Abstract
Salmonella enterica serovar Typhimurium invasion genes are necessary for bacterial invasion of intestinal epithelial cells and are thought to allow salmonellae to enter and cross the intestinal epithelium during infection. Many invasion genes are encoded on Salmonella pathogenicity island 1 (SPI1), and their expression is activated by HilA, a transcription factor also encoded on SPI1. We have studied the role of Salmonella invasion genes during infection of mice following intragastric inoculation. We have found that strains containing a mutation in hilA or invG were recovered from the intestinal contents, intestinal tissues, and systemic tissues at a lower frequency than their parental wild-type strain. In contrast, a strain in which SPI1 is deleted was recovered from infected mice at a frequency similar to that of its parental wild-type strain. The ΔSPI1 phenotype indicates that S. enterica does not require invasion genes to cross the intestinal epithelium and infect systemic tissues. This result has forced us to reconsider the long-held belief that invasion genes directly mediate bacterial infection of the intestinal mucosa and traversion of the intestinal barrier during infection. Instead, our results suggest that hilA is required for bacterial colonization of the host intestine. The seemingly contradictory phenotype of the ΔSPI1 mutant suggests that deletion of another gene(s) encoded on SPI1 suppresses the hilA mutant defect. We propose a model for S. enterica pathogenesis in which hilA and invasion genes are required for salmonellae to overcome a host clearance response elicited by another SPI1 gene product(s).
Infection with Salmonella enterica serovar Typhimurium can cause a systemic, typhoid-like disease in mice. Following ingestion, bacteria can colonize the intestinal tract, penetrate the intestinal epithelium, and access systemic sites such as the spleen and liver through the lymphatic and blood circulation (7). Passage of the bacteria through the intestinal wall is believed to be initiated by bacterial invasion into enterocytes and M cells (7, 22, 26, 43). The ability of salmonellae to penetrate the intestinal mucosa has been correlated with their ability to invade cultured nonphagocytic cells (14, 17, 26). Salmonella invasion into cultured epithelial cells is mediated by a bacterial type III secretion system (25). Secretion of bacterial proteins such as SopE and SptP into the host cell cytosol reorganizes the cytoskeleton, leading to membrane ruffling and bacterial uptake (12, 13, 19, 28, 46). The type III secretion system is encoded by genes on Salmonella pathogenicity island 1 (SPI1) (33). Expression of the SPI1 secretion system as well as many of its secreted effectors are coordinately regulated by HilA, a transcriptional activator encoded on SPI1 (1–3, 11).
Invasion genes appear to be important for serovar Typhimurium infection of mice via the gastric route. Ligated loop assays have shown that bacterial strains with mutations in hilA and other invasion genes have a reduced ability to enter and disrupt M cells compared to their wild-type (WT) parental strain (10, 26, 36). Studies of the dose of bacteria required to kill 50% of infected mice (50% lethal dose [LD50]) also suggest that invasion genes are important for infection via the gastric route. The LD50 of invasion mutants administered intraperitoneally (i.p.) is the same as that of the WT, whereas the LD50 of invasion mutants administered intragastrically (i.g.) is increased at least 20-fold over that of the WT (1, 4, 14, 26, 36). In these studies, i.p. inoculation is thought to deliver bacteria directly to systemic sites and bypass the need for invasion genes to traverse the intestinal wall. The decrease in virulence of a hilA mutant and other invasion mutants when inoculated i.g. but not i.p. suggests that invasion genes are important for entering and crossing the intestinal epithelium.
Invasion genes also contribute to the ability of salmonellae to induce migration of neutrophils (PMNs) into and across the intestinal epithelium (15, 27, 31). A hallmark of gastroenteritis, this transmigration response requires Salmonella adhesion to the epithelial apical membrane. Bacterial contact causes the epithelial cells to express and secrete chemokines that are chemotactic for PMNs (30). In vitro studies have shown that hilA and other SPI1 invasion genes are required for serovar Typhimurium to induce cytokine expression and transepithelial migration of PMNs (16). Invasion genes also appear to be involved in nitric oxide production in cultured colonic epithelial cells. A Salmonella enterica serovar Dublin mutant defective in invA, which encodes a component of the SPI1 secretion system, does not induce NO production compared to a WT strain. NO production increases in response to stimulation with a combination of gamma interferon and interleukin-1 or tumor necrosis factor alpha (45 and references within). Thus, invasion factors may enhance NO production by inducing the expression of these cytokines. Interestingly, serovar Typhimurium may induce invasion and the production of certain cytokines via the same mechanism. The delivery of SopE, one of the type III secretion effectors, into COS-1 cells activates a CDC42-dependent signal transduction cascade (19) which appears to stimulate both membrane ruffling and expression of interleukin-8 (21). Invasion genes have also been shown to be important for serovar Typhimurium to induce macrophage apoptosis in vitro (8, 34). SipB, an SPI1-secreted protein, directly interacts with and activates the proapoptosis enzyme caspase-1 in cultured macrophages (20).
