Abstract
Neonatal Escherichia coli meningitis remains a devastating disease, with unacceptably high morbidity and mortality despite advances in supportive care measures and bactericidal antibiotics. To further our ability to improve the outcome of affected neonates, a better understanding of the pathogenesis of the disease is necessary. To identify potential bacterial genes which contribute to E. coli invasion of the blood-brain barrier, a cerebrospinal fluid isolate of E. coli K1 was mutagenized with TnphoA. TnphoA mutant 27A-6 was found to have a significantly decreased ability to invade brain microvascular endothelial cells compared to the wild type. In vivo, 32% of the animals infected with mutant 27A-6 developed meningitis, compared to 82% of those infected with the parent strain, despite similar levels of bacteremia. The DNA flanking the TnphoA insertion in 27A-6 was cloned and sequenced and determined to be homologous to E. coli K-12 aslA (arylsulfatase-like gene). The deduced amino acid sequence of the E. coli K1 aslA gene product shows homology to a well-characterized arylsulfatase family of enzymes found in eukaryotes, as well as prokaryotes. Two additional aslA mutants were constructed by targeted gene disruption and internal gene deletion. Both of these mutants demonstrated decreased invasion phenotypes, similar to that of TnphoA mutant 27A-6. Complementation of the decreased-invasion phenotypes of these mutants was achieved when aslA was supplied in trans. This is the first demonstration that this locus contributes to invasion of the blood-brain barrier by E. coli K1.
K1 encapsulated Escherichia coli bacteria are responsible for the majority of cases of gram-negative meningitis among neonates. Mortality from this disease remains substantial, despite improvements in antibiotics and supportive care. In addition, the neurologic morbidity of surviving infants is approximately 50%. It is clear from these data that this disease continues to offer considerable clinical challenges for improved mortality and morbidity, and a more complete understanding of its pathogenesis is necessary if there is to be improved patient outcome.
From animal studies, as well as studies of infants with E. coli meningitis, it appears that a high level of bacteremia is closely associated with the development of meningitis (14, 22). While the achievement of this level of bacteremia is necessary for subsequent bacterial invasion of the central nervous system, it in itself is not sufficient for the bacteria to cross the blood-brain barrier; successful invasion of brain microvascular endothelial cells (BMEC), which constitute the blood-brain barrier, has been shown to require multiple bacterial determinants. The availability of BMEC and a neonatal rat model of hematogenous meningitis is integral to our ability to dissect the pathogenesis of this disease and relate our in vitro data to relevant biological systems.
Previous studies have identified factors which mediate complex and multifactorial interactions between bacterial gene products and BMEC for E. coli K1 traversal of the blood-brain barrier, including S fimbriae (34, 45), outer membrane protein A (OmpA) (36, 37), and the K1 capsule (18, 22). TnphoA mutagenesis has identified several genes (ibeA, ibeB, and yijP) which contribute to E. coli K1 invasion of the blood-brain barrier in vitro and in vivo (19, 20, 48).
Recent studies demonstrate that ibeA encodes a protein with multiple transmembrane domains and binds to a novel binding molecule-receptor on BMEC, Ibe10R (35). The addition of purified Ibe10R protein, or a polyclonal antibody raised against Ibe10R, to in vitro invasion assays significantly inhibits invasion by E. coli K1 (35). yijP encodes a putative membrane protein which is a homologue of the gene product termed L7028 or ecf3, encoded on virulence plasmid p0157 of E. coli O157:H7 (48). Additional work suggests that not all meningitis-causing E. coli K1 bacteria possess all of these identified virulence factors (8). Therefore, despite the body of work, the invasion process has yet to be fully elucidated.
In a previous communication, we reported the isolation of several E. coli K1 TnphoA mutants that were deficient in the ability to invade BMEC (19). For this study, we further characterized one mutant, 27A-6.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Strains used in this study were derived from E44, a spontaneous rifampin-resistant mutant of a cerebrospinal fluid (CSF) isolate of K1-encapsulated E. coli RS218 (O18:K1:H7) which has been previously characterized (19, 49). 27A-6 is a single-insertion TnphoA mutant of E44. JH3 is an E44 aslA gene disruption mutant. JH6 is an E44 aslA internal gene deletion mutant, and HB101 is a laboratory E. coli K-12 strain (Table 1). For invasion assays, bacteria were grown under microaerophilic conditions (3) for 14 to 16 h at 37°C in brain heart infusion broth (Difco Laboratories, Detroit, Mich.) under rifampin (100 μg/ml), kanamycin (50 μg/ml), chloramphenicol (25 μg/ml) and/or ampicillin (100 μg/ml) selection, as needed. All of the mutant and wild-type strains grown to stationary phase attained similar cell densities.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| E44 | Rifampin-resistant RS218 (O18:K1:H7) | 19 |
| 27A-6 | E44 (aslA::TnphoA) | 19 |
| JH3 | E44 (aslA::pEP185.2) | This study |
| JH6 | E44 (ΔaslA) | This study |
| HB101 | E. coli K-12 | 2 |
| Plasmids | ||
| p7B8 | pSupercos with 20- to 40-kb insert containing aslA | This study |
| p5C10 | pTM100 with 7-kb insert containing aslA | This study |
| pJAH44 | aslA PCR product cloned in pCR2.1 | This study |
| pSupercos | Cloning vector, Apr | Stratagene |
| pTM100 | Cloning vector, pACYC derivative, Cmr Tetr | 4 |
| pCR2.1 | Cloning vector, Apr | Invitrogen |
Cloning and sequencing of DNA adjacent to the TnphoA insertion.
