Abstract
The lack of a long-term in vitro culture method has severely restricted the study of Plasmodium vivax, in part because it limits genetic manipulation and reverse genetics. We used the recently optimized Plasmodium cynomolgi Berok in vitro culture model to investigate the putative P. vivax drug resistance marker MDR1 Y976F. Introduction of this mutation using clustered regularly interspaced short palindromic repeats–CRISPR-associated protein 9 (CRISPR-Cas9) increased sensitivity to mefloquine, but had no significant effect on sensitivity to chloroquine, amodiaquine, piperaquine, and artesunate. To our knowledge, this is the first reported use of CRISPR-Cas9 in P. cynomolgi, and the first reported integrative genetic manipulation of this species.
Keywords: CRISPR-Cas9, Plasmodium cynomolgi, antimalarial drug resistance, chloroquine, mefloquine, molecular markers, Plasmodium vivax
The first integrative genetic manipulation of Plasmodium cynomolgi (cause of zoonotic malaria and P. vivax model) suggests mdr1 Y976F is not a marker of chloroquine-resistant vivax malaria; however, it increases the sensitivity of P. cynomolgi and possibly P. vivax to mefloquine.
(See the Editorial Commentary by Sibley on pages 1119–20.)
Chloroquine resistance (CQR) was first reported in Plasmodium vivax in 1989, 30 years after being observed in Plasmodium falciparum. Chloroquine-resistant P. vivax is particularly prevalent in New Guinea and eastern Indonesia, but it has also emerged in or spread to many other endemic areas, including Thailand, Ethiopia, and South America [1, 2]. Drug resistance in P. vivax is less well studied and understood than in P. falciparum, but is an equally pressing issue, as infection with multidrug-resistant parasites has been associated with increased severity and mortality, particularly in young children [3].
The genetic basis of P. vivax CQR remains elusive. Variations in 2 genes, crt (chloroquine resistance transporter) and mdr1 (multidrug resistance), incriminated for P. falciparum CQR, have been analyzed in P. vivax clinical isolates but yielded inconclusive results [2]. Moreover, although increased expression of Pvcrt has been clinically linked to CQR, the relationship was not confirmed in ex vivo assays using parasites from patient samples [4]. One polymorphism, Y976F, in the Pvmdr1 gene has been linked to CQR in some studies but not others [2, 5]. The unavailability of continuous in vitro cultivation of P. vivax clearly precludes efforts to validate candidate genes.
Plasmodium cynomolgi, a simian malaria parasite of Southeast Asian macaques, has long served as an excellent model for P. vivax, with which it is closely related phylogenetically, the 2 species being nearly indistinguishable morphologically and biologically [6]. We have recently adapted one of the P. cynomolgi strains, Berok, to routine in vitro cultivation, from which a cloned line (K4A7) was derived [7], thus providing parasites amenable to modern genetic manipulation that can be uniquely employed for investigation pertinent to P. vivax. We have applied clustered regularly interspaced short palindromic repeats–CRISPR-associated protein 9 (CRISPR-Cas9) to introduce the Y976F point mutation in the Pcymdr1 gene to P. cynomolgi Berok to establish whether it influences drug susceptibility.
METHODS
Macaque Blood
Nonhuman primate blood and serum were sourced from Macaca fascicularis (cynomolgus monkeys), which were maintained at the Monash Animal Research Platform, in Gippsland, Australia. Leukocytes were removed through filtration with preequilibrated nonwoven filters, and red blood cells (RBCs) were adjusted to 50% hematocrit with Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with GlutaMAX.
Sequence Alignments
Plasmodium mdr1 sequences were sourced for homology analysis from PlasmoDB (PVX_080100, PCYB_101870, and PCYM_1008800). Amino and nucleic acid sequence alignments and pairwise comparisons were completed with MegAlign Pro (DNASTAR) using MUSCLE software. Phylogenetic trees were compiled using the Kimura model with MegAlign Pro using BIONJ clustering and pairwise gap removal.
