Summary
Cardiac pericytes are a critical yet enigmatic cell type within the coronary microvasculature. Since primary human cardiac pericytes are not readily accessible, we present a protocol to generate them from human induced pluripotent stem cells (hiPSCs). Our protocol involves several steps, including the generation of intermediate cell types such as mid-primitive streak, lateral plate mesoderm, splanchnic mesoderm, septum transversum, and epicardium, before deriving cardiac pericytes. With hiPSC-derived cardiac pericytes, researchers can decipher the mechanisms underlying coronary microvascular dysfunction.
For complete details on the use and execution of this protocol, please refer to Shen et al.1
Subject areas: Developmental Biology, Stem Cells, Cell Differentiation
Graphical abstract

Highlights
-
•
A 7-stage stepwise protocol to generate hiPSC-derived epicardial cells
-
•
Highly efficient differentiation of epicardial cells into cardiac pericytes
-
•
Derived cardiac pericytes exhibit typical morphology, gene markers, and cell functions
Cardiac pericytes are a critical yet enigmatic cell type within the coronary microvasculature. Since primary human cardiac pericytes are not readily accessible, we present a protocol to generate them from human induced pluripotent stem cells (hiPSCs). Our protocol involves several steps, including the generation of intermediate cell types such as mid-primitive streak, lateral plate mesoderm, splanchnic mesoderm, septum transversum, and epicardium, before deriving cardiac pericytes. With hiPSC-derived cardiac pericytes, researchers can decipher the mechanisms underlying coronary microvascular dysfunction.
Before you begin
The protocol below describes the specific steps for using human induced pluripotent stem cells (hiPSCs) to derive cardiac pericytes. However, human embryonic stem cells (hESCs) can also be used. The utilization of either hiPSCs or hESCs must be approved by the institutional Stem Cell Research Oversight (SCRO) Committee. In this protocol, hiPSCs and their derivatives are maintained in a humidified incubator at 37°C with 5% CO2. Cell culture media and matrix-coated culture vessels should be prepared before initiating the differentiation protocol. Cytokines and small molecules should be prepared as stock aliquots and added freshly at desired concentrations to the basal chemically defined medium (CDM) right before use. Culture media should be warmed to 37°C before use. All cell culture procedures should be performed in a Class II biosafety cabinet, using strict aseptic techniques.
Preparation of matrix-coated culture vessels for hiPSC maintenance and differentiation
Timing: 30–60 min
-
1.Prepare Matrigel-coated culture vessels.
-
a.Thaw a bottle of Growth Factor Reduced (GFR) Matrigel (10 mL) overnight at 4°C.
-
b.Prechill 1000 μL filtered tips, 1.5 mL Eppendorf tubes, and a cooling block for Eppendorf tubes at −20°C.
-
c.Put the thawed Matrigel bottle on ice to avoid solidification.
-
d.Transfer the prechilled tip box, Eppendorf tubes, and cooling block together with the Matrigel bottle kept on ice to a biosafety cabinet. Keep everything sterile.
-
e.Place Eppendorf tubes in the cooling block and quickly aliquot 2 mg of Matrigel per Eppendorf tube and store at −80°C for further use.Note: Matrigel concentrations vary from lot to lot. Consult the manufacturer’s datasheet to calculate the precise volume of the aliquot containing 2 mg Matrigel.
-
f.Dilute one aliquot (2 mg) of Matrigel in 25 mL of ice-cold DMEM/F12 medium to make 80 μg/mL Matrigel coating solution.Note: Matrigel coating solution is good for one month when kept at 4°C.
-
g.Add 1 mL or 8 mL of Matrigel coating solution to each well of a 6-well plate or a T75 flask (∼8.7 μg/cm2), respectively.Note: If the coated culture vessels need to be used immediately, incubate them in a humidified incubator at 37°C at least for 30 min. Otherwise, coat them with Matrigel and store at 4°C with sealing parafilm.Note: The coated vessels can be safely stored at 4°C for up to two weeks.
 CRITICAL: Make sure culture vessels are always fully covered by Matrigel coating solution before use. Each batch of Matrigel should be tested to ensure consistent growth performance of hiPSCs.Alternatives: Matrigel is a product extracted from mouse sarcoma and therefore is not chemically defined. In cases where a chemically defined system is required, Vitronectin or other recombinant human matrix proteins can be used as alternatives.
CRITICAL: Make sure culture vessels are always fully covered by Matrigel coating solution before use. Each batch of Matrigel should be tested to ensure consistent growth performance of hiPSCs.Alternatives: Matrigel is a product extracted from mouse sarcoma and therefore is not chemically defined. In cases where a chemically defined system is required, Vitronectin or other recombinant human matrix proteins can be used as alternatives. -
h.When cells are ready to be seeded, aspirate Matrigel coating solution completely from culture vessels and briefly wash them with DPBS.
-
i.Add the desired volumes of cell culture medium to the culture vessels and return them back to the incubator for equilibration.
-
a.
-
2.Prepare gelatin-coated culture vessels.
-
a.Warm up gelatin stock solution (2%) in water bath until a transparent liquid phase is achieved.
-
b.Add 25 mL of gelatin stock solution to 500 mL of tissue culture grade water in a biosafety cabinet and mix thoroughly to make 0.1% gelatin coating solution.Note: Gelatin coating solution is good for two months when kept at 4°C.Alternatives: 0.1% gelatin solution can also be purchased from different vendors to minimize batch-to-batch variations.
-
c.Add 1 mL or 8 mL of gelatin coating solution to each well of a 6-well plate or a T75 flask, respectively.Note: Incubate the coated culture vessels in the hood for at least 15 min before use.Note: Gelatin coating solution should be prewarmed at 37°C before use.
 CRITICAL: Make sure culture vessels are always fully covered by gelatin coating solution before use.
CRITICAL: Make sure culture vessels are always fully covered by gelatin coating solution before use.
-
a.
Preparation of cytokine and small molecule stock aliquots for hiPSC differentiation
Timing: 1–2 h
-
3.Prepare 0.1% bovine serum albumin (BSA) solution.
-
a.Dissolve 500 mg of BSA powder in 5 mL of DPBS to make 10% stock solution and pass through a sterile 0.22 μm syringe filter unit.
-
b.Store the 10% BSA stock solution at −80°C as 100 μL aliquots for up to 24 months.
-
c.Dilute 10% BSA stock aliquots into 0.1% working solution using 9.9 mL of sterile DPBS before use.
-
a.
-
4.Reconstitute and aliquot recombinant growth factors.
-
a.Add 1 mL of 0.1% BSA solution to 100 μg of basic fibroblast growth factor (FGF2) to make 100 μg/mL stock solution.
-
b.Add 1 mL of 0.1% BSA solution to 100 μg of bone morphogenic protein 4 (BMP4) to make 100 μg/mL stock solution.
-
c.Add 1 mL of 0.1% BSA solution to 10 μg of activin A to make 10 μg/mL stock solution.
-
d.Add 1 mL of 0.1% BSA solution to 10 μg of platelet-derived growth factor-BB (PDGF-BB) to make 10 μg/mL stock solution.