In summary, invasion genes have the potential to contribute to many aspects of Salmonella pathogenesis by allowing the bacteria to directly and indirectly alter the behavior of host cells, such as epithelial cells, PMNs, and macrophages. Previous studies have primarily focused on investigating the role of invasion genes in allowing salmonellae to enter and cross intestinal epithelial cells during infection. We have explored the role of invasion genes during Salmonella pathogenesis by studying three mutant strains—one mutant containing a deletion of SPI1, one mutant containing an insertion mutation in hilA, and one mutant containing an insertion mutation in invG, a component of the type III secretion apparatus encoded on SPI1. We have compared the levels of mutant and WT bacteria recovered from different tissues following intragastric inoculation of mice. Results from these infection assays together with LD50 determinations and other published results suggest that invasion genes may contribute to Salmonella pathogenesis by mediating bacterial interactions with nonepithelial cell types. We propose a model in which SPI1 is involved in a complex cross-talk scheme between the bacteria and the host.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
RM12, a clone of SL1344, was tested for virulence in mice and used as the WT serovar Typhimurium strain in this study (23). P22 was used to transduce mutations into the parental RM12 strain. RM40 contains the hilA::Tn5lacZY-080 (tet) mutation which was transduced from EE658 (3). RM69 contains the Δspi1::kan mutation in which SPI1 was deleted and replaced by a kanamycin resistance marker (41). The Δspi1::kan mutation was transduced from SD11 (41). RM50 contains the invG::cam mutation, which was transduced from SB201 (gift of Jorge Galán). All strains were grown in Luria-Bertani (LB) according to Sambrook et al. (40) without pH adjustment to saturation by rolling culture tubes overnight at 37°C. CFU were determined on LB-agar plates (40) containing 5 g of NaCl per liter plus streptomycin (100 mg/liter), tetracycline (25 mg/liter), kanamycin (50 mg/liter), or chloramphenicol (25 mg/liter) when appropriate.
Animal infections.
Female BALB/c mice 6 to 8 weeks old (Taconic Farms, Inc.) were given drinking water containing streptomycin (5 mg/ml) for 24 h prior to infection. Fresh drinking water containing antibiotic was supplied every 4 days for the duration of the experiment. Animals were starved overnight prior to inoculation. Bacterial inocula were prepared by centrifuging broth cultures and resuspending the bacteria in phosphate-buffered saline (PBS); 0.1 ml of the inoculum was delivered i.g. using a feeding needle (Harvard Instruments). The CFU in each inoculum was determined by plating dilutions on agar plates containing appropriate antibiotics.
LD50 and mean day to death.
To determine LD50s, groups of four animals were infected with the specified inocula (10, 100, 1,000, and 10,000 CFU) for each strain. Animal deaths were recorded for 3 weeks, and the LD50 was estimated by the Reed and Muench method (39). The mean day to death refers to the average time it took animals to die.
C.I.'s and single infections.
For competitive indices (C.I.'s) and total bacterial counts (CFU), infected tissues were harvested and homogenized in PBS, and dilutions were plated on LB-agar plates containing streptomycin, tetracycline, kanamycin, or chloramphenicol. Specifically, a fragment of the intestine approximately corresponding to the ileum, three Peyer's patches proximal to the cecum, mesenteric lymph nodes, and the spleen were removed from infected animals. The intestinal segment was further divided into tissue (intestine) and nontissue (contents) fractions by extruding the luminal material within each segment. All bacterial strains used are streptomycin resistant. All mutant strains used are resistant to an additional antibiotic, such as kanamycin, tetracycline, or chloramphenicol. Therefore, the WT CFU was estimated by subtracting the sum of CFU on kanamycin, chloramphenicol, and tetracycline (sum of CFU for mutant strains) from the CFU on streptomycin (CFU for all strains). C.I.'s were determined for each tissue sample by dividing mutant CFU by WT CFU.
Statistical analysis.
All statistical tests were done using the Prism statistical analysis package (GraphPad Software, San Diego, Calif.). C.I.'s were logarithmically transformed, and arithmetic means were calculated and used for analysis. The 95% confidence interval for the log10 C.I. of the mutants was compared to 0 (one-sample t test). For the CFU comparisons, paired and unpaired Student t tests were done for each tissue (mixed and single infections, respectively). Mean day to death for the WT and hilA::tet strains was compared by the generalized Wilcoxon test.
RESULTS
The ability of salmonellae to infect experimental animals via the i.g. route can be inconsistent (4). One explanation for this unreliability is inhibition of Salmonella colonization by the resident intestinal flora. Germ-free mice have been seen to have an LD50 1,000-fold lower than non-germ-free mice when infected with serovar Typhimurium (35). Reduction of microflora by treatment with streptomycin also increases the susceptibility of mice to infection by increasing the ability of salmonellae to colonize the intestinal tract and translocate to the mesentery, spleen, and liver (5, 32, 38). Similarly, we found that streptomycin treatment of mice prior to i.g. inoculation increases the reproducibility of infection and dramatically reduces the dose required for infection (data not shown).
The numbers of bacteria recovered from mice following infection can vary greatly from animal to animal. One way of reducing this variability is to perform mixed infections (4, 14). This approach allows direct comparison of mutant strains and the WT strain in the same animal. In addition to using the WT strain as an internal standard, mixed infections have the advantage of reducing the number of animals necessary for determining the infection phenotype of a mutant bacterial strain.
Mixed infections.
