Abstract
The endothelium is the frontline target of multiple metabolic stressors and pharmacological agents. As a consequence, endothelial cells (ECs) display highly dynamic and diverse proteome profiles. We describe here the culture of human aortic ECs from healthy and type 2 diabetic donors, the treatment with a small molecular coformulation of trans-resveratrol and hesperetin (tRES+HESP), followed by proteomic analysis of whole-cell lysate. A number of 3666 proteins were presented in all of the samples and thus further analyzed. We found that 179 proteins had a significant difference between diabetic ECs vs. healthy ECs, while 81 proteins had a significant change upon the treatment of tRES+HESP in diabetic ECs. Among them, 16 proteins showed a difference between diabetic ECs and healthy ECs and the difference was reversed by the tRES+HESP treatment. Follow-up functional assays identified activin A receptor-like type 1 and transforming growth factor β receptor 2 as the most pronounced targets suppressed by tRES+HESP in protecting angiogenesis in vitro. Our study has revealed the global differences in proteins and biological pathways in ECs from diabetic donors, which are potentially reversible by the tRES+HESP formula. Furthermore, we have identified the TGFβ receptor as a responding mechanism in ECs treated with this formula, shedding light on future studies for deeper molecular characterization.
Introduction
The endothelium is a vital organ lining the entire vascular system, controlling the passage of materials, including nutrients, cytokines, metabolites, toxins, and treatment agents, into and out of the bloodstream.1 The deterioration of endothelial health is the primary step in the onset and progression of atherosclerosis and is closely related to long-term cardiovascular outcomes.2 Meanwhile, the endothelium is one of the primary targets of detrimental factors or therapeutic agents. Ample evidence from experimental and clinical studies has shown that numerous pharmacological agents used for metabolic and cardiovascular diseases impact endothelial function despite different structures and mechanisms of action.3 However, it is challenging to perform a thorough assessment of the molecular response of endothelium to these interventions due to the anatomic locations of the endothelium and the complexity of endothelial activities regulating vascular homeostasis.
Proteomics has emerged as a powerful tool to assay endpoints of the proteome to reveal information on the underlying biochemistry and physiologic state of tissue or cells.4 Notably, limited studies have reported the proteomic findings as tied to human aortic endothelial response to pharmacological agents, and fewer have used it to understand the interactions of treatments under pathologies. We have adopted the tandem mass tags (TMT) approach, which offers the capability to quantify multiple samples (each sample labeled with a unique TMT tag) under different conditions in a single proteomics experiment. This enables us to quantify thousands of proteins simultaneously and understand their regulatory network under normal physical and pathological conditions with or without treatments. The proteomics data can then be used to select relevant markers and pathways and further analyzed by functional assays. The proteomic-based approach provides an ideal platform for mechanistic studies on environmental stimuli and pharmacological interventions.
A recent report demonstrated the coformulation of two dietary compounds, trans-resveratrol (trans-3,5,4′-trihydroxystilbene, tRES) and hesperetin (HESP) (structures were shown in Figure 1), acted synergistically to improve glucose metabolic profile and arterial function in overweight and obese subjects.5 The coformulation was screened out from a pool of ∼100 dietary bioactive compounds with the ability to induce the gene expression of glyoxalase 1 (GLO1), an enzyme that detoxifies reactive metabolites during glycolysis and protects cells against glycation stress.6 What has not been addressed was that tRES+HESP displayed disproportionately potent vaso-protective effects, as demonstrated by improved arterial function in recipients. Indeed, we have previously observed that topical application of tRES-HESP accelerates skin wound closure with a concomitant augmentation of angiogenesis in a mouse model of type 2 diabetes.7 The protection of this coformulation on EC function is possibly beyond its induction of GLO1, as shown in a previous proteomic analysis of tRES-treated human umbilical vein ECs under normal conditions.8 However, no information is available on the proteome profile shift in dermal microvascular ECs from diabetic donors or whether this shift can be reversed by tRES+HESP treatment. In this study, we aim to establish the connection between the efficacy of tRES+HESP on EC function and the altered proteomic profiles. We believe this study gains substantial knowledge of the signaling mechanisms underlying the beneficial effects of tRES+HESP treatment on endothelial function in diabetes.
Figure 1.
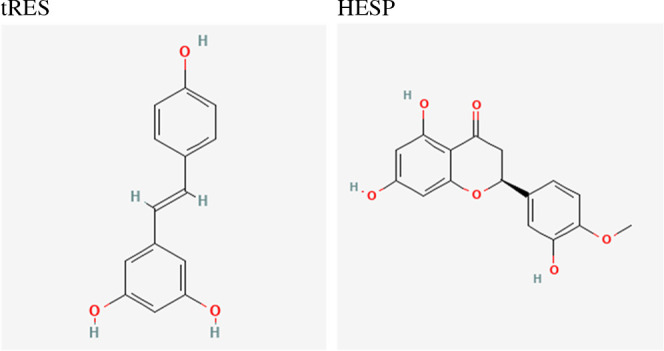
Structure of trans-resveratrol (tRES) and hesperetin (HESP). The two-dimensional (2D) chemical structures of tRES and HESP are shown above. The source images are from the National Library of Medicine National Center for Biotechnology Information (tRES: https://pubchem.ncbi.nlm.nih.gov/compound/445154; HESP: https://pubchem.ncbi.nlm.nih.gov/compound/72281).
Research Design and Methods
Human Aortic Endothelial Cells (HAECs) In Vitro Culture and Treatments
HAECs from healthy (cc-2535) or diabetic (cc-2920) donors were purchased from Lonza and cultured in endothelial growth media (EGM-2, Lonza CC-3162) at 37 °C and 5% CO2. HAECs were used between passages 4 and 6, and all experiments were performed in HAECs from 11 donors (donor information: healthy n = 6, age 52.17 ± 3.301 years, male/female = 4:2, diabetic n = 5, age 60.6 ± 2.821 years, male/female = 3:2). HAECs were treated with 5 μM tRES plus 5 μM HESP, using 0.002% dimethylsulfoxide (DMSO) as vehicle control for 48 h. For transfection of plasmids (all purchased from Origene), in each well in a six-well plate, 1μg of plasmids carrying activin A receptor-like type 1 (ACVRL1) (#RC213389), a disintegrin and metalloprotease domain metallopeptidase domain 9 (ADAM9) (#RC222453), integrin, α V (ITGAV) (#RC227470), propionyl CoA carboxylase, β polypeptide (PCCB) (#RG202019), transforming growth factor β receptor 2 (TGFBR2) (#RC219855), and vector control (#PS100001) were transfected into HAECs with MegaTran 2.0 (#TT210002) for 48 h according to the protocol of the manufacturer. After the transfection, The HAECs were treated with tRES+HESP or vehicle control as described above.
Functional Assays (Proliferation, Network Formation, Apoptosis)
In the network formation assay, HAECs were plated in a 48-well cell culture plate (5 × 104 cells per well) precoated with 160 μL of growth factor-reduced Matrigel matrix (Corning) as described previously.9 Cell proliferation was evaluated using the BrdU Cell Proliferation Assay Kit (Cell Signaling, #6813). Cell apoptosis was determined by Annexin V-PI staining using a Countess II FL Automated Cell Counter (Invitrogen). Approximately 1 × 106 cells from each sample were stained with 20 μL of FITC-conjugated Annexin V (Invitrogen Cat No: A13199) and 2 μL of the 100 μg/mL propidium iodide (PI) (Invitrogen Cat No-P1304MP). The cells were incubated at room temperature in the dark for 15 min. The cell suspension was centrifuged, and the supernatant was removed. The cells were then washed and resuspended in 1× Annexin-binding buffer (Invitrogen Cat No-V13246). 10 μL of this suspension was loaded on the countess slide, and the fluorescence was measured using appropriate filters (Annexin V-Ex/Em: 485/535 nm; PI-Ex/Em: 535/617 nm). The Annexin Vlow-PI-cells were defined as live cells; the Annexin Vhigh/PI– were defined as apoptotic cells; and the Annexin Vhigh/PIhigh cells were defined as dead cells. The fraction of the apoptotic cells was calculated as the percentage of apoptotic cells (Annexin Vhigh/PI–) in the total cell numbers counted.