-
e.Add 1 mL of 0.1% BSA solution to 100 μg of epidermal growth factor (EGF) to make 100 μg/mL stock solution.
-
f.Add 1.67 mL of IMDM medium and 1.67 mL of Ham’s F12 medium to 100 mg of transferrin to make 30 mg/mL stock solution.
-
a.
Note: Aliquot all the above recombinant growth factors at the volume of 10 μL per vial, except for transferrin at 250 μL per vial, and store at −80°C for up to 12 months.
CRITICAL: Avoid repeated freeze and thaw cycles to prevent recombinant growth factors from degradation. Once aliquots are thawed, keep leftovers at 4°C for up to a week.
-
5.Reconstitute and aliquot small molecules.
-
a.Add 1.993 mL of dimethyl sulfoxide (DMSO) to 10 mg of CHIR99021 to make 10 mM stock solution.
-
b.Add 2.602 mL of DMSO to 10 mg of SB431542 to make 10 mM stock solution.
-
c.Add 11.862 mL of DMSO to 5 mg of A83-01 to make 1 mM stock solution.
-
d.Add 13.177 mL of DMSO to 5 mg of Wnt-C59 to make 1 mM stock solution.
-
e.Add 3.254 mL of DMSO to 10 mg of LY294002 to make 10 mM stock solution.
-
f.Add 16.642 mL of DMSO to 50 mg of retinoic acid (RA) to make 10 mM stock solution.
-
g.Add 3.873 mL of DMSO to 10 mg of thalidomide to make 1 mM stock solution.
-
h.Add 3.123 mL of sterile culture grade water to 10 mg of Y27632 dihydrochloride to make 10 mM stock solution.
-
i.Add 4 mL of sterile culture grade water to 100 mg of L-ascorbic acid 2-phosphate sesquimagnesium salt hydrate (AA) to make 25 mg/mL stock solution.
-
a.
Note: Aliquot all the above small molecule stocks at the volume of 25 μL per vial and store at −20°C for up to 12 months.
CRITICAL: Protect RA and AA stock aliquots from light.
Maintenance of hiPSCs
Timing: 1 week
-
6.Prepare complete StemMACS iPS-Brew XF medium.
-
a.Thaw StemMACS iPS-Brew XF, 50× Supplement at 4°C prior to use.
-
b.Add 10 mL of StemMACS iPS-Brew XF, 50× Supplement to 500 mL of StemMACS iPS-Brew XF, Basal Medium.
-
c.Mix thoroughly.
-
a.
Note: Use the complete medium within 2 weeks when stored at 4°C. For longer storage, prepare 50 mL aliquots and store at −20°C for up to 2 months. Thaw aliquots of complete medium overnight at 4°C. Once thawed, keep aliquots at 4°C and use within 2 weeks.
CRITICAL: Avoid warming up the complete Brew medium to 37°C as this can potentially impact its quality. Instead, bring the cold complete Brew medium to 25°C by allowing the bottle to sit on the bench for at least 30 min.
Alternatives: hiPSC maintenance media from other vendors, such Essential 8 or mTeSR, can be used as alternatives to StemMACS iPS-Brew XF medium.
-
7.
Thaw a cryopreserved vial of hiPSCs and transfer the cells to a Matrigel-coated 6-well plate containing complete StemMACS iPS-Brew XF (Brew) medium supplemented with 10 μM of Y27632 dihydrochloride (a ROCK inhibitor).
Note: For a detailed protocol on thawing hiPSCs, please refer to https://www.stemcell.com/cryopreservation-and-thawing-of-es-ips-cells.html.
-
8.
Add fresh Brew medium the next day to remove the ROCK inhibitor.
-
9.
Refresh Brew medium every other day until the confluency of hiPSCs reaches ∼85%.
Note: We do not recommend using antibiotics during hiPSC maintenance.
CRITICAL: It is important to avoid overgrowth of hiPSCs, as it may lead to karyotype abnormality.
-
10.
Rinse hiPSCs with DPBS (without Ca2+ and Mg2+) followed by cell dissociation using 0.5 mL of Gentle Cell Dissociation Reagent for 5–7 min at 37°C.
-
11.
Terminate cell dissociation when clear boundaries can be seen between individual hiPSCs.
Alternatives: We routinely use Gentle Cell Dissociation Reagent from STEMCELL Technologies for hiPSC dissociation to minimize batch-to-batch variations. However, in-house made EDTA dissociation solution also works well. The recipe is as follows: add 500 μL of 0.5 M EDTA (pH 8.0) and 0.9 g of NaCl into 500 mL of DPBS (without Ca2+ and Mg2+). Sterilize the EDTA solution by filtration through a 0.22-μm pore size Stericup system and store at 4°C–25°C for up to 12 months.
Note: Optimal dissociation time may vary among different hiPSC lines. In most cases, hiPSCs should still loosely attach to culture plates. If a majority of the hiPSCs detach from the culture plates, collect the cell suspension and centrifuge it at 300× g for 3 min at 16°C–25°C. Decant the supernatant and resuspend the cell pellet using Brew medium, adding 10 μM ROCK inhibitor.
-
12.
Aspirate the Gentle Cell Dissociation Reagent and add 1 mL of Brew medium plus 10 μM ROCK inhibitor to each well.
-
13.
Pipette cells up and down for about 4–5 times using a P1000 pipette tip and add an appropriate volume of cell suspension (split ratio normally ranges from 1:6 to 1:12) to a new well of Matrigel-coated plates.
-
14.
Move the plate in a left-to-right and back-to-forth motion to distribute hiPSCs evenly over the surface of each well.
-
15.
Return the plate to the incubator.
-
16.
Repeat the procedures starting from step 9.