Genomic DNA from mutant 27A-6 was digested with MluI, which does not cut TnphoA, and was ligated into the MluI site of pCVD433 as previously described (19). The constructs were electroporated into DH5α, and transformants were selected for kanamycin resistance, indicating the presence of Tn5. Sequence data of the DNA flanking the transposon were obtained by using oligonucleotide primers Tnp5 (5′-GCACGATGAAGAGCAGAAG-3′) and Tnp3 (5′-TGCTAAGAGAATCACGCAGAG-3′), which are complementary to the 5′ and 3′ ends, respectively of TnphoA. The remaining sequence information was obtained by primer walking of a plasmid that contained the wild-type aslA sequence. Sequencing was determined by the dideoxy-chain termination method described by Sanger et al. (39) with Sequenase kit version 2.0 from U.S. Biochemicals or using an Applied Biosystems (Foster City, Calif.) automated sequencer at the Childrens Hospital Los Angeles Research Institute Biotechnology Core Facility. Sequences obtained were analyzed using the BLAST program (National Center for Biotechnology Information at the National Library of Medicine). Both strands of the entire E. coli K1 aslA locus were sequenced.
Gene disruption mutant.
JH3 was constructed by targeted disruption of the aslA gene via suicide plasmid integration into the wild-type E. coli K1 locus. An internal fragment of aslA was cloned by PCR using oligonucleotide primers based on the E. coli K-12 aslA sequence with SacI and KpnI restriction sites incorporated at the 5′ and 3′ ends of the primers, respectively. Primer sequences were as follows: ORF2, 5′-CGGTGTCTGATATGTACACCG-3′; ORF1, 5′-CATCCCTTTCCAGTAACG-3′. These primers anneal at bp 905 and 1453 of aslA, respectively. The internal open reading frame (ORF) fragment was digested with SacI and KpnI and ligated into suicide vector pEP185.2 (23). The ligation mixtures were transformed into S17-1λ pir (43), and the resulting plasmids were transferred into wild-type E. coli K1 strain E44 by conjugation. Integration of the suicide plasmid via homologous recombination at the site of the cloned internal ORF fragment resulted in a gene disruption. Transconjugants were selected on rifampin and chloramphenicol. Gene disruption was confirmed by Southern blot analysis.
Deletion mutant.
To construct an aslA deletion mutant (JH6), gene splicing by the overlap extension method was used (17). PCR product 1 was produced from oligonucleotide primers aslA-S (5′-GATATCCGCTCTGGGTATAG-3′), containing a SacI tag annealing to bp 1733, and aslA-SΔ (5′-CGGTCAGCTTGATAACACGC-3′), annealing at bp 1299 and containing overlap sequence 5′-GCCGCCCGCGCTCTG-3′. PCR product 2 was produced from oligonucleotide primers aslA-K (5′-CTGCCTTACATATCAAC-3′), annealing to bp 80 and containing a KpnI tag, and aslA-KΔ (5′-CACATCGTCCAGCAAG-3′), annealing at bp 543 and containing overlap sequence 5′-CAGAGCGCGGGCGGC-3′. For PCR product 3, we used PCR products 1 and 2, which anneal at their overlap sequences, as a template and primers aslA-S and aslA-K to produce a 900-bp PCR product, resulting in an approximately 750-bp deletion (bp 543 to 1299) in aslA. PCR product 3 was digested with SacI and KpnI and ligated into suicide vector pEP185.2 (23). The ligation mixtures were transformed into S17-1λ pir (43), and the resulting plasmids were transferred into wild-type E. coli K1 strain E44 by conjugation. Initial transconjugants were selected on rifampin and chloramphenicol, indicating integration of the suicide plasmid via homologous recombination. Colonies were then subject to cycloserine enrichment to induce plasmid loss (32). Colonies that had lost the plasmid and successfully integrated the deletion-containing aslA gene (double-crossover event) were identified by chloramphenicol sensitivity and confirmed by PCR and Southern analysis.
Phenotypic characterization of mutants.
Mutants were examined for retention of important phenotypic characteristics of the wild-type strain, including growth rates, biochemistries, antibiograms, and the presence of the K1 capsule, O18 lipopolysaccharide (LPS), S fimbriae, and OmpA. Growth curves were determined for mutants and the wild type in experimental medium containing the appropriate antibiotics. The presence of the K1 capsule was verified by the antiserum agar technique with antiserum to group B meningococci as previously described (19, 22). O18 LPS was detected by colony blot analysis with an anti-O18 monoclonal antibody (19). S fimbriae were detected by mannose-sensitive and mannose-resistant hemagglutination tests, as well as by colony blot assays with anti-type 1 fimbrial antibody and anti-S fimbrial monoclonal antibodies (19, 34). Members of the outer membrane complex were isolated by sonication and selective detergent solubilization, and their patterns were examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (TnphoA mutant 27A-6) (19). OmpA was detected by colony blot assay and Western blotting of Omp complex members with an anti-OmpA antibody. TnphoA mutant 27A-6 was examined for insertions in ORFs of some known virulence factors by Southern hybridization with probes for K1 and OmpA as previously described (19).
Arylsulfatase activity assay.
To detect arylsulfatase activity, indoxysulfate indicator agar plates were prepared by combining 3 mM tyramine, 0.25 mg of indoxysulfate per ml, 0.25% succinate (E. coli) or xylose (Klebsiella pneumoniae), 1 mM MgSO4, 0.00005% vitamin B, 0.1% Casamino Acids, and 5 μg of nicotine per ml in M63 minimal-medium agar (29). Bacteria were plated for isolated colonies. Arylsulfatase activity was indicated by blue colonies. The positive and negative controls used were K. pneumoniae and E. coli K-12 strain HB101, respectively.
Cloning of wild-type loci for complementation.
