Escape Variants of Neutralizing Influenza Virus Monoclonal Antibodies. J Vis Exp, (2017); published online EpubAug 29 ( 10.3791/56067). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Crooks GE, Hon G, Chandonia JM, Brenner SE, WebLogo: a sequence logo generator. Genome Res 14, 1188–1190 (2004); published online EpubJun ( 10.1101/gr.849004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data to understand the conclusions of this research are available in the main text, supplementary materials, and the following repositories: Heavy and light chain mAb sequences specific to this study are deposited in GenBank (MW857987–MW858070) and all mAb accession numbers are summarized in Data file S2. Electron microscopy maps are deposited on to Electron Microscopy Data Bank (EMD-23792 – EMD23978) and the 045–09 2B05 + HA model to is deposited on Protein Data Bank (PDB: 7MEM).


