Significance
Narcolepsy with cataplexy is characterized by excessive daytime sleepiness and sudden loss of muscle tone (cataplexy) triggered by strong emotions. The best marker of the disease is low/absent hypocretin (HCRT), a hypothalamic neuropeptide. An autoimmune attack targeting these neurons is believed to cause HCRT deficiency, despite any direct evidence. We demonstrate that a substantial number of HCRT neurons are present in the brain of patients, but HCRT gene is silenced by methylation. The epigenetic silencing is not restricted to HCRT but also CRH and PDYN genes are methylated in the hypothalamus of patients. Our results strongly suggest that HCRT and CRH neurons are not destroyed but epigenetically deactivated, and possibilities exist to reactivate them to treat or even cure narcolepsy.
Keywords: QRFP, HCRT, CRH, gene silencing, methylation
Abstract
Narcolepsy with cataplexy is a sleep disorder caused by deficiency in the hypothalamic neuropeptide hypocretin/orexin (HCRT), unanimously believed to result from autoimmune destruction of hypocretin-producing neurons. HCRT deficiency can also occur in secondary forms of narcolepsy and be only temporary, suggesting it can occur without irreversible neuronal loss. The recent discovery that narcolepsy patients also show loss of hypothalamic (corticotropin-releasing hormone) CRH-producing neurons suggests that other mechanisms than cell-specific autoimmune attack, are involved. Here, we identify the HCRT cell-colocalized neuropeptide QRFP as the best marker of HCRT neurons. We show that if HCRT neurons are ablated in mice, in addition to Hcrt, Qrfp transcript is also lost in the lateral hypothalamus, while in mice where only the Hcrt gene is inactivated Qrfp is unchanged. Similarly, postmortem hypothalamic tissues of narcolepsy patients show preserved QRFP expression, suggesting the neurons are present but fail to actively produce HCRT. We show that the promoter of the HCRT gene of patients exhibits hypermethylation at a methylation-sensitive and evolutionary-conserved PAX5:ETS1 transcription factor-binding site, suggesting the gene is subject to transcriptional silencing. We show also that in addition to HCRT, CRH and Dynorphin (PDYN) gene promoters, exhibit hypermethylation in the hypothalamus of patients. Altogether, we propose that HCRT, PDYN, and CRH are epigenetically silenced by a hypothalamic assault (inflammation) in narcolepsy patients, without concurrent cell death. Since methylation is reversible, our findings open the prospect of reversing or curing narcolepsy.
Narcolepsy with cataplexy (also called narcolepsy type 1 or NT1, hereafter referred to as narcolepsy) is a sleep disorder characterized by excessive daytime sleepiness and cataplexy (loss of skeletal muscle tone triggered by strong emotions) (1). Shortly after the discovery of hypocretin (HCRT) neuropeptides, the postmortem examination of hypothalamic tissue from narcolepsy patients revealed a near absence of hypocretin (HCRT) gene transcript, prompting the hypothesis of destruction of HCRT-producing neurons in narcolepsy (2, 3). HCRT-1 deficiency (cerebrospinal fluid [CSF] HCRT-1 levels <110 pg/mL) is presently the best biological marker of the disease (4). It is currently widely, if not unanimously, believed that HCRT neuronal loss results from an immune or autoimmune process, based on the strong association of narcolepsy with the HLA-DQB1*06:02 allele (5), as well as the finding of a T cell receptor alpha gene polymorphism (6), and the acute increase in childhood narcolepsy that followed the 2009 to 2010 H1N1 vaccination (7). Although autoreactive T cells against various antigens expressed by HCRT neurons were reported (8, 9), direct evidence of HCRT cell death, lateral hypothalamic inflammation, or T cell infiltration, is lacking.
Based on several observations, we hypothesized that HCRT deficiency could result from the absence of HCRT expression, rather than from neuronal death. Evidence from patients with secondary narcolepsy (10–13) has indicated that CSF HCRT deficiency can be transient and normalize following treatment, suggesting that hypothalamic inflammatory reactions can cause reversible HCRT deficiency. Up to 48% of patients with traumatic brain injury exhibit HCRT deficiency, which recovers within 6 mo (14). Intriguingly, a 28-y-old female patient diagnosed with narcolepsy with cataplexy and undetectable CSF HCRT-1, who was placed under intravenous immunoglobulins starting as soon as 15 d after her first cataplexy attack, saw her CSF HCRT-1 level normalized, and her symptoms improved (15). Furthermore, lower CSF HCRT-1 levels, and even HCRT deficiency, were reported during hypersomnia episodes in patients with Kleine–Levin syndrome, suggesting expression levels can change without permanent cell loss (16). Aligned with these findings, evidence suggests that HCRT gene expression is finely tuned in response to both local and external environmental changes in humans and animal models.
Very recently, the mapping of hypothalamic neuropeptides in postmortem samples of narcolepsy patients led to the observation of a massive loss (88%) of corticotropin-releasing hormone (CRH) neurons in the paraventricular hypothalamic nucleus, while vasopressin, oxytocin, tyrosine hydroxylase and thyrotropin-releasing hormone neurons were unchanged (17). The cause of CRH deficiency in narcolepsy is unknown but it seems unlikely that an autoimmune attack would target both HCRT and CRH neurons, two highly specialized and unrelated cell types, while leaving other hypothalamic cells intact.
If HCRT deficiency is not caused by cellular destruction, then either loss-of-function mutations or epigenetic gene silencing are involved. Extensive genetic analyses of narcolepsy have failed to uncover causative gene mutations, with the exception of a few cases (2, 18, 19), and the disease is thought to be primarily sporadic. Epigenome-wide association studies have revealed sites of differential DNA methylation in blood and hypothalamic samples of narcolepsy patients (20, 21), but no site-specific hypermethylation associated with HCRT deficiency was found.
Results
Preserved Hypothalamic Expression of the HCRT Neuron-Coexpressed Neuropeptide QRFP in Narcolepsy.
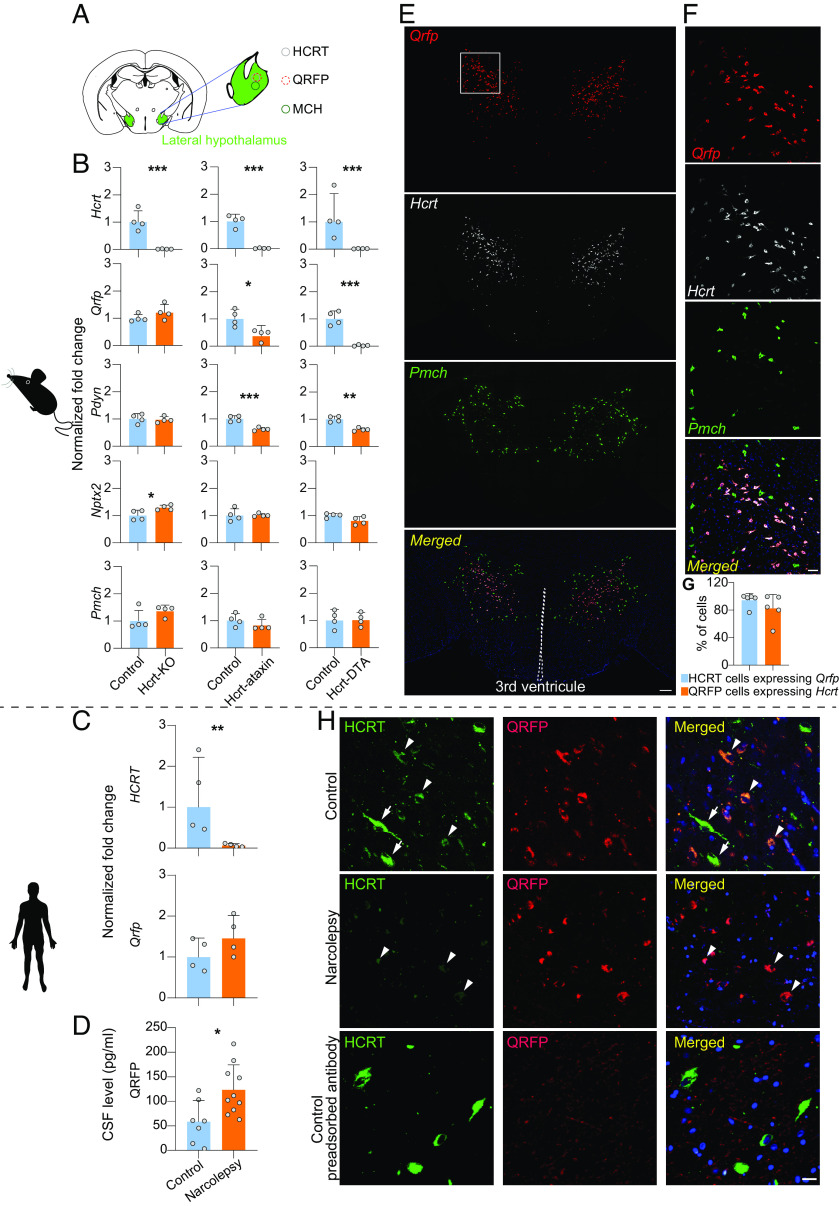
Several mouse models phenocopy narcolepsy symptoms with astonishing precision, including the Hcrt gene KO mouse, whose hypothalamus is known to contain cells with “HCRT-neuronal” identity (notably because they express HCRT promoter-driven transgenes), but they do not express Hcrt and several HCRT-neuron-ablated transgenic models, whose hypothalamus has lost HCRT-neurons due to expression of a neurotoxin. We previously performed RNA sequencing to identify differentially expressed genes in HCRT-neuron-ablated (Hcrt-ataxin3 transgenic) mice and in Hcrt-KO mice and their respective controls throughout several brain regions. This analysis revealed a gene, called Qrfp (26RFa: RFamide family 26 amino acid peptide), which displayed a striking hypothalamus-specific higher than a twofold decrease in expression in Hcrt-ataxin-3 Tg mice, whereas levels were unchanged in Hcrt-KO mice (22), and tended even to be up-regulated (Fig. 1B). To confirm loss of Qrfp expression in another HCRT-neuron-ablated model, we quantified Qrfp transcript in the hypothalamus of Hcrt-DTA mice, in which a bigenic system drives conditional expression of diphtheria toxin A in HCRT cells (23). The Qrfp transcript was found to be nearly abolished (up to 10-fold downregulation) in these mice (Fig. 1B), while no change was observed for Pmch, the MCH peptide-encoding gene, which is expressed in cells that are distinct, but intermingled with the HCRT and QRFP-producing neurons in the lateral hypothalamus (Fig. 1A). Similar analysis of two other neuropeptides known to colocalize with Hcrt, namely Pdyn (dynorphin) and Nptx2 (Narp) indicated no change for Nptx2 but around 50% decrease for Pdyn in neuron-ablated models (Fig. 1B). These findings suggested that Qrfp is either strongly coexpressed with Hcrt (and more specifically than Pdyn and Nptx2) or is dependent on HCRT cells and support the utility of Qrfp downregulation as a proxy for HCRT neuronal loss.