Parasite Culture
The experiments in this study used the single-cell cloned line Berok (K4A7), derived from the continuous culture of P. cynomolgi Berok (K4) [7], which was cultured as described by Christensen et al (2022) [8]. Base medium was prepared with RPMI 1640 medium with HEPES containing 2 mM L-glutamine, supplemented with 4 g/L D-glucose, 360 μM hypoxanthine, 5 g/L Albumax II lipid rich bovine serum albumin, 25 μg/mL penicillin G potassium salt, and 0.5 μg/mL cefquinome sulphate. Growth medium was supplemented with 10% (v/v) heat-inactivated horse serum. Parasites were maintained at a hematocrit of 2.5%, and between 0.5% and 5% parasitemia, and cultured at 37°C in a mixed gas chamber (5% CO2, 5% O2, and 90% N2).
Plasmid Constructs
DNA constructs for CRISPR manipulation of P. cynomolgi were adapted from the plasmids pDC2_vCam_GFP_hDHFR and pDC2_Cam_Cas9_hDHFR. A Streptococcus pyogenes Cas9 and a U6 promotor expressing a chimeric guide RNA from the plasmid pDC2_Cam_Cas9_U6_hDHFR were inserted into pDC2_vCam_GFP_hDHFR through restriction cloning, following digestion with XhoI and AvrII, and BamHI respectively. The guide RNA g1 was constructed by annealing primers p1 and p2 (Supplementary Table 1) and inserted following digestion of the plasmid with BbsI as described by Ng and Fidock (2013) [9].
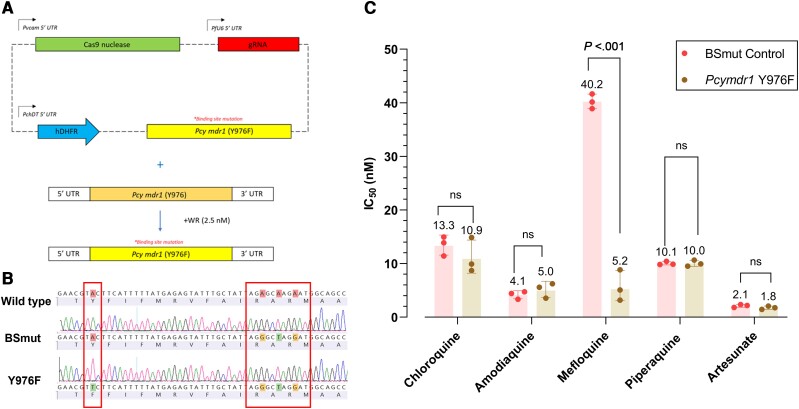
An 859-bp donor template was polymerase chain reaction (PCR) amplified from P. cynomolgi Berok K4A7 genomic DNA using primers p3 and p4 (Supplementary Table 1) and was inserted into the cloning plasmid pGEM. The Y976F mutation was introduced at amino acid position 976 through site-directed mutagenesis using primers p5 and p6 (Figure 1 and Supplementary Table 1). Silent mutations were also introduced at positions 986–988 using primers p7 and p8 (Supplementary Table 1) to restrict cleavage following successful donor insertion and mutation. The donor sequence was cloned into pDC2_vCam_Cas9_hDHFR through restriction cloning following digestion with SfoI and AatII to generate the transfection plasmid pDC2_vCam_Cas9_mdr1_hDHFR (Figure 1A). A separate transfection plasmid was assembled containing the silent binding site mutations with a wild-type Y976.
Figure 1.
The genetic modification of MDR1 Y976F in Plasmodium cynomolgi Berok K4A7 using CRISPR-Cas9 and its corresponding association with in vitro sensitivity to 5 antimalarial drugs. A, An all-in-one approach [8] was used for the CRISPR-Cas9 strategy for editing the mdr1 locus, consisting of a Streptococcus pyogenes Cas9 gene with a Plasmodium vivax calmodulin promotor, a U6-driving gRNA, a human dhfr selection cassette, and an mdr1-specific donor template for homology-directed repair. Donors coding for MDR1 Y976F and silent binding-site mutations were generated by site-directed mutagenesis of the wild-type P. cynomolgi mdr1 donor sequence. B, Sequencing results showing the introduction of individual Pcymdr1 mutations in recombinant parasites (Y976F), with wild-type amino and nucleic acid sequences for reference. The mutation Y976F was introduced into Pcymdr1, along with silent binding-site mutations, and a separate control BSmut line was produced with the same silent binding-site mutations and wild-type Y976. C, The geometric mean (3 biological replicates) of IC50 values are shown for chloroquine, artesunate, mefloquine, piperaquine, and amodiaquine against P. cynomolgi K4A7 Pcymdr1 Y976F and the BSmut control. Significance was determined by ordinary 2-way analysis of variance and a Šídák multiple comparisons test. Abbreviations: Cas9, CRISPR-associated protein 9; CRISPR, clustered regularly interspaced short palindromic repeats; IC50, 50% inhibitory concentration; ns, not significant; UTR, untranslated region.
