Abstract
Growth and productivity of crop plants worldwide are often adversely affected by anthropogenic and natural stresses. Both biotic and abiotic stresses may impact future food security and sustainability; global climate change will only exacerbate the threat. Nearly all stresses induce ethylene production in plants, which is detrimental to their growth and survival when present at higher concentrations. Consequently, management of ethylene production in plants is becoming an attractive option for countering the stress hormone and its effect on crop yield and productivity. In plants, ACC (1-aminocyclopropane-1-carboxylate) serves as a precursor for ethylene production. Soil microorganisms and root-associated plant growth promoting rhizobacteria (PGPR) that possess ACC deaminase activity regulate growth and development of plants under harsh environmental conditions by limiting ethylene levels in plants; this enzyme is, therefore, often designated as a “stress modulator.” TheACC deaminase enzyme, encoded by the AcdS gene, is tightly controlled and regulated depending upon environmental conditions. Gene regulatory components of AcdS are made up of the LRP protein-coding regulatory gene and other regulatory components that are activated via distinct mechanisms under aerobic and anaerobic conditions. ACC deaminase-positive PGPR strains can intensively promote growth and development of crops being cultivated under abiotic stresses including salt stress, water deficit, waterlogging, temperature extremes, and presence of heavy metals, pesticides and other organic contaminants. Strategies for combating environmental stresses in plants, and improving growth by introducing the acdS gene into crop plants via bacteria, have been investigated. In the recent past, some rapid methods and cutting-edge technologies based on molecular biotechnology and omics approaches involving proteomics, transcriptomics, metagenomics, and next generation sequencing (NGS) have been proposed to reveal the variety and potential of ACC deaminase-producing PGPR that thrive under external stresses. Multiple stress-tolerant ACC deaminase-producing PGPR strains have demonstrated great promise in providing plant resistance/tolerance to various stressors and, therefore, it could be advantageous over other soil/plant microbiome that can flourish under stressed environments.
Keywords: environnemental stress, ethylene, plants, PGPR, ACC deaminase, mode of action
Introduction
Plant growth and productivity are affected by myriad complex factors, both physiological and environmental, including plant genotype, soil physical and chemical characteristics, availability of nutrients, and other variables (Schwachtje et al., 2019). In addition, crop growth and yield may be stressed by several biotic and abiotic factors, i.e., salinity, drought, temperature, mechanical wounding, waterlogging, organic contaminants, heavy metals and other xenobiotics (Gupta et al., 2013; Gull et al., 2019). As a consequence of these factors, ~35–50% yield loss has beed reported so far in major crops globally (Stallworth et al., 2020). Abiotic stresses are therefore considered as a primary influence affecting agricultural production worldwide.
Global food supplies must be increased to fulfil the increasing demands of rapidly-growing populations (Place et al., 2017). Response to several biotic and nutritional challenges in plant husbandry can be resolved using chemical pesticides, fertilizers, and other agrochemicals. However, using non-biological methods to address problems posed by abiotic stressershas its share of difficulties. Plants respond to external challenges by altering production of certain hormones, which promotes the synthesis of stress-related proteins that afford protection against the negative effects of stressors (Gupta et al., 2020). In this regard, ethylene is considered as the most common phytohormone mediating stress response in many crop plants (Tiwari et al., 2020). In contrast, when ethylene production exceeds a certain threshold, it becomes “stress ethylene.” Excessive levels of ethylene adversely affect proliferation of roots, shoots, and other yield parameters and, thus, hamper overall plant performance (Klay et al., 2018; Mog et al., 2018). The detrimental impacts of the high ethylene levels can be reduced by various soil/plant-colonizing microbiomes that contain the essential enzyme ACC deaminase (Glick, 2014; Saikia et al., 2018). ACC deaminase (ACCD) converts the harmful form of ethylene to a non-toxic state (Das and Osborne, 2018). The ACCD decreases ethylene levels in plants by breaking down ACC into α-ketobutyrate (C4H6O3) and ammonia (Bharti and Barnawal, 2019) which in turn allow roots/shoots or entire plants to grow normally (Glick, 2014). Thus, ACCD permits plants to thrive in challenging environments by reducing harmful concentrations of ethylene (Han et al., 2015; Ravanbakhsh et al., 2017; Sarkar et al., 2018a,b). ACC serves as the originator of ethylene in plants (Ouaked et al., 2003). “Induced systemic tolerance” refers to the inherent characteristics of assigning tolerance to abiotic stressors through ACCD activity and some redundant PGPR processes to alleviate stresses in host plants (Arya et al., 2018; Carlson et al., 2020). Therefore, PGPR equipped with ACCD activity are essential organisms that play a major role in the reduction/mitigation of the toxic effects of several environmental stressors such as salinity, drought, heavy metals, and organic pollutants (Table 1). The production of the stress hormone, ethylene, and its impact on plants while growing under stress has previously been explained. Taking relevant papers into account, the present review describes the importance of ethylene in plant physiology and the function of bacterial ACC deaminase in reducing stress-induced ethylene levels in plants, thereby circumventing the negative effects of environmental stressors.
Table 1.
Examples of ACC deaminase producing PGPR.
| PGPR strains | Source | ACC deaminase activity (nmol α-ketobutyrate mg protein−1 h−1) | Reference |
|---|---|---|---|
| Achromobacter xylosooxidans A551 | Pisum sativum rhizosphere | 400 ± 4.0 | Belimov et al. (2009) |
| Burkholderia cepacia PSBB1 | Vicia faba rhizosphere | – | Shahid and Khan (2018) |
| Pseudomonas putida UW4 | – | 3,030 ± 60 | Hontzeas et al. (2006) |
| Rhizobium leguminosarum 128C53K | Pisum sativum rhizosphere | 5.0 ± 1.0 | Belimov et al. (2005) |
| Serratia proteamaculans | Rhizosphere region of salt-affected T. aestivum (L.) | 276 ± 00 | Zahir et al. (2009) |
| Serratia marcescens BC-3 | Rhizosphere soils of salt and petroleum amended E. crusgali plants | 38,520 ± 00 | Liu et al. (2015) |
| Bacillus pumilus SB1-ACC3 | Oryza sativa (L.) rhizosphere | 1,460 ± 00 | Bal et al. (2013) |
| Bacillus licheniformis B2r | Salinity-stressed rhizosphere soils | 860 ± 00 | Chookietwattana and Maneewan (2012) |
| Bacillus sp. MR4 | Rhizosphere soils of Arabidopsis thaliana (L.) and Festuca rubra (L.) | 15,920 ± 00 | Grobelak et al. (2018) |
| Bacillus cereus LB1 | Tissue of Carthamus tinctorius (L.) | 2,400 ± 00 | Hemida and Reyad (2018) |
| Bacillus aerius SB1 | Tissue of Carthamus tinctorius (L.) | 1,800 ± 00 | Hemida and Reyad (2018) |
| Pseudomonas sp. R3 | Rhizosphere soil of Arabidopsis thaliana (L.) and Festuca rubra (L.) | 23,490 ± 00 | Grobelak et al. (2018) |
| Mesorhizobium ciceri strain LMS-1 (pRKACC) | – | 2,305 ± 00 | Nascimento et al. (2012) |
| Sinorhizobium meliloti KYA71 and KYA40 | Soil and Water Research Institute (Iran) | 326,136 ± 00 | Khosravi et al. (2014) |
| Serratia ficaria | Salinity-stressed rhizosphere soils | 326 ± 00 | Nadeem et al. (2010) |
| Pseudomonas sp. ST3 | Rhizosphere soil of Ipomoea aquatica (L.) | 900 ± 00 | Trung et al. (2016) |
| Enterobacter aerogenes | Rhizosphere soil of salt-treated Zea mays (L.) | 341 ± 00 | Nadeem et al. (2007) |
| Enterobacter sp. CS1 | - | 170 ± 00 | Huang et al. (2013) |
| Enterobacter cloacae ZNP-4 | Ziziphus nullifera (L.) rhizosphere soil | 188.90 ± 9.3 | Singh et al. (2022) |
| Pseudomonas sp. TR15a | Contaminated rhizosphere of Trifoliumrepens (L.) | 53.74 ± 00 | Kumar et al. (2021) |
| Bacillus amyloliquefaciens | Pearl millet rhizosphere | 2196.23 ± 00 | Murali et al. (2021) |
| Ensifer adhaerens KS23 | Rhizosphere soil of leguminous crop | 174.2 ± 00 | Katiyar et al. (2021) |
| Achromobacter sp. | Rhizosphere soil | 4.90 ± 00 | Sun et al. (2022) |
| Streptomyces hydrogenans DH16 | 363 ± 00 | Kaur and Manhas (2022) |
Ethylene: Biosynthesis, physiology, regulation, and stress response in plants
Ethylene, the smallest and simplest gaseous phytohormone produced by plants, regulates a suite of biological and functional processes in plants (Light et al., 2016; Fernandez-Moreno and Stepanova, 2019). Processes regulated by ethylene include seedling germination, ripening/maturation of fruit, senescence, development of root hairs and nodules, elongation of roots, and epinasty (Zhang et al., 2016; Zhu et al., 2016; Sun et al., 2019; Figure 1).
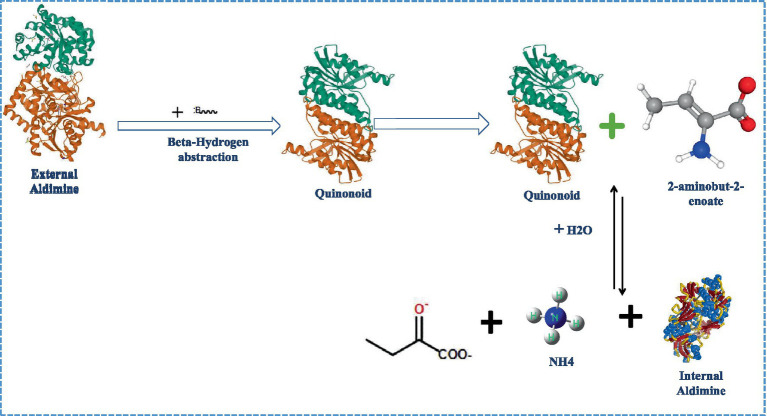
Figure 1.
Elucidation of route1 (Direct β-hydrogen extraction) for ACC metabolism by ACC deaminase.
Ethylene production in plants is primarily influenced by environmental factors and depends on the degree and intensity of environmental variables. The identification of ethylene as a plant growth regulator was revealed by early leaf shedding, geotropism of etiolated pea seedlings when exposed to lighting gas, and the ripening/maturation of plant organs when exposed to kerosene combustion gas (Pierik et al., 2006; Glick et al., 2007a).
A wide array of biotic and abiotic factors (e.g., salinity, drought, waterlogging, flooding, agrochemicals, pesticides, heavy metals, organic and inorganic pollutants, phytopathogens) inducesethyleneproduction in plants (Gontia-Mishra et al., 2014). Henceforth, the ethylene produced under such environmental stresses is regarded as “stress ethylene” (Glick, 2014). The stress ethylene triggers genes to be transcribed and further expressed, resulting in plant senescence. Ethylene biosynthesis in plants follows a relatively straightforward system where methionine is converted to S-adenosyl methionine (SAM) by the enzyme SAM synthetase that is subsequently used as a substrate by ACC synthase to generate 1-aminocyclopropane-1-carboxylic acid (ACC). The ACC generated in this process acts as precursor for ethylene production by the action of enzyme ACC oxidase.
ACCD: Biochemical properties and mode of action
When ACC deaminase was identified in soil microorganisms for the first time, it was demonstrated to transform ACC to ammonia (NH3) and α-ketobutyrate, which were subsequently metabolized by microbes (Honma and Shimomura, 1978). ACCD is a pyridoxal PO₄3−-dependent enzyme. In order to activate the enzyme, about 3.0 mol of pyridoxal PO₄3− (enzyme bound) mol−1 of enzyme or 1.0 mol trimeric−1subunitis required (Honma, 1985; Karthikeyan et al., 2004). This enzyme was first purified from Pseudomonas sp. strain ACP; however, strains of P. chloroaphis 6G5 (Klee et al., 1991) and P. putida GR12-2 (Jacobson et al., 1994) have also been utilized for partial purification of ACCD. The molecular mass and shape of enzyme isolated from all three sources appear to be identical. Pseudomonas sp. strain ACP was found to have a native size of 110–112 KDa, while P. putida GR12-2 had a native size of 105 KDa. In nature, this enzyme is found in the trimeric form with ~36,500 Da mass subunit.
At pH 6.0 and pH 9.0, the absorption maxima of pure ACC deaminase from Pseudomonas sp. were 416 and 326 nm, respectively (Honma, 1985). The 326 nm band observed at pH 9.0 could represent the activation form of ACCD to which inhibitors and substrates strongly bind (Jacobson et al., 1994). The published range of Km values for enzyme extracts from various bacteria at pH 8.5 is 1.5–17.4 mM, indicating that the enzyme has a low affinity for ACC. Following second-order kinetics, the total efficiency (kcat/km) of ACC deaminase is around 690 M−1S−1. The ACC deaminase Km value for 1-amino cyclopropane 1-carboxylate has been established using enzyme extracts from microorganisms at pH 8.5 (Klee et al., 1991). Several bacterial species produced ACCD enzyme and their activity was evaluated over a broad pH range and at pH 8.0 to 8.5 showing highest activity. The optimal temperature for ACC deaminase activity is 30°C (Glick et al., 1998).
Because ACC deaminase is an inducible enzyme, its production is triggered when its substrate, ACC, is present. In P. putida strain GR12-2 and Pseudomonas sp. strain ACP GR12-2, the lowest level of substrate for induction was determined to be 100 nM. ACCD induction is a lengthy and complex procedure. Within a few hours of incubation with the substrate, the enzyme expresses its activity which, steadily declines thereafter (Jha et al., 2012). In a minimal medium supplied with (NH₄)₂SO₄ (ammonium sulfate) as Nsource, the basal level of enzyme activity was observed. It was further demonstrated that growing bacteria in a minimal medium that contained ACC as the only N source led to increased enzyme activity, suggesting that the substrate ACC had a direct relationship with induction of enzyme activity (Honma, 1983). Expression of ACCD and the activation of other amino acids such as L-alanine, DL-alanine, and D-serine, increase to a lesser degree than in the case of ACC. Furthermore, both ACC and amino-isobutyric acid (C4H9NO2) produced a similar degree of enzyme activity in Pseudomonas sp. strain ACP (Honma, 1983). According to Glick et al. (1998), ACC is released from plant roots or seeds, ingested by soil microbiota, and hydrolysed to ammonia and α-ketobutyrate. The quantum of ACC outside the plant root, however, decreases due to ACC absorption and hydrolysis. The equilibrium between levels of internal and external ACC is also maintained by the exudation of excess ACC into the rhizosphere. As a result, a reduction in ACC levels reduces the production of stress hormone ethylene in host plants and stimulating growth of the plant (Glick et al., 1998).
L-isomers of amino acids such as L-alanine, L-serine, L-homoserine, and L-aminobutyric acid inhibit ACC deaminase competitively, with L-alanine and L-serine exhibiting greatest inhibition. ACC deaminase isolated from Pseudomonas sp. can also use ACC-related compounds like 2-alkyl-ACC and vinyl-ACC as substrates. Strain ACP, although the enzyme has a peculiar preference for D-amino acids, being inactive with any L-amino acids or derivatives. According to NMR research, a proton is removed from the β-carbon of D-alanine but not from the L-isomer. These findings support the stero-specific breakage of the cyclopropane ring during ACC deamination, which explains the deamination of D-amino acids and many substituted D-alanines. The iodoacetamide derivative 1,5 N-iodoacetamidoethyl-1-aminonapthalene-5-sulfonic acid (1,5-I-AEDANS) inactivates ACC deaminase more effectively in the presence of D-alanine than iodoacetamide. During inactivation, a thiol group in cysteine residue 162 is altered, as it is the aldimine connection between pyridoxal phosphate and lysine residue 51 (Honma et al., 1993). The primary feature of the ACC deaminase-catalyzed process is the opening of the ACC cyclopropane ring. The most likely method for cleaving the cyclopropane bond appear to be nucleophilic addition and elimination, although the full reaction mechanism is unknown (Thibodeaux and Liu, 2011).
Enzymology of ACC deaminase
The deamination of ACC, the precursor of the gaseous phytohormone ethylene, is carried out by the tryptophan synthase beta (β) superfamily enzyme ACC deaminase (EC 3.5.99.7), which is dependent on the pyridoxal 5′-phosphate (PLP) molecule. To initiate the ACC deaminase enzyme activity, 1 mol pyridoxal phosphate (vitamin B6) works as a firmly bound cofactor (Singh et al., 2015). It is found in the cytoplasm of bacterial cells and has a molecular mass of 35–42 kDa (Gamalero and Glick, 2015). PLP is thought to be an inducible enzyme that requires a substrate, ACC, at a concentration of <100 nM to activate the process. By switching ACC deaminase-producing bacterial strains from nutrient-rich growth media to minimal media containing ACC as its sole N source, the induction of enzymatic activity by substrate, ACC, is proven. Other amino acids such as D-alanine, L-alanine, D-valine, 2-alkyl-ACC, vinyl-ACC, and 2-aminoisobutyric acid, all of which are similar to ACC in structure and behavior, can also activate ACC deaminase. Furthermore, 2-aminoisobutyric acid has the same ability to stimulate activity as ACC (Malerba et al., 1996).
Activation of ACCD has been observed at various pH levels. The pH range 8.5–9.0 has, however, been found to impart the highest efficiency for the substrate and competing inhibitors. The L-amino acids or their derivatives decrease the activation of ACC deaminase. At pH 9.0, the ACC deaminase absorption spectra showed the strongest band at 326 nm. The activity of Pseudomonas putida strain GR12-2 ACC deaminase was reported to be highest at 30°C (Jacobson et al., 1994). At pH 8.5, enzyme Km value ranged from 1.5 to ~17.4 mM, indicating that it does not have a strong affinity for ACC (Hontzeas et al., 2004). The enzyme has a catalytic efficiency of roughly 690 M−1 S−1 (kcat/km) (Klee et al., 1994). Because ACC oxidase has a stronger affinity for ACC than ACC deaminase, the lower Kmvalues indicate that ACC deaminase should be present in higher concentrations (100–1,000 fold) in order to utilize the ACC substrate before ACC oxidase and hence reduce ethylene levels (Glick et al., 1998).
Mechanism of ACC deaminase enzymatic reaction
Stressed plants generate ACC, which is hydrolyzed by the microbial enzyme ACC deaminase to α-ketobutyrateand ammonia, thus reducing stress-induced ethylene and related growth inhibition. The elimination reaction and addition of nucleophiles that breaks the cyclopropane ring is the fundamental feature of the ACC deaminase-catalyzed second-order process (Glick et al., 2007a,b). Two possible mechanisms by which ACC deaminase carried out the deamination of its substrate ACC (Walsh et al., 1981; Zhao et al., 2003) include: (i) Direct β-hydrogen extraction in whichLys-mediated hydrolytic reactions break the cyclopropane ring when a hydrogen atom is extracted from the ACC substrate (Figure 1); and (ii) Nucleophilic addition followed by β-hydrogen extraction where ACCcarbon is attacked nucleophilically, and the cyclopropane ring is opened via Lys51-mediated hydrogen abstraction. The internal aldimine (imine analogue of aldehyde group) is located between the ACC deaminase lysine residue and pyridoxal phosphate cofactor. The trans-aldimination process occurs when the ACC amino group displaces the L-lysine residue from the enzyme active site, leading to the production of external aldimine via an aminyl intermediate that is present in both proposed pathways (Hontzeas et al., 2006). In route 1, a Lys basic residue on an external aldimine removes the methylene proton directly, forming quinonoid, which results in the formation of a new quinonoid molecule by protonation and electronic configuration (Joshi et al., 2012). The process continues with quinonoid nucleophilic attack by basic lysine amino residues, yielding another quinonoid and 2-aminobut-2-enoate, which is then reversibly hydrolyzed to provide 2-oxobutanoate and an ammonium ion, restoring the internal aldimine (Ose et al., 2003). Following the formation of external aldimine, route 2 departs from route 1 by performing a nucleophilic attack on the proton of the β-carbon of ACC (pro-S), resulting in the synthesis of quinonoid, followed by hydrogen removal from the carbon of ACC (pro-R). Following quinonoid production, the steps are identical to route 1 (Ose et al., 2003).
