ABSTRACT
Increased mitochondrial reactive oxygen species (mROS) and glycolysis have been established in pulmonary hypertension (PH). However, the effect of elevated mROS on glycolytic shift and how increased glycolysis promotes hypoxic pulmonary artery smooth muscle cell (PASMC) proliferation and vascular remodeling remain elusive. Here, we reported that hypoxia-induced mROS inhibit HIF-1α hydroxylation and further trigger PASMC glycolytic switch through the upregulated HIF-1α/PDK1&PDK2/p-PDH-E1α axis, which facilitates lactate accumulation and histone lactylation. Through H3K18la and HIF-1α ChIP–seq analysis, we found that the enhanced histone lactylation of HIF-1α targets, such as Bmp5, Trpc5, and Kit, promotes PASMC proliferation. Knockdown of Pdk1&2 blunts lactate production, histone lactylation marks, and PASMC proliferation. Moreover, pharmacological intervention with lactate dehydrogenase inhibitor diminishes histone lactylation and ameliorates PASMC proliferation and vascular remodeling in hypoxic PH rats. Taken together, this study provides proof of concept for anti-remodeling therapy through lactate manipulation.
Keywords: pulmonary hypertension, hypoxia, reactive oxygen species, cell proliferation, glycolysis, histone lactylation
Introduction
A striking pathological alteration of pulmonary hypertension (PH) is the obstructive remodeling of pulmonary arterioles, principally manifested by the progressive proliferation of pulmonary artery smooth muscle cells (PASMCs) (Rabinovitch, 2012; Humbert et al., 2019). Recently, numerous molecular abnormalities have been identified in proliferating PASMCs (Morrell et al., 2009; Rabinovitch, 2012). Glycolytic shift, as a common denominator in PH, has aroused widespread attention (Paulin and Michelakis, 2014; Sutendra and Michelakis, 2014; Ryan and Archer, 2015). The cancer-like metabolic abnormality is ubiquitous but has not been well studied. Generally, metabolism is adjusted immediately in response to various pathophysiological stimuli. As important stressful mediators (Shadel and Horvath, 2015), mitochondrial reactive oxygen species (mROS) have recently been identified as essential sensors for hypoxia in PH onset (Adesina et al., 2015; Pak et al., 2018; Sommer et al., 2020). We, therefore, postulated that increased mROS might be involved in the glycolytic switch of proliferating PASMCs under hypoxia.
The central issue in this uncoupled glycolysis is the inhibition of pyruvate dehydrogenase (PDH) catalysis of pyruvate to acetyl-CoA (Sutendra and Michelakis, 2013). As a gate-keeping checkpoint, PDH is typically inactivated by phosphorylation at serine 293 mediated by PDH kinase 1 (PDK1) and PDK2 in vascular cells (Bowker-Kinley et al., 1998; Michelakis et al., 2017). PDK could be upregulated by an established transcriptional factor, hypoxia-inducible factor 1-alpha (HIF-1α) (Manalo et al., 2005). It has been suggested that HIF-1α stabilization occurs partly from increased mROS (Movafagh et al., 2015). Hence, we further hypothesized that HIF-1α accumulation mediated by elevated mROS might trigger a glycolytic switch of proliferating PASMCs under hypoxia.
Recently, it has emerged that epigenetic mechanisms could initiate the activated phenotypes of pulmonary vascular cells, including PASMC proliferation (Cheng et al., 2019; Hu et al., 2019). As a fundamental regulation of chromatin accessibility, metabolism has been implicated in the epigenetic control of gene transcription and cellular identities (Reid et al., 2017). During this metabolon–epigenetics interaction, diverse cofactors and metabolites are utilized by epigenetic enzymes to modify chromatin structure, such as acetyl-CoA required for histone acetylation (Berger and Sassone-Corsi, 2016). Lactate, as the end-product in uncoupled glycolysis, is widely known as a metabolic by-product. However, it has recently been reported that increased lactate could facilitate lactylation of histone lysine residues, which functions as a novel epigenetic modification (Zhang et al., 2019). Whether enhanced histone lactylation induced by glycolysis promotes PASMC proliferation remains unknown.
Here, we demonstrate that increased mROS orchestrate glycolytic shift with PASMC proliferation in a HIF-1α-dependent manner under hypoxia. The uncovered intermediate mechanism highlights the lactate-derived histone lactylation of HIF-1α targets associated with cell proliferation. Disruption of lactate production and histone lactylation blunts PASMC proliferation and vascular remodeling. These findings elucidate a novel epigenetic modification in PH and suggest that lactate manipulation might represent a suitable avenue for developing therapies against PH.
Results
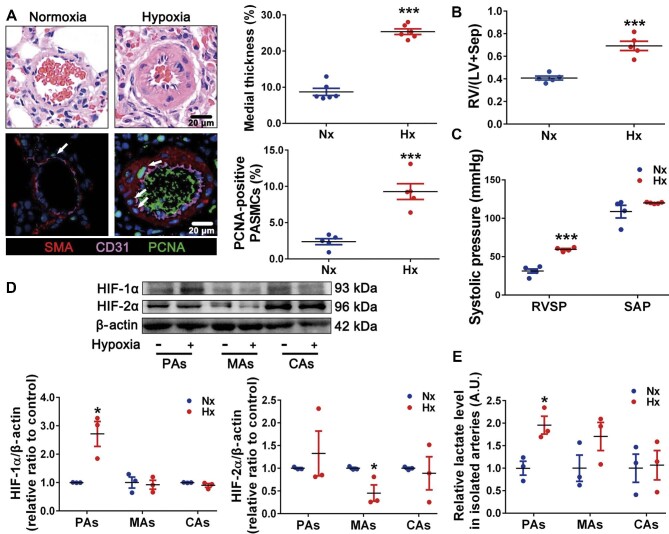
Hypoxia specifically increases the levels of HIF-1α and lactate in remodeled pulmonary arteries
To explore the potential effect of HIF-1α and HIF-2α on glycolytic switch during pulmonary arteries (PAs) remodeling, we examined the levels of HIF-1α, HIF-2α, and lactate in pulmonary and systemic circulatory arteries. Firstly, hypoxia increased PA medial thickness and PASMC proliferation (Figure 1A). Meanwhile, hypoxia enhanced the ratio right ventricle/(left ventricle plus septum) (RV/(LV + Sep); Figure 1B) and right ventricular systolic pressure (RVSP) rather than systemic artery pressure (SAP) (Figure 1C; Supplementary Figure S1). Moreover, levels of HIF-1α protein (Figure 1D) and lactate (Figure 1E), as two important elements in PH pathogenesis, were increased in hypoxic PAs rather than in mesenteric arteries (MAs) and carotid arteries (CAs). Furthermore, immunofluorescence staining demonstrated that the upregulated HIF-1α was mainly accumulated in PASMC nuclei and co-localized well with the increased proliferating cell nuclear antigen (PCNA)-positive nuclei (Supplementary Figure S2). HIF-2α protein was reduced in hypoxic MAs but unchanged in PAs and CAs (Figure 1D).
Figure 1.
The levels of HIF-1α and lactate are increased in remodeled PAs from hypoxic PH rats. (A) Top, representative H&E images of lung sections, with quantification of the ratio of medial thickness to diameter. Bottom, representative immunofluorescence images of lung sections stained for CD31 (pink), SMA (red), and PCNA (green, white arrows), with quantification of the percentage of PCNA-positive nuclei. Scale bar, 20 μm. (B) Changes in RV/(LV + Sep). (C) Changes in RVSP and SAP. (D) Immunoblots for HIF-1α, HIF-2α, and β-actin in isolated PAs, MAs, and CAs. (E) Relative lactate levels in freshly isolated PAs, MAs, and CAs. Data represent mean ± SEM; *P < 0.05, ***P < 0.001 vs. Nx; two-tailed unpaired Student's t-test. Nx, normoxia; Hx, hypoxia; A.U., arbitrary unit.
