Abstract
Amebae have an Hsp60-associated, mitochondrion-derived organelle (crypton). In this study, the crypton was stained with multiple DNA-binding fluorochromes and a monoclonal anti-double-stranded DNA antibody. Transmission microscopy of partially purified cryptons revealed organelles bound by a double membrane.
The endosymbiont hypothesis, one of the great ideas in cell biology, suggests that mitochondria derive from a phagocytosed α-purple bacterium (11). Evidence for this idea includes sequences of rRNA genes, which are present in the mitochondrial genome, and sequences of heat shock proteins such as Hsp60, which are targeted to mitochondria (12, 31). Although the lumenal parasites (amebae, giardia, and trichomonads) appear to be “amitochondriate,” they each contain an hsp60 gene encoding a homologue of the mitochondrial Hsp60 (5, 7, 8, 24). The trichomonad mitochondrion has been converted into a fermentation factory, which is called a hydrogenosome because it produces hydrogen gas via an iron-dependent hydrogenase (Fe-hydrogenase) (2, 6, 19). The amebic mitochondrion-derived organelle was called “mitosome” or “crypton,” because it is small and rare and its function remains unclear (17, 28). The crypton contains Hsp60, which is induced by heat shock and is functional in a groEL mutant of Escherichia coli (17). Like nucleus-encoded mitochondrial and hydrogenosomal proteins, the amebic Hsp60 has a presequence rich in Leu and Ser, which is cleaved at Arg-2 (4, 9, 17). In contrast, the crypton lacks enzymes of oxidative phosphorylation and lacks fermentation proteins including ferredoxin and alcohol dehydrogenase 1 (ADH1) (15, 17, 21).
An amebic cytosolic structure, which was stained green with acridine orange, was called EhKO (Entamoeba histolytica kinetoplastid organelle) by Esther Orozco and colleagues, because it contains circular DNAs (20). They also localized the 24-kb rRNA episome, the pyruvate:ferredoxin oxidoreductase (POR), and a TATA-binding protein to the EhKO (16, 23). The goals in this study were to determine whether the amebic crypton/mitosome is the same as or different from the EhKO and to determine whether the organelle is bound by a double membrane as is present around mitochondria and hydrogenosomes (1–3, 11, 19).
The crypton contains 2.2% of the amount of DNA in the amebic nucleus.
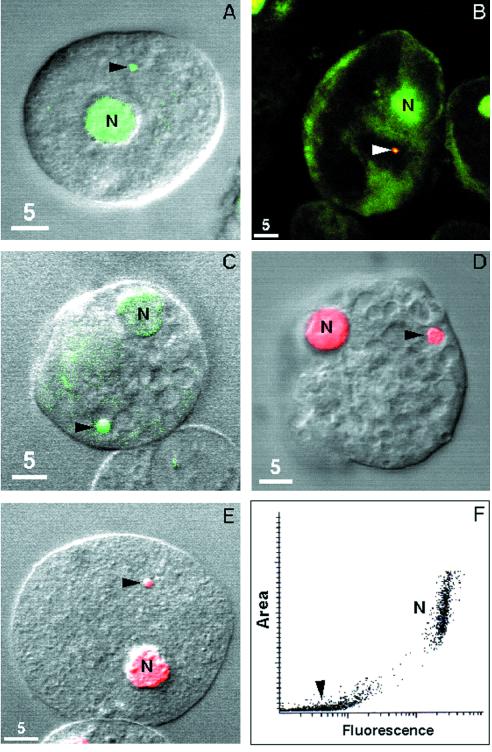
E. histolytica HM-1 strain trophozoites were fixed in 2% paraformaldehyde, permeabilized with 0.1% Triton X-100, treated with 20 μg of RNAse per ml, stained with DNA-binding fluorochromes (1 μM sytox green or 1 μg of propidium iodide per ml), and viewed with a Leica NT-TCS confocal microscope (27). One or at most two cryptons per amebae stained well with sytox green (Fig. 1A and B), 1 μM acridine orange (unfixed cells) (Fig. 1C), and propidium iodide (Fig. 1E). The DNA-associated organelles were the crypton, because they also stained with rabbit antibodies to the C terminus of Hsp60 (Fig. 1B), which were detected with Texas red-conjugated goat anti-rabbit antibodies (8, 12, 17). The crypton was also stained with a 2C10 monoclonal antibody to anti-double-stranded DNA from a Lupus mouse (Fig. 1D), which was diluted to 1 μg/ml and detected with Texas red-conjugated goat anti-mouse antibodies, as previously described (13). Western blots showed that the anti-DNA monoclonal antibody bound to amebic DNA but not to amebic RNA or protein (data not shown). Anti-DNA antibodies have been used to identify the hydrogenosomal genome of Nyctotherus ovalis, a protozoan parasite of the cockroach hindgut (1). Interestingly, cryptons were also visible as convex spheres by differential interference contrast microscopy (Fig. 1A and C to E). Nuclei, which have a double membrane, also appeared convex, while cytosolic vacuoles, which have a single membrane, appeared concave (see below).
