Summary
Expansion microscopy of millimeter-large mechanically heterogeneous tissues, such as whole vertebrate embryos, has been limited, particularly when combined with post-expansion immunofluorescence. Here, we present a protocol to perform ultrastructure expansion microscopy of whole vertebrate embryos, optimized to perform post-expansion labeling. We describe steps for embedding and denaturing zebrafish larvae or mouse embryos. We then detail procedures for hydrogel handling and mounting. This protocol is particularly well suited for super-resolution imaging of macromolecular protein complexes in situ but does not preserve lipids.
For complete details on the use and execution of this protocol, please refer to Steib et al.1
Subject areas: Developmental Biology, Microscopy, Model Organisms, Molecular Biology
Graphical abstract

Highlights
-
•
TissUExM for expansion microscopy of whole vertebrate embryos
-
•
Procedures for embedding and denaturing for post-expansion labeling
-
•
Procedures for gel handling and mounting
-
•
Applicable to study protein complexes at super resolution in situ
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Expansion microscopy of millimeter-large mechanically heterogeneous tissues, such as whole vertebrate embryos, has been limited, particularly when combined with post-expansion immunofluorescence. Here, we present a protocol to perform ultrastructure expansion microscopy of whole vertebrate embryos, optimized to perform post-expansion labeling. We describe steps for embedding and denaturing zebrafish larvae or mouse embryos. We then detail procedures for hydrogel handling and mounting. This protocol is particularly well suited for super-resolution imaging of macromolecular protein complexes in situ but does not preserve lipids.
Before you begin
We divided this section into four parts. First, we list the different pre-TissUExM embryo fixation options and explain how to plan the number of embryos to prepare. Next, we describe the rationale for implementing an optional collagenase digestion between the gelation and denaturation steps. Lastly, we comment on time needed to prepare all the solutions required.
Embryos fixation
The fixative should be chosen before TissUExM. We suggest performing a regular immunofluorescence to identify the fixative giving the best result for the antibody and organelle of interest.
The protocol uses 4% PFA-fixed two-days post-fertilization (2 dpf) zebrafish (ZF) embryos. We dilute commercially available 16% PFA in PBS. TissUExM can also be used on ZF up to 5 dpf or fixed with Dent’s fixative (−20°C; Methanol 80%, DMSO 20%). PFA-fixed embryos can be gradually dehydrated in methanol and stored at −20°C. Similarly, this protocol is applicable for 4% PFA-fixed and dehydrated mouse embryos.
-
1.48 hpf embryos fixations.
-
a.Place embryos in a 2 mL tube filled with 4% PFA. Do not fix more than ∼30 embryos per tube. Use multiple tubes if needed.
-
b.Incubate 6 h at RT or overnight at 4°C.
-
c.Replace the 4% PFA solution with 0.1% PBS-Tween 20 (PBS-T).
-
a.
-
2.Embryos dehydration in the same 2 mL tube, at RT, with orbital agitation.
-
a.5 min in 500 μL 25%–75% methanol-PBS.
-
b.5 min in 500 μL 50%–50% methanol-PBS.
-
c.5 min in 500 μL 75%–55% methanol-PBS.
-
d.Replace with 100% methanol. Store at −20°C.
-
a.
Number of embryos per experiment
Plan the number of embryos based on the number of antibody combinations you will need, aiming for one whole embryo/gel/antibody combination. We suggest duplicating the number of embryos to start with, as loss can happen up to the labeling step. Although experienced users may observe 0–5% loss, beginners could lose up to 50% of their embryos/gels. We do not recommend processing more than twelve gels per experiment. The protocol below describes the preparation of two gels.
Conditions for additional collagenase VII digestion
We found that collagenase treatment is necessary from 3 dpf onward to allow non-disruptive expansion. We recommend performing the collagenase VII digestion between gelation (after embedding) and denaturation (prior to expansion). In this way, tissues are anchored and crosslinked to the gel prior to the digestion of internal myotendinous regions.
Solutions preparation
Considering the number of solutions needed, we recommend users to plan 1 or 2 days to prepare solutions in advance. Note that most solutions are stable for 6–12 months.
