Abstract
In the human gut, the growth of the pathogen Clostridioides difficile is impacted by a complex web of interspecies interactions with members of human gut microbiota. We investigate the contribution of interspecies interactions on the antibiotic response of C. difficile to clinically relevant antibiotics using bottom-up assembly of human gut communities. We identify 2 classes of microbial interactions that alter C. difficile’s antibiotic susceptibility: interactions resulting in increased ability of C. difficile to grow at high antibiotic concentrations (rare) and interactions resulting in C. difficile growth enhancement at low antibiotic concentrations (common). Based on genome-wide transcriptional profiling data, we demonstrate that metal sequestration due to hydrogen sulfide production by the prevalent gut species Desulfovibrio piger increases the minimum inhibitory concentration (MIC) of metronidazole for C. difficile. Competition with species that display higher sensitivity to the antibiotic than C. difficile leads to enhanced growth of C. difficile at low antibiotic concentrations due to competitive release. A dynamic computational model identifies the ecological principles driving this effect. Our results provide a deeper understanding of ecological and molecular principles shaping C. difficile’s response to antibiotics, which could inform therapeutic interventions.
This study examines how interactions between constituent members of the human gut microbiome influence the pathogen C. difficile’s response to clinically relevant antibiotics, providing key insights into ecological principles and molecular mechanisms that influence antibiotic susceptibility in this health-relevant system.
Introduction
The bacterial pathogen Clostridioides difficile can infect the human gastrointestinal tract, an environment teeming with a dense microbiota. Gut microbiota can inhibit C. difficile’s growth and ability to persist over time in the human gut, a phenomenon known as colonization resistance [1]. The key role of colonization resistance is illustrated by the increased risk of C. difficile infection after treatment with antibiotics that decimate the microbiota [2] and by the efficacy of fecal microbiota transplants from healthy human donors in eliminating recurrent C. difficile infections [3]. Previous studies have provided a deeper understanding of interactions between gut microbiota and C. difficile. For example, interspecies interactions between individual gut microbes and C. difficile have been studied in vitro and analyses of human microbiome data have identified gut microbes whose presence or absence is associated with altered outcomes of C. difficile infection [4–6]. Multiple mechanisms of interaction have been determined, such as inhibition of C. difficile germination by Clostridium scindens via production of secondary bile acids [6] and promotion of C. difficile growth by Bacteroides thetaiotaomicron via succinate cross-feeding [7]. While much is known about how the microbiota impacts C. difficile growth, how the microbiota impacts C. difficile antibiotic susceptibility is largely unknown.
Similar to other pathogens, C. difficile antibiotic susceptibility has been studied using in vitro experiments of monoculture growth. However, monoculture experiments do not consider how interactions with resident community members can modify the antibiotic susceptibility of a pathogen. For example, monospecies antibiotic susceptibility of the pathogen Pseudomonas aeruginosa did not always correlate with the efficacy of treatment for polymicrobial infections [8]. If microbial interactions substantially decrease the pathogen’s antibiotic susceptibility, treatments based on monoculture susceptibility to antibiotics may not be effective in eradicating the pathogen. Alternatively, if communities increase the pathogen’s antibiotic susceptibility, the standard antibiotic dosage may exceed the dose needed to eradicate the pathogen, yielding unnecessary and avoidable disruption to the native microbiota. Understanding how constituent members of microbiota alter a pathogen’s susceptibility to an antibiotic could be used to guide the design of treatments to eradicate the pathogen.
Previous studies have shown that interspecies interactions can alter a given microbe’s response to antibiotics by increasing or decreasing susceptibility compared to monoculture [9]. One example is exposure protection, where susceptible microbes are protected from an antibiotic by species that degrade the antibiotic [10]. In addition, a previous study showed that an increase in antibiotic susceptibility occurred when the growth of a resistant microbe depended on cross-feeding with another organism that was susceptible to the antibiotic [11].
While previous studies have identified specific types of interspecies interactions that impact antibiotic susceptibility, the prevalence of susceptibility-altering microbial interactions across different microbial communities is not well understood. In a human urinary tract infection community, around a third of total interactions between species were estimated to yield a change in the susceptibility to 2 different antibiotics based on spent media experiments [12]. Studies of a fruit fly microbiome, a multispecies wound infection biofilm, and a multispecies brewery biofilm each identified a change in antibiotic susceptibility of a given species in the community compared to monospecies [13–15]. By contrast, no significant changes in antibiotic susceptibility were observed for 15 characterized species in a community of human gut microbes or an Escherichia coli pathogen introduced into a porcine microbiome [16,17]. Based on this observed variation in the contribution of interspecies interactions to antibiotic susceptibility across different systems, it is not known whether microbial interactions can impact C. difficile’s antibiotic susceptibility.
We used a diverse human gut community [4] to study the impact of microbial interactions on C. difficile’s antibiotic susceptibility. We focused on 2 antibiotics, vancomycin and metronidazole, that are used to treat C. difficile infections. Vancomycin is a glycopeptide that inhibits cell wall synthesis whose activity is specific to gram-positive bacteria [18]. Metronidazole is a DNA-damaging agent that is effective against both gram-positive and gram-negative anaerobic bacteria. Metronidazole is a prodrug which is inactive until its nitro group is reduced to nitroso radicals in the cytoplasm of anaerobic bacteria [19]. The proposed mechanism of metronidazole reduction is due to the cofactors ferredoxin and/or flavodoxin in reactions catalyzed by multiple enzymes including reductases, hydrogenases, and pyruvate ferredoxin/flavodoxin oxidoreductase (PFOR) [20–22]. Metronidazole, previously a recommended first-line treatment, is now only recommended in rare cases due to an observed decrease in its clinical effectiveness [23,24].
Using a bottom-up approach, we perform a detailed and quantitative characterization of how human gut microbes impact C. difficile’s response to these antibiotics. We show that gut microbes infrequently alter C. difficile’s minimum inhibitory concentration (MIC), but gut microbes frequently impact C. difficile’s response to subinhibitory concentrations of antibiotics. In communities with antibiotic-sensitive species that also compete with C. difficile, we observe that C. difficile’s growth is enhanced in the presence of low concentrations of antibiotics due to competitive release. A dynamic ecological model representing the antibiotic recapitulates these trends. In addition, we demonstrate that Desulfovibrio piger substantially increases C. difficile’s MIC for metronidazole. We investigate the mechanism determining the increased MIC of C. difficile using transcriptional profiling and media perturbations. Our data suggest D. piger’s impact on the environment induces a metal starvation transcriptional response in C. difficile, leading to down-regulation of enzymes required to reduce metronidazole to its active form. In sum, biotic interactions shape C. difficile antibiotic susceptibility at both subinhibitory and minimal inhibitory concentration regimes via distinct mechanisms. These results highlight the need to consider biotic interactions in the design of future therapeutic treatments to eradicate pathogens.
Results
The minimum inhibitory concentration of C. difficile to metronidazole or vancomycin is modified by a subset of gut microbes in pairwise communities
C. difficile infections occur in the context of complex resident gut communities. However, antibiotic treatments to eliminate C. difficile infection are designed based on the susceptibility of C. difficile in monoculture, which neglect the role of interspecies interactions in shaping antibiotic susceptibility. Understanding how the gut microbiota alters C. difficile antibiotic susceptibility could inform the treatment of C. difficile by antibiotics. To investigate this question, we evaluated C. difficile’s response to antibiotics in the presence of synthetic communities of gut microbes (Fig 1A). The 13-member human gut community was designed to span the phylogenetic diversity of the human gut microbiome. The interactions between these gut microbes and C. difficile have been deciphered in the absence of antibiotics [4] (Fig 1B). We selected the antibiotics metronidazole and vancomycin for their clinical relevance in the treatment of C. difficile infections and for the differences in their activity spectra. Metronidazole has broad spectrum activity against the 13 gut microbes, whereas the activity of vancomycin is specific for gram-positive species. To determine how C. difficile would respond to both low and high antibiotic concentrations in the human gut, we characterized C. difficile’s MIC and its growth in the presence of subinhibitory concentrations across a range of ecological contexts (Fig 1C).
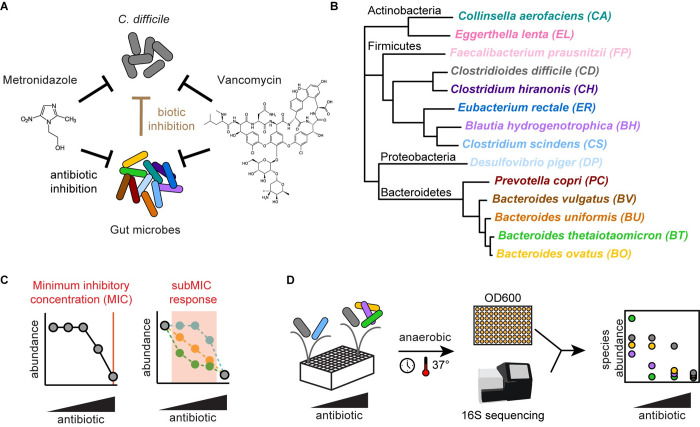
Fig 1. Investigating the effects of interspecies interactions on the clinically relevant antibiotic response of C. difficile in human gut communities.
(A) Schematic of factors affecting C. difficile growth in synthetic gut communities in the presence of antibiotics metronidazole or vancomycin. The DNA-damaging antibiotic metronidazole and the cell wall inhibitor vancomycin can inhibit C. difficile and gut microbes (black arrows). Biotic inhibition by gut microbes (brown arrow) inhibits C. difficile. (B) Phylogenetic tree of 14-member human gut community, spanning 4 major phyla of gut microbiota and pathogen C. difficile. Black text indicates phylum name. Colored text indicates species name. Phylogeny based on a concatenated alignment of 37 marker genes [25]. (C) Schematic of 2 aspects of the response of a microbial population to antibiotics: the MIC and the subMIC response. (D) Schematic of methods to determine the antibiotic susceptibility of multispecies communities. Communities are incubated anaerobically in microtiter plates. Absolute abundance is determined by multiplying optical density (OD600) by relative abundance based on 16S rRNA gene sequencing. MIC, minimum inhibitory concentration; subMIC, sub-minimum inhibitory concentration.
To determine the antibiotic susceptibility of C. difficile in communities, we used a broth dilution method based on the clinical method detailed by the Clinical and Laboratory Standards Institute [26] (Methods). We inoculated communities in liquid culture containing a 2-fold dilution series of antibiotics. After incubation, we determined the absolute abundance of each species in the communities at a fixed time point (48 h) by multiplying the community optical density at 600 nm (OD600) by its relative abundance determined via 16S rRNA gene sequencing (Fig 1D). The MIC for each species in the community was determined as the lowest antibiotic concentration where growth at 48 h was below a threshold (Methods).
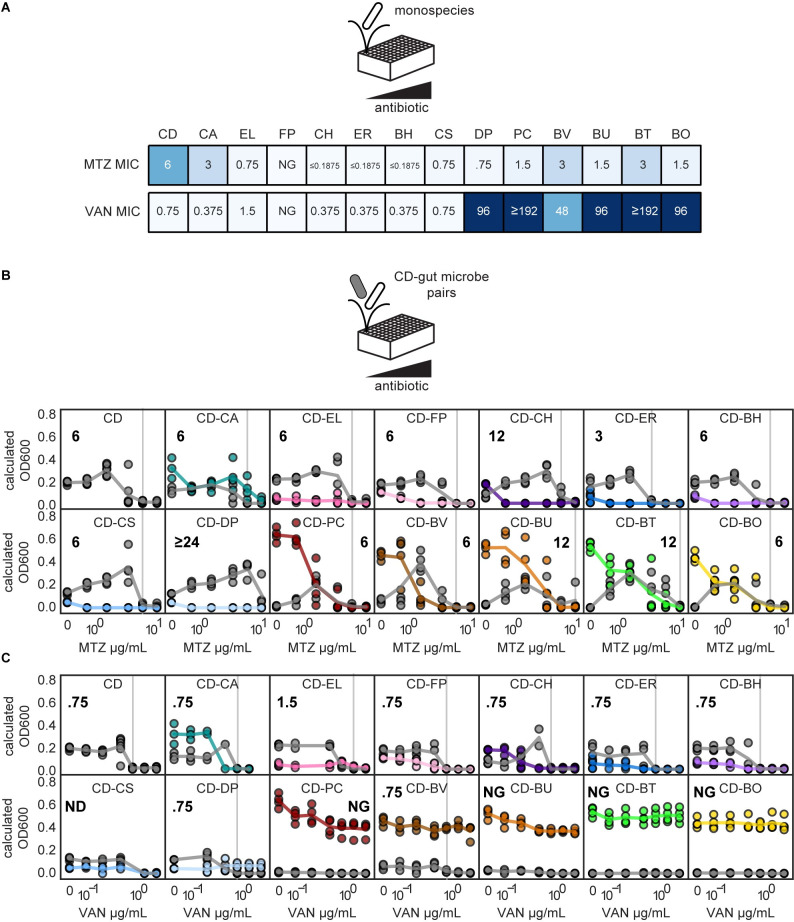
Using this method, we determined the MIC of each species in monoculture (Fig 2A) and in each pairwise community containing C. difficile (Fig 2B and 2C). Consistent with the mechanism of the antibiotics, all species were susceptible to metronidazole in the range of antibiotics tested, whereas gram-negative species (Bacteroidetes and Proteobacteria phyla, Fig 1D) were not susceptible to vancomycin (Figs 2A and S1A and S1B).
Fig 2. C. difficile response to metronidazole or vancomycin is modified in specific pairwise communities.
(A) Heatmaps of MIC of monocultures in the presence of metronidazole (MTZ) or vancomycin (VAN). Species indicated by 2-letter species code (Fig 1B). (B, C) Line plots of species absolute abundance at 48 h as a function of antibiotic concentration for C. difficile in monoculture or C. difficile in pairwise communities. Each x-axis is semi-log scale. The values on the y-axis are calculated OD600 (OD600 multiplied by relative abundance from 16S rRNA gene sequencing). Data points indicate individual biological replicates. Lines indicate the average of n = 1 to n = 8 biological replicates. Vertical gray line and bold number indicate MIC of C. difficile in each condition. Color indicates species (Fig 1B). The data underlying all panels in this figure can be found in DOI: 10.5281/zenodo.7626486. MIC, minimum inhibitory concentration.
In most pairwise communities, the metronidazole and vancomycin MICs of C. difficile were unchanged compared to monoculture. Across metronidazole and vancomycin conditions, the MIC of C. difficile was unchanged in 15 pairwise communities and displayed a moderate difference (2-fold) in 5 pairwise communities compared to monospecies (Fig 2B and 2C). However, C. difficile had a large change in MIC (≥4-fold) in coculture with D. piger (metronidazole MIC increased from 6 μg/mL to greater than or equal to 24 μg/mL) (Fig 2B). In the remaining 4 pairwise communities, C. difficile’s growth was strongly inhibited in all concentrations and no MIC could be calculated (Fig 2C, indicated as “NG”). In sum, the MIC of C. difficile was infrequently impacted by pairwise interspecies interactions in the human gut community.
For select pairwise communities, we quantified C. difficile abundance using both OD600 multiplied by 16S rRNA gene sequencing relative abundance and count of colony-forming units (CFUs) on C. difficile selective agar plates to validate the OD600-based absolute abundance method (S2A Fig). Our results showed that CFU counting and the OD600-based absolute abundance method yielded the same MIC value in 11 out of 12 conditions (S2B and S2C Fig). In 1 condition, C. difficile’s growth was strongly inhibited below the MIC threshold for all concentrations. Therefore, an MIC could not be determined using the OD600-based method, although an MIC could be revealed using the CFU method. Overall, these trends demonstrate that the OD600-based absolute abundance method is a reliable way to measure C. difficile MIC, although the 2 methods can vary in sensitivity.
Enhancement of C. difficile abundance in pairwise communities in the presence of low antibiotic concentrations
Pathogens may encounter antibiotic concentrations lower than the MIC during antibiotic treatments. If the pathogen has gained resistance to the antibiotic, the entire dose regimen could be subinhibitory. Alternatively, even if the target dose is greater than the MIC, sub-minimum inhibitory concentrations (subMICs) can occur at the beginning and end of dosing regimens and between daily dosages [27]. Therefore, we investigated the role of microbial interactions on C. difficile’s response to subinhibitory antibiotic concentrations (concentrations lower than C. difficile’s MIC or subMICs).
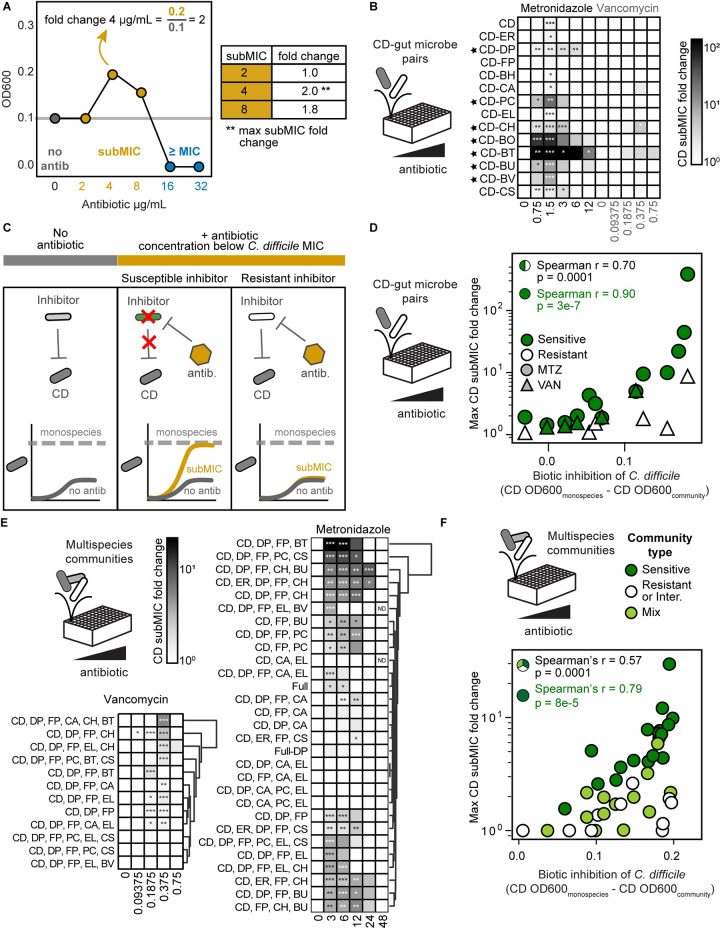
In many pairwise communities, C. difficile’s abundance was similar or reduced in the subMIC range than in the absence of antibiotic (Fig 2B and 2C). However, in a subset of pairwise communities, C. difficile’s abundance in the subMIC range was enhanced compared to the absence of antibiotic (Fig 2B and 2C). For example, C. difficile monospecies abundance was similar in the presence of 0.75 μg/mL metronidazole and the absence of antibiotic. However, in the presence of B. thetaiotaomicron, the abundance of C. difficile was significantly higher in the presence of 0.75 μg/mL metronidazole than in the absence of antibiotic (Fig 2B). To provide further insights, we computed the subMIC fold change at each subMIC, defined as the species absolute abundance at the given subMIC divided by the species absolute abundance in the absence of antibiotic (Fig 3A). Using this metric, C. difficile’s growth was enhanced for at least 1 subMIC in 1 pairwise community in the presence of vancomycin and 11 pairwise communities in the presence of metronidazole (Figs 3B and S3A and S3B). In 7 pairwise communities, the growth enhancement of C. difficile in the presence of metronidazole was significantly greater than the enhancement observed for the C. difficile monoculture in the presence of metronidazole (Fig 3B). In sum, our results illustrate that approximately half of the species altered C. difficile’s response to low concentrations of metronidazole.
Fig 3. C. difficile growth is enhanced in communities with antibiotic-sensitive biotic inhibitors in the presence of antibiotic concentrations lower than the MIC.