FIG. 1.
Confocal micrographs of nuclei (N) and cryptons (arrows) of E. histolytica trophozoites stained with DNA-binding fluorochromes, including sytox green (green [A and B]), acridine orange (green [C]), and propidium iodide (red [E]) (27). The crypton was also stained with anti-Hsp60 antibodies (yellow [B]) and a mouse monoclonal antibody to double-stranded DNA (red [D]) (13, 17). (F) Scatter diagram shows area and propidium iodide staining of the crypton and nuclei (14).
The relative amounts of propidium iodide that bound to the amebic crypton and nucleus were measured using an Olympus microscope equipped with an argon laser and an attached camera and image analysis system (CompuCyte, Cambridge, Mass.) (Fig. 1F) (14). The propidium iodide-stained organelle contained 2.2% as much DNA as the amebic nucleus, although there was marked variability in the amount of crypton DNA. Assuming that amebae are diploid and have a 20-Mb haploid genome, the DNA content of the nucleus is 40 Mb and the crypton is 880 kb (29; S. Ghosh and J. Samuelson, unpublished observations).
The 24-kb rRNA episome is present in the nucleus, while Fe-dependent hydrogenase is present in the cytosol.
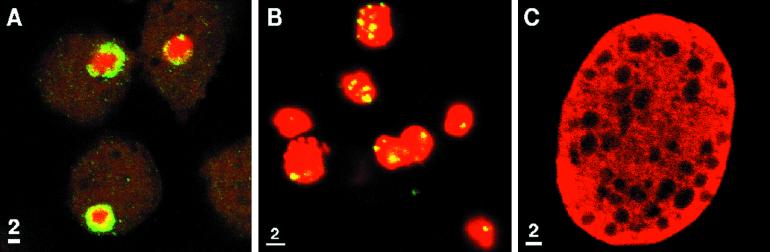
Amebae were allowed to adhere to polyLys-coated slides, were fixed in methanol-acetic acid (3:1), air-dried, and denatured in 70% formamide at 70°C (18). Plasmids, which contained ∼40% of the 24-kb rRNA episome, were labeled with biotin, hybridized to denatured amebae in 30% formamide and 2× SSC (1× SSC is 300 mM sodium chloride and 30 mM sodium citrate [pH 7]) at 37°C, and washed with the same buffer (26). Amebae were incubated with fluorescein isothiocyanate-avidin, treated with 200 μg of RNAse per ml, and counterstained with propidium iodide. Numerous copies of the 24-kb rRNA episome were present in the periphery of amebic nuclei but not within the cytosol (Fig. 2A). On two previous occasions the rRNA episomes have been shown to be present at the periphery of amebic nuclei (26, 30). Episomal plasmids based upon pBluescript, which were selected in transfected amebae with G418, were present in clumps throughout the amebic nucleus and were absent from the cytosol (Fig. 2B) (10). This clumping is consistent with concatenation of the foreign plasmids in transfected amebae. Negative controls with pBluescript in nontransfected parasites showed no fluorescence in situ hybridization (FISH) signal. Because the morphology of the crypton was not well preserved in the methanol-acetic acid fixative used for FISH, it was impossible to rule out the possibility that small numbers of rRNAs or episomal plasmids were present in the crypton. However, in no cases were cytosolic structures heavily stained with the rRNA probes as recently described by autoradiographic methods (20).
FIG. 2.
Confocal micrographs of FISH of amebae with probes to the 24-kb rRNA episome (green [A]) and to foreign episomes in transfected parasites (green [B]) (10, 18, 26). Confocal micrograph of epitope-tagged Fe-dependent hydrogenase (red [C]) in the cytosol of transfected amebae (10).
We recently cloned an E. histolytica Fe-hydrogenase gene, which encodes a protein with 34% amino acid identity with an Fe-hydrogenase of Trichomonas vaginalis (6; J. Field and J. Samuelson, unpublished observations). Amebae were transfected with a plasmid containing a modified amebic Fe-hydrogenase gene, which encodes a protein with amebic chitinase repeats as an epitope tag at the C terminus (10). Parasites were fixed, permeabilized, and immunostained with rabbit antibodies to the chitinase repeats exactly as previously described (10). The amebic Fe-dependent hydrogenase, which lacks an organelle-targeting presequence, was present in the cytosol (Fig. 2C) (4, 9, 17). Previously, ferredoxin, ADH1, and Hsp60 less its presequence were identified in the parasite cytosol (17). Further, amebic fermentation enzymes (ferredoxin, ADH1, ADHE, ADH3, and POR) all lack presequences, which might target them to the crypton (15, 22, 25, 32).