Fig. 1.
Expression of the HCRT-neuron coexpressed QRFP neuropeptide is intact in the hypothalamus of narcolepsy patients. (A) Schematic presentation of the lateral hypothalamus with major neuropeptidergic cell groups. (B) Expression levels of Hcrt, Qrfp, Pdyn, Nptx2, and Pmch transcripts in the hypothalamus of Hcrt-KO mice and two HCRT neuron-ablated transgenic lines, Hcrt-ataxin and Hcrt-DTA mice (n = 4 in each group). As expected, Hcrt expression is dramatically reduced in all three narcolepsy mouse models as compared to their respective controls. Qrfp expression is unchanged (or tends to increase) in Hcrt-KO mice, but is profoundly decreased in both neuron-ablated models. Pdyn expression is also substantially decreased in cell-ablated mice, although less than Qrfp, while Nptx2 does not seem to be affected. Also, Pmch is unchanged in all three mouse models. (C) The HCRT transcript is also dramatically decreased in the hypothalamus of narcolepsy patients, as expected, while QRFP expression tends to be increased, similar to Hcrt-KO mice. *P < 0.05, **P < 0.01, and ***P < 0.001, t tests on log transformed normalized fold change of mutated mice compared to their respective controls, n = 4. Bars represent geometric mean + geometric SD. (D) Quantification by RIA of the QRFP peptide in the CSF of narcolepsy (n = 10) and control subjects (n = 7) indicates increased QRFP levels in narcolepsy patients. *P < 0.05, t test. Bars represent mean + SD. (E) RNAscope in situ hybridization analysis of coronal sections through the mouse hypothalamus depicts a dense population of Qrfp, Hcrt, and Pmch expressing neurons in the lateral hypothalamic area. The merge photomicrograph shows extensive coexpression of Qrfp and Hcrt mRNA, while Pmch-expressing neurons surround but do not overlap with Qrfp+Hcrt neurons. (F) Higher magnification images (corresponding to the region highlighted with the white rectangle on top left) show coexpression of Qrfp and Hcrt mRNAs (pink), and absence of coexpression with the Pmch transcript, which is expressed in independent neurons (green). (Scale bars: A = 200 µm; B = 50 µm.) (G) Cell number quantification of Hcrt neurons expressing Qrfp and Qrfp neurons expressing Hcrt throughout the entire lateral hypothalamus (n = 5). (H) Postmortem human hypothalamic sections subjected to immunofluorescence staining for HCRT, QRFP, and merged immunofluorescence images are shown for a control subject (Upper), a narcolepsy patient (Middle), or a control subject after preadsorption of the antiserum with QRFP peptide (Lower). Note that neurons with low HCRT immunoreactivity (arrowheads) appear QRFP-positive, while neurons with high HCRT immunoreactivity (arrows) do not. In the patient, again neurons with residual, low-intensity staining for HCRT show strong QRFP staining. The number of QRFP-positive neurons in the patient and control subject appears similar. Lower panels of a control subject with QRFP peptide-preadsorbed QRFP antibody confirm QRFP antibody specificity (scale bars: 20 μm).
To determine whether QRFP expression is lost in the hypothalamus of narcolepsy patients as it is in HCRT neuron-ablated mice, RNA was extracted from the hypothalamic region of four narcolepsy and four control postmortem fresh frozen brains (24). Strikingly, the QRFP expression was not only preserved in the hypothalamus of narcolepsy patient (Fig. 1C), but also showed a very similar tendency to increase in patients (45% higher than controls), as we had found in Hcrt-KO mice (Fig. 1B). This may suggest competition between the two genes for the recruitment of similar transcription factors. Note that Qrfp and Hcrt are structurally and functionally related genes, their cDNAs share 50% sequence similarity and their seven-transmembrane receptors can heterodimerize (25). To next investigate QRFP peptide, we measured QRFP expression level by radioimmunoassay (RIA) in the CSF of 10 narcolepsy patients and seven control subjects. CSF-QRFP showed a significant twofold increase in patients (Fig. 1D), suggesting that CSF QRFP measurement might be a biomarker for narcolepsy. Altogether, our results suggest that HCRT neurons may be present in narcolepsy brains, but HCRT gene might be inactive.
There are conflicting reports on whether Qrfp and Hcrt are coexpressed in the same neurons in mice. One study reported two distinct hypothalamic neuronal populations in adult mice (26), while another study using single-cell RNA-sequencing of hypothalamic neurons of juvenile mice indicated colocalization (27). Hierarchical clustering defined by molecular fingerprints suggested that Hcrt- and Qrfp-expressing neurons share a similar neuronal lineage (27). Since no commercially available QRFP antibody shows specificity in mice, we tested if Hcrt and Qrfp transcripts are colocalized in the mouse hypothalamus by triple fluorescent in situ hybridization using RNAscope® (28) with probe sets covering the full-length Hcrt, Qrfp, and Pmch transcripts (Fig. 1 E and F). Quantification of RNAscope data indicated that 94.3 ± 10% of Hcrt-expressing neurons coexpressed Qrfp, while 82.5 ± 20.4% of Qrfp-expressing neurons coexpressed Hcrt (N = 5), with no colocalization with Pmch (Fig. 1G). Also, recently, Takahashi et al. reported the generation of a knockin mouse line expressing iCre from the endogenous Qrfp gene and observed low levels of iCre expression in HCRT immunoreactive neurons, supporting Hcrt/Qrfp coexpression (29).
To test whether HCRT and QRFP peptides colocalize in humans, we performed double immunostaining of postmortem hypothalamic sections (Fig. 1H). Immunostaining of four lateral hypothalamic sections from four control subjects and four sections from 2 narcolepsy patients revealed, as expected, over 90% reduction in the number of HCRT-immunoreactive neurons (n = 553 in controls versus n = 50 in patients). The same analysis for QRFP-immunoreactive neurons indicated no difference between patients and controls (n = 328 in controls and n = 278 in patients). In controls, we found that among QRFP-positive neurons, 62.0% were also HCRT-positive, and 38.7% of HCRT-positive cells appeared QRFP-positive, indicating significant colocalization. Next, among the very few and low-intensity HCRT-positive neurons found in patients (N = 50), 82% (n = 41) were QRFP-positive. Affecting these neuronal counts, we made the additional observation that the intensity of HCRT and QRFP immunostaining appeared inversely related, i.e., neurons with low HCRT staining displayed high QRFP signal, and vice versa (Fig. 1H, arrowheads). This finding aligns with our observations above at the mRNA level, showing that QRFP transcript levels tend to increase in narcolepsy patients, and suggests the existence of an HCRT–QRFP gene interaction. Altogether these data confirm that QRFP mRNA and protein are preserved in narcolepsy and largely colocalize with HCRT, supporting survival of a substantial number of these cells.
HCRT Gene Promoter Shows Site-Specific Epigenetic Alteration in Narcolepsy.
If indeed human narcolepsy brains contain HCRT neurons but similarly to Hcrt-KO mice do not express HCRT, and because HCRT gene mutations are not found in human narcolepsy, lack of expression may be the result of epigenetic dysregulation. Epigenetic control of gene expression can operate via different mechanisms, including DNA methylation, histone modification, chromatin structural change, and microRNAs. DNA methylation and histone modification are major mechanisms for gene silencing, which mainly occur in regulatory regions of genes such as promoters and enhancers.
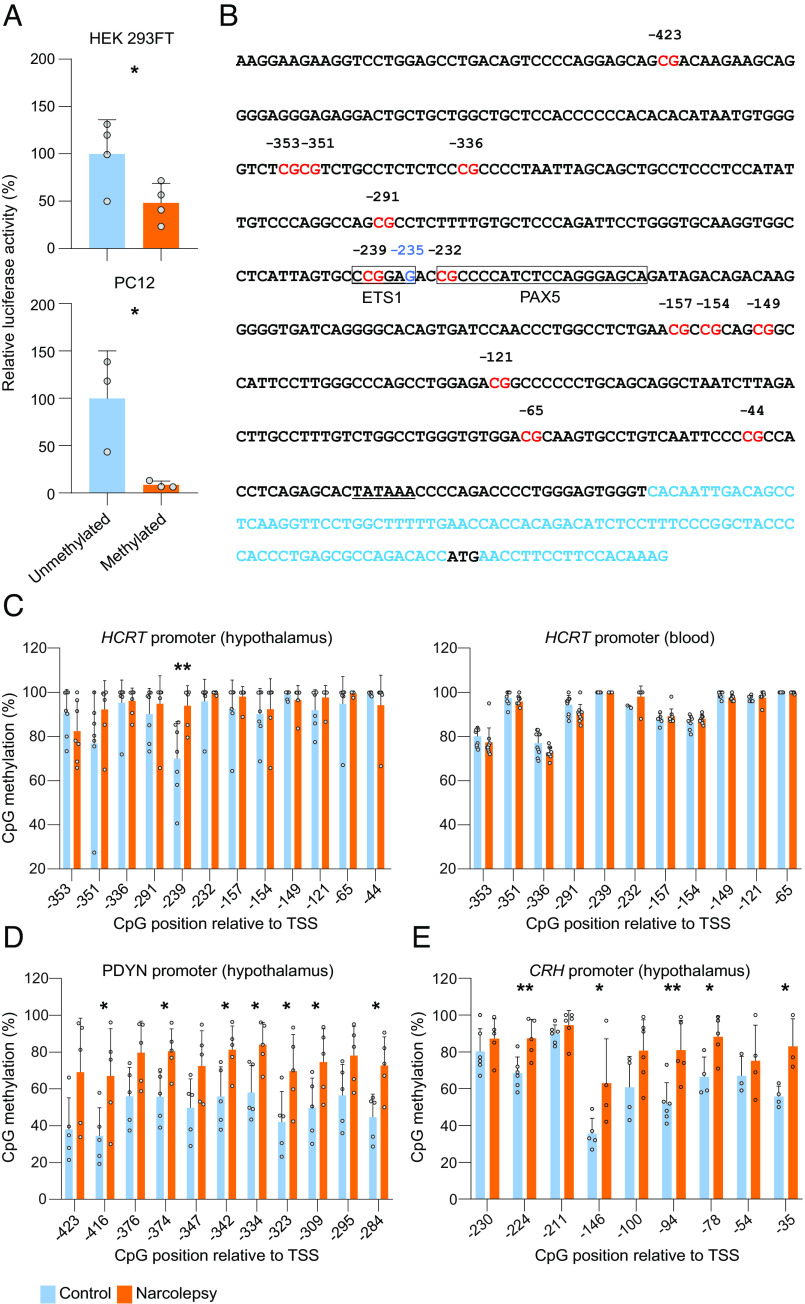
Transgenic analysis of the human HCRT promoter using lacZ reporter mice identified a 3.2-kb fragment upstream of the transcription start site that fully recapitulates expression in mice (30). Extensive deletion analysis of this fragment revealed that a 57-bp core region in the proximal promoter is critical for HCRT expression and represents the main regulatory region for the binding of transcription factors (31). To determine the methylation sensitivity of the human HCRT gene, we cloned a 1,800-bp proximal promoter in a CpG-free luciferase reporter vector (pCpGL) and performed luciferase assays. Two different cell lines (HEK 293FT and PC12) were transfected with intact unmethylated human HCRT promoter-luciferase pCpGL construct, or the same construct that had been preexposed to M.Sssl methyltransferase in vitro. Methylation of the HCRT promoter decreased the expression of luciferase by 60 to 90% (P < 0.05) (Fig. 2A), confirming that methylation of the HCRT promoter inhibits its expression.