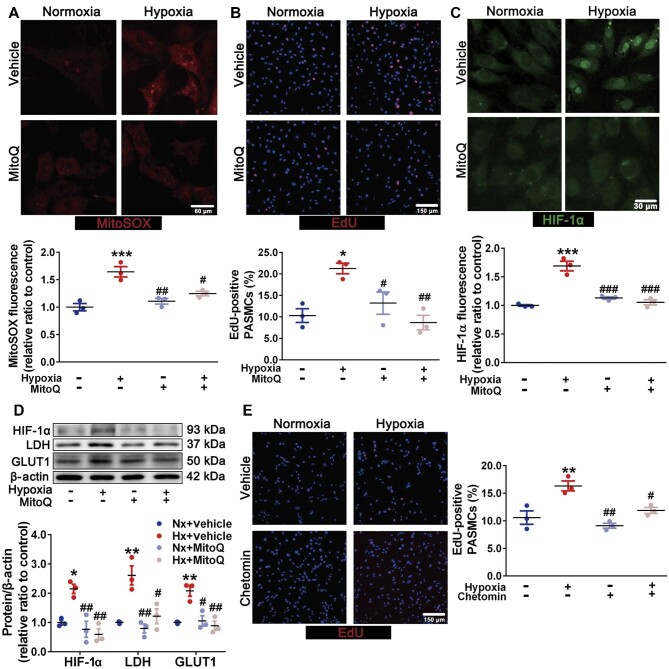
Hypoxia facilitates PASMC proliferation in a mROS-dependent manner
mROS, the important hypoxia mediators, are known to play important roles in adapting to hypoxia (Diebold and Chandel, 2016). Consequently, we further evaluated the mROS levels and their roles in PASMC proliferation under hypoxia. Indeed, hypoxia upregulated mROS levels (Figure 2A) and proliferative capacity of PASMCs (Figure 2B). Mitoquinone mesylate (MitoQ), as a mitochondria-targeted antioxidant, significantly attenuated hypoxia-induced mROS accumulation (Figure 2A). Meanwhile, the intracellular hydrogen peroxide (H2O2) was also decreased by MitoQ incubation (Supplementary Figure S3). Furthermore, MitoQ alleviated the increased PASMC viability (Supplementary Figure S4) and proliferation under hypoxia (Figure 2B).
Figure 2.
Increased mROS facilitate PASMC proliferation in a HIF-1α-dependent manner. (A–D) Primary PASMCs were exposed to normoxia and hypoxia with or without MitoQ (0.5 μM) incubation. (A) Representative MitoSOX images of PASMCs (scale bar, 60 μm) with quantification of relative fluorescent density. (B) Representative EdU (red) images of PASMCs (scale bar, 150 μm) with quantification of the percentage of EdU-positive nuclei. (C) Representative immunofluorescence images of PASMCs stained for HIF-1α (scale bar, 30 μm) with quantification of the relative fluorescence density of HIF-1α. (D) Immunoblots for HIF-1α, GLUT1, LDH, and β-actin. (E) Representative EdU (red) images of PASMCs exposed to normoxia and hypoxia with or without 50 nM chetomin incubation, with quantification of the percentage of EdU-positive nuclei. Scale bar, 150 μm. Data represent mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001 vs. Nx; #P < 0.05, ##P < 0.01, ###P < 0.001 vs. Hx; two-way ANOVA and Bonferroni's post-test.
mROS-dependent HIF-1α accumulation promotes the PASMC proliferative phenotype
As another key regulator during oxygen fluctuation, HIF could transcriptionally modulate metabolic status and cell proliferation (Choudhry and Harris, 2018). We, therefore, examined the potential roles of HIF-1α and HIF-2α in PASMC proliferation. Firstly, hypoxia promoted HIF-1α accumulation (Figure 2C and D) rather than HIF-2α accumulation (Supplementary Figure S5) in isolated PASMCs, which was inhibited by MitoQ incubation (Figure 2C and D). Additionally, the protein levels of HIF-1α targets, such as glucose transporter 1 (GLUT1) and lactate dehydrogenase (LDH), were also increased, which were attenuated by MitoQ (Figure 2D). Meanwhile, the glucose consumption and nicotinamide adenine dinucleotide (reduced)/nicotinamide adenine dinucleotide (NADH/NAD+) ratio were elevated under hypoxia, which were mitigated by MitoQ intervention (Supplementary Figure S6A and B). Nevertheless, the adenosine triphosphate (ATP) production was decreased, which could not be improved by MitoQ (Supplementary Figure S6C). Chetomin, a HIF-1α inhibitor (Tian et al., 2020), could also blunt the enhanced proliferative capacity of hypoxic PASMCs (Figure 2E). Taken together, our findings indicate that under hypoxia, mROS are critical for HIF-1α stabilization to promote a PASMC proliferative phenotype with metabolic abnormality.
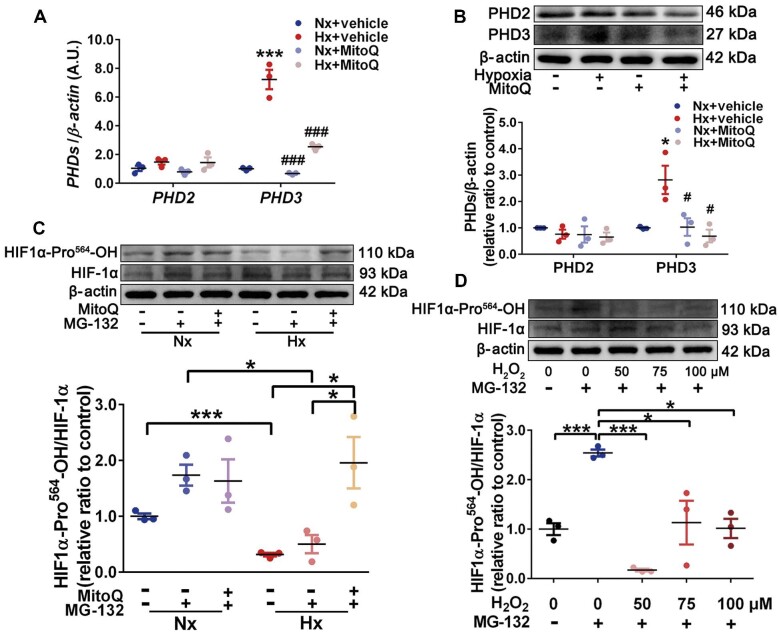
Increased mROS enhance HIF-1α stabilization by inhibiting HIF-1α hydroxylation
Given that the levels of Hif1α mRNA were unchanged under hypoxia (Supplementary Figure S7), we posited that a mROS-dependent post-translational mechanism might be involved in HIF-1α accumulation. Hence, we investigated whether mROS-mediated inhibition of proline hydroxylases (PHDs), which are required for HIF-1α proteasomal degradation (Chee et al., 2019), facilitated HIF-1α stabilization under hypoxia. Interestingly, hypoxia boosted the mRNA and protein levels of PHD3 rather than PHD2, which could be reversed by MitoQ (Figure 3A and B). In view of the increased HIF-1α protein level under hypoxia, we postulated that PHD3 upregulation was driven by a negative feedback mechanism due to its inactivation mediated by mROS. To explore this speculation, we examined the levels of hydroxylated HIF-1α (HIF1α-Pro564-OH). With MG-132 (a proteasome inhibitor) incubation, the ratio of HIF1α-Pro564-OH to total HIF-1α was decreased under hypoxia, indicating an inactivated PHD3, which was improved by MitoQ (Figure 3C). Additionally, exogenous H2O2 could also reduce the ratio of HIF1α-Pro564-OH to total HIF-1α (Figure 3D). Collectively, our data suggest that hypoxia-induced mROS inhibit HIF-1α hydroxylation possibly through inactivating PHD3, resulting in HIF-1α stabilization.
Figure 3.
mROS enhance HIF-1α stabilization by inhibiting HIF-1α hydroxylation. (A and B) PASMCs were exposed to normoxia and hypoxia with or without MitoQ (0.5 μM) incubation. (A) mRNA levels of PHD2 and PHD3 relative to β-actin determined by RT-qPCR. (B) Immunoblots for PHD2, PHD3, and β-actin. Data represent mean ± SEM; *P < 0.05, ***P < 0.001 vs. Nx; #P < 0.05, ###P < 0.001 vs. Hx; two-way ANOVA and Bonferroni's post-test. (C) Immunoblots for HIF1α-Pro564-OH, total HIF-1α, and β-actin in PASMCs exposed to normoxia and hypoxia with or without MitoQ and MG-132 (10 μM) incubation. Data represent mean ± SEM; *P < 0.05, ***P < 0.001. (D) Immunoblots for HIF1α-Pro564-OH, total HIF-1α, and β-actin in PASMCs incubated with exogenous H2O2 (0, 50, 75, and 100 μM) and MG-132 (10 μM). Data represent mean ± SEM; *P < 0.05, ***P < 0.001.
