Summary:
Interferon-γ (IFN-γ) is a key cytokine in response to viral or intracellular bacterial infection in mammals. While a number of enhancers are described to promote IFN-γ responses, to our knowledge, no silencers for the Ifng gene have been identified. By examining H3K4me1 histone modification in naïve CD4+ T cells within Ifng locus, we identified a silencer (CNS–28) which restrains Ifng expression. Mechanistically, CNS–28 maintains Ifng silence by diminishing enhancer-promoter interactions within Ifng locus in a GATA3-dependent but T-bet independent manner. Functionally, CNS–28 restrains Ifng transcription in NK cells, CD4+ and CD8+ T cells during both innate and adaptive immune responses. Moreover, CNS–28 deficiency resulted in repressed type 2 responses due to elevated IFN-γ expression, shifting Th1 and Th2 paradigm. Thus, CNS–28 activity ensures immune cell quiescence by cooperating with other regulatory cis elements within the Ifng gene locus to minimize autoimmunity.
eTOC blurb
There are multiple enhancer regions for the Ifng locus, but Ifng restraint is unknown. Cui et al. identify a silencer CNS–28 that diminishes enhancer-promoter interactions within Ifng locus in a GATA3-dependent but T-bet-independent manner. Together with other regulatory elements, CNS–28 activity ensures immune cell quiescence and minimizes autoimmunity.
Graphical Abstract
Introduction
Cytokines are key regulators for immune homeostasis and host defense. The expression of cytokine genes requires precise control by different regulatory elements.1,2 While interactions between the transcription factors (TFs) with promoters and proximal regulatory elements are critical for gene expression, functions of distal regulatory elements such as enhancers, silencers and boundary elements are now appreciated for their role in controlling gene activation and suppression in a cell type-specific manner. CD4+ T helper (Th) cells are ideal for studying these distal regulatory elements due to their ease in isolation, expansion, and differentiation.
Th cell subtypes, including Th1, Th2, Th9, Th17, and Treg cells are extensively studied during adaptive immunity in mammals.3 Naïve CD4+ T cells can be readily differentiated into different Th cell subtypes upon encountering various antigens and microenvironments.4 Among these cells, Th1 cell-derived IFN-γ exerts a broad spectrum of functions, including host defense, autoimmune responses, tumor surveillance and neuronal function.5–8 The mechanisms underlying the dynamic regulation of IFN-γ expression have been extensively studied. T-bet (T-box gene, TBX21) is a key TF in regulating type 1 immune responses, including determining Th1 cell differentiation and Ifng expression.1 T-bet directly binds to the Ifng promoter for its activation.9 Further studies suggest that the interactions between T-bet and multiple distal elements of conserved non-coding sequence (CNSs) also critically contribute to Ifng expression.10–12 Identification of the binding of other TFs, such as STAT4, Runx3, and NFAT, to CNS across the Ifng locus facilitated the functional mapping of the cis-regulatory elements.11,13–15 All these identified upstream and downstream CNSs, including CNS–54, CNS–34, CNS–22, CNS–6 and CNS+18–20, have been shown to act as enhancers for Ifng transcription.16 However, to our knowledge, no negative regulatory elements have been found for the expression of Ifng.
T-bet and GATA3 are known as the master TFs for Th1 and Th2 cells, respectively.17,18 The mutual exclusive expression of IFN-γ and IL-4 in CD4+ T cells indicates the antagonistic nature of T-bet and GATA3. Meanwhile, interactions of T-bet or GATA3 with distal regulatory elements lead to the suppression of Il4 or Ifng transcription in the alternate lineage.19–21 T-bet and GATA3 can be co-expressed within Th1 cells both in vitro and in vivo,20,22,23 implying the potential plasticity in genomic regulation and functionality between Th1 and Th2 cells.4,24 Indeed, during Th1 cell differentiation, GATA3 can bind at genomic regions harboring Th1 genes, including the Ifng locus.25,26 Despite the mapping of GATA3 binding sites in Th1 cells, the interaction between GATA3 and distal elements in the fate decision of Th1 cells and transcription of Ifng remains unclear.
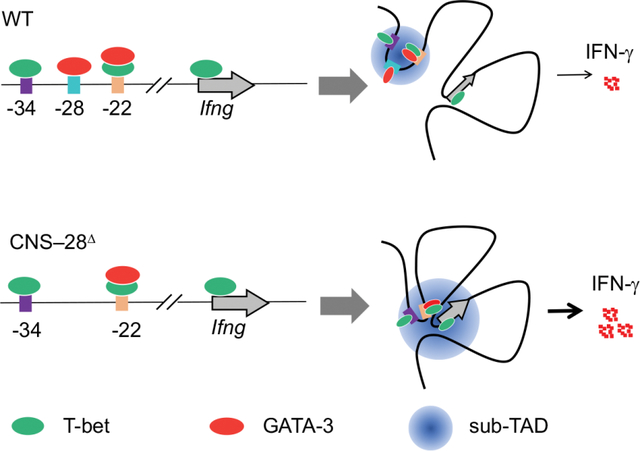
Epigenomic modifications encode necessary information on chromatin required for cellular differentiation.27 Chromatin modifications play critical roles in T cell development, plasticity and memory formation.28–30 The histone H3 lysine 4 (H3K4)-methylation is considered a permissive epigenetic mark for the assessment of active and primed enhancers.27 The MLL family of histone methyltransferases, including SETD1A, MLL1 (KMT2A), MLL2 (KMT2B), MLL3 (KMT2C) and MLL4 (KMT2D), catalyzes the methylation of H3K4.31 Our previous work demonstrates that MLL4 plays a key role for shaping the enhancer landscape in Treg cells by catalyzing the methylation of H3K4 via both direct binding and long-distance chromatin looping.32 However, it is still unclear whether such effects by MLL4 also impact the differentiation and function of other T cell subsets. In the current study, we found that loss of MLL4 led to substantial increase of Ifng expression during Th1 cell differentiation. Through genetic deletion of Mll4 and H3K4me1 binding profile analysis, we identify CNS–28, a distal element for the Ifng locus. MLL4 facilitates H3K4 monomethylation of CNS–28 and promotes the formation of a mini chromatin domain containing CNS–22, CNS–28 and CNS–34. Further, we demonstrate that CNS–28 functions as a silencer for Ifng expression via repression of CNS–22-promoter interactions within the Ifng locus. Mice missing CNS–28 exhibited elevated systemic IFN-γ from both adaptive and innate immune cells. Therefore, we present here a negative mechanism for proper Ifng expression, restraining the IFN-γ overreaction to ensure tissue homeostasis.
Results
Loss of Mll4 induces enhanced IFN-γ expression and chromatin interaction frequencies within Ifng domain.
Our previous study indicates that loss of MLL4 led to dysregulated Treg cell function and disrupted immune homeostasis.32 To further investigate the role of MLL4 in type 1 immune responses, we polarized naïve CD4+ T cells from Cd4creMll4fl/fl (Mll4 KO) and control mice under Th1 conditions. We observed elevated IFN-γ production from cultured Th1 cells by flow cytometry and ELISA analyses (Figures 1A, 1B, and S1A). These results suggest that MLL4 represses the IFN-γ expression during Th1 differentiation. It is well documented that Th1 differentiation requires IL-12 signaling via STAT4, which induces T-bet expression.7,33,34 However, our data indicated that neither STAT4 nor T-bet were involved in MLL4-mediated IFN-γ repression (Figures S1B–S1C). MLL4 deletion resulted in elevated Ifng mRNA expression levels in CD4+ T cells during Th1 cell differentiation (Figures 1C, and S1D). Consistently, the enriched Gene Ontology terms in the upregulated genes included immune responses (Figure S1E).
Fig 1. Loss of Mll4 induces enhanced IFN-g expression and interaction frequencies within the Ifng domain.
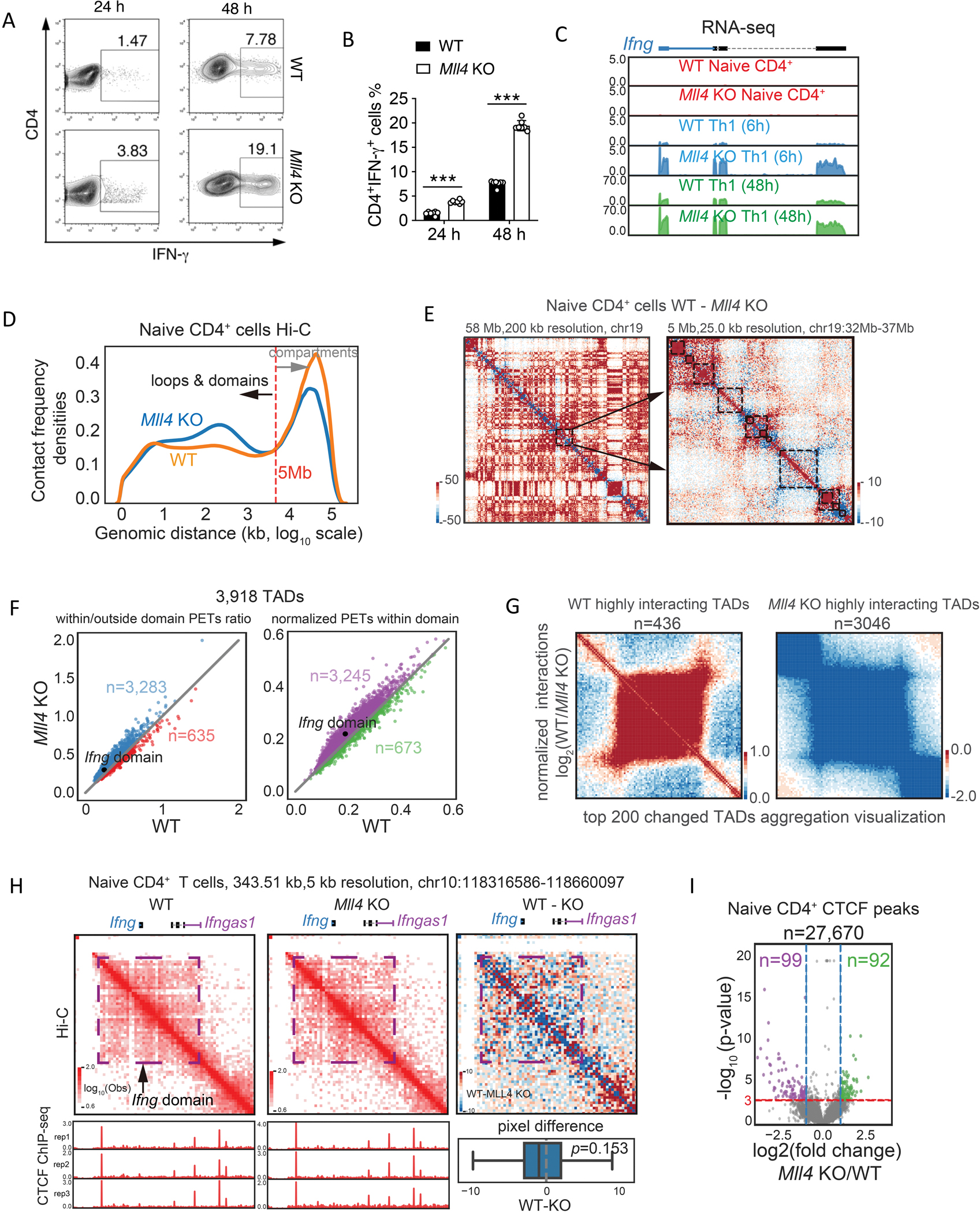
(A) Naive CD4+ T cells from WT and Mll4 KO mice were stimulated under Th1 condition and harvested at 24 and 48 hours. Intracellular expression levels of IFN-γ produced in these cells were determined by flow cytometry.
(B) Quantification of IFN-γ producing cells from multiple experiments as measured in (A) above.
(C) Genome browser images of RNA-seq analysis of Ifng expression in WT and Mll4 KO CD4+ T cells under Th1 condition. The RNA-seq data were generated in this study.
(D) Distribution of interacting paired-end tags (PETs) frequencies against genomic distances of Hi-C data in wild-type and Mll4 KO naïve CD4+ T cells. Hi-C data were obtained from 32 and down-sampled to 300 million for a fair comparison. Only PETs with distance longer than 1 kb were used to draw the density plot.
(E) Examples of chromatin interaction changes measured by Hi-C after Mll4 KO in naïve CD4+ T cells. The changes of Hi-C interaction frequency were visualized by subtracting the number of PETs detected in KO cells from the number of PETs detected in WT cells. The blue color indicates decreased interaction frequencies, and the red color indicates increased interaction frequencies. Left panel shows the interaction difference heatmap for Chromosome 19 with 200 kb resolution, and arrows indicate a random selected region for zoom-in visualization as the right panel. The right panel shows the interaction changes in a 5 Mb genomic region with 25 kb resolution; the black rectangles mark the TADs called by Juicer with the WT Hi-C data.
(F) Quantitation of Hi-C interaction changes for TADs comparing Mll4 KO and WT mice in naïve CD4+ T cells. The left panel shows relative changes regarding the TAD compactness, and the right panel shows changes of interaction densities within the TADs. Only PETs with a distance longer than 1 kb were used for the calculation. The numbers indicate the TADs with higher interacting densities in KO or WT cells. The TAD contains the Ifng locus was indicated.
(G) Hi-C data aggregation analysis of highly interacting TADs in WT or Mll4 KO cells. WT or KO highly interacting domains were obtained by overlapping the consistently changed domains from compactness and interaction densities within TADs in (F). Only the top 200 changed TADs were used to draw the aggregation heatmaps.
(H) Hi-C interaction frequencies of Ifng domain were increased in Mll4 KO naïve CD4+ T cells (middle panel) compared to WT cells (left panel). The changes of interaction frequency were visualized by subtracting the number of PETs detected in KO cells from the number of PETs detected in WT cells (right panel). The blue color indicates decreased interaction frequencies, and the red color indicates increased interaction frequencies. CTCF ChIP-seq profiles for WT and KO naïve CD4+ T cells were shown below the red heatmaps for corresponding cell types. Distribution of pixel level (5kb resolution) difference for the Ifng domain was shown below the red/blue heatmap. Two sided Wilcoxon signed-rank test P-value was shown for the statistical difference of interactions within the Ifng domain. CTCF ChIP-seq data were generated in this study.
(I) Volcano plots of significantly changed CTCF peaks for the naïve CD4+ T cells affected by Mll4 KO. Mean values from three replicates were used to calculate fold changes (KO/WT) and Poisson P-values. Fold change > 2 (or < 0.5) and P-value smaller than 0.001 were set as the significant cutoffs. Numbers of total peaks, WT specific peaks and KO specific peaks were shown.
Data are representative of at least two independent experiments (A, C-I) or pooled from two independent experiments (B). ***p < 0.001. (Two-way ANOVA with Tukey’s multiple comparison test, error bars represent SD).