Fig. 2.
Selective hypermethylation of the HCRT gene promoter at a PAX5:ETS1-binding site in the hypothalamus of narcolepsy patients. (A) Expression of a human HCRT gene promoter-driven luciferase reporter in two different cell lines is suppressed by methylation. Both cultures were transfected with a HCRT-promoter-driven luciferase construct either methylated or unmethylated. Values are averages of three wells measured in duplicate over three different transfections, and reported as a percentage of the reference unmethylated vector. *P < 0.05, t tests of methylated compared to unmethylated, n = 4 (HEK 293FT) and n = 3 (PC12). Bars represent mean + SD. (B) Sequence of the human HCRT promoter. Shown are 500 nucleotides upstream of the transcription start site, together with the first exon (in blue), with 5′ untranslated region and the translation start site (ATG, black). CpGs are highlighted in red. The CCCGAG sequence in the PAX5:ETS1 binding site is boxed with a G (instead of A in the consensus ETS1 sequence) in blue (−235). The TATA box is underlined. (C, Left) Methylation status at the 12 most proximal CpGs of the HCRT promoter (see B) in hypothalamic sections in seven narcolepsy patients and seven controls. One of the 12 CpGs is significantly hypermethylated in narcolepsy patients relative to controls (−239 CpG). (C, Right) Methylation status of 11 out of the 12 most proximal CpGs of the HCRT promoter from peripheral blood samples of nine narcolepsy and 10 control subjects. (D) Methylation status of the PDYN promoter from hypothalamic sections of five narcolepsy and five control subjects shows significant hypermethylation at 7 out of the 11 CpGs relative to controls. (E) Methylation status of the CRH promoter from brain hypothalamic sections in five narcolepsy and five control subjects shows significant hypermethylation at 5 out of the 9 CpGs relative to controls. Bisulfite sequencing of hypothalamic HCRT and PDYN promoters was performed by targeted next-generation sequencing, while data for blood and CRH derive from Sanger sequencing (percentage + SD). *P < 0.05 and **P < 0.005, t tests between narcolepsy patients and controls. CpG positions are indicated relative to the transcriptional start site (TSS).
To determine whether promoter methylation is involved in HCRT deficiency of narcolepsy patients, we next evaluated HCRT gene promoter methylation in DNA samples extracted from postmortem lateral hypothalami of seven patients and seven control subjects. The proximal human HCRT promoter contains 13 CpGs (Fig. 2B). We applied bisulfite treatment, followed by either of two sequencing methods to quantify the CpG methylation status (SI Appendix, Table S1). The first method relies on direct Sanger sequencing of PCR amplicons and measures the average methylation of DNA molecules in a tissue. When sufficient high-quality DNA was available, we also performed targeted next-generation bisulfite sequencing. Twelve of the 13 CpG sites within the proximal HCRT promoter (all except the most 5′ CpG in Fig. 2B) were tested. A higher methylation level was observed in narcolepsy patients as compared to controls at eight of these twelve CpGs, and one of them was significantly hypermethylated both by Sanger and targeted sequencing (CpG −239, methylation 93.9% in patients vs. 69.93% in controls, P < 0.01, Fig. 2 C, Left). Interestingly, the 57-bp core regulatory region of the proximal human HCRT promoter contains only 2 CpGs: CpG −291 and the patient-hypermethylated CpG-239. This core region was shown to be one of the 2 subregions essential for correct HCRT expression and transcription factor binding in vivo (31). Therefore, the patient differentially methylated residue is positioned at a functionally critical region.
Previous postmortem analysis of narcolepsy patients’ brains indicated that immunostaining for two other neuropeptides, PDYN (Dynorphin) and NPTX2 (Narp), which partially colocalize with HCRT, are also lost in the lateral hypothalamus (32, 33). In silico analysis of the promoter regions of these two genes revealed that both harbor a CpG island, suggesting that DNA methylation may affect their expression. The NPTX2 promoter contains a very high-density CpG island hindering bisulfite sequencing. We therefore focused on the methylation status of the PDYN promoter by applying the same methods as for the HCRT promoter (SI Appendix, Table S1). All 11 CpGs tested showed higher methylation in the hypothalamus of patients than in controls, and 7 from these 11 were significantly hypermethylated (N = 5 in each group, P < 0.05; Fig. 2D). Therefore, loss of PDYN signal in narcolepsy might also be due to promoter methylation rather than cell loss.
To determine whether differential methylation in narcolepsy patients compared to controls is specific to HCRT neurons, we tested the methylation status of the HCRT receptor 2 gene (HCRTR2). HCRTR2 is widely expressed in the hypothalamus including the lateral hypothalamus but we have shown that HCRT neurons do not express HCRTR2 (34). The HCRTR2 promoter contains a CpG island, in which we evaluated 10 CpGs for methylation level (SI Appendix, Table S1). Surprisingly, we found very low methylation (all CpGs had a methylation level below 35%), and no difference between narcolepsy patients and controls was observed (n = 4 narcolepsy patients and 6 controls, SI Appendix, Fig. S1A). This suggests that promoter hypermethylation within the lateral hypothalamus of narcolepsy patients has cell- and gene-specificity.
As DNA methylation patterns are tissue-specific, we tested methylation of the HCRT and PDYN promoters in DNA samples extracted from the peripheral blood cells of narcolepsy patients and controls. Eleven out of the same 12 CpGs of HCRT as tested in the brain samples were also tested by Sanger sequencing. Similar to the hypothalamus DNA, methylation at these 11 CpGs varied between 68 and 100% in the blood DNA (Fig. 2 C, Right). We however found that the CpG -239 that was hypermethylated in the hypothalamus of narcolepsy patients compared to controls was methylated at 100% in peripheral blood of all subjects, in patients (n = 9) or controls (n = 10) (Fig. 2 C, Right), consistent with the brain-specific expression of these genes. The fact that HCRT methylation level is not dramatically different in the brain of patients (although the pattern is), likely reflects that our hypothalamic samples contain heterogeneous populations of cells and the fraction of the relevant HCRT-producing neurons is small, leading to dilution of the signal. The same analysis was also performed for PDYN DNA from blood samples, which likewise indicated high methylation and no difference between narcolepsy patients and controls (n = 9 narcolepsy patients and 10 controls, SI Appendix, Fig. S1B). We next examined the CpG methylation levels of the QRFP gene promoter, which features 9 CpGs (SI Appendix, Table S1). We found no changes either in the hypothalamus or blood of narcolepsy patients as compared to controls (n = 7 narcolepsy patients and 6 controls for the hypothalamus, n = 9 narcolepsy and 10 controls for the blood, SI Appendix, Fig. S1 C and D), consistently with our observation above that QRFP transcript and protein are intact or increased in narcolepsy. Altogether these results suggest that the DNA-methylation alterations we find in narcolepsy are both cell type- and gene specific.
During the preparation of this manuscript, we made an unexpected discovery in an independent study aimed at mapping expression of hypothalamic neuropeptides in human narcolepsy. Analysis of CRH-expressing neurons revealed a severe loss of CRH signal in the paraventricular hypothalamus area of narcolepsy patients (17). Since CRH and HCRT are not coexpressed, we hypothesized that similar to HCRT, the CRH gene expression might be inhibited by promoter methylation, rather than CRH neuron-specific autoimmune attack. The CRH promoter is known to contain a CpG island whose expression is sensitive to methylation, specifically at a cAMP response element (CRE) bound by the CREB protein (35, 36). We thus performed bisulfite DNA sequencing of the proximal CRH promoter (SI Appendix, Table S1) using postmortem hypothalamic samples from 6 narcolepsy patients and 6 controls and found hypermethylation in narcolepsy patients at several CpGs including CpG −224, which is located in the CREB-binding site (Fig. 2E). Our finding of CRH promoter hypermethylation in narcolepsy is consistent with our hypothesis that expression loss is due to CRH gene silencing. The alternate hypothesis that an autoimmune attack electively targets HCRT and CRH neurons, two very different cell types, seems less likely.
A Methylation-Sensitive PAX5:ETS1 Transcription Factor Complex Binds and Regulates the HCRT Gene Promoter.