Cryptons appear to be bound by a double membrane.
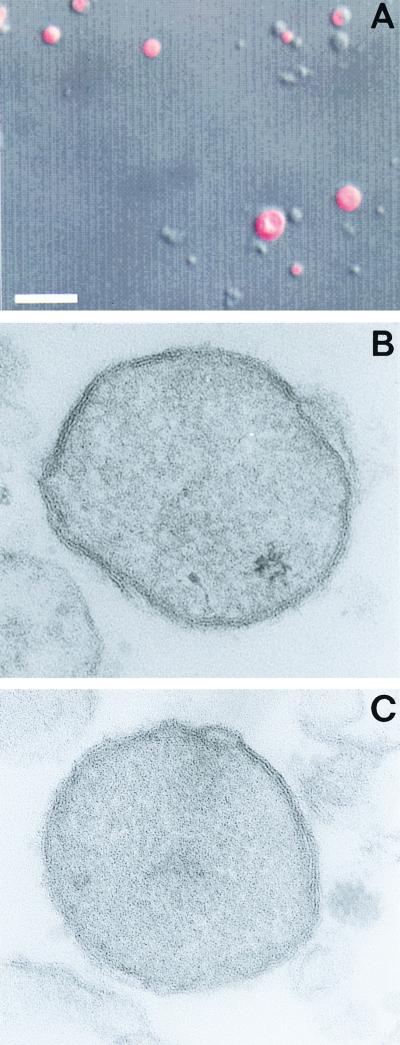
The crypton is small and rare, so parasites were disrupted by gentle homogenization, nuclei were removed by low-speed centrifugation, and an enriched fraction of propidium iodide-stained cryptons was obtained using a 20% Percoll gradient (Fig. 3A). These fractions were fixed in 2% paraformaldehyde, postfixed in 1% osmium tetroxide, stained in block with uranyl acetate, dehydrated in graded ethanols, and embedded in Epon. Although there was contamination with vesicles and vacuoles, all of which were bound by a single membrane, putative cryptons were present as 0.5- to 1-μm-diameter circles that were filled with electron-dense material and bound by two closely applied membranes (Fig. 3B and C). Putative cryptons, which were abundant and easy to identify, resembled hydrogenosomes of T. vaginalis and N. ovalis, which are also bound by two closely applied membranes (1, 3). Because the partially purified cryptons fell apart when ultrathin frozen sections were cut, we were unable to use immunoelectron microscopy to demonstrate binding of antibodies to Hsp60 or double-stranded DNA.
FIG. 3.
(A) Confocal micrograph of partially purified cryptons stained red with propidium iodide and (B and C) transmission micrographs of cryptons, which are bound by a double membrane. (B and C) Magnification, ×125,000.
Conclusions.
These results, which are summarized in Table 1, suggest that the amebic crypton/mitosome has genomic DNA and so likely is the same organelle as the EhKO (17, 20, 28). This conclusion was unexpected because the amebic crypton appears atrophic and most hydrogenosomes lack a genome (1, 17, 19). Here and in our previous studies, the crypton did not contain rRNA genes and fermentation enzymes (hydrogenase, ferredoxin, and ADH1), which have been attributed to the EhKO (15, 17, 20, 23). Further, there are no examples of nuclear rRNA genes in mitochondrial genomes (11), while fermentation enzymes of hydrogenosomes all have organelle-targeting sequences (1, 4–6). Finally, the crypton appears to be bound by a double membrane, which was expected as the crypton is descended from mitochondria and appears to have similar machinery for binding and transporting Hsp60 to the matrix (8, 17).
TABLE 1.
Staining of nucleus and crypton with various probes
| Probe | Result for:
|
|
|---|---|---|
| Nucleus | Crypton | |
| Sytox green | + | + |
| Acridine orange | + | + |
| Propidium iodide | + | + |
| Anti-ds DNA antibodiesa | + | + |
| Anti-Hsp60 antibodies | − | + |
| 24-kb rRNA episomes | + | − |
| Foreign episomes | + | − |
| Hydrogenase | − | − |
| Double-membrane | + | + |
ds, double stranded.
Acknowledgments
This work was supported in part by NIH grants AI-33492 and GM-31318 (to J.S.) and HL-33009 and HL-43510 (to R.R.).