A custom cosmid library was constructed using wild-type E44 Sau3AI-digested DNA fragments ligated into the BamHI site of the Supercos 1 cosmid vector (Stratagene Cloning Systems, La Jolla, Calif.). Insert sizes ranged from 20 to 40 kb. Transformants were subjected to colony hybridization using a PCR-derived internal fragment of aslA as a probe. Colony hybridization was performed on approximately 1,200 clones. Seven strongly hybridizing colonies were recovered and used for complementation analysis. One clone, p7B8, was found to consistently complement the decreased-invasion mutants and was chosen for further characterization. p7B8 was digested with Sau3AI, and the resulting subgenomic fragments were ligated into the BamHI site of pTM100. Transformants were probed with the probe described above. One hybridizing subclone, p5C10, was identified and determined to complement the mutant phenotypes.
The wild-type E44 aslA ORF was cloned using primers aslA4 (5′-CATCTACCGCCATATCTTAAAGCAATGGCTG-3′) and aslA3 (5′-CAGGTCTGTATGACAACAAG-3′), which anneal at bp 2007 of E. coli K1 aslA and bp 39713 of the E. coli genomic sequence of the region between 84.5 and 86.5 min (accession no. M87049). The PCR product was ligated into vector pCR2.1-topo, resulting in plasmid pJAH44. DNA inserts were confirmed by sequencing. All plasmids were transferred to E44-derived strains by electroporation.
mRNA slot blot analysis.
Total cellular RNA was purified from stationary-phase bacteria grown in brain heart infusion broth using the TRIzol reagent (Gibco BRL). RNA samples were treated with RNase-free DNase, and the integrity of each sample was assessed by examination of an electrophoresed aliquot. We applied 5.0 and 0.5 μg of purified RNA to nylon membranes (Pro-Nytran; Schleicher & Schuell) using a slot blot apparatus (Bio-Rad). Membranes were prepared and subjected to hybridization as previously described (3). Membranes were probed with the internal ORF DNA fragment of aslA. In addition, a 526-bp internal fragment of the E. coli 23S rRNA gene, generated by PCR using oligonucleotide primers 23s1 (5′-GACTAAGCGTACACGGTG-3′) and 23s2 (5′-GCCTTGTACGTACACGG-3′), was used to confirm equivalent RNA loading.
HBMEC invasion phenotypes.
Tissue culture invasion assays were performed with human BMEC (HBMEC). These cells were isolated and cultured as described previously (45). Approximately 107 bacteria were added to confluent HBMEC monolayers and 500 μl of experimental medium (Ham's F12: Medium 199–1× Earle salts [1:1], 5% heat-inactivated fetal bovine serum, 1% Na pyruvate, 0.5% glutamine) (Irvine Scientific) for a multiplicity of infection of 100. The cells and bacteria were incubated for 1.5 h at 37°C in 5% CO2 without shaking. The monolayers were then washed with RPMI medium and reincubated with experimental medium containing gentamicin (100 μg/ml) for 1 h at 37°C to kill extracellular bacteria. This step was omitted in experiments performed to determine the total number of cell-associated bacteria. To determine the number of viable intracellular bacteria, the monolayers were washed with RPMI medium and lysed with 150 μl of sterile water for 20 min. The number of CFU in each well was determined by dilutions plated on Luria-Bertani agar with antibiotic (mutants) or on blood agar plates (wild type). Invasion frequency was calculated as follows: number of recovered bacteria/number of bacteria inoculated × 100. Wild-type E44 had an invasion frequency of 0.1 to 0.8%. HB101, a noninvasive E. coli K-12 strain that was used as a negative control, had an invasion frequency of 0.001% ± 0.007%. All invasion experiments were conducted in duplicate and performed a minimum of three times.
Animal model of E. coli bacteremia and meningitis.
E. coli bacteremia and meningitis, defined as a positive CSF culture, were induced in 5-day-old rats by a previously described method (36). Briefly, outbred, specific-pathogen-free, pregnant Sprague-Dawley rats with timed conception were purchased from Charles River Breeding Laboratories (Wilmington, Mass.); the rats delivered in our vivarium 5 to 7 days after arrival. At 5 days of age, all members of each litter were randomly assigned to one of two groups, which received intracardiac injections of E. coli strain E44 or 27A-6. Pilot experiments were performed with each bacterial strain to determine the inoculum size that would induce a level of bacteremia (105 to 108 CFU/ml of blood) found to be necessary for hematogenous bacteria to enter the central nervous system (22). Inoculum sizes were as follows for two experiments: E44, 5.5 × 106 and 6.5 × 106 CFU; 27A-6, 1 × 107 and 8 × 106 CFU. At 1 to 2 h after bacterial inoculation, blood and CSF specimens were obtained as described previously for quantitative cultures (36).
Nucleotide sequence accession number.
The sequence of aslA from E. coli K1 strain RS218 has been submitted to the GenBank database under accession number AF263456.
RESULTS
In vitro and in vivo phenotype of TnphoA mutant 27A-6.
As previously reported, E44 was subjected to TnphoA mutagenesis to identify mutants with a decreased ability to invade the blood-brain barrier in vitro (19). Despite having the same multiplicity of infection, mutant 27A-6 was determined to be significantly less invasive in HBMEC than parent strain E44 (clinical CSF isolate), 0.026% ± 0.011% versus 0.40% ± 0.08% invasion, respectively (P < 0.01) (Table 2). Total cell-associated counts of mutant and wild-type E. coli K1 bacteria were similar (data not shown). This suggests that the mutation leading to the decreased-invasion phenotype is not the result of decreased association with HBMEC. In addition, 27A-6 was tested in the neonatal rat meningitis model to determine the percentage of animals developing meningitis, in animals achieving similar levels of bacteremia (E44, 7.50 ± 0.32 mean log CFU/ml of blood; 27A-6, 7.60 ± 0.49 mean log CFU/ml of blood). Meningitis, defined as a positive CSF culture, was observed in 82% (14 of 17) of the animals infected with wild-type strain E44 but in only 32% (7 of 22) of those infected with mutant strain 27A-6 (P < 0.001). These data suggest that the locus disrupted by TnphoA in mutant 27A-6 contributes to penetration of the blood-brain barrier in vitro and in vivo.
