Abstract
Synthetic biology aims to design or assemble existing bioparts or bio-components for useful bioproperties. During the past decades, progresses have been made to build delicate biocircuits, standardized biological building blocks and to develop various genomic/metabolic engineering tools and approaches. Medical and pharmaceutical demands have also pushed the development of synthetic biology, including integration of heterologous pathways into designer cells to efficiently produce medical agents, enhanced yields of natural products in cell growth media to equal or higher than that of the extracts from plants or fungi, constructions of novel genetic circuits for tumor targeting, controllable releases of therapeutic agents in response to specific biomarkers to fight diseases such as diabetes and cancers. Besides, new strategies are developed to treat complex immune diseases, infectious diseases and metabolic disorders that are hard to cure via traditional approaches. In general, synthetic biology brings new capabilities to medical and pharmaceutical researches. This review summarizes the timeline of synthetic biology developments, the past and present of synthetic biology for microbial productions of pharmaceutics, engineered cells equipped with synthetic DNA circuits for diagnosis and therapies, live and auto-assemblied biomaterials for medical treatments, cell-free synthetic biology in medical and pharmaceutical fields, and DNA engineering approaches with potentials for biomedical applications.
Subject terms: Biotechnology, Nanobiotechnology
Introduction
The concept of synthetic biology was proposed in 1910s by Stephane Le Duc.1 In this field, research strategies have been changed from the description and analysis of biological events to design and manipulate desired signal/metabolic routes, similar to the already defined organic synthesis. Unlike organic synthesis successfully developed in the early 19th century,2 synthetic biology is restricted by DNA, RNA and protein technology within the complexity of biological systems. Today, synthetic biology has been developed extensively. It becomes a multidisciplinary field aims to develop new biological parts, systems, or even individuals based on existing knowledge. Researchers can apply the engineering paradigm to produce predictable and robust systems with novel functionalities that do not exist in nature. Synthetic biology is tightly connected with many subjects including biotechnology, biomaterials and molecular biology, providing methodology and disciplines to these fields.
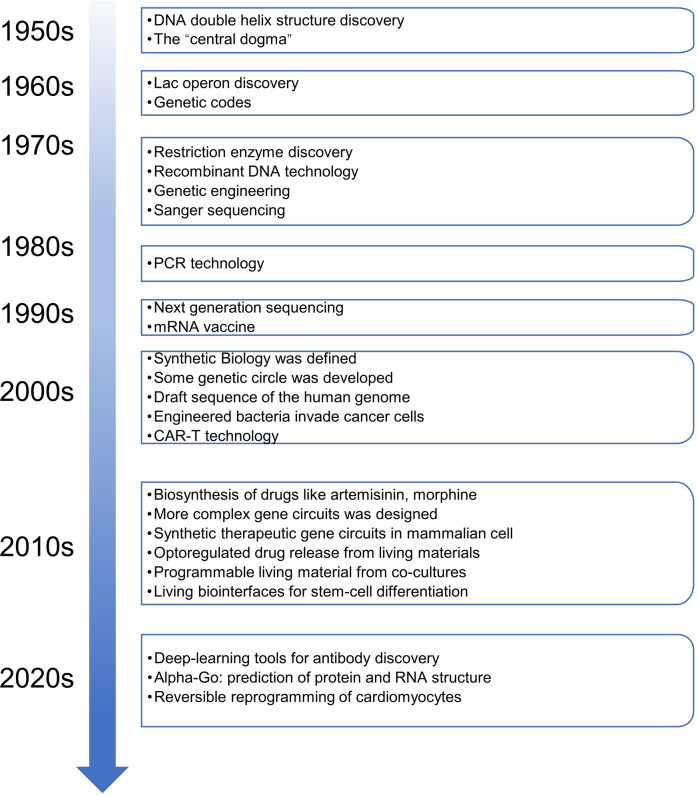
The timeline of synthetic biology developments is summarized here (Fig. 1). In general, the history of synthetic biology can be divided into three stages. The initial stage was found across the 20th century. Although the simplest organisms such as virus particles, bacteria, archaea and fungi were hard to engineer in the 20th century, some achievements were still acquired in the early explorations including the synthesis of crystalline bovine insulin,3 chemical synthesis of DNA and RNA,4 decoding of genetic codes5 and the establishment of central dogma of molecular biology.6 Synthetic biology has been accumulating its strengths in this period, as knowledge of genome biology and molecular biology are developed rapidly at the end of the 20th century (Fig. 1).
Fig. 1.
Timeline of major milestones in synthetic biology. The timeline begins at 1950s and expands to 2020s. Important events are listed in the right panels
The development stage begins in the 21st century. In the first decade of the new millennium, synthetic biology is known to every biological researcher to include inventions of bioswitches,7 gene circuits based on quorum sensing signals,8 yeast cell-factory for amorphadiene synthesis9 (Table 1), BioBrick standardized assembly10 and the iGEM conferences11 (Fig. 1). Two principles in synthetic biology designs have been considered in this stage including bottle-up12 and top-down13 ones, referring to the de novo creations of artificial lives by assembling basic biological molecules and engineering natural-existed cells to meet actual demands, respectively. However, most circuits are well-designed but still not enough for producing complex metabolites or sensing multiple signals, especially the applications are not well prepared for medical and pharmaceutical usages. Anyhow, synthetic biology is gradually becoming a most topical area, on the eve of rapid developments.
Table 1.
Applications and yields of biosynthesized pharmaceuticals
| Application | Classification | Production host | Titer (g/L) | Year | Ref. | |
|---|---|---|---|---|---|---|
| Artemisic acid | A precursor of anti-malarial drug artemisinin | Terpenoids | Saccharomyces cerevisiae | 25 | 2013 | 23 |
| Thebaine | Pain management and palliative care | Alkaloids | E. coli/Saccharomyces cerevisiae | 2.1 × 10−3/6.6 × 10−5 | 2016/2015 | 323,324 |
| Hydrocodone | Pain management and palliative care | Alkaloids | E. coli/Saccharomyces cerevisiae | 4 × 10−5/3 × 10−7 | 2016/2015 | 323,324 |
| Codeine | Treat severe pain | Alkaloids | E. coli | 304 µg L−1 OD−1 | 2019 | 396 |
| Sitagliptin | Increased insulin secretion | Amines | In vitro | N.A.a | 2010 | 337 |
| Ginsenoside Rh2 | Cancer prevention and therapy | Terpenoids | Saccharomyces cerevisiae | 2.2 | 2019 | 317 |
| Ginsenoside compound K | Increased resistance to stress and aging | Terpenoids | Saccharomyces cerevisiae | 1.4 × 10−3 / 1.17/5.0 | 2014/2020/2021 | 318,397,398 |
| Guaia-6,10(14)-diene | A precursor of kidney cancer drug Englerin A | Terpenoids | Saccharomyces cerevisiae | 0.8 | 2020 | 399 |
| Taxadiene | A precursor of cancer drug Taxol | Terpenoids | E. coli / Saccharomyces cerevisiae | 1.0 / 8.7 × 10−3 | 2010/2008 | 315,400 |
| Adenosylcobalamin (vitamin B12) | Vital cofactor for human | Corrinoids | E. coli | 307 µg g−1 DCWb | 2018 | 401 |
| Baicalein | Neuroprotective agent | Flavonoids | E. coli | 0.02 | 2019 | 402 |
| Miltiradiene | A precursor of cardiovascular diseases drug tanshinone | Terpenoids | Saccharomyces cerevisiae | 0.3 / 3.5 | 2012/2020 | 403,404 |
| Catharanthine | A precursor of anti-cancer drug vinblastine and vincristine | Alkaloids | Saccharomyces cerevisiae | 2.7 × 10−5 | 2022 | 405 |
| Breviscapine | Treat cardio- and cerebrovascular diseases | Flavonoids | Saccharomyces cerevisiae | 0.2 | 2018 | 406 |
| Scopolamine | Treat neuromuscular disorders | Alkaloids | Saccharomyces cerevisiae | 6 × 10−2 | 2020 | 407 |
| (S)-tetrahydropalmatine | Use as an analgesic and anxiolytic drug | Alkaloids | Saccharomyces cerevisiae | 3.6 × 10−6 | 2021 | 328 |
| Cannabigerolic acid | A precursor of various cannabinoids; reduce pain without hallucination | Alkaloids | Saccharomyces cerevisiae | 0.1 | 2019 | 327 |
| Triptolide | Treatment of rheumatoid arthritis | Terpenoids | Saccharomyces cerevisiae | 30.5 μg g−1 | 2020 | 408 |
| Psilocybin | Treatment of addiction, depression and post-traumatic stress disorder. | Amino acid derivatives | E. coli / Saccharomyces cerevisiae | 1.2 / 0.6 | 2019 / 2020 | 330,331 |
| Monacolin J | A precursor for simvastatin (Zocor), an important drug for treating hypercholesterolemia. | Polyketides | Aspergillus terreus | 4.7 | 2017 | 409 |
| Acarbose | Clinically used to treat patients with type 2 diabetes | oligosaccharide | Actinoplanes sp. | 7.4 | 2020 | 410 |
| α-Tocoterienol | Natural vitamin E, used as a valuable supplementation | Terpenoids | Saccharomyces cerevisiae | 0.3 | 2020 | 411 |
| Avermectin B1a | Widely used in the field of animal health, agriculture and human health | Polyketides | Streptomyces avermitilis | 6.4 | 2010 | 412 |
| Carnosic acid | Potent antioxidant and anticancer agents | Terpenoids | Saccharomyces cerevisiae | 1 × 10−3 | 2016 | 413 |
| Noscapine | Anticancer drug | Alkaloids | Saccharomyces cerevisiae | 2.2 × 10−3 | 2018 | 414 |
| Farnesene | Widely used in industry, a precursor of vitamin E | Terpenoids | Saccharomyces cerevisiae | 55.4 | 2022 | 415 |
| (−)-Deoxypodophyllotoxin | A precursor to anti-cancer drug etoposide | Alkaloids | Nicotiana benthamiana | 4.3 mg/g dry plant weight | 2019 | 416 |
| Dencichine | Promote aggregation of platelets | Amino acid derivatives | E. coli | 1.29 | 2022 | 334 |
aN.A. not applicable
bDCW the abbreviation of dry cell mass
The fast-growing stage begins from the 2010s, the emergences of genome editing technologies especially CRISPR/Cas9,14 low-cost DNA synthesis,15 next-generation DNA sequencing16 and high-throughput screening methods,17 workflows of design-build-test-learn (DBTL)18 and progresses in engineering biology19 (Fig. 1), have allowed synthetic biology to enter a fast-growing period,20 both in the lab-scale discoveries and industry-scale productions. Typically, Venter et al. assembled an artificial chromosome of Mycoplasma mycoides and transplanted it to M. capricolum to create new living cells.21 Besides, new methods have accelerated the discovery and engineering of metabolite biosynthesis pathways, microbial artemisinic acid synthesis has been made possible,22,23 which is the first industrialized plant metabolite produced by microbial cells. To realize the ultimate goal of design bio-systems similar to design electronic or mechanical systems, this is just the beginning. More efforts are needed to generate complex and stable biocircuits for various applications in the present of synthetic biology.
Besides scientists, investors also have realized the potentials in this field. Financial investments help establish synthetic-biology-related companies encouraged by the prediction that the global market of synthetic biology valued 9.5 billion dollars by 2021, including synthetic biology products (e.g., BioBrick parts, synthetic cells, biosynthesized chemicals) and enabling technologies (e.g., DNA synthesis, gene editing),24 they are expected to reach 37 billion dollars by 2026. Most investments focus on medical applications.25 Scientists and capital market are all optimistic about the future.
Started from chemical biosynthesis, synthetic biology has been expanded to cover areas in medical treatments, pharmaceutical developments, chemical engineering, food and agriculture, and environmental preservations. This paper focuses on the advances of synthetic biology in medical and pharmaceutical fields, including cell therapies, bacterial live diagnosis and therapeutics, production of therapeutic chemicals, nanotechnology and nanomaterial applications and targeted gene engineering.
Genetic engineering of therapeutic chassis
Engineered mammalian cells for medical applications
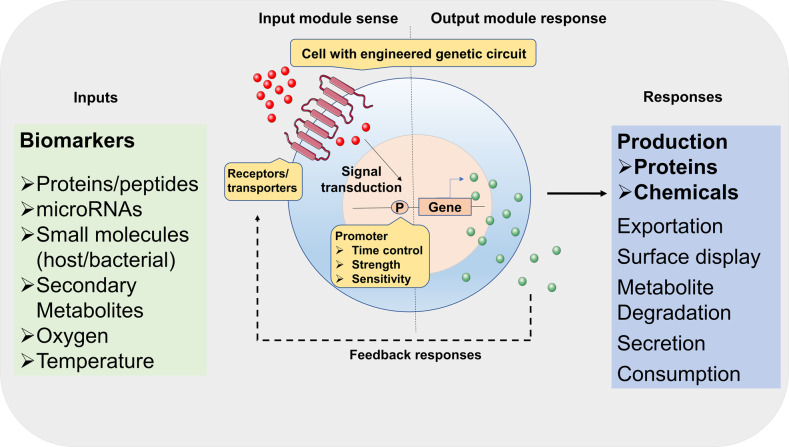
With the advances in synthetic biology, researchers created various novel therapies using living cell chassis rationally designed from existing signaling networks with new constructs for their purposes, including e.g., production of medical biomolecules, synthetic gene networks for sensing or diagnostics, and programmable organisms, to handle mechanisms underlying disease and related organism/individual events (Fig. 2). We highlighted here synthetic biology strategies in mammalian cell engineering for metabolic disorders, tissue engineering and cancer treatments, as well as approaches in cell therapy and the design of gene circuits.
Fig. 2.
Development of smart living cells based on synthetic biology strategies. Smart cells can sense various environmental biomarkers, from chemicals to proteins. External signals are transducted into cells to trigger downstream responses. The products are also in the form of chemicals to proteins for customized demands. The sensing-reponsing system is endowing cells with new or enhanced abilities. P represents promoters
Therapies based on chimeric antigen receptor (CAR)-T cells
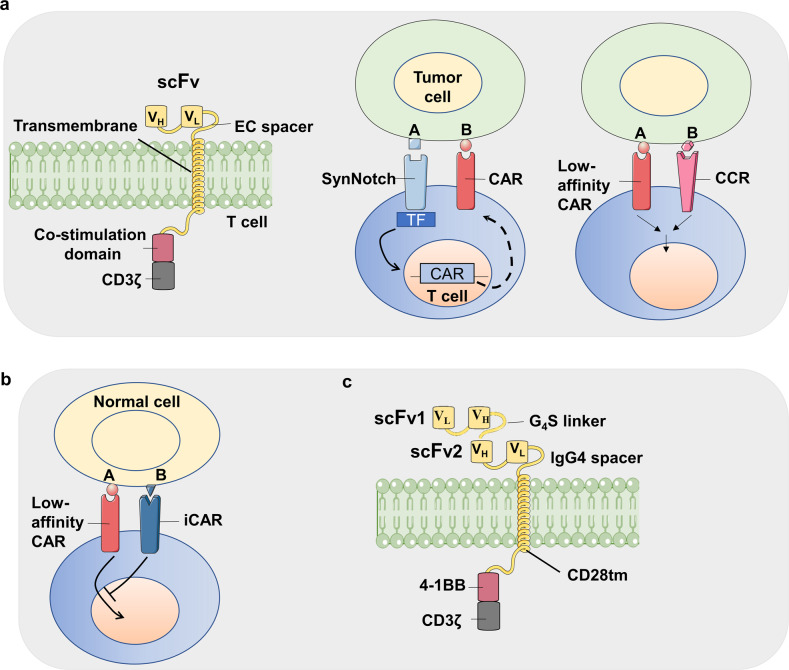
CARs are engineered receptors containing both antigen-binding and T cell-activating domains. T cells are acquired from patients and engineered ex vivo to express a specific CAR, and followed by transferring into the original donor patient, where they eliminate cancer cells that surface-displayed the target antigen.26 CAR-T is a novel cell therapy began from 2000s.27 The first generation of CARs are single-chain variable fragments (scFv) targeting CD19.28 The development of artificial CARs comprises three generations. The first-generation CARs only contain a CD3ζ intracellular domain, while the second-generation CARs also possess a co-stimulatory domain, e.g., 4-1BB or CD3ζ (Fig. 3). Studies with the third-generation chimeric antigen receptors with multiple co-stimulatory signaling domains are also under investigation (Fig. 3).29 Because scFvs have the ability to recognize cell surface proteins, the targeting of tumors mediated via CAR-T cell is neither restricted nor dependent on antigen processing and presentation. CAR-T cells are therefore not limited to tumor escaping from MHC loss. For cancer immunotherapy, the main advantage of employing CAR-based methods is attributed to that the scFv derived from antibodies with affinities several orders of magnitude higher than conventional TCRs.30 In addition, CARs can target glycolipids, abnormal glycosylated proteins and conformational variants that cannot be easily recognized by TCRs. Based on clinical trial results, there is an increasing evidence that CAR-T cells have the ability to deliver powerful anti-tumor therapeutic effects, leading to the recent FDA approval of CAR-T therapies directed against the CD19 protein for the treatment of acute lymphoblastic leukemia (ALL) and large B-cell lymphoma (DLBCL).
Fig. 3.
Synthetic biology in the designs of chimeric antigen receptors (CAR). a The AND gate used in artificial CARs. Three typical CARs i.e. Costimulation domain-based second-generation CAR, synNotch receptor-assisted CAR with multiple recognization mechanisms and chimeric costimulation receptor (CCR)-based CAR are exhibited from left to right. b The artificial CARs with inhibitory CAR (iCAR) system. The system can prevent recognizing self-antigens on somatic cells. c The artificial CARs sensing different tumor antigens. Two ScFvs recognizing different targets are tandemly fused, the engineered CAR can be triggered by multiple antigens. The figure is inspired by the paper468
In addition, CAR-T applications are stepping into commercialization. The first approved CAR T-cell therapy was Kymriah which is CD19-targeted for treating DLBCL developed by Novartis and University of Pennsylvania.31 DLBCL is a typical form of non-hodgkin lymphomas (NHL) that consist of 40% of total lymphomas.32 The FDA also approved Yescarta (axicabtagene ciloleucel) in 2017 for DLBCL treatments.33 In the clinical studies, patients with DLBCL were treated with the CD19-targeted CAR T-cells, with 25% partial responders and more than 50% complete responders.34,35 Durable responses of over two years were observed, indicating the therapeutic effects of the CAR-T cells. However, cytokine storm, an excessive release of pro-inflammatory cytokines, was observed in Yescarta treated patients (13%),36 indicating the safety needs to be improved.
The selection of target antigen is the determinant in CAR-T cell therapies.37–39 If CAR-T cells can recognize protein expressed on non-malignant cells, severe cell toxicities could occur with the off-target activities.40 The optimal target antigen is the one that is consistently expressed on the surface of cancer cells but not on the surface of normal cells.37,41,42 Multiple myeloma is hard to treat via chemicals or stem cell transplantation.43,44 CAR-T cell therapies are effective for multiple myeloma in preclinical studies.45 However, to date, no antigen has been characterized that is strongly and constantly expressed on multiple myeloma cells but not on somatic cells. Among the antigens used so far, a member of the TNF superfamily proteins, B cell maturation antigen (BCMA), is the most favorable candidate for a multiple myeloma cell-directed CAR-T therapy target.42,46,47 BCMA is expressed in cancer cells in almost all multiple myeloma patients, the expression of this antigen on somatic cells is limited to plasma cells and some kinds of B cells.42,48 BCMA was the first antigen for multiple myeloma to be used in a clinical trial via a CAR-T cell approach leading to systematic responses in patients with this cancer.40,42,49 Twelve patients received BCMA CAR-T cells in the dose-gradient clinical trial. Two patients treated with 9 × 106 CAR-T cells/kg body weight were obtained with good remissions, though the treatment had toxicity related to cytokine storms.49 Many clinical trials investigating the safety and/or efficacy of anti-BCMA CAR-T cells are currently ongoing or finished.
Idecabtagene vicleucel (Abecma, also abbreviated as Ide-cel) is developed by Bristol-Myers Squibb, uses the anti-BCMA 11D5-3 scFv, the same as the 11D5-3-CD828Z CAR tested at the NCI.49 However, the co-stimulatory domain is different, the CAR used in idecabtagene vicleucel is delivered using a lentivirus vector and has a 4-1BB co-stimulatory domain instead of a CD28 one.50 In a multicenter phase I trial for idecabtagene vicleucel,50,51 the therapy is highly effective for treating multiple myeloma patients. A phase II trial named KarMMa, designed to further evaluate the safety and ability of idecabtagene vicleucel, is undergoing.52 The initial results of KarMMa demonstrates its deep, durable responses in heavily pretreated multiple myeloma patients.52 Efficacy and safety were reflected in early reports, supporting a favorable idecabtagene vicleucel clinical benefit-risk profile across the target dose range in primary clinical results.
Receptor engineering in medical therapies
SynNotch receptors are a class of artificially engineered receptors that are used in medical applications (Fig. 3).53 Notch receptors are transmembrane receptors participating in signal transductions,54 comprising an extracellular domain, a transmembrane and an intracellular domain.55 The transmembrane and intracellular domains are usually retained in synNotch architects,56 whereas the signal-input extracellular domain is engineered to sense scFvs and nanobodies,57 providing possibilities of recognizing agents to initiate signaling in living cells.
Also, the modular extracellular sensor architecture (MESA) was developed intending to detect extracellular free ligands54,58 based on the synNotch idea. MESA designs have two membrane proteins each containing an extracellular ligand-binding domain which senses the chemicals or proteins and can be a small molecule-binding domain or antibody based sensing module, a transmembrane domain and either an intracellular transcriptional factor with relasing ability from the complex, protease recognition sequence or a protease. After ligand binding to the extracellular domain, MESA receptors dimerize and induce an intracellular proteolytic cleavage that allows the transcriptional factor dissociate for downstream regulations. The method allows more flexible sensor designs without limiting to Notch receptors. This system has also been remade recently to signal transduction via a split protease59 or split transcriptional factor patterns.60 The synNotch design has been constructed with a series of receptors called synthetic intramembrane proteolysis receptors (SNIPRs) containing domains from other natural receptors other than mouse Notch protein that are also cleavable by endogenous proteases.61 Similiar to synNotch, SNIPRs bind to their antigens and function via dissociating a transcriptional factor to sense cell and immune factors.62 For synNotch, SNIPR and MESA, the choice of ligand-binding domains and transcriptional factor domains enables customization of both sensing (signal input) and function (signal output) steps when using the systems. SNIPR and MESA also enrich the available engineering tools for the artificial receptor-effectors. However, some limitations still remain such as high background signals, off-target effects, the immunogenicity from the murine Notch protein, the large size of artificial receptors and transcriptional regulators.56,61,63 Many efforts are needed to improve the system.
Receptor engineering applications are commonly related to CAR-T therapies. The receptors can be designed to target two specific antigens, one using the synNotch and the other via a traditional CAR. In preclinical models, T-cells engineered for targeting dual-antigen expressing cells are established.64 TEV protease can be fused to MESA receptors, cleaving the transcriptional factor off for functionalization.58 A humanized synthetic construct can reduce immunogenicity and minimize off-target effects. Zhu et al. constructed a framework for human SNIPRs with future applications in CAR-T therapies, preventing CAR-T cells from being activated via non-tumor signals.61 Besides the above synthetic receptors, based on the same idea, Engelowski et al. designed a synthetic cytokine receptor sensing nanobodies by the fusion of GFP/mCherry nanobodies to native IL-23 intracellular domains.65 Another receptor engineering strategy is to rewire receptor-transduced signals to novel effector genes. Using a scFv complementary to VEGF, the engineered receptor senses VEGF and released dCas9 protein, then the IL-2 expression are up-regulated. The system is successfully explored in Jurkat T cells.58
The HEK-β cells used for diabetes treatments
β-cells are existing in pancreatic islets that synthesize and secrete insulin.66 As the only site of insulin synthesis in mammals, β-cells sense blood glucose using a signal transduction pathway that comprises glycolysis and the stimulus-sensing-secretion coupling process.67,68 The secretion of insulin is consisted with the following steps. Blood glucose is transported into β-cells and metabolized via glycolysis inside the cell, resulting in cell membrane depolarization, energy generation and closing of K+ATP channels, which activates the calcium channel Cav1.3 to induce calcium influx with the secretion of insulin granules. The excessive blood-glucose concentration in diabetes patients is from the deficiency of insulin-producing β cells for type 1 diabetes, or from low insulin sensitivity of body cells for type 2 diabetes.69 Using a synthetic biology-based multiple screening approach, Xie et al. engineered human kidney cells HEK-293 to sense blood glucose levels for insulin secretion.70 The design combines automatic diagnosis and treatment in diabetes therapy. The researchers demonstrated that overexpression of Cav1.3 provided the pathway for constructing a β-cell-like glucose-sensing module in somatic cells.70 The combination of Cav1.3-controlled calcium and a synthetic Ca2+-inducible promoter allowed the monitoring of glucose levels using a tuned in vivo transcriptional response. After the construction of artificial HEK-293-β cells, the cell line HEK-293-β for glucose-response insulin production which maintained glucose homeostasis for over 3 weeks, via implanting the cells intraperitoneally to mice, also auto-corrected diabetic hyperglycemia within 3 days in T1D mice in this study.
The advantages of HEK-293-β cells are clear. Compared to primate pancreatic islets, HEK-293-β cells were adequately efficient in stabilizing postprandial glucose metabolism in T1D mice. Moreover, HEK-β cells are more easily for cultivation in vitro. It is expected that the engineered human cells have the prospect to be produced easily, cost-effectively and robustly, following current rules and regulations for pharmaceutical manufacturing, allowing the production of ready-to-use commercials with good properties for product purity, stability and quality. This highly innovative engineered cell raises the possibility that any cell type could be rationally reprogramming to achieve customized abilities such as blood glucose control.
