Abstract
Invasin-mediated invasion of host cells by the pathogen Yersinia enterocolitica was shown to be affected by flagellar-dependent motility. Motility appears to be required to ensure the bacterium migrates to and contacts the host cell. Nonmotile strains of Y. enterocolitica were less invasive than motile strains, but the reduction in invasion could be overcome by artificially bringing the bacteria into host cell contact by centrifugation. Mutations in known regulatory genes of the flagellar regulon, flhDC and fliA, resulted in less inv expression but did not have a significant effect on invasin levels. However, invasin levels were reduced for strains that harbored flhDC on a multicopy plasmid, apparently as a result of increased proteolysis of invasin.
Gastroenteritis caused by Yersinia enterocolitica is a disease that requires the coordinated expression of many bacterial genes for pathogenesis (3–5, 19, 20). Recently, it became clear that virulence genes can be regulated as part of the flagellar regulon, indicating that this regulon contributes to Y. enterocolitica pathogenesis (20). One virulence gene that is regulated as part of the flagellar regulon is yplA. It has been shown that yplA is required for survival of Y. enterocolitica in the Peyer's patches and for stimulation of the acute inflammatory response of the host to the infection (18). The mechanism by which yplA influences the host inflammatory response is not known, but it has been demonstrated to encode a phospholipase that is secreted by the type III flagellum secretion system (20). These results raise the possibility that other virulence genes belong to the flagellar regulon and that components of this regulon contribute to pathogenesis in ways that have not been previously recognized. Another virulence gene that may be regulated as part of the flagellar regulon is inv. The inv gene encodes invasin, a 92-kDa outer membrane protein which mediates invasion of the host (17), primarily at the M cells overlying the lymphoid follicles (Peyer's patches) lining the ileum (8, 10). Recently, two Y. enterocolitica mutants were isolated that exhibited increased levels of motility and decreased levels of invasin (2). It was postulated that these effects were due to mutations that coordinately affected regulation of the flagellar transcriptional regulon and inv. In this study, we determined the role of flagellum-dependent motility of Y. enterocolitica in host cell invasion and examined the influence of the flagellar regulon on inv expression.
Motility is required for efficient cellular invasion.
Y. enterocolitica is highly motile when cultivated in T medium (1% [wt/vol] tryptone) and exhibit very little motility when cultivated in Luria-Bertani (LB) medium (21). Invasion of HEp-2 cells for bacteria cultivated in these media showed that growth in LB medium resulted in a severe reduction in Y. enterocolitica invasion compared to growth in T medium (Table 1, without centrifugation). This suggested motility might be required for efficient host cell invasion. To more directly determine if bacterial motility contributes to cellular invasion, we examined the invasion phenotype of motility mutants, which carried mutations in either flhDC or fliA. These genes are known to encode transcriptional regulators of the flagellar regulon and are required for the expression of motility (11, 21). Although these mutants were grown in T medium, these strains exhibited low levels of invasion (Table 1, without centrifugation). The amount of invasion observed with the flhDC and fliA mutants was comparable to the inv mutant (Table 1, without centrifugation). Complementation of the mutations with a plasmid encoded copy of either flhDC or fliA, respectively, restored motility-dependent invasion (Table 1, without centrifugation). These results indicated that the flagellar regulon is required for host cell invasion by Y. enterocolitica.
TABLE 1.
Expression of motility affects Y. enterocolitica invasion of HEp-2 cells
| Strain genotypea | Plasmidb | % Relative invasion of HEp-2 cellsc:
|
|
|---|---|---|---|
| Without centrifugation | With centrifugation | ||
| Wild type | 100 | 115 | |
| Wild type (LB medium) | 16 | 138 | |
| Wild type | pTM100 | 69 | 88 |
| Wild type | pGY10 | 40 | 60 |
| Wild type | pGY10* | 117 | 177 |
| flhDC | 5 | 88 | |
| flhDC | pTM100 | 7 | 106 |
| flhDC | pGY10 | 65 | 36 |
| fliA | 6 | 195 | |
| fliA | pTM100 | 5 | 203 |
| fliA | pJB222 | 117 | 278 |
| inv | 8 | 3 | |
Strains represented are JB580v (wild type) (12), GY460v (flhDC) (21), VK1v (fliA) (11), and JP273v (inv) (17). A “v” designation indicates the presence of the virulence plasmid.