We acknowledge the expert technical support of Jean Lai for confocal microscopy and Maria Ericsson for electron microscopy. We also thank Leona Samson of the Department of Cancer Cell Biology, Harvard School of Public Health, for use of the CompuCyte image analysis system and David Stollar of the Department of Biochemistry, Tufts University School of Medicine, for the monoclonal anti-double-stranded DNA antibody.
REFERENCES
- 1.Akhmanova A, Voncken F, van Alen T, van Hoek A, Boxma B, Vogels G, Veenhuis M, Hackstein J H. A hydrogenosome with a genome. Nature. 1998;396:527–528. doi: 10.1038/25023. [DOI] [PubMed] [Google Scholar]
- 2.Andersson S G E, Kurland C G. Origins of mitochondria and hydrogenosomes. Curr Opin Microbiol. 1999;2:535–541. doi: 10.1016/s1369-5274(99)00013-2. [DOI] [PubMed] [Google Scholar]
- 3.Benchimol M, Johnson P J, Desouza W. Morphogenesis of the hydrogenosome—an ultrastructural study. Biol Cell. 1996;87:197–205. [PubMed] [Google Scholar]
- 4.Bradley P J, Lahti C J, Plumper E, Johnson P J. Targeting and translocation of proteins into the hydrogenosome of the protist Trichomonas: similarities with mitochondrial protein import. EMBO J. 1997;16:3484–3493. doi: 10.1093/emboj/16.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bui E T N, Bradley P J, Johnson P J. A common evolutionary origin for mitochondria and hydrogenosomes. Proc Natl Acad Sci USA. 1996;93:9651–9656. doi: 10.1073/pnas.93.18.9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bui E T, Johnson P J. Identification and characterization of [Fe]-hydrogenases in the hydrogenosome of Trichomonas vaginalis. Mol Biochem Parasitol. 1996;76:305–310. doi: 10.1016/0166-6851(96)02567-4. [DOI] [PubMed] [Google Scholar]
- 7.Cavalier-Smith T. Eukaryotes with no mitochondria. Nature. 1987;326:332–333. doi: 10.1038/326332a0. [DOI] [PubMed] [Google Scholar]
- 8.Clark C G, Roger A J. Direct evidence for secondary loss of mitochondria in Entamoeba histolytica. Proc Natl Acad Sci USA. 1995;92:6518–6521. doi: 10.1073/pnas.92.14.6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claros M G, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh S K, Field J, Frisardi M, Rosenthal B, Mai Z, Rogers R, Samuelson J. Chitinase secretion by encysting Entamoeba invadens and transfected Entamoeba histolytica trophozoites: localization of secretory vesicles, endoplasmic reticulum, and Golgi apparatus. Infect Immun. 1999;67:3073–3081. doi: 10.1128/iai.67.6.3073-3081.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray M W, Burger G, Land B F. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 12.Hartl F U. Molecular chaperones in cellular protein folding. Nature. 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 13.Jang Y-J, Sanford D, Chung H Y, Baek S Y, Stollar B D. The structural basis for DNA binding by an anti-DNA autoantibody. Mol Immunol. 1998;35:1207–1217. doi: 10.1016/s0161-5890(98)00095-9. [DOI] [PubMed] [Google Scholar]
- 14.Kamentsky L A, Burger D E, Gershman R J, Kamentsky L D, Luther E. Slide-based laser scanning cytometry. Acta Cytolog. 1997;41:123–143. doi: 10.1159/000332315. [DOI] [PubMed] [Google Scholar]
- 15.Kumar A, Shen P-S, Descoteaux S, Pohl J, Bailey G, Samuelson J. Cloning and expression of an NADP+-dependent alcohol dehydrogenase gene of Entamoeba histolytica. Proc Natl Acad Sci USA. 1992;85:1782–1786. doi: 10.1073/pnas.89.21.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luna-Arias J P, Hernandez-Rivas R, de Dios-Bravo G, Garcia J, Mendoza L, Orozco E. The TATA-binding protein of Entamoeba histolytica: cloning of the gene location of the protein by immunofluorescence and confocal microscopy. Microbiol. 1999;145:33–40. doi: 10.1099/13500872-145-1-33. [DOI] [PubMed] [Google Scholar]