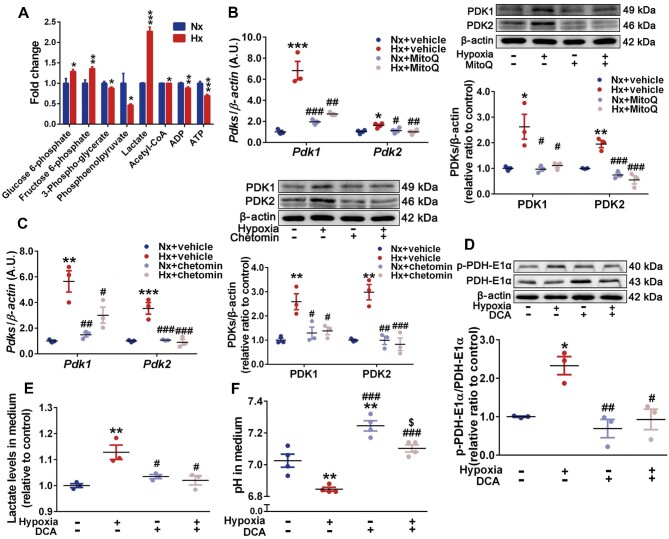
PDK1 and PDK2 induced by the mROS–HIF-1α axis drive PASMC glycolytic shift
Considering the essential role of glycolysis in PASMC fate, we performed a targeted analysis of PASMC metabolites in the glycolytic pathway through liquid chromatography tandem–mass spectrometry (LC–MS/MS). A total of eight metabolites were analyzed. The levels of three metabolites significantly increased under hypoxia, including glucose 6-phosphate (increased to 1.3-fold), fructose 6-phosphate (increased to 1.4-fold), and lactate (increased to 2.3-fold) (Figure 4A). The levels of the other five metabolites, including 3-phospho-glycerate, phosphoenolpyruvate, acetyl-CoA, adenosine diphosphate (ADP), and ATP, were decreased. These findings suggest an increased glycolysis in hypoxic PASMCs.
Figure 4.
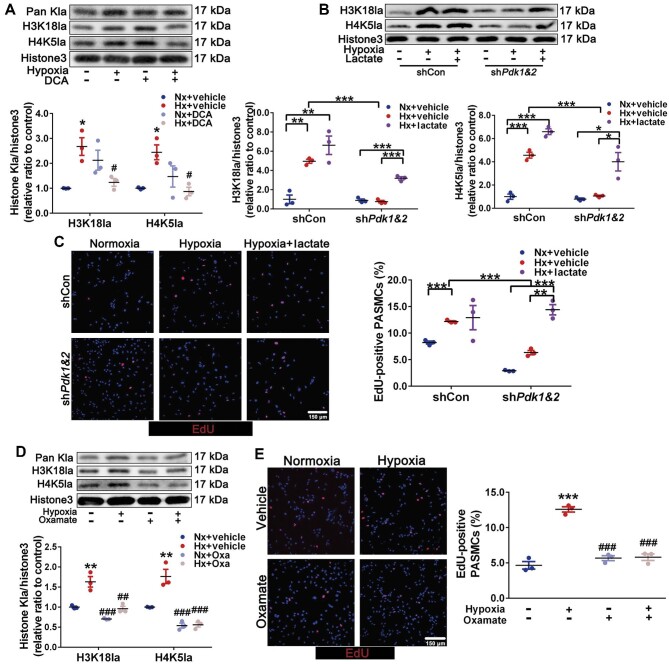
HIF-1α-induced PDK1 and PDK2 promote PASMC glycolytic shift. (A) Significant fold changes of PASMC metabolites in the glycolytic pathway determined by targeted analysis via LC–MS/MS. (B and C) PASMCs were exposed to normoxia and hypoxia with or without MitoQ (0.5 μM, B)/chetomin (50 nM, C) incubation. Left, mRNA levels of Pdk1 and Pdk2 relative to β-actin determined by RT-qPCR. Right, immunoblots for PDK1, PDK2, and β-actin. (D–F) PASMCs were exposed to normoxia and hypoxia with or without DCA (10 mM) incubation. (D) Immunoblots for p-PDH-E1α, total PDH-E1α, and β-actin, with quantification of the ratio of p-PDH-E1α to total PDH-E1α. (E and F) Relative lactate concentrations (E) and pH (F) in PASMC culture medium. Data represent mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001 vs. Nx; #P < 0.05, ##P < 0.01, ###P < 0.001 vs. Hx; two-tailed unpaired Student's t-test (A) or two-way ANOVA and Bonferroni's post-test (B–F).
We further demonstrated that the levels of PDK1 and PDK2, two crucial mediators of glycolysis, were increased in hypoxic PASMCs in a mROS- and HIF-1α-dependent manner (Figure 4B and C). Increased PDK1 and PDK2 promoted phosphorylation of the PDH-E1α component (p-PDH-E1α), which was attenuated by sodium dichloroacetate (DCA), a PDK inhibitor (Figure 4D). Moreover, increased pyruvate (Supplementary Figure S8A) and lactate (Figure 4E) as well as decreased pH (Figure 4F) in PASMC culture medium were also mitigated by DCA. However, the elevated mitochondrial acetyl-CoA and NADH/NAD+ could continually increase with DCA incubation (Supplementary Figure S8B and C). The decreased ATP production and mitochondrial membrane potential (ΔΨ) hyperpolarization were not improved significantly by DCA (Supplementary Figures S8D and S9). As an additional glycolytic mechanism (Zhang et al., 2017), pyruvate kinase muscle isoform 1 (PKM1) and PKM2 were not increased (Supplementary Figure S10). Taken together, our results indicate that accompanied by mitochondrial dysfunction, mROS-dependent HIF-1α accumulation enhances PASMC glycolysis by PDK1 and PDK2-mediated phosphorylation of PDH-E1α under hypoxia.
Histone lactylation induced by lactate accumulation promotes PASMC proliferation
Due to the fact that lactate accumulation could trigger histone lactylation (Zhang et al., 2019), we further investigated whether PDK1 and PDK2-induced glycolysis promotes hypoxic PASMC proliferation by enhancing lysine lactylation (Kla) of histone. Indeed, we showed that hypoxia increased L-lactyl lysine (Pan Kla), Kla of histone H3 (Lys18) (H3K18la), and Kla of histone H4 (Lys5) (H4K5la), which were reversed by DCA (Figure 5A). Moreover, the clearance of mROS with MitoQ could also attenuate the hypoxia-induced elevation of H3K18la and H4K5la (Supplementary Figure S11). However, histone acetylation marks, including Pan Kac, H3K18ac, and H4K5ac, were unchanged (Supplementary Figure S12). Furthermore, Pdk1&2 silencing, manifested by their reduced mRNA and protein levels, decreased the level of p-PDH-E1α (Supplementary Figure S13) and reduced the levels of H3K18la and H4K5la that were reversed by exogenous L-lactate incubation (Figure 5B). Correspondingly, hypoxia-induced PASMC proliferation was also inhibited by Pdk1&2 silencing, which was offset by exogenous L-lactate (Figure 5C). Meanwhile, oxamate, an LDH inhibitor, abrogated lactate production (Supplementary Figure S14), histone lactylation marks (Figure 5D), and PASMC proliferation (Figure 5E) under hypoxia. Consequently, PDK1 and PDK2-induced lactate accumulation promotes PASMC proliferation through upregulating histone lactylation under hypoxia.
Figure 5.
Histone lactylation induced by lactate accumulation promotes PASMC proliferation. (A) Immunoblots for Pan Kla, H3K18la, H4K5la, and histone H3 in PASMCs exposed to normoxia and hypoxia with or without DCA (10 mM) incubation. (B and C) PASMCs with shCon or shPdk1&2 were exposed to normoxia, hypoxia, and hypoxia + lactate (10 mM). (B) Immunoblots for H3K18la, H4K5la, and histone H3. (C) Representative EdU (red) images, with quantification of the percentage of EdU-positive nuclei. Scale bar, 150 μm. Data represent mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001. (D and E) PASMCs were exposed to normoxia and hypoxia with or without oxamate (50 μM) incubation. (D) Immunoblots for H3K18la, H4K5la, and histone H3. (E) Representative EdU (red) images, with quantification of the percentage of EdU-positive nuclei. Scale bar, 150 μm. Data represent mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001 vs. Nx; #P < 0.05, ##P < 0.01, ###P < 0.001 vs. Hx; two-way ANOVA and Bonferroni's post-test. Oxa, oxamate; histone3, histone H3.