The ACC deaminase gene and its expression
ACC deaminase gene
As previously mentioned, the AcdS gene encoding ACC deaminase has been identified in various bacterial and fungal species. ACC deaminase has recently been discovered in a variety of Gram-negative bacteria (Gontia-Mishra et al., 2017), fungi (Rauf et al., 2021), endophytes (Sofy et al., 2021) and rhizobia (Saghafi et al., 2019). An ACC deaminase gene has been identified in several species, notably R. leguminosarum bv. Trifoli and Mesorhizobium loti MAFF303099. The degree of ACC deaminase expression, however, differs from one organism to another. A portion of the AcdS gene was amplified and examined in a variety of environmental isolates using a universal pair of primers. Various workers have developed several pairs of primers to identify the presence of the bacterial AcdS gene. Only a few bacterial species have had the entire genetic makeup and function of the ACCD gene described (Duan et al., 2013). It has also been discovered that the nucleotide sequences of the AcdS gene are very similar to those of two other genes, i.e., dcyD and yedO, which encode for another PLP-dependent enzyme, D-cysteine sulfhydralase. Earlier studies have shown that certain genes previously thought to code for ACC deaminase activity also code for D-cysteine desulfhydrase (Riemenschneider et al., 2005). Nascimento et al. (2014) used Pseudomonas sp. strain UW-4 as a reference to evaluate the key protein residues recognized to be crucial for ACC deaminase function, including Leu322, Glu296, Ser78, Tyr295 and Lys51. Any alteration in residues at certain sites was considered to indicate D-cysteine desulfhydrase.
With few exceptions, the AcdS gene in the majority of bacterial species is chromosomal DNA-borne. In symbiotic bacteria M. loti (symbiont of lotus spp.), the ACC deaminase gene is associated with nitrogen fixation genes and might be regulated by NifA, which is known to activate nif gene expression in association with the product of rpoN gene (Ma et al., 2003a). Only a small fraction of the putative AcdS gene has been shown to encode active enzyme (Glick et al., 2013).
Regulation of ACC deaminase
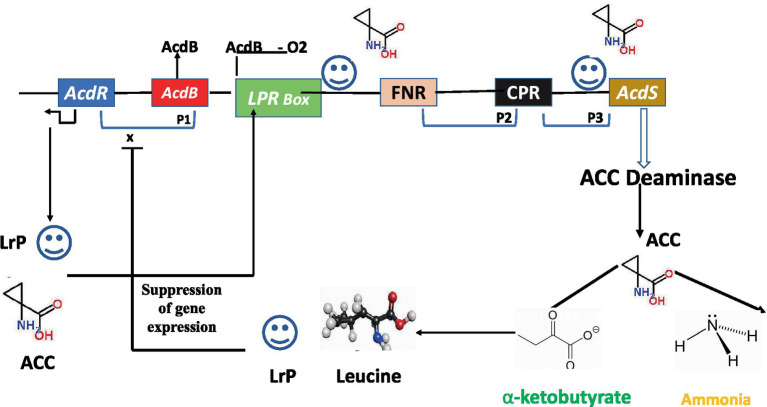
AcdS is highly controlled, whose expression varies with O2level, quantity of substrate, and product accumulation. With few exceptions, regulation of the AcdSgene in various bacterial taxa is poorly understood. Li et al. (2000) presented the model for regulating ACC deaminase genes in P. putida strain UW-4. Regulatory elements for the expression of ACC deaminase gene consist of regulatory gene AcdR located 5′ upstream of ACC deaminase structural gene (AcdS), promoter regions for binding of regulatory proteins like Lrp box for binding of Lrp protein, AcdB box for binding regulatory protein AcdB, FNR box for binding of fumarate and nitrate reductase protein, and CRP box for binding of cAMP receptor protein (Figure 2). The LRP creates an active octamer in the presence of ACC, which binds to an ACC-AcdBcomplex (Gupta and Pandey, 2019). Glycerophosphoryl diester phosphodiesterase is encoded by the gene dB, which forms a complex with ACC. By attaching to the promoter region of AcdS, this triparental complex promotes its transcription. In other bacteria studied for AcdS gene expression, AcdBhas not been demonstrated to play a function. Leucine, which is generated from α-ketobutyrate, a breakdown product of the ACCD-catalyzed process, inhibits expression of the ACC deaminase gene. As the quantity of leucine rises, it favors creation of inactive LRP dimers, which prevents the AcdS gene from being transcribed (Figure 2).
Figure 2.
Regulatory circuits of AcdS gene expression in Pseudomonas putida UW4 and related bacteria. AcdR, regulatory gene for ACC deaminase; AcdB, encoding for glycerophosphoryl diester phosphodiester; LRP, leucine responsive protein; FNR, fumarate nitrate reductase protein; CRP, c-AMP receptor protein; AcdS, gene for encoding ACC deaminase.
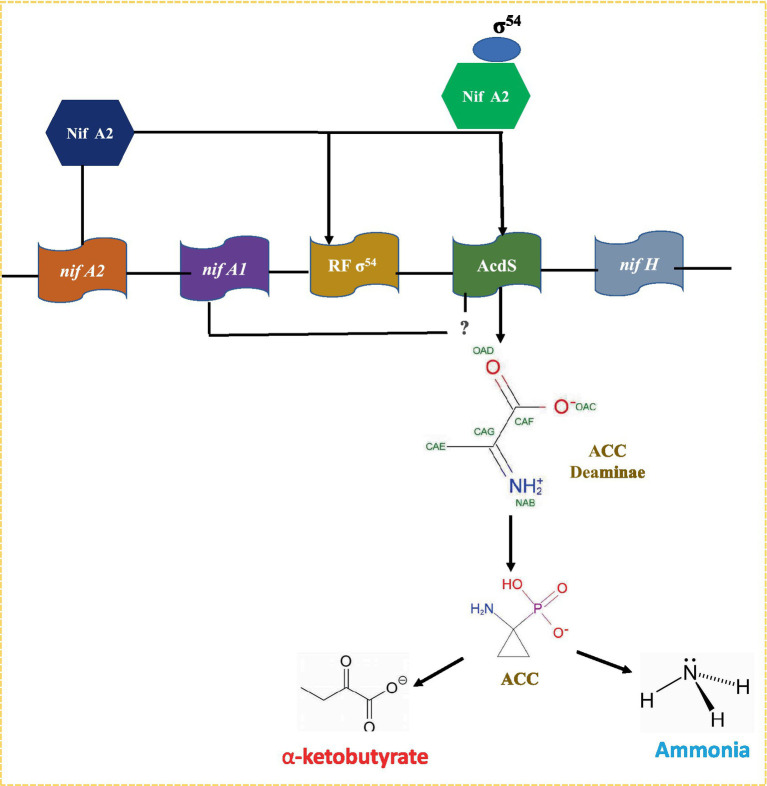
The regulatory mechanism that controls AcdS expression differs from bacterial species to species. The majority of bacteria have AcdR encoding LRP or related sequences, according to results of the IMG database analysis. LRP-like protein and the 70 promoters are also implicated in the regulation of the AcdS gene in Bradyrhizobium japonicum USDA 110 and Rhizobium leguminosarum bv. Viciae 128 C53K (Kaneko et al., 2002; Ma et al., 2003a). According to the evolutionary analysis of the AcdS and AcdRgene evolved in a similar fashion. Instead of the AcdR gene, Burkholderia sp. CCGE 1002 and B. phymatum STM 815 have two copies (megaplasmid and the other on the second chromosome) of the AcdS gene. In smaller replicons, these shards of evidence point to chromosomal rearrangement or gene insertion events. Some bacteria, such as Achromobacterxylosooxidans A-551 and Variovoraxparrdoxus 5C2, lack all the regulatory components as observed in the model bacterium P. putida UW4.In M. loti, the upstream elements of AcdS and nifH contain nifA1 and nifA2 (regulatory N2 fixing unit) and σ54 RNA polymerase sigma recognition site. It was hypothesized that expression of ACC deaminase in M. loti required the symbiotic nitrogen fixing regulatory gene nifA2 (Nukui et al., 2006).
The nifA2 encoded protein NifA2 interacts with σ54 RNA polymerase, favoring AcdS transcription. The nifA1 also affects transcription of the AcdS gene to some extent; however, its role in expression of AcdS is not fully understood (Nukui et al., 2006; Figure 3). The AcdS gene is expressed in root nodules, which minimizes the negative effects of ethylene-induced senescence and increases the concentration of fixed nitrogen in nodules. The activity of ACC deaminase is commonly measured in free-living organisms; however, in M. loti, it was only found in symbiotic nodules (Uchiumi et al., 2004).
Figure 3.
A model for acds gene regulation in nitrogen fixing Mesorhizobium sp. Expression of acds is positively regulated by NIFA2 protein which binds to σ54 and switch on transcription of AcdS gene. Nifa1 is also required in regulation of AcdS but its role is not well-understood.
It must be emphasized that, unlike free-living bacteria, ACC deaminase among nodule-forming rhizobia does not reduce ethylene levels throughout the plant and, hence, cannot be employed to protect plants from various stresses (Ferguson and Mathesius, 2014; Vargas et al., 2017). Furthermore, the amount of ACCD produced in the nodule is only 2–10% of the amount produced by free-living bacteria.
The GntR protein coding gene is presentadjacent to theAcdS gene in various Meiothermus and Actinobacteria. This suggests that some downstream components may be involved in ACC deaminase expression control as well. The lack of a promoter region in some members of these genera clearly suggests that control of AcdS gene transcription is mediated by the interaction of the AcdSgene with a downstream element close to thatgene. Brenneria sp. EniD312, Burkholderiaxenovorans LB4000, and Pantoea sp. are examples of Actinobacteria and Proteobacteria. At-9B, a transcription regulatory element belonging to the LysR family was identified near the AcdS gene. However, it is still not clear howACC deaminase specifically functions in such organisms. Therefore, to fully comprehend the mechanism of ACC deaminase regulation and function in various bacterial genera, additional genetic and biochemical research is required.
When triggered by ACC, the putative ACC deaminase gene in M. loti MAFF303099 contains no regulatory elements and shows no enzyme activity (Ma et al., 2003b). ACC concentrations as low as 1 M promote ACC deaminase expression in R. leguminosarum bv. Viciae 128C53K. The introduction of the ACC deaminase and its regulatory gene from R. leguminosarum bv. Viciae 128C53K to a S. meliloti strain resulted in an increase in Medicago sativa nodulation efficiency (Ma et al., 2004). Furthermore, in terms of nodulation, the latter strain outperformed the wild type (Ma et al., 2004).
ACC deaminase producing PGPR: Ecological significance
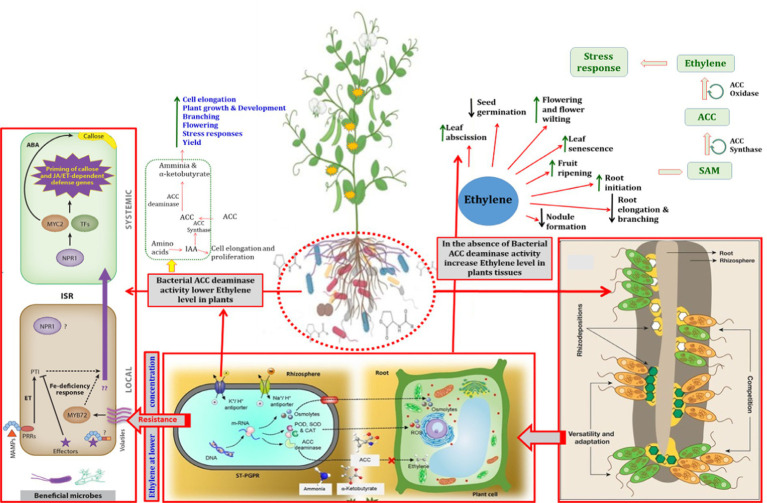
The relevance of PGPR having ACCD activity in reducing the effects of stress ethylene has been extensively studied. When ACCD-producing bacteria are present on the root surface of a stressed plant, they function as ACC reservoirs, reducing ethylene levels in the plant and promoting root development. Because of their extensive root growth, plants inoculated with ACCD harboring PGPR may have better tolerance to a variety of environmental challenges. Several environmental stresses (salinity, flooding, extreme temperatures, heavy metal toxicity, water deficit, nutrient deficiency, and pathogenicity) are the key limiting factors for agricultural production and productivity across the globe. It is presumed that global climate change might augment the occurrence and magnitude of environmental stresses, i.e., abiotic and biotic in the near future (Saleem et al., 2007; Timmusk et al., 2011). These stresses cause significant reduction in the crop growth and yield of stressed plants. It is well established that ethylene production increased significantly under environmental stressed condition especially in stress-sensitive crop varieties. This is commonly known as “stress ethylene” produced as a consequence of abiotic and biotic stresses. On the other hand it is well known that the ACC deaminase-producing organisms were much abundant in the rhizosphere of wild barley (Hordeum spontaneum) growing in a stressed environment than they were in a similar (nearby) less stressed environment (Timmusk et al., 2011). Under stresses conditions, rhizospheric and endophytic bacterial/microorganisms produces ACC deaminase which break the ACC (prerequisite of ethylene production) to α-ketobutyrate and ammonia and thereby diminishes level of “stresses ethylene” in the stressed host plants. Few reports indicated that Methylobacterium spp. (phytopathogenic in nature) modulate plant growth and development by decreasing environmental stress, immobilizing heavy metals, degrading toxic organic compounds and even inhibiting plant pathogens (Reinhold-Hurek and Hurek, 2011; Brader et al., 2014; Santoyo et al., 2016; Shahzad et al., 2017; Ek-Ramos et al., 2019). A number of bacteria have been discovered in soil/rhizosphere that can utilize ACC as a sole source of nitrogen, are capable of alleviating different environmental stresses, and can support improved growth and overall performance of agricultural crops (Chauhan et al., 2017; Figure 4). Plant synthesis of ethylene is also regarded as a stress response, and is closely linked to a variety of stress factors including as waterlogging, salinity, presence of heavy metals, and nutrient deficiencies (Dimkpa et al., 2009). It may be possible to apply phytoremediation at contaminated sites by taking advantage of the variation in ACC deaminase activity among microbial species under extreme environmental conditions (Glick, 2005). By biotransforming toxic substances, rhizodegradation mediated by root exudates, and/or detoxification of heavy metals, ACC deaminase-producing bacteria support plants in phytoremediation and enable host plants to thrive under challenging conditions (Qin et al., 2014). By expanding the plant root system and improving root access to soil, ACC deaminase rhizospheric bacterial populations can accelerate rhizo-remediation (Kalsoom et al., 2022). With modified root structure and architecture, inorganic pollutants are more effectively absorbed by the plant. According to Belimov et al. (2005), increased root growth was positively correlated with increased bacterial ACC deaminase activity when cadmium accumulated in plant tissue. Synthesis of minimal quantities of ethylene in leguminous plants has been shown to disrupt the Nod factor involved in the signal transduction pathway, which was prevented by rhizobial inoculation (Guinel, 2015). As a result, PGPR-produced ACC deaminase shields plants from the detrimental effects of ethylene when exposed to abiotic stress (Sapre et al., 2018a,b). Some widely acclaimed bacterial genera synthesizing ACC deaminase include Achromobacter (Sun et al., 2022), Brevibacterium linens (Choi et al., 2022), Bacillus amyloliquefaciens (Murali et al., 2021), Ensiferadhaerens (Katiyar et al., 2021), Variovorax sp. (Bessadok et al., 2020), Enterobacter sp. (Sagar et al., 2020), Rhizobium (Saghafi et al., 2019), Bradyrhizobium (Greetatorn et al., 2019), Pseudomonas (Nascimento et al., 2019), Bacillus (Din et al., 2019), Burkholderia, Enterobacter, Serratia (Zafar-ul-Hye et al., 2019), Azotobacter (Rizvi and Khan, 2018), Achromobacter (Shahid et al., 2019a,b), and Acinetobacter, Alcaligenes (Gontia-Mishra et al., 2017). Table 1 lists certain PGPR-containing ACC deaminase activity (ACCD) positive bacteria.
Figure 4.
Representation of the direct and indirect roles of bacterial ACC deaminase in plant growth and development. MAMPs represent microbe-associated molecular patterns; ET, ethylene; PTI, PAMP triggered immunity; ISR, induced systemic resistance; TFs, transcription factors; ABA, abscisic acid; POD, peroxidase; SOD, superoxide dismutase; CAT, catalase; PGPRs, plant growth promoting rhizobacteria; ROS, reactive oxygen species; JA: jasmonic acid.
Biochemistry of ACC deaminase
ACC deaminase is a multimeric enzyme in the tryptophan synthase β-superfamily of pyridoxal phosphate-binding proteins (Glick et al., 2007a,b; Gamalero and Glick, 2015) and is cytoplasmically localized. It has a subunit of mass of ~35–42kD, whereas its natural size is between 100 and 112 kD (Raghuwanshi and Prasad, 2018). This enzyme does not have high affinity for the substrate (1.5–6.0 mM). As a co-factor, pyridoxal phosphate is required for ACC deaminase activity (Glick et al., 2007a,b), and is required for activity of ACC synthase, which catalyzes the synthesis of ACC. Enzyme ACCD exists in the microbial community in very low quantities, and in comparison, to ACC deaminase, ACC oxidase has a substantially higher affinity for ACC (Singh et al., 2015). The level of ethylene in bacterial species depends primarily on activities of ACC oxidase and ACC deaminase (Glick, 2014). Amino acids such as L-alanine, DL-alanine, and DL-valine also stimulate enzyme activity to a modest degree, whereas 4-aminobutanoic acid can stimulate enzyme activity to about the same degree as ACC (Honma, 1983; Raghuwanshi and Prasad, 2018). At pH 8.5, the substrate ACC, as well as the competing inhibitors L-alanine and L-serine has maximum affinity (Hontzeas et al., 2006; Stress, 2018). The acdS genes present in certain bacteria and numerous fungi belonging to different genera are thought to have originated from a collective progenitor (Nascimento et al., 2014). Vertical gene transfer is widespread in many bacteria, while horizontal gene transfer, such as inter-kingdom transfer, also occurs occasionally. The structural genes (acdS) and regulatory genes (acdR) of ACC deaminase genes have been reported in numerous rhizobacterial groups including endophytic, rhizospheric and root nodulating rhizobia such as Rhizobium spp. (Kumar et al., 2016), Bradyrhizobium spp. (Greetatorn et al., 2019), Mesorhizobium spp. (Senthilkumar et al., 2016) and non-rhizobial groups such as Burkholderia spp. (Sarkar et al., 2018a,b), Pseudomonas spp. (Azadikhah et al., 2019), Achromobacter spp. (Chandra et al., 2020), Enterobacter spp. (Kruasuwan and Thamchaipenet, 2018), Azotobacter spp. (Viscardi et al., 2016), Bacillus spp. (Din et al., 2019), and Leclercia spp. (Kang et al., 2019). Regardless, however, even if certain strains of a genus and species possess an acdS gene, not all strains of that genus and species have ACCD.
Bioinoculation impact of ACC deaminase-producing PGPR: Management of biotic and abiotic stresses
Plants may be exposed to a wide range of environmental stresses, both biotic and biotic. ACCD-containing bacterial species safeguards plants from the deleterious impacts of environmental stresses including drought, salinity, high temperature, waterlogging, excess pesticides, heavy metals, and other xenobiotic contaminants by decreasing the activity of stressor-induced ethylene (Figure 4; Ali and Kim, 2018; Danish et al., 2020; Misra and Chauhan, 2020). The utilization of ACCD-positive PGPR for mitigating multiple abiotic stresses and their positive response on plants appears in Table 2.
Table 2.