Transfection
Transfections were carried out using the P3 Primary cell 4D Nucleofector X Kit L (Lonza), using the protocol described by Mohring et al (2019) [10] for the transfection of Plasmodium knowlesi. Briefly, synchronous schizont-stage parasites were enriched through gradient centrifugation on a 55% Nycodenz cushion, followed by a 3-hour incubation with 1 μM of 4-[7-[(dimethylamino)methyl]-2-(4-fluorophenyl)imidazo[1,2-a]pyridin-3-yl]pyrimidin-2-amine (compound 2), which inhibits parasite egress [11]. Parasites were washed in prewarmed complete media and pelleted through centrifugation at 627g, and the supernatant was discarded.
Five to ten microliters of pelleted schizonts were added to 100 μL of P3 Primary Cell solution (Lonza), along with 20 μg of plasmid DNA eluted in 10 μL of sterile Tris and Ethylenediaminetetraacetic acid buffer. The transfection solution was transferred to a 4D Nucleofector X Kit L cuvette and transfected using program FP158 on the Amaxa 4D Nucleofector. Transfected cells were immediately transferred to a 1.4-mL Eppendorf tube with 500 mL of prewarmed media and 150 μL fresh RBCs. Cells were incubated at 37°C on a shaking incubator at 650 rpm for 30–40 minutes, and were subsequently transferred to a 6-well plate containing 5 mL of prewarmed complete media, gassed, and incubated in a gas chamber at 37°C.
Successfully transfected parasites were selected through the use of 2.5 nM WR99210 (Jacobus Pharmaceuticals) after 24 hours, and were kept under drug pressure for 6 days with daily media changes. Media was changed every 48 hours, and cultures were split 1:2 with fresh RBCs every 2 weeks. Editing was confirmed by PCR and Sanger sequencing with primers p9 and p10 (Supplementary Table 1).
Drug Assays
P. cynomolgi sensitivity to antimalarial drugs was measured by incubating synchronous ring-stage parasites with a 10-point 2-fold serial dilution of drug concentrations at 0.8% parasitemia and 2% hematocrit in 96-well plates. Assay plates were incubated at 37°C for 72 hours under normal culturing conditions. Final parasitemia was determined through flow cytometry using a BD FACSCanto II following staining for 20 minutes with 1× SYBR Green I nucleic acid and 100 nM Mitotracker Deep Red. Experiments were carried out in duplicate, with 3 biological replicates. Fifty percent inhibitory concentration (IC50) values were calculated through nonlinear regression analysis, and statistical significance was determined through an ordinary 2-way analysis of variance and a Šídák multiple comparisons test using GraphPad Prism version 9.3.1.
RESULTS
Pairwise comparisons between the P. cynomolgi Berok and P. vivax mdr1 genes showed a similarity of 88.7% and 93.9% at the nucleotide and amino acid levels, respectively (Supplemental Figure 1). Interestingly, P. cynomolgi Berok, B, and M strains all carry T958M and F1076L, which have previously been linked to CQR in P. vivax [2].
We constructed a single-plasmid CRISPR vector expressing a S. pyogenes Cas9 gene with a P. vivax calmodulin promotor (1.15 kb), a U6-driven gRNA, an hDHFR selection cassette driven by P. chabaudi DHFR-TS promoter (0.6 kb), and a repair template (Figure 1A). Transfections with this system successfully introduced a Y976F mutation in the Pcymdr1 gene (Figure 1B), as well as silent binding-site mutations to prevent recleavage following homology-directed repair. We also introduced the same silent binding-site mutations into a line with wild-type Y976 to function as a control (the BSmut control) and to ensure that these silent mutations did not alter the phenotype. We genotyped transfected parasites with primers p9 and p10 (Supplementary Table 1), which were located outside of the repair template to avoid plasmid amplification.
