Abstract
Vibrio vulnificus is an opportunistic gram-negative pathogen that commonly contaminates oysters. Predisposed individuals who consume raw oysters can die within days from sepsis, and even otherwise healthy people are susceptible to serious wound infection after contact with contaminated seafood or seawater. Numerous secreted and cell-associated virulence factors have been proposed to account for the fulminating and destructive nature of V. vulnificus infections. Among the putative virulence factors is an elastolytic metalloprotease. We cloned and sequenced the vvpE gene encoding an elastase of V. vulnificus ATCC 29307. The functions of the elastase were assessed by constructing vvpE insertional knockout mutants and evaluating phenotypic changes in vitro and in mice. Although other types of protease activity were still observed in vvpE mutants, elastase activity was completely absent in the mutants and was restored by reintroducing the recombinant vvpE gene. In contrast to previous characterization of elastase as a potential virulence factor, which was demonstrated by injecting the purified protein into animals, inactivation of the V. vulnificus vvpE gene did not affect the ability of the bacteria to infect mice and cause damage, either locally in subcutaneous tissues or systemically in the liver, in both iron-treated and normal mice. Furthermore, a vvpE mutant was not affected with regard to cytolytic activity toward INT407 epithelial cells or detachment of INT407 cells from culture dishes in vitro. Therefore, it appears that elastase is less important in the pathogenesis of V. vulnificus than would have been predicted by examining the effects of administering purified proteins to animals. However, V. vulnificus utilizes a variety of virulence factors; hence, the effects of inactivation of elastase alone could be masked by other compensatory virulence factors.
The pathogenic marine bacterium Vibrio vulnificus is the causative agent of food-borne diseases such as life-threatening septicemia and possibly gastroenteritis in individuals with underlying predisposing conditions such as liver damage, excess levels of iron, and immunocompromised conditions (2, 14). Wound infections result from exposure to seawater or from the handling of shellfish contaminated with V. vulnificus. Mortality from septicemia is very high (>50%), and death may occur within 1 to 2 days after the first signs of illness (14, 47). Several potential virulence factors including an endotoxin, a polysaccharide capsule (46, 55, 57), iron-sequestering systems (19, 54), a cytolytic hemolysin (43, 53), an elastase (16, 24, 36), a phospholipase A2 (48), and other exotoxins have been identified for V. vulnificus. However, to date, only the capsule (55) and iron-sequestering systems (19) have been confirmed as virulence factors by using the molecular version of Koch's postulates (6, 11), in which mutations are constructed in genes encoding putative virulence factors, followed by complementation of any observed attenuating phenotypes. It is interesting that a mutation in the cytolytic hemolysin of V. vulnificus exhibited no attenuating effect in mouse models of disease (52). Most recently, a pleiotropic mutation in a gene encoding prepilin peptidase was shown to significantly attenuate the virulence of V. vulnificus in mice (37). The prepilin peptidase mutant was defective in the secretion of cytolysin, elastase, chitinase, and probably other proteins, so it is difficult to assign the attenuation to a particular effector protein.
The proteolytic activity of V. vulnificus has been characterized as elastase, collagenase, and caseinase (16, 24). The V. vulnificus elastase activity is from a neutral metalloprotease, and the characteristics of the protease as a potential virulence factor have been studied primarily using the purified protein in animal models (17, 23, 25–28, 32). Injection of purified elastase could reproduce many of the observed aspects of disease caused by V. vulnificus, including dermonecrosis, destruction of tissues, edema, and ulceration. These diverse activities are believed to be caused by the proteolytic degradation or inactivation of biologically important host proteins and immune system components such as collagen, fibrin, and complement. Conversely, increased vascular permeability is stimulated by the activation of Hagemann factor and prekallikrein directly by the elastase, leading to the production of bradykinin (23, 30, 32). Additionally, the activity of the elastase toward the host iron-binding proteins is involved in the utilization of heme and iron (34, 35). More direct evidence for the relevance of these diverse biological and biochemical activities during infection is based on injecting biochemical inhibitors of protease activity or neutralizing antibodies during infection of experimental animals (20, 27). Additionally, some data have been reported on undefined chemically induced mutants deficient in the production of elastase (27). However, no definitive analysis of the role of the V. vulnificus elastase by means of the construction of a defined mutation has been reported.
Recently the gene that encodes a V. vulnificus protease was cloned and sequenced (4, 5). The deduced gene product was predicted to be a 609-amino-acid polypeptide, with the mature protease having a molecular mass of 45 kDa and consisting of 413 amino acids generated by deletion of the N-terminal 196 amino acids. By using the mature protease purified from recombinant Escherichia coli, two functional domains, a 35-kDa N-terminal domain required for catalytic activity and a 10-kDa domain required for attachment to the substrate, were identified (31).
To study the role of the elastase in the pathogenesis of infection, we constructed by allelic exchange two V. vulnificus mutants that no longer produced elastase. Using both iron-treated and normal mice, we observed no alteration in virulence as determined by levels of local and systemic infection or histopathology. Furthermore, the ability of V. vulnificus to lyse or cause the detachment of cultured epithelial cells was not affected by the protease mutation.
MATERIALS AND METHODS
Strains, plasmids, media, and culture conditions.
The strains and plasmids used in this study are listed in Table 1. E. coli strains used for plasmid DNA replication or conjugational transfer of plasmids were grown in Luria-Bertani (LB) broth or on LB broth containing 1.5% (wt/vol) agar. Nutrient agar plates supplemented with 1.5% (wt/vol) skim milk were used for screening E. coli transformants carrying and expressing the recombinant V. vulnificus elastase gene. Unless noted otherwise, V. vulnificus strains were grown in LB medium supplemented with 2.0% (wt/vol) NaCl (LBS). For mouse and cell culture experiments, V. vulnificus strains were grown in LB broth containing 0.85% (wt/vol) NaCl (LBN). For 50% lethal dose (LD50) experiments, vibrios were grown in brain heart infusion broth containing 2.5% (wt/vol) NaCl (BHI-N). When required, appropriate antibiotics were added to the media as follows: ampicillin at 100 μg/ml, chloramphenicol at 10 μg/ml, kanamycin at 50 μg/ml, and tetracycline at 10 μg/ml. All medium components were purchased from Difco (Detroit, Mich.), and chemicals were purchased from Sigma (St. Louis, Mo.).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| Bacterial strains | ||
| V. vulnificus | ||
| ATCC 29307 | Clinical isolate; virulent | Laboratory collection |
| MO6-24/O | Clinical isolate; virulent | 55 |
| KC64 | ATCC 29307 vvpE::pKC9844; elastase deficient | This study |
| CMM111 | MO6-24/O vvpE::pKC9844; elastase deficient | This study |
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZ ΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Laboratory collection |
| SY327λpir | Δ(lac pro) argE(Am) recA56 gyrA rpoB λpir; host for π-requiring plasmids | 21 |
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu λpir; Kmr; host for π-requiring plasmids; conjugal donor | 21 |
| Plasmids | ||
| pNQ705 | Cloning vector; R6K γ ori (requires π); oriT of RP4; Cmr | 22 |
| pRK415 | IncP ori; broad-host-range vector; oriT of RP4; Tcr | 13 |
| pKC980 | pUC18 with 2.5-kb partial Sau3A fragment encoding vvpE; Apr | This study |
| pKC9844 | pNQ705 with 0.6-kb HindIII-PstI fragment encoding internal coding sequence of vvpE; for allelic exchange; Cmr | This study |
| pRKY980 | pRK415 with 2.5-kb EcoRI-SalI fragment carrying vvpE from pKC980; Tcr | This study |
Apr, ampicillin resistant; Cmr, chloramphenicol resistant; Kmr, kanamycin resistant; Tcr, tetracycline resistant.
Measurement of cell growth and enzyme activities.
For comparison of the growth rates and protease activities of parental, wild-type V. vulnificus ATCC 29307 and its elastase mutant, KC64, 50-ml cultures of nutrient broth in 250-ml Erlenmeyer flasks were inoculated with an initial cell density (measured as the optical density at 600 nm [OD600]) of approximately 0.005 and were incubated at 30°C with shaking. The inocula were from late-exponential-phase cultures in LBS. Samples of 5 ml were removed at the indicated times for determination of cell density, total protease activity, and elastase activity. Bacterial growth was monitored by measuring the OD600 of cultures. For measurements of enzyme activities in supernatants, bacterial cells were removed from cultures by centrifugation at 10,000 × g for 5 min and the supernatants were filtered through 0.45-μm-pore-size filters. Minor modifications of procedures described previously (8, 44) were used for determination of total protease activity, defined as a casein-hydrolyzing activity, and of elastase activity. For total protease activity, the reaction was initiated by addition of 100 μl of filtered supernatant as an enzyme source to 2 ml of 0.25-mg/ml azocasein in 10 mM Tris-HCl buffer (pH 7.5). After incubation for 1 h at 30°C, the reaction was stopped by addition of 2 ml of 8% (wt/vol) trichloroacetic acid. The reaction mixture was clarified by centrifugation, and 2 ml of the supernatant was transferred to a new tube. Color development was enhanced by addition of 2 ml of 0.5 M sodium hydroxide, and the absorbance at 400 nm was measured. For elastase activity, 100 μl of enzyme source was added to 1 ml of a solution containing 20 mg of elastin-Congo red/ml in 10 mM sodium phosphate (pH 7.0). The resulting reaction mixture was incubated for 4 h at 37°C, and the absorbance at 495 nm was determined. One unit of enzyme activity is defined as an increase in absorbance of 0.001 per hour for total protease activity and of 0.01 per hour for elastase activity. The mean of triplicate results was used.