Institutional permissions (if applicable)
All experiments using ZF were performed following the European directive 2010/63/EU and Home Office guidelines under the project license PP6020928.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| 1-Phenyl-2-thiourea (PTU) | Sigma | Cat#P7629 |
| Paraformaldehyde (PFA, 16%) | Fisher Scientific | Cat#28908 |
| Methanol | Sigma | Cat#34860 |
| DMSO | Sigma | Cat#D2650 |
| Triton X100 | Fisher Scientific | Cat#10102913 |
| Formaldehyde (FA, 36.5%–38%) | Sigma | Cat#F8775 |
| Acrylamide (AA, 40%) | Sigma | Cat#A4058 |
| N,N′-methylbisacrylamide (bis-AA, 2%) | Sigma | Cat#M1533 |
| Sodium acrylate (SA, 97%–99%) | Sigma | Cat#408220 |
| 4-Hydroxy-TEMPO | Sigma | Cat#176141 |
| Ammonium persulfate (APS) | Thermo Fisher | Cat#17874 |
| Tetramethylethylenediamine (TEMED) | Thermo Fisher | Cat#17919 |
| Sodium dodecyl sulfate (SDS) | Sigma | Cat#L3771 |
| Tris | Merck | Cat#T1503 |
| Tween 20 | Merck | Cat#P1379 |
| Phosphate buffer saline | Merck | Cat#D8537 |
| Poly-D-Lysine (PDL, 1 mg/mL) | Gibco | Cat#A3890401 |
| 12 mm coverslip, Menzel | Thermo Fisher | Cat#11846933 |
| 24 mm coverslip, Menzel | Thermo Fisher | Cat#11817892 |
| Attofluor chamber | Thermo Fisher | Cat#A7816 |
| ATTO 647N-NHS | Sigma | Cat#18373 |
| Experimental models: Organisms/strains | ||
| Zebrafish: WT/AB | Zebrafish International Resource Center (ZIRC) | ZFIN:ZDB-GENO-960809-7 |
Materials and equipment
All solutions are prepared similarly to Gambarotto at al.,2 except for the monomer solution that we modified to add Triton-X-100. Furthermore, we use a solution containing Triton-X-100 to supplement each step prior to embedding, a solution containing 4-hydroxy-TEMPO (4-OH) to slow down the gelation kinetic, a denaturation-like buffer for gel equilibration, and a calcium-containing buffer for optional collagenase digestion. Note that most solutions can be stored over several weeks or months, and that the monomer solution containing sodium acrylate should be prepared at least 24 h prior to gelation.
Sodium Acrylate
| Reagent | Final concentration | Amount |
|---|---|---|
| Sodium acrylate (99%) | 38% | 19 g |
| ddH2O | N/A | 31 mL |
| Total | N/A | 50 mL |
Dissolve slowly at 4°C with agitation then filter (0.45μm). Store at 4°C for 6 months.
CRITICAL: Use personal protective equipment (PPE), including gloves and surgical mask. Work under a fume hood if possible. The solution should be beige and not display a strong yellow color, associated with low batch purity. Contact the manufacturer if needed.
Triton X-100
| Reagent | Final concentration | Amount |
|---|---|---|
| Triton (99%) | 10% | 10 g |
| ddH2O | N/A | ∼80 mL |
| Total | N/A | 100 mL |
Dissolve with agitation then filter (0.45μm). Store at RT for 12 months.
Inactivated monomer solution (gelation solution)
| Reagent | Final concentration | Amount |
|---|---|---|
| Sodium acrylate (38%) | 21% | 500 μL |
| Acrylamide (40%) | 10% | 250 μL |
| Bis-Acrylamide (2%) | 0,1% | 50 μL |
| Triton 10% | 0,1% | 10 μL |
| PBS 10 × | 1 × | 90 μL |
| Total | N/A | 900 μL |
Work on ice or at 4°C. Aliquot in PCR tubes containing exactly 90 μL. Store at −20°C. The solution should not freeze.
CRITICAL: Use PPE, including gloves and surgical mask. Work under a fume hood if possible. The solution should be prepared at least 24 h before use.
4-hydroxy-TEMPO
| Reagent | Final concentration | Amount |
|---|---|---|
| 4-hydroxyTEMPO | 0.5% | 0.25 g |
| ddH2O | N/A | ∼49,5 mL |
| Total | N/A | 50 mL |
Prepare intermediate stocks of 10 mL. Aliquot in PCR tubes containing ∼3.5 μL each. Store at −20°C.
CRITICAL: Use PPE, including gloves and surgical mask. Work under a fume hood if possible.
TEMED
| Reagent | Final concentration | Amount |
|---|---|---|
| TEMED | 10% | 10 μL |
| ddH2O | N/A | 90 μL |
| Total | N/A | 100 μL |
Aliquot in PCR tubes containing ∼3.5 μL each. Store at −20°C.
CRITICAL: Use PPE, including gloves and surgical mask. Work under a fume hood if possible.
APS
| Reagent | Final concentration | Amount |
|---|---|---|
| APS (99%) | 10% | 0.01 g |
| ddH2O | N/A | ∼90 μL |
| Total | N/A | 100 μL |
Aliquot in PCR tubes containing ∼3.5 μL each. Store at −20°C.
CRITICAL: Use PPE, including gloves and surgical mask. Work under a fume hood if possible.
SDS
| Reagent | Final concentration | Amount |
|---|---|---|
| SDS (99%) | 350 mM | 10 g |
| ddH2O | N/A | ∼80 mL |
| Total | N/A | 100 mL |
Add little by little with agitation. Filter (0.45 μm) and keep at RT for 6 months. Place at 37°C for 10 min if crystals form.
CRITICAL: Use PPE, including gloves and surgical mask. Work under a fume hood if possible.
NaCl
| Reagent | Final concentration | Amount |
|---|---|---|
| Sodium Chloride (99%) | 5 M | 29.2 g |
| ddH2O | N/A | ∼95 mL |
| Total | N/A | 100 mL |
Filter (0.45 μm) and store at RT for 12 months.