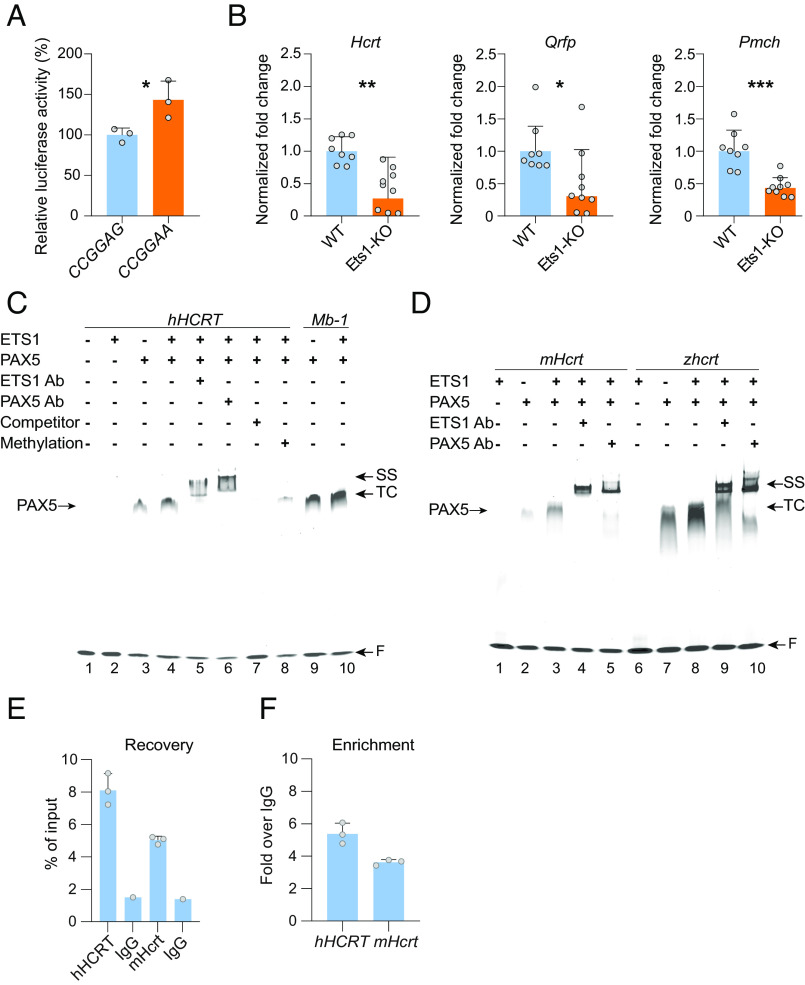
The hypermethylated CpG −239 residue of the HCRT promoter is located within a suboptimal ETS1-binding site (5′-CCGGAG-3′), a sequence originally identified in the promoter of the early B cell-specific MB-1 (CD79A) gene, encoding the Ig-alpha protein (37). ETS1 binding at this site was reported to be repressed by methylation (38, 39). Monomeric ETS1 is expressed in an autoinhibited form with very low affinity for its binding site, but it can bind either as a dimer at two head-to-head ETS1-binding sites, or cooperatively with another transcription factor, such as PAX5 or RUNX1. To determine whether this site in the human HCRT promoter recruits ETS1, we mutated the binding sequence to create an optimal ETS1-binding site (5′-CCGGAA-3′) in our HCRT promoter-driven luciferase reporter plasmid. This mutation caused a significantly increase in luciferase activity (Fig. 3A), indicating that the identified binding site is important for HCRT promoter-driven expression.
Fig. 3.
A PAX5:ETS1 complex regulates HCRT expression. (A) Mutagenesis of a suboptimal 5′-CCGGAG-3′ site of the human HCRT promoter into the optimized ETS1 binding site (5′-CCGGAA-3′) leads to a significant increase in expression of a luciferase reporter. *P < 0.05, t tests on CCGGAA compared to CCGGAG, n = 3. (B) The expression of the Hcrt transcript is significantly decreased in the hypothalamus of Ets1-KO mice. Levels of the Pmch and Qrfp transcripts are also significantly decreased, suggesting ETS1 may regulate expression of several lateral hypothalamic neuropeptide genes. *P < 0.05, and ***P < 0.001, t tests on log transformed normalized fold change of Ets1-KO mice compared to wild-type controls, n = 8 wild-type and n = 9 Ets1-KO. Bars represent geometric mean + geometric SD. (C) Inverted grayscale fluorescence micrograph of an EMSA gel showing the cooperative binding of PAX5 and ETS1 to the human HCRT (lanes 1 to 8) and Mb-1 (lane 9 to 10) promoters. In lane 8, the HCRT probe was synthesized to contain a methylated Cytosine at the −239 CpG residue of the sense strand, resulting in severely reduced binding. For HCRT: recombinant human ETS1 at 200 nM, PAX5 at 100 nM, for Mb-1: ETS1 and PAX5 at 100 nM, ETS1 and PAX5 antibodies (Ab) at 1:20, unlabeled specific competitor at 200X. (D) EMSA evidence of conserved cooperative binding of PAX5 and ETS1 to the mouse (lanes 1 to 5) and zebrafish proximal Hcrt/hcrt promoters (lanes 6 to 10). ETS1 at 200 nM, PAX5 at 100 nM, ETS1 and PAX5 antibodies at 1:20. TC: ternary complex, SS: super-shift, F: free probe. (E) ChIP-qPCR performed on hypothalamic tissue of 17 Hcrt-Cre transgenic mice and an anti-ETS1 antibody yielded substantial recovery (as % of input) of both the human and mouse hypocretin promoters, with fold enrichments (F) over the control IgG IP.
To assess the significance of the ETS1-binding site for endogenous Hcrt gene-expression in vivo, we next quantified Hcrt transcripts in the hypothalamus of Ets1-KO mice. A significant decrease in Hcrt gene expression was observed as compared to their wild-type littermates (Fig. 3B), suggesting that ETS1 controls the expression of the mouse Hcrt gene in vivo, consistently with our in vitro data above using the human HCRT promoter sequence. The 57-bp core regulatory region of the human HCRT promoter is well conserved in the mouse (82.5% base identity), although the mouse promoter shows an optimal ETS1-binding site (5′-CGGAA-3′) instead of the suboptimal human sequence (5′-CCGGAG-3′). Qrfp and Pmch expressions were also significantly decreased in the hypothalamus of Ets1-KO mice (Fig. 3B), suggesting that the ETS1 transcription factor controls the expression of multiple hypothalamic neuropeptides in mice. Consistently, both Qrfp and Pmch promoters were found to contain an optimal ETS1-binding site, similar to the mouse Hcrt promoter, at a similar position relative to the transcription start site.
Interestingly, we found that methylation levels of the last 2 CpG residues of the QRFP promoter were substantially lower than the one of all other CpGs of the QRFP promoter, in hypothalamus and blood of narcolepsy patients, or control subjects (SI Appendix, Fig. S1 C and D). These 2 CpGs overlap a 5′-CGGAAGCCG-3′ sequence, which contains an ETS1-binding site (underlined). This is consistent with regulation of the QRFP promoter by ETS1, and the relative hypomethylation of its ETS1-binding site may indicate ETS1 occupancy, reflecting active QRFP expression in the hypothalamus of narcolepsy patients.
To test if, similarly to what was reported in the MB-1 promoter, PAX5 recruits ETS1 at the CpG −239 of the human HCRT promoter, we performed in vitro electrophoretic mobility shift assays (EMSA) with full-length human recombinant PAX5 protein and a probe including 43 bp (−255 to −213) of the human HCRT promoter, or a 35-bp MB-1 probe (37) (SI Appendix, Table S2). PAX5 protein titration with both probes confirmed binding, although with lower affinity for the HCRT than for the MB-1 probe. Additionally, at concentrations higher than 100 nM, PAX5 was found to bind both probes as a dimer. As expected, the full-length human recombinant ETS1 showed very limited binding (trace binding at over 1-μg protein concentration). However, when PAX5 was added in the binding reaction, a ternary complex was detected (Fig. 3C, lane 4). Interestingly, although PAX5 alone binds HCRT probe as a dimer, in the presence of ETS1, the ternary complex seems to contain monomeric PAX5, leading to a similarly sized complex. The addition of ETS1 or PAX5 antibody to the binding reaction confirmed that the complex contains both proteins (Fig. 3C, lanes 5 and 6). The ternary complex formation could be completely removed with 200-fold excess unlabeled HCRT probe as a specific competitor (Fig. 3C, lane 7). Next, we tested the effect on binding of site-specific methylation by using the same HCRT probe but with the CpG −239 residue being methylated on the sense strand (SI Appendix, Table S2). Methylation dramatically reduced the ternary complex formation (Fig. 3C, lane 8), confirming the methylation-sensitive binding of ETS1. These results confirm that, similar to the MB-1 promoter (Fig. 3C, lanes 9 and 10), the HCRT promoter can cooperatively bind PAX5 and ETS1 in a site-specific and methylation-dependent manner. Our data further evidence structural differences in the PAX5:ETS1 complex at the HCRT and MB-1 promoters, with PAX5 binding the HCRT sequence as a dimer and at higher concentrations. PAX5 was first identified as a B cell-specific transcription factor, but it also plays important functions in the brain. Full-body Pax5 KO in mice leads to severe structural defects in the embryonic brain (40). The function of Pax5 in the postnatal brain is less well understood, but the gene was recently reported to be up-regulated in specific hypothalamic neuronal subtypes shortly after birth (41), and a role in the adult brain was suggested by the observation that its expression is down-regulated in the hippocampus of patients with bipolar disorder (42).
To determine whether PAX5:ETS1 cooperative binding is conserved across species, we performed EMSA with probes targeting the promoter of the mouse Hcrt, and the zebrafish hcrt gene (SI Appendix, Table S2), both containing putative PAX5 and ETS1-binding sites, with zebrafish hcrt gene containing several ETS1-binding sites. Both the mouse and the zebrafish probes showed PAX5 and PAX5:ETS1 cooperative binding using the human recombinant proteins (Fig. 3D), and the ternary complex could be supershifted with ETS1 and PAX5 antibodies, confirming that the cooperative PAX5:ETS1 binding to the Hcrt gene is evolutionary conserved.
To confirm recruitment of ETS1 at both the human HCRT and mouse Hcrt promoters in vivo, we performed chromatin immunoprecipitation (ChIP) with hypothalamic extracts of Hcrt-Cre transgenic mice (43), which contain both the endogenous Hcrt gene and the human HCRT-driven Cre-expressing transgene, and an anti-ETS1 antibody. ChIP-qPCR demonstrated efficient immunoprecipitation of both human and mouse promoters by the ETS1 antibody, with fold enrichments of 5.4 ± 0.6 for the human and 3.6 ± 0.2 for the mouse promoter (Fig. 3 E and F), indicating ETS1 protein is bound at both promoters in vivo.