Given that MLL4 is critical in modifying the chromatin structure for gene expression,32 we examined the Ifng locus by Hi-C analysis. MLL4 deletion led to substantial increases in chromatin interactions between 10 Kbp to 5 Mbp while it decreased interactions between 5Mbp to 100 Mbp (Figure 1D). Consistently, increased interactions in the MLL4 deletion cells were often detected along the diagonal of the interaction matrix, as represented by the blue color, while the red color far away from the diagonal indicates decreased long-distance interactions (Figure 1E). Globally, a total of 3,918 topologically associating domains (TADs) were identified from WT naïve CD4+ T cells; and the Mll4 deletion resulted in more TADs with increased intradomain interactions, including the Ifng locus (Figure 1F). Further aggregate analysis of the top 200 changed TADs indicated a difference in the interaction intensities between the WT and Mll4 KO cells (Figure 1G). Specific examination of the Hi-C paired-end tags (PETs) at the Ifng locus revealed an increase in intra-TADs interaction in the Mll4 KO cells despite with only a modest statistical difference (P-value=0.153, two-sided Wilcoxon signed-rank test) (Figure 1H), suggesting that MLL4 regulates Ifng expression by altering chromatin organization. Meanwhile, there were only a small number of significantly changed CTCF peaks genome-widely in the Mll4 KO cells, and the CTCF binding patterns did not show significant changes in the Ifng locus (Figures 1H and 1I), suggesting that the changes in chromatin interaction in the loss of MLL4 are not caused by CTCF binding alterations.
Identification of the Ifng silencer CNS–28.
Since MLL4 is responsible for generating the H3K4me1 mark on chromatin,32 we further analyzed the distribution of H3K4me1 modification in naïve CD4+ T cells from Mll4 KO and control mice (Figure 2A). There are several known enhancers including CNS–22, CNS–34, CNS+18–20, and CNS+55 in the Ifng genomic region that positively contribute to Ifng expression during Th1 cell differentiation.35 Although Ifng was not expressed in the naïve CD4+ T cells, these enhancer elements were associated with the active histone mark H3K4me1 (Figure 2A), consistent with the notion that at least a fraction of naïve CD4+ T cells are primed for Th1 differentiation.36 In addition to these known enhancers observed in both WT and Mll4 KO cells, we also detected a H3K4me1 peak at the −28Kb region upstream of the Ifng gene in WT but not in the absence of MLL4 (Figure 2A). These data suggest that the decreased H3K4me1 signal at −28Kb region upon deletion of Mll4 may be linked to the enhanced IFN-γ induction during Th1 cell polarization, raising the possibility that this site acts as a regulatory element for Ifng expression.
Fig 2. Identification of the Ifng silencer CNS–28.
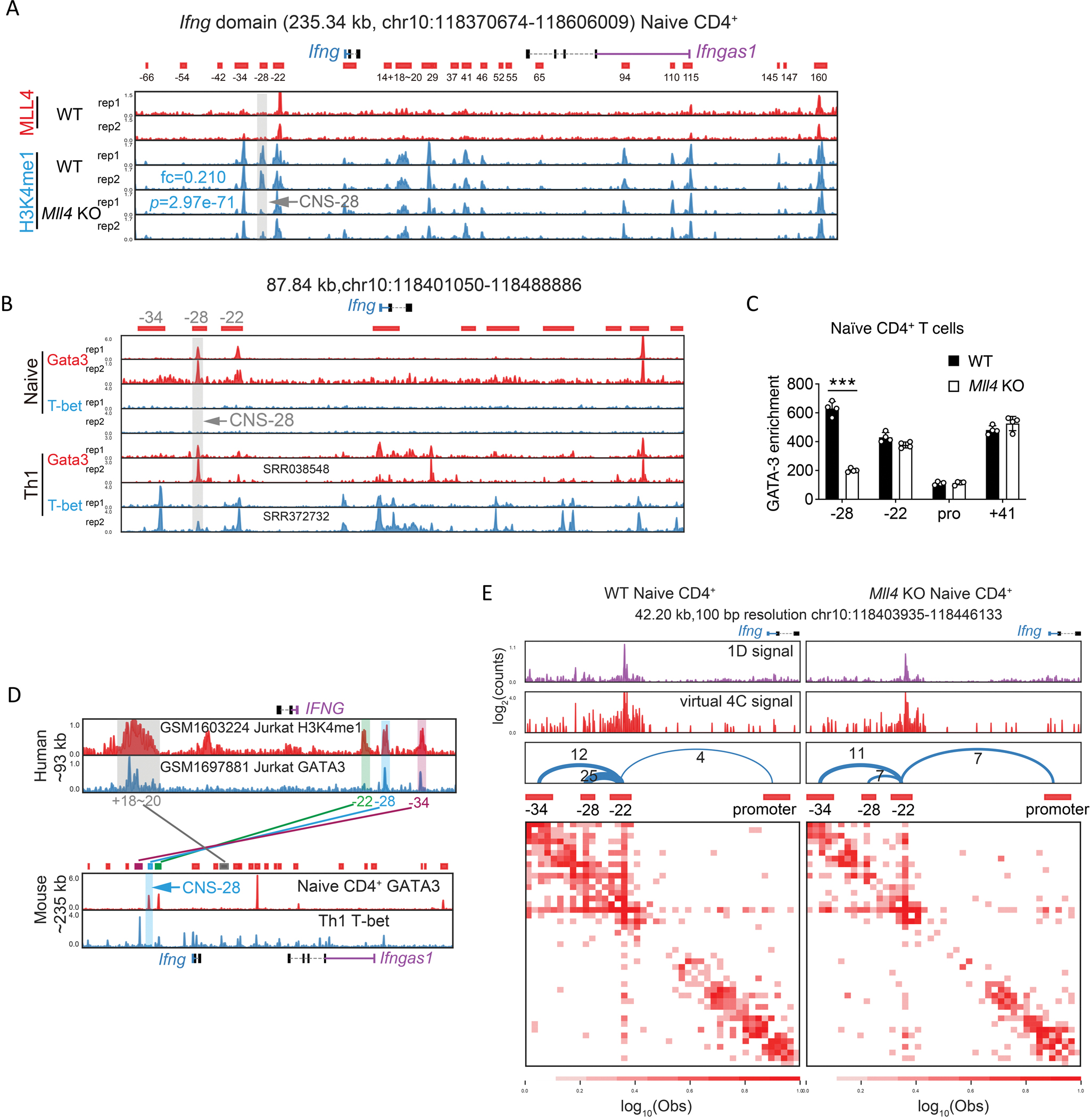
(A) Genome browser images of MLL4 and H3K4me1 ChIP-seq profiles in the Ifng domain. The ChIP-seq data were obtained from 32. Peaks were called and combined from H3K4me1 ChIP-seq data in WT naïve CD4+ and Th1 cells by cLoops2 and shown as the red blocks below the genes as putative cis-regulatory elements, annotated according to the relative distance to Ifng promoter, with – standing for upstream and + indicating downstream of Ifng TSS. The gray bar and arrow highlighted the absence of an H3K4me1 peak located at CNS-28 after the deletion of MLL4. The mean of normalized reads counts (to a total of 10 million reads) from two replicates of H3K4me1 ChIP-seq in the CNS-28 locus were used to calculate fold change (KO/WT) and Poisson P-value.
(B) Genome browser images of GATA-3 and T-bet ChIP-seq binding profiles around Ifng in naïve CD4+ T cells and Th1 differentiated cells. Th1 GATA-3 ChIP-seq data annotated with SRR038548 was obtained from 26, Th1 T-bet ChIP-seq data annotated with SRR372732 was obtained from 59, and the other ChIP-seq data shown in this panel were generated in this study.
(C) ChIP-qPCR analysis of GATA-3 binding to CNS–28, CNS–22, Ifng promoter and CNS+41 at the Ifng locus in WT and Mll4 KO naïve CD4+ T cells.
(D) Histone modification mark H3K4me1 at CNS–34, CNS–28, CNS–22 and CNS+18–20 is conserved between human T lymphocyte cell line Jurkat and mouse CD4+ T cells, while binding of GATA-3 is only conserved at CNS–28. CNS–28 is not bound by T-bet in mice Th1 cells. Human IFNG gene is located in negative strand and mouse Ifng gene is located in positive strand. GATA-3 ChIP-seq data in naïve CD4+ cells and T-bet ChIP-seq data in Th1 cells were generated in this study and were also shown in panel B. Human ChIP-seq data were obtained from 60 and 61 with the annotation of GSM number from GEO in the figure.
(E) The interaction matrix heatmaps from Hi-TrAC data around CNS–34, –28, –22, and Ifng promoter in WT and Mll4 KO naïve CD4+ T cells. CNS–22 was set as the viewpoint for the virtual 4C plots. The number of interacting reads for each chromatin loop is shown above the arches. The cLoops2 plot module generated the plots.
Data are representative of at least two independent experiments (A-E). ***p < 0.001. NS: not statistically significant. (Two-way ANOVA with Tukey’s multiple comparison test, error bars represent SD).
To further characterize the Ifng –28Kb region, we examined T-bet binding patterns using ChIP-seq and found that T-bet bound to CNS–34, CNS–22, CNS–6 and the promoter region of Ifng but no binding was detected at −28Kb (Figure 2B). Another key TF for Th2 cells, GATA3, is also known to be associated with the Ifng locus and plays a critical role for Th1/2 differentiation balance.25,37 Loss of MLL4 resulted in a reduced binding of GATA3 at −28Kb site in naïve CD4+ T cells, while no changes of GATA3 binding were detected at CNS–22, CNS+41 and Ifng promoter regions (Figures 2B and 2C). We further discovered that loss of GATA3 caused only a very modest decrease in H3K4me1 modification at CNS–28 (Figure S2A), and the regulation of defense response was among the top five enriched GO terms for the changed H3K4me1 peaks in GATA3 deficient cells (Figures S2B and S2C). Moreover, only a limited number of significantly changed CTCF peaks genome-widely were identified and no substantial changes in CTCF binding were detected in the Ifng locus in the Gata3 KO naïve CD4+ T cells (Figures S2D and S2E). Altogether, these data indicate that GATA3 acts downstream of MLL4 on CNS–28. Moreover, while −28Kb site is not within a highly conserved region, we observed shared features that both human and mouse −28Kb sites were associated with H3K4me1 modifications and bound by GATA3 (Figure 2D). Thus, we defined −28Kb region as CNS–28 site despite lacking high sequence conservation.
To better understand the effects of MLL4 deficiency on the three-dimensional organization of the Ifng locus, we employed Hi-TrAC to examine the interactions of all regulatory elements at high-resolution.38 By applying Hi-TrAC to naïve CD4+ T cells, we obtained about 35 million unique PETs and a total of 27,963 chromatin loops, with 204 wildtype (WT) and 115 Mll4 KO-specific loops (Figures S2F and S2G). While we did not find any enriched GO terms in the WT-specific loops, pathways involved in leukocyte activation or cell activation were identified from the target genes of Mll4 KO-specific loops (Figures S2H and S2I). MLL4 deletion reduced the interaction among CNS–22, CNS–28 and CNS–34, while it increased the interaction between CNS–22 and Ifng promoter regions (Figure 2E), despite the overall increased interactions in the domain in Mll4 KO cells measured by Hi-C (Figures 1H and 1I). Given the known enhancer activities of CNS–22,11 our data implicate that CNS–28 restrains Ifng expression via locally trapping cis elements of CNS–22 and affecting chromatin looping within the Ifng locus.
Loss of CNS–28 results in tissue inflammation and enhanced IFN-γ production.
To directly test whether CNS–28 contributes to IFN-γ expression, we first generated G28Δ mice, which lost a 1kb fragment of the CNS–28 region (Figure S3A). G28Δ mice exhibited normal T cell development in the periphery (Figures S3B–S3G). In vitro differentiation showed elevated Th1 response from G28Δ mice, implicating that CNS–28 plays a role in repressing Ifng expression (Figures S3H and S3I). To pinpoint the function of CNS–28 more specifically, we next generated CNS–28Δ mice with precise deletion of a 156 bp fragment containing two GATA3 binding motifs within CNS–28 (Figure S4A). ChIP-qPCR assays confirmed that GATA3 binding at CNS–28 was compromised in naïve CD4+ T cells from CNS–28Δ compared to the control mice, while its bindings were not affected at CNS–22, CNS+41 and Ifng promoter regions (Figure S4B). Similar to G28Δ mice, we also found no defect in Th17 or Treg cells from CNS–28Δ mice relative to WT mice (Figures 3A and 3B). However, both Th0 and Th1 cells from CNS–28Δ mice exhibited elevated IFN-γ compared to WT cells (Figure 3A and 3B). Considering the enhanced IFN-γ expression, we asked whether CNS–28Δ mice exhibited any symptoms regarding tissue inflammation. We examined CNS–28Δ mice over time and found that while young (8-week-old) mice displayed normal tissue homeostasis (data not shown), non-lymphoid tissues from the 40-week-old CNS–28Δ mice showed enhanced immune-cell infiltration in multiple organs compared to WT mice (Figures 3C and 3D). Additionally, we noticed elevated IFN-γ levels in 40-week-old CNS–28Δ compared to WT mice but not in young (8-week-old) WT and CNS–28Δ mice (Figure 3E). These data suggest that loss of CNS–28 results in disrupted systemic immune tolerance and chronic inflammation via elevated IFN-γ responses. We further examined lymphatic organs in CNS–28Δ mice and found comparable CD4+ and CD8+ T cell proportion in spleen and lymph nodes (Figures S4C, S4D, 3F, and 3G). Young CNS–28Δ mice also showed similar levels of Foxp3+ Treg cells in spleen and lymph nodes compared to WT mice (Figure S4F). Further, while young WT and CNS–28Δ mice exhibited similar CD4+ T cell composition in spleens and lymph nodes, we observed a greater proportion of activated (CD62LloCD44+) CD4+ T cells in 40-week-old CNS–28Δ mice than in their WT counterparts (Figures S4G, S4H, 3H, and 3I), consistent with hyperactivation of immune responses in CNS–28Δ mice.
Fig 3. Loss of CNS–28 results in tissue inflammation and enhanced IFN-γ production.