The induced pluripotent stem cells (iPSCs) for medical applications
Synthetic biology also helps in generating human stem cells via overexpressing certain de-differentiation-related genes. One of the applications is the induced pluripotent stem cells. iPSCs are pluripotent stem cells generated from somatic cells.71 Pioneered by Yamanaka’s lab, the introduction of four transcriptional factors including Oct3/4, Sox2, c-Myc, and Klf4, resulted in changing fibroblasts to embryonic stem (ES)-like cells,72 which can re-differentiate into blood cells, bone cells or neurons for possible treatment of damages to various tissues and organs.73 iPSCs are not created using human embryos, circumvented ethical concerns in contrast with ES cells.74 Additionally, autologous somatic cell-derived iPSCs avoid immunological rejections.75
iPSCs are self-renewable with continuous subculture properties.76 The somatic cell samples from patients are induced into iPSCs able to serve as an unlimited repository for medical researches. The iPSC cell lines for Down’s syndrome and polycystic kidney disease are established.77,78 An project termed StemBANCC calls for collections of iPSC cell lines for drug screening.79 Various applications combined with therapeutic chemicals and iPSC cell lines are undergoing high-throughput drug screening and analysis.80,81
iPSCs are aimed to be used for tissue regeneration and therapy developments. Type O red blood cells can be derived from iPSCs to meet demands for blood transfusion.82 When cancer patients require large quantities of NK cells in immunotherapies, the cells can be manufactured using iPSCs to circumvent their low availabilities.83 The anti-aging effects of iPSCs are observed during mouse studies.84 The chemical-induced differentiation of iPSCs to cardiomyocytes has been commonly used.85 These iPSC-cardiomyocytes are recapitulated with genetic codes in patients whom they derived, allowing the establishment of models of long QT syndrome and ischemic heart disease.85,86 Cord-blood cells can be induced into pluripotent stem cells for treating malfunctional mice retina,87 re-differentiated iPSCs are employed to cure brain lesions in mice with their motor abilities regained after the therapy.88
iPSCs are successfully used for organ regeneration, for example, ex vivo cardiomyocytes can be used to regenerate fetal hearts to normal hearts via the Yamanaka’s method.89 Human “liver buds” can be generated from three different cells including iPSCs, endothelial stem cells and mesenchymal stem cells.90 The bio-mimicking processes made the liver buds self-packaging into a complex organ for transplanting into rodents. It functions well for metabolizing drugs.91
Some iPSC applications are advanced to clinical stages. For example, a group in Osaka University made “myocardial sheets” from iPSCs, transplanted them into patients with severe heart failure, the clinical research plan was approved in Japan,92 patients are under recruiting. Additionally, two men in China received iPSC-differentiated cardiomyocytes treatments.93 They were reported to be in good condition although no detailed data are revealed.93 iPSCs derived from skin cells from six patients are reprogrammed to retinal epithelial cells (RPCs) to replace degenerated RPCs in an ongoing phase I clinical trial.94 Similarly, phase I clinical trials are also undergoing for thalassemia treatment using autologous iPSCs differentiated hematopoietic stem cells,95 patients are recruiting. Till now, no Phase III study on stem cell-related therapy has been conducted. The major concern is the safety of iPSCs with the carcinogenic possibilities: teratoma has been observed in iPSCs injected mice,96 low-induction efficiency, incomplete reprogramming of genomes, immunogenicity and vector genomic integrations are also issues of concerns.97,98 More efforts are required for clinical applications.
Synthetic biology in tissue engineering
Tissue engineering aims to repair damaged tissues and restoring their normal functions. The use of synthetic biology in tissue engineering allows control of cell behaviors. Artificial genetic constructs can regulate cell functions by rewiring cellular signals. As engineered cells are building blocks in tissues with special properties to achieve smarter functions, synthetic biology allows complex tissue engineering for new medical studies.
By overexpression of functional genes or transcriptional factors, stem cells can differentiate to generate specific tissue cells successfully.99 This is a simple and common way in stem cell-based tissue engineering. However, the gene overexpression lacks feedback control mechanisms to avoid excess nutrient consumption or cell toxicity.100 For an instance, constitutive overexpression of the anti-apoptotic factor Bcl-2 leads to tumorigenesis risks.101,102 CRISPR/dCas9 bioswitches or synthetic mRNAs are found able to solve the problem via time and spatial-specific expression of genes.103,104 Moreover, introductions of genetic circuits sensing small molecules or cell-surface proteins are well studied, especially Tet repressor-based system.105 Gersbach et al. designed a Tet-off system controlling Runx2 factors that can regulate the in vivo osteogenic processes.106 Yao et al. employed a Tet-on system to express Sox9 specifically in engineered rat chondrocytes, Sox9 is a key factor maintaining chondrocyte viability, activating the protein expressions for type II collagen and aggrecan in cartilage tissue engineering.107 Chondrocyte degradation was inhibited after Dox (Tet system inducer) injection in implanted cell scaffolds.107 The Tet-on system is also used for overexpressing interleukin-1 receptor antagonist (IL-1Ra) gene to modulate inflammatory cytokines during the chondrogenesis processes in cartilage repairs108 (Table 2). Tet-switches have aided elapsed time controllable gene expressions for tissue engineering.
Table 2.
Synthetic biology in mammalian cell-based therapies
| Main features | Cell host /cell type | Genetic manipulations | Applications | Stages | Reference | |
|---|---|---|---|---|---|---|
| Tisagenlecleucel (Kymriah) | CD19-targeted CAR-T cancer immunotherapy | Patient’s own T-cells | The chimeric antigen receptor (CAR) is composed of a murine single‐chain antibody fragment that recognizes CD19, fused to intracellular signaling domains from 4‐1BB and CD3‐ζ. | Acute lymphoblastic leukemia and diffuse large B‐cell lymphoma | Approved | 417 |
| Axicabtagene ciloleucel (Yescarta) | CD19-targeted CAR-T cancer immunotherapy | Patient’s own T-cells | Expressing a CAR comprising an anti‐CD19 single chain variable fragment linked to CD28 and CD3‐ζ costimulatory domains. | Diffuse large B‐cell lymphoma | Approved | 418 |
| Idecabtagene vicleucel (Abecma) | B-cell maturation antigen (BCMA)-directed CAR-T cell therapy | Patient’s own T-cells | Comprises an anti-BCMA single-chain variable fragment (scFv) fused to a CD8 linker region, the 4-1BB co-stimulatory and the CD3‐ζ signaling domains | Relapsed and refractory multiple myeloma | Approved | 419 |
| SynNotch | An engineered Notch receptor to construct Multi-antigen prime-and-kill recognition circuits to induce effective proteins | Patient’s own T-cells | A synNotch receptor that recognizes EGFRvIII or MOG, induces expression of a CAR. | glioblastoma | Pre-Clinical | 56,420 |
| HEK-β cells | Engineering a synthetic circuit into human cells that can sense the glucose concentration and to correct blood sugar concentration | HEK-293 cells | Ectopic expression of a calcium channel, expression of insulin under control of elements of the calcium-responsive NFAT promoter, expression of both a short version of the glucagon-like peptide (GLP-1), a known insulin secretagogue, and its receptor (GLP1R). | Diabetes mellitus | Pre-Clinical | 70,421 |
| Caffeine-stimulated advanced Regulators (C-STAR) system | Sensing caffeine to produce peptides for treating diabetes | HEK-293T or hMSC-hTERT cells | Overexpression of T2D-treating peptide shGLP-1 under STAT3 promoter | Type 2 diabetes | Pre-Clinical | 422 |
| Guanabenz-controlled genetic circuits | Guanabenz activates the secretion of peptides GLP-1 and leptin. | Hana3A cells | The cTAAR1 signal transduction is rewired to dose-dependently control expression of the glucagon-like peptide 1 (GLP-1) and leptin via an IgG-Fc linker under the induction of guanabenz. | The metabolic syndrome | Pre-Clinical | 423 |
| Green tea-triggered genetic control system | Engineering cells to respond to protocatechuic acid (PCA), a metabolite in green tea to treat diabetes in mouse and nonhuman primate models. | HEK-293 cells | Using PCA-ON sensor (artificial KRAB-PcaV fusion repressor) to overproduce insulin and shGLP-1 | Diabetes mellitus | Pre-Clinical | 424 |
| Synthetic optogenetic transcription device | Light-controlled expression of the shGLP-1 peptide to attenuate glycemic excursions in type II diabetic mice | HEK-293 cells | Overexpression of melanopsin; PNFAT-driven expression of shGLP-1 | Type 2 diabetic mice | Pre-Clinical | 425 |
| Red/far-red light-mediated and miniaturized Δphytochrome A (ΔPhyA)-based photoswitch (REDMAP) system | Small and highly sensitive light-inducible switch in mammalian cells | HEK-293 cells | The PhyA interaction domain FHY1 is fused to the VP64 to create a light-dependent transactivator (FHY1-VP64), the DNA-binding domain Gal4 is fused to phytochrome ΔPhyA to create a fusion light sensor domain (ΔPhyA-Gal4), the transactivator can bind to its synthetic promoter P5 × UAS to initiate transgene expression, following exposure to far-red light (730 nm), the transactivator terminates transgene expression. | Type 1 diabetic (T1D) mice and rats | Pre-Clinical | 387 |
| Gene expression by radiowave heating | Heating of iron oxide nanoparticles by radiowaves remotely activated insulin gene expression in cultured cells or mouse models | HEK-293T cells | Heated iron oxide nanoparticles activate TRPV1 channel to pump calcium, the insulin gene is driven by a Ca2+-sensitive promoter. | Lowers blood glucose in mice | Pre-Clinical | 192 |
| Electronic control of designer mammalian cells | Using wireless-powered electrical stimulation of cells to trigger the release of insulin | Human β cells | Coupling ectopic expression of the L-type voltage-gated channel CaV1.2 and the rectifying potassium channel Kir2.1 to the desired output through endogenous calcium signaling, the insulin gene is overexpressed by the system. | Type 1 diabetic mice | Pre-Clinical | 421 |
| Self-sufficient control of urate homeostasis | Senses uric acids levels and triggers dose-dependent derepression of a urate oxidase that eliminates uric acid | HeLa cells | The HucR start codon was modified (GTG→ATG) and fused to a Kozak consensus sequence for maximum expression, fusing it to the C terminus of the KRAB protein, eight tandem hucO modules downstream of the PSV40 promoter to drive the signal peptide-uricase cassette SSIgk-mUox. | Acute hyperuricemia in mice | Pre-Clinical | 426 |
| Dopamine sensors for hypertension control | A synthetic dopamine-sensitive transcription controller to produce the atrial natriuretic peptide to reduce blood pressure under pleasure situations | HEK-293 cells | Rewiring the human dopamine receptor D1 (DRD1) via cAMP to synthetic promoters containing cAMP response element-binding protein 1(CREB1)-specific cAMP-responsive operator modules to express atrial natriuretic peptide | Hypertension in mice | Pre-Clinical | 427 |
| Insulin self-regulation circuit for correcting insulin resistance | A self-adjusting synthetic gene circuit to reverse insulin resistance in diabetes and obesity animal models | HEK-293 cells | Ectopically express the human insulin receptor (IR) via the rewiring of the MAPK pathway, expression of adiponectin transgene (consisting of three tandem adiponectin molecules fused to a human IgG-Fc fragment), regulated by a synthetic promoter specific to the hybrid transcription factor TetR-ELK1 | Insulin resistance in mice | Pre-Clinical | 428 |
| Smartphone-controlled optogenetically engineered cells | Remotely control release of glucose-decreasing proteins by engineered mammalian cells implanted diabetic mice under the control of far-red light | HEK-293 cells | The bacterial light-activated c-di-GMP synthase BphS and the c-di-GMP-specific phosphodiesterase YhjH are designed to regulate drug production according to user-defined glycemic thresholds. | Diabetes mellitus in mice | Pre-Clinical | 186 |
| Modified rapamycin-induced CAR-T cells | Engineered T cell “on” or “off” by administering small molecule rapalog | K562 cells | The part I constructs of the ON-switch are similar to conventional CAR, in addition to the FKBP domain for heterodimerization; part II variants contained the additional DAP10 ectodomain for homodimerization and the CD8α transmembrane domain for membrane anchoring. | Xenografted matched cancer cells | Pre-Clinical | 429 |
| Synthetic RNA regulatory systems for T-cell proliferation | Linking rationally designed circuit to growth cytokine targets to control mouse and primary human T-cell proliferation | CTLL-2 and TCM cells | Using tetracycline-responsive switches to inactivate ribozyme and cell-proliferation associated cytokines that are expressed to promote T-cell growth. | N.A. | N.A. | 430 |
| Nonimmune cell cancer therapies | A new class of synthetic T-cell receptor-like signal-transduction device to kill target cells | HEK-293T and hMSC cells | Employs JAK-STAT signaling mediated by the IL4 and IL13 receptor, with STAT6 as a signaling scaffold, and uses CD45-mimetic molecule upon specific cell contact as an OFF/ON switching mechanism. | N.A. | Pre-Clinical | 431 |
| PD-1 and CTLA-4 based inhibitory chimeric antigen receptors (iCARs) | Designed antigen-specific inhibitory receptors to block these unwanted “on-target, off-tumor” responses. | Mice’s own T-cells | The cytoplasmic domain of CTLA-4 or PD-1 was fused to the human prostate-specific membrane antigen (PSMA) transmembrane domain. | Leukemia in mice | Pre-Clinical | 432 |
| Resveratrol-triggered regulation devices in CAR-T cells | Allow precise control over T cell activity through adjustment of resveratrol dosage | Mice’s own T-cells | RESrep device consists of a resveratrol-dependent transactivator ResA3 that fused to a synthetic activator VPR via the C terminus of TtgR, the chimeric transactivator can bind to the resveratrol-dependent promotor PResA1, positioned in front of a promoter PhCMVmin. | Mouse tumor model of B cell leukemia | Pre-Clinical | 433 |
| CAR-transduced natural killer cells (CAR-NT) in CD19-positive lymphoid tumors | CD19-targeted CAR-NK cancer immunotherapy | HLA-mismatched anti-CD19 CAR-NK cells | NK cells were transduced with a retroviral vector expressing genes that encode anti-CD19 CAR, IL-15, and inducible caspase 9 as a safety switch. | Non-Hodgkin’s lymphoma or chronic lymphocytic leukemia | Phase 1 and 2 trial | 434 |
| CAR-macrophages (CAR-M) for solid cancer immunotherapy | CD19-targeted CAR-M cancer immunotherapy | Human macrophage THP-1 cells | First-generation anti-CD19 CAR encoding the CD3ζ intracellular domain, targeting the solid tumor antigens mesothelin or HER2 | Mice bearing SKOV3 lung or peritoneal metastases | Phase 1 | 435 |
| In vivo gene editing to treat Duchenne muscular dystrophy (DMD) | CRISPR-Cas9 can correct disease-causing mutations in dog models of DMD. | Systemic delivery in skeletal muscle or vein | Using Streptococcus pyogenes Cas9 coupled with a sgRNA to target a region adjacent to the exon 51 splice acceptor site to correct the skipping of exon 51 | Duchenne muscular dystrophy in dog | Phase 1 and 2 trial | 436 |
| In vivo gene editing to treat Leber congenital amaurosis 10 (EDIT-101) | Targeted genomic deletion using the CRISPR/Cas9 system for the treatment of mice and cynomolgus monkeys with LCA10 bearing the CEP290 splice mutation | Subretinal delivered in mice | A combination of specific pairs of sgRNAs and Cas9 to excise the intronic fragment containing the IVS26 splice mutation in CEP290 gene | Leber congenital amaurosis 10 in mice or cynomolgus monkeys | Phase 1 and 2 trial | 437 |
| CRISPR-edited stem cells to treat human diseases | Transplanted CRISPR-edited CCR5-ablated HSPCs into a patient for acute lymphoblastic leukemia with HIV-1 infection | Edited CD34+ cells | Cells were transfected with a ribonucleoprotein complex comprising Cas9 protein and two designed guiding RNAs targeting CCR5. | Acute lymphoblastic leukemia patient with HIV-1 infection | Clinical | 438 |
| Mammalian synthetic cellular recorders integrating biological events (mSCRIBE) | Recording of molecular events into mammalian cellular genomic DNA | HEK-293T cells | This device consists of a self-targeting guide RNA (stgRNA) that repeatedly directs Streptococcus pyogenes Cas9 nuclease activity toward the DNA that encodes the RNA, when cellular sensors regulate the Cas9 activity, the device enabling localized, continuous DNA mutagenesis as a function of stgRNA expression. | N.A. | N.A. | 439 |
| Synthetic gene network for thyroid hormone homeostasis for Graves’ disease | A gene circuit that monitor increased thyroid hormone levels and drive the expression of a validated TSH receptor antagonist. | CHO-K1 cells | This synthetic control device consists of a synthetic thyroid-sensing receptor (TSR), a yeast Gal4 protein/human thyroid receptor-α fusion, which reversibly triggers expression of the TSHAntag gene from TSR-dependent promoters. | Graves’ disease in mouse models | Pre-Clinical | 440 |
| Aroma-triggered pain relief based on synthetic cell engineering | Spearmint (R-carvone) induced analgesic peptide production in mice | Hana 3A cells | Ectopic expression of the R-carvone-responsive olfactory receptor OR1A1 rewired via an artificial G-protein deflector to induce the expression of a secretion-engineered and stabilized huwentoxin-IV variant | Relief chronic pain in mice | Pre-Clinical | 441 |
| Synthetic gene circuit controls human IPSCs differentiation | Using vanillic acid as the inducer for cell-fate gene expressions in the transition of hIPSCs to beta-like cells | hIPSC cells | vanillic acid-triggered expression switches for the transcription factors Ngn3 and Pdx1 with the concomitant induction of MafA | N.A. | Pre-Clinical | 442 |
| Cytokine-induced anti-inflammatory factors to treat experimental psoriasis | Designed and engineered human cells that sequentially detected elevated TNF and IL22 levels from a psoriatic flare and produced therapeutic doses of IL4 and IL10. | HEK-293T cells | Rewired TNFR-signaling through NF-κB to a synthetic NF-κB-responsive promoter that controlled the expression of human IL22 receptor α which enables IL22-mediated activation of the JAK signal transducer and activator of transcription STAT signaling cascade, driving expression of the cytokines IL4 and IL10. | Psoriasis in mice | Pre-Clinical | 443 |
| Designer exosomes to deliver therapeutic cargo into brain (EXOtic devices) | The device enhances exosome biogenesis, packaging of specific RNAs into exosomes, secretion of exosomes, targeting, and delivery of mRNA into the cytosol of target cells to treat Parkinson’s disease. | HEK-293T cells | Overexpressing STEAP3, SDC4, NadB and Cx43 variant S368A, targeting CHRNA7 receptor | Parkinson’s disease in mice | Pre-Clinical | 444 |
| Genetic-code-expanded cell-based therapy for treating diabetes in mice | A genetic code expansion-based therapeutic system, to achieve fast therapeutic protein expression in response to cognate ncAAs at the translational level | HEK-293T cells | A ncAA-triggered therapeutic switch (NATS) system composed of a bacterial aaRS-tRNA pair and an insulin gene carrying an ectopic amber codon | Diabetes in mice | Pre-Clinical | 445 |
| Synthetic mammalian cell-based microbial-control device | Detects microbial chemotactic formyl peptides through a formyl peptide sensor (FPS) and responds by releasing AI-2 to inhibit pathogens | HEK-293T cells | A FPS module that detects formylated peptides by FPR1, the adapter protein Gα16 redirects receptor signaling to the Ca2+ transduction pathway, constitutively expressed 5′-methylthioadenosine nucleosidase MTAN cleaving endogenous SAH and the LuxS under the control of a Ca2+-responsive promoter to produce AI-2. | Inhibit Vibrio harveyi and Candida albicans | Pre-Clinical | 446 |
| Human liver buds | Self-packaging into a complex organ using three stem cells | iPSCs, endothelial stem cells and mesenchymal stem cells | N.A. | Generation of a functional human organ from pluripotent stem cells | Pre-Clinical | 90 |
| Reprogramming of cardiomyocytes drives heart regeneration | Uses of ex vitro cardiomyocytes for regeneration of fetal hearts to normal hearts by the Yamanaka’s method | Patient’s own cardiomyocyte cells | Cardiomyocytes specific expression of OSKM (Oct4, Sox2, Klf4, and c-Myc) is enabled by administration of doxycycline. | Heart failure in mice | Pre-Clinical | 89 |
N.A. not applicable
The optogenetic induction systems are also used in the control of cell behaviors in tissue engineering. Light inducible proteins are able to respond to UV and far-infrared lights, making light induction applicable.109 Various optogenetic circuits are constructed by fusing light-sensitive motifs to well-characterized transcriptional factors.110,111 Spatial-specific gene activation has been successfully employed to guide the arrangement of cells.112 Sakar et al. used blue light-induced channel rhodopsin-2 to achieve dynamic and region-specific contractions of tissues.113 The optogenetic control of engineered murine-derived muscle cells offers remote gene activation or silencing via the light-sensitive membrane Na+ channel and ion-inducible downstream elements for tissue engineering.
Inspired by successes of CAR-T cells, G protein-coupled receptors (GPCRs) are engineered to sense artificial ligands for tissue engineering.114 Park et al. successfully designed and used a GPCR sensing clozapine-N-oxide (CNO) in primary cells for the control of cell migration in response to CNO concentration gradients.115 This technology could make a valuable module for wound healing and cell regeneration. Synthetic biology makes possible to program cells to multicellular structures in a self-assembly manner.116 Toda et al. employed synNotch methods to engineer cell adhesion signals in a population of mouse fibroblasts that were turned into multilayers and polarized according to the synNotch receptor types.117
Besides cells, biomaterials are commonly used in tissue engineering, served as scaffolds and bio-mimicked organs.118 the auto-modulation characteristics of biomaterials in response to stimuli or chemical compounds are useful in biomaterial-based tissue engineering. Baraniak et al. engineered the B16 cell line with a green fluorescent protein (GFP) reporter induced by RheoSwitch Ligand 1 (RSL1), which was coated on poly(ester urethane) films, allowing GFP activation for up to 300 days on the film.119 Deans et al. constructed an isopropyl-β-d-thiogalactoside (IPTG)-induced Lac-off system in Chinese hamster ovary (CHO) cells, and IPTG encapsulated in poly(lactide-co-glycolide) (PLGA) scaffolds or PEG beads was released in a sustainable manner. The reporter gene indicated that the induction lasted over 10 days in mouse models implanted subcutaneously into the dorsal region,120 the GFP fluorescence level was observed to be controlled by its locations.121 The spatial-induced gene expression regulation has become a design-of-concept in many applications like cartilage repair and in vivo 3D cell scaffolds.
In summary, expressions of biological circuits could generate functionalized cells for tissue engineering. Multiple synthetic biology designs e.g. time and spatial-dependent gene expression, induction and autoregulation systems and smart biomaterials are available in this field. The state-of-the-art development still remains with many obstacles from moving truly synthetic tissues into clinic, but at least some foundations are settled for future studies.
Engineered bacterial cells for therapeutical applications
Synthetic biology approaches have promoted genetically engineered bacteria for novel live therapeutics (Fig. 2).122 Bacteria containing synthetic gene circuits can control the timing, localization and dosages of bacterial therapeutic activities sensing specific disease biomarkers and thus develop a powerful new method against diseases.123 Synthetic biology-based engineering methods allow to program living bacterial cells with unique therapeutic functions, offering flexibility, sustainability and predictability, providing novel designs and toolkits to conventional therapies.124 Here some advances are presented for engineered bacterial cells harboring gene circuits capable of sensing and transduction of signals derived from intracellular or extracellular biomarkers, also the treatments and diagnosis based on these signaling pathways. The concept of bacterial cell-based live therapeutics and diagnostics are rapidly growing strategies with promises for effective treatments of a wide variety of human diseases.
Engineered bacterial cells in cancer diagnosis and treatments
Some anaerobic/facultative anaerobic bacterial cells are good candidates for tumor treatments. They can target the anaerobic microenvironment of tumors, they also have the tumor lysis-inducing and trigger inflammation abilities useful in fighting against solid tumors.125 Engineered microbes can become suitable tools for cancer in vivo diagnosis. Danino et al. engineered E. coli with LacZ reporter gene, the bacterium produces LacZ when in contact with tumor cells. Subsequently, mice were injected with chemiluminescence substrates for LacZ (Table 3). The luminescence is enriched in the urine to generate red color.126 The method is more sensitive than microscopes as it can detect tumors smaller than 1 cm. Similarly, Royo et al. constructed a salicylic acid-induced circuit converting 5-fluorocytosine to toxic products in attenuated Salmonella enterica for tumor killing.127 Salmonella enterica localized in tumor tissues after the injection, with the additional providing of salicylic acid (inducer) and 5-fluorocytosine (substrate), tumor cells were eliminated via the formation of 5-fluorouracil from the bacterial cells.
Table 3.