(A) Schematic of subMIC fold change metric using example data. The x-axis is semi-log scale. Table indicates subMIC fold changes for the 3 subMICs (2, 4, and 8 μg/mL) in this example. Maximum subMIC fold change (indicated by **) is defined as the largest fold change across all subMICs. (B) Heatmap of subMIC fold changes for C. difficile in pairwise communities at each concentration at a 48 h. Shading indicates subMIC fold change. SubMIC fold change is calculated as the average C. difficile absolute abundance at the subMIC divided by the average C. difficile absolute abundance in the absence of antibiotic, where the average is of n = 1 to n = 8 biological replicates. Asterisks indicate significant difference (*P < 0.05, **P < 0.01, ***P < 0.001) between C. difficile OD600 at subMIC and C. difficile OD600 in the absence of antibiotic according to an unpaired t test. Black stars indicate pairs with a maximum subMIC fold change significantly greater than the maximum subMIC fold change of C. difficile monospecies (p < 0.05 based on an unpaired t test). (C) Schematic of C. difficile growth in the presence of antibiotic sensitive and resistant inhibitors. (D) Scatterplot of maximum C. difficile subMIC fold change in pairwise communities as a function of the degree of biotic inhibition of C. difficile in pairwise communities for metronidazole (MTZ, circles) and vancomycin (VAN, triangles). Biotic inhibition of C. difficile (x-axis) is defined as the average C. difficile OD600 in monospecies minus the average C. difficile OD600 in the pairwise community in no antibiotic conditions. The maximum C. difficile subMIC fold change (y-axis) is the maximum of the C. difficile subMIC fold change across all subMICs at 48 h. The subMIC fold change is calculated as in panel B, with n = 1 to n = 8 biological replicates. Species are classified as sensitive if the species monospecies MIC was less than the monospecies MIC of C. difficile and resistant if the species monospecies MIC was greater than or equal to the monospecies MIC of C. difficile. Each point represents 1 pairwise community in metronidazole or vancomycin. The Spearman correlation is annotated for all data points (black) and for pairwise communities with sensitive gut microbes only (green). (E) Heatmap of subMIC fold changes for C. difficile in multispecies communities at each concentration of metronidazole and vancomycin at 48 h. Shading indicates subMIC fold change. SubMIC fold change is calculated as in panel B, with n = 1 to n = 4 biological replicates. Asterisks indicate significant difference (*P < 0.05, **P < 0.01, ***P < 0.001) between C. difficile OD600 at subMIC and C. difficile OD600 in the absence of antibiotic according to an unpaired t test. Communities clustered by subMIC fold change using Euclidean distance hierarchical clustering. “Full” indicates 14-member community, “Full-DP” indicates 13-member community containing all species except DP. (F) Scatterplot of maximum C. difficile subMIC fold change in multispecies communities as a function of the degree of biotic inhibition of C. difficile in multispecies communities for metronidazole (circles) and vancomycin (triangles). Biotic inhibition of C. difficile (x-axis) is calculated as in panel D. The maximum C. difficile subMIC fold change (y-axis) is calculated as in panel B, with n = 1 to n = 4 biological replicates. Community type is sensitive (“Sens.”) if all species excluding C. difficile are sensitive inhibitors. Community type is resistant or intermediate (“Resis. or Intermed.”) if all species excluding C. difficile are either intermediate-inhibitors or resistant-inhibitors. Community type is considered “mix” if the given community contains both sensitive inhibitors and resistant- or intermediate-inhibitors. Species are categorized as biotic inhibitors if the absolute abundance of C. difficile in the presence of this species was significantly lower than the absolute abundance of C. difficile in monospecies, in the absence of antibiotics, as determined by an unpaired t test. Species are classified as sensitive and resistant as in panel D. C. aerofaciens was classified as intermediate (see text). Each point represents 1 community in the presence of metronidazole or vancomycin. The Spearman correlation is annotated for all data points (black) and for sensitive communities only (green). The data underlying panels BDEF in this figure can be found in DOI: 10.5281/zenodo.7626486. MIC, minimum inhibitory concentration; subMIC, sub-minimum inhibitory concentration.
In monoculture, C. difficile’s growth was moderately enhanced in the presence of a single subMIC (1.5 μg/mL metronidazole, subMIC fold change of 1.6, S3A Fig). Time-series OD600 measurements of C. difficile monoculture revealed that this moderate growth enhancement was due to a difference in growth phase in the presence and absence of the antibiotic at the measured time point (48 h) (S3C and S3D Fig). By contrast, C. difficile’s growth was enhanced in 2 pairwise communities in the presence of subMICs compared to no treatment over multiple time points that was not due to differences in growth phase (S3E and S3F Fig). Thus, while differences in monoculture growth phases yielded a modest enhancement of C. difficile in response to a single subMIC, a different mechanism determined the observed larger and sustained growth enhancements in pairwise communities.
We hypothesized the action of the antibiotic in certain pairwise communities relieved the biotic inhibition of C. difficile, which would in turn enhanced C. difficile growth, representing competitive release [28,29]. This competitive release would occur in pairwise communities with competitors (i.e., biotic inhibitors) with a higher susceptibility to the antibiotic than C. difficile. By contrast, if a biotic inhibitor displayed a higher resistance to the antibiotic than C. difficile, the degree of biotic inhibition would remain unchanged at subMICs, and thus, C. difficile’s growth would not change substantially compared to its growth in absence of the antibiotic (Fig 3C). Supporting this hypothesis, the maximum subMIC fold change of C. difficile displayed a strong positive correlation with the degree of biotic inhibition in the presence of the coculture partner (Fig 3D). In addition, the correlation was enhanced in pairwise communities containing gut microbes that were sensitive to the antibiotic (Fig 3D). In the presence of both individual antibiotics, the maximum subMIC fold change of C. difficile was significantly higher when paired with biotic inhibitors with higher sensitivity to the antibiotic than C. difficile (S3G Fig). In sum, antibiotic induced inhibition of susceptible biotic inhibitors enhanced C. difficile’s growth in response to subMICs. This suggests that C. difficile’s growth in the gut could be enhanced in the subMIC regime if patients harbor antibiotic-sensitive biotic inhibitors.
We investigated whether the C. difficile subMIC growth enhancements measured using the OD600-based absolute abundance method were due to increases in C. difficile abundance as opposed to other factors such as changes in cell morphology. To this end, we determined the subMIC fold changes of C. difficile in select pairwise communities that were characterized using CFU counting (S2B and S2C Fig). Our results showed that the CFU method and OD600-based absolute abundance method identified a significant subMIC fold change in the C. difficile-B. thetaiotaomicron and C. difficile-D. piger pairs in presence of metronidazole (S2D Fig). The methods also agreed in detecting no significant subMIC fold changes in 10 other pairwise communities in metronidazole or vancomycin (S2D Fig). However, the CFU method did not detect significant subMIC fold changes in 3 pairwise communities for which the OD600-based method detected significant fold changes (C. difficile-C. hiranonis vancomycin and metronidazole conditions and C. difficile-C. scindens metronidazole condition, S2D Fig). These discrepancies may be due to differences in sensitivities of the 2 methods, as the spot dilution CFU method (Methods) cannot distinguish small fold changes (average variation of technical replicates is 2-fold).
To determine if subMIC fold changes could be resolved using the lower sensitive CFU method, we increased the resolution of subinhibitory concentrations tested. The finer resolution may capture subMICs with larger fold changes that are large enough to be detectable by the CFU method. Additionally, the finer resolution may result in multiple subMICs with significant fold changes, increasing confidence in the results. We used the CFU method with a finer resolution of antibiotic concentrations to measure growth enhancement in the C. difficile-B. vulgatus pairwise community in the presence of metronidazole, which exhibited a significant C. difficile growth enhancement using the OD600-based method (Fig 3B). Our results demonstrated a significant C. difficile subMIC fold change at multiple concentrations in the pairwise community, while no significant fold changes were observed in the C. difficile monoculture (S4 Fig). This demonstrates that the observed C. difficile growth enhancement using the OD600-based method (Fig 3B) was consistent with CFU counting. Overall, our results demonstrate that C. difficile subMIC growth enhancements can also be detected by the CFU-based absolute abundance method.
Enhancement of C. difficile abundance in multispecies communities in the presence of low antibiotic concentrations
We investigated if the OD600-based absolute abundance trends observed in pairwise communities persisted in multispecies communities that are more representative of human gut microbiota. The communities consisted of 2- and 3-member core communities (CC) predicted to display minimal biotic inhibition of C. difficile guided by a previously developed dynamic computational model of our system [4] (CC-2: D. piger and Faecalibacterium prausnitzii. CC-3: D. piger, F. prausnitzii, and Eggerthella lenta). In addition to this core community, we introduced at least 1 antibiotic-sensitive inhibitor, antibiotic-resistant inhibitor, or a combination of species in these 2 groups, creating 10 communities for metronidazole and 12 communities for vancomycin (3- to 6-members). To further explore the behavior of communities in response to antibiotic perturbations, we characterized the response of 3-, 4-, 5-, 13-, or 14-member communities (19 total) with no consistent core members to metronidazole.
In 6 of 29 communities characterized in the presence of metronidazole, C. difficile’s MIC was 4-fold greater than in monoculture, increasing from 6 μg/mL to 24 μg/mL. The remaining communities displayed moderate (2-fold, 14 communities) or no change (9 communities) (S5A Fig, S1 Table). In 4 of 12 communities examined in the presence of vancomycin, C. difficile’s growth was strongly inhibited at all concentrations and no MIC could be calculated (S5B Fig, S2 Table, indicated as “NG”). The remaining 8 communities displayed either a moderate increase in MIC (2-fold, 2 communities) or no change (6 communities) (S5B Fig, S2 Table). Consistent with the pairwise community data, C. difficile’s metronidazole MIC was altered in certain communities, but C. difficile’s vancomycin MIC was not substantially altered in any of the characterized communities.
We tested if the multispecies communities containing antibiotic-sensitive biotic inhibitors displayed an enhancement in C. difficile’s growth at subMICs. In response to metronidazole or vancomycin, C. difficile’s growth was enhanced in response to at least 1 subMIC in most communities (21 of 29 communities for metronidazole, 9 of 12 communities for vancomycin, Figs 3E and S6). Consistent with the pairwise community data, the degree of biotic inhibition of C. difficile was positively correlated with the magnitude of the maximum subMIC fold change (Fig 3F). In addition, the correlation was stronger for communities composed of only antibiotic-sensitive biotic inhibitors (Fig 3F). Consistent with these trends, the number of sensitive inhibitors in the community and the absolute abundance of sensitive inhibitors displayed a positive correlation with the maximum subMIC fold change (S7A Fig). Communities containing antibiotic-sensitive biotic inhibitors had a significantly higher average subMIC fold change than communities containing only resistant inhibitors or a mix of both inhibitor types (S7B Fig).
Communities containing a mix of antibiotic sensitive and resistant inhibitors, mirroring the makeup of the gut microbiota, displayed low subMIC fold changes similar in magnitude to resistant biotic inhibitor communities (S7B Fig). In communities with a non-zero abundance of resistant biotic inhibitors, the correlation between the growth enhancement of C. difficile and the number of sensitive inhibitors or abundance of sensitive inhibitors vanished (S7A Fig). These data suggest that resistant biotic inhibitors can suppress the C. difficile growth enhancement caused by sensitive biotic inhibitors. We analyzed if the growth enhancement of C. difficile in multispecies communities could be predicted based on the sum of the growth enhancements of C. difficile in pairwise communities. While the relationship displayed a moderate correlation, it was not additive (S7C and S7D Fig). This demonstrates that community effects, such as interspecies interactions between constituent community members and growth enhancement suppression by resistant inhibitors, play key roles in determining the magnitude of growth enhancement of C. difficile at subMICs. Overall, these results demonstrate that C. difficile growth enhancement at subMICs can occur in multispecies communities, suggesting this response may occur in the human gut microbiome.
A dynamic ecological model representing pairwise interactions and monospecies antibiotic susceptibilities can capture the response of C. difficile to subMICs
We hypothesized that a model capturing the dynamics of species growth, interspecies interactions, and monospecies antibiotic susceptibilities could recapitulate the observed trends in C. difficile growth enhancement at subMICs. We assumed that inferred interspecies interactions in the absence of antibiotics contributed to the community response to antibiotics. We tested whether a model that neglects complex antibiotic-dependent interspecies interactions could predict the qualitative trends of C. difficile growth at subMICs.
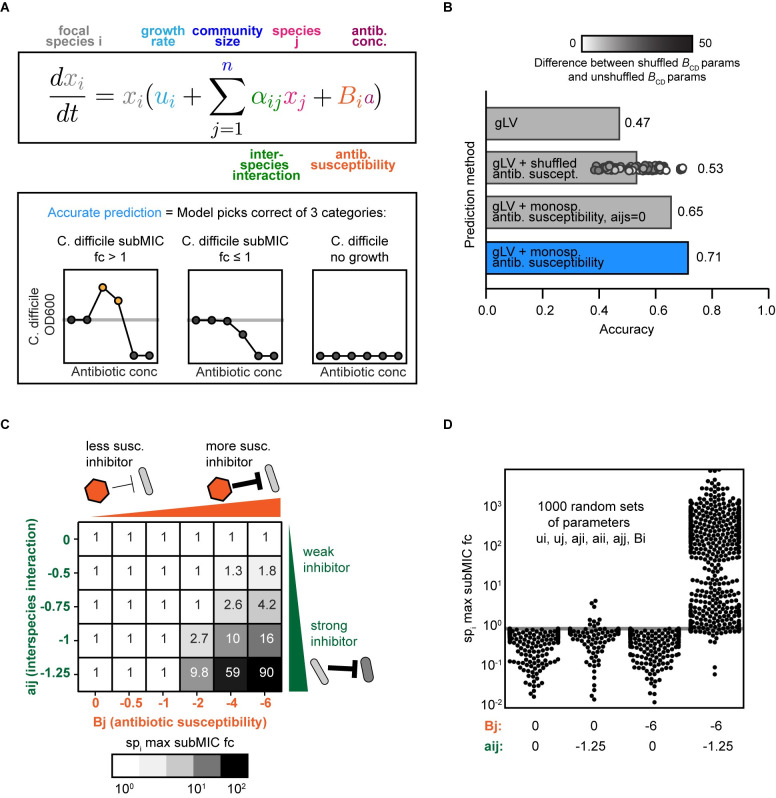
The gLV model is a system of coupled ordinary differential equations that captures individual species’ growth rate and pairwise interactions with each community member. We use an expansion of the generalized Lotka–Volterra model (gLV) that captures antibiotic perturbations [30] (Fig 4A). In this expanded model, the growth of each species is modified by an antibiotic term consisting of the concentration of antibiotic and the susceptibility of each species (Bi) (Methods, Fig 4A).
Fig 4. A modified generalized Lotka–Volterra model of community dynamics in response to antibiotics captures trends in antibiotic concentrations lower than the MIC in the presence of antibiotic sensitive biotic inhibitors.
(A) Top: Schematic of the antibiotic expansion of the gLV model. Bottom: Schematic of accuracy metric used in panel B. (B) Qualitative accuracy of multiple models for pairwise and multispecies communities. Models include (1) standard gLV model lacking an antibiotic term; (2) gLV model with randomly shuffled antibiotic susceptibility parameters (average of 100 permutations), and 100 shuffled values shown as points, shaded according to the absolute value of the difference between the shuffled C. difficile antibiotic susceptibility parameter and the original C. difficile antibiotic susceptibility parameter; (3) gLV model with antibiotic susceptibility terms inferred from monospecies with all interaction parameters set to zero (aij = 0, where i! = j); (4) full gLV model with antibiotic susceptibility terms inferred from monospecies data. (C) Simulated maximum subMIC fold changes for a focal species in a pairwise community for 48 h for a representative parameter set. Growth rates of the 2 species are equal (ui = uj = 0.25), intraspecies interactions are equal (aii = ajj = −0.8), and initial ODs are equal (xi(0) = xj(0) = 0.0022). The interspecies interaction coefficients aji are set to 0 and Bi is set to −2. (D) Simulated max subMIC fold change for a focal species in a pairwise community for 1,000 randomly sampled parameter sets. Parameters Bj and aij are set to constant values of 0 or −6 and 0 or −1.25, respectively. Other parameters were randomly sampled between upper and lower bounds. The lower and upper bounds on each parameter value are aji (−1.25, 1.25), growth rates (0, 1), intraspecies interactions (−1.25, 0), Bi (−6, 0). Gray horizontal line at y = 1 indicates no change in growth compared to the no antibiotic condition. The data and modeling scripts underlying panels BCD in this figure can be found in DOI: 10.5281/zenodo.7726490. MIC, minimum inhibitory concentration; subMIC, sub-minimum inhibitory concentration.
The monospecies antibiotic susceptibility parameters were inferred from measurements of individual species growth in the presence of a range of antibiotic concentrations (2-fold dilutions) (Methods, S1 Fig). We used growth rate and interspecies interaction parameters that were previously inferred based on absolute abundance measurements of 159 monospecies and communities (2- to 14-member) in the absence of antibiotics [4].
We evaluated whether this model could qualitatively predict the trends in growth enhancement of C. difficile in the subMIC range (S8 and S9 Figs). C. difficile’s response to each antibiotic concentration was classified into 3 categories (C. difficile subMIC fold-change > 1, C. difficile subMIC fold-change ≤ 1, or no growth of C. difficile, Fig 4A). The expanded gLV model correctly predicts C. difficile’s qualitative response to 71% of antibiotic conditions (Figs 4B and S10A). We designed a set of null models to evaluate the contribution of different terms of the expanded gLV model to model performance. The full model displays higher accuracy than a null model that lacked antibiotic susceptibility terms or has randomly shuffled antibiotic susceptibility parameters (47% and 53% accuracy, Fig 4C). These data highlight that the fitted monospecies susceptibilities play a major role in the model’s predictive performance. The full model also outperforms a null model that lacks interspecies interaction terms (65%, Fig 4C), indicating that the inferred interspecies interactions contribute to the growth enhancement of C. difficile in response to subMICs. Taken together, these findings indicate that biotic inhibition and monospecies antibiotic susceptibility are major variables determining the growth of C. difficile in response to subMICs in microbial communities.
We explored model simulations to determine if the enhancement of C. difficile growth at subMICs required a sensitive biotic inhibitor in a wide variety of communities, beyond those that were experimentally characterized. To this end, we simulated 1,000 pairwise communities with a wide range of growth rates, interspecies interactions, and antibiotic susceptibilities. When paired with a sensitive biotic inhibitor, growth is enhanced at subMICs, consistent with the trends observed in our experiments (Fig 4D). By contrast, growth enhancements are not observed in simulated communities that lack an antibiotic-sensitive biotic inhibitor of the focal species (Fig 4D). This trend is also present in larger communities (S10B Fig). The focal species’ growth enhancement at subMICs increases with biotic inhibition and antibiotic susceptibility of the inhibitor species (Fig 4E). Overall, these model simulations suggest that the antibiotic sensitive inhibitor trend we observe with C. difficile in human gut communities is generalizable to other species and communities with variable richness, interaction networks, and antibiotic susceptibilities.
D. piger substantially increases C. difficile’s minimum inhibitory concentration to metronidazole
Changes in antibiotic susceptibility of a pathogen due to significant microbial interactions could reduce the efficacy of antibiotic treatments. In the presence of D. piger, we observed a substantial increase in C. difficile MIC, in addition to a moderate growth enhancement of C. difficile at subMICs (Fig 2B). Consistent with the proposed antibiotic-sensitive biotic inhibition mechanism, D. piger was a weak biotic inhibitor of C. difficile and was more sensitive to metronidazole than C. difficile (Figs 2A and 3C). However, the substantial increase in C. difficile MIC in the presence versus absence of D. piger (≥24 μg/mL compared to 6 μg/mL) was not explained by the proposed antibiotic-sensitive biotic inhibition mechanism and the expanded gLV antibiotic model failed to predict this trend (S8A Fig). While competitive interactions (i.e., resource competition or biological warfare) contributed to the observed subMIC growth response, the mechanism of increased metronidazole MIC in the presence of D. piger was unknown.