Proteomic Analysis
Diabetic ECs from four donors were treated with tRES+HESP (both at 5 μM) or vehicle control for 24 h. At the time this experiment was carried out, the maximum number of samples one TMT labeling experiment can handle was 11; therefore, we optioned to use three healthy ECs without tRES+HESP treatment (healthy controls, three samples) and four diabetic ECs (four with tRES+HESP treatment, diabetic tRES+HESP; and four with vehicle treatment, diabetic controls, total eight samples). The cells were lysed with CellLytic buffer (Sigma) mixed with Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific, 1:100 dilution) and 250 μg of proteins from each of the 11 samples were subsequently digested in-solution by trypsin. The concentration of the resulting tryptic peptides was measured, and 100 μg peptides from each sample were labeled with one of the 11 unique TMT tags purchased from Thermo Fisher Scientific. Labeling efficiency for each sample was measured by high-performance liquid chromatography–electrospray ionization tandem mass spectrometry (HPLC-ESI-MS/MS), which was all >97%. An aliquot of the combined TMT-labeled peptides (100 μg) was separated using high pH reverse phase fractionation purchased from Thermo Fisher Scientific, and each fraction of the peptide mixture was separated with a gradient of 2–35% buffer B (100% ACN and 0.1% FA) in 180min, starting with 2% B and increase to 5% B in 3 min, to 10% B in 50 min, to 25% B in 92 min, to 35% B in 30 min, to 90% B in 3 min, hold at 90% B for 10 min, and then 2% buffer B at a flow rate of 300 nl/min on a C18-reversed-phase column (75 μm ID), 35 cm length packed in-house with ReproSil-Pur C18-AQ μm resin (Dr. Maisch GmbH) in buffer A (0.1% FA). MS spectra were generated from an Orbitrap Fusion Lumos equipped with a nano-LC-based electrospray ionization source and coupled online to a Dionex Ultimate 3000 UPLC system (Thermo Fisher Scientific).
A “top 3 second” data-dependent tandem mass spectrometry approach was utilized to identify peptides in the samples. In a top 3-second scan protocol, a full scan spectrum (survey scan, 400–1400 Th) is acquired, followed by collision-induced dissociation (CID) mass spectra of the most abundant ions in the survey scan for 3 s. The survey scan was acquired using the Orbitrap mass analyzer to obtain high mass accuracy and high mass resolution data (120,000 resolution), and the top intense peptides were selected and subjected to fragmentation in the linear ion trap (LTQ). Dynamic exclusion was set at 30 s. The charge state rejection function was enabled with “unassigned” and “single” charge states rejected. By knowing the accurate mass and fragmentation pattern of the peptide, the peptide’s amino acid sequence can be reliably inferred.
The RAW mass spectrometry files were analyzed using MaxQuant, a popular quantitative proteomics software package10 (ver.1.6.2.1), to identify proteins and to obtain the intensity for the 11 TMT reporter ions for each peptide and protein. The RAW mass spectrometry files were searched against a database with forward and reversed Uniprot Human protein sequences downloaded from www.uniprot.org. Standard settings in the MaxQuant were applied. Parent mass tolerance was 5 p.p.m., and fragment mass tolerance was 0.5 Da. Two missing trypsin cleavage sites were allowed. Methionine oxidation (M), phosphorylation (STY), and acetyl (protein N-term) were allowed as variable modifications. Carbamidomethyl (C) as fixed modification. The false discovery rate (FDR) for proteins and peptides (with minimum seven amino acids) was set to 0.01. Only proteins identified with a minimum of two unique peptides were considered.
The TMT reporter ion intensities were used to calculate the relative abundance for each protein in these 11 samples. We first normalized the reporter ion intensity for each of the proteins against the reporter ion intensity for the 11th TMT tag to obtain the ratio for each protein. We then calculated the median for the resulting ratios of proteins for each TMT tag (i.e., each of the 11 samples used for the preliminary study). The median ratio for each 11 TMT tag should be identical since an equal number of TMT-labeled peptides were mixed before HPLC-ESI-MS/MS. Most proteins are expected to have a similar abundance among these 11 samples. Therefore, for each TMT tag, the protein ratio was normalized against its median for the 3666 proteins to correct for errors in TMT tag mixing. This normalization is commonly used in TMT or other labeling techniques that require mixing.
The resulting normalized protein ratios were log 2 transformed and compared among the three groups of samples (healthy controls, diabetic controls, and diabetic tRES+HESP). Independent t-test was used to assess the statistical significance of the effects of type 2 diabetes on protein abundance between healthy controls and diabetic controls, while paired t-test was used to assess the statistical significance of the effects of tRES+HESP treatment on protein abundance between diabetic controls and diabetic tRES+HESP treatment groups.
Experimental Design and Statistical Rationale
The EC functions were tested after treatment of tRES+HESP coformulation. The proteomics analyses were performed to assess the global profiles of protein abundance in diabetic ECs after tRES+HESP treatment. Based on the DAVID bioinformatics results and literature search, the proteins selected were then validated in tRES+HESP-treated EC by functional assays. All values are expressed as mean ± standard deviation (SD). For continuous variables that failed Shiparo–Wilk normality tests, such as mRNA expression, protein levels, staining quantifications, and functional assays, the statistical significance of differences between the two groups was determined using the Mann–Whitney U test. When more than two groups of treatments were performed, the Kruskal–Wallis test was applied across all of the groups, and if significant, the pairs of primary interest, based on scientific rationale, were assessed using the Mann–Whitney U test with Benjamini Krieger, and Yekutieli’s adjustment for multiple comparisons.11 These gatekeeping approaches and the adjustments preserved α spending and controlled false positive rate inflation due to multiple hypothesis testing. The significant differences that came from post hoc comparisons of groups were noted. A value of p < 0.05 was considered statistically significant. All of these statistical analyses were performed using GraphPad Prism 9 (GraphPad Software).
Data and Resource Availability
The mass spectrometry proteomics dataset generated in this study are available at PRoteomics IDEntifications Database (PRIDE) (PXD036094), submitted through proteomexchange.org. The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Results
tRES+HESP Augments EC Functions In Vitro
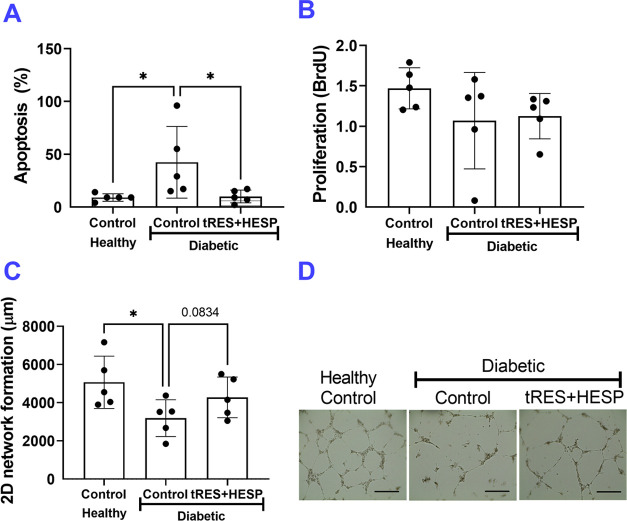
We tested the EC functions of diabetic ECs with or without tRES+HESP coformulation treatment in vitro, using healthy ECs as controls. The tRES+HESP (both at 5 μM) or vehicle control were added to the cell culture media for 24 h before the functional tests were performed. The vessel-like network formation by ECs on growth factor-reduced Matrigel matrix was visualized. As shown in Figure 2A, the cell apoptosis in diabetic ECs was significantly higher than that in healthy ECs but was reversed by tRES+HESP treatment. The proliferation was comparable among treatment groups (Figure 2B). The diabetic ECs formed less network than healthy ECs (Figure 2C,D). This was somewhat rescued upon tRES+HESP treatment (p = 0.0834 vs. diabetic ECs receiving control treatment). These observations suggested that the tRES+HESP stimulated the tube of diabetic ECs.
Figure 2.