Plasmids represented include pTM100, which served as the cloning vector (14); pGY10, which is a clone of flhDC in pTM100 (21); and pGY10*, which is a flhDC::TnMax2 derivative of pGY10 (this study) (9).
Invasion assays were done in duplicate as described previously, but with or without centrifugation to bring the bacteria into contact with the HEp-2 cells (15). Except where indicated, all cultures were grown overnight in T medium at 26°C. Values for invasion were calculated as described previously (15) and normalized to the wild-type strain JB580v, which was arbitrarily set at 100. The actual percent invasion for the wild-type strain, JB580v, was 24 ± 5% when grown in T medium and assayed for invasion under conditions without centrifugation.
The role of the flagellar regulon in invasion by Y. enterocolitica could be to ensure migration of the bacteria to host cells or could be due to regulatory effects on invasin levels. To determine if bacterial migration to host cells was important, the levels of invasion were determined for each Y. enterocolitica strain when brought into contact with the HEp-2 cell monolayer by centrifugation (Table 1, with centrifugation). These results demonstrated that all of the bacterial strains tested, except the inv mutant, retained the ability to invade HEp-2 cells once the bacteria were brought into contact with the HEp-2 cells. This indicates motility is required to ensure that the bacteria reach host cells but is not essential for invasion once the bacteria contact the host cell. Furthermore, these data indicate why previous studies, which routinely involved invasion assay protocols that employ centrifugation, did not reveal a role for motility in host cell invasion. When the flhDC mutant was complemented with a plasmid-encoded copy of flhDC (pGY10), invasion levels increased but not to wild-type levels. The negative effect was also observed for the wild-type strain harboring pGY10 (Table 1, with or without centrifugation). In contrast, invasion was not significantly affected by the presence of the parental plasmid (pTM100) or when flhDC of pGY10 was inactivated by insertion of the transposon TnMax2 (pGY10*) (Table 1). The fliA mutant, when centrifuged onto the monolayer, consistently showed increased invasion.
inv expression is affected by expression of the flagellar regulon.
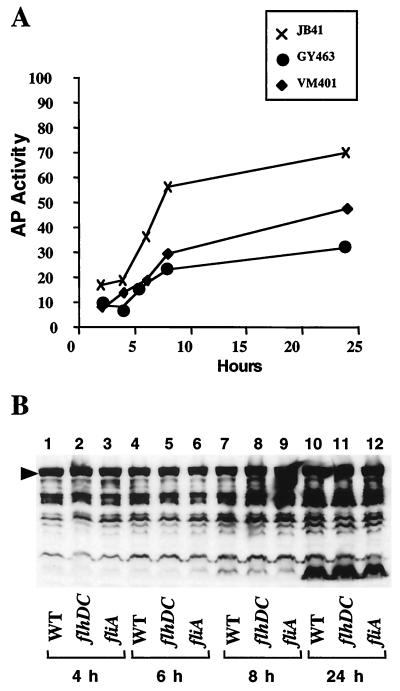
Results from cellular invasion assays indicated that motility was important for the migration of the bacteria to the host cell. However, these results do not eliminate the possibility that inv expression is modulated in response to expression of the flagellar regulon. Therefore, levels of inv expression were analyzed using an inv-phoA translational fusion (2, 16). The inv-phoA fusion was introduced into strains carrying a mutation in either flhDC or fliA to test whether mutations in genes encoding regulators of flagellar genes affect levels of inv expression. The results showed that, compared to the wild-type strain (JB41v), the presence of a mutation in either flhDC (strain GY463v) or fliA (strain VM401v) decreased inv expression two- to threefold (Fig. 1A). However, this effect on inv transcription did not result in a reduction in the amount of invasin produced by these strains, as determined by immunoblot analysis with an anti-invasin polyclonal antibody (Fig. 1B). This suggests decreased invasion by motility mutants might be slightly affected by expression of inv, but the loss of motility per se is predominantly responsible for reduced cell invasion by these mutants. This is consistent with the data showing that centrifugation to bring bacteria into contact with the host cell eliminates the need for flhDC or fliA.