Synthetic biology in microbe-based therapies
| Main features | Microorganism type | Genetic manipulations | Applications | Stages | Reference | |
|---|---|---|---|---|---|---|
| SYNB1020 | Transform ammonia into L-arginine to treat hyperammonia | E. coli Nissle 1917 | Deleted the gene argR, thyA and integrated the gene argA215, under the control of the fnrS promoter (PfnrS) | Hyperammonia in mice and cynomolgus monkeys | Phase 1b/2a | 150 |
| SYNB1618 | Engineered Escherichia coli Nissle to express genes encoding Phe-metabolizing enzymes to treat phenylketonuria | E. coli Nissle 1917 | Two chromosomally integrated copies of pheP and three copies of stlA under the regulatory control of PfnrS, two additional copies of stlA were placed under the control of the Ptac promoter. | Phenylketonuria | Phase 1/2 | 151 |
| Probiotic-associated therapeutic curli hybrids (PATCH) | Genetically engineer Escherichia coli Nissle 1917 (EcN) to create fibrous matrices that promote gut epithelial integrity in situ | E. coli Nissle 1917 | CsgA fused to TFF3, under the control of an inducible promoter (PBAD) | Dextran sodium sulfate (DSS)-induced colitis in mice | Pre-Clinical | 145 |
| Engineered bacteria for colorectal-cancer chemoprevention | The engineered Escherichia coli bound specifically to colorectal cancer cells and secreted myrosinase to transform small molecule form broccoli to anticancer agents. | E. coli Nissle 1917 | Expressed and secreted YebF-I1 myrosinase catalyzes the glucosinolate hydrolysis to sulforaphane, while the expression of INP-HlpA facilitates bacterial CRC cell binding. | Colorectal-cancer in mice | Pre-Clinical | 447 |
| Bacteria engineered to reduce ethanol-induced liver disease | Bacteria engineered to produce IL-22 induce expression of REG3G to reduce ethanol-induced steatohepatitis | Lactobacillus reuteri | L. reuteri EF-Tu promoter drives murine IL-22 gene. | Ethanol-induced liver disease in mice | Pre-Clinical | 154 |
| Bacteria-mediated tumor therapy triggered via photothermal nanoparticles | Bacteria are coated with nanogold particles (or indocyanine green-loaded nanoparticles) able to receive light for heat generation, inducing therapeutic protein TNF-α in tumor sites. | E. coli MG1655/ attenuated Salmonella typhimurium | A widely used temperature-sensitive plasmid pBV220 containing TcI repression and tandem pR-pL operator-promoter was introduced to express human TNF-α. | Breast tumor in mice | Pre-Clinical | 448,449 |
| Engineered bacteria overexpressing anti-inflammatory cytokines | Therapeutic dose of IL-10 can be reduced by localized delivery of a bacterium genetically engineered to secrete the cytokine. | Lactococcus lactis MG1363 | lactococcal P1 promoter driving usp45 secretion leader fused to the mIL-10 gene. | Murine colitis | Pre-Clinical | 450 |
| Modified bacteria producing peptides to inhibit obesity | Engineered bacteria that express the therapeutic factor N-acylphosphatidylethanolamines (NAPEs) into the gut microbiota | E. coli Nissle 1917 | Overexpressing N-acyltransferase At1g78690 under the intrinsic promoter from Arabidopsis thaliana | Obesity in mice | Pre-Clinical | 149 |
| Synthetic genetic system to eliminate gut pathogens | A gene encoding an anti-biofilm enzyme induced by P. aeruginosa-specific quorum sensing signal | E. coli Nissle 1917 | Genes alr and dadX are deleted, the 3OC12-HSL-inducible promoter drives the expression of DspB and E7. | Pseudomonas aeruginosa gut infection in Caenorhabditis elegans and mice | Pre-Clinical | 146 |
| Bacteria synchronized for drug delivery | Engineer a clinically relevant bacterium to lyse synchronously and routinely at a threshold population density to release genetically encoded cargo | attenuated Salmonella enterica serovar Typhimurium | The luxI promoter regulates production of autoinducer (AHL), which binds LuxR and enables it to transcriptionally activate the promoter, negative feedback arises from cell death that is triggered by a bacteriophage lysis gene (φX174 E) which is also under control of the luxI promoter. | Subcutaneous liver metastasis in mice | Pre-Clinical | 451 |
| Engineering bacteria to serve as whole-cell diagnostic biosensors | Sensing abnormal glucose concentrations in human urine samples | E. coli DH5α | Combinatorial of pCpxP and pYeaR promoters driving expression of integrase TP901 and BxB1 | Detection of urine glucose in human samples | Pre-Clinical | 135 |
| Engineered bacteria as live diagnostics of inflammation | Engineered a commensal murine Escherichia coli strain to detect tetrathionate, which is produced during inflammation. | E. coli strain NGF-1 | ttrR/S genes and the PttrBCA promoter from S. typhimurium to drive Cro expression, and inserted it into the genome, containing the phage lambda cI/Cro genetic switch can sense and record environmental perturbations. | Detection of gut inflammations | Pre-Clinical | 452,453 |
| Recording of cellular events over time using engineered bacteria | A multiplexing strategy to simultaneously record the temporal availability of three metabolites (copper, trehalose, and fucose) in the environment of a cell population over time. | E. coli BL21 | The E. coli cas1-cas2 cassette is downstream of the PLTetO-1 promoter, the CopA/GalS or TreR sensors drive RepL. | Recording specific environmental factors surrounding the cells | Pre-Clinical | 454 |
| Ingestible micro-bio-electronic device (IMBED) for in situ biomolecular detection | Heme-sensitive probiotic biosensors demonstrate accurate diagnosis of gastrointestinal bleeding in swine via miniaturized luminescence readout electronics that wirelessly communicate with an external device. | E. coli Nissle 1917 | The heme biosensor PL(HrtO), overexpressing HrtO and ChuA, luxCDABE was used as the output of the genetic circuit to generate luminescence captured by electronic devices. | Gastrointestinal bleeding in swine | Pre-Clinical | 455 |
| Probiotics detect and suppress cholera | Engineered an L. lactis strain that specifically detects quorum-sensing signals of V. cholerae in the gut and triggers expression of an enzymatic reporter that is readily detected in fecal samples. | L. lactis subsp. cremoris MG1363 | Designed an L. lactis hybrid receptor that combines the transmembrane ligand binding domain of CqsS with the signal transduction domain of NisK, placed the gene tetR downstream of the chimeric repressor-controlled nisA promoter to enable constitutive repression of an engineered Bacillus subtilis xylA-tetO promoter. | Detection of cholera infection in mice | Pre-Clinical | 456 |
| Engineering probiotics for detection of cancer in urine | Orally administered diagnostic that can noninvasively indicate the presence of liver metastasis by producing easily detectable signals in urine. | E. coli Nissle 1917 | Genomic expression of luxCDABE, IPTG-inducible lacZ in plasmid | Indicate the presence of liver metastasis | Pre-Clinical | 126 |
| Underwater adhesives made by bacterial self-assembling multi-protein nanofibers | Fusing mussel foot proteins of Mytilus galloprovincialis with CsgA proteins | E. coli C3016 strain | Overexpression of the fused protein in E. coli | Novel bio-adhesive | N.A. | 248,250 |
| Engineered modularized receptors activated via ligand-induced dimerization (EMeRALD) to detect pathological biomarkers | Build EMeRALD receptor detecting bile salts in E. coli by rewiring bile salt-sensing modules from Vibrio cholerae and Vibrio parahaemolyticus | E. coli strain NEB10β | Synthetic bile acid sensor TcpP-TcpH for taurocholic acid; synthetic bile acid sensor VtrA-VtrC for taurodeoxycholic acid, driving sfGFP as the reporter | Detection of bile acid concentration in serum | Pre-Clinical | 457 |
| Kill tumor cells via salicylic acid-induced circuit | Salmonella spp., carrying an expression module encoding the 5-fluorocytosine-converting enzyme cytosine deaminase in the bacterial chromosome or in a plasmid, to mice with tumors | attenuated Salmonella enterica | Carries an expression module with a gene of interest (cytosine deaminase) under control of the XylS2-dependent Pm promoter | Eliminate xenografted tumor in mice | Pre-Clinical | 127 |
| Ultrasound-controllable engineered bacteria for cancer immunotherapy | Engineer therapeutic bacteria to be controlled by focused ultrasound to release of immune checkpoint inhibitors | E. coli Nissle 1917 | TcI42-containing thermal switch to express αCTLA-4 and αPD-L1 nanobodies in high temperature generated from the ultrasound | Eliminate xenografted tumor in mice | Pre-Clinical | 458 |
| Living bacterial polymer materials in gastrointestinal tract | Auto-lysis bacteria contain self-assembly materials to glue microbes up for stabilizing gut microbiota. | E. coli MC4100Z1 | Cells carrying the ePop circuit produce ELPs fused with either multiple SpyCatcher or SpyTag sequences | Maintain gut microbes under perturbations by antibiotics | Pre-Clinical | 459 |
| Ketone-producing probiotics as a colitis treatment | Develop a sustainable approach to treat chronic colitis using engineered EcN that can sustainably release 3-hydroxybutyrate | E. coli Nissle 1917 | The ldhA gene is knocked-out, phaB, phaA and tesB genes are overexpressed under fnrS promoter in the genome. | Acute colitis in mice | Pre-Clinical | 147 |
| Optotheranostic nanosystem for ulcerative colitis via engineered bacteria | Developed diagnosis and treatment kits containing two parts: the optical diagnosis sensor to smartphone processing and (ii) treatment based on optogenetic probiotics | E. coli Nissle 1917 | A light-responsive EcN strain containing light-inducing promoter pDawn to drive mil-10 gene for IL-10 production | Ulcerative colitis in mice | Pre-Clinical | 389 |
| SYNB1891 | Targets STING-activation to phagocytic antigen-presenting cells (APCs) in the tumor and activates complementary innate immune pathways. | E. coli Nissle 1917 | The CDA-producing enzyme DacA from Listeria monocytogenes was expressed in EcN under PfnrS promoter, both dapA and thyA deleted in the genome. | Murine melanoma tumors and A20 B cell lymphoma tumors | Phase 1 | 460 |
| Bacterial flagellin triggered enhanced cancer immunotherapy | Engineered a Salmonella typhimurium producing the flagellin B protein from another bacterium Vibrio vulnificus to induce an effective antitumor immune response | S. typhimurium | relA and spoT genes were deleted in the genome, the pelB leader sequence was fused to the upstream of flaB to guide extracellular secretion, under the control of a PBAD promoter. | Mice colon tumors | Pre-Clinical | 461 |
| Quorum-sensing Salmonella spatial-selectively trigger protein expression within tumors | Integrated Salmonella with a quorum-sensing (QS) switch that only initiates drug expression in the tightly packed colonies present within tumors | attenuated Salmonella enterica | The pluxI promoter controls one operon consisting of genes encoding for proteins LuxR, GFP, and LuxI, LuxI produces the communication molecule 3OC6HSL. | Controlled therapy for mammary cancer in mice | Pre-Clinical | 462 |
| Tumor-specific lysis and releasing anti-cancer agents | Engineered a non-pathogenic Escherichia coli strain to specifically lyse within the tumor microenvironment and release an encoded nanobody antagonist of CD47 | E. coli Pir1+ | A stabilized plasmid that drives constitutive expression of a hemagglutinin (HA)-tagged variant of CD47nb, the strain overexpresses luxI and lyses at a critical threshold owing to the production of ϕX174E, resulting in bacterial death and therapeutic release. | Eliminating planted melanoma, mammary tumor in mice | Pre-Clinical | 463 |
| Engineered probiotics for regularly self-lysis to release nanobodies | Engineered a probiotic bacteria system to release nanobodies targeting the immune checkpoints | E. coli Nissle 1917 | The PD-L1nb and CTLA-4nb sequences were cloned onto separate plasmids downstream of a strong constitutive tac promoter on a high-copy plasmid, an HA protein tag was added to the 3′ end of the nanobody sequences. | Colorectal cancer and B cell lymphoma in mice | Pre-Clinical | 464 |
| Engineering of symbiont bacteria in mosquitos to control malaria | Serratia AS1 was genetically engineered for secretion of anti-Plasmodium effector proteins, and the recombinant strains inhibit development of Plasmodium falciparum in mosquitoes. | Serratia AS1 | The five effector genes were cloned in a single construct, (MP2)2-scorpine-(EPIP)4-Shiva1-(SM2)2, under the control of a single promoter. | Malaria prevention | N.A. | 465 |
| Engineered bacterial communication prevents Vibrio cholerae virulence | Escherichia coli Nissle 1917 to express the auto inducer molecule cholera autoinducer 1(CAI-1) to increase the mice’s survival in cholera infections | E. coli Nissle 1917 | Express the gene cqsA, under control of the native constitutive promoter PfliC | Prevents Vibrio cholerae virulence | Pre-Clinical | 466 |
| Noninvasive assessment of gut function: Record-seq | A CRISPR-based recording method (Record-seq) to capture the transcriptional changes that occur in Escherichia coli bacteria as they pass through the intestines | E. coli MG1655 | An anhydrotetracycline (aTc)-inducible transcriptional recording plasmid consisted of FsRT-Cas1–Cas2 and CRISPR arrays | N.A. | N.A. | 467 |
N.A. not applicable
To improve the effects of bacteria-based cancer therapies, some studies aim to further enhance bacterial tumor tropism.128 Some bacteria have natural affinity for the anaerobic environment of solid tumors, like E. coli or attenuated Vibrio cholerae, Salmonella typhimurium, and Listeria monocytogenes.128 However, the affinity is not sufficient for targeted therapies, bacterial cells in vivo are still dispersed in general. They can be augmented by introducing synthetic surface adhesins targeted to bind cancer-specific molecules like neoantigens or other chemicals or proteins that are enriched in cancer cells, not accumulated in somatic cells. Engineering of adhesins are demonstrated to be effective in enhancing bacterial tumor reactions. The adhesins are membrane-displayed proteins with extracellular immunoglobulin domains that can be engineered via library directed evolution screens. Piñero-Lambea et al. constructed a constitutive genetic circuit in E. coli with an artificial adhesin targeting green fluorescent protein (GFP) as the evidence of a proof of concept, it demonstrated the abilities from that binding of the cell membrane-engineered bacteria to GFP-expressing HeLa cells are successful both in vitro and in mice.129 Importantly, the intravenous delivery of this engineered bacteria to mice resulted in effective and efficient colonization in xenografted solid tumors of HeLa cells at a dose 100 times lower than that for a bacterial strain expressing an irrelevant control adhesin, or for the wild-type strain, suggesting that similarly engineered bacteria can be used to carry therapeutic agents to tumors at low doses with marginal potential systemic basal toxicities.130,131 However, few tumor-targeting bacteria have entered clinical stages. The facultative anaerobe Salmonella typhimurium VNP2000, has been engineered for safety with anti-tumor abilities in pre-clinical studies,132 yet it failed in the phase I clinical trial for marginal anti-tumor effects and dose-dependent side effects.133 Some other clinical investigations based on bacteria Clostridia novyi-NT or Bifidobacterium longum APS001F are ongoing or recruited for their phase I trials.134
Engineered bacterial cells for diabetes diagnosis and treatments
Bacteria have been engineered to detect glucose concentrations for diabetes. Courbet et al. described an approach in sensing abnormal glucose concentrations in human urine samples.135 They encapsulated the bacterial sensors in hydrogel beads, glucose in urine will change the color to red in beads. The in vitro bacterial glucometer has found outperforming the detection limit of urinary dipsticks by one order of magnitude.
Some proteins and peptides are biosynthesized in engineered gut bacteria for diabetes treatments. The engineered probiotic L. gasseri ATCC 33323 produced GLP-1 protein, the bacterium is orally delivered to diabetic rats,136 demonstrating a down-regulation of blood glucose levels to 33%. Similarly, engineered L. lactis FI5876 was reconstructed to biosynthesize and deliver incretin hormone GLP-1 to stimulate β-cell insulin secretion under conditions of high glucose concentrations. Results showed the glucose tolerance is improved in high-fat diet mice.137 The probiotic L. paracasei ATCC 27092 is engineered to secret angiotensin (1-7) [Ang-(1-7)], increasing the concentrations of Ang-(1-7) (an anti-inflammatory, vasodilator and angiogenic peptide phamarceutical), and reduced the side effects on retina and kidney in diabetic mice, as the insulin production level is increased after oral administration of the bacteria. Following the design, oral uptake of engineered B. longum HB15 which produces penetratin (a cell-penetrating peptide with the ability of enhancing delivery of insulin), and GLP-1 fusion protein also enhanced the production of GLP-1 in the colorectal tract.138–140 L. paracasei BL23 was also successfully designed to produce monomer GLP-1 analogs displayed to the bacterial membrane via fusing GLP-1 to peptidoglycan-anchor protein PrtP, the engineered bacteria enhanced glycemic control in rats with diabetes. However, the efficacy is still limited and needed further investigations.141 In addition to GLP-1, some other proteins like the immunomodulatory cytokine IL-10 along with human proinsulin were simultaneously introduced to engineered L. lactis MG1363, the combination therapy with low-dose systemic anti-CD3 allowing reversal of irregulated self-autoimmune triggered diabetes in non-obese diabetic mice.142,143 This design could possibly be effective for the treating of type 1 diabetes in human.
Engineered bacterial cells for diagnosis and treatments of gastrointestinal diseases
Probiotics can be used to treat inflammatory bowel disease (IBD).144 IBD is chronic inflammation of tissues in the digestive tract, including ulcerative colitis and Crohn’s disease. Patients are suffering from diarrhea, pain and weight loss. Synthetic biology approaches and ideas help bacteria acquire more powerful abilities against gastrointestinal diseases. Praveschotinunt et al. designed an engineered E. coli Nissle 1917 (EcN) that produces extracellular fibrous matrices to enhance gut mucosal healing abilities for alleviating IBD in mice.145 Curli fibrous proteins (CsgA) were fused with trefoil factor (TFF) domains to promote the reconstruction of cell surface, and the bacterium could produce fibrous matrices via the in situ protein self-assembly of the modified curli fibers. The results revealed that the designed EcN significantly inhibited the production of pro-inflammatory cytokines, alleviated the weight loss of mice, maintained colon length, demonstrating its anti-inflammation ability in the dextran sodium sulfate (DSS)-induced acute colitis mouse model. The design could be expanded to a general approach for probiotic-based live therapeutics in IBD treatments.
Bacteria are feasible to be engineered to directly eliminate pathogens for preventing infectious diseases in gastrointestinal tracts. Pseudomonas aeruginosa is a common multidrug-resistant pathogen difficult to treat. Engineered EcN has been employed for the detection, prevention and treatment of gut infections by P. aeruginosa.146 The designed EcN was able to sense the biomarker N-acyl homoserine lactone produced by P. aeruginosa, and autolyzed to release a biofilm degradation enzyme dispersin and pyocin S5 bacteriocin to eliminate the pathogen in the intestine. Moreover, the reprogrammed bacteria displayed long-term (over 15 days) prophylactic abilities against P. aeruginosa and was demonstrated to be more useful than treating a pre-established infection in mouse models. 3-Hydroxybutyrate (3HB) is a component of human ketone bodies with therapeutic effects in colitis. Yan et al. constructed an EcN overexpressing 3HB biosynthesis pathway.147 Compared to wild-type EcN, the engineered E. coli demonstrated better effects on mouse weights, colon lengths, occult blood levels, gut tissue myeloperoxidase activity and proinflammatory cytokine concentrations.147 However, the studies are the preliminary results in mice, they have not reached clinical trials yet. Further efforts are needed to evaluate their applications in human.
Engineered bacterial cells for metabolic disorders
Engineered gut microbes also have been used to target metabolic disorders.148 E. coli was designed to treat obesity synthesizing anorexigenic lipids precursors in mice with high-fat diet.149 Some efforts are made to degrade toxic compounds accumulated in patients via live bacteria. Kurtz et al. engineered an E. coli Nissle 1917 strain for converting ammonia to L-arginine in the intestine and reducing systemic hyperammonemia in both mouse and monkey models.150 Isabella et al. reprogrammed E. coli Nissle 1917 to overexpress phenylalanine degradation pathway to metabolize excess phenylalanine in phenylketonuria (PKU) patients. In the Pahenu2/enu2 PKU mouse model, oral uptake of the engineered bacterium significantly down-regulated blood phenylalanine concentration by 38%.151
Alcoholic liver disease is the major cause of liver disorders, widely risking the health of heavy drinkers.152 The engineered Bacillus subtilis and L. lactis could be employed to express ethanol degradation pathway (alcohol dehydrogenase and aldehyde dehydrogenase) for the detoxification of alcohol and alleviate liver injury from alcohol overconsumption.153 Moreover, the lectin regenerating islet-derived 3 gamma (REG3G) protein is decreased in the gastrointestinal tract during chronic ethanol uptake. L. reuteri was designed to overexpress the interleukin-22 (IL-22) gene, which increased REG3G abundance in the intestine, reduced inflammation and damage in liver using an alcoholic liver disease mouse model.154
Synthetic biology approaches have allowed the construction and design of engineered live biotherapeutics. Many cases are targeting future clinical applications. The examples discussed here indicate that, with the development of circuit designs and understanding in microorganism hosts, researchers can construct live biotherapeutics that function in a precise, systematic, inducible and robust manner. However, many efforts are still needed to weaken bacterial toxicity and increase the controllability in vivo.
Synthetic biology in the fabrication of emerging therapeutic materials
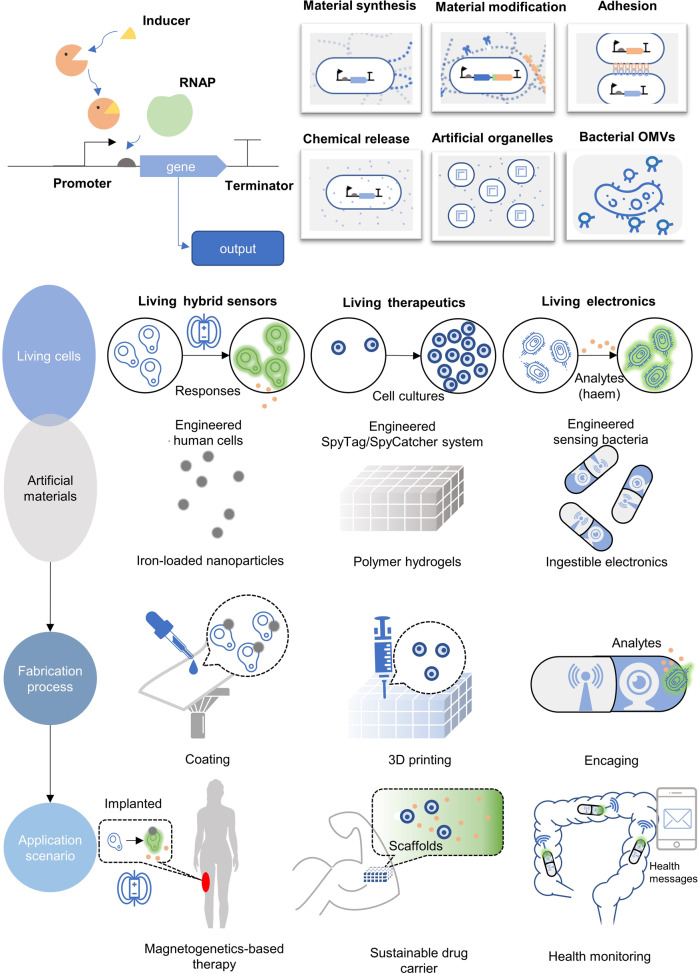
Besides engineered cells, engineered nanomaterials are also commonly used in medical fields. Nanobiotechnology aims to solve important biological concerns similar to drug delivery, disease diagnosis and treatment based on its unique physical, chemical and biological properties of micro-nano scale materials155,156 (Fig. 4). Nanomaterials possess unique mechanical, magnetic and electronic properties, able to respond to external signals, controlling their downstream circuits.157 However, traditional nanomaterials are generated from physical and chemical processes, the solvents and modifying molecules are frequently causing bio-safety issues.158 Recently, biological nanomaterials have been developed exhibiting their advantages in environmentally friendly, enhanced biocompatibility and bioactivity, and low tissue toxicity under the guidance of synthetic biology.159 Based on synthetic biology concepts and approaches, the genetic engineered bacteria,160 yeast161 and tobacco mosaic virus162 (TMV) can serve as bio-factories for nanomaterials.163 Mammalian cell-derived vesicles and nanoparticles have suitable biocompatibility, also commonly used as nanomedicines.164 Biological materials can be constructed and engineered with the help of synthetic biology, extending their application scenarios in modern disease treatments.
Fig. 4.
The designs and applications in synthetic material biology. Generally, a genetic circuit is constructed to synthesize biological materials or sense environments. The engineered bacteria are endowed with new characteristics like color change and unique surface properties. The applications for cells with excellular matrices are diverse including magnet field induced therapies, development of novel drug carrier or health monitoring via sophiscated biofabrication processes. This figure is partially inspired by the paper469
Synthetic biology in the artificial organelles
Following the principles of synthetic biology, biocatalysis or trigger-sensing modulus nanoparticles can be processed to self-assembly organelles,165,166 which are biomimicry of characteristics of living cells like enzyme reaction compartmentalization and stimuli-responses (Fig. 4). The design also provides new inputs for constructing artificial cells.167 Additionally, combinations of artificial organelles and engineered living cell chassis including CAR-T cells and engineered bacteria, the nano-living hybrid system can exert its dual effects to enhance therapeutic results or more strictly control of artificial systems.
Polymersomes are artificial hollow vesicles made by amphiphilic polymers, using as shells of artificial organelles. van Oppen et al. employed a polymersome-based system that was anchored with cell-penetrating peptides on its outer membrane. The artificial organelles possess inside catalase, allowing degradation of external reactive oxidative molecules, perform as a synthetic organelle, protecting the cells from ROS damages triggered via H2O2, which showed abilities in uptaking by human primary fibroblasts and human embryonic kidney cells.168 A similar design relying on polymersomes equipped with two enzymes and related transmembrane channels, was used to mimic cell peroxisomes. These organelles were able to deal with both H2O2 and superoxide radicals. The results further demonstrated the feasibility of artificial organelle with catalase activity. Based on similar ideas, engineered polymersomes may play a role in treating medical conditions including Parkinson’s, Alzheimer’s, Huntington’s, metabolic diseases, cancers and acatalasemia via harboring various therapeutic proteins inside of the artificial organelles.169,170
Moreover, the fusion of nanobiotechnology and synthetic biology may achieve novel functions. First, researchers can create “artificial lives” via assembling nanoparticles following the “bottle-up” principle. The idea can be applied in constructing biological components using inorganic scaffolds and functional nanomaterials with nucleic acids and protein inside of the nanoparticles.171,172 The “top-down” principle, or engineering natural cells for actual demands, can be used as a guidance when using nanomaterials in living cells for chimeric biological systems to increase the robustness, stability and sensitivity in specific medical applications.
Constructing nanoparticle-mediated genetic circuits
Auto-responses can be achieved via internal environmental stimulus to induce genetic switch ON/OFF173 (Fig. 4). However, the irreversible situation of genetic switches is a common and difficult problem.174,175 To circumvent the weakness of genetic constructs, nanoparticles are employed to sense signals for the transductions in vivo. Light, sound, heat and magnet stimuli are easy to respond for nanoparticles, they can be used as inducer systems for solid tumor and diabetes treatments. Yet the spatial-specific induction is hard for physical stimulus.176 Overall, via combining the advantages of genetic sensor and nanoparticles, it is feasible to convert physical stimuli into genetic switch with specified input signals by introducing nanoparticles for signal transduction, and the time-spatial control of gene expressions are realized.177
Near-infrared (NIR) light-responsible gene circuits are feasible for in vivo therapeutical applications for their better transmission of NIR light able to penetrate tissues and lower toxicity.178 NIR-sensing protein is identified in plants and bacteria, like the bacterial phytochromes (BphPs).179 However, NIR-sensing proteins are generally with low brightness.180 Also, the lacking of structural information hindered their rational engineering.180 To circumvent the disadvantages of NIR light-responsible protein, researchers have use nanomaterials converting NIR light into visible light. For example, Chen et al. employed nanoparticles doped with lanthanide to derive 980 nm NIR light into visible light, controlling genetic gates of opsin-expressing neurons in mice models.181,182 Another design uses plasmonic gold nanorods or photothermal responsible nanoparticles to transduct NIR light into up-regulation of temperature, then the promoters of heat-shock protein are activated for downstream gene expression.183,184 One disadvantage for nanoparticles is that they must be injected into human body, it could be solved by developing genetically engineered nanoparticles.185 Similar to magnetogenetics, in which biosynthesized ferritin can be used as a tool to prepare exogenous paramagnetic nanoparticles. However, the penetration depth needs much improvements in these samples (less than 1 cm), which is not enough for the applications of cell therapy demands in humans. Some researchers couple light-generating microdevices with photosensitive engineered therapeutic cells to address the problem (Fig. 4),186–188 patients can control the release of drugs via applications of their own smartphone or real-time monitoring their health. Besides, some genetic-encoded luminescent module can produce light in situ with a protein like various luciferases, all emit the desired wavelength with corresponding substrates. The in vivo light induces the photosensitive proteins that trigger transgene expressions for customized demands.189
In addition to optogenetics, magnetogenetics emerges for regulating the cell activities and has been applied for controlling of nanomaterial therapies remotely and non-invasively (Fig. 4).190–193 Magnetic fields can penetrate human body without losses, which is a preferred characteristic in deep-tissue targeted therapies. Previous magnetogenetics tools are mainly externally injected magnetic nanoparticles.190,192,194 The nanoparticles are usually with radius of <10 nm, toxicity free and water-soluble.190 Heating of nanoparticles using remote magnetic fields can activate temperature-sensitive cation channels in cells. the next-generation tools are heterologously expressed receptor-targeted ferritin proteins in the form of nanoparticles (iron-loaded particles) in engineered cells, which could sense and transduce magnetic signals to cell membrane-anchored receptors like transient receptor potential channel 1 (TRPV1) or TRPV4.191,193 The membrane receptors are ion channels allowing calcium influx with the magnet stimuli. The described gene circuit can be manipulated to control NFAT-dependent transcriptional regulators for downstream functional genes. Implanted engineered therapeutic cells can achieve target-specific treatments and precise control of therapeutic dosage, time and location under magnetic fields.
However, the mechanisms of the magnetic activation of the sensor channels are still not clear, the theories proposed are under debate for a long time.195 TRPV channels are activated by a variety of signals including but not limited to mechanical forces and heat. Recently, a new mechanism is raised to solve the problem that how radio-frequency weak magnetic fields (1 mT) could trigger transient responses in living cells with ferritin-anchored TRPV channels.196 The mechanism is the dissociation of free Fe3+ from ferritin protein, resulting in an enhanced oxidation of membrane lipids via increased production of reactive oxygen species (ROS).196 These oxidized lipids have the ability to turn on the TRPV channels, resulting in calcium influx.196–198 Recently, ROS is reported to be involved in the treatment of combined electric and static magnetic fields in type 2 diabetic mice to increase their insulin sensitivity.199 In this research, low-energy fields can induce the expression of nuclear factor erythroid 2-related factor 2 (Nrf2), a transcriptional regulator controlling ROS levels.199 Moreover, the local ROS accumulation does not have side effects in mice, it is promising to induce gene expression via electromagnetic fields mediated by redox states.200 Magnetogenetics are exhibiting its potentials in remote control and targeted therapies. However, more efforts are needed to establish the magnetogenetic platform. Despite improvements in recent years, the cell toxicity and biocompatibility are two main obstacles of magnetic nanoparticles that still challenges their in vivo applications.