Diabetic EC functions are compromised compared with healthy ECs. Cell functions between healthy and type 2 diabetic human aorta ECs were compared using in vitro assays, including cell apoptosis (Annexin V+/PI– staining), proliferation (BrdU incorporation), and two-dimensional (2D) network formation. (A) Apoptosis was evaluated by Annexin V-PI staining and counted as the percentage of the cells that were Annexin V+/PI–; n = 5 per group, *p < 0.05. (B) Proliferation was evaluated by the BrdU assay. Optic density values were recorded; n = 5 per group. (C) Quantification of network formation on Matrigel; n = 5 per group, *p < 0.05. (D) Representative images of network formation; bar = 400 μm. In all figures, horizontal lines show mean ± SD.
tRES+HESP Rescues Abnormalities in Protein Abundance of Multiple Proteins in Diabetic ECs In Vitro
We performed comparative proteomics studies on the effect of tRES+HESP on the global profiles of protein abundance in diabetic ECs. The characteristics of the participants are listed in Table 1. There is no significant difference in any characteristics provided by the vendor (Lonza) between the healthy and type 2 diabetic participants (P > 0.05). Based on the proteomics analysis, 3666 proteins with minimal 2 unique peptides and >99.99% confidence were identified.
Table 1. Characteristics of Participants for the Proteomics Analysisa.
| group | gender (M/F) | race (African Americans/Caucasian) | ethnicity (hispanic/nonhispanic) | age (years) | BMI (kg/m2) |
|---|---|---|---|---|---|
| healthy | 2/1 | 1/2 | 0/3 | 54.3 ± 2.3 | 23.7 ± 0.9 |
| diabetic | 2/2 | 1/3 | 1/3 | 58.2 ± 2.0 | 33.2 ± 4.6 |
Data are given as means ± SEM when applicable.
We have analyzed the dispersion of the resulting normalized protein ratios using the coefficient of variation (CV), which was defined as the ratio of the standard deviation to the mean. The CV for the normalized protein ratio for each of the 3666 proteins for the three healthy controls was obtained, and the mean and medium for these 3666 CVs, along with those for the four diabetic controls, and four diabetic controls with tRES+HESP treatment, are listed in the Supporting Information, Table S1. The results indicated that the average CV for normalized protein ratios for the same set of samples was ∼6%, which is much better than that for the label-free HPLC-ESI-MS/MS analysis and western blotting, suggesting high reproducibility of the TMT labeling approach.
Although a large number of proteins were identified, a series of filters were used to narrow the number of proteins that were used in comparisons between groups to minimize false positives: (1) identified in all 11 samples and with TMT reporter ion intensity, and then, 3666 proteins met this criterion; (2) with a fold change greater than 1.15 (i.e., 1.15-fold increase) or less than 0.87 (i.e., 1.15-fold decrease) between the comparisons, and then, 324 (diabetic controls vs. healthy controls) and 149 (diabetic tRES+HESP treatment vs. diabetic controls) proteins met this criterion; and (3) with a P-value < 0.05, and 179 (diabetic controls vs. healthy controls) and 81 (diabetic tRES+HESP treatment vs. diabetic controls) proteins met this criterion. Therefore, 179 proteins have a significant difference between diabetic controls vs. healthy controls (Supporting Information, Figure S1), while 81 proteins have a significant change upon tRES+HESP treatment in diabetic ECs (Supporting Information, Figure S2). The false discovery rate (FDR) has emerged as a powerful method to correct for multiple statistical comparisons, which offers an excellent balance between false positives and false negatives (Bonferroni type of corrections tend to have excessive false negatives).12 FDR is defined as12
There were 324 and 149 t-tests performed to assess the effects of type 2 diabetes or tRES+HESP treatment on protein abundance, respectively. In total, 179 out of the 324 proteins showed significant differences between diabetic controls vs. healthy controls, and 81 out of the 149 proteins showed significant differences between diabetic tRES+HESP treatment vs. diabetic controls (P < 0.05); among these 179 and 81 proteins with P < 0.05, there may be 16.2 and 7.5 false positives (324*0.05 = 16.2 and 179*0.05 = 7.5), respectively. Thus, the FDR is 0.09 and 0.09 (16.2/324 = 0.09 and 7.5/81 = 0.09) for this study, respectively, which is less than the reported FDR values for many omics studies in various journals (FDR ranges from 0.1 to 0.25).13
We have performed gene ontology (GO) and pathway enrichment analyses using the Database for Annotation, Visualization, and Integrated Discovery (DAVID)14 for the 179 proteins with a significant difference between diabetic controls vs. healthy controls. Categorical annotation was provided in the form of GO biological process, cellular component, and molecular function, as well as Kyoto Encyclopedia of Genes and Genomes pathways and Reactome (reactome.org).14 DAVID analysis for the 179 proteins with significant differences between healthy and diabetic ECs indicated that multiple diseases, biological processes, molecular functions, and pathways are significantly enriched (Supporting Information, Table S2, P < 0.05). Meanwhile, DAVID analysis for the 81 proteins with significant differences between diabetic tRES+HESP treatment vs. diabetic controls indicated that multiple diseases, biological processes, molecular functions, and pathways are significantly enriched (Supporting Information, Table S3, P < 0.05). Please note that one protein can be assigned multiple diseases, biological processes, molecular functions, and pathways.
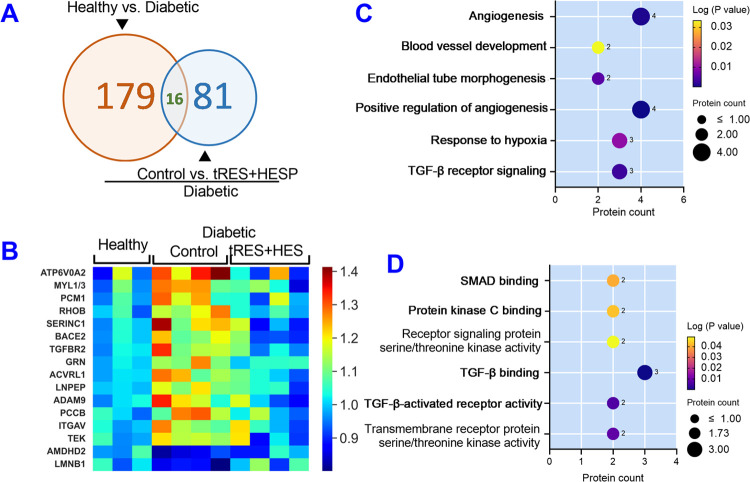
In addition to analyzing the proteomics data in an unbiased fashion as described above, we searched for proteins that were (1) different between diabetic and healthy controls (fold change for diabetic control vs. healthy control is >1.15-fold and P < 0.05) and (2) the difference was reversed by the tRES+HESP treatment (fold change for diabetic tRES+HESP treatment vs. healthy control is <1.05-fold, i.e., diabetic tRES+HESP treatment is within 5% of the healthy control). In this proteomics study, a total of 16 proteins reached our criteria (Figure 3A and Table 2). The heatmap of these proteins is shown in Figure 3B.
Figure 3.
Differentially expressed proteins in diabetic ECs reversed by tRES+HESP treatment and their distribution in biological processes and molecular functions. (A) Schema of significantly altered proteins in healthy controls, diabetic controls, and diabetics with tRES+HESP treatment. (B) Heatmap of 16 differentially expressed proteins and their abundance pattern in diabetic controls (n = 4) compared with healthy controls (n = 3) but reversed by tRES+HESP treatment (n = 4). These proteins were analyzed for their distribution in enriched biological processes and molecular functions revealed by DAVID analysis. The enriched biological processes (C) and molecular functions (D) are shown on Y-axis. The greater the −Log (P-value) value (e.g., the smaller P-value) in the Y-axis, the more likely a pathway is significantly enriched just by chance. The size of the dots indicated protein numbers (listed nearby).
