Abstract
Global deployment of vaccines that can provide protection across several age groups is still urgently needed to end the COVID-19 pandemic, especially in low- and middle-income countries. Although vaccines against SARS-CoV-2 based on mRNA and adenoviral-vector technologies have been rapidly developed, additional practical and scalable SARS-CoV-2 vaccines are required to meet global demand. Protein subunit vaccines formulated with appropriate adjuvants represent an approach to address this urgent need. The receptor-binding domain (RBD) is a key target of SARS-CoV-2 neutralizing antibodies but is poorly immunogenic. We therefore compared pattern recognition receptor (PRR) agonists alone or formulated with aluminum hydroxide (AH) and benchmarked them against AS01B and AS03-like emulsion-based adjuvants for their potential to enhance RBD immunogenicity in young and aged mice. We found that an AH and CpG adjuvant formulation (AH:CpG) produced an 80-fold increase in anti-RBD neutralizing antibody titers in both age groups relative to AH alone and protected aged mice from the SARS-CoV-2 challenge. The AH:CpG-adjuvanted RBD vaccine elicited neutralizing antibodies against both wild-type SARS-CoV-2 and the B.1.351 (beta) variant at serum concentrations comparable to those induced by the licensed Pfizer-BioNTech BNT162b2 mRNA vaccine. AH:CpG induced similar cytokine and chemokine gene enrichment patterns in the draining lymph nodes of both young adult and aged mice and enhanced cytokine and chemokine production in human mononuclear cells of younger and older adults. These data support further development of AH:CpG-adjuvanted RBD as an affordable vaccine that may be effective across multiple age groups.
One Sentence Summary:
An aluminum hydroxide:CpG adjuvant improves SARS-CoV-2 RBD immunogenicity in aged mice and activates leukocytes isolated from older humans.
INTRODUCTION
The coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a serious threat to humanity. Rapid deployment of safe and effective vaccines is proving key to reducing morbidity and mortality of COVID-19, especially in high-risk populations, such as older adults (those ≥65 years of age) (1). Vaccine technologies such as mRNA and adenoviral vector vaccines have dramatically accelerated the process of vaccine development and have shown high efficacy in preclinical and clinical studies. They have therefore been granted Emergency Use Authorization and, in one example, licensure by the Food and Drug Administration (2–9). Unfortunately, worldwide access to these vaccines may be limited by the need for ultra-cold storage of mRNA vaccines, cost, and concerns regarding global production capacity and scalability especially in low- and middle-income countries (1). This situation represents a major ethical problem related to inequity and may also promote the emergence of vaccine-resistant SARS-CoV-2 strains due to high infection rates in unvaccinated regions (10). Thus, there is an unmet need to develop additional affordable, easily scalable, and effective vaccine approaches against SARS-CoV-2. To this end, alternative platforms, such as inactivated and protein subunit SARS-CoV-2 vaccines, have entered various stages of clinical development and in some cases have already been deployed at the population level (11–17). These approaches may play an essential role in the global fight against COVID-19 since they utilize well-established technologies, do not require low-temperature storage, and have proven records of safety and effectiveness in various age groups, including young children and older adults.
With the exception of inactivated viruses, most SARS-CoV-2 vaccine candidates aim to target the SARS-CoV-2 spike glycoprotein, as it is required for binding to the human receptor angiotensin-converting enzyme 2 (ACE2) and subsequent cell fusion. In particular, the receptor-binding domain (RBD) of the spike protein plays a key role in ACE2 binding and is targeted by neutralizing antibodies that exert a protective role against SARS-CoV-2 infection (18–20). The RBD is an attractive candidate for a SARS-CoV-2 subunit vaccine and is relatively easy to produce at scale (21, 22); however, it is poorly immunogenic on its own. Structural biology-based vaccine design has been employed to overcome this limitation and has generated encouraging results in preclinical and clinical studies (22–30). A complementary approach to increase the immunogenicity of vaccine antigens is the use of adjuvants, which can enhance antigen immunogenicity by activating receptors of the innate immune system called pattern-recognition receptors (PRRs) or by modulating antigen pharmacokinetics (31, 32). Adjuvant formulations of aluminum salts and PRR agonists have been shown to enhance vaccine immune responses compared to aluminum salts or PRR agonists alone (33). AS04 was the first adjuvant system composed of aluminum salts and a PRR agonist, specifically the TLR4 agonist monophosphoryl lipid A (MPLA), to be included in licensed vaccines for human papillomavirus and hepatitis B vaccines (33). Thus, combinations of aluminum salts and PRR agonists represent a promising adjuvant platform to enhance RBD immunogenicity.
Here, we evaluated several combinations of PRR agonists and aluminum hydroxide (AH) and found that formulating the TLR9 agonists CpG oligodeoxynucleotides with AH and RBD dramatically enhanced immune responses toward RBD in young mice in a prime-boost immunization schedule. The AH:CpG-adjuvanted RBD vaccine also elicited a robust anti-RBD immune response in aged mice, with the administration of an additional boost dose generating an anti-RBD antibody response comparable to young adult mice and providing complete protection from live SARS-CoV-2 challenge. Furthermore, we discovered unique immunological properties of the AH:CpG adjuvant formulation that demonstrated in vitro activation of human leukocytes. Overall, our comprehensive, head-to-head adjuvant comparison study demonstrates that AH:CpG co-adjuvantation can overcome both the poor immunogenicity of RBD and immunosenescence, supporting this approach for the development of a scalable, affordable, and safe global SARS-CoV-2 vaccine suitable for older adults.
RESULTS
Evaluation of multiple AH:PRR agonist formulations in young adult mice identifies AH:CpG as an effective adjuvant.
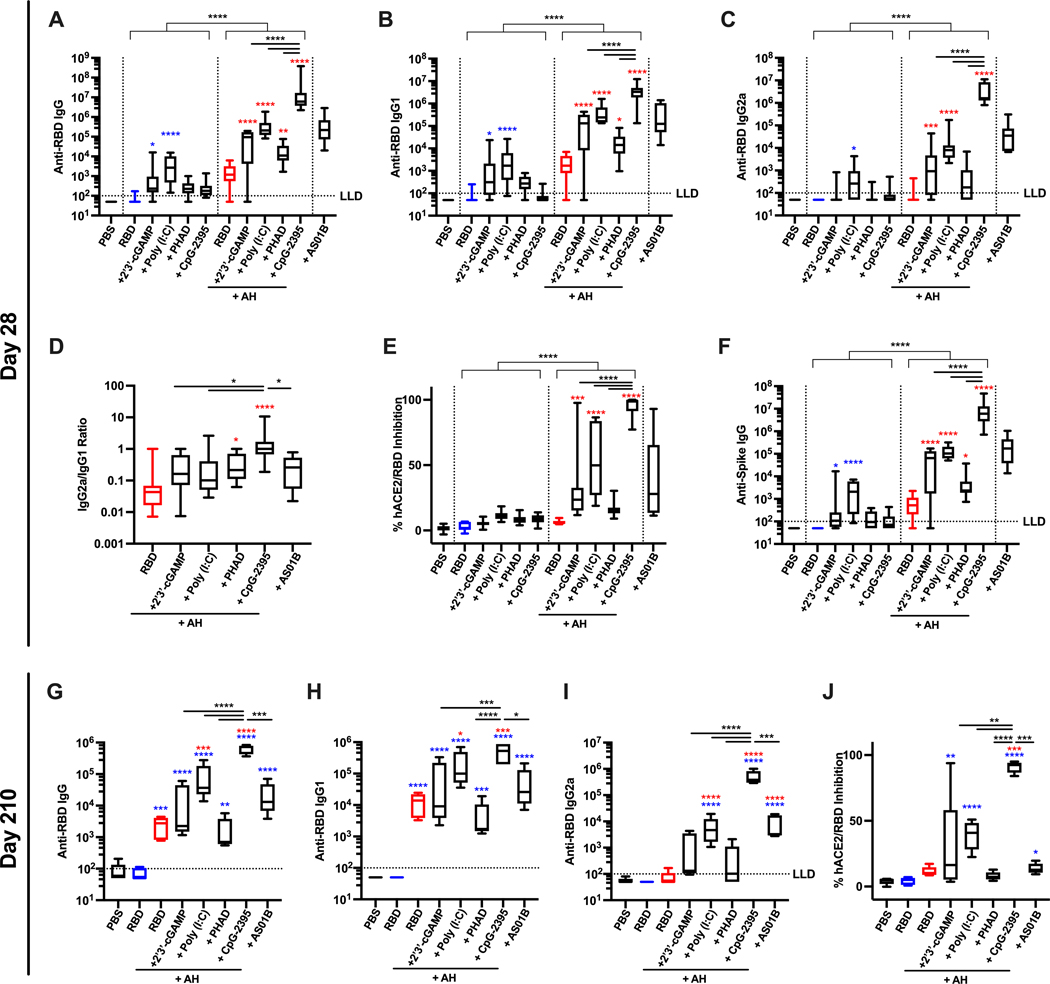
We first evaluated whether distinct AH:PRR agonist formulations can overcome the low immunogenicity of monomeric RBD proteins. To this end, we performed a comprehensive comparison of PRR agonists, including 2’3’-cGAMP, a ligand of the stimulator of interferon (IFN) genes (STING), Poly (I:C) (TLR3 ligand), PHAD (synthetic MPLA, TLR4 ligand), and CpG-ODN 2395 (TLR9 ligand). Each PRR agonist was formulated with and without AH. We also included AS01B (a liposome-based adjuvant containing MPLA and the saponin QS-21) as a clinical-grade benchmark adjuvant with potent immunostimulatory activity. The immunogenicity of vaccine formulations was first evaluated in young (3-month-old) adult mice. Mice were immunized intramuscularly twice with 10 μg of monomeric RBD protein formulated with or without adjuvant, in a two-dose prime-boost regimen (Days 0 and 14). Two weeks after the boost immunization, humoral immune responses were evaluated. AH:PRR agonist formulations enhanced both anti-RBD antibody titers and inhibition of RBD binding to human ACE2 (hACE2) as compared to their respective non-AH adjuvanted formulations (Fig. 1A to E). The antibody response elicited by AH alone was highly skewed to IgG1, with minimal inhibition of hACE2-RBD binding (Fig. 1D and E). Among various AH:PRR agonist formulations, AH:CpG demonstrated the highest induction of total IgG, IgG1, and IgG2a along with a balanced IgG2a:IgG1 ratio (Fig. 1A to D). Furthermore, the AH:CpG formulation significantly enhanced hACE2-RBD binding inhibition compared to all the other AH:PRR agonist formulations (P < 0.0001, Fig. 1E). Antibodies induced by monomeric RBD immunization recognized the native trimeric spike protein, as demonstrated by a binding enzyme-linked immunosorbent assay (ELISA) with a prefusion stabilized form of spike trimer (Fig. 1F). To assess long-term immunogenicity, we then evaluated antibody responses and hACE2-RBD binding inhibition on Day 210 (Fig. 1G to J). Of note, AH:CpG formulation maintained high hACE2-RBD binding inhibition whereas animals vaccinated with other adjuvant formulations exhibited waning immune responses (Fig. 1E and J). To evaluate sex-related differences in vaccine immunogenicity, young adult male and female mice were immunized with RBD proteins formulated with AH and AH:CpG (fig. S1). Similar to female mice, AH:CpG formulation elicited robust anti-RBD humoral immune responses after prime-boost immunization in male mice (fig. S1).