Selected examples of ACCD synthesizing PGPR strains in alleviation of abiotic and biotic stress.
| S. No. | ACC deaminase producing PGPR | Source | Used against/host plant | Stress | Application response | References |
|---|---|---|---|---|---|---|
| Salinity stress | ||||||
| 1 | Bacillus mycoides PM-35 | Rhizosphere soil | Zea mays (L.) | – | Enhanced chlorophyll, soluble sugar and protein content and capacity to scavenge radical ions | Ali et al. (2022a) |
| 2 | Enterobacter cloacae ZNP-4 | Ziziphus nummularia | T. aestivum (L.) | – | Increased growth parameters like shoot (41%) and root length (31%), fresh plant weight (28%), dry biomass (29%) and leaf chlorophyll | Singh et al. (2022) |
| 3 | Enterobacter cloacae PM23 | Rhizosphere soil | Zea mays (L.) | – | Enhanced the power of radical scavenging, relative water content (RWC), soluble sugars, proteins, phenolic content, total flavonoid content in salt-treated Z. mays plants | Ali et al. (2022b) |
| 4 | Bacillus marisflavi CHR JH 203 and Bacillus cereus (BST YS1-42) | Leguminous crop | Pisum sativum (L.) | – | Increased dry biomass, biochemical constituents (carbohydrates, protein, reducing soluble sugars, leaf chlorophyll, phenolics and flavonoids) | Gupta et al. (2021) |
| 5 | Glutamicibacter sp. YD01 | Rhizosphere of Oryza sativa | Oryza sativa (L.) | – | Decreased levels of Na+ ion buildup and, electrolyte leakage; improved plant development | Ji et al. (2020) |
| 6 | Bacillus aryabhattai EWR29 | Wheat rhizosphere soil | T. aestivum (L.) | – | Mitigated the negative impact of NaCl, significantly enhanced growth, and reduced proline content | Farahat et al. (2020) |
| 7 | Paenibacillus sp. ACC-06 and Aneurinibacillusaneurinilyticus ACC-02 | Allium sativum (L.) rhizosphere soil | Phaseolus vulgaris (L.) | – | Negatively affected NaCl-induced pressure and enhanced biological properties (length, fresh weight, biomass) and photosynthetic capability of plant | Gupta and Pandey (2019) |
| 8 | Serratia grimesii BXF1 | Rhizosphere soil | Phaseolus vulgaris (L.) | – | Promoted formation of early root nodules and growth; improved the symbiotic attributes of plants | Tavares et al. (2018) |
| 9 | Bacillus, Acinetobacter and Enterobacter | Soil | Medicago sativa (L.) | – | Height, leaf-to-stem ratio, fresh weight, dry biomass, pigments used for photosynthetic energy, nitrogen, phosphorus and potassium content all increased in the plants. | Daur et al. (2018) |
| 10 | Enterobacter sp. | Soil | Oryza sativa (L.) | – | Lowered antioxidative enzymatic responses and NaCl-induced ethylene in bacteria-treated plants; improved plant yield and productivity | Sarkar et al. (2018a,b) |
| 11 | Klebsiella sp. | Rhizosphere of T. aestivum | Avena sativa (L.) | – | Reduced salt stress and boosted plant development in salt-stressed soil. Expression profiles of the rbcL and WRKY1 genes were positively regulated | Sapre et al. (2018a,b) |
| 12 | Pseudomonas sp., Bacillus cereus and Bacillus sp. | Brassica napus rhizosphere | Festuca rubra and Brassica napus (L.) | – | Potentially ameliorated the salinity and enhanced the physiological and biochemical traits of plants | Grobelak et al. (2018) |
| 13 | Bacillus cereus LB1 and Bacillus aerius SB1 | Rhizosphere soil | Carthamus tinctorus | – | Mitigated toxicity of NaCl and promoted vegetative growth of plant | Hemida and Reyad (2018) |
| 14 | Pseudomonas frederiksbergensis | Soil | Capsicum annum (L.) | – | Increased resistance of plants to NaCl stress observed in bacterial treated plants, as evidenced by increased antioxidant enzymatic activity responsiveness in leaf tissue and lowered hydrogen ion concentrations | Chatterjee et al. (2017) |
| 15 | Bacillus licheniformis HSW-16 | Rhizosphere of T. aestivum | Triticum aestivum (L.) | – | ACCD-positive PGPR strain positively influenced plant growth by relieving toxic effect of salts | Singh and Jha (2016) |
| 16 | Paenibacilluslentimorbus B-30488 | Rhizosphere soil | Lycopersicon esculentum | – | Suppressed growth of phytopathogens and inhibited southern blight disease in tomato; improved overall plant growth | Dixit et al. (2016) |
| 17 | Dietzianatronolimnaea | Rhizosphere soil | Triticum aestivum (L.) | – | Halotolerant PGPR strain increased different antioxidant defensive enzymes and stressor metabolites thus improving salt tolerance ability of plant | Bharti et al. (2016) |
| 18 | Pseudomonas putida | Desert regions of Rajasthan | C. arietinum (L.) | – | Relieved salt-induced toxicity and modulated the growth, physiology, biochemical properties and expression of various stress-related genes | Tiwari et al. (2016) |
| 19 | Variovorax paradoxus 5C-2 | Soil | Pisum sativum (L.) | – | Loweredthe proline and MDA content and antioxidant enzymes and enhanced the plant growth | Wang C. et al. (2016), Wang P. et al. (2016), and Wang Q. et al. (2016) |
| 20 | Pseudomonas sp. ST3 | Root nodule of Vigna unguiculata | Vigna unguiculata (L.) | – | Improved the plant water-relation status, ionic balance, biological attributes, and photosynthetic machinery of peas by relieving the NaCl-induced toxic effect | Trung et al. (2016) |
| 21 | Bacillus sp., Zhihengliuellahalotolerans and Staphylococcus succinus | Root nodule of T. aestivum | Triticum estivum (L.) | – | Improved ion balance, nutritional content and homeostasis | Orhan (2016) |
| 22 | Variovorax paradoxus 5C-2 | Root nodule of P. sativum | P. sativum (L.) | – | Water uptake, ionic homeostasis, overall growth, dry phyto-mass accumulation, leaf chlorophyll and grain yield of pea plants significantly improved | Wang C. et al. (2016), Wang P. et al. (2016), and Wang Q. et al. (2016) |
| 23 | Pseudomonas stutzeri A1501 | Rhizosphere of O. sativa | Oryza sativa (L.) | – | Restricted level of salts and improved the development and yield features of plant | Han et al. (2015) |
| 24 | Pseudomonas fluorescens YsS6 | Soil | Lycopersicum esculentum (L.) | – | Augmented seedling germination, vigor index (SVI), plant length (root and shoot) and plant dry biomass | Ali et al. (2014) |
| 25 | Bacillus flexus, Isoptericola dokdonensis and Arthrobacter soli | Inner tissues of Limonium sinense | L. sinense (L.) | – | Protected against salinity effects; increased the flavenoid accumulation | Qin et al. (2014) |
| 26 | Rhizobium leguminosarum | Pea root nodule | P. sativum (L.) | – | Augmented lengths of shoots and roots, dry biomass, chlorophyll synthesis, LHb content and nutrient uptake of plants | Ahmad et al. (2013) |
| 27 | Pseudomonas putida UW4 | Soil | Lycopersicum esculentum (L.) | – | Increased expression of mRNA in different ROS-scavenging enzymes and stressor metabolites, i.e., proline | Yan et al. (2013) |
| Drought stress | ||||||
| 28 | Bacillus megaterium (MU2) | Maize rhizosphere soil | T. aestivum (L.) | – | Potentially increased germination indices, vigor indices (SVI), plant fresh weight and dry biomass | Rashid et al. (2022) |
| 29 | Pseudomonas sp. | Rhizosphere soil of cereal crop | Arabidopsis thaliana (L.) | – | Increased plant survival, LRWC, chlorophyll, glycine betaine, stressor proline, and malondialdehyde content in drought-induced A. thaliana plants by 95, 59, 30, 38, 23, and 43%, respectively | Yasmin et al. (2022) |
| 30 | Serratia marcescens and Pseudomonas sp. | Rhizosphere of cereal crops | T. aestivum (L.) | – | Both strains potentially improved ROS, water status, osmolyte accumulation, chlorophyll and carotenoids content in plant leaves | Khan and Singh (2021) |
| 31 | Enterobacter cloacae 2WC2 | Withaniacoagulans plant | Zea mays (L.) | – | Morpho-biological parameters, RWC and antioxidant defence enzymes of PEG-treated plants increased following application of E. cloacae strain 2WC2 | Maqbool et al. (2021) |
| 32 | Bacillus velezensis strain D3 | Rhizosphere soil of rain-fed area | – | Photosynthetic capacity, stomatal conductance, vapor pressure, water-use efficiency, and transpiration rate all improved | Nadeem et al. (2021) | |
| 33 | Enterobacter HS9 and Bacillus G9 | Soil | Mucuna pruriens (L.) | – | Improved water uptake, rate of respiration and synthesis of chlorophyll | Saleem et al. (2018) |
| 34 | Ochrobactrumpseudogrignonense RJ12, Pseudomonas sp. RJ-15 and Bacillus subtilis RJ-46 | Drought-affected rhizosphere soils | Vigna mungo and P. sativum (L.) | – | Germination attributes, plant length (root and shoot) and dry biomass enhanced | Saikia et al. (2018) |
| 35 | Mitsuaria sp. and Burkholderia | Arabidopsis thaliana | A. thaliana and Zea mays (L.) | – | Lowered evapotranspiration; altered proline, MDA, and levels of plant hormones. | Huang et al. (2017) |
| 36 | Bacillus pumilus and Bacillus firmus | Rhizosphere of Solanum tuberosum | S. tuberosum (L.) | – | Enhanced proline content in tubers; greater mRNA expression levels of several ROS scavenging enzymes responsible for increased plant tolerance to salt and drought stress. | Gururani et al. (2013) |
| 37 | Bacillus cereus AR156, Bacillus subtilis SM21 and Serratia sp. XY21 | Soil | Cucumis sativus (L.) | – | Root:shoot ratio and vegetative growth increased | Wang et al. (2012) |
| 38 | Pseudomonas fluorescens ACC-5 | Nodule | Pisum sativum (L.) | – | Increased water uptake by plants | Zahir et al. (2008) |
| 39 | Pseudomonas sp. | Drought-stressed soil | Pisum sativum (L.) | – | Increased plant height, leaf-to-stem ratio, fresh plant weight, dry biomass, chlorophyll a, b, and total chlorophyll; increased N, P, and K contents. | Arshad et al. (2008) |
| Heavy metal stress | ||||||
| 40 | Bacillus gibsonii (PM11) and Bacillus xiamenensis (PM14) | Industrially polluted rhizosphere | Linumusitatissimum (L.) | – | Incresed fresh and dry biomass, chlorophyll content, proline concentration, and antioxidant enzymatic activity of plants | Zainab et al. (2020) |
| 41 | Agrobacterium fabrum and Leclercia adecarboxylata | Metal-contaminated rhizosphere | Zea mays (L.) | – | Potentially alleviated Cr toxicity and improved the overall growth of plants by reducing metal uptake | Danish et al. (2019) |
| 42 | Rhizobium leguminosarum bv. viciae 1066S | Metal-contaminated rhizosphere | Pisum sativum (L.) | – | Increased shoot biomass, nodulation, nitrogen fixation, water usage efficiency (WUE), and nutritional mineral uptake | Belimov et al. (2019) |
| 43 | Agrobacterium fabrum (CdtS5) and Stenotrophomonas maltophilia (CdtS7) | Cd-contaminated wheat rhizophere | Tritium estivum (L.) | Cd | Alleviated Cd toxicity and lowered uptake of Cd; improved growth, chlorophyll content and yield attributes of wheat | Zafar-Ul-Hye et al. (2018) |
| 44 | Combination of Pseudomonas sp., Bacillus cereus and Bacillus sp. | Rhizosphere soil | Festuca rubra and Brassica napus (L.) | Heavy metals | Sequestered the metal, reduced proline, MDA and antioxidant enzymes, reduced metal levels within the plant | Grobelak et al. (2018) |
| 45 | Azotobacter chroococcum | Metal-contaminated rhizosphere | Zea mays (L.) | Heavy metals | Detoxified the metals and increased biological and physiological parameters of the plant | Rizvi and Khan (2018) |
| 46 | Pseudomonas aeruginosa | Metal-polluted soil | C. arietinum (L.) | Heavy metals | Enhanced root length, shoot length, biomass, chlorophyll formation, nodulation, symbiotic attributes and seed yield of plant | Saif and Khan (2018) |
| 47 | Enterobacter aerogenes MCC 3092 | Rhizosphere of Oryza sativa | Oryza sativa (L.) | Cd | Alleviated phytotoxicity of Cd, reduced level of ethylene, antioxidant enzymes (CAT, SOD, POD), increased growth and chlorophyll content of plants | Pramanik et al. (2018) |
| 48 | Enterobacter ludwigii (HG 2) and Klebsiella pneumonia | Alternanthera sessilis and Cyperus esculentus rhizosphere | T. aestivum (L.) | Cr | Much improved growth promotion of wheat seedlings. | Gontia-Mishra et al. (2016) |
| 49 | Enterobacter sp., Serratia sp. and Klebsiella sp. | Rhizospheres of plants growing in mining waste | Helianthus annuus (L.) | Pb | Lowered toxicity of Cd, promoted growth features of plants | Carlos et al. (2016) |
| 50 | Pseudomonas fluorescens and Bacillus thuringiensis | Rhizosphere of Zea mays | T. aestivum (L.) | Cr | Improved plant growth and decreased Cr accumulation in roots and shoots | Shahzadi et al. (2013) |
| 51 | Pseudomonas stutzeri A1501 | – | Oryza sativa (L.) | Ni | increased metal tolerance of plants | Han et al. (2015) |
| 52 | Azotobacter sp. | Metal-contaminated rhizosphere | Zea mays (L.) | Pb | Lowered Pb toxicity and enhanced plant biometric parameters, biomass production, chlorophyll a and b and carotenoids, protein, proline, glutathione S-transferase and enzymes of POD and CAT | Hassan et al. (2014) |
| 53 | Ochrobactrum sp. and Bacillus spp. | Slag disposal site | Oryza sativa (L.) | Heavy metals | Mitigated toxicity of heavy metals, reduced ethylene level and enhanced overall growth of plants | Pandey et al. (2013) |
| Organic pollutant stress | ||||||
| 54 | Burkholderia sp. | Soil | Assorted vegetables | Organic pollutant | Lowered phenol toxicity, thus increasing overall functioning of plants | Chen et al. (2017) |
| 55 | Enterobacter intermedius, Bacillus circulans and Serratia carnosus | Z. mays and per nigrum Rhizosphere soil | Z. mays (L.) | Organic pollutant | Improvement in vegetative development of plant was quite noticeable | Ajuzieogu et al. (2015) |
| 56 | Pseudomonas aeruginosa SLC-2and Serratia marcescens BC-3 | Contaminated soil | Avena sativa (L.) | Organic pollutant | Degraded/detoxified the pollutant and improved biological properties and yield of plants even in petroleum-contaminated soil | Liu et al. (2015) |
| 57 | Acinetobacter sp. | Ployscyclic aromatic hydrocarbon (PAHs)-contaminated soil | A. sativa (L.) | Organic pollutant | DegradedPAHs and hydrocarbons; decreased level of MDA, free proline content and ROS-scavenging enzymes; increased overall performance of plants | Xun et al. (2015) |
| 58 | Pseudomonas aeruginosa and Serratia marcescens | Rhizosphere of Echinochloa | A. Sativa (L.) | Organic pollutant | A pronounced increase in A. sativa plants | Liu et al. (2015) |
| Agrochemicals stress | ||||||
| 59 | Burkholderiacepacia | Cabbage rhizosphere | C. arietinum (L.) | Pesticide | Alleviated toxicity of glyphosate; enhanced overall plant growth and performance | Shahid and Khan (2018) |
| 60 | Rhizobium leguminosarum | Root nodules of pea | P. sativum (L.) | Pesticide | Improved length, biomass, symbiotic features, nutrient uptake and seed attributes of plants under kitazin stress | Shahid et al. (2019a,b) |
| Biotic stress | ||||||
| 61 | Pseudomonas putida | Withaniasomnifera rhizosphere soil | Papaver somniferum (L.) | Peronospora sp. causing downy mildew disease | Biochemical and physiological (stomatal behavior and rate of transpiration) parameters significantly increased | Barnawal et al. (2017) |
| 62 | Bacillus xiamenensis PM14 | Sugarcane rhizosphere | Saccharum officinarum L. | Colletotrichum falcatum causing red rot disease | Potentially suppressed symptoms of disease, enhanced plant growth, enhanced production of defensive enzymes and content of proline | Xia et al. (2020) |
| 63 | Pseudomonas sp. strain S3 | rhizospheric soil of turmeric (Curcuma longa) | Solanum lycopersicum (L.) | Rhizoctonia solani | Improved morphological features, photosynthetic attributes and osmolytes in plants | Pandey and Gupta (2020) |
| 64 | Paenibacilluslentimorbus B-30488 | rhizospheric soil of tomato | Solanum lycopersicum (L.) | Scelerotiumrolfsii causing southern blight diseases | Controlled the disease, increased defense enzymes and improved plant growth attributes | Dixit et al. (2016) |
| 65 | Delftiatsuruhatensis WGR–UOM–BT1 | Rauwolfia serpentina Rhizosphere | Solanum lycopersicum (L.) | Fusarium oxysporum | Protected plant from fungal disease; significantly improved characteristic growth features of tomato | Prasannakumar et al. (2015) |
In general, every plant has innate ability to withstand the adverse effects of the environment. However, under such stressed conditions, a number of physio-biochemical cascades activated and deactivated upon sensing the type of stresses. Among them, certain phytohormones play important role in stresses plants (Babalola et al., 2003; Glick et al., 2012). However, a number of microorganisms present either in rhizosphere, phyllosphere or endosphere of the plants play crucial role in the sensing and transducing signal to the plants under stressed conditions in coordinated manner. It is well established that ethylene at lower concentration worked as signaling molecules and regulate several gene expression, transcription and translation lead to overall plant development (Shaharoona et al., 2006; Yim et al., 2012; Bal et al., 2013; Ek-Ramos et al., 2019). In contrast, ethylene at higher concentration causes programme cell death, accelerating abscission, aging, inhibiting root elongation, senescence, leaf and fruit drop, etc. Under such circumstances, ACC deaninase either produced by plant or microorganisms cleave the ACC and lowering down the production of excess amount of ethylene even under stressed condition (Glick et al., 1998). Further, microorganisms synthesizing IAA along with endogenous plant IAA could accelerate the amalgamation of the enzyme ACC synthase translating the compound S-adenosyl methionine to ACC being the immediate precursor of ethylene in higher plants (Glick, 2012). It was revealed that phyllosphere methylobacteria distributed in the rice leaves produce the enzyme ACC deaminase, which control the ethylene concentrations level in the rice plant (Chinnadurai et al., 2009). The beneficial impact of ACCD-positive PGPR in the alleviation of various stresses is briefly discussed in the following sections.
Salinity stress
Salinity is a critical environmental stress that strongly influences plant productivity worldwide (Pirasteh-Anosheh et al., 2016; Hussain et al., 2019; Singh S. et al., 2021; Singh U. B. et al., 2021). Among the total global cultivable area, ~20% of area suffer from salinity stress; as a direct result of irrigation, this situation is becoming more serious (Kataria and Verma, 2018; Singh U. B. et al., 2021). Globally, the land area affectted by salinity/sodicity is estimated to be over 800 million hectares (MH) (FAO, 2008; Rengasamy, 2010; Dixit et al., 2015). Salinity affects plant physiology via differing mechanisms including disruption of chlorophyll synthesis, increased levels of photorespiration and transpiration, and fluctuation in homeostasis in plant cells (Miller et al., 2010; Sahu et al., 2021). Nutrient imbalance due to salinity stress is another variable that adversely affects plant growth and yield (Singh U. B. et al., 2021). This imbalance interrupts proper uptake and transport of nutrients to growing shoots and that ultimately causes mineral deficiencies in the plant (Panda et al., 2017; Singh U. B. et al., 2021). High levels of salt result in oxidative burst of cellular organelles. Increased production of ROS follows, which damages the plasma membrane and adversely affectscellular metabolism and homeostasis. Salinity causes overproduction of ethylene which increases abscission of leaves and petals, and accelerates organ senescence that ultimately leads to premature death of the plant (Zahir et al., 2009; Singh S. et al., 2021). ACCD-containing PGPR have been used to resolve salinity stress in several crops including vegetables and legumes (Shahid et al., 2021a, 2022a,b,c). These PGPR transform ACC to NH3 and α-ketobutyrate, which the plant uses as a source of nitrogen, while also mitigating the deleterious effects of salt stress (Siddikee et al., 2012; Barnawal et al., 2014). Even in rather saline environment, salt-tolerant and ACCD-producing bacteria can thrive, and their beneficial characteristics assist plants in overcoming the impacts of stress (Thijs et al., 2014; Han et al., 2021; Sagar et al., 2022).
Microorganisms that survive and flourish in media containing sodium chloride (NaCl) up to 1–33% are known as halotolerant bacteria (Arora et al., 2017; Kumar M. et al., 2019; Singh et al., 2020b). Substantial literature is available on salt-tolerant ACCD-producing PGPR strains that can safeguard plants against the harmful effects of salt. Wang C. et al. (2016), Wang P. et al. (2016), and Wang Q. et al. (2016) found that the ACCD-synthesizing V. paradoxus 5C-2 reduced the negative effects of NaCl in pea by enhancing water relations and ion homeostasis, as well as increasing plant growth, dry biomass, chlorophyll synthesis, and yield when pea was grown in a saline environment. Halotolerant strains of Enterobacter, Bacillus and Acinetobacter containing ACCD genes increased plant height, biomass, leaf-to-stem ratio, leaf relative water content (LRWC), production of leaf chlorophyll and nutrient status of Medicago sativa (L.) plants cultivated in salinity-stressed agricultural soil (Daur et al., 2018). The early nodulation process and growth of common beans cultivated under high levels of salt stress have been shown to be stimulated by the endophytic bacterium Serratia grimesii BXF-1 (Tavares et al., 2018). In a similar study, Ji et al. (2020) reported that Glutamicibacter sp. strain YD-01 tolerated exceedingly high salt levels. When treated as a biological inoculant to Oryza sativa (L.), this strain exhibited low levels of Na+ buildup and decreased electrolyte leakage (EL) during salt treatment, as well as increased plant productivity. In a similar study, two NaCl-tolerant and ACCD-positive PGPR, Aneurinibacillusaneurinilyticus ACC-02 and Paenibacillus sp. ACC-06, imparted a positive response to morphological attributes (length and biomass), biochemical features, and yield of salt-treated Phaseolus vulgaris (L.) by limiting the negative effects of NaCl (Gupta and Pandey, 2019). Wheat (Triticum aestivum L.) plants cultivated in saline-sodic soil treated with fertilizer and ACCD positive strains of S. succinus, Zhihengliuella halotolerans and Bacillus sp., either alone or in combination, grew and yielded better than those cultivated in soil treated solely with NaCl (Orhan, 2016; Singh U. B. et al., 2021).