DNA techniques, cloning, and sequencing.
Isolation of genomic or plasmid DNA and transformation of E. coli strains were carried out according to the procedures outlined by Sambrook et al. (42). Restriction enzymes and DNA-modifying enzymes were used as recommended by the supplier (Promega, Madison, Wis.). Primary DNA manipulations were carried out in E. coli DH5α, and restriction mapping was used to confirm that transformants contained the appropriate plasmids.
For cloning the protease gene, genomic DNA isolated from V. vulnificus ATCC 29307 was partially digested with Sau3AI. DNA fragments in the 2- to 6-kb range were purified from agarose gels by using the Geneclean II kit (Bio 101, Inc., Vista, Calif.) and used for ligation into pUC18 digested with BamHI. Competent cells of E. coli DH5α were transformed with the ligation products, and the transformants exhibiting clear zones around colonies on nutrient agar-skim milk plates were identified. One such clone contained a 2.5-kb insert and was named pKC980.
The nucleotide sequence of the entire 2.5-kb insert of pKC980 was determined by primer walking using the dideoxy-chain termination method with Top DNA polymerase (Bioneer, Seoul, Korea). Comparisons of nucleotide and amino acid sequences were conducted using BLAST (basic local alignment search tool) (1, 9).
Insertional inactivation of the elastase gene by allelic exchange.
To construct mutant V. vulnificus strains deficient in the metalloprotease, a mutation was introduced into the vvpE gene carried by pKC980. The 5′ end and 3′ end of the DNA insert of pKC980, corresponding to the regions encoding the N-terminal 24 amino acids and the C-terminal 388 amino acids of the elastase, respectively, were removed by digestion with PstI and HindIII. The overhang ends were blunt ended using the Klenow fragment of DNA polymerase I. The resulting 0.6 kb-DNA fragment was isolated from an agarose gel and was ligated into the allelic exchange suicide vector pNQ705 (22), which had been linearized with EcoRV. The resulting plasmid, pKC9844, encodes the 5′- and 3′-truncated vvpE gene as depicted in Fig. 1A. Since pNQ705 has the R6K γ origin for replication requiring the π protein (15) and can replicate only in bacterial hosts carrying the pir gene encoding the π protein, the pKC9844 allelic exchange recombinant was transformed into E. coli SY327λpir (22).
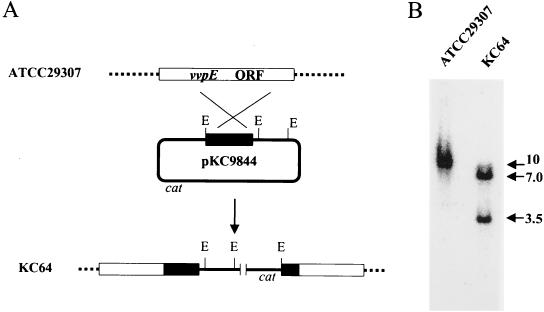
FIG. 1.
Construction of vvpE isogenic mutant by insertional mutagenesis. (A) Homologous recombination between strain ATCC 29307 and plasmid pKC9844 resulted in insertional mutation of vvpE in strain KC64. Dashed lines, the V. vulnificus chromosome; solid line, the plasmid sequence; open rectangles, the target vvpE gene; solid rectangles, the truncated vvpE gene; large X, genetic crossover. Abbreviations: E, EcoRI; cat, chloramphenicol acetyltransferase gene. (B) Southern blot analysis of strain KC64, generated by homologous recombination. Genomic DNAs from strain ATCC 29307 and strain KC64 were digested with EcoRI and hybridized to a 32P-labeled DNA probe consisting of a HindIII-PstI fragment internal to the vvpE coding sequence. The positions of hybridization are indicated in kilobases at the right. Details of the procedure are described in Materials and Methods.
The truncated vvpE gene was then introduced into the V. vulnificus chromosome by homologous recombination as follows. pKC9844 was transformed into E. coli SM10λpir, which supplies tra gene products to mobilize pKC9844 for conjugation via the RP4 origin of transfer, oriT (21, 45). E. coli SM10λpir(pKC9844) was used as a conjugational donor to deliver pKC9844 into the wild-type V. vulnificus strains ATCC 29307 and MO6-24/O. Conjugation was carried out by methods described previously (45), with slight modifications. Briefly, the recipient strains, V. vulnificus ATCC 29307 and MO6-24/O, and the donor strain, SM10λpir(pKC9844), were grown overnight on agar plates, and cells were recovered with sterile cotton swabs. Both cell masses were spotted and mixed on LB agar. The mixture of cells was incubated for at least 8 h, suspended in 1 ml of saline, and then spread on TCBS (thiosulfate citrate bile salts) agar (to select against the E. coli donor) supplemented with skim milk (as an indicator for protease activity) and chloramphenicol (to select for pKC9844). The desired transconjugants were selected by chloramphenicol resistance and screened for the inability to exhibit clearing of the skim milk. Potential mutants were subsequently tested for lack of elastase activity. The V. vulnificus elastase mutants chosen for further analysis were named KC64 and CMM111, derived from the parental strains ATCC 29307 and MO6-24/O, respectively.
Southern blot analysis.
Approximately 10 μg of genomic DNA isolated from V. vulnificus ATCC 29307 and its protease mutant, KC64, was digested completely with EcoRI and separated on a 0.7% (wt/vol) agarose gel. After transfer to a nitrocellulose membrane (Schleicher and Schuell, Keene, N.H.), the DNA was fixed by UV irradiation and hybridized with a 0.6-kb PstI-HindIII DNA probe representing the internal sequences of the vvpE coding region. The probe was labeled with [α-32P]dCTP using the Prime-a-gene labeling system (Promega). The prehybridization and hybridization solutions consisted of 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt's reagent, 0.5% (wt/vol) sodium dodecyl sulfate (SDS), and 100 μg of denatured, fragmented salmon sperm DNA/ml. Prehybridization and hybridization were done for 2 and 16 h, respectively, at 65°C. After hybridization, the membranes were washed at room temperature for 2 h in 0.1× SSC–0.4% SDS and then at 68°C for 1 h in the same solution. The blot was exposed using a phosphorimage analyzer (BAS1500 model; Fuji Photo Film Co. Ltd., Tokyo, Japan).
Analysis of expression of vvpE mRNA and VvpE protein from insertion mutants. (i) Northern blot analysis of vvpE mRNA.
For Northern blot analysis, a 1.2-kb HindIII DNA fragment derived from the region of vvpE encoding the C-terminal 389 amino acids was labeled and used as a probe. Total cellular RNA from cultures of KC64 and ATCC 29307 grown to an OD600 of 1.5 was isolated by using the Trizol reagent kit according to the manufacturer's specifications (GIBCO-BRL, Gaithersburg, Md.) and was suspended in diethyl pyrocarbonate-treated water. RNA was separated by agarose gel electrophoresis, transferred to a nylon membrane, and hybridized as described previously (33), with slight modifications. Briefly, 5.5 μl of RNA (approximately 20 μg) was mixed with 1 μl of 10× MOPS buffer [1× MOPS buffer is 0.02 M 3-(N-morpholino)propanesulfonic acid (pH 7.0), 5 mM sodium acetate (pH 5.2), and 1 mM EDTA], 3.5 μl of 37% (wt/vol) formaldehyde, and 10 μl of deionized formamide. The mixture was heated for 10 min at 60°C and then mixed with 10 μl of loading buffer (50% [vol/vol] glycerol, 0.2% [wt/vol] xylene cyanole, 0.2% [wt/vol] bromophenol blue). The RNA was separated by electrophoresis in a 1.2% (wt/vol) agarose gel containing 1.1% (wt/vol) formaldehyde and 1× MOPS buffer. The gel was soaked for 45 min in 10× SSC, and the separated RNA was transferred to a nylon membrane (Nytran; Schleicher and Schuell) and fixed by UV irradiation. The membrane was hybridized and developed by the same procedure as that used for Southern blot analysis, except that the 32P-labeled 1.2-kb HindIII DNA probe was used.
(ii) Western blot analysis of VvpE protein.