Note: For better cell survival and differentiation efficiency, it is recommended to passage hiPSCs at least twice before initiating epicardial cell differentiation if they have been recently thawed from cryovials.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-α-SMA (1:250) | MilliporeSigma | Cat#A5228 |
| Anti-Calponin (1:800) | MilliporeSigma | Cat#C2687 |
| Anti-Fibroblast antibody, clone TE-7 (1:100) | Novus Biologicals | Cat#NBP2-50082 |
| Anti-GATA4 (1:500) | Santa Cruz Biotechnology | Cat#sc-25301 |
| Anti-NG2 (1:100) | Millipore | Cat#MAB2029 |
| Anti-PDGFRβ (1:250) | R&D Systems | Cat#AF385 |
| Anti-Smooth muscle myosin heavy chain (1:200) | Abcam | Cat#Ab53219 |
| Anti-TBX18 (1:200) | Abcam | Cat#Ab115262 |
| Anti-TCF21 (1:250) | MilliporeSigma | Cat#HPA013189 |
| Anti-TBX20 (1:250) | MilliporeSigma | Cat# HPA008192 |
| Anti-WT1 (1:500) | Abcam | Cat#ab89901 |
| Anti-ZO1 (1:250) | Thermo Fisher Scientific | Cat#33-9100 |
| Donkey anti-Mouse IgG, Alexa Fluor Plus 594 (1:500) | Thermo Fisher Scientific | Cat#A32744 |
| Donkey anti-Rabbit IgG, Alexa Fluor Plus 594 (1:500) | Thermo Fisher Scientific | Cat#A32754 |
| Donkey anti-Mouse IgG, Alexa Fluor Plus 488 (1:500) | Thermo Fisher Scientific | Cat#A32766 |
| Donkey anti-Rabbit IgG, Alexa Fluor Plus 488 (1:500) | Thermo Fisher Scientific | Cat#A32790 |
| Donkey anti-Goat IgG, Alexa Fluor Plus 647 (1:500) | Thermo Fisher Scientific | Cat#A32849 |
| Chemicals, peptides, and recombinant proteins | ||
| 4′,6-Diamidino-2-phenylindole (DAPI) | Thermo Fisher Scientific | Cat#D1306 |
| A83-01 (1 mM) | Tocris | Cat#2939 |
| Advanced DMEM/F12 | Gibco | Cat#12634010 |
| Bambanker | Wako Chemicals USA | Cat#30214681 |
| Bovine serum albumin (BSA) | MilliporeSigma | Cat#A7906 |
| Culture-grade water | Thermo Fisher Scientific | Cat#15230162 |
| CHIR99021 (10 mM) | Selleck Chemicals | Cat#S2924 |
| Chemically defined lipid concentrate | Gibco | Cat#11905-031 |
| Dimethyl sulfoxide (DMSO) | MilliporeSigma | Cat#D2650 |
| DMEM/F-12, HEPES | Gibco | Cat#11330057 |
| DPBS without calcium and magnesium | Gibco | Cat#14190250 |
| Gelatin solution (2%) | MilliporeSigma | Cat#G1393 |
| Gentle Cell Dissociation Reagent | STEMCELL Technologies | Cat#07174 |
| Glutamax™-I | Gibco | Cat#35050-061 |
| Ham’s F12 Nutrient Mix | Gibco | Cat#11765-054 |
| IMDM | Gibco | Cat#12440-053 |
| LY294002 (10 mM) | Selleck Chemicals | Cat#S1105 |
| L-Ascorbic acid 2-phosphate sesquimagnesium salt hydrate (25 mg/mL) | MilliporeSigma | Cat#A8960 |
| Matrigel Growth Factor Reduced (GFR) Basement Membrane Matrix | Corning | Cat#356231 |
| Monothioglycerol | MilliporeSigma | Cat#M6145 |
| Paraformaldehyde (20%) | Electron Microscopy Sciences | Cat#15713-S |
| Pericyte medium | ScienCell | Cat#1201 |
| Polyvinyl alcohol (PVA, 100 mg/mL) | MilliporeSigma | Cat#P8136 |
| Recombinant Human/Murine/Rat Activin A (10 μg/mL) | PeproTech | Cat#120-14E |
| Recombinant Human BMP4 (100 μg/mL) | PeproTech | Cat#120-05ET |
| Recombinant Human EGF (100 μg/mL) | PeproTech | Cat#AF-100-15 |
| Recombinant Human FGF2 (100 μg/mL) | PeproTech | Cat# |
| Recombinant Human PDGF-BB (10 μg/mL) | PeproTech | Cat#100-14B |
| Retinoic acid (10 mM) | MilliporeSigma | Cat#R2625 |
| SB431542 (10 mM) | Selleck Chemicals | Cat#S1067 |
| StemMACS™ iPS-Brew XF | Miltenyi Biotech | Cat#130-104-368 |
| Thalidomide (1 mM) | Selleck Chemicals | Cat#S1193 |
| Transferrin (30 mg/mL) | MilliporeSigma | Cat#T8158 |
| TrypLE™ Select Enzyme (1×) | Gibco | Cat#12605036 |
| TrypLE™ Select Enzyme (10×) | Gibco | Cat#A1217701 |
| Wnt-C59 (1 mM) | Selleck Chemicals | Cat#S7037 |
| Y27632 dihydrochloride (10 mM) | Selleck Chemicals | Cat#S1049 |
| Experimental models: Cell lines | ||
| Human induced pluripotent stem cells (hiPSCs) | Stanford Cardiovascular Institute Biobank (SCVI) | SCVI15, male; SCVI100, female; SCVI480, female; SCVI1098, female |
Materials and equipment
Chemically defined medium (CDM)
| Reagent | Final concentration | Amount |
|---|---|---|
| Ham’s F-12 Nutrient Mix | 50% | 240 mL |
| IMDM | 50% | 240 mL |
| Chemically defined lipid concentrate | 1% | 5 mL |
| Glutamax™-I (200 mM) | 2 mM | 5 mL |
| PVA (100 mg/mL)∗ | 1 mg/mL | 5 mL |
| Transferrin (30 mg/mL) | 15 μg/mL | 250 μL |
| Monothioglycerol | 450 μM | 20 μL |
Note:∗To prepare 100 mg/mL PVA solution, weigh 10 g of PVA and add the powder slowly into a sterile flask containing 100 mL of culture grade water with constant gentle agitation until the PVA is completely dissolved. The solution can be stored at 4°C for 2 months.
Note: Filter the CDM using a Stericup filtration system with a pore size of 0.22 μm and store it at 4°C for up to 2 months.
Mid-primitive streak (MPS) induction medium
| Reagent | Final concentration | Amount |
|---|---|---|
| CDM | N/A | 10 mL |
| Activin A (10 μg/mL) | 10 ng/mL | 10 μL |
| BMP4 (100 μg/mL) | 50 ng/mL | 5 μL |
| FGF2 (100 μg/mL) | 20 ng/mL | 2 μL |
| CHIR99021 (10 mM) | 6 μM | 6 μL |
| LY294002 (1 mM) | 2 μM | 2 μL |
Lateral plate mesoderm (LPM) induction medium
| Reagent | Final concentration | Amount |
|---|---|---|
| CDM | N/A | 10 mL |
| BMP4 (100 μg/mL) | 30 ng/mL | 3 μL |
| A83-01 (1 mM) | 1 μM | 10 μL |
| Wnt-C59 (1 mM) | 1 μM | 10 μL |
Splanchnic mesoderm (SM) induction medium
| Reagent | Final concentration | Amount |
|---|---|---|
| CDM | N/A | 10 mL |
| BMP4 (100 μg/mL) | 30 ng/mL | 3 μL |
| FGF2 (100 μg/mL) | 20 ng/mL | 2 μL |
| A83-01 (1 mM) | 1 μM | 10 μL |
| Wnt-C59 (1 mM) | 1 μM | 10 μL |
| Retinoic acid (10 mM) | 2 μM | 2 μL |
Septum transversum (ST) induction medium
| Reagent | Final concentration | Amount |
|---|---|---|
| CDM | N/A | 10 mL |
| BMP4 (100 μg/mL) | 40 ng/mL | 4 μL |
| Retinoic acid (10 mM) | 2 μM | 2 μL |
Note: All the above intermediate cell type induction media can be stored at 4°C for up to a week. However, we encourage to prepare them fresh. Warm up the media to 37°C before use.