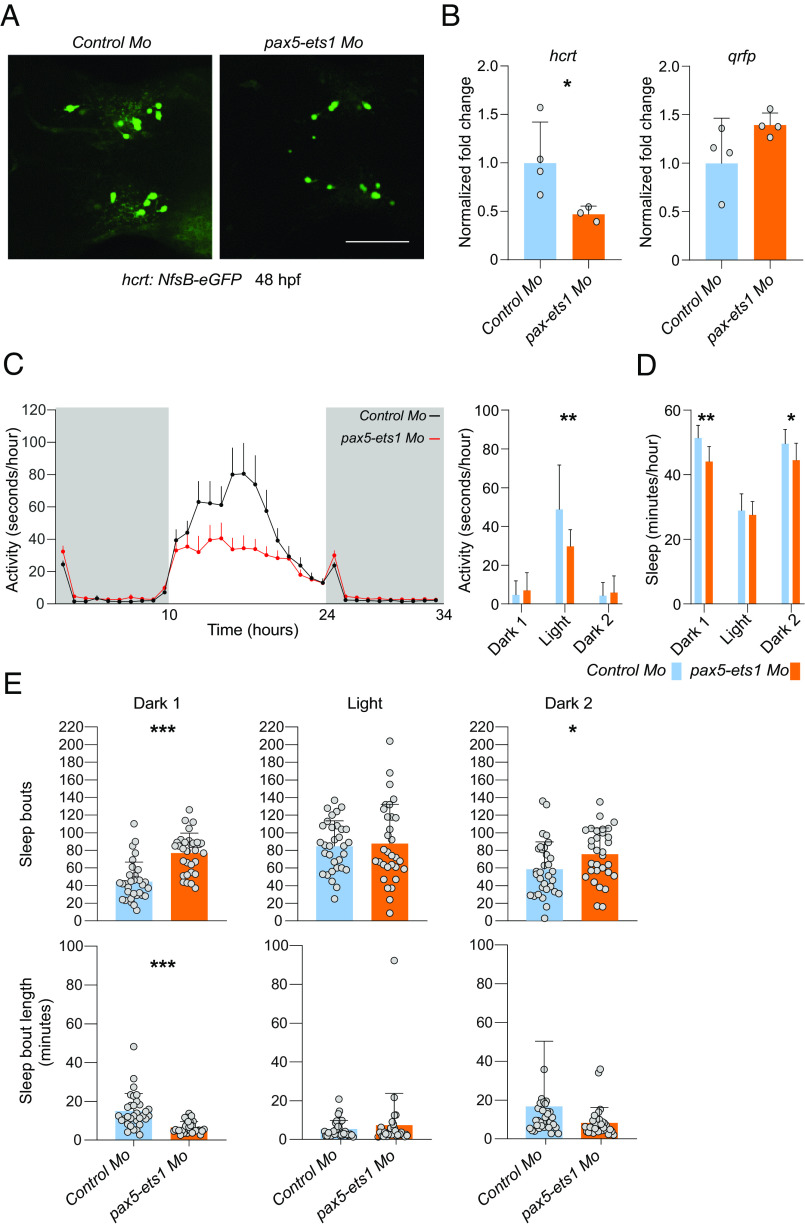
We next knocked down both pax5 and ets1 genes by injecting morpholino oligonucleotides in zebrafish embryos at the one-cell stage and assessed fish endogenous hcrt transcript levels. Fish with normal morphogenetic development (SI Appendix, Table S3) were selected for gene expression and activity/sleep analysis. Use of a transgenic line expressing a zebrafish hcrt promoter-driven eGFP reporter (44) allowed us to show that while hcrt-reporter expression was diminished by pax5-ets1 knockdown, the number of hcrt-positive cells was unchanged (Fig. 4A). Fish with pax5 and ets1 knockdown showed decrease hcrt transcript levels, relative to fish injected with a control morpholino oligonucleotide, while the expression of qrfp was unchanged (Fig. 4B). Furthermore, rest-activity analysis revealed that pax5 and ets1 knockdown fish exhibited a significant decrease in spontaneous locomotor activity during the light period (Fig. 4C), while sleep was decreased (Fig. 4D) and largely fragmented during the major sleep period (dark) (Fig. 4E), a trait commonly seen in narcolepsy (45). Prober et al., (46) showed that overexpression of hcrt in zebrafish leads to increased locomotor activity and reduced sleep, while Elbaz et al., (44) produced a zebrafish model where hcrt neurons are ablated, and found increased sleep and sleep fragmentation similar to our observations, although they did not find decreased locomotor activity. Altogether, our results suggest that PAX5:ETS1 may have a conserved function in controlling HCRT gene expression in man, mouse, and fish.
Fig. 4.
PAX5:ETS1 regulates the zebrafish hcrt expression and sleep. (A) Zebrafish larvae injected with morpholinos (Mo) against both ets1 and pax5 exhibit a substantial decrease in hcrt:nfs-eGFP expression (GFP immunoreactivity 48 h postfertilization (hpf), scale bar: 50 μm), and hcrt mRNA level (B), relative to fish injected with a control morpholinowhile the level of expression of qrfp is not changed or tends to increase. *P < 0.05, t test on log transformed normalized fold change of pax5 and ets1 morpholinos-injected fish compared to control morpholino-injected fish (n = 4 RNA samples extracted from pooled 60 fish at 5dpf each). (C) Knockdown of ets1 and pax5 also leads to decreased spontaneous locomotor activity during the light period (dark periods are indicated in gray), (D) decreased sleep during the dark period, and (E) increased sleep fragmentation in the dark period (i.e., a higher number of sleep bouts, of shorter average duration). Recording corresponds to fish at 5 to 6 dpf. N = 32, mean + SD; *P < 0.05, **P < 0.01, ***P < 0.001, t tests.
Discussion
In this study, we demonstrate the use of the coexpressed QRFP neuropeptide as a marker of a subpopulation of HCRT neurons, that is independent of concomitant HCRT expression, and has better specificity than previously identified coexpressed genes. Several studies sought to discover gene products enriched in HCRT neurons, with the aim of identifying potential self-antigens acting as targets of a hypothetical autoimmune process. Different approaches were used and yielded a number of candidate HCRT cell coexpressed genes (24, 47, 48). However, none of these genes are specific to HCRT neurons and are expressed in other forebrain cell types. We have used RNA-sequencing as a sensitive gene expression analytical tool to differentiate the hypothalamus of HCRT cell-ablated with the one of control mice, and did not find the previously reported genes but discovered Qrfp as the most significant differentially expressed gene, lost specifically within the hypothalamus of Hcrt-ataxin mice but not in other brain areas (22). We here confirm and extend this observation by showing that over 95% of Qrfp transcript is lost in HCRT neuron-ablated Hcrt-DTA mice. Loss of Qrfp expression when HCRT neurons are destroyed may be due to Qrfp transcript colocalization, or alternatively indicate that HCRT neurons are necessary for Qrfp gene expression. Because there is no mechanistic evidence that HCRT neurons would control Qrfp expression in other cells, colocalization of the two transcripts is the most parsimonious explanation. Additionally, we found that loss of Hcrt expression in Hcrt-KO mice leads to upregulation of Qrfp expression, possibly reflecting an interaction between the two genes within the same cells, for instance by competition for recruitment of common transcription factors. Most directly, we used RNAscope in mice to demonstrate that over 94% of Hcrt-expressing neurons in the lateral hypothalamus coexpress Qrfp. Reciprocally 82% of Qrfp-expressing neurons express Hcrt. The 18% of Qrfp-expressing neurons that do not express Hcrt are mainly located outside the lateral hypothalamus. Although we cannot directly transpose mouse findings to humans, or two different detection techniques (RNAscope versus immunohistochemistry), we show that 38.7% of HCRT-immunoreactive cells in the brain of human control subjects are immunoreactive for QRFP, and 62% of QRFP-positive neurons are HCRT-positive. The lower figures in humans as compared to mice may stem from several factors, most notably the lower sensitivity of immunohistochemistry compared to in-situ RNA hybridization. Additionally, these data are confounded by our observation that when HCRT-immunoreactivity is low, QRFP-immunoreactivity is high. Consistently we observe that among the few remaining and low-expressing HCRT neurons in the brain of narcolepsy patients, 82% are QRFP-positive.
Even if only 38.7% of HCRT neurons appear to coexpress QRFP in controls, the loss of HCRT neurons by a hypothetical immune attack would be expected to lead to a significant decrease in QRFP. In contrast, we show that not only it is not the case but QRFP expression is increased in the brain of patients. Altogether, these results support that QRFP is a marker of HCRT neurons and because its expression is not decreased but increased, a substantial number of HCRT neurons must be preserved in narcolepsy patients but do not productively express HCRT.
The second important finding of this study is the mechanism by which HCRT neurons are silenced in narcolepsy. If these neurons are not destroyed but do not express HCRT, then either there is a physiological downregulation or epigenetic mechanisms are involved. In contrast to Hcrt-KO mice, human narcolepsy patients carry an intact HCRT gene. In addition, none of the narcolepsy transcriptome studies found loss of any critical gene that could control HCRT expression, such as a signaling molecule or a transcription factor. We do not exclude that at disease onset, a local immune reaction (inflammation) caused a certain level of cell loss. Accordingly, both epigenetic gene silencing and cell death may occur in narcolepsy. We show here that not only the human HCRT (as well as Pdyn and CRH) promoter is hypermethylated in the hypothalamus of narcolepsy patients but that the methylation occurs at a highly critical and phylogenetically conserved CpG within the PAX5:ETS1 binding site. One may argue that if HCRT neurons are destroyed then our methylation analysis is based on non-HCRT-expressing neurons, that could be hypermethylated. First, we clearly show in the first part that HCRT-neurons could not be absent in the brain of narcolepsy patients. Second, we show that the pattern of methylation in the hypothalamus is different from that of blood cells, which do not express HCRT. For instance, CpG-336 is nearly 100% methylated in the lateral hypothalamus while its methylation is below 80% in blood cells (Fig. 2C) and the differences are even larger for Pdyn promoter methylation between the hypothalamus and blood cells (Fig. 2D and SI Appendix, Fig. S1B). Finally, non-HCRT hypothalamic cells may not be fully methylated but only partially leading to undetectable HCRT peptide (for instance by immunohistochemistry), and these neurons can express detectable HCRT under certain circumstances, as previously reported (see below) (49). The % DNA methylation values we obtain from our hypothalamic samples need here to be put in context. Clearly our samples contain heterogeneous populations of cells, and HCRT neurons (estimated to number about 70,000 in humans) represent only a small fraction of them (evaluated to grossly represent about 2%). Yet the difference in methylation level between patients and controls that we observe (with ~95% methylation in patients vs. ~70% in controls at CpG −239, i.e., approximatively 25%) appears about an order of magnitude greater than the number of DNA molecules plausibly contributed by HCRT neurons in the tissue sample. We find that the CpG −239 of the HCRT promoter is overall about 70% methylated in the hypothalamus of control subjects, indicating that this site is not always methylated in cells that do not express HCRT, and mechanisms other than DNA methylation are also implicated in the regulation of HCRT expression in different cell types. In the hypothalamus of patients, methylation at this site approaches 95%. A 25% HCRT DNA methylation increase in narcolepsy patients strongly suggests that methylation has occurred within HCRT neurons, as well as a fraction of non-HCRT cells. Thus, the HCRT gene methylation changes that our values reflect must be accounted by a sizable population of non-HCRT cells in addition to HCRT neurons, and the same applies to CRH gene DNA methylation.