Histone lactylation of HIF-1α targets boosts PASMC proliferation
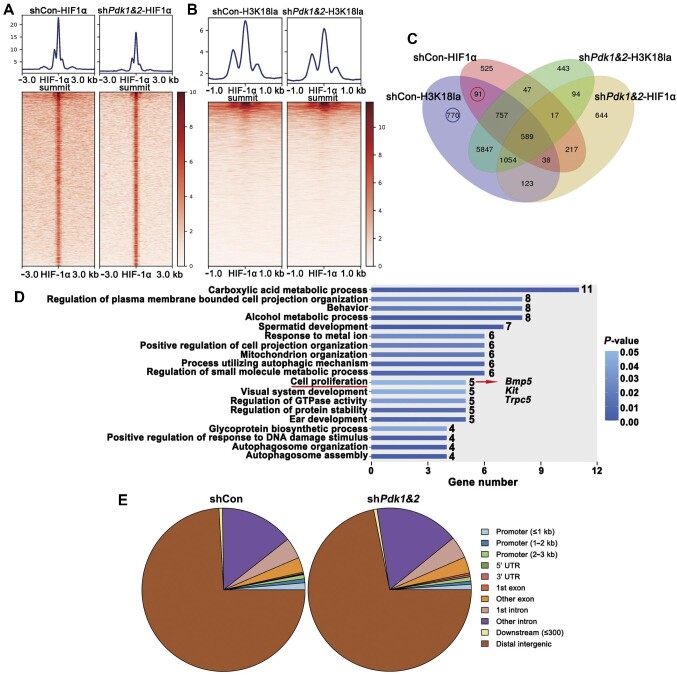
To explore the underlying regulation of histone lactylation on HIF-1α targets, hypoxic PASMCs with or without Pdk1&2 silencing were subjected to HIF-1α and H3K18la ChIP–seq analysis. There were fewer HIF-1α ChIP–seq peaks under Pdk1&2 silencing (Figure 6A), the majority of which were distributed on distal intergenic regions (Figure 6E), possibly indicating HIF-1α function related to distal enhancers. Meanwhile, we showed a mild reduction in H3K18la density around HIF-1α peaks (Figure 6B) with Pdk1&2 silencing. Moreover, we demonstrated Pdk1&2-independent H3K18la peaks at Pdk1 and Pdk2 promoter region (Supplementary Figure S15), suggesting histone lactylation-independent expression of Pdk1 and Pdk2. The 91 HIF-1α targets that overlapped with H3K18la peaks in a Pdk1&2-dependent manner (red circle in Figure 6C) were subjected to gene ontology (GO) enrichment analysis, and five genes were significantly enriched in cell proliferation (Figure 6D). The mRNA levels of Bmp5, Kit, and Trpc5 were validated to be elevated under hypoxia, which were attenuated by Pdk1&2 knockdown (Supplementary Figure S16). Additionally, we performed an analysis of other 770 H3K18la specifically regulating genes (blue circle in Figure 6C) that did not overlap with HIF-1α targets. The results demonstrated some essential catabolic processes, mitochondrion organization, glucose import, and mitochondrial electron transport process (Supplementary Figure S17), which might suggest that metabolism-mediated histone lactylation modification could, in turn, regulate the metabolic processes.
Figure 6.
PDK1 and PDK2-dependent histone lactylation of HIF-1α targets associated with cell proliferation. (A) Peak plots of HIF-1α ChIP–seq ± 3 kb around HIF-1α summit. (B) Heatmaps of H3K18la density ± 1 kb around the HIF-1α peaks. (C) Venn diagram of genes from HIF-1α and H3K18la ChIP–seq peaks with shCon vs. shPdk1&2. (D) Red-circled 91 genes in venn diagram were picked for GO enrichment analysis. P < 0.05 was set as the threshold of significance. (E) Distribution of HIF-1α peaks in the genome.
Inhibition of LDH attenuates histone lactylation and PAs remodeling in hypoxic PH rats
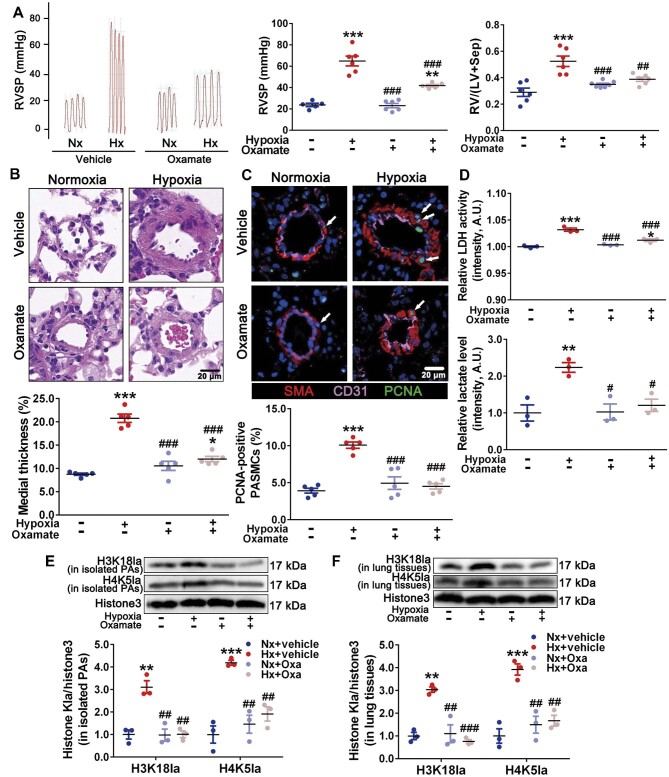
In vivo, oxamate attenuated hypoxia-induced elevation of RVSP and RV/(LV + Sep) (Figure 7A). Meanwhile, oxamate mitigated PA medial thickness (Figure 7B) and PASMC proliferation (Figure 7C). The elevated LDH activity and lactate levels in PA homogenates from hypoxic rats were decreased by oxamate intervention (Figure 7D). Moreover, the increased histone lactylation marks, H3K18la and H4K5la, in homogenates of isolated PAs (Figure 7E) and lung tissues (Figure 7F) from hypoxia rats were reduced by oxamate. To further investigate the potential roles of Bmp5, Kit, and Trpc5 in PAs remodeling, we examined their mRNA levels in isolated pulmonary arterioles. The results demonstrated that the expression levels of Bmp5, Kit, and Trpc5 in hypoxic pulmonary arterioles were increased, which were mitigated by oxamate treatment (Supplementary Figure S18). Taken together, these findings suggest that inhibition of LDH might attenuate PH and PAs remodeling by decreasing lactate production and histone lactylation of HIF-1α targets associated with cell proliferation.
Figure 7.
Inhibition of LDH attenuates histone lactylation as well as PH and PAs remodeling in hypoxic PH model. (A) Representative images of tracing of RVSP (left), with quantification (middle). Changes in RV/(LV + Sep) (right). (B) Representative H&E images of lung sections, with quantification of the ratio of medial thickness to diameter. Scale bar, 20 μm. (C) Representative immunofluorescence images of lung sections stained for CD31 (pink), SMA (red), and PCNA (green, white arrows), with quantification of the percentage of PCNA-positive nuclei. Scale bar, 20 μm. (D) Relative levels of LDH activity (top) and lactate (bottom) in freshly isolated PAs. (E and F) Immunoblots for H3K18la, H4K5la, and histone H3 in isolated PAs (E) and lung tissues (F). Data represent mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001 vs. Nx; #P < 0.05, ##P < 0.01, ###P < 0.001 vs. Hx; two-way ANOVA and Bonferroni's post-test.