Wild-type and VvpE− mutant V. vulnificus strains were grown in LBN broth to an OD420 of approximately 3.25. Cells were harvested by centrifugation, and supernatants were filter sterilized through 0.22-μm-pore-size filters. Supernatants were concentrated 200-fold by precipitation with trichloroacetic acid and suspension in water. Cells and concentrated supernatants were boiled in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer (18). Equivalent amounts of cells and supernatants of the various strains were electrophoresed by SDS-PAGE (4 to 15% [wt/vol] acrylamide, Tris-HCl precast Ready Gel; Bio-Rad, Hercules, Calif.), and proteins were transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore Corp., Bedford, Mass.) using Towbin transfer buffer (49). One of the duplicate membranes was stained with Coomassie brilliant blue, and the second was examined for the presence of VvpE protein by Western blot analysis using the immunoglobulin G (IgG) fraction of rabbit anti-V. vulnificus elastase serum, kindly provided by Shin-Ichi Miyoshi (27). Briefly, the gel was presoaked with phosphate-buffered saline (PBS) containing 0.05% (wt/vol) Tween 20 (PBS-T) and then incubated with a 1:500 dilution of the anti-elastase serum in PBS-T for 1.5 h at room temperature. The membrane was washed three times with PBS-T and then incubated with goat anti-rabbit IgG conjugated with alkaline phosphatase (Sigma). After three more washes, the antibody-antigen complexes were visualized by using the 5-bromo-4-chloro-3-indolylphosphate (BCIP)-nitroblue tetrazolium (NBT) substrate (Sigma).
Subcutaneous infection of mice.
We examined the virulence of V. vulnificus ATCC 29307 and KC64 using both iron dextran-treated and normal ICR mice (specific pathogen free; Harlan Sprague-Dawley, Indianapolis, Ind.), as described elsewhere (46a). Briefly, female mice from 7 to 11 weeks of age were housed under specific-pathogen-free conditions. For experiments involving iron treatment, mice were injected intraperitoneally with 250 μg of iron dextran (Sigma)/g of body weight immediately before subcutaneous (s.c.) injection into the lower back with bacterial cells suspended in buffered saline containing 0.01% (wt/vol) gelatin (BSG). Between 15 and 24 h later, the mice were euthanized with carbon dioxide, the s.c. lesion was removed for enumeration of bacterial CFU and histology, and a portion of the liver was removed for enumeration of CFU. To enumerate CFU in tissues, samples were weighed, suspended in BSG, and homogenized in glass tissue homogenizers. Homogenates were diluted in BSG and plated on LBN. When mice died before harvest, they were assigned the highest value obtained for the same tissue and the same strain in the same experiment. Occasionally, mice were unaffected by the particular V. vulnificus strains examined. These apparent inoculation failures are reported as such. Bacterial cultures were initially grown as overnight, static LBN cultures at room temperature. On the day of the infection, the overnight cultures were diluted 1:20 into fresh prewarmed (37°C) LBN and incubated with shaking until the OD420 reached 1.0 to 1.5. The cells were harvested by centrifugation and then suspended and diluted in BSG for inoculation. For iron dextran-treated mice, bacterial inocula of ATCC 29307 and KC64 ranged from 84 to 206 CFU per mouse for one experiment and from 530 to 560 CFU per mouse for a repetition. For normal mice, inocula were approximately 2 × 107 CFU per mouse for one experiment and 2 × 106 to 4 × 106 CFU per mouse for a repetition. For MO6/24-O and CMM111, inocula for iron dextran-treated mice ranged from 100 to 900 CFU/mouse among three different experiments. All manipulations of mice were approved by the University of Florida Institutional Animal Care and Use Committee.
Histological analysis.
Skin lesions were fixed in buffered 10% (vol/vol) formalin, embedded in paraffin, and sectioned at 5 μm at the University of Florida Department of Pathology, Immunology, and Laboratory Medicine Diagnostic Referral Laboratory. Sections were stained with hematoxylin and eosin. Initial examination by the veterinary pathologist for lesion characteristics and severity was conducted in a blinded manner.
LD50 determination.
Bacteria were grown in BHI-N broth overnight at 25°C. The following day, 0.1 ml of the culture was inoculated into 100 ml of BHI-N broth and shaken at 25°C. After 4 h of cultivation, bacterial cells were harvested by centrifugation and suspended in PBS to appropriate concentrations. Groups of six to eight normal female CD-1 mice (specific pathogen free; 8 weeks old; Daehan Animal Co., Taejon, Korea) were injected intraperitoneally with 0.2 ml of serial dilutions of bacterial suspensions. The infected mice were observed for 48 h, and the LD50s were calculated by the method of Reed and Muench (40).
Cell culture experiments.
To examine the effects of the vvpE mutation on the ability of V. vulnificus to damage epithelial cells, we performed three different assays using INT407 intestinal epithelial cells. Bacterial cells for infection were grown as described above for animal infection, except that the bacteria were ultimately suspended in cell culture medium, Dulbecco's modified Eagle medium (DMEM) containing 10% (vol/vol) fetal calf serum (Life Technologies/BRL), instead of BSG. INT407 cells grown in DMEM containing 10% fetal calf serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml were seeded to achieve approximately 70% confluence in 24-well culture plates (Costar) for the day of infection. One hour before infection, antibiotic-containing medium was replaced with antibiotic-free medium. The INT407 cells were infected at a multiplicity of infection of 5 and allowed to incubate at 37°C for 1 h, at which time gentamicin was added to a final concentration of 100 μg/ml to kill the bacteria. This was done to prevent the bacteria from rapidly multiplying in the cell culture medium and destroying the monolayers before cytopathology could be examined. Bacterial cells and any extracellular products remained in the cell culture wells for the duration of the experiment. Twenty-four hours later, the infected cell cultures were assayed for lysis of host cells by using the Cytotoxicity Detection Kit (Boehringer Mannheim), killing and/or destruction of the cell monolayer with the CellTiter 96 AQueous Cell Proliferation Assay kit (Promega), and detachment of host cells by a modification of a crystal violet staining technique (41).
The Cytotoxicity Detection Kit measures lactate dehydrogenase (LDH) activity released into the culture supernatant by lysed cells. At harvest time, a sample of supernatant from experimental wells (either infected or uninfected) was centrifuged at 250 × g for 10 min to remove cells and debris. To measure total LDH activity in either infected or uninfected wells, Triton X-100 was added to the wells containing host cells and the remaining supernatant at a final concentration of 1% (vol/vol) to lyse host cells. The colorimetric assay to measure LDH activity was then performed according to the manufacturer's instructions, by reading the A490. For each well, we computed the LDH activity in the supernatant divided by the total LDH activity in the well. The percent LDH release by uninfected cells, approximately 15%, was subtracted from the percent LDH release in V. vulnificus-infected wells. Triplicate wells were run for each sample, and the experiment was performed at least twice.
The CellTiter Proliferation Assay measures nonspecific dehydrogenase activity of metabolically active cells that remain attached to the culture dish. After the 24-h treatment period, the wells were rinsed twice with Hanks balanced salt solution (HBSS) to remove dead and detached cells. DMEM (0.5 ml) containing 100 μg of gentamicin/ml was added back, followed by 0.1 ml of the tetrazolium reagent. The reaction was developed and the A490 was read according to the manufacturer's instructions. We report the percent loss of viable and attached cells in infected wells compared to that in uninfected wells. Triplicate wells were run for each sample, and the experiment was performed three times.
The crystal violet staining assay measures the amount of cell mass remaining attached to culture dishes after infection. This assay was originally developed to measure the effects of tumor necrosis factor alpha on cell lines (41). After the 24-h treatment period, wells were rinsed twice with PBS, followed by staining with 0.167% (wt/vol) crystal violet diluted in PBS. After 10 min the wells were rinsed with PBS until no further leaching of crystal violet was observed. The crystal violet stain in attached cells was released by rinsing the wells with 95% (vol/vol) ethanol. The relative concentration of crystal violet was measured by the A490. As for the CellTiter Proliferation Assay, we calculated the percentage of INT407 cells detached from infected wells compared with that for uninfected wells. Triplicate wells were run for each sample, and the experiment was performed twice.
Nucleotide sequence accession number.
The nucleotide sequence of V. vulnificus ATCC 29307 vvpE was submitted to GenBank and was assigned accession number AF102028.
RESULTS
Cloning and sequencing analysis of vvpE from V. vulnificus ATCC 29307.
A fragment of genomic DNA from V. vulnificus ATCC 29307 that conferred elastolytic activity on E. coli DH5α was cloned as described in Materials and Methods, yielding plasmid pKC980. The nucleotide sequence of the 2.5-kb DNA insert containing the vvpE structural gene and the upstream regulatory region was determined, revealing a coding region consisting of 1,830 nucleotides. The deduced amino acid sequence revealed a protein of 609 amino acids with a theoretical molecular mass of 65,965 Da and a pI of 6.07. The nucleotide sequence of V. vulnificus ATCC 29307 vvpE was submitted to GenBank and was assigned accession number AF102028.