Other epigenetic mechanisms (histone modifications and chromatin remodeling enzymes, microRNAs and long noncoding RNAs), were not analyzed in our limited postmortem material in this study, but we recently reported that the evolutionary-conserved miRNA-137 is also involved in Hcrt expression (50). Whether in addition to methylation, histone modifications (acetylation, methylation, or phosphorylation) are involved cannot be excluded, especially because of a crosstalk exists between the two mechanisms. For instance, DNA methylation can guide histone modifications and DNA methylation can also take its cues from histone modifications (51, 52).
Epigenetic silencing of HCRT neurons might be facilitated by genetic predispositions (the most significant being the HLA-DQB1*06:02 allele) that make individuals particularly susceptible to immune-related processes triggered by an inflammatory insult within the lateral hypothalamus, for instance a viral infection that causes local release of proinflammatory cytokines within the lateral hypothalamus. Systemic inflammation has been linked to reduced HCRT CSF levels in patients and rats (53), through nonidentified mechanisms. Further supporting a link between inflammation and reduced HCRT expression, a polymorphism in the proinflammatory TNF-alpha gene was reported to be significantly associated with narcolepsy (54, 55), and narcolepsy patients are reported to have increased serum TNF-alpha levels (56, 57). Moreover, TNF-alpha was found to dramatically decrease HCRT expression in vitro (58). Importantly, prolonged exposure to TNF-alpha was reported to induce promoter methylation in cell lines (59). The unexplained recent discovery of a marked increase in the number of HCRT-immunoreactive neurons in the hypothalamus of heroin addicts, later reproduced in mice and shown to be independent of neurogenesis (49), is another example of changes in HCRT gene expression that could be explained by epigenetic mechanisms operating at the HCRT gene promoter. HCRT gene expression malleability in response to various external cues would be consistent with HCRT prime role as a global environmental integrator, and therefore susceptible to epigenetic modifications as in narcolepsy. Altogether we propose that human narcolepsy is not a neurodegenerative disease, but rather results from an epigenetic remodeling of a number of hypothalamic tissue- and cell-specific genes. The disease is therefore neither appropriately modeled by the Hcrt-KO mice, since in addition to loss of HCRT expression it also features loss of hypothalamic NPTX2 and PDYN transcripts, nor by HCRT-neuron ablation, since other hypothalamic cell types (e.g., CRH-producing cells) are also affected.
Intriguingly, a rare condition called autosomal dominant cerebellar ataxia, deafness and narcolepsy, is caused by dominant mutations in the gene encoding the maintenance DNA methyltransferase 1 (DNMT1) (60, 61). Dominant mutations in DNMT1 were associated with increased methylation at normally unmethylated regions of promoters and CpG islands, leading to gene silencing (62), strengthening the notion of a causal link between DNA hypermethylation and narcolepsy. Partial or transient epigenetic modulation of HCRT expression might also be involved in narcolepsy without cataplexy, idiopathic hypersomnia or Kleine–Levin syndrome. Furthermore, the gradual decrease in the number of HCRT-immunoreactive neurons with aging (63, 64) might also be due to epigenetic downregulation of HCRT gene expression, leading to profound changes in sleep and sleepiness in aged subjects.
Narcolepsy is increasingly recognized as a hypothalamic disorder, rather than exclusively a HCRT disease, involving not only sleep-wake, but also motor, psychiatric, emotional, cognitive, metabolic, and autonomic abnormalities (1). Together with other studies, our results strongly suggest that narcolepsy results from dysregulations, including epigenetic alterations, in several other neuropeptidergic systems (PDYN, NPTX2, CRH, and QRFP) with diverse functions.
Methylation being reversible, our findings open the avenue for therapeutic interventions based on DNA demethylation, leading to treatment or even cure of narcolepsy. Although DNA methylation/demethylation events were initially thought to be restricted to dividing cells, recent evidence indicates that they also occur in postmitotic cells, including in neurons (65). Epigenetic editing tools, including pharmacological compounds, or site-specific demethylating genetic reagents, are becoming available (66). Beyond narcolepsy, epigenetic silencing may represent a key causative factor in several other immune, autoimmune, neurodegenerative, or neuropsychiatric diseases.
Materials and Methods
Animals.
Hcrt-KO and Hcrt-ataxin mice were bred locally on C57BL/6J background. Brains of Hcrt-DTA (23), Ets1-KO (67) and their controls were provided by international collaborators. Brains of Hcrt-DTA mice were sampled 4 wk after doxycycline removal from the diet, while their controls were maintained under doxycycline. All animal procedures followed Swiss federal laws and were approved by the State of Vaud Veterinary Office. At all times, care was taken to minimize animal discomfort and avoid pain.
Human Tissue Samples.
Hypothalamic RNA samples from four narcolepsy patients and four controls were available in the laboratory of M. Honda (24) and were tested locally by qPCR with TaqMan assays Hs01891339_s1 (HCRT), Hs01650960_s1 (QRFP), and reference genes Hs00744842_sH (TUBA1B), Hs01375212_g1 (RPS18), and Hs04185005_g1 (RPS27). Formalin fixed sections (40 μm, 2 to 4 sections/subject) through the lateral hypothalamus from narcolepsy patients and controls were obtained through international collaborators. All tissues samples were from postmortem brains previously published (17, 49, 68–71). The research protocol and the use of human biological material was approved by the local ethics committee (SwissEthics). Formalin fixed sections were washed with PBS, and genomic DNA was extracted with the AllPrep DNA FFPE Tissue kit (Qiagen). CSF samples from narcolepsy and control subjects were provided by the laboratory of GJ. Lammers.
RT-qPCR.
RNA extraction, quantification, and processing are detailed in SI Appendix. Amplification was carried out with TaqMan assay kits Mm01964030_s1 (Hcrt), Mm01701538_m1 (Qrfp), Mm0124886_g1 (Pmch), Mm00457573_m1 (Pdyn), Mm00479438_m1 (Nptx2), and reference genes Mm01973893_g1 (Eef1a1), Mm00850060_s1 (Rps9). qPCR for fish samples were performed by primers in combination with Power SYBR Green (Thermo Fisher Scientific) using a ViiA7 real-time PCR system (Thermo Fisher Scientific). Primers were: hcrt-F 5′-CTCCTGCAAACTCTACGAGATG-3′, hcrt-R 5′-GTCGTTGTTGAGATGCACTAAA-3′, qrfp-F 5′-AATGCTGCCACACCAACCAA-3′, qrfp-R 5′-TCCCAGGTCCTGAAACAAAGC-3′, and the reference gene 18s-F 5′- AGCGTGCGGGAAACCACGAG-3′ and 18s-R 5′- AAGCCGCAGGCTCCACTCCT-3′. Normalized fold expression was calculated according to Taylor et al. (72).
QRFP RIA.
Quantification of QRFP in human CSF samples was carried out using a specific RIA, previously described in detail (73). CSF samples were pumped at a flow rate of 1.5 mL/min through one Sep-Pak C18 cartridge. Bound material was eluted with acetonitrile/water/TFA (50:49.9:0.1; v/v/v), and acetonitrile was evaporated under reduced pressure. Finally, the dried extracts were resuspended in PBS 0.1M and assayed for QRFP. Data represent the mean of duplicates for each sample.
RNAscope-In Situ Hybridization Experiments.
Tissue processing is detailed in SI Appendix. Fluorescent in situ hybridization for the simultaneous detection of the Qrfp, Hcrt, and Pmch transcripts was performed using RNAscope (ACD; Advanced Cell Diagnostics). The Hcrt probe targeted the region 2 to 577 (accession number NM_010410.2; ACD, Cat. No. 490461-C2). The Qrfp probe targeted the region 112 to 1,081 (accession number NM_183424.4; ACD, Cat No. 464341-C3). The Pmch probe targeted the region 4 to 652 (accession number NM_029971.2; ACD, Cat. No. 478721-C1). Negative and positive control probes recognizing dihydrodipicolinate reductase, DapB (a bacterial transcript) and PolR2A (C1 channel), PPIB (C2 channel) and UBC (C3 channel), respectively, were processed in parallel with the target probes to ensure tissue RNA integrity and optimal assay performance.
Immunofluorescence and Confocal Microscopy of Human Tissues.
Tissue processing is detailed in SI Appendix. QRFP antibody (73) at 1:400 was applied overnight at room temperature, followed by a second incubation with QRFP antibody 1:400 and HCRT antibody 1:500 (goat HCRT anti HCRT, Santa Cruz Biotechnology, SC-8070) at 4 °C for 24 h. After rinsing with 1xPBS for 3 times (10 min each), secondary antibodies were applied (donkey IgGs coupled to Alexa-594, or -488 fluorophores) in 1:500 dilutions for 2 h at room temperature followed by rinsing with 1xPBS for 3 times (10 min each). To test the specificity of QRFP antibody, QRFP antibody was blocked with QRFP (NM_198180) Human Over-expression Lysate (2 μg/mL, 1%BSA) for 2 h at room temperature before use in the above-described immunofluorescence assay. Images were acquired on an inverted Zeiss LSM780 confocal laser-scanning microscope (405-, 488-, and 561-nm lasers) using a 40× oil objective (EC plan-Neofluar 40×/1.30 Oil DIC M 27). For each human section used for cell quantification, confocal images covering the HCRT positive cell field were acquired at 8-bit image depth and a frame of 1,024 × 1,024 pixels and tiled together at size of 2,125.48 × 2,125.48 μm using ZEN software. Immunoreactive cell counts were evaluated within that frame using ImageJ software.