Table 2. Fold Changes of the 16 Proteins among Three Groups of Samplesa.
| gene name | protein name | healthy controls | diabetic controls | diabetic tRES+HESP |
|---|---|---|---|---|
| LMNB1 | lamin-B1 | 1.00 ± 0.04 | 0.86 ± 0.02 | 1.03 ± 0.05 |
| ADAM9 | disintegrin and metalloproteinase domain-containing protein 9 | 1.00 ± 0.02 | 1.19 ± 0.06 | 0.99 ± 0.07 |
| ACVRL1 | activin A receptor-like type 1; receptor protein serine/threonine kinase; serine/threonine-protein kinase receptor; serine/threonine-protein kinase receptor R3; TGF-B superfamily receptor type I | 1.00 ± 0.01 | 1.19 ± 0.05 | 1.02 ± 0.04 |
| AMDHD2 | putative N-acetylglucosamine-6-phosphate deacetylase | 1.00 ± 0.04 | 0.86 ± 0.02 | 0.95 ± 0.04 |
| PCM1 | pericentriolar material 1 protein | 1.00 ± 0.04 | 1.19 ± 0.05 | 1.03 ± 0.06 |
| TEK | angiopoietin-1 receptor | 1.00 ± 0.03 | 1.16 ± 0.02 | 1.00 ± 0.07 |
| PCCB | propionyl CoA carboxylase β chain, mitochondrial | 1.00 ± 0.03 | 1.20 ± 0.05 | 1.02 ± 0.05 |
| ATP6V0A2 | V-type proton ATPase 116 kDa subunit a isoform 2 | 1.00 ± 0.09 | 1.31 ± 0.05 | 1.02 ± 0.08 |
| GRN | acrogranin; granulin-1; granulin-2; granulin-3; granulin-4; granulin-5; granulin-6; granulin-7; granulins; paragranulin | 1.00 ± 0.01 | 1.16 ± 0.04 | 1.04 ± 0.01 |
| ITGAV | integrin α-V | 1.00 ± 0.02 | 1.15 ± 0.04 | 1.04 ± 0.06 |
| RHOB | Rho-related GTP-binding protein RhoB | 1.00 ± 0.04 | 1.19 ± 0.03 | 1.02 ± 0.02 |
| MYL1; MYL3 | myosin light chain 1/3, skeletal muscle isoform | 1.00 ± 0.06 | 1.22 ± 0.04 | 1.02 ± 0.06 |
| SERINC1 | serine incorporator 1 | 1.00 ± 0.02 | 1.24 ± 0.06 | 0.98 ± 0.07 |
| LNPEP | leucyl-cystinyl aminopeptidase; leucyl-cystinyl aminopeptidase, pregnancy serum form | 1.00 ± 0.01 | 1.15 ± 0.03 | 1.02 ± 0.04 |
| BACE2 | β-secretase 2 | 1.00 ± 0.02 | 1.16 ± 0.03 | 0.97 ± 0.05 |
| TGFBR2 | TGF-β receptor type 2 | 1.00 ± 0.02 | 1.19 ± 0.05 | 1.01 ± 0.04 |
Healthy controls, diabetic controls, and diabetic controls with tRES+HESP.
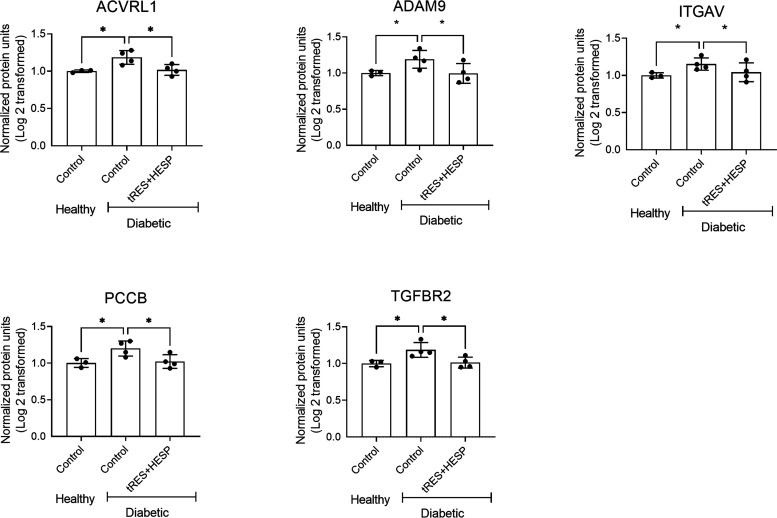
DAVID analysis for these 16 proteins indicated that 11 of them were assigned to the cytoplasm, 9 to the plasma membrane, 4 to the nucleus, 4 to the cytoskeleton, 1 to the endoplasmic reticulum (ER), 1 to the Golgi apparatus, and 1 to the mitochondrion. In addition, multiple biological processes are significantly enriched for these 16 proteins (Table 2, P < 0.05), such as angiogenesis, blood vessel development, endothelial tube morphogenesis, positive regulation of angiogenesis, response to hypoxia, and transforming growth factor-β receptor signaling, etc. (Figure 3C). Moreover, multiple molecular functions are significantly enriched for these 16 proteins (P < 0.05), such as SMAD binding, protein kinase C binding, receptor signaling protein serine/threonine kinase activity, transforming growth factor-β binding, transforming growth factor β-activated receptor activity, and transmembrane receptor protein serine/threonine kinase activity, etc. (Figure 3D). Please note that one protein can be assigned multiple cellular localization, biological processes, or molecular functions. Based on the DAVID bioinformatics results and literature search, we have selected the top five proteins ACVRL1, ADAM9, ITGAV, PCCB, and TGFBR2 (Figure 4). Their functions and activities are demonstrated in Table 3. These proteins are the likely mediators contributing to the beneficial effect of tRES+HESP on angiogenesis.
Figure 4.
Five proteins whose abundance was different between diabetic and healthy controls and reversed by the tRES+HESP treatment. Data are given as fold changes. The mean value of the normalized intensity for TMT reporter ion for each protein in the healthy ECs was set to 1.00, and all of the fold changes were relative to healthy ECs. A fold change of 2 indicates a 2-fold increase, and a fold change of 0.5 indicates a 2-fold decrease. *P < 0.05 diabetic controls vs. healthy controls. In all figures, horizontal lines show mean ± SD.
Table 3. Functions and Activities of the Five Proteins Altered by tRES+HESP.
| gene name | protein name | function, activity and pathways |
|---|---|---|
| ACVRL1 | activin A receptor-like type 1, aka, TGF-B superfamily receptor type I | an important regulator of normal blood vessel development. Its mutations in mice with the presence of oxidative stress are associated with decoupling eNOS.40 |
| ADAM9 | disintegrin and metalloproteinase domain-containing protein 9 | cleaves and releases cytokines such as TNF and epidermal growth factor receptor ligands and regulate inflammatory or regenerative processes. May mediate cell–cell, cell–matrix interactions.41 |
| ITGAV | integrin α-V | receptor for fibronectin, MMP2, and thrombospondins. Binds with fractalkine (CX3CL1).42 Participate in TGFB-1 release.43 |
| PCCB | propionyl CoA carboxylase β chain, mitochondrial | mitochondrial fatty acid β-oxidation and metabolism of water-soluble vitamins and cofactors.44 |
| TGFBR2 | TGF-β receptor type 2 | gatekeeper receptor for TGFbeta signaling during embryonic hematopoiesis and vasculogenesis has both Ser/Thr kinase activity and Tyr kinase activity.45 |
Validation of Causal Roles of ACVRL1, ADAM9, ITGAV, PCCB, and TGFBR2 in the tRES+HESP-Induced Improvement of EC Function
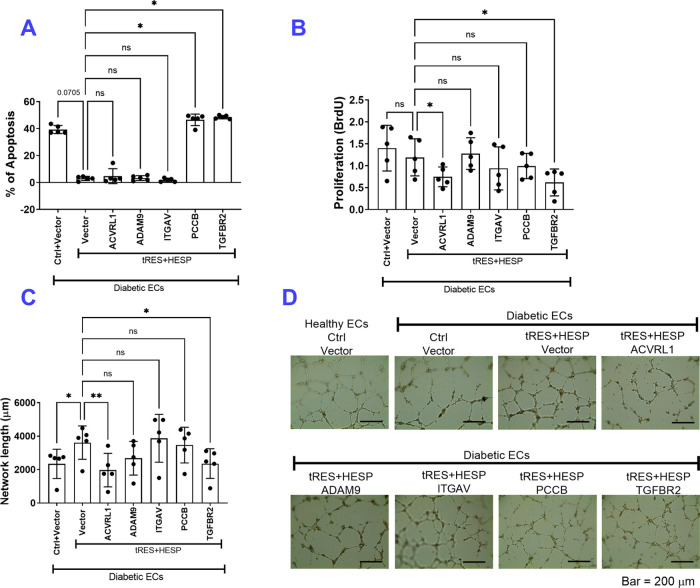
We then performed functional validations in EC validations on these five molecules and tested their essentiality in tRES+HESP-promoted EC angiogenesis in vitro (Figure 5). ECs were transfected with plasmids carrying ACVRL1, ADAM9, ITGAV, PCCB, and TGFBR2 gene or empty vector, together with 5 μM tRES and HESP or 0.002% DMSO control for 48 h. The apoptosis, proliferation, and network formation assessment were performed to evaluate the EC angiogenesis. As shown in Figure 5, under the control treatment and empty vector transfection, diabetic ECs showed increased apoptosis (Figure 5A), comparable proliferation (Figure 5B), and decreased network formation (Figure 5C,D). PCCB and TGFBR2 showed a significant role in tRES+HESP-induced protection against cell apoptosis (Figure 5A), while ACVRL1 and TGFBR2 participated in tRES+HESP’s protection of cell proliferation (Figure 5B) and network formation (Figure 5C). Through these functional tests, we believe that at least three molecules are playing a contributing role, PCCB, ACVRL1, and TGFBR2. Given that migration and network formation are comprehensive measurements of cellular behavior of generating new vessels, these results suggest that ACVRL1 and TGFBR2 are two vital molecules regulated by tRES+HESP treatment in ECs.