Discussion
In this study, we elucidate the previously uncharacterized effect of the mROS–HIF-1α axis on PASMC glycolytic shift, a crucial process in driving PASMCs towards a more proliferative phenotype. As a novel epigenetic modification, glycolysis-induced histone lactylation of HIF-1α targets associated with cell proliferation facilitates vascular remodeling and fuels PH development. Based on these findings, we add to many others that demonstrate how increased glycolysis contributes to vascular remodeling and propose that lactate manipulation might serve as a potential intervention for PH.
It has been indicated that PASMC proliferation could rely on increased glycolysis accompanied by either minimal or enhanced oxidative phosphorylation (Michelakis et al., 2017; Kovacs et al., 2019; Miyagawa et al., 2019). The differences might derive from different initial stimuli or molecular abnormalities. In this study, we demonstrated a marked increase of lactate and a mild decrease of ATP and acetyl-CoA under hypoxia. In addition, the levels of PDK1, PDK2, and p-PDH-E1α were increased in proliferating PASMCs, but the levels of PKM1 and PKM2 were not. Moreover, increased lactate secretion from PASMCs could be reversed by DCA incubation. These findings suggest that an uncoupled glycolysis mediated by increased PDK1 and PDK2 has been involved in PASMC proliferation, which reinforces that most proliferating cells adopt a less efficient glycolysis (Vander et al., 2009).
Most metabolic alterations could indirectly determine cell identities through remodeling chromatin and regulating gene expression (Lu and Thompson, 2012; Klemm et al., 2019). This metabolon–epigenetics interaction has been involved in persistently activated PASMC phenotype and vascular remodeling (Cheng et al., 2019; Hu et al., 2019; Miyagawa et al., 2019). A recent study has reported a lactate-dependent histone lactylation under hypoxia (Zhang et al., 2019). Here, we demonstrate that PDK1 and PDK2-mediated increase in lactate pool facilitates histone lactylation and that histone lactylation plays an important role in defining HIF-1α binding according to H3K18la and HIF-1α ChIP–seq analysis. Furthermore, of the 91 HIF-1α targets that overlapped with H3K18la peaks in a PDK1 and PDK2-dependent manner, cell proliferation item could be significantly enriched, indicating the regulation of histone lactylation on PASMC proliferation. Although more direct modulation on histone lactylation could not be carried out, LDH inhibitor significantly attenuated histone lactylation, PASMC proliferation, and PAs remodeling. We clarify, for the first time to our knowledge, that glycolysis-induced histone lactylation promotes PASMC proliferation.
Consistent with previous studies (Pisarcik et al., 2013; Ball et al., 2014), we also elucidate that hypoxia promotes PAs remodeling through enhancing HIF-1α rather than HIF-2α in PASMCs. Additionally, it has been demonstrated that the upregulation of HIF-2α mainly occurs in endothelial cells (Dai et al., 2018). Actually, the roles of HIF-1α and HIF-2α in PH might depend on the cytological context (Waypa and Schumacker, 2019). A recent study demonstrated that HIF-2αMyh11Cre mice were not protected from hypoxia-induced PH, suggesting that HIF-2α in SMCs might not be involved in the development of PH (Tang et al., 2018). Moreover, some other studies have revealed that the relevance of HIF-2α in pulmonary vascular remodeling depends on its expression in endothelial cells (Cowburn et al., 2016; Kapitsinou et al., 2016). For instance, HIF-2αTie2Cre mice exhibited a normal pulmonary phenotype under hypoxia (Dai et al., 2016). A previous study has suggested that most cells respond to certain conditions through either HIF-1α or HIF-2α (Rosenberger et al., 2002). In this study, the levels of HIF-1α and HIF-2α were examined in isolated PAs, the adventitia and endothelium of which had been stripped off. This indicates the accumulation of HIF-1α in medial SMCs. All these suggest that the cell-specific expression of HIF-1α and HIF-2α seems to reflect its intrinsic property. Nevertheless, this certainly warrants further investigations.
HIF-1α is an essential regulator in adapting to hypoxia, which is principally modulated by PHDs-mediated hydroxylation of proline residues (Pro402 and Pro564 in HIF-1α, Pro405 and Pro531 in HIF-2α) followed by VHL-directed proteasomal degradation (Tian et al., 2011; Ivan and Kaelin, 2017). In this study, HIF-1α protein level was specifically upregulated in PAs rather than in MAs and CAs under identical hypoxia. Meanwhile, hypoxia enhanced the mRNA and protein levels of PHD3. We, therefore, postulated that in the initial phase of hypoxia sensing, an intermediate mechanism might be implicated in PHD3 inactivation and subsequent HIF-1α accumulation. Several studies have identified increased mROS in PAs rather than in systemic arteries under hypoxia (Wang and Zheng, 2010; Adesina et al., 2015). Through modulating protein kinases and transcription factors, mROS could trigger hypoxic signal transduction (Sommer et al., 2017). Thus, we ultimately elucidate that the mROS–HIF-1α crosstalk orchestrated by inactivated PHD3 initiates oxygen sensing and glycolytic shift in hypoxic PASMCs.
Currently, there are plenty of debates on the sources and quantities of intracellular ROS under hypoxia. As the metabolic center, mitochondria are recognized as the most important sources of ROS (Mittal et al., 2007; Dikalova et al., 2010). With regard to mROS quantities, numbers of researches suggest that the duration of hypoxia plays a critical role. Increased mROS primarily occur in early rather than sustained hypoxia (Pak et al., 2018; Sommer et al., 2020). Generally, metabolic status tends to be adjusted timely to oxygen alterations, and the notable upregulation of histone lactylation marks has been revealed within hours under hypoxia. Therefore, we mainly focused on the initial effect of increased mROS on HIF-1α stabilization and glycolytic switch under subacute hypoxia, despite that the total superoxides are decreased during chronic hypoxia (Pak et al., 2018). As for HIF-1α accumulation and metabolic alteration under sustained hypoxia, two important researches indicate that the CD146–HIF-1α and RASSF1A–HIF-1α loops might play essential roles (Dabral et al., 2019; Luo et al., 2019).
Limitations also exist in this study. Although human PASMCs or PAs from PH patients are most suitable for investigation, they are expensive and very difficult to obtain. The undefinition of ‘writer’, ‘reader’, and ‘eraser’ of histone lactylation has hampered the exploration of more direct effect of histone lactylation on PASMC proliferation. Owing to the lack of PASMCs-targeted antioxidants, the effects of mROS clearance on downstream mechanisms could not be evaluated in vivo. The unavailable information on lactylation modifications of other histone lysine residues, enzymes, and transcriptional factors has confounded us whether they are also involved in PASMC proliferation to some extent. Future studies on ‘lactyl-proteome’ are needed to characterize lactylation modifications in PH pathogenesis.
In summary, we demonstrate that glycolytic shift driven by mROS-mediated HIF-1α stabilization promotes hypoxic PASMC proliferation and PAs remodeling. The proposed mechanism reveals that lactate accumulation facilitates histone lactylation of HIF-1α targets associated with proliferative phenotype. These findings elucidate a hitherto more novel initiator and downstream effector of increased glycolysis in PH onset and provide proof of concept for anti-remodeling therapy through lactate manipulation.
Materials and methods
Rodent PH models
The animal experiments were approved by Animal Care and Use Committee of Fourth Military Medical University and performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Adult male Sprague Dawley (SD) rats (250–300 g) were obtained from the Experimental Animal Center of Fourth Military Medical University. All the rats were maintained in a specific pathogen-free facility. For hypoxia-induced PH, SD rats (n = 6) were exposed to 10% O2 (hypoxia) in a hypobaric hypoxia chamber (a custom-made chamber) or room air (normoxia) for 4 weeks (Lahm et al., 2012). The chamber was subjected to 12-h light/dark cycle and opened for 30 min once a day for replenishing water and food. For preventive purpose, rats were randomly divided into four groups (n = 7): Nx + vehicle, intraperitoneal injection with an equal volume of vehicle (0.9% NaCl) daily for 4 weeks under normoxia; Hx + vehicle, intraperitoneal injection with an equal volume of vehicle daily for 4 weeks under hypoxia; Nx + Oxa: intraperitoneal injection with the LDH inhibitor (oxamate, 750 mg/kg) (Zhao et al., 2011) daily for 4 weeks under normoxia; Hx + Oxa: intraperitoneal injection with oxamate daily for 4 weeks under hypoxia.