Figure 1. BD formulated with AH:CpG induced robust production of anti-RBD antibodies in young adult mice.
Young adult, 3-month-old BALB/c mice were immunized intramuscularly on Days 0 and 14 with 10 μg of monomeric SARS-CoV-2 RBD protein with indicated adjuvants. Each PRR agonist was administered alone or formulated with aluminum hydroxide (AH). (A to F) Serum samples were collected on Day 28, and (A) Anti-RBD IgG, (B) IgG1, (C) IgG2a, (D) IgG2a:IgG1 ratio, (E) hACE2-RBD inhibition rate, and (F) anti-spike protein IgG were assessed. n=10 per group. Data were combined from two individual experiments. (G to J) Serum samples were collected on Day 210, and (G) Anti-RBD IgG, (H) IgG1, (I) IgG2a and (J) hACE2-RBD inhibition rate were assessed. n=5 per group. Data were analyzed by two-way (A to C, E and F) (AH and PRR agonist) or one-way (D, G to J) ANOVAs followed by post-hoc Tukey’s test for multiple comparisons. *P <0.05, **P <0.01, ***P <0.001, **** P <0.0001. Blue and red colored asterisks indicate comparisons to RBD and AH-adjuvanted RBD groups, respectively. Box-and-whisker plots represent the minimum, first quartile, median, third quartile, and maximum value. LLD, lower limit of detection.
AH:CpG-formulated RBD vaccine is immunogenic in aged mice.
To assess the vaccine response in the context of aging, the immunogenicity of RBD vaccines adjuvanted with AH:PRR agonists was further studied in aged mice (14-month-old). Similar to young mice, the AH:CpG formulation also elicited the highest humoral immune response after prime-boost immunization in aged mice (Fig. 2A to F). Of note, the vaccine adjuvanted with AH:CpG produced significantly higher hACE2-RBD inhibition (P < 0.001) and neutralizing titers (P < 0.01) compared to the vaccine adjuvanted with AS01B, which is known as a potent adjuvant in older adult human (34, 35) (Fig. 2E and F). However, antibody concentrations were generally lower in aged mice, and the magnitude of the immune response of aged mice receiving the AH:CpG vaccine was significantly lower than that of young mice (P < 0.01), suggesting an impaired vaccine response due to immunosenescence in the older adults (fig. S2). To determine whether an additional dose can improve vaccine immunogenicity in aged mice, we administered a second booster dose two weeks after the last immunization. On Day 42 (two weeks after the 2nd boost), an enhancement in humoral responses was observed in AH:PRR agonist formulations (Fig. 2G to L). Notably, a significant enhancement of hACE2-RBD inhibition was observed in aged mice receiving the two-boost AH:CpG vaccination regimen (P = 0.01), with inhibition reaching the degree observed in young mice that had received AH:CpG in a prime-boost regimen (fig. S2). High serum neutralizing antibody titers were observed in the AH:CpG and AS01B adjuvanted groups after the 2nd boost but not in the non-adjuvanted nor AH alone-adjuvanted RBD groups. Assessment of cytokine production by splenocytes isolated from immunized mice and restimulated in vitro with spike peptides demonstrated high Th1 (IFN-γ and interleukin (IL)-2) and low Th2 (IL-4) polarizing cytokine production in the AH:CpG and AS01B groups (Fig. 2M). These results demonstrate that the AH:CpG-adjuvanted RBD vaccine is highly immunogenic in aged mice and that an additional booster dose can further enhance anti-RBD humoral responses to match those observed in young mice.
Figure 2. AH:CpG adjuvant formulation elicited a robust anti-RBD response in aged mice.
Aged, 14-month-old BALB/c mice were immunized intramuscularly on Days 0, 14, and 28 with 10 μg of monomeric SARS-CoV-2 RBD protein with indicated adjuvants. Each PRR agonist was formulated with aluminum hydroxide (AH). Serum samples were collected and analyzed on day 28 prior to the 2nd boost (A to F), and day 42 (G to L). (A, G) Anti-RBD IgG, (B, H) IgG1, (C, I) IgG2a, (D, J) IgG2a:IgG1 ratio, (E, K) hACE2-RBD inhibition rate, and (F, L) neutralizing titers were assessed. n=9 to 10 per group. (M) Splenocytes were collected 2 weeks after the final immunization and stimulated with a SARS-CoV 2 spike peptide pool in the presence of anti-CD28 antibody. After 24 (for IL-2 and IL-4) and 96 (for IFN-γ) hours, supernatants were harvested and cytokine concentrations were measured by ELISA. n=4 to 5 per group. Data were log-transformed and analyzed by one-way ANOVAs followed by post-hoc Tukey’s test for multiple comparisons. *P <0.05, **P <0.01, ***P <0.001, **** P <0.0001. Blue and red colored asterisks indicate comparisons to RBD and AH-adjuvanted RBD groups, respectively. Box-and-whisker plots represent the minimum, first quartile, median, third quartile, and maximum value. LLD, lower limit of detection.
AH:CpG adjuvant formulation induces robust germinal center responses in young and aged mice.
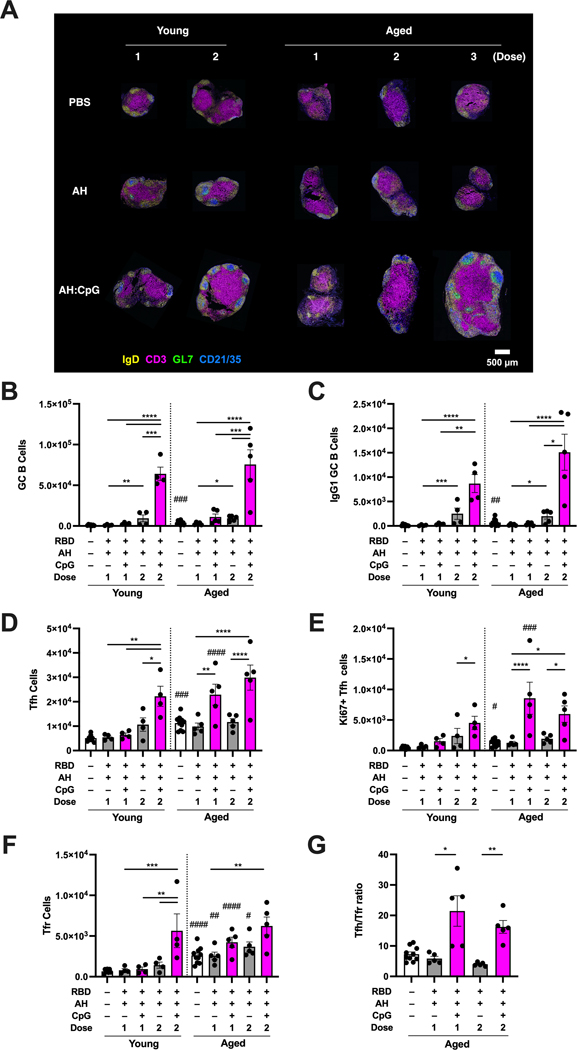
Germinal center (GC) responses in draining lymph nodes (dLNs) are fundamental for the induction of high-quality long-lasting B cell responses, and T follicular helper (Tfh) cells are key regulators of GC responses (36, 37). To further assess how vaccine adjuvants, immunization regimens, and age modulate the GC response, we immunized young and aged mice with either single or a multiple dose regimen of RBD vaccines formulated with AH or AH:CpG. In line with the humoral response profiles observed in previous experiments (Fig. 1A to E, Fig. 2A to L, fig. S2), dLN sections analyzed by immunofluorescence and confocal microscopy revealed a robust induction of GC structures in the draining inguinal LN, as indicated by increased area of GL7+ cells and CD21/35+ follicular dendritic cells (FDCs) surrounded by a mantle of IgD+ naive B cells, after multiple doses of AH:CpG adjuvanted vaccine in young and aged mice (Fig. 3A). To perform quantitative immunophenotyping analysis of dLNs, frequencies and absolute numbers of GC B cells, Tfh cells, and T follicular regulatory (Tfr) cells in dLNs were evaluated by flow cytometry 7 days after the final immunization. Robust GC B cell, class-switched IgG1+ GC B cell, Tfh cell, and proliferating Ki67+ Tfh cell responses were observed in the dLN after 2 doses of AH:CpG in immunized young and aged mice (Fig. 3B to E, fig. S3 and S4). Notably, the absolute number of Tfh cells was higher in aged mice than in young mice after the first dose of AH:CpG immunization (Fig. 3D and E). Defective T cell-dependent B cell responses due to aging have been associated with the overabundance of suppressive Tfr cells and impairment of antigen-specific Tfh cell responses (38). In line with those findings, we observed an increased number of Tfr cells in aged mice compared to young mice in each treatment group (Fig. 3F). Furthermore, a higher Tfh:Tfr ratio was observed in aged mice after AH:CpG immunization compared to AH adjuvanted vaccine, suggesting that overcoming overabundant Tfr cells through expanding Tfh cells is a strategy to enhance vaccine responses in aged mice (Fig. 3G).