LaSR basal medium
| Reagent | Final concentration | Amount |
|---|---|---|
| Advanced DMEM/F-12 medium | N/A | 500 mL |
| Glutamax (200 mM) | 2.5 mM | 6.25 mL |
| L-Ascorbic acid (25 mg/mL) | 100 μg/mL | 2 mL |
Note: LaSR basal media should be protected from light and is stable for up to 2 months at 4°C.
Proepicardial organ (PEO) specification medium
| Reagent | Final concentration | Amount |
|---|---|---|
| LaSR basal medium | N/A | 50 mL |
| SB431542 (10 mM) | 10 μM | 50 μL |
| Retinoic acid (10 mM) | 2 μM | 10 μL |
Note: We do not recommend storing EPI specification medium. Therefore, prepare it right before use. A83-01 at the final concentration of 1 μM can be used as an alternate to 10 μM SB431542.
Epicardial cell (EPI) maintenance medium
| Reagent | Final concentration | Amount |
|---|---|---|
| LaSR basal medium | N/A | 100 mL |
| SB431542 (10 mM) | 10 μM | 100 μL |
Note: The EPI maintenance medium can be stored at 4°C in dark for 2 weeks. A83-01 at the final concentration of 1 μM can be used as an alternate to 10 μM SB431542.
Cardiac pericyte induction medium, 100 mL
| Reagent | Final concentration | Amount |
|---|---|---|
| ScienCell Pericyte medium | N/A | 100 mL |
| PDGF-BB (10 μg/mL) | 10 ng/mL | 100 μL |
| EGF (100 μg/mL) | 10 ng/mL | 10 μL |
| Thalidomide (1 mM) | 1 μM | 100 μL |
Note: We have observed that thalidomide can significantly facilitate the differentiation and proliferation of cardiac pericytes without causing any noticeable abnormalities in gene expression or cell functions. However, if users are concerned that thalidomide may interfere with their targeted pathways of interest, they may choose to omit it from the cardiac pericyte induction medium recipe. In such cases, it may take up to three weeks to obtain fully differentiated cardiac pericytes from EPI cells.
CRITICAL: Thalidomide is a well-known teratogen, and its exposure should be avoided during pregnancy.
Step-by-step method details
Differentiation of human induced pluripotent stem cells into epicardial cells
Timing: 2 weeks
Steps 1–12 describe the process of differentiating hiPSCs into epicardial cells (EPI) through a stepwise approach that mimics the developmental trajectory of their primary counterpart (Figure 1). The purity of intermediate cell types at each stage, including mid-primitive streak (MPS), lateral plate mesoderm (LPM), splanchnic mesoderm (SM), septum transversum (ST), and EPI, can be confirmed by immunofluorescence staining of cell type-specific markers.2 The induction media for intermediate cell types at different stages should be prepared according to the tables in the “materials and equipment” section.
Note: While all stage-specific induction media can be stored at 4°C for up to a week, it is advisable to prepare them fresh for best results. It is, therefore, recommended to make only the required amount of induction media for each batch of differentiation. Make sure to warm up induction medium to 37°C in a water bath before use.
CRITICAL: To ensure high differentiation efficiency, verify the quality of hiPSCs prior to differentiation through examination of cell morphology and key pluripotency markers such as OCT4, NANOG, SSEA4, and TRA-1-60 using immunofluorescence staining or flow cytometry analysis. Additionally, hiPSCs should have a normal karyotype, be free of differentiated cells, and test negative for mycoplasma.
-
1.On day 0, when hiPSCs reach 85% confluency, remove Brew medium from one well of a 6-well plate.
-
a.Wash cells with 1 mL of DPBS.
-
b.Add 0.5 mL of 1× TrypLE™ Select Enzyme to the well and incubate for 4–5 min at 37°C.
-
c.When majority of hiPSCs come off the plate, add 0.5 mL of Brew medium followed by gentle pipetting for 5 times.
-
d.Transfer the hiPSC suspension to a 5 mL Eppendorf tube and centrifuge at 300× g for 3 min at 16°C–25°C.
-
e.Resuspend the hiPSC pellet in 1 mL of Brew medium containing 10 μM ROCK inhibitor and count cells.
-
f.Seed hiPSCs at 2–3 densities, with one density per well, to ensure at least one well has an optimal density of cells (Figure 2i) for the differentiation process to continue.
-
a.
Note: As the MPS induction medium can lead to significant cell death, we normally prepare 2 wells by adding 1.5 mL of the complete Bew medium containing 10 μM ROCK inhibitor to each well of a Matrigel-coated 6-well plate, and then seed 2×104 cells/cm2 in one well and 2.5×104 cells/cm2 in the other well as single cells.
Note: While either 2×104 cells/cm2 or 2.5×104 cells/cm2 worked well for most hiPSC lines, some hiPSC lines may require a lower seeding density (e.g., 1.5×104 cells/cm2) to avoid over-confluency by day 2, while others may need a higher density (e.g., 3×104 cells/cm2) if 2.5×104 cells/cm2 do not result in sufficient viable MPS cells. For a new hiPSC line that has never been tested, the users may seed hiPSCs in 3 densities (e.g., 1.5×104 cells/cm2, 2×104 cells/cm2, and 2.5 × 104 cells/cm2) to start.
Note: The users should also take the proliferation rate of hiPSCs into consideration when determine the optimal seeding density. It is recommended to aim for less than 30% confluency of LPM cells by the end of day 2 to avoid 100% confluency of ST cells on day 5. It is also important to note that since EPI cells need to be seeded at a very low density on day 8 due to their high proliferation rate, choosing one well of ∼30% confluent cells starting on day 3 for continued differentiation will be sufficient. It will significantly reduce the reagent cost of EPI differentiation. troubleshooting 1.
Note: While we have optimized the seeding densities of hiPSCs for EPI cell differentiation in a 6-well format, users may choose to adapt this protocol to a 12-well format to further reduce differentiation reagent costs. However, the optimal seeding densities should be tested empirically.
CRITICAL: To ensure successful differentiation, the viability of hiPSCs should be greater than 90% before cell seeding. It is also important to distribute hiPSCs within the well evenly by gently moving the plate in a left-to-right and back-to-forth motion. Then allow the cells to rest in the hood for 10 min before returning to the incubator.
CRITICAL: Make sure to perform the following differentiation procedures at the same time each day to ensure that the cells are exposed to stage-specific induction medium for 22–26 h on each day.
-
2.
On day 1, wash hiPSCs with DPBS and add 1.5 mL of prewarmed MPS induction medium per well (Figure 2ii). troubleshooting 2.
-
3.
On day 2, wash MPS cells with DPBS and add 1.5 mL of prewarmed LPM induction medium per well (Figure 2iii). troubleshooting 3.
-
4.
On day 3, evaluate the cell confluency of all wells, and choose the well that has roughly 30% confluent LPM cells to continue the differentiation process. Wash LPM cells with DPBS and add 1.5 mL of pre-warmed SM induction medium to the well.
-
5.
On day 4, refresh 1.5 mL of pre-warmed SM induction medium (Figure 2iv).
-
6.
On day 5, wash SM cells with DPBS and add 2 mL of pre-warmed ST induction medium.