We investigated the robustness of this trend across different environmental conditions and for different C. difficile isolates. C. difficile displayed an increased MIC in coculture with D. piger and in monoculture in D. piger’s spent media (≥8-fold increase in MIC) (Fig 5A). These data indicate that the increase in C. difficile metronidazole MIC was not dependent on cell-to-cell contact with D. piger or prior exposure of D. piger to metronidazole. C. difficile displayed a higher metronidazole MIC in D. piger spent media harvested at late exponential phase/early stationary phase than in spent media harvested at earlier time points in exponential phase (S11A and S11B Fig). Further, multiple C. difficile clinical isolates had an increased MIC in D. piger spent media (S11C Fig). Finally, C. difficile exhibited a higher metronidazole MIC in coculture with D. piger in a different chemically defined growth medium (S11D Fig). These data demonstrate that the protective effect of D. piger on C. difficile’s response to metronidazole was robust to variations in C. difficile’s strain background, media composition, and the growth phase of D. piger.
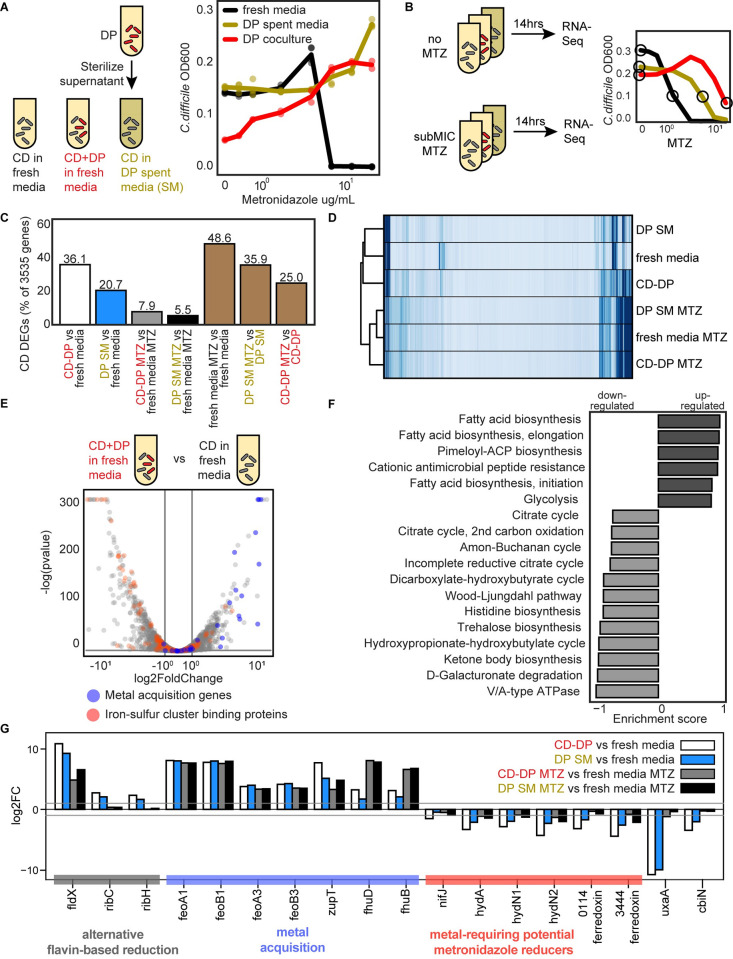
Fig 5. D. piger increases the metronidazole MIC of C. difficile and induces metal starvation genome-wide transcriptional response.
(A) Abundance of C. difficile at 41 h in response to different metronidazole concentrations. The OD600 value for C. difficile in the presence of D. piger is calculated OD600 (OD600 multiplied by relative abundance from 16S rRNA gene sequencing). The x-axis is semi-log scale. Data points represent biological replicates. Lines indicate average of n = 2 to n = 4 biological replicates. (B) Schematic of genome-wide transcriptional profiling experiment. The x-axis is semi-log scale of metronidazole (MTZ) concentration. Lines indicate average C. difficile OD600 at 14 h for n = 4 biological replicates. Circles indicate conditions sampled for RNA-Seq. (C) C. difficile DEGs between culture conditions. DEGs are defined as genes that displayed greater than 2-fold change and a p-value less than 0.05. (D) Clustered heatmap of RPKM for each gene (rows) and in each sample (columns) for C. difficile. Each column represents the average of n = 2 biological replicates. Hierarchical clustering was performed based on Euclidean distance using the single linkage method of the Python SciPy clustering package. (E) Volcano plot of log transformed transcriptional fold changes for C. difficile in the presence of D. piger. Gray vertical lines indicate 2-fold change and the gray horizontal line indicates the statistical significance threshold (p = 0.05). Blue indicates genes annotated to be involved in metal import. Red indicates genes predicted to contain iron-sulfur clusters by MetalPredator [31]. (F) Enriched gene sets in C. difficile grown in the presence of D. piger compared to C. difficile grown in fresh media. All gene sets with significant enrichment scores from GSEA are shown. Gene sets are defined using modules from the KEGG. (G) Bar plot of the log transformed fold changes of a set of genes across different conditions. Gray horizontal lines indicate 2-fold change. The data underlying panels AB in this figure can be found in DOI: 10.5281/zenodo.7626486 and the data underlying panels CDEFG in this figure can be found in DOI 10.5281/zenodo.7049035 and 10.5281/zenodo.7049027 and S3 and S4 Tables. DEG, differentially expressed gene; GSEA, Gene Set Enrichment Analysis; KEGG, Kyoto Encyclopedia of Genes and Genomes; MIC, minimum inhibitory concentration; RPKM, reads per kilobase million.
D. piger could alter C. difficile’s metronidazole MIC via a direct chemical interaction with metronidazole which reduces its efficacy (e.g., degradation or sequestration) or an indirect effect that modifies the activities of C. difficile’s intracellular networks which in turn yields an increase in metronidazole MIC. To test for a direct chemical interaction, we incubated metronidazole in either D. piger spent media or fresh media and characterized C. difficile’s growth response to each of these conditions in fresh media. C. difficile’s metronidazole MIC was equal in these conditions, indicating that compounds present in D. piger’s spent media do not directly interact with metronidazole to reduce its activity (S11E Fig).
C. difficile displays a genome-wide transcriptional signature of metal limitation in response to D. piger
We considered indirect effects of D. piger on C. difficile’s intracellular network activities which in turn could alter C. difficile’s response to metronidazole. We first considered resource competition as a possible indirect effect because resource competition is a common mechanism driving interspecies interactions in microbial communities. C. difficile’s abundance was not substantially lower in D. piger’s spent media than monoculture (Fig 5A), suggesting that resource competition was not a major driver of the increased MIC. In addition, supplementing concentrated fresh media into the D. piger spent media, which would alleviate any resource competition, did not alter C. difficile’s metronidazole MIC (S11F Fig). Therefore, these data suggest that D. piger did not affect the metronidazole MIC of C. difficile via resource competition.
We considered if D. piger affected the metronidazole MIC of C. difficile by altering patterns in gene expression. We performed genome-wide transcriptomic profiling of C. difficile in late exponential phase in the following environments: (1) monoculture in fresh media; (2) monoculture in D. piger spent media; and (3) coculture with D. piger. Each of these cultures was exposed to a subMIC of metronidazole that reduced growth by approximately 50% or no treatment was applied (Fig 5B). The antibiotic concentration was chosen to allow sufficient growth for transcriptional profiling.
C. difficile’s gene expression profile in D. piger conditions (i.e., D. piger spent media and coculture with D. piger) was significantly altered compared to C. difficile monoculture (Tables 1 and S3). Differentially expressed genes (DEGs) were defined as genes with >2-fold change and a p-value less than 0.05. In the absence of antibiotics, 36% and 21% of C. difficile’s 3,535 genes were differentially expressed in the D. piger coculture and spent media, respectively, compared to the C. difficile monoculture in fresh media (Fig 5C, white and blue bars). Of these genes, 72 in the D. piger coculture and 47 in the D. piger spent media exhibited >8-fold change, demonstrating that the presence of D. piger had a large impact on C. difficile’s gene expression profile. In the presence of metronidazole, a smaller percentage of genes were differentially expressed between the D. piger and fresh media conditions (8% and 6%, Fig 5C, gray and black bars). Large shifts in C. difficile gene expression occurred due to the addition of metronidazole, with 25% to 49% of genes differentially expressed (Fig 5C, brown bars). The 3 metronidazole conditions clustered closely together, indicating that the antibiotic induced similar changes in gene expression regardless of the media condition or ecological context (Fig 5D).
Table 1. C. difficile gene expression in D. piger conditions compared to fresh media for genes with >16-fold differential expression.
| D. piger coculture vs. fresh media | D. piger spent media vs. fresh media | |||||
|---|---|---|---|---|---|---|
| Locus_tag | Gene name | log2 FC | padj | log2 FC | padj | |
| CDR20291_1925 | fldX | flavodoxin | 10.86 | 0.0E+00 | 9.27 | 0.0E+00 |
| CDR20291_2391 | none | hypothetical protein | 9.54 | 0.0E+00 | 8.57 | 9.0E-197 |
| CDR20291_0517 | none | putative membrane protein | 8.73 | 0.0E+00 | 8.87 | 0.0E+00 |
| CDR20291_0516 | none | putative cation transporting ATPase | 8.10 | 0.0E+00 | 8.36 | 0.0E+00 |
| CDR20291_1327 | feoA1 | putative ferrous iron transport protein A | 8.09 | 9.0E-167 | 8.03 | 3.0E-140 |
| CDR20291_1328 | feoB1 | ferrous iron transport protein B | 7.79 | 0.0E+00 | 8.00 | 0.0E+00 |
| CDR20291_0946 | zupT | zinc transporter | 7.71 | 1.0E-51 | 5.14 | 3.0E-20 |
| CDR20291_1329 | none | putative exported protein | 7.60 | 1.0E-105 | 7.49 | 1.0E-81 |
| CDR20291_1326 | none | putative ferrous iron transport protein | 7.12 | 2.0E-227 | 6.96 | 2.0E-157 |
| CDR20291_2009 | none | putative Na(+)/H(+) antiporter | 5.46 | 5.7E-221 | 3.93 | 1.0E-87 |
| CDR20291_2825 | none | ABC transporter, ATP-binding protein | 5.30 | 1.6E-136 | 3.81 | 5.0E-51 |
| CDR20291_2826 | none | putative ABC transporter, permease protein | 5.14 | 3.4E-130 | 3.58 | 4.0E-45 |
| CDR20291_2827 | none | hypothetical protein | 4.91 | 7.4E-126 | 3.52 | 1.0E-44 |
| CDR20291_0455 | none | putative membrane protein | 4.84 | 2.0E-54 | 1.94 | 5.0E-06 |
| CDR20291_1545 | none | putative iron compound ABC transporter, permease protein | 4.66 | 9.0E-106 | 3.59 | 3.0E-46 |
| CDR20291_1546 | none | putative iron compound ABC transporter, permease protein | 4.52 | 5.0E-61 | 3.40 | 4.0E-28 |
| CDR20291_1547 | none | putative iron compound ABC transporter, ATP-binding protein | 4.46 | 4.0E-67 | 3.03 | 2.0E-23 |
| CDR20291_0687 | plfB | formate acetyltransferase | 4.38 | 5.0E-203 | 2.51 | 5.0E-46 |
| CDR20291_1021 | acpP | acyl carrier protein | 4.30 | 9.0E-101 | 2.00 | 9.0E-16 |
| CDR20291_1548 | none | putative iron compound ABC transporter, substrate-binding protein | 4.23 | 8.0E-117 | 2.77 | 3.0E-28 |
| CDR20291_2437 | none | putative sugar transporter, substrate-binding lipoprotein | 4.20 | 1.1E-165 | 2.16 | 3.0E-28 |
| CDR20291_3135 | feoB3 | putative ferrous iron transport protein B | 4.14 | 1.9E-189 | 4.26 | 2.0E-148 |
| CDR20291_2824 | none | ABC transporter, substrate-binding protein | 4.11 | 9.9E-129 | 2.83 | 4.0E-40 |
| CDR20291_1022 | fabF | 3-oxoacyl-[acyl-carrier-protein] synthase II | 4.00 | 1.0E-121 | 1.58 | 2.0E-16 |
| CDR20291_0363 | none | Radical SAM-superfamily protein | −4.00 | 2.0E-134 | −3.26 | 9.0E-71 |
| CDR20291_0368 | hadB | subunit of oxygen-sensitive 2-hydroxyisocaproyl-CoA dehydratase | −4.01 | 3.0E-183 | −1.72 | 5.0E-25 |
| CDR20291_2064 | gabT | 4-aminobutyrate aminotransferase | −4.04 | 3.0E-89 | −3.07 | 4.0E-41 |
| CDR20291_0364 | none | putative membrane protein | −4.09 | 6.0E-164 | −3.50 | 2.0E-78 |
| CDR20291_3178 | fdhD | formate dehydrogenase accessory protein | −4.09 | 8.0E-127 | −2.45 | 4.0E-37 |
| CDR20291_0523 | cotJC1 | putative spore-coat protein | −4.11 | 5.0E-107 | −1.18 | 2.0E-09 |
| CDR20291_0755 | rbr | rubrerythrin | −4.11 | 2.0E-155 | −2.40 | 2.0E-29 |
| CDR20291_2227 | abfH | NAD-dependent 4-hydroxybutyrate dehydrogenase | −4.12 | 3.9E-181 | −2.00 | 4.0E-30 |
| CDR20291_2509 | none | putative membrane protein | −4.14 | 1.8E-164 | −0.67 | 2.0E-04 |
| CDR20291_0731 | crt1 | 3-hydroxybutyryl-CoA dehydratase (crotonase) | −4.20 | 1.0E-08 | −2.81 | 1.0E-05 |
| CDR20291_2794 | ntpI | V-type sodium ATP synthase subunit I | −4.27 | 3.2E-174 | −1.79 | 8.0E-27 |
| CDR20291_0177 | none | putative oxidoreductase, NAD/FAD binding subunit | −4.29 | 1.0E-147 | −3.29 | 2.0E-66 |
| CDR20291_1478 | none | hypothetical protein | −4.30 | 8.0E-122 | −2.80 | 2.0E-39 |
| CDR20291_3176 | hydN2 | electron transport protein | −4.31 | 2.7E-160 | −2.32 | 4.0E-41 |
| CDR20291_2566 | ctfB | butyrate—acetoacetate CoA-transferase subunit B | −4.32 | 5.2E-11 | −2.31 | 2.0E-05 |
| CDR20291_0011 | none | putative translation elongation factor | −4.33 | 2.0E-135 | −1.72 | 1.0E-16 |
| CDR20291_1511 | none | hypothetical protein | −4.33 | 5.0E-128 | −1.88 | 7.0E-22 |
| CDR20291_0175 | none | putative oxidoreductase, acetyl-CoA synthase subunit | −4.39 | 2.0E-183 | −3.02 | 1.0E-59 |
| CDR20291_2416 | none | conserved hypothetical protein | −4.41 | 5.7E-102 | −3.53 | 5.0E-59 |
| CDR20291_2787 | ntpD | V-type sodium ATP synthase subunit D | −4.41 | 3.9E-158 | −2.18 | 1.0E-31 |
| CDR20291_2228 | abfT | 4-hydroxybutyrate CoA transferase | −4.41 | 5.6E-189 | −2.08 | 1.0E-32 |
| CDR20291_0176 | none | putative oxidoreductase, electron transfer subunit | −4.42 | 1.0E-181 | −3.37 | 6.0E-61 |
| CDR20291_3444 | none | ferredoxin | −4.45 | 7.0E-182 | −2.61 | 4.0E-25 |
| CDR20291_2132 | asrA | anaerobic sulfite reductase subunit A | −4.51 | 1.7E-10 | −2.81 | 3.0E-06 |
| CDR20291_1905 | none | putative transcriptional regulator | −4.51 | 4.1E-52 | −1.29 | 3.0E-07 |
| CDR20291_2788 | ntpB | V-type sodium ATP synthase subunit B | −4.56 | 5.8E-199 | −1.91 | 1.0E-26 |
| CDR20291_2800 | adhE | aldehyde-alcohol dehydrogenase | −4.69 | 9.4E-15 | −3.76 | 1.0E-09 |
| CDR20291_0521 | none | hypothetical protein | −4.71 | 6.0E-93 | −1.15 | 5.0E-07 |
| CDR20291_3177 | none | hypothetical protein | −4.71 | 5.1E-59 | −2.74 | 2.0E-25 |
| CDR20291_2492 | none | adp-ribosyltransferase binding component | −4.75 | 3.0E-208 | −1.78 | 2.0E-20 |
| CDR20291_2789 | ntpA | V-type sodium ATP synthase subunit A | −4.75 | 1.4E-213 | −2.15 | 3.0E-36 |
| CDR20291_2491 | none | cdta (adp-ribosyltransferase enzymatic component) | −4.79 | 9.4E-219 | −1.66 | 5.0E-20 |
| CDR20291_2792 | ntpE | V-type sodium ATP synthase subunit E | −4.80 | 6.0E-145 | −1.81 | 7.0E-23 |
| CDR20291_1008 | etfB3 | electron transfer flavoprotein beta-subunit | −4.81 | 2.0E-92 | −4.13 | 5.0E-60 |
| CDR20291_0522 | cotJB1 | putative spore-coat protein | −4.83 | 4.0E-107 | −1.38 | 3.0E-09 |
| CDR20291_0728 | none | putative hydroxymethylglutaryl-CoA lyase | −4.83 | 7.0E-14 | −3.85 | 2.0E-10 |
| CDR20291_2793 | ntpK | V-type sodium ATP synthase subunit K | −4.91 | 1.2E-184 | −1.78 | 4.0E-24 |
| CDR20291_2790 | ntpG | V-type sodium ATP synthase subunit G | −4.93 | 8.4E-144 | −1.92 | 2.0E-21 |
| CDR20291_2230 | abfD | gamma-aminobutyrate metabolism dehydratase/isomerase | −4.96 | 4.5E-250 | −2.15 | 4.0E-33 |
| CDR20291_0709 | none | conserved hypothetical protein | −4.99 | 2.0E-251 | −3.38 | 1.0E-65 |
| CDR20291_1007 | none | conserved hypothetical protein | −5.03 | 2.0E-118 | −4.47 | 1.0E-69 |
| CDR20291_2229 | none | conserved hypothetical protein | −5.09 | 9.9E-189 | −2.74 | 3.0E-27 |
| CDR20291_2791 | ntpC | V-type sodium ATP synthase subunit C | −5.10 | 2.4E-183 | −2.03 | 2.0E-27 |
| CDR20291_0706 | none | putative membrane protein | −5.20 | 9.0E-267 | −3.43 | 4.0E-68 |
| CDR20291_0191 | none | putative membrane protein | −5.26 | 3.0E-254 | −4.21 | 4.0E-127 |
| CDR20291_0708 | none | putative amidohydrolase | −5.32 | 2.0E-226 | −3.72 | 2.0E-63 |
| CDR20291_0707 | none | putative membrane protein | −5.64 | 1.0E-286 | −3.77 | 3.0E-87 |
| CDR20291_0760 | none | putative membrane protein | −5.72 | 3.0E-291 | −4.08 | 2.0E-113 |
| CDR20291_3092 | none | conserved hypothetical protein | −6.15 | 0.0E+00 | −4.44 | 4.0E-77 |
| CDR20291_1689 | none | hypothetical protein | −6.48 | 1.0E-39 | −4.14 | 6.0E-31 |
| CDR20291_2074 | hcp | hydroxylamine reductase | −6.92 | 0.0E+00 | −0.87 | 7.0E-06 |
| CDR20291_2417 | none | conserved hypothetical protein | −7.27 | 0.0E+00 | −7.54 | 0.0E+00 |
| CDR20291_1690 | none | conserved hypothetical protein | −7.84 | 2.0E-158 | −3.50 | 5.0E-68 |
| CDR20291_2075 | none | iron-sulfur binding protein | −8.29 | 0.0E+00 | −1.62 | 7.0E-14 |
| CDR20291_2419 | none | putative aminotransferase | −9.21 | 0.0E+00 | −8.46 | 0.0E+00 |
| CDR20291_2418 | none | putative membrane protein | −9.43 | 0.0E+00 | −8.94 | 0.0E+00 |
| CDR20291_2766 | kdgT | 2-keto-3-deoxygluconate permease | −10.13 | 0.0E+00 | −9.66 | 0.0E+00 |
| CDR20291_2768 | uxaA | putative altronate hydrolase | −10.81 | 0.0E+00 | −10.01 | 0.0E+00 |
| CDR20291_1692 | none | putative pyridine nucleotide-disulfide oxidoreductase | −11.08 | 0.0E+00 | −2.96 | 9.0E-67 |
| CDR20291_2767 | uxaA’ | altronate hydrolase (N-terminus) | −11.44 | 3.4E-41 | −15.26 | 3.0E-21 |
| CDR20291_1691 | none | putative nitrite and sulfite reductase subunit | −12.01 | 0.0E+00 | −3.97 | 9.0E-67 |
To provide deeper insights into specific genes that varied across conditions, we analyzed the genes with the largest expression changes in D. piger conditions. Many of the genes up-regulated in D. piger and D. piger antibiotic conditions compared to fresh media had annotated roles in metal acquisition (blue points in Figs 5E and S12). To acquire iron, C. difficile uses iron permeases to import free ferrous iron into the cell [32,33]. We observed >10-fold up-regulation in 2 ferrous transporters (feoAB1, feoAB3) in D. piger and D. piger metronidazole conditions (Table 1, Fig 5G). Alternative transporters were also up-regulated, such as uptake genes for ferrichrome siderophores (fhuB, fhuD) and catecholate siderophores (CDR20291_1545–1548, orthologs of yclNOPQ in CD630) as well as a zinc transporter gene (zupT). These genes (feo, fhu, ycl, and zupT) are regulated by the iron regulator fur [32] and have been shown to be up-regulated in response to iron starvation and zinc starvation [34–36], suggesting that metal limitation in D. piger conditions was responsible for the observed changes. Consistent with the metal limitation signature, ferrous transporters that were not up-regulated (feoAB2) have been documented as unresponsive to iron limitation [32,34].