TABLE 2.
In vitro HBMEC invasion phenotype of wild-type and mutant E. coli strains
| Strain | Mean invasion frequency (%)a ± SD |
|---|---|
| E44 | 0.40 ± 0.08 |
| HB101 | 0.003 ± 0.004 |
| 27A-6 | 0.026 ± 0.011 |
| JH3 | 0.014 ± 0.020 |
| JH6 | 0.025 ± 0.014 |
All invasion experiments were conducted in duplicate and performed a minimum of three times.
Characterization of transposon mutant 27A-6.
An extensive investigation was undertaken to examine mutant 27A-6 to determine if the expression of known virulence factors was altered. Phenotypic studies of the mutant revealed that there were no alterations in the K1 capsule, O18 LPS, type 1 and S fimbriae, outer membrane protein profile, and OmpA as determined by methods previously described (19, 22, 34). In addition, the biochemistries, antibiograms, and growth curves were identical to those of the wild-type strain (data not shown).
Identification and characterization of gene locus aslA.
To determine the genetic basis for the observed decreased-invasion phenotype of mutant 27A-6, we cloned the DNA region flanking the TnphoA insertion site. A 500-bp segment of flanking DNA was cloned and sequenced and found to be homologous (94% nucleotide and 97% amino acid homology) to E. coli K-12 aslA, a putative arylsulfatase-like gene. This E. coli K-12 gene was named because the deduced amino acid sequence contains sulfatase consensus motifs I and II, which are homologous (55 and 70% amino acid identity, respectively) to those of K. pneumoniae (previously named K. aerogenes) AtsA, an arylsulfatase which is involved in sulfate metabolism. No definitive gene product or function has been identified for E. coli K-12 aslA (30, 40).
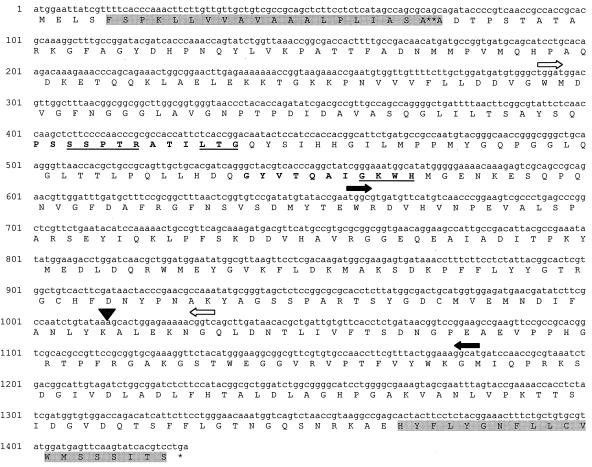
The entire E. coli K1 aslA gene was cloned and sequenced. Similar to E. coli K-12, aslA in E. coli K1 was found adjacent to and divergently oriented with respect to aslB, which encodes a putative regulatory protein. Figure 1 depicts the nucleotide and predicted amino acid sequences of aslA, along with the several important features of its predicted protein structure. The TnphoA insertion site was determined to be at the 3′ end of aslA, at bp 1283. aslA encodes a polypeptide of 475 amino acids with a predicted molecular mass of 52 kDa (10). Computer analysis predicts that AslA contains two membrane-spanning segments (24) and an amino-terminal signal sequence with a putative cleavage site between amino acids 24 and 25 (31). As in K. pneumoniae AtsA and the predicted amino acid sequence of E. coli K-12 AslA, two sulfatase motifs, designated I and II, were identified in the deduced amino acid sequence (7). Both are located in the N terminus of the protein (Fig. 1). Sulfatase motif I, containing highly conserved amino acid residues (S/CXPXRXXXXLTGX), and motif II (GKWH) have been shown to be essential for the catalytic activity of the enzyme (38, 46, 47).
FIG. 1.
Nucleotide and deduced amino acid sequences of the E. coli K1 aslA locus. ▾, TnphoA insertion site; ∗∗, cleavage site for putative signal sequence. Boldface amino acid sequences denote sulfatase motifs I and II, and important amino acid residues are underlined. Putative transmembrane domains are shaded. ➧ , disruption of JH3; ➭ , deletion of JH6.
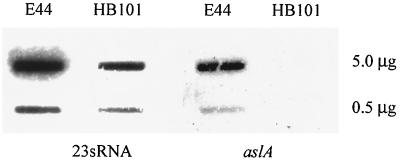
Wild-type E. coli K1 and E. coli K-12 were examined for in vitro arylsulfatase activity by using indoxysulfate indicator plates, and there was no detectable activity for either E. coli K1 or E. coli K-12, compared to a K. pneumoniae positive control (data not shown). Slot blot assays were performed on RNA isolated from wild-type E. coli K1 and E. coli K-12, and the blots were probed with an internal DNA fragment of E. coli K1 aslA. As shown in Fig. 2, the aslA transcript was detectable in E. coli K1 but not in E. coli K-12 strain HB101. These data suggest that although aslA is expressed in E. coli K1 but not in E. coli K-12, no arylsulfatase activity is detectable by this method in vitro.
FIG. 2.
RNA slot blot analysis of aslA transcript levels. mRNA was isolated from strains E44 (K1) and HB101 (K-12) (5.0 and 0.5 μg in the top and bottom slots, respectively) and probed with an internal fragment of aslA. Transcript levels for a constitutive expressing gene, 23sRNA, were determined in parallel to confirm equivalent loading.
Complementation of transposon mutant 27A-6.