Figure 3. AH:CpG adjuvant formulation elicited robust Tfh cell responses in aged mice.
Young (3-month-old) and aged (14-month-old) BALB/c mice were immunized with 10 μg of monomeric SARS-CoV-2 RBD protein with indicated adjuvants following either a 1, 2 or 3 dose (14 days apart) regimen. Draining inguinal lymph nodes were collected. (A) Representative confocal images of whole LN sections collected 10 days after final immunization are shown. LN sections were stained with monoclonal antibodies against IgD (yellow), CD3 (magenta), GL7 (green), and CD21/35 (blue). Scale bar, 500 μm. (B to G) Immune cells from lymph nodes collected 7 days after final immunization were phenotyped by flow cytometry. Absolute numbers of (B) germinal center (GC) B cells (CD19+GL7+FAS+CD4−), (C) IgG1+ GC B cells, (D) T follicular helper (Tfh) cells (CD4+CXCR5+ICOS+Foxp3−CD19−), (E) Ki67+ proliferating Tfh cells, (F) T follicular regulatory (Tfr) cells (CD4+ CXCR5+ICOS+Foxp3+CD19−), and (G) the Tfh:Tfr ratio were assessed. n=4 to 5 per group. Results are presented as mean ± SEM. Statistical significance was determined by two-way repeated measures ANOVA corrected for multiple comparisons. * indicates comparisons of vaccine regimens among same age groups. # indicates comparisons between young and aged mice that received same immunization. */#P <0.05, **/##P <0.01, ***/###P <0.001, ****/#### P <0.0001.
AH:CpG-formulated RBD vaccine protects aged mice from lethal SARS-CoV-2 challenge.
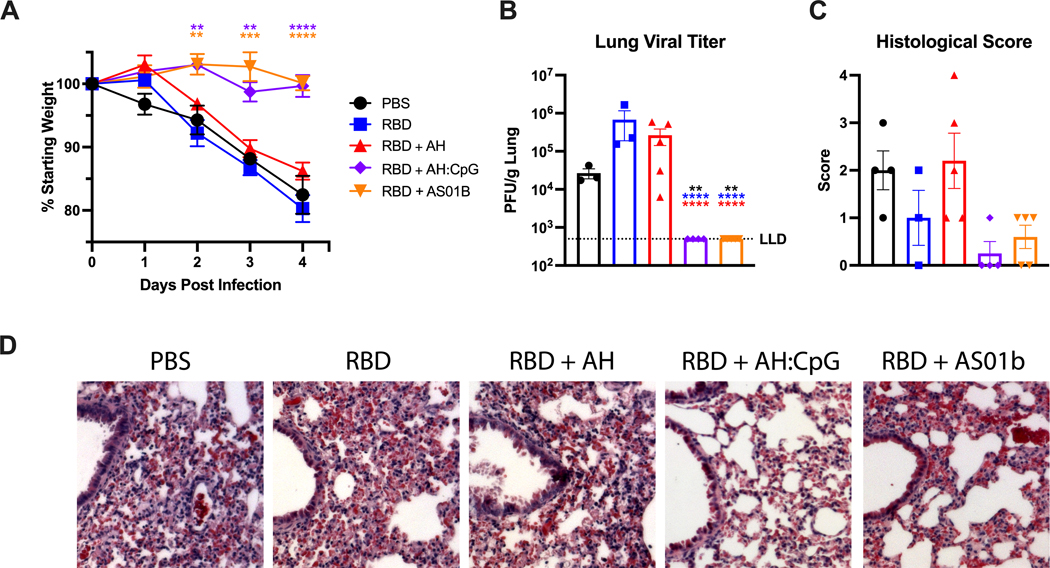
Neutralizing antibodies are key to protecting from SARS-CoV-2 infection. Since RBD formulated with AH:CpG elicited high titers of neutralizing antibodies, we assessed the protection of immunized mice in a challenge model. To this end, we employed the mouse-adapted SARS-CoV-2 MA10 virus strain (39). When tested in young (3-month-old) and aged (14-month-old) BALB/c mice, SARS-CoV-2 MA10 elicited dose-dependent weight loss (fig. S5A and B). Notably, aged mice challenged with 103 plaque forming units (PFU) or more of MA10 exhibited dose-dependent mortality by 4 days post-infection (dpi) (fig. S5C). Considerable titers of infectious virus accompanied with histological manifestations such as interstitial congestion, immune cell infiltration, and edema were observed in the lungs of young and aged mice at 4 dpi (fig. S5D and E). None of the young mice died by 4 dpi, including those that received the highest viral dose, in contrast with aged mice. Next, immunized aged mice were challenged with SARS-CoV-2 MA10 six weeks after the second boost of AH:CpG adjuvanted RBD vaccine. Body weight changes were assessed daily up to 4 dpi when the mice were euthanized for viral titer and histopathology analyses. Aged mice immunized with the AH:CpG and AS01B adjuvanted vaccines showed no weight loss up to 4 dpi, whereas aged mice immunized with non-adjuvanted or AH-adjuvanted RBD showed rapid body weight loss of greater than 10% through 4 dpi (Fig. 4A). Lung tissues were harvested and tested for SARS-CoV-2 viral titer in lung. No detectable live virus was observed in lung tissues of the AH:CpG and AS01B adjuvanted groups, whereas viral titers were detectable in the vehicle, non-adjuvanted, and AH-adjuvanted groups (Fig. 4B). Histopathological analysis conducted in lung tissues further confirmed the reduced SARS-CoV-2 infection in aged animals vaccinated with AH:CpG and AS01B adjuvants (Fig. 4C and D).
Figure 4. AH:CpG-adjuvanted vaccine protected aged mice from SARS-CoV-2 challenge.
Aged, 14-month-old BALB/c mice were immunized as in Fig. 2. On Day 70 (6 weeks post 2nd boost), mice were challenged intranasally with 103 PFU of mouse-adapted SARS-CoV-2 (MA10). (A) Body weight changes were assessed daily up to 4 days post-infection. Data are presented as mean ± SEM with body weights shown for surviving mice at each time-point (one mouse in the RBD group died at 4 days post infection). Data were analyzed by one-way ANOVA followed by Dunnett’s Test for comparisons to the PBS group. (B) Viral titers in lung homogenates at 4 days post SARS-CoV-2 challenge are shown. Results are presented as mean ± SEM. Data were analyzed by one-way ANOVA followed by post-hoc Tukey’s test for multiple comparisons. **P <0.01, **** P <0.0001. Black, blue, and red colored asterisks respectively indicate comparisons to PBS, RBD, and RBD + aluminum hydroxide (AH) groups. LLD, lower limit of detection. (C) Lung interstitial inflammation was evaluated and converted to a score of 0 to 4 with 0 being no inflammation and 4 being most severe. (D) Representative H&E stained lung images at 4 days post-challenge are shown. Scale bar, 500 μm. n=4 to 5 animals per group.
AH:CpG-formulated RBD and spike mRNA vaccines elicit comparable concentrations of neutralizing antibodies against wild type SARS-CoV-2 and variants.
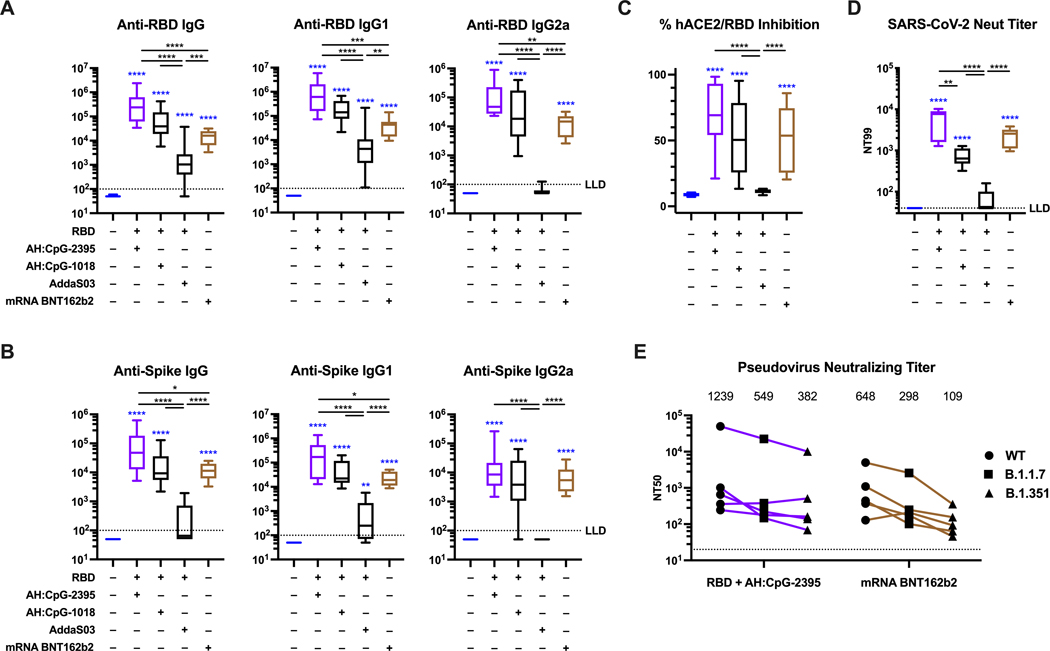
Recently, it has been reported that SARS-CoV-2 mRNA vaccines are more immunogenic than RBD adjuvanted with oil-in-water emulsions (40). To assess whether this is a general feature of RBD protein vaccines, we used the clinical-grade authorized BNT162b2 spike mRNA vaccine (Pfizer-BioNTech) as a benchmark and compared it to RBD formulated with AddaS03 (a commercially available version of the oil-in-water emulsion AS03) and to AH:CpG in aged mice. Along with class C CpG-2395, we also tested class B CpG-1018, which is included in the Heplisav-B vaccine and has also been tested in combination with spike or RBD and AH in SARS-CoV-2 studies, including human vaccine trials (12, 16, 41). In accordance with previously published data, the mRNA vaccine was highly immunogenic, whereas RBD formulated with AddaS03 failed to induce robust neutralizing antibodies (Fig. 5A to D). Of note, both AH:CpG formulations elicited concentrations of anti-RBD (Fig. 5A), anti-spike protein (Fig. 5B) and neutralizing antibodies (Fig. 5C and D) comparable to or greater than those elicited by mRNA vaccination.