Hemodynamics measurement and tissue preparation
Hemodynamics in all rats was evaluated as described previously (Luo et al., 2019). Briefly, anesthesia was performed through intraperitoneal injection with pentobarbital sodium (30 mg/kg) and chloral hydrate (350 mg/kg). Then, rats were placed in a supine position on a homeothermic plate. RVSP was continuously monitored for 10–15 min by inserting a polyethylene catheter, connected to a pressure transducer (AD Instruments), into right ventricle through right external jugular vein. Subsequently, the catheter was inserted into aorta through left CA to measure SAP. All data were recorded and analyzed using PowerLab data-acquisition system (AD Instruments) and LabChart 6 for Windows software. Ultimately, all rats were immediately exsanguinated and infused with chilled phosphate-buffered saline (PBS) to remove the blood in pulmonary circulation. Then, lung tissues, CAs, and MAs were isolated and removed. Right ventricle free wall was dissected from left ventricle plus septum and weighed, and right ventricle hypertrophy was assessed by the ratio RV/(LV + Sep). The dissected peripheral zone of left lungs along with CAs and MAs were snap-frozen in liquid nitrogen for preparation of homogenates. Lower lobes of right lungs were fixed in 4% paraformaldehyde solution for 48 h and embedded with paraffin. The remaining lung lobes were utilized to isolate the peripheral PAs.
Histologic analysis
After paraffin-embedded and sliced, the lung tissue slides (5 μm thick) were stained with hematoxylin and eosin (H&E) for vascular morphometry analysis. To quantitate medial thickness, PAs with diameter of 50–100 μm were randomly collected by light microscope (Leica). The degree of medial thickness, defined as medial thickness (%) = 100 × (medial thickness/diameter), was calculated using Image-Pro Plus.
Immunofluorescence on tissues
To detect proliferative PASMCs and HIF-1α expression levels, the paraffin sections of lung tissues were triple immuno-stained for PCNA/smooth muscle α-actin (SMA)/CD31 and SMA/HIF-1α/PCNA, respectively. After deparaffinized and antigen retrieval as described previously (Luo et al., 2019), all slides were stained using a TSAPLus Staining Kit (G1236, Servicebio). Briefly, slides were incubated with the first primary antibody to PCNA (1:50, 10205-2-AP, Proteintech) or SMA (1:50, 14395-1-AP, Proteintech) overnight at 4°C. After washing, slides were covered with corresponding secondary antibody labeled with horseradish peroxidase (HRP) at room temperature for 50 min. After another washing, all slides were incubated with FITC-Tyramide solution for 10 min and immersed in EDTA antigen retrieval buffer for 15 min to remove the first primary and secondary antibodies. Subsequently, all slides were incubated with the second primary antibody to SMA (1:50, 14395-1-AP, Proteintech) or HIF-1α (1:50, NB100-105, Novus Biologicals) and corresponding secondary antibody labeled with HRP as described above. All slides were then incubated with CY3-Tyramide solution and subjected to antigen retrieval. Finally, all slides were incubated with the third primary antibody to CD31 (1:50, 11265-1-AP, Proteintech) or PCNA (1:50, 10205-2-AP, Proteintech) and corresponding secondary antibody labeled with CY5, and then counterstained with DAPI. After incubated with spontaneous fluorescence quenching reagent for 5 min, all slides were sealed with coverslip and anti-fade mounting medium. The fluorescent images for proliferative PASMCs (PCNA [glowing green], SMA [glowing red], and CD31 [glowing pink]) and HIF-1α expression (SMA [glowing white], HIF-1α [glowing red], and PCNA [glowing green]) were taken using PANNORAMIC Diagnostic Scanners, a digital slide scanner, and CaseViewer software (3DHISTECH). The percentage of PCNA-positive nuclei was calculated.
Lactate assay
The levels of lactate in the homogenates of isolated arterial medial layer and PASMC culture medium were determined using commercially available Lactate Assay Kit (MAK064, Sigma–Aldrich) following the manufacturer's instructions.
LDH activity assay
The homogenates of freshly isolated PA medial layer were prepared. Then, the LDH activity was measured using commercially available Lactate Dehydrogenase Activity Assay Kit (BC0685, Solarbio) following the manufacturer's instructions.
PASMC isolation and culture
Primary PASMCs were isolated from adult male SD rats using a modified enzymatic disaggregation (Luo et al., 2019). In brief, pulmonary arteriole was dissected away from lung lobes under stereomicroscope. After stripping off the adventitia and endothelium, the nude medial explants were cut into 1–2 mm3 sections and placed in Hank's balanced salt solution containing 0.1% collagenase (Worthington Biochemical Corporation) to be digested into single cells for 1 h at 37°C. Subsequently, cell suspensions were transferred into centrifugal tubes containing fetal bovine serum. PASMCs were collected through centrifugation at 90–140× g for 5 min and re-suspended in Dulbecco's modified Eagle's medium containing 15% fetal bovine serum and antibiotics. The purity and identity were confirmed through immunofluorescence staining against SMA. PASMCs from passages 2–5 were used for experiments.
Treatment with compounds and drugs
Primary PASMCs were quiesced in serum-free medium for 24 h and then exposed to normoxia (21% O2) and hypoxia (5% O2) in the absence or presence of mitochondria-targeted antioxidant MitoQ (0.5 μM, HY-100116A, MCE), chetomin (50 nM, GC17405, GlpBio), MG-132 (10 μM, S2619, Selleck), H2O2 (50, 75, and 100 mM, Beyotime), DCA (10 mM, 347795, Sigma–Aldrich), lactate (10 mM, 71718, Sigma–Aldrich), and sodium oxamate (50 μM, GC44911, GlpBio). After indicated time periods, the relative changes in mRNA, protein, metabolites, and proliferation capacity were assessed through corresponding methods. The concentrations of these compounds were chosen according to previous studies.
Measurement of mROS
mROS were measured using a highly selective molecular probe, MitoSOXTM Red mitochondrial superoxide indicator, for live-cell imaging (M36008, ThermoFisher Scientific). Briefly, PASMCs (1.0 × 105) were seeded on chamber slides. When PASMCs grew to 50% confluence, all chambers were exposed to normoxia (21% O2) and hypoxia (5% O2) with or without MitoQ (0.5 μM) incubation. After 12 h, they were stained with MitoSOXTM working solution (5 μM) within a hypoxic glove chamber for 10 min. Following washing, all chambers were observed immediately under confocal laser scanning microscope (Olympus FLUOVIEW FV 1000). The density of fluorescent signal for mROS was quantified using Image-Pro Plus.
Measurement of H2O2
PASMCs were exposed to normoxia (21% O2) and hypoxia (5% O2) with or without MitoQ (0.5 μM) incubation for 12 h. The levels of H2O2 in cultured PASMCs were examined using a commercially available Hydrogen Peroxide Assay Kit (S0038, Beyotime) following the manufacturer's instructions.
Immunofluorescence on cultured PASMCs
PASMCs (1.0 × 105) seeded on chamber slides were exposed to normoxia (21% O2) and hypoxia (5% O2) with or without MitoQ (0.5 μM) incubation for 12 h and then fixed with 4% paraformaldehyde. Subsequently, they were subjected to permeabilization using QuickBlockTM Blocking Buffer for Immunol Staining (P0260, Beyotime) and incubated with the primary antibody to HIF-1α (1:50, NB100-105, Novus Biologicals) at 4°C overnight. The next day, slides were washed and incubated with Alexa 488-conjugated donkey anti-mouse IgG (1:2000, A21202, Invitrogen) for 60 min at room temperature. All slides were counterstained with DAPI. Images were taken using confocal laser scanning microscope (Olympus FLUOVIEW FV 1000). The density of fluorescent signal was quantified using Image-Pro Plus.
EdU incorporation assay
The proliferative capacity of PASMCs was evaluated by 5-ethynyl-2′-deoxyuridine (EdU) incorporation assay using Cell-Light EdU Apollo567 In Vitro Kit (C10310, RIBOBIO). Briefly, PASMCs (1.0 × 105) were seeded on chamber slides and then quiesced in serum-free medium for 24 h. After different interventions for 24 h, all chambers were incubated with complete medium containing 50 μM EdU for 2 h. Fixation, Apollo staining, and counterstaining were performed according to the manufacturer's instructions. The EdU-positive PASMCs were assessed under confocal laser scanning microscope (Olympus FLUOVIEW FV 1000).