Sapre et al. (2018a,b) reported that a salt-tolerant and ACCD-producing PGPR strain of Klebsiella sp. was inoculated to Avena sativa plants treated with varying levels of NaCl. The PGPR strain improved plant development under salt stress and progressively regulated the rbcL and WRKY1 gene expression profiles.
Drought stress
Insufficient availability of water, referred to as drought, unfavorably affects crop productivity. Under drought stress many plants physiological and biochemical effects including reduction in water potential, turgor loss, wilting, stomatal closure, and alteration in structures of membranes and proteins are reported (Kaushal and Wani, 2016). Drought stress is documented to slow plant growth, resulting in lower yields, necessitating the use of drought-resistant plant growth techniques. Several researchers have utilized ACCD-producing and drought-tolerant PGPR strains for ameliorating water stress. ACCD-positive PGPR strains Ochrobactrumpseudogrignonense RJ-12, Pseudomonas sp. RJ-15 and B. subtilis RJ-46 were isolated from drought-stressed rhizosphere soil and utilized as bioinoculants to Vigna mungo and Pisum sativum cultivated under drought stress. ThePGPR strains increased the germination attributes, morphological features and dry weight accumulation in plants (Saikia et al., 2018). Saleem et al. (2018) reported that two ACCD-containing drought-resistant Enterobacter HS-9 and Bacillus G-9 strains improved overall growth of Mucuna pruriens cultivated in drought-stressed conditions. In another crop-based study, two strains of Bacillus (B. pumilus and B. firmus) were reported to enhance the expression levels of mRNA of several ROS scavenging enzymes, and decreased proline concentration in drought-stressed tubers (Gururani et al., 2013). Additionally, the inoculation of ACCD-producing drought-tolerant PGPR strains of Burkholderiaand Mitsuaria sp. recovered from the rhizosphere of Arabidopsis thaliana were reported to lower evapotranspiration rate as well as levels of proline and malondialdehyde. Levels of phytohormones were also altered (Huang et al., 2017).
Waterlogging stress
Flooding is a common abiotic stress that impacts a wide range of plants. During flooding, plant roots experience anoxia (lack of oxygen), prompting production of ACC that oxidizes ethylene as it moves within the plant. The secreted ethylene has negative consequences on leaves, such as epinasty (rapid nastic motions), chlorosis, necrosis, and lower fruit output (Paul et al., 2016). To eliminate the epinasty response in plants, ethylene production inhibitors likeCO2, cobalt chloride, 7-chloro-4-ethoxycarbonylmethoxy-5-methyl-2,1,3-benzothiadiazole, L-α-(2-aminoethoxyvinyl)-glycine (AVG), silver nitrate, and 1-methylcyclopropene (1-MCP) have been used (Jackson, 2008). In addition to these, ACCD-synthesizing PGPR operate as an ACC sink, and their application reduces ethylene levels significantly, protecting plants from flooding stress (Ali and Kim, 2018). Tolerance against waterlogging stress in rice seedlings was enhanced by ACC deaminase-synthesizing Streptomyces sp. GMKU 336. The bacteria reduced levels of ethylene and improved root elongation, biomass production, leaf area and chlorophyll content (Jaemsaeng et al., 2018). Etesami et al. (2014) reported that ACC deaminase-positive endophytic P. fluorescens strain REN1 significantly elongated rice roots, endophytically colonized plants and promoted development of seedlings under waterlogged conditions. Barnawal et al. (2012) observed that ACC deaminase PGPR strains protected Ocimum sanctum (L.) plants against waterlogging. Compared to waterlogged plants without bacterial inoculation, the selected bacteria modulated the negative alterations in stress-induced ethylene production, decreased the lipid peroxidation and proline content, and substantially increased the chlorophyll concentration and foliar nutrient uptake in O. sanctum plant. Furthermore, ACCD-containing PGPR strains (P. putida ATCC17399/pRK415, Enterobacter cloacae UW4 and E. cloacae CAL2) enhanced various physiological reactions of S. lycopersicum (L.) under flooding stress (Grichko and Glick, 2001).
Agrochemical stress
Agrochemicals including pesticides, herbicides and fungicides are among the most significant anthropogeniccompounds that adversely affect microbial physiology (Shahid et al., 2019a,b, 2020), composition and functions (Ataikiru et al., 2019; Shahid et al., 2021b; Shahid and Khan, 2022a,b), soil fertility (Sanchez-Hernandez, 2019) and crop productivity (Shahid et al., 2018a,b; Khan et al., 2020). Stress ethylene production causes the agrochemical to obstruct plant development viaunknown mechanisms. Several beneficial pesticide-tolerant soil microbes (PGPR) are reported which can degrade pesticides (Shahid et al., 2019a,b, 2021c; Shahid and Khan, 2019). In addition, a plentiful ACC deaminase-positive and pesticide-tolerant PGPR has been shown to support legumes grown in degraded or stressed soils (Zaidi et al., 2016; Ahmed et al., 2017; Rizvi et al., 2017; Zaidi et al., 2017). Shahid and Khan (2018) reported that glyphosate-tolerant PGPR strain Burkholderiacepacia PSBB1 isolated from the contaminated rhizosphere of Vicia faba produced considerable ACC deaminase and alleviated the toxicity of the herbicide, and enhanced overall growth and performance of chickpea plants raised in herbicide-amended soil.
Heavy metal stress
Soil pollution by heavy metals has become one of the greatest environmental and agronomic challenges worldwide (Ashraf et al., 2019). Certain heavy metals including Zn, Cu, and Co are used by plants in trace quantities; however, they become toxic at higher concentrations and cause deleterious effects to plant growth and development (Dixit et al., 2015). Roots are primarily responsible for nutritient (including metal) uptake by plants. Stress ethylene is produced in soils having high concentrations of heavy metals, which limits root morphogenesis (Saif et al., 2017). Numerous reports existin the literature regarding utilization of metal-tolerant and ACCD-generating PGPR strains capable of optimizing plant growth under heavy metal-stressed conditions (Płociniczak et al., 2014; Pramanik et al., 2018; Manoj et al., 2020). ACCD-positive PGPR support phytoremediation by increasing the uptake of harmful metals by enlarging/improving root growth under metal stress (Santos et al., 2019). In this regard, several agronomists and microbiologists have isolated metal-tolerant and ACCD-producing PGPR strains from different contaminated sites for use as potent bioinoculants for various crops grown in soils contaminated with heavy metals. For instance, single or co-inoculation of metal-tolerating ACCD-producing PGPR strains such as Bacillus sp., B. cereus and Pseudomonas sp. to Festuca rubra and Brassica napus plants resulted in substantial increases in plant growth and yield (Grobelak et al., 2018). Pandey et al. (2013) reported that metal-tolerant ACCD-positive PGPR strains of Ochrobactrum sp. and Bacillus spp., when used with rice plantsgrown in heavy metal-contaminated soils, mitigate the toxic effect of metals, reduced ethylene levels and enhanced overall growth of plants. Similarly, two Cr-tolerant PGPR strains, Enterobacter ludwigii and Klebsiella pneumonia, significantly reduced the toxicity of Cr and promoted seedling germination, and increased protein and carbohydrate content of wheat plants even in the presence of high concentrations of Cr (Gontia-Mishra et al., 2016). Other PGPR strains like Pseudomonas fluorescens and Bacillus thuringiensis (Shahzadi et al., 2013), Achromobacter xylosoxidans and Bacillus pumilus (Chandra et al., 2019), Enterobacter sp., Serratia sp. and Klebsiella sp. (Carlos et al., 2016), and Enterobacter aerogenes MCC 3092 (Pramanik et al., 2018) are also reported to alleviate toxic ethylene levels vis-à-vis enhanced growth of crops.
Temperature (chilling and heat) stress
Extreme (low or high) temperatures cause substantial losses in yield and productivity of crops (Lesk et al., 2016; Wang C. et al., 2016; Wang P. et al., 2016; Wang Q. et al., 2016). Temperature extremes cause plants to modify many metabolic processes (Yadav, 2010). Temperature changes cause drastic alteration in membrane shape, catalytic characteristics, enzyme performance, and nutrient transport (Subramanian et al., 2016). Low temperatures (between 0 and 15°C) cause yield losses in a variety of tropical and subtropical crops. Cold stress generally slows rate of germination, reduces growth, causes yellowing (chlorosis) of leaves, and reduces tiller formation (Yadav, 2010). Chilling causes lesions on leaf surfaces, discoloration, and rapid senescence in horticultural crops due to reduced chlorophyll production. Chilling, like other environmental stresses, results in production of ethylene which inhibits overall plant development. The use of ACCD-synthesizing bacterial strains in Vitis vinifera (L.) and Solanum lycopersicum (L.) was reported to alleviate chilling stress (Theocharis et al., 2012; Subramanian et al., 2016). Some cold-tolerant and ACCD-negative PGPR strains, viz., P. frederiksbergensis, Sphingomonasfaeni and Flavobacterium sp. were transformed with a plasmid pRKACCharboring the acdS gene from Pseudomonas putida UW4. The role of these altered PGPRs that overexpressed the acdS gene in alleviating chilling stress in S. lycopersicum (L.), Setariaitalica (L.) and Eleusine coracanawas investigated (Subramanian et al., 2015; Srinivasan et al., 2017).
Air pollution stress
Sulfur dioxide (SO2), ozone (O3), nitrogen oxides (NOx), and volatile organic compounds (VOCs) are anthropogenicand naturally-occurring pollutants that impart negative impacts to human health and ecosystems (Sharma et al., 2013). Atmospheric pollutants deleteriously affect plants by inhibiting enzyme systems and metabolic activities (Saxena and Kulshrestha, 2016). The increased synthesis of ethylene in plants in response to air pollutants is well documented, and is thought to be one of the key regulators in plant tolerance to air pollution stress, particularly O3 exposure (Rao and Davis, 2001). According to one study, inhibition of the ethylene expressing gene resulted in considerable reduction of O3-induced leaf damage in tomato plants (Moeder et al., 2002). As a result, bacteria that produce ACC deaminase have received greater attention as a stress management tool for plants suffering from air pollution.
Nutrient deficiency
Excessive application of chemical fertilizers in agriculture is costly, andosols considered a potential source of soil and water pollution (Kumar R. et al., 2019). A variety of beneficial ACC deaminase-synthesizing bacteria are known to boost productivity and efficiency of fertilized crops, either directly or indirectly. At low fertilizer application rates, PGPR ACC deaminase activity may reduce ethylene concentrations in wheat plants exposed to nutritional stress by hydrolyzing ACC to α-ketobutyrate and NH3 (Hemissi et al., 2019). The authorsfurther claim that PGPR, which comprise ACCD-generating bacteria, might be used in concert with fertilizers to boost nutrient intake and plant development. Multiple studies have demonstrated the critical role of microbially-synthesized ACC deaminase in promoting plant growth, which allows them to withstand abiotic stress and ultimately create a symbiotic interaction between plants and the native rhizosphere (Tahir et al., 2006).
Stress from other organic contaminants
Rapid worldwide industrial development and modernization has resulted in the manufacture and release of significant volumes of hazardous organic pollutants into natural habitats. Polycyclic aromatic hydrocarbons (PAHs), petroleum, and other xenobiotics based on hydrocarbons are known to limit crop productivity (Jajoo, 2017; Li et al., 2019). Most plants are stressed by the presence of organic pollutants in soil, which causes them to produce more ethylene. However, the exact mechanisms of excessive ethylene production remain unknown. Organic contaminants such as refrigerants and organic solvents are reported to be degraded by several bacterial species belonging to different genera. In the presence of organic pollutants, ACCD-producing PGPRs have consistently improved plant development (Xun et al., 2015). PGPR can also aid in plant-mediated remediation (phytoremediation) by bio-transforming harmful substances to innocuous forms. ACCD-producing PGPR is known to play a significant role in elongation of roots and overall plant growth, which explains why host plants are superior at phytoremediating organic chemicals. Phenol-degrading PGPR strain Burkholderia sp. isolated from phenol-contaminated soil was reported to reduce the phytotoxicity of phenol and improve growth and biochemical activities in plants (Chen et al., 2017). Similarly, ACCD-producing and petroleum-degrading PGPR strains S. marcescens BC-3 and P. aeruginosa SLC-2 augmented the growth and physiological properties of Avena sativa grown in petroleum-contaminated soil (Liu et al., 2015). An ACCD-producing and PAHs-tolerant soil bacterium Acinetobacter sp., when applied to A. sativa plants cultivated in hydrocarbon-contaminated soil, decreased the MDA, antioxidant enzymes, and free proline contents of shoot tissues and increased yield, photosynthetic pigments, and protein content of plants (Xun et al., 2015). In, another study, two PGPR-degrading P. aeruginosa and S. marcescens strains isolated from the rhizosphere of Echinochloa promoted the growth of Ascophyllum sativum (Liu et al., 2015). Application to polluted soil of Microbacterium sp. strain F10a-R containing ACC deaminase enzymes and other multifarious PGP features resulted in elimination of pyrene and phenanthrene, both hazardous PAHs, and boosted wheat growth (Sheng et al., 2009).
Biotic stress
Pathogen attack
Plants often respond to attack/infection of bacterial pathogens, fungal pathogens, viruses, and nematodes by increasing ethylene levels in their tissue (Van Loon et al., 2006). Soil application of potent ACCD-producing PGPR strains may reduce injuries from induced ethylene triggered by numerous pathogenic bacteria such as Agrobacterium tumefaciens (Toklikishvili et al., 2010), Pseudomonas syringae pv. tomato (Indiragandhi et al., 2008), and Erwinia spp. (Wang et al., 2000), and those caused by phytopathogenic fungi such as Pythium aphanidermatum (El-Tarabily, 2013), P. ultimum (Wang et al., 2000), and Pyriculariaoryzae (Amutharaj et al., 2012). The PGPR either directly or indirectly inhibit pathogen development by synthesizing a variety of antimicrobial metabolites (Singh et al., 2016a, 2020a). The efficiency and efficacy of varying species and genera of ACC deaminase-producing PGPR strains have demonstrated a positive effect in the suppression of different diseases caused by phytopathogens (Singh et al., 2016b; Shahid et al., 2017). Bursaphelenchusxylophilus is a pathogenic nematode commonly known as pine/wood nematode and is associated with by pine wilt disease. This nematode was suppressed by ACC deaminase-containing B. subtilis (Nascimento et al., 2013). In an in-vitro study, Al-Shwaiman et al. (2022) reported that multi-stress tolerant and biocontrol agent Beijerinckiafluminensis supressed the growth of major fungal phytopathogens (Aletrnariaalternata, Rhizoctonia solani, Fusariumoxysporum, Ustilaginoidea virens) by producing defensive extracellular enzymes. Dixit et al. (2016) assessed the plant growth-regulating and biocontrol efficiency of ACC deaminase-producing strain Paenibacilluslentimorbus B-30488, which suppresses the growth of fungal pathogens and inhibits southern blight disease in tomatoes. Additionally, ACCD containing Pseudomonas putida recovered from Withaniasomnifera (L.) rhizosphere soil and applied toPeronospora sp. causing downy mildew disease infected Papaver somniferum (L.) plants. It was observed that the potential ACCD candidate significantly modulated the biochemical and physiological (stomatal behavior and rate of transpiration) parameters by reducing the incidence of disease in plant (Barnawal et al., 2017; Malviya et al., 2020). Based on these data, inoculation of ACC deaminase-containing bacteria to crops suffering from pathogenic stress can protect the plants effectively. In addition, ACCD-synthesizing PGPR strains lower the quantity of ethylene generated in plants infected with soil-borne and foliar disease (Glick, 2014).
Certain plant growth-promoting microorganisms produce the enzyme ACC deaminase, which indirectly promote plant growth by lowering down the ethylene level in plants (Glick, 1995). Under biotic stressed condition, ACC deaminase transcriptionally regulated differently by several biotic factors (Gontia-Mishra et al., 2014). Few reports indicated that Methylobacterium spp. (phytopathogenic in nature) modulate plant growth by inhibiting plant pathogens indirectly. ACCD producting Methylobacterium spp. synthesized certain polymer degrading pectinase and cellulase, suggesting that they can indirectly induce systemic resistance during pathogen attack (Chinnadurai et al., 2009; Tsolakidou et al., 2019). Under biotic stressed condition, PGPMs produce ACC deaminase which modulates the level of ethylene by hydrolyzing ACC, a precursor of ethylene, in ammonia and a-ketobutyrate (Babalola et al., 2003; Nascimento et al., 2014). The lower concentration of ethylene induced jasmonate dependent pathways in plants which further modulate synthesis of antioxidative biomolecules which in turn reduce the synthesis of reactive oxygen species and superoxide radicals and protect plants from programme cell death against invasion caused by hemi-biotroph and necrotroph. In contrast, ET dependent pathways lead to PCD in the plants attacked by obligate and biotrophs which restrict colonization and invasion of the pathogen. Some time, elevated ET cause premature leaf and fruit drop in the plants attacked by biotrophs (Etesami et al., 2020).
Concluding remarks and future prospects
In agricultural systems worldwide, environmentally-benign management approaches are necessary to improve food security in the face of constantly changing agro-climatic conditions. The current review focuses on the interaction and mechanistic action of ACCD-synthesizing rhizobacteria on abiotic and biotic stress tolerance induction. It is well recognized that multiple stress-tolerant ACC deaminase-synthesizing bacterial strains are advantageous over other conventional bacterial strains, and can thrive in sufficient numbers in new and stressful environments to impart favorable impacts to crop plants. Under abiotic- and biotic-stressed situations, powerful PGPR strains enhance crop growth and production. Keeping in mind the many significant environmental hazards encountered in agronomic practices from anthropogenic and natural factors, there is an urgent need for a major paradigm shift in agricultural practices. The costs associated with generating and modifying transgenic plants capable of tolerating biotic and abiotic stresses are substantial. To overcome this problem, focus has shifted to the identification and development of ACCD-containing PGPR formulations that support plants in combatting stressed environmental conditions. The survival of such beneficial PGPR strains under harsh circumstances poses a challenge for their large-scale production, yet the exploitation of a powerful PGPR strain is likely to provide wide-ranging solutions to problems in modern agriculture. Research has demonstrated that ethylene balance is crucial for plant growth and development under abiotic stress conditions, and application of PGPR bacteria may be useful in protecting plants from such stresses. Therefore, rhizobacteria should be screened for ACC deaminase production. Utilizing ACC deaminase-synthesizing bacterial strains as biological inoculants for abiotic stress management could be critical for long-term sustainability of agriculture. Furthermore, uncovering the essential mechanistic action of these PGPR strains will help to expand the applicability of this technology.
Author contributions
MS and MK conceived and designed the study. MS, MK, US, PS, and HS performed the literature search. MS wrote the first draft of the manuscript. MS and US prepared the figures and artwork. MS, MK, US, RK, RS, and AK edited the manuscript. MS, PS, and AM formatted the reference as per Journal’s style. All authors contributed to the article and approved the submitted version.
Funding
This work is funded by Network Project on Application of Microorganisms in Agriculture and Allied Sectors (AMAAS), Indian Council of Agricultural Research, New Delhi.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer RK declared a shared affiliation with the authors MS, US, and HS to the handling editor at the time of review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
MS is thankful to DST-SERB for the National Post-Doctoral Fellowship (PDF/2022/000970). The authors MS and US would like to thank the ICAR-NBAIM for providing research facilities. Thanks to John Pichtel, Ball State University (United States) for assistance with the manuscript. Authors have picked-up some of the figures/artwork to prove their concept and acknowledged the Social cites for valuable help. We would like to extend their sincere thanks to the Network Project on Application of Microorganisms in Agriculture and Allied Sectors (AMAAS), Indian Council of Agricultural Research, New Delhi, for providing financial support to carry out the research.