Figure 5. AH:CpG-adjuvanted RBD vaccines and an authorized spike mRNA vaccine elicit comparable concentrations of neutralizing antibodies in aged mice.
Aged, 14-month-old BALB/c mice were immunized intramuscularly on Days 0 and 14 with monomeric SARS-CoV-2 RBD protein with indicated adjuvants or with BNT162b2 spike mRNA vaccine as described in Methods. Serum samples were collected and analyzed on day 28. (A) Anti-RBD binding ELISA, (B) anti-spike protein binding ELISA, (C) hACE2-RBD inhibition rate, and (D) SARS-CoV-2 virus neutralizing (Neut) titer were assessed. n=9 to 10 per group. Data were combined from two individual experiments and analyzed by one-way ANOVAs followed by post-hoc Tukey’s test for multiple comparisons. (E) Pseudovirus neutralizing titers against wild-type spike or the B.1.17 or B.1.351 spike variants were assessed. n=5 per group. The numbers indicate geometric mean titers (GMT). Each symbol represents an animal. *P <0.05, **P <0.01, ***P <0.001, **** P <0.0001. Blue colored asterisks indicate comparisons to the PBS group. Box-and-whisker plots represent the minimum, first quartile, median, third quartile, and maximum value. LLD, lower limit of detection.
SARS-CoV-2 variants of concern (VOCs) such as B.1.1.7 (alpha variant) and B.1.351 (beta variant) have emerged and have exhibited reduced neutralization from serum samples of convalescent or vaccinated individuals (42–45). A recent report showed that the mRNA BNT162b2 vaccine maintained its effectiveness against severe COVID-19 with the B.1.351 variant at greater than 90% (46). We therefore evaluated whether RBD + AH:CpG and mRNA BNT162b2 vaccines elicit neutralizing antibodies against these VOCs. As expected, we observed reduced titers against the variants, especially against B.1.351 (Fig. 5E). The neutralization titers of RBD + AH:CpG decreased by 3.2-fold against B.1.351, and the mRNA BNT162b2 decreased by 6.0-fold. Neutralizing titers against the B.1.351 were comparable between RBD + AH:CpG and mRNA BNT162b2 with geometric mean titers (GMT) of 382 and 109, respectively.
Variant SARS-CoV-2 RBD vaccines formulated with AH:CpG confer broad neutralization in aged mice.
As described previously, currently available vaccines, such as the mRNA BNT162b2 vaccine, have demonstrated high vaccine effectiveness against severe COVID-19 due to VOCs such as the beta (B.1.351) variant (46). However, lower vaccine effectiveness against B.1.351 infection compared to the original strain has been reported, along with increased breakthrough infections among those who had received two doses of BNT162b2 vaccines (46–48). Therefore, we developed a modified RBD protein derived from B.1.351 variant (B.1.351-RBD) and evaluated whether the AH:CpG adjuvant can enhance broad neutralization responses when formulated with variant antigens. High abundance of binding antibodies against wild-type (WT)-RBD and B.1.351-RBD were observed in both young and aged mice after two doses of AH:CpG-adjuvanted vaccines formulated with WT-RBD, B.1.351-RBD, or the 1:1 mixture of the two antigens (bivalent-RBD) (Fig. 6A and B). Next, we evaluated neutralization titers against WT and B.1.351 strains. Although we observed 3.3- to 5.2-fold reduced titers against the B.1.351 compared to the WT strain in mice immunized with WT-RBD + AH:CpG, neutralizing titers against the B.1.351 were comparable to WT strain in mice which received either B.1.351-RBD or bivalent-RBD formulated with AH:CpG (Fig. 6C and D). Thus, these data suggest that the AH:CpG adjuvant can elicit high concentrations of neutralizing antibodies against VOCs when formulated with modified antigens.
Figure 6. Variant SARS-CoV-2 RBD vaccines formulated with AH:CpG confer broad neutralization in aged mice.
Young (3-month-old) and aged (14-month-old) BALB/c mice were immunized intramuscularly on days 0 and 14 with 10 μg of monomeric SARS-CoV-2 RBD protein derived from wild type (WT-RBD), B.1.351 beta variant (B.1.351-RBD), or a 1:1 mixture (Bivalent-RBD) with indicated adjuvants. Serum samples were collected on day 28. (A and B) Anti-WT and B.1.351-RBD binding ELISA were assessed in young (A) and aged (B) mice. n=10 per group. Box-and-whisker plots represent the minimum, first quartile, median, third quartile, and maximum value. (C and D) Pseudovirus neutralizing titers (NT50) values against wild-type (circle) and B.1.351 (triangle) variant SARS-CoV-2 strains were assessed. n=5 per group. Fold differences in titers are shown. The numbers indicate GMT. Each symbol represents an animal. Data were analyzed by one-way ANOVAs corrected for multiple comparisons. **P <0.01, **** P <0.0001. LLD, lower limit of detection.
Innate immune signaling potentiated by AH:CpG formulation is preserved in aged mice.
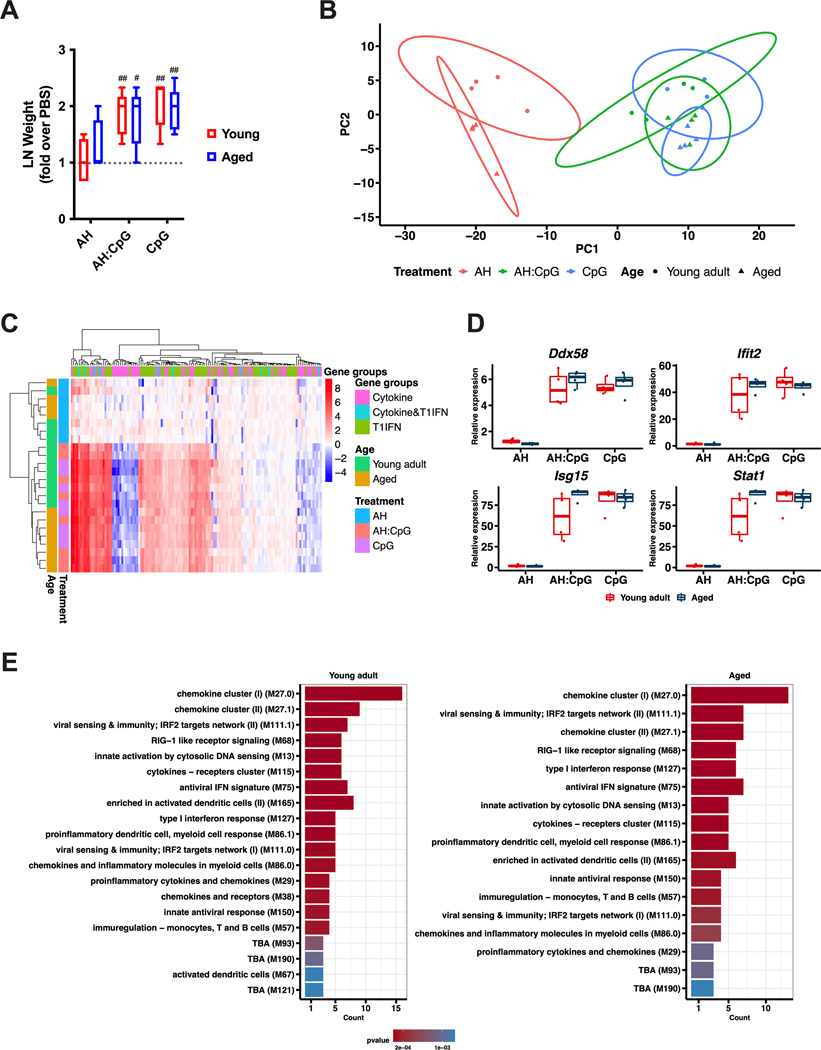
LNs are critical sites for the interaction between innate and adaptive immune systems and orchestrate the development of vaccine immune responses (49, 50). Specifically, activation of the innate immune system can induce a rapid response in the LN characterized by LN expansion, which is driven by lymphocyte accrual and expression of proinflammatory molecules (51, 52). To gain further insights into the mechanism of action of the AH:CpG formulation, we collected dLNs 24 hours post-injection of AH:CpG or either adjuvant alone. CpG and AH:CpG induced comparable dLN expansion in both age groups (Fig. 7A). To characterize the molecular events associated with these treatments further, RNA isolated from dLNs after injection of vehicle, CpG, or AH:CpG was subjected to a quantitative real-time PCR array comprised of 157 genes related to cytokines, chemokines, and type 1 IFN responses. Principal component analysis and hierarchical clustering analysis demonstrated a marked separation between AH and CpG-containing treatments, whereas similar patterns were observed between groups treated with AH:CpG and CpG alone in both age groups (Fig. 7B and C). Generalized linear model analysis comparing gene expressions of the significantly differentially expressed genes (Benjamini-Hochberg corrected, adjusted P ≤ 0.05) after AH, CpG, and AH:CpG treatments further revealed over-representation of similar pathways between young and aged mice (Fig. 7D and E). These results suggest that CpG and AH:CpG activate similar pathways in young and aged mice to elicit an innate immune response in the dLN.