Measurement of culture medium pH
The culture medium of PASMCs exposed to normoxia (21% O2) and hypoxia (5% O2) with or without DCA (10 mM) incubation for 24 h was collected. Extracellular pH was immediately measured using a pH meter (FiveEasy Plus, METTLER TOLEDO).
Targeted analysis of metabolites in glycolysis
PASMCs isolated from seven rats were subjected to normoxia (21% O2) and hypoxia (5% O2) for 24 h. They were then collected, washed with cold PBS, pelleted (5 min at 14000× g, 4°C), and snap-frozen in liquid nitrogen. Targeted analysis of the metabolites in glycolysis was promptly performed using a LC–MS/MS system (Applied Protein Technology). Briefly, PASMCs (1.0 × 107) were vortexed in 1 ml of cold methanol/acetonitrile/H2O (2:2:1, v/v/v) and sonicated for 30 min under ice bath. The homogenates were placed in –20°C for 1 h and centrifuged for 20 min (15500× g, 4°C). The supernatant was dried by a vacuum drying system. The dried samples were dissolved in 100 μl acetonitrile/H2O (1:1, v/v), completely mixed, and centrifuged (15500× g, 4°C) for 15 min. The supernatants were subjected to targeted metabolite analysis using an Agilent 1290 Infinity chromatography system and AB Sciex QTRAP 5500 mass spectrometer.
Lentivirus transduction
For Pdk1 and Pdk2 double silencing, PASMCs at 50%–70% confluence were transduced with lentiviral particles (GenePharma) that packaged Pdk1 shRNA (5′-GGTTTATGTTCCGTCCCATCT-3′) or scramble shRNA (5′-TTCTCCGAACGTGTCACGT-3′) (LV10N, MOI = 50) and Pdk2 shRNA (5′-GCTGTCCATGAAGCAGTTTCT-3′) or scramble shRNA (5′-TTCTCCGAACGTGTCACGT-3′) (LV3, MOI = 60) in the presence of 6 μg/ml polybrene. After transduction for 72 h, PASMCs were selected with puromycin (2 μg/ml). The efficacy of Pdk1&2 silencing was evaluated by immunoblots or real-time quantitative PCR (RT-qPCR).
ChIP–seq analysis
ChIP–seq analysis was performed as previously described (Miyagawa et al., 2019). PASMCs with or without shPdk1&2 were subjected to hypoxia for 24 h and fixed for protein–DNA cross-linking with 1% formaldehyde (F8775, Sigma) for 10 min at room temperature. Glycine solutions were then added to neutralize formaldehyde for 5 min. The collected cells were incubated with DTT solutions for 10 min on ice and centrifuged (2000× g, 4°C) for 5 min to extract cell nuclei. Chromatin was sheared with sonifier, and 10 μl of chromatin fragments was removed as input sample. The remaining chromatin lysates were immunoprecipitated with specific antibodies for HIF-1α (NB100-105, Novus Biologicals) and H3K18la (PTM-1406, PTM BIO) at 4°C overnight on a nutator. The chromatin–antibody complex was pulled down using Protein G Magnetic beads (9006, Cell Signaling Technology), and then chromatin fragments were eluted from beads. The cross-linking of ChIP-DNA and input sample was reversed by proteinase K incubation (10012, Cell Signaling Technology) at 65°C for 2 h and purified with purification columns. The ChIP-DNA was subjected to end repair and A-tailing. The adapters were ligated and size-selected from the gel. Subsequently, the DNA fragments were amplified by PCR. PCR products were purified, size-selected, and subjected to quality and quantity evaluation by Agilent 2100 Bioanalyzer and ABI StepOnePlus RT-qPCR System, respectively. The final samples were sequenced on Illumina Novaseq6000. Reads of HIF-1α and H3K18la ChIP–seq were trimmed with FASTQ groomer (Blankenberg et al., 2010) and aligned to rat genome (UCSC, rn6) with Soap (Li et al., 2009). Peaks were called utilizing MACS2 (Zhang et al., 2008) and aligned peaks were visualized through Integrated Genome Browser. The pie charts of peak distribution were visualized utilizing ChIPseeker (Yu et al., 2015). Peak plots and heatmaps were performed using deeptools (Ramirez et al., 2016). For annotated genes, the biological pathways were identified by GO enrichment analyses with the R package ClusterProfiler (Yu et al., 2012).
Statistical analysis
All summary data are presented as mean ± standard error of the mean (SEM). The significance of difference in statistical data was determined by two-tailed unpaired Student's t-test analysis, one-way analysis of variance (ANOVA), or two-way ANOVA followed by Bonferroni's test using GraphPad Prism software. Difference of P < 0.05 was considered to be significant.
Supplementary Material
Acknowledgements
The authors thank Xiaohui Di (Key Laboratory of Biomedical Information Engineering, Ministry of Education, Xi'an Jiaotong University) for her technical support.
Contributor Information
Jian Chen, Department of Respiratory and Critical Care Medicine, Tangdu Hospital, Fourth Military Medical University, Xi'an 710038, China.
Meiling Zhang, College of Life Science and Medicine, Northwest University, Xi'an 710069, China.
Yanjie Liu, Department of Medicinal Chemistry, School of Pharmacy, Xi'an Jiaotong University, Xi'an 710061, China.
Shihong Zhao, Department of Respiratory and Critical Care Medicine, Tangdu Hospital, Fourth Military Medical University, Xi'an 710038, China.
Yanxia Wang, Department of Pathology, Xijing Hospital and School of Basic Medicine, Fourth Military Medical University, Xi'an 710032, China.
Meng Wang, College of Life Science and Medicine, Northwest University, Xi'an 710069, China.
Wen Niu, Department of pathophysiology, School of Basic Medicine, Fourth Military Medical University, Xi'an 710032, China.
Faguang Jin, Department of Respiratory and Critical Care Medicine, Tangdu Hospital, Fourth Military Medical University, Xi'an 710038, China.
Zhichao Li, Department of Respiratory and Critical Care Medicine, Tangdu Hospital, Fourth Military Medical University, Xi'an 710038, China.
Funding
This work was supported by the National Natural Science Foundation of China (82070054, 31671186, 81970076, and 81800046) and the Achievement Cultivation Project of Tangdu Hospital (2020CGPY001).
Conflict of interest: none declared.