-
7.
On days 6 and 7, refresh 2 mL of pre-warmed ST induction medium per day.
Note: ST cells normally become sub-confluent (∼85%) on day 5. However, for some hiPSC lines, ST cells may reach only 50%–70% confluency on day 8, which does not compromise the differentiation efficiency.
-
8.On day 8, wash ST cells (Figure 2v) with DPBS and incubate with 1 mL of 10× TrypLE™ Select Enzyme for 5–7 min at 37°C.
-
a.When majority of ST cells come off the plate, add 1mL of LaSR basal medium followed by gentle pipetting for 10 times.
-
b.Transfer the ST cell suspension to a 5 mL Eppendorf tube and centrifuge at 300× g for 5 min at 16°C–25°C.
-
c.Resuspend the ST cell pellet in 1 mL of LaSR basal medium containing 10 μM ROCK inhibitor and 10 μM of SB431542.
-
d.Count ST cells and seed at 2.5 × 103 cells/cm2 as single cells in Matrigel-coated T75 flasks.
-
e.Add 10 mL of LaSR basal medium containing 10 μM ROCK inhibitor to each flask.
-
f.Evenly distribute ST cells by gently moving the flasks in a left-to-right and back-to-forth motion.
-
g.Re-pellet the remaining ST cells and cryopreserve with Bambanker freezing medium as back up.
-
a.
CRITICAL: Because EPI cells are susceptible to epithelial-to-mesenchymal transition (EMT),3 particularly when forming clusters, it is essential to dissociate ST cells into single cells and seed them at a low density before initiating PEO and EPI differentiation.
Note: ST cells can be cryopreserved and subsequently used as a starting point for continuing EPI differentiation upon thawing. As ST cells can produce a large amount of tight junction proteins, it is more efficient to obtain single cells using 10× TrypLE™ Select Enzyme rather than 1× TrypLE™ Select Enzyme.
Pause point: hiPSC-derived ST cells can be cryopreserved indefinitely in liquid nitrogen tank before being further differentiated into PEO and EPI cells.
Note: We recommend using LaSR basal medium containing ROCK inhibitor to replate ST cells, as we have observed significant cell death in some hiPSC lines when using the EPO specification medium for ST cell replating.
-
9.
On day 9, wash ST cells with DPBS and add 12 mL of PEO specification medium per flask. Leave the cells undisturbed for 2 days (Figure 2vi). troubleshooting 4 and 5.
-
10.
On day 11, wash PEO cells with DPBS and add 12 mL of EPI maintenance medium per flask. Change the medium every other day until EPI cells reach approximately 90% confluency (Figure 2vii). troubleshooting 6.
Note: Due to their fast proliferation rate and the epithelial cell-like morphology, EPI cells typically reach sub-confluency on days 10–12.
CRITICAL: It is important to avoid overgrowing EPI cells, as they can easily undergo EMT when they reach full confluency.
-
11.Once EPI cells become sub-confluent, wash flasks with DPBS.
-
a.Add 4 mL of 10× TrypLE™ Select Enzyme per flask and incubate for 7–10 min until all cells come off the flasks.
-
b.Add 4 mL of LaSR basal medium to each flask and collect EPI cell suspension to a 15 mL Falcon tube for centrifugation at 300× g for 5 min at 16°C–25°C.
-
c.Decant the supernatant by careful aspiration and resuspend the cell pellet using 10 mL of LaSR maintenance medium containing 10 μM of ROCK inhibitor per flask.
-
d.Seed EPI cells at 5×103 cells/cm2. EPI cells at this stage are labeled as passage 1 (Figure 2viii).
-
a.
CRITICAL: It is crucial to dissociate EPI cells using 10× TrypLE™ Select Enzyme for a sufficient amount of time to obtain single cells. Otherwise, the EPI cells may be difficult to spin down and may appear as a fluffy clump floating in the Falcon tube.
-
12.On the following day, aspirate the spent medium.
-
a.Add 12 mL of fresh EPI maintenance medium per flask.
-
b.Refresh the medium every other day until EPI cells reach approximately 90% confluency.
-
c.Repeat step 11 for further cell passaging or cell cryopreservation. Otherwise, proceed with cardiac pericyte differentiation.
-
a.
Note: a detailed protocol on hiPSC cryopreservation and thawing https://www.stemcell.com/cryopreservation-and-thawing-of-es-ips-cells.html is also applicable to EPI cells.
Pause point: hiPSC-derived EPI cells can be cryopreserved indefinitely in liquid nitrogen tank.
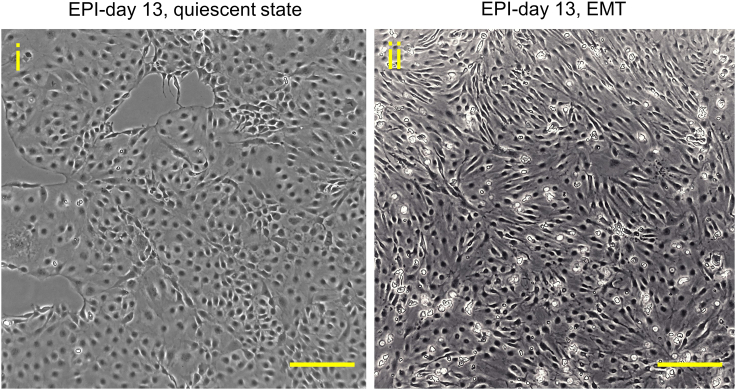
Note: In our experience, EPI cells can be expanded for up to 25 passages without losing their cell type markers or epithelial cell-like morphology. Since serial passaging improves the maturity of EPI cells, it is recommended to initiate cardiac pericyte differentiation using EPI cells that have undergone at least three passages. It is crucial to prevent overgrowth of EPI cells during cell maintenance, as they may undergo EMT (Figure 3) and compromise the purity of the resulting cardiac pericytes.
Note: Before initiating cardiac pericyte differentiation, it is recommended to perform immunofluorescence staining of EPI markers. To do this, seed 6×103 EPI cells/cm2 in 12-well plates and grow them to sub-confluency in EPI maintenance medium. Fix the EPI cells using 4% paraformaldehyde and proceed with immunostaining (dilutions for primary and secondary antibodies shown in the key resources table) for ZO1, WT1, TBX18, and TCF21 (Figure 4).
CRITICAL: PFA is hazardous. Perform the above staining step in a fume hood and properly dispose of waste.
Figure 1.
A schematic showing a stepwise protocol to generate epicardial cells from human induced pluripotent stem cells (hiPSCs)
To generate intermediate mesodermal cell types and EPI cells, stage-specific inhibition and activation of morphogens were achieved using various combinations of cytokines and small molecules. CDM, chemically defined medium; MPS, mid-primitive streak; LPM, lateral plate mesoderm; SM, splanchnic mesoderm; ST, septum transversum; PEO, pre-epicardial organ; EPI, epicardial cell.
Figure 2.
