Abstract
The PALB2 gene is a breast cancer (BC) and ovarian cancer (OC) predisposition gene involved in the homologous recombination repair pathway. However, the prevalence and clinicopathological association of PALB2 pathogenic/likely pathogenic (PV/LPV) variants in Middle East is still not fully explored. Total 918 BC/OC patients from Saudi Arabia were selected for PALB2 mutations screening using capture sequencing technology. Five heterozygous PVs or LPVs were identified in six cases, accounting for 0.65% (6/918) of entire cohort. Two cases (33.3%) harbored PVs and four cases (66.7%) carried LPVs. Four PVs/LPVs (80%) were frameshift along with one novel splicing LPV (c.2835-2_2835-1delinsTT). One recurrent LPV (c.3425delT: p.L1142fs) was identified in two cases. All six affected carriers have breast cancer diagnosis with median age of 39.5 years (range 34–49 years). Only two cases (33%) have documented family history of cancer. Breast cancer phenotype was invasive ductal unilateral cancer in all cases with 66.7% of hormone receptor positive and 16% of triple negative tumors. Germline PVs/LPVs in the PALB2 gene were observed in low frequency of 0.65% in Saudi BC and/or OC. Our study confirms one recurrent LPV and one novel LPV in Saudi breast cancer patients.
Subject terms: Cancer genetics, Breast cancer, Cancer genomics
Introduction
Breast cancer is the leading cause of cancer mortality in women worldwide1. In Saudi Arabia, breast cancer is commonest cancer affecting Saudi females2. 5–10% of breast cancer and 10–15% ovarian cancer are thought to be hereditary3–6.
BRCA1/2 germline pathogenic variants (PVs) and likely pathogenic variants (LPVs) are known to be the main susceptibility genes in hereditary breast and ovarian cancer (HBOC)5,7,8. The Partner and Localizer of BRCA2 (PALB2) gene plays a critical role in homologous recombination repair of double standard DNA breaks for checkpoint control function through its ability to recruit BRCA2 and RAD51 to DNA breaks9–14.
Recently, PALB2 has been identified as one of the common predisposing genes for breast cancer after BRCA1/2 with penetrance estimated at 33–70% depending on age at diagnosis and family history15–18. Germline PVs/LPVs in PALB2 have also been identified in ovarian cancer and pancreatic cancer patients19,20. The prevalence of germline PALB2 PV/LPV in hereditary HBOC patients ranges between 0.8 and 1.5%21–23.
However, data about the prevalence of germline PALB2 variants in breast and ovarian cancers from Middle Eastern ethnicity is still scarce. Therefore, we have conducted this study in a cohort of 918 breast and ovarian cancer patients from Middle Eastern ethnicity to investigate the prevalence of germline PALB2 PVs and LPVs in breast cancer and ovarian cancer patients, and their molecular and clinicopathological characteristics in this population.
In addition, identifying the presence of recurrent and/or none PALB2 PVs/LPVs will help in understanding the contribution to breast cancer and ovarian cancer risk and tailor the best preventive and treatment option of PALB2 carriers from Middle Eastern ethnicity.
Materials and methods
Study population
The cases comprised 918 patients, 791 with breast cancer and 127 with epithelial ovarian cancer. All patients were of Saudi origin, diagnosed and treated at King Faisal Specialist Hospital and Research Centre (KFSH&RC) from 1989 to 2016. Relaxed criteria were used to select high-risk patients. For breast cancer, the criteria used were age ≤ 50 years, positive family history of cancer, triple negative breast cancers or bilateral tumors. Similarly, age ≤ 50 years, along with positive family history of cancer were considered high risk for ovarian cancer24. In our cohort of 918 breast and ovarian cancer patients, 271 patients had a positive family history of cancer in first and/or second-degree relatives. Among the patients with positive family history, 84.5% (229/271) had a relative with one of the well-known hereditary cancers (breast, ovarian, uterine, gastrointestinal, pancreatic, brain or soft tissue sarcomas).