The N-terminal 20-amino-acid sequence previously determined from purified metalloprotease of V. vulnificus by Kothary and Kreger (17) is present within the deduced 609-amino-acid polypeptide encoded by vvpE. The two amino acid sequences are nearly identical, with the initial residue of the Kothary and Kreger sequence corresponding to our Ala residue at deduced position 197. There was a single difference at amino acid residue 200, with an Asn in the Kothary and Kreger sequence and an Asp in VvpE. Thus the 609-amino-acid VvpE polypeptide is presumed to be processed by deleting the N-terminal 196 amino acids to form the mature protein. Furthermore, the N-terminal amino acids show the typical pattern of putative leader peptides, as observed in other metalloproteases (12). The signal peptide is predicted to be cleaved between Ala24 and Ala25 based on the von Heijne method (51). The mature elastase protein is therefore deduced to consist of 413 amino acids, with a calculated molecular weight of 45,430 Da. This result was not unexpected, since many other bacterial metalloproteases have been shown to rely on signal and leader sequences to aid in transport across the bacterial membrane, in addition to undergoing other stages of maturation. Furthermore, the calculated molecular weight and that observed by Western blot analysis (see below) are in good agreement with those observed for the purified metalloprotease of V. vulnificus (17, 24).
A database search for similar sequences revealed two other protease genes, vvp and empV, cloned from V. vulnificus strains YJ061 and CKM-1, respectively, whose nucleotide sequences showed high levels of identity with the vvpE sequence. The vvpE coding sequence is between 97 and 99% identical with those of empV and vvp, respectively. The deduced amino acid sequence of VvpE was 98% (599 of 609 amino acids) and 96% (570 of 589 amino acids) identical to those of the EmpV and Vvp proteases of V. vulnificus, respectively. Furthermore, the amino acid sequence of VvpE contains the conserved regions, such as zinc-binding residues (H-343-X-V-S-H-347) and active-site residues (V-425-H-X-X-S-G-X-X-N-X-A-X-Y-437), observed in many other extracellular zinc-containing metalloproteases of Vibrio spp. (12). All of this information confirmed that the vvpE gene encodes the metalloprotease gene of V. vulnificus ATCC 29307.
Construction of V. vulnificus isogenic mutants deficient in elastase activity.
We used standard suicide vector methods to insertionally inactivate the vvpE genes of the wild-type V. vulnificus strains ATCC 29307 and MO6-24/O. Plasmid pKC9844, encoding 5′- and 3′-truncated vvpE, was integrated into the genomes of V. vulnificus ATCC 29307 and V. vulnificus MO6-24/O by a single homologous recombination event, leading to a partial diploid of the vvpE gene consisting of two mutant genes truncated in the N terminus- or C terminus-coding regions separated by intervening vector DNA (Fig. 1A). The insertional disruption of vvpE in the mutants was confirmed by Southern blot analysis (Fig. 1B). When wild-type V. vulnificus ATCC 29307 genomic DNA digested with EcoRI was hybridized with the internal coding sequence probe, a 10-kb band was observed. A representative strain of the caseinase-negative transconjugants, KC64, was chosen from those insertional mutants whose EcoRI-digested genomic DNA produced bands of approximately 3.5 and 7.0 kb hybridizing with the 32P-labeled vvpE probe, as shown in Fig. 1B. Since plasmid pKC9844 carries three EcoRI sites from the vector, pNQ705, this pattern of hybridization confirms that the vvpE gene of KC64 was disrupted by insertion of vector DNA of pKC9844, as depicted in Fig. 1A. Similarly, the vvpE mutant strain CMM111 was constructed from parental strain MO6-24/O.
To determine the stability of the insertional mutation, V. vulnificus KC64 was grown overnight without chloramphenicol selection. The inserted plasmid DNA was stably maintained, as determined by maintenance of chloramphenicol resistance (all of more than 500 colonies tested) and Southern analysis (data not shown). The mutation was also stably maintained during growth in mice, since most, if not all, colonies of KC64 isolated from infected mice retained chloramphenicol resistance and lack of elastase activity (data not shown).
Total protease and elastase activities of V. vulnificus KC64.
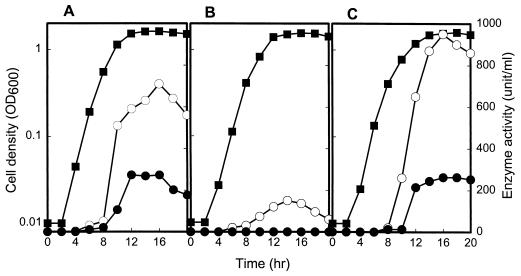
The mutant strain KC64 was characterized for total protease and elastase activities relative to those of the wild-type strain ATCC 29307. For ATCC 29307 both total protease activity and elastase activity were produced during growth at mid-exponential phase and reached a maximum during stationary phase (Fig. 2A). The disruption of vvpE in KC64 resulted in a complete loss of elastase activity and also reduced the production of total protease activity. The residual level of total protease activity in KC64 corresponded to approximately one-third that in the wild type (Fig. 2B). These data demonstrated that the vvpE gene encoded the elastase activity of V. vulnificus. The observation that the mutant still exhibited protease activity revealed the existence of at least one more protease being produced by V. vulnificus ATCC 29307. Therefore, we have designated vvpE to represent V. vulnificus elastase, in order to differentiate it from the other genes encoding other potential proteases of V. vulnificus.
FIG. 2.
Growth kinetics and activities of total protease and elastase in V. vulnificus strains. V. vulnificus strains were grown in nutrient broth as described in Materials and Methods. Samples removed at the indicated times from cultures of strains ATCC 29307 (A), KC64 (B), and KC64(pRKY980) (C) were analyzed. ■, cell density; ○, total protease activity; ●, elastolytic protease activity.
Although it seemed unlikely that the decrease in protease activity by two-thirds resulted from polar effects of the vvpE insertional mutation on downstream genes, this possibility could not be ruled out a priori. Therefore, we investigated whether reintroduction of recombinant vvpE could complement the lack of elastase activity of KC64 cells. A 2.5-kb EcoRI-SalI fragment carrying vvpE was isolated from pKC980 and was subcloned into vector pRK415 (13) digested with HindIII. Since pRK415 has an IncP1 origin and RP4 oriT, the resulting plasmid, pRKY980, was mobilizable into KC64 by conjugation. The protease activity of KC64(pRKY980) was restored to a level comparable to the wild-type level of ATCC 29307 (Fig. 2C). Therefore, the decreased protease activity of KC64 resulted from inactivation of functional vvpE rather than from any polar effects on any genes downstream of vvpE.
Confirmation of lack of vvpE mRNA and VvpE protein in mutant strains.
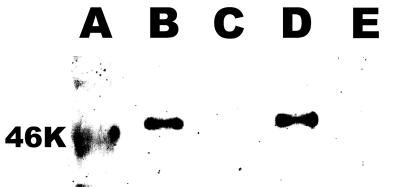
Because the vvpE mutants were created by insertional inactivation of the wild-type gene by using a suicide plasmid encoding a DNA fragment internal to the coding sequence, it was possible, though unlikely, that one of the two resulting partial genes, that encoding the C terminus, could still be expressed and yield a mature peptide. To examine this possibility, we performed both Northern and Western blot analyses of the parental and mutant strains. Northern blot analysis using a probe to the 3′ end of the vvpE coding sequence identified a vvpE transcript of approximately 2.0 kb produced by the wild-type strain ATCC 29307 but not by the mutant strain KC64 (data not shown). Western blot analysis of parental and vvpE mutants for both sets of V. vulnificus strains demonstrated that no anti-elastase-reactive proteins were produced by the mutants in either culture supernatants (Fig. 3) or whole cells (data not shown). In addition to the lack of elastase activity (Fig. 2), these combined results confirm that the vvpE gene disruptions eliminate all detectable expression of the vvpE gene and VvpE protein. This result was not unexpected given that the mutant copy of the vvpE gene encoding the C-terminal portion of VvpE had no promoter, ribosome-binding site, or start codon.
FIG. 3.
Lack of secretion of VvpE by mutant V. vulnificus. Parental and vvpE mutant V. vulnificus strains were grown in LBN to late-exponential-phase growth. Cells (not shown) and supernatants were then examined for the presence of VvpE elastase protein by Western blot analysis using a rabbit anti-V. vulnificus elastase antibody (27), as described in Materials and Methods. Lane A, 46-kDa molecular mass marker; lanes B and D, the wild-type parental strains ATCC 29307 and MO6-24/0, respectively; lanes C and E, the corresponding VvpE− mutant strains KC64 and CMM111, respectively.
vvpE is not essential for virulence of V. vulnificus in mice.