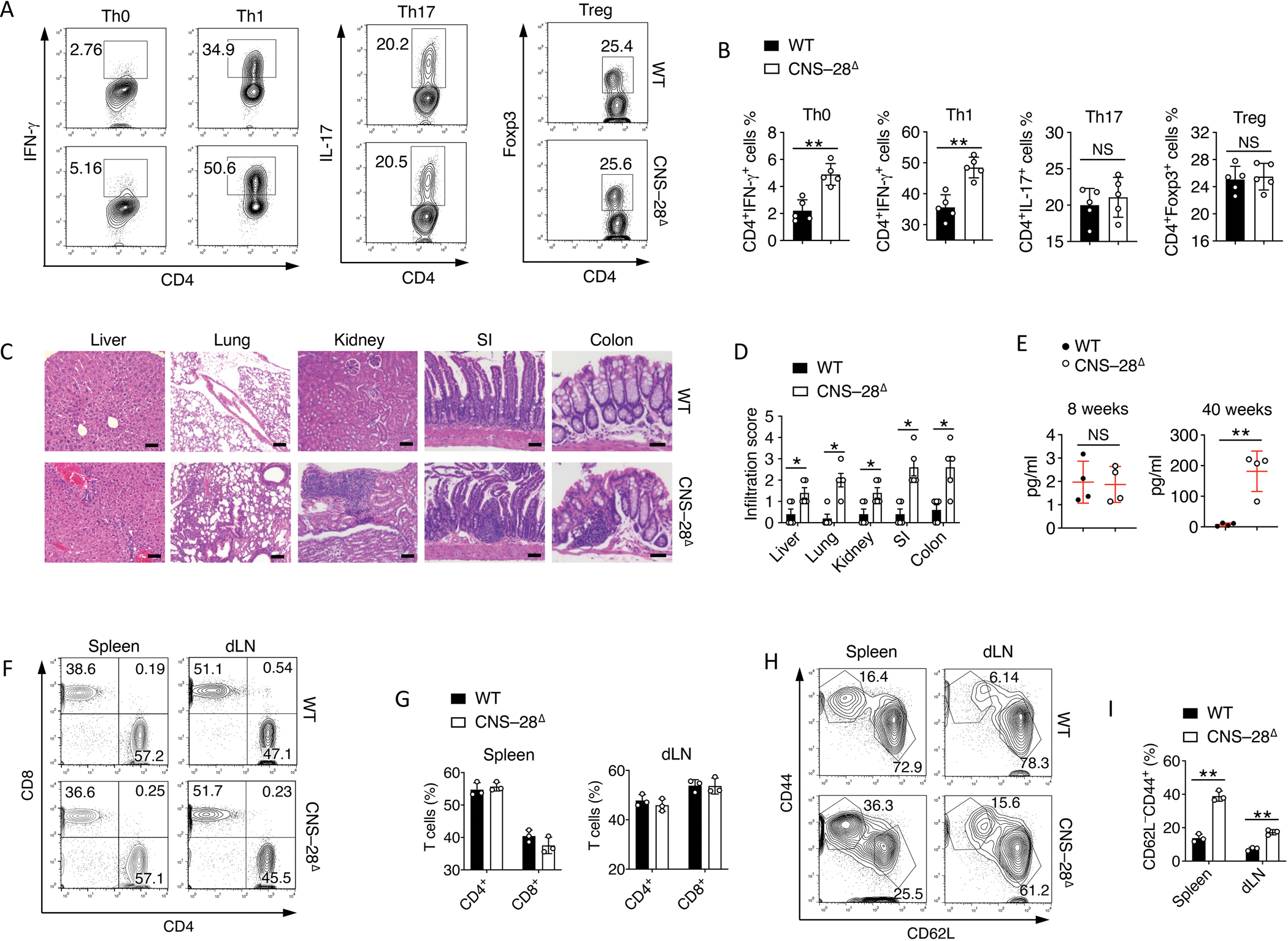
(A-B) Naive CD4+ T cells from WT and CNS–28Δ mice were stimulated under Th0, Th1, Th17 and pTreg conditions and harvested at 72 hours. Intracellular staining of indicated cytokines produced by different polarized T cells from WT and CNS–28Δ mice was analyzed by (A) flow cytometry and (B) Quantification.
(C) Representative hematoxylin and eosin staining of liver, lung, kidney, small intestine (SI) and colon sections. Scale bar: liver, lung, 100 μm; kidney, SI, colon, 50 μm.
(D) Quantification of histological analysis from Fig. 3C of mice aged 40 weeks old. 0–3 score: 0, no mononuclear infiltration; 3, high degree of mononuclear infiltration.
(E) ELISA of IFN-γ in the serum in WT and CNS–28Δ mice at 8 or 40 weeks of age.
(F-G) (F) Flow cytometry analysis and (G) quantification of CD4 and CD8 expression on CD45+ cells from spleen and peripheral lymph nodes (dLN) of WT and CNS–28Δ mice at 40 weeks of age.
(H-I) (H) Flow cytometry and (I) quantification of CD44 and CD62L expression on CD4+ T cells from spleen and peripheral lymph nodes (dLN) of WT and CNS–28Δ mice at 40 weeks of age.
Data are representative of at least two independent experiments (A, C-I) and pooled from two independent experiments (B). *p < 0.05, **p<0.01. NS: not statistically significant. (Student’s t-test, error bars represent SD).
CNS–28 represses intrachromosomal interactions within Ifng locus.
To investigate the molecular mechanism of how CNS–28 is involved in the modulation of Ifng expression, we analyzed kinetics of Ifng and Tbx21 expression during Th1 cell differentiation and found that both genes exhibited higher levels in CNS–28Δ than in control T cells. Furthermore, Ifng expression started to elevate as early as 12 hours after the differentiation while Tbx21 expression starts to increase at 48 hours in CNS–28Δ compared to the control T cells (Figure 4A), indicating CNS–28 regulates Ifng via a T-bet independent manner, at least at the early phase of expression. We confirmed the conclusion with RNA-seq data (Figure S5A). Further, only limited numbers of genes were significantly changed in CNS–28Δ cells during in vitro Th1 differentiation (Figure S5B). We only found enriched GO terms for the 72hr up-regulated genes in the CNS–28Δ Th1 cells, which included inflammatory response and cytokine production (Figure S5C). These data suggest that the loss of CNS–28 has a local effect rather than a systemic effect.
Fig 4. CNS–28 represses interaction between CNS-22 and Ifng promoter.
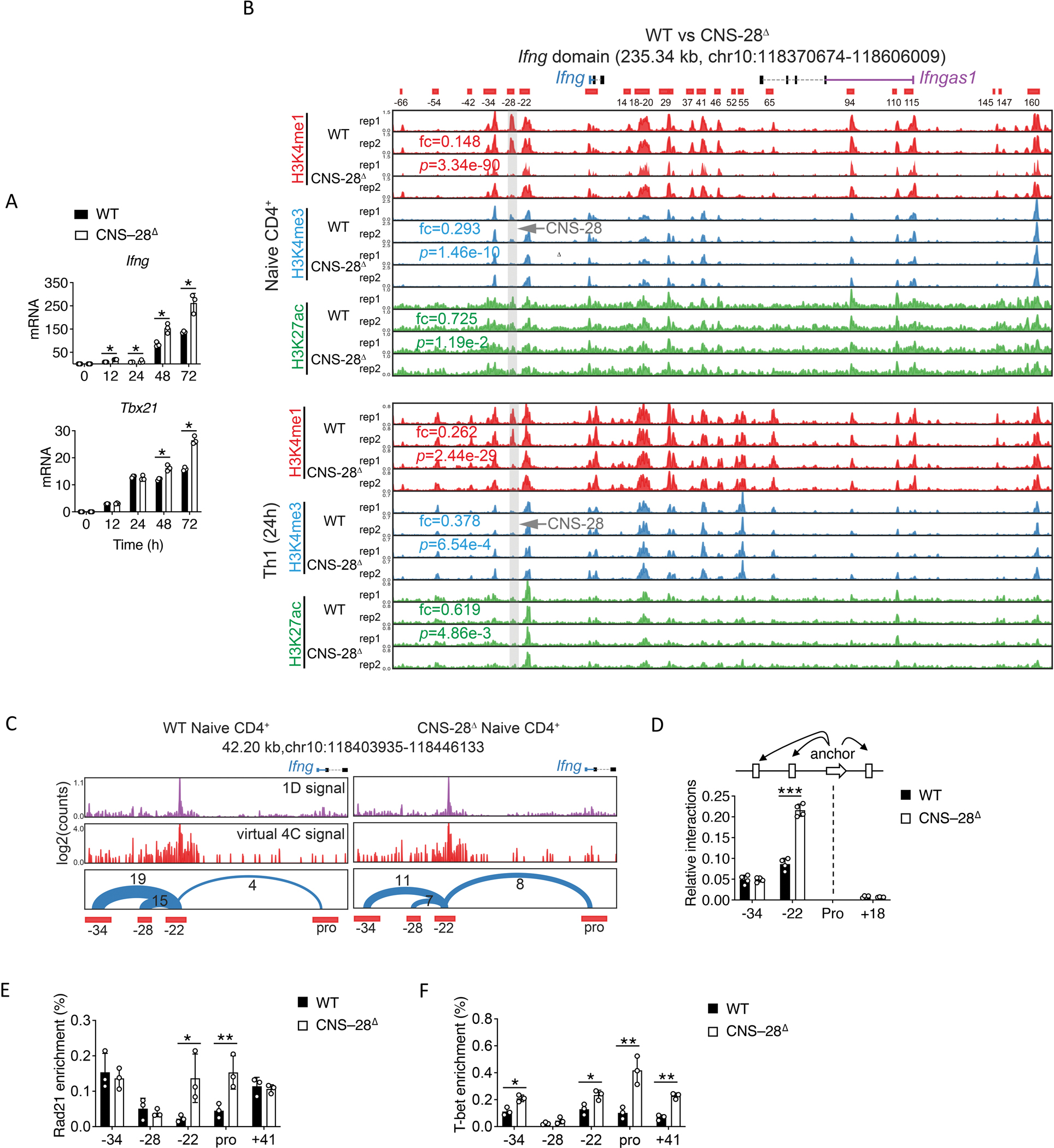
(A) qPCR analysis of mRNA in WT and CNS–28Δ naïve CD4+ T cells stimulated for 12, 24, 48 and 72 h (horizontal axis) under Th1 conditions; results are presented relative to those of Gapdh.
(B) Genome browser images of H3K4me1, H3K4me3 and H3K27ac ChIP-seq profiles in the Ifng domain. The ChIP-seq data were generated in this study. The mean of normalized reads counts (to a total of 10 million reads) from two replicates of ChIP-seq data in the CNS-28 locus were used to calculate fold changes (KO/WT) and Poisson P-values.
(C) The chromatin interactions originating from CNS-22 detected by Hi-TrAC in WT and CNS–28Δ naïve CD4+ T cells. The high-quality unique PETs from Hi-TrAC libraries were down-sampled to 37 million for a fair comparison between WT and CNS–28Δ cells. CNS–22 was set as the viewpoint for the virtual 4C plots. The number of interacting PETs for each chromatin loop is shown above the arches. The cLoops2 plot module generated the plots.
(D) 3C-qPCR analysis of interaction intensity between the Ifng promoter and other indicated elements in WT and CNS–28Δ Th1 cells. The cartoon above the data indicates the fixed anchor fragment (dashed black lines) and other Hpa I fragments used for the assay.
(E-F) ChIP-qPCR analysis of the binding of (E) Rad21 or (F) T-bet to the Ifng locus in WT or CNS–28Δ Th1 cells, presented relative to input.
Data are representative of at least two independent experiments (A-F). *p < 0.05, **p < 0.01, ***p<0.001, NS: not statistically significant. (Two-way ANOVA with Tukey’s multiple comparison test, error bars, SD).
Given the early induction of Ifng in CNS–28Δ Th1 cells, we performed ChIP-seq assays to examine different histone modifications in naïve CD4+ T cells and 24h differentiated Th1 cells. We noticed that CNS-28 was associated with high levels of H3K4me1, but low levels of H3K4me3 and H3K27ac, and loss of CNS–28 led to diminished H3K4me1 in both naïve and Th1 conditions while H3K4me3 and H3K27ac histone marks were not affected (Figure 4B). These data suggest that CNS-28 does not function as an either direct or indirect enhancer for Ifng or other genes. Moreover, we found no changes of histone modification at Tbx21 locus between CNS–28Δ and control mice (Figure S5D), suggesting that T-bet may not be directly involved in the function of CNS–28. Analysis of these ChIP-seq data revealed that the global patterns of these histone modifications are very similar between wildtype and CNS–28Δ cells (Figures S5E–S5G). Only a limited number of changed peaks were detected and no enriched GO terms were found for the changed peaks, implicating that the effect of CNS–28 is mainly limited to the Ifng locus.
On the other hand, while similar to naïve and in vitro differentiated Th1 cells, significantly decreased H3K4me1 signals at the CNS-28 region were also observed in CNS–28Δ CD4+ memory T cells (Figure S6A). Globally, 722 and 3,813 H3K4me1 peaks were decreased and increased, respectively, in CNS–28Δ CD4+ memory T cells (Figure S6B). The increased peaks were associated with pathways involved in immune functions (Figure S6C). Deletion of CNS–28 resulted in 55 down-regulated and 167 up-regulated genes in CD4+ memory T cells (Figure S6D). The enhanced genes were enriched in pathways involved in the regulation of immune responses (Figure S6E). CNS–28Δ CD4+ memory T cells exhibited higher Ifng expression than WT cells (Figure S6F), indicating the role of CNS–28 in controlling IFN-γ response in vivo.
Next, considering the potential role of CNS–28 in modulating the spatial organization of Ifng locus, we performed Hi-TrAC assay on naïve CD4+ T cells (Figure S6G). Among a total of 27,873 chromatin loops genome-wide, CNS–28 deletion resulted in only 43 decreased and 47 increased loops, suggesting the effect of CNS–28 is mainly limited to the Ifng locus (Figure S6H). We noticed that deletion of CNS–28 led to decreased interactions among CNS–22, CNS–28 and CNS–34 meanwhile leading to increased interactions between CNS–22 and Ifng promoter (Figure 4C). Further, chromosome conformation capture (3C) assays in Th1 cells confirmed that enhanced interactions between CNS–22 and Ifng promoter in CNS–28Δ Th1 cells, while CNS–34-promoter and CNS+18-promoter interactions were not changed (Figure 4D). Cohesin (Rad21) is important for spatial chromatin organization and is involved in enhancer-promoter interactions.39,40 We assessed the binding of Rad21 at the Ifng locus in the Th1 cell setting using ChIP-qPCR assays. We found increased binding of Rad21 at CNS–22 and Ifng promoter in CNS–28Δ Th1 cells than in WT Th1 cells (Figure 4E). Furthermore, while no change of Tbx21 expression was observed at 24h of Th1 cell differentiation between WT and CNS–28Δ mice, there was greater binding of T-bet to CNS–34, CNS–22, Ifng promoter and CNS+18 regions in the CNS–28Δ than in WT Th1 cells, supporting the elevated Ifng expression in CNS–28Δ Th1 cells (Figure 4F). Altogether, our data indicate that CNS–28 served a critical role in silencing the IFN-γ response, via a chromatin-looping mechanism, repressing T-bet interaction with distal cis enhancer elements within Ifng locus.
CNS–28 is critical for type 1 responses during host defense and inflammation
To further investigate the role of CNS–28 in CD4+ T cells in vivo, we carried out chronic autoimmune inflammation by using a T cell transfer colitis model. Either WT or CNS–28Δ CD4+CD25−CD45RBhi cells were transferred into Rag2−/− mice to induce colitis. Mice that received CNS–28Δ CD4+CD25−CD45RBhi cells exhibited more severe disease with worse colonic tissue damage (Figures 5A–5C). Moreover, recovered CNS–28Δ T cells from colonic tissues showed higher IFN-γ expression levels than did WT T cells (Figures 5D and 5E), indicating that CNS–28 suppressed Th1 responses during inflammatory reactions.
Fig 5. CNS–28 is critical for type 1 responses during host defense and inflammation.