References
- Adesina S.E., Kang B.Y., Bijli K.M.et al. (2015). Targeting mitochondrial reactive oxygen species to modulate hypoxia-induced pulmonary hypertension. Free Radical Biol. Med. 87, 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball M.K., Waypa G.B., Mungai P.T.et al. (2014). Regulation of hypoxia-induced pulmonary hypertension by vascular smooth muscle hypoxia-inducible factor-1α. Am. J. Respir. Crit. Care Med. 189, 314–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S.L., Sassone-Corsi P. (2016). Metabolic signaling to chromatin. Cold Spring Harb. Perspect. Biol. 8, a019463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenberg D., Gordon A., Von Kuster G.et al. (2010). Manipulation of FASTQ data with Galaxy. Bioinformatics 26, 1783–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowker-Kinley M.M., Davis W.I., Wu P.et al. (1998). Evidence for existence of tissue-specific regulation of the mammalian pyruvate dehydrogenase complex. Biochem. J. 329, 191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee N.T., Lohse I., Brothers S.P. (2019). mRNA-to-protein translation in hypoxia. Mol. Cancer 18, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Wang Y., Du L. (2019). Epigenetic modulation in the initiation and progression of pulmonary hypertension. Hypertension 74, 733–739. [DOI] [PubMed] [Google Scholar]
- Choudhry H., Harris A.L. (2018). Advances in hypoxia-inducible factor biology. Cell Metab. 27, 281–298. [DOI] [PubMed] [Google Scholar]
- Cowburn A.S., Crosby A., Macias D.et al. (2016). HIF2α–arginase axis is essential for the development of pulmonary hypertension. Proc. Natl Acad. Sci. USA 113, 8801–8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabral S., Muecke C., Valasarajan C.et al. (2019). A RASSF1A–HIF1α loop drives Warburg effect in cancer and pulmonary hypertension. Nat. Commun. 10, 2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z., Li M., Wharton J.et al. (2016). Prolyl-4 hydroxylase 2 (PHD2) deficiency in endothelial cells and hematopoietic cells induces obliterative vascular remodeling and severe pulmonary arterial hypertension in mice and humans through hypoxia-inducible factor-2α. Circulation 133, 2447–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z., Zhu M.M., Peng Y.et al. (2018). Therapeutic targeting of vascular remodeling and right heart failure in pulmonary arterial hypertension with a HIF-2α inhibitor. Am. J. Respir. Crit. Care Med. 198, 1423–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold L., Chandel N.S. (2016). Mitochondrial ROS regulation of proliferating cells. Free Radical Biol. Med. 100, 86–93. [DOI] [PubMed] [Google Scholar]
- Dikalova A.E., Bikineyeva A.T., Budzyn K.et al. (2010). Therapeutic targeting of mitochondrial superoxide in hypertension. Circ. Res. 107, 106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C.J., Zhang H., Laux A.et al. (2019). Mechanisms contributing to persistently activated cell phenotypes in pulmonary hypertension. J. Physiol. 597, 1103–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert M., Guignabert C., Bonnet S.et al. (2019). Pathology and pathobiology of pulmonary hypertension: state of the art and research perspectives. Eur. Respir. J. 53, 1801887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M., Kaelin W.J. (2017). The EGLN–HIF O2-sensing system: multiple inputs and feedbacks. Mol. Cell 66, 772–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitsinou P.P., Rajendran G., Astleford L.et al. (2016). The endothelial prolyl-4-hydroxylase domain 2/hypoxia-inducible factor 2 axis regulates pulmonary artery pressure in mice. Mol. Cell. Biol. 36, 1584–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm S.L., Shipony Z., Greenleaf W.J. (2019). Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 20, 207–220. [DOI] [PubMed] [Google Scholar]
- Kovacs L., Cao Y., Han W.et al. (2019). PFKFB3 in smooth muscle promotes vascular remodeling in pulmonary arterial hypertension. Am. J. Respir. Crit. Care Med. 200, 617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahm T., Albrecht M., Fisher A.J.et al. (2012). 17β-estradiol attenuates hypoxic pulmonary hypertension via estrogen receptor-mediated effects. Am. J. Respir. Crit. Care Med. 185, 965–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Yu C., Li Y.et al. (2009). SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967. [DOI] [PubMed] [Google Scholar]
- Lu C., Thompson C.B. (2012). Metabolic regulation of epigenetics. Cell Metab. 16, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Teng X., Zhang L.et al. (2019). CD146–HIF-1α hypoxic reprogramming drives vascular remo659_deling and pulmonary arterial hypertension. Nat. Commun. 10, 3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manalo D.J., Rowan A., Lavoie T.et al. (2005). Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood 105, 659–669. [DOI] [PubMed] [Google Scholar]
- Michelakis E.D., Gurtu V., Webster L.et al. (2017). Inhibition of pyruvate dehydrogenase kinase improves pulmonary arterial hypertension in genetically susceptible patients. Sci. Transl. Med. 9, eaao4583. [DOI] [PubMed] [Google Scholar]
- Mittal M., Roth M., Konig P.et al. (2007). Hypoxia-dependent regulation of nonphagocytic NADPH oxidase subunit NOX4 in the pulmonary vasculature. Circ. Res. 101, 258–267. [DOI] [PubMed] [Google Scholar]
- Miyagawa K., Shi M., Chen P.I.et al. (2019). Smooth muscle contact drives endothelial regeneration by BMPR2–Notch1-mediated metabolic and epigenetic changes. Circ. Res. 124, 211–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell N.W., Adnot S., Archer S.L.et al. (2009). Cellular and molecular basis of pulmonary arterial hypertension. J. Am. Coll. Cardiol. 54, S20–S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movafagh S., Crook S., Vo K. (2015). Regulation of hypoxia-inducible factor-1a by reactive oxygen species: new developments in an old debate. J. Cell. Biochem. 116, 696–703. [DOI] [PubMed] [Google Scholar]
- Pak O., Scheibe S., Esfandiary A.et al. (2018). Impact of the mitochondria-targeted antioxidant MitoQ on hypoxia-induced pulmonary hypertension. Eur. Respir. J. 51, 1701024. [DOI] [PubMed] [Google Scholar]
- Paulin R., Michelakis E.D. (2014). The metabolic theory of pulmonary arterial hypertension. Circ. Res. 115, 148–164. [DOI] [PubMed] [Google Scholar]
- Pisarcik S., Maylor J., Liu J.et al. (2013). Activation of hypoxia-inducible factor-1 in pulmonary arterial smooth muscle cells by endothelin-1. Am. J. Physiol. Lung Cell Mol. Physiol. 304, L549–L561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovitch M. (2012). Molecular pathogenesis of pulmonary arterial hypertension. J. Clin. Invest. 122, 4306–4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F., Ryan D.P., Gruning B.et al. (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid M.A., Dai Z., Locasale J.W. (2017). The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat. Cell Biol. 19, 1298–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberger C., Mandriota S., Jurgensen J.S.et al. (2002). Expression of hypoxia-inducible factor-1α and -2α in hypoxic and ischemic rat kidneys. J. Am. Soc. Nephrol. 13, 1721–1732. [DOI] [PubMed] [Google Scholar]
- Ryan J.J., Archer S.L. (2015). Emerging concepts in the molecular basis of pulmonary arterial hypertension: part I: metabolic plasticity and mitochondrial dynamics in the pulmonary circulation and right ventricle in pulmonary arterial hypertension. Circulation 131, 1691–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadel G.S., Horvath T.L. (2015). Mitochondrial ROS signaling in organismal homeostasis. Cell 163, 560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer N., Alebrahimdehkordi N., Pak O.et al. (2020). Bypassing mitochondrial complex III using alternative oxidase inhibits acute pulmonary oxygen sensing. Sci. Adv. 6, a694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer N., Huttemann M., Pak O.et al. (2017). Mitochondrial complex IV subunit 4 isoform 2 is essential for acute pulmonary oxygen sensing. Circ. Res. 121, 424–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutendra G., Michelakis E.D. (2013). Pyruvate dehydrogenase kinase as a novel therapeutic target in oncology. Front. Oncol. 3, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutendra G., Michelakis E.D. (2014). The metabolic basis of pulmonary arterial hypertension. Cell Metab. 19, 558–573. [DOI] [PubMed] [Google Scholar]
- Tang H., Babicheva A., Mcdermott K.M.et al. (2018). Endothelial HIF-2α contributes to severe pulmonary hypertension due to endothelial-to-mesenchymal transition. Am. J. Physiol. Lung Cell. Mol. Physiol. 314, L256–L275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Wu D., Dasgupta A.et al. (2020). Epigenetic metabolic reprogramming of right ventricular fibroblasts in pulmonary arterial hypertension: a pyruvate dehydrogenase kinase-dependent shift in mitochondrial metabolism promotes right ventricular fibrosis. Circ. Res. 126, 1723–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y.M., Yeoh K.K., Lee M.K.et al. (2011). Differential sensitivity of hypoxia inducible factor hydroxylation sites to hypoxia and hydroxylase inhibitors. J. Biol. Chem. 286, 13041–13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander H.M., Cantley L.C., Thompson C.B. (2009). Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.X., Zheng Y.M. (2010). Role of ROS signaling in differential hypoxic Ca2+ and contractile responses in pulmonary and systemic vascular smooth muscle cells. Respir. Physiol. Neurobiol. 174, 192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waypa G.B., Schumacker P.T. (2019). Roles of HIF1 and HIF2 in pulmonary hypertension: it all depends on the context. Eur. Respir. J. 54, 1901929. [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y.et al. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.G., He Q.Y. (2015). ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383. [DOI] [PubMed] [Google Scholar]
- Zhang D., Tang Z., Huang H.et al. (2019). Metabolic regulation of gene expression by histone lactylation. Nature 574, 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Wang D., Li M.et al. (2017). Metabolic and proliferative state of vascular adventitial fibroblasts in pulmonary hypertension is regulated through a microRNA-124/PTBP1 (polypyrimidine tract binding protein 1)/pyruvate kinase muscle axis. Circulation 136, 2468–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A.et al. (2008). Model-based analysis of ChIP–Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Liu H., Liu Z.et al. (2011). Overcoming trastuzumab resistance in breast cancer by targeting dysregulated glucose metabolism. Cancer Res. 71, 4585–4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.