Denaturation buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris (99%) | 38% | 19 g |
| NaCl 5 M | 200 mM | 4 mL |
| SDS 350 mM | 200 mM | 57,14 mL |
| Adjust pH to 9 with HCl | ||
| ddH2O | N/A | ∼25 mL |
| Total | N/A | 100 mL |
Filter (0.45 μm) and keep at RT. Place at 37°C for 10 min if crystals form.
CRITICAL: Use PPE, including gloves and surgical mask, as SDS is an irritant.
Denaturation-like buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris (99%) | 38% | 19 g |
| NaCl 5 M | 200 mM | 4 mL |
| ddH2O | N/A | ∼80 mL |
| Total | N/A | 100 mL |
Filter (0.45 μm) and keep at RT for 6 months.
Tween 20
| Reagent | Final concentration | Amount |
|---|---|---|
| Tween 20 (99%) | 10% | 10 mL |
| PBS | N/A | ∼90 mL |
| Total | N/A | 100 mL |
Filter (0.45 μm) and keep at RT for 12 months.
PBS-T
| Reagent | Final concentration | Amount |
|---|---|---|
| Tween 20 (10%) | 0,1% | 500 μL |
| PBS | N/A | 49,5 mL |
| Total | N/A | 50 mL |
Filter (0.45 μm) and keep at RT for 12 months.
PBS-BSA
| Reagent | Final concentration | Amount |
|---|---|---|
| BSA (99%) | 2% | 1 g |
| PBS | N/A | ∼48 mL |
| Total | N/A | 50 mL |
Filter (0.45 μm) and keep at 4°C for 3 months.
CRITICAL: Do not vortex.
CaCl2
| Reagent | Final concentration | Amount |
|---|---|---|
| CaCl2 (99%) | 5 M | 7.35 g |
| ddH2O | N/A | ∼ 40 mL |
| Total | N/A | 50 mL |
The solution will heat, so dissolve it little by little in an open beaker. Once the solution cooled, filter (0.45 μm) and keep at RT for 12 months.
CRITICAL: Do not let the solution heat and do not close the container, due to the risk of explosion.
Calcium-containing buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| CaCl2 (99%) | 40 mM | 2 mL |
| Tris | 50 mM | 0.3 g |
| NaCl | 200 mM | 2 mL |
| H20 | N/A | ∼45 mL |
| Total | N/A | 50 mL |
Filter (0.45 μm) and keep at RT for 6 months.
Gelation chamber
Using a clear plastic box or Petri Dish, layer the bottom part with multiple sheets of tissue paper. Use parafilm to maintain the tissues paper stuck to the plastic of the box. Leave the top part clear, as it will allow monitoring the gelation step under a binocular microscope.
The gelation chamber can be used multiple times. To avoid contamination, the tissue paper should be air-dried between each use.
Step-by-step method details
The steps from embryo rehydration to gelation are specific to TissUExM.1 From the denaturation step, gel handling is similar to U-ExM2,3 but the incubations time and solution volumes are scaled up for whole embryos.
Embryos rehydration and anchoring – Day 1
Timing: 1 h preparation, 3 d incubation
Stored embryos are first rehydrated to reach a near-native fixed state in PBS. The anchoring step consists in anchoring acrylamide (AA) to primary amines from the sample.
-
1.Embryos rehydration in a 2 mL tube, at RT, with orbital agitation.
-
a.5 min in 500 μL 75%–25% methanol-PBS.
-
b.5 min in 500 μL 50%–50% methanol-PBS.
-
c.5 min in 500 μL 25%–75% methanol-PBS.
-
d.5 min in 500 μL PBS.
-
a.
-
2.Individual embryo anchoring.
-
a.Under a fume hood, prepare 1 mL of the anchoring solution in a 1.5 mL Epi-tube, aiming for 500 μL/embryo. Vortex.Anchoring solution master mix for two gels
Reagent Amount PBS 1× 453 μL Triton 10% 10 μL AA 40% 500 μL FA 38% 37 μL -
b.Coat a plastic pipette with PBS-Tween 0.1% (PBS-T).
-
c.Transfer each embryo to an empty 2 mL tube. Remove the excess of liquid.
-
d.Add 500 μL of crosslinking solution to each embryo.
-
e.Seal the tube with parafilm. Invert multiple times.
-
f.Place at 37°C for 72 h. Gently invert the tube occasionally.
 CRITICAL: It is crucial that AA is evenly distributed and reaches all the tissues of the embryo. This will affect the embedding homogeneity in the following next step. The volume of anchoring solution should be in excess compared to embryo size. If larger samples are used, do not hesitate to scale up the volume and incubation time.
CRITICAL: It is crucial that AA is evenly distributed and reaches all the tissues of the embryo. This will affect the embedding homogeneity in the following next step. The volume of anchoring solution should be in excess compared to embryo size. If larger samples are used, do not hesitate to scale up the volume and incubation time.
-
a.
Gelation preparation – Day 4
Timing: 30 min preparation, overnight incubation
This step allows a first penetration of the gelation components in the tissues, prior to the rigidification of the gel matrix.