HCRT Promotor Cloning.
1,800 base pairs of the human HCRT promoter upstream of the transcription start site were cloned in multiple cloning sites of the pCpG-basic vector (74) upstream of the firefly luciferase sequence. The cloned plasmids were transformed in a Pir1 bacterial strain and sequenced for verification. The bacterial colonies were cultured with Zeocin selection at large scale and maxiplasmid preparations were carried out using the HiSpeed Plasmid Maxi Kit (Qiagen, 12662). CpG Methyltransferase, M.SssI (New England Biolabs, M0226s) was used to methylate the CpG sites on the HCRT promoter.
Luciferase Assay.
HEK 293FT and PC12 cell lines were cultured at 1.5-2*105 density in 24-well plates in DMEM medium containing 10% FBS. Cultures were transfected with methylated and unmethylated plasmids. The transfection was performed using X-tremeGENE™ HP DNA Transfection Reagent (Sigma, 6366244001) with the DNA to reagent ration of 1:3. The transfected plasmids were mixed with Firefly and Renilla luciferases with ratio of 19:1. Then, 48 h after transfection the cells were lysed and prepared for the luciferase activity measurement according to Promega kit (E1910). Luciferase activity was measured using the GloMax 96 microplate luminometer from Promega. The firefly luciferase activity was normalized to the Renilla activity.
DNA Methylation Analysis.
Using the EpiTect Fast DNA Bisulfite kit (Qiagen), 500 ng DNA was treated with sodium bisulfite. Primers for nested PCR (2 fragments for HCRT, 1 for PDYN and HCRTR2) were designed with MethPrimer (75) (SI Appendix, Table S1). Methods for methylation quantification are detailed in SI Appendix.
Electrophoretic Mobility Shift Assay (EMSA).
Oligonucleotides probes (SI Appendix, Table S2) were purchased from Microsynth AG (Balgach, Switzerland). To test the effect on binding of selective methylation at the CpG-239 residue, the HCRT probe was synthesized with a methyl-Cytosine at this position on the sense strand (Fig. 3C, lane 8). Probes were 3′ labeled with Dyomics 781. Unlabeled oligos were used for competition assays. Assay procedure is detailed in SI Appendix.
ChIP-qPCR.
ChIP was performed using iDeal ChIP-seq kit for Transcription Factors (Diagenode, C01010054) following the provided protocol. Tissue processing and chromatin preparation is detailed in SI Appendix. Sheared chromatin was subjected to magnetic immunoprecipitation using 1 μg anti-ETS1 (Cell Signaling Technology, D8O8A) and anti-IgG in ChIP reaction provided by the kit. Quantification of ChIP was performed by qPCR using Power SYBR™ Green PCR Master Mix (Fisher Scientific, 4367659) using primers as listed in SI Appendix, Table S2. The % recovery was determined as 2^((Ct(input)-6.64)-Ct(IP))*100, where 6.64 is the adjusting factor to correct for the input dilution (1:100), and the fold enrichment of target over the control sequences were calculated as 2^(Ct(target)-Ct(IgG)).
Morpholino Study in Zebrafish.
Morpholino antisense oligos targeting ets1 and pax5 transcripts were designed according to Pham et al. (76) and Kwak et al. (77), respectively. Subsequently, 0.25 pmol ets1 (5′-GTCATGGTCACGCATTCAAACGTAC-3′) or a combination of 0.375 pmol pax5 TB1 (5′-CAGTGGATTTCCATCTGTTTTAAA-3′) and 0.375 pmol pax5 TB2 (5′- CTCGGATCTCCCAGGCAACATGGT-3′) was injected in 1 to 2 cell zygotes in a total volume of 1 nL. The control morpholino corresponds to the standard sequence proposed by the manufacturer (5′-CCTCTTACCTCAGTTACAATTTATA-3′). Morpholino synthesis was performed by Gene-tools (http://www.gene-tools.com). Visualization of Hcrt neurons was performed using the Tg(hcrt:nfsB-EGFP) transgenic line, in which a 2-kb fragment of the zebrafish hcrt promoter drives expression of a nfsB-EGFP fusion protein (44). Imaging was performed on homozygous embryos, using a LSM710 confocal microscope (Zeiss). Locomotion tracking is detailed in SI Appendix. Larvae exhibiting morphological defects (SI Appendix, Table S3) were excluded from analysis to control against motor impairments.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank T. Scammell, D. Swaab, Y. Dauvilliers, and C. Peyron for providing human brain tissues. We thank the Lausanne Genomic Technologies Facility for help in Next-Generation-methylation sequencing. We thank T. Kilduff and A. Yamanake for providing Hcrt-DTA mice brains, H. Okamoto for providing Tg(hcrt:nfsB-eGFP) fish, and M. Rehli for providing the CpG-free vector. We also thank T. Miyagawa and M. Shimada from Tokyo Metropolitan Institute of Medical Science for assistance. We thank all the persons at the zebrafish facility of the University of Lausanne. We thank also the Genomic Technology Facility, the Protein Analysis Facility, and the Cellular Imaging Facility of the University of Lausanne. We are grateful to U. Schibler for scientific advice and with R. Jaenisch for their comments on our draft manuscript. This work was supported by E-RARE NARCOMICS (Swiss NSF grant 185655 to M.T.), Swiss NSF (grant 173126 to M.T.), and the State of Vaud (Faculty of Biology and Medicine, University of Lausanne). The zebrafish work was supported by the Swiss NSF (grant 188789 to F.A.). L.S. has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant (Agreement No. 707404).
Author contributions
A.S., R.F., F.A., N.C., R.L., G.J.L., A.V., and M.T. designed research; A.S., M.R., L.S., S.L., M.-L.P., C.P., L.A.G.-S., D.F., M.H., Y.A., D.G., M.P., A.A., C.I., A.V., and M.T. performed research; L.S. and L.A.G.-S. contributed new reagents/analytic tools; A.S., M.R., S.L., M.-L.P., C.P., D.F., M.H., Y.A., D.G., M.P., A.A., C.I., N.C., A.V., and M.T. analyzed data; and A.S., M.R., R.F., F.A., N.C., R.L., G.J.L., A.V., and M.T. wrote the paper.
Competing interests
M.T. reports consulting for NLS Pharmaceutics and an unrestricted research grant from NLS Pharmaceutics and Jazz Pharmaceuticals. G.J.L. reports consulting for Jazz Pharmaceuticals, UCB, Bioprojet, and NLS Pharmaceutics and is a member of the advisory board of Jazz Pharmaceuticals, UCB, and Bioprojet. R.F. reports consulting for Takeda Pharmaceutical and Bioprojet/AOP Orphan and grant support from Jazz Pharmaceuticals and Bioprojet. L.S. reports consulting for Takeda Pharmaceutical. University of Lausanne has a patent, with M.T. and A.S. as inventors, on epigenetic treatments of narcolepsy. All other authors declare no competing interests.
Footnotes
This article is a PNAS Direct Submission. L.d.L. is a guest editor invited by the Editorial Board.
Contributor Information
Ali Seifinejad, Email: ali.seifinejad@mail.ch.
Mehdi Tafti, Email: mehdi.tafti@unil.ch.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Bassetti C. L. A., et al. , Narcolepsy–clinical spectrum, aetiopathophysiology, diagnosis and treatment. Nat. Rev. Neurol. 15, 519–539 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Peyron C., et al. , A mutation in a case of early onset narcolepsy and a generalized absence of hypocretin peptides in human narcoleptic brains. Nat. Med. 6, 991–997 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Thannickal T. C., et al. , Reduced number of hypocretin neurons in human narcolepsy. Neuron 27, 469–474 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishino S., Ripley B., Overeem S., Lammers G. J., Mignot E., Hypocretin (orexin) deficiency in human narcolepsy. Lancet 355, 39–40 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Tafti M., et al. , DQB1 locus alone explains most of the risk and protection in narcolepsy with cataplexy in Europe. Sleep 37, 19–25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hallmayer J., et al. , Narcolepsy is strongly associated with the T-cell receptor alpha locus. Nat. Genet. 41, 708–711 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dauvilliers Y., et al. , Post-H1N1 narcolepsy-cataplexy. Sleep 33, 1428–1430 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Latorre D., et al. , T cells in patients with narcolepsy target self-antigens of hypocretin neurons. Nature 562, 63–68 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Jiang W., et al. , In vivo clonal expansion and phenotypes of hypocretin-specific CD4(+) T cells in narcolepsy patients and controls. Nat. Commun. 10, 5247 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kashiwagi T. T., et al. , Sleepiness due to Wernicke’s encephalopathy with bilateral hypothalamic lesion in a 5-year-old girl. Sleep 27, A315 (2004). [Google Scholar]
- 11.Oka Y., et al. , Low CSF hypocretin-1/orexin-A associated with hypersomnia secondary to hypothalamic lesion in a case of multiple sclerosis. J. Neurol. 251, 885–886 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Nozaki H., et al. , A patient with anti-aquaporin 4 antibody who presented with recurrent hypersomnia, reduced orexin (hypocretin) level, and symmetrical hypothalamic lesions. Sleep Med. 10, 253–255 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Dauvilliers Y., et al. , Reversal of symptomatic tumoral narcolepsy, with normalization of CSF hypocretin level. Neurology 69, 1300–1301 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Baumann C. R., Werth E., Stocker R., Ludwig S., Bassetti C. L., Sleep-wake disturbances 6 months after traumatic brain injury: A prospective study. Brain 130, 1873–1883 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Dauvilliers Y., Abril B., Mas E., Michel F., Tafti M., Normalization of hypocretin-1 in narcolepsy after intravenous immunoglobulin treatment. Neurology 73, 1333–1334 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Wang J. Y., et al. , Cerebrospinal fluid orexin a levels and autonomic function in kleine-levin syndrome. Sleep 39, 855–860 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shan L., Balesar R., Swaab D. F., Lammers G. J., Fronczek R., Reduced numbers of corticotropin-releasing hormone neurons in narcolepsy type 1. Ann. Neurol. 91, 282–288 (2022), 10.1002/ana.26300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Degn M., et al. , Rare missense mutations in P2RY11 in narcolepsy with cataplexy. Brain 140, 1657–1668 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Hor H., et al. , A missense mutation in myelin oligodendrocyte glycoprotein as a cause of familial narcolepsy with cataplexy. Am. J. Hum. Genet. 89, 474–479 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimada M., Miyagawa T., Toyoda H., Tokunaga K., Honda M., Epigenome-wide association study of DNA methylation in narcolepsy: An integrated genetic and epigenetic approach. Sleep 41, zsy019 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Shimada M., et al. , Epigenome-wide association study of narcolepsy-affected lateral hypothalamic brains, and overlapping DNA methylation profiles between narcolepsy and multiple sclerosis. Sleep 43, zsz198 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Seifinejad A., et al. , Molecular codes and in vitro generation of hypocretin and melanin concentrating hormone neurons. Proc. Natl. Acad. Sci. U.S.A. 116, 17061–17070 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tabuchi S., et al. , Conditional ablation of orexin/hypocretin neurons: A new mouse model for the study of narcolepsy and orexin system function. J. Neurosci. 