In addition to genes directly related to metal transport, many of the DEGs in C. difficile were linked to iron metabolism. Flavodoxin (fldX) was the most up-regulated gene in the D. piger conditions (Table 1, Fig 5G). Flavodoxin can replace iron-requiring electron transfer proteins (such as ferredoxin) in iron-limiting conditions [37]. Flavodoxin uses a nonmetal cofactor, flavin, whose biosynthesis genes (ribACH) were up-regulated in D. piger conditions (Table 1). Previous studies have shown that flavodoxin was regulated by fur and de-repressed in iron-limiting and zinc-limiting conditions [32,34–36], and riboflavin biosynthesis genes were up-regulated in iron-limiting conditions [35]. Iron-containing proteins have been shown to be down-regulated in iron-limited conditions [35]. Using a bioinformatic analysis, we found that 9% of C. difficile’s down-regulated genes (62 of 678 genes) in the D. piger coculture condition were predicted to contain iron-sulfur clusters (red points in Fig 5E, S4 Table). In sum, genes for the metal-independent flavodoxin were up-regulated, whereas transcripts for iron-requiring proteins were down-regulated, consistent with a metal-limited environment for C. difficile created by D. piger.
To identify other biological pathways that were significantly altered in D. piger conditions, we performed a gene set enrichment analysis (GSEA) using Kyoto Encyclopedia of Genes and Genomes (KEGG) modules (Figs 5F and S12). Many of the identified pathways could be connected to metals. For example, many of the enzymes in the down-regulated Wood–Ljungdahl pathway have iron-sulfur clusters, namely carbon monoxide dehydrogenase (cooCS, CDR20291_0653, CDR20291_0655) and formate dehydrogenase H (fdhF) [38–40]. In addition, the [NiFe] hydrogenase (hydAN1N2) in the Wood–Ljungdahl pathway contains a nickel-iron cluster. These enzymes have been shown to be down-regulated under iron-limited conditions [35]. Pathways for cationic antimicrobial peptide resistance and fatty acid biosynthesis were up-regulated, which has been previously attributed to iron limitation [35]. Similarly, the down-regulation of pathways for V-type ATPases, D-galacturonate degradation, and trehalose biosynthesis has been connected to iron limitation (Fig 5F) [35]. D. piger also affected the expression of C. difficile toxins (tcdA, tcdB, and binary toxin genes CDR20291_2491 and CDR20291_2492) that were down-regulated between 4- and 27-fold in the D. piger conditions (S3 Table).
To provide further insights into the contribution of iron limitation on the patterns of gene expression in C. difficile, we evaluated the relationship between gene expression changes in C. difficile in the presence of D. piger and gene expression changes in C. difficile in iron-limited media previously characterized in a separate study [35]. For all DEGs, we compared the log2 fold changes between C. difficile in the D. piger coculture and C. difficile in monoculture in the absence of metronidazole with the log2 fold changes observed between C. difficile in iron-limited and iron-rich media in the previous study. These fold changes displayed an informative relationship and were qualitatively consistent for 89% of genes between the 2 studies (S13A Fig, Pearson r = 0.61, p = 8*10−54). Similarly, the fold changes of C. difficile cultured in D. piger spent media showed 87% qualitative agreement with the fold changes in the C. difficile iron-limitation study (S13B Fig). The informative relationship between these datasets suggests that the shifts in gene expression for the majority of C. difficile’s genes in the D. piger conditions can be explained by metal limitation.
Overall, these data suggest that D. piger created a metal-limited environment for C. difficile, as metal acquisition and alternative genes involved in flavin metabolism were up-regulated, whereas transcripts for metal-requiring proteins were down-regulated (Fig 5G). These trends were observed in both D. piger coculture and D. piger spent media and were also consistent with the trends in the D. piger conditions with metronidazole (Fig 5G).
Differentially expressed genes in C. difficile in the presence of D. piger are linked to metronidazole resistance
Since the majority of C. difficile’s DEGs could be explained by metal limitation, we investigated the connection between metal limitation and C. difficile metronidazole MIC. To identify potential connections, we compared the set of DEGs in the presence of D. piger to genes previously shown to play a role in metronidazole resistance in C. difficile.
In 2 studies of metronidazole-resistant C. difficile mutants, iron-related genes were implicated in metronidazole resistance. In a study of C. difficile strain ATCC 700057, multiple mutants with increased metronidazole resistance acquired a truncation in feoB1, which resulted in reduced intracellular iron and a shift to flavodoxin [41]. Similarly, in a metronidazole-resistant mutant of a NAP1 strain, iron-uptake genes were down-regulated and a shift to flavodoxin was observed [42]. In each of these studies, the proposed mechanism of metronidazole resistance was attributed to down-regulation of enzymes predicted to reduce metronidazole to its active form. In the D. piger conditions, enzymes hypothesized to reduce metronidazole were also down-regulated, namely ferredoxin genes (fdxA, CDR20291_0114, CDR20291_3444), pyruvate-ferredoxin/flavodoxin oxidoreductase (PFOR) (nifJ), and hydrogenases (hydA, hydN1, hydN2) (Table 1, Fig 5G) [20,21]. Down-regulation of these enzymes in D. piger conditions may reduce the rate of conversion of metronidazole into its active form, thus increasing the tolerance of C. difficile. The down-regulated enzymes are each predicted to contain iron clusters (S4 Table), and hydrogenases are known to contain nickel cofactors [39], suggesting that their down-regulation and the subsequent decrease in metronidazole susceptibility could be attributed to metal limitation.
This mechanism of resistance has been proposed in other species beyond C. difficile. For example, Bacteroides fragilis metronidazole-resistant mutants displayed reduced PFOR expression [21]. Therefore, if this mechanism was responsible for the increase in C. difficile’s metronidazole MIC by D. piger, Bacteroides species should also display a higher MIC. Indeed, B. thetaiotaomicron displayed a higher metronidazole MIC in D. piger spent media compared to fresh media (S14 Fig).
We also identified that cbiN, a putative cobalt transporter that was down-regulated in the D. piger conditions (Table 1, Fig 5G) and in iron-limited media [35] has been previously implicated in metronidazole resistance. A single SNP present in cbiN distinguished a metronidazole-resistant R010 isolate of C. difficile from a metronidazole sensitive R010 isolate isolated from the same patient [43]. In our data, cbiN was down-regulated by 10-fold in the D. piger coculture and 4-fold in D. piger spent media.
Another enzyme that was down-regulated in the D. piger conditions, altronate hydrolase (uxaA), has potential connections with metronidazole resistance. Notably, altronate hydrolase was substantially down-regulated in the coculture with D. piger, where the magnitude of this decrease in transcript abundance was the second largest in the transcriptome (>1,000-fold reduction, Table 1, Fig 5G). Altronate hydrolase catalyzes the dehydration of the six-carbon altronate as part of galacturonate degradation [44]. One of the 17 mutations that distinguished a metronidazole-resistant NAP1 C. difficile strain from the metronidazole sensitive C. difficile reference strain occurred in the altronate hydrolase gene. The mutation in altronate hydrolase was one of 3 frameshift mutations in the mutant and likely rendered altronate hydrolase nonfunctional [45]. Studies in E. coli have demonstrated that this enzyme requires iron or manganese for its catalytic activity [46], consistent with its strong down-regulation in metal-limited media [35].
The mutations in cbiN and uxaA in metronidazole-resistant C. difficile isolates suggests that the observed down-regulation of cbiN and uxaA in response to metal limitation may contribute to C. difficile’s increased metronidazole MIC. These genes are potential links between metal limitation and metronidazole susceptibility, in addition to the down-regulation of metal-containing oxidoreductases, hydrogenases, and ferredoxins predicted to convert metronidazole into its active form.
Hydrogen sulfide production by D. piger promotes metal sequestration
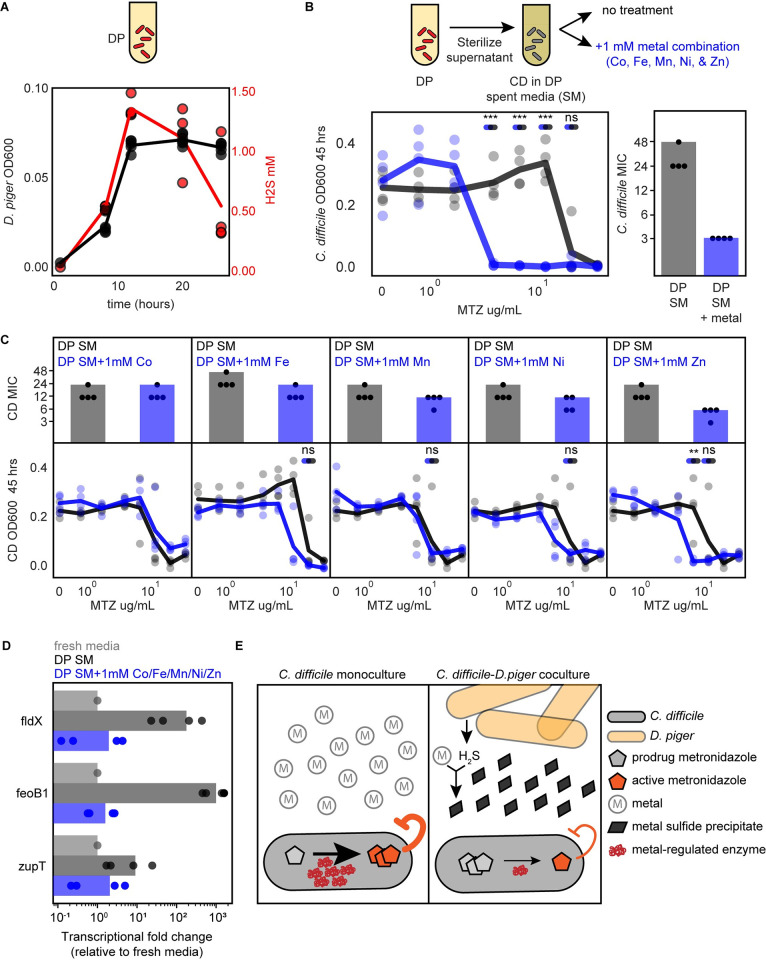
The global changes in C. difficile’s gene expression profile suggest that D. piger created a metal-limited environment. We hypothesized that these metal limitations could have been caused by hydrogen sulfide produced by D. piger [47]. In D. piger cultures, we observed a characteristic black precipitate (ferrous sulfide) that forms when iron combines with produced hydrogen sulfide [48]. Other divalent metals can precipitate with sulfide as well [49,50] and may be precipitating in addition to ferrous sulfide in the presence of D. piger.
To estimate how much metal is precipitated by D. piger produced sulfide, we quantified the amount of sulfide in a monoculture of D. piger over time (Methods). The amount of sulfide peaked in late exponential phase at 1.4 mM (Fig 6A). Hydrogen sulfide is volatile and escapes during growth. Therefore, the total amount of produced hydrogen sulfide was likely higher than the measured concentration. While C. difficile also produces a small amount of hydrogen sulfide, the amount of hydrogen sulfide in the C. difficile and D. piger coculture was similar to the amount in the D. piger monoculture (S15A Fig). The produced hydrogen sulfide in the D. piger spent media and D. piger coculture (>1.4 mM) was in excess of the total divalent metal concentration in the media (1.2 mM iron and micromolar concentrations of other metals, S15B Fig). Therefore, the produced hydrogen sulfide could precipitate all divalent metals in the media, creating a metal-limited environment for C. difficile. This would explain the metal-limited signature in C. difficile’s gene expression profile. Supporting this hypothesis, iron depletion due to the precipitation with excess sulfide has been previously shown to induce Fur-regulated genes in C. difficile [51].
Fig 6. Supplementation of D. piger spent media with metals eliminates the protective effect on C. difficile in the presence of metronidazole.
(A) Line plot of absolute abundance (black) and hydrogen sulfide production (red) of D. piger in monoculture. Data points represent biological replicates. Each biological replicate is the average of 2 technical replicates. Line indicates average of n = 4 biological replicates. (B) Line plot and bar plot of C. difficile metronidazole (MTZ) susceptibility in D. piger spent media (SM) with and without metal supplementation at 48 h. Metal supplementation condition contained 1 mM of Co, Mn, Ni, Zn, and Fe. The x-axis is semi-log scale (line plot). Data points represent biological replicates. Lines indicate average of n = 4 biological replicates. Asterisks indicate significant difference between conditions with and without metal supplementation (*P < 0.05, **P < 0.01, ***P < 0.001) according to an unpaired t test, “ns” indicates not significant. Statistical significance was performed at the lower of the 2 MICs and concentrations between the MICs of the 2 conditions. Bar plot displays MIC of data shown in line plot. Data points represent the MIC of n = 4 biological replicates. Bar represents the MIC determined based on average OD600 of n = 4 biological replicates. (C) Line plots and bar plots of C. difficile antibiotic susceptibility in D. piger spent media (SM) with and without supplementation of individual metals at 48 h. Each x-axis is semi-log scale. Data points represent biological replicates. Lines indicate the average of n = 4 biological replicates. Bar plot displays MIC of data shown in line plot. Data points represent MIC of n = 4 biological replicates. Bar represents the MIC determined based on an average OD600 of n = 4 biological replicates. Statistical significance as described in panel B. For metals with a change in MIC between the 2 conditions, statistical significance was tested at the lower of the 2 MICs and any concentrations between the MICs of the 2 conditions. (D) Bar plot of relative gene expression of 3 genes in C. difficile grown in fresh media, D. piger spent media (DP SM), and D. piger spent media with metal supplementation as detected by qRT-PCR. Fold change was calculated using the 2−ΔΔCt method (Methods). Data points represent biological replicates, with each point calculated as the average of 3 technical replicates. Bar indicates the average of n = 4 biological replicates. Statistical significance was determined based on ΔCt values (S15 Fig). (E) Schematic of proposed mechanism for D. piger alteration of C. difficile metronidazole susceptibility. (Left) In monoculture, metals are available in the environment, and metal containing enzymes in C. difficile are expressed and reduce the prodrug metronidazole to its active form. (Right) In coculture with D. piger, hydrogen sulfide produced by D. piger sequesters metals, which forms metal sulfide precipitates. In response to metal limitation, the expression of metal binding proteins in C. difficile that reduce the conversion of metronidazole from prodrug to its active form is reduced. This in turn reduces the rate of conversion of metronidazole from its inactive to active form and increases the tolerance of C. difficile. The data underlying panels ABCD in this figure can be found in DOI: 10.5281/zenodo.7626486. MIC, minimum inhibitory concentration.
Supplementation of D. piger spent media with combination of metals eliminates the protective effect of C. difficile from metronidazole
Based on the hypothesis that metal precipitation by hydrogen sulfide leads to an increase in C. difficile’s metronidazole MIC, we tested whether removing hydrogen sulfide from D. piger spent media eliminates this protective effect. The majority of hydrogen sulfide was eliminated from D. piger spent media by purging with nitrogen gas for 15 min (S16A Fig). We visually observed that nitrogen-purged spent media formed less ferrous sulfide precipitate than the untreated spent media (S16B Fig). Consistent with the proposed mechanism, the metronidazole MIC of C. difficile was reduced in the nitrogen-purged spent media compared to the untreated spent media (S16C and S16D Fig). This suggests that hydrogen sulfide contributed to the increase in C. difficile metronidazole MIC via metal precipitation.
Additionally, we hypothesized that supplementing D. piger spent media with metals that have precipitated would reduce the protective effect on C. difficile’s metronidazole MIC. We characterized C. difficile’s metronidazole MIC in media supplemented with cobalt, iron, manganese, nickel, and zinc since multiple divalent metals can form sulfide precipitates [49,50] and limitation of multiple divalent metals can lead to similar gene expression changes in C. difficile [35,36]. We introduced these metals in high concentrations (millimolar range) into the D. piger spent media to account for unreacted hydrogen sulfide that could precipitate the supplemented metals. Our results showed that C. difficile’s metronidazole MIC was substantially reduced in D. piger spent media supplemented with the five-metal combination compared to D. piger spent media without this addition (MIC decreased from 48 μg/mL to 3 μg/mL, Fig 6B). By contrast, C. difficile’s antibiotic MIC was not altered in fresh media supplemented with the five-metal combination, demonstrating that this effect was dependent on environmental modification by D. piger (S17A Fig). Supplementation of D. piger spent media with individual metals revealed that none of the individual metals decreased metronidazole MIC as substantially as the combination of metals (Fig 6C).
Consistent with these results, the expression of 3 genes that indicate metal limitation (fldX, feoB1, and zupT) was significantly reduced in C. difficile cultured in D. piger spent media supplemented with the metal combination compared to the spent media without metal supplementation (Figs 6D and S17B). Overall, this data suggests that metal limitation in D. piger spent media caused a global shift in C. difficile’s transcriptome and substantially reduced metronidazole susceptibility. Notably, both of these effects were eliminated by metal supplementation. Combining our data together, we propose a biological mechanism for the effect of D. piger on C. difficile’s metronidazole susceptibility (Fig 6E). In this proposed mechanism, hydrogen sulfide produced by D. piger sequesters divalent metals in the media. This in turn creates a metal limited environment for C. difficile, which leads to down-regulation of enzymes requiring metal cofactors for their activities. The down-regulated genes include enzymes that perform redox reactions that reduce metronidazole to its active form. Therefore, in coculture with D. piger, the activated concentration of metronidazole is reduced, yielding a substantially higher antibiotic MIC as a consequence of a reduction in DNA damage.
Discussion
A fundamental question is uncovering the role of biotic interactions in shaping antibiotic susceptibility in microbiomes. In particular, understanding how microbial interactions alter the antibiotic susceptibility of major human pathogens such as C. difficile could enable tailored antibiotic treatments informed by ecological context. This understanding could inform new microbiome interventions that selectively eradicate human pathogens while minimizing disruption of healthy gut microbiota and minimize the acquisition of antibiotic resistance. We investigated the contribution of interspecies interactions to the antibiotic susceptibility of a major human gut pathogen C. difficile. We observed 2 types of alterations in C. difficile antibiotic susceptibility: changes in C. difficile MIC and changes in C. difficile abundance at subinhibitory concentrations. Substantial changes in C. difficile MIC were rare in our conditions, only occurring in a small fraction of communities (S18A Fig). By contrast, enhancements in the abundance of C. difficile at subMICs were more frequently observed, occurring in 52% of characterized communities (S18B Fig). Our work demonstrates that pathogen growth can be altered by interspecies interactions across a wide range of antibiotic concentrations, which should be considered in the design of antibiotic treatments.