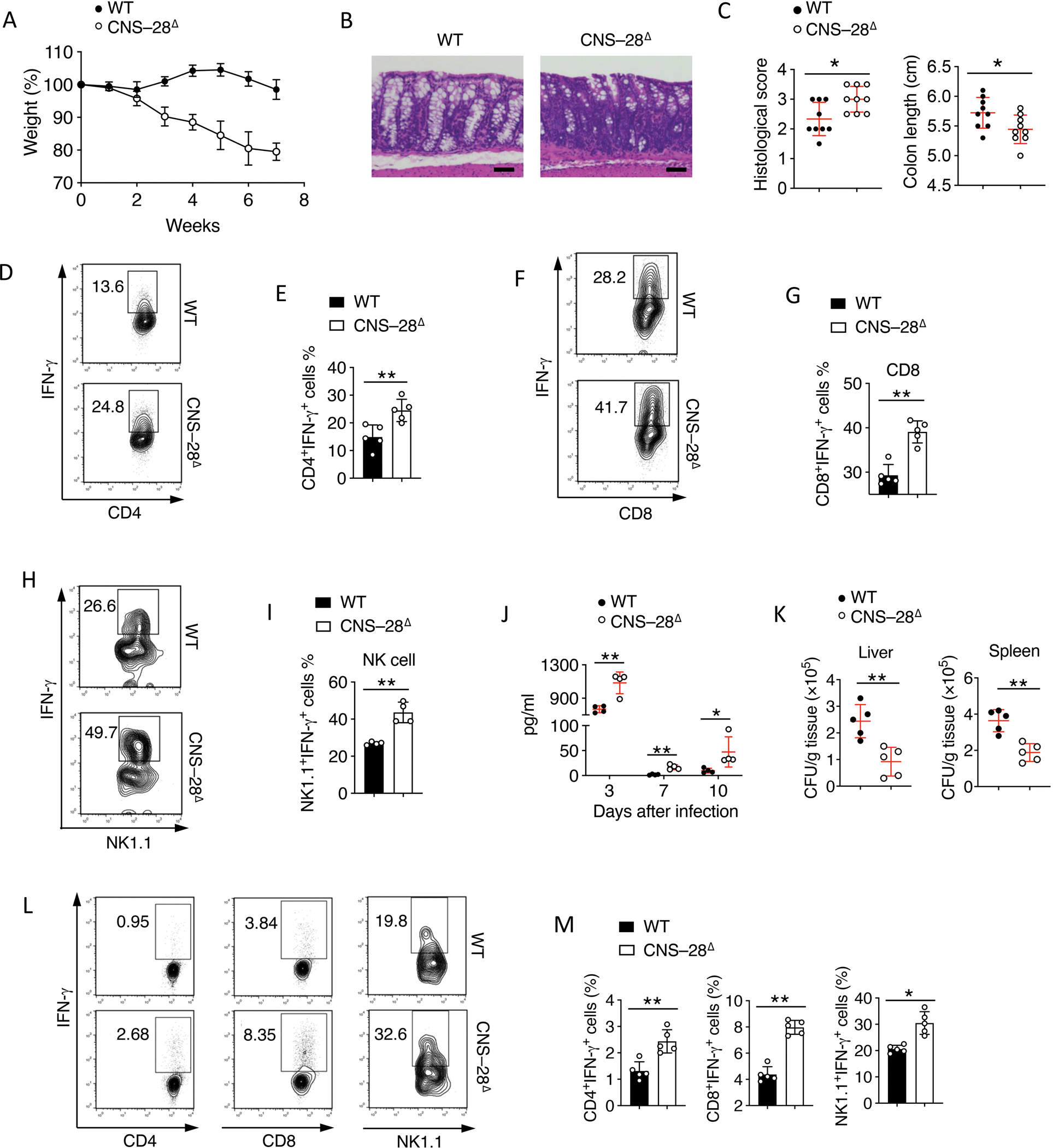
(A) Body weight of Rag2−/− mice transferred i.p. with CD4+CD25−CD62L+ cells from WT or CNS–28Δ mice.
(B) Hematoxylin and eosin staining of colonic tissue from the different groups as in (A) 10 weeks after colitis induction, scale bar, 50μm.
(C) Left: Quantification of pathological changes in the colon of mice as in (A); Right: Colon lengths of Rag2−/− mice which had received the indicated cells for transfer as in (A), measured from the colocecal junction to the anal verge
(D-E) (D)Flow cytometry analysis and (E) Quantification of IFN-γ expression by CD4+ T cells isolated from LP from indicated groups as in (A) 10 weeks after colitis induction.
(F-I) Splenocytes from WT and CNS–28Δ mice were isolated and cultured in the presence of IL-12 and IL-2 for 6 h. IFN-γ expression in (F, G) CD8+ T cells or (H, I) NK1.1+ NK cells were measured by (F, H) flow cytometry or (G, I) Quantification.
(J-M) Listeria were inoculated into WT and CNS–28Δ mice by oral gavage. The mice were sacrificed on day 14 for further tests.
(J) IFN-γ level in serum was assessed by ELISA at indicated day.
(K) Bacteria CFU was counted at day 7 after infection in liver and spleen.
(L) Flow cytometry analysis and (M) Quantification of IFN-γ expression in CD4+, CD8+ T cells from spleen 7 days after infection and NK cells isolated from spleen 1 day after infection.
Data are representative of at least two independent experiments (A, B, C-M) or pooled from two independent experiments (C). *p < 0.05, **p < 0.01 (Student’s t test, error bars represent SD).
We next investigated whether CNS–28 also modulated Ifng expression during innate immune responses. We observed that both CD8+ T cell and NK cell exhibited elevated IFN-γ after in vitro activation (Figures 5F–5I). By utilizing a Listeria infection model in which innate immune cell-derived IFN-γ plays a key role for host defense,41 we observed CNS–28Δ mice exhibited elevated serum IFN-γ level during acute infection (Figure 5J), accompanied with reduced bacteria CFU in both spleen and liver at day 7 compared to control mice (Figure 5K). Meanwhile, although CD4+ T cells were found to mildly increase IFN-γ during infection, both CD8+ and NK cells displayed higher IFN-γ in CNS–28Δ mice than in control mice (Figures 5L and 5M). Taken together, consistent with our in vitro data, these results suggest that CNS–28 is a key genomic element which is critical for IFN-γ restriction in both innate and adaptive immune cells.
CNS–28 represses type 2 responses via enhanced IFN-γ expression.
In addition to elevated Th1 response in vitro (Figures 3A and 3B), we further found dampened Th2 differentiation in the CNS–28 deletion cells (Figures S7A, S7B, 6A, and 6B). We then evaluated whether deletion of CNS–28 affected type 2 response in vivo using a model of house dust mite (HDM)-induced asthma in which type 2 cytokines dominate during tissue inflammation 42. After HDM challenge for 10 days, we found CNS–28Δ mice were protected from mononuclear cell infiltration into the lung tissue compared to WT mice (Figure 6C). Loss of CNS–28 resulted in reduced immunoglobulin E (IgE) in the serum and bronchoalveolar lavage fluid (Figure 6D and 6E). The lung tissue of CNS–28Δ mice exhibited less inflammatory leukocytes, including eosinophils and T cells, than did WT mice (Figures S7C, S7D, and 6F). Moreover, recovered infiltrated CD4+ T cells from lung tissue of CNS–28Δ mice also showed less type 2 cytokines compared to WT T cells (Figures 6G and 6H). Altogether, these results indicated that deletion of CNS–28 represses type 2 response during both in vitro Th2 differentiation and in vivo mucosal allergic responses.
Fig 6. Reduced type 2 responses by deletion of CNS–28.
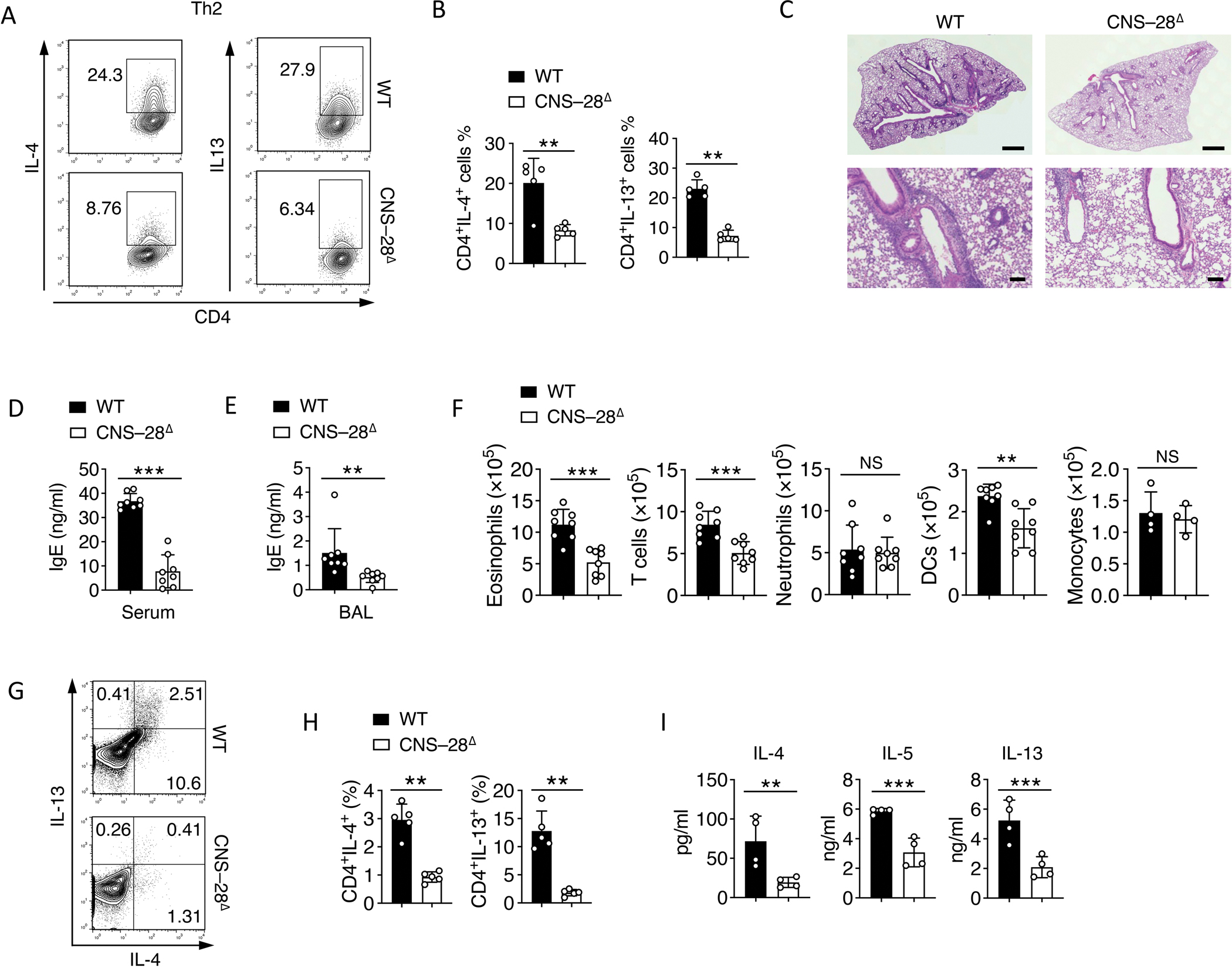
(A-B) Naïve CD4+ T cells from WT and CNS–28Δ mice were stimulated under Th2 condition and harvested at 72 hours. Intracellular staining of indicated cytokines was measured by (A) flow cytometry and (B) Quantification.
(C) Hematoxylin-and-eosin staining of lung-tissue sections of WT and CNS–28Δ mice, assessed after 10 days of HDM challenge. Scale bar, top row, 0.5 mm; bottom row, 100 μm.
(D-E) ELISA of IgE in the (D) serum and (E) bronchoalveolar lavage (BAL) fluid of WT and CNS–28Δ mice 10 days after HDM challenge.
(F) Frequency of inflammatory cells in the lung tissue of WT and CNS–28Δ mice, assessed at 10 days after HDM challenge.
(G) Flow cytometry analysis and (H) Quantification of type 2 cytokines in CD4+ T cells isolated from the lung 10 days after HDM challenge.
(I) Recovered cells from the lung 10 days after HDM challenge were restimulated by HDM. The levels of type 2 cytokines levels in medium were assessed by ELISA after 3 days of restimulation.
Data are representative of at least two independent experiments (A-C, G-I) or pooled from two independent experiments (D-F). **p < 0.01, ***p < 0.001, NS: not statistically significant. (Student’s t test, error bars represent SD).
There is reciprocal regulation between Th1 and Th2 cells.25,43 We examined the kinetics of Ifng and Th2-cell-associated gene expression during Th2 cell differentiation. CNS–28Δ T cells exhibited lower expression of Il4, Il5 and Il13, but higher expression of Ifng at 12 h after Th2 cell differentiation than did the WT T cells, suggesting a role of CNS–28 in suppressing the early stage Ifng expression during Th2 responses (Figure 7A). We also found elevated IFN-γ production from CNS–28Δ Th2 cells during in vitro differentiation (Figure 7B). To determine whether the suppression of type 2 responses in CNS–28Δ Th2 cells was due to elevated expression of IFN-γ, we supplied a neutralizing antibody to IFN-γ during Th2 cell differentiation and observed full restoration of type 2 cytokines expression in CNS–28Δ Th2 cells (Figures 7C and 7D). Therefore, these data suggested that IFN-γ induced in Th2 cells was fully responsible for the reduction in the type 2 cytokines.
Fig 7. CNS–28 represses type 2 responses due to enhanced IFN-γ production.
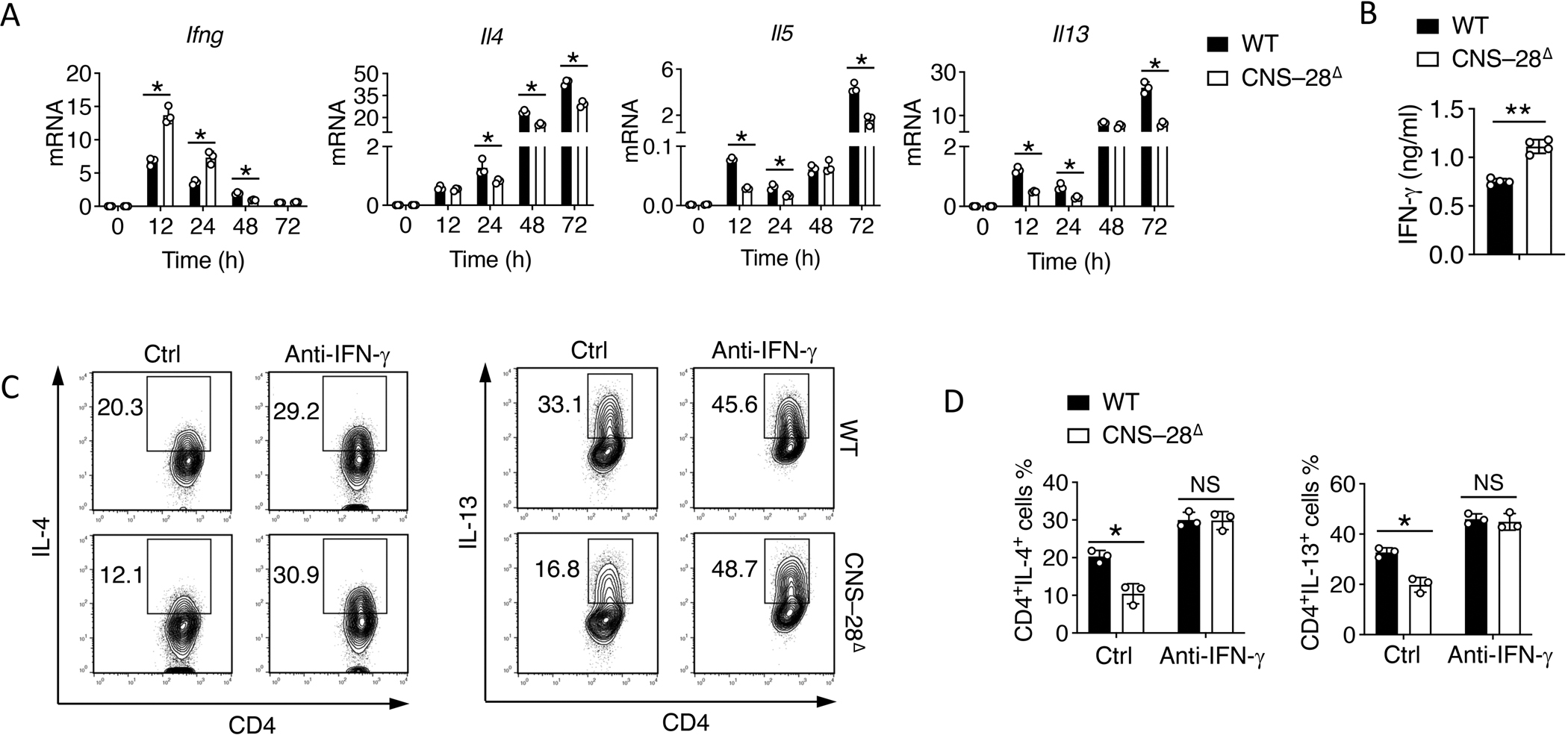
(A) qPCR analysis of mRNA during differentiation of WT and CNS–28Δ naïve CD4+ T cells under Th2 conditions for 12, 24, 48 or 72 h (horizontal axis); results are presented relative to those of Gapdh.