Figure 5.
Functional validation of five proteins in tRES+HESP-induced angiogenesis. Human aortic endothelial cells (ECs) from healthy or diabetic donors received plasmids carrying ACVRL1, ADAM9, ITGAV, PCCB, and TGFBR2 gene or vector control, with tRES+HESP (both at 5 μM) or DMSO control treatment for 48 h. Cell functions, including apoptosis, proliferation, and network formation, were performed. (A) Cell apoptosis was determined by Annexin V-PI staining using a Countess II FL Automated Cell Counter; n = 5 per group. *P < 0.05 vs. D-HAEC + vector + tRES+HESP. (B) The proliferation of the ECs evaluated using the BrdU assay; n = 5 per group. *P < 0.05 vs. D-HAEC + vector + tRES+HESP. (C) Quantification of network on Matrigel formed by the ECs; n = 5 per group. *P < 0.05 vs. D-HAEC + vector + tRES+HESP. (D) Representative images of network formation; bar = 200 μm. In all figures, horizontal lines show mean ± SD.
Discussion
This study tested the functional impact of tRES+HESP coformulation in human aortic ECs. Through the proteomic studies and follow-up functional tests, we compared the EC protein profiles between healthy donors and type 2 diabetes donors and identified potentially critical molecules mediating the protection of this small molecule coformulation on EC function (Figure 6). Our study has shed light on the molecular changes induced by this formula outside its action on GLO1.
Figure 6.
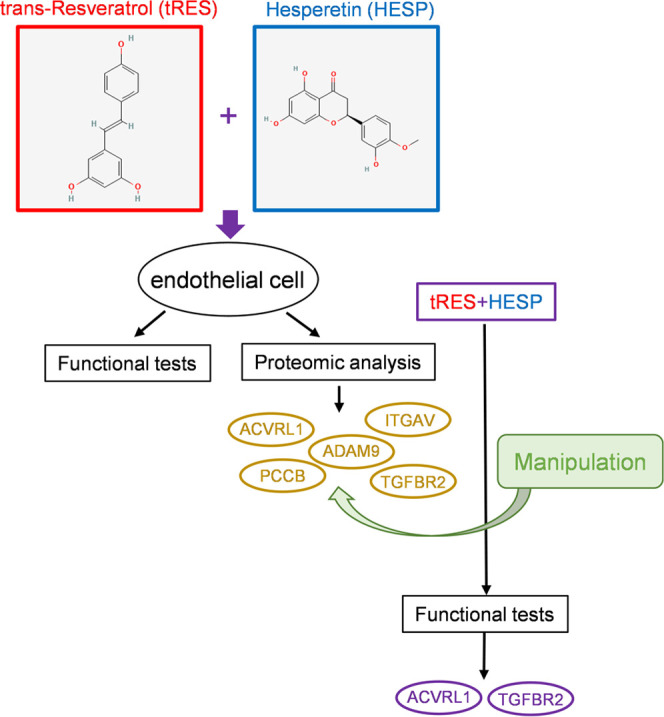
Schema of the workflow and key findings. Human endothelial cells received tRES+HESP treatment, followed by functional tests and proteomic analysis. The proteins ACVRL1, ADAM9, ITGAV, PCCB, and TGFBR2 were chosen. After the genetic manipulation of the five proteins, together with tRES+HESP treatment, functional assays identified that ACVRL1 and TGFBR2 are the most pronounced mediators for tRES+HESP-induced protection of angiogenesis.
Glycation of lipids, DNA, and proteins contributes to an antiproliferative and apoptotic response to highly cytotoxic stress. Methylglyoxal is one of the most extensively studied glycating agents.15 GLO1 of the glyoxalase system has a major role in the metabolism of methylglyoxal, and thus it is plausible to develop GLO1 inducers to alleviate glycation stress as a prospective regimen to prevent and reverse early-stage type 2 diabetes and vascular complications.16 The coformulation of tRES+HESP was established previously to reduce insulin resistance in overweight and obese subjects due to its ability to alleviate glycation stress by inducing GLO1.5,17 Our previous report has recaptured the induction of GLO1 by this formula in endothelial progenitor cells,7 and we have shown that the topical application of the formula augmented wound angiogenesis in a mouse model of type 2 diabetes. The glycation stress induces chronic inflammation and interferes with angiogenesis, both of which are responsible for poor wound healing in individuals with hyperglycemia. Therefore, it is conceivable that correcting the glycation stress is a working mechanism by which the tRES+HESP formula accelerates wound healing in diabetic animals. The tRES+HESP coformulation was proposed to ameliorate insulin resistance in obese individuals because it showed strong inductions of two potent molecules in response to metabolic turbulence, glyoxalase 1, which eliminates glycation stress, and Nrf2, which activates antioxidant genes. In fact, each of tRES and HESP can activate enzymes with an antioxidant nature to scavenge reactive oxygen species and stimulate anti-inflammatory cytokines,18 and each tRES and HESP have unique but related pathways in augmenting EC functions. tRES, the dominant isoform of resveratrol, exhibits antimicrobiome and antitumor activities.19 In addition, tRES has been reported to work by inhibiting ET-1 synthesis,20 activating the estrogen receptor,21 sirtuin 1,22 phosphorylation of AMP-activated protein kinase,23 and potassium channels.24 HESP, a flavonoid found in oranges, mandarins, and lemons, exerts neuroprotective effects in experimental models of neurogenerative diseases.25 Together with its 7-O-glycoside hesperidin, HESP has been reported to inhibit VCAM1 expression and adhesion of monocytes to ECs,26 thrombogenic plasminogen activator inhibitor-1 levels,27 and activate the estrogen receptor.28 While the mechanism by which these two bioactive polyphenols synergize their signaling in ECs is still elusive, all of these abovementioned pathways are potent signals to directly or indirectly promote eNOS enzymatic activity and preserve NO bioavailability by counteracting oxidative stress. Consistently, our observations suggest the proangiogenic and antiapoptotic activities of tRES+HESP in ECs from type 2 diabetes donors (Figure 2A,B), supporting the protection of this formula on cardiometabolic health.
It was the limitation of our study that we did not include the experiments of healthy ECs being exposed to tRES+HESP. However, numerous clinical trials have shown that tRES intake, with or without other diet modifications, showed some improvement of EC functions in healthy subjects.29−31 This is related to its antioxidant, anti-inflammatory, and antitumor activities as mentioned above. However, marginal benefits or unfavorable outcomes from clinical trials have also been reported.19,32 Similar to tRES, HESP (and hesperidin) showed benefits in EC functions in healthy subjects and obese subjects through its antioxidant, anti-inflammatory, or antibacterial actions,26,33,34 but marginal/no effects on cardiovascular health have also been reported.35 The limited solubility and bioavailability through diet is a barrier to translating the expected health benefits of polyphenols like tRES and HESP, while high doses of individual use often lead to adverse effects. The coformulation of tRES-HESP through oral administration has advantages because HESP inhibits intestinal glucuronosyl transferase and facilitates the uptake of these two compounds, increasing their bioavailability in vivo.5,36 Nonetheless, whether tRES+HESP cast benefits endothelial function in healthy subjects without risk factors or pathologies (including aging) remains inconclusive, which warrants future investigations in the healthy population.