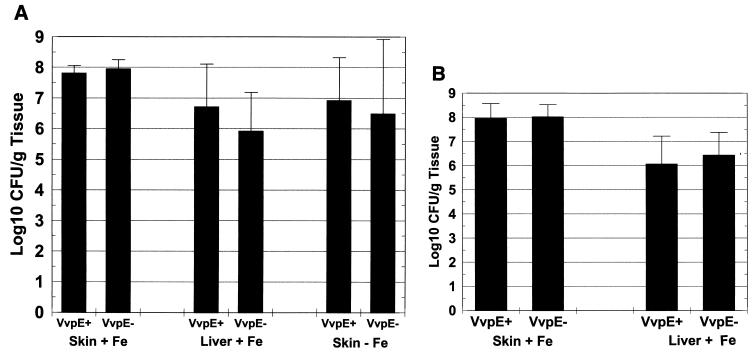
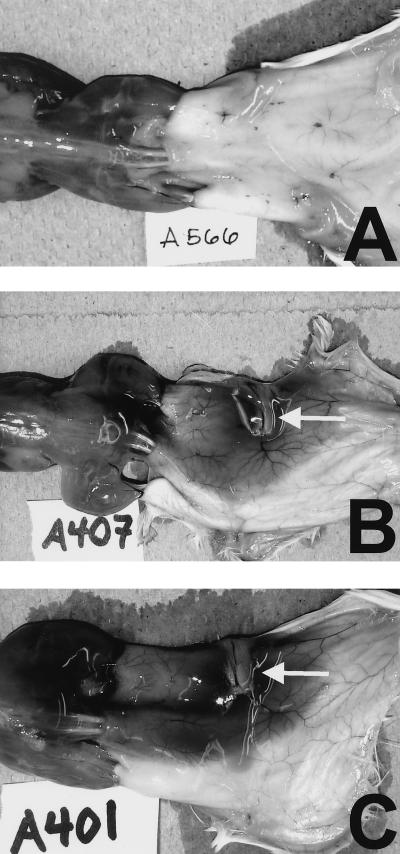
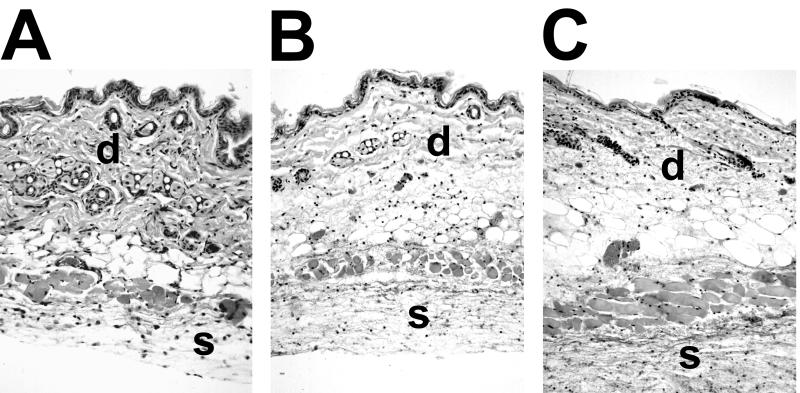
Several studies have demonstrated numerous activities of metalloproteases of V. vulnificus when they are injected into animals (17, 23, 25–28, 32). The effects observed were consistent with the pathogenesis of infection in animals, including vasodilation, increased vascular permeability, edema, and necrosis. To examine the effects of the lack of elastase activity on the virulence of V. vulnificus during infection, we used both iron dextran-treated and normal mice inoculated s.c. with either wild-type V. vulnificus ATCC 29307 or the VvpE− strain KC64. In iron dextran-treated mice, s.c. injection of approximately 102 CFU of either strain resulted in extremely ill mice and occasional death within 24 h postinoculation. There was no difference between the counts of ATCC 29307 and KC64 bacteria recovered from s.c. lesions, which were as high as 108 CFU/g of tissue for both strains (Fig. 4A). The s.c. lesions produced by the parent and metalloprotease mutant strains were indistinguishable at the gross (Fig. 5) and histological (Fig. 6) levels from those observed in our analysis of other virulent V. vulnificus strains in this model (46a). In Fig. 5 it is clear that both ATCC 29307 and KC64 produced extensive hemorrhagic and edematous lesions, with dilation of the associated vasculature. The regional lymphatics were also inflamed (Fig. 5). Histopathological findings included extensive edema of the dermis and edema and necrosis of s.c. tissue (Fig. 6), with extension of infection through the dermis up to the epidermis. It is notable that, despite the red coloration of the lesions, intact red blood cells were rarely observed outside of blood vessels, most likely because they had been lysed by the hemolytic activity of the vibrios. Bacteria were easily observed in the s.c. lesion, with necrotic or degenerating polymorphonuclear leukocytes nearby. We also occasionally observed perivascular infection by either of the V. vulnificus strains localized to dilated blood vessels. To examine the effects of the vvpE mutation on the spreading of the infection beyond the site of inoculation, we examined the CFU in the livers of the s.c. inoculated mice. As was seen for infection of s.c. tissues, there was no significant difference in CFU per gram of liver tissue between the wild-type strain ATCC 29307 and the VvpE− mutant KC64, with yields of approximately 106 CFU/g of tissue (Fig. 4A). Therefore, the elastase also appeared to be dispensable for vibrios reaching and multiplying in the liver.
FIG. 4.
Lack of effect of the vvpE mutation on infection of mice by V. vulnificus. Mice were either injected intraperitoneally with iron dextran (+ Fe) or not (−Fe) immediately before s.c. inoculation of the wild-type strain V. vulnificus ATCC 29307 (VvpE+) or the elastase mutant strain KC64 (VvpE−) (A) or the wild-type V. vulnificus strain MO6-24/O (VvpE+) or the elastase mutant strain CMM111 (VvpE−) (B). Fifteen to 24 h later, CFU in skin lesion and liver were quantified as described in the text. Error bars, standard deviations. (A) For VvpE+ infection of iron-treated mice, n = 8, plus 1 unaffected mouse; for VvpE− infection of iron-treated mice, n = 10; for infections of non-iron-treated mice, n = 10. The differences in CFU per gram of tissue between the VvpE+ and VvpE− strains were not significant in any pair (P > 0.2). (B) Combined results of three experiments are shown. For VvpE+ infection of iron-treated mice, n = 10, plus 2 unaffected mice; for VvpE− infection of iron-treated mice, n = 10, plus 3 unaffected mice. The differences in CFU per gram of tissue between the VvpE+ and VvpE− strains were not significant in any pair (P > 0.75).
FIG. 5.
Dorsal view of gross pathology of s.c. lesions caused by infection with VvpE+ and VvpE− V. vulnificus strains. Mice were injected intraperitoneally with iron dextran. Immediately afterwards mice were injected s.c. in the lower right dorsal quadrant with 102 CFU of either the parental V. vulnificus strain ATCC 29307 (B) or the VvpE− mutant KC64 (C). Control mice (A) received no further injections after iron dextran. Between 15 and 24 h later, mice were euthanized, and the skin was peeled back from head to tail to reveal s.c. tissues. In panels B and C a large, edematous, hemorrhagic lesion is visible unilaterally in the area surrounding the injection site. The regional lymph node is inflamed (white arrows), and the localized vasculature is dilated. Some lesion material is adherent to the underlying musculature over the lower back, as well.
FIG. 6.
Histopathology of s.c. lesions caused by infection with VvpE+ and VvpE− V. vulnificus strains. Mice were injected intraperitoneally with iron dextran. Immediately afterwards, mice were injected s.c. in the lower right dorsal quadrant with 102 CFU of either the parental V. vulnificus strain ATCC 29307 (B) or the VvpE− mutant KC64 (C). Control mice (A) received no further injections after iron dextran. Between 15 and 24 h later, mice were euthanized, and a tissue sample from the lesion was fixed in formalin, embedded in paraffin, sectioned, and stained with hematoxylin and eosin, as described in the text. In all panels, the dermis is labeled with the latter “d.” In panel A the dermis is intact, with thick, intensely staining, and dense collagen fibers. In panels B and C the dermis is edematous, as evidenced by the less-intense, open staining. Infection by both strains of V. vulnificus extended into the dermis. The s.c. region beneath the cutaneous muscle layer is labeled with the letter “s.” In panel A the tissue is populated with healthy cells with well-defined nuclei. In panels B and C the tissues contain necrotic cells with condensed nuclei, fibrin deposition, and numerous vibrios.
Since it had been proposed that the metalloprotease could remove iron from transferrin (35) and increase the utilization of heme (34), we also examined the effects of the metalloprotease mutation on disease in non-iron-treated mice. To cause disease in this model, 106 to 107 CFU of either strain had to be injected s.c.; however, s.c. lesions were still observed with levels of infection indistinguishable between the parent and protease mutant (Fig. 4A). Recovery of bacteria from livers was mostly undetectable for either strain in non-iron-treated mice, in spite of the relatively high levels of s.c. infections. Therefore, in the mouse model of infection by s.c. inoculation, in which both local and systemic aspects of infection are measured, the VvpE metalloprotease was completely dispensable for disease.
To examine the effects of the vvpE mutation in the MO6-24/O background, we examined infection of s.c. inoculated, iron dextran-treated mice. As shown in Fig. 4B, there was no significant difference in either skin lesion or liver CFU between the parental strain MO6-24/O and the vvpE mutant CMM111. We also examined the effects of the vvpE mutation on the lethality of both the ATCC 29307 and MO6-24/O strain backgrounds in non-iron-treated mice (Table 2). The intraperitoneal LD50s of the vvpE mutant strains were very similar to those of the respective parent strains. Therefore, the lack of a significant change in the mouse virulence phenotype caused by the vvpE mutation is observed in both V. vulnificus strain backgrounds examined.
TABLE 2.