Detailed clinico-pathological data, including follow-up data, were noted from case records and summarized in Table 1 (breast cancer) and Table 2 (epithelial ovarian cancer). The Institutional Review Board of King Faisal Specialist Hospital and Research Centre approved this study and the Research Advisory Council (RAC) of King Faisal Specialist Hospital and Research Centre provided waiver of informed consent under project RAC # 2140 008, since only retrospective patient data were analyzed. It is confirmed that all methods were performed in accordance with the relevant guidelines and regulations.
Table 1.
Summary of clinico-pathological variables in breast cancer patients (n = 791).
| Clinico-pathological variable | n (%) |
|---|---|
| Age at diagnosis, years | |
| Mean ± SD | 40.9 ± 9.8 |
| Median (range) | 39 (13–84) |
| ≤ 30 | 77 (9.8) |
| 31–40 | 387 (48.9) |
| 41–50 | 220 (27.8) |
| 51–60 | 68 (8.6) |
| > 60 | 39 (4.9) |
| Family history of cancer | |
| No | 529 (66.9) |
| Yes | 262 (33.1) |
| Bilateral breast cancer | |
| Yes | 15 (1.9) |
| No | 776 (98.1) |
| Histologic grade | |
| Well differentiated | 50 (6.3) |
| Moderately differentiated | 342 (43.2) |
| Poorly differentiated | 360 (45.5) |
| Unknown | 40 (5.0) |
| Tumor size | |
| T1 | 208 (26.3) |
| T2 | 329 (41.6) |
| T3 | 131 (16.6) |
| T4 | 101 (12.8) |
| Unknown | 22 (2.7) |
| Lymph node status | |
| Negative | 294 (37.2) |
| Positive | 475 (60.1) |
| Unknown | 22 (2.7) |
| Distant metastasis | |
| Absent | 697 (88.1) |
| Present | 72 (9.2) |
| Unknown | 22 (2.7) |
| Stage | |
| I | 111 (14.0) |
| II | 317 (40.1) |
| III | 269 (34.0) |
| IV | 72 (9.2) |
| Unknown | 22 (2.7) |
| ER | |
| Positive | 439 (55.5) |
| Negative | 352 (44.5) |
| PR | |
| Positive | 401 (50.7) |
| Negative | 390 (49.3) |
| Her-2 neu | |
| Positive | 254 (32.1) |
| Negative | 537 (67.9) |
| Triple negative breast cancer | |
| Yes | 199 (25.2) |
| No | 592 (74.8) |
Table 2.
Summary of clinico-pathological variables in epithelial ovarian cancer patients (n = 127).
| n (%) | |
|---|---|
| Age | |
| Mean ± SD | 39.7 ± 9.8 |
| Median (range) | 41.3 (17–66) |
| ≤ 30 | 30 (23.6) |
| 31–40 | 29 (22.8) |
| 41—50 | 59 (46.5) |
| 51–60 | 8 (6.3) |
| > 60 | 1 (0.8) |
| Family history of cancer | |
| No | 118 (92.9) |
| Yes | 9 (7.1) |
| Histopathology | |
| High grade serous | 56 (44.1) |
| Low grade serous | 27 (21.3) |
| Mucinous | 20 (15.7) |
| Endometrioid | 16 (12.6) |
| Clear cell | 3 (2.4) |
| Undifferentiated | 5 (3.9) |
| Histological grade | |
| Grade 1 | 33 (26.0) |
| Grade 2 | 53 (41.7) |
| Grade 3 | 40 (31.5) |
| Unknown | 1 (0.8) |
| pT | |
| T1 | 29 (22.8) |
| T2 | 13 (10.2) |
| T3 | 85 (67.0) |
| pN | |
| N0 | 120 (94.5) |
| N1 | 7 (5.5) |
| pM | |
| M0 | 105 (82.7) |
| M1 | 22 (17.3) |
| Stage | |
| I | 29 (22.8) |
| II | 9 (7.1) |
| III | 67 (52.8) |
| IV | 22 (17.3) |
DNA extraction
DNA samples were extracted from formalin-fixed and paraffin-embedded (FFPE) normal tissues of breast cancer or ovarian cancer patients utilizing Gentra DNA Isolation Kit (Gentra, Minneapolis, MN, USA) according to the manufacturer’s protocols as described in the previous study25. Two pathologists examined the histopathology slides to ensure that Normal tissues were obtained from different FFPE blocks such as uninvolved lymph nodes or non-cancerous breast tissue away from the tumor in order to minimize somatic contamination.