Figure 7. AH:CpG induced comparable innate immune responses in lymph nodes isolated from young and aged mice.
Young and aged mice were subcutaneously injected with aluminum hydroxide (AH), CpG, or AH:CpG. Twenty-four hours later, draining lymph nodes (dLNs) were collected and RNA was extracted. (A) Weights of dLNs were measured and expressed as fold over contralateral, PBS-injected dLNs. n=5 per group. # and ## respectively indicate P <0.05 and 0.01 when comparing each group against the value 1 (which represents the contralateral control sample expressed as fold). (B to E) RNA isolated from dLNs was subjected to a quantitative real-time PCR array comprised of 157 genes related to cytokines, chemokines, and type 1 IFN (T1IFN) responses. n=4 animals per group. Principal component analysis (B) and unsupervised hierarchical clustering (C) revealed major differences between treatments and highlighted the marked difference between AH and CpG-containing treatments. (D) A generalized linear model comparing treatment and age with each gene was performed. The top 4 significant genes were selected and plotted with their relative expression values by age and treatment. Statistical analysis of the plots employed the Kruskal-Wallis test to compare mean differences across groups and Wilcoxon test to compare between ages, followed by Benjamini-Hochberg correction for multiple comparisons. (E) Over-representation analysis of differentially expressed genes (Benjamini-Hochberg corrected, adjusted P ≤ 0.05) using the blood transcriptional modules (89) was performed from the significant gene results after the generalized linear model by treatment. The top 20 modules are summarized per age.
AH:CpG synergistically enhances proinflammatory cytokines from human PBMCs and MoDC-dependent memory T cell activation.
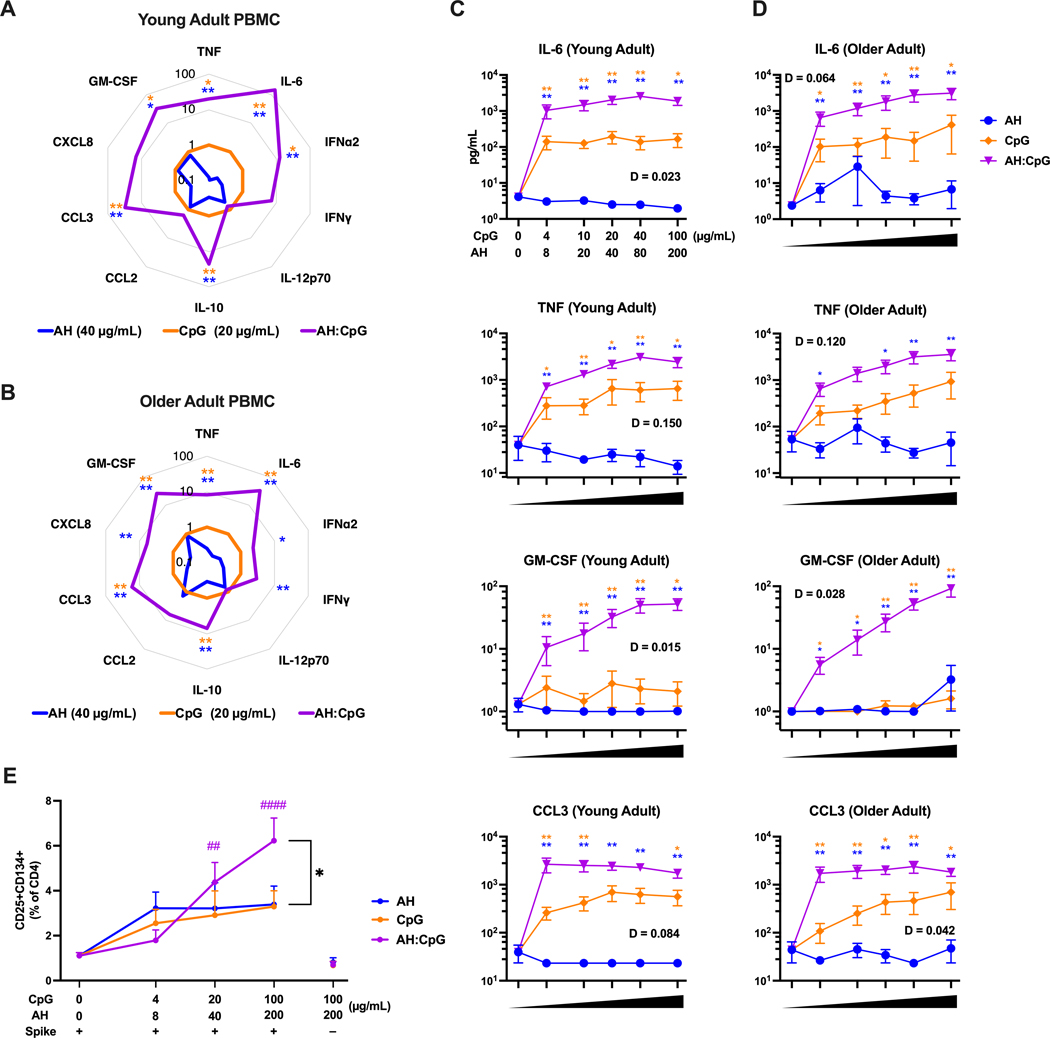
In order to assess the translational relevance of an adjuvanted vaccine formulations, especially those indicated for use in the very young and older adults, it is key to confirm its ability to activate human immune cells in an age-dependent manner. To this end, we stimulated human peripheral blood mononuclear cells (PBMCs) isolated from young adults (18 to 40 years old) and older adults (≥65 years old) with CpG, AH, and the admixed AH:CpG formulation and measured cytokine and chemokine production. Whereas AH induced limited or no cytokine production, both CpG alone and AH:CpG activated young and older adult PBMCs in a concentration-dependent manner (Fig. 8A to D, fig. S6). PBMCs of both age groups treated with AH:CpG produced higher concentrations of various proinflammatory cytokines and chemokines than those treated with CpG alone (Fig. 8A to D). Of note, CpG and AH synergistically induced IL-6, IL-10, tumor necrosis factor (TNF), CCL3, and granulocyte-macrophage colony-stimulating factor (GM-CSF) production in both young and older adult PBMCs, as defined mathematically (D value, see Methods) (Fig. 8C and D, fig. S6). There were no consistent sex-related differences regarding proinflammatory cytokines and chemokines production in either age group (fig. S7).
Figure 8. AH:CpG synergistically enhanced proinflammatory cytokine production from human PBMCs and enhanced MoDC-dependent memory T cell activation.
(A to D) Human PBMCs collected from young adults (A, C) and older adults (B, D) were cultured in vitro for 24 hours with CpG alone, aluminum hydroxide (AH) alone, or a combination of both. Supernatants were collected for multiplexing bead array. n=6 per age group. (A and B) Radar plot analysis presented as a fold-change over the CpG alone group for the 20 μg/mL CpG and 40 μg/mL AH conditions. (C and D) Cytokine and chemokine concentrations are shown at the indicate doses of CpG and AH. Results are presented as mean ± SEM. Unpaired Mann-Whitney tests were applied at each concentration. Blue and yellow colored asterisks indicate comparisons of AH:CpG formulation to AH and CpG alone groups, respectively. Degree of synergy was calculated using an adapted Loewe definition of additivity (D <1: synergy, D=1: additivity, D >1: antagonism). (E) Human monocyte-derived dendritic cells from young adults were stimulated with spike protein and indicated adjuvants and co-cultured with CD4+ T cells for 5 days. The percent of CD25+CD134+ CD4+ T cells of total CD4+ T cells was assessed. n=5. Results are presented as mean ± SEM. Statistical significance was determined by two-way repeated measures ANOVA corrected for multiple comparisons. * indicates comparisons among AH, CpG, and AH:CpG at the same concentration. # indicates comparisons among same adjuvant groups to the baseline control (antigen only). Comparisons among experimental groups are indicated by color code. * P <0.05, **/ ## P <0.01, #### P <0.0001.
To detect the effect of CpG and AH adjuvants on adaptive immunity in a human in vitro system, we developed a human monocyte-derived dendritic cell (MoDC) and T cell co-culture system. Briefly, peripheral blood was collected from SARS-CoV-2 vaccinated young adult donors, and monocytes were isolated and differentiated into MoDCs. After treatment with recombinant spike protein and adjuvants as indicated (Fig. 8E), MoDCs were then cocultured with autologous CD4+ T cells for 5 days, and activated spike protein-specific T cells were quantified by co-expression of CD25 (IL-2 receptor α) and CD134 (OX40 receptor). CD25 is present on activated memory T cells, whereas CD134 plays a critical role in Th-mediated immune responses in vivo. This system enabled us to detect the effect of adjuvants on antigen-presenting cells by measuring activation of antigen-specific T cells. In agreement with murine in vivo data, we observed that AH:CpG significantly enhanced activation of antigen-specific T cells compared to CpG or AH alone (P = 0.02, Fig. 8E). Collectively, in human in vitro systems, AH:CpG formulation elicited proinflammatory cytokine and chemokine production from PBMCs, and enhanced MoDC-dependent memory T cell activation.