Bright-field phase contrast images showing differentiation of hiPSCs into mesodermal cell types and epicardial cells at different developmental stages
Representative phase contrast images showing the chronological morphology change from day 0 (hiPSCs) to day 12 (epicardial cells) under the current differentiation protocol. Images on days 0, 9, and 12 were captured within 12–18 h of cell dissociation and in the presence of 10 μM of ROCK inhibitor. Scale bars, 500 μm.
Figure 3.
Bright-field phase contrast images showing epicardial cells at a quiescent state and undergoing epithelial-to-mesenchymal transition (EMT)
Representative phase contrast images showing quiescent (i) and EMT process-activated (ii) epicardial cells from two independent batches. Scale bars, 500 μm.
Figure 4.
Immunofluorescent images showing high expression levels of epicardial cell-specific markers
Immunostaining of epicardial markers WT1, TCF21, TBX18 and epithelial maker ZO1 was performed on high quality (A) and low quality (B) epicardial cells harvested on day 13 of differentiation. DAPI staining was included in merged images. Note that a large portion of epicardial cells in (B) are undergoing epithelial-to-mesenchymal transition. Scale bars, 100 μm.
Differentiation of epicardial cells into cardiac pericytes
Timing: 12 days
Steps 13–17 describe the process of differentiating EPI cells into nascent cardiac pericytes.
Day 0: Seed EPI cells for cardiac pericyte differentiation
-
13.
Dissociate EPI cells into single cells using 10× TrypLE™ Select Enzyme and seed at the density of 2×104 cells/cm2 in one Matrigel-coated 6-well plate using 2 mL of EPI maintenance medium containing 10 μM of ROCK inhibitor (Figure 5).
Note: EPI cells seeded at this density typically form a complete layer the following day. If not, grow the EPI cells in EPI maintenance medium until they reach 100% confluency.
Figure 5.
A schematic showing the generation of cardiac pericyte from epicardial cells
Seeded epicardial cells need to form a monolayer before cardiac pericyte induction medium is added. Pericyte medium is refreshed every other day. One 6-well plate of early cardiac pericytes is recommended to be replated in two T75 flasks at day 7. Fully differentiated cardiac pericytes are recommended to be replated in the regular ScienCell Pericyte Medium at the density of 1×104 cells/cm2. Typically, hiPSC-derived cardiac pericytes show a senescent phenotype by days 45–50 of differentiation and should not be used for biochemical assays. PDGF-BB, platelet-derived growth factor-BB; EGF, epidermal growth factor.
Days 1–6: Refresh cardiac pericyte induction medium every other day
-
14.
On day 1, wash EPI cells with DPBS and add 2 mL of cardiac pericyte induction medium per well. Leave cell undisturbed for 2 days (Figure 5).
-
15.
On days 3 and 5, refresh 2 mL of cardiac pericyte induction medium per well (Figure 5).
Note: In our experience, it is crucial to keep early cardiac pericytes at a very high confluency, as it can facilitate a complete EMT and cardiac pericyte differentiation via a paracrine effect.
Day 7: Replate early cardiac pericytes to gelatin-coated flasks
-
16.On day 7, wash cells with DPBS and add 1 mL of 10× TrypLE™ Select Enzyme to each well and incubate for 5 min at 37°C.
-
a.Add 0.5 mL of pericyte medium to each well and pipette up and down for 4–5 times.
-
b.Spin cells down at 300 ×g for 5 min at 16°C–25°C and resuspend them in 1 mL of cardiac pericyte induction medium.
-
c.Split cells into 2 gelatin-coated T75 flasks using 12 mL of cardiac pericyte induction medium per flask (Figure 5). troubleshooting 7.
-
a.
Days 8–12: Refresh cardiac pericyte induction medium every other day
-
17.
On days 9 and 11, refresh 12 mL of cardiac pericyte induction medium per flask (Figure 5).
Note: During the initial 6 days of differentiation, early cardiac pericytes display a uniform morphology (Figure 6i). However, upon replating, cells exhibit a heterogeneous morphology (Figure 6ii). Nevertheless, after an additional week of maintenance in ScienCell pericyte medium, the cells can regain a relatively uniform morphology (Figure 6iii). When maintained in a low serum pericyte medium (with serum concentration reduced from 2% to 0.1%) after becoming sub-confluent, the cells can form a more uniform and aligned morphology (Figure 6iv). Flow cytometry and quantitative real-time PCR data from our previous study suggest that by day 12 of differentiation, epicardial cell-derived cardiac pericytes already express a broad range of cell type-selective markers, including PDGFRβ, NG2, CD13, and CD146.1 This indicates that the cardiac pericytes are developing and maturing as expected.
Pause point: hiPSC-derived nascent cardiac pericytes can be cryopreserved indefinitely in liquid nitrogen tank for future expansion.
Figure 6.
Bright-field phase contrast images showing the morphology changes of cardiac pericytes at differentiation and maintenance stages
At day 6 of differentiation, cardiac pericytes display a uniform morphology (i), but become more heterogeneous after replating (ii). During cardiac pericyte maintenance, a predominant stellate-shaped morphology is achieved, as shown on day 21 of differentiation (iii). When day 21 cardiac pericytes were cultured in a low serum medium for 2 days, they adopted an elongated, well-aligned, spindle-shaped morphology (iv). Scale bars, 200 μm.
Cardiac pericyte expansion and maturation
Timing: Variable durations (up to 32 days) depending on experimental plans
Steps 18–20 describe the process of prompting nascent cardiac pericytes to become mature while expanding the cell population for downstream functional assays.
Days 13–45: Refresh ScienCell pericyte medium every other day; replate or cryopreserve sub-confluent cardiac pericytes
-
18.From day 13 onwards, refresh 12 mL of ScienCell pericyte medium per flask every other day until cardiac pericytes become fully confluent.
-
a.To dissociate the cardiac pericytes, treat the cells with 1× TrypLE™ Select Enzyme for 3 min at 37°C.
-
b.Replate the cells in gelatin-coated 6-well plates at a density of 1 × 104 cells/cm2 using ScienCell pericyte medium.
-
c.Refresh the medium every other day.
-
d.When the cells reach 95% confluency, dissociate them again using the same procedure (Figure 5). troubleshooting 8.
-
a.
-
19.Cryopreserve hiPSC-derived cardiac pericytes for future use.
-
a.We recommend collecting cells at day 18–21 of differentiation and suspend them in Bambanker freezing medium at a density of 1 × 106 cells per vial.
-
b.Put cryovials in a Corning CoolCell™ LX Cell freezing container and store at −80°C overnight before transferring them into liquid nitrogen.
-
c.The remaining cardiac pericytes can be further expanded or used for molecular and cellular assays as needed.
-
d.Refer to step 12 to get details on how to properly thaw hiPSC-derived cardiac pericytes for future use.
-
a.
-
20.
For a quality control purpose, check the expression levels of cardiac pericyte markers (e.g., PDGFRβ, NG2, CD13, and CD146), smooth muscle cell markers (e.g., smooth muscle-myosin heavy chain and calponin), and cardiac fibroblast markers (e.g., anti-fibroblast antibody, clone TE-7) using either flow cytometry1 or immunofluorescence staining (Figure 7).