34, 6495–6509 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Honda M., et al. , IGFBP3 colocalizes with and regulates hypocretin (orexin). PLoS One 4, e4254 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies J., et al. , Orexin receptors exert a neuroprotective effect in Alzheimer’s disease (AD) via heterodimerization with GPR103. Sci. Rep. 5, 12584 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okamoto K., et al. , QRFP-deficient mice are hypophagic, lean, hypoactive and exhibit increased anxiety-like behavior. PLoS One 11, e0164716 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romanov R. A., et al. , Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nat. Neurosci. 20, 176–188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang F., et al. , RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 14, 22–29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi T. M., et al. , A discrete neuronal circuit induces a hibernation-like state in rodents. Nature 583, 109–114 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Sakurai T., et al. , Structure and function of human prepro-orexin gene. J. Biol. Chem. 274, 17771–17776 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Moriguchi T., Sakurai T., Takahashi S., Goto K., Yamamoto M., The human prepro-orexin gene regulatory region that activates gene expression in the lateral region and represses it in the medial regions of the hypothalamus. J. Biol. Chem. 277, 16985–16992 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Blouin A. M., et al. , Narp immunostaining of human hypocretin (orexin) neurons: Loss in narcolepsy. Neurology 65, 1189–1192 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crocker A., et al. , Concomitant loss of dynorphin, NARP, and orexin in narcolepsy. Neurology 65, 1184–1188 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vassalli A., Li S., Tafti M., Comment on “Antibodies to influenza nucleoprotein cross-react with human hypocretin receptor 2”. Sci. Transl. Med. 7, 314le312 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Zhang X., et al. , Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. U.S.A. 102, 4459–4464 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elliott E., Ezra-Nevo G., Regev L., Neufeld-Cohen A., Chen A., Resilience to social stress coincides with functional DNA methylation of the Crf gene in adult mice. Nat. Neurosci. 13, 1351–1353 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Fitzsimmons D., et al. , Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promoter. Genes Dev. 10, 2198–2211 (1996). [DOI] [PubMed] [Google Scholar]
- 38.Polansky J. K., et al. , Methylation matters: Binding of Ets-1 to the demethylated Foxp3 gene contributes to the stabilization of Foxp3 expression in regulatory T cells. J. Mol. Med. (Berl). 88, 1029–1040 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maier H., Colbert J., Fitzsimmons D., Clark D. R., Hagman J., Activation of the early B-cell-specific mb-1 (Ig-alpha) gene by Pax-5 is dependent on an unmethylated Ets binding site. Mol. Cell Biol. 23, 1946–1960 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Urbánek P., Wang Z. Q., Fetka I., Wagner E. F., Busslinger M., Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell 79, 901–912 (1994). [DOI] [PubMed] [Google Scholar]
- 41.Kim D. W., et al. , Gene regulatory networks controlling differentiation, survival, and diversification of hypothalamic Lhx6-expressing GABAergic neurons. Commun. Biol. 4, 95 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benes F. M., et al. , Regulation of the GABA cell phenotype in hippocampus of schizophrenics and bipolars. Proc. Natl. Acad. Sci. U.S.A. 104, 10164–10169 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsuki T., et al. , Selective loss of GABA(B) receptors in orexin-producing neurons results in disrupted sleep/wakefulness architecture. Proc. Natl. Acad. Sci. U.S.A. 106, 4459–4464 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elbaz I., Yelin-Bekerman L., Nicenboim J., Vatine G., Appelbaum L., Genetic ablation of hypocretin neurons alters behavioral state transitions in zebrafish. J. Neurosci. 32, 12961–12972 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Middelkoop H. A., et al. , Circadian distribution of motor activity and immobility in narcolepsy: Assessment with continuous motor activity monitoring. Psychophysiology 32, 286–291 (1995). [DOI] [PubMed] [Google Scholar]
- 46.Prober D. A., Rihel J., Onah A. A., Sung R. J., Schier A. F., Hypocretin/orexin overexpression induces an insomnia-like phenotype in zebrafish. J. Neurosci. 26, 13400–13410 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cvetkovic-Lopes V., et al. , Elevated Tribbles homolog 2-specific antibody levels in narcolepsy patients. J. Clin. Invest. 120, 713–719 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dalal J., et al. , Translational profiling of hypocretin neurons identifies candidate molecules for sleep regulation. Genes Dev. 27, 565–578 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thannickal T. C., et al. , Opiates increase the number of hypocretin-producing cells in human and mouse brain and reverse cataplexy in a mouse model of narcolepsy. Sci. Transl. Med. 10, eaao4953 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holm A., et al. , The evolutionarily conserved miRNA-137 targets the neuropeptide hypocretin/orexin and modulates the wake to sleep ratio. Proc. Natl. Acad. Sci. U.S.A. 119, e2112225119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richards E. J., Elgin S. C., Epigenetic codes for heterochromatin formation and silencing: Rounding up the usual suspects. Cell 108, 489–500 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Eden S., Hashimshony T., Keshet I., Cedar H., Thorne A. W., DNA methylation models histone acetylation. Nature 394, 842 (1998). [DOI] [PubMed] [Google Scholar]
- 53.Ogawa Y., et al. , Reduced CSF orexin levels in rats and patients with systemic inflammation: A preliminary study. BMC Res. Notes 15, 221 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hohjoh H., Terada N., Miki T., Honda Y., Tokunaga K., Haplotype analyses with the human leucocyte antigen and tumour necrosis factor-alpha genes in narcolepsy families. Psychiatry Clin. Neurosci. 55, 37–39 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Hohjoh H., et al. , Significant association of a single nucleotide polymorphism in the tumor necrosis factor-alpha (TNF-alpha) gene promoter with human narcolepsy. Tissue Antigens 54, 138–145 (1999). [DOI] [PubMed] [Google Scholar]
- 56.Himmerich H., et al. , Plasma levels of tumor necrosis factor alpha and soluble tumor necrosis factor receptors in patients with narcolepsy. Arch. Intern. Med. 166, 1739–1743 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Chen Y. H., Huang Y. S., Chen C. H., Increased plasma level of tumor necrosis factor alpha in patients with narcolepsy in Taiwan. Sleep Med. 14, 1272–1276 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Zhan S., et al. , Tumor necrosis factor-alpha regulates the Hypocretin system via mRNA degradation and ubiquitination. Biochim. Biophys. Acta 1812, 565–571 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu Y., Starzinski-Powitz A., Guo S. W., Prolonged stimulation with tumor necrosis factor-alpha induced partial methylation at PR-B promoter in immortalized epithelial-like endometriotic cells. Fertil. Steril. 90, 234–237 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Moghadam K. K., et al. , Narcolepsy is a common phenotype in HSAN IE and ADCA-DN. Brain 137, 1643–1655 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Baets J., et al. , Defects of mutant DNMT1 are linked to a spectrum of neurological disorders. Brain 138, 845–861 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kernohan K. D., et al. , Identification of a methylation profile for DNMT1-associated autosomal dominant cerebellar ataxia, deafness, and narcolepsy. Clin. Epigenetics 8, 91 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hunt N. J., Rodriguez M. L., Waters K. A., Machaalani R., Changes in orexin (hypocretin) neuronal expression with normal aging in the human hypothalamus. Neurobiol. Aging 36, 292–300 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Li S. B., et al. , Hyperexcitable arousal circuits drive sleep instability during aging. Science 375, eabh3021 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo J. U., Su Y., Zhong C., Ming G. L., Song H., Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145, 423–434 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu X. S., et al. , Rescue of fragile X syndrome neurons by DNA methylation editing of the FMR1 gene. Cell 172, 979–992.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bories J. C., et al. , Increased T-cell apoptosis and terminal B-cell differentiation induced by inactivation of the Ets-1 proto-oncogene. Nature 377, 635–638 (1995). [DOI] [PubMed] [Google Scholar]
- 68.Crocker A., et al. , Concomitant loss of dynorphin, NARP, and orexin in narcolepsy. Neurology 65, 1184–1188 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Valko P. O., et al. , Increase of histaminergic tuberomammillary neurons in narcolepsy. Ann. Neurol. 74, 794–804 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Scammell T. E., Matheson J. K., Honda M., Thannickal T. C., Siegel J. M., Coexistence of narcolepsy and Alzheimer’s disease. Neurobiol. Aging 33, 1318–1319 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dauvilliers Y., et al. , Hypothalamic immunopathology in anti-Ma-associated diencephalitis with narcolepsy-cataplexy. JAMA Neurol. 70, 1305–1310 (2013). [DOI] [PubMed] [Google Scholar]
- 72.Taylor S. C., et al. , The ultimate qPCR experiment: Producing publication quality, reproducible data the first time. Trends Biotechnol. 37, 761–774 (2019). [DOI] [PubMed] [Google Scholar]
- 73.Bruzzone F., et al. , Anatomical distribution and biochemical characterization of the novel RFamide peptide 26RFa in the human hypothalamus and spinal cord. J. Neurochem. 99, 616–627 (2006). [DOI] [PubMed] [Google Scholar]
- 74.Klug M., Rehli M., Functional analysis of promoter CpG methylation using a CpG-free luciferase reporter vector. Epigenetics 1, 127–130 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Li L. C., Dahiya R., MethPrimer: Designing primers for methylation PCRs. Bioinformatics 18, 1427–1431 (2002). [DOI] [PubMed] [Google Scholar]
- 76.Pham V. N., et al. , Combinatorial function of ETS transcription factors in the developing vasculature. Dev. Biol. 303, 772–783 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kwak S. J., et al. , Zebrafish pax5 regulates development of the utricular macula and vestibular function. Dev. Dyn. 235, 3026–3038 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.