Eight mice were infected with a total of 104 CFU containing a mixture of WT and three mutant strains, a hilA::tet, a Δspi1::kan, and an invG::cam mutant, in an approximately 1:1:1:1 ratio (Fig. 1). Tissues were harvested 5 days postinoculation, homogenized, diluted, and plated on LB-agar plates containing antibiotics. All strains are streptomycin resistant; therefore, to determine the WT CFU, the sum of the mutant CFUs, as determined on agar plates containing tetracycline, kanamycin, or chloramphenicol, was subtracted from the total streptomycin-resistant CFU. The C.I. was determined as mutant CFU/WT CFU and compared to 1 (one-sample t test).
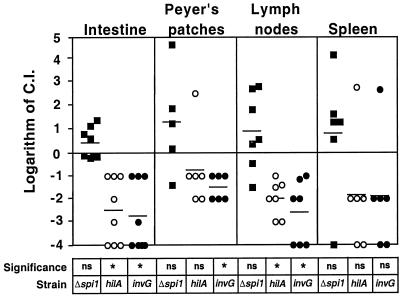
FIG. 1.
Mean C.I. of mutant strains for different tissues at 5 days postinoculation. Eight mice were orally infected with a mixture of WT and mutant strains in equal proportions. Tissues were harvested at 5 days postinoculation, homogenized, and plated on streptomycin, tetracycline, kanamycin, or chloramphenicol. Mean C.I.'s were determined as described in Materials and Methods. Each symbol is the logarithm of C.I. Solid squares, C.I.'s for the Δspi1::kan mutant; open circles, C.I.'s for the hilA::tet mutant; solid circles, C.I.'s for the invG::cam mutant. Horizontal lines are the mean of the logarithm of C.I.'s. ns, mean C.I. not significantly different from 1 (log C.I. = 0) (P > 0.05, one-sample t test). An asterisk indicates that a mean C.I. is significantly different from 1 (log C.I. = 0) (P < 0.05, one-sample t test).
The mean C.I. for the hilA::tet and invG::cam mutants was lower than 1 for all of the tissues analyzed. However, the mean C.I. was not statistically significantly different from 1 for the invG::cam mutant in the spleen and for the hilA::tet mutant in the Peyer's patches and spleen (P > 0.05, one-sample t test). Our explanation for why statistical significance was not achieved is that these three groups of C.I.'s included a single outlier that represents tissues from the same animal.
The Δspi1::kan mutant and WT strains were recovered at a similar frequency from all tissues at all times postinoculation (P > 0.05, one-sample t test). Although the recovery of the Δspi1::kan mutant was not significantly different from that of the WT, the calculated mean C.I. was greater than 1 in the intestine, Peyer's patches, mesenteric lymph nodes, and spleen.
Mixed versus single infections.
We were concerned that the presence of different bacterial strains in the infected mice could alter their virulence phenotypes. For example, the presence of WT bacteria might suppress a virulence defect of the Δspi1::kan mutant. In fact, complementation of the invasion defect of an invE mutant strain by WT bacteria has been reported to occur in vitro (18). In addition, the WT and/or the Δspi1::kan strain could compete for host factors and reduce the recovery of the hilA::tet mutant from a mixed infection. To investigate these possibilities, we compared results from mixed and single infections. Groups of four animals were inoculated with 105 CFU of a single strain or 105 CFU of a 1:1 mixture of strains. Tissues were harvested 5 days postinoculation.
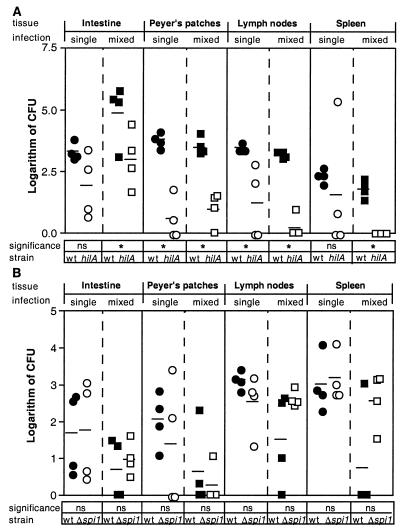
Statistical analyses showed that the hilA::tet mutant was recovered at a reduced frequency compared to WT from Peyer's patches and mesenteric lymph nodes in both the single and mixed infections (P < 0.05, unpaired t test and paired t test, respectively) (Fig. 2A). The hilA::tet mutant was also recovered at a reduced frequency from the intestine and spleen in both infections; however, this reduction was only statistically significant in the case of mixed infections (P < 0.05, paired t test). The Δspi1::kan mutant, on the other hand, was recovered at a similar frequency to the WT from all tissues in both single and mixed infections (P > 0.05, unpaired t test and paired t test, respectively) (Fig. 2B). These results suggest there is no trans-complementation of the Δspi1::kan mutant by WT bacteria during a mixed infection and that the reduced recovery of the hilA::tet mutant is not dependent on coinfection with the WT strain.
FIG. 2.
Comparison of mixed and single infections. (A) Infection with WT and hilA mutant strains; (B) infection with WT and Δspi1 mutant strains. Groups of four mice were orally infected with 105 total CFU. Circles represent single infections with WT or mutant strains. Squares represent mixed infections in a 1:1 ratio of WT to mutant. Tissues were recovered 5 days postinoculation, homogenized, and plated on streptomycin, tetracycline, or kanamycin. Each symbol is the logarithm of CFU for each animal tissue. Solid symbols, WT CFU; open symbols, mutant CFU. Horizontal lines represent means of the logarithm of CFU. ns, no significant difference between the mutant and the WT logarithm of CFU (P > 0.05, nonpaired t test for single infections, paired t test for mixed infections). An asterisk indicates that there is a significant difference between the mutant and the WT logarithm of CFU (P < 0.05, nonpaired t test for single infections, paired t test for mixed infections).