References
- Ahmad E., Khan M. S., Zaidi A. (2013). ACC deaminase producing pseudomonas putida strain PSE3 and rhizobium leguminosarum strain RP2 in synergism improves growth, nodulation and yield of pea grown in alluvial soils. Symbiosis (Philadelphia, PA) 61, 93–104. doi: 10.1007/s13199-013-0259-6 [DOI] [Google Scholar]
- Ahmed B., Zaidi A., Khan M. S., Rizvi A., Saif S., Shahid M. (2017). “Perspectives of plant growth promoting rhizobacteria in growth enhancement and sustainable production of tomato” in Microbial strategies for vegetable production, eds. Zaidi, A., Khan, M. (Cham: Springer; ), 125–149. [Google Scholar]
- Ajuzieogu C. A., Ibiene A. A., Stanley H. O. (2015). Laboratory study on influence of plant growth promoting rhizobacteria (PGPR) on growth response and tolerance of Zea mays to petroleum hydrocarbon. Afr. J. Biotechnol. 14, 2949–2956. doi: 10.5897/AJB2015.14549 [DOI] [Google Scholar]
- Ali S., Charles T. C., Glick B. R. (2014). Amelioration of high salinity stress damage by plant growth-promoting bacterial endophytes that contain ACC deaminase. Plant Physiol. Biochem. 80, 160–167. doi: 10.1016/j.plaphy.2014.04.003, PMID: [DOI] [PubMed] [Google Scholar]
- Ali S., Kim W. C. (2018). Plant growth promotion under water: decrease of waterlogging-induced ACC and ethylene levels by ACC deaminase-producing bacteria. Front. Microbiol. 9:1096. doi: 10.3389/fmicb.2018.01096, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali B., Wang X., Saleem M. H., Azeem M. A., Afridi M. S., Nadeem M., et al. (2022a). Bacillus mycoides PM35 reinforces photosynthetic efficiency, antioxidant defense, expression of stress-responsive genes, and ameliorates the effects of salinity stress in maize. Life 12:219. doi: 10.3390/life12020219, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali B., Wang X., Saleem M. H., Sumaira, Hafeez A., Afridi M. S., et al. (2022b). PGPR-mediated salt tolerance in maize by modulating plant physiology, antioxidant defense, compatible solutes accumulation and bio-surfactant producing genes. Plan. Theory 11:345. doi: 10.3390/plants11030345, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shwaiman H. A., Shahid M., Elgorban A. M., Siddique K. H., Syed A. (2022). Beijerinckiafluminensis BFC-33, a novel multi-stress-tolerant soil bacterium: deciphering the stress amelioration, phytopathogenic inhibition and growth promotion in Triticum aestivum (L.). Chem 295:133843. doi: 10.1016/j.chemosphere.2022.133843Get [DOI] [PubMed] [Google Scholar]
- Amutharaj P., Sekar C., Natheer S. E. (2012). Intergeneric microbial co aggregates: bio inoculation effect of ACC deaminase positive wild type strains of pseudomonas and Paenibacillus, as co aggregates, on the maximization of ISR against Pyriculariaoryzae in upland rice cv. ASD-19. CIB Tech. J. Microbiol. 1, 57–66. [Google Scholar]
- Arora S., Singh A. K., Sahni D. (2017). “Bioremediation of salt-affected soils: challenges and opportunities” in Bioremediation of salt affected soils: an Indian perspective, eds. Arora, S., Singh, A., Singh, Y. (Cham: Springer; ), 275–301. [Google Scholar]
- Arshad M., Shaharoona B., Mahmood T. (2008). Inoculation with pseudomonas spp. containing ACC deaminase partially eliminates the effects of drought stress on growth, yield, and ripening of pea (Pisum sativum L.). Pedosphere 18, 611–620. doi: 10.1016/S1002-0160(08)60055-7 [DOI] [Google Scholar]
- Arya B., Komala B. R., Sumalatha N. T., Surendra G. M., Gurumurthy P. R. (2018). PGPR induced systemic tolerance in plant. Int. J.Curr. Microbiol. Appl. Sci. 7, 453–462. [Google Scholar]
- Ashraf S., Ali Q., Zahir Z. A., Ashraf S., Asghar H. N. (2019). Phytoremediation: environmentally sustainable way for reclamation of heavy metal polluted soils. Ecotoxicol. Environ. Safe 174, 714–727. doi: 10.1016/j.ecoenv.2019.02.068, PMID: [DOI] [PubMed] [Google Scholar]
- Ataikiru T. L., Okpokwasili G. S. C., Okerentugba P. O. (2019). Impact of pesticides on microbial diversity and enzymes in soil. Public Administration Series--Bibliography, 4, 1–16. doi: 10.9734/sajrm/2019/v4i230104 [DOI] [Google Scholar]
- Azadikhah M., Jamali F., Nooryazdan H. R., Bayat F. (2019). Growth promotion and yield enhancement of barley cultivars using ACC deaminase producing Pseudomonas fluorescens strains under salt stress. Spanish J. Agric. Res. 17:16. doi: 10.5424/sjar/2019171-13828 [DOI] [Google Scholar]
- Babalola O. O., Osir E. O., Sanni A. I., Odhiambo G. D., Bulimo W. D. (2003). Amplification of 1-amino-cyclopropane-1-carboxylic (ACC) deaminase from plant growth promoting rhizobacteria in Striga-infested soil. African J. Biotechnol. 2, 157–160. doi: 10.5897/AJB2003.000-1032 [DOI] [Google Scholar]
- Bal H. B., Nayak L., Das S., Adhya T. K. (2013). Isolation of ACC deaminase PGPR from rice rhizo-sphere and evaluating their plant growth promoting activity under salt stress. Plant Soil 366, 93–105. doi: 10.1007/s11104-012-1402-5 [DOI] [Google Scholar]
- Barnawal D., Bharti N., Maji D., Chanotiya C. S., Kalra A. (2012). 1-Aminocyclopropane-1-carboxylic acid (ACC) deaminase-containing rhizobacteria protect Ocimum sanctum plants during waterlogging stress via reduced ethylene generation. Plant Physiol. Biochem. 58, 227–235. doi: 10.1016/j.plaphy.2012.07.008, PMID: [DOI] [PubMed] [Google Scholar]
- Barnawal D., Bharti N., Maji D., Chanotiya C. S., Kalra A. (2014). ACC deaminase-containing Arthrobacter protophormiae induces NaCl stress tolerance through reduced ACC oxidase activity and ethylene production resulting in improved nodulation and mycorrhization in Pisum sativum. J. Plant Physiol. 171, 884–894. doi: 10.1016/j.jplph.2014.03.007, PMID: [DOI] [PubMed] [Google Scholar]
- Barnawal D., Pandey S. S., Bharti N., Pandey A., Ray T., Singh S., et al. (2017). ACC deaminase-containing plant growth-promoting rhizobacteria protect Papaversomniferum from downy mildew. J. Appl. Microbiol. 122, 1286–1298. doi: 10.1111/jam.13417, PMID: [DOI] [PubMed] [Google Scholar]
- Belimov A. A., Dodd I. C., Hontzeas N., Theobald J. C., Safronova V. I., Davies W. J. (2009). Rhizosphere bacteria containing 1-aminocyclopropane1-carboxylate deaminase increase yield of plants grown in drying soil via both local and systemic hormone signalling. New Phytol. 181, 413–423. doi: 10.1111/j.1469-8137.2008.02657.x, PMID: [DOI] [PubMed] [Google Scholar]
- Belimov A. A., Hontzeas N., Safronova V. I., Demchinskaya S. V., Piluzza G., Bullitta S., et al. (2005). Cadmium-tolerant plant growth-promoting bacteria associated with the roots of Indian mustard (Brassica juncea L. Czern.). Soil Biol. Biochem. 37, 241–250. doi: 10.1016/j.soilbio.2004.07.033 [DOI] [Google Scholar]
- Belimov A. A., Zinovkina N. Y., Safronova V. I., Litvinsky V. A., Nosikov V. V., Zavalin A. A., et al. (2019). Rhizobial ACC deaminase contributes to efficient symbiosis with pea (Pisum sativum L.) under single and combined cadmium and water deficit stress. Environ. Exp. Bot. 167:103859. doi: 10.1016/j.envexpbot.2019.103859 [DOI] [Google Scholar]
- Bessadok K., Navarro-Torre S., Pajuelo E., Mateos-Naranjo E., Redondo-Gómez S., Caviedes M. Á., et al. (2020). The ACC-deaminase producing bacterium Variovorax sp. CT7. 15 as a tool for improving Calicotomevillosa nodulation and growth in arid regions of Tunisia. Microorganisms 8:541. doi: 10.3390/microorganisms8040541, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti N., Barnawal D. (2019). “Amelioration of salinity stress by PGPR: ACC deaminase and ROS scavenging enzymes activity” in PGPR amelioration in sustainable agriculture 85–10, eds. Amit Kishore Singh, Ajay Kumar and Pawan Kumar Singh (Woodhead Publishing. Elsevier; ). [Google Scholar]
- Bharti N., Pandey S. S., Barnawal D. (2016). Plant growth promoting rhizobacterial Dietzianatronolimnaea modulates the expression of stress responsive genes providing protection of wheat from salinity stress. Sci. Rep. 6:34768. doi: 10.1038/srep34768, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brader G., Compant S., Mitter B., Trognitz F., Sessitsch A. (2014). Metabolic potential of endophytic bacteria. Curr. Opin. Biotechnol. 27, 30–37. doi: 10.1016/j.copbio.2013.09.012, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlos M. H. J., Stefani P. V. Y., Janette A. M. (2016). Assessing the effects of heavy metals in ACC deaminase and IAA production on plant growth-promoting bacteria. Microbiol. Res. 188-189, 53–61. doi: 10.1016/j.micres.2016.05.001, PMID: [DOI] [PubMed] [Google Scholar]
- Carlson R., Tugizimana F., Steenkamp P. A., Dubery I. A., Hassen A. I., Labuschagne N. (2020). Rhizobacteria-induced systemic tolerance against drought stress in Sorghum bicolor (L.) Moench. Microbiol. Res. 232:126388. doi: 10.1016/j.micres.2019.126388, PMID: [DOI] [PubMed] [Google Scholar]
- Chandra D., Srivastava R., Glick B. R., Sharma A. K. (2020). Rhizobacteria producing ACC deaminase mitigate water-stress response in finger millet (Eleusine coracana (L.) Gaertn.). 3 Biotech 10:65. doi: 10.1007/s13205-019-2046-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra D., Srivastava R., Gupta V. V., Franco C. M., Sharma A. K. (2019). Evaluation of ACC-deaminase-producing rhizobacteria to alleviate water-stress impacts in wheat (Triticumaestivum L.) plants. Canadian J. Microbiol. 65, 387–403. doi: 10.1139/cjm-2018-0636 [DOI] [PubMed] [Google Scholar]
- Chatterjee P., Samaddar S., Anandham R., Kang Y., Kim K., Selvakumar G., et al. (2017). Beneficial soil bacterium pseudomonas frederiksbergensis OS261 augments salt tolerance and promotes red pepper plant growth. Front. Plant Sci. 8:705. doi: 10.3389/fpls.2017.00705, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan P. S., Anandham R., Han G. H., Sa T. (2017). Erratum to: isolation, characterization, and use for plant growth promotion under salt stress, of ACC deaminase-producing halotolerant bacteria derived from coastal soil. J. Microbiol. Biotechnol. 20, 1577–1584. doi: 10.4014/jmb.1007.07011 [DOI] [PubMed] [Google Scholar]
- Chen J., Li S., Xu B. (2017). Characterization of Burkholderia sp. XTB-5 for phenol degradation and plant growth promotion and its application in bioremediation of contaminated soil. Land Degrad. Dev. 28, 1091–1099. doi: 10.1002/ldr.2646 [DOI] [Google Scholar]
- Chinnadurai C., Balachandar D., Sundaram S. P. (2009). Characterization of 1-aminocyclopropane-1-carboxylate deaminase producing methylobacteria from phyllosphere of rice and their role in ethylene regulation. World J. Microbiol. Biotechnol. 25, 1403–1411. doi: 10.1007/s11274-009-0027-1 [DOI] [Google Scholar]
- Choi J., Roy Choudhury A., Walitang D. I., Lee Y., Sa T. (2022). ACC deaminase-producing Brevibacterium linens RS16 enhances heat-stress tolerance of rice (Oryza sativa L.). Physiol. Planta. 174:e13584. doi: 10.1111/ppl.13584 [DOI] [PubMed] [Google Scholar]
- Chookietwattana K., Maneewan K. (2012). Selection of efficient salt-tolerant bacteria containing ACC deaminase for promotion of tomato growth under salinity stress. Soil Environ. 31, 30–36. [Google Scholar]
- Danish S., Kiran S., Fahad S., Ahmad N., Ali M. A., Tahir F. A., et al. (2019). Alleviation of chromium toxicity in maize by Fe fortification and chromium tolerant ACC deaminase producing plant growth promoting rhizobacteria. Ecotoxicol. Environ. Saf 185:109706. doi: 10.1016/j.ecoenv.2019.109706, PMID: [DOI] [PubMed] [Google Scholar]
- Danish S., Zafar-ul-Hye M., Mohsin F., Hussain M. (2020). ACC-deaminase producing plant growth promoting rhizobacteria and biochar mitigate adverse effects of drought stress on maize growth. PLos One 15:e0230615. doi: 10.1371/journal.pone.0230615, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Das A., Osborne J. W. (2018). “Bioremediation of heavy metals” in Nanotechnology, food security and water treatment, eds. Gothandam, K., Ranjan, S., Dasgupta, N., Ramalingam, C., Lichtfouse, E. (Cham: Springer; ), 277–311. [Google Scholar]
- Daur I., Saad M. M., Eida A. A., Ahmad S., Shah Z. H., Ihsan M. Z., et al. (2018). Boosting alfalfa (Medicago sativa L.) production with rhizobacteria from various plants in Saudi Arabia. Front. Microbiol. 9:477. doi: 10.3389/fmicb.2018.0047, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimkpa C., Weinand T., Asch F. (2009). Plant–rhizobacteria interactions alleviate abiotic stress conditions. Plant Cell Environ. 32, 1682–1694. doi: 10.1111/j.1365-3040.2009.02028.x, PMID: [DOI] [PubMed] [Google Scholar]
- Din B. U., Sarfraz S., Xia Y., Kamran M. A., Javed M. T., Sultan T., et al. (2019). Mechanistic elucidation of germination potential and growth of wheat inoculated with exopolysaccharide and ACC-deaminase producing bacillus strains under induced salinity stress. Ecotoxicol. Environ. Saf. 183:109466. doi: 10.1016/j.ecoenv.2019.109466, PMID: [DOI] [PubMed] [Google Scholar]
- Dixit R., Agrawal L., Gupta S., Kumar M., Yadav S., Chauhan P. S., et al. (2016). Southern blight disease of tomato control by 1-aminocyclopropane-1-carboxylate (ACC) deaminase producing Paenibacilluslentimorbus B-30488. Plant Sign. Behav. 11, 1–36. doi: 10.1080/15592324.2015.1113363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R., Malaviya D., Pandiyan K., Singh U. B., Sahu A., Shukla R., et al. (2015). Bioremediation of heavy metals from soil and aquatic environment: an overview of principles and criteria of fundamental processes. Sustain. 7, 2189–2212. doi: 10.3390/su7022189 [DOI] [Google Scholar]
- Duan J., Jiang W., Cheng Z., Heikkila J. J., Glick B. R. (2013). The complete genome sequence of the plant growth-promoting bacterium pseudomonas sp. UW4. PLoS One 8:e58640. doi: 10.1371/journal.pone.0058640, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ek-Ramos M. J., Gomez-Flores R., Orozco-Flores A. A., Rodríguez-Padilla C., González-Ochoa G., Tamez-Guerra P. (2019). Bioactive products from plant-endophytic gram-positive bacteria. Front. Microbiol. 10:463. doi: 10.3389/fmicb.2019.00463, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Tarabily K. A. (2013). Biocontrol of damping-off and root and crown rots of cucumber caused by Pythium aphanidermatum by ACC deaminase producing endophytic actinomycetes. Phytopathology 103:40. [Google Scholar]
- Etesami H., Mirseyed Hosseini H., Alikhani H. A. (2014). Bacterial biosynthesis of 1-aminocyclopropane-1-caboxylate (ACC) deaminase, a useful trait to elongation and endophytic colonization of the roots of rice under constant flooded conditions. Physiol. Mol. Biol. Plants 20, 425–434. doi: 10.1007/s12298-014-0251-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etesami H., Noori F., Ebadi A., Reiahi Samani N. (2020). “Alleviation of stress-induced ethylene-mediated negative impact on crop plants by bacterial ACC deaminase: perspectives and applications in stressed agriculture management” in Plant microbiomes for sustainable agriculture by A. eds. Meena R., et al. (Springer, Singapore: Springer Nature Switzerland AG; ), 287–315. [Google Scholar]
- FAO (2008). FAO land and plant nutrition management service. Available at: http://www.fao.org/ag/agl/agll/spush
- Farahat M. G., Mahmoud M. K., Youseif S. H., Saleh S. A., Kamel Z. (2020). Alleviation of salinity stress in wheat by ACC deaminase-producing bacillus aryabhattai EWR29 with multifarious plant growth-promoting attributes. Plant Arch. 20, 417–429. [Google Scholar]
- Ferguson B. J., Mathesius U. (2014). Phytohormone regulation of legume-rhizobia interactions. J. Chemical Ecol. 40, 770–790. doi: 10.1007/s10886-014-0472-7, PMID: [DOI] [PubMed] [Google Scholar]
- Fernandez-Moreno J. P., Stepanova A. N. (2019). Monitoring ethylene in plants: genetically encoded reporters and biosensors. Small Methods 4:1900260. doi: 10.1002/smtd.201900260 [DOI] [Google Scholar]
- Gamalero E., Glick B. R. (2015). Bacterial modulation of plant ethylene levels. PlantPhysiol. 169, 13–22. doi: 10.1104/pp.15.00284, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B. R. (1995). The enhancement of plant growth by free-living bacteria. Can. J. Microbiol. 41, 109–117. doi: 10.1139/m95-015 [DOI] [Google Scholar]
- Glick B. R. (2005). Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase. FEMS Microbiol. Lett. 251, 1–7. doi: 10.1016/j.femsle.2005.07.030, PMID: [DOI] [PubMed] [Google Scholar]
- Glick B. R. (2012). Plant growth-promoting bacteria: mechanisms and applications. Scientifica 2012:963401, 1–15. doi: 10.6064/2012/963401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B. R. (2014). Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 169, 30–39. doi: 10.1016/j.micres.2013.09.009, PMID: [DOI] [PubMed] [Google Scholar]
- Glick B. R., Cheng Z., Czarny J., Duan J. (2007a). “Promotion of plant growth by ACC deaminase-producing soil bacteria” in New perspectives and approaches in plant growth-promoting Rhizobacteria research, eds. Bakker, P. A. H. M., Raaijmakers, J. M., Bloemberg, G., Höfte, M., Lemanceau, P., Cooke, B. M. (Dordrecht: Springer; ), 329–339. [Google Scholar]
- Glick B. R., Penrose D. M., Li J. (1998). A model for the lowering of plant ethylene concentrations by plant growth-promoting bacteria. J. Theoretical Biol. 190, 63–68. doi: 10.1006/jtbi.1997.0532 [DOI] [PubMed] [Google Scholar]
- Glick B. R., Todorovic B., Czarny J., Cheng Z., Duan J., McConkey B. (2007b). Promotion of plant growth by bacterial ACC deaminase. Critical Review Plant Sci. 26, 227–242. doi: 10.1080/07352680701572966 [DOI] [Google Scholar]
- Glick B. R., Nascimento F. X., Vicente C. S. L., Barbosa P., Espada M., Mota M., et al. (2013). Evidence for the involvement of ACC deaminase from Pseudomonas putida UW4 in the biocontrol of pine wilt disease. caused by Bursaphelenchus xylophilus. Biocontrol 58, 427–433. doi: 10.1007/s10526-012-9500-0 [DOI] [Google Scholar]
- Gontia-Mishra I., Sapre S., Kachare S., Tiwari S. (2017). Molecular diversity of 1-aminocyclopropane-1-carboxylate (ACC) deaminase producing PGPR from wheat (Triticum aestivum L.) rhizosphere. Plant Soil 414, 213–227. doi: 10.1007/s11104-016-3119-3 [DOI] [Google Scholar]
- Gontia-Mishra I., Sapre S., Sharma A. (2016). Alleviation of mercury toxicity in wheat by the interaction of mercury-tolerant plant growth-promoting rhizobacteria. J. Plant Growth Reg. 35, 1000–1012. doi: 10.1007/s00344-016-9598-x [DOI] [PubMed] [Google Scholar]
- Gontia-Mishra I., Sasidharan S., Tiwari S. (2014). Recent developments in use of 1-aminocyclopropane-1-carboxylate (ACC) deaminase for conferring tolerance to biotic and abiotic stress. Biotechnol. Lett. 36, 889–898. doi: 10.1007/s10529-014-1458-9, PMID: [DOI] [PubMed] [Google Scholar]
- Greetatorn T., Hashimoto S., Sarapat S., Tittabutr P., Boonkerd N., Uchiumi T., et al. (2019). Empowering rice seedling growth by endophytic Bradyrhizobium sp. SUTN 9-2. Lett. Appl. Microbiol. 68, 258–266. doi: 10.1111/lam.13114, PMID: [DOI] [PubMed] [Google Scholar]
- Grichko V. P., Glick B. R. (2001). Amelioration of flooding stress by ACC deaminase-containing plant growth-promoting bacteria. Plant Physiol. Biochem. 39, 11–17. doi: 10.1016/S0981-9428(00)01212-2 [DOI] [Google Scholar]
- Grobelak A., Kokot P., Świątek J., Jaskulak M., Rorat A. (2018). Bacterial ACC deaminase activity in promoting plant growth on areas contaminated with heavy metals. J. Ecol. Eng. 19, 150–157. doi: 10.12911/22998993/89818 [DOI] [Google Scholar]
- Guinel F. C. (2015). Ethylene, a hormone at the center-stage of nodulation. Front. Plant Sci. 6, 1–21. doi: 10.3389/fpls.2015.01121, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gull A., Lone A. A., Wani N. U. I. (2019). “Biotic and abiotic stresses in plants” in Abiotic and Biotic Stress in Plants (London, United Kingdom: Intech Open; ). [Google Scholar]
- Gupta A., Bano A., Rai S., Kumar M., Ali J., Sharma S., et al. (2021). ACC deaminase producing plant growth promoting rhizobacteria enhance salinity stress tolerance in Pisumsativum. 3 Biotech 11, 514–517. doi: 10.1007/s13205-021-03047-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K., Dey A., Gupta B. (2013). Plant polyamines in abiotic stress responses. Acta Physiol.Plant. 35, 2015–2036. doi: 10.1007/s11738-013-1239-4 [DOI] [Google Scholar]
- Gupta S., Pandey S. (2019). Unravelling the biochemistry and genetics of ACC deaminase-an enzyme alleviating the biotic and abiotic stress in plants. Plant Gene. 18:100175. doi: 10.1016/j.plgene.2019.100175 [DOI] [Google Scholar]
- Gupta A., Sinha R., Fernandes J. L., Abdelrahman M., Burritt D. J., Tran L. S. P. (2020). Phytohormones regulate convergent and divergent responses between individual and combined drought and pathogen infection. Critical Reviews Biotech. 40, 320–340. doi: 10.1080/07388551.2019.1710459, PMID: [DOI] [PubMed] [Google Scholar]
- Gururani M. A., Upadhyaya C. P., Baskar V., Venkatesh J., Nookaraju A., Park S. W. (2013). Plant growth-promoting rhizobacteria enhance abiotic stress tolerance in Solanum tuberosum through inducing changes in the expression of ROS-scavenging enzymes and improved photosynthetic performance. J. Plant Growth Regul. 32, 245–258. doi: 10.1007/s00344-012-9292-6 [DOI] [Google Scholar]