No significant change was observed in the chloroquine sensitivity of P. cynomolgi for the recombinant Y976F line as compared to that of the BSmut control, with geometric mean IC50 values of 10.9 nM (SD 1.3 nM) and 13.3 nM (SD 1.2 nM), respectively (Figure 1C). Similarly, the Y976F mutation did not result in any significant changes to the sensitivity profile of P. cynomolgi to the antimalarial drugs artesunate, amodiaquine, or piperaquine (Figure 1C). However, we observed that the Y976F mutant led to a significant sensitization to mefloquine, with a mean IC50 of 5.2 nM (SD 1.7 nM) compared to 40.2 nM (SD 1.0 nM) in the BSmut control (P < .0001) (Figure 1C).
DISCUSSION
A fundamental understanding of resistance mechanisms notwithstanding, deciphering the genetic basis of drug resistance provides the ability to monitor and restrict the spread of resistant parasites, thus averting morbidity and mortality and optimizing control measures. For P. falciparum this has been possible through the combined availability of routine in vitro cultivation and genetic manipulation, two avenues lacking for P. vivax.
A combination of clinical studies, in vitro phenotyping, and reverse genetics has provided the community with verified molecular markers of drug resistance for P. falciparum, which additionally enhanced target optimization for drug development against resistant parasite populations [12, 13]. The emergence of P. vivax lines resistant to chloroquine, still the first-line treatment for this parasite, requires similar investigations. However, these are limited to clinical drug efficacy trials and ex vivo sensitivity assays [14], both of which are subject to biological and technical variability. In vivo nonhuman primate models are prohibitively expensive and cannot provide a tenable alternative to the extensive in vitro reverse genetic investigations conducted for P. falciparum. To date, only one prior study has successfully edited P. vivax, as a proof of concept showing the introduction of a selectable marker with parasites selected in vivo in Saimiri monkeys [15]. While recent reports of a humanized mouse model for blood-stage P. vivax are impressive, this model has not yet shown to be amenable to transfection [16].
We have exploited a cultured P. cynomolgi cloned line to introduce the Pcymdr1 Y976F mutation, which is possibly associated with CQR in P. vivax [5], by an integrative genetic manipulation through CRISPR-Cas9. To our knowledge, this is the first time gene editing has been achieved with P. cynomolgi. We observed no significant change in chloroquine sensitivity, which strongly suggests that this mutation is not directly linked to a resistant phenotype. Similarly, no change in sensitivity to the related first-line drug amodiaquine resulted from the introduction of the Y976F mutation, although it had previously been linked to amodiaquine and sulfadoxine-pyrimethamine in P. vivax alongside Pvdhfr mutations [17]. These observations suggest that the Pvmdr1 Y976F mutation, which is predominant in Papua Indonesia but also present in P. vivax populations spanning the globe [2, 18], is not being selected for by exposure to chloroquine.
Interestingly, we did note an increase in sensitivity to mefloquine, of a magnitude similar to that reported earlier in ex vivo studies of Thai and Indonesian P. vivax isolates [19]. Our results demonstrated a 7.7-fold reduction in mefloquine IC50 values, from 40 to 5 nM for P. cynomolgi, which compares favorably with the 8.6-fold reduction, from 121 to 14 nM, for P. vivax. While one should be cautious in making comparisons to orthologous P. falciparum MDR1 (Pfmdr1), a recent mechanistic study by Shafik et al (2022) [20] supports earlier observations [21] showing that mutant isoforms of Pfmdr1 almost universally increase the sensitivity of P. falciparum to mefloquine. Mutant isoforms were reported to have a reduced capacity to transport mefloquine into the digestive vacuole, resulting in an increased concentration in the parasite cytosol [20]. In contrast, increased expression of wild-type Pfmdr1 [22] and Pvmdr1 [19] increase the accumulation of mefloquine in the digestive vacuole, thereby sequestering it away from its cytosolic target. It is important to note that the P. cynomolgi Berok K4A7 clone used here has a single copy of Pcymdr1.