We observed a ≥4-fold increase in C. difficile’s MIC compared to monospecies in 4% of pairwise communities (Fig 2) and 16% of multispecies communities in response to single antibiotics (S1 and S2 Tables). This is qualitatively consistent with the small fraction of interspecies interactions shown to modify susceptibility in other microbial communities [12–15]. While we focused on antibiotics used to treat C. difficile, future work could apply our approach to study the impact of gut microbes on the response of C. difficile to antibiotics that are risk factors for initiating C. difficile infections such as clindamycin, cephalosporins, and fluoroquinolones [52].
The observed increase in C. difficile metronidazole MIC in the presence of specific gut may contribute to the ineffectiveness of metronidazole in treating C. difficile infections in the human colon. Due to the low achieved metronidazole concentration in the human colon [53], even modest increases in C. difficile metronidazole MIC could allow the pathogen to survive. Based on our results, future testing of C. difficile antibiotic susceptibility could include conditions of C. difficile cultured with physiologically relevant microbial communities. For example, C. difficile could be cultured with a panel of resuspended fecal samples from multiple donors with disparate human gut microbiome compositions. Antibiotic treatments could be scored by balancing minimization of the disruption of healthy gut microbiota with minimization of the variability of C. difficile’s MIC across donor samples.
We propose a mechanism wherein D. piger depletes bioavailable divalent metals in the environment, resulting in transcriptional down-regulation of enzymes in C. difficile requiring metal cofactors. These enzymes are proposed to reduce metronidazole to its active form. This implies that a lower expression level of these enzymes protects C. difficile from the action of the antibiotic (Fig 6E). Iron limitation has been shown to increase microbial resistance to antibiotics with proton motive force (PMF)-dependent uptake due to the decrease in iron-sulfur containing complexes crucial for generating the PMF [54]. Because metronidazole passively diffuses into the cell independent of PMF [55], iron limitation likely impacts not the initial uptake rate of metronidazole but instead the conversion rate from prodrug to activated inhibitor. Our results are supported by previous studies that have identified a link between diminished intracellular iron levels and C. difficile metronidazole resistance [41,42]. Interestingly, heme limitation has previously been shown to sensitize C. difficile to metronidazole [56]. This is in contrast to the metal limitation induced metronidazole protection we observed here, suggesting a complex relationship between heme and non-heme iron and metronidazole susceptibility.
In our metal supplementation experiments, the cocktail of 5 metals caused a substantial decrease in C. difficile MIC, whereas the addition of individual metals, including iron alone, did not have this effect. We hypothesize that multiple metal cofactors are required for the enzymatic activities that contribute to metronidazole reduction. While some enzymes proposed to contribute to metronidazole reduction, such as PFOR, only require iron cofactors, others such as hydrogenase require both iron and nickel cofactors, supporting this hypothesis [20,21,39,57]. An alternative hypothesis is that iron is the major cofactor needed by enzymes that reduce metronidazole, but both iron and non-iron divalent metals are involved in the transcriptional regulation of these enzymes due to cross-regulation of iron and other divalent metals. A cross-regulation of the zinc and iron starvation responses has been demonstrated in Staphylococcus aureus [58].
Our proposed mechanism of metronidazole protection has implications beyond D. piger and C. difficile. There are multiple commensal species in addition to D. piger that can produce hydrogen sulfide [59]. These species may increase C. difficile metronidazole MIC through a similar mechanism. Beyond C. difficile, metronidazole is a widely used antibiotic to treat anaerobic bacterial infections [19], suggesting hydrogen sulfide producing bacteria could protect other pathogens from the action of metronidazole via divalent metal sequestration (S14 Fig).
Estimates of sulfide concentration in the colon (10−3 M range [60,61]) are higher than estimates of bioavailable iron (approximately 10−3 M based on fecal concentrations, with 30% estimated to be present in a bioavailable form [62,63]). The excess of sulfide suggests physiological concentrations of iron may be precipitated by sulfide in the colon. However, during C. difficile infection, iron concentrations vary significantly as damage of epithelial cells can release heme into the environment [64], leading to an increase in available iron. However, metal sequestration by the host immune system [65] could simultaneously reduce available iron. Thus, the effects of sulfide on bioavailable metal pools in mammalian gut infections are unknown. Future studies could investigate the effect of hydrogen sulfide on C. difficile metronidazole susceptibility in an infection model in the murine gut. If the mechanism persists in in vivo, reduction of dietary protein could be considered during metronidazole treatment, as lower dietary protein has been shown to reduce hydrogen sulfide production by gut microbiota [60,66].
We observed alterations in C. difficile’s subMIC response in approximately half of the characterized communities (Fig 3B and 3E). Our CFU experiments demonstrated that the growth enhancement in a subset of conditions could be explained by changes in C. difficile abundance (S2D and S4 Figs). This does not exclude the possibility that some fraction of the observed C. difficile growth enhancements based on the OD600 method could be impacted by changes in cell morphology. Nevertheless, the relationship between the degree of biotic inhibition by susceptible competitors and focal strain growth enhancements at subinhibitory antibiotic concentrations is consistent in our computational model. This model does not capture changes in cell morphology, yet is able to recapitulate the overall trends in C. difficile growth enhancements (Fig 4).
Our observed growth enhancement of C. difficile at subMICs is an example of the ecological principle of competitive release, where resistant species expand after removal of antibiotic-sensitive competitors [28]. Competitive release has been previously observed in a 10-member model community of a lung infection due to cystic fibrosis [29] and a 2-member and 4-member brewery community [14]. Here, we systematically studied competitive release in both pairwise interactions and multispecies communities and its impact on the growth of a major human gut pathogen C. difficile. We demonstrated that the magnitude of the focal strain’s growth enhancement depends on the degree of biotic inhibition (Fig 3D and 3F). In addition, we showed that resistant inhibitors can suppress the growth enhancement caused by antibiotic sensitive biotic inhibitors (Figs 3D, 3F and S7A and S7B). Additionally, our computational model identified general ecological principles driving subMIC growth enhancements, which can be applied to other antibiotic treated communities.
Our findings suggest that growth enhancement of pathogens could occur in high richness communities such as the human gut microbiome in the presence of subinhibitory concentrations of antibiotics in the absence of resistant inhibitors. Metronidazole and vancomycin concentrations in the colon are estimated based on fecal concentrations. Therefore, it is unknown whether subinhibitory concentrations of these antibiotics occur in the colon. Based on fecal concentrations, it has been suggested that C. difficile may encounter subinhibitory concentrations during metronidazole treatment [67]. To avoid growth enhancement of C. difficile or other pathogens at subinhibitory concentrations, antibiotic treatments could be combined with bacterial therapeutics consisting of cocktails of resistant biotic inhibitors to suppress pathogen growth enhancement. This benefit of resistant commensal species has been suggested previously [68]. Our data demonstrates that for vancomycin treatment, Bacteroides thetaiotaomicron and Prevotella copri are good candidates for such a cocktail as they are resistant biotic inhibitors that inhibit C. difficile in communities across all antibiotic concentrations (S5A Fig). Use of narrow spectrum antibiotics or selective bacteriophages [69] that specifically inhibit a pathogen could also reduce the risk of pathogen growth enhancement at subMICs by minimizing disruption of the biotic competitors.
Overall, we found 2 classes of interactions that yield enhanced C. difficile growth in the presence of antibiotics. These results suggest that the discovery of interactions that sensitize C. difficile to antibiotics may be challenging. Therefore, future ecologically informed therapeutic strategies could exploit the strong biotic competition of C. difficile by diverse human gut species [4] that are resistant to the antibiotics as opposed to interactions that sensitize C. difficile to antibiotics.
Methods
Strain and culturing information
Information on strains used in this study is found in S5 Table. Single use glycerol stocks for each strain were created as described previously [70]. All cultures were grown in anaerobic Basal Broth (ABB, Oxoid) in an anaerobic chamber with an atmosphere of 2.5 ± 0.5% H2, 15 ± 1% CO2 and balance N2 (Coy Lab products). Starter cultures were prepared by inoculating 100 μL of single use glycerol into 5 mL of ABB. D. piger starter cultures were supplemented with 28 mM sodium lactate (Sigma-Aldrich) and 2.7 mM magnesium sulfate (Sigma-Aldrich). E. rectale starter cultures were supplemented with 33 mM sodium acetate (Sigma-Aldrich). To ensure organisms were at similar growth stages at experimental set up, starter cultures were inoculated either 16 or 41 h prior, depending on the organism’s growth rate (see S5 Table). Cultures were incubated at 37°C with no shaking.
Antibiotic titrations
Antibiotic susceptibility was determined following the Clinical and Laboratory Standards Institute protocol for antibiotic susceptibility of monospecies anaerobic organisms [26], with modifications to apply the method to multispecies communities. The protocol was modified to use broth microdilution method for all conditions, because some communities contained Bacteroides species for which broth microdilution method is recommended. The broth microdilution protocol was modified to use ABB media in order to support growth of all members of the community. Lastly, the protocol was modified to determine species absolute abundance as the product of optical density and relative abundance from 16S rRNA sequencing because our conditions contained multiple species.
Metronidazole (Sigma-Aldrich M1547) and vancomycin (Sigma-Aldrich V1130) stocks of 1 mg/mL were made in water, filter sterilized with 0.2 μM filters, and stored at −20°C as single use aliquots. Stocks were diluted in 2-fold dilution series at 10× the desired concentration. Cultures were inoculated into ABB in 96 deep well plates. Monospecies were inoculated with a starting OD600 of 0.0022. Pairs were inoculated with a starting OD600 of 0.00022 for C. difficile and 0.00198 for other species. Multispecies communities were inoculated with a total OD600 of 0.0066 with evenness of 1. Immediately after inoculation, the 10× antibiotic dilution series was diluted 1:10 into the communities. The plates were covered with gas-permeable seals (Breathe-Easy) and incubated at 37°C with no shaking.
After 48 h, cultures were mixed by pipetting and 200 μL aliquots were removed for sequencing and measuring OD600. Sequencing aliquots were spun down at 1,739 g for 15 min after which the supernatant was removed and the pellets were stored at −80°C. OD600 was measured for 2 dilutions of each sample and the dilution in the linear range of the instrument (Tecan Infinite Pro 200) was selected, and 16S rRNA sequencing was performed to determine relative abundances as described below. Species absolute abundance was calculated by multiplying the relative abundance of each species by the community OD600. The MIC was defined as the lowest concentration for which all concentrations greater to and equal restrict organism growth to significantly less (p < 0.05) than 0.05 OD600 or 5,000 CFU/mL as determined by a one-sample t test.
Determination of community composition
DNA extraction, library preparation, and sequencing were performed as described previously [4]. Briefly, cell pellets were genome extracted using the Qiagen DNEasy protocol with gram-positive lysozyme pretreatment modified for 96-well plates. Genomic DNA was normalized to 2 ng/μL in water and the 16S v3-v4 region was amplified using dual-indexed primers arrayed in 96-well plates. Samples were cleaned with DNA Clean and Concentrator kit (Zymo) and sequenced on an Illumina MiSeq.
Sequencing data was analyzed as described previously [4]. Briefly, reads were demultiplexed with Basespace FastQ Generation, paired ends were merged with PEAR v0.90 [71], and mapped to a custom database of our species using the mothur v1.40.5 command classify.seqs with the Wang method with bootstrap cutoff value of 60% [72]. Relative abundance of an organism was calculated by dividing the number of reads mapped to that organism by the total number of reads for that sample. Absolute abundance was calculated by multiplying the relative abundance by the OD600 of that sample. Samples were removed from further analysis if >1% of the reads were mapped to species not expected to be in the sample (indicating contamination).
C. difficile colony-forming unit counting
C. difficile selective plates were created by autoclaving C. difficile agar (Oxoid CM0601) and adding horse blood (Lampire 7233401, 70 mL/1L media), norfloxacin (Santa Cruz 215586, 120 μg/mL), moxalactam (Santa Cruz 250419, 320 μg/mL), and erythromycin (Santa Cruz 204742, 100 μg/mL) after media cooled to 55°C. After 48 h of incubation, cultures were mixed and diluted into PBS. Four dilutions of each sample were spotted on C. difficile selective agar plates, with 2 technical replicates per sample. Plates were incubated at 37°C for 48 h at which point colonies were counted in the dilution spot containing between 5 and 50 colonies. The CFU/mL for each sample was calculated as the average of the 2 technical replicates times the dilution factor. The lower limit of detection for the assay was 5,000 CFU/mL.
Generalized Lotka–Volterra model with antibiotic perturbation
The antibiotic perturbation extension of the generalized Lotka–Volterra model [30] is a set of N-coupled first-order ordinary differential equations (Eq 1):
where N is the number of species, Xi is the abundance of species i, ri is the basal growth rate of species i, αij is the growth modification of species i by species j, and Xj is the abundance of species j. The parameter αij is constrained to be negative when i = j, representing intraspecies competition. In the antibiotic term, Bi is the sensitivity of species i to antibiotic, and a is the concentration of the antibiotic. We modify the model to make the antibiotic concentration modified constant over time.
Values for growth rates parameters ri and interaction parameters aij come from our previous work [4]. Values for antibiotic sensitivity parameters Bi were inferred in this study from time-series measurements of monospecies antibiotic titrations using a custom python script. The equation used to infer Bi from monospecies data was Eq 1, with αij = 0 for all j! = i. The minimize function of the Scipy optimize package was used to determine the optimal Bi for each species xi, that resulted in the lowest cost between the antibiotic gLV model and the time-series measurements across all the antibiotic concentrations in the titration. The antibiotic concentrations were scaled so that the concentrations range between 0 and 1. The cost in the optimization was computed by simulating the species abundance in each antibiotic condition with an ODE solver and summing the mean-squared error between the abundance of the species in the simulation and the abundance of the species in the data at each time point in each antibiotic concentration.
Development of null computational models
Null model 1 (“gLV”) is the generalized Lotka–Volterra model. This is the same as the generalized Lotka–Volterra model with antibiotic perturbation (Eq 1) without the antibiotic term. This can also be thought of as the generalized Lotka–Volterra with antibiotic perturbation with all species antibiotic susceptibility set to zero (not susceptible). Null model 1 is Eq 1 with Bi = 0 for all species.
Null model 2 (“gLV + shuffled antib. suscept.”) is the generalized Lotka–Volterra model with antibiotic perturbation (Eq 1) with a shuffled set of antibiotic susceptibility parameters. For metronidazole conditions, the 14 metronidazole susceptibility parameters were shuffled and for the vancomycin conditions, the 14 vancomycin susceptibility parameters were shuffled (the shuffled Bi for species i is equal to the unshuffled B of any one of the 14 species). The accuracy of Null model 2 was averaged across 1,000 sets of shuffled parameters. The distance between the shuffled B for C. difficile and the unshuffled B for C. difficile was calculated as the absolute abundance of the difference between shuffled BCD and unshuffled BCD.
Null model 3 (“gLV + monosp. Antib. Susceptibility, aijs = 0”) is the generalized Lotka–Volterra model with antibiotic perturbation (Eq 1) with no interspecies interactions. Null model 3 is Eq 1 with αij = 0 for all j! = i.
Spent media preparation
D. piger starter culture was incubated for 48 h. The culture was then diluted into ABB to an OD600 of 0.0022. The cultures were incubated at 37°C with no shaking. Unless otherwise indicated, cultures were incubated for 12 h. After incubation, cultures were spun down at 1,739 g for 15 min. Supernatant was removed and filter sterilized with Steriflip 0.2 μM filters.
Metronidazole incubation experiment
D. piger spent media was prepared following the procedure described above. An aliquot of metronidazole was incubated in D. piger spent media or rich media (ABB) at 37°C. After 12 h of incubation, the incubated metronidazole was diluted into a C. difficile culture in fresh media (ABB) following the antibiotic titration method described above.
Iron-sulfur cluster prediction
The protein FASTA sequence from the C. difficile R20291 reference genome (GenBank assembly accession GCF_000027105.1) was run on the Metal Predator web-server [31].
Metal quantification and supplementation
The amount of metal in fresh media (Anaerobic Basal Broth) was analyzed via Inductively Coupled Plasma Mass Spectroscopy by the Wisconsin State Lab of Hygiene.
In metal supplementation experiments, supernatants were supplemented with iron (II) sulfate (Alfa Aesar), manganese (II) sulfate monohydrate (Sigma-Aldrich), nickel sulfate (II) hexahydrate (Sigma-Aldrich), zinc (II) sulfate heptahydrate (Alfa Aesar), and/or cobalt (II) chloride hexahydrate (Sigma-Aldrich). Metal compounds were prepared as 100× stocks in water and filter sterilized with 0.2 μM filters.
Hydrogen sulfide quantification
Hydrogen sulfide was quantified using the Cline Assay. Immediately before performing the assay, sodium sulfide (Alfa Aesar) was added to nitrogen purged water into a sealed vial. The stock was then diluted into 1% zinc acetate to 1 mM and further diluted into 1% zinc acetate to desired concentrations for a standard curve. Samples were removed from the anaerobic chamber and immediately diluted 5-fold into 1% zinc acetate. Cline reagent was prepared in advance (1.6 g N,N-dimethyl-p-phenylenediamine sulfate (Acros Organics), 2.4 g iron (III) chloride (Spectrum Chemical), 50 mL concentrated hydrochloric acid, 50 mL water, stored in the dark) of which 3 μL was added to the standards and samples. Samples were mixed by pipetting and incubated for 20 min in the dark before measuring absorbance at 670 nm. If the samples contained cells, after incubation the samples were spun down at 1,739 g for 10 min and the supernatant was transferred to a new plate to measure OD670.
Whole-genome transcriptomics
D. piger spent media was prepared following the procedure described above. C. difficile and D. piger starter cultures were incubated for 48 h. C. difficile monoculture and CD-DP coculture conditions were inoculated from starter cultures into 96 deep well plates. For monoculture conditions, C. difficile was inoculated to an OD600 of 0.0022. For CD-DP coculture, C. difficile was inoculated to an OD600 of 0.0022 and D. piger was inoculated to an OD600 of 0.0198. Immediately after inoculation, antibiotics were added as a 1:10 dilution of 10× antibiotic stocks. The plates were covered with gas-permeable seals (BreathEasy) and were incubated at 37°C with no shaking. After 14 h, 800 μL of RNAprotect (Qiagen) was added to 400 μL of culture after which the cultures were mixed by pipetting and then incubated for 5 min at room temperature. Cultures were then centrifuged at room temperature for 10 min at 1,739 g and supernatant was carefully removed. Pellets were stored at −80°C.
RNA was extracted using acidic phenol bead-beating method. Pellets were resuspended in 500 μL 2× Buffer B (200 mM sodium chloride, 20 mM ethylenediaminetetraacetic acid) and transferred to 2 mL microcentrifuge tubes containing 500 μL Phenol:Chloroform:IAA (125:24:1, pH 4.5) and 210 μL 20% sodium dodecyl sulfate and were bead-beated with acid washed beads (Sigma G1277) for 3 min. All solutions were RNAse-free. Samples were centrifuged at 4°C for 5 min at 8,000 rpm, and 600 μL of the upper aqueous phase was added to 60 μL 3 M sodium acetate and 660 μL cold isopropanol and chilled on ice for 5 min before freezing for 5 min at −80°C. Samples were centrifuged at 4°C for 15 min at 18,200 g, the supernatant was decanted, and the pellet was washed with cold 100% ethanol. The pellets were dried in a biosafety cabinet for 10 min and then resuspended in 100 μL RNAse-free water. Samples were purified using RNeasy Mini Kit (Qiagen) and genomic DNA was removed using RNAse-Free DNase Set (Qiagen). Two replicates of each condition were sent to GENEWIZ (New Jersey, United States of America) for sequencing. GENEWIZ depleted rRNA with Ribozero rRNA Removal Kit (Illumina) before cDNA library preparation using NEBNext Ultra RNA Library Prep (NEB). GENEWIZ sequenced the libraries on Illumina HiSeq. Data was de-multiplexed using Illumina’s bcl2fastq 2.17 software, where one mismatch was allowed for index sequence identification.