- Han Y., Wang R., Yang Z., Zhan Y., Ma Y., Ping S., et al. (2015). 1-aminocyclopropane-1-carboxylate deaminase from pseudomonas stutzeri A1501 facilitates the growth of rice in the presence of salt or heavy metals. J. Microbiol. Biotechnol. 25, 1119–1128. doi: 10.4014/jmb.1412.12053 [DOI] [PubMed] [Google Scholar]
- Han L., Zhang H., Xu Y., Li Y., Zhou J. (2021). Biological characteristics and salt-tolerant plant growth-promoting effects of an ACC deaminase-producing Burkholderiapyrrocinia strain isolated from the tea rhizosphere. Arch. Microbiol. 203, 2279–2290. doi: 10.1007/s00203-021-02204-x, PMID: [DOI] [PubMed] [Google Scholar]
- Hassan W., Bano R., Bashir F., David J. (2014). Comparative effectiveness of ACC-deaminase and/or nitrogen-fixing rhizobacteria in promotion of maize (Zea mays L.) growth under lead pollution. Environ. Sci. Pollut. Res. 21, 10983–10996. doi: 10.1007/s11356-014-3083-5 [DOI] [PubMed] [Google Scholar]
- Hemida K. A., Reyad A. M. M. (2018). Improvement salt tolerance of safflower plants by endophytic bacteria. J. Hort. Plant Res. 5, 38–56. doi: 10.18052/www.scipress.com/JHPR.5.38 [DOI] [Google Scholar]
- Hemissi I., Hammami R., Hachana A., Arfaoui H., Sifi B. (2019). Fertilizer-dependent efficiency of Mesorhizobium strain for improving growth, nutrient uptake and grain yield of durum wheat (Triticum turgidium L.) variety. J. New Sci. 61, 3885–3891. [Google Scholar]
- Honma M. (1983). Enzymatic determination of 1-aminocyclopropane-1-carboxylic acid. Agric. Biol. Chem. 47, 617–618. [Google Scholar]
- Honma M. (1985). Chemically reactive sulfhydryl groups of 1-aminocyclopropane-1-carboxylate deaminase. Agric. Biol. Chem. 49, 567–571. doi: 10.1080/00021369.1985.10866774 [DOI] [Google Scholar]
- Honma M. (1993). “Stereospecific reaction of 1-aminocyclopropane-1-carboxylate deaminase” in Cellular and molecular aspects of the plant hormone ethylene (Dordrecht: Springer; ), 111–116. [Google Scholar]
- Honma M., Kawai J., Yamada M. (1993). Identification of the reactive sulfhydryl group of 1-aminocyclopropane-1-carboxylate deaminase. Biosci. Biotechnol. Biochem. 57, 2090–2093. doi: 10.1271/bbb.57.2090, PMID: [DOI] [PubMed] [Google Scholar]
- Honma M., Shimomura T. (1978). Metabolism of 1-aminocyclopropane-1-carboxylic acid. Agric. Biol. Chem. 42, 1825–1831. doi: 10.1080/00021369.1978.10863261 [DOI] [Google Scholar]
- Hontzeas N., Hontzeas C. E., Glick B. R. (2006). Reaction mechanisms of the bacterial enzyme 1-aminocyclopropane-1-carboxylate deaminase. Biotechnol. Adv. 24, 420–426. doi: 10.1016/j.biotechadv.2006.01.006, PMID: [DOI] [PubMed] [Google Scholar]
- Hontzeas N., Zoidakis J., Glick B. R., Abu-Omar M. M. (2004). Expression and characterization of 1-aminocyclopropane-1-carboxylate deaminase from the rhizobacterium pseudomonas putida UW4: a key enzyme in bacterial plant growth promotion. Biochim. Biophysica Acta-Proteins Proteom. 1703, 11–19. doi: 10.1016/j.bbapap.2004.09.015, PMID: [DOI] [PubMed] [Google Scholar]
- Huang C. H., Hsiang T., Trevors J. T. (2013). Comparative bacterial genomics: defining the minimal core genome. Anton. Leeuw. 103, 385–398. doi: 10.1007/s10482-012-9819-7, PMID: [DOI] [PubMed] [Google Scholar]
- Huang X. F., Zhou D., Lapsansky E. R., Reardon K. F., Guo J., Andales M. J., et al. (2017). Mitsuaria sp. and Burkholderia sp. from Arabidopsis rhizosphere enhance drought tolerance in Arabidopsis thaliana and maize (Zea mays L.). Plant Soil 419, 523–539. doi: 10.1007/s11104-017-3360-4 [DOI] [Google Scholar]
- Hussain S., Shaukat M., Ashraf M., Zhu C., Jin Q., Zhang J. (2019). Salinity stress in arid and semi-arid climates: effects and management in field crops. Climate change and agriculture. Intech Open. [Google Scholar]
- Indiragandhi P., Anandham R., Kim K., Yim W. J., Madhaiyan M., Sa T. M. (2008). Induction of defense responses in tomato against pseudomonas syringaepv. Tomato by regulating the stress ethylene level with Methylobacteriumoryzae CBMB20 containing 1-aminocyclopropane-1carboxylate deaminase. World J. Microbiol. Biotechnol. 24, 1037–1045. doi: 10.1007/s11274-007-9572-7 [DOI] [Google Scholar]
- Jackson M. B. (2008). Ethylene-promoted elongation: an adaptation to submergence stress. Ann. Bot. 101, 229–248. doi: 10.1093/aob/mcm237, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson C. B., Pasternak J. J., Glick B. R. (1994). Partial purification and characterization of 1-aminocyclopropane-1-carboxylate deaminase from the plant growth promoting rhizobacterium pseudomonas putida GR12-2. Canadian J. Microbiol. 40, 1019–1025. doi: 10.1139/m94-16 [DOI] [Google Scholar]
- Jaemsaeng R., Jantasuriyarat C., Thamchaipenet A. (2018). Positive role of 1-aminocyclopropane-1-carboxylate deaminase-producing endophytic Streptomyces sp. GMKU 336 on flooding resistance of mung bean. Agric. Natural Res. 52, 330–334. doi: 10.1016/j.anres.2018.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jajoo A. (2017). “Effects of environmental pollutants polycyclic aromatic hydrocarbons (PAH) on photosynthetic processes” in Photosynthesis: structures, mechanisms, and applications (Cham, eds. Hou, H., Najafpour, M., Moore, G., Allakhverdiev, S. Springer; ), 249–259. [Google Scholar]
- Jha C. K., Annapurna K., Saraf M. (2012). Isolation of Rhizobacteria from Jatrophacurcas and characterization of produced ACC deaminase. J. Basic Microbiol. 52, 285–295. doi: 10.1002/jobm.201100113 [DOI] [PubMed] [Google Scholar]
- Ji J., Yuan D., Jin C., Wang G., Li X., Guan C. (2020). Enhancement of growth and salt tolerance of rice seedlings (Oryza sativa L.) by regulating ethylene production with a novel halotolerant PGPR strain Glutamicibacter sp. YD01 containing ACC deaminase activity. Acta Physiol. Plant. 42:42. doi: 10.1007/s11738-020-3034-3 [DOI] [Google Scholar]
- Joshi M., Srivastava R., Sharma A. K., Prakash A. (2012). Screening of resistant varieties and antagonistic fusarium oxysporum for biocontrol of fusarium wilt of chilli. J. Plant Pathol. Microbiol. 03, 1–6. doi: 10.4172/2157-7471.1000134 [DOI] [Google Scholar]
- Kalsoom A., Batool R., Jamil N. (2022). Beneficial rhizospheric associated traits of chromate resistant bacteria for remediation of Cr (VI) contaminated soil. Biorem. J., 16, 1–19. doi: 10.1080/10889868.2022.2054930 [DOI] [Google Scholar]
- Kaneko T., Nakamura Y., Sato S., Minamisawa K., Uchiumi T., Sasamoto S., et al. (2002). Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9, 189–197. doi: 10.1093/dnares/9.6.189, PMID: [DOI] [PubMed] [Google Scholar]
- Kang S. M., Shahzad R., Bilal S., Khan A. L., Park Y. G., Lee K. E., et al. (2019). Indole-3-acetic-acid and ACC deaminase producing Leclerciaadecarboxylata MO1 improves Solanum lycopersicum L. growth and salinity stress tolerance by endogenous secondary metabolites regulation. BMC Microbiol. 19:80. doi: 10.1186/s12866-019-1450-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan S., Zhou Q., Zhao Z., Kao C. L., Tao Z., Robinson H., et al. (2004). Structural analysis of pseudomonas 1-aminocyclopropane-1-carboxylate deaminase complexes: insight into the mechanism of a unique pyridoxal-5′-phosphate dependent cyclopropane ring-opening reaction. Biochemist 43, 13328–13339. doi: 10.1021/bi048878g, PMID: [DOI] [PubMed] [Google Scholar]
- Kataria S., Verma S. K. (2018). “Salinity stress responses and adaptive mechanisms in major glycophytic crops: the story so far” in Salinity responses and tolerance in plants, eds. Kumar, V., Wani, S., Suprasanna, P., Tran, LS. (Cham: Springer; ), 1, 1–39. [Google Scholar]
- Katiyar P., Dubey R. C., Maheshwari D. K. (2021). ACC deaminase-producing Ensiferadhaerens KS23 enhances proximate nutrient of Pisum sativum L. cultivated in high altitude. Arch. Microbiol. 203, 2689–2698. doi: 10.1007/s00203-021-02250-5, PMID: [DOI] [PubMed] [Google Scholar]
- Kaur T., Manhas R. K. (2022). Evaluation of ACC deaminase and indole acetic acid production by Streptomyces hydrogenans DH16 and its effect on plant growth promotion. Biocat. Agric. Biotech. 42:102321. doi: 10.1016/j.bcab.2022.102321 [DOI] [Google Scholar]
- Kaushal M., Wani S. P. (2016). Plant growth-promoting rhizobacteria: drought stress alleviators to ameliorate crop production in drylands. Ann. Microbiol. 66, 35–42. doi: 10.1007/s13213-015-1112-3 [DOI] [Google Scholar]
- Khan S., Shahid M., Khan M. S., Syed A., Bahkali A. H., Elgorban A. M., et al. (2020). Fungicide-tolerant plant growth-promoting rhizobacteria mitigate physiological disruption of white radish caused by fungicides used in the field cultivation. Int. J. Environ. Res. Public Health 17:7251. doi: 10.3390/ijerph17197251, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Singh A. V. (2021). Multifarious effect of ACC deaminase and EPS producing pseudomonas sp. and Serratia marcescens to augment drought stress tolerance and nutrient status of wheat. World J.Microbiol. Biotech. 37, 198–117. doi: 10.1007/s11274-021-03166-4, PMID: [DOI] [PubMed] [Google Scholar]
- Khosravi H., Alikhani H. A., Yakhchali B., Kharkhane A. A. (2014). Isolation, cloning and sequence analysis of 1-aminocyclopropane-1-carboxylate deaminase gene from native Sinorhizobiummeliloti. Iran. J. Biotech. 12, 50–56. doi: 10.15171/ijb.1000 [DOI] [Google Scholar]
- Klay I., Gouia S., Liu M., Mila I., Khoudi H., Bernadac A., et al. (2018). Ethylene response factors (ERF) are differentially regulated by different abiotic stress types in tomato plants. Plant Sci. 274, 137–145. doi: 10.1016/j.plantsci.2018.05.023, PMID: [DOI] [PubMed] [Google Scholar]
- Klee H. J., Hayford M. B., Kretzmer K. A., Barry G. F., Kishore G. M. (1991). Control of ethylene synthesis by expression of a bacterial enzyme in transgenic tomato plants. Plant Cell 3, 1187–1193. doi: 10.1105/tpc.3.11.1187, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee H. J., Romano C. P., Binns A. (1994). The roles of phytohormones in development as studied in transgenic plants. Critic. Review Plant Sci. 13, 311–324. doi: 10.1080/07352689409701918 [DOI] [Google Scholar]
- Kruasuwan W., Thamchaipenet A. (2018). 1-aminocyclopropane-1-carboxylate (ACC) deaminase-producing endophytic diazotrophic Enterobacter sp. EN-21 modulates salt–stress response in sugarcane. J. Plant Growth Regul. 37, 849–858. doi: 10.1007/s00344-018-9780-4 [DOI] [Google Scholar]
- Kumar P., Dubey R. C., Maheshwari D. K., Bajpai V. (2016). ACC deaminase producing rhizobium leguminosarum rpn5 isolated from root nodules of Phaseolus vulgaris L. Bangladesh J. Bot. 45, 477–484. [Google Scholar]
- Kumar M., Etesami H., Kumar V. (2019). Saline soil-based agriculture by halotolerant microorganisms Springer. [Google Scholar]
- Kumar R., Kumar R., Prakash O. (2019). Chapter-5 the impact of chemical fertilizers on our. Environ. Ecosyst. 35:69. [Google Scholar]
- Kumar A., Maleva M., Bruno L. B., Rajkumar M. (2021). Synergistic effect of ACC deaminase producing pseudomonas sp. TR15a and siderophore producing Bacillus aerophilus TR15c for enhanced growth and copper accumulation in Helianthus annuus L. Chemosphere 276:130038. doi: 10.1016/j.chemosphere.2021.130038, PMID: [DOI] [PubMed] [Google Scholar]
- Lesk C., Rowhani P., Ramankutty N. (2016). Influence of extreme weather disasters on global crop production. Nature 529, 84–87. doi: 10.1038/nature16467, PMID: [DOI] [PubMed] [Google Scholar]
- Li J., Ovakim D. H., Charles T. C., Glick B. R. (2000). An ACC deaminase minus mutant of Enterobactercloacae UW4No longer promotes root elongation. Cur. Microbiol. 41, 101–105. doi: 10.1007/s002840010101, PMID: [DOI] [PubMed] [Google Scholar]
- Li Z., Wang W., Zhu L. (2019). Effects of mixed surfactants on the bioaccumulation of polycyclic aromatic hydrocarbons (PAHs) in crops and the bioremediation of contaminated farmlands. Sci. Total Environ 646, 1211–1218. doi: 10.1016/j.scitotenv.2018.07.349, PMID: [DOI] [PubMed] [Google Scholar]
- Light K. M., Wisniewski J. A., Vinyard W. A., Kieber-Emmons M. T. (2016). Perception of the plant hormone ethylene: known-knowns and known-unknowns. J. Biol. Inorganic Chem. 21, 715–728. doi: 10.1007/s00775-016-1378-3, PMID: [DOI] [PubMed] [Google Scholar]
- Liu J. L., Xie B. M., Shi X. H., Ma J. M., Guo C. H. (2015). Effects of two plant growth-promoting rhizobacteria containing 1-aminocyclopropane-1carboxylate deaminase on oat growth in petroleum-contaminated soil. Int. J. Environ. Sci. Technol. 12, 3887–3894. doi: 10.1007/s13762-015-0798-x [DOI] [Google Scholar]
- Ma W., Charles T. C., Glick B. R. (2004). Expression of an exogenous 1-aminocyclopropane-1-carboxylate deaminase gene in Sinorhizobiummeliloti increases its ability to nodulate alfalfa. Appl. Environ. Microbiol. 70, 5891–5897. doi: 10.1128/AEM.70.10.5891-5897.2004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Guinel F. C., Glick B. R. (2003a). Rhizobium leguminosarum biovar viciae 1-aminocyclopropane-1-carboxylate deaminase promotes nodulation of pea plants. Appl. Environ. Microbiol. 69, 4396–4402. doi: 10.1128/AEM.69.8.4396-4402.2003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Sebestianova S. B., Sebestian J., Burd G. I., Guinel F. C., Glick B. R. (2003b). Prevalence of 1-aminocyclopropane-1-carboxylate deaminase in rhizobium spp. Antonie Van Leeuwenhoek 83, 285–291. doi: 10.1023/A:1023360919140, PMID: [DOI] [PubMed] [Google Scholar]
- Malerba M., Crosti P., Bianchetti R. (1996). 2-Aminoisobutyric acid prevents the substrate-induced inactivation of ACC-synthase without inhibiting the enzyme. Plant Sci. 113, 131–138. doi: 10.1016/0168-9452(95)04294-6 [DOI] [Google Scholar]
- Malviya D., Singh U. B., Singh S., Sahu P. K., Pandiyan K., Kashyap A. S., et al. (2020). “Microbial interactions in the rhizosphere contributing crop resilience to biotic and abiotic stresses” in Rhizosphere microbes. eds. Sharma, et al. (Singapore: Springer; ), 1–33. [Google Scholar]
- Manoj S. R., Karthik C., Kadirvelu K., Arulselvi P. I., Shanmugasundaram T., Bruno B., et al. (2020). Understanding the molecular mechanisms for the enhanced phytoremediation of heavy metals through plant growth promoting rhizobacteria: a review. J. Environ.Manag. 254:109779. doi: 10.1016/j.jenvman.2019.109779, PMID: [DOI] [PubMed] [Google Scholar]
- Maqbool S., Amna A., Mehmood S., Suhaib M., Sultan T., Munis M. (2021). Interaction of acc deaminase and antioxidant enzymes to induce drought tolerance in Enterobacter cloacae 2wc2 inoculated maize genotypes. Pak. J. Bot. 53. doi: 10.30848/PJB2021-3(28) [DOI] [Google Scholar]
- Miller G., Susuki N., Ciftci-Yilmaz S. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 33, 453–467. doi: 10.1111/j.1365-3040.2009.02041.x [DOI] [PubMed] [Google Scholar]
- Misra S., Chauhan P. S. (2020). ACC deaminase-producing rhizosphere competent bacillus spp. mitigate salt stress and promote Zea mays growth by modulating ethylene metabolism. 3 Biotech 10, 119–114. doi: 10.1007/s13205-020-2104-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeder W., Barry C. S., Tauriainen A. A., Betz C., Tuomainen J., Utriainen M., et al. (2002). Ethylene synthesis regulated by biphasic induction of 1-aminocyclopropane-1-carboxylic acid synthase and 1-aminocyclopropane-1-carboxylic acid oxidase genes is required for hydrogen peroxide accumulation and cell death in ozone-exposed tomato. Plant Physiol. 130, 1918–1926. doi: 10.1104/pp.009712, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mog B., Adiga D., Nayak M. G. (2018). Role of plant growth hormones in cashew: key strategy for modifying crop performance. Int. J. Curr. Microbiol. App. Sci. 7, 1470–1484. doi: 10.20546/ijcmas.2018.707.174 [DOI] [Google Scholar]
- Murali M., Singh S. B., Gowtham H. G., Shilpa N., Prasad M., Aiyaz M., et al. (2021). Induction of drought tolerance in Pennisetum glaucum by ACC deaminase producing PGPR-bacillus amyloliquefaciens through antioxidant defense system. Microbiol. Res. 253:126891. doi: 10.1016/j.micres.2021.126891, PMID: [DOI] [PubMed] [Google Scholar]
- Nadeem S. M., Ahmad M., Tufail M. A., Asghar H. N., Nazli F., Zahir Z. A. (2021). Appraising the potential of EPS-producing rhizobacteria with ACC-deaminase activity to improve growth and physiology of maize under drought stress. Physiol. Plant. 172, 463–476. doi: 10.1111/ppl.13212, PMID: [DOI] [PubMed] [Google Scholar]
- Nadeem S. M., Zahair Z. A., Naveed M., Arshad M. (2007). Preliminary investigations on inducing salt tolerance in maize through inoculation with rhizobacteria containing ACC deaminase activity. Can. J. Microbiol. 53, 1141–1149. doi: 10.1139/W07-081, PMID: [DOI] [PubMed] [Google Scholar]
- Nadeem S. M., Zahir Z. A., Naveed M., Asghar H. N., Arshad M. (2010). Rhizobacteria capable of producing ACC deaminase may mitigate salt stress in wheat. Soil Sci. Soc. Am. J. 74, 533–542. doi: 10.2136/sssaj2008.0240 [DOI] [Google Scholar]
- Nascimento F., Brígido C., Alho L., Glick B. R., Oliveira S. (2012). Enhanced chickpea growth-promotion ability of a Mesorhizobium strain expressing an exogenous ACC deaminase gene. Plant Soil 353, 221–230. doi: 10.1007/s11104-011-1025-2 [DOI] [Google Scholar]
- Nascimento F. X., Rossi M. J., Soares C. R., McConkey B. J., Glick B. R. (2014). New insights into 1-aminocyclopropane-1-carboxylate (ACC) deaminase phylogeny, evolution and ecological significance. PLoS One 9:e99168. doi: 10.1371/journal.pone.0099168, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento F. X., Tavares M. J., Franck J., Ali S., Glick B. R., Rossi M. J. (2019). ACC deaminase plays a major role in Pseudomonas fluorescens YsS6 ability to promote the nodulation of alpha-and beta proteobacteria rhizobial strains. Arch.Microbiol. 201, 817–822. doi: 10.1007/s00203-019-01649-5 [DOI] [PubMed] [Google Scholar]
- Nascimento F. X., Vicente C. S., Barbosa P., Espada M., Glick B. R., Mota M., et al. (2013). Evidence for the involvement of ACC deaminase from pseudomonas putida UW4 in the biocontrol of pine wilt disease caused by Bursaphelenchusxylophilus. Biol. Control 58, 427–433. doi: 10.1007/s10526-012-9500-0 [DOI] [Google Scholar]
- Nukui N., Minamisawa K., Ayabe S. I., Aoki T. (2006). Expression of the 1-aminocyclopropane-1-carboxylic acid deaminase gene requires symbiotic nitrogen-fixing regulator gene nifA2 in Mesorhizobium loti MAFF303099. Appl. Environ. Microbiol. 72, 4964–4969. doi: 10.1128/AEM.02745-05, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orhan F. (2016). Alleviation of salt stress by halotolerant and halophilic plant growth-promoting bacteria in wheat (Triticum aestivum). Brazilian J. Microbiol. 47, 621–627. doi: 10.1016/j.bjm.2016.04.001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ose T., Fujino A., Yao M., Watanabe N., Honma M., Tanaka I. (2003). Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction. J. Biol. Chem. 278, 41069–41076. doi: 10.1074/jbc.M305865200, PMID: [DOI] [PubMed] [Google Scholar]
- Ouaked F., Rozhon W., Lecourieux D., Hirt H. (2003). A MAPK pathway mediates ethylene signaling in plants. EMBO J. 22, 1282–1288. doi: 10.1093/emboj/cdg131, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda A., Rangani J., Kumari A., Parida A. K. (2017). Efficient regulation of arsenic translocation to shoot tissue and modulation of phytochelatin levels and antioxidative defense system confers salinity and arsenic tolerance in the halophyte Suaeda maritima. Environ. Exp. Bot. 143, 149–171. doi: 10.1016/j.envexpbot.2017.09.007 [DOI] [Google Scholar]
- Pandey S., Ghosh P. K., Ghosh S., De T. K., Maiti T. K. (2013). Role of heavy metal resistant Ochrobactrum sp. and bacillus spp. strains in bioremediation of a rice cultivar and their PGPR like activities. J. Microbiol. 51, 11–17. doi: 10.1007/s12275-013-2330-7, PMID: [DOI] [PubMed] [Google Scholar]
- Pandey S., Gupta S. (2020). Evaluation of pseudomonas sp. for its multifarious plant growth promoting potential and its ability to alleviate biotic and abiotic stress in tomato (Solanumlycopersicum) plants. Sci. Rep. 10, 1–15. doi: 10.1038/s41598-020-77850-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul M. V., Iyer S., Amerhauser C., Lehmann M., van Dongen J. T., Geigenberger P. (2016). Oxygen sensing via the ethylene response transcription factor RAP2. 12 affects plant metabolism and performance under both normoxia and hypoxia. Plant Physiol. 172, 141–153. doi: 10.1104/pp.16.00460, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R., Tholen D., Poorter H., Visser E. J., Voesenek L. A. (2006). The Janus face of ethylene: growth inhibition and stimulation. Trends Plant Sci. 11, 176–183. doi: 10.1016/j.tplants.2006.02.006, PMID: [DOI] [PubMed] [Google Scholar]
- Pirasteh-Anosheh H., Ranjbar G., Pakniyat H., Emam Y. (2016). Physiological mechanisms of salt stress tolerance in plants: an overview. Plant-Environ. Interact. Responses Approaches Mitigate Stress, 141–160. doi: 10.1002/9781119081005.ch8 [DOI] [Google Scholar]
- Place F., Meybeck A., Colette L., de Young C., Gitz V., Dulloo E., et al. (2017). Food security and sustainable resource use? What are the resource challenges to food security.