DISCUSSION
The risk of COVID-19-related morbidity and mortality increases with age (53, 54). Currently authorized SARS-CoV-2 vaccines have proven effective at preventing severe COVID-19 (2–4). Nevertheless, there is still the need to develop affordable and accessible vaccines that can provide protection across several age groups, especially for low- and middle-income countries (1, 10, 55). Protein subunit vaccines formulated with appropriate adjuvants represent a promising strategy to address this urgent need. Here, we performed a comprehensive head-to-head comparison of multiple adjuvants in age-specific in vivo and ex vivo animal models, along with age-specific human in vitro screening. We determined an appropriate adjuvant for a SARS-CoV-2 RBD vaccine in young and older adults, focusing on the innate and humoral immune response reported to align best with known correlates of protection (56, 57). We found that the AH:CpG adjuvant formulation enhances anti-RBD neutralizing antibody titers and type 1 immunity (IgG2a switching, Th1 polarization) and induced robust Tfh responses in dLNs in both age groups. Aged mice immunized with AH:CpG were protected from mouse-adapted SARS-CoV-2 challenge. Of note, RBD adjuvanted with AH:CpG elicited concentrations of neutralizing antibodies comparable to the clinical-grade BNT162b2 spike mRNA vaccine. The translational relevance of our findings is also highlighted by the synergistic activation of human PBMCs and enhanced MoDC-dependent memory T cell activation upon stimulation with AH:CpG. Overall, our results expand upon recent preclinical and clinical studies on the enhanced immunogenicity of spike protein formulated with AH:CpG by showing that a vaccine composed of RBD and AH:CpG can also induce a robust anti-SARS-CoV-2 immune response across different age groups. Since an RBD antigen is amenable to high-yield manufacturability (58–60), our study also supports the development of RBD formulated with AH:CpG as an affordable and accessible vaccine.
Among various AH:PRR agonist formulations, AH:CpG elicited the highest immune responses in both young and aged mice. We observed that vaccine immune responses were generally lower in aged mice than in young adult mice, even in the group receiving RBD formulated with AH:CpG. Although the lower concentrations of anti-RBD antibodies observed in aged mice are likely sufficient for protection, we found that an additional booster dose in the aged mice overcame the observed age-dependent reductions in vaccine response and protected these mice from SARS-CoV-2 challenge. We employed AH, which has been used for over 90 years with a firmly established record of safety and efficacy (33) and AS01B, which recently demonstrated excellent adjuvant effects among older adults (34, 35), as “benchmarking” adjuvants to compare the exploratory adjuvant formulations with more established adjuvants. In this context, we demonstrated that the AH:CpG adjuvanted vaccine was superior to a vaccine adjuvanted only with AH and was not inferior to AS01B. In the context of the aged mice prime-boost setting, AH:CpG-adjuvanted SARS-CoV-2 RBD outperformed AS01B with respect to functional anti-RBD inhibition (Geometric mean with SD, 57±2% vs. 14±3%) and neutralizing antibodies titers (2344 ± 7 versus 117± 4).
In this study, AH:CpG dramatically enhanced vaccine immune responses compared to vaccines adjuvanted with AH or CpG alone in both young and aged mice. AH:PRR agonist formulations have shown promising adjuvanticity in preclinical models, and AS04 (a formulation of aluminum salts and MPLA) is employed in several licensed vaccines (33). Although the precise mechanism of action of AH:PRR agonist formulations has not been completely uncovered and is potentially influenced by the degree of adsorption of PRR ligands onto AH, the effects of these formulations are at least in part mediated by enhanced activation of innate immune cells at the injection site (32, 61). In our murine model, we also showed that AH:CpG and CpG alone induce comparable proinflammatory gene expression profiles in dLNs. To gain additional mechanistic insight and increase the translational relevance of our findings, we tested the activity of AH:CpG on human PBMCs isolated from young adults and older individuals and found that this adjuvant formulation synergistically enhances cytokine and chemokine production compared to AH or CpG. We also employed a human MoDC:T cell co-culture system to study the effect of adjuvants on adaptive immunity in an in vitro system and demonstrated that the AH:CpG formulation markedly enhanced MoDC-dependent memory T cell activation. These results might be explained by either synergistic activation by AH and CpG of distinct molecular pathways or by adsorption of CpG onto AH leading to the formation of macromolecular complexes that are more efficiently internalized or that promote enhanced TLR9 activation. Further work is required to define the underlying molecular mechanism of action of AH:CpG in vivo and in vitro.
The rationale for use of a synthetic TLR9 agonist CpG as an adjuvant for SARS-CoV-2 subunit vaccine is multi-fold. First, CpG has been used as a vaccine adjuvant in licensed vaccines with well-known mechanisms, substantial safety data, and confirmed effectiveness (62, 63). Second, CpG has demonstrated adjuvant effects in older adults. CpG enhanced vaccinal antigen immunogenicity in aged murine and porcine models (64–69). Several human trials demonstrated that older individuals had a higher seroprotection rate when immunized with the CpG-adjuvanted hepatitis B vaccine compared to the conventional alum-adjuvanted vaccine (70, 71). Further, AH:CpG-adjuvanted SARS-CoV-2 spike protein vaccines have demonstrated safety, immunogenicity, and efficacy in young adult animal models (57, 72, 73), and in a human clinical study involving an older population (12). Finally, Biological E. Limited has recently completed early phase (1 and 2) trials of a AH:CpG-adjuvanted SARS-CoV-2 RBD protein vaccine candidate (trial # CTRI/2020/11/029032) which was intended for low- and middle-income countries and are currently advancing through manufacturing (17). In September 2021, The Drugs Controller General of India (DCGI) approved plans for Biological E to conduct a phase 3 trial in adults and a phase 2/3 trial in children and adolescents (CTRI/2021/06/034014 and CTRI/2021/08/036074) of its vaccine, Corbevax, with plans for its emergency authorization before the end of 2021 (74).
CpG is classified into 4 major classes, with distinct activation profiles of human cells (75). Class B CpG-1018 has been extensively evaluated in clinical trials. We observed that CpG-1018 and the class C CpG-2395 formulated with AH elicit comparable concentrations of neutralizing antibodies, resulting in adjuvanted RBD formulations that were both comparable to the clinical-grade BNT162b2 spike mRNA vaccine. Studies of TLR7/8 agonists as precision adjuvants with robust activity in early life (76), including in enhancing spike protein immunogenicity in the young (77), further support the use of adjuvants to enhance vaccine immunogenicity in target populations. Together, and in light of our results in the older individuals, these studies suggest that precision adjuvant approaches hold substantial promise to generate scalable adjuvanted SARS-CoV-2 vaccine formulations that do not require freezing and afford robust protection to vulnerable populations across the lifespan (78).
Recently, several SARS-CoV-2 VOCs have emerged harboring mutations in the RBD region and showing various degrees of reduced neutralization by serum samples obtained from convalescent or vaccinated individuals (42–44). Current data suggest that an additional booster dose of the original vaccine is useful to enhance the durability of immunity and reduce immune evasion due to VOCs in vulnerable populations such as older adults (79, 80). However, it is possible that booster doses that account for mutations in the spike protein will be required in order to achieve complete immunity against such variants (81). Several vaccines composed of multiple protein antigens adsorbed onto aluminum salts alone or co-formulated with MPLA have been produced (61, 82). We demonstrated that, when formulated with B.1.351-RBD or bivalent-RBD (mixture of WT- and B.1.351-RBD), the AH:CpG adjuvant formulation robustly enhanced induction of neutralizing antibody against B.1.351, suggesting that AH:CpG can be used as an adjuvant platform for multi-valent vaccines composed of RBDs from multiple SARS-CoV-2 strains or potentially from other coronaviruses.
Our study features several strengths, including (i) defining a combination adjuvantation system based on the common AH backbone that demonstrated mathematical synergy in its ability to activate human mononuclear cells; (ii) accounting for age-specific immunity that can play major roles in vaccine immunogenicity and is often overlooked in vaccine discovery; (iii) accounting for species-specificity by assessing the activity of the adjuvant formulation in human in vitro and murine in vivo platforms; (iv) testing the ability of the adjuvanted formulation to protect in a SARS-CoV-2 challenge model; and (v) benchmarking to the authorized BNT162b2 spike mRNA vaccine to place our studies in context. As with any research our study also has some limitations, including that (i) we performed in vivo analysis only in mice, establishing the need for future translational research in additional animal models and humans, (ii) all adjuvants and antigens were compared in a single dose and further analysis should be performed in multiple doses to evaluate both efficacy and reactogenicity, and (iii) the analysis of the ratio of Tfh:Tfr cells relies on surface markers which, although informative for the study of immune ontogeny, do not fully account for antigen specificity. Nevertheless, since we used standard doses of adjuvants and antigens in mouse systems (1/30 and 1/18th of the human dose for CpG (12) and BNT162b2 (3) respectively, to compare the CpG-adjuvanted RBD subunit vaccine to the mRNA vaccine), it should be underscored that the results in this study hold promise from a translational perspective.
Overall, the current study aimed to evaluate an optimal adjuvant formulation to enhance immunogenicity and efficacy of RBD-based subunit vaccines in older adults, which is otherwise reduced as an effect of aging. We demonstrated that an AH:CpG adjuvant formulation induces potent anti-RBD responses in both young and aged mice and overcomes both the poor immunogenicity of the antigen and impaired immune responses in the aged. We discovered unique immunological properties of the AH:CpG adjuvant formulation that demonstrated in vitro synergistic enhancement of human leukocyte activation. These observations indicate that formulating RBD with AH:CpG represents a promising approach to develop a practical, thermostable, scalable, effective, and affordable vaccine that may be effective across multiple age groups and could potentially incorporate multiple RBD proteins to achieve cross-strain protection.
MATERIALS AND METHODS
Study design.
The aim of this study was to assess optimal combinations of RBD antigen and adjuvants in pre-clinical models that take age-dependent vaccine immune responses and COVID-19 susceptibility into account. To this end, we used age-specific mouse in vivo and human in vitro models. Sample size and age criteria were chosen empirically based on the results of previous studies. Mouse experiments aimed to include in total 10 mice per group and were combined from two individual experiments. Mice were randomly assigned to different treatment groups. In order to assess the translational relevance and potential mechanism of an adjuvant formulation, we designed a human in vitro study with peripheral blood collected from healthy young adults, aged 18 to 40 years (n = 6), and older participants, aged ≥ 65 years (n = 6), with approval from the Ethics Committee of the Boston Children’s Hospital (protocol number X07–05-0223) and Institutional Review Board of Brigham and Women’s Hospital, Boston (protocol number 2013P002473). All participants signed an informed consent form prior to enrollment. Investigators were not blinded. No data outliers were excluded.
Animals.