Capture sequencing
Targeted capture sequencing was performed on 918 samples using Illumina platform with the custom designed panel and all the quality metrics were applied as described previously26. ACMG/AMP 2015 guideline and ClinVar were utilized for interpretation of pathogenicity of variants.
Haplotype analysis
Haplotype analysis was performed using PHASE version 2.1.1 algorithm27,28. Number of variant positions, variant nucleotide positions and genotypes for each sample at those positions were supplied as an input in the algorithm. Following default parameter were set: number of iterations = 100, thinning interval = 1, burn-in = 100.
Results
Molecular results
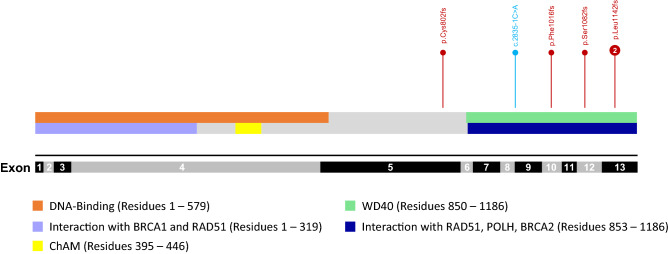
In the entire cohort, five different types of PVs or LPVs were identified in six cases, accounting for 0.65% (6/918) of all analyzed cases. Among these five variants, four (80%) were frameshift variants while one (20%) was splicing variant. All four frameshift variants were reported previously as PV or LPV in breast cancer cases while the splicing LPV was novel (Table 3). The PV/LPV distribution in the PALB2 gene was presented in the Fig. 1.
Table 3.
Pathogenic/ Likely pathogenic PALB2 (NM_024675.4) variants in breast cancer.
| Chr | Start | End | Ref | Alt | HGVS | ClinVar | Mutation type | |
|---|---|---|---|---|---|---|---|---|
| 1 | chr16 | 23,614,916 | 23,614,916 | A | – | c.3425delT:p.L1142fs | Likely pathogenic | Frameshift deletion |
| 2 | chr16 | 23,641,068 | 23,641,069 | CA | – | c.2406_2407del:p.C802fs | Pathogenic | Frameshift deletion |
| 3 | chr16 | 23,619,290 | 23,619,291 | CT | – | c.3244_3245del:p.S1082fs | Pathogenic | Frameshift deletion |
| 4 | chr16 | 23,634,452 | 23,634,453 | CT | AA | c.2835-2_2835-1delinsTT | NA | Splicing variant |
| 5 | chr16 | 23,632,748 | 23,632,748 | A | – | c.3048delT:p.F1016fs | Likely pathogenic | Frameshift deletion |
Figure 1.
Schematic representation of the PALB2 protein with variant positions and domains. Pins represent mutation position. Number represents the number of cases with the mutation (unmarked pin represents 1 case). Red pins represent frameshift mutations; blue pin represents splice-site mutation.
In the six mutant cases, all cases were breast cancer. None of ovarian cancer cases was found to carry any PVs or LPVs in PALB2. Only one recurrent LPV, NM_024675.4:c.3425del:p.Leu1142fs was identified. This variant was present in two unrelated cases. Interestingly, this homozygous PALB2 p.Leu1142fs variant was also previously reported in two Fanconi Anemia families in our population with strong family history of multiple cancer types including Wilms tumor, neruoblastoma and acute myeloid leukemia29. Therefore, haplotype analysis was performed for these two cases utilizing Capture Sequencing data. However, the founder mutation could not be confirmed due to the limited available SNPs information extracted from capture sequencing data. In addition, it was first time that the splicing variant c.2835-2_2835-1delinsTT was reported as LPV in cancer patients. However, other PALB2 PVs or LPVs detected in our cohort were previously reported in breast cancer or ovarian cancer patients in the literature30 (Table 3).