While the introduction of this mutation did not result in decreased sensitivity to chloroquine, it is possible that it may alter the phenotype in different backgrounds. Indeed, initial reports of this mutation correlating with CQR noted significant differences in sensitivity between Y976F lines from Thailand and Indonesia [5]. Isolation and culture adaption of strains from human infections of P. cynomolgi [23, 24] could allow for further investigation into the role of the parasite genetic background in contributing to P. vivax CQR. Interestingly, the Y976F mutation did not affect the sensitivity of P. cynomolgi to piperaquine (the first-line partner drug used with dihydroartemisinin to effectively treat CQR P. vivax malaria [25, 26]), a finding also supported in earlier field work on PvMDR1 polymorphisms [19]. This is important as piperaquine can be preferred to lumefantrine for the treatment of vivax malaria, primarily due to piperaquine's longer half-life kinetics and thus longer period of posttreatment prophylaxis [27]. Further studies are required to assess whether the Y976F mutation alters P. cynomolgi sensitivity to lumefantrine.
This study demonstrates that P. cynomolgi genetic manipulation offers an excellent opportunity to investigate the contribution of various candidate genetic mutations, alone or in defined combinations, to the susceptibility of P. vivax to a given antimalarial drug. This model also has the potential to improve our broader understanding of P. vivax biology. However, a notable caveat is its reliance on nonhuman primate blood, which is both expensive and of limited supply. It should also be noted that the transfection methodology used in this study still requires considerable optimization. Our successful editing of Pcymdr1 Y976F was achieved after 2 years of considerable effort, and we were unable to edit a range of other orthologous markers of drug resistance such as Pcydhfr (dihydrofolate reductase) or Pcycrt (chloroquine resistance transporter). Other simian models for P. vivax such as P. knowlesi provide a greater transfection efficiency and the ability to culture adapted parasites in human RBCs [10], and at this stage P. knowlesi is preferred for reverse engineering P. vivax orthologs. Nonetheless, the very close similarity of P. cynomolgi with P. vivax at the genomic and phenotypic levels warrants further investment into optimizing P. cynomolgi genetic manipulation and leveraging this model to gain important insights into P. vivax drug resistance.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Kurt E Ward, Department of Microbiology and Immunology, Columbia University Irving Medical Center, New York, New York, USA; Department of Microbiology and Immunology, University of Otago, Dunedin, New Zealand.
Peter Christensen, Department of Microbiology and Immunology, University of Otago, Dunedin, New Zealand.
Annie Racklyeft, Department of Microbiology and Immunology, University of Otago, Dunedin, New Zealand.
Satish K Dhingra, Department of Microbiology and Immunology, Columbia University Irving Medical Center, New York, New York, USA.
Adeline C Y Chua, Department of Microbiology and Immunology, University of Otago, Dunedin, New Zealand; A*STAR Infectious Diseases Laboratory, Agency for Science, Technology, and Research, Singapore, Singapore.
Caroline Remmert, Department of Microbiology and Immunology, University of Otago, Dunedin, New Zealand.
Rossarin Suwanarusk, Department of Microbiology and Immunology, University of Otago, Dunedin, New Zealand; Department of Protozoology, Institute of Tropical Medicine, Nagasaki University, Nagasaki, Japan.
Jessica Matheson, Department of Microbiology and Immunology, University of Otago, Dunedin, New Zealand.
Michael J Blackman, Malaria Biochemistry Laboratory, Francis Crick Institute, London, United Kingdom; Faculty of Infectious and Tropical Diseases, London School of Hygiene and Tropical Medicine, London, United Kingdom.
Osamu Kaneko, Department of Protozoology, Institute of Tropical Medicine, Nagasaki University, Nagasaki, Japan.
Dennis E Kyle, Center for Tropical and Emerging Global Diseases, University of Georgia, Athens, Georgia, USA.
Marcus C S Lee, Parasites and Microbes Programme, Wellcome Sanger Institute, Hinxton, United Kingdom.
Robert W Moon, Faculty of Infectious and Tropical Diseases, London School of Hygiene and Tropical Medicine, London, United Kingdom.