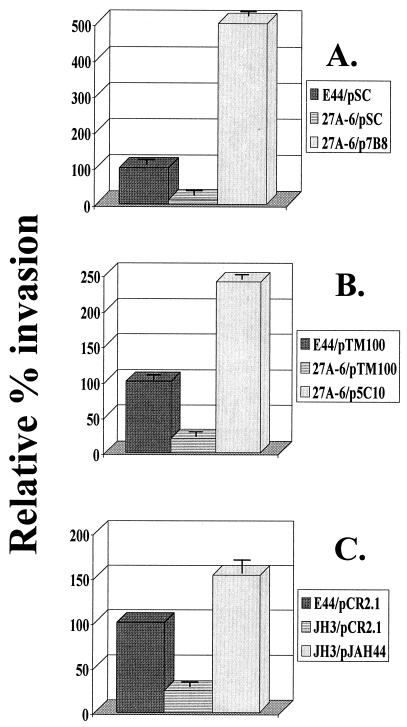
An important step in the investigation of the genetic basis of a mutant phenotype is demonstration of the ability of wild-type DNA supplied in trans to restore the wild-type phenotype. To approach this goal, we screened a wild-type E44 cosmid genomic library using the TnphoA chromosomal junction clone as a probe. A hybridizing clone (p7B8) was identified and assayed for the ability to complement 27A-6's mutant invasion phenotype. p7B8 was found to restore the invasiveness phenotype to mutant 27A-6 by raising its invasion frequency fivefold higher than that of E44 containing a cloning vector (Fig. 3A).
FIG. 3.
Complementation of HBMEC invasion phenotype of aslA mutant strains with wild-type DNA supplied in trans. (A) Complementation of TnphoA mutant 27A-6 with cosmid clone p7B8. (B) Complementation of TnphoA mutant 27A-6 with subclone p5C10. (C) Complementation of aslA disruption mutant JH3 with pJAH44 (aslA ORF). Results are presented as relative percent invasion, with that of strain E44 containing the respective cloning vector set at 100%. The data in panels A and B are from a single experiment conducted in duplicate and are representative of numerous experiments performed with similar results. The experiments whose results are shown in panel C were conducted in duplicate and performed three times. The error bars represent the standard errors of the means.
Because the complementing cosmid clone contains a large DNA insert of 20 to 40 kb, we endeavored to define a smaller subclone of DNA that would confer the wild-type invasiveness phenotype on the mutant. Subcloning of p7B8 and complementation analysis of resultant clones identified a subclone (p5C10) with a 7-kb insert containing aslA. Similar to p7B8, p5C10 was found to raise the transposon mutant invasion frequency to a level 2.4-fold above that of the wild-type strain containing the cloning vector (Fig. 3B).
Characterization of an aslA gene disruption mutant and a gene deletion mutant.
To exclude the possibility that an unidentified second-site mutation and/or a polar effect was responsible for the observed decreased-invasion phenotype of transposon mutant 27A-6, an additional aslA gene disruption mutant was generated. As shown in Table 2, this mutant, JH3, demonstrated a phenotype like that of 27A-6 as measured by the in vitro HBMEC invasion assay; JH3 was approximately 30-fold less invasive than E44. In a complementary approach to confirm the genetic basis for the decreased-invasion mutant, we generated an internal deletion mutant. This mutant, JH6, has a 750-bp (50%) deletion of the internal region of aslA. JH6 was found to be approximately 16-fold less invasive than wild-type E44 (Table 2), supporting the importance of this gene locus for in vitro invasion of HBMEC. Phenotypic studies of JH3 and JH6 revealed that there were no alterations in the K1 capsule, O18 LPS, type 1 and S fimbriae, and OmpA as determined by methods previously described (19, 22, 34). In addition, the biochemistries, antibiograms, and growth curves were identical to those of the wild-type strain (data not shown).
The decreased-invasion phenotype of JH6 was complemented by cosmid p7B8 and subclone p5C10 (data not shown). In addition, the decreased-invasion phenotype of JH3 was complemented by cosmid clone p7B8 (data not shown). Concomitantly, a 2-kb clone containing aslA alone (pJAH44) was constructed. pJAH44 was found to complement the decreased-invasion phenotype of mutant JH3 by fully restoring the mutant invasion frequency to that of the wild type containing the cloning vector (Fig. 3C).
DISCUSSION
We have previously identified multiple bacterial and environmental factors which contribute to E. coli K1's ability to cross the blood-brain barrier in vitro and in vivo, including the K1 capsule (18, 22), S fimbriae (34, 45), OmpA (36, 37), IbeA (19), IbeB (20), YijP (48), the finP traJ locus (5), environmental growth conditions (3), and the level of bacteremia (22). This process appears to be mediated by complex interactions between multiple bacterial, host, and environmental determinants.
In this report, we present data on E. coli K1 aslA and its contributions to the invasion equation. Our earlier communication on TnphoA mutagenesis identified several mutant strains containing TnphoA insertions in ibeA, ibeB, and yijP with decreased HBMEC invasion phenotypes compared to that of parental wild-type strain E44 (19, 20, 48). One such mutant, 27A-6, was further characterized in this study. 27A-6 was shown to have significantly decreased HBMEC invasion ability in vitro and an attenuated ability to cause meningitis in newborn rats compared to the wild type. Two additional aslA mutants, an aslA disruption mutant (JH3) and an aslA internal deletion mutant (JH6), also demonstrated significantly decreased invasion phenotypes. We were able to complement the decreased HBMEC invasion phenotypes of all of the mutant strains with constructs containing the aslA gene locus. These data, taken together, strongly suggest that aslA plays a role in E. coli K1 invasion of the blood-brain barrier.