FIG. 1.
Effect of flhDC and fliA on inv transcription and invasin protein levels. (A) Units of alkaline phosphatase activity (AP activity) were measured for strains containing a previously described inv-phoA fusion (16). Alkaline phosphatase activity was measured for Y. enterocolitica strain JB41v (wild type) (×), strain GY463v (flhDC) (●), and strain VM401v (fliA) (⧫) grown at 26°C in T medium. Strain JB41v was previously described (2), GY463v is an inv-phoA derivative of strain GY460v (21), and VM401v is an inv-phoA derivative of strain JB400v. Strain JB400v has a chromosomal deletion removing the 5′ end of the fliA locus (1). Transcription of inv is induced as cultures exit the logarithmic growth phase (16). To be sure these strains did not exhibit differences in growth phase induction, inv-phoA expression was measured at several time points after subculture. Values represent the calculated mean for assays performed in duplicate and had a standard deviation of less than 10%. All strains exhibited a similar growth rate. (B) Levels of invasin protein were determined by immunoblot analysis using an anti-invasin polyclonal antibody (16). Cultures were grown as described in part A and samples were normalized by examining equivalent units of optical density. The arrow indicates the position of full-length invasin.
Expression of flhDC from a plasmid results in decreased steady state levels of full-length invasin protein.
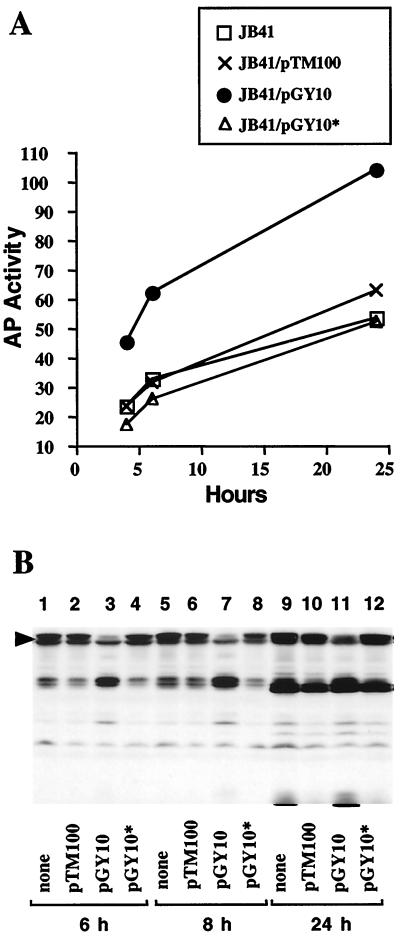
As noted above, the presence of flhDC on a multicopy plasmid resulted in a slight decrease in Y. enterocolitica invasion of HEp-2 cells. This decrease in invasion was similar to the reduction in invasion previously described for strains harboring mutations that affect motility and levels of invasin (2). To clarify the nature of the negative effect of plasmid encoded flhDC on invasion observed in this study, we determined whether reduced invasion was due to reduced inv expression by examining the effect of plasmid encoded flhDC on inv-phoA expression in the wild-type strain JB41v (Fig. 2A). When a plasmid-encoded copy of flhDC was introduced into strain JB41v, inv expression did not decrease but increased approximately twofold (Fig. 2A). The presence of the cloning vector (pTM100) or the transposon-inactivated derivative of flhDC (pGY10*) did not increase inv expression (Fig. 2A). These results indicate that the presence of multiple copies of flhDC causes increased inv expression. This finding is consistent with the previous conclusion that inv expression is affected by the flagellar regulon, but these results did not explain the negative effect of multicopy flhDC on cell invasion. An alternative explanation for the reduction of cellular invasion by Y. enterocolitica strains harboring pGY10 is that steady-state levels of invasin are reduced as a result of increased proteolysis. Therefore, invasin levels were determined for these same samples by immunoblot analysis using anti-invasin polyclonal rabbit antibody (Fig. 2B). The data revealed that levels of full-length invasin decreased for strain JB41v when containing pGY10 but were unchanged when containing other plasmids. A similar decrease in invasin levels was observed for strains JB580v (wild type) and GY460v (flhDC) when containing pGY10 (data not shown).