Intestinal wall versus content.
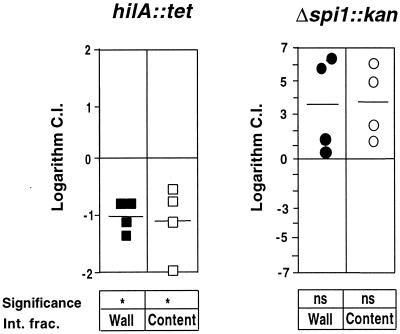
We were interested in knowing if the reduced recovery of the hilA::tet mutant in the intestinal wall (Fig. 1 and 2A) might be due to reduced colonization in the luminal fraction of the intestine. We also wanted to know if the Δspi1::kan mutant was able to colonize the intestinal lumen as well as it could colonize the intestinal tissue. Groups of four animals were infected with a mixture of 105 CFU of WT and a mutant (the hilA::tet or Δspi1::kan mutant) in an approximately 1:1 ratio. A fragment of the small intestine corresponding to the ileum was harvested at 5 days postinoculation. The intestinal contents and wall were homogenized separately and plated. Figure 3 shows the C.I.'s for both the hilA::tet and Δspi1::kan mutants in these samples. The mean C.I. for the hilA::tet mutant is significantly less than 1 for both the intestinal wall and contents (P < 0.05, one-sample t test). The mean C.I. for the Δspi1::kan mutant is not significantly different from 1 for both the intestinal wall and contents (P > 0.05, one-sample t test). There was no significant difference between the C.I.'s for each mutant strain in the intestinal wall versus the intestinal contents (P > 0.05, paired t test).
FIG. 3.
C.I.'s for intestinal wall and contents of the hilA::tet and Δspi1::kan mutant strains from individual animals, 5 days after oral inoculation. Symbols represent the logarithm of C.I., which was determined as described in Materials and Methods. Solid symbols, C.I.'s for the intestinal wall for both mutant strains; open symbols, C.I.'s for the intestinal contents for both mutant strains. Horizontal lines represent means of logarithm of C.I.'s. ns, mean C.I. not significantly different from 1 (log C.I. = 0) (P > 0.05, one-sample t test); an asterisk indicates that the mean C.I. is significantly different from 1 (log C.I. = 0) (P < 0.05, one-sample t test). Int. frac., intestinal fraction.
LD50 and mean day to death.
We obtained LD50 and mean day to death values for the WT, Δspi1::kan, and hilA::tet strains using the Reed-Muench method (39) (Table 1). We infected four groups of four animals with 10, 100, 1,000, and 10,000 CFU and recorded deaths for 3 weeks. Mean day to death was determined as the average time it took mice to die at each dose.
TABLE 1.
LD50 and mean day to death for WT and mutant strainsa
| Strain | Genotype | LD50 (CFU) | Mean day of death |
|---|---|---|---|
| RM12 | WT | <10 | 9 |
| RM40 | hilA::tet | <10 | 14 |
| RM69 | Δspi1::kan | 3.4 × 103 | 13 |
Groups of four animals were infected with 10, 100, 1,000, or 10,000 CFU of each strain and followed for 3 weeks. LD50 was determined by the Reed-Muench formula.
As shown in Table 1, the LD50s for the WT and the hilA::tet strains are similar (<10 CFU). It is possible that the LD50 for the hilA mutant is actually higher than that for the WT strain, but 8 to 10 CFU was the lowest dose tested. In contrast, the LD50 for the Δspi1::kan mutant is at least 1,000-fold higher than the LD50 for the WT strain. This result was surprising in light of our C.I. data showing that the Δspi1::kan mutant is able to infect mice as efficiently as WT at 5 days postinoculation.
The mean day to death at the LD50 for the hilA::tet mutant was higher than that for the WT strain (14 versus 9 days), although this difference was not statistically significant (P > 0.05, generalized Wilcoxon test). The mean day to death at the LD50 for the Δspi1::kan mutant was approximately the same as that for the hilA::tet mutant.
DISCUSSION
We have shown that invasion genes encoded on SPI1 are not necessary for serovar Typhimurium to enter and cross the intestinal wall. In addition, we have shown that invasion genes are important for bacterial colonization of the intestinal lumen. Our results suggest that deletion of a gene(s) on SPI1 suppresses the infection defect of the hilA and invG mutants. As discussed below, we propose that an SPI1-encoded factor stimulates host clearance responses in the intestine. In this model, invasion genes allow salmonellae to colonize the intestine by counteracting this host clearance response, possibly by causing the apoptosis of luminal phagocytes.
After i.g. inoculation, hilA and invG mutants are less able than a WT strain to infect intestinal and systemic tissues. By examining the contents of the intestinal lumen, we found that a mutation in hilA also reduces the recovery of salmonellae from the nontissue fraction of the ileum. Thus, hilA appears to be important for serovar Typhimurium to colonize the extracellular, luminal compartment of the intestine. Therefore, the reduced numbers of the hilA mutant found in the intestinal wall and systemic tissues might be the direct result of the reduced numbers of mutant bacteria present in the intestinal lumen.