We do not know how AslA contributes to E. coli K1 invasion. The E. coli K-12 gene was named because of the sequence homology of the deduced protein sequence to AtsA, a K. pneumoniae arylsulfatase. However, aslA does not appear to be expressed in noninvasive E. coli K-12, as determined by measurement of RNA transcript levels. Furthermore, not unexpectedly, and in fitting with our hypothesis of the multifactorial nature of the invasion process, a plasmid containing E. coli K1 aslA did not confer invasiveness on noninvasive E. coli K-12 strain DH5α (data not shown). The E. coli K1 sequence is highly homologous to the E. coli K-12 sequence (94% nucleotide and 97% amino acid homology), but in contrast to E. coli K-12, we were able to detect the aslA transcript in E. coli K1. Analysis of the deduced protein sequence indicates that E. coli K1 AslA is also a member of the arylsulfatase family of enzymes that contain highly conserved primary amino acid sequence motifs and the predicted three-dimensional structure (13, 16, 33, 47, 50). These enzymes, which are necessary for the cleavage of sulfate esters, are found in humans, lower eukaryotes, and prokaryotes (11, 29, 44). In bacteria, these genes are expressed under conditions of sulfur starvation, where the enzymes function in sulfate scavenging from exogenous substrates (15, 46). The genes are regulated, repressed or derepressed, by the availability of preferred sources of inorganic sulfate or sulfur (1, 28, 30, 51) and by posttranslation modification (12, 13, 27, 41, 42, 46, 47). We were not able to detect arylsulfatase activity in E. coli K1 by the indoxylsulfate method. Studies are in progress to determine whether or not E. coli K1 AslA has sulfatase activity.
Sulfatases of all origins share a unique amino acid residue, α-formylglycine, which is the result of posttranslational modification of a cysteine in eukaryotes and some prokaryotes, for example, Pseudomonas aeruginosa and Mycobacterium tuberculosis (6, 12, 13, 40, 41, 42), or a serine in K. pneumoniae and E. coli (27, 40, 46). The α-formylglycine, which is contained within motif I, and downstream motif II (GKWH) are located in the catalytic site of the enzyme (9, 25, 38, 46, 47).
There are several similarities between E. coli K1 AslA and K. pneumoniae AtsA which further strengthen the hypothesis that AslA in E. coli K1 is a bacterial arylsulfatase. For example, both contain a serine instead of a cysteine, as do most other known sulfatases, in the conserved amino acid sequence described above, which has been shown to be modified to α-formylglycine in K. pneumoniae (27, 40). When AtsA is overexpressed in E. coli K-12 via an expression vector, a catalytically active enzyme containing the α-formylglycine residue is produced, suggesting that E. coli can modify serine residues to α-formylglycine. Both K. pneumoniae AtsA and E. coli K1 AslA contain putative signal sequences and transmembrane regions, suggesting a periplasmic or membrane cellular location (40). Additionally, both are found in a gene cluster with a putative regulator gene, B. In K. pneumoniae, atsB has recently been found to be required for posttranslational modification of the serine residue and is now called arylsulfatase-activating protein (40, 46). E. coli K1 aslA is found in a cluster with aslB, which has been sequenced (data not shown) and is an atsB homologue.
In summary, our data support the idea that AslA plays a role in the penetration of the blood-brain barrier by E. coli K1 in vitro and in vivo. Experiments are in progress to further elucidate the mechanism by which AslA contributes to HBMEC invasion and the potential interactions between AslA and AslB.
ACKNOWLEDGMENTS
This work was supported by The Infectious Diseases Society of America Procter and Gamble Young Investigator Award in Bacterial Infections, The American Heart Association Clinician-Scientist Award (1122-CS), and U.S. Public Health Service grants K08-AI01457-01 (to J.A.H.) and R01 NS 26310 (to K.S.K.).
We thank Carol A. Wass for excellent technical assistance.
REFERENCES
- 1.Adachi R, Murooka Y, Harada T. Derepression of arylsulfatase synthesis in Aerobacter aerogenesby tyramine. J Bacteriol. 1973;116:19–24. doi: 10.1128/jb.116.1.19-24.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachmann B J. Pedigrees of some mutant strains of Escherichia coliK-12. Bacteriol Rev. 1972;36:525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Badger J L, Kim K S. Environmental growth conditions influence the ability of Escherichia coliK1 to invade brain microvascular endothelial cells and confer serum resistance. Infect Immun. 1998;66:5692–5697. doi: 10.1128/iai.66.12.5692-5697.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badger J L, Miller V L. Expression of invasin and motility are coordinately regulated in Yersinia enterocolitica. J Bacteriol. 1998;180:793–800. doi: 10.1128/jb.180.4.793-800.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Badger J L, Wass C A, Kim K S. Identification of Escherichia coliK1 genes contributing to human brain microvascular endothelial cell (HBMEC) invasion by differential fluorescence induction (DFI) Mol Microbiol. 2000;36:174–182. doi: 10.1046/j.1365-2958.2000.01840.x. [DOI] [PubMed] [Google Scholar]
- 6.Beil S, Kehrli H, James P, Staudenmann W, Cook A M, Leisinger T, Kertesz M A. Purification and characterization of the arylsulfatase synthesized by Pseudomonas aerogenosa PAO during growth in sulfate-free medium and cloning of the arylsulfatase gene (atsA) Eur J Biochem. 1995;229:385–394. doi: 10.1111/j.1432-1033.1995.0385k.x. [DOI] [PubMed] [Google Scholar]