(B) ELISA assessment of IFN-γ in the culture medium in WT and CNS–28Δ differentiated Th2 cells.
(C-D) Activated WT and CNS–28Δ naive CD4+ T cells were stimulated with combinations of IL-4 with anti-IFN-γ antibody for 3 days. Intracellular staining of type 2 cytokines was measured by (C) flow cytometry and (D) Quantification.
Data are representative of at least two independent experiments (A-D). *p < 0.05, ***p < 0.01, NS: not statistically significant. (A, D, Two-way ANOVA with Tukey’s multiple comparison test; B, Student’s t test, error bars represent SD).
Discussion
While multiple cis-regulatory elements surrounding Ifng, including enhancers and boundary elements, promote Ifng expression,16 the contributions of negative regulatory elements to Ifng regulation have not been resolved. Here, we identified CNS–28 as a distal silencer for Ifng expression. In naïve CD4+ T cells, MLL4 mediates remote H3K4 methylation at CNS–28 which is bound by GATA3, forming a unique local chromatin configuration to inhibit Ifng expression. Deletion of CNS–28 promoted interactions between CNS–22 with Ifng promoter without interfering with Tbx21 activation. Functionally, we demonstrated that CNS–28 restrains Ifng transcription in Th1, Tc1 and NK cells during both innate and adaptive immune responses, protecting the tissues from autoimmunity and immune hyperreaction. Moreover, we found that loss of CNS–28 led to disrupted Th1/Th2 balance, repressing type 2 responses due to elevated IFN-γ expression.
Early studies have characterized the function of nearby cis-regulatory elements at the Ifng gene locus, such as CNS1, CNS2, HS1, HS2, and HS3.33 The dynamic interplay of histone modification and TF binding at these regions coordinate to regulate the developmental specificity of IFN-γ expression.33 Both CNS–22 and CNS–34 function as enhancers to promote Ifng expression during Th1 cell differentiation.16 Consistently, our data showed that both CNS–22 and CNS–34 are associated with active histone modification marks. Moreover, CNS–22 and CNS–34 can reorganize the chromatin structure of the extended Ifng locus by recruiting CTCF and cohesin.44,45 Our data in this study indicated that CNS–28 also contributes to chromatin organization by suppressing the enhancer-promoter interactions within the Ifng locus. Such impacts only occurred at CNS–22 but not CNS–34. While some studies have shown that CNS–34 acts as T-bet dependent enhancer in adaptive immunity,10,46 other studies suggest that CNS–34 has enhancer blocking activity to ensure a poised state of the Ifng locus in naïve CD4+ T cells.47 The different TFs bound at CNS–22 and CNS–34 may lead to the distinct modes of how CNS–28 interacts with these two elements in regulating Ifng transcription subsequent to differentiation-driven local chromatin remodeling.16 In addition, the H3K4 mono-methylation at CNS–28 is catalyzed in trans by MLL4 bound at CNS–22 via long-distance chromatin looping in naïve CD4+ T cells, indicating a preferential interaction between CNS–22 and CNS–28.32
GATA3 inhibits type 1 responses.15,48,49 It serves as a key factor for fate decision during Th1 and Th2 differentiation and switch by repressing T-bet target genes and promoting Th2 genes.50 GATA3 is also expressed in different cell types, such as T cells and NK cells, which are essential for immune function,51,52 and regulating immune responses in a cell type specific manner. Loss of GATA3 leads to hyper-reaction of type 1 immune responses.50 Further, GATA3 deficiency in Treg cells could impair Treg cell function and Foxp3 expression for the generation of immune tolerance.53 Thus, in addition to such extrinsic effect via Treg cells, our findings complemented the previous studies and provided additional evidence that GATA3 may repress type 1 responses by intrinsic control of IFN-γ through CNS–28.
T-bet and GATA3 collaborate in regulating many genes by binding to the same sites in the genome.25 During Th1 cell differentiation, GATA3 binding is detected at the sites not associated with the expression of Th2 specific genes. Instead, GATA3 occupies the sites co-bound by T-bet in Th1 cells. More importantly, the presence of T-bet is responsible for GATA3 binding at Th1 specific sites.16,25 Our work shows that MLL4 deficiency compromised GATA3 binding to CNS-28 but did not change the CTCF binding patterns at the Ifng locus. Thus GATA3 acts downstream of MLL4 and binds to CNS–28 independent of T-bet to repress Ifng activation and expression, which is crucial in maintaining the naïve state of T cells and preventing autoimmunity. Th1 differentiation shuffles the divergent interactions of T-bet and GATA3 on the chromatin. In particular, the dynamic switch of decreased GATA3 binding at CNS–28 with increased T-bet binding at CNS–22 leads to chromatin reconfiguration for the upregulation of IFN-γ.11,44,54 While it has been suggested that GATA3 expression and redistribution in Th1 cells may contribute to plasticity of T cells by associating with T-bet, our results provided an alternative mechanism by which GATA3 alone exerts its function to secure T cell quiescence from hyperactivation via modulating chromatin configuration.
Numerous studies support that high order chromatin organization plays important roles in Ifng gene regulation.16,45,55–58 Here we identified a CNS–28 distal regulatory element which maintains quiescent chromatin structure to repress Ifng gene in a T-bet independent manner. We demonstrated that CNS–28 plays a crucial role in restraining IFN-γ response during both innate and adaptive immunity, protecting the body from autoimmunity. Therefore, our findings provided insights into how cis elements cooperate with each other for gene silencing to confine cellular quiescence, revealing a T-bet independent mechanism for a tight control of Ifng expression.
Study Limitation
We suggested that CNS–28 functions independently from T-bet, but this conclusion is based on circumstantial evidence. Crossing CNS–28Δ mice with T-bet deletion mice would provide more conclusive evidence. Additionally, our data do not fully address whether the regulation of CNS-28 function in repressing Ifng expression requires the enzymatic activity of MLL4. Thus, it would be interesting to test whether the point mutation, which inactivates the enzymatic activity but retains the chromatin binding of MLL4, compromises the function of CNS-28. Although we showed that GATA-3 acts downstream of MLL4, it is still unclear how GATA-3 regulates the function of CNS–28 in repressing Ifng expression. Further investigation is required to assess the chromatin conformation changes in Ifng locus in GATA-3 deletion cells, focusing on the looping of CNS–34, CNS–28, CNS–22 and promoter region, to understand whether GATA3 participates in repression of Ifng expression by impacting CNS–22-promoter interactions via CNS–28.
STAR★Methods
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Keji Zhao ( zhaok@nhlbi.nih.gov ).
Materials availability
All unique reagents generated in this study will be made available upon request. An agreement with our institute’s Materials Transfer Agreement (MTA) may be required.
Data and Code availability.
The sequencing data including ChIP-seq, RNA-seq and Hi-TrAC data have been deposited in the Gene Expression Omnibus database with accession number: GSE204946.
All codes used for data analysis in this paper are public and listed in the key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FITC anti-mouse CD4 | Biolegend | Cat# 130308, RRID: AB_1279237 |
| APC anti-mouse CD8a | Biolegend | Cat# 100711, RRID: AB_312750 |
| Brilliant Violet 785™ anti-mouse CD45 | Biolegend | Cat# 103149, RRID: AB_2564590 |
| PE anti-mouse CD62L | Biolegend | Cat# 104407, RRID: AB_313094 |
| APC anti-mouse CD44 | Biolegend | Cat# 103012, RRID: AB_312963 |
| PE/Cy7 anti-mouse IL-17A | Biolegend | Cat# 506922, RRID: AB_2125010 |
| Pacific Blue™ anti-mouse IFN-γ | Biolegend | Cat# 505818, RRID: AB_893526 |
| APC anti-mouse IL-4 | Biolegend | Cat# 504106, RRID: AB_315320 |
| PE/Cy7 anti-mouse CD11b | Biolegend | Cat# 101216, RRID: AB_312799 |
| PerCP/Cyanine5.5 anti-mouse CD3 | Biolegend | Cat# 100217, RRID: AB_1595597 |
| PE anti-mouse CD170 (Siglec-F) | Biolegend | Cat# 155506, RRID: AB_2750235 |
| APC anti-mouse F4/80 | Biolegend | Cat# 123116, RRID: AB_893481 |
| APC/Fire™ 750 anti-mouse Ly-6C | Biolegend | Cat# 128045, RRID: AB_2616730 |
| Brilliant Violet 421™ anti-mouse I-A/I-E (MHC-II) | Biolegend | Cat# 107631, RRID: AB_10900075 |
| FITC anti-mouse Ly-6G | Biolegend | Cat# 127606, RRID: AB_1236494 |
| APC anti-mouse CD11c | ThermoFisher | Cat# 17-0114-81, RRID: AB_469345 |
| APC anti-mouse FOXP3 | ThermoFisher | Cat# 77-5775-40, RRID: AB_469981 |
| PE anti-mouse CD45RB | Biolegend | Cat# 103308, RRID: AB_313015 |
| anti-mouse CD3ε | Bio X Cell | Cat# BE0001-1, RRID: AB_1107634 |
| anti-mouse CD28 | Bio X Cell | Cat# BE0015-5, RRID: AB_1107628 |
| anti-mouse IFNγ | Bio X Cell | Cat# BE0055, RRID: AB_1107694 |
| anti-pSTAT4 | R and D Systems | Cat# PA-ST4, RRID: AB_2302639 |
| anti-STAT4 | R and D Systems | Cat# MAB5287, RRID: AB_2302638 |
| anti-H3 | Abcam | Cat# 10799, RRID: AB_470239 |
| anti-H3K4me1 | Abcam | Cat# ab8895, RRID: AB_306847 |
| anti-H3K4me3 | Millipore | Cat# 17-614, RRID: AB_11212770 |
| anti-H3K27ac | Abcam | Cat# ab4729, RRID: AB_2118291 |
| anti-GATA3 | BD bioscience | Cat# 558686, RRID: AB_2108590 |
| anti-T-bet | BD bioscience | Cat# 561263, RRID: AB_10563082 |
| anti-MLL4 | 1 | N/A |
| Bacterial strains | ||
| Listeria monocytogenes | ATCC | Cat# 35152™ |
| Chemicals, peptides, and recombinant proteins | ||
| House Dust mite protein | Greer Laboratories | Cat# XPB82D3A2.5 |
| LB Broth | Sigma | Cat# L3522 |
| LB Broth with agar | Sigma | Cat# L3147 |
| PMA | Sigma | Cat# P8139 |
| Ionomycin | Sigma | Cat# I0634 |
| Recombinant Human TGF-beta 1 Protein | R&D | Cat# 240-B-002 |
| Recombinant Mouse IL-12 Protein | R&D | Cat# 419-ML-010 |
| Recombinant Mouse IL-4 Protein | R&D | Cat# 404-ML-010 |
| Recombinant Mouse IL-6 Protein | R&D | Cat# 406-ML-005 |
| formaldehyde | Thermo Scientific | Cat# 28908 |
| T4 DNA polymerase | New England Biolabs | Cat#M0203L |
| rCutSmart buffer | New England Biolabs | Cat# B6004S |
| Hpa I | New England Biolabs | Cat# R0105L |
| MluCI | New England Biolabs | Cat# R0538L |
| NlaIII | New England Biolabs | Cat# R0125L |
| Proteinase K | Roche | Cat# 03115828001 |
| T4 DNA Ligase | New England Biolabs | Cat# M0202L |
| Critical commercial assays | ||
| ELISA MAX™ Standard Set Mouse IFN-γ | Biolegend | Cat# 430801 |
| Mouse IgE ELISA MAX™ Deluxe | Biolegend | Cat# 432404 |
| Fixation/Permeabilization Solution Kit with GolgiPlug™ | BD Biosciences | Cat# 555028 |
| Foxp3 / Transcription Factor Fixation/Permeabilization Concentrate and Diluent | ThermoFisher | Cat# 00-5521-00 |
| iScript RT Supermix for RT-qPCR | Biorad | Cat# 1708841 |
| RNeasy Plus Mini Kit | Qiagen | Cat# 74136 |
| End-It DNA-Repair kit | Epicentre | Cat#ER0720 |
| Deposited data | ||
| Hi-C data from wild-type and MLL4 KO mice naïve CD4+ cells | 2 | GEO accession#: GSE69162 |
| MLL4 and H3K4me1 ChIP-seq data from wild-type and MLL4 KO mice naïve CD4+ cells | 2 | GEO accession#: GSE69162 |
| GATA3 ChIP-seq data from mice Th1 cells | 3 | SRA accession#: SRR038548 |
| T-bet ChIP-seq data from mice Th1 cells | 4 | SRA accession#: SRR372732 |
| H3K4me1 ChIP-seq data from human Jurkat cells | 5 | GEO accession#: GSM1603224 |
| GATA3 ChIP-seq data from human Jurkat cells | 6 | GEO accession#: GSE68976 |
| RNA-seq | This paper | GEO accession#: GSE204946 |
| ChIP-seq | This paper | GEO accession#: GSE204946 |
| Hi-TrAC | This paper | GEO accession#: GSE204946 |
| Experimental models: Organisms/strains | ||
| Mouse: Mll4fl/fl | 1 | N/A |
| Mouse: CD4cre | Taconic | Model#4196 |
| Mouse: hCD2Cre | (Gurram et al., 2023) | N/A |
| Mouse: Gata3fl/fl | (Gurram et al., 2023) | N/A |
| Mouse: G28∆ | This paper | N/A |
| Mouse: CNS-28∆ | This paper | N/A |
| Oligonucleotides | ||
| CNS–28∆ sgRNA-1: ATTAAGACCTCGTTGAAGGC | IDT | N/A |
| CNS–28∆ sgRNA-2: GAGATCTTATCATGCCGTCT | IDT | N/A |
| G28∆ sgRNA-1: AATTAAGTCTTAACAGAAGG | IDT | N/A |
| G28∆ sgRNA-2: ACTCTGCATGGTTCCCATT | IDT | N/A |
| Cas9 mRNA | TriLink Biotechnologies | L-6125 |
| CNS-22/F: ACCTGCACTTCTGTGAGCACAT | IDT | N/A |
| CNS-22/R: AGGCGCTGACATCATGCTT | IDT | N/A |
| CNS-22/Probe: CACGCATCGCCCCGCCCTAT | IDT | N/A |
| CNS-28/F: GTTGAAGGCAGGTACTGTGATA | IDT | N/A |
| CNS-28/R: CCATCCTAGACGGCATGATAAG | IDT | N/A |
| CNS-28/Probe: ACTCATGTCCATGTGCTA | IDT | N/A |
| CNS+41/F: GCAAAGGCTCAGACTGAAGATA | IDT | N/A |
| CNS+41/R: ACAGAGTTTCTGGAGAGAGTAGA | IDT | N/A |
| CNS+41/Probe: CGGAGTCAGAAGCAGGGTTACAGC | IDT | N/A |
| Ifng promoter/F: CAGCCGTCCCCAACCA | IDT | N/A |
| Ifng promoter/R: GCCCTTGTAATGTGAATTTCTCATC | IDT | N/A |
| Ifng promoter/Probe: CAAAGGCTCCCTGTGCTGTGCTCTG | IDT | N/A |
| Foxp3 probe | ThermoFisher | Mm00475162 |
| Ifng probe | ThermoFisher | Mm01168134 |
| Tbx21 probe | ThermoFisher | Mm00450960 |
| Gata3 probe | ThermoFisher | Mm00484683 |
| Il4 probe | ThermoFisher | Mm00445259 |
| Il5 probe | ThermoFisher | Mm00439646 |
| Il13 probe | ThermoFisher | Mm00434204 |
| Actb probe | ThermoFisher | 4352341E |
| Software and algorithms | ||
| GraphPad Prism version 8 for MacOS | Graphpad Software | https://www.graphpad.com |
| FlowJo | TreeStar | https://www.flowjo.com/ |
| Python (v3) | Python Community | https://www.python.org |
| seaborn (v0.11.0) | 7 | https://seaborn.pydata.org |
| Bowtie2 (v.2.3.5) | 8 | https://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| STAR (v2.7.3a) | 9 | https://github.com/alexdobin/STAR |
| Cuffdiff (v2.2.1) | 10 | https://github.com/cole-trapnell-lab/cufflinks |
| deepTools2 (v3.3.0) | 11 | https://deeptools.readthedocs.io/en/develop/ |
| HiC-Pro (v2.11.1) | 12 | https://github.com/nservant/HiC-Pro |
| Juicer (v1.11.04) | 13 | https://github.com/aidenlab/juicer |
| cLoops2 (v0.0.3) | 14 | https://github.com/YaqiangCao/cLoops2 |
| HOMER (v4.10.4) | 15 | http://homer.ucsd.edu/homer/ |
| Other | ||
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Mice
Mll4fl/fl mice on mixed C57BL/6 and 129 backgrounds were provided by K.Ge in NIDDK/NIH, and have been previously described 1,2. CD4-Cre mice on the C57BL/6 background were purchased from Taconic. The Ifng −28kb mutant mice were generated using the CRISPR/Cas9 method 16. Briefly two single guide RNAs (sgRNA-1: ATTAAGACCTCGTTGAAGGC; sgRNA-2: GAGATCTTATCATGCCGTCT for CNS–28Δ mice), (sgRNA-1:AATTAAGTCTTAACAGAAGGAGG; sgRNA-2:ACTCTGCATGGTTCCCATTTGG for G28Δ mice ) were designed and in vitro transcribed using Thermo Fisher’s sgRNA synthesis services. These two sgRNAs (20 ng/ul each) were co-microinjected with Cas9 mRNA (50 ng/ul) into the cytoplasm of fertilized eggs collected from C57BL/6N mice. The injected embryos were cultured overnight in M16 medium, and those embryos which reached 2-cell stage of development were implanted into the oviducts of pseudo pregnant foster mothers (CD-1 mice from Charles River Laboratory). Offspring born to the foster mothers were genotyped by PCR and Sanger sequencing. Founder mice with desired deletions were bred with C57BL/6 mice to establish the mouse line. Mice were bred and maintained in NHLBI animal facility. All experiments were performed on 6 to 10 weeks old mice according to a protocol approved by the NHLBI and NCI Animal Care and Use Committee.