Ligated loop assays have shown that invasion genes can be important for Salmonella entry into enterocytes, M cells, and Peyer's patches (9, 26, 36). Unfortunately, because ligated loop assays are so short, certain processes that affect bacterial colonization in vivo, such as bacterial replication and host clearance responses, cannot be assessed in these assays. Therefore, our finding that a hilA mutant exhibits an intestinal lumen colonization defect suggests that further work must be done to determine the relevance of the events observed during ligated loop assays for Salmonella pathogenesis in the murine model.
Several mechanisms might account for the hilA colonization defect. The hilA mutant might not adhere to or replicate at the mucosal surface. Another possibility is that the hilA mutant does not colonize the intestine because it is susceptible to bactericidal host factors such as phagocytes. Invasion genes have been found to be important for the ability of salmonellae to kill phagocytes in vitro (8, 20, 34). As discussed below, we favor the idea that the hilA mutant is defective in colonization because it is killed by phagocytes in the intestine.
Since we observed an infection defect in the hilA::tet mutant 5 days postinoculation, it was surprising that the LD50 for this mutant was indistinguishable from that for the WT strain. However, although the hilA mutant is defective for infection at early times postinoculation, we have observed that after a delay, the hilA mutant can be recovered from systemic tissues at levels similar to those of the WT strain (data not shown). Invasion gene-independent mechanisms may ultimately allow the hilA mutant to enter nonphagocytic cells, infect systemic sites, and kill mice. In vitro studies suggest that salmonellae possess invasion factors in addition to those encoded on SPI1. Stone et al. have described two classes of mutations that reduce Salmonella enterica serovar Enteritidis entry into nonphagocytic cells and that do not map to SPI1 (42). Hong and Miller have reported that SigD, which is normally secreted by the SPI1 type III secretion system, contributes to a serovar Typhimurium invasion mechanism that can operate independently of invA (24). Ligated loop assays with invA and invG mutant strains suggest that under certain circumstances, invasion genes are not necessary for invasion into M cells (10). In addition, serovar Typhimurium might enter enterocytes and M cells via a receptor, in a similar fashion to CFTR-dependent entry of Salmonella enterica serovar Typhi into epithelial cells (37). Alternatively, we favor the idea that bacterial entry into epithelial and M cells may be unnecessary for infection of the intestinal mucosa and systemic sites. In fact, in the absence of invasion genes, salmonellae appear to be shuttled from the intestine to the bloodstream within CD18-expressing phagocytes (44).
Our LD50 results contrast with other findings showing that the i.g. LD50 for a hilA mutant strain is 10- to 50-fold higher than that for the WT strain (1, 36). One possible explanation for the difference between our LD50 results and previous reports is that we used a streptomycin-treated mouse model. While the LD50 for WT salmonellae has been reported to be between 104 and 105 CFU (1, 14, 36), streptomycin treatment reduces the LD50 to 10. We speculate that killing of resident intestinal microflora by streptomycin might enhance phagocyte migration into the intestinal lumen and therefore shuttling of bacteria from the intestine to systemic sites (44). Thus, streptomycin treatment may affect Salmonella infection in two ways—by reducing competition with the intestinal microflora and by increasing the contribution of phagocytes to systemic infection.
The most intriguing results we have obtained are from our infection studies with the Δspi1 mutant. This mutant strain was recovered at a frequency similar to that of the WT strain from all tissues. This result is surprising because the Δspi1 mutant lacks the hilA gene and so was expected to have an infection defect similar to that of the hilA mutant. Our results suggest that deletion of an SPI1 gene(s) in the Δspi1 strain suppresses the hilA infection defect and allows the salmonellae to colonize and infect their host in an SPI1-independent manner. We favor a model in which a hilA-independent SPI1 factor(s) stimulates a host clearance response that the bacteria can overcome by expressing a hilA-dependent factor(s).
It is also surprising that the Δspi1 strain can infect the intestinal mucosa and spread to systemic sites efficiently because, like other invasion mutants, the Δspi1 strain is not able to enter HEp-2 cells in vitro (S. Damrauer and C. Lee, unpublished results). However, as discussed above, invasion gene-independent mechanisms for bacterial penetration of the intestinal epithelium might account for the ability of the Δspi1 mutant to infect intestinal and systemic tissues.
Our result showing an increased LD50 for the Δspi1 mutant was surprising considering the high recovery of this mutant in systemic tissues 5 days postinoculation. The number of Salmonella bacteria present at systemic sites is usually directly correlated with disease and death. One explanation for our results is that the Δspi1 strain might not induce certain host responses and consequently might not damage the host to the same degree as the WT strain. Interestingly, Khan et al. have reported that a waaN mutant strain defective in lipid A acylation is recovered at very high numbers in the spleens and livers of infected mice but does not induce cytokine expression, tissue damage, or death (29). In addition, the waaN mutant strain is ultimately cleared from the spleen and liver of infected animals. The Δspi1 mutant might similarly be cleared at later times postinoculation. In this case, SPI1 genes might be important for persistence of salmonellae at systemic sites.