- 7.Biaroch A, Bucher P, Hofmann K. The PROSITE database, its status in 1997. Nucleic Acids Res. 1997;25:217–221. doi: 10.1093/nar/25.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bingen E, Bonacorsi S, Brahimi N, Denamur E, Elion J. Virulence patterns of Escherichia coliK1 strains associated with neonatal meningitis. J Clin Microbiol. 1997;35:2981–2982. doi: 10.1128/jcm.35.11.2981-2982.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bond C S, Clements P R, Ashby S J, Collyer C A, Harrop S J, Hopwood Guss J M. Structure of a human lysosomal sulfatase. Structure. 1997;5:277–289. doi: 10.1016/s0969-2126(97)00185-8. [DOI] [PubMed] [Google Scholar]
- 10.Combet C, Blanchet C, Geourjon C, Deléage G. Network protein sequence analysis. Trends Biochem Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 11.de Hostos E L, Schilling J, Grossman A R. Structure and expression of the gene encoding the periplasmic arylsulfatase of Chlamydomonas reinhardtii. Mol Gen Genet. 1989;218:229–239. doi: 10.1007/BF00331273. [DOI] [PubMed] [Google Scholar]
- 12.Dierks R, Miech C, Hummerjohann J, Bernhard S, Kertesz M A, von Figura K. Posttranslational formation of formylglycine in prokaryotic sulfatases by modification of either cysteine or serine. J Biol Chem. 1998;273:25560–25564. doi: 10.1074/jbc.273.40.25560. [DOI] [PubMed] [Google Scholar]
- 13.Dierks R, Lecca M R, Schlotterhose P, Schmidt B, von Figura K. Sequence determinants directing conversion of cysteine to formylglycine in eukaryotic sulfatases. EMBO J. 1999;18:2084–2091. doi: 10.1093/emboj/18.8.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dietzman D E, Fischer G W, Schoenknecht F D. Neonatal Escherichia coli septicemia—bacterial counts in blood. J Pediatr. 1974;85:128–130. doi: 10.1016/s0022-3476(74)80308-2. [DOI] [PubMed] [Google Scholar]
- 15.Dogeson K S, White G F, Fitzgerald J W. Sulfatases of microbial origin. Boca Raton, Fla: CRC Press, Inc.; 1982. [Google Scholar]
- 16.Franco B, Germana M, Giancarlo P, Levilliers J, Bernard L, Gebbia M, Cox L, Maroteaux P, Sheffield L, Rappold G A, Andria G, Petit C, Ballabio A. A cluster of sulfatase genes on Xp22.3: mutations in chondrodysplasia punctata (CDPX) and implications for warfarin embryopathy. Cell. 1995;81:15–25. doi: 10.1016/0092-8674(95)90367-4. [DOI] [PubMed] [Google Scholar]
- 17.Ho S N, Junt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 18.Hoffman J A, Wass C, Stins M F, Kim K S. The capsule supports survival but not traversal of Escherichia coliK1 across the blood-brain barrier. Infect Immun. 1999;67:3566–3570. doi: 10.1128/iai.67.7.3566-3570.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang S-H, Wass C, Fu Q I, Prasadarao N V, Stins M, Kim K S. Escherichia coli invasion of brain microvascular endothelial cells in vitro and in vivo: molecular cloning and characterization of invasion gene ibe10. Infect Immun. 1995;63:4470–4475. doi: 10.1128/iai.63.11.4470-4475.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang S-H, Chen Y-H, Fu Q I, Stins M, Wang Y, Wass C, Kim K S. Identification and characterization of an Escherichia coli invasion gene locus, ibeB, required for penetration of brain microvascular endothelial cells. Infect Immun. 1999;67:2103–2109. doi: 10.1128/iai.67.5.2103-2109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim K S. Comparison of cefotaxime, imipenem-cilastatin, ampicillin-gentamicin, and ampicillin-chloramphenicol in the treatment experimental Escherichia colibacteremia and meningitis. Antimicrob Agents Chemother. 1985;28:433–436. doi: 10.1128/aac.28.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim K S, Itabashi H, Gemski P, Sadoff J, Warren R L, Cross A S. The K1 capsule is the critical determinant in the development of Escherichia coli meningitis in the rat. J Clin Investig. 1992;90:897–905. doi: 10.1172/JCI115965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kinder S A, Badger J L, Bryant G O, Pepe J C, Miller V L. Cloning of the YenI restriction endonuclease and methyltransase from Yersinia enterocolitica serotype O8 and construction of transformable R-M+mutant. Gene. 1993;136:271–275. doi: 10.1016/0378-1119(93)90478-l. [DOI] [PubMed] [Google Scholar]
- 24.Klein P, Kanehisa M, DeLisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985;815:468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- 25.Lukatela G, Krauss N, Theis K, Selmer T, Gieselmann V, Von Figura K, Saenger W. Crystal structure of human arylsulfatase A: the aldehyde function and the metal ion at the active site suggest a novel mechanism for sulfate ester hydrolysis. Biochemistry. 1998;37:3654–3664. doi: 10.1021/bi9714924. [DOI] [PubMed] [Google Scholar]
- 26.Michiels T, Cornelis G R. Secretion of hybrid proteins by the YersiniaYop export system. J Bacteriol. 1991;173:1677–1685. doi: 10.1128/jb.173.5.1677-1685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miech C, Dierks T, Selmer T, von Figura K, Schmidt B. Arylsulfatase from Klebsiella pneumoniaecarries a formylglycine generated from a serine. J Biol Chem. 1998;273:4835–4837. doi: 10.1074/jbc.273.9.4835. [DOI] [PubMed] [Google Scholar]