-
3.Embryos wash.
-
a.Under a fume hood, remove the crosslinking solution.
-
b.Rinse 3 × 5 min with 500 μL of PBS-T at RT with orbital agitation.
-
a.
-
4.Gelation preparation.
-
a.Using a plastic pipette coated with PBS-T, transfer each embryo to a clean 2 mL tube.
-
b.Add 90 μL of inactivated monomer solution (MS).
-
c.Incubate overnight at 4°C.
-
a.
- 5.
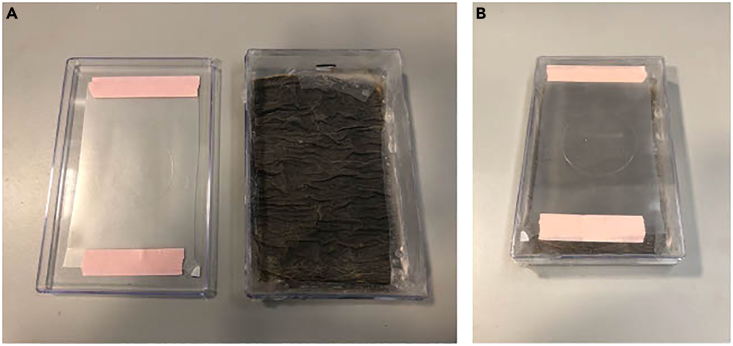
Figure 1.
How to prepare the gelation chamber
(A) Place a clean parafilm on the lid and humidify the paper with water on the bottom part.
(B) Put the closed box at −20°C overnight.
Gelation – Day 5
Timing: 4 h
This step requires careful handling of the sample, to determine embryo orientation and ensure gel integrity. The processing should not take more than 30 min. Every step should be performed rapidly and precisely. We suggest preparing the gelation station (binocular, gelation chamber, tweezers, coverslips, …) in advance, with every tool ready to minimize waiting times (Figure 2A). The inactivated MS should be thawed shortly before use. During activation, keep the MS tube open and vortex between each step. Rapidly place the activated solution onto the dried embryos (Methods video S1, Embedding, related to step 2).
-
6.Activators preparation.
-
a.To prepare two gels, thaw one aliquot of ∼3.5 μL of 4OH-TEMPO, one aliquot of ∼3.5 μL TEMED and one aliquot of ∼3.5 μL APS.
-
a.
-
7.Embryos wash.
-
a.From the tubes at 4°C, remove the inactivated MS solution.
-
b.Rinse 3× with 500 μL PBS-T with orbital agitation.
-
a.
-
8.Embryos orientation.
-
a.Take the gelation chamber out of −20°C. Place the bottom part with parafilm under a binocular microscope.
-
b.With a plastic pipette coated with PBS-T, transfer each embryo onto the parafilm, in the smallest volume possible (Figure 2B).
-
a.
-
9.Monomer solution activation.
-
a.Thaw one aliquot of inactivated MS.
-
b.Rapidly add directly to the tube and in this order: 3 μL PBS, 2 μL 4OH-TEMPO, 2.5 μL TEMED, and 2.5 μL APS. Vortex between each addition (Figure 2C).
-
a.
-
10.Embryos gelation.
-
a.Rapidly remove the excess liquid from the embryo.
-
b.Add 45 μL of activated gelation solution to each embryo.
-
c.Reorient the embryo with a lash if necessary. Place it in the center of the drop. Pop the bubble if any. Let the embryo stabilize.
-
d.Flip a 12 mm coverslip on each embryo, making sure it did not slide to the edge. Replace the coverslip if necessary (Figure 2D).
-
e.Wait 5 min for stabilization. Close the gelation chamber and gently place it at 4°C.
-
f.Incubate 1 h.
-
g.Gently transfer the entire box from 4°C to 37°C.
-
h.Incubate 2 h, troubleshooting 1.
-
a.
Optional: Collagenase VII digestion.
Figure 2.
How to prepare the gelation
(A) Gelation station with binocular, lid of the gelation chamber, pair of tweezers and coverslips ready-to-use.
(B) Inset on the two embryos placed directly on the parafilm.
(C) Ice box to thaw gelation activators. On the left side, PBS and 4OH-tempo do not require fast addition. On the right side, TEMED and APS must be pipetted fast. Vortex with tube open after adding each component.
(D) Inset on two embryos covered by a gelation drop and coverslips, to be incubated for gel polymerization.
For ZF larvae older than 3 dpf, place the gel in an Epi-tube containing 1 mL of 1000 U of Collagenase VII, diluted in calcium-containing buffer. Incubate 24 h at 37°C. The next day, wash 15 min in calcium-containing buffer and 15 min in PBS before proceeding to denaturation.
Note: Individual embryos should not be on the edge of the coverslip, but rather in the center.
CRITICAL: Embryo orientation in the gel is determined from the moment the coverslip flattens the gelation drop. It will be impossible to change the orientation of the embryo after that step. Bubbles should be avoided. If bubbles are visible, you can remove the coverslip, place the embryo back in the center of the gelation solution drop and add a new coverslip.