Effect of the vvpE mutation on the lethality of V. vulnificus to normal mice
| Straina | Intraperitoneal LD50 (CFU)
|
|
|---|---|---|
| Wild type | vvpE mutantb | |
| ATCC 29307 (n = 8) | 1.8 × 107 | 5.8 × 107 |
| MO6-24/O (n = 10) | 2.8 × 106 | 3.8 × 106 |
Parental V. vulnificus strain background. n, number of mice for each inoculation group, ranging from 104 to 109 CFU in 10-fold increments.
The vvpE mutants of ATCC 29307 and MO6-24/O are KC64 and CMM111, respectively (see Table 1).
The VvpE metalloprotease has no phenotype in interactions of V. vulnificus with epithelial cells in vitro.
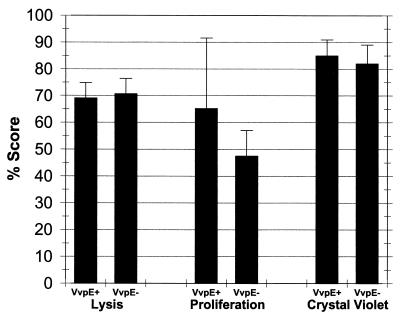
Since many of the biological activities of protease from V. vulnificus are associated with effects on host cells, we examined the parental strain, ATCC 29307, and the protease mutant, KC64, for different effects on INT407 intestinal epithelial cells. We measured the abilities of the strains to lyse the epithelial cells by release of LDH from the host cells. As shown in Fig. 7, KC64 exhibited cytolytic activity similar to that of ATCC 29307, with approximately 70% LDH release higher than the background of uninfected cells, which release approximately 15% of total LDH (P < 0.00005 compared with either V. vulnificus strain). To examine the ability of the bacteria to cause the detachment of host cells from culture dishes without necessarily lysing them, we used the CellTiter Proliferation Assay on vibrio-infected cell cultures. This assay produced somewhat variable results; however, there was no consistent difference between the abilities of the protease mutant and the wild-type parent to detach INT407 cells after 24 h of interaction (Fig. 7). In other repetitions of the experiment (not shown), the protease mutant showed either lower or higher activity than the parent strain in causing detachment of INT407 cells. Finally, we examined the abilities of the V. vulnificus strains to cause the detachment or release of the host cells from the cell culture plates, known effects of proteases, using an assay that measures crystal violet staining of the INT407 cells remaining attached to culture dishes (41). In agreement with results obtained using the CellTiter Proliferation Assay, ATCC 29307 and KC64 caused similar levels of detachment after 24 h of treatment, approximately 80 to 85% (Fig. 7). Therefore, using these in vitro cell culture assays, we could not identify a virulence-associated phenotype for the VvpE elastolytic protease by comparing the wild-type parent, ATCC 29307, with its isogenic metalloprotease mutant, KC64.
FIG. 7.
Lack of effect of the vvpE mutation on the cytopathology of V. vulnificus in INT407 cells. INT407 intestinal epithelial cells were infected at a multiplicity of infection of 5 for 1 h before addition of gentamicin at 100 μg/ml. Twenty-four hours later, lysis was measured using LDH release into culture supernatant; values are expressed as percent release from vibrio-infected cells compared with total LDH (Triton X-released) of vibrio-infected cells, with the background lysis (15%) of uninfected cells subtracted. The proliferation assay measures metabolically active cells remaining attached to culture dishes; values are expressed as percent loss compared with uninfected wells. The crystal violet assay measures percent loss of crystal violet-staining material for vibrio-infected cell cultures compared with uninfected wells. Triplicate wells of each experiment were run, and these assays were repeated at least once. The values for the VvpE+ and VvpE− strains were not significantly different in any of the three assays (P > 0.2).
DISCUSSION
Disease caused by infection with V. vulnificus is remarkable for the invasive nature of the infection, ensuing severe tissue damage, and rapidly fulminating course. Understanding the molecular pathogenesis of this multifaceted host-pathogen interaction is critical for the development of improved treatment and prevention, as well as elucidating how certain bacteria can circumvent host defenses, multiply in the host, and cause such extensive damage. The characterization of somatic as well as secreted products of V. vulnificus has yielded a large list of putative virulence attributes, whose known functions are easily imagined to explain the pathology of disease. These putative virulence factors include a carbohydrate capsule, lipopolysaccharide, a cytolysin/hemolysin, an elastolytic metalloprotease, iron-sequestering systems, a lipase, and pili. However, only the capsule and iron acquisition systems have been confirmed to be essential for virulence by the use of the molecular version of Koch's postulates (19, 55). Notably, a null mutation in the gene encoding hemolysin had no effect on virulence in mice (52). A prepilin peptidase, whose function is essential for the secretion of numerous proteins by V. vulnificus including elastase, hemolysin, and chitinase, has also been shown to be essential for virulence in mice (37). However, which of the observed or as yet unidentified secreted proteins affected by the prepilin peptidase mutation are responsible for the attenuation is unknown. In the present study we focused on analysis of the elastase, since numerous studies have shown that injection of the protease into experimental animals can reproduce many aspects of the disease process observed during experimental infection with viable V. vulnificus.
Elastase, a metalloprotease with a broad substrate specificity, including biologically important host molecules, has been suggested to be an important virulence factor of various human-pathogenic bacteria (12). The well-characterized elastase of Pseudomonas aeruginosa is capable of degrading or inactivating elastin, collagen, immunoglobulins, serum complement factors, and some plasma proteins (7, 39). This enzyme is important for massive tissue destruction, which may aid the bacteria in invading the host (7, 39). There have been several different lines of evidence leading to the hypothesis that elastase is an important, if not an essential, component of virulence for V. vulnificus during infection of animals. Many of these studies involved injection of purified protein into animals, with the production of specific symptoms or pathology observed during infection. For example, injection of elastase resulted in dermonecrosis and toxicity in mice (17). Edema can be caused by combined increases in vasodilation and vascular permeability. V. vulnificus elastase can affect vascular permeability by stimulating histamine release from mast cells (27) and by stimulating the production of bradykinin (23), which acts on the vascular endothelium. Interestingly, elastase stimulates the bradykinin pathway in two different steps: first in the initiation events by activation of Hagemann factor and later by the cleavage of prekallikrein to kallikrein (30, 32). Most recently, Miyoshi and colleagues (26) examined in great detail the pathology associated with the injection of elastase in order to understand the mechanism of edema and hemorrhage. They concluded that the elastase degraded type IV collagen of the basement membrane underlying the vascular endothelium. The lack of support for the endothelium, coupled with the increase in vascular permeability due to stimulation of histamine release and bradykinin production, the decrease in clotting due to stimulation of fibrinolysis, and necrosis of the vascular endothelium, could all contribute to the massive edema and hemorrhage observed during infection. Consistent with this hypothesis, administration of antibodies to collagen decreased the effects of infection on the vasculature.
In contrast to the reported effects of administering V. vulnificus protease in experimental animals, the analysis of down-regulation of elastase activity by host protease inhibitors has complicated the overall picture. For example, elastase is inhibited by α2-macroglobulin, normally found in plasma (16, 28, 30). It was originally proposed that the presence of α2-macroglobulin in plasma would restrict the biological action of the protease to the interstitial tissues. However, elastase activity could not be detected in interstitial fluids, even in the presence of high levels of V. vulnificus infection (3). It was later found that leakage of plasma into tissues, i.e., edema formation, caused the inhibition of elastase by α2-macroglobulin (28, 29). Therefore, it appeared that the success of the protease in causing edema could ultimately result in elimination of its activity.
Another likely unrelated function of elastase is the release of or acquisition of heme from heme-binding host proteins and iron from transferrin (34, 35). This was shown directly by demonstrating that the protease could degrade hemoglobin. In contrast to the wild-type parent, protease-deficient mutants were unable to grow in synthetic medium containing haptoglobin-hemoglobin unless V. vulnificus protease was added. Similarly, protease-deficient mutants could not acquire iron from transferrin (35).
As opposed to the studies described above, in which the biochemical activities of elastase were examined in vitro or in which the biological activities were examined in animal models, further evidence supporting a role for elastase in virulence was provided by inhibiting its function in vivo by administration of biochemical inhibitors of the enzyme, antibodies neutralizing the protease, or mutant vibrios. For example, administration of soybean trypsin inhibitor or anti-metalloprotease antibodies inhibited increased vascular permeability during infection (27). Furthermore, Miyoshi and Shinoda (27) constructed nitrosoguanidine-induced mutant V. vulnificus strains that were defective in protease activity. These mutant strains also caused less vascular permeability in rat skin and exhibited lower levels of virulence, as measured by CFU in tissues or LD50. However, the exact nature of the mutations was not determined, and, as described above for the prepilin peptidase which exhibits numerous effects on protein secretion, it is possible that factors other than metalloprotease were affected by the mutations. What was needed was the construction of a defined mutation in the elastase gene.