Figure 7.
High expression levels of cardiac pericyte markers but no smooth muscle cell or fibroblast markers in high quality hiPSC-derived cardiac pericytes
(A) Cardiac pericytes are positive for PDGFRβ and α-SMA.
(B) Cardiac pericytes are positive for NG2 and negative for an anti-fibroblast antibody (TE-7).
(C) Cardiac pericytes are partially positive for calponin and negative for a mature smooth muscle cell marker SM-MHC.
(D) Cells fail to acquire a cardiac pericyte fate are weakly positive for α-SMA and are negative for PDGFRβ.
(E) Cardiac fibroblasts derived from epicardial cells are positive for TE-7 and negative for NG2.
(F) Cardiac smooth muscle cells derived from epicardial cells are positive for both SM-MHC and calponin. hiPSC, human induced pluripotent stem cell; SMA, smooth muscle actin; NG2, Neuron-glial antigen 2; PDGFR, platelet-derived growth factor; SM-MHC, smooth muscle myosin heavy chain. Scale bars, 50 μm.
Expected outcomes
This protocol outlies a robust method for generating bona fide EPI cells, followed by cardiac pericytes that have potential utility in modeling genetic variants or environmental risk factor-induced coronary microvascular dysfunction. The first stage of differentiation toward EPI cells is accomplished using a stepwise protocol that recapitulates the developmental trajectory of this cell type in vivo. This approach leads to an almost 100% differentiation efficiency of EPI cells, which have epigenetic and transcriptional characteristics similar to their primary counterparts.1 hiPSC-derived EPI cells are highly proliferative and can be expanded for up to 25 passages. However, it is still recommended to use EPI cells between passages 3–8 for cardiac pericyte differentiation, as these cells can readily undergo EMT and give rise to fibroblast-like cells.4 EPI cells have been shown to aid in heart regeneration,5 so they may be a limitless source for cell therapy following ischemic myocardial injury.
In our experience, it takes at least 12 days to generate cardiac pericytes from EPI cells. While cell type-specific markers for pericytes have not yet been established,6 we expect cardiac pericytes to express high levels of PDGFRβ and NG2, low levels of smooth muscle cell-specific markers such as calponin and SM-MHC, and negligible levels of fibroblast markers (Figure 7). Cardiac pericytes should also express cardiac transcription factors, such as TBX20 and GATA4 (Figure 8). Our iterative differentiation of cardiac pericytes from 10 hiPSC lines has resulted in >95% differentiation efficiency consistently.1
Figure 8.
High expression levels of cardiac transcription factors in hiPSC-derived cardiac pericytes
Cardiac pericytes are positive for cardiac transcription factors TBX20 (A’) and GATA4 (A’’). Cells are counterstained with PDGFRβ. hiPSC, human induced pluripotent stem cell; PDGFR, platelet-derived growth factor. Scale bars, 50 μm.
hiPSC-derived cardiac pericytes normally show a moderate proliferation rate and exhibit a heterogeneous morphology, particularly in the early stage of differentiation. However, as cardiac pericytes become confluent, they tend to show a homogeneous, well-aligned morphology. Our previous study showed that hiPSC-derived cardiac pericytes exhibited a potent pro-angiogenic capacity both in vitro and in vivo,1 making them another promising cell source to facilitate neovascularization in ischemic myocardium.7 Moreover, cardiac pericytes can be used to recapitulate chemotherapy-induced coronary microvascular dysfunction, as demonstrated by both in vivo8 and in vitro studies.1
Limitations
Although lineage-tracing studies suggest that cardiac pericytes originate from EPI cells,9 it is still under debate whether other progenitor cell types, such as cardiac neural crest cells, also contribute to cardiac pericytes, which may contribute to cell lineage-specific functional heterogeneity. Emerging single-cell RNA sequencing data have identified several subclusters of cardiac pericytes with distinct transcriptional profiles,10,11 suggesting functional heterogeneity within this cell type. Therefore, it is important to acknowledge that our EPI cell-derived cardiac pericytes may not fully capture the functional complexity of this poorly characterized cell type under pathophysiological conditions. A better understanding of the developmental program of cardiac pericytes will aid in optimizing the current differentiation protocol.4
Cardiac pericytes generated using this protocol are fully differentiated but relatively immature. Extended culture does not seem to be a viable option to significantly improve cell maturity, as these cells tend to exhibit senescence-related changes around days 45–50 of differentiation. We, therefore, recommend using cardiac pericytes from days 21–35 of differentiation for experiments, provided that a sufficient number of cells can be obtained. If more mature cardiac pericytes are required, co-culturing them with endothelial cells or generating three-dimensional cardiac or vascular tissues may help achieve functional maturation.
Troubleshooting
Problem 1
hiPSCs fail to distribute evenly in cell culture plates but form patchy clusters instead (step 1).
Potential solution
Gently pipette hiPSCs into single cells. Make sure to mix the cell suspension right before adding the hiPSCs into the wells to avoid cells from clustering together. Once hiPSCs are seeded, shake the plates immediately in a left-to-right and back-to-forth motion. Leave the cell culture plates at 16°C–25°C for 10 min before transferring them back to the incubator.
Problem 2
Excessive cell death and detachment happens after changing to MPS induction medium (step 2).
Potential solution
-
•
A substantial loss of cells is expected at this point. However, if most of the cells are detached, the differentiation will fail. Since the concentrations of cytokines and small molecules present in the MPS induction medium has been extensively titrated, we do not recommend adjusting the concentration of any component in the medium first. Instead, try to increase the hiPSC seeding density for differentiation. If the problem persists, users may consider reducing the CHIR99021 concentration to 4–5 μM.
-
•
If multiple hiPSC lines experience the same issue, it is recommended to test whether cells are contaminated with mycoplasma.
-
•
Renewing hiPSCs every 3–4 days is crucial. If hiPSCs require at least 5–7 days to reach 85% confluency, it is a sign that the cells are not in good condition to start differentiation. In this scenario, we recommend making one more passage and adjusting the seeding density of hiPSCs, provided that the cells are mycoplasma-free.
Problem 3
LPM cells reach over 70% confluency by the end of day 2 of differentiation (step 3).
Potential solution
Terminate the current batch of differentiation and start a new one by seeding hiPSCs at least 3 different densities. Continue differentiation on the well that yields approximately 20%–30% LPM cells.
Problem 4
ST cells fail to attach to Matrigel-coated flasks or form small spheroid-shaped clusters (step 9).
Potential solution
-
•
Users may forget to add ROCK inhibitor to the LaSR basal medium before replating cells. In such a case, thaw a vial of cryopreserved ST cells and reseed cells in a new Matrigel-coated T75 flask with LaSR basal medium containing a ROCK inhibitor.
-
•
ST cells may be over-digested in 10× TrypLE™ Select Enzyme or damaged due to extensive pipetting. Thaw a vial of cryopreserved ST cells and check cell viability. If it is below 50%, it is recommended to start a new batch of differentiation. Otherwise, users may try to seed the thawed cells in a new Matrigel-coated T75 flask to see whether this solves the problem.