Denaturation – Day 5
Timing: 15 min preparation, 3 d incubation
This step inhibits mechanical resistance generated by the specimen when the gel will expand. It is based on temperature and pH to denature the tertiary structures of proteins in the embryo. Only AA-anchored amines will remain, maintaining the relative position of each protein to one another. This step also includes delipidating, leading to the optical clearing of the embryo.
-
11.Gel detachment.
-
a.Set a Thermoblock at 70°C.
-
b.In a 35 mm dish or a 6-well plate, add ∼2 mL of the denaturation buffer.
-
c.Using forceps, gently detach the coverslip and the gel from the parafilm.
-
d.Transfer the coverslip and gel to the well with the denaturation buffer. The coverslip is placed downward so that the gel can detach and float in the denaturation buffer (Methods video S2, Gel detachment, related to step 3).
-
e.Incubate 10 min at RT with 100 rpm orbital agitation, troubleshooting 2.
-
a.
-
12.Embryo denaturation.
-
a.For each embryo, fill a 1.5 mL Epi-tube with ∼1.2 mL of the denaturation buffer.
-
b.Using a spatula, transfer each gel detached from its coverslip and place it in the tube. Complete with ∼100 μL to fill the tube (Methods video S3, Transfer to tube, related to step 3).
-
c.Incubate at 70°C for 72 h.
-
a.
Note: Denaturation buffer contains SDS, which is an irritant. Handle tubes with gloves and avoid spilling when closing the tube.
Gel equilibration and control – Day 8
Timing: 3 h
This step marks the end of gel preparation. We provide tips to control gel integrity and trimming.
-
13.Gel equilibration.
-
a.Take tubes out of the Thermoblock.
-
b.Prepare 35 mm dishes or a 6-well plate and add ∼2 mL of denaturation-like buffer.
-
c.Using a plastic pipette, gently remove all the denaturation buffer, pipetting from the bottom of the tube, limiting contacts with the gel (Methods video S4, Transfer from tube, related to step 3).
-
d.Transfer the gel to the well.
-
e.Incubate 15 min at RT at 100 rpm with orbital agitation.
-
f.Remove the denaturation-like buffer and add ∼2 mL of PBS.
-
g.Incubate 15 min at RT at 100 rpm with orbital agitation, troubleshooting 3.
-
a.
Pause point: Gels can be stored in PBS at 4°C for a month from now. Consider adding 0.05% sodium azide to prevent from contamination.
-
14.Gel control.
-
a.Option1, for beginners: ATTO 647N NHS-ester.
-
i.Prepare a solution of ATTO 647N NHS-ester/PBS (1:2000).
-
ii.In the gel-containing well, replace PBS with ATTO647N NHS-ester/PBS. Protect from light with aluminum foil.
-
iii.Incubate 2 h at RT at 100 rpm with orbital agitation (or 16–72 h at 4°C, with optional shaking).
-
iv.Replace with PBS and verify embryo integrity within the gel, with or without a binocular, troubleshooting 4.
-
v.If validated, proceed to trim, as described in section 17.Note: This step is not compatible with further antibody staining, as most epitopes will be masked by the dye. The fish will become blue and visible directly by eye in the gel.Optional: Blocking.If future antibody staining requires a blocking step, it is possible to perform it along DAPI labeling, for 2 h at RT. By default, we use the following blocking solution:
Reagent Final concentration Amount DMSO 0.1% 500 μL Triton-X-100 (10%) 0.5% 250 μL Fetal calf or goat serum 5% 2.5 mL BSA 2% 1 g PBS N/A ∼45 mL Total N/A 50 mL Filter (0.45μm) and keep at 4°C for 6 months. Pause point: Gels can be stored in PBS at 4°C and protected from light, for 2–3 weeks.
Pause point: Gels can be stored in PBS at 4°C and protected from light, for 2–3 weeks.
-
i.
-
b.Option2, routine.
-
i.Prepare a solution of DAPI/PBS (1:2000).
-
ii.In the gel-containing well, replace PBS with DAPI/PBS, Protect from light with aluminum foil.
-
iii.Incubate 2 h at RT at 100 rpm with orbital agitation.
-
iv.Replace with PBS.
-
v.Measure the gel diameter. The diameter should consistently be between 26-30 mm.
-
vi.Optional: Place the gel in an Attofluor chamber with a 24 mm coverslip if controlled is performed with an epifluorescence microscope,
-
vii.Control embryo positioning and orientation in the gel, troubleshooting 5.Optional: Trimming.If antibody concentration or cost is a limiting factor, it is possible to trim the gel at that step and reduce its size to fit in a smaller well (2 mL in a 6-well plate, 1 mL in a 12-well plate). However, we recommend beginners to not trim, as the gel can be more difficult to extract from a 12-well format). If trimming is chosen, please refer to section 17.
 Pause point: Gels can be stored in PBS at 4°C and protected from light, for 1–2 weeks.
Pause point: Gels can be stored in PBS at 4°C and protected from light, for 1–2 weeks.
-
i.