Georges Snounou, 11-INSERM U1184, Immunology of Viral Infections and Autoimmune Diseases, Infectious Disease Models and Innovative Therapies Department, Institut de biologie François Jacob, Direction de Recherche Fondamentale, Commissariat à l'énergie atomique et aux énergies alternatives-Université Paris Sud, Fontenay-aux-Roses, France.
Laurent Rénia, A*STAR Infectious Diseases Laboratory, Agency for Science, Technology, and Research, Singapore, Singapore; Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore, Singapore; School of Biological Sciences, Nanyang Technological University, Singapore, Singapore.
David A Fidock, Department of Microbiology and Immunology, Columbia University Irving Medical Center, New York, New York, USA; Center for Malaria Therapeutics and Antimicrobial Resistance, Division of Infectious Diseases, Department of Medicine, Columbia University Irving Medical Center, New York, New York, USA.
Bruce Russell, Department of Microbiology and Immunology, University of Otago, Dunedin, New Zealand; Department of Protozoology, Institute of Tropical Medicine, Nagasaki University, Nagasaki, Japan.
Pablo Bifani, A*STAR Infectious Diseases Laboratory, Agency for Science, Technology, and Research, Singapore, Singapore; Faculty of Infectious and Tropical Diseases, London School of Hygiene and Tropical Medicine, London, United Kingdom; Department of Microbiology and Immunology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore.
Notes
Financial support. This work was supported by the Agency for Science, Technology, and Research, Singapore (A*STAR) (core funding to A*STAR Infectious Diseases Laboratory, and grant number JCO-DP BMSI/15-800006-SIGN to L. R.); Health Research Council, New Zealand (grant number 17/678 e-ASIA Research Program to B. R.); University of Otago (Deans Bequest Grant to B. R.); Japanese Society for the Promotion of Science (Invitational Fellowship for Research in Japan to B. R.); US National Institutes of Health (grant number R01 AI050234 to D. A. F.); Agence Nationale de la Recherche, France (grant number ANR-17-CE13-0025-01 to G. S.); Francis Crick Institute (support to M. J. B.), which receives its core funding from Cancer Research UK (grant number FC001043), UK Medical Research Council (MRC) (grant number FC001043), and Wellcome (grant number FC001043); Wellcome (grant number 206194/Z/17/Z to M. C. S. L.); MRC and UK Department for International Development (grant number MR/M021157/1 MRC Career Development Award to R. W. M.); and Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan (grant number 22K19441 Grants-in-Aid for Scientific Research to O. K.). Funding to pay the Open Access publication charges for this article was provided by CAUL – Council of Australian University Librarians.
References
- 1. Price RN, von Seidlein L, Valecha N, et al. Global extent of chloroquine-resistant Plasmodium vivax: a systematic review and meta-analysis. Lancet Infect Dis 2014; 14:982–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Buyon LE, Elsworth B, Duraisingh MT. The molecular basis of antimalarial drug resistance in Plasmodium vivax. Int J Parasitol Drugs Drug Resist 2021; 16:23–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tjitra E, Anstey NM, Sugiarto P, et al. Multidrug-resistant Plasmodium vivax associated with severe and fatal malaria: a prospective study in Papua, Indonesia. PLoS Med 2008; 5:e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pava Z, Handayuni I, Wirjanata G, et al. Expression of Plasmodium vivax crt-o is related to parasite stage but not ex vivo chloroquine susceptibility. Antimicrob Agents Chemother 2016; 60:361–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Suwanarusk R, Russell B, Chavchich M, et al. Chloroquine resistant Plasmodium vivax: in vitro characterisation and association with molecular polymorphisms. PLoS One 2007; 2:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tachibana S, Sullivan SA, Kawai S, et al. Plasmodium cynomolgi genome sequences provide insight into Plasmodium vivax and the monkey malaria clade. Nat Genet 2012; 44:1051–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chua ACY, Ong JJY, Malleret B, et al. Robust continuous in vitro culture of the Plasmodium cynomolgi erythrocytic stages. Nat Commun 2019; 10:3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Christensen P, Racklyeft A, Ward KE, et al. Improving in vitro continuous cultivation of Plasmodium cynomolgi, a model for P. vivax. Parasitol Int 2022; 89:102589. [DOI] [PubMed] [Google Scholar]