Synthetic biology in drug delivery
The synthetic biology constructs are usually encapsulated in carriers for their functions in vivo. The safety concerns of viral vectors restrict their applications for editing human genome.201 Therefore, non-viral carriers are attracting more and more attentions. Nanotechnology can aid to deliver therapeutic agents including genetic circuits and genome engineering tools.202,203 With the advances in nanotechnology, more choices are available for targeted and controllable-release in DNA/RNA delivery system.204
One of the examples, the DNA/RNA delivery system based on liposome nanomaterials, has become an effective and potential gene therapy method, with a variety of artificial lipid vectors approved for clinical uses. For example, an RNAi therapeutic agent under the trade name Onpattro, has been developed by Alnylam Pharmaceuticals. The drug was approved in 2018 for the treatment of polyneuropathy.205 Liposomes are small lipid vesicles, the size is between 50 nm and 1 μm.206 Liposome are generally amphiphilic consisted with a hydrophobic tail and a hydrophilic head, employed for delivering drugs in various treatments.207 Because liposomes reduce drug toxicity, deliver drugs directly to targets via site-specific injections, and envelope drugs free from degradation, they have advantages over traditional drug therapies in delivery. CRISPR/Cas9-aided gene therapies are commonly using lipid-based nanoparticles integrating negatively charged mRNA, gRNA scaffolds and CRISPR genes with positively charged liposomes via electrostatic interactions.208 Felgner et al. first designed and used liposomes by enveloping DNA and delivered it to target mammalian cells in the plasma membrane, leading to DNA expression after its endocytosis.209 The liposome vector not only helps therapeutic DNAs to pass through the cell membrane barrier, but also protects them from DNase degradation and immune responses to maintain their activities. Partially inspired by the results that liposomes can be applied in human therapies, liposomes also have delivered mRNA encoding SARS-CoV-2 antigens to humans as vaccines. Both the Moderna mRNA-1273 and BioNTech/Pfizer BNT162b2 vaccines are encapsulated in liposomes, with their clinical use approvals.210
Nanotechnology can also aid synthetic biology to deliver chemicals.211,212 Nanocarriers deliver chemicals minimize off-target effects,213,214 enhancing therapeutic results215,216 compared to traditional drug administrations. External physical stimuli can also initiate the release of chemicals to make the system sustainable and controllable.217 Here, we discuss the application of synthetic biology-guided biological chemical carriers.
The genetically encoded post-translational modified protein can self-assemble to carry hydrophobic drugs.218 The protein with different structure and material properties can be easily manipulated at the amino sequence level. Based on synthetic biology approaches, Mozhdehi et al. designed and co-expressed an elastin-like polypeptide and an N-myristoyl transferase in E. coli.219 The N-myristoyl transferase enzyme modified the polypeptide with myristoyl groups in bacteria, generating a temperature-induced self-assembly behavior.219 The lipid core of the purified recombinant protein can carry hydrophobic compounds with a prolonged drug half-life.220 The protein can form complex assembly systems encapsulated with chemicals. Li et al. used an in silico designed cationic chimera near-infrared fluorescent protein and anionic carboxylate-terminated PEG to prepare a protein-PEG nanocarrier.221 The nanoprotein is amphiphilic, resulting in the aggregation and phase separation in aqueous solutions to form nanoparticles.221 The engineered nanoparticle achieved imaging of solid tumor and metastasis in vivo without transfections for the fluorescent nature of the protein,221 as well as the nanoprotein served as the long-term drug carrier, which can improve half-life and therapeutic effects of IL1-Ra significantly.222
Engineered bacterial outer-membrane vesicles (OMVs) as nanocarriers
Bacterial outer membrane vesicles (OMVs) are lipid spheres released from Gram-negative bacterial outer membranes, they can be used for trafficking biochemicals to other cells in the environment.223 The gene manipulation methods from synthetic biology can improve bio-originated nanoparticle abilities,224 expanding the application scenarios of outer-membrane vesicles (OMV) and engineered cells.225,226
Engineered OMV anchored with recombinant proteins are potentially used in medical and clinical fields (Fig. 4). The general strategy to surface display proteins in the engineering of OMV is to fuse their genes together in the OMV expression system. Many studies have employed the E. coli Cytolysin A (ClyA) protein as the fusion chassis to anchor exogenous proteins to OMV membranes.227–230 In recent studies, ClyA has been reported to successfully fuse to the domain 4 of Bacillus anthracis protective antigen, to extracellular domain of the influenza A matrix protein 2 (M2), and to GFP without influences OMV formation.231 The alternative strategy is to express proteins to the periplasm and assembly to the OMV when the fusion step hampers protein functions.232 However, the heterologous protein is enveloped inside of the OMV, which is a main disadvantage of the strategy. Bartolini et al. also employed the method to carry Chlamydia muridarum protein HtrA in OMVs as a vaccine against Chlamydia infections.233,234 Some proteins from Streptococcus spp. are expressed to the periplasm with the E. coli OmpA signal peptide to packed them into OMVs.235 Even though these proteins are located inside of the OMV, they were able to activate the immune responses,232,233,235 the generated IgG antibodies had strong activity to specific pathogens in murine models.225,232,235 The results indicated that antigen location is not a decisive factor in OMV-elicited immune responses.
Besides proteins, OMVs can be engineered to carry chemicals. LPS and capsular polysaccharides (CPS) decorating the cell membrane of pathogens are also vaccine candidates.236 However, polysaccharides trigger immune responses apart from T-cells, the immunological memory cannot be established.237 To circumvent the problem, polysaccharides are anchored to nanocarriers to elicit immunological memories. Polysaccharide and capsule synthesis genes are expressed in E. coli, packed into OMVs using the mentioned methods. The designed OMVs are potentially used as vaccines after further optimizations. Chen et al. employed the O-antigen polysaccharide from Francisella tularensis, the genes were heterologous expressed in E. coli to produce the glyco-modified OMVs.238,239 Mice injected with the engineered OMVs were protected against F. tularensis strains.238 Another similar design uses Streptococcus pneumoniae CPS (Sp-CPS) biosynthesis genes. They were overexpressed in E. coli, located both on the membrane of engineered OMVs and bacterial cells.240,241 After the vaccination via injecting these collected OMVs, the vaccine was effective in opsonophagocytosis assays and IgG antibodies were triggered against Sp-CPS.240 In general, synthetic biology approaches have developed better engineered OMVs for immunotherapies,242,243 with bright prospects in drug targeted-delivery and combined therapies.
Biomimetic medical adhesive materials
Traditional medical adhesive materials are limited in underwater uses, which hampered their applications in body fluids. Recently, some biomimetic designs are conducted to solve the problem based on synthetic biology ideas (Fig. 4).244 Many marine organisms (e.g. mussel and barnacle) have extraordinary adhesive capacities to rock surfaces,245,246 as they produce L-3,4-dihydroxyphenylalanine (DOPA) as an important component of the adhesion proteins in underwater surfaces.247 Zhong et al. reported a strong underwater adhesive by fusion of CsgA curli protein and mussel foot proteins.248 The excellent design reconciled the biocompatibility and adhesion activity, with the prospect of in vivo applications like tissue repairs. Zhang et al. is inspired by natural biomaterials like bones and mussel foots,249 they developed a Bacillus spp. extracellular matrix-based living glue.250 The live material is adhesive with regeneration abilities. Engineered mammalian cells could be constructed with adhesive proteins, serving as in vivo live functional glues. As summarized above, the novel live biomedical adhesives are hotspots in medical synthetic biology. However, most studies are focused in the material properties rather than their biocompatibility and biodegradability, adequate efforts are needed to promote the material for clinical applications.
Genetically encoded click chemistry in medical applications
Inspired by click chemistry, isopeptide bond was engineered for the establishment of protein-protein linkages.251 The genetic-encoded click chemistry is more applicable in living organisms compared with traditional click chemistry. The SpyTag/SpyCatcher system is an application of the natural click-like reaction among Gram-positive bacterial pilus,252,253 using biological ways to form stable chemical bonds between amino acids, additional modifications of biomacromolecules are not needed in click chemistry-oriented proteins (Fig. 4).254 Genetically encoded click chemistry (or Spy chemistry) is a powerful tool for materials made via synthetic biology.255
Hydrogels are cross-linked hydrophilic polymer networks,256 serving as carriers for biomacromolecules and stem cells due to their biocompatibilities and extracellular matrix (ECM) like properties.257 Hydrogel materials synthesized using chemical polymerizations are facing bioactivity problems.258 The protein characteristics are decided by amino acid sequences. Protein hydrogels are easier to synthesize and be controlled using various DNA sequences. Yang et al. employs the SpyTag/SpyCatcher system to synthesize a 4-arm star-like light-sensing protein. The protein can form rapid sol-gel and gel-sol phase transitions in response to AdoB12 and light, respectively.259 Biofilm-degrading glycosyl hydrolase PslG can be enveloped into the hydrogel, endowing the material with abilities against multidrug-resistant bacteria in chronic infections. Sun et al. designed a Spy-network containing multiple SpyTags and SpyCatchers in elastin-like proteins and the leukemia inhibitory factor. The proteins were turned into a high-mechanical strength hydrogel, allowing mouse embryonic stem cells to maintain pluri-potentials without adding other cytokines in the gel.260
Genetically encoded click chemistry has also used in the vaccine development. Some designed proteins can self-assembly into virus-like particles (VLPs) to surface display antigens for mimicking pathogens.261 Synthetic vaccines are causing more and more attentions for their efficiency and safety compared to canonical vaccines developed from dead or attenuated microorganisms. Genetically encoded click chemistry is a useful approach to modify the surface with heterologous antigens to enhance their immunogenicity.262,263 The easy formation of chemical bonds based on Spy chemistry provide a customized and convenient method to design synthetic vaccines via encoded protein self-assembly. Liu et al. developed a synthetic vaccine using the SpyCatcher/SpyTag chemistry via covalently ligating specific antigens and chemicals. The result demonstrates this engineered vaccine targets dendritic cells successfully.264 The generated protein-chemical hybrid vaccine remained the individual functions and had the ability to trigger B and T cell responses. Brune et al. engineered virus-like particles (VLPs) via exhibiting SpyCatcher on material surfaces, further enabling the modification of VLPs with SpyTag-expressing malarial antigens to develop novel vaccines.265 The VLP-antigen vaccine can trigger immune responses rapidly and efficiently via only one single immunization, indicating the potential of this effective, simple, and modular modification method.
Genetic code expansion for medical and pharmaceutical applications
A protein usually consists of 20 natural amino acids. To add non-canonical amino acids (ncAAs) into proteins, the genetic code expansion technology has been developed.266 ncAAs can be used to modify proteins via conjugation with peptides or chemicals depending on actual demands. Employing a termination codon (UAG/UGA/UAA), the heterologous bioorthogonal aminoacyl-tRNA synthase (aaRS)-tRNA pairs can add ncAAs to any site in a protein.267 Many different aaRS/tRNA pairs have been developed.268–270 The high-efficiency genetic code expansion devices allow the production of ncAA-containing protein and multiple ncAA-inserted proteins.271,272 The ncAA insertions are succeed in all main model organisms.273,274 Applications of the genetic code expansion system in medical fields are summarized here.
Genetic code expansion for antibody-drug conjugates
The antibody-drug conjugates (ADC) combine antigen-recognizing abilities of antibodies and tumor-killing capacities of chemicals commonly used in tumor therapies.275 Traditional ADC drugs are chemical modification of cysteines or lysines in the antibodies, which may affect the immunogenicity, stability and half-life.276 With the development of genetic code expansion technology, the introduction of a functional ncAA in the antibodies are feasible.277 The site-specific, high-efficiency conjugation between antibodies and chemicals can be achieved. Oller-Salvia et al. developed a novel genetic code expansion system incorporating a cyclopropene derivative of lysine into antibodies.278 The antibody conjugates to monomethyl auristatin E (MMAE) via a rapid Diels-Alder reaction.278 The resulting ADC was stable and effective in serum. Wang et al. conjugated the Lck inhibitor dasatinib to monoclonal antibody CXCR4 using genetic code expansion methods.279 The ADC avoids the side reactions during the chemical modification. The resulting dasatinib-antibody conjugate inhibited T-cell activation with low EC50 with negligible effects on cell viability.
Genetic code expansion in the bispecific antibodies
Bispecific antibodies (BsAb) possess two specific antigen binding sites with enhanced tumor-killing abilities.280 Some BsAbs have been approved by FDA.281 The traditional BsAb production method relies on fusions of proteins, resulting in steric hindrance in the ligand-binding domains.282 Additionally, the antibody production is at a low level with short half-life.283 Synthesis of BsAbs via chemical modifications meets similar questions to ADC productions.284 Genetic code expansion methods can conjugate two antibodies via a PEG linker to circumvent the challenges. Kim et al. introduced a ncAA (pAcF) to the antigen-binding fragment Fab region of anti-HER2 and anti-CD3 antibodies to form BsAb via two-step reactions.285 Picomolar concentrations of the BsAb induced effector-cell mediated cytotoxicity in vitro. Employing the Diels-Alder reaction between tetrazine-containing ncAA and bicyclononyne- containing ncAA, a BsAb recognizing BCMA was developed to treat multiple myeloma,286 successfully overcoming the drug-resistances in patients with multiple myeloma.
Genetic code expansion for engineering adeno-associated viruses (AAV)
AAVs are small parvovirus infecting human and primates.287 AAVs are commonly used in gene therapies to achieve non-pathogenic, broad host range and high transfection and expression efficiencies.288 However, the controllability and targeting ability are limited, hampering their applications. Zhang et al. used genetic code expansio to enhance the targeting ability of AAVs, conjugating cyclic arginyl-glycyl-aspartic acid (cRGD) to the shell protein of AAVs for targeting integrin.289 Erickson et al. engineered AAVs for opto-control of the infection.290 The R585 and R588 residues in vp1 protein of AAV2 were replaced by a light-sensitive ncAA, which hampered the interaction of vp1 and HSPG protein, resulting in inhibiting the infection of AAV. Exposed to UV light would remove the light-sensing moiety, recovered the infecting abilities of AAVs.290 The method enhances time-spatial controllability of AAV vectors.
Genetic code expansion for prolonging a protein half-life
PEG is commonly used in prolonging the half-life of therapeutic proteins.291 However, the random-modified PEG usually influences binding sites of therapeutic agents.292 Thus, genetic code expansion may provide advantages in modifying proteins. Cho et al. used genetic code expansion to site-specifically modify PEG in human growth hormone, which is highly instable in clinical applications.293 The modified human growth hormone is also with good batch to batch repeatability during the manufacturing processes. Some ncAAs increase protein stabilities per se. Xuan et al. demonstrated incorporation of a reactive isothiocyanate group into proteins to improve the heat-stability of myoglobin. Stable thiourea crosslinks were formed between the proteins.294 Similar designs using long chain thiol-containing or fluorinated ncAAs were also verified.295,296
Genetic code expansion for developing novel vaccines
ncAAs provide a wide variety of modifications of potential antigens that are candidates for vaccines. Gauba et al. inserted ncAAs containing nitrophenyl moiety into murine TNF-α protein for strong antibody response even with adjuvants.297 ncAA-addicted genetically modified organism (GMO) is useful for vaccine developments.298 The inactivated or attenuated pathogen-based vaccines usually have reduced effectiveness.299 Construction of a GMO strain that relies on ncAA to survive has been conducted to amplify live-virus vaccines. By introducing a termination codon in the genome of influenza A virus, HIV-1 or hepatitis D virus, the viruses can only replicate in engineered cells with specific aaRS/tRNA pairs and ncAAs. Si et al. inserted a termination codon in the NP protein of influenza A viruses, leading to a stronger immunogenicity and triggering broader immune responses.300 Based on the same idea, more and more live bacterial vaccines are under development.298 However, bacteria are more complex compared to viruses. Many mutation mechanisms can help bacteria to escape from expression terminations.301 The termination escapes restrict further applications with genetic code expansion in bacteria. Mandell et al. constructed a bacterium that metabolically dependent on ncAAs for survival.302 The bacterium exhibited unprecedented resistance to evolutionary escapes, providing a hint to the development of live bacteria vaccines.
Other medical applications of genetic code expansion
The genetic code expansion technology can be applied for the construction of controllable CAR-T cells. Incorporation of p-azidophenylalanine (pAzF) into the Fab allows the identification and conjugation of fluorescein isothiocyanate (FITC), activating the antibody for cancer treatments.303 Changing the inducer FITC to a short peptide was also proven applicable in cancer therapies.304 FITC or peptides were used as inducers of CAR-T cells that provide a more safety-control approach for immunotherapies. The genetic code expansion has also been applied for biosynthesis of peptide natural products. Nisin is a complex lanthipeptide with broad-spectrum of anti-bacterial activities. Zambaldo et al. introduced a number of ncAAs into nisin, equipping it with novel macrocyclic topologies with enhanced activities.305
The genetic code expansion methods are developing rapidly, modifying proteins both in vivo and site-specifically. The most sophisticated organism for this method is zebrafish and mouse.306 The method should be improved to apply in more higher species. Although more than 200 different ncAAs have been used for genetic code expansion, most ncAAs are based on similar structural units. Enriching structure types is another direction for developments. In the future, genetic code expansion technology will bring more delicate treatments for mankinds.
Synthetic biology in the biosynthesis of therapeutic drugs
In the recent years, synthetic biology approaches has become promising in sustainable and cost-effective production of phamarceuticals. Synthetic biology designs (Fig. 5) and constructs biological circuits or chassis including bacteria, yeasts, cell cultures or whole plants, for effectively producing high-value added phamarceutical products or phamarceutical intermediates. It offers a scalable and sustainable way for productions of bioproducts using CO2 based substrates, the production is rapid and robust, feasible for the large-scale industrial production, bioproducts can be manufactured without excessive cultivating and harvesting of medicinal plants (Table 1).
Fig. 5.
Technologies commonly used in synthetic biology. Various synthetic biology methods and tools have been developed to promote the design-build-test-learn cycle of cell factory construction, and these technologies are reforming the medical uses for synthetic biology. Pathway design is the first step, primary results are acquired via the constructed genetic circuits. Some optimizations are needed before next-round of tests, and the characteristics of the system is better understood from preliminary data. The design-build-test-learn cycles are iterative processes to improve robustness and efficacy of synthetic biology systems
As a classical field in synthetic biology, synthesis of pharmarceuticals is different from other medical applications. it generally uses yeast or bacteria as the production chassis. Synthetic biology concepts are extensively used in microorganisms, especially the DBTL (design-build-test-learn) (Fig. 5). DBTL cycle comprises the molecular biology designs and constructs in the beginning, and the experimental results are the basis for the new cycles of designs. The single-cell systems are easier to be manipulated than mammalian cells, In manmalian systems, the DBTL cycle can take very long, which is also an obstacle for mammalian synthetic biology. In the microbial synthesis of drugs, high-throughput screening and directed evolution are commonly used to accelerate experimental paces. Synthetic biology in microbes points to the direction of manmalian synthetic biology in a sense.
Biosynthesis of terpenoid drugs
Terpenoids are 5-carbon compound isoprene derivatives, also the largest group of plant secondary metabolites comprising approximately 60% of identified natural products.307 Many of them are bioactive medical ingradients.308 The anti-malaria drug, artemisinin, is sesquiterpene lactone containing an endoperoxide bridge.309 Initially, artemisinin was extracted from the plant Artemisia annua310 with a very low (0.01%-1%) content,311 much less than the actual medical demands. The chemical route to artemisinin is difficult and inefficient mainly due to the multiple-chiral centers of this molecule.312 The microbial synthesis of artemisinin prodrugs lowered drug cost. Biosynthesis of amorphadiene was a milestone in synthetic biology. The recombinant E. coli synthesized initially only 24 µg caryophyllene equivalent/ml.9 After continuous optimizations, another artemisinin prodrug, namely, artemisinic acid, reached 25 g/L produced by engineered yeast.22,23 The biosynthesis of artemisinic acid is a successful example of synthetic biology.
Taxol is a diterpene extracted from Pacific yew trees, serving as an anti-cancer agent.313 Its production mainly relies on laborious and low-efficiency plant cell cultures.314 Ajikumar et al. engineered E. coli cells to produce a taxol precursor, taxadiene, at a titer of 1 g/L.315
The ginsenosides are triterpene saponins found in the plant genus Panax with cancer prevention and anti-aging effects.316 Using the yeast cell-factory, various ginsenosides including ginsenoside Rh2 and ginsenoside compound K are synthesized with the titers of 2.2 g/L and 5.0 g/L, respectively.317,318 Microbial approach reduces the shortage of ginsenoside for clinical uses.
Biosynthesis of alkaloid drugs
Alkaloids are a variety of organic compounds containing at least one nitrogen atom.319 As a natural product, alkaloids are commonly used as they have pharmacological activities.320 Biosynthesis of alkaloids circumvent the bans on growing certain plants like poppy and marijuana.321 The formation of chiral centers during biosynthesis also outcompetes chemical synthesis for most chiral alkaloid compounds.322 Galanie et al. employed engineered yeast cells to produce thebaine and hydrocodone.323 Overexpression of 21 genes (for thebaine) or 23 genes (for hydrocodone) led to their formations of 6.6 × 10−5 g/L and 3 × 10−7 g/L, respectively. Nakagawa et al. improved the process using E. coli chassis.324 The titers for thebaine and hydrocodone were enhanced to 2.1 × 10−3 and 4 × 10−5 g/L, respectively. The production of opiates reached miligram level. Subsequent metabolic engineering are needed to promote biosynthesized opiates to meet market demands.
Similar to the biosynthesis of artemisinic acid, cannabinoids are natural products from cannabis, commonly used for pain killing and anxiolytic actions.325 (S)-Tetrahydropalmatine and cannabigerolic acid are two well-known cannabinoid hard to extract from plants.326 The biosynthesis processes for cannabigerolic acid were established by Luo et al. The yield from yeast reached 0.1 g/L.327 (S)-Tetrahydropalmatine biosynthesized by yeast by Hafner et al. reached 3.6 × 10−6 g/L, a successful concept-of-proof for microbial production of complicated cannabinoids.328
Biosynthesis of amino acid-derivative drugs
Using amino acids as building blocks, amino acid derivatives are also played an important role in human health.329 This class of compounds is usually synthesized via biological routes rather than chemical synthesis for their multiple chirality moieties. Compared with alkaloid and terpenoids, amino acid-derivatives are more simple in structures with diversity.329 Psilocybin is a L-tryptophan derivative with effects of anti-drug-addiction, relieving depression and anti-post-traumatic stress disorder effects.330 E. coli or Saccharomyces cerevisiae have been engineered to heterologously express the synthetic pathways, forming 1.2 g/L and 0.6 g/L psilocybin, respectively.330,331 Dencichine, also known as β-N-oxalyl-L-α,β-diaminopropionic acid (β-ODAP), is a plant metabolite first isolated from Lathyrus sativus seeds. Dencichine can induce platelet aggregation in human blood, and it is the main effective component of the Chinese medicine Yunnan Baiyao.332,333 The authors optimized metabolic flux to dencichine in E. coli to the production with final titer reaching 1.29 g L−1 and a yield of 0.28 g g−1 glycerol.334 Microbial production of dencichine exhibits an example of employing artificial enzymes and pathways to produce a desired chemical in synthetic biology applications.
Biocatalytic of asymmetric synthesis
Synthetic biology can assist multiple chiral-center chemical developments. Sitagliptin (Januvia) is a commonly used diabetes treatment, inhibiting DPP-4 enzyme in a competitive manner, reducing the cleavage of GLP-1 to increase the secretion of insulin.335 The market of Januvia reached 1.4 billion dollars by 2021.336 For chemical synthesis of sitagliptin, the chiral amine is transferred via a rhodium-based chiral catalyst with a low stereoselectivity and the product contaminated with rhodium.337 A transaminase and synthetic-biology-based engineering approach based on homologous modeling and saturation mutagenesis, a process was developed that substantially improved the efficiency and purity for sitagliptin synthesis.337
Cell-free synthetic biology in medical applications
Till now, efforts in synthetic biology have mainly focused on reprogramming organisms, development of genetic circuits and biological modules. However, because our knowledge on how life works is limited, the complex feature of creatures hindered progresses in synthetic biology. User-defined systems can solve the problem. Cell-free system is prepared to perform in vitro biological activities free from living cells (e.g. transcription and translation).338 As it is open, easy to control, flexible and high tolerance to cytotoxicity,339,340 the system has been used in synthesizing proteins that are difficult to express or toxic in cells (Fig. 6).341 Moreover, cell-free systems fit well to high-throughput screening.342 Recently, with the development of cell-free biosensing diagnosis343 and the advances in lyophilization,344 the applications of cell-free synthetic biology have expanded into medical and pharmaceutical fields.345
Fig. 6.
The charasteristics of cell-free synthetic biology. The types, advantages, products, and bottlenecks of cell-free systems are summarized in this figure. Generally, cell-free systems are used to produce pharmaceuticals or served as in vitro sensors. The main advantages are convenient, flexible and high tolerance to cytotoxicity. After solving the problems like high cost and instabilities, the system is promising for actual medical applications
Cell-free synthetic biology in pharmaceutical protein synthesis
Protein and peptide drugs are target-specific mostly with high activities and low toxicity for medical uses.346–348 Many well-known drugs are proteins or peptides like Trastuzumab (Herceptin),349 Adalimumab (Humira),350 Insulin Glargine (Lantus)351 and 13-valent pneumococcal conjugate vaccine (PCV13).352 70% of the protein drugs are produced using the CHO cells.353 However, some proteins are toxic for growth of cell hosts.354 Cell-free protein synthesis (CFPS) provides a solution to the toxicity problems.355 Additionally, screening of intracellular proteins are feasible in CFPS systems,356 also lyophilization technologies allow the cell-free system to maintain highly active after one-year preservation.357
The cell-lysate based- and purified component systems are two commonly used CFPS systems.358 Theoretically, any organism could be used as the source in cell-lysate based system. The most common cell extract is from E. coli, wheat germ and yeast.359 E. coli lysate is frequently used for protein synthesis,360 wheat germ lysates for construction of protein arrays,361,362 yeast lysates for synthesis of glycoproteins.363 The purified component system comprises all purified translational-elements. Shimizu et al. developed a cell-free system using 36 transcription/translation related enzymes with highly purified ribosomes.364 The system is efficient although minimum. However, the high cost of purified components hampers its applications. The cell-lysate based system is the first choice of CFPS systems.
Vaccination is the most effective way for pandemic prevention.365 Cell-free systems provide a platform for rapid production of vaccines. Kanter et al. developed a cell-free system for highly effective production of a fusion protein consisting of a single chain Fv antibody fragment (scFv) connected to granulocyte-macrophage colony-stimulating factor (GM-CSF), a vaccine of B-cell lymphoma.366 Lu et al. described a CFPS overexpressing a domain of pandemic H1N1 influenza virus for potentially and broadly protective influenza vaccines.367 Besides bacterial systems, eukaryotic cell-free systems can express complex vaccines. Tsuboi et al. successfully expressed three malarial proteins in yeast lysate based cell-free systems, which is hard to produce in recombinant cells.368
Antibodies are important for disease treatments and diagnosis.369 CFPS is commonly used during the synthesis of antibodies. Ryabova et al. successfully produced functional scFv fragments in E. coli lysate-based cell-free system.370 Post-translational modification (PTM) is the final maturation step of proteins.371 Glycosylation is the main form of PTM important for maintaining the half-life and activity of protein drugs including some antibodies.372,373 CFPS can also introduce functional PTM to proteins. Jaroentomeechai et al. used CFPS to synthesize N-glycosylated scFv using E. coli cell-free systems.374 Overall, cell-free systems are useful complements to recombinant expressing systems for their rapid and on-demand properties.