-
a.
Immuno-labeling – Day 8
Timing: 1 week, including 2 × 4 h of preparation and 4 + 3 d of incubation
This step consists of the immunofluorescence, as for cleared samples. We use passive labeling, but other groups have described active labeling options based on the electrophoresis of the antibodies.4,5
-
15.Antibody staining.
-
a.Transfer individual gels in a 6-well plate or individual 2 mL tubes, depending on incubation limitations.
-
b.Incubate each gel in 2 mL PBS-BSA 2% or blocking buffer with primary antibodies. The default dilution is 1:100. Incubate 4 d at 37°C and 100 rpm orbital agitation. We suggest using a shaking incubator with temperature controlled.
-
c.Wash 3 × 45 min with PBS-T at RT at 100 rpm with orbital agitation.
-
d.Incubate each gel in 2 mL PBS-BSA 2% with secondary antibodies. The default dilution is 1:250. Incubate 3 d at 37°C and 100 rpm orbital agitation. We suggest using a shaking incubator with temperature controlled.
-
e.Wash 3 × 45 min with PBS-T at RT with 100 rpm with orbital agitation.
-
f.Replace the last washing solution with 2 mL of PBS.
-
a.
-
16.Staining control.
-
a.Place individual gels in an Attofluor chamber.
-
b.Control staining efficiency using an epifluorescence microscope, troubleshooting 6.
-
c.If necessary, try to retain which orientation of the gel is optimal for visualization of the tissue of interest (= which side of the gel should be closer to the objective, if the working distance is limiting).
-
a.
Note: Samples will be brighter in PBS-state gels (∼2,2× expansion) than in water (4× expansion). We recommend beginners to stain for acetylated-tubulin or polyglutamylated-tubulin as centriolar markers, which are used as landmarks to control intracellular isotropic expansion.
Secondary antibodies should be coupled to fluorophores that remain stable in water, such as Alexa 488, 555, or 568. For example, Rhodamine fluorescence is lost when placed in water.
Optional: ATTO 647N NHS ester labeling, as in section 14.
We recommend beginners to add this step to visualize the embryo in the gel and trim it easily. Note that NHS ester labeling provides limited subcellular information combined with 4× expansion of whole embryos.
-
17.Trimming.
-
a.Place individual gels on a parafilm.
-
b.Measure their individual diameter (value should be ∼26–30 mm) (Figure 3A).
-
c.Trim the gel excess to obtain a rectangle of ∼12–15 mm (Figure 3B) (Methods video S5, trimming, related to step 5).
-
d.Measure the new size for future calculation of the expansion factor.
-
a.
Note: As the gel is deformable and soft, trimming is not very precise. Prefer keeping an excess of gel rather than cutting too close to the embryo. Performing the trimming on a parafilm will ease the gel manipulation.
Pause point: Gels can be stored in PBS at 4°C and protected from light, for 1–2 weeks.
Figure 3.
How to handle the gel
(A) ATTO 647N NHS-ester labeled gel, in PBS-state, measured prior to trimming.
(B) Same gel measured after trimming.
(C) Mounting set up on a plastic box covered with a parafilm and Attofluor chamber. Plastic scalpels and spatula allow gel handling. Kimwipe should be used to blot the excess water from the gel.
(D) Same gel after water immersion, measured when fully expanded.
Expansion – Day 15
Timing: 3 h minimum, overnight preferred
This step involves the physical and maximal expansion of the gel, to reach 4× expansion compared to its size at gelation.
-
18.Water exchanges.
-
a.Use a 6-well plate for trimmed gels smaller than 13 mm. For larger gels, use a bigger container, like a 100 mL or a 250 mL beaker.
-
b.Transfer each gel and cover it with an excess of water.
-
c.Incubate 1 h at RT, protected from light.
-
d.Replace the water. Incubate between 1 h and 16 h, protected from light. We recommend overnight incubation whenever possible.
-
e.Replace the water one last time. Incubate 1 h.
-
f.Place the gel on a parafilm and measure its final size (Figure 3C).
-
a.
Note: Water-rehydrated is the most unstable state for antibodies and fluorophores. Expand only prior to imaging and prefer PBS immersion for storage.
CRITICAL: Do not let the gel dry, otherwise it will fragilize its structure, make it brittle, and eventually break. If the gel becomes opaque and porous during one of the transfer steps, rehydrate with a few drops of water. The gel should become translucent again instantly.
Mounting for imaging
Timing: 10 min
This step is usually performed at the microscope once the optimal orientation of the gel has been defined. You will have to consider the working distance of the objectives, which can be a major limitation. We detail the mounting for inverted microscopes here. We recommend using Attofluor chambers for two reasons: 1/ PDL-coated coverslips can easily be prepared and replaced, 2/ the weight of the chamber helps stabilize the sample, compared to a plastic confocal dish. Mounting is a crucial step to prevent the gel from drifting while imaging.
-
19.Mounting.
-
a.Prepare the mounting station with a parafilm, Kimwipes, and a spatula Figure 3D).
-
b.Place a PDL-coated coverslip in the Attofluor chamber.