- 28.Murooka Y, Higashiura K T, Harada T. Genetic mapping of tyramine oxidass and arylsulfatase genes and their regulation in intergenic hybrids of enteric bacteria. J Bacteriol. 1978;136:714–722. doi: 10.1128/jb.136.2.714-722.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murooka Y, Ishibashi K, Yasumoto M, Sasaki M, Sugino H, Azakami H, Yamashita M. A sulfur- and tyramine-regulated Klebsiella aerogenes operon containing the arylsulfatase (atsA) gene and the atsBgene. J Bacteriol. 1990;172:2131–2140. doi: 10.1128/jb.172.4.2131-2140.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murooka Y, Yamada T, Tanabe S, Harada T. Immunological study of the regulation of cellular arylsulfatase synthesis in Klebsiella aerogenes. J Bacteriol. 1977;132:247–253. doi: 10.1128/jb.132.1.247-253.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 32.Pepe J C, Badger J L, Miller V L. Growth phase and low pH affect the thermal regulation of the Yersinia enterocolitica invgene. Mol Microbiol. 1994;11:123–135. doi: 10.1111/j.1365-2958.1994.tb00295.x. [DOI] [PubMed] [Google Scholar]
- 33.Peters C, Schmidt B, Rommerskirch W, Rupp K, Zühlsdorf M, Vingron M, Meyer H E, Pohlmann R, von Figura K. Phylogenetic conservation of arylsulfatases. cDNA cloning and expression of human arylsulfatase B. J Biol Chem. 1990;265:3374–3381. [PubMed] [Google Scholar]
- 34.Prasadarao N V, Wass C A, Hacker J, Jann K, Kim K S. Adhesion of S-fimbriated Escherichia colito brain glycolipids mediated by sfaA gene-encoded protein of S-fimbriae. J Biol Chem. 1993;268:10356–10363. [PubMed] [Google Scholar]
- 35.Prasadarao N V, Wass C A, Huang S-H, Kim K S. Identification and characterization of a novel Ibe10 binding protein that contributes to Escherichia coliinvasion of brain microvascular endothelial cells. Infect Immun. 1999;67:1131–1138. doi: 10.1128/iai.67.3.1131-1138.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prasadarao N V, Wass C A, Kim K S. Endothelial cell GlcNAcβ1-4GlcNAc epitopes for outer membrane protein A enhance traversal of Escherichia coliacross the blood-brain barrier. Infect Immun. 1996;64:154–160. doi: 10.1128/iai.64.1.154-160.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prasadarao N V, Wass C A, Weiser J N, Stins M F, Huang S-H, Kim K S. Outer membrane protein A of Escherichia colicontributes to invasion of brain microvascular endothelial cells. Infect Immun. 1996;64:146–153. doi: 10.1128/iai.64.1.146-153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Recksiek M, Selmer T, Dierks T, Schmidt B, von Figura K. Sulfatases, trapping of the sulfated enzyme intermediate by substituting the active site formylglycine. J Biol Chem. 1998;273:6096–6103. doi: 10.1074/jbc.273.11.6096. [DOI] [PubMed] [Google Scholar]
- 39.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schirmer A, Kolter R. Computational analysis of bacterial sulfatases and their modifying enzymes. Chem Biol. 1998;5:181–186. doi: 10.1016/s1074-5521(98)90154-5. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt B, Selmer T, Ingendoh A, von Figura K. A novel amino acid modification in sulfatases that is defective in multiple sulfatase deficiency. Cell. 1995;82:271–278. doi: 10.1016/0092-8674(95)90314-3. [DOI] [PubMed] [Google Scholar]
- 42.Selmer T, Hallmann A, Schmidt B, Sumper M, von Figura K. The evolutionary conservation of a novel of a protein modification, the conversion of cysteine to serinesemialdehyde in arylsulfatase from Volvox carteri. Eur J Biochem. 1996;238:341–345. doi: 10.1111/j.1432-1033.1996.0341z.x. [DOI] [PubMed] [Google Scholar]
- 43.Simon M, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 44.Stein C, Gieselmann V, Kreysing J, Schmidt B, Pohlmann R, Waheed A, Meyer H E, O'Brien J S, von Figura K. Cloning and expression of human arylsulfatase A. J Biol Chem. 1989;264:1252–1259. [PubMed] [Google Scholar]
- 45.Stins M F, Prasadarao N V, Ibric L, Wass C A, Luckett P, Kim K S. Binding characteristics of S fimbriated Escherichia colito isolated brain microvascular endothelial cells. Am J Pathol. 1994;145:1228–1236. [PMC free article] [PubMed] [Google Scholar]
- 46.Szameit C, Miech C, Balleininger M, Schmidt B, von Figura K, Dierks T. The iron sulfur protein AtsB is required for posttranslational formation of formylglycine in the Klebsiellasulfatase. J Biol Chem. 1999;274:15375–15381. doi: 10.1074/jbc.274.22.15375. [DOI] [PubMed] [Google Scholar]
- 47.Waldow A, Schmidt B, Dierks T, von Bülow R, von Figura K. Amino acid residues forming the active site of arylsulfatase A. J Biol Chem. 1999;274:12284–12288. doi: 10.1074/jbc.274.18.12284. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Huang S-H, Wass C A, Stins M F, Kim K S. The gene locus yijP contributes to Escherichia coliK1 invasion of brain microvascular endothelial cells. Infect Immun. 1999;67:4751–4756. doi: 10.1128/iai.67.9.4751-4756.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiser J N, Gotschlich E C. Outer membrane protein A (OmpA) contributes to serum resistance and pathogenicity of Escherichia coliK-1. Infect Immun. 1991;59:2252–2258. doi: 10.1128/iai.59.7.2252-2258.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson P J, Morris C P, Anson D S, Occhiodoro T, Bielicki J, Clements P R, Hopwood J J. Hunter syndrome: isolation of an iduronate-2-sulfatase cDNA clone and analysis of patient DNA. Proc Natl Acad Sci USA. 1990;87:8531–8535. doi: 10.1073/pnas.87.21.8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamada T, Murooka Y, Harada T. Comparative immunological studies on arylsulfatase in bacteria of the family Enterobacteriaceae: occurrence of latent arylsulfatase protein regulated by sulfur compounds and tyramine. J Bacteriol. 1978;133:536–541. doi: 10.1128/jb.133.2.536-541.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]