Methods
In vitro T cell differentiation
CD4+ Naive (CD44loCD62L+CD25−) T cells were obtained from spleens and lymph nodes of indicated mice by flow cytometric cell sorting. The purity of isolated T cell populations routinely exceeded 98%. Naive T cells were stimulated with plate-bound anti-CD3 (145–2C11, 1μg/ml) and anti-CD28 (PV-1, 1μg/ml) and polarizing cytokines (Th1: 20 ng/ml IL-12; Th2: 20 ng/ml IL-4; Th17: 20 ng/ml IL-6, 2 ng/ml TGF-β; Treg: 4 ng/ml TGF-β; all cytokines from R&D). The antibody anti-IFN-g (XMG1.2, 10ng/ml) was added in Th2 differentiation condition as indicated.
Flow cytometry
For intracellular cytokine staining, cells were cultured as described above and stimulated for 4 h at 37°C in culture medium containing PMA (50ng/ml; Sigma), ionomycin (1μg/ml; Sigma) and monensin (GolgiStop; 1μg/ml; BD Biosciences). Surface markers were stained in PBS with 1% FCS for 20 minutes at room temperature, then subsequently fixed in Cytoperm/Cytofix (BD Biosciences), permeabilized with Perm/Wash Buffer (BD Biosciences), and stained with cytokine antibodies diluted in Perm/Wash. For Foxp3 staining, cells were fixed and permeabilized with the Foxp3 Staining Buffer Set, according to the manufacturer’s instructions (eBiosciences). All flow cytometry data were acquired on a FACS X-20 (Becton Dickinson) and analyzed with FlowJo software (TreeStar).
Immunoblot analysis
Th1 cells were lysed in RIPA buffer and proteins were resolved by the Novex NuPage SDS-PAGE gel system (Life Technologies). Proteins were transferred to Supported Nitrocellulose Membrane (Bio-Rad) and were incubated with anti-pSTAT4 (R&D, PA-ST4), anti-STAT4 (R&D, MAB5287) and anti-H3 (Abcam, 10799). Blots were visualized with Pierce ECL Western Blotting Substrate (Thermo Scientific).
Histology
Mouse tissues were fixed in 10% formalin for 48 hours, preserved in 70% ethanol, and embedded in paraffin. The paraffin blocks were cut into 10 um longitudinal sections and stained with hematoxylin and eosin (H&E) by Histoserv Inc. (Germantown, MD). Scoring of the infiltrates was performed by blinded assessment of at least three sections of each tissue per mouse.
Assessment of intestinal inflammation
Tissues were graded semiquantitatively from 0 to 5 in a blinded fashion. Score 0: No changes observed; Score 1: Minimal scattered mucosal inflammatory cell infiltrates, with or without minimal epithelial hyperplasia; Score 2: Mild scattered to diffuse inflammatory cell infiltrates, sometimes extending into the submucosa and associated with erosions, with minimal to mild epithelial hyperplasia and minimal to mild mucin depletion from goblet cells; Score 3: mild to moderate inflammatory cell infiltrates that were sometimes transmural, often associated with ulceration, with moderate epithelial hyperplasia and mucin depletion; Score 4: marked inflammatory cell infiltrates that were often transmural and associated with ulceration, with marked epithelial hyperplasia and mucin depletion; Score 5: marked transmural inflammation with severe ulceration and loss of intestinal glands.
ELISAs
The cell supernatants or mouse serum were collected, and IFN-γ or IgE measurement was performed according to the manufacturer’s recommendations (Biolegnd).
Quantitative RT-PCR
For gene expression detection, total RNA was isolated from whole cells using the Qiagen miniRNA extraction kit following the manufacturer’s instructions. RNA was quantified and complementary DNA was reverse-transcribed using the iScript kit (Bio-rad) following the manufacturer’s instructions. The cDNA samples were used at 20ng/well in a 384 well plate and run in triplicate. PCR reactions were set up using TaqMan Universal PCR Master Mix (Applied Biosystems) on a QuantStudio™ 6 Flex. Quantification of relative mRNA expression was normalized to the expression of Actb. Primers-probe mixtures were purchased from Applied Biosystems: Foxp3 (Mm00475162), Ifng (Mm01168134), Tbx21 (Mm00450960), Gata3 (Mm00484683), Il4 (Mm00445259), Il5 (Mm00439646), Il13 (Mm00434204), Actb (4352341E).
T Cell-Induced Colitis
Naive (CD44loCD45RBhi) CD4+ T cells were purified from WT or IfngΔCNS28 mice. 7 × 105 naive T cells were injected intraperitoneally (i.p.) were injected into age- and sex-matched Rag2−/− mice, and weight loss was monitored over 6–8 weeks.
HDM antigen–induced allergic asthma
To generate HDM antigen–induced allergic asthma, 25μg house dust mites (HDM) protein (Greer Laboratories) in 25μl PBS was injected into each mouse nasally on day 0, day 7, day 8 and day 9. The injections were performed in anesthetized mice (2.5% Isoflurane) using a VetEquip Inhalation Anesthesia System (VetEquip, Inc). The mice were sacrificed on day 10.
Listeria infection model
Mice were infected with 2 × 104 CFU/200 ml Listeria monocytogenes (ATCC® 35152™) and weighed daily. On day 7 after infection, spleen and liver were homogenized in PBS, and serial dilutions of the homogenates were plated on LB plates incubated at 37 °C and CFU were counted.
Chromosome-conformation capture (3C)
3C assays were performed as described previously 17. In brief, 1 × 106 cells were resuspended in 1ml of medium and fixed with 1% formaldehyde for 10 minutes at room temperature. Fixed cells were resuspended in 1ml of cold lysis buffer on ice for 10 min. The nuclei were pelleted at 400g at 4°C for 5 min. 1 x rCutSmart buffer with 0.1% SDS was added to the nuclei and the samples incubated at 65°C for 10 min. Then Triton X-100 was added to final 2% and incubated at 37°C for 1h while shaking at 750rpm. 50U of Hpa I were used to digest the nuclei overnight at 37°C while shaking at 750rpm. The nuclei pellet was precipitated and washed with washing buffer (PBS+10mM EDTA+10mM NaCL+0.1% Triton X-100). The pellet was resuspended in 1x ligation buffer + 10U Ligase and incubate overnight at 16°C. Reverse cross-links the reaction by adding 15μl Proteinase K (20mg/ml) and incubating overnight at 65°C. DNA was purified using a standard phenol/chloroform extraction.
ChIP-seq and RNA-seq assays
ChIP-seq assays were performed as described previously 3,18. In brief, cells for ChIP-seq were fixed for 10 minutes in 1% formaldehyde and sonicated and chromatin immunoprecipitation was performed with anti-H3K4me1(ab8895, Abcam), anti-H3K4me2 (ab32356, Abcam), anti-H3K4me3 (17–614, Millipore), anti-H3K9me3 (ab8898, Abcam), anti-H3K27ac (ab4729, Abcam), anti-H3K27me3 (07–449, Millipore), anti-MLL4 19,20, anti-GATA3 (558686, BD bioscience), anti-T-bet (561263, BD bioscience). DNA was end-repaired using an End-It DNA-Repair kit (Epicentre), and was indexed, amplified and sequenced on an Illumina HiSeq-2500. RNA-seq libraries using poly-A RNA isolated from both the WT and MLL4 KO cells were prepared as previous described 21 and sequenced by Illumina HiSeq-2500.
ChIP-qPCR assays
The chromatin immunoprecipitation (ChIP) assays were performed as described previously 22. The immunoprecipitated DNA was analyzed by multiplex q-PCR with following primers: CNS-22/F 5’ ACCTGCACTTCTGTGAGCACAT 3’, R 5’ AGGCGCTGACATCATGCTT 3’, Probe 5’ CACGCATCGCCCCGCCCTAT 3’; CNS-28/F 5’ GTTGAAGGCAGGTACTGTGATA 3’, R 5’ CCATCCTAGACGGCATGATAAG 3’, Probe 5’ ACTCATGTCCATGTGCTA 3’; CNS+41/ F 5’ GCAAAGGCTCAGACTGAAGATA 3’, R 5’ ACAGAGTTTCTGGAGAGAGTAGA 3’, Probe 5’ CGGAGTCAGAAGCAGGGTTACAGC 3’; Ifng promoter/F 5’ CAGCCGTCCCCAACCA 3’, R 5’ GCCCTTGTAATGTGAATTTCTCATC, Probe 5 CAAAGGCTCCCTGTGCTGTGCTCTG 3’.
Sequencing data bioinformatics analysis
Mouse reference genome mm10 and annotations from GENCODE (M21) 23 were used for all analysis. RNA-seq data, ChIP-seq data and Hi-TrAC data were pre-processed similarly with previous our study 24.
Hi-C data generated previously 2 were re-analyzed by processing into paired-end tags (PETs) by HiC-Pro (v2.11.1) 12; specifically, restriction fragments were generated by digest_genome.py -r ĜTAC CÂTG ĈTAG and all other parameters were kept default. Hi-C replicates were pooled together and down-sampled to equal 300 million unique intra-chromosomal interacting PETs both for wild-type and knockout sample for all following analyses. Only PETs with a distance greater than 1 kb were used to plot the decay of interaction densities with genomic distance to show the genome-wide effect of Mll4 knockout. Hi-C domains were called by the arrowhead algorithm implemented in Juicer (v1.11.04) 13 with the resolution of 5 kb, 10 kb, and 20 kb and KR normalization, all other parameters kept defaults. For each domain, only PETs with a distance greater than 1 kb were quantified for the intra-domain PETs number and the PETs with only one end located in the domain to compare the effect of Mll4 knockout. Heatmap visualizations and domain aggregation analysis were performed by cLoops2 (v0.0.3) 14. 5 kb resolution contact matrix for the Ifng domain region were obtained by the cLoops2 dump module, box plot, and two-sided Wilcoxon signed-rank test were used to check the pixel/bin-wise difference of Hi-C interaction densities.
Briefly, raw reads of RNA-seq data were mapped by STAR (v2.7.3a) 9. BigWig files were also generated by STAR and normalized into Reads per Million (RPM) for genome browser images with the cLoops2 plot module. Cuffdiff (v2.2.1) 10 was used to perform differential expressed genes (DEGs) analysis and generate fold changes and P-values with settings of --no-js-tests -- compatible-hits-norm. Fold change >= 2, P-value < 1e-3, and gene expression level >1 RPKM either in wild-type or knockout sample were used as cutoffs for DEGs for all RNA-seq data generated in this study.
Raw reads of public and this study generated ChIP-seq data were mapped by Bowtie2 (v2.3.5) 8. Mapped unique paired-end reads with MAPQ ≥ 10 were used for the following analysis. Sample-wise genome-wide correlation analysis was performed with multiBigwigSummary bins and parameter -bs 1000 in the deepTools2 (v3.3.0) package 11. ChIP-seq signals were also quantified into RPM as BigWig files and shown genome browser images by the cLoops2 plot module. ChIP-seq peaks were called by the cLoops2 callPeaks module (v0.0.3) 14 for each replicate first, with the settings of -eps 300,500 -minPts 10,20 for H3K4me1, -eps 150,300 -minPts 10,20 for H3K4me3 and H3K27ac, and -eps 150 -minPts 10,20 for CTCF ChIP-seq data. Overlapped peak regions from the same antibody biological replicates, both wild-type and knockout conditions, were compiled together as union sets for statistical analysis of highly changed peaks. For each peak, reads were quantified and scaled to a total depth of 10 million for each replicate, then the mean value from replicates was used for the Poisson test. Fold change >= 2 and P-value < 1e-3 were used as cutoffs for peaks for all ChIP-seq data generated in this study. CNS region annotations around Ifng gene locus were based on the H3K4me1 peaks from Naïve the Th1 cells.