Model for the contribution of SPI1 genes to virulence.
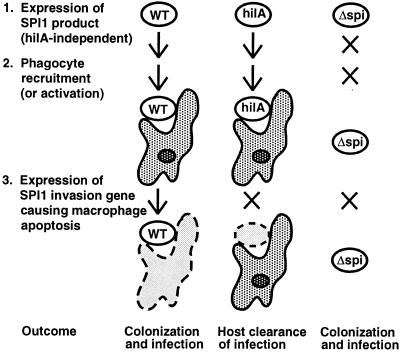
We propose a working model to explain why invasion genes are important for bacterial colonization and persistence in the intestine (Fig. 4). Our model attempts to consider current ideas about the role of invasion genes in protein translocation and stimulation of host cell signal transduction as well as their potential role in altering epithelial cell, neutrophil, and macrophage biology. The infection phenotypes of the WT and invG, hilA, and Δspi1 mutant strains can be explained by speculating that three important events occur when serovar Typhimurium infects its murine host (Fig. 4). Analogous to elicitins and avirulence factors of plant pathogens (6, 47), Salmonella spp. express a hilA-independent SPI1 product that elicits a defense response in its host. The SPI1 elicitin may recruit or activate intestinal phagocytes which can clear the infection. However, expression of SPI1 invasion genes allows the bacteria to overcome the host clearance response, possibly by triggering apoptosis of the phagocytes in the intestinal lumen. Our results suggest that the host response is stimulated by the expression of an SPI1 elicitin gene(s) that is present in the WT, invG, and hilA strains but deleted in the Δspi1 strain. Although the WT strain and the hilA and invG mutants trigger the host defense mechanism, only the WT strain can successfully infect the host because it can express invasion genes and counteract the host response in the third step. In this scenario, the hilA and invG mutants are cleared by the recruited or activated phagocytes. In contrast, even in the absence of hilA, the Δspi1 strain can infect the host because it expresses no SPI1 elicitin and does not trigger host clearance mechanisms.
FIG. 4.
Model for the role of SPI1 in colonization of the intestine by serovar Typhimurium. The left-hand column lists the three events that lead to colonization and infection by the WT strain. The bottom row shows the outcome for each bacterial strain. X's show which processes (arrows) are blocked for the mutant strains. Live bacteria (white) and host cells (shaded) are outlined in solid lines, whereas dashed outlines around bacteria or host cells (pale grey) indicate that they have been killed.
Although it may not seem beneficial to serovar Typhimurium to stimulate a host clearance response, there may be situations in which such an activity promotes pathogenesis, for example, by inducing inflammatory diarrhea and thus enhancing bacterial transmission to new hosts. Future experiments will be aimed at identifying the SPI1 elicitin gene(s) that must be deleted to suppress the hilA infection defect. Such studies may shed light on the mechanisms by which mammalian hosts sense and defend against Salmonella infections.
Our in vivo studies of three SPI1 mutants have revealed new details about the interaction between serovar Typhimurium and its murine host. First, the contribution of SPI1 genes to pathogenesis may be more complex than previously thought. Our results indicate that invasion genes play a role in serovar Typhimurium colonization of the intestinal wall and lumen in mice. Second, our results suggest that an SPI1 gene product induces a host clearance response during Salmonella infection. Previous in vivo analyses had not revealed the possible contributions of SPI1 genes to bacterial colonization of the ileal lumen or to induction and evasion of a host clearance response. Our work emphasizes the importance of using several in vivo approaches and bacterial mutants to study the role of bacterial genes in pathogenesis.
ACKNOWLEDGMENTS
We thank members of the Lee lab, M. Starnbach, D. Schauer, N. Mantis, and J. Mekalanos for critical reading and helpful discussion of the manuscript. We thank S. D'Orazio, K. Klose, D. Schauer, M. Starnbach, and L. Steele for providing R.M. with technical advice. We thank S. Damrauer for technical assistance with the LD50 experiments. We thank Jorge Galán for providing us with strain SB201.
This work was funded by NIH (R01-AI33444). R.M. was supported by a Graduate Prize Fellowship scholarship and funds from the Harvard School of Public Health Division of Biological Sciences-Biological Sciences in Public Health.