Cell-free synthetic biology for diagnosis
Generally, detection of pathogens are based-on biosensors.375 The sensing elements include enzymes, transcriptional factors, antibodies, organelles, whole-cells and tissues.376–380 Although many biosensors are rapid and sensitive, the disadvantages are including the instability of enzymes, biosafety concerns of whole-cell biosensors and the complexity in preparing microfluidic sensors.293,381 Therefore, cell-free sensors are developed. Pellinen et al. used luciferase as the reporter, Tet repressor and MerR regulatory proteins as the sensing elements, for the detection of tetracycline and the toxic mercury in cell-free systems.382 Davies et al. constructed a cell-free protein array to screen high-immunogenicity proteins in human serums after virus infections, for the prophylactic uses and diagnosis.383 In remote regions or harsh environments, cell-free systems lyophilized and attached on papers (or other matrices) are convenient and stable.384 Pardee et al. employed lyophilized cell-free sensors to rapid determination of Ebola and Zika virus.385,386 Future cell-free synthetic biology may lead to sophisticated design and synthesis of more complicated therapeutic agents, or rapid and sensitive biosensors for chronic disease diagnostics.
Discussion and future perspectives
Since the rapid developments started from more than a decade ago, synthetic biology has grown substantially and has emerged with many achievements, both in science and application aspects (Fig. 1). In this review, we summarized the advanced strategies and designs in synthetic biology for traditional pharmaceutical and medical applications, such as engineered smart cells (Fig. 2),387 live probiotic therapeutics,151 diagnostics,388 stem cells,83 drug production,23 nanocarriers389 and artificial vaccine developments.300 The novel approach will enrich clinical regimens, shorten drug development cycle and lower pharmaceutical prices.
Synthetic biology approaches that most probably bring (or has brought) dramatic changes in biomedical fields include: the use of light for time-spartial controllable precise cell therapeutics (optigenetics), designed bacteria to target cancer cells, engineered cells rewiring metabolic flux in human or engineer the gut-brain-liver axis (engineered live therapeutics). Recent studies have shown possibilities that biosystems mentioned above are functioning well in manmalian and exhibiting considerable therapeutic effects in animal models or even volunteers.70 However, they are just developed in their early stages. Many efforts are still needed to translate the lab findings to commercial products for patients.
The personalized engineered medicine is the next-gneration treatment strategy in the future. Smart therapeutics based on genetic-encoded circuits that can intepret environmental signal into effector activities will be commonly used. The auto-regulated therapeutic cells that sense diagnostic inputs for therapeutic outputs are one-station solutions for diagnosis, disease prevention and treatments (Fig. 2). Some applictions like CAR-T therapies have entered clinical stages, but most of the smart cells are not. Many attempts have failed in the early clinical, mainly for the low therapeutical abilities and unexpected side effects in human. Future works should emphasis on their safety as well as the efficacy and stability in treatments.
The combination of synthetic biology and artificial intelligence (AI) is promising to accelerate the advances both in medical and pharmaceutical fields, although the field is in initial stage. AI is a hit not only in computer science, but also in biology research.390 The AI prediction of protein structures ranks as the top one in ten scientific breakthroughs in 2021.391 The era of AI and big data is arriving, in-depth learning technique is advantageous in the characterization of complex objects,392 fusion of multimodal features393 and auto-sample generations.394 AI can be applied in the synthetic biology field. At present, the combined applications of AI and synthetic biology have mainly been focused on the following three aspects, including, firstly, foresight of future research directions; collection of related synthetic biology data, then distinguish the casual link to analyze and evaluate the application and development directions. This is very helpful in analysis of numerous clinical datasets. Secondly, in the pharmaceutical applications, screening effective drugs based on AI and bioinformatic big data, testing candidate chemicals and simulating the therapeutic processes in disease models. It is a high-throughput method saving much manpower. Thirdly, development of novel drugs via reconstruction or modification the genomes by in-depth AI learning models, synthesizing novel compounds for drug discoveries. In the future, AI is promising to assist medical synthetic biology in designing more complicated systems (engineered cells or tissues) based on actual demands, substantially decreasing labor amounts of researchers.
However, some shortages and bottlenecks are to tackle for medical synthetic biology. Much effort is needed before the synthetic biology-based therapy become an available clinical option (Fig. 7). Although engineered cells containing genetic circuits are one of the most exciting designs in recent decades, they have limitations in actual uses of extracellular, signal-transduction free diseases which can be treated via traditional ways.395 Tissue-specific engineered therapeutics are not succeed till now. The interferences of manmalian metabolisms are remain unknown. Solving these problems will be helpful for synthetic biology-based clinical applications.
Fig. 7.
The present situations, technical bottlenecks and future developments of synthetic biology based gene therapies. Some diagnosis and therapeutical approaches are available via rewiring metabolic and (or) signaling pathways in present synthetic biology. However, some bottlenecks like safety, versatility and efficacy are needing to tackle. Besides, novel designs such as AI-aided synthetic biology and rationally constructed live organisms and proteins are progressing
The majority of synthetic biology is still applied in microbes. However, most of the major issues, especially in solving human health problems, are needed for mammalian systems. Therefore, much efforts must be made for advancing mammalian synthetic biology to the next-generation therapeutic treatments, including the engineering of synthetic gene networks for disease treatments, tissue engineering or stem-cell generation and differentiation.
Additionally, synthetic biology-based therapeutics are still facing same social problems in ethical and legal fields similar to transgenic foods and stem cell therapies, although they can be imposed of better control from stringent pathways.
Even so, the future for synthetic biology-based therapeutics are promising, with new tools and applications developed in biomedical fields and highly-efficient microbial pharmaceutical production in the twenty-first century.
Acknowledgements
This study was supported by grants from the Ministry of Science and Technology of China [Grant number 2018YFA0900200], National Natural Science Foundation of China [Grant number 32130001], Center of Life Sciences of Tsinghua-Peking University, the Shuimu Tsinghua Scholar Program and Chunfeng Foundation. This project is also funded by the National Natural Science Foundation of China [Grant numbers 31961133017, 31961133018]. These grants are part of MIX-UP, a joint NSFC and EU H2020 collaboration. In Europe, MIX-UP has received funding from the European Union’s Horizon 2020 research and innovation program [grant agreement Number 870294].
Author contributions
Y.X. made the charts and wrote the original draft. Y.X, L.X., and Z.C. sketched the figures. The work is supervised by G.Q.C. and the paper was reviewed and edited by G.Q.C. All authors have read and approved the article.
Competing interests
The authors declare no competing interests.
References
- 1.Le Duc, S. (eds) The Mechanism of Life (Rebman Company, 1914).
- 2.Hopkins FG. The centenary of Wöhler’s synthesis of urea (1828–1928) Biochem. J. 1928;22:1341. doi: 10.1042/bj0221341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun Y. The creation of synthetic crystalline bovine insulin. Protein Cell. 2015;6:781–783. doi: 10.1007/s13238-015-0221-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caruthers MH. Gene synthesis machines: DNA chemistry and its uses. Science. 1985;230:281–285. doi: 10.1126/science.3863253. [DOI] [PubMed] [Google Scholar]
- 5.Crick FH. The genetic code-yesterday, today, and tomorrow. Cold Spring Harb. Symp. Quant. Biol. 1966;31:3–9. doi: 10.1101/SQB.1966.031.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Crick F. Central dogma of molecular biology. Nature. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 7.Slepchenko BM, Terasaki M. Bio-switches: what makes them robust? Curr. Opin. Genet. Dev. 2004;14:428–434. doi: 10.1016/j.gde.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Jayaraman A, Wood TK. Bacterial quorum sensing: signals, circuits, and implications for biofilms and disease. Annu. Rev. Biomed. Eng. 2008;10:145–167. doi: 10.1146/annurev.bioeng.10.061807.160536. [DOI] [PubMed] [Google Scholar]
- 9.Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 2003;21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 10.Knight, T. Idempotent vector design for standard assembly of biobricks. MIT’s DSpace (2003). https://dspace.mit.edu/handle/1721.1/21168;jsessionid=98E77285D00B9BAFF807B2B0268D7DE6.
- 11.Smolke CD. Building outside of the box: iGEM and the BioBricks Foundation. Nat. Biotechnol. 2009;27:1099–1102. doi: 10.1038/nbt1209-1099. [DOI] [PubMed] [Google Scholar]
- 12.Breithaupt H. The engineer’s approach to biology. EMBO Rep. 2006;7:21–23. doi: 10.1038/sj.embor.7400607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts M, Cranenburgh R, Stevens M, Oyston P. Synthetic biology: biology by design. Microbiology. 2013;159:1219. doi: 10.1099/mic.0.069724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian J, Ma K, Saaem I. Advancing high-throughput gene synthesis technology. Mol. Biosyst. 2009;5:714–722. doi: 10.1039/b822268c. [DOI] [PubMed] [Google Scholar]
- 16.Mardis ER. Next-generation sequencing platforms. Annu. Rev. Anal. Chem. 2013;6:287–303. doi: 10.1146/annurev-anchem-062012-092628. [DOI] [PubMed] [Google Scholar]
- 17.Zeng W, Guo L, Xu S, Chen J, Zhou J. High-throughput screening technology in industrial biotechnology. Trends Biotechnol. 2020;38:888–906. doi: 10.1016/j.tibtech.2020.01.001. [DOI] [PubMed] [Google Scholar]
- 18.Gurdo N, Volke DC, Nikel PI. Merging automation and fundamental discovery into the design-build-test-learn cycle of nontraditional microbes. Trends Biotechnol. 2022;40:1148–1159. doi: 10.1016/j.tibtech.2022.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 20.Cameron DE, Bashor CJ, Collins JJ. A brief history of synthetic biology. Nat. Rev. Microbiol. 2014;12:381–390. doi: 10.1038/nrmicro3239. [DOI] [PubMed] [Google Scholar]
- 21.Gibson DG, et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 22.Ro DK, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 23.Paddon CJ, et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496:528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- 24.Kitney R, Freemont P. Synthetic biology-the state of play. FEBS Lett. 2012;586:2029–2036. doi: 10.1016/j.febslet.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Si T, Zhao H. A brief overview of synthetic biology research programs and roadmap studies in the United States. Synth. Syst. Biotechnol. 2016;1:258–264. doi: 10.1016/j.synbio.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feins S, Kong W, Williams EF, Milone MC, Fraietta JA. An introduction to chimeric antigen receptor (CAR) T-cell immunotherapy for human cancer. Am. J. Hematol. 2019;94:S3–S9. doi: 10.1002/ajh.25418. [DOI] [PubMed] [Google Scholar]
- 27.Sadelain M, Rivière I, Brentjens R. Targeting tumours with genetically enhanced T lymphocytes. Nat. Rev. Cancer. 2003;3:35–45. doi: 10.1038/nrc971. [DOI] [PubMed] [Google Scholar]
- 28.Sadelain M, Brentjens R, Rivière I. The basic principles of chimeric antigen receptor designmaking better chimeric antigen receptors. Cancer Discov. 2013;3:388–398. doi: 10.1158/2159-8290.CD-12-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Der Stegen SJ, Hamieh M, Sadelain M. The pharmacology of second-generation chimeric antigen receptors. Nat. Rev. Drug Discov. 2015;14:499–509. doi: 10.1038/nrd4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stone JD, Kranz DM. Role of T cell receptor affinity in the efficacy and specificity of adoptive T cell therapies. Front. Immunol. 2013;4:244. doi: 10.3389/fimmu.2013.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perica K, Curran KJ, Brentjens RJ, Giralt SA. Building a CAR garage: preparing for the delivery of commercial CAR T cell products at Memorial Sloan Kettering Cancer Center. Biol. Blood Marrow Transplant. 2018;24:1135–1141. doi: 10.1016/j.bbmt.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li S, Young KH, Medeiros LJ. Diffuse large B-cell lymphoma. Pathology. 2018;50:74–87. doi: 10.1016/j.pathol.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 33.Locke FL, et al. Long-term safety and activity of axicabtagene ciloleucel in refractory large B-cell lymphoma (ZUMA-1): a single-arm, multicentre, phase 1-2 trial. Lancet Oncol. 2019;20:31–42. doi: 10.1016/S1470-2045(18)30864-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haslauer T, Greil R, Zaborsky N, Geisberger R. CAR T-cell therapy in hematological malignancies. Int. J. Mol. Sci. 2021;22:8996. doi: 10.3390/ijms22168996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selim AG, et al. CAR-T cell therapy: practical guide to routine laboratory monitoring. Pathology. 2021;53:408–415. doi: 10.1016/j.pathol.2021.02.002. [DOI] [PubMed] [Google Scholar]
- 36.Viardot A, Wais V, Sala E, Koerper S. Chimeric antigen receptor (CAR) T-cell therapy as a treatment option for patients with B-cell lymphomas: perspectives on the therapeutic potential of Axicabtagene ciloleucel. Cancer Manag. Res. 2019;11:2393. doi: 10.2147/CMAR.S163225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sadelain M, Rivière I, Riddell S. Therapeutic T cell engineering. Nature. 2017;545:423–431. doi: 10.1038/nature22395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei J, Han X, Bo J, Han W. Target selection for CAR-T therapy. J. Hematol. Oncol. 2019;12:1–9. doi: 10.1186/s13045-019-0758-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei J, et al. The model of cytokine release syndrome in CAR T-cell treatment for B-cell non-Hodgkin lymphoma. Signal Transduct. Target. Ther. 2020;5:1–9. doi: 10.1038/s41392-020-00256-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brudno JN, Kochenderfer JN. Recent advances in CAR T-cell toxicity: mechanisms, manifestations and management. Blood Rev. 2019;34:45–55. doi: 10.1016/j.blre.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srivastava S, Riddell SR. Engineering CAR-T cells: design concepts. Trends Immunol. 2015;36:494–502. doi: 10.1016/j.it.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carpenter RO, et al. B-cell maturation antigen is a promising target for adoptive T-cell therapy of multiple myeloma anti-BCMA chimeric antigen receptor. Clin. Cancer Res. 2013;19:2048–2060. doi: 10.1158/1078-0432.CCR-12-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fisher SA, et al. Mesenchymal stromal cells as treatment or prophylaxis for acute or chronic graft‐versus‐host disease in haematopoietic stem cell transplant (HSCT) recipients with a haematological condition. Cochrane Database Syst. Rev. 2019;1:1. doi: 10.1002/14651858.CD009768.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Costa LJ, Usmani SZ. Defining and managing high-risk multiple myeloma: current concepts. J. Natl Compr. Canc. Netw. 2020;18:1730–1737. doi: 10.6004/jnccn.2020.7673. [DOI] [PubMed] [Google Scholar]
- 45.Ding L, Hu Y, Huang H. Novel progresses of chimeric antigen receptor (CAR) T cell therapy in multiple myeloma. Stem Cell Invest. 2021;8:1. doi: 10.21037/sci-2020-029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laabi Y, et al. A new gene, BCM, on chromosome 16 is fused to the interleukin 2 gene by at (4; 16)(q26; p13) translocation in a malignant T cell lymphoma. EMBO J. 1992;11:3897–3904. doi: 10.1002/j.1460-2075.1992.tb05482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novak AJ, et al. Expression of BCMA, TACI, and BAFF-R in multiple myeloma: a mechanism for growth and survival. Blood. 2004;103:689–694. doi: 10.1182/blood-2003-06-2043. [DOI] [PubMed] [Google Scholar]
- 48.Ng LG, et al. B cell-activating factor belonging to the TNF family (BAFF)-R is the principal BAFF receptor facilitating BAFF costimulation of circulating T and B cells. J. Immunol. 2004;173:807–817. doi: 10.4049/jimmunol.173.2.807. [DOI] [PubMed] [Google Scholar]
- 49.Ali SA, et al. T cells expressing an anti-B-cell maturation antigen chimeric antigen receptor cause remissions of multiple myeloma. Blood. 2016;128:1688–1700. doi: 10.1182/blood-2016-04-711903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raje N, et al. Anti-BCMA CAR T-cell therapy bb2121 in relapsed or refractory multiple myeloma. N. Engl. J. Med. 2019;380:1726–1737. doi: 10.1056/NEJMoa1817226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Friedman KM, et al. Effective targeting of multiple B-cell maturation antigen-expressing hematological malignances by anti-B-cell maturation antigen chimeric antigen receptor T cells. Hum. Gene Ther. 2018;29:585–601. doi: 10.1089/hum.2018.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Munshi NC, et al. Idecabtagene vicleucel (ide-cel; bb2121), a BCMA-targeted CAR T-cell therapy, in patients with relapsed and refractory multiple myeloma (RRMM): Initial KarMMa results. J. Clin. Oncol. 2020;38:15. doi: 10.1200/JCO.2020.38.15_suppl.8503. [DOI] [Google Scholar]
- 53.Wang Z, et al. Using apelin-based synthetic Notch receptors to detect angiogenesis and treat solid tumors. Nat. Commun. 2020;11:1–13. doi: 10.1038/s41467-020-15729-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Z, Yu Z, Cai Y, Du R, Cai L. Engineering of an enhanced synthetic Notch receptor by reducing ligand-independent activation. Commun. Biol. 2020;3:1–7. doi: 10.1038/s42003-020-0848-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang H, Zang C, Liu XS, Aster JC. The role of Notch receptors in transcriptional regulation. J. Cell. Physiol. 2015;230:982–988. doi: 10.1002/jcp.24872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choe JH, et al. SynNotch-CAR T cells overcome challenges of specificity, heterogeneity, and persistence in treating glioblastoma. Sci. Transl. Med. 2021;13:eabe7378. doi: 10.1126/scitranslmed.abe7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He L, Huang J, Perrimon N. Development of an optimized synthetic Notch receptor as an in vivo cell-cell contact sensor. Proc. Natl Acad. Sci. USA. 2017;114:5467–5472. doi: 10.1073/pnas.1703205114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwarz KA, Daringer NM, Dolberg TB, Leonard JN. Rewiring human cellular input-output using modular extracellular sensors. Nat. Chem. Biol. 2017;13:202–209. doi: 10.1038/nchembio.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dolberg TB, et al. Computation-guided optimization of split protein systems. Nat. Chem. Biol. 2021;17:531–539. doi: 10.1038/s41589-020-00729-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baeumler TA, Ahmed AA, Fulga TA. Engineering synthetic signaling pathways with programmable dCas9-based chimeric receptors. Cell Rep. 2017;20:2639–2653. doi: 10.1016/j.celrep.2017.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu I, et al. Modular design of synthetic receptors for programmed gene regulation in cell therapies. Cell. 2022;185:1431–1443. doi: 10.1016/j.cell.2022.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Manhas J, Edelstein HI, Leonard JN, Morsut L. The evolution of synthetic receptor systems. Nat. Chem. Biol. 2022;18:244–255. doi: 10.1038/s41589-021-00926-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Daringer NM, Dudek RM, Schwarz KA, Leonard JN. Modular extracellular sensor architecture for engineering mammalian cell-based devices. ACS Synth. Biol. 2014;3:892–902. doi: 10.1021/sb400128g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roybal KT, et al. Precision tumor recognition by T cells with combinatorial antigen-sensing circuits. Cell. 2016;164:770–779. doi: 10.1016/j.cell.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Engelowski E, et al. Synthetic cytokine receptors transmit biological signals using artificial ligands. Nat. Commun. 2018;9:1–15. doi: 10.1038/s41467-018-04454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ren J, et al. Pancreatic islet cell therapy for type I diabetes: understanding the effects of glucose stimulation on islets in order to produce better islets for transplantation. J. Transl. Med. 2007;5:1–15. doi: 10.1186/1479-5876-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ashcroft FM, Rorsman P. KATP channels and islet hormone secretion: new insights and controversies. Nat. Rev. Endocrinol. 2013;9:660–669. doi: 10.1038/nrendo.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hashimoto N, et al. Ablation of PDK1 in pancreatic β cells induces diabetes as a result of loss of β cell mass. Nat. Genet. 2006;38:589–593. doi: 10.1038/ng1774. [DOI] [PubMed] [Google Scholar]
- 69.Ozougwu J, Obimba K, Belonwu C, Unakalamba C. The pathogenesis and pathophysiology of type 1 and type 2 diabetes mellitus. J. Physiol. Pathophysiol. 2013;4:46–57. doi: 10.5897/JPAP2013.0001. [DOI] [Google Scholar]
- 70.Xie M, et al. β-cell–mimetic designer cells provide closed-loop glycemic control. Science. 2016;354:1296–1301. doi: 10.1126/science.aaf4006. [DOI] [PubMed] [Google Scholar]
- 71.Patel M, Yang S. Advances in reprogramming somatic cells to induced pluripotent stem cells. Stem Cell Rev. Rep. 2010;6:367–380. doi: 10.1007/s12015-010-9123-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 73.Yu Y, Alkhawaji A, Ding Y, Mei J. Decellularized scaffolds in regenerative medicine. Oncotarget. 2016;7:58671. doi: 10.18632/oncotarget.10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Araki R, et al. Negligible immunogenicity of terminally differentiated cells derived from induced pluripotent or embryonic stem cells. Nature. 2013;494:100–104. doi: 10.1038/nature11807. [DOI] [PubMed] [Google Scholar]
- 75.Liu X, Li W, Fu X, Xu Y. The immunogenicity and immune tolerance of pluripotent stem cell derivatives. Front. Immunol. 2017;8:645. doi: 10.3389/fimmu.2017.00645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu M, et al. Stable expression of a truncated TLX variant drives differentiation of induced pluripotent stem cells into self-renewing neural stem cells for production of extracellular vesicles. Stem Cell. Res. Ther. 2022;13:1–17. doi: 10.1186/s13287-022-03131-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Park IH, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Freedman BS, et al. Reduced ciliary polycystin-2 in induced pluripotent stem cells from polycystic kidney disease patients with PKD1 mutations. J. Am. Soc. Nephrol. 2013;24:1571–1586. doi: 10.1681/ASN.2012111089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morrison M, et al. StemBANCC: governing access to material and data in a large stem cell research consortium. Stem Cell Rev. Rep. 2015;11:681–687. doi: 10.1007/s12015-015-9599-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sharma A, et al. High-throughput screening of tyrosine kinase inhibitor cardiotoxicity with human induced pluripotent stem cells. Sci. Transl. Med. 2017;9:eaaf2584. doi: 10.1126/scitranslmed.aaf2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shaheen N, et al. Human induced pluripotent stem cell-derived cardiac cell sheets expressing genetically encoded voltage indicator for pharmacological and arrhythmia studies. Stem Cell Rep. 2018;10:1879–1894. doi: 10.1016/j.stemcr.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim C. Disease modeling and cell based therapy with iPSC: future therapeutic option with fast and safe application. Blood Res. 2014;49:7. doi: 10.5045/br.2014.49.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cichocki F, et al. iPSC-derived NK cells maintain high cytotoxicity and enhance in vivo tumor control in concert with T cells and anti-PD-1 therapy. Sci. Transl. Med. 2020;12:eaaz5618. doi: 10.1126/scitranslmed.aaz5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sarkar TJ, et al. Transient non-integrative expression of nuclear reprogramming factors promotes multifaceted amelioration of aging in human cells. Nat. Commun. 2020;11:1–12. doi: 10.1038/s41467-020-15174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pang L. Toxicity testing in the era of induced pluripotent stem cells: A perspective regarding the use of patient-specific induced pluripotent stem cell-derived cardiomyocytes for cardiac safety evaluation. Curr. Opin. Toxicol. 2020;23:50–55. doi: 10.1016/j.cotox.2020.04.001. [DOI] [Google Scholar]