- Płociniczak T., Sinkkonen A., Romantschuk M. (2014). Characterization of Enterobacterintermedius MH8b and its use for the enhancement of heavy metals uptake by Sinapis alba L. Appl. Soil Ecol. 63, 1–7. doi: 10.1016/j.apsoil.2012.09.009 [DOI] [Google Scholar]
- Pramanik K., Mitra S., Sarkar A., Maiti T. K. (2018). Alleviation of phytotoxic effects of cadmium on rice seedlings by cadmium resistant PGPR strain Enterobacter aerogenes MCC 3092. J. Hazard. Mater. 351, 317–329. doi: 10.1016/j.jhazmat.2018.03.009, PMID: [DOI] [PubMed] [Google Scholar]
- Prasannakumar S. P., Gowtham H. G., Hariprasad P., Shivaprasad K., Niranjana S. R. (2015). Delftiatsuruhatensis WGR–UOM–BT1, a novel rhizobacterium with PGPR properties from Rauwolfia serpentina (L.) Benth. Ex Kurz also suppresses fungal phytopathogens by producing a new antibiotic—AMTM. Lettt. Appl. Microbiol. 61, 460–468. doi: 10.1111/lam.12479, PMID: [DOI] [PubMed] [Google Scholar]
- Qin S., Zhang Y. J., Yuan B., Xu P. Y., Xing K., Wang J., et al. (2014). Isolation of ACC deaminase-producing habitat-adapted symbiotic bacteria associated with halophyte Limonium sinense (Girard) Kuntze and evaluating their plant growth-promoting activity under salt stress. Plant Soil 374, 753–766. doi: 10.1007/s11104-013-1918-3 [DOI] [Google Scholar]
- Raghuwanshi R., Prasad J. K. (2018). “Perspectives of rhizobacteria with ACC deaminase activity in plant growth under abiotic stress” in Root biology, eds. Giri, B., Prasad, R., Varma, A. (Cham: Springer; ), 303–321. [Google Scholar]
- Rao M. V., Davis K. R. (2001). The physiology of ozone induced cell death. Planta 213, 682–690. doi: 10.1007/s004250100618 [DOI] [PubMed] [Google Scholar]
- Rashid U., Yasmin H., Hassan M. N., Naz R., Nosheen A., Sajjad M., et al. (2022). Drought-tolerant bacillus megaterium isolated from semi-arid conditions induces systemic tolerance of wheat under drought conditions. Plant Cell Rep. 41, 549–569. doi: 10.1007/s00299-020-02640-x [DOI] [PubMed] [Google Scholar]
- Rauf M., Awais M., Ud-Din A., Ali K., Gul H., Rahman M. M., et al. (2021). Molecular mechanisms of the 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase producing Trichoderma asperellum MAP1 in enhancing wheat tolerance to waterlogging stress. Front. Plant Sci. 11:2213. doi: 10.3389/fpls.2020.614971, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanbakhsh M., Sasidharan R., Voesenek L. A., Kowalchuk G. A., Jousset A. (2017). ACC deaminase-producing rhizosphere bacteria modulate plant responses to flooding. J. Ecol. 105, 979–986. doi: 10.1111/1365-2745.12721 [DOI] [Google Scholar]
- Reinhold-Hurek B., Hurek T. (2011). Living inside plants: bacterial endophytes. Curr. Opin. Plant Biol. 14, 435–443. doi: 10.1016/j.pbi.2011.04.004 [DOI] [PubMed] [Google Scholar]
- Rengasamy P. (2010). Soil processes affecting crop production in salt-affected soils. FunctionalPlant Biol. 37, 613–620. doi: 10.1071/FP09249 [DOI] [Google Scholar]
- Riemenschneider A., Wegele R., Schmidt A., Papenbrock J. (2005). Isolation and characterization of ad-cysteine desulfhydrase protein from Arabidopsis thaliana. FEBS J. 272, 1291–1304. doi: 10.1111/j.1742-4658.2005.04567.x, PMID: [DOI] [PubMed] [Google Scholar]
- Rizvi A., Khan M. S. (2018). Heavy metal induced oxidative damage and root morphology alterations of maize (Zea mays L.) plants and stress mitigation by metal tolerant nitrogen fixing Azotobacter chroococcum. Ecotoxicol. Environ. Safe. 157, 9–20. doi: 10.1016/j.ecoenv.2018.03.063, PMID: [DOI] [PubMed] [Google Scholar]
- Rizvi A., Zaidi A., Khan M. S., Saif S., Ahmed B., Shahid M. (2017). Growth improvement and management of vegetable diseases by plant growth-promoting rhizobacteria. In Microbial Strategies for Vegetable Production. eds. Zaidi, A., Khan, M. Springer, Cham.. doi: 10.1007/978-3-319-54401-4_5 [DOI] [Google Scholar]
- Sagar A., Rai S., Ilyas N., Sayyed R. Z., Al-Turki A. I., El Enshasy H. A., et al. (2022). Halotolerant rhizobacteria for salinity-stress mitigation: diversity, mechanisms and molecular approaches. Sustainability 14:490. doi: 10.3390/su14010490 [DOI] [Google Scholar]
- Sagar A., Sayyed R. Z., Ramteke P. W., Sharma S., Marraiki N., Elgorban A. M., et al. (2020). ACC deaminase and antioxidant enzymes producing halophilic Enterobacter sp. PR14 promotes the growth of rice and millets under salinity stress. Physiol. Molecular Biol. Plants 26, 1847–1854. doi: 10.1007/s12298-020-00852-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghafi D., Ghorbanpour M., Ajirloo H. S., Lajayer B. A. (2019). Enhancement of growth and salt tolerance in Brassica napus L. seedlings by halotolerant rhizobium strains containing ACC-deaminase activity. Plant Physiol. Rep. 24, 225–235. doi: 10.1007/s40502-019-00444-0 [DOI] [Google Scholar]
- Sahu P. K., Singh S., Singh U. B., Chakdar H., Sharma P. K., Sarma B. K., et al. (2021). Inter-genera colonization of Ocimumtenuiflorum endophytes in tomato and their complementary effects on Na+/K+ balance, oxidative Stress regulation, and root architecture under elevated soil salinity. Front. Microbiol. 12:744733. doi: 10.3389/fmicb.2021.744733, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saif S., Khan M. S. (2018). Assessment of toxic impact of metals on proline, antioxidant enzymes, and biological characteristics of Pseudomonas aeruginosa inoculated Cicer arietinum grown in chromium and nickel-stressed sandy clay loam soils. Environ.Monit. Assess. 190, 290–218. doi: 10.1007/s10661-018-6652-0, PMID: [DOI] [PubMed] [Google Scholar]
- Saif S., Khan M. S., Zaidi A., Rizvi A., Shahid M. (2017). Metal toxicity to certain vegetables and bioremediation of metal-polluted soils. In Microbial Strategies for Vegetable Production. eds. Zaidi, A., Khan, M. Springer, Cham. doi: 10.1007/978-3-319-54401-4_8 [DOI] [Google Scholar]
- Saikia J., Sarma R. K., Dhandia R., Yadav A., Bharali R., Gupta V. K., et al. (2018). Alleviation of drought stress in pulse crops with ACC deaminase producing rhizobacteria isolated from acidic soil of Northeast India. Sci. Rep. 8:3560. doi: 10.1038/s41598-018-21921-w, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem M., Arshad M., Hussain S., Bhatti A. S. (2007). Perspective of plant growth promoting rhizobacteria (PGPR) containing ACC deaminase in stress agriculture. J. Ind. Microbiol. Biotechnol. 34, 635–648. doi: 10.1007/s10295-007-0240-6, PMID: [DOI] [PubMed] [Google Scholar]
- Saleem A. R., Brunetti C., Khalid A., Della Rocca G., Raio A., Emiliani G. (2018). Drought response of Mucuna pruriens (L.) inoculated with ACC deaminase and IAA producing rhizobacteria. PLoS One 13, 1–18. doi: 10.1371/journal.pone.0191218, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Hernandez J. C. (2019). Biochar mitigates the impact of pesticides on soil enzyme activities. Biorem. Agric. Soils, 193–216. doi: 10.1201/9781315205137-10 [DOI] [Google Scholar]
- Santos E., Pires F. R., Ferreira A. D., Egreja Filho F. B., Madalão J. C., Bonomo R., et al. (2019). Phytoremediation and natural attenuation of sulfentrazone: mineralogy influence of three highly weathered soils. Int. J. Phytoremediation 21, 652–662. doi: 10.1080/15226514.2018.1556583 [DOI] [PubMed] [Google Scholar]
- Santoyo G., Moreno-Hagelsieb G., del Carmen Orozco-Mosqueda M., Glick B. R. (2016). Plant growth-promoting bacterial endophytes. Microbiol. Res. 183, 92–99. doi: 10.1016/j.micres.2015.11.008 [DOI] [PubMed] [Google Scholar]
- Sapre S., Gontia-Mishra I., Tiwari S. (2018a). Klebsiella sp. confers enhanced tolerance to salinity and plant growth promotion in oat seedlings (Avena sativa). Microbiol. Res. 206, 25–32. doi: 10.1016/j.micres.2017.09.009, PMID: [DOI] [PubMed] [Google Scholar]
- Sapre S., Gontia-Mishra I., Tiwari S. (2018b). “ACC deaminase-producing bacteria: a key player in alleviating abiotic stresses in plants” in Plant growth promoting Rhizobacteria for agricultural sustainability (Singapore: Springer; ), 267–291. [Google Scholar]
- Sarkar A., Ghosh P. K., Pramanik K., Mitra S., Soren T., Pandey S., et al. (2018a). A halotolerant Enterobacter sp. displaying ACC deaminase activity promotes rice seedling growth under salt stress. Res.Microbiol. 169, 20–32. doi: 10.1016/j.resmic.2017.08.005, PMID: [DOI] [PubMed] [Google Scholar]
- Sarkar A., Pramanik K., Mitra S., Soren T., Maiti T. K. (2018b). Enhancement of growth and salt tolerance of rice seedlings by ACC deaminase-producing Burkholderia sp. MTCC 12259. J. Plant Physiol. 231, 434–442. doi: 10.1016/j.resmic.2017.08.005, PMID: [DOI] [PubMed] [Google Scholar]
- Saxena P., Kulshrestha U. (2016). “Biochemical effects of air pollutants on plants” in Plant responses to air pollution, Kulshrestha, U., Saxena, P. (eds) (Singapore: Springer; ), 59–70. [Google Scholar]
- Schwachtje J., Whitcomb S. J., Firmino A. A., Zuther E., Hincha D. K., Kopka J. (2019). Induced, imprinted and primed metabolic responses of plants to changing environments: do plants store and process information by metabolic imprinting and metabolic priming? Front. Plant Sci. 10:106. doi: 10.3389/fpls.2019.00106, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthilkumar M., Paulraj S., Alagupalamuthirsolai M., Singh M., Singh J. (2016). Synergistic effect of Mesorhizobiumciceri and 1-amino cyclopropane 1-carboxylate (ACC) deaminase producing rhizobacteria on chickpea growth and yield. J. Food Legumes 29, 37–42. [Google Scholar]
- Shaharoona B., Arshad M., Zahir Z. A. (2006). Effect of plant growth promoting rhizobacteria containing ACC-deaminase on maize (Zea mays L.) growth under axenic conditions and on nodulation in mung bean (Vigna radiata L.). Lett. Appl. Microbiol. 42, 155–159. doi: 10.1111/j.1472-765X.2005.01827.x, PMID: [DOI] [PubMed] [Google Scholar]
- Shaharoona B., Jamro G. M., Zahir Z. A., Arshad M., Memon K. S. (2007). Effectiveness of various pseudomonas spp. and Burkholderiacaryophylli containing ACC-deaminase for improving growth and yield of wheat (Triticum aestivum L.). J. Microbiol. Biotechnol. 17, 1300–1307. doi: 10.1016/j.soilbio.2006.03.024, PMID: [DOI] [PubMed] [Google Scholar]
- Shahid M., Ahmed B., Khan M. S. (2018a). Evaluation of microbiological management strategy of herbicide toxicity to greengram plants. Biocat. Agric. Biotech. 14, 96–108. doi: 10.1016/j.bcab.2018.02.009 [DOI] [Google Scholar]
- Shahid M., Ahmed B., Zaidi A., Khan M. S. (2018b). Toxicity of fungicides to Pisum sativum: a study of oxidative damage, growth suppression, cellular death and morpho-anatomical changes. RSC Adv. 8, 38483–38498. doi: 10.1039/C8RA03923B, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid M., Al-Khattaf F. S., Danish M., Atef Hatamleh A., Mohamed A., Ali S. (2022a). PGPR Kosakoniaradicincitans KR-17 increases the salt tolerance of radish by regulating ion-homeostasis, photosynthetic molecules, redox potential, and stressor metabolites. Front. Plant Sci. 13:2270. doi: 10.3389/fpls.2022.919696, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid M., Ameen F., Maheshwari H. S., Ahmed B., AlNadhari S., Khan M. S. (2021a). Colonization of Vigna radiata by a halotolerant bacterium Kosakoniasacchari improves the ionic balance, stressor metabolites, antioxidant status and yield under NaCl stress. Appl. Soil Ecol. 158:103809. doi: 10.1016/j.apsoil.2020.103809 [DOI] [Google Scholar]
- Shahid M., Khan M. S. (2018). Glyphosate induced toxicity to chickpea plants and stress alleviation by herbicide tolerant phosphate solubilizing Burkholderiacepacia PSBB1 carrying multifarious plant growth promoting activities. 3 Biotech 8, 131–117. doi: 10.1007/s13205-018-1145-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid M., Khan M. S. (2019). Fungicide tolerant Bradyrhizobium japonicum mitigate toxicity and enhance greengram production under hexaconazole stress. J. Environ. Sci. 78, 92–108. doi: 10.1016/j.jes.2018.07.007, PMID: [DOI] [PubMed] [Google Scholar]
- Shahid M., Khan M. S. (2022a). Tolerance of pesticides and antibiotics among beneficial soil microbes recovered from contaminated rhizosphere of edible crops. Cur. Res. Microbial Sci. 3:100091. doi: 10.1016/j.crmicr.2021.100091, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid M., Khan M. S. (2022b). Ecotoxicological implications of residual pesticides to beneficial soil bacteria: a review. Pesticide Biochem. Physiol 188:105272. doi: 10.1016/j.pestbp.2022.105272, PMID: [DOI] [PubMed] [Google Scholar]
- Shahid M., Khan M. S., Syed A., Marraiki N., Elgorban A. M. (2021b). Mesorhizobiumciceri as biological tool for improving physiological, biochemical and antioxidant state of Cicer aritienum (L.) under fungicide stress. Sci. Rep. 11, 1–18. doi: 10.1038/s41598-021-89103-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid M., Khan M. S., Zaidi A. (2020). Fungicide toxicity to legumes and its microbial remediation: a current perspective. Pestic. Crop Prod. Physiol. Biochem. Action, 15–33. doi: 10.1002/9781119432241.ch2 [DOI] [Google Scholar]
- Shahid M., Manoharadas S., Altaf M., Alrefaei A. F. (2021c). Organochlorine pesticides negatively influenced the cellular growth, morphostructure, cell viability, and biofilm-formation and phosphate-solubilization activities of Enterobacter cloacae strain EAM 35. ACS Omega. 6, 5548–5559. doi: 10.1021/acsomega.0c05931, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid M., Shah A. A., Basit F., Noman M., Zubair M., Ahmed T., et al. (2019a). Achromobacter sp. FB-14 harboring ACC deaminase activity augmented rice growth by upregulating the expression of stress-responsive CIPK genes under salinity stress. Brazilian J. Microbiol. 51, 719–728. doi: 10.1007/s42770-019-00199-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid M., Singh U. B., Ilyas T., Malviya D., Vishwakarma S. K., Shafi Z., et al. (2022c). Bacterial inoculants for control of fungal diseases in Solanum lycopersicum L. (tomatoes): a comprehensive overview. In Rhizosphere Microbes. Microorganisms for Sustainability, eds. Singh, U.B., Sahu, P.K., Singh, H.V., Sharma, P.K., Sharma, S. K. vol 40. Springer, Singapore., 311–339. doi: 10.1007/978-981-19-5872-4_1 [DOI] [Google Scholar]
- Shahid M., Zaidi A., Ehtram A., Khan M. S. (2019b). In vitro investigation to explore the toxicity of different groups of pesticides for an agronomically important rhizosphere isolate Azotobacter vinelandii. Pesticide Biochem. Physiol 157, 33–44. doi: 10.1016/j.pestbp.2019.03.006, PMID: [DOI] [PubMed] [Google Scholar]
- Shahid M., Zaidi A., Khan M., Rizvi A., Saif S., Ahmed B. (2017). Recent advances in management strategies of vegetable diseases. In Microbial Strategies for Vegetable Production. eds. Zaidi, A., Khan, M. Springer, Cham. doi: 10.1007/978-3-319-54401-4_9 [DOI] [Google Scholar]
- Shahid M., Zeyad M. T., Syed A., Singh U. B., Mohamed A., Bahkali A. H., et al. (2022b). Stress-tolerant endophytic isolate Priestiaaryabhattai BPR-9 modulates physio-biochemical mechanisms in wheat (Triticum aestivum L.) for enhanced salt tolerance. Int. J. Environ. Res. Public Health 19:10883. doi: 10.3390/ijerph191710883, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahzad R., Khan A. L., Bilal S., Asaf S., Lee I. J. (2017). Plant growth-promoting endophytic bacteria versus pathogenic infections: an example of bacillus amyloliquefaciens RWL-1 and fusarium oxysporum f. sp. lycopersici in tomato. Peer J. 5:e3107. doi: 10.7717/peerj.3107, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahzadi I., Khalid A., Mahmood S., Arshad M., Mahmood T., Aziz I. (2013). Effect of bacteria containing ACC deaminase on growth of wheat seedlings grown with chromium contaminated water. Pak. J. Bot. 45, 487–494. [Google Scholar]
- Sharma S. B., Jain S., Khirwadkar P., Kulkarni S. (2013). The effects of air pollution on the environment and human health. Indian J. Res. Pharmacy Biotech. 1, 391–396. [Google Scholar]
- Sheng X. F., He L. Y., Zhou L., Shen Y. Y. (2009). Characterization of microbacterium sp. F10a and its role in polycyclic aromatic hydrocarbon removal in low-temperature soil. Can. J. Microbiol. 55, 529–535. doi: 10.1139/W09-005, PMID: [DOI] [PubMed] [Google Scholar]
- Siddikee M. A., Chauhan P. S., Sa T. (2012). Regulation of ethylene biosynthesis under salt stress in red pepper (Capsicum annuum L.) by 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase-producing halotolerant bacteria. J. Plant Growth Regul. 31, 265–272. doi: 10.1007/s00344-011-9236-6 [DOI] [Google Scholar]