FIG. 2.
Expression of inv and invasin protein levels in strain JB41v when flhDC is present on a multicopy plasmid. Units of alkaline phophatase activity (AP activity) were measured and cultures were grown as described in Fig. 1. (A) Alkaline phosphatase activity was measured for strain JB41v alone (□) and when containing the parental cloning vector (pTM100) (×), a plasmid-encoded clone of flhDC (pGY10) (●), or a plasmid-encoded flhDC::TnMax2 derivative (pGY10*) (▵). (B) Levels of invasin protein were determined by immunoblot analysis using an anti-invasin polyclonal antibody (16). Cultures were grown as described in panel A, and samples were normalized by examining equivalent units of optical density. The arrow indicates the position of full-length invasin.
Based on these results, we hypothesized that the degradation phenotype was due to aberrant expression of the flagellar regulon. Consistent with this hypothesis, examination of flagellin preparations from cultures of strains harboring pGY10 confirmed that these strains overproduce flagellar proteins (data not shown). In addition, a mutation in fliA was found to suppress the degradation phenotype (data not shown). These results suggest that invasin degradation occurs in response to aberrant overexpression of the flagellar regulon. The degradation phenotype was also observed when E. coli harboring inv was transformed with plasmids encoding flhDC (data not shown). This may indicate invasin is degraded by a conserved protease that is activated in these bacteria in response to increased levels of flagellar proteins. The physiological significance of proteolytic digestion of invasin is not known, but this observation does account for the reduced invasion of HEp-2 cells observed when flhDC is present in multicopy. In this case, the degradation of invasin could be a regulatory mechanism that allows the bacterium to restrict the conditions leading to bacterial invasion of epithelial cells. Alternatively, increased production of flagellar proteins, which are localized to the cell envelope, might interfere with proper targeting of invasin to the outer membrane. Improper localization of invasin could stimulate degradation, which would affect bacterial invasion.
Conclusions.
The role of the flagellar regulon in Y. enterocolitica pathogenesis appears to be multifactorial. The bacterial type III flagellum secretion system is required for the appropriate localization of specific virulence factors such as YplA (20). Motility also may have a role in host invasion by allowing the bacterium to efficiently migrate to host cells to initiate contact. In some cases virulence genes, such as yplA, will be regulated coordinately at the transcriptional level as part of the flagellar regulon (20) (D. H. Schmiel, G. M. Young, and V. L. Miller, unpublished data). For other virulence genes, such as inv, expression and protein levels may be modulated in response to expression of the flagellar regulon.
Acknowledgments
We thank the members of the Miller lab for constructive discussions and suggestions.
This work was supported by National Institutes of Health Grants AI27342 to V.L.M. and 5 T AI07172 to G.M.Y. J.L.B. is a recipient of the UCPF award.
REFERENCES
- 1.Badger J. Ph.D. thesis. Los Angeles: University of California; 1996. [Google Scholar]
- 2.Badger J L, Miller V L. Expression of invasin and motility are coordinately regulated in Yersinia enterocolitica. J Bacteriol. 1998;180:793–800. doi: 10.1128/jb.180.4.793-800.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bottone E J. Yersinia enterocolitica: the charisma continues. Clin Microbiol Rev. 1997;10:257–276. doi: 10.1128/cmr.10.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornelis G R, Boland A, Boyd A P, Geuijen C, Iriate M, Neyt C, Sory M-P, Stainer I. The virulence plasmid of Yersinia, an antihost genome. Microbiol Mol Biol Rev. 1998;62:1315–1352. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darwin A J, Miller V L. Identification of Yersinia enterocolitica genes affecting survival in an animal host using signature-tagged transposon mutagenesis. Mol Microbiol. 1999;32:51–62. doi: 10.1046/j.1365-2958.1999.01324.x. [DOI] [PubMed] [Google Scholar]