The data was quality checked using FastQC. The BBDuk, BBSplit, and BBMap tools from BBTools suite were used to trim adapters, deplete rRNA, and map the remaining mRNA reads to the reference genomes (C. difficile: GenBank assembly accession GCF_000027105.1. D. piger: Genbank assembly accession GCA_000156375.1). FeatureCounts was used to map reads to features on the genome. Reads per kilobase million (RPKM) values were computed using a custom python script. The DESeq2 Bioconductor library v4.0.3 was used in R v4.0.4 to quantify differential gene expression using a negative binomial generalized linear models with apeglm shrinkage estimator. When calculating RPKM of C. difficile genes in the C. difficile-D. piger coculture, the “reads mapped” in the denominator was the number of reads mapped to the C. difficile genome. Similarly, when quantifying differential gene expression for C. difficile genes in the C. difficile-D. piger coculture, only reads mapped to the C. difficile genome were provided to DeSeq2.
Real-time quantitative reverse transcription PCR (qRT-PCR)
C. difficile cultures were prepared the same as the transcriptomic samples and were incubated for the same duration (14 h). RNA was extracted using the same method as the transcriptomic samples and was purified using RNeasy Mini Kit (Qiagen). Genomic DNA was removed using RNAse-Free DNase Set (Qiagen). We performed cDNA synthesis with 1 ug of total RNA using the iScript Select cDNA Synthesis Kit (Bio-rad). We performed qRT-PCR (Bio-rad CFX Connect) with SsoAdvanced Universal SYBR Green SuperMix (Bio-rad). We used previously published primers for the fldX, feoB1, zupT, and rrs genes [32,73]. We computed the fold changes using the 2−ΔΔCt method [74], where:
where “targ” refers to target gene fldX, feoB1, or zupT and rrs is the reference gene [73]. Sample refers to media type. Fresh media was used as control condition for Ctcontrol. Each Ct value was the average of 3 technical replicates.
Gene set enrichment analysis
GSEA was performed using the GSEA method of the ClusterProfiler R package (v4.2.2) [75]. KEGG modules for C. difficile R20291 (KEGG T number T00998) were used as gene sets and were supplied as a user defined annotation with the TERM2GENE field. The analysis was run with the log2FCs calculated by DeSeq2. The p-value cutoff used was 0.05 and the minimum gene set size used was 3.
Supporting information
(A, B) Line plots of monospecies OD600 across antibiotic concentrations for metronidazole (MTZ) and vancomycin (VAN) at 48 h. Each x-axis is semi-log scale. Colored data points indicate biological replicates. Colored lines indicate the average of n = 2 to n = 4 biological replicates. Bold number indicates MIC of data. Black lines indicate model fit. (C) Table of scaled antibiotic susceptibility parameter B for each species. The data underlying panels AB in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A) Schematic of 2 methods for determining absolute abundance: calculated absorbance at 600 nm (OD600) or CFU counting. (B, C) Double axis line plots of absolute abundance at 48 h of C. difficile in the presence of (B) metronidazole (MTZ) and (C) vancomycin (VAN). Left axis (black) is calculated OD600 (OD600 multiplied by relative abundance from 16S sequencing) and right axis (red) is CFU per mL. The x-axis is semi-log scale. Black points indicate biological replicates. Red points indicate biological replicates, each the average of 2 technical replicates. Lines represent the mean of n = 4 biological replicates. Bold number indicates the MIC based on calculated OD600 (black) or CFU (red). “NG” indicates no growth above the MIC threshold for any conditions, and no MIC was able to be determined. Blue outlines indicate conditions for which the MIC was consistent between the 2 methods. Data collected on a separate day from Fig 2. (D) Heatmaps of C. difficile subMIC fold change in data from panels BC. Black indicates significant fold change (greater than 1) in at least 1 concentration, white indicates no significant fold change at any concentration. Significance was determined using unpaired t test between the abundance of C. difficile at a given subMIC and the abundance of C. difficile in the absence of antibiotic, P < 0.05. C. difficile abundance determined by CFU counting (left heatmap) or calculated OD600 method (right heatmap). The data underlying panels BCD in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A, B) Line plots of subMIC fold change for pairwise communities in metronidazole (MTZ) or vancomycin (VAN) at 48 h. SubMIC fold change is calculated as the species absolute abundance at the subMIC divided by the species absolute abundance in the absence of the antibiotic. Black horizontal line at y = 1 indicates no change in growth compared to the no antibiotic condition. Each x-axis is semi-log scale. Data points indicate biological replicates. Line represents mean of n = 3 to n = 8 biological replicates in panel A and n = 1 to n = 8 biological replicates in panel B. (C) Line plots of C. difficile monospecies over time. (Top) C. difficile OD600 at 0 μg/mL and 1.5 μg/mL metronidazole. (Bottom) C. difficile subMIC fold change μg/mL metronidazole. Gray horizontal line at y = 1 indicates no change in growth compared to no antibiotic condition. SubMIC fold change calculated as in panels AB. Points represent biological replicates. Line represents average of n = 4 biological replicates. Statistical significance analyzed for all time points where C. difficile OD600 was greater in the presence of antibiotic than the absence of antibiotic (“ns” P > 0.05, according to an unpaired t test). (D) Microscopy images of select C. difficile conditions from panel C: no antibiotic at 24 and 48 h and 1.5 μg/mL metronidazole at 48 h. Exponential phase C. difficile cells in presence of subMIC of metronidazole do not show morphological difference from exponential phase C. difficile cells grown in absence of antibiotic. (E) Line plots of C. difficile in pair with Bacteroides thetaiotaomicron over time. (Top) C. difficile OD600 in the presence of 0 μg/mL or 1.5 μg/mL metronidazole. (Bottom) C. difficile subMIC fold change in the presence of 1.5 μg/mL metronidazole. Gray horizontal line at y = 1 indicates no change in growth compared to no antibiotic condition. SubMIC fold change calculated as in panels AB. Points represent biological replicates. Line represents average of n = 4 biological replicates. Statistical significance analyzed for all time points where C. difficile OD600 was greater in the presence of antibiotic than the absence of antibiotic (“ns” P > 0.05, according to an unpaired t test). (F) Line plots of C. difficile in pair with Clostridium hiranonis over time. (Top) C. difficile OD600 in the presence of 0 μg/mL or 0.1875 μg/mL vancomycin. (Bottom) C. difficile subMIC fold change in the presence of 0.1875 μg/mL vancomycin. Gray horizontal line at y = 1 indicates no change in growth compared to the no antibiotic condition. SubMIC fold change calculated as in panels AB. Points represent biological replicates. Line represents average of n = 4 biological replicates. Statistical significance was analyzed for all time points where C. difficile OD600 was greater in the presence of antibiotic than the absence of antibiotic (*P < 0.05, **P < 0.01, ***P < 0.001, ns P > 0.05, according to an unpaired t test). (G) Box plot of maximum subMIC fold change for C. difficile in pairwise communities at 48 h. The maximum subMIC fold change is the maximum of the average subMIC fold change across all subMICs. The average subMIC fold change was calculated for each subMIC concentration by taking the average C. difficile absolute abundance at the subMIC divided by the average C. difficile absolute abundance in the no antibiotic condition (n = 1 to n = 4 biological replicates). C. difficile did not grow at any concentrations of vancomycin in CD-BO, CD-BT, CD-BU, and CD-PC pairs so these communities were not included in the analysis. Each data point represents 1 pairwise community. Gray horizontal line at y = 1 indicates no change in growth compared to the no antibiotic condition. Species are categorized as sensitive inhibitors if (1) the absolute abundance of C. difficile in a given pairwise community was significantly lower than the monoculture C. difficile abundance, in the absence of antibiotics, as determined by an unpaired t test; and (2) if the monospecies MIC was less than the monospecies MIC of C. difficile. Asterisks indicate significant difference (*P < 0.05, **P < 0.01, ***P < 0.001) according to a one-sided Mann–Whitney U test. The data underlying panels ABCEFG in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
Line plot of C. difficile abundance in presence of metronidazole in (black) monoculture or in (brown) coculture with B. vulgatus. Data points indicate biological replicates. Lines indicate the average of n = 4 biological replicates. Asterisks indicate significance between C. difficile CFU/mL at given concentration and C. difficile CFU/mL in the absence of antibiotic (*P < 0.05, **P < 0.01, ***P < 0.001, ns P > 0.05). The data underlying this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A, B) Line plots of absolute abundance across A metronidazole (MTZ) and B vancomycin (VAN) concentrations at 48 h. Each x-axis is semi-log scale. Y-axis is calculated OD600 (OD600 multiplied by relative abundance based on 16S sequencing). Data points indicate biological replicates. Lines indicate the average of n = 1 to n = 4 biological replicates in panel A and n = 3 to n = 4 biological replicates in panel B. Color indicates species, see Fig 1C. Red borders indicate communities with ≥4-fold change in C. difficile MIC (see S1 Table). The data underlying all panels in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A, B) Line plots of subMIC fold change in metronidazole (MTZ) or vancomycin (VAN) at 48 h. SubMIC fold change is calculated as the species absolute abundance in a given subMIC divided by the species absolute abundance in the absence of the antibiotic. Black horizontal line at y = 1 indicates no change in growth compared to no antibiotic condition. Each x-axis is semi-log scale. Data points indicate biological replicates. Lines indicate the average of n = 1 to n = 4 biological replicates in panel A and n = 3 to n = 4 biological replicates in panel B. Color indicates species, see Fig 1C. The data underlying all panels in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A) Scatterplots of maximum C. difficile subMIC fold change in multispecies communities at 48 h as a function of the number (left) or abundance (right) of sensitive biotic inhibitors for both metronidazole and vancomycin. The x-axis represents the number of sensitive biotic inhibitors in a given community in the beginning of the experiment (left) or summed absolute abundance of sensitive biotic inhibitors in a given community at 48 h in the absence of the antibiotic (right). The average subMIC fold change was calculated for each subMIC concentration by computing the average C. difficile absolute abundance in a given subMIC divided by the average C. difficile absolute abundance in the absence of the antibiotic, average of n = 1 to n = 4 biological replicates. The maximum subMIC fold change (y-axis) is the maximum of the average subMIC fold change across all subMICs. The sensitive and resistant inhibitors are determined as in Fig 3F. Red points indicate communities where the sum of absolute abundance of resistant biotic inhibitors at 48 h was greater than 0.05 OD600. Spearman correlation annotated for all data points (black) and for only communities with resistant inhibitors >0.05 OD600 (red). (B) Box plot of the maximum subMIC fold change for C. difficile in multispecies communities. The maximum subMIC fold change (y-axis) is calculated as in panel A. Each data point represents 1 community. Community type (x-axis) determined as in Fig 3F. Gray horizontal line at y = 1 indicates no change in growth compared to the no antibiotic condition. Asterisks indicate a significant difference (*P < 0.05, **P < 0.01, ***P < 0.001) according to a one-sided Mann–Whitney U test. (C) Schematic of subMIC difference metric. The x-axis is semi-log scale. Gray horizontal line at y = 1 indicates no change in growth compared to the no antibiotic condition. (D) Scatterplots comparing the predicted value of the C. difficile subMIC difference in a given community (x-axis) with average experimental value of the C. difficile subMIC difference in the same community (y-axis). The predicted value was calculated by summing the C. difficile subMIC differences of each CD-SpX pairwise community for all SpX. SubMIC differences greater than 0 have subMIC fold changes greater than 1. SubMIC differences less than 0 have subMIC fold changes less than 1. Each point represents a given community at given concentration of metronidazole or vancomycin. Number of sensitive inhibitors (color) was determined as in Fig 3F. Black line indicates best fit linear regression for all data points. Gray y = x line represents a perfect prediction of community subMIC difference. Annotated Pearson correlation values are based on all data points. Annotated percentages indicate the percentage of total data points in a given quadrant. The data underlying panels ABD in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A, B) Line plots of model prediction of absolute abundance of C. difficile and gut microbe pairwise communities at 48 h in presence of A metronidazole (MTZ) or B vancomycin (VAN). Each x-axis is semi-log scale. Y-axis is calculated OD600 (OD600 multiplied by relative abundance from 16S sequencing). Points indicate experimental data. Lines indicate model simulations. Color indicates species, see Fig 1C. Bold number is the sum of squared errors for C. difficile (square of the difference between model OD600 and the average experimental OD600, summed across all concentrations). The data underlying all panels in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A, B) Line plots of model prediction of absolute abundance of species in multispecies communities at 48 h in the presence of A metronidazole (MTZ) or B vancomycin (VAN). Each x-axis is semi-log scale. The y-axis denotes calculated OD600 (OD600 multiplied by relative abundance from 16S sequencing). Data points indicate experimental data. Lines indicate model simulations. Color indicates species, see Fig 1C. Bold number is sum of squared errors for C. difficile (square of difference between model OD600 and average experimental OD600, summed across all concentrations). The data underlying all panels in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A) Antibiotic gLV model prediction of pairwise and multispecies community data in the presence of each antibiotic concentration. These predictions are summarized in the blue “gLV + monospecies antib susceptibility” bar in Fig 4B. (B) Simulated maximum subMIC fold change for a focal species cocultured with a 4 other species for 1,000 randomly generated parameter sets. All parameters were randomized between a set of bounds. Bounds: aji (−1.25, 1.25), growth rates (0, 1), intraspecies interactions (−1.25, 0), and antibiotic susceptibility (−6, 0). Color of the data point indicates the abundance of focal species in the community in the absence of antibiotics (where light colored data points indicate inhibition of a focal species). A community is classified as containing a susceptible inhibitor if for any non-focal species j, Bj < −0.1 and afocal,j < −0.1. Gray horizontal line at y = 1 indicates no change in growth compared to the no antibiotic condition. The data and modeling scripts underlying all panels in this figure can be found in DOI: 10.5281/zenodo.7726490.
(TIF)
(A) Line plot of the growth curve of D. piger. Colored data points indicate the time at which 5 separate D. piger spent media samples were harvested. Data points represent biological replicates. The line represents the mean of n = 5 biological replicates. (B) Line plot of C. difficile abundance at 41 h in 5 different D. piger spent medias in the presence of metronidazole (MTZ). The x-axis is semi-log scale. Data points indicate biological replicates. Line represents the mean of n = 4 biological replicates. (C) Line plot of the abundance of 4 clinical C. difficile isolates in the presence of metronidazole. Each x-axis is semi-log scale. The y-axis denotes the integral of C. difficile OD600 from 0 to 48 h. Data points indicate biological replicates. The line represents the mean of n = 3 biological replicates. Asterisks indicate a significant difference (*P < 0.05, **P < 0.01, ***P < 0.001) according to an unpaired t test. (D) Line plot of C. difficile abundance at 48 h in a rich chemically defined media (“DM38”) in the presence of metronidazole. The x-axis is semi-log scale. For the D. piger coculture condition, OD600 denotes calculated OD600 (OD600 multiplied by relative abundance from 16S rRNA gene sequencing). Data points indicate biological replicates. The line represents the mean of n = 6 (fresh media) or n = 3 (DP SM) biological replicates. Asterisks indicate a significant difference (*P < 0.05, **P < 0.01, ***P < 0.001) according to an unpaired t test. (E) Line plot of C. difficile abundance at 48 h in fresh media in the presence of metronidazole that was incubated in fresh media or D. piger spent media. The x-axis is semi-log scale. Data points indicate biological replicates. Line represents the mean of n = 4 biological replicates. No significant difference (“ns”, p > 0.05) according to an unpaired t test. (F) Line plot of C. difficile abundance at 48 h in fresh media, D. piger spent media, or D. piger spent media with a fresh media spike. The x-axis is semi-log scale. Data points indicate biological replicates. The line represents the mean of n = 4 biological replicates. Asterisks indicate significant difference (*P < 0.05, **P < 0.01, ***P < 0.001, “ns” P > 0.05) according to an unpaired t test. The data underlying panels ABD in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A–E) Left: Schematic of 2 conditions being compared. Middle: Volcano plot of the fold change in C. difficile transcript abundance. Gray vertical lines indicate 2-fold change (1 in log2) and gray horizontal line indicates statistical significance (p = 0.05). Blue indicates genes annotated to be involved in metal import. Red indicates genes predicted to contain iron-sulfur clusters by MetalPredator. Right: Enriched KEGG modules in C. difficile. All KEGG pathways with significant enrichment scores from Gene Set Enrichment Analysis (GSEA) are shown. The data underlying all panels in this figure can be found in S3 and S4 Tables.
(TIF)
(A, B) Scatterplots to evaluate the patterns in gene expression for gene orthologs between CDR20291 (this study) and CD630Δerm (Berges and colleagues [35]). Genes only shown if differentially expressed in both studies. X-axis: log2 fold change between low iron (0.2 μM) and high iron (15 μM) media in Berges and colleagues [35]. Y-axis: log2 fold change between C. difficile in coculture with D. piger (panel A) or C. difficile in D. piger spent media (SM) (panel B) and C. difficile in fresh media in this study. Each data point indicates a gene. Percentages indicate percentage of genes in each quadrant. Black line indicates best fit linear regression for all data points. Gray line is y = x. The data underlying all panels in this figure can be found in S3 Table.
(TIF)
Line plot of B. thetaiotaomicron abundance at 48 h in fresh media or in 75% D. piger spent media, 25% fresh media. The x-axis is semi-log scale. Data points represent biological replicates. Lines indicate the average of n = 3 biological replicates. The data underlying this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A) Bar plot of the amount of hydrogen sulfide in multiple conditions after 10 h of incubation. “Blank” represents a fresh media control. The y-axis denotes the percentage of the amount of hydrogen sulfide in the D. piger monoculture. Data points represent the mean of n = 2 technical replicates. Bar represents the mean of n = 8 biological replicates. Asterisks indicate significant difference (*P < 0.05, **P < 0.01, ***P < 0.001, “ns” P > 0.05) according to an unpaired t test. (B) Bar plot of the concentration of divalent transition metals in fresh media. Points represent the mean of n = 2 technical replicates. Bar represents the mean of n = 3 biological replicates. The data underlying all panels in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A) Bar plot of hydrogen sulfide concentration in different medias. Data points represent n = 4 biological replicates. Bar represents the average of biological replicates. (B) Image of D. piger spent media 2 h after nitrogen purge (right) or no treatment (left). Precipitation of a black ferrous sulfide turns the media a darker color. (C) Line plot of C. difficile OD600 at 41 h in different medias in the presence of metronidazole. The x-axis is semi-log scale. Data points represent biological replicates. Lines indicate the average of n = 4 to n = 8 biological replicates. (D) Bar plot displaying MIC of data shown in C. Data points represent the MIC of n = 4 to n = 8 biological replicates. Bar represents the MIC determined based on an average OD600 of n = 4 to n = 8 biological replicates. The data underlying panels ACD in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A) Line plots and bar plots of C. difficile metronidazole susceptibility in fresh media with and without metal supplementation. (Top) Bar plots of C. difficile metronidazole MIC in untreated fresh media (gray) and media with 1 mM metal supplementation (blue). In the metal combination condition (“Combo”), all 5 metals were supplemented at 1 mM. Data points represent the MIC of n = 4 biological replicates. Bar represents the MIC determined based on an average OD600 of n = 4 biological replicates. (Bottom) Line plots of C. difficile OD600 at 45 h in untreated fresh media (gray) or media supplemented with 1 mM metal (blue) in the presence of metronidazole (MTZ). Each x-axis is semi-log scale. Data points represent biological replicates. Lines indicate the average of n = 4 biological replicates. (B) Bar plot of delta Ct (ΔCt) values of 3 genes in C. difficile grown in fresh media, D. piger spent media (DP SM), or D. piger spent media with metal supplementation based on qRT-PCR. Data points represent biological replicates, with each point calculated as the average of 3 technical replicates. Bar indicates the average of n = 4 biological replicates. Asterisks indicate significant difference (*P < 0.05, **P < 0.01, ***P < 0.001, “ns” P > 0.05) according to an unpaired t test. The data underlying all panels in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A) Pie charts of the change in C. difficile MIC in pairwise and multispecies communities compared to monoculture. (B) Pie charts of the growth enhancement of C. difficile in pairwise and multispecies communities. Growth enhancement is defined as a maximum subMIC fold change that is greater than 1 and is greater than the maximum subMIC fold change in C. difficile monoculture for that antibiotic (i.e., greater than 1.61 for metronidazole). The data underlying all panels in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
MICs in μg/mL. C. difficile monoculture MIC from same experimental day given for comparison, as monoculture MIC can vary due to preculture conditions and glycerol stock batches.