We therefore undertook this study to identify the function of the metalloprotease during an infectious process, rather than the artificial system of injecting purified protein, by constructing isogenic metalloprotease mutants of V. vulnificus ATCC 29307 and MO6-24/O and applying the molecular version of Koch's postulates (6, 11). When the isogenic vvpE mutants were compared with the parental strain for virulence in s.c. inoculated mice, the mutants did not show any significant differences in any aspect of the disease process (Fig. 4 through 6; Table 2). The morbidity of iron dextran-treated mice inoculated with <103 CFU of the VvpE+ and VvpE− strains was identical, and the gross pathology (Fig. 5) and histopathology (Fig. 6) of the s.c. lesions were indistinguishable. Consistent with our other studies (Starks et al., submitted), s.c. lesions were extremely edematous, with extensive necrosis of host tissue and death of phagocytes in regions of infection. The extension of infection upwards into the dermis from the s.c. inoculation site was the same for the wild type and the VvpE− mutant, suggesting that the collagenase activity of the metalloprotease was not necessary for the spread of the infection through the skin layers. These results argue that previously characterized effects on host cells and vasculature observed during V. vulnificus infection and reproduced by injection of purified metalloprotease do not, in fact, require the production of elastolytic metalloprotease in vivo during an infection. Furthermore, the quantitation of infection in s.c. tissues, which most likely reflects the growth of V. vulnificus due to degradation of host tissues, leakage of fluids from the vasculature into tissues, and putative destruction of proteinaceous antimicrobial effectors such as complement, was not significantly different between the VvpE+ and VvpE− strains (Fig. 4). In addition to the putative role of the metalloprotease in localized infection, we also examined the role of VvpE in the systemic spread of V. vulnificus to deeper tissues such as the liver. As shown in Fig. 4, there was again no significant difference in CFU recovered between wild-type and VvpE− strains. Importantly, colonies of the VvpE− mutant recovered from all tissues of mice retained their caseinase-negative phenotype, as observed upon plating on skim milk agar media. This control was essential, since the mutation we constructed could revert by excision of the suicide plasmid pKC9844 from the vvpE gene.
The metalloprotease has been reported to aid in the acquisition of iron and heme from host proteins (34, 35). We considered the possibility that using the iron dextran-treated mouse model could preclude these iron-associated functions from being important due to excess levels of iron. Therefore, we repeated s.c. inoculation of the ATCC 29307 pair of strains using normal mice. Lack of iron treatment required an increase in inocula to 106 to 107 CFU, and the resulting pathology was greatly reduced compared with that in iron-treated mice. However, s.c. lesions still contained equivalent bacterial counts regardless of whether the parent or the mutant strain was used (Fig. 4A). The added pressure of normal levels of iron did not enable the detection of a virulence phenotype for the VvpE elastase in s.c. inoculated mice.
In agreement with our results obtained by infection of mice, no significant differences were observed between the effects of wild-type and VvpE− mutant V. vulnificus ATCC 29307 on INT407 cells in culture (Fig. 7). The experiments were designed to assay effects on the host cells by vibrio products produced during an initial 1-h infection period, in which the bacteria rapidly multiply in the tissue culture medium. Twenty-four hours after infection, similar levels of lysis, death, and detachment for the epithelial cells were caused by the wild-type and VvpE− strains (Fig. 7). It should be noted that we examined effects on epithelial cells, not vascular endothelial cells, which could be more relevant during infection. However, the in vivo infection data, both quantitative CFU and qualitative pathology, failed to support a differential effect of the elastase on virulence. It is likely that the effects on epithelial cells that we observed for both strains were due to the action of the hemolysin/cytolysin. These results demonstrate that the vvpE gene is not essential for virulence of V. vulnificus in these animal and cell culture models.
The major problem to be addressed is the discrepancy between our infection experiments and those studies that relied primarily on the injection of proteins into animals. First, it is noteworthy that our vvpE mutant, which lacked elastase activity, exhibited residual total-protease activity, revealing the existence of at least one more gene for protease. Although other explanations are possible, the lack of significant difference in virulence between the vvpE mutant and the wild-type parent could be related to the presence of this other protease(s) of V. vulnificus. We find it difficult to imagine that the effects of inactivation of elastase were completely compensated by expression of the other protease(s) in V. vulnificus; however, analysis of this hypothesis awaits identification and mutation of the gene(s) encoding the other protease(s). We therefore caution that our results demonstrate that the metalloprotease encoded by the vvpE gene of V. vulnificus is dispensable for virulence in mice; however, we cannot make any conclusions as to the role of proteases in general. Additionally, the cytolysin/hemolysin has the potential of duplicating several aspects of the activity of the elastase, including stimulation of histamine release, vascular permeability, and toxicity (10, 38, 56). The virulence of most organisms is multifactorial, and backup or redundant virulence factors have often been identified. The best-known examples involve adherence factors, such that in order to observe effects of inactivation of specific adhesins, the specific mutations must be examined in the background of mutations in the redundant systems (50). Whether the elastolytic protease is completely dispensable in the mouse model of infection or whether redundant and fully compensatory virulence factors exist in V. vulnificus remains to be determined. However, our results strongly underscore the necessity of examining putative virulence attributes by using the molecular version of Koch's postulates (6, 11) in addition to injecting purified bacterial products into animal or cell culture systems.
ACKNOWLEDGMENTS
We are indebted to D. Milton for providing the plasmid pNQ705 and the E. coli strains with λ pir. We thank Shin-Ichi Miyoshi, Okayama University, for kindly providing rabbit anti-V. vulnificus elastase antibody. We thank Trenton R. Schoeb for his expertise and assistance in examining histopathological results and Thomas J. Doyle for his expert technical assistance with animal and cell culture experiments.
This study was supported by a grant to J.H.R. and S.H.C. from the KRF (1998-019-F00032), Republic of Korea. Research in the laboratory of P.A.G. was supported by the USDA (USDA-NRICGP 9802757).
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D L. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Blake P A, Weaver R E, Hollis D G. Diseases of humans (other than cholera) caused by vibrios. Annu Rev Microbiol. 1980;34:341–367. doi: 10.1146/annurev.mi.34.100180.002013. [DOI] [PubMed] [Google Scholar]
- 3.Bowdre J H, Poole M D, Oliver J D. Edema and hemoconcentration in mice experimentally infected with Vibrio vulnificus. Infect Immun. 1981;32:1193–1199. doi: 10.1128/iai.32.3.1193-1199.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng J C, Shao C P, Hor L I. Cloning and nucleotide sequencing of the protease gene of Vibrio vulnificus. Gene. 1996;183:255–257. doi: 10.1016/s0378-1119(96)00488-x. [DOI] [PubMed] [Google Scholar]
- 5.Chuang Y C, Chang T M, Chang M C. Cloning and characterization of the gene (empV) encoding extracellular metalloprotease from Vibrio vulnificus. Gene. 1997;189:163–168. doi: 10.1016/s0378-1119(96)00786-x. [DOI] [PubMed] [Google Scholar]
- 6.Falkow S. Molecular Koch's postulates applied to microbial pathogenicity. Rev Infect Dis. 1988;10:S274–S276. doi: 10.1093/cid/10.supplement_2.s274. [DOI] [PubMed] [Google Scholar]
- 7.Galloway D R. Pseudomonas aeruginosa elastase and elastolysis revisited: recent developments. Mol Microbiol. 1991;5:2315–2321. doi: 10.1111/j.1365-2958.1991.tb02076.x. [DOI] [PubMed] [Google Scholar]
- 8.Gambello M J, Iglewski B H. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gish W, States D J. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 10.Gray L D, Kreger A S. Mouse skin damage caused by cytolysin from Vibrio vulnificus and by V. vulnificus infection. J Infect Dis. 1987;155:236–241. doi: 10.1093/infdis/155.2.236. [DOI] [PubMed] [Google Scholar]
- 11.Gulig P A. Use of isogenic mutants to study bacterial virulence factors. J Microbiol Methods. 1993;18:275–287. [Google Scholar]
- 12.Hase C C, Finkelstein R A. Bacterial extracellular zinc-containing metalloproteases. Microbiol Rev. 1993;57:823–837. doi: 10.1128/mr.57.4.823-837.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 14.Klontz K C, Lieb S, Schreiber M, Janowski H T, Baldy L M, Gunn R A. Syndromes of Vibrio vulnificus infections. Clinical and epidemiologic features in Florida cases, 1981–1987. Ann Intern Med. 1988;109:318–323. doi: 10.7326/0003-4819-109-4-318. [DOI] [PubMed] [Google Scholar]