- 6.de Lorenzo V, Eltis L, Kessler B, Timmis K N. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene. 1993;123:17–24. doi: 10.1016/0378-1119(93)90533-9. [DOI] [PubMed] [Google Scholar]
- 7.Givskov M, Ebert L, Christiansen G, Benedik M J, Molin S. Induction of phospholipase and flagellar synthesis in Serratia liquefaciens is controlled by expression of the master operon flhD. Mol Microbiol. 1995;15:445–454. doi: 10.1111/j.1365-2958.1995.tb02258.x. [DOI] [PubMed] [Google Scholar]
- 8.Grutzkau A, Hanski C, Hahn H, Riecken E O. Involvement of M cells in the bacterial invasion of Peyer's patches: a common mechanism shared by Yersinia enterocolitica and other enteroinvasive bacteria. Gut. 1990;31:1011–1015. doi: 10.1136/gut.31.9.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haas R, Kahrs A F, Facius D, Allmeier H, Schmitt R, Meyer T F. TnMax—a versatile mini-transposon for the analysis of cloned genes and shuttle mutagenesis. Gene. 1993;130:23–31. doi: 10.1016/0378-1119(93)90342-z. [DOI] [PubMed] [Google Scholar]
- 10.Hanski C, Naumann M, Hahn H, Riecken E O. Determinants of invasion and survival of Yersinia enterocolitica in intestinal tissue: an in vivo study. Med Microbiol Immunol. 1989;178:289–296. doi: 10.1007/BF00191063. [DOI] [PubMed] [Google Scholar]
- 11.Kapatral V, Olson J W, Pepe J C, Miller V L, Minnich S A. Temperature-dependent regulation of Yersinia enterocolitica class III flagellar genes. Mol Microbiol. 1996;19:1061–1071. doi: 10.1046/j.1365-2958.1996.452978.x. [DOI] [PubMed] [Google Scholar]
- 12.Kinder S A, Badger J L, Bryant G O, Pepe J C, Miller V L. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O:8 and construction of a transformable R−M+ mutant. Gene. 1993;136:271–275. doi: 10.1016/0378-1119(93)90478-l. [DOI] [PubMed] [Google Scholar]
- 13.Michaelis S, Inouye H, Oliver D, Beckwith J. Mutations that alter the signal sequence of alkaline phosphatase in Escherichia coli. J Bacteriol. 1983;154:366–374. doi: 10.1128/jb.154.1.366-374.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michiels T, Cornelis G R. Secretion of hybrid proteins by the Yersinia Yop export system. J Bacteriol. 1991;173:1677–1685. doi: 10.1128/jb.173.5.1677-1685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller V L, Falkow S. Evidence for two genetic loci from Yersinia enterocolitica that can promote invasion of epithelial cells. Infect Immun. 1988;56:1242–1248. doi: 10.1128/iai.56.5.1242-1248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pepe J C, Badger J L, Miller V L. Growth phase and low pH affect the thermal regulation of the Yersinia enterocolitica inv gene. Mol Microbiol. 1994;11:123–135. doi: 10.1111/j.1365-2958.1994.tb00295.x. [DOI] [PubMed] [Google Scholar]
- 17.Pepe J C, Miller V L. Yersinia enterocolitica invasin: a primary role in the initiation of infection. Proc Natl Acad Sci USA. 1993;90:6473–6477. doi: 10.1073/pnas.90.14.6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmiel D H, Wagar E, Karamanou L, Weeks D, Miller V L. Phospholipase A of Yersinia enterocolitica contributes to pathogenesis in a mouse model. Infect Immun. 1998;66:3941–3951. doi: 10.1128/iai.66.8.3941-3951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young G M, Miller V L. Identification of novel chromosomal loci affecting Yersinia enterocolitica pathogenesis. Mol Microbiol. 1997;25:319–328. doi: 10.1046/j.1365-2958.1997.4661829.x. [DOI] [PubMed] [Google Scholar]
- 20.Young G M, Schmiel D H, Miller V L. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc Natl Acad Sci USA. 1999;96:6456–6461. doi: 10.1073/pnas.96.11.6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young G M, Smith M, Minnich S A, Miller V L. The Yersinia enterocolitica motility master regulatory operon, flhDC, is required for flagellin production, swimming motility and swarming motility. J Bacteriol. 1998;181:2823–2833. doi: 10.1128/jb.181.9.2823-2833.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]