An important finding from the proteomic analysis was the activation of TGFβ signaling in diabetic ECs that can be reversed by tRES+HESP treatment (Supporting Information, Figure S1), as evidenced by multiple molecules in the TGFβ signaling pathway, including the TGFβ ligands, TGFβ binding, TGFβ receptor activity, Smad activity, and subsequently transmembrane receptor protein serine/threonine kinase activity (Supporting Information, Figure S2). In fact, TGFβ has been suggested to have complicated roles in modulating angiogenesis. TGFβ was initially considered as a proproliferative factor because the TGFβ1 or TGFβR2 mutant mice are partly embryonic lethal due to multiorgan inflammatory cell infiltration and necrosis, delayed vasculogenesis, and disorganized primary vessels.37 As our understanding of TGFβ evolves, it is now known that while low levels of TGFβ promote angiogenesis, higher levels of TGFβ result in growth inhibition of EC proliferation and vascular maturation.38 The diabetic ECs possessed higher levels of TGFβ signaling as indicated by protein abundance. ACVRL1 (TGFβ superfamily receptor type 1) and TGFβ receptor type 2 were among the top five altered molecules (Table 3) that have been tested further for functional essentiality in tRES+HESP-induced protection on ECs. Our results indicated that inhibition of ACVRL1 and TGFBR2 mediates the tRES+HESP’s protection in both proliferation and network formation tests (Figure 5B,D). Additionally, inhibition of TGFBR2 also contributes to the tRES+HESP’s protection of ECs against apoptosis (Figure 5A). Based on these observations, we propose that tRES+HESP inhibits the TGFβ signal in ECs to modulate angiogenesis. Nonetheless, multiple signaling pathways might be involved. A recent report has shown that Nrf2 activation also mediates the beneficial effects of tRES+HESP.16 Nrf2 was not picked up in our proteomic study based on our filtering criteria. However, crosstalks have been found between TGFβ and Nrf signaling. Particularly, TGFβ1 interacts with Nrf2-mediated antioxidant responses in various cell types,39 but this action seems to be predominantly limited to the process of mesothelial–mesenchymal transition. Moreover, though ADAM9 and ITGAV did not show a role in apoptosis, proliferation, and network formation, they might participate in the action of tRES and HESP through modulation of inflammatory and immune responses given their roles in inflammation. Future study is needed to gain more insight into the precise mechanism(s) for the protective benefit of tRES and HESP on EC functions.
It is noted that proteomics can provide comparable quantification results to western blotting and offer the capability to assess thousands of proteins simultaneously, offering the advantage of unbiased discovery and not requiring prior knowledge on which proteins to probe for. Recently, we have adopted the TMT approach. Since the TMT-labeled samples are combined before HPLC-ESI-MS/MS analysis, the experimental variation on protein quantification is significantly reduced. Using TMT labeling, the average standard deviation of protein abundance for the same set of samples was ∼6% of the mean, which is much better than that for the label-free HPLC-ESI-MS/MS analysis and western blotting. This will enable the detection of small differences among different conditions reproducibly.
In conclusion, our study has provided substantial knowledge of the signaling mechanisms, identifying TGFβR as one of the critical targets, which will help the translation of preclinical findings into clinical settings. This new small-molecule formula offers an inexpensive, easily administered solution. It has the capability of further advantages in combining systemic administration and local tissue application to prevent vascular complications associated with diabetes.
Glossary
Abbreviations
- ACVRL1
activin A receptor-like type 1
- ADAM9
a disintegrin and metalloprotease domain metallopeptidase domain 9
- CV
coefficient of variation
- DAVID
the Database for Annotation, Visualization, and Integrated Discovery
- DMSO
dimethylsulfoxide
- EC
endothelial cell
- ER
endoplasmic reticulum
- FDR
false discovery rate
- GO
gene ontology
- HAEC
human aortic endothelial cells
- HESP
hesperetin
- HPLC-ESI-MS/MS
high-performance liquid chromatography–electrospray ionization tandem mass spectrometry
- ITGAV
integrin, α V
- PVDF
poly(vinylidene fluoride)
- PCCB
propionyl CoA carboxylase, β polypeptide
- TGFBR2
transforming growth factor β receptor 2
- TMT
tandem mass tags
- tRES
trans-resveratrol
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c00678.
Heatmap of differentially expressed proteins and their abundance pattern in healthy and diabetic endothelial cells (ECs) (Figure S1); heatmap of differentially expressed proteins and their abundance pattern in diabetic ECs with control or tRES+HESP treatment (Figure S2); the mean and medium for the coefficient of variation for the normalized protein ratio (Table S1); DAVID analysis for the proteins with significant differences between healthy and diabetic ECs (Table S2); and DAVID analysis for the proteins with significant differences between diabetic tRES+HESP treatment vs. diabetic controls (Table S3) (PDF)
Author Contributions
⊥ A.M. and H.L. contributed equally to the manuscript. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript. A.M., H.L., X.Z., and R.J.: methodology. A.M., X.Z., R.J., and F.-S.Y.: formal analysis. A.M., X.Z., R.J., F.-S.Y., and Z.Y.: data curation. A.M., H.L., and J.-M.W.: writing—original draft. A.M., H.L., S.P., M.V., R.J., F.-S.Y., Z.Y., and J.-M.W.: writing—review and editing. Z.Y. and J.-M.W.: supervision. Z.Y. and J.-M.W.: project administration. Z.Y. and J.-M.W.: funding acquisition.
This work was supported in part by the NIH/NIDDK R01 DK109036 (To J.-M.W.), R01 DK119222 (to J.-M.W.), and R01 DK128937 (to Z.Y. and J.-M.W.).
The authors declare no competing financial interest.