- 15.Kolter R M, Inuzuka M, Helinski D R. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell. 1978;15:1199–1208. doi: 10.1016/0092-8674(78)90046-6. [DOI] [PubMed] [Google Scholar]
- 16.Kothary M H, Kreger A S. Production and partial characterization of an elastolytic protease of Vibrio vulnificus. Infect Immun. 1985;50:534–540. doi: 10.1128/iai.50.2.534-540.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kothary M H, Kreger A S. Purification and characterization of an elastolytic protease of Vibrio vulnificus. J Gen Microbiol. 1987;133:1783–1791. doi: 10.1099/00221287-133-7-1783. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Litwin C M, Rayback T W, Skinner J. Role of catechol siderophore synthesis in Vibrio vulnificus virulence. Infect Immun. 1996;64:2834–2838. doi: 10.1128/iai.64.7.2834-2838.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maruo K, Akaike T, Ono T, Maeda H. Involvement of bradykinin generation in intravascular dissemination of Vibrio vulnificus and prevention of invasion by a bradykinin antagonist. Infect Immun. 1998;66:866–869. doi: 10.1128/iai.66.2.866-869.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milton D L, Norqvist A, Wolf-Watz H. Cloning of a metalloprotease gene involved in the virulence mechanism of Vibrio anguillarum. J Bacteriol. 1992;174:7235–7244. doi: 10.1128/jb.174.22.7235-7244.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyoshi N, Miyoshi S, Sugiyama K, Suzuki Y, Furuta H, Shinoda S. Activation of the plasma kallikrein-kinin system by Vibrio vulnificus protease. Infect Immun. 1987;55:1936–1939. doi: 10.1128/iai.55.8.1936-1939.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyoshi N, Shimizu C, Miyoshi S, Shinoda S. Purification and characterization of Vibrio vulnificus protease. Microbiol Immunol. 1987;31:13–25. doi: 10.1111/j.1348-0421.1987.tb03064.x. [DOI] [PubMed] [Google Scholar]
- 25.Miyoshi S, Hirata Y, Tomochika K, Shinoda S. Vibrio vulnificus may produce a metalloprotease causing an edematous skin lesion in vivo. FEMS Microbiol Lett. 1994;121:321–325. doi: 10.1111/j.1574-6968.1994.tb07120.x. [DOI] [PubMed] [Google Scholar]
- 26.Miyoshi S, Nakazawa H, Kawata K, Tomochika K, Tobe K, Shinoda S. Characterization of the hemorrhagic reaction caused by Vibrio vulnificus metalloprotease, a member of the thermolysin family. Infect Immun. 1998;66:4851–4855. doi: 10.1128/iai.66.10.4851-4855.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyoshi S, Shinoda S. Role of the protease in the permeability enhancement by Vibrio vulnificus. Microbiol Immunol. 1988;32:1025–1032. doi: 10.1111/j.1348-0421.1988.tb01467.x. [DOI] [PubMed] [Google Scholar]
- 28.Miyoshi S, Shinoda S. Inhibitory effect of α2-macroglobulin on Vibrio vulnificus protease. J Biochem (Tokyo) 1989;106:299–303. doi: 10.1093/oxfordjournals.jbchem.a122848. [DOI] [PubMed] [Google Scholar]
- 29.Miyoshi S, Shinoda S. Alpha-macroglobulin-like plasma inactivator for Vibrio vulnificus metalloprotease. J Biochem (Tokyo) 1991;110:548–552. doi: 10.1093/oxfordjournals.jbchem.a123617. [DOI] [PubMed] [Google Scholar]
- 30.Miyoshi S, Shinoda S. Activation mechanism of human Hageman factor-plasma kallikrein-kinin system by Vibrio vulnificus metalloprotease. FEBS Lett. 1992;308:315–319. doi: 10.1016/0014-5793(92)81301-2. [DOI] [PubMed] [Google Scholar]
- 31.Miyoshi S, Wakae H, Tomochika K, Shinoda S. Functional domains of a zinc metalloprotease from Vibrio vulnificus. J Bacteriol. 1997;179:7606–7609. doi: 10.1128/jb.179.23.7606-7609.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molla A, Yamamoto T, Akaike T, Miyoshi S, Maeda H. Activation of hageman factor and prekallikrein and generation of kinin by various microbial proteinases. J Biol Chem. 1989;264:10589–10594. [PubMed] [Google Scholar]
- 33.Mytelka D S, Chamberlin M J. Escherichia coli fliAZY operon. J Bacteriol. 1996;178:24–34. doi: 10.1128/jb.178.1.24-34.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishina Y, Miyoshi S, Nagase A, Shinoda S. Significant role of an exocellular protease in utilization of heme by Vibrio vulnificus. Infect Immun. 1992;60:2128–2132. doi: 10.1128/iai.60.5.2128-2132.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okujo N, Akiyama T, Miyoshi S, Shinoda S, Yamamoto S. Involvement of vulnibactin and exocellular protease in utilization of transferrin- and lactoferrin-bound iron by Vibrio vulnificus. Microbiol Immunol. 1996;40:595–598. doi: 10.1111/j.1348-0421.1996.tb01114.x. [DOI] [PubMed] [Google Scholar]
- 36.Oliver J D, Wear J E, Thomas M B, Warner M, Linder K. Production of extracellular enzymes and cytotoxicity by Vibrio vulnificus. Diagn Microbiol Infect Dis. 1986;5:99–111. doi: 10.1016/0732-8893(86)90112-4. [DOI] [PubMed] [Google Scholar]
- 37.Paranjpye R N, Lara J C, Pepe J C, Pepe C M, Strom M S. The type IV leader peptidase/N-methyltransferase of Vibrio vulnificus controls factors required for adherence to HEp-2 cells and virulence in iron-overloaded mice. Infect Immun. 1998;66:5659–5668. doi: 10.1128/iai.66.12.5659-5668.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park J W, Ma S N, Song E S, Song C H, Chae M R, Park B H, Rho R W, Park S D, Kim H R. Pulmonary damage by Vibrio vulnificus cytolysin. Infect Immun. 1996;64:2873–2876. doi: 10.1128/iai.64.7.2873-2876.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parmely M J. Pseudomonas metalloproteases and host-microbe relationship. In: Fick R B, editor. Pseudomonas aeruginosa the opportunist: pathogenesis and disease. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 79–94. [Google Scholar]
- 40.Reed L J, Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 41.Ruff M R, Gifford G E. Purification and physico-chemical characterization of rabbit tumor necrosis factor. J Immunol. 1980;125:1671–1676. [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 43.Shinoda S, Miyoshi S, Yamanaka H, Miyoshi-Nakahara N. Some properties of Vibrio vulnificus hemolysin. Microbiol Immunol. 1985;29:583–590. doi: 10.1111/j.1348-0421.1985.tb00862.x. [DOI] [PubMed] [Google Scholar]
- 44.Simbert R M, Krieg N R. Phenotypic characterization. In: Gerhardt P, editor. Methods for general and molecular bacteriology. Washington, D.C.: American Society for Microbiology; 1994. pp. 607–654. [Google Scholar]
- 45.Simon R, Priefer U, Pühler A. A broad-host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 46.Simpson L M, White V K, Zane S F, Oliver J D. Correlation between virulence and colony morphology in Vibrio vulnificus. Infect Immun. 1987;55:269–272. doi: 10.1128/iai.55.1.269-272.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46a.Starks, A. M., T. R. Schoeb, M. L. Tamplin, S. Parveen, T. J. Doyle, P. E. Bomeisl, G. L. Escudero, and P. A. Gulig. Pathogenesis of infection by clinical and environmental strains of Vibrio vulnificus in iron dextran-treated mice. Infect. Immun., in press. [DOI] [PMC free article] [PubMed]
- 47.Tacket C O, Brenner F, Blake P A. Clinical features and an epidemiological study of Vibrio vulnificus infections. J Infect Dis. 1984;149:558–561. doi: 10.1093/infdis/149.4.558. [DOI] [PubMed] [Google Scholar]
- 48.Testa J, Daniel L W, Kreger A S. Extracellular phospholipase A2 and lysophospholipase produced by Vibrio vulnificus. Infect Immun. 1984;45:458–463. doi: 10.1128/iai.45.2.458-463.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4355. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Velden A W, Baumler A J, Tsolis R M, Heffron F. Multiple fimbrial adhesins are required for full virulence of Salmonella typhimurium in mice. Infect Immun. 1998;66:2803–2808. doi: 10.1128/iai.66.6.2803-2808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wright A C, Morris J G., Jr The extracellular cytolysin of Vibrio vulnificus: inactivation and relationship to virulence in mice. Infect Immun. 1991;59:192–197. doi: 10.1128/iai.59.1.192-197.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wright A C, Morris J G, Jr, Maneval D R, Jr, Richardson K, Kaper J B. Cloning of the cytotoxin-hemolysin gene of Vibrio vulnificus. Infect Immun. 1985;50:922–924. doi: 10.1128/iai.50.3.922-924.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wright A C, Simpson L M, Oliver J D. Role of iron in the pathogenesis of Vibrio vulnificus infections. Infect Immun. 1981;34:503–507. doi: 10.1128/iai.34.2.503-507.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wright A C, Simpson L M, Oliver J D, Morris J G., Jr Phenotypic evaluation of acapsular transposon mutants of Vibrio vulnificus. Infect Immun. 1990;58:1769–1773. doi: 10.1128/iai.58.6.1769-1773.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamanaka H, Sugiyama K, Furuta H, Miyoshi S, Shinoda S. Cytolytic action of Vibrio vulnificus haemolysin on mast cells from rat peritoneal cavity. J Med Microbiol. 1990;32:39–43. doi: 10.1099/00222615-32-1-39. [DOI] [PubMed] [Google Scholar]
- 57.Yoshida S, Ogawa M, Mizuguchi Y. Relation of capsular materials and colony opacity to virulence of Vibrio vulnificus. Infect Immun. 1985;47:446–451. doi: 10.1128/iai.47.2.446-451.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]