Clinico-pathological characteristics of breast cancer patients with PALB2 PV/LPVs
Breast cancer phenotype was invasive ductal (100%) with 66.7% of hormone-receptor positive and 16.7% of triple negative tumors. All six patients had either grade 2 (66.7%) or grade III (33.3%) tumors, with majority being stage II (66.7%). Family history was positive in 33.3% (2/6) of germline PALB2 PV/LPV carriers. One of the patients had a sister with breast cancer, whereas another patient’s mother (liver cancer) and sister (breast cancer) had history of cancer. None of the patients had a personal history of other cancers. One of the patient died due to disease progression (metastasis to lung, liver, bone and brain) (Table 4).
Table 4.
Clinico-pathological details of PALB2 mutated cases in breast cancer (n = 6).
| S no | Age | pT | pN | pM | Stage | Grade | Histology | ER | PR | Her-2 | TNBC | BRCA 1/2 mutation | Other tumors in patient | Family history | Status |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 40 | T4 | N2 | M0 | III | 2 | IDC | Positive | Positive | Positive | No | Negative | No | Negative | Alive |
| 2 | 38 | T2 | N2 | M1 | IV | 3 | IDC | Negative | Negative | Positive | No | Negative | No | Negative | Dead |
| 3 | 39 | T1 | N1 | M0 | II | 2 | IDC | Positive | Positive | Negative | No | Negative | No | Positive—sister had breast cancer | Alive |
| 4 | 41 | T2 | N0 | M0 | II | 2 | IDC | Negative | Negative | Negative | Yes | Negative | No | Positive—mother had liver cancer and sister had breast cancer | Alive |
| 5 | 34 | T3 | N0 | M0 | II | 3 | IDC | Positive | Positive | Negative | No | Negative | No | Negative | Alive |
| 6 | 49 | T2 | N0 | M0 | II | 2 | IDC | Positive | Positive | Positive | No | Negative | No | Negative | Alive |
Discussion
We identified six cases harboring PALB2 PVs or LPVs among 918 breast and ovarian cancer patients. The previously reported frequency rate range of PALB2 PV/LPV is from 0.1 to 1.5% depending on the population, cohort size and method of testing22,31–34. The frequency of PALB2 PV/LPV in this cohort is 0.65%, and therefore lies within the lowest frequencies reported worldwide.
In current study, all the PALB2 PVs/LPVs were identified in breast cancer cases and no PALB2 PVs or LPVs were found in ovarian cases. However, this should be interpreted with caution since the ovarian cancer cohort is small in comparing to breast cancer cohort included in this study.
The one recurrent frameshift LPV p.Leu1142fs was found in two unrelated young patients with breast cancer. Both patients have a family history of breast cancer. This recurrent PALB2 LPV identified in our cohort is particularly interesting since it was previously reported in two Fanconi anemia families with strong family history of multiple cancer types from Saudi Arabia 201629,35. We also found one submission of this alteration in the ClinVar database from different laboratories reporting germline testing of hereditary breast cancer (undisclosed ancestry)30. However, haplotype analysis based on the limited available SNPs data of two cases harboring this LPV did not confirm founder effect. Therefore, further analysis to confirm the founder effect of this LVP is needed. In addition, the patient carrying the novel LPV c.2835-2_2835-1delinsTT did not have family history of breast cancer.
Molecular and clinicopathological characteristics of breast cancer cases carrying PALB2 PVs/LPVs showed that all breast cancer tumors were invasive ductal carcinoma. Majority of the tumors were histologically intermediate to high grade, positive for HER2, ER and PR expression. Young onset was apparent in all the PALB2 PV or LPV carries with all of them under the age of 50 years of age, including 4 under the age of 40.
This study has limitation; first, it is retrospective single center study where selection bias can’t be excluded. Second, the ovarian cancer cohort is relatively small to draw a conclusion regarding the absence of PALB2 PV/LPV in ovarian cancer from this ethnicity. Third, germline filtering by peripheral blood sample could not be performed, due to unavailability of such samples.