The script tracPre2.py in the cLoops2 package pre-processed raw reads of Hi-TrAC data generated in this study. To compare CNS-28/Gata3/Mll4 knockout and wild-type control, PETs from replicates were pooled and downsampled to the same total depth for all downstream analysis and visualization. Pooled PETs in naïve cells were used to call loops with the cLoops2 callLoops module by settings of -eps 1000 -minPts 10,20 -cut 1000 -mcut 5000000, requiring at least 10 PETs supporting a confident loop. Differentially enriched loops analysis was performed by the cLoops2 callDiffLoops module with the default parameters. Visualizations of Hi-TrAC data around the CNS-34, CNS-28, CNS-22, and Ifng promoter region with 1D signal, virtual 4C signal (CNS_22 set as the viewpoint), arches showing the number of PETs, and heatmaps were performed by the cLoops2 plot module.
Gene Ontology (GO) terms enrichment analysis for DEGs from RNA-seq was performed by findGO.pl in the HOMER package 15, and annotatePeaks.pl with the option of -go was used for significantly different peaks, both requiring more than ten overlapping genes in the terms, and there are fewer than 500 genes in the terms. Only top enriched terms sorted by ascending P-values were shown, and redundant terms were removed manually.
Supplementary Material
Identification of the Ifng silencer CNS–28
CNS–28 represses interaction between CNS-22 and Ifng promoter
CNS–28 maintains tissue quiescence by restraining IFN-γ production
CNS–28 deletion represses type 2 responses via enhanced IFN-γ expression
Acknowledgements
We thank NHLBI DNA Sequencing Core facility for sequencing the NGS libraries; the NHLBI Flow Cytometry Core facility and NCI CCR flow cytometry core facility for sorting the cells; and NHLBI Transgenic Core facility for generating the knockout mouse lines. The authors thank Yolanda L. Jones, National Institutes of Health Library and the NIH Fellows Editorial Board (FEB), for editing assistance. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). This research was supported by the Intramural Research Programs of National Heart, Lung and Blood Institute, National Cancer Institute, and National Institute of Allergy and Infectious Diseases of National Institutes of Health.
Footnotes
Declaration of interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference:
- 1.Lazarevic V, Glimcher LH, and Lord GM (2013). T-bet: a bridge between innate and adaptive immunity. Nature reviews. Immunology 13, 777–789. 10.1038/nri3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bustamante J, Boisson-Dupuis S, Abel L, and Casanova JL (2014). Mendelian susceptibility to mycobacterial disease: genetic, immunological, and clinical features of inborn errors of IFN-gamma immunity. Semin Immunol 26, 454–470. 10.1016/j.smim.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bluestone JA, Mackay CR, O’Shea JJ, and Stockinger B (2009). The functional plasticity of T cell subsets. Nature reviews. Immunology 9, 811–816. 10.1038/nri2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Shea JJ, and Paul WE (2010). Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science 327, 1098–1102. 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filiano AJ, Xu Y, Tustison NJ, Marsh RL, Baker W, Smirnov I, Overall CC, Gadani SP, Turner SD, Weng Z, et al. (2016). Unexpected role of interferon-gamma in regulating neuronal connectivity and social behaviour. Nature 535, 425–429. 10.1038/nature18626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schroder K, Hertzog PJ, Ravasi T, and Hume DA (2004). Interferon-gamma: an overview of signals, mechanisms and functions. Journal of leukocyte biology 75, 163–189. 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 7.Zhu J, and Paul WE (2010). Peripheral CD4+ T-cell differentiation regulated by networks of cytokines and transcription factors. Immunological reviews 238, 247–262. 10.1111/j.1600-065X.2010.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abbas AK, Murphy KM, and Sher A (1996). Functional diversity of helper T lymphocytes. Nature 383, 787–793. 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 9.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, and Glimcher LH (2000). A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100, 655–669. 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 10.Balasubramani A, Shibata Y, Crawford GE, Baldwin AS, Hatton RD, and Weaver CT (2010). Modular utilization of distal cis-regulatory elements controls Ifng gene expression in T cells activated by distinct stimuli. Immunity 33, 35–47. 10.1016/j.immuni.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatton RD, Harrington LE, Luther RJ, Wakefield T, Janowski KM, Oliver JR, Lallone RL, Murphy KM, and Weaver CT (2006). A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity 25, 717–729. 10.1016/j.immuni.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 12.Shnyreva M, Weaver WM, Blanchette M, Taylor SL, Tompa M, Fitzpatrick DR, and Wilson CB (2004). Evolutionarily conserved sequence elements that positively regulate IFN-gamma expression in T cells. Proceedings of the National Academy of Sciences of the United States of America 101, 12622–12627. 10.1073/pnas.0400849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee DU, Avni O, Chen L, and Rao A (2004). A distal enhancer in the interferon-gamma (IFN-gamma) locus revealed by genome sequence comparison. The Journal of biological chemistry 279, 4802–4810. 10.1074/jbc.M307904200. [DOI] [PubMed] [Google Scholar]
- 14.Chang S, and Aune TM (2005). Histone hyperacetylated domains across the Ifng gene region in natural killer cells and T cells. Proceedings of the National Academy of Sciences of the United States of America 102, 17095–17100. 10.1073/pnas.0502129102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yagi R, Junttila IS, Wei G, Urban JF Jr., Zhao K, Paul WE, and Zhu J (2010). The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-gamma. Immunity 32, 507–517. 10.1016/j.immuni.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balasubramani A, Mukasa R, Hatton RD, and Weaver CT (2010). Regulation of the Ifng locus in the context of T-lineage specification and plasticity. Immunological reviews 238, 216–232. 10.1111/j.1600-065X.2010.00961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mosmann TR, and Coffman RL (1989). TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annual review of immunology 7, 145–173. 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 18.Oestreich KJ, and Weinmann AS (2012). Master regulators or lineage-specifying? Changing views on CD4+ T cell transcription factors. Nature reviews. Immunology 12, 799–804. 10.1038/nri3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ansel KM, Greenwald RJ, Agarwal S, Bassing CH, Monticelli S, Interlandi J, Djuretic IM, Lee DU, Sharpe AH, Alt FW, and Rao A (2004). Deletion of a conserved Il4 silencer impairs T helper type 1-mediated immunity. Nature immunology 5, 1251–1259. 10.1038/ni1135. [DOI] [PubMed] [Google Scholar]
- 20.Jenner RG, Townsend MJ, Jackson I, Sun K, Bouwman RD, Young RA, Glimcher LH, and Lord GM (2009). The transcription factors T-bet and GATA-3 control alternative pathways of T-cell differentiation through a shared set of target genes. Proceedings of the National Academy of Sciences of the United States of America 106, 17876–17881. 10.1073/pnas.0909357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naoe Y, Setoguchi R, Akiyama K, Muroi S, Kuroda M, Hatam F, Littman DR, and Taniuchi I (2007). Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf beta binding to the Il4 silencer. The Journal of experimental medicine 204, 1749–1755. 10.1084/jem.20062456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hegazy AN, Peine M, Helmstetter C, Panse I, Frohlich A, Bergthaler A, Flatz L, Pinschewer DD, Radbruch A, and Lohning M (2010). Interferons direct Th2 cell reprogramming to generate a stable GATA-3(+)T-bet(+) cell subset with combined Th2 and Th1 cell functions. Immunity 32, 116–128. 10.1016/j.immuni.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 23.Cousins DJ, Lee TH, and Staynov DZ (2002). Cytokine coexpression during human Th1/Th2 cell differentiation: direct evidence for coordinated expression of Th2 cytokines. Journal of immunology 169, 2498–2506. 10.4049/jimmunol.169.5.2498. [DOI] [PubMed] [Google Scholar]
- 24.Messi M, Giacchetto I, Nagata K, Lanzavecchia A, Natoli G, and Sallusto F (2003). Memory and flexibility of cytokine gene expression as separable properties of human T(H)1 and T(H)2 lymphocytes. Nature immunology 4, 78–86. 10.1038/ni872. [DOI] [PubMed] [Google Scholar]
- 25.Kanhere A, Hertweck A, Bhatia U, Gokmen MR, Perucha E, Jackson I, Lord GM, and Jenner RG (2012). T-bet and GATA3 orchestrate Th1 and Th2 differentiation through lineage-specific targeting of distal regulatory elements. Nat Commun 3, 1268. 10.1038/ncomms2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei G, Abraham BJ, Yagi R, Jothi R, Cui K, Sharma S, Narlikar L, Northrup DL, Tang Q, Paul WE, et al. (2011). Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity 35, 299–311. 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Schones DE, and Zhao K (2009). Characterization of human epigenomes. Curr Opin Genet Dev 19, 127–134. 10.1016/j.gde.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidl C, Delacher M, Huehn J, and Feuerer M (2018). Epigenetic mechanisms regulating T-cell responses. The Journal of allergy and clinical immunology 142, 728–743. 10.1016/j.jaci.2018.07.014. [DOI] [PubMed] [Google Scholar]
- 29.Rothenberg EV (2014). The chromatin landscape and transcription factors in T cell programming. Trends in immunology 35, 195–204. 10.1016/j.it.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. (2009). Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30, 155–167. S1074–7613(08)00555–4 [pii] 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hess JL (2004). Mechanisms of transformation by MLL. Crit Rev Eukaryot Gene Expr 14, 235–254. 10.1615/critreveukaryotgeneexpr.v14.i4.10. [DOI] [PubMed] [Google Scholar]
- 32.Placek K, Hu G, Cui K, Zhang D, Ding Y, Lee JE, Jang Y, Wang C, Konkel JE, Song J, et al. (2017). MLL4 prepares the enhancer landscape for Foxp3 induction via chromatin looping. Nature immunology 18, 1035–1045. 10.1038/ni.3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee GR, Kim ST, Spilianakis CG, Fields PE, and Flavell RA (2006). T helper cell differentiation: regulation by cis elements and epigenetics. Immunity 24, 369–379. 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 34.Thierfelder WE, van Deursen JM, Yamamoto K, Tripp RA, Sarawar SR, Carson RT, Sangster MY, Vignali DA, Doherty PC, Grosveld GC, and Ihle JN (1996). Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature 382, 171–174. 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- 35.Aune TM, Collins PL, Collier SP, Henderson MA, and Chang SJ (2013). Epigenetic activation and silencing of the gene that encodes IFN-gamma. Front Immunol 4. ARTN 112 10.3389/fimmu.2013.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai B, Gao W, Cui K, Xie W, Tang Q, Jin W, Hu G, Ni B, and Zhao K (2018). Principles of nucleosome organization revealed by single-cell micrococcal nuclease sequencing. Nature 562, 281–285. 10.1038/s41586-018-0567-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei G, Abraham BJ, Yagi R, Jothi R, Cui KR, Sharma S, Narlikar L, Northrup DL, Tang QS, Paul WE, et al. (2011). Genome-wide Analyses of Transcription Factor GATA3-Mediated Gene Regulation in Distinct T Cell Types. Immunity 35, 299–311. 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu S, Cao Y, Cui K, Tang Q, and Zhao K (2022). Hi-TrAC reveals division of labor of transcription factors in organizing chromatin loops. Nat Commun 13, 6679. 10.1038/s41467-022-34276-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanyal A, Lajoie BR, Jain G, and Dekker J (2012). The long-range interaction landscape of gene promoters. Nature 489, 109–113. 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren G, Jin W, Cui K, Rodrigez J, Hu G, Zhang Z, Larson DR, and Zhao K (2017). CTCF-Mediated Enhancer-Promoter Interaction Is a Critical Regulator of Cell-to-Cell Variation of Gene Expression. Molecular cell 67, 1049–1058 e1046. 10.1016/j.molcel.2017.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thale C, and Kiderlen AF (2005). Sources of interferon-gamma (IFN-gamma) in early immune response to Listeria monocytogenes. Immunobiology 210, 673–683. 10.1016/j.imbio.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 42.Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, and Wahl SM (2003). Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. The Journal of experimental medicine 198, 1875–1886. 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou M, and Ouyang W (2003). The function role of GATA-3 in Th1 and Th2 differentiation. Immunol Res 28, 25–37. 10.1385/IR:28:1:25. [DOI] [PubMed] [Google Scholar]
- 44.Sekimata M, Perez-Melgosa M, Miller SA, Weinmann AS, Sabo PJ, Sandstrom R, Dorschner MO, Stamatoyannopoulos JA, and Wilson CB (2009). CCCTC-binding factor and the transcription factor T-bet orchestrate T helper 1 cell-specific structure and function at the interferon-gamma locus. Immunity 31, 551–564. 10.1016/j.immuni.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, and Merkenschlager M (2009). Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460, 410–413. 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, Stamatoyonnapoulos JA, and Wilson CB (2007). Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat Immunol 8, 732–742. 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schoenborn JR, and Wilson CB (2007). Regulation of interferon-gamma during innate and adaptive immune responses. Adv Immunol 96, 41–101. 10.1016/S0065-2776(07)96002-2. [DOI] [PubMed] [Google Scholar]
- 48.Ouyang W, Ranganath SH, Weindel K, Bhattacharya D, Murphy TL, Sha WC, and Murphy KM (1998). Inhibition of Th1 development mediated by GATA-3 through an IL-4-independent mechanism. Immunity 9, 745–755. 10.1016/s1074-7613(00)80671-8. [DOI] [PubMed] [Google Scholar]
- 49.Usui T, Nishikomori R, Kitani A, and Strober W (2003). GATA-3 suppresses Th1 development by downregulation of Stat4 and not through effects on IL-12Rbeta2 chain or T-bet. Immunity 18, 415–428. 10.1016/s1074-7613(03)00057-8. [DOI] [PubMed] [Google Scholar]