- 17.Mai Z, Ghosh S, Frisardi M, Rosenthal B, Rogers R, Samuelson J. Hsp60 is targeted to a cryptic mitochondrion-derived organelle (“crypton”) in the microaerophilic protozoan parasite Entamoeba histolytica. Mol Cell Biol. 1999;19:2198–2205. doi: 10.1128/mcb.19.3.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNeil J A, Johnson C V, Carter K C, Singer R H, Lawrence J B. Localizing DNA and RNA within nuclei and chromosomes by fluorescence in situ hybridization. Genet Anal Tech Applic. 1991;8:41–58. doi: 10.1016/1050-3862(91)90049-w. [DOI] [PubMed] [Google Scholar]
- 19.Muller M. The hydrogenosome. J Gen Microbiol. 1993;139:2879–2889. doi: 10.1099/00221287-139-12-2879. [DOI] [PubMed] [Google Scholar]
- 20.Orozco E, Gharaibeh R, Riveron A M, Delgadillo D M, Mercado M, Sanchez T, Gomez Conde E, Vargas M A, Lopez-Revilla R. A novel cytoplasmic structure containing DNA networks in Entamoeba histolytica trophozoites. Mol Gen Genet. 1997;254:250–257. doi: 10.1007/s004380050413. [DOI] [PubMed] [Google Scholar]
- 21.Reeves R E. Metabolism of Entamoeba histolytica Schaudinn, 1903. Adv Parasitol. 1984;23:105–142. doi: 10.1016/s0065-308x(08)60286-9. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez M A, Baez-Camargo M, Delgadillo D M, Orozco E. Cloning and expression of an Entamoeba histolytica NADP+-dependent alcohol dehydrogenase gene. Biochim Biophys Acta. 1996;1306:23–26. doi: 10.1016/0167-4781(96)00014-0. [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez M A, Garcia-Perez R M, Mendoza L, Sanchez T, Guillen N, Orozco E. The pyruvate:ferredoxin oxidoreductase enzyme is located in the plasma membrane and in a cytoplasmic structure in Entamoeba. Microb Pathog. 1998;25:1–10. doi: 10.1006/mpat.1998.0202. [DOI] [PubMed] [Google Scholar]
- 24.Roger A J, Svard S G, Tovar J, Clark C G, Smith M W, Gillin F D, Sogin M L. A mitochondrial-like chaperonin 60 gene in Giardia lamblia: evidence that diplomonads once harbored an endosymbiont related to the progenitor of mitochondria. Proc Natl Acad Sci USA. 1998;95:229–234. doi: 10.1073/pnas.95.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenthal B, Mai Z, Caplivski D, Ghosh S, de la Vega H, Graf T, Samuelson J. Evidence for the bacterial origin of genes encoding fermentation enzymes of the amitochondriate protozoan parasite Entamoeba histolytica. J Bacteriol. 1997;179:3736–3745. doi: 10.1128/jb.179.11.3736-3745.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sehgal D, Mittal V, Ramachandran S, Dhar S K, Bhattacharya A. Nucleotide sequence organization and analysis of the nuclear ribosomal DNA circle of the protozoan parasite Entamoeba histolytica. Mol Biochem Parasitol. 1994;67:205–214. doi: 10.1016/0166-6851(94)00129-4. [DOI] [PubMed] [Google Scholar]
- 27.Shapiro, H. Practical flow cytometry. Alton R. Liss, New York, N.Y.
- 28.Tovar J, Fischer A, Clark C G. The mitosome, a novel organelle related to mitochondria in the amitochondrial parasite Entamoeba histolytica. Mol Microbiol. 1999;32:1013–1021. doi: 10.1046/j.1365-2958.1999.01414.x. [DOI] [PubMed] [Google Scholar]
- 29.Willhoeft U, Tannich E. The electrophoretic karyotype of Entamoeba histolytica. Mol Biochem Parasitol. 1999;99:41–53. doi: 10.1016/s0166-6851(98)00178-9. [DOI] [PubMed] [Google Scholar]
- 30.Willhoeft U, Tannich E. Fluorescence microscopy and fluorescence in situ hybridization of Entamoeba histolytica nuclei to analyse mitosis and the localization of repetitive DNA. Mol Biochem Parasitol. 2000;105:291–296. doi: 10.1016/s0166-6851(99)00181-4. , 2000. [DOI] [PubMed] [Google Scholar]
- 31.Yang D, Oyaizu Y, Oyaizu H, Olsen G J, Woese C R. Mitochondrial origins. Proc Natl Acad Sci USA. 1985;82:4443–4447. doi: 10.1073/pnas.82.13.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang W, Li E, Kairong T, Stanley S L., Jr Entamoeba histolytica has an alcohol dehydrogenase homologous to the multifunctional adhE gene product of Escherichia coli. Mol Biochem Parasitol. 1994;64:253–260. doi: 10.1016/0166-6851(93)00020-a. [DOI] [PubMed] [Google Scholar]