REFERENCES
- 1.Ahmer B M, van Reeuwijk J, Watson P R, Wallis T S, Heffron F. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol Microbiol. 1999;31:971–982. doi: 10.1046/j.1365-2958.1999.01244.x. [DOI] [PubMed] [Google Scholar]
- 2.Bajaj V, Hwang C, Lee C A. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol. 1995;18:715–727. doi: 10.1111/j.1365-2958.1995.mmi_18040715.x. [DOI] [PubMed] [Google Scholar]
- 3.Bajaj V, Lucas R L, Hwang C, Lee C A. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol. 1996;22:703–714. doi: 10.1046/j.1365-2958.1996.d01-1718.x. [DOI] [PubMed] [Google Scholar]
- 4.Bäumler A J, Tsolis R M, Valentine P J, Ficht T A, Heffron F. Synergistic effect of mutations in invA and lpfC on the ability of Salmonella typhimurium to cause murine typhoid. Infect Immun. 1997;65:2254–2259. doi: 10.1128/iai.65.6.2254-2259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohnhoff M, Drake B L, Miller C P. Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Proc Soc Exp Biol Med. 1954;86:132–139. doi: 10.3181/00379727-86-21030. [DOI] [PubMed] [Google Scholar]
- 6.Bonas U, Van den Ackerveken G. Gene-for-gene interactions: bacterial avirulence proteins specify plant disease resistance. Curr Opin Microbiol. 1999;2:94–98. doi: 10.1016/s1369-5274(99)80016-2. [DOI] [PubMed] [Google Scholar]
- 7.Carter P B, Collins F M. The route of enteric infection in normal mice. J Exp Med. 1974;139:1189–1203. doi: 10.1084/jem.139.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L M, Kaniga K, Galán J E. Salmonella spp. are cytotoxic for cultured macrophages. Mol Microbiol. 1996;21:1101–1115. doi: 10.1046/j.1365-2958.1996.471410.x. [DOI] [PubMed] [Google Scholar]
- 9.Clark M A, Hirst B H, Jepson M A. Inoculum composition and Salmonella pathogenicity island 1 regulate M-cell invasion and epithelial destruction by Salmonella typhimurium. Infect Immun. 1998;66:724–731. doi: 10.1128/iai.66.2.724-731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark M A, Reed K A, Lodge J, Stephen J, Hirst B H, Jepson M A. Invasion of murine intestinal M cells by Salmonella typhimurium inv mutants severely deficient for invasion of cultured cells. Infect Immun. 1996;64:4363–4368. doi: 10.1128/iai.64.10.4363-4368.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eichelberg K, Galán J E. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI1)-encoded transcriptional activators InvF and HilA. Infect Immun. 1999;67:4099–4105. doi: 10.1128/iai.67.8.4099-4105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu Y, Galán J E. A Salmonella protein antagonizes Rac-1 and Cdc 42 to mediate host-cell recovery after bacterial invasion. Nature. 1999;401:293–297. doi: 10.1038/45829. [DOI] [PubMed] [Google Scholar]
- 13.Fu Y, Galán J E. The Salmonella typhimurium tyrosine phosphatase SptP is translocated into host cells and disrupts the actin cytoskeleton. Mol Microbiol. 1998;27:359–368. doi: 10.1046/j.1365-2958.1998.00684.x. [DOI] [PubMed] [Google Scholar]
- 14.Galan J E, Curtiss R., III Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galyov E E, Wood M W, Rosqvist R, Mullan P B, Watson P R, Hedges S, Wallis T S. A secreted effector protein of Salmonella dublin is translocated into eukaryotic cells and mediates inflammation and fluid secretion in infected ileal mucosa. Mol Microbiol. 1997;25:903–912. doi: 10.1111/j.1365-2958.1997.mmi525.x. [DOI] [PubMed] [Google Scholar]
- 16.Gewirtz A T, Siber A M, Madara J L, McCormick B A. Orchestration of neutrophil movement by intestinal epithelial cells in response to Salmonella typhimurium can be uncoupled from bacterial internalization. Infect Immun. 1999;67:608–617. doi: 10.1128/iai.67.2.608-617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giannella R A, Washington O, Gemski P, Formal S B. Invasion of HeLa cells by Salmonella typhimurium: a model for study of invasiveness of Salmonella. J Infect Dis. 1973;128:69–75. doi: 10.1093/infdis/128.1.69. [DOI] [PubMed] [Google Scholar]
- 18.Ginocchio C, Pace J, Galán J E. Identification and molecular characterization of a Salmonella typhimurium gene involved in triggering the internalization of salmonellae into cultured epithelial cells. Proc Natl Acad Sci USA. 1992;89:5976–5980. doi: 10.1073/pnas.89.13.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hardt W D, Chen L M, Schuebel K E, Bustelo X R, Galán J E. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell. 1998;93:815–826. doi: 10.1016/s0092-8674(00)81442-7. [DOI] [PubMed] [Google Scholar]
- 20.Hersh D, Monack D M, Smith M R, Ghori N, Falkow S, Zychlinsky A. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc Natl Acad Sci USA. 1999;96:2396–2401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hobbie S, Chen L M, Davis R J, Galán J E. Involvement of mitogen-activated protein kinase pathways in the nuclear responses and cytokine production induced by Salmonella typhimurium in cultured intestinal epithelial cells. J Immunol. 1997;159:5550–5559. [PubMed] [Google Scholar]