- 9. Ng CL, Fidock DA. Plasmodium falciparum in vitro drug resistance selections and gene editing. Methods Mol Biol 2019; 2013:123–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mohring F, Hart MN, Rawlinson TA, et al. Rapid and iterative genome editing in the malaria parasite Plasmodium knowlesi provides new tools for P. vivax research. eLife 2019; 8:e45829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Collins CR, Hackett F, Strath M, et al. Malaria parasite cGMP-dependent protein kinase regulates blood stage merozoite secretory organelle discharge and egress. PLoS Pathog 2013; 9:e1003344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yuthavong Y, Tarnchompoo B, Vilaivan T, et al. Malarial dihydrofolate reductase as a paradigm for drug development against a resistance-compromised target. Proc Natl Acad Sci U S A 2012; 109:16823–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim J, Tan YZ, Wicht KJ, et al. Structure and drug resistance of the Plasmodium falciparum transporter PfCRT. Nature 2019; 576:315–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Russell B, Suwanarusk R, Malleret B, et al. Human ex vivo studies on asexual Plasmodium vivax: the best way forward. Int J Parasitol 2012; 42:1063–70. [DOI] [PubMed] [Google Scholar]
- 15. Moraes Barros RR, Straimer J, Sa JM, et al. Editing the Plasmodium vivax genome, using zinc-finger nucleases. J Infect Dis 2015; 211:125–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Luiza-Batista C, Thiberge S, Serra-Hassoun M, et al. Humanized mice for investigating sustained Plasmodium vivax blood-stage infections and transmission. Nat Commun 2022; 13:4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marfurt J, Monbrison F, Brega S, et al. Molecular markers of in vivo Plasmodium vivax resistance to amodiaquine plus sulfadoxine-pyrimethamine: mutations in Pvdhfr and Pvmdr1. J Infect Dis 2008; 198:409–17. [DOI] [PubMed] [Google Scholar]
- 18. Ferreira MU, Nobrega de Sousa T, Rangel GW, et al. Monitoring Plasmodium vivax resistance to antimalarials: persisting challenges and future directions. Int J Parasitol Drugs Drug Resist 2021; 15:9–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Suwanarusk R, Chavchich M, Russell B, et al. Amplification of pvmdr1 associated with multidrug-resistant Plasmodium vivax. J Infect Dis 2008; 198:1558–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shafik SH, Richards SN, Corry B, Martin RE. Mechanistic basis for multidrug resistance and collateral drug sensitivity conferred to the malaria parasite by polymorphisms in PfMDR1 and PfCRT. PLoS Biol 2022; 20:e3001616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sidhu AB, Valderramos SG, Fidock DA. Pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol Microbiol 2005; 57:913–26. [DOI] [PubMed] [Google Scholar]
- 22. Price RN, Uhlemann AC, Brockman A, et al. Mefloquine resistance in Plasmodium falciparum and increased Pfmdr1 gene copy number. Lancet 2004; 364:438–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sai-ngam P, Pidtana K, Suida P, et al. Case series of three malaria patients from Thailand infected with the simian parasite, Plasmodium cynomolgi. Malar J 2022; 21:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Putaporntip C, Kuamsab N, Pattanawong U, et al. Plasmodium cynomolgi co-infections among symptomatic malaria patients, Thailand. Emerg Infect Dis 2021; 27:590–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kenangalem E, Poespoprodjo JR, Douglas NM, et al. Malaria morbidity and mortality following introduction of a universal policy of artemisinin-based treatment for malaria in Papua, Indonesia: a longitudinal surveillance study. PLoS Med 2019; 16:e1002815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Phyo AP, Lwin KM, Price RN, et al. Dihydroartemisinin-piperaquine versus chloroquine in the treatment of Plasmodium vivax malaria in Thailand: a randomized controlled trial. Clin Infect Dis 2011; 53:977–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Commons RJ, Simpson JA, Thriemer K, et al. The efficacy of dihydroartemisinin-piperaquine and artemether-lumefantrine with and without primaquine on Plasmodium vivax recurrence: a systematic review and individual patient data meta-analysis. PLoS Med 2019; 16:e1002928. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.