Problem 5
PEO cells become almost 100% confluent by the end of day 9 (step 9).
Potential solution
For certain hiPSC lines, PEO cells can proliferate from 10% to approximately 100% confluency within 24 h. In this case, if cryopreserved backup ST cells are available, it is recommended to reseed these cells at a lower density in a new Matrigel-coated T75 flask. However, if backup cells are not available, day 10 PEO cells should be dissociated using 10× TrypLE™ Select Enzyme and re-plated at 1-2.5×103 cells/cm2 in PEO specification medium containing ROCK inhibitor. The following day, switch to EPI maintenance medium.
Problem 6
EPI cells fail to show clear boundaries and lose the epithelial cell morphology in some regions of the flasks (step 10).
Potential solution
This indicates that EPI cells have initiated the EMT process. To address this issue, dissociated EPI cells using 10× TrypLE™ Select Enzyme and replate cells in a new Matrigel-coated T75 flask at the density of 2.5×103 cells/cm2 in EPI maintenance medium containing a ROCK inhibitor. Switch to regular EPI maintenance medium the following day and check cell morphology after 2 days. If a uniform epithelial cell morphology is observed, continue with the culture. If not, redo step 9 using cryopreserved ST cells at the density of 1×103 cells/cm2.
Problem 7
A typical epithelial cell morphology is still observed among early cardiac pericytes at day 6 of differentiation (step 16).
Potential solution
EPI cells derived from certain hiPSC lines may take longer than 6 days to completely lose their epithelial cell-like morphology. If the cells are dissociated for replating on gelatin-coated plates at this point, significant cell death could occur. In such cases, we recommend continuing to refresh the cardiac pericyte induction medium in the same plates for an additional 2–4 days until a typical morphology is observed, as shown in Figure 6i. This will allow more time for the EMT process to complete and minimize cell death.
Problem 8
Cardiac pericyte proliferate slow in ScienCell pericyte medium (step 18).
Potential solution
In our experience, cardiac pericytes have a lower proliferation rate compared to EPI cells. To enhance the proliferation of cardiac pericytes, we recommend replating the cells at a low dilution ratio of 1:2-1:3. If a larger number of cardiac pericytes are required within a short time frame, an alternative approach would be to differentiate additional EPI cells and pool them together to minimize batch-to-batch variabilities. Another option is to supplement the ScienCell pericyte medium with thalidomide, provided that it does not interfere with the signaling pathways under investigation. However, to mitigate any potential impact of thalidomide on the functional properties of cardiac pericytes, it is advisable to wash out the compound by culturing the cells in ScienCell pericyte medium containing reduced serum (e.g., 0.1%–0.5%) for at least 2–3 days.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Joseph C. Wu (joewu@stanford.edu).
Materials availability
This study did not generate novel unique reagents. Materials associate with this protocol are available upon reasonable request from the lead contact.
Data and code availability
This protocol did not generate any new datasets or original codes.
Acknowledgments
This study was supported by research grants from Tobacco-Related Disease Research Program (TRDRP) 30FT0852 (M.S.); American Heart Association (AHA) Career Development Award 19CDA34760019 (C.L.); and the National Institutes of Health (NIH) R01 HL113006, R01 HL126527, R01 HL130020, R01 HL163680, and P01 HL141084 (J.C.W).
Author contributions
M.S. developed this protocol and wrote the manuscript. J.C.W. supervised the project and revised the manuscript. S.R.Z., Y.K., L.L., Y.Z., and C.L. validated this protocol independently and revised the manuscript.
Declaration of interests
J.C.W. is a cofounder of Greenstone Biosciences but has no competing interests, as the work presented was performed independently.
Contributor Information
Mengcheng Shen, Email: mcshen@stanford.edu.
Joseph C. Wu, Email: joewu@stanford.edu.
References
- 1.Shen M., Liu C., Zhao S.R., Manhas A., Sundaram L., Ameen M., Wu J.C. Stepwise generation of human induced pluripotent stem cell-derived cardiac pericytes to model coronary microvascular dysfunction. Circulation. 2023;147:515–518. doi: 10.1161/CIRCULATIONAHA.122.061770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen M., Liu C., Wu J.C. Generation of embryonic origin-specific vascular smooth muscle cells from human induced pluripotent stem cells. Methods Mol. Biol. 2022;2429:233–246. doi: 10.1007/978-1-0716-1979-7_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knight-Schrijver V.R., Davaapil H., Bayraktar S., Ross A.D.B., Kanemaru K., Cranley J., Dabrowska M., Patel M., Polanski K., He X., et al. A single-cell comparison of adult and fetal human epicardium defines the age-associated changes in epicardial activity. Nat. Cardiovasc. Res. 2022;1:1215–1229. doi: 10.1038/s44161-022-00183-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ameen M., Sundaram L., Shen M., Banerjee A., Kundu S., Nair S., Shcherbina A., Gu M., Wilson K.D., Varadarajan A., et al. Integrative single-cell analysis of cardiogenesis identifies developmental trajectories and non-coding mutations in congenital heart disease. Cell. 2022;185:4937–4953.e23. doi: 10.1016/j.cell.2022.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xia Y., Duca S., Perder B., Dündar F., Zumbo P., Qiu M., Yao J., Cao Y., Harrison M.R.M., Zangi L., et al. Activation of a transient progenitor state in the epicardium is required for zebrafish heart regeneration. Nat. Commun. 2022;13:7704. doi: 10.1038/s41467-022-35433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee L.L., Chintalgattu V. Pericytes in the heart. Adv. Exp. Med. Biol. 2019;1122:187–210. doi: 10.1007/978-3-030-11093-2_11. [DOI] [PubMed] [Google Scholar]
- 7.Chen W.C.W., Baily J.E., Corselli M., Díaz M.E., Sun B., Xiang G., Gray G.A., Huard J., Péault B. Human myocardial pericytes: multipotent mesodermal precursors exhibiting cardiac specificity. Stem Cell. 2015;33:557–573. doi: 10.1002/stem.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chintalgattu V., Rees M.L., Culver J.C., Goel A., Jiffar T., Zhang J., Dunner K., Jr., Pati S., Bankson J.A., Pasqualini R., et al. Coronary microvascular pericytes are the cellular target of sunitinib malate-induced cardiotoxicity. Sci. Transl. Med. 2013;5:187ra69. doi: 10.1126/scitranslmed.3005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai C.L., Martin J.C., Sun Y., Cui L., Wang L., Ouyang K., Yang L., Bu L., Liang X., Zhang X., et al. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baek S.H., Maiorino E., Kim H., Glass K., Raby B.A., Yuan K. Single cell transcriptomic analysis reveals organ specific pericyte markers and identities. Front. Cardiovasc. Med. 2022;9:876591. doi: 10.3389/fcvm.2022.876591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Litviňuková M., Talavera-López C., Maatz H., Reichart D., Worth C.L., Lindberg E.L., Kanda M., Polanski K., Heinig M., Lee M., et al. Cells of the adult human heart. Nature. 2020;588:466–472. doi: 10.1038/s41586-020-2797-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This protocol did not generate any new datasets or original codes.