In summary, this study was able to identify the prevalence of PALB2 PV/LPV and describe the molecular and clinical characteristics of PALB2 PV/LPV carriers in Saudi Arabia. Frequency of PALB2 PV/LPV in breast cancer seems to be lower than in other population31–34. PALB2 c.3425delT:p.Leu1142fs is a recurrent LPV that seems to be responsible for one third of PALB2 mutant cases. Recent studies demonstrated that PARP inhibitor is effective in treating breast cancer patients carrying PALB2 PV or LPV, opening a new avenue of target treatment that might benefit breast cancer patients36,37. Genetic testing for relevant genes such as PALB2 must be included in molecular and genetic evaluation of breast cancer patient from this population, which could contribute, to a better understanding of breast cancer risk and implementing preventive and therapeutic strategies for breast cancer patients from Middle Eastern ethnicity.
Author contributions
A.K.S. and K.S.A. designed the study and wrote the manuscript. I.A.A., A.T., F.A.D. and D.A. coordinated sample acquisition. K.I., S.A., Z.Q., M.A., W.H., M.D. and I.G. prepared and performed experiments. S.K.P contributed in acquisition of clinical data. S.K.P., K.I., S.A. and Z.Q. analyzed clinical and experimental data. All authors reviewed and approved the manuscript.
Data availability
All the data generated or analyzed during this study are included in this published article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Abdul K. Siraj and Rong Bu.
References
- 1.Ferlay, J. et al. Global cancer observatory: cancer today. Lyon, France: Int. Agency Res. Cancer3, 2019 (2018).
- 2.Sanyour RA, Abdullah M. Real time data analysis and visualization for the breast cancer disease. Period. Eng. Nat. Sci. 2019;7:395–407. [Google Scholar]
- 3.Yoshida R. Hereditary breast and ovarian cancer (HBOC): review of its molecular characteristics, screening, treatment, and prognosis. Breast Cancer. 2021;28:1167–1180. doi: 10.1007/s12282-020-01148-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamauchi H, Takei J. Management of hereditary breast and ovarian cancer. Int. J. Clin. Oncol. 2018;23:45–51. doi: 10.1007/s10147-017-1208-9. [DOI] [PubMed] [Google Scholar]
- 5.El Ansari FZ, et al. Screening of BRCA1/2 genes mutations and copy number variations in patients with high risk for hereditary breast and ovarian cancer syndrome (HBOC) BMC Cancer. 2020;20:747. doi: 10.1186/s12885-020-07250-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura, S., Aoki, D. & Miki, Y. in Hered. Breast Ovarian Cancer (ed Daisuke Aoki Seigo Nakamura, Yoshio Miki) VIII, 324 (Springer Singapore, 2022).
- 7.Hodgson, A. & Turashvili, G. Pathology of hereditary breast and ovarian cancer. Front Oncol.10, 531790. 10.3389/fonc.2020.531790 (2020). [DOI] [PMC free article] [PubMed]
- 8.Miki Y, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 9.Xia B, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 10.Sy SMH, Huen MSY, Zhu YY, Chen JJ. PALB2 regulates recombinational repair through chromatin association and oligomerization. J. Biol. Chem. 2009;284:18302–18310. doi: 10.1074/jbc.M109.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sy SMH, Huen MSY, Chen JJ. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc. Natl. Acad. Sci. 2009;106:7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buisson R, Masson JY. PALB2 self-interaction controls homologous recombination. Nucleic Acids Res. 2012;40:10312–10323. doi: 10.1093/nar/gks807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park JY, et al. Breast cancer-associated missense mutants of the PALB2 WD40 domain, which directly binds RAD51C, RAD51 and BRCA2, disrupt DNA repair. Oncogene. 2014;33:4803–4812. doi: 10.1038/onc.2013.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foo TK, et al. Compromised BRCA1–PALB2 interaction is associated with breast cancer risk. Oncogene. 2017;36:4161–4170. doi: 10.1038/onc.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antoniou AC, et al. Breast-cancer risk in families with mutations in PALB2. N. Engl. J. Med. 2014;371:497–506. doi: 10.1056/NEJMoa1400382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X, et al. Cancer risks associated with germline PALB2 pathogenic variants: an international study of 524 families. J. Clin. Oncol. 