- 86.Wei H, Wang C, Guo R, Takahashi K, Naruse K. Development of a model of ischemic heart disease using cardiomyocytes differentiated from human induced pluripotent stem cells. Biochem. Biophys. Res. Commun. 2019;520:600–605. doi: 10.1016/j.bbrc.2019.09.119. [DOI] [PubMed] [Google Scholar]
- 87.Park TS, et al. Vascular progenitors from cord blood-derived induced pluripotent stem cells possess augmented capacity for regenerating ischemic retinal vasculature. Circulation. 2014;129:359–372. doi: 10.1161/CIRCULATIONAHA.113.003000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tang H, et al. Tracking induced pluripotent stem cells-derived neural stem cells in the central nervous system of rats and monkeys. Cell. Reprogram. 2013;15:435–442. doi: 10.1089/cell.2012.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen Y, et al. Reversible reprogramming of cardiomyocytes to a fetal state drives heart regeneration in mice. Science. 2021;373:1537–1540. doi: 10.1126/science.abg5159. [DOI] [PubMed] [Google Scholar]
- 90.Takebe T, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 91.Grompe M, Strom S. Mice with human livers. Gastroenterology. 2013;145:1209–1214. doi: 10.1053/j.gastro.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 92.Guo R, et al. Stem cell-derived cell sheet transplantation for heart tissue repair in myocardial infarction. Stem Cell. Res. Ther. 2020;11:1–13. doi: 10.1186/s13287-019-1536-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li J, et al. Engineered tissue for cardiac regeneration: current status and future perspectives. Bioengineering. 2022;9:605. doi: 10.3390/bioengineering9110605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maeda T, Mandai M, Sugita S, Kime C, Takahashi M. Strategies of pluripotent stem cell-based therapy for retinal degeneration: update and challenges. Trends Mol. Med. 2022;28:388–404. doi: 10.1016/j.molmed.2022.03.001. [DOI] [PubMed] [Google Scholar]
- 95.Deinsberger J, Reisinger D, Weber B. Global trends in clinical trials involving pluripotent stem cells: A systematic multi-database analysis. NPJ Regen. Med. 2020;5:1–13. doi: 10.1038/s41536-020-00100-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Knoepfler PS. Deconstructing stem cell tumorigenicity: a roadmap to safe regenerative medicine. Stem Cells. 2009;27:1050–1056. doi: 10.1002/stem.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.de Almeida PE, Ransohoff JD, Nahid A, Wu JC. Immunogenicity of pluripotent stem cells and their derivatives. Circ. Res. 2013;112:549–561. doi: 10.1161/CIRCRESAHA.111.249243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhao X, et al. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461:86–90. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
- 99.Hodgkinson CP, Gomez JA, Mirotsou M, Dzau VJ. Genetic engineering of mesenchymal stem cells and its application in human disease therapy. Hum. Gene Ther. 2010;21:1513–1526. doi: 10.1089/hum.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hoffman T, et al. Synthetic biology and tissue engineering: toward fabrication of complex and smart cellular constructs. Adv. Funct. Mater. 2020;30:1909882. doi: 10.1002/adfm.201909882. [DOI] [Google Scholar]
- 101.Reed JC, Cuddy M, Slabiak T, Croce CM, Nowell PC. Oncogenic potential of bcl-2 demonstrated by gene transfer. Nature. 1988;336:259–261. doi: 10.1038/336259a0. [DOI] [PubMed] [Google Scholar]
- 102.Kirkin V, Joos S, Zörnig M. The role of Bcl-2 family members in tumorigenesis. Biochim. Biophys. Acta Mol. Cell Res. 2004;1644:229–249. doi: 10.1016/j.bbamcr.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 103.Cheng AW, et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Warren L, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Slusarczyk AL, Lin A, Weiss R. Foundations for the design and implementation of synthetic genetic circuits. Nat. Rev. Genet. 2012;13:406–420. doi: 10.1038/nrg3227. [DOI] [PubMed] [Google Scholar]
- 106.Gersbach CA, Le Doux JM, Guldberg RE, Garcia AJ. Inducible regulation of Runx2-stimulated osteogenesis. Gene Ther. 2006;13:873–882. doi: 10.1038/sj.gt.3302725. [DOI] [PubMed] [Google Scholar]
- 107.Jo A, et al. The versatile functions of Sox9 in development, stem cells, and human diseases. Genes Dis. 2014;1:149–161. doi: 10.1016/j.gendis.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Glass KA, et al. Tissue-engineered cartilage with inducible and tunable immunomodulatory properties. Biomaterials. 2014;35:5921–5931. doi: 10.1016/j.biomaterials.2014.03.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mansouri M, Strittmatter T, Fussenegger M. Light‐controlled mammalian cells and their therapeutic applications in synthetic biology. Adv. Sci. 2019;6:1800952. doi: 10.1002/advs.201800952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Polstein LR, Gersbach CA. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat. Chem. Biol. 2015;11:198–200. doi: 10.1038/nchembio.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yamada M, Suzuki Y, Nagasaki SC, Okuno H, Imayoshi I. Light control of the Tet gene expression system in mammalian cells. Cell Rep. 2018;25:487–500. e486. doi: 10.1016/j.celrep.2018.09.026. [DOI] [PubMed] [Google Scholar]
- 112.Sauers DJ, et al. Light-activated gene expression directs segregation of co-cultured cells in vitro. ACS Chem. Biol. 2010;5:313–320. doi: 10.1021/cb9002305. [DOI] [PubMed] [Google Scholar]
- 113.Sakar MS, et al. Formation and optogenetic control of engineered 3D skeletal muscle bioactuators. Lab Chip. 2012;12:4976–4985. doi: 10.1039/c2lc40338b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Conklin BR, et al. Engineering GPCR signaling pathways with RASSLs. Nat. Methods. 2008;5:673–678. doi: 10.1038/nmeth.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Park JS, et al. Synthetic control of mammalian-cell motility by engineering chemotaxis to an orthogonal bioinert chemical signal. Proc. Natl Acad. Sci. USA. 2014;111:5896–5901. doi: 10.1073/pnas.1402087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gilbert C, Ellis T. Biological engineered living materials: growing functional materials with genetically programmable properties. ACS Synth. Biol. 2018;8:1–15. doi: 10.1021/acssynbio.8b00423. [DOI] [PubMed] [Google Scholar]
- 117.Toda S, Blauch LR, Tang SK, Morsut L, Lim WA. Programming self-organizing multicellular structures with synthetic cell-cell signaling. Science. 2018;361:156–162. doi: 10.1126/science.aat0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen F, Liu X. Advancing biomaterials of human origin for tissue engineering. Prog. Polym. Sci. 2016;53:86–168. doi: 10.1016/j.progpolymsci.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Baraniak PR, et al. Spatial control of gene expression within a scaffold by localized inducer release. Biomaterials. 2011;32:3062–3071. doi: 10.1016/j.biomaterials.2010.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Deans TL, Singh A, Gibson M, Elisseeff JH. Regulating synthetic gene networks in 3D materials. Proc. Natl Acad. Sci. USA. 2012;109:15217–15222. doi: 10.1073/pnas.1204705109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Singh A, Deans TL, Elisseeff JH. Photomodulation of cellular gene expression in hydrogels. ACS Macro. Lett. 2013;2:269–272. doi: 10.1021/mz300591m. [DOI] [PubMed] [Google Scholar]
- 122.Lu Y, et al. Engineering bacteria‐activated multifunctionalized hydrogel for promoting diabetic wound healing. Adv. Funct. Mater. 2021;31:2105749. doi: 10.1002/adfm.202105749. [DOI] [Google Scholar]
- 123.Charbonneau MR, Isabella VM, Li N, Kurtz CB. Developing a new class of engineered live bacterial therapeutics to treat human diseases. Nat. Commun. 2020;11:1–11. doi: 10.1038/s41467-020-15508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cubillos-Ruiz A, et al. Engineering living therapeutics with synthetic biology. Nat. Rev. Drug Discov. 2021;20:941–960. doi: 10.1038/s41573-021-00285-3. [DOI] [PubMed] [Google Scholar]
- 125.Xu J, Liu XS, Zhou S-F, Wei MQ. Combination of immunotherapy with anaerobic bacteria for immunogene therapy of solid tumours. Gene Ther. Mol. Biol. 2009;13:36–52. [Google Scholar]
- 126.Danino T, et al. Programmable probiotics for detection of cancer in urine. Sci. Transl. Med. 2015;7:289ra284. doi: 10.1126/scitranslmed.aaa3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Royo JL, et al. In vivo gene regulation in Salmonella spp. by a salicylate-dependent control circuit. Nat. Methods. 2007;4:937–942. doi: 10.1038/nmeth1107. [DOI] [PubMed] [Google Scholar]
- 128.Yu YA, et al. Visualization of tumors and metastases in live animals with bacteria and vaccinia virus encoding light-emitting proteins. Nat. Biotechnol. 2004;22:313–320. doi: 10.1038/nbt937. [DOI] [PubMed] [Google Scholar]
- 129.Piñero-Lambea C, et al. Programming controlled adhesion of E. coli to target surfaces, cells, and tumors with synthetic adhesins. ACS Synth. Biol. 2015;4:463–473. doi: 10.1021/sb500252a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pinero-Lambea C, Ruano-Gallego D, Fernandez LA. Engineered bacteria as therapeutic agents. Curr. Opin. Biotechnol. 2015;35:94–102. doi: 10.1016/j.copbio.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 131.Bertrand N, Wu J, Xu X, Kamaly N, Farokhzad OC. Cancer nanotechnology: the impact of passive and active targeting in the era of modern cancer biology. Adv. Drug Del. Rev. 2014;66:2–25. doi: 10.1016/j.addr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Thamm DH, et al. Systemic administration of an attenuated, tumor-targeting Salmonella typhimurium to dogs with spontaneous neoplasia: phase I evaluation. Clin. Cancer Res. 2005;11:4827–4834. doi: 10.1158/1078-0432.CCR-04-2510. [DOI] [PubMed] [Google Scholar]
- 133.Toso JF, et al. Phase I study of the intravenous administration of attenuated Salmonella typhimurium to patients with metastatic melanoma. J. Clin. Oncol. 2002;20:142. doi: 10.1200/JCO.2002.20.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhou S, Gravekamp C, Bermudes D, Liu K. Tumour-targeting bacteria engineered to fight cancer. Nat. Rev. Cancer. 2018;18:727–743. doi: 10.1038/s41568-018-0070-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Courbet A, Endy D, Renard E, Molina F, Bonnet J. Detection of pathological biomarkers in human clinical samples via amplifying genetic switches and logic gates. Sci. Transl. Med. 2015;7:289ra283. doi: 10.1126/scitranslmed.aaa3601. [DOI] [PubMed] [Google Scholar]
- 136.Duan FF, Liu JH, March JC. Engineered commensal bacteria reprogram intestinal cells into glucose-responsive insulin-secreting cells for the treatment of diabetes. Diabetes. 2015;64:1794–1803. doi: 10.2337/db14-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Arora T, et al. Microbially produced glucagon-like peptide 1 improves glucose tolerance in mice. Mol. Metab. 2016;5:725–730. doi: 10.1016/j.molmet.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wei P, Yang Y, Li T, Ding Q, Sun H. A engineered Bifidobacterium longum secreting a bioative penetratin-Glucagon-like peptide 1 fusion protein enhances Glucagon-like peptide 1 absorption in the intestine. J. Microbiol. Biotechnol. 2015;24:10. [PubMed] [Google Scholar]
- 139.Carter CS, et al. Therapeutic delivery of Ang (1–7) via genetically modified probiotic: a dosing study. J. Gerontology: Ser. A. 2020;75:1299–1303. doi: 10.1093/gerona/glz222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Verma A, et al. Angiotensin-(1–7) expressed from Lactobacillus bacteria protect diabetic retina in mice. Transl. Vis. Sci. Technol. 2020;9:20. doi: 10.1167/tvst.9.13.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lin Y, et al. Oral delivery of pentameric glucagon-like peptide-1 by recombinant Lactobacillus in diabetic rats. PLoS ONE. 2016;11:e0162733. doi: 10.1371/journal.pone.0162733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Takiishi T, et al. Reversal of autoimmune diabetes by restoration of antigen-specific tolerance using genetically modified Lactococcus lactis in mice. J. Clin. Invest. 2012;122:1717–1725. doi: 10.1172/JCI60530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang M, Gao Z, Zhang Y, Pan L. Lactic acid bacteria as mucosal delivery vehicles: a realistic therapeutic option. Appl. Microbiol. Biotechnol. 2016;100:5691–5701. doi: 10.1007/s00253-016-7557-x. [DOI] [PubMed] [Google Scholar]
- 144.Jakubczyk D, Leszczyńska K, Górska S. The effectiveness of probiotics in the treatment of inflammatory bowel disease (IBD)-a critical review. Nutrients. 2020;12:1973. doi: 10.3390/nu12071973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Praveschotinunt P, et al. Engineered E. coli Nissle 1917 for the delivery of matrix-tethered therapeutic domains to the gut. Nat. Commun. 2019;10:5580. doi: 10.1038/s41467-019-13336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hwang IY, et al. Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nat. Commun. 2017;8:15028. doi: 10.1038/ncomms15028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yan X, et al. Construction of a sustainable 3-hydroxybutyrate-producing probiotic Escherichia coli for treatment of colitis. Cell. Mol. Immunol. 2021;18:2344–2357. doi: 10.1038/s41423-021-00760-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.He M, Shi B. Gut microbiota as a potential target of metabolic syndrome: the role of probiotics and prebiotics. Cell Biosci. 2017;7:1–14. doi: 10.1186/s13578-017-0183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chen Z, et al. Incorporation of therapeutically modified bacteria into gut microbiota inhibits obesity. J. Clin. Invest. 2014;124:3391–3406. doi: 10.1172/JCI72517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kurtz CB, et al. An engineered E. coli Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans. Sci. Transl. Med. 2019;11:aau7975. doi: 10.1126/scitranslmed.aau7975. [DOI] [PubMed] [Google Scholar]
- 151.Isabella VM, et al. Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat. Biotechnol. 2018;36:857–864. doi: 10.1038/nbt.4222. [DOI] [PubMed] [Google Scholar]
- 152.Cusi K, et al. American Association of Clinical Endocrinology clinical practice guideline for the diagnosis and management of nonalcoholic fatty liver disease in primary care and endocrinology clinical settings: co-sponsored by the American Association for the Study of Liver Diseases (AASLD) Endocr. Pract. 2022;28:528–562. doi: 10.1016/j.eprac.2022.03.010. [DOI] [PubMed] [Google Scholar]
- 153.Lu J, et al. Co-expression of alcohol dehydrogenase and aldehyde dehydrogenase in Bacillus subtilis for alcohol detoxification. Food Chem. Toxicol. 2020;135:110890. doi: 10.1016/j.fct.2019.110890. [DOI] [PubMed] [Google Scholar]
- 154.Hendrikx T, et al. Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut. 2019;68:1504–1515. doi: 10.1136/gutjnl-2018-317232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Orive G, Gascón AR, Hernández RM, Domínguez-Gil A, Pedraz JL. Techniques: new approaches to the delivery of biopharmaceuticals. Trends Pharmacol. Sci. 2004;25:382–387. doi: 10.1016/j.tips.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 156.Razzacki SZ, Thwar PK, Yang M, Ugaz VM, Burns MA. Integrated microsystems for controlled drug delivery. Adv. Drug Del. Rev. 2004;56:185–198. doi: 10.1016/j.addr.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 157.Patra JK, et al. Nano based drug delivery systems: recent developments and future prospects. J. Nanobiotechnol. 2018;16:1–33. doi: 10.1186/s12951-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Choi Y, Park TJ, Lee DC, Lee SY. Recombinant Escherichia coli as a biofactory for various single-and multi-element nanomaterials. Proc. Natl Acad. Sci. USA. 2018;115:5944–5949. doi: 10.1073/pnas.1804543115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Mishra RK, Ha SK, Verma K, Tiwari SK. Recent progress in selected bio-nanomaterials and their engineering applications: an overview. J. Sci.: Adv. Mater. Devices. 2018;3:263–288. [Google Scholar]
- 160.Azizi S, Ahmad MB, Namvar F, Mohamad R. Green biosynthesis and characterization of zinc oxide nanoparticles using brown marine macroalga Sargassum muticum aqueous extract. Mater. Lett. 2014;116:275–277. doi: 10.1016/j.matlet.2013.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Shaheen, T. I., Salem, S. & Fouda, A. in Microbial Nanobiotechnology. Materials Horizons: From Nature to Nanomaterials (eds. Lateef, A., Gueguim-Kana, E. B., Dasgupta, N., Ranjan, S.) (Springer, 2021).
- 162.Shankar PD, et al. A review on the biosynthesis of metallic nanoparticles (gold and silver) using bio-components of microalgae: Formation mechanism and applications. Enzym. Microb. Technol. 2016;95:28–44. doi: 10.1016/j.enzmictec.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 163.Iravani S, Varma RS. Biofactories: engineered nanoparticles via genetically engineered organisms. Green. Chem. 2019;21:4583–4603. doi: 10.1039/C9GC01759C. [DOI] [Google Scholar]
- 164.Jahromi LP, Fuhrmann G. Bacterial extracellular vesicles: understanding biology promotes applications as nanopharmaceuticals. Adv. Drug Del. Rev. 2021;173:125–140. doi: 10.1016/j.addr.2021.03.012. [DOI] [PubMed] [Google Scholar]
- 165.Kumar M, et al. Amino-acid-encoded biocatalytic self-assembly enables the formation of transient conducting nanostructures. Nat. Chem. 2018;10:696–703. doi: 10.1038/s41557-018-0047-2. [DOI] [PubMed] [Google Scholar]
- 166.van Rijn P, et al. Challenges and advances in the field of self-assembled membranes. Chem. Soc. Rev. 2013;42:6578–6592. doi: 10.1039/c3cs60125k. [DOI] [PubMed] [Google Scholar]
- 167.Gaur D, Dubey NC, Tripathi BP. Biocatalytic self-assembled synthetic vesicles and coacervates: From single compartment to artificial cells. Adv. Colloid Interface Sci. 2022;299:102566. doi: 10.1016/j.cis.2021.102566. [DOI] [PubMed] [Google Scholar]
- 168.Van Oppen LM, et al. Biodegradable synthetic organelles demonstrate ROS shielding in human-complex-I-deficient fibroblasts. ACS Cent. Sci. 2018;4:917–928. doi: 10.1021/acscentsci.8b00336. [DOI] [Google Scholar]
- 169.Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov. 2013;12:931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 170.Fruehauf JP, Meyskens FL., Jr Reactive oxygen species: a breath of life or death? Clin. Cancer Res. 2007;13:789–794. doi: 10.1158/1078-0432.CCR-06-2082. [DOI] [PubMed] [Google Scholar]
- 171.Prindle A, et al. A sensing array of radically coupled genetic ‘biopixels’. Nature. 2012;481:39–44. doi: 10.1038/nature10722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jewett MC, Patolsky F. Nanobiotechnology: synthetic biology meets materials science. Curr. Opin. Biotechnol. 2013;24:551–554. doi: 10.1016/j.copbio.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 173.Xie M, Fussenegger M. Designing cell function: assembly of synthetic gene circuits for cell biology applications. Nat. Rev. Mol. Cell Biol. 2018;19:507–525. doi: 10.1038/s41580-018-0024-z. [DOI] [PubMed] [Google Scholar]
- 174.Zhang C, Zhuang Q, Liu J, Liu X. Synthetic Biology in Chimeric Antigen Receptor T (CAR T) Cell Engineering. ACS Synth. Biol. 2022;11:1–15. doi: 10.1021/acssynbio.1c00256. [DOI] [PubMed] [Google Scholar]
- 175.Aratboni HA, et al. LED control of gene expression in a nanobiosystem composed of metallic nanoparticles and a genetically modified E. coli strain. J. Nanobiotechnol. 2021;19:1–12. doi: 10.1186/s12951-021-00937-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hasty J, Pradines J, Dolnik M, Collins JJ. Noise-based switches and amplifiers for gene expression. Proc. Natl Acad. Sci. USA. 2000;97:2075–2080. doi: 10.1073/pnas.040411297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Shin T-H, Cheon J. Synergism of nanomaterials with physical stimuli for biology and medicine. Acc. Chem. Res. 2017;50:567–572. doi: 10.1021/acs.accounts.6b00559. [DOI] [PubMed] [Google Scholar]
- 178.Yan Z, et al. Visible-light-responsive reduced graphene oxide/g-C3N4/TiO2 composite nanocoating for photoelectric stimulation of neuronal and osteoblastic differentiation. RSC Adv. 2022;12:8878–8888. doi: 10.1039/D2RA00282E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Chernov KG, Redchuk TA, Omelina ES, Verkhusha VV. Near-infrared fluorescent proteins, biosensors, and optogenetic tools engineered from phytochromes. Chem. Rev. 2017;117:6423–6446. doi: 10.1021/acs.chemrev.6b00700. [DOI] [PubMed] [Google Scholar]
- 180.Piatkevich KD, Subach FV, Verkhusha VV. Engineering of bacterial phytochromes for near-infrared imaging, sensing, and light-control in mammals. Chem. Soc. Rev. 2013;42:3441–3452. doi: 10.1039/c3cs35458j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chen S, et al. Near-infrared deep brain stimulation via upconversion nanoparticle-mediated optogenetics. Science. 2018;359:679–684. doi: 10.1126/science.aaq1144. [DOI] [PubMed] [Google Scholar]
- 182.Patel M, Meenu M, Pandey JK, Kumar P, Patel R. Recent development in upconversion nanoparticles and their application in optogenetics: a review. J. Rare Earths. 2021;40:847–861. doi: 10.1016/j.jre.2021.10.003. [DOI] [Google Scholar]
- 183.Miller IC, Gamboa Castro M, Maenza J, Weis JP, Kwong GA. Remote control of mammalian cells with heat-triggered gene switches and photothermal pulse trains. ACS Synth. Biol. 2018;7:1167–1173. doi: 10.1021/acssynbio.7b00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wang Y, et al. Photothermal‐responsive conjugated polymer nanoparticles for remote control of gene expression in living cells. Adv. Mater. 2018;30:1705418. doi: 10.1002/adma.201705418. [DOI] [PubMed] [Google Scholar]
- 185.Feliu N, Neher E, Parak WJ. Toward an optically controlled brain. Science. 2018;359:633–634. doi: 10.1126/science.aar7379. [DOI] [PubMed] [Google Scholar]
- 186.Shao J, et al. Smartphone-controlled optogenetically engineered cells enable semiautomatic glucose homeostasis in diabetic mice. Sci. Transl. Med. 2017;9:eaal2298. doi: 10.1126/scitranslmed.aal2298. [DOI] [PubMed] [Google Scholar]
- 187.Folcher M, et al. Mind-controlled transgene expression by a wireless-powered optogenetic designer cell implant. Nat. Commun. 2014;5:1–11. doi: 10.1038/ncomms6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhang XA, Chen I, Chang C. Recent progress in near-field nanolithography using light interactions with colloidal particles: from nanospheres to three-dimensional nanostructures. Nanotechnology. 2019;30:352002. doi: 10.1088/1361-6528/ab2282. [DOI] [PubMed] [Google Scholar]
- 189.Li T, et al. A synthetic BRET-based optogenetic device for pulsatile transgene expression enabling glucose homeostasis in mice. Nat. Commun. 2021;12:1–10. doi: 10.1038/s41467-021-20913-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Huang H, Delikanli S, Zeng H, Ferkey DM, Pralle A. Remote control of ion channels and neurons through magnetic-field heating of nanoparticles. Nat. Nanotechnol. 2010;5:602–606. doi: 10.1038/nnano.2010.125. [DOI] [PubMed] [Google Scholar]
- 191.Wheeler MA, et al. Genetically targeted magnetic control of the nervous system. Nat. Neurosci. 2016;19:756–761. doi: 10.1038/nn.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Stanley SA, et al. Radio-wave heating of iron oxide nanoparticles can regulate plasma glucose in mice. Science. 2012;336:604–608. doi: 10.1126/science.1216753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Stanley SA, Sauer J, Kane RS, Dordick JS, Friedman JM. Remote regulation of glucose homeostasis in mice using genetically encoded nanoparticles. Nat. Med. 2015;21:92–98. doi: 10.1038/nm.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Tong S, Zhu H, Bao G. Magnetic iron oxide nanoparticles for disease detection and therapy. Mater. Today. 2019;31:86–99. doi: 10.1016/j.mattod.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Meister M. Physical limits to magnetogenetics. Elife. 2016;5:e17210. doi: 10.7554/eLife.17210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Brier MI, et al. Uncovering a possible role of reactive oxygen species in magnetogenetics. Sci. Rep. 2020;10:1–13. doi: 10.1038/s41598-020-70067-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hernández-Morales M, Shang T, Chen J, Han V, Liu C. Lipid oxidation induced by RF waves and mediated by ferritin iron causes activation of ferritin-tagged ion channels. Cell Rep. 2020;30:3250–3260. doi: 10.1016/j.celrep.2020.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Miller, B. A. & Zhang, W. in Transient Receptor Potential Channels. Advances in Experimental Medicine and Biology (ed Islam, M.) (Springer, 2011).