- 22.Hohmann A W, Schmidt G, Rowley D. Intestinal colonization and virulence of Salmonella in mice. Infect Immun. 1978;22:763–770. doi: 10.1128/iai.22.3.763-770.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoiseth S K, Stocker B A. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 24.Hong K H, Miller V L. Identification of a novel Salmonella invasion locus homologous to Shigella ipgDE. J Bacteriol. 1998;180:1793–1802. doi: 10.1128/jb.180.7.1793-1802.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones B D, Ghori N, Falkow S. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J Exp Med. 1994;180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones M A, Wood M W, Mullan P B, Watson P R, Wallis T S, Galyov E E. Secreted effector proteins of Salmonella dublin act in concert to induce enteritis. Infect Immun. 1998;66:5799–5804. doi: 10.1128/iai.66.12.5799-5804.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaniga K, Uralil J, Bliska J B, Galán J E. A secreted protein tyrosine phosphatase with modular effector domains in the bacterial pathogen Salmonella typhimurium. Mol Microbiol. 1996;21:633–641. doi: 10.1111/j.1365-2958.1996.tb02571.x. [DOI] [PubMed] [Google Scholar]
- 29.Khan S A, Everest P, Servos S, Foxwell N, Zahringer U, Brade H, Rietschel E T, Dougan G, Charles I G, Maskell D J. A lethal role for lipid A in Salmonella infections. Mol Microbiol. 1998;29:571–579. doi: 10.1046/j.1365-2958.1998.00952.x. [DOI] [PubMed] [Google Scholar]
- 30.McCormick B A, Colgan S P, Delp-Archer C, Miller S I, Madara J L. Salmonella typhimurium attachment to human intestinal epithelial monolayers: transcellular signalling to subepithelial neutrophils. J Cell Biol. 1993;123:895–907. doi: 10.1083/jcb.123.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCormick B A, Hofman P M, Kim J, Carnes D K, Miller S I, Madara J L. Surface attachment of Salmonella typhimurium to intestinal epithelia imprints the subepithelial matrix with gradients chemotactic for neutrophils. J Cell Biol. 1995;131:1599–1608. doi: 10.1083/jcb.131.6.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller C P, Bohnhoff M. Changes in the mouse's enteric microflora associated with enhanced susceptibility to Salmonella infection following streptomycin treatment. J Infect Dis. 1963;113:59–66. doi: 10.1093/infdis/113.1.59. [DOI] [PubMed] [Google Scholar]
- 33.Mills D M, Bajaj V, Lee C A. A 40 kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol Microbiol. 1995;15:749–759. doi: 10.1111/j.1365-2958.1995.tb02382.x. [DOI] [PubMed] [Google Scholar]
- 34.Monack D M, Raupach B, Hromockyj A E, Falkow S. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc Natl Acad Sci USA. 1996;93:9833–9838. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nardi R M, Silva M E, Vieira E C, Bambirra E A, Nicoli J R. Intragastric infection of germfree and conventional mice with Salmonella typhimurium. Braz J Med Biol Res. 1989;22:1389–1392. [PubMed] [Google Scholar]
- 36.Penheiter K L, Mathur N, Giles D, Fahlen T, Jones B D. Non-invasive Salmonella typhimurium mutants are avirulent because of an inability to enter and destroy M cells of ileal Peyer's patches. Mol Microbiol. 1997;24:697–709. doi: 10.1046/j.1365-2958.1997.3741745.x. [DOI] [PubMed] [Google Scholar]
- 37.Pier G B, Grout M, Zaidi T, Meluleni G, Mueschenborn S S, Banting G, Ratcliff R, Evans M J, Colledge W H. Salmonella typhi uses CFTR to enter intestinal epithelial cells. Nature. 1998;393:79–82. doi: 10.1038/30006. [DOI] [PubMed] [Google Scholar]
- 38.Que J U, Hentges D J. Effect of streptomycin administration on colonization resistance to Salmonella typhimurium in mice. Infect Immun. 1985;48:169–174. doi: 10.1128/iai.48.1.169-174.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reed L J, Muench H. A simple method of estimating fifty per cent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T, editors. Molecular cloning: a laboratory manual. 2nd ed. Vol. 3. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Schechter L M, Damrauer S M, Lee C A. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol Microbiol. 1999;32:629–642. doi: 10.1046/j.1365-2958.1999.01381.x. [DOI] [PubMed] [Google Scholar]
- 42.Stone B J, García C M, Badger J L, Hassett T, Smith R I, Miller V L. Identification of novel loci affecting entry of Salmonella enteritidis into eukaryotic cells. J Bacteriol. 1992;174:3945–3952. doi: 10.1128/jb.174.12.3945-3952.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeuchi A. Electron microscope studies of experimental Salmonella infection. I. Penetration into the intestinal epithelium by Salmonella typhimurium. Am J Pathol. 1967;50:109–136. [PMC free article] [PubMed] [Google Scholar]
- 44.Vázquez-Torres A, Jones-Carson J, Bäumler A J, Falkow S, Valdivia R, Brown W, Le M, Berggren R, Parks W T, Fang F C. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature. 1999;401:804–808. doi: 10.1038/44593. [DOI] [PubMed] [Google Scholar]
- 45.Witthoft T, Eckmann L, Kim J M, Kagnoff M F. Enteroinvasive bacteria directly activate expression of iNOS and NO production in human colon epithelial cells. Am J Physiol. 1998;275:G564–G571. doi: 10.1152/ajpgi.1998.275.3.G564. [DOI] [PubMed] [Google Scholar]
- 46.Wood M W, Rosqvist R, Mullan P B, Edwards M H, Galyov E E. SopE, a secreted protein of Salmonella dublin, is translocated into the target eukaryotic cell via a sip-dependent mechanism and promotes bacterial entry. Mol Microbiol. 1996;22:327–338. doi: 10.1046/j.1365-2958.1996.00116.x. [DOI] [PubMed] [Google Scholar]
- 47.Yu L M. Elicitins from Phytophthora and basic resistance in tobacco. Proc Natl Acad Sci USA. 1995;92:4088–4094. doi: 10.1073/pnas.92.10.4088. [DOI] [PMC free article] [PubMed] [Google Scholar]