- 50.Zhu J, Min B, Hu-Li J, Watson CJ, Grinberg A, Wang Q, Killeen N, Urban JF Jr., Guo L, and Paul WE (2004). Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nature immunology 5, 1157–1165. 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
- 51.Samson SI, Richard O, Tavian M, Ranson T, Vosshenrich CA, Colucci F, Buer J, Grosveld F, Godin I, and Di Santo JP (2003). GATA-3 promotes maturation, IFN-gamma production, and liver-specific homing of NK cells. Immunity 19, 701–711. 10.1016/s1074-7613(03)00294-2. [DOI] [PubMed] [Google Scholar]
- 52.Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, and Ho IC (2003). Critical roles for transcription factor GATA-3 in thymocyte development. Immunity 19, 863–875. 10.1016/s1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Su MA, and Wan YY (2011). An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity 35, 337–348. 10.1016/j.immuni.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Balasubramani A, Winstead CJ, Turner H, Janowski KM, Harbour SN, Shibata Y, Crawford GE, Hatton RD, and Weaver CT (2014). Deletion of a conserved cis-element in the Ifng locus highlights the role of acute histone acetylation in modulating inducible gene transcription. PLoS Genet 10, e1003969. 10.1371/journal.pgen.1003969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Balasubramani A, Winstead CJ, Turner H, Janowski KM, Harbour SN, Shibata Y, Crawford GE, Hatton RD, and Weaver CT (2014). Deletion of a Conserved cis-Element in the Ifng Locus Highlights the Role of Acute Histone Acetylation in Modulating Inducible Gene Transcription. Plos Genet 10. ARTN e1003969 10.1371/journal.pgen.1003969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Collier SP, Henderson MA, Tossberg JT, and Aune TM (2014). Regulation of the Th1 Genomic Locus from Ifng through Tmevpg1 by T-bet. Journal of Immunology 193, 3959–3965. 10.4049/jimmunol.1401099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collins PL, Henderson MA, and Aune TM (2012). Lineage-specific adjacent IFNG and IL26 genes share a common distal enhancer element. Genes Immun 13, 481–488. 10.1038/gene.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pham D, Vincentz JW, Firulli AB, and Kaplan MH (2012). Twist1 Regulates Ifng Expression in Th1 Cells by Interfering with Runx3 Function. Journal of Immunology 189, 832–840. 10.4049/jimmunol.1200854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakayamada S, Kanno Y, Takahashi H, Jankovic D, Lu KT, Johnson TA, Sun HW, Vahedi G, Hakim O, Handon R, et al. (2011). Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity 35, 919–931. 10.1016/j.immuni.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reeder JE, Kwak YT, McNamara RP, Forst CV, and D’Orso I (2015). HIV Tat controls RNA Polymerase II and the epigenetic landscape to transcriptionally reprogram target immune cells. Elife 4. 10.7554/eLife.08955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA, et al. (2016). Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351, 1454–1458. 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Reference:
- 1.Lee JE, Wang C, Xu S, Cho YW, Wang L, Feng X, Baldridge A, Sartorelli V, Zhuang L, Peng W, and Ge K (2013). H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife 2, e01503. 10.7554/eLife.01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Placek K, Hu G, Cui K, Zhang D, Ding Y, Lee JE, Jang Y, Wang C, Konkel JE, Song J, et al. (2017). MLL4 prepares the enhancer landscape for Foxp3 induction via chromatin looping. Nature immunology 18, 1035–1045. 10.1038/ni.3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei G, Abraham BJ, Yagi R, Jothi R, Cui K, Sharma S, Narlikar L, Northrup DL, Tang Q, Paul WE, et al. (2011). Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity 35, 299–311. 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakayamada S, Kanno Y, Takahashi H, Jankovic D, Lu KT, Johnson TA, Sun HW, Vahedi G, Hakim O, Handon R, et al. (2011). Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity 35, 919–931. 10.1016/j.immuni.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reeder JE, Kwak YT, McNamara RP, Forst CV, and D’Orso I (2015). HIV Tat controls RNA Polymerase II and the epigenetic landscape to transcriptionally reprogram target immune cells. Elife 4. 10.7554/eLife.08955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA, et al. (2016). Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351, 1454–1458. 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waskom ML (2021). Seaborn: statistical data visualization. Journal of Open Source Software 6, 3021. [Google Scholar]
- 8.Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, and Pachter L (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature Biotechnology 31, 46-+. 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–165. 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Servant N, Varoquaux N, Lajoie BR, Viara E, Chen CJ, Vert JP, Heard E, Dekker J, and Barillot E (2015). HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol 16, 259. 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durand NC, Shamim MS, Machol I, Rao SS, Huntley MH, Lander ES, and Aiden EL (2016). Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst 3, 95–98. 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao Y, Liu S, Ren G, Tang Q, and Zhao K (2022). cLoops2: a full-stack comprehensive analytical tool for chromatin interactions. Nucleic Acids Res 50, 57–71. 10.1093/nar/gkab1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589. 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang HY, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, and Jaenisch R (2013). One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell 153, 910–918. 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, and Forne T (2007). Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat Protoc 2, 1722–1733. 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 18.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, and Zhao K (2007). High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837. 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Lee JE, Wang CC, Xu SLY, Cho YW, Wang LF, Feng XS, Baldridge A, Sartorelli V, Zhuang LN, Peng WQ, and Ge K (2013). H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. Elife 2. ARTN e01503 10.7554/eLife.01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ang SY, Uebersohn A, Spencer CI, Huang Y, Lee JE, Ge K, and Bruneau BG (2016). KMT2D regulates specific programs in heart development via histone H3 lysine 4 di-methylation. Development 143, 810–821. 10.1242/dev.132688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren G, Jin W, Cui K, Rodrigez J, Hu G, Zhang Z, Larson DR, and Zhao K (2017). CTCF-Mediated Enhancer-Promoter Interaction Is a Critical Regulator of Cell-to-Cell Variation of Gene Expression. Molecular cell 67, 1049–1058 e1046. 10.1016/j.molcel.2017.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu R, Liu H, Chen X, Kirby M, Brown PO, and Zhao K (2001). Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell 106, 309–318. 10.1016/s0092-8674(01)00446-9. [DOI] [PubMed] [Google Scholar]
- 23.Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, et al. (2019). GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 47, D766–D773. 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao Y, Liu S, Cui K, Tang Q, and Zhao K (2022). Hi-TrAC reveals fractal nesting of super-enhancers. bioRxiv, 2022.2007.2013.499926. 10.1101/2022.07.13.499926. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data including ChIP-seq, RNA-seq and Hi-TrAC data have been deposited in the Gene Expression Omnibus database with accession number: GSE204946.
All codes used for data analysis in this paper are public and listed in the key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FITC anti-mouse CD4 | Biolegend | Cat# 130308, RRID: AB_1279237 |
| APC anti-mouse CD8a | Biolegend | Cat# 100711, RRID: AB_312750 |
| Brilliant Violet 785™ anti-mouse CD45 | Biolegend | Cat# 103149, RRID: AB_2564590 |
| PE anti-mouse CD62L | Biolegend | Cat# 104407, RRID: AB_313094 |
| APC anti-mouse CD44 | Biolegend | Cat# 103012, RRID: AB_312963 |
| PE/Cy7 anti-mouse IL-17A | Biolegend | Cat# 506922, RRID: AB_2125010 |
| Pacific Blue™ anti-mouse IFN-γ | Biolegend | Cat# 505818, RRID: AB_893526 |
| APC anti-mouse IL-4 | Biolegend | Cat# 504106, RRID: AB_315320 |
| PE/Cy7 anti-mouse CD11b | Biolegend | Cat# 101216, RRID: AB_312799 |
| PerCP/Cyanine5.5 anti-mouse CD3 | Biolegend | Cat# 100217, RRID: AB_1595597 |
| PE anti-mouse CD170 (Siglec-F) | Biolegend | Cat# 155506, RRID: AB_2750235 |
| APC anti-mouse F4/80 | Biolegend | Cat# 123116, RRID: AB_893481 |
| APC/Fire™ 750 anti-mouse Ly-6C | Biolegend | Cat# 128045, RRID: AB_2616730 |
| Brilliant Violet 421™ anti-mouse I-A/I-E (MHC-II) | Biolegend | Cat# 107631, RRID: AB_10900075 |
| FITC anti-mouse Ly-6G | Biolegend | Cat# 127606, RRID: AB_1236494 |
| APC anti-mouse CD11c | ThermoFisher | Cat# 17-0114-81, RRID: AB_469345 |
| APC anti-mouse FOXP3 | ThermoFisher | Cat# 77-5775-40, RRID: AB_469981 |
| PE anti-mouse CD45RB | Biolegend | Cat# 103308, RRID: AB_313015 |
| anti-mouse CD3ε | Bio X Cell | Cat# BE0001-1, RRID: AB_1107634 |
| anti-mouse CD28 | Bio X Cell | Cat# BE0015-5, RRID: AB_1107628 |
| anti-mouse IFNγ | Bio X Cell | Cat# BE0055, RRID: AB_1107694 |
| anti-pSTAT4 | R and D Systems | Cat# PA-ST4, RRID: AB_2302639 |
| anti-STAT4 | R and D Systems | Cat# MAB5287, RRID: AB_2302638 |
| anti-H3 | Abcam | Cat# 10799, RRID: AB_470239 |
| anti-H3K4me1 | Abcam | Cat# ab8895, RRID: AB_306847 |
| anti-H3K4me3 | Millipore | Cat# 17-614, RRID: AB_11212770 |
| anti-H3K27ac | Abcam | Cat# ab4729, RRID: AB_2118291 |
| anti-GATA3 | BD bioscience | Cat# 558686, RRID: AB_2108590 |
| anti-T-bet | BD bioscience | Cat# 561263, RRID: AB_10563082 |
| anti-MLL4 | 1 | N/A |
| Bacterial strains | ||
| Listeria monocytogenes | ATCC | Cat# 35152™ |
| Chemicals, peptides, and recombinant proteins | ||
| House Dust mite protein | Greer Laboratories | Cat# XPB82D3A2.5 |
| LB Broth | Sigma | Cat# L3522 |
| LB Broth with agar | Sigma | Cat# L3147 |
| PMA | Sigma | Cat# P8139 |
| Ionomycin | Sigma | Cat# I0634 |
| Recombinant Human TGF-beta 1 Protein | R&D | Cat# 240-B-002 |
| Recombinant Mouse IL-12 Protein | R&D | Cat# 419-ML-010 |
| Recombinant Mouse IL-4 Protein | R&D | Cat# 404-ML-010 |
| Recombinant Mouse IL-6 Protein | R&D | Cat# 406-ML-005 |
| formaldehyde | Thermo Scientific | Cat# 28908 |
| T4 DNA polymerase | New England Biolabs | Cat#M0203L |
| rCutSmart buffer | New England Biolabs | Cat# B6004S |
| Hpa I | New England Biolabs | Cat# R0105L |
| MluCI | New England Biolabs | Cat# R0538L |
| NlaIII | New England Biolabs | Cat# R0125L |
| Proteinase K | Roche | Cat# 03115828001 |
| T4 DNA Ligase | New England Biolabs | Cat# M0202L |
| Critical commercial assays | ||
| ELISA MAX™ Standard Set Mouse IFN-γ | Biolegend | Cat# 430801 |
| Mouse IgE ELISA MAX™ Deluxe | Biolegend | Cat# 432404 |
| Fixation/Permeabilization Solution Kit with GolgiPlug™ | BD Biosciences | Cat# 555028 |
| Foxp3 / Transcription Factor Fixation/Permeabilization Concentrate and Diluent | ThermoFisher | Cat# 00-5521-00 |
| iScript RT Supermix for RT-qPCR | Biorad | Cat# 1708841 |
| RNeasy Plus Mini Kit | Qiagen | Cat# 74136 |
| End-It DNA-Repair kit | Epicentre | Cat#ER0720 |
| Deposited data | ||
| Hi-C data from wild-type and MLL4 KO mice naïve CD4+ cells | 2 | GEO accession#: GSE69162 |
| MLL4 and H3K4me1 ChIP-seq data from wild-type and MLL4 KO mice naïve CD4+ cells | 2 | GEO accession#: GSE69162 |
| GATA3 ChIP-seq data from mice Th1 cells | 3 | SRA accession#: SRR038548 |
| T-bet ChIP-seq data from mice Th1 cells | 4 | SRA accession#: SRR372732 |
| H3K4me1 ChIP-seq data from human Jurkat cells | 5 | GEO accession#: GSM1603224 |
| GATA3 ChIP-seq data from human Jurkat cells | 6 | GEO accession#: GSE68976 |
| RNA-seq | This paper | GEO accession#: GSE204946 |
| ChIP-seq | This paper | GEO accession#: GSE204946 |
| Hi-TrAC | This paper | GEO accession#: GSE204946 |
| Experimental models: Organisms/strains | ||
| Mouse: Mll4fl/fl | 1 | N/A |
| Mouse: CD4cre | Taconic | Model#4196 |
| Mouse: hCD2Cre | (Gurram et al., 2023) | N/A |
| Mouse: Gata3fl/fl | (Gurram et al., 2023) | N/A |
| Mouse: G28∆ | This paper | N/A |
| Mouse: CNS-28∆ | This paper | N/A |
| Oligonucleotides | ||
| CNS–28∆ sgRNA-1: ATTAAGACCTCGTTGAAGGC | IDT | N/A |
| CNS–28∆ sgRNA-2: GAGATCTTATCATGCCGTCT | IDT | N/A |
| G28∆ sgRNA-1: AATTAAGTCTTAACAGAAGG | IDT | N/A |
| G28∆ sgRNA-2: ACTCTGCATGGTTCCCATT | IDT | N/A |
| Cas9 mRNA | TriLink Biotechnologies | L-6125 |
| CNS-22/F: ACCTGCACTTCTGTGAGCACAT | IDT | N/A |
| CNS-22/R: AGGCGCTGACATCATGCTT | IDT | N/A |
| CNS-22/Probe: CACGCATCGCCCCGCCCTAT | IDT | N/A |
| CNS-28/F: GTTGAAGGCAGGTACTGTGATA | IDT | N/A |
| CNS-28/R: CCATCCTAGACGGCATGATAAG | IDT | N/A |
| CNS-28/Probe: ACTCATGTCCATGTGCTA | IDT | N/A |
| CNS+41/F: GCAAAGGCTCAGACTGAAGATA | IDT | N/A |
| CNS+41/R: ACAGAGTTTCTGGAGAGAGTAGA | IDT | N/A |
| CNS+41/Probe: CGGAGTCAGAAGCAGGGTTACAGC | IDT | N/A |
| Ifng promoter/F: CAGCCGTCCCCAACCA | IDT | N/A |
| Ifng promoter/R: GCCCTTGTAATGTGAATTTCTCATC | IDT | N/A |
| Ifng promoter/Probe: CAAAGGCTCCCTGTGCTGTGCTCTG | IDT | N/A |
| Foxp3 probe | ThermoFisher | Mm00475162 |
| Ifng probe | ThermoFisher | Mm01168134 |
| Tbx21 probe | ThermoFisher | Mm00450960 |
| Gata3 probe | ThermoFisher | Mm00484683 |
| Il4 probe | ThermoFisher | Mm00445259 |
| Il5 probe | ThermoFisher | Mm00439646 |
| Il13 probe | ThermoFisher | Mm00434204 |
| Actb probe | ThermoFisher | 4352341E |
| Software and algorithms | ||
| GraphPad Prism version 8 for MacOS | Graphpad Software | https://www.graphpad.com |
| FlowJo | TreeStar | https://www.flowjo.com/ |
| Python (v3) | Python Community | https://www.python.org |
| seaborn (v0.11.0) | 7 | https://seaborn.pydata.org |
| Bowtie2 (v.2.3.5) | 8 | https://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| STAR (v2.7.3a) | 9 | https://github.com/alexdobin/STAR |
| Cuffdiff (v2.2.1) | 10 | https://github.com/cole-trapnell-lab/cufflinks |
| deepTools2 (v3.3.0) | 11 | https://deeptools.readthedocs.io/en/develop/ |
| HiC-Pro (v2.11.1) | 12 | https://github.com/nservant/HiC-Pro |
| Juicer (v1.11.04) | 13 | https://github.com/aidenlab/juicer |
| cLoops2 (v0.0.3) | 14 | https://github.com/YaqiangCao/cLoops2 |
| HOMER (v4.10.4) | 15 | http://homer.ucsd.edu/homer/ |
| Other | ||
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