-
c.With a spatula, dry the gel on a Kimwipe by blotting it several times on a Kimwipe. It is essential to remove as much water as possible to favor grip on the coverslip.
-
d.Place the gel in the chamber. Press very gently if air bubbles are between the coverslip and the gel, troubleshooting 7.
-
e.Optional: trim the corners of the gel if needed to close the chamber.
-
f.Add 1–2 drops of water on top of the gel. The gel is now ready to be imaged (Methods video S6, mounting, related to step 5).
-
a.
CRITICAL: Make sure the gel stays hydrated by adding a drop of water every ∼30 min during the imaging session. Artifacts such as sudden drift or fluorescent increase may mean that the gel is shrinking due to dehydration. In this case, rehydrate the gel and wait 5 min until the gel stabilizes before imaging more.
Note: Once adherent to the PDL-coated coverslip, the gel will be difficult to detach. You can use a pipet to add water between the gel and the coverslip. When detaching, be very gentle not to break the gel.
Expected outcomes
Embedded embryos should have retained their morphology, be cleared, and 4× bigger than their initial size. Fluorescent labeling should be homogeneous within the embryo. With a 10× objective on a confocal microscope, tiling (12–16 tiles) should be used to image the entire embryo. Images are described in Steib et al.1
Limitations
The protocol has not been tested on fish older than 5 dpf, so more optimizations might be required when expanding older fish.
Although TissUExM is a versatile approach, the gel preparation is not compatible with mineralized structures, such as C.elegans and D. melanogaster cuticles, A. thaliana roots, or gastrulated sea urchin shells. In general, any external mineralized layer should be removed to allow the penetration of chemicals. Thus, each of these samples should be treated accordingly prior to the TissUExM process.
Internal mineralized structures could be treated enzymatically between gelation and denaturation, similarly to the collagenase VII digestion.
Importantly, TissUExM is not adapted for lipid visualization because the denaturation step solubilizes all lipids.
Because the sample increases its size 4 folds, the working distance of each objective should be considered when designing the experiment when performing confocal microscopy.
Troubleshooting
Troubleshooting 1: embryo gelation problems & solutions
We list potential gelation problems, from minor to major:
-
•
Small hole in the gel: this means that a bubble has formed between the gelation solution and the coverslip. It has no consequence if the bubble is away from the embryo. Always burst bubbles with a pair of tweezers before placing the coverslip.
-
•
Embryo on the edge of the gel: this means that the embryo moved under the pressure of the coverslip. Always let the embryo stabilize in the center of the gelation drop prior to adding the coverslip. Alternatively, remove the coverslip, place the embryo in the center of the gelation drop and add a new coverslip. Only proceed with polymerization when the embryo is far enough from the coverslip edge.
-
•
Gel polymerized before adding the coverslip: this means that manipulation of the embryo is too slow. The presence of 4-OH TEMPO should allow 20–25 min manipulation at RT.
-
•
Gel never polymerized: Like for SDS-page, it would mean that either TEMED or APS was forgotten. Consider using half of a monomer solution aliquot for gelation and the other half as blank. Monitor the polymerization at 4°C and 37°C in the tube to reflect the polymerization state of the embryo in the gelation chamber.
Troubleshooting 2: gel detachment from the parafilm problems & solutions
A potential start of denaturation problem is that the gel may stick more to the parafilm rather than to the coverslips after total polymerization. If you detach the coverslip without the gel, use a pair of tweezers or a spatula and gently glide between the parafilm and the gel. The gel will then detach.
Troubleshooting 3: fragile gels after equilibration & solutions
We list potential gel integrity control problems, from minor to major:
-
•
Thinner gel on one side: this occurs when the sample is not perfectly placed in the center of the drop. Trim the thin fragile part on the edge of the gel to avoid gel breakage and embryo damage. Keep processing the solid part of the gel containing the embryo.
-
•
Irregular gel, breaking at transfer: this happens when the monomer solution is used fresh. Always store the aliquots of MS at least one night at −20°C before use.
Troubleshooting 4: problems after NHS-Ester staining & solutions
We list potential gel NHS staining control problems, from minor to major:
-
•
Heterogeneous or superficial staining: make sure to use a sufficient concentration of NHS-Ester 647N Dye and incubate with agitation at 100 rpm on an orbital shaker. Re-incubate 2 h at RT with agitation.
-
•
Sample too bright for imaging: repeat washes, 3 × 1 h at RT in PSB-T. Control in PBS, then in water.
-
•
Broken embryo: this correlates with poor anchoring and/or poor denaturation. Please start over, ensuring that the volume of the anchoring solution is at least 3× the volume of the sample. If anchoring is the problem, the damage should occur in the first 10 min of denaturation, already at RT, and should be visible by the naked eye. For fish older than 3 dpf or samples with cuticles, proceed to enzymatic digestion prior to anchoring. If damages are detected only after expansion, this means denaturation should be performed over a longer period.
Troubleshooting 5: problems after DAPI staining & solutions
We list potential gel DAPI staining control problems, from minor to major:
-
•
Heterogeneous or superficial staining: make sure to incubate with agitation at 100 rpm. Re-incubate 2 h at RT with agitation.