(XLSX)
MICs in μg/mL. C. difficile monoculture MIC from same experimental day given for comparison, as monoculture MIC can vary due to preculture conditions and glycerol stock batches. “No growth” represents no growth of C. difficile across all concentrations and therefore no MIC could be determined.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
The authors would like to thank Dr. Tricia Kiley for helpful discussions.
Abbreviations
- CFU
colony-forming unit
- DEG
differentially expressed gene
- GSEA
Gene Set Enrichment Analysis
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MIC
minimum inhibitory concentration
- PFOR
pyruvate ferredoxin/flavodoxin oxidoreductase
- PMF
proton motive force
- RPKM
reads per kilobase million
- subMIC
sub-minimum inhibitory concentration
Data Availability
The datasets and computer code used in this study are available in the following databases: • Species abundance data, CFU data, hydrogen sulfide data and qRT-PCR data are available from Zenodo: https://doi.org/10.5281/zenodo.7626486 • RNA-seq data is available from Zenodo: https://doi.org/10.5281/zenodo.7049035 and https://doi.org/10.5281/zenodo.7049027 • Modeling computer scripts are available from Zenodo: https://doi.org/10.5281/zenodo.7726490.
Funding Statement
Research was sponsored by the National Institutes of Health and was accomplished under grant numbers R35GM124774 and R21AI159980. S.H. was supported in part by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number T32GM008349. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. doi: 10.1038/nri3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown KA, Khanafer N, Daneman N, Fisman DN. Meta-analysis of antibiotics and the risk of community-associated Clostridium difficile infection. Antimicrob Agents Chemother. 2013;57:2326–2332. doi: 10.1128/AAC.02176-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dowle C. Faecal microbiota transplantation: a review of FMT as an alternative treatment for Clostridium difficile infection. Biosci Horizons Int J Student Res. 2016;9:hzw007. [Google Scholar]
- 4.Hromada S, Qian Y, Jacobson TB, Clark RL, Watson L, Safdar N, et al. Negative interactions determine Clostridioides difficile growth in synthetic human gut communities. Mol Syst Biol. 2021;17:e10355. doi: 10.15252/msb.202110355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schubert AM, Rogers MAM, Ring C, Mogle J, Petrosino JP, Young VB, et al. Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy controls. MBio. 2014;5:e01021–e01014. doi: 10.1128/mBio.01021-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreyra JA, Wu KJ, Hryckowian AJ, Bouley DM, Weimer BC, Sonnenburg JL. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe. 2014;16:770–777. doi: 10.1016/j.chom.2014.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith AL, Fiel SB, Mayer-Hamblett N, Ramsey B, Burns JL. Susceptibility Testing of Pseudomonas aeruginosa Isolates and Clinical Response to Parenteral Antibiotic Administration: Lack of Association in Cystic Fibrosis. Chest. 2003;123:1495–1502. doi: 10.1378/chest.123.5.1495 [DOI] [PubMed] [Google Scholar]
- 9.Bottery MJ, Pitchford JW, Friman V-P. Ecology and evolution of antimicrobial resistance in bacterial communities. ISME J. 2021;15:939–948. doi: 10.1038/s41396-020-00832-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perlin MH, Clark DR, McKenzie C, Patel H, Jackson N, Kormanik C, et al. Protection of Salmonella by ampicillin-resistant Escherichia coli in the presence of otherwise lethal drug concentrations. Proceedings Biol Sci. 2009;276:3759–3768. [DOI] [PMC free article] [PubMed]
- 11.Adamowicz EM, Muza M, Chacón JM, Harcombe WR. Cross-feeding modulates the rate and mechanism of antibiotic resistance evolution in a model microbial community of Escherichia coli and Salmonella enterica. PLoS Pathog. 2020;16:e1008700. doi: 10.1371/journal.ppat.1008700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Vos MGJ, Zagorski M, McNally A, Bollenbach T. Interaction networks, ecological stability, and collective antibiotic tolerance in polymicrobial infections. Proc Natl Acad Sci U S A. 2017;114:10666–10671. doi: 10.1073/pnas.1713372114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aranda-Díaz A, Obadia B, Dodge R, Thomsen T, Hallberg ZF, Güvener ZT, et al. Bacterial interspecies interactions modulate pH-mediated antibiotic tolerance. Elife. 2020;9:e51493. doi: 10.7554/eLife.51493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parijs I, Steenackers HP. Competitive inter-species interactions underlie the increased antimicrobial tolerance in multispecies brewery biofilms. ISME J. 2018;12:2061–2075. doi: 10.1038/s41396-018-0146-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalton T, Dowd SE, Wolcott RD, Sun Y, Watters C, Griswold JA, et al. An In Vivo Polymicrobial Biofilm Wound Infection Model to Study Interspecies Interactions. PLoS ONE. 2011;6:e27317. doi: 10.1371/journal.pone.0027317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klümper U, Recker M, Zhang L, Yin X, Zhang T, Buckling A, et al. Selection for antimicrobial resistance is reduced when embedded in a natural microbial community. ISME J. 2019;13:2927–2937. doi: 10.1038/s41396-019-0483-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aranda-Díaz A, Ng KM, Thomsen T, Real-Ramírez I, Dahan D, Dittmar S, et al. Establishment and characterization of stable, diverse, fecal-derived in vitro microbial communities that model the intestinal microbiota. Cell Host Microbe. 2022;30:260–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler MS, Hansford KA, Blaskovich MAT, Halai R, Cooper MA. Glycopeptide antibiotics: Back to the future. J Antibiot (Tokyo). 2014;67:631–644. doi: 10.1038/ja.2014.111 [DOI] [PubMed] [Google Scholar]
- 19.Löfmark S, Edlund C, Nord CE. Metronidazole Is Still the Drug of Choice for Treatment of Anaerobic Infections. Clin Infect Dis. 2010;50:S16–S23. doi: 10.1086/647939 [DOI] [PubMed] [Google Scholar]
- 20.Church DL, Rabin HR, Lalshley EJ. Role of hydrogenase 1 of clostridium pasteurianum in the reduction of metronidazole. Biochem Pharmacol. 1988;37:1525–1534. doi: 10.1016/0006-2952(88)90014-7 [DOI] [PubMed] [Google Scholar]
- 21.Diniz CG, Farias LM, Carvalho MAR, Rocha ER, Smith CJ. Differential gene expression in a Bacteroides fragilis metronidazole-resistant mutant. J Antimicrob Chemother. 2004;54:100–108. doi: 10.1093/jac/dkh256 [DOI] [PubMed] [Google Scholar]
- 22.Leitsch D, Kolarich D, Binder M, Stadlmann J, Altmann F, Duchêne M. Trichomonas vaginalis: metronidazole and other nitroimidazole drugs are reduced by the flavin enzyme thioredoxin reductase and disrupt the cellular redox system. Implications for nitroimidazole toxicity and resistance. Mol Microbiol. 2009;72:518–536. doi: 10.1111/j.1365-2958.2009.06675.x [DOI] [PubMed] [Google Scholar]
- 23.Johnson S, Lavergne V, Skinner AM, Gonzales-Luna AJ, Garey KW, Kelly CP, et al. Clinical Practice Guideline by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA): 2021 Focused Update Guidelines on Management of Clostridioides difficile Infection in Adults. Clin Infect Dis. 2021;73:e1029–e1044. doi: 10.1093/cid/ciab549 [DOI] [PubMed] [Google Scholar]
- 24.McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, et al. Clinical Practice Guidelines for Clostridium difficile Infection in Adults and Children: 2017 Update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis. 2018;66:e1–e48. doi: 10.1093/cid/cix1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darling AE, Jospin G, Lowe E, Matsen FA IV, Bik HM, Eisen JA. PhyloSift: phylogenetic analysis of genomes and metagenomes. PeerJ. 2014;2:e243. doi: 10.7717/peerj.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.CLSI. Methods for antimicrobial susceptibility testing of anaerobic bacteria. Approved standard. CLSI Doc. 2018:M11–A9. [Google Scholar]
- 27.Paterson IK, Hoyle A, Ochoa G, Baker-Austin C, Taylor NGH. Optimising Antibiotic Usage to Treat Bacterial Infections. Sci Rep. 2016;6:37853. doi: 10.1038/srep37853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Roode JC, Culleton R, Bell AS, Read AF. Competitive release of drug resistance following drug treatment of mixed Plasmodium chabaudi infections. Malar J. 2004;3:33. doi: 10.1186/1475-2875-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varga JJ, Zhao CY, Davis JD, Hao Y, Farrell JM, Gurney JR, et al. Antibiotics Drive Expansion of Rare Pathogens in a Chronic Infection Microbiome Model. mSphere 2022;7:e00318–22. doi: 10.1128/msphere.00318-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stein RR, Bucci V, Toussaint NC, Buffie CG, Rätsch G, Pamer EG, et al. Ecological Modeling from Time-Series Inference: Insight into Dynamics and Stability of Intestinal Microbiota. PLoS Comput Biol. 2013;9:31–36. doi: 10.1371/journal.pcbi.1003388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valasatava Y, Rosato A, Banci L, Andreini C. MetalPredator: a web server to predict iron–sulfur cluster binding proteomes. Bioinformatics. 2016;32:2850–2852. doi: 10.1093/bioinformatics/btw238 [DOI] [PubMed] [Google Scholar]
- 32.Ho TD, Ellermeier CD. Ferric uptake regulator fur control of putative iron acquisition systems in Clostridium difficile. J Bacteriol. 2015;197:2930–2940. doi: 10.1128/JB.00098-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lau CKY, Krewulak KD, Vogel HJ. Bacterial ferrous iron transport: the Feo system. FEMS Microbiol Rev. 2016;40:273–298. doi: 10.1093/femsre/fuv049 [DOI] [PubMed] [Google Scholar]
- 34.Hastie JL, Hanna PC, Carlson PE. Transcriptional response of Clostridium difficile to low iron conditions. Pathog Dis. 2018;76:fty009. doi: 10.1093/femspd/fty009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berges M, Michel A-M, Lassek C, Nuss AM, Beckstette M, Dersch P, et al. Iron Regulation in Clostridioides difficile. Front Microbiol. 2018;9:3183. doi: 10.3389/fmicb.2018.03183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez CA, Beavers WN, Weiss A, Knippel RJ, Zackular JP, Chazin W, et al. The Immune Protein Calprotectin Impacts Clostridioides difficile Metabolism through Zinc Limitation. MBio. 2019;10:e02289–e02219. doi: 10.1128/mBio.02289-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoch DC, Valentine RC. Ferredoxins and flavodoxins of bacteria. Annu Rev Microbiol. 1972;26:139–162. doi: 10.1146/annurev.mi.26.100172.001035 [DOI] [PubMed] [Google Scholar]
- 38.Dobbek H, Svetlitchnyi V, Gremer L, Huber R, Meyer O. Crystal Structure of a Carbon Monoxide Dehydrogenase Reveals a [Ni-4Fe-5S] Cluster. Science. 2001;293:1281–1285. [DOI] [PubMed] [Google Scholar]
- 39.Shafaat HS, Rüdiger O, Ogata H, Lubitz W. [NiFe] hydrogenases: A common active site for hydrogen metabolism under diverse conditions. Biochim Biophys Acta—Bioenerg. 2013;1827:986–1002. doi: 10.1016/j.bbabio.2013.01.015 [DOI] [PubMed] [Google Scholar]
- 40.Constanze P, Gary SR, Valley S. Anaerobic Formate and Hydrogen Metabolism. EcoSal Plus. 2016:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aditi D, Xiaoqian W, Wenwen H, Palmer KL, Hurdle JG. Chromosomal Resistance to Metronidazole in Clostridioides difficile Can Be Mediated by Epistasis between Iron Homeostasis and Oxidoreductases. Antimicrob Agents Chemother. 2020;64:e00415–e00420. doi: 10.1128/AAC.00415-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chong PM, Lynch T, McCorrister S, Kibsey P, Miller M, Gravel D, et al. Proteomic Analysis of a NAP1 Clostridium difficile Clinical Isolate Resistant to Metronidazole. PLoS ONE. 2014;9:e82622. doi: 10.1371/journal.pone.0082622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boekhoud IM, Hornung BVH, Sevilla E, Harmanus C, Bos-Sanders IMJG, Terveer EM, et al. Plasmid-mediated metronidazole resistance in Clostridioides difficile. Nat Commun. 2020;11:598. doi: 10.1038/s41467-020-14382-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smiley JD, Ashwell G. Uronic Acid Metabolism in Bacteria: III. PURIFICATION AND PROPERTIES OF d-ALTRONIC ACID AND d-MANNONIC ACID DEHYDRASES IN ESCHERICHIA COLI. J Biol Chem. 1960;235:1571–1575. [PubMed] [Google Scholar]
- 45.Lynch T, Chong P, Zhang J, Hizon R, Du T, Graham MR, et al. Characterization of a stable, metronidazole-resistant Clostridium difficile clinical isolate. PLoS ONE. 2013;8:e53757. doi: 10.1371/journal.pone.0053757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dreyer JL. The role of iron in the activation of mannonic and altronic acid hydratases, two Fe-requiring hydro-lyases. Eur J Biochem. 1987;166:623–630. doi: 10.1111/j.1432-1033.1987.tb13559.x [DOI] [PubMed] [Google Scholar]
- 47.Loubinoux J, Valente FMA, Pereira IAC, Costa A, Grimont PAD, Le Faou AE. Reclassification of the only species of the genus Desulfomonas, Desulfomonas pigra, as Desulfovibrio piger comb. nov. Int J Syst Evol Microbiol. 2002;52:1305–1308. doi: 10.1099/00207713-52-4-1305 [DOI] [PubMed] [Google Scholar]
- 48.Loubinoux J, Bronowicki J-P, Pereira IAC, Mougenel J-L, Le Faou AE. Sulfate-reducing bacteria in human feces and their association with inflammatory bowel diseases. FEMS Microbiol Ecol. 2002;40:107–112. doi: 10.1111/j.1574-6941.2002.tb00942.x [DOI] [PubMed] [Google Scholar]
- 49.Azabou S, Mechichi T, Sayadi S. Zinc precipitation by heavy-metal tolerant sulfate-reducing bacteria enriched on phosphogypsum as a sulfate source. Miner Eng. 2007;20:173–178. [Google Scholar]
- 50.Foucher S, Battaglia-Brunet F, Ignatiadis I, Morin D. Treatment by sulfate-reducing bacteria of Chessy acid-mine drainage and metals recovery. Chem Eng Sci. 2001;56:1639–1645. [Google Scholar]
- 51.Dubois T, Dancer-Thibonnier M, Monot M, Hamiot A, Bouillaut L, Soutourina O, et al. Control of Clostridium difficile Physiopathology in Response to Cysteine Availability. Infect Immun. 2016;84:2389–2405. doi: 10.1128/IAI.00121-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eze P, Balsells E, Kyaw MH, Nair H. Risk factors for Clostridium difficile infections—an overview of the evidence base and challenges in data synthesis. J Glob Health. 2017;7:10417. doi: 10.7189/jogh.07.010417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bolton RP, Culshaw MA. Faecal metronidazole concentrations during oral and intravenous therapy for antibiotic associated colitis due to Clostridium difficile. Gut. 1986;27:1169 LP– 1172. doi: 10.1136/gut.27.10.1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ezraty B, Barras F. The ‘liaisons dangereuses’ between iron and antibiotics. FEMS Microbiol Rev. 2016;40:418–435. doi: 10.1093/femsre/fuw004 [DOI] [PubMed] [Google Scholar]
- 55.Müller M, Lindmark DG. Uptake of metronidazole and its effect on viability in trichomonads and Entamoeba invadens under anaerobic and aerobic conditions. Antimicrob Agents Chemother. 1976;9:696–700. doi: 10.1128/AAC.9.4.696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boekhoud IM, Sidorov I, Nooij S, Harmanus C, Bos-Sanders IMJG, Viprey V, et al. Haem is crucial for medium-dependent metronidazole resistance in clinical isolates of Clostridioides difficile. J Antimicrob Chemother. 2021;76:1731–1740. doi: 10.1093/jac/dkab097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chabrière E, Charon M, Volbeda A, Pieulle L, Hatchikian EC, Fontecilla–Camps J. Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate. Nat Struct Biol. 1999;6:182–190. doi: 10.1038/5870 [DOI] [PubMed] [Google Scholar]
- 58.Fojcik C, Arnoux P, Ouerdane L, Aigle M, Alfonsi L, Borezée-Durant E. Independent and cooperative regulation of staphylopine biosynthesis and trafficking by Fur and Zur. Mol Microbiol. 2018;108:159–177. doi: 10.1111/mmi.13927 [DOI] [PubMed] [Google Scholar]
- 59.Wolf PG, Cowley ES, Breister A, Matatov S, Lucio L, Polak P, et al. Diversity and distribution of sulfur metabolic genes in the human gut microbiome and their association with colorectal cancer. Microbiome. 2022;10:64. doi: 10.1186/s40168-022-01242-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Magee EA, Richardson CJ, Hughes R, Cummings JH. Contribution of dietary protein to sulfide production in the large intestine: an in vitro and a controlled feeding study in humans. Am J Clin Nutr. 2000;72:1488–1494. doi: 10.1093/ajcn/72.6.1488 [DOI] [PubMed] [Google Scholar]
- 61.Deplancke B, Finster K, Graham WV, Collier CT, Thurmond JE, Gaskins HR. Gastrointestinal and Microbial Responses to Sulfate-Supplemented Drinking Water in Mice. Exp Biol Med. 2003;228:424–433. doi: 10.1177/153537020322800413 [DOI] [PubMed] [Google Scholar]
- 62.Kortman GAM, Raffatellu M, Swinkels DW, Tjalsma H. Nutritional iron turned inside out: intestinal stress from a gut microbial perspective. FEMS Microbiol Rev. 2014;38:1202–1234. doi: 10.1111/1574-6976.12086 [DOI] [PubMed] [Google Scholar]
- 63.Lobo AR, Gaievski EHS, De Carli E, Alvares EP, Colli C. Fructo-oligosaccharides and iron bioavailability in anaemic rats: the effects on iron species distribution, ferroportin-1 expression, crypt bifurcation and crypt cell proliferation in the caecum. Br J Nutr. 2014;112:1286–1295. doi: 10.1017/S0007114514002165 [DOI] [PubMed] [Google Scholar]
- 64.Knippel RJ, Zackular JP, Moore JL, Celis AI, Weiss A, Washington MK, et al. Heme sensing and detoxification by HatRT contributes to pathogenesis during Clostridium difficile infection. PLoS Pathog. 2018;14:e1007486. doi: 10.1371/journal.ppat.1007486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zygiel EM, Nolan EM. Transition Metal Sequestration by the Host-Defense Protein Calprotectin. Annu Rev Biochem. 2018;87:621–643. doi: 10.1146/annurev-biochem-062917-012312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lobel L, Cao YG, Fenn K, Glickman JN, Garrett WS. Diet posttranslationally modifies the mouse gut microbial proteome to modulate renal function. Science. 2020;369:1518–1524. doi: 10.1126/science.abb3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kuijper EJ, Wilcox MH. Decreased Effectiveness of Metronidazole for the Treatment of Clostridium difficile Infection? Clin Infect Dis. 2008;47:63–65. doi: 10.1086/588294 [DOI] [PubMed] [Google Scholar]
- 68.Waldetoft KW, Sundius S, Kuske R, Brown SP. Defining the Benefits of Antibiotic Resistance in Commensals and the Scope for Resistance Optimization. MBio. 2022;14:e01349–e01322. doi: 10.1128/mbio.01349-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nale JY, Spencer J, Hargreaves KR, Buckley AM, Trzepiński P, Douce GR, et al. Bacteriophage Combinations Significantly Reduce Clostridium difficile Growth In Vitro and Proliferation In Vivo. Antimicrob Agents Chemother. 2016;60:968–981. doi: 10.1128/AAC.01774-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Clark RL, Connors BM, Stevenson DM, Hromada SE, Hamilton JJ, Amador-Noguez D, et al. Design of synthetic human gut microbiome assembly and butyrate production. Nat Commun. 2021;12:3254. doi: 10.1038/s41467-021-22938-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Metcalf D, Sharif S, Weese JS. Evaluation of candidate reference genes in Clostridium difficile for gene expression normalization. Anaerobe. 2010;16:439–443. doi: 10.1016/j.anaerobe.2010.06.007 [DOI] [PubMed] [Google Scholar]