2020;38:674–685. doi: 10.1200/JCO.19.01907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorling L, et al. Breast cancer risk genes—association analysis in more than 113,000 women. N. Engl. J. Med. 2021;384:428–439. doi: 10.1056/NEJMoa1913948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu C, et al. A population-based study of genes previously implicated in breast cancer. N. Engl. J. Med. 2021;384:440–451. doi: 10.1056/NEJMoa2005936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dansonka-Mieszkowska A, et al. A novel germline PALB2 deletion in Polish breast and ovarian cancer patients. BMC Med. Genet. 2010;11:20. doi: 10.1186/1471-2350-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peterlongo P, et al. PALB2 germline mutations in familial breast cancer cases with personal and family history of pancreatic cancer. Breast Cancer Res. Treat. 2011;126:825–828. doi: 10.1007/s10549-010-1305-1. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen-Dumont T, et al. Mutation screening of PALB2 in clinically ascertained families from the Breast Cancer Family Registry. Breast Cancer Res. Treat. 2015;149:547–554. doi: 10.1007/s10549-014-3260-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kluska A, et al. PALB2 mutations in BRCA1/2-mutation negative breast and ovarian cancer patients from Poland. BMC Med. Genom. 2017;10:14. doi: 10.1186/s12920-017-0251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hauke J, et al. Gene panel testing of 5589 BRCA 1/2-negative index patients with breast cancer in a routine diagnostic setting: results of the German Consortium for Hereditary Breast and Ovarian Cancer. Cancer Med. 2018;7:1349–1358. doi: 10.1002/cam4.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petrucelli N, Daly MB, Feldman GL. Hereditary breast and ovarian cancer due to mutations in BRCA1 and BRCA2. Genet. Med. 2010;12:245–259. doi: 10.1097/GIM.0b013e3181d38f2f. [DOI] [PubMed] [Google Scholar]
- 25.Abubaker J, et al. Clinicopathological analysis of papillary thyroid cancer with PIK3CA alterations in a Middle Eastern population. J. Clin. Endocrinol. Metab. 2008;93:611–618. doi: 10.1210/jc.2007-1717. [DOI] [PubMed] [Google Scholar]
- 26.Siraj AK, et al. Expanding the spectrum of germline variants in cancer. Hum. Genet. 2017;136:1431–1444. doi: 10.1007/s00439-017-1845-0. [DOI] [PubMed] [Google Scholar]
- 27.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am. J. Hum. Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghazwani Y, et al. Clinical characteristics and genetic subtypes of Fanconi anemia in Saudi patients. Cancer Genet. 2016;209:171–176. doi: 10.1016/j.cancergen.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Landrum MJ, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tischkowitz, M. et al. Contribution of the PALB2 c.2323C>T [p.Q775X] founder mutation in well-defined breast and/or ovarian cancer families and unselected ovarian cancer cases of French Canadian descent. BMC Med. Genet.14, 5. 10.1186/1471-2350-14-5 (2013). [DOI] [PMC free article] [PubMed]
- 32.Damiola F, et al. Mutation analysis of PALB2 gene in French breast cancer families. Breast Cancer Res. Treat. 2015;154:463–471. doi: 10.1007/s10549-015-3625-7. [DOI] [PubMed] [Google Scholar]
- 33.Couch FJ, et al. Associations between cancer predisposition testing panel genes and breast cancer. JAMA Oncol. 2017;3:1190–1196. doi: 10.1001/jamaoncol.2017.0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buys SS, et al. A study of over 35,000 women with breast cancer tested with a 25-gene panel of hereditary cancer genes. Cancer. 2017;123:1721–1730. doi: 10.1002/cncr.30498. [DOI] [PubMed] [Google Scholar]
- 35.Reid S, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat. Genet. 2007;39:162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 36.Barchiesi, G. et al. Emerging role of PARP inhibitors in metastatic triple negative breast cancer. Current scenario and future perspectives. Front. Oncol.11, 769280 (2021). [DOI] [PMC free article] [PubMed]
- 37.Collet, L. et al. PARP inhibitors: A major therapeutic option in endocrine-receptor positive breast cancers. Cancers (Basel)14, 599 (2022). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data generated or analyzed during this study are included in this published article.