- 199.Carter CS, et al. Exposure to static magnetic and electric fields treats type 2 diabetes. Cell Metab. 2020;32:561–574. doi: 10.1016/j.cmet.2020.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Raghunath A, et al. Antioxidant response elements: Discovery, classes, regulation and potential applications. Redox Biol. 2018;17:297–314. doi: 10.1016/j.redox.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.David RM, Doherty AT. Viral vectors: the road to reducing genotoxicity. Toxicol. Sci. 2017;155:315–325. doi: 10.1093/toxsci/kfw220. [DOI] [PubMed] [Google Scholar]
- 202.Chen Y, Groves B, Muscat RA, Seelig G. DNA nanotechnology from the test tube to the cell. Nat. Nanotechnol. 2015;10:748–760. doi: 10.1038/nnano.2015.195. [DOI] [PubMed] [Google Scholar]
- 203.Deng H, Huang W, Zhang Z. Nanotechnology based CRISPR/Cas9 system delivery for genome editing: Progress and prospect. Nano Res. 2019;12:2437–2450. doi: 10.1007/s12274-019-2465-x. [DOI] [Google Scholar]
- 204.Angell C, Xie S, Zhang L, Chen Y. DNA nanotechnology for precise control over drug delivery and gene therapy. Small. 2016;12:1117–1132. doi: 10.1002/smll.201502167. [DOI] [PubMed] [Google Scholar]
- 205.Tenchov R, Bird R, Curtze AE, Zhou Q. Lipid nanoparticles-from liposomes to mRNA vaccine delivery, a landscape of research diversity and advancement. ACS Nano. 2021;15:16982–17015. doi: 10.1021/acsnano.1c04996. [DOI] [PubMed] [Google Scholar]
- 206.Yanar F, Mosayyebi A, Nastruzzi C, Carugo D, Zhang X. Continuous-flow production of liposomes with a millireactor under varying fluidic conditions. Pharmaceutics. 2020;12:1001. doi: 10.3390/pharmaceutics12111001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Sercombe L, et al. Advances and challenges of liposome assisted drug delivery. Front. Pharmacol. 2015;6:286. doi: 10.3389/fphar.2015.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Liu C, Zhang L, Liu H, Cheng K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J. Control. Release. 2017;266:17–26. doi: 10.1016/j.jconrel.2017.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Felgner PL, et al. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl Acad. Sci. USA. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Dobrowolski C, Paunovska K, Hatit MZ, Lokugamage MP, Dahlman JE. Therapeutic RNA delivery for COVID and other diseases. Adv. Healthc. Mater. 2021;10:2002022. doi: 10.1002/adhm.202002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ma W, et al. The biological applications of DNA nanomaterials: current challenges and future directions. Signal Transduct. Target. Ther. 2021;6:1–28. doi: 10.1038/s41392-021-00727-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Huang X, Liu Y, Yung B, Xiong Y, Chen X. Nanotechnology-enhanced no-wash biosensors for in vitro diagnostics of cancer. ACS Nano. 2017;11:5238–5292. doi: 10.1021/acsnano.7b02618. [DOI] [PubMed] [Google Scholar]
- 213.Fan J, et al. A multifunction lipid-based CRISPR-Cas13a genetic circuit delivery system for bladder cancer gene therapy. ACS Synth. Biol. 2019;9:343–355. doi: 10.1021/acssynbio.9b00349. [DOI] [PubMed] [Google Scholar]
- 214.Elzoghby AO, Samy WM, Elgindy NA. Protein-based nanocarriers as promising drug and gene delivery systems. J. Control. Release. 2012;161:38–49. doi: 10.1016/j.jconrel.2012.04.036. [DOI] [PubMed] [Google Scholar]
- 215.Loh XJ, Lee T, Dou Q, Deen GR. Utilising inorganic nanocarriers for gene delivery. Biomater. Sci. 2016;4:70–86. doi: 10.1039/C5BM00277J. [DOI] [PubMed] [Google Scholar]
- 216.Lin Y, et al. Exosome-liposome hybrid nanoparticles deliver CRISPR/Cas9 system in MSCs. Adv. Sci. 2018;5:1700611. doi: 10.1002/advs.201700611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Mirkhani N, Gwisai T, Schuerle S. Engineering cell-based systems for smart cancer therapy. Adv. Intell. Syst. 2022;4:2100134. doi: 10.1002/aisy.202100134. [DOI] [Google Scholar]
- 218.Milligan JJ, Saha S, Jenkins IC, Chilkoti A. Genetically encoded elastin-like polypeptide nanoparticles for drug delivery. Curr. Opin. Biotechnol. 2022;74:146–153. doi: 10.1016/j.copbio.2021.11.006. [DOI] [PubMed] [Google Scholar]
- 219.Mozhdehi D, et al. Genetically encoded lipid-polypeptide hybrid biomaterials that exhibit temperature-triggered hierarchical self-assembly. Nat. Chem. 2018;10:496–505. doi: 10.1038/s41557-018-0005-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Luginbuhl KM, et al. Recombinant synthesis of hybrid lipid–peptide polymer fusions that self‐assemble and encapsulate hydrophobic drugs. Angew. Chem. Int. Ed. 2017;56:13979–13984. doi: 10.1002/anie.201704625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Li J, et al. Engineered near-infrared fluorescent protein assemblies for robust bioimaging and therapeutic applications. Adv. Mater. 2020;32:2000964. doi: 10.1002/adma.202000964. [DOI] [PubMed] [Google Scholar]
- 222.Ma C, et al. Significantly improving the bioefficacy for rheumatoid arthritis with supramolecular nanoformulations. Adv. Mater. 2021;33:2100098. doi: 10.1002/adma.202100098. [DOI] [PubMed] [Google Scholar]
- 223.Schwechheimer C, Kuehn MJ. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat. Rev. Microbiol. 2015;13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Shao J, Zaro J, Shen Y. Advances in exosome-based drug delivery and tumor targeting: from tissue distribution to intracellular fate. Int. J. Nanomed. 2020;15:9355. doi: 10.2147/IJN.S281890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Micoli F, MacLennan CA. Outer membrane vesicle vaccines. Semin. Immunol. 2020;50:101433. doi: 10.1016/j.smim.2020.101433. [DOI] [PubMed] [Google Scholar]
- 226.Nguyen PQ, Courchesne NMD, Duraj-Thatte A, Praveschotinunt P, Joshi NS. Engineered living materials: prospects and challenges for using biological systems to direct the assembly of smart materials. Adv. Mater. 2018;30:1704847. doi: 10.1002/adma.201704847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Chen DJ, et al. Delivery of foreign antigens by engineered outer membrane vesicle vaccines. Proc. Natl Acad. Sci. USA. 2010;107:3099–3104. doi: 10.1073/pnas.0805532107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Galen JE, et al. Adaptation of the endogenous Salmonella enterica serovar Typhi clyA-encoded hemolysin for antigen export enhances the immunogenicity of anthrax protective antigen domain 4 expressed by the attenuated live-vector vaccine strain CVD 908-htrA. Infect. Immun. 2004;72:7096–7106. doi: 10.1128/IAI.72.12.7096-7106.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Wai SN, et al. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell. 2003;115:25–35. doi: 10.1016/S0092-8674(03)00754-2. [DOI] [PubMed] [Google Scholar]
- 230.Liu H, et al. Bacterial extracellular vesicles-based therapeutic strategies for bone and soft tissue tumors therapy. Theranostics. 2022;12:6576. doi: 10.7150/thno.78034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Rappazzo CG, et al. Recombinant M2e outer membrane vesicle vaccines protect against lethal influenza A challenge in BALB/c mice. Vaccine. 2016;34:1252–1258. doi: 10.1016/j.vaccine.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 232.Muralinath M, Kuehn MJ, Roland KL, Curtiss R., III Immunization with Salmonella enterica serovar Typhimurium-derived outer membrane vesicles delivering the pneumococcal protein PspA confers protection against challenge with Streptococcus pneumoniae. Infect. Immun. 2011;79:887–894. doi: 10.1128/IAI.00950-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Bartolini E, et al. Recombinant outer membrane vesicles carrying Chlamydia muridarum HtrA induce antibodies that neutralize chlamydial infection in vitro. J. Extracell. Vesicles. 2013;2:20181. doi: 10.3402/jev.v2i0.20181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Backert S, Bernegger S, Skórko‐Glonek J, Wessler S. Extracellular HtrA serine proteases: an emerging new strategy in bacterial pathogenesis. Cell. Microbiol. 2018;20:e12845. doi: 10.1111/cmi.12845. [DOI] [PubMed] [Google Scholar]
- 235.Fantappiè L, et al. Antibody-mediated immunity induced by engineered Escherichia coli OMVs carrying heterologous antigens in their lumen. J. Extracell. Vesicles. 2014;3:24015. doi: 10.3402/jev.v3.24015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Micoli F, Costantino P, Adamo R. Potential targets for next generation antimicrobial glycoconjugate vaccines. FEMS Microbiol. Rev. 2018;42:388–423. doi: 10.1093/femsre/fuy011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Avci FY, Kasper DL. How bacterial carbohydrates influence the adaptive immune system. Annu. Rev. Immunol. 2009;28:107–130. doi: 10.1146/annurev-immunol-030409-101159. [DOI] [PubMed] [Google Scholar]
- 238.Chen L, et al. Outer membrane vesicles displaying engineered glycotopes elicit protective antibodies. Proc. Natl Acad. Sci. USA. 2016;113:E3609–E3618. doi: 10.1073/pnas.1518311113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Weyant KB, Mills DC, DeLisa MP. Engineering a new generation of carbohydrate-based vaccines. Curr. Opin. Chem. Eng. 2018;19:77–85. doi: 10.1016/j.coche.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Price NL, et al. Glycoengineered outer membrane vesicles: a novel platform for bacterial vaccines. Sci. Rep. 2016;6:1–9. doi: 10.1038/srep24931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Pumpens P, et al. The true story and advantages of RNA phage capsids as nanotools. Intervirology. 2016;59:74–110. doi: 10.1159/000449503. [DOI] [PubMed] [Google Scholar]
- 242.Liu C, et al. Engineered extracellular vesicles and their mimetics for cancer immunotherapy. J. Control. Release. 2022;349:679–698. doi: 10.1016/j.jconrel.2022.05.062. [DOI] [PubMed] [Google Scholar]
- 243.Long Q, et al. Engineered bacterial membrane vesicles are promising carriers for vaccine design and tumor immunotherapy. Adv. Drug Del. Rev. 2022;186:114321. doi: 10.1016/j.addr.2022.114321. [DOI] [PubMed] [Google Scholar]
- 244.Meredith HJ, Jenkins CL, Wilker JJ. Enhancing the adhesion of a biomimetic polymer yields performance rivaling commercial glues. Adv. Funct. Mater. 2014;24:3259–3267. doi: 10.1002/adfm.201303536. [DOI] [Google Scholar]
- 245.Petrone L, et al. Mussel adhesion is dictated by time-regulated secretion and molecular conformation of mussel adhesive proteins. Nat. Commun. 2015;6:1–12. doi: 10.1038/ncomms9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Liang C, et al. Biochemistry of barnacle adhesion: an updated review. Front. Mar. Sci. 2019;6:565. doi: 10.3389/fmars.2019.00565. [DOI] [Google Scholar]
- 247.Zhang X, Liu H, Yue L, Bai Y, He J. Mussel-mimetic polymer underwater adhesives with L-Dopa functionality: influencing adhesion properties and simplified operation procedures. J. Mater. Sci. 2020;55:7981–7997. doi: 10.1007/s10853-020-04572-z. [DOI] [Google Scholar]
- 248.Zhong C, et al. Strong underwater adhesives made by self-assembling multi-protein nanofibres. Nat. Nanotechnol. 2014;9:858–866. doi: 10.1038/nnano.2014.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Chen AY, Zhong C, Lu TK. Engineering living functional materials. ACS Synth. Biol. 2015;4:8–11. doi: 10.1021/sb500113b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Zhang C, et al. Engineered Bacillus subtilis biofilms as living glues. Mater. Today. 2019;28:40–48. doi: 10.1016/j.mattod.2018.12.039. [DOI] [Google Scholar]
- 251.Sun F, Zhang W. Unleashing chemical power from protein sequence space toward genetically encoded “click” chemistry. Chin. Chem. Lett. 2017;28:2078–2084. doi: 10.1016/j.cclet.2017.08.052. [DOI] [Google Scholar]
- 252.Kang HJ, Coulibaly F, Clow F, Proft T, Baker EN. Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science. 2007;318:1625–1628. doi: 10.1126/science.1145806. [DOI] [PubMed] [Google Scholar]
- 253.Zakeri B, Howarth M. Spontaneous intermolecular amide bond formation between side chains for irreversible peptide targeting. J. Am. Chem. Soc. 2010;132:4526–4527. doi: 10.1021/ja910795a. [DOI] [PubMed] [Google Scholar]
- 254.Lin Z, et al. Spy chemistry‐enabled protein directional immobilization and protein purification. Biotechnol. Bioeng. 2020;117:2923–2932. doi: 10.1002/bit.27460. [DOI] [PubMed] [Google Scholar]
- 255.Liu Y, Ba F, Liu W, Wu C, Li J. Plug-and-play functionalization of protein-polymer conjugates for tunable catalysis enabled by genetically encoded “click” chemistry. ACS Catal. 2022;12:4165–4174. doi: 10.1021/acscatal.2c00846. [DOI] [Google Scholar]
- 256.Lu L, et al. The formation mechanism of hydrogels. Curr. Stem Cell Res. Ther. 2018;13:490–496. doi: 10.2174/1574888X12666170612102706. [DOI] [PubMed] [Google Scholar]
- 257.Ren K, et al. Injectable polypeptide hydrogels with tunable microenvironment for 3D spreading and chondrogenic differentiation of bone-marrow-derived mesenchymal stem cells. Biomacromolecules. 2016;17:3862–3871. doi: 10.1021/acs.biomac.6b00884. [DOI] [PubMed] [Google Scholar]
- 258.DeForest CA, Polizzotti BD, Anseth KS. Sequential click reactions for synthesizing and patterning three-dimensional cell microenvironments. Nat. Mater. 2009;8:659–664. doi: 10.1038/nmat2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Yang Z, et al. Dynamically tunable, macroscopic molecular networks enabled by cellular synthesis of 4-arm star-like proteins. Matter. 2020;2:233–249. doi: 10.1016/j.matt.2019.09.013. [DOI] [Google Scholar]
- 260.Sun F, Zhang W, Mahdavi A, Arnold FH, Tirrell DA. Synthesis of bioactive protein hydrogels by genetically encoded SpyTag-SpyCatcher chemistry. Proc. Natl Acad. Sci. USA. 2014;111:11269–11274. doi: 10.1073/pnas.1401291111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Ding X, Liu D, Booth G, Gao W, Lu Y. Virus‐like particle engineering: from rational design to versatile applications. Biotechnol. J. 2018;13:1700324. doi: 10.1002/biot.201700324. [DOI] [PubMed] [Google Scholar]
- 262.Berti F, Adamo R. Antimicrobial glycoconjugate vaccines: an overview of classic and modern approaches for protein modification. Chem. Soc. Rev. 2018;47:9015–9025. doi: 10.1039/C8CS00495A. [DOI] [PubMed] [Google Scholar]
- 263.Wu F, Liu J. Decorated bacteria and the application in drug delivery. Adv. Drug Del. Rev. 2022;188:114443. doi: 10.1016/j.addr.2022.114443. [DOI] [PubMed] [Google Scholar]
- 264.Liu Z, et al. A stable three-dimensional topological Dirac semimetal Cd3As2. Nat. Mater. 2014;13:677–681. doi: 10.1038/nmat3990. [DOI] [PubMed] [Google Scholar]
- 265.Brune KD, et al. Plug-and-Display: decoration of Virus-Like Particles via isopeptide bonds for modular immunization. Sci. Rep. 2016;6:1–13. doi: 10.1038/srep19234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Lemke EA. The exploding genetic code. ChemBioChem. 2014;15:1691–1694. doi: 10.1002/cbic.201402362. [DOI] [PubMed] [Google Scholar]
- 267.Huang Y, Liu T. Therapeutic applications of genetic code expansion. Synth. Syst. Biotechnol. 2018;3:150–158. doi: 10.1016/j.synbio.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Chin JW, et al. An expanded eukaryotic genetic code. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 269.Srinivasan G, James CM, Krzycki JA. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science. 2002;296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 270.Bryson DI, et al. Continuous directed evolution of aminoacyl-tRNA synthetases. Nat. Chem. Biol. 2017;13:1253–1260. doi: 10.1038/nchembio.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Schmied WH, Elsässer SJ, Uttamapinant C, Chin JW. Efficient multisite unnatural amino acid incorporation in mammalian cells via optimized pyrrolysyl tRNA synthetase/tRNA expression and engineered eRF1. J. Am. Chem. Soc. 2014;136:15577–15583. doi: 10.1021/ja5069728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Amiram M, et al. Evolution of translation machinery in recoded bacteria enables multi-site incorporation of nonstandard amino acids. Nat. Biotechnol. 2015;33:1272–1279. doi: 10.1038/nbt.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Chen Y, et al. Heritable expansion of the genetic code in mouse and zebrafish. Cell Res. 2017;27:294–297. doi: 10.1038/cr.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Vargas-Rodriguez O, Sevostyanova A, Söll D, Crnković A. Upgrading aminoacyl-tRNA synthetases for genetic code expansion. Curr. Opin. Chem. Biol. 2018;46:115–122. doi: 10.1016/j.cbpa.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Hallam TJ, Wold E, Wahl A, Smider VV. Antibody conjugates with unnatural amino acids. Mol. Pharm. 2015;12:1848–1862. doi: 10.1021/acs.molpharmaceut.5b00082. [DOI] [PubMed] [Google Scholar]
- 276.Sievers EL, Senter PD. Antibody-drug conjugates in cancer therapy. Annu. Rev. Med. 2013;64:15–29. doi: 10.1146/annurev-med-050311-201823. [DOI] [PubMed] [Google Scholar]
- 277.Kim CH, Axup JY, Schultz PG. Protein conjugation with genetically encoded unnatural amino acids. Curr. Opin. Chem. Biol. 2013;17:412–419. doi: 10.1016/j.cbpa.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Olle-Salvia B, Kym G, Chin JW. Rapid and efficient generation of stable antibody-drug conjugates via an encoded cyclopropene and an inverse‐electron‐demand Diels-Alder reaction. Angew. Chem. Int. Ed. 2018;57:2831–2834. doi: 10.1002/anie.201712370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Wang RE, et al. An immunosuppressive antibody-drug conjugate. J. Am. Chem. Soc. 2015;137:3229–3232. doi: 10.1021/jacs.5b00620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Yu S, et al. Recent advances of bispecific antibodies in solid tumors. J. Hematol. Oncol. 2017;10:1–16. doi: 10.1186/s13045-017-0522-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Byrne H, Conroy PJ, Whisstock JC, O’Kennedy RJ. A tale of two specificities: bispecific antibodies for therapeutic and diagnostic applications. Trends Biotechnol. 2013;31:621–632. doi: 10.1016/j.tibtech.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Wu C, et al. Simultaneous targeting of multiple disease mediators by a dual-variable-domain immunoglobulin. Nat. Biotechnol. 2007;25:1290–1297. doi: 10.1038/nbt1345. [DOI] [PubMed] [Google Scholar]
- 283.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 284.Chames P, Baty D. Bispecific antibodies for cancer therapy: the light at the end of the tunnel? MAbs. 2009;1:539–547. doi: 10.4161/mabs.1.6.10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Kim CH, et al. Synthesis of bispecific antibodies using genetically encoded unnatural amino acids. J. Am. Chem. Soc. 2012;134:9918–9921. doi: 10.1021/ja303904e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Ramadoss NS, et al. An anti-B cell maturation antigen bispecific antibody for multiple myeloma. J. Am. Chem. Soc. 2015;137:5288–5291. doi: 10.1021/jacs.5b01876. [DOI] [PubMed] [Google Scholar]
- 287.Wu Z, Yang H, Colosi P. Effect of genome size on AAV vector packaging. Mol. Ther. 2010;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Gardlík R, et al. Vectors and delivery systems in gene therapy. Med. Sci. Monit. 2005;11:110–121. [PubMed] [Google Scholar]
- 289.Zhang C, et al. Development of next generation adeno-associated viral vectors capable of selective tropism and efficient gene delivery. Biomaterials. 2016;80:134–145. doi: 10.1016/j.biomaterials.2015.11.066. [DOI] [PubMed] [Google Scholar]
- 290.Erickson SB, et al. Precise photoremovable perturbation of a virus-host interaction. Angew. Chem., Int. Ed. 2017;129:4298–4301. doi: 10.1002/ange.201700683. [DOI] [PubMed] [Google Scholar]
- 291.Hu Y, Hou Y, Wang H, Lu H. Polysarcosine as an alternative to PEG for therapeutic protein conjugation. Bioconjugate Chem. 2018;29:2232–2238. doi: 10.1021/acs.bioconjchem.8b00237. [DOI] [PubMed] [Google Scholar]
- 292.Harris JM, Chess RB. Effect of pegylation on pharmaceuticals. Nat. Rev. Drug discov. 2003;2:214–221. doi: 10.1038/nrd1033. [DOI] [PubMed] [Google Scholar]
- 293.Cho H, et al. Optimized clinical performance of growth hormone with an expanded genetic code. Proc. Natl Acad. Sci. USA. 2011;108:9060–9065. doi: 10.1073/pnas.1100387108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Xuan W, Li J, Luo X, Schultz PG. Genetic incorporation of a reactive isothiocyanate group into proteins. Angew. Chem., Int. Ed. 2016;128:10219–10222. doi: 10.1002/ange.201604891. [DOI] [PubMed] [Google Scholar]
- 295.Liu T, et al. Enhancing protein stability with extended disulfide bonds. Proc. Natl Acad. Sci. USA. 2016;113:5910–5915. doi: 10.1073/pnas.1605363113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Yang CY, et al. Improved stability and half-life of fluorinated phosphotriesterase using Rosetta. ChemBioChem. 2014;15:1761–1764. doi: 10.1002/cbic.201402062. [DOI] [PubMed] [Google Scholar]
- 297.Gauba V, et al. Loss of CD4 T-cell-dependent tolerance to proteins with modified amino acids. Proc. Natl Acad. Sci. USA. 2011;108:12821–12826. doi: 10.1073/pnas.1110042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Kang M, Lu Y, Chen S, Tian F. Harnessing the power of an expanded genetic code toward next-generation biopharmaceuticals. Curr. Opin. Chem. Biol. 2018;46:123–129. doi: 10.1016/j.cbpa.2018.07.018. [DOI] [PubMed] [Google Scholar]
- 299.Jang YH, Seong B. Principles underlying rational design of live attenuated influenza vaccines. Clin. Exp. Vaccine Res. 2012;1:35–49. doi: 10.7774/cevr.2012.1.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Si L, et al. Generation of influenza A viruses as live but replication-incompetent virus vaccines. Science. 2016;354:1170–1173. doi: 10.1126/science.aah5869. [DOI] [PubMed] [Google Scholar]
- 301.Ko J, et al. Pyrrolysyl‐tRNA synthetase variants reveal ancestral aminoacylation function. FEBS Lett. 2013;587:3243–3248. doi: 10.1016/j.febslet.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Mandell DJ, et al. Biocontainment of genetically modified organisms by synthetic protein design. Nature. 2015;518:55–60. doi: 10.1038/nature14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Cao Y, et al. Design of switchable chimeric antigen receptor T cells targeting breast cancer. Angew. Chem., Int. Ed. 2016;55:7520–7524. doi: 10.1002/anie.201601902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Ma JS, et al. Versatile strategy for controlling the specificity and activity of engineered T cells. Proc. Natl Acad. Sci. USA. 2016;113:E450–E458. doi: 10.1073/pnas.1524193113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Zambaldo C, Luo X, Mehta AP, Schultz PG. Recombinant macrocyclic lanthipeptides incorporating non-canonical amino acids. J. Am. Chem. Soc. 2017;139:11646–11649. doi: 10.1021/jacs.7b04159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Zheng Y, Lewis TL, Jr, Igo P, Polleux F, Chatterjee A. Virus-enabled optimization and delivery of the genetic machinery for efficient unnatural amino acid mutagenesis in mammalian cells and tissues. ACS Synth. Biol. 2017;6:13–18. doi: 10.1021/acssynbio.6b00092. [DOI] [PubMed] [Google Scholar]
- 307.Firn, R. (eds) Nature’s chemicals: the natural products that shaped our world. (OUP Oxford, 2009).
- 308.Ashour, M., Wink, M. & Gershenzon, J. Biochemistry of terpenoids: monoterpenes, sesquiterpenes and diterpenes. in Annual plant reviews volume 40: biochemistry of plant secondary metabolism, (eds Michael W.) (Blackwell, 2010).
- 309.Liu C, Zhao Y, Wang Y. Artemisinin: current state and perspectives for biotechnological production of an antimalarial drug. Appl. Microbiol. Biotechnol. 2006;72:11–20. doi: 10.1007/s00253-006-0452-0. [DOI] [PubMed] [Google Scholar]
- 310.Hao J, Han W, Xue B, Deng X. Microwave-assisted extraction of artemisinin from Artemisia annua L. Sep. Purif. Technol. 2002;28:191–196. doi: 10.1016/S1383-5866(02)00043-6. [DOI] [Google Scholar]
- 311.Hong G, Hu W, Li J, Chen X, Wang L. Increased accumulation of artemisinin and anthocyanins in Artemisia annua expressing the Arabidopsis blue light receptor CRY1. Plant Mol. Biol. Rep. 2009;27:334–341. doi: 10.1007/s11105-008-0088-6. [DOI] [Google Scholar]
- 312.Dhingra V, Rao KV, Narasu ML. Artemisinin: present status and perspectives. Biochem. Educ. 1999;27:105–109. doi: 10.1016/S0307-4412(98)00282-9. [DOI] [Google Scholar]
- 313.Li F, Weng J. Demystifying traditional herbal medicine with modern approach. Nat. Plants. 2017;3:1–7. doi: 10.1038/nplants.2017.109. [DOI] [PubMed] [Google Scholar]
- 314.Zhang C, Wu J, He G. Effects of inoculum size and age on biomass growth and paclitaxel production of elicitor-treated Taxus yunnanensis cell cultures. Appl. Microbiol. Biotechnol. 2002;60:396–402. doi: 10.1007/s00253-002-1130-5. [DOI] [PubMed] [Google Scholar]
- 315.Ajikumar PK, et al. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science. 2010;330:70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Yang Y, Ren C, Zhang Y, Wu X. Ginseng: an nonnegligible natural remedy for healthy aging. Aging Dis. 2017;8:708. doi: 10.14336/AD.2017.0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Wang P, et al. Synthesizing ginsenoside Rh2 in Saccharomyces cerevisiae cell factory at high-efficiency. Cell Discov. 2019;5:1–14. doi: 10.1038/s41421-018-0075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Shi Y, et al. Engineering yeast subcellular compartments for increased production of the lipophilic natural products ginsenosides. Metab. Eng. 2021;67:104–111. doi: 10.1016/j.ymben.2021.06.002. [DOI] [PubMed] [Google Scholar]
- 319.Ma X, Gang DR. The lycopodium alkaloids. Nat. Prod. Rep. 2004;21:752–772. doi: 10.1039/b409720n. [DOI] [PubMed] [Google Scholar]
- 320.He M, Qu C, Gao O, Hu X, Hong X. Biological and pharmacological activities of Amaryllidaceae alkaloids. RSC Adv. 2015;5:16562–16574. doi: 10.1039/C4RA14666B. [DOI] [Google Scholar]
- 321.Rubio-Piña J, Vázquez-Flota F. Pharmaceutical applications of the benzylisoquinoline alkaloids from Argemone mexicana L. Curr. Top. Med. Chem. 2013;13:2200–2207. doi: 10.2174/15680266113139990152. [DOI] [PubMed] [Google Scholar]
- 322.Chrzanowska M, Rozwadowska MD. Asymmetric synthesis of isoquinoline alkaloids. Chem. Rev. 2004;104:3341–3370. doi: 10.1021/cr030692k. [DOI] [PubMed] [Google Scholar]
- 323.Galanie S, Thodey K, Trenchard IJ, Filsinger Interrante M, Smolke CD. Complete biosynthesis of opioids in yeast. Science. 2015;349:1095–1100. doi: 10.1126/science.aac9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Nakagawa A, et al. Total biosynthesis of opiates by stepwise fermentation using engineered Escherichia coli. Nat. Commun. 2016;7:1–8. doi: 10.1038/ncomms10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Haroutounian S, et al. Societal issues and policy implications related to the use of cannabinoids, cannabis, and cannabis-based medicines for pain management. Pain. 2021;162:S110–S116. doi: 10.1097/j.pain.0000000000002001. [DOI] [PubMed] [Google Scholar]
- 326.Pereira SR, Hackett B, O’Driscoll DN, Sun MC, Downer EJ. Cannabidiol modulation of oxidative stress and signalling. Neuronal Signal. 2021;5:NS20200080. doi: 10.1042/NS20200080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Luo X, et al. Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature. 2019;567:123–126. doi: 10.1038/s41586-019-0978-9. [DOI] [PubMed] [Google Scholar]
- 328.Hafner J, Payne J, MohammadiPeyhani H, Hatzimanikatis V, Smolke C. A computational workflow for the expansion of heterologous biosynthetic pathways to natural product derivatives. Nat. Commun. 2021;12:1–14. doi: 10.1038/s41467-021-22022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Barrett, G. (eds) Chemistry and Biochemistry Of The Amino Acids (Springer Science & Business Media, 2012).