- 74.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- 75.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation. 2021;2:100141. doi: 10.1016/j.xinn.2021.100141 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A, B) Line plots of monospecies OD600 across antibiotic concentrations for metronidazole (MTZ) and vancomycin (VAN) at 48 h. Each x-axis is semi-log scale. Colored data points indicate biological replicates. Colored lines indicate the average of n = 2 to n = 4 biological replicates. Bold number indicates MIC of data. Black lines indicate model fit. (C) Table of scaled antibiotic susceptibility parameter B for each species. The data underlying panels AB in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A) Schematic of 2 methods for determining absolute abundance: calculated absorbance at 600 nm (OD600) or CFU counting. (B, C) Double axis line plots of absolute abundance at 48 h of C. difficile in the presence of (B) metronidazole (MTZ) and (C) vancomycin (VAN). Left axis (black) is calculated OD600 (OD600 multiplied by relative abundance from 16S sequencing) and right axis (red) is CFU per mL. The x-axis is semi-log scale. Black points indicate biological replicates. Red points indicate biological replicates, each the average of 2 technical replicates. Lines represent the mean of n = 4 biological replicates. Bold number indicates the MIC based on calculated OD600 (black) or CFU (red). “NG” indicates no growth above the MIC threshold for any conditions, and no MIC was able to be determined. Blue outlines indicate conditions for which the MIC was consistent between the 2 methods. Data collected on a separate day from Fig 2. (D) Heatmaps of C. difficile subMIC fold change in data from panels BC. Black indicates significant fold change (greater than 1) in at least 1 concentration, white indicates no significant fold change at any concentration. Significance was determined using unpaired t test between the abundance of C. difficile at a given subMIC and the abundance of C. difficile in the absence of antibiotic, P < 0.05. C. difficile abundance determined by CFU counting (left heatmap) or calculated OD600 method (right heatmap). The data underlying panels BCD in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A, B) Line plots of subMIC fold change for pairwise communities in metronidazole (MTZ) or vancomycin (VAN) at 48 h. SubMIC fold change is calculated as the species absolute abundance at the subMIC divided by the species absolute abundance in the absence of the antibiotic. Black horizontal line at y = 1 indicates no change in growth compared to the no antibiotic condition. Each x-axis is semi-log scale. Data points indicate biological replicates. Line represents mean of n = 3 to n = 8 biological replicates in panel A and n = 1 to n = 8 biological replicates in panel B. (C) Line plots of C. difficile monospecies over time. (Top) C. difficile OD600 at 0 μg/mL and 1.5 μg/mL metronidazole. (Bottom) C. difficile subMIC fold change μg/mL metronidazole. Gray horizontal line at y = 1 indicates no change in growth compared to no antibiotic condition. SubMIC fold change calculated as in panels AB. Points represent biological replicates. Line represents average of n = 4 biological replicates. Statistical significance analyzed for all time points where C. difficile OD600 was greater in the presence of antibiotic than the absence of antibiotic (“ns” P > 0.05, according to an unpaired t test). (D) Microscopy images of select C. difficile conditions from panel C: no antibiotic at 24 and 48 h and 1.5 μg/mL metronidazole at 48 h. Exponential phase C. difficile cells in presence of subMIC of metronidazole do not show morphological difference from exponential phase C. difficile cells grown in absence of antibiotic. (E) Line plots of C. difficile in pair with Bacteroides thetaiotaomicron over time. (Top) C. difficile OD600 in the presence of 0 μg/mL or 1.5 μg/mL metronidazole. (Bottom) C. difficile subMIC fold change in the presence of 1.5 μg/mL metronidazole. Gray horizontal line at y = 1 indicates no change in growth compared to no antibiotic condition. SubMIC fold change calculated as in panels AB. Points represent biological replicates. Line represents average of n = 4 biological replicates. Statistical significance analyzed for all time points where C. difficile OD600 was greater in the presence of antibiotic than the absence of antibiotic (“ns” P > 0.05, according to an unpaired t test). (F) Line plots of C. difficile in pair with Clostridium hiranonis over time. (Top) C. difficile OD600 in the presence of 0 μg/mL or 0.1875 μg/mL vancomycin. (Bottom) C. difficile subMIC fold change in the presence of 0.1875 μg/mL vancomycin. Gray horizontal line at y = 1 indicates no change in growth compared to the no antibiotic condition. SubMIC fold change calculated as in panels AB. Points represent biological replicates. Line represents average of n = 4 biological replicates. Statistical significance was analyzed for all time points where C. difficile OD600 was greater in the presence of antibiotic than the absence of antibiotic (*P < 0.05, **P < 0.01, ***P < 0.001, ns P > 0.05, according to an unpaired t test). (G) Box plot of maximum subMIC fold change for C. difficile in pairwise communities at 48 h. The maximum subMIC fold change is the maximum of the average subMIC fold change across all subMICs. The average subMIC fold change was calculated for each subMIC concentration by taking the average C. difficile absolute abundance at the subMIC divided by the average C. difficile absolute abundance in the no antibiotic condition (n = 1 to n = 4 biological replicates). C. difficile did not grow at any concentrations of vancomycin in CD-BO, CD-BT, CD-BU, and CD-PC pairs so these communities were not included in the analysis. Each data point represents 1 pairwise community. Gray horizontal line at y = 1 indicates no change in growth compared to the no antibiotic condition. Species are categorized as sensitive inhibitors if (1) the absolute abundance of C. difficile in a given pairwise community was significantly lower than the monoculture C. difficile abundance, in the absence of antibiotics, as determined by an unpaired t test; and (2) if the monospecies MIC was less than the monospecies MIC of C. difficile. Asterisks indicate significant difference (*P < 0.05, **P < 0.01, ***P < 0.001) according to a one-sided Mann–Whitney U test. The data underlying panels ABCEFG in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
Line plot of C. difficile abundance in presence of metronidazole in (black) monoculture or in (brown) coculture with B. vulgatus. Data points indicate biological replicates. Lines indicate the average of n = 4 biological replicates. Asterisks indicate significance between C. difficile CFU/mL at given concentration and C. difficile CFU/mL in the absence of antibiotic (*P < 0.05, **P < 0.01, ***P < 0.001, ns P > 0.05). The data underlying this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A, B) Line plots of absolute abundance across A metronidazole (MTZ) and B vancomycin (VAN) concentrations at 48 h. Each x-axis is semi-log scale. Y-axis is calculated OD600 (OD600 multiplied by relative abundance based on 16S sequencing). Data points indicate biological replicates. Lines indicate the average of n = 1 to n = 4 biological replicates in panel A and n = 3 to n = 4 biological replicates in panel B. Color indicates species, see Fig 1C. Red borders indicate communities with ≥4-fold change in C. difficile MIC (see S1 Table). The data underlying all panels in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A, B) Line plots of subMIC fold change in metronidazole (MTZ) or vancomycin (VAN) at 48 h. SubMIC fold change is calculated as the species absolute abundance in a given subMIC divided by the species absolute abundance in the absence of the antibiotic. Black horizontal line at y = 1 indicates no change in growth compared to no antibiotic condition. Each x-axis is semi-log scale. Data points indicate biological replicates. Lines indicate the average of n = 1 to n = 4 biological replicates in panel A and n = 3 to n = 4 biological replicates in panel B. Color indicates species, see Fig 1C. The data underlying all panels in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A) Scatterplots of maximum C. difficile subMIC fold change in multispecies communities at 48 h as a function of the number (left) or abundance (right) of sensitive biotic inhibitors for both metronidazole and vancomycin. The x-axis represents the number of sensitive biotic inhibitors in a given community in the beginning of the experiment (left) or summed absolute abundance of sensitive biotic inhibitors in a given community at 48 h in the absence of the antibiotic (right). The average subMIC fold change was calculated for each subMIC concentration by computing the average C. difficile absolute abundance in a given subMIC divided by the average C. difficile absolute abundance in the absence of the antibiotic, average of n = 1 to n = 4 biological replicates. The maximum subMIC fold change (y-axis) is the maximum of the average subMIC fold change across all subMICs. The sensitive and resistant inhibitors are determined as in Fig 3F. Red points indicate communities where the sum of absolute abundance of resistant biotic inhibitors at 48 h was greater than 0.05 OD600. Spearman correlation annotated for all data points (black) and for only communities with resistant inhibitors >0.05 OD600 (red). (B) Box plot of the maximum subMIC fold change for C. difficile in multispecies communities. The maximum subMIC fold change (y-axis) is calculated as in panel A. Each data point represents 1 community. Community type (x-axis) determined as in Fig 3F. Gray horizontal line at y = 1 indicates no change in growth compared to the no antibiotic condition. Asterisks indicate a significant difference (*P < 0.05, **P < 0.01, ***P < 0.001) according to a one-sided Mann–Whitney U test. (C) Schematic of subMIC difference metric. The x-axis is semi-log scale. Gray horizontal line at y = 1 indicates no change in growth compared to the no antibiotic condition. (D) Scatterplots comparing the predicted value of the C. difficile subMIC difference in a given community (x-axis) with average experimental value of the C. difficile subMIC difference in the same community (y-axis). The predicted value was calculated by summing the C. difficile subMIC differences of each CD-SpX pairwise community for all SpX. SubMIC differences greater than 0 have subMIC fold changes greater than 1. SubMIC differences less than 0 have subMIC fold changes less than 1. Each point represents a given community at given concentration of metronidazole or vancomycin. Number of sensitive inhibitors (color) was determined as in Fig 3F. Black line indicates best fit linear regression for all data points. Gray y = x line represents a perfect prediction of community subMIC difference. Annotated Pearson correlation values are based on all data points. Annotated percentages indicate the percentage of total data points in a given quadrant. The data underlying panels ABD in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A, B) Line plots of model prediction of absolute abundance of C. difficile and gut microbe pairwise communities at 48 h in presence of A metronidazole (MTZ) or B vancomycin (VAN). Each x-axis is semi-log scale. Y-axis is calculated OD600 (OD600 multiplied by relative abundance from 16S sequencing). Points indicate experimental data. Lines indicate model simulations. Color indicates species, see Fig 1C. Bold number is the sum of squared errors for C. difficile (square of the difference between model OD600 and the average experimental OD600, summed across all concentrations). The data underlying all panels in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A, B) Line plots of model prediction of absolute abundance of species in multispecies communities at 48 h in the presence of A metronidazole (MTZ) or B vancomycin (VAN). Each x-axis is semi-log scale. The y-axis denotes calculated OD600 (OD600 multiplied by relative abundance from 16S sequencing). Data points indicate experimental data. Lines indicate model simulations. Color indicates species, see Fig 1C. Bold number is sum of squared errors for C. difficile (square of difference between model OD600 and average experimental OD600, summed across all concentrations). The data underlying all panels in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A) Antibiotic gLV model prediction of pairwise and multispecies community data in the presence of each antibiotic concentration. These predictions are summarized in the blue “gLV + monospecies antib susceptibility” bar in Fig 4B. (B) Simulated maximum subMIC fold change for a focal species cocultured with a 4 other species for 1,000 randomly generated parameter sets. All parameters were randomized between a set of bounds. Bounds: aji (−1.25, 1.25), growth rates (0, 1), intraspecies interactions (−1.25, 0), and antibiotic susceptibility (−6, 0). Color of the data point indicates the abundance of focal species in the community in the absence of antibiotics (where light colored data points indicate inhibition of a focal species). A community is classified as containing a susceptible inhibitor if for any non-focal species j, Bj < −0.1 and afocal,j < −0.1. Gray horizontal line at y = 1 indicates no change in growth compared to the no antibiotic condition. The data and modeling scripts underlying all panels in this figure can be found in DOI: 10.5281/zenodo.7726490.
(TIF)
(A) Line plot of the growth curve of D. piger. Colored data points indicate the time at which 5 separate D. piger spent media samples were harvested. Data points represent biological replicates. The line represents the mean of n = 5 biological replicates. (B) Line plot of C. difficile abundance at 41 h in 5 different D. piger spent medias in the presence of metronidazole (MTZ). The x-axis is semi-log scale. Data points indicate biological replicates. Line represents the mean of n = 4 biological replicates. (C) Line plot of the abundance of 4 clinical C. difficile isolates in the presence of metronidazole. Each x-axis is semi-log scale. The y-axis denotes the integral of C. difficile OD600 from 0 to 48 h. Data points indicate biological replicates. The line represents the mean of n = 3 biological replicates. Asterisks indicate a significant difference (*P < 0.05, **P < 0.01, ***P < 0.001) according to an unpaired t test. (D) Line plot of C. difficile abundance at 48 h in a rich chemically defined media (“DM38”) in the presence of metronidazole. The x-axis is semi-log scale. For the D. piger coculture condition, OD600 denotes calculated OD600 (OD600 multiplied by relative abundance from 16S rRNA gene sequencing). Data points indicate biological replicates. The line represents the mean of n = 6 (fresh media) or n = 3 (DP SM) biological replicates. Asterisks indicate a significant difference (*P < 0.05, **P < 0.01, ***P < 0.001) according to an unpaired t test. (E) Line plot of C. difficile abundance at 48 h in fresh media in the presence of metronidazole that was incubated in fresh media or D. piger spent media. The x-axis is semi-log scale. Data points indicate biological replicates. Line represents the mean of n = 4 biological replicates. No significant difference (“ns”, p > 0.05) according to an unpaired t test. (F) Line plot of C. difficile abundance at 48 h in fresh media, D. piger spent media, or D. piger spent media with a fresh media spike. The x-axis is semi-log scale. Data points indicate biological replicates. The line represents the mean of n = 4 biological replicates. Asterisks indicate significant difference (*P < 0.05, **P < 0.01, ***P < 0.001, “ns” P > 0.05) according to an unpaired t test. The data underlying panels ABD in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A–E) Left: Schematic of 2 conditions being compared. Middle: Volcano plot of the fold change in C. difficile transcript abundance. Gray vertical lines indicate 2-fold change (1 in log2) and gray horizontal line indicates statistical significance (p = 0.05). Blue indicates genes annotated to be involved in metal import. Red indicates genes predicted to contain iron-sulfur clusters by MetalPredator. Right: Enriched KEGG modules in C. difficile. All KEGG pathways with significant enrichment scores from Gene Set Enrichment Analysis (GSEA) are shown. The data underlying all panels in this figure can be found in S3 and S4 Tables.
(TIF)
(A, B) Scatterplots to evaluate the patterns in gene expression for gene orthologs between CDR20291 (this study) and CD630Δerm (Berges and colleagues [35]). Genes only shown if differentially expressed in both studies. X-axis: log2 fold change between low iron (0.2 μM) and high iron (15 μM) media in Berges and colleagues [35]. Y-axis: log2 fold change between C. difficile in coculture with D. piger (panel A) or C. difficile in D. piger spent media (SM) (panel B) and C. difficile in fresh media in this study. Each data point indicates a gene. Percentages indicate percentage of genes in each quadrant. Black line indicates best fit linear regression for all data points. Gray line is y = x. The data underlying all panels in this figure can be found in S3 Table.
(TIF)
Line plot of B. thetaiotaomicron abundance at 48 h in fresh media or in 75% D. piger spent media, 25% fresh media. The x-axis is semi-log scale. Data points represent biological replicates. Lines indicate the average of n = 3 biological replicates. The data underlying this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A) Bar plot of the amount of hydrogen sulfide in multiple conditions after 10 h of incubation. “Blank” represents a fresh media control. The y-axis denotes the percentage of the amount of hydrogen sulfide in the D. piger monoculture. Data points represent the mean of n = 2 technical replicates. Bar represents the mean of n = 8 biological replicates. Asterisks indicate significant difference (*P < 0.05, **P < 0.01, ***P < 0.001, “ns” P > 0.05) according to an unpaired t test. (B) Bar plot of the concentration of divalent transition metals in fresh media. Points represent the mean of n = 2 technical replicates. Bar represents the mean of n = 3 biological replicates. The data underlying all panels in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A) Bar plot of hydrogen sulfide concentration in different medias. Data points represent n = 4 biological replicates. Bar represents the average of biological replicates. (B) Image of D. piger spent media 2 h after nitrogen purge (right) or no treatment (left). Precipitation of a black ferrous sulfide turns the media a darker color. (C) Line plot of C. difficile OD600 at 41 h in different medias in the presence of metronidazole. The x-axis is semi-log scale. Data points represent biological replicates. Lines indicate the average of n = 4 to n = 8 biological replicates. (D) Bar plot displaying MIC of data shown in C. Data points represent the MIC of n = 4 to n = 8 biological replicates. Bar represents the MIC determined based on an average OD600 of n = 4 to n = 8 biological replicates. The data underlying panels ACD in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A) Line plots and bar plots of C. difficile metronidazole susceptibility in fresh media with and without metal supplementation. (Top) Bar plots of C. difficile metronidazole MIC in untreated fresh media (gray) and media with 1 mM metal supplementation (blue). In the metal combination condition (“Combo”), all 5 metals were supplemented at 1 mM. Data points represent the MIC of n = 4 biological replicates. Bar represents the MIC determined based on an average OD600 of n = 4 biological replicates. (Bottom) Line plots of C. difficile OD600 at 45 h in untreated fresh media (gray) or media supplemented with 1 mM metal (blue) in the presence of metronidazole (MTZ). Each x-axis is semi-log scale. Data points represent biological replicates. Lines indicate the average of n = 4 biological replicates. (B) Bar plot of delta Ct (ΔCt) values of 3 genes in C. difficile grown in fresh media, D. piger spent media (DP SM), or D. piger spent media with metal supplementation based on qRT-PCR. Data points represent biological replicates, with each point calculated as the average of 3 technical replicates. Bar indicates the average of n = 4 biological replicates. Asterisks indicate significant difference (*P < 0.05, **P < 0.01, ***P < 0.001, “ns” P > 0.05) according to an unpaired t test. The data underlying all panels in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
(A) Pie charts of the change in C. difficile MIC in pairwise and multispecies communities compared to monoculture. (B) Pie charts of the growth enhancement of C. difficile in pairwise and multispecies communities. Growth enhancement is defined as a maximum subMIC fold change that is greater than 1 and is greater than the maximum subMIC fold change in C. difficile monoculture for that antibiotic (i.e., greater than 1.61 for metronidazole). The data underlying all panels in this figure can be found in DOI: 10.5281/zenodo.7626486.
(TIF)
MICs in μg/mL. C. difficile monoculture MIC from same experimental day given for comparison, as monoculture MIC can vary due to preculture conditions and glycerol stock batches.
(XLSX)
MICs in μg/mL. C. difficile monoculture MIC from same experimental day given for comparison, as monoculture MIC can vary due to preculture conditions and glycerol stock batches. “No growth” represents no growth of C. difficile across all concentrations and therefore no MIC could be determined.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
The datasets and computer code used in this study are available in the following databases: • Species abundance data, CFU data, hydrogen sulfide data and qRT-PCR data are available from Zenodo: https://doi.org/10.5281/zenodo.7626486 • RNA-seq data is available from Zenodo: https://doi.org/10.5281/zenodo.7049035 and https://doi.org/10.5281/zenodo.7049027 • Modeling computer scripts are available from Zenodo: https://doi.org/10.5281/zenodo.7726490.