Female and male, 3-month-old BALB/c mice were purchased from Jackson Laboratory. Female, 12 to 13-month-old BALB/c mice purchased from Taconic Biosciences were used for aged mice experiments. Mice were housed under specific pathogen-free conditions at Boston Children’s Hospital, and all the procedures were approved under the Institutional Animal Care and Use Committee (IACUC) and operated under the supervision of the Department of Animal Resources at Children’s Hospital (ARCH) (Protocol number 19–02-3897R). At the University of Maryland School of Medicine, mice were housed in a biosafety level 3 (BSL3) facility for all SARS-CoV-2 infections with all the procedures approved under the IACUC (Protocol number #1120004) to MBF.
SARS-CoV-2 spike protein and RBD expression and purification.
Full-length SARS-CoV-2 spike glycoprotein (M1-Q1208, GenBank MN90894) and RBD constructs (amino acid residues R319-K529, GenBank MN975262.1), both with an HRV3C protease cleavage site, a TwinStrepTag and an 8XHisTag at C-terminus were obtained from Barney S. Graham (NIH Vaccine Research Center) and Aaron G. Schmidt (Ragon Institute), respectively. Mammalian expression construct for RBD domain (N331-P528) of B.1.351 (beta variant), carrying K417N, E484K, and N501Y mutations, was synthesized (Gene Universal Inc.). These mammalian expression vectors were used to transfect Expi293F suspension cells (Thermo Fisher Scientific) using polyethylenimine (Polysciences). Cells were allowed to grow at 37°C, 8% CO2 for an additional 5 days before harvesting for purification. Protein was purified in phosphate-buffered saline (PBS) (pH 7.4) from filtered supernatants by using either StrepTactin resin (IBA) or Cobalt-TALON resin (Takara). Affinity tags were cleaved off from eluted protein samples by HRV 3C protease, and tag-removed proteins were further purified by size-exclusion chromatography using a Superose 6 10/300 column (Cytiva) for full-length spike protein and a Superdex 75 10/300 Increase column (Cytiva) for RBD domain in a PBS buffer (pH 7.4).
Adjuvants and immunization.
The adjuvants and their doses used were: Alhydrogel adjuvant 2% (100 μg), 2’3’-cGAMP (10 μg), Poly (I:C) HMW (50 μg), CpG-ODN 2395 (50 μg), AddaS03 (25 μL) (all from InvivoGen), CpG-ODN 1018 (50 μg, 5’ TGA CTG TGA ACG TTC GAG ATG A 3’) (Integrated DNA Technologies), PHAD (50 μg) (Avanti Polar Lipids), and AS01B (40 μL) (obtained from the Shingrix vaccine, GSK Biologicals SA). Mice were injected with 10 μg of recombinant monomeric SARS-CoV-2 RBD protein with or without adjuvants. Each PRR agonist was formulated with and without aluminum hydroxide. Mock treatment mice received phosphate-buffered saline (PBS) alone. BNT162b2 spike mRNA vaccine (Pfizer-BioNTech) was obtained as residual volumes in used vials from the Boston Children’s Hospital employee vaccine clinic, strictly using material that would only otherwise be discarded, and was used within 6 hours from the time of reconstitution. BNT162b2 suspension (100 μg/mL) was diluted 1:3 in PBS, and 50 μL (1.67 μg) was injected. Injections (50 μL) were administered intramuscularly in the caudal thigh on days 0 and 14 (both age groups) and day 28 (aged mice only, where relevant). Blood samples were collected 2 weeks post-immunization.
ELISA.
RBD- and spike protein-specific antibody concentrations were quantified in serum samples by ELISA by modification of a previously described protocol(83). Briefly, high-binding flat-bottom 96-well plates (Corning) were coated with 50 ng per well RBD or 25 ng per well spike protein and incubated overnight at 4 °C. Plates were washed with 0.05% Tween 20 PBS and blocked with 1% bovine serum albumin (BSA) PBS for 1 hour at room temperature. Serum samples were serially diluted 4-fold from 1:100 up to 1:1.05 × 108 and then incubated for 2 hours at room temperature. Plates were washed three times and incubated for 1 hour at room temperature with horseradish peroxidase (HRP)-conjugated anti-mouse IgG, IgG1, IgG2a, or IgG2c (Southern Biotech). Plates were washed five times and developed with tetramethylbenzidine (1-Step Ultra TMB-ELISA Substrate Solution, Thermo Fisher Scientific, for RBD-ELISA, and BD OptEIA Substrate Solution, BD Biosciences, for spike protein ELISA) for 5 minutes, then stopped with 2 N H2SO4. Optical densities (ODs) were read at 450 nm with SpectraMax iD3 microplate reader (Molecular Devices). End-point titers were calculated as the dilution that emitted an optical density exceeding a 3× background. An arbitrary value of 50 was assigned to the samples with OD values below the limit of detection for which it was not possible to interpolate the titer.
hACE2-RBD inhibition assay.
The hACE2-RBD inhibition assay employed a modification of a previously published protocol(84). Briefly, high-binding flat-bottom 96-well plates (Corning) were coated with 100 ng per well recombinant human ACE2 (hACE2) (Sigma-Aldrich) in PBS, incubated overnight at 4°C, washed three times with 0.05% Tween 20 PBS, and blocked with 1% BSA PBS for 1 hour at room temperature. Each serum sample was diluted 1:160, pre-incubated with 3 ng of RBD-Fc in 1% BSA PBS for 1 hour at room temperature, and then transferred to the hACE2-coated plate. RBD-Fc without pre-incubation with serum samples was added as a positive control, and 1% BSA PBS without serum pre-incubation was added as a negative control. Plates were then washed three times and incubated with HRP-conjugated anti-human IgG Fc (Southern Biotech) for 1 hour at room temperature. Plates were washed five times and developed with tetramethylbenzidine (BD OptEIA Substrate Solution, BD Biosciences) for 5 min, then stopped with 2 N H2SO4. The optical density was read at 450 nm with SpectraMax iD3 microplate reader (Molecular Devices). Percentage inhibition of RBD binding to hACE2 was calculated with the following formula: Inhibition (%) = [1 – (Sample OD value – Negative Control OD value)/(Positive Control OD value – Negative Control OD value)] x 100.
SARS-CoV-2 neutralization titer determination.
All serum samples were heat-inactivated at 56°C for 30 minutes to remove complement and allowed to equilibrate to room temperature prior to processing for neutralization titers. Samples were diluted in duplicate to an initial dilution of 1:5 or 1:10 followed by 1:2 serial dilutions (vaccinated sample), resulting in a 12-dilution series with each well containing 100 μL. All dilutions were performed in Dulbecco’s Modified Eagle Medium (Quality Biological), supplemented with 10% (v/v) FBS (heat-inactivated, Sigma-Aldrich), 1% (v/v) penicillin/streptomycin (Gemini Bio-products) and 1% (v/v) L-glutamine (2 mM final concentration, Gibco). Dilution plates were then transported into the BSL3 laboratory and 100 μL of diluted SARS-CoV-2 (WA-1, courtesy of Natalie Thornburg/CDC) inoculum was added to each well to result in a multiplicity of infection (MOI) of 0.01 upon transfer to titering plates. A non-treated, virus-only control and mock infection control were included on every plate. The sample-virus mixture was then incubated at 37°C (5.0% CO2) for 1 hour before transferring to 96-well titer plates with confluent VeroE6 cells. Titer plates were incubated at 37°C (5.0% CO2) for 72 hours, followed by cytopathic effect (CPE) determination for each well in the plate. The first sample dilution to show CPE was reported as the minimum sample dilution required to neutralize >99% of the concentration of SARS-CoV-2 tested (NT99).
Pseudovirus neutralization assay.
The SARS-CoV-2 pseudoviruses expressing a luciferase reporter gene were generated in an approach similar to as described previously (85, 86). Briefly, the packaging plasmid psPAX2 (AIDS Resource and Reagent Program), luciferase reporter plasmid pLenti-CMV Puro-Luc (Addgene), and spike protein expressing pcDNA3.1-SARS CoV-2 SΔCT of variants were co-transfected into HEK293T cells by lipofectamine 2000 (Thermo Fisher Scientific). Pseudoviruses of SARS-CoV-2 variants were generated by using WA1/2020 strain (Wuhan/WIV04/2019, Global Initiative on Sharing Avian Influenza Data (GISAID) accession ID: EPI_ISL_402124), B.1.1.7 variant (GISAID accession ID: EPI_ISL_601443), or B.1.351 variant (GISAID accession ID: EPI_ISL_712096). The supernatants containing the pseudotype viruses were collected 48 hours post-transfection, which were purified by centrifugation and filtration with 0.45 μm filter. To determine the neutralization activity of the plasma or serum samples from participants, HEK293T-hACE2 cells were seeded in 96-well tissue culture plates at a density of 1.75 × 104 cells per well overnight. Three-fold serial dilutions of heat-inactivated serum or plasma samples were prepared and mixed with 50 μL of pseudovirus. The mixture was incubated at 37°C for 1 hour before adding to HEK293T-hACE2 cells. 48 hours after infection, cells were lysed in Steady-Glo Luciferase Assay (Promega) according to the manufacturer’s instructions. SARS-CoV-2 neutralization titers were defined as the sample dilution at which a 50% reduction in the relative light unit (RLU) was observed relative to the average of the virus control wells.
Splenocyte restimulation assay.
Immunized mice were euthanized 2 weeks after the final immunization, and spleens were collected. To isolate splenocytes, spleens were mashed through a 70 μm cell strainer, and the resulting cell suspensions were washed with PBS and incubated with 2 mL of ACK lysis buffer (Gibco) for 2 minutes at room temperature to lyse erythrocytes. Splenocytes were washed again with PBS and plated in flat-bottom 96-well plates (2 × 106 cells per well). Then, SARS-CoV-2 spike peptides (PepTivator SARS-CoV-2 Prot_S, Miltenyi Biotec) were added at a final concentration of 0.6 nmol/ml in the presence of 1 μg/mL anti-CD28 antibody (total cell culture volume, 200 μL per well). After 24 (for IL-2 and IL-4) and 96 (for IFN-γ) hours, supernatants were harvested, and cytokine concentrations were measured by ELISA (Invitrogen) according to the manufacturer’s protocol.