Supplementary Material
References
- Rajendran P.; Rengarajan T.; Thangavel J.; Nishigaki Y.; Sakthisekaran D.; Sethi G.; Nishigaki I. The vascular endothelium and human diseases. Int. J. Biol. Sci. 2013, 9, 1057–1069. 10.7150/ijbs.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez E.; Flammer A. J.; Lerman L. O.; Elizaga J.; Lerman A.; Fernandez-Aviles F. Endothelial dysfunction over the course of coronary artery disease. Eur. Heart J. 2013, 34, 3175–3181. 10.1093/eurheartj/eht351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Kiseleva R. Y.; Glassman P. M.; Greineder C. F.; Hood E. D.; Shuvaev V. V.; Muzykantov V. R. Targeting therapeutics to endothelium: are we there yet. Drug Delivery Transl. Res. 2018, 8, 883–902. 10.1007/s13346-017-0464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Su J. B. Vascular endothelial dysfunction and pharmacological treatment. World J. Cardiol. 2015, 7, 719–741. 10.4330/wjc.v7.i11.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa Rojo A.; Heylen D.; Aerts J.; Thas O.; Hooyberghs J.; Ertaylan G.; Valkenborg D. Towards Building a Quantitative Proteomics Toolbox in Precision Medicine: A Mini-Review. Front. Physiol. 2021, 12, 723510 10.3389/fphys.2021.723510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M.; Weickert M. O.; Qureshi S.; Kandala N. B.; Anwar A.; Waldron M.; Shafie A.; Messenger D.; Fowler M.; Jenkins G.; et al. Improved Glycemic Control and Vascular Function in Overweight and Obese Subjects by Glyoxalase 1 Inducer Formulation. Diabetes 2016, 65, 2282–2294. 10.2337/db16-0153. [DOI] [PubMed] [Google Scholar]
- Rabbani N.; Thornalley P. J. Glyoxalase 1 Modulation in Obesity and Diabetes. Antioxid. Redox Signaling 2018, 30, 354–374. 10.1089/ars.2017.7424. [DOI] [PubMed] [Google Scholar]
- Li H.; O’Meara M.; Zhang X.; Zhang K.; Seyoum B.; Yi Z.; Kaufman R. J.; Monks T. J.; Wang J. M. Ameliorating Methylglyoxal-Induced Progenitor Cell Dysfunction for Tissue Repair in Diabetes. Diabetes 2019, 68, 1287–1302. 10.2337/db18-0933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao B.; Tang M.; Li Z.; Zhou R.; Deng Y.; Nie C.; Yuan Z.; Zhou L.; Tang M.; Tong A.; et al. Proteomics analysis of human umbilical vein endothelial cells treated with resveratrol. Amino Acids 2012, 43, 1671–1678. 10.1007/s00726-012-1248-4. [DOI] [PubMed] [Google Scholar]
- Wang J. M.; Tao J.; Chen D. D.; Cai J. J.; Irani K.; Wang Q.; Yuan H.; Chen A. F. MicroRNA miR-27b rescues bone marrow-derived angiogenic cell function and accelerates wound healing in type 2 diabetes mellitus. Arterioscler., Thromb., Vasc. Biol. 2014, 34, 99–109. 10.1161/atvbaha.113.302104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Cox J.; Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]; b Neuhauser N.; Nagaraj N.; McHardy P.; Zanivan S.; Scheltema R.; Cox J.; Mann M. High performance computational analysis of large-scale proteome data sets to assess incremental contribution to coverage of the human genome. J. Proteome Res. 2013, 12, 2858–2868. 10.1021/pr400181q. [DOI] [PubMed] [Google Scholar]; c Weisser H.; Nahnsen S.; Grossmann J.; Nilse L.; Quandt A.; Brauer H.; Sturm M.; Kenar E.; Kohlbacher O.; Aebersold R.; et al. An Automated Pipeline for High-Throughput Label-Free Quantitative Proteomics. J. Proteome Res. 2013, 12, 1628–1644. 10.1021/pr300992u. [DOI] [PubMed] [Google Scholar]
- Benjamini Y.; Krieger A. M.; Yekutieli D. Adaptive linear step-up procedures that control the false discovery rate. Biometrika 2006, 93, 491–507. 10.1093/biomet/93.3.491. [DOI] [Google Scholar]
- a Glickman M. E.; Rao S. R.; Schultz M. R. False discovery rate control is a recommended alternative to Bonferroni-type adjustments in health studies. J. Clin. Epidemiol. 2014, 67, 850–857. 10.1016/j.jclinepi.2014.03.012. [DOI] [PubMed] [Google Scholar]; b Storey J. D.; Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 9440–9445. 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Pounds S. B. Estimation and control of multiple testing error rates for microarray studies. Briefings Bioinf. 2006, 7, 25–36. 10.1093/bib/bbk002. [DOI] [PubMed] [Google Scholar]
- a Steele M. A.; Schiestel C.; AlZahal O.; Dionissopoulos L.; Laarman A. H.; Matthews J. C.; McBride B. W. The periparturient period is associated with structural and transcriptomic adaptations of rumen papillae in dairy cattle. J. Dairy Sci. 2015, 98, 2583–2595. 10.3168/jds.2014-8640. [DOI] [PubMed] [Google Scholar]; b Zheng D.; Kille P.; Feeney G. P.; Cunningham P.; Handy R. D.; Hogstrand C. Dynamic transcriptomic profiles of zebrafish gills in response to zinc supplementation. BMC Genomics 2010, 11, 553. 10.1186/1471-2164-11-553. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Rahimov F.; King O. D.; Leung D. G.; Bibat G. M.; Emerson C. P. Jr.; Kunkel L. M.; Wagner K. R. Transcriptional profiling in facioscapulohumeral muscular dystrophy to identify candidate biomarkers. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 16234–16239. 10.1073/pnas.1209508109. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Geetha T.; Langlais P.; Luo M.; Mapes R.; Lefort N.; Chen S. C.; Mandarino L. J.; Yi Z. Label-free proteomic identification of endogenous, insulin-stimulated interaction partners of insulin receptor substrate-1. J. Am. Soc. Mass Spectrom. 2011, 22, 457–466. 10.1007/s13361-010-0051-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W.; Sherman B. T.; Tan Q.; Kir J.; Liu D.; Bryant D.; Guo Y.; Stephens R.; Baseler M. W.; Lane H. C.; et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007, 35, W169–W175. 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalkwijk C. G.; Stehouwer C. D. A. Methylglyoxal, a Highly Reactive Dicarbonyl Compound, in Diabetes, Its Vascular Complications, and Other Age-Related Diseases. Physiol. Rev. 2020, 100, 407–461. 10.1152/physrev.00001.2019. [DOI] [PubMed] [Google Scholar]
- Rabbani N.; Thornalley P. J. Emerging Glycation-Based Therapeutics-Glyoxalase 1 Inducers and Glyoxalase 1 Inhibitors. Int. J. Mol. Sci. 2022, 23, 2453 10.3390/ijms23052453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani N.; Xue M.; Weickert M. O.; Thornalley P. J. Reversal of Insulin Resistance in Overweight and Obese Subjects by trans-Resveratrol and Hesperetin Combination-Link to Dysglycemia, Blood Pressure, Dyslipidemia, and Low-Grade Inflammation. Nutrients 2021, 13, 2374 10.3390/nu13072374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Moussa C.; Hebron M.; Huang X.; Ahn J.; Rissman R. A.; Aisen P. S.; Turner R. S. Resveratrol regulates neuro-inflammation and induces adaptive immunity in Alzheimer’s disease. J. Neuroinflammation 2017, 14, 1. 10.1186/s12974-016-0779-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Parhiz H.; Roohbakhsh A.; Soltani F.; Rezaee R.; Iranshahi M. Antioxidant and anti-inflammatory properties of the citrus flavonoids hesperidin and hesperetin: an updated review of their molecular mechanisms and experimental models. Phytother. Res. 2015, 29, 323–331. 10.1002/ptr.5256. [DOI] [PubMed] [Google Scholar]
- Salehi B.; Mishra A. P.; Nigam M.; Sener B.; Kilic M.; Sharifi-Rad M.; Fokou P. V. T.; Martins N.; Sharifi-Rad J. Resveratrol: A Double-Edged Sword in Health Benefits. Biomedicines 2018, 6, 91 10.3390/biomedicines6030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W. F.; Yang H. Y.; Liu J. C.; Cheng T. H.; Chen C. H.; Shih C. M.; Lin H.; Wang T. C.; Lian W. S.; Chen J. J.; et al. Inhibition of cyclic strain-induced endothelin-1 secretion by tetramethylpyrazine. Clin. Exp. Pharmacol. Physiol. 2005, 32, 536–540. 10.1111/j.1440-1681.2005.04227.x. [DOI] [PubMed] [Google Scholar]
- Bowers J. L.; Tyulmenkov V. V.; Jernigan S. C.; Klinge C. M. Resveratrol acts as a mixed agonist/antagonist for estrogen receptors alpha and beta. Endocrinology 2000, 141, 3657–3667. 10.1210/endo.141.10.7721. [DOI] [PubMed] [Google Scholar]