- 330.Adams AM, et al. In vivo production of psilocybin in E. coli. Metab. Eng. 2019;56:111–119. doi: 10.1016/j.ymben.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 331.Milne, N. et al. Metabolic engineering of Saccharomyces cerevisiae for the de novo production of psilocybin and related tryptamine derivatives. Metab. Eng.60, 25–36 (2020).. [DOI] [PMC free article] [PubMed]
- 332.Wang C, et al. Ionic liquid-microemulsions assisting in the transdermal delivery of Dencichine: preparation, in-vitro and in-vivo evaluations, and investigation of the permeation mechanism. Int. J. Pharm. 2018;535:120–131. doi: 10.1016/j.ijpharm.2017.10.024. [DOI] [PubMed] [Google Scholar]
- 333.Zhao G, Wang X. The hemostatic component of Panax notoginseng: dencichine. Chin. Tradit. Herb. Drugs. 1986;17:34–36. [Google Scholar]
- 334.Li W, et al. Biosynthesis of plant hemostatic dencichine in Escherichia coli. Nat. Commun. 2022;13:5492. doi: 10.1038/s41467-022-33255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Pratley RE, Salsali A. Inhibition of DPP-4: a new therapeutic approach for the treatment of type 2 diabetes. Curr. Med. Res. Opin. 2007;23:919–931. doi: 10.1185/030079906X162746. [DOI] [PubMed] [Google Scholar]
- 336.Sova M, Frlan R, Gobec S, Časar Z. Efficient and straightforward syntheses of two United States pharmacopeia sitagliptin impurities: 3-desamino-2, 3-dehydrositagliptin and 3-desamino-3, 4-dehydrositagliptin. ACS Omega. 2020;5:5356–5364. doi: 10.1021/acsomega.9b04393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Savile CK, et al. Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science. 2010;329:305–309. doi: 10.1126/science.1188934. [DOI] [PubMed] [Google Scholar]
- 338.Katzen F, Chang G, Kudlicki W. The past, present and future of cell-free protein synthesis. Trends Biotechnol. 2005;23:150–156. doi: 10.1016/j.tibtech.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 339.Lu Y. Cell-free synthetic biology: Engineering in an open world. Synth. Syst. Biotechnol. 2017;2:23–27. doi: 10.1016/j.synbio.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Liu J, et al. Research progress on the application of cell-free synthesis systems for enzymatic processes. Crit. Rev. Biotechnol. 2022;1:1–18. doi: 10.1080/07388551.2022.2090314. [DOI] [PubMed] [Google Scholar]
- 341.Chong S. Overview of cell‐free protein synthesis: historic landmarks, commercial systems, and expanding applications. Curr. Protoc. Mol. Biol. 2014;108:16–30. doi: 10.1002/0471142727.mb1630s108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Zhang Y, et al. Accurate high-throughput screening based on digital protein synthesis in a massively parallel femtoliter droplet array. Sci. Adv. 2019;5:eaav8185. doi: 10.1126/sciadv.aav8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Zhang L, Guo W, Lu Y. Advances in cell‐free biosensors: principle, mechanism, and applications. Biotechnol. J. 2020;15:2000187. doi: 10.1002/biot.202000187. [DOI] [PubMed] [Google Scholar]
- 344.Pardee K. Perspective: Solidifying the impact of cell-free synthetic biology through lyophilization. Biochem. Eng. J. 2018;138:91–97. doi: 10.1016/j.bej.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Zhang P, Yang J, Cho E, Lu Y. Bringing light into cell-free expression. ACS Synth. Biol. 2020;9:2144–2153. doi: 10.1021/acssynbio.0c00211. [DOI] [PubMed] [Google Scholar]
- 346.Oh EJ, et al. Target specific and long-acting delivery of protein, peptide, and nucleotide therapeutics using hyaluronic acid derivatives. J. Control. Release. 2010;141:2–12. doi: 10.1016/j.jconrel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 347.Ayoub M, Scheidegger D. Peptide drugs, overcoming the challenges, a growing business. Chim. Oggi. 2006;24:46. [Google Scholar]
- 348.Torchilin VP, Lukyanov AN. Peptide and protein drug delivery to and into tumors: challenges and solutions. Drug Discov. Today. 2003;8:259–266. doi: 10.1016/S1359-6446(03)02623-0. [DOI] [PubMed] [Google Scholar]
- 349.Barok M, Joensuu H, Isola J. Trastuzumab emtansine: mechanisms of action and drug resistance. Breast Cancer Res. 2014;16:1–12. doi: 10.1186/bcr3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Bartelds GM, et al. Clinical response to adalimumab: relationship to anti-adalimumab antibodies and serum adalimumab concentrations in rheumatoid arthritis. Ann. Rheum. Dis. 2007;66:921–926. doi: 10.1136/ard.2006.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Rosenstock J, Park G, Zimmerman J, Group UIGTDI. Basal insulin glargine (HOE 901) versus NPH insulin in patients with type 1 diabetes on multiple daily insulin regimens. US Insulin Glargine (HOE 901) Type 1 Diabetes Investigator Group. Diabetes Care. 2000;23:1137–1142. doi: 10.2337/diacare.23.8.1137. [DOI] [PubMed] [Google Scholar]
- 352.Moore MR, et al. Effect of use of 13-valent pneumococcal conjugate vaccine in children on invasive pneumococcal disease in children and adults in the USA: analysis of multisite, population-based surveillance. Lancet Infect. Dis. 2015;15:301–309. doi: 10.1016/S1473-3099(14)71081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Bosques CJ, et al. Chinese hamster ovary cells can produce galactose-α-1, 3-galactose antigens on proteins. Nat. Biotechnol. 2010;28:1153–1156. doi: 10.1038/nbt1110-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Dumon-Seignovert L, Cariot G, Vuillard L. The toxicity of recombinant proteins in Escherichia coli: a comparison of overexpression in BL21 (DE3), C41 (DE3), and C43 (DE3) Protein Expr. Purif. 2004;37:203–206. doi: 10.1016/j.pep.2004.04.025. [DOI] [PubMed] [Google Scholar]
- 355.Carlson ED, Gan R, Hodgman CE, Jewett MC. Cell-free protein synthesis: applications come of age. Biotechnol. Adv. 2012;30:1185–1194. doi: 10.1016/j.biotechadv.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Chi CW, et al. High‐throughput tumor‐on‐a‐chip platform to study tumor-stroma interactions and drug pharmacokinetics. Adv. Health Mater. 2020;9:2000880. doi: 10.1002/adhm.202000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Salehi AS, et al. Cell‐free protein synthesis of a cytotoxic cancer therapeutic: Onconase production and a just‐add‐water cell‐free system. Biotechnol. J. 2016;11:274–281. doi: 10.1002/biot.201500237. [DOI] [PubMed] [Google Scholar]
- 358.Hong SH, Kwon Y, Jewett MC. Non-standard amino acid incorporation into proteins using Escherichia coli cell-free protein synthesis. Front. Chem. 2014;2:34. doi: 10.3389/fchem.2014.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Gan R, Jewett MC. A combined cell‐free transcription‐translation system from Saccharomyces cerevisiae for rapid and robust protein syntheis. Biotechnol. J. 2014;9:641–651. doi: 10.1002/biot.201300545. [DOI] [PubMed] [Google Scholar]
- 360.He M. Cell-free protein synthesis: applications in proteomics and biotechnology. N. Biotechnol. 2008;25:126–132. doi: 10.1016/j.nbt.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 361.Zhu H, Snyder M. Protein chip technology. Curr. Opin. Chem. Biol. 2003;7:55–63. doi: 10.1016/S1367-5931(02)00005-4. [DOI] [PubMed] [Google Scholar]
- 362.Merkel JS, Michaud GA, Salcius M, Schweitzer B, Predki PF. Functional protein microarrays: just how functional are they? Curr. Opin. Biotechnol. 2005;16:447–452. doi: 10.1016/j.copbio.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 363.Laukens, B., Wachter, C. D. & Callewaert, N. in Glyco-Engineering,Part of the Methods in Molecular Biology Book Series (eds Alexandra, C.) (Springer, 2015).
- 364.Shimizu Y, et al. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 365.Rappuoli R, Pizza M, Del Giudice G. & De Gregorio, E. Vaccines, new opportunities for a new society. Proc. Natl Acad. Sci. USA. 2014;111:12288–12293. doi: 10.1073/pnas.1402981111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Kanter G, et al. Cell-free production of scFv fusion proteins: an efficient approach for personalized lymphoma vaccines. Blood. 2007;109:3393–3399. doi: 10.1182/blood-2006-07-030593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Lu Y, Welsh JP, Swartz JR. Production and stabilization of the trimeric influenza hemagglutinin stem domain for potentially broadly protective influenza vaccines. Proc. Natl Acad. Sci. USA. 2014;111:125–130. doi: 10.1073/pnas.1308701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Tsuboi T, et al. Wheat germ cell-free system-based production of malaria proteins for discovery of novel vaccine candidates. Infect. Immun. 2008;76:1702–1708. doi: 10.1128/IAI.01539-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Padlan EA. Anatomy of the antibody molecule. Mol. Immunol. 1994;31:169–217. doi: 10.1016/0161-5890(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 370.Ryabova LA, Desplancq D, Spirin AS, Plückthun A. Functional antibody production using cell-free translation: effects of protein disulfide isomerase and chaperones. Nat. Biotechnol. 1997;15:79–84. doi: 10.1038/nbt0197-79. [DOI] [PubMed] [Google Scholar]
- 371.Foley TL, Burkart MD. Site-specific protein modification: advances and applications. Curr. Opin. Chem. Biol. 2007;11:12–19. doi: 10.1016/j.cbpa.2006.11.036. [DOI] [PubMed] [Google Scholar]
- 372.Jia W, et al. Polypeptide N-acetylgalactosaminyltransferase 18 retains in endoplasmic reticulum depending on its luminal regions interacting with ER resident UGGT1, PLOD3 and LPCAT1. Glycobiology. 2021;31:947–958. doi: 10.1093/glycob/cwab031. [DOI] [PubMed] [Google Scholar]
- 373.Guarino C, DeLisa MP. A prokaryote-based cell-free translation system that efficiently synthesizes glycoproteins. Glycobiology. 2012;22:596–601. doi: 10.1093/glycob/cwr151. [DOI] [PubMed] [Google Scholar]
- 374.Jaroentomeechai T, et al. Single-pot glycoprotein biosynthesis using a cell-free transcription-translation system enriched with glycosylation machinery. Nat. Commun. 2018;9:1–11. doi: 10.1038/s41467-018-05110-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Yu T, X Y. Array‐based biosensors for bacteria detection: from the perspective of recognition. Small. 2021;17:2006230. doi: 10.1002/smll.202006230. [DOI] [PubMed] [Google Scholar]
- 376.Karube I, Nomura Y. Enzyme sensors for environmental analysis. J. Mol. Catal. B: Enzym. 2000;10:177–181. doi: 10.1016/S1381-1177(00)00125-9. [DOI] [Google Scholar]
- 377.Li B, et al. Recent advance in the sensing of biomarker transcription factors. TrAC Trends Anal. Chem. 2020;132:116039. doi: 10.1016/j.trac.2020.116039. [DOI] [Google Scholar]
- 378.Kimura H, Asano R, Tsukamoto N, Tsugawa W, Sode K. Convenient and universal fabrication method for antibody-enzyme complexes as sensing elements using the SpyCatcher/SpyTag system. Anal. Chem. 2018;90:14500–14506. doi: 10.1021/acs.analchem.8b04344. [DOI] [PubMed] [Google Scholar]
- 379.Mandl J, Mészáros T, Bánhegyi G, Hunyady L, Csala M. Endoplasmic reticulum: nutrient sensor in physiology and pathology. Trends Endocrinol. Metab. 2009;20:194–201. doi: 10.1016/j.tem.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 380.Ziółkowski R, Jarczewska M, Górski Ł, Malinowska E. From small molecules toward whole cells detection: Application of electrochemical aptasensors in modern medical diagnostics. Sensors. 2021;21:724. doi: 10.3390/s21030724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Zhang, X., Ju, H. & Wang, J. (eds.) Electrochemical Sensors, Biosensors and Their Biomedical Applications (Academic Press, 2011).
- 382.Pellinen T, Huovinen T, Karp M. A cell-free biosensor for the detection of transcriptional inducers using firefly luciferase as a reporter. Anal. Biochem. 2004;330:52–57. doi: 10.1016/j.ab.2004.03.064. [DOI] [PubMed] [Google Scholar]
- 383.Davies DH, et al. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc. Natl Acad. Sci. USA. 2005;102:547–552. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Smith MT, Berkheimer SD, Werner CJ, Bundy BC. Lyophilized Escherichia coli-based cell-free systems for robust, high-density, long-term storage. BioTechniques. 2014;56:186–193. doi: 10.2144/000114158. [DOI] [PubMed] [Google Scholar]
- 385.Pardee K, et al. Paper-based synthetic gene networks. Cell. 2014;159:940–954. doi: 10.1016/j.cell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Pardee K, et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 387.Zhou Y, et al. A small and highly sensitive red/far-red optogenetic switch for applications in mammals. Nat. Biotechnol. 2022;40:262–272. doi: 10.1038/s41587-021-01036-w. [DOI] [PubMed] [Google Scholar]
- 388.Cao Z, Cheng S, Wang X, Pang Y, Liu J. Camouflaging bacteria by wrapping with cell membranes. Nat. Commun. 2019;10:3452. doi: 10.1038/s41467-019-11390-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Cui M, et al. Optotheranostic nanosystem with phone visual diagnosis and optogenetic microbial therapy for ulcerative colitis at-home care. ACS Nano. 2021;15:7040–7052. doi: 10.1021/acsnano.1c00135. [DOI] [PubMed] [Google Scholar]
- 390.Patel VL, et al. The coming of age of artificial intelligence in medicine. Artif. Intell. Med. 2009;46:5–17. doi: 10.1016/j.artmed.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Baek M, et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373:871–876. doi: 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Chen J, Sun J, Wang G. From unmanned systems to autonomous intelligent systems. Engineering. 2022;12:16–19. doi: 10.1016/j.eng.2021.10.007. [DOI] [Google Scholar]
- 393.Zhang W, Yu J, Zhao W, Ran C. DMRFNet: deep multimodal reasoning and fusion for visual question answering and explanation generation. Inf. Fusion. 2021;72:70–79. doi: 10.1016/j.inffus.2021.02.006. [DOI] [Google Scholar]
- 394.Kumar M, et al. Secure video communication using firefly optimization and visual cryptography. Artif. Intell. Rev. 2022;55:2997–3017. doi: 10.1007/s10462-021-10070-8. [DOI] [Google Scholar]
- 395.Zhou B, et al. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct. Target. Ther. 2020;5:1–14. doi: 10.1038/s41392-020-00258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Dastmalchi M, et al. Neopinone isomerase is involved in codeine and morphine biosynthesis in opium poppy. Nat. Chem. Biol. 2019;15:384–390. doi: 10.1038/s41589-019-0247-0. [DOI] [PubMed] [Google Scholar]
- 397.Yan X, et al. Production of bioactive ginsenoside compound K in metabolically engineered yeast. Cell Res. 2014;24:770–773. doi: 10.1038/cr.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Wang D, et al. Elucidation of the complete biosynthetic pathway of the main triterpene glycosylation products of Panax notoginseng using a synthetic biology platform. Metab. Eng. 2020;61:131–140. doi: 10.1016/j.ymben.2020.05.007. [DOI] [PubMed] [Google Scholar]
- 399.Siemon T, et al. Semisynthesis of plant-derived englerin A enabled by microbe engineering of guaia-6, 10 (14)-diene as building block. J. Am. Chem. Soc. 2020;142:2760–2765. doi: 10.1021/jacs.9b12940. [DOI] [PubMed] [Google Scholar]
- 400.Engels B, Dahm P, Jennewein S. Metabolic engineering of taxadiene biosynthesis in yeast as a first step towards Taxol (Paclitaxel) production. Metab. Eng. 2008;10:201–206. doi: 10.1016/j.ymben.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 401.Fang, H. et al. Metabolic engineering of Escherichia coli for de novo biosynthesis of vitamin B12. Nat. Commun.9, 1–12 (2018). [DOI] [PMC free article] [PubMed]
- 402.Li J, et al. Production of plant-specific flavones baicalein and scutellarein in an engineered E. coli from available phenylalanine and tyrosine. Metab. Eng. 2019;52:124–133. doi: 10.1016/j.ymben.2018.11.008. [DOI] [PubMed] [Google Scholar]
- 403.Hu T, et al. Engineering chimeric diterpene synthases and isoprenoid biosynthetic pathways enables high-level production of miltiradiene in yeast. Metab. Eng. 2020;60:87–96. doi: 10.1016/j.ymben.2020.03.011. [DOI] [PubMed] [Google Scholar]
- 404.Zhou YJ, et al. Modular pathway engineering of diterpenoid synthases and the mevalonic acid pathway for miltiradiene production. J. Am. Chem. Soc. 2012;134:3234–3241. doi: 10.1021/ja2114486. [DOI] [PubMed] [Google Scholar]
- 405.Zhang J, et al. A microbial supply chain for production of the anti-cancer drug vinblastine. Nature. 2022;609:341–347. doi: 10.1038/s41586-022-05157-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Liu X, et al. Engineering yeast for the production of breviscapine by genomic analysis and synthetic biology approaches. Nat. Commun. 2018;9:1–10. doi: 10.1038/s41467-018-02883-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Srinivasan P, Smolke CD. Biosynthesis of medicinal tropane alkaloids in yeast. Nature. 2020;585:614–619. doi: 10.1038/s41586-020-2650-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Tu L, et al. Genome of Tripterygium wilfordii and identification of cytochrome P450 involved in triptolide biosynthesis. Nat. Commun. 2020;11:1–12. doi: 10.1038/s41467-020-14776-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Huang X, Liang Y, Yang Y, Lu X. Single-step production of the simvastatin precursor monacolin J by engineering of an industrial strain of Aspergillus terreus. Metab. Eng. 2017;42:109–114. doi: 10.1016/j.ymben.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 410.Zhao Q, et al. A severe leakage of intermediates to shunt products in acarbose biosynthesis. Nat. Commun. 2020;11:1–15. doi: 10.1038/s41467-020-15234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Shen B, et al. Fermentative production of Vitamin E tocotrienols in Saccharomyces cerevisiae under cold-shock-triggered temperature control. Nat. Commun. 2020;11:1–14. doi: 10.1038/s41467-020-18958-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Zhuo Y, et al. Reverse biological engineering of hrdB to enhance the production of avermectins in an industrial strain of Streptomyces avermitilis. Proc. Natl Acad. Sci. USA. 2010;107:11250–11254. doi: 10.1073/pnas.1006085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Ignea C, et al. Carnosic acid biosynthesis elucidated by a synthetic biology platform. Proc. Natl Acad. Sci. USA. 2016;113:3681–3686. doi: 10.1073/pnas.1523787113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Li Y, et al. Complete biosynthesis of noscapine and halogenated alkaloids in yeast. Proc. Natl Acad. Sci. USA. 2018;115:E3922–E3931. doi: 10.1073/pnas.1721469115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Ye Z, et al. Revolution of vitamin E production by starting from microbial fermented farnesene to isophytol. Innovation. 2022;3:100228. doi: 10.1016/j.xinn.2022.100228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Schultz BJ, Kim SY, Lau W, Sattely ES. Total biosynthesis for milligram-scale production of etoposide intermediates in a plant chassis. J. Am. Chem. Soc. 2019;141:19231–19235. doi: 10.1021/jacs.9b10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Yip A, Webster RM. The market for chimeric antigen receptor T cell therapies. Nat. Rev. Drug Discov. 2018;17:161–162. doi: 10.1038/nrd.2017.266. [DOI] [PubMed] [Google Scholar]
- 418.Neelapu SS, et al. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N. Engl. J. Med. 2017;377:2531–2544. doi: 10.1056/NEJMoa1707447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Munshi NC, et al. Idecabtagene vicleucel in relapsed and refractory multiple myeloma. N. Engl. J. Med. 2021;384:705–716. doi: 10.1056/NEJMoa2024850. [DOI] [PubMed] [Google Scholar]
- 420.Williams JZ, et al. Precise T cell recognition programs designed by transcriptionally linking multiple receptors. Science. 2020;370:1099–1104. doi: 10.1126/science.abc6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Krawczyk K, et al. Electrogenetic cellular insulin release for real-time glycemic control in type 1 diabetic mice. Science. 2020;368:993–1001. doi: 10.1126/science.aau7187. [DOI] [PubMed] [Google Scholar]
- 422.Bojar D, Scheller L, Hamri GC, Xie M, Fussenegger M. Caffeine-inducible gene switches controlling experimental diabetes. Nat. Commun. 2018;9:1–10. doi: 10.1038/s41467-018-04744-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Ye H, et al. Pharmaceutically controlled designer circuit for the treatment of the metabolic syndrome. Proc. Natl Acad. Sci. USA. 2013;110:141–146. doi: 10.1073/pnas.1216801110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Yin J, et al. A green tea-triggered genetic control system for treating diabetes in mice and monkeys. Sci. Transl. Med. 2019;11:eaav8826. doi: 10.1126/scitranslmed.aav8826. [DOI] [PubMed] [Google Scholar]
- 425.Ye H, Baba MD, Peng R, Fussenegger M. A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science. 2011;332:1565–1568. doi: 10.1126/science.1203535. [DOI] [PubMed] [Google Scholar]
- 426.Kemmer C, et al. Self-sufficient control of urate homeostasis in mice by a synthetic circuit. Nat. Biotechnol. 2010;28:355–360. doi: 10.1038/nbt.1617. [DOI] [PubMed] [Google Scholar]
- 427.Rössger K, Charpin-El Hamri G, Fussenegger M. Reward-based hypertension control by a synthetic brain-dopamine interface. Proc. Natl Acad. Sci. USA. 2013;110:18150–18155. doi: 10.1073/pnas.1312414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Ye H, et al. Self-adjusting synthetic gene circuit for correcting insulin resistance. Nat., Biomed. Eng. 2016;1:1–9. doi: 10.1038/s41551-016-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Wu C, Roybal KT, Puchner EM, Onuffer J, Lim WA. Remote control of therapeutic T cells through a small molecule-gated chimeric receptor. Science. 2015;350:aab4077. doi: 10.1126/science.aab4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Chen YY, Jensen MC, Smolke CD. Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proc. Natl Acad. Sci. USA. 2010;107:8531–8536. doi: 10.1073/pnas.1001721107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Kojima R, Scheller L, Fussenegger M. Nonimmune cells equipped with T-cell-receptor-like signaling for cancer cell ablation. Nat. Chem. Biol. 2018;14:42–49. doi: 10.1038/nchembio.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Fedorov VD, Themeli M, Sadelain M. PD-1-and CTLA-4-based inhibitory chimeric antigen receptors (iCARs) divert off-target immunotherapy responses. Sci. Transl. Med. 2013;5:215ra172. doi: 10.1126/scitranslmed.3006597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Yang L, et al. Engineering genetic devices for in vivo control of therapeutic T cell activity triggered by the dietary molecule resveratrol. Proc. Natl Acad. Sci. USA. 2021;118:e2106612118. doi: 10.1073/pnas.2106612118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Liu E, et al. Use of CAR-transduced natural killer cells in CD19-positive lymphoid tumors. N. Engl. J. Med. 2020;382:545–553. doi: 10.1056/NEJMoa1910607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Klichinsky M, et al. Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat. Biotechnol. 2020;38:947–953. doi: 10.1038/s41587-020-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Amoasii L, et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science. 2018;362:86–91. doi: 10.1126/science.aau1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Maeder ML, et al. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat. Med. 2019;25:229–233. doi: 10.1038/s41591-018-0327-9. [DOI] [PubMed] [Google Scholar]
- 438.Xu L, et al. CRISPR-edited stem cells in a patient with HIV and acute lymphocytic leukemia. N. Engl. J. Med. 2019;381:1240–1247. doi: 10.1056/NEJMoa1817426. [DOI] [PubMed] [Google Scholar]
- 439.Perli SD, Cui CH, Lu TK. Continuous genetic recording with self-targeting CRISPR-Cas in human cells. Science. 2016;353:aag0511. doi: 10.1126/science.aag0511. [DOI] [PubMed] [Google Scholar]
- 440.Saxena P, Charpin-El Hamri G, Folcher M, Zulewski H, Fussenegger M. Synthetic gene network restoring endogenous pituitary-thyroid feedback control in experimental Graves’ disease. Proc. Natl Acad. Sci. USA. 2016;113:1244–1249. doi: 10.1073/pnas.1514383113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Wang H, Xie M, Charpin-El Hamri G, Ye H, Fussenegger M. Treatment of chronic pain by designer cells controlled by spearmint aromatherapy. Nat. Biomed. Eng. 2018;2:114–123. doi: 10.1038/s41551-018-0192-3. [DOI] [PubMed] [Google Scholar]
- 442.Saxena P, et al. A programmable synthetic lineage-control network that differentiates human iPSCs into glucose-sensitive insulin-secreting beta-like cells. Nat. Commun. 2016;7:1–14. doi: 10.1038/ncomms11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Schukur L, Geering B, Charpin-El Hamri G, Fussenegger M. Implantable synthetic cytokine converter cells with AND-gate logic treat experimental psoriasis. Sci. Transl. Med. 2015;7:318ra201. doi: 10.1126/scitranslmed.aac4964. [DOI] [PubMed] [Google Scholar]
- 444.Kojima R, et al. Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson’s disease treatment. Nat. Commun. 2018;9:1–10. doi: 10.1038/s41467-018-03733-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Chen C, et al. Genetic-code-expanded cell-based therapy for treating diabetes in mice. Nat. Chem. Biol. 2022;18:47–55. doi: 10.1038/s41589-021-00899-z. [DOI] [PubMed] [Google Scholar]
- 446.Sedlmayer F, Hell D, Müller M, Ausländer D, Fussenegger M. Designer cells programming quorum-sensing interference with microbes. Nat. Commun. 2018;9:1–13. doi: 10.1038/s41467-018-04223-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Ho CL, et al. Engineered commensal microbes for diet-mediated colorectal-cancer chemoprevention. Nat. Biomed. Eng. 2018;2:27–37. doi: 10.1038/s41551-017-0181-y. [DOI] [PubMed] [Google Scholar]
- 448.Fan J, et al. Bacteria-mediated tumor therapy utilizing photothermally-controlled TNF-α expression via oral administration. Nano Lett. 2018;18:2373–2380. doi: 10.1021/acs.nanolett.7b05323. [DOI] [PubMed] [Google Scholar]
- 449.Chen F, et al. Nanophotosensitizer-engineered Salmonella bacteria with hypoxia targeting and photothermal-assisted mutual bioaccumulation for solid tumor therapy. Biomaterials. 2019;214:119226. doi: 10.1016/j.biomaterials.2019.119226. [DOI] [PubMed] [Google Scholar]
- 450.Steidler L, et al. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science. 2000;289:1352–1355. doi: 10.1126/science.289.5483.1352. [DOI] [PubMed] [Google Scholar]
- 451.Din MO, et al. Synchronized cycles of bacterial lysis for in vivo delivery. Nature. 2016;536:81–85. doi: 10.1038/nature18930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Kotula JW, et al. Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc. Natl Acad. Sci. USA. 2014;111:4838–4843. doi: 10.1073/pnas.1321321111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Riglar DT, et al. Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nat. Biotechnol. 2017;35:653–658. doi: 10.1038/nbt.3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Sheth RU, Yim SS, Wu FL, Wang HH. Multiplex recording of cellular events over time on CRISPR biological tape. Science. 2017;358:1457–1461. doi: 10.1126/science.aao0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Mimee M, et al. An ingestible bacterial-electronic system to monitor gastrointestinal health. Science. 2018;360:915–918. doi: 10.1126/science.aas9315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Mao N, Cubillos-Ruiz A, Cameron DE, Collins JJ. Probiotic strains detect and suppress cholera in mice. Sci. Transl. Med. 2018;10:eaao2586. doi: 10.1126/scitranslmed.aao2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Chang H, et al. Programmable receptors enable bacterial biosensors to detect pathological biomarkers in clinical samples. Nat. Commun. 2021;12:1–12. doi: 10.1038/s41467-021-25538-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Abedi MH, et al. Ultrasound-controllable engineered bacteria for cancer immunotherapy. Nat. Commun. 2022;13:1–11. doi: 10.1038/s41467-022-29065-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Dai Z, et al. Living fabrication of functional semi-interpenetrating polymeric materials. Nat. Commun. 2021;12:1–9. doi: 10.1038/s41467-021-23812-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Leventhal DS, et al. Immunotherapy with engineered bacteria by targeting the STING pathway for anti-tumor immunity. Nat. Commun. 2020;11:2739. doi: 10.1038/s41467-020-16602-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Zheng JH, et al. Two-step enhanced cancer immunotherapy with engineered Salmonella typhimurium secreting heterologous flagellin. Sci. Transl. Med. 2017;9:eaak9537. doi: 10.1126/scitranslmed.aak9537. [DOI] [PubMed] [Google Scholar]
- 462.Swofford CA, Van Dessel N, Forbes NS. Quorum-sensing Salmonella selectively trigger protein expression within tumors. Proc. Natl Acad. Sci. USA. 2015;112:3457–3462. doi: 10.1073/pnas.1414558112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Chowdhury S, et al. Programmable bacteria induce durable tumor regression and systemic antitumor immunity. Nat. Med. 2019;25:1057–1063. doi: 10.1038/s41591-019-0498-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Gurbatri CR, et al. Engineered probiotics for local tumor delivery of checkpoint blockade nanobodies. Sci. Transl. Med. 2020;12:eaax0876. doi: 10.1126/scitranslmed.aax0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Wang S, et al. Driving mosquito refractoriness to Plasmodium falciparum with engineered symbiotic bacteria. Science. 2017;357:1399–1402. doi: 10.1126/science.aan5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Duan F, March JC. Engineered bacterial communication prevents Vibrio cholerae virulence in an infant mouse model. Proc. Natl Acad. Sci. USA. 2010;107:11260–11264. doi: 10.1073/pnas.1001294107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Schmidt F, et al. Noninvasive assessment of gut function using transcriptional recording sentinel cells. Science. 2022;376:eabm6038. doi: 10.1126/science.abm6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Wu M, Jusiak B, Lu TK. Engineering advanced cancer therapies with synthetic biology. Nat. Rev. Cancer. 2019;19:187–195. doi: 10.1038/s41568-019-0121-0. [DOI] [PubMed] [Google Scholar]
- 469.Tang T, et al. Materials design by synthetic biology. Nat. Rev. Mater. 2021;6:332–350. doi: 10.1038/s41578-020-00265-w. [DOI] [Google Scholar]