- Singh R. P., Jha P. N. (2016). A halotolerant bacterium bacillus licheniformis HSW-16 augments induced systemic tolerance to salt stress in wheat plant (Triticum aestivum). Front. Plant Sci. 7:1890. doi: 10.3389/fpls.2016.01890, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh U. B., Malviya D., Singh S., Imran M., Pathak N., Alam M., et al. (2016a). Compatible salt-tolerant rhizosphere microbe-mediated induction of phenylpropanoid cascade and induced systemic responses against Bipolarissorokiniana (Sacc.) shoemaker causing spot blotch disease in wheat (Triticum aestivum L.). Appl. Soil Ecol. 108, 300–306. doi: 10.1016/j.apsoil.2016.09.014 [DOI] [Google Scholar]
- Singh U. B., Malviya D., Singh S., Pradhan J. K., Singh B. P., Roy M., et al. (2016b). Bio-protective microbial agents from rhizosphere eco-systems trigger plant defense responses provide protection against sheath blight disease in rice (Oryza sativa L.). Microbiol. Res. 192, 300–312. doi: 10.1016/j.micres.2016.08.007, PMID: [DOI] [PubMed] [Google Scholar]
- Singh U. B., Malviya D., Singh S., Singh P., Ghatak A., Imran M., et al. (2021). Salt-tolerant compatible microbial inoculants modulate physio-biochemical responses enhance plant growth, Zn biofortification and yield of wheat grown in saline-sodic soil. Int. J. Environ. Res. Public Health 18:9936. doi: 10.3390/ijerph18189936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. P., Pandey D. M., Jha P. N., Ma Y. (2022). ACC deaminase producing rhizobacterium Enterobactercloacae ZNP-4 enhance abiotic stress tolerance in wheat plant. PLoS One 17:e0267127. doi: 10.1371/journal.pone.0267127, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. P., Shelke G. M., Kumar A., Jha P. N. (2015). Biochemistry and genetics of ACC deaminase: a weapon to “stress ethylene” produced in plants. Front. Microbiol. 6:937. doi: 10.3389/fmicb.2015.00937, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Singh U. B., Malviya D., Paul S., Sahu P. K., Trivedi M., et al. (2020a). Seed biopriming with microbial inoculant triggers local and systemic defense responses against Rhizoctonia solani causing banded leaf and sheath blight in maize (Zea mays L.). Int. J. Environ. Res. Public Health 17:1396. doi: 10.3390/ijerph17041396, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Singh U. B., Trivdi M., Malviya D., Sahu P. K., Roy M., et al. (2021). Restructuring the cellular responses: connecting microbial intervention with ecological fitness and adaptiveness to the maize (Zea mays L.) grown in saline–sodic soil. Front. Microbiol. 11:568325. doi: 10.3389/fmicb.2020.568325, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Singh U. B., Trivedi M., Sahu P. K., Paul S., Paul D., et al. (2020b). Seed biopriming with salt-tolerant endophytic Pseudomonas geniculata-modulated biochemical responses provide ecological fitness in maize (Zea mays L.) grown in saline sodic soil. Int. J. Environ. Res. Public Health 17:253. doi: 10.3390/ijerph17010253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofy M. R., Aboseidah A. A., Heneidak S. A., Ahmed H. R. (2021). ACC deaminase containing endophytic bacteria ameliorate salt stress in Pisum sativum through reduced oxidative damage and induction of antioxidative defense systems. Environ. Sci. Pollut. Res. 28, 40971–40991. doi: 10.1007/s11356-021-13585-3, PMID: [DOI] [PubMed] [Google Scholar]
- Srinivasan R., Mageswari A., Subramanian P. (2017). Exogenous expression of ACC deaminase gene in psychrotolerant bacteria alleviates chilling stress and promotes plant growth in millets under chilling conditions. Indian J. Exp. Biol. 55, 463–468. [Google Scholar]
- Stallworth S., Schumaker B., Fuller M. G., Tseng T. M. (2020). “Consequences and mitigation strategies of biotic and abiotic Stress in Rice (Oryza sativa L.)” in Plant Stress physiology (eds. Akbar Hossain Intech Open; ) [Google Scholar]
- Stress A. (2018). Perspectives of Rhizobacteria with ACC deaminase activity in plant growth under. Root Biology 52:303. doi: 10.1007/978-3-319-75910-4_12 [DOI] [Google Scholar]
- Subramanian P., Kim K., Krishnamoorthy R. (2016). Cold stress tolerance in psychrotolerant soil bacteria and their conferred chilling resistance in tomato (Solanum lycopersicum mill.) under low temperatures. PLoS One 11:e0161592. doi: 10.1371/journal.pone.0161592, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian P., Krishnamoorthy R., Chanratana M. (2015). Expression of an exogenous 1-aminocyclopropane-1-carboxylatedeaminase gene in psychrotolerant bacteria modulates ethylene metabolism and cold induced genes in tomato under chilling stress. Plant Physiol. Biochem. 89, 18–23. doi: 10.1016/j.plaphy.2015.02.003, PMID: [DOI] [PubMed] [Google Scholar]
- Sun L., Zhang X., Ouyang W., Yang E., Cao Y., Sun R. (2022). Lowered cd toxicity, uptake and expression of metal transporter genes in maize plant by ACC deaminase-producing bacteria Achromobacter sp. J. Hazard. Mater. 423:127036. doi: 10.1016/j.jhazmat.2021.127036, PMID: [DOI] [PubMed] [Google Scholar]
- Sun L. R., Zhao Z. J., Hao F. S. (2019). NADPH oxidases, essential players of hormone signalings in plant development and response to stresses. Plant Signal. Behav. 14:1657343. doi: 10.1080/15592324.2019.1657343, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahir M., Arshad M., Naveed M., Zahir Z. A., Shaharoona B., Ahmad R. (2006). Enrichment of recycled organic waste with N fertilizer and PGPR containing ACC-deaminase for improving growth and yield of tomato. Soil Environ. 31, 23–15. [Google Scholar]
- Tavares M. J., Nascimento F. X., Glick B. R., Rossi M. J. (2018). The expression of an exogenous ACC deaminase by the endophyte Serratia grimesii BXF 1 promotes the early nodulation and growth of common bean. Letter Appl. Microbiol. 66, 252–259. doi: 10.1111/lam.12847 [DOI] [PubMed] [Google Scholar]
- Theocharis A., Bordiec S., Fernandez O., Paquis S., Dhondt-Cordelier S., Baillieul F., et al. (2012). BurkholderiaphytofirmansPsJN primes Vitis vinifera L. and confers a better tolerance to low non-freezing temperatures. Mol. Plant-Microbe Interact. 25, 241–249. doi: 10.1094/MPMI-05-11-0124, PMID: [DOI] [PubMed] [Google Scholar]
- Thibodeaux C. J., Liu H. W. (2011). Mechanistic studies of 1-aminocyclopropane-1-carboxylate deaminase: characterization of an unusual pyridoxal 5′-phosphate-dependent reaction. Biochemist 50, 1950–1962. doi: 10.1021/bi101927s, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thijs S., Weyens N., Sillen W., Gkorezis P., Carleer R., Vangronsveld J. (2014). Potential for plant growth promotion by a consortium of stress-tolerant 2, 4-dinitrotoluene-degrading bacteria: isolation and characterization of a military soil. Microbial Biotech. 7, 294–306. doi: 10.1111/1751-7915.12111, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmusk S., Paalme V., Pavlicek T., Bergquist J., Vangala A., Danilas T., et al. (2011). Bacterial distribution in the rhizosphere of wild barley under contrasting microclimates. PLoS One 6:e17968. doi: 10.1371/journal.pone.0017968, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S., Gupta D., Fatima A., Singh S., Prasad S. M. (2020). “Phytohormonal metabolic engineering for abiotic stress in plants: new avenues and future prospects” in Plant life under changing environment, eds. Durgesh Kumar Tripathi, Vijay Pratap Singh, Naleeni Ramawat (Academic Press; ), 543–576. [Google Scholar]
- Tiwari S., Lata C., Chauhan P. S., Nautiyal C. S. (2016). Pseudomonas putida attunes morphophysiological, biochemical and molecular responses in Cicer arietinum L. during drought stress and recovery. Plant Physiol. Biochem. 99, 108–117. doi: 10.1016/j.plaphy.2015.11.001, PMID: [DOI] [PubMed] [Google Scholar]
- Toklikishvili N., Dandurishvili N., Vainstein A., Tediashvili M., Giorgobiani N., Lurie S. (2010). Inhibitory effect of ACC deaminase producing bacteria on crown gall formation in tomato plants infected by agrobacterium tumefaciens or A. vitis. Plant Pathol. 59, 1023–1030. doi: 10.1111/j.1365-3059.2010.02326.x [DOI] [Google Scholar]
- Trung N. T., Hieu H. V., Thuan N. H. (2016). Screening of strong 1-aminocyclopropane-1-carboxylate deaminase producing bacteria for improving the salinity tolerance of cowpea. Appl. Micro Open Access 2:1000111. doi: 10.4172/amoa.1000111 [DOI] [Google Scholar]
- Tsolakidou M. D., Pantelides L. S., Tzima A. K., Kang S., Paplomatas E. J., Tsaltas D. (2019). Disruption and overexpression of the gene encoding ACC (1-aminocyclopropane-1-carboxylic acid) deaminase in soil-borne fungal pathogen Verticillium dahliae revealed the role of ACC as a potential regulator of virulence and plant defense. Mol. Plant-Microbe Interact. 32, 639–653. doi: 10.1094/MPMI-07-18-0203-R [DOI] [PubMed] [Google Scholar]
- Uchiumi T., Ohwada T., Itakura M., Mitsui H., Nukui N., Dawadi P., et al. (2004). Expression islands clustered on the symbiosis island of the Mesorhizobium loti genome. J. Bacteriol. 186, 2439–2448. doi: 10.1128/JB.186.8.2439-2448.2004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loon L. C., Geraats B. P. J., Linthorst H. J. M. (2006). Ethylene as a modulator of disease resistance in plants. Trends Plant Sci. 11, 184–191. doi: 10.1016/j.tplants.2006.02.005 [DOI] [PubMed] [Google Scholar]
- Vargas L. K., Volpiano C. G., Lisboa B. B., Giongo A., Beneduzi A., Passaglia L. M. P. (2017). Potential of rhizobia as plant growth-promoting rhizobacteria Springer International Publishing, 153–174. [Google Scholar]
- Viscardi S., Ventorino V., Duran P., Maggio A., De Pascale S., Mora M. L., et al. (2016). Assessment of plant growth promoting activities and abiotic stress tolerance of Azotobacter chroococcum strains for a potential use in sustainable agriculture. J. Soil Sci. Plant Nut. 16, 848–863. doi: 10.4067/S0718-95162016005000060 [DOI] [Google Scholar]
- Walsh C., Pascal R. A., Jr., Johnston M., Raines R., Dikshit D., Krantz A., et al. (1981). Mechanistic studies on the pyridoxal phosphate enzyme 1-aminocyclopropane-1-carboxylate deaminase from pseudomonas sp. Biochemist 20, 7509–7519. doi: 10.1021/bi00529a028, PMID: [DOI] [PubMed] [Google Scholar]
- Wang Q., Dodd I. C., Belimov A. A., Jiang F. (2016). Rhizosphere bacteria containing 1-aminocyclopropane-1-carboxylate deaminase increase growth and photosynthesis of pea plants under salt stress by limiting Na1 accumulation. Funct. Plant Biol. 43, 161–172. doi: 10.1071/FP15200, PMID: [DOI] [PubMed] [Google Scholar]
- Wang C., Knill E., Glick B. R., Défago G. (2000). Effect of transferring 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase genes into Pseudomonas fluorescens strain CHA0 and its gacA derivative CHA96 on their growth-promoting and disease-suppressive capacities. Can. J. Microbiol. 46, 898–907. doi: 10.1139/w00-07 [DOI] [PubMed] [Google Scholar]
- Wang C., Wang F., Zhang Q., Liang W. (2016). Individual and combined effects of tebuconazole and carbendazim on soil microbial activity. Eur. J. Soil Bio. J. 72, 6–13. doi: 10.1016/j.ejsobi.2015.12.005 [DOI] [Google Scholar]
- Wang C. J., Yang W., Wang C., Gu C., Niu D. D., Liu H. X., et al. (2012). Induction of drought tolerance in cucumber plants by a consortium of three plant growth-promoting rhizobacterium strains. PLoS One 7:e52565. doi: 10.1371/journal.pone.0052565, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Zhang Z., Chen Y., Wei X., Feng B., Tao F. (2016). How much yield loss has been caused by extreme temperature stress to the irrigated rice production in China? Clim. Chang. 134, 635–650. doi: 10.1007/s10584-015-1545-5 [DOI] [Google Scholar]
- Xia Y., Farooq M. A., Javed M. T., Kamran M. A., Mukhtar T., Ali J., et al. (2020). Multi-stress tolerant PGPR Bacillusxiamenensis PM14 activating sugarcane (Saccharum officinarum L.) red rot disease resistance. Plant Physiol. Biochem. 151, 640–649. doi: 10.1016/j.plaphy.2020.04.016 [DOI] [PubMed] [Google Scholar]
- Xun F., Xie B., Liu S. (2015). Effect of plant growth-promoting bacteria (PGPR) and arbuscular mycorrhizal fungi (AMF) inoculation on oats in saline-alkali soil contaminated by petroleum to enhance phytoremediation. Environ. Sci. Pollut. Res. 22, 598–608. doi: 10.1007/s11356-014-3396-4, PMID: [DOI] [PubMed] [Google Scholar]
- Yadav S. K. (2010). Cold stress tolerance mechanisms in plants: a review. Agron. Sustain. Dev. 30, 515–527. doi: 10.1051/agro/2009050 [DOI] [Google Scholar]
- Yan K., Shao H., Shao C., Chen P., Zhao S., Brestic M. (2013). Physiological adaptive mechanisms of plants grown in saline soil and implications for sustainable saline agriculture in coastal zone. Acta Physiol. Plant. 35, 2867–2878. doi: 10.1007/s11738-013-1325-7 [DOI] [Google Scholar]
- Yasmin H., Bano A., Wilson N. L., Nosheen A., Naz R., Hassan M. N., et al. (2022). Drought-tolerant pseudomonas sp. showed differential expression of stress-responsive genes and induced drought tolerance in Arabidopsis thaliana. Physiol. Planta. 174:e13497. doi: 10.1111/ppl.13497, PMID: [DOI] [PubMed] [Google Scholar]
- Yim W. J., Woo S. M., Kim K. Y., Sa T. M. (2012). Regulation of ethylene emission in tomato (Lycopersicon esculentum mill.) and red pepper (Capsicum annuum L.) inoculated with ACC deaminase producing Methylobacterium spp. Korean J. Soil Sci. Fertilizer 45, 37–42. [Google Scholar]
- Zafar-ul-Hye M., Danish S., Abbas M., Ahmad M., Munir T. M. (2019). ACC deaminase producing PGPR bacillus amyloliquefaciens and agrobacterium fabrum along with biochar improve wheat productivity under drought stress. Agronomy 9:343. doi: 10.3390/agronomy9070343 [DOI] [Google Scholar]
- Zafar-Ul-Hye M. U., Shahjahan A., Danish S. U., Abid M. U., Qayyum M. F. (2018). Mitigation of cadmium toxicity induced stress in wheat by acc-deaminase containing PGPR isolated from cadmium polluted wheat rhizosphere. Pak. J. Bot. 50, 1727–1734. [Google Scholar]
- Zahir Z. A., Ghani U., Naveed M., Nadeem S. M., Asghar H. N. (2009). Comparative effectiveness of pseudomonas and Serratia sp. containing ACC deaminase for improving growth and yield of wheat (Triticum aestivum L.) under salt-stressed conditions. Arch. Microbiol. 191, 415–424. doi: 10.1007/s00203-009-0466-Y, PMID: [DOI] [PubMed] [Google Scholar]
- Zahir Z. A., Munir A., Asghar H. N., Shaharoona B., Arshad M. (2008). Effectiveness of rhizobacteria containing ACC deaminase for growth promotion of peas (Pisum sativum) under drought conditions. J. Microbiol. Biotechnol. 18, 958–963. [PubMed] [Google Scholar]
- Zaidi A., Khan M. S., Ahmad E., Saif S., Rizvi A., Shahid M. (2016). Growth stimulation and management of diseases of ornamental plants using phosphate solubilizing microorganisms: current perspective. Acta Physiol. Planta. 38, 1–21. doi: 10.1007/s11738-016-2133-7 [DOI] [Google Scholar]
- Zaidi A., Khan M. S., Saif S., Rizvi A., Ahmed B., Shahid M. (2017). “Role of nitrogen-fixing plant growth-promoting rhizobacteria in sustainable production of vegetables: current perspective” in Microbial strategies for vegetable production, Zaidi, A., Khan, M. (eds) (Cham: Springer; ), 49–79. [Google Scholar]
- Zainab N., Din B. U., Javed M. T., Afridi M. S., Mukhtar T., Kamran M. A., et al. (2020). Deciphering metal toxicity responses of flax (Linumusitatissimum L.) with exopolysaccharide and ACC-deaminase producing bacteria in industrially contaminated soils. Plant Physiol. Biochem. 152, 90–99. doi: 10.1016/j.plaphy.2020.04.039, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang H., Li A., Zhang Z., Huang Z., Lu P., Zhang D., et al. (2016). Ethylene response factor TERF1, regulated by ethylene-INSENSITIVE3-like factors, functions in reactive oxygen species (ROS) scavenging in tobacco (Nicotiana tabacum L.). Sci. Rep. 6:29948. doi: 10.1038/srep29948, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Chen H., Li K., Du W., He S., Liu H. W. (2003). Reaction of 1-amino-2-methylenecyclopropane-1-carboxylate with 1-aminocyclopropane-1-carboxylate deaminase: analysis and mechanistic implications. Biochemist 42, 2089–2103. doi: 10.1021/bi020567n, PMID: [DOI] [PubMed] [Google Scholar]
- Zhu T., Deng X., Zhou X., Zhu L., Zou L., Li P., et al. (2016). Ethylene and hydrogen peroxide are involved in brassinosteroid-induced salt tolerance in tomato. Sci. Rep. 6:35392. doi: 10.1038/srep35392, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]