- Hubbard B. P.; Gomes A. P.; Dai H.; Li J.; Case A. W.; Considine T.; Riera T. V.; Lee J. E.; Lamming D. W.; et al. Evidence for a common mechanism of SIRT1 regulation by allosteric activators. Science 2013, 339, 1216–1219. 10.1126/science.1231097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. J.; Ahmad F.; Philp A.; Baar K.; Williams T.; Luo H.; Ke H.; Rehmann H.; Taussig R.; Brown A. L.; et al. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell 2012, 148, 421–433. 10.1016/j.cell.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protić D.; Radunović N.; Spremović-Rađenović S.; Živanović V.; Heinle H.; Petrović A.; Gojković-Bukarica L. The Role of Potassium Channels in the Vasodilatation Induced by Resveratrol and Naringenin in Isolated Human Umbilical Vein. Drug Dev. Res. 2015, 76, 17–23. 10.1002/ddr.21236. [DOI] [PubMed] [Google Scholar]
- Evans J. A.; Mendonca P.; Soliman K. F. A. Neuroprotective Effects and Therapeutic Potential of the Citrus Flavonoid Hesperetin in Neurodegenerative Diseases. Nutrients 2022, 14, 2228 10.3390/nu14112228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizza S.; Muniyappa R.; Iantorno M.; Kim J. A.; Chen H.; Pullikotil P.; Senese N.; Tesauro M.; Lauro D.; Cardillo C.; et al. Citrus polyphenol hesperidin stimulates production of nitric oxide in endothelial cells while improving endothelial function and reducing inflammatory markers in patients with metabolic syndrome. J. Clin. Endocrinol. Metab. 2011, 96, E782–E792. 10.1210/jc.2010-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giménez-Bastida J. A.; González-Sarrías A.; Vallejo F.; Espín J. C.; Tomás-Barberán F. A. Hesperetin and its sulfate and glucuronide metabolites inhibit TNF-α induced human aortic endothelial cell migration and decrease plasminogen activator inhibitor-1 (PAI-1) levels. Food Funct. 2016, 7, 118–126. 10.1039/c5fo00771b. [DOI] [PubMed] [Google Scholar]
- Liu L.; Xu D. M.; Cheng Y. Y. Distinct effects of naringenin and hesperetin on nitric oxide production from endothelial cells. J. Agric. Food Chem. 2008, 56, 824–829. 10.1021/jf0723007. [DOI] [PubMed] [Google Scholar]
- Gresele P.; Pignatelli P.; Guglielmini G.; Carnevale R.; Mezzasoma A. M.; Ghiselli A.; Momi S.; Violi F. Resveratrol, at concentrations attainable with moderate wine consumption, stimulates human platelet nitric oxide production. J. Nutr. 2008, 138, 1602–1608. 10.1093/jn/138.9.1602. [DOI] [PubMed] [Google Scholar]
- Wong R. H.; Berry N. M.; Coates A. M.; Buckley J. D.; Bryan J.; Kunz I.; Howe P. R. Chronic resveratrol consumption improves brachial flow-mediated dilatation in healthy obese adults. J. Hypertens. 2013, 31, 1819–1827. 10.1097/HJH.0b013e328362b9d6. [DOI] [PubMed] [Google Scholar]
- Ozemek C.; Hildreth K. L.; Blatchford P. J.; Hurt K. J.; Bok R.; Seals D. R.; Kohrt W. M.; Moreau K. L. Effects of resveratrol or estradiol on postexercise endothelial function in estrogen-deficient postmenopausal women. J. Appl. Physiol. 2020, 128, 739–747. 10.1152/japplphysiol.00488.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Gonçalinho G. H. F.; Roggerio A.; Goes M.; Avakian S. D.; Leal D. P.; Strunz C. M. C.; Mansur A. P. Comparison of Resveratrol Supplementation and Energy Restriction Effects on Sympathetic Nervous System Activity and Vascular Reactivity: A Randomized Clinical Trial. Molecules 2021, 26, 3168 10.3390/molecules26113168. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Mankowski R. T.; You L.; Buford T. W.; Leeuwenburgh C.; Manini T. M.; Schneider S.; Qiu P.; Anton S. D. Higher dose of resveratrol elevated cardiovascular disease risk biomarker levels in overweight older adults—A pilot study. Exp. Gerontol. 2020, 131, 110821 10.1016/j.exger.2019.110821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morand C.; Dubray C.; Milenkovic D.; Lioger D.; Martin J. F.; Scalbert A.; Mazur A. Hesperidin contributes to the vascular protective effects of orange juice: a randomized crossover study in healthy volunteers. Am. J. Clin. Nutr. 2011, 93, 73–80. 10.3945/ajcn.110.004945. [DOI] [PubMed] [Google Scholar]
- Li L.; Lyall G. K.; Martinez-Blazquez J. A.; Vallejo F.; F A. T.-B.; Birch K. M.; Boesch C. Blood Orange Juice Consumption Increases Flow-Mediated Dilation in Adults with Overweight and Obesity: A Randomized Controlled Trial. J. Nutr. 2020, 150, 2287–2294. 10.1093/jn/nxaa158. [DOI] [PubMed] [Google Scholar]
- Schär M. Y.; Curtis P. J.; Hazim S.; Ostertag L. M.; Kay C. D.; Potter J. F.; Cassidy A. Orange juice-derived flavanone and phenolic metabolites do not acutely affect cardiovascular risk biomarkers: a randomized, placebo-controlled, crossover trial in men at moderate risk of cardiovascular disease. Am. J. Clin. Nutr. 2015, 101, 931–938. 10.3945/ajcn.114.104364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani N.; Thornalley P. J. Glyoxalase 1 Modulation in Obesity and Diabetes. Antioxid. Redox Signaling 2019, 30, 354–374. 10.1089/ars.2017.7424. [DOI] [PubMed] [Google Scholar]
- a Akhurst R. J.; Lehnert S. A.; Faissner A.; Duffie E. TGF beta in murine morphogenetic processes: the early embryo and cardiogenesis. Development 1990, 108, 645–656. 10.1242/dev.108.4.645. [DOI] [PubMed] [Google Scholar]; b Oshima M.; Oshima H.; Taketo M. M. TGF-beta receptor type II deficiency results in defects of yolk sac hematopoiesis and vasculogenesis. Dev. Biol. 1996, 179, 297–302. 10.1006/dbio.1996.0259. [DOI] [PubMed] [Google Scholar]
- a Li C.; Guo B.; Bernabeu C.; Kumar S. Angiogenesis in breast cancer: the role of transforming growth factor beta and CD105. Microsc. Res. Tech. 2001, 52, 437–449. . [DOI] [PubMed] [Google Scholar]; b Hofer E.; Schweighofer B. Signal transduction induced in endothelial cells by growth factor receptors involved in angiogenesis. Thromb. Haemostasis 2007, 97, 355–363. 10.1160/TH06-08-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Churchman A. T.; Anwar A. A.; Li F. Y. L.; Sato H.; Ishii T.; Mann G. E.; Siow R. C. M. Transforming growth factor-beta1 elicits Nrf2-mediated antioxidant responses in aortic smooth muscle cells. J. Cell. Mol. Med. 2009, 13, 2282–2292. 10.1111/j.1582-4934.2009.00874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Yazaki K.; Matsuno Y.; Yoshida K.; Sherpa M.; Nakajima M.; Matsuyama M.; Kiwamoto T.; Morishima Y.; Ishii Y.; Hizawa N. ROS-Nrf2 pathway mediates the development of TGF-beta1-induced epithelial-mesenchymal transition through the activation of Notch signaling. Eur. J. Cell Biol. 2021, 100, 151181 10.1016/j.ejcb.2021.151181. [DOI] [PubMed] [Google Scholar]
- Jerkic M.; Kabir M. G.; Davies A.; Yu L. X.; McIntyre B. A.; Husain N. W.; Enomoto M.; Sotov V.; Husain M.; Henkelman M.; et al. Pulmonary hypertension in adult Alk1 heterozygous mice due to oxidative stress. Cardiovasc. Res. 2011, 92, 375–384. 10.1093/cvr/cvr232. [DOI] [PubMed] [Google Scholar]
- Dreymueller D.; Uhlig S.; Ludwig A. ADAM-family metalloproteinases in lung inflammation: potential therapeutic targets. Am. J. Physiol.: Lung Cell. Mol. Physiol. 2015, 308, L325–L343. 10.1152/ajplung.00294.2014. [DOI] [PubMed] [Google Scholar]
- Fujita M.; Takada Y. K.; Takada Y. Integrins alphavbeta3 and alpha4beta1 act as coreceptors for fractalkine, and the integrin-binding defective mutant of fractalkine is an antagonist of CX3CR1. J. Immunol. 2012, 189, 5809–5819. 10.4049/jimmunol.1200889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annes J. P.; Chen Y.; Munger J. S.; Rifkin D. B. Integrin alphaVbeta6-mediated activation of latent TGF-beta requires the latent TGF-beta binding protein-1. J. Cell. Biol. 2004, 165, 723–734. 10.1083/jcb.200312172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongkittichote P.; Ah Mew N.; Chapman K. A. Propionyl-CoA carboxylase - A review. Mol. Genet. Metab. 2017, 122, 145–152. 10.1016/j.ymgme.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Ark A.; Cao J.; Li X. TGF-beta receptors: In and beyond TGF-beta signaling. Cell. Signalling 2018, 52, 112–120. 10.1016/j.cellsig.2018.09.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.