-
•
Cytoplasmic dots in addition to nuclear labeling: the additional dots are due to mitochondrial DNA, exposed when DAPI is too concentrated or when denaturation temperature has been increased above 70°C.
-
•
Broken embryo: this correlates with poor anchoring and/or poor denaturation. Please start over, ensuring that the volume of the anchoring solution is at least 3× the volume of the sample. If anchoring is the problem, the damage should occur in the first 10 min of denaturation, already at RT, and should be visible by the naked eye. For fish older than 3 dpf or samples with cuticles, proceed to enzymatic digestion prior to anchoring. If damages are detected only after expansion, this means denaturation should be performed for a longer period.
Troubleshooting 6: staining problems & solutions
We list potential immuno-labeling problems, from minor to major:
-
•
Staining limited to superficial external layers of the embryo: This represents a penetration problem. Make sure to incubate antibodies at 37°C to relax the gel and favor antibody penetration. If possible, use small antibodies purified from sera. Eventually, consider active labeling.4,5
-
•
Dotty staining at super-resolution: this likely represents epitope degradation, often due to chemical fixation prior to the TissUExM process. As for immunofluorescence, test different chemical fixations.
-
•
Unspecific staining: add a blocking step prior to antibody labeling. As for immunofluorescence, adapt the blocking buffer to specific antibody requirements.
-
•
Faint staining in one color: Increase antibody concentration.
-
•
No staining in one color: Ensure to use a fluorophore that is stable in water. For example, Rhodamine loses fluorescence properties in the water, while Alexa fluorophores are stable.
-
•
Loss of fluorescence over days: this is due to epitope destabilization and unbinding in the gel. Make sure to store gels in PBS at 4°C between imaging sessions.
Troubleshooting 7: mounting problems & solutions
We list potential mounting problems, from minor to major:
-
•
The gel is gliding on the PDL-coverslip/The sample is drifting under the microscope: Make sure to dry the gel enough before adhering it to the coverslip, by wiping it on a Kimwipe until no water marks the tissue. Alternatively, prep new fresh PDL-coated coverslips.
-
•
There is a bubble between the gel and the coverslip: press very gently on the gel with a spatula, moving the bubble to one side of the gel.
-
•
The gel is mounted on the wrong side: add plenty of water to detach the gel from the coverslip, very gently. Proceed to mount again. In general, always confirm gel orientation on a non-coated coverslip before proceeding to final mounting.
-
•
Fluorescence rapidly increases during the imaging: this means the gel is drying and currently shrinking. Stop the imaging, add 2–3 drops of water, let the gel stabilize for 5 min and image again.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Julien Vermot (j.vermot@imperial.ac.uk).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate new datasets or codes.
Acknowledgments
We thank our collaborators for their feedback on the protocol. E.S. is funded by the Marie Sklodowska Curie Action Program (H2020- MSCA-IF-2020, GA101028893). J.V. is funded by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (GA682938).
Author contributions
Conceptualization, E.S., J.V.; Investigation, E.S.; Writing, E.S., C.V.-P., J.V.; Review & Editing, E.S., C.V.-P., J.V.; Funding Acquisition, E.S., J.V.; Supervision, J.V.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2023.102257.
Contributor Information
Emmanuelle Steib, Email: e.steib@imperial.ac.uk.
Julien Vermot, Email: j.vermot@imperial.ac.uk.
References
- 1.Steib E., Tetley R., Laine R.F., Norris D.P., Mao Y., Vermot J. TissUExM enables quantitative ultrastructural analysis in whole vertebrate embryos by expansion microscopy. Cell Rep. Methods. 2022;2 doi: 10.1016/j.crmeth.2022.100311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gambarotto D., Hamel V., Guichard P. In: Methods in Cell Biology Expansion Microscopy for Cell Biology. Guichard P., Hamel V., editors. Academic Press; 2021. Chapter 4 - ultrastructure expansion microscopy (U-ExM) pp. 57–81. [DOI] [PubMed] [Google Scholar]
- 3.Gambarotto D., Zwettler F.U., Le Guennec M., Schmidt-Cernohorska M., Fortun D., Borgers S., Heine J., Schloetel J.-G., Reuss M., Unser M., et al. Imaging cellular ultrastructures using expansion microscopy (U-ExM) Nat. Methods. 2019;16:71–74. doi: 10.1038/s41592-018-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim S.-Y., Cho J.H., Murray E., Bakh N., Choi H., Ohn K., Ruelas L., Hubbert A., McCue M., Vassallo S.L., et al. Stochastic electrotransport selectively enhances the transport of highly electromobile molecules. Proc. Natl. Acad. Sci. USA. 2015;112:E6274–E6283. doi: 10.1073/pnas.1510133112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ku T., Swaney J., Park J.-Y., Albanese A., Murray E., Cho J.H., Park Y.-G., Mangena V., Chen J., Chung K. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat. Biotechnol. 2016;34:973–981. doi: 10.1038/nbt.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate new datasets or codes.