Lymph node immunophenotyping by flow cytometry.
Draining lymph nodes were collected, passed through 70 micron filters, and resuspended in PBS supplemented with 1% FBS and 1 mM EDTA. Cell suspensions were split for staining with the following antibodies to B and T cell antigens for 30 minutes at 4°C: anti-CD4 (RM4–5, allophycocyanin (APC)-Cy7, BioLegend, 1 μg/ml), anti-ICOS (15F9, phycoerythrin (PE), BioLegend, 1 μg/ml), anti-CXCR5 (2G8, biotin, BD Biosciences, 5 μg/ml), anti-CD19 (6D5, Brilliant Violet 510, BioLegend, 0.5 μg/ml), T- and B- cell activation antigen (GL-7, fluorescein isothiocyanate (FITC), BD Biosciences, 2.5 μg/ml), anti-PD-1 (RMP 1–30, PE-Cy7, BioLegend, 1 μg/ml), CD38 (90, Pacific Blue, BioLegend, 2.5 μg/ml), CD138 (281–2, APC, BioLegend, 0.5 μg/ml), and anti-CD95 (Jo2, PE-Cy7, BD Biosciences, 1 μg/ml). For further CXCR5 detection, streptavidin (Brilliant Violet 421, BioLegend, 1.25 μg/ml) was used at 4°C. For intracellular staining, samples were fixed with the Foxp3 Fix/Perm buffer set according to the manufacturer’s instructions (eBioscience). Samples were then intracellularly stained with anti-Foxp3 (FJK-16s, Alexa Fluor 488, Invitrogen, 2.5 μg/ml) and anti-Ki67 (B56, Alexa Fluor 647, BD Biosciences, 1:200 dilution) or anti-IgG1 (A85–1, PE, BD Biosciences, 1 μg/ml). Samples were analyzed on a Cytek Aurora flow cytometer. Data analyses were performed using FlowJo v10 (FlowJo LLC).
Lymph node immunofluorescence stain and microscopy.
Mouse inguinal LNs were harvested and fixed with a mixture of 4% paraformaldehyde and 10% sucrose for 4 hours at 4°C. Following three washes with PBS, samples were equilibrated in 30% sucrose overnight. Samples were placed in molds with OCT compound (Tissue Tek) and then frozen using a 2-methylbutane bath submerged in liquid nitrogen. 5 μm sections were obtained on a cryostat (Leica CM1850). The frozen sections were air-dried at room temperature for 10 minutes and then rehydrated in Tris Buffered Saline (TBS) for 5 minutes at room temperature. Tissue sections were then permeabilized in 0.05% Triton X-100 TBS for 10 minutes at room temperature. Sections were blocked with 5% normal donkey serum (Jackson ImmunoResearch Lab Inc) for 1 hour at room temperature. Slides were then incubated with Alexa 594 conjugated anti-mouse CD3 (1:200, BioLegend, Cat#: 100240), Brilliant Violet 421 conjugated anti-mouse CD21/CD35 (1:100, BioLegend, Cat#: 123422), Alexa 647 conjugated anti-mouse IgD (1:200, BioLegend, Cat#: A05708) and Alexa 488 conjugated anti-mouse/human GL7 (1:200, BioLegend, Cat#: 144612) for 2 hours at room temperature. The tissue sections were then washed three times with TBS and slides were mounted with Prolong Gold anti-fade mounting media (Invitrogen). The fluorescent images were taken with a Zeiss LSM 880 confocal microscope using Zen black software version 2.3 SP1. Tile scan images were taken with a 20× 0.8 N.A. objective lens. The images were represented with the following false colors: Alexa 594-CD3 with red, BV421-CD21/CD35 with blue, Alexa 647-IgD with yellow and Alexa 488-GL7 with green.
SARS-CoV-2 mouse challenge study.
Mice were anesthetized by intraperitoneal injection 50 μl of a mix of xylazine (0.38 mg/mouse) and ketamine (1.3 mg/mouse) diluted in PBS. Mice were then intranasally inoculated with 1 × 103 PFU of mouse-adapted SARS-CoV-2 (MA10, courtesy of Dr. Ralph Baric (UNC)) in 50 μl divided between nares(39). Different doses of SARS-CoV-2 were used where indicated. Challenged mice were weighed on the day of infection and daily for up to 4 days post-infection. At 4 days post-infection, mice were euthanized and lungs were harvested to determine virus titer by a plaque assay and prepared for histological scoring.
SARS-CoV-2 plaque assay.
SARS-CoV-2 lung titers were quantified by homogenizing harvested lungs in PBS (Quality Biological Inc.) using 1.0 mm glass beads (Sigma-Aldrich) and a Beadruptor (Omni International Inc.). Homogenates were added to Vero E6 cells. SARS-CoV-2 virus titers determined by counting plaque-forming units (PFU) using a 6-point dilution curve.
Histopathology analysis.
Slides were prepared as 5 μm sections and stained with hematoxylin and eosin. A pathologist was blinded to information identifying the treatment groups and fields were examined by light microscopy and analyzed. The severity of interstitial inflammation was evaluated and converted to a score of 0 to 4 with 0 being no inflammation and 4 being most severe. Interstitial inflammation was evaluated for the number of neutrophils present in the interstitial space as well as the extent of neutrophilic apoptosis. Once scoring was complete, scores for each group were averaged and the standard deviation for the scoring was computed.
Murine LN gene expression analysis by quantitative real-time PCR array.
Mice were subcutaneously injected on Day 0 with the indicated treatments in a volume of 50 μL on each side of the back (one side for the compound and the contralateral side for saline of vehicle control). Twenty-four hours post-injection, draining (brachial) LNs were collected for subsequent analysis. LNs were transferred to a beadbeater and homogenized in TRI Reagent (Zymo Research). Samples were then centrifuged, and the clear supernatant was transferred to a new tube for subsequent RNA isolation. RNA was isolated from TRI Reagent samples using phenol-chloroform extraction or column-based extraction systems (Direct-zol RNA Miniprep, Zymo Research) according to the manufacturer’s protocol. RNA concentration and purity (260/280 and 260/230 ratios) were measured by NanoDrop (Thermo Fisher Scientific). cDNA was prepared from RNA with RT2 First Strand Kit, according to the manufacturer’s instructions (Qiagen). cDNA was quantified using 96-well PCR array analysis on a PAMM-150ZA plate (Cytokines & Chemokines) and PAMM-016ZA plate (Type I Interferon Response) (both Qiagen). Quantitative real time-PCR (QRT-PCR) was run on a 7300 real-time PCR system (Applied Biosystems – Life Technologies). mRNA abundance was normalized to three housekeeping genes: Actb, Gapdh, and Gusb. Relative quantification of gene expression was calculated by the ΔΔCt (relative expression over PBS treatment group).
Human PBMC isolation.
PBMCs were isolated based on previously described protocols (87). Briefly, heparinized whole blood was centrifuged at 500 g for 10 minutes, then the upper layer of platelet-rich plasma was removed. Plasma was centrifuged at 3000 g for 10 minutes, and platelet-poor plasma (PPP) was collected and stored on ice. The remaining blood was reconstituted to its original volume with heparinized Dulbecco’s PBS and layered on Ficoll-Paque gradients (Cytiva) in Accuspin tubes (Sigma-Aldrich). PBMCs were collected after centrifugation and washed twice with PBS.
Human PBMCs stimulation.
PBMCs were resuspended at a concentration of 200,000 cells per well in a 96-well U-bottom plate (Corning) in 200 μL RPMI-1640 media (Gibco) supplemented with 10% autologous PPP, 100 IU/mL penicillin, 100 μg/mL streptomycin, and 2 mM L-glutamine. PBMCs were incubated for 24 hours at 37ºC in a humidified incubator at 5% CO2 with indicated treatments. After culture, plates were centrifuged at 500 g and supernatants were removed by pipetting without disturbing the cell pellet. Cytokine expression profiles in cell culture supernatants were measured using customized Milliplex human cytokine magnetic bead panels (Milliplex). Assays were analyzed on the Luminex FLEXMAP 3D employing xPONENT software (Luminex) and Millipore Milliplex Analyst. Cytokine measurements were excluded from analysis if fewer than 30 beads were recovered. Synergy was evaluated using the Loewe definition of additivity, with D > 1 indicating antagonism, D = 1 additivity, and D < 1 synergy (88). In order to fit regression curves more closely to the data, higher concentrations were excluded from linear regressions when calculating D values if the cytokine concentrations plateaued or decreased.
Human MoDC and T cell co-culture.
Monocytes were isolated from cryopreserved PBMCs by positive selection with magnetic CD14+ beads on Day 1 according to manufacturer protocols (Miltenyi Biotec). CD14+ monocytes were then differentiated to monocyte-derived dendritic cells (MoDCs) in RPMI-1640 media supplemented with 500 IU/mL IL-4, 1000 IU/mL GM-CSF (both Miltenyi), penicillin, streptomycin, 10% autologous plasma, and 2 mM L-glutamine at a concentration of 0.67 to 1 × 106 per ml for 5 days. On Day 6, MoDCs were collected and resuspended at 25,000 cells per well in a 96-well U-bottom plate and stimulated with the indicated concentrations of CpG 2395 (Invivogen) or Alhydrogel (Invivogen), and 1 μg/mL recombinant spike protein (Invitrogen). CD4+ T cells were isolated from cryopreserved PBMCs by positive selection with magnetic CD4+ beads (Miltenyi) on day 6 and rested overnight in RPMI-1640 supplemented with 10% autologous plasma. On Day 7, CD4+ T cells were added at a concentration of 250,000 cells per well to the MoDC plates, and the co-cultured plates were incubated for 4 days. Cells were then stained with 20 μg/ml each of anti–CD4-V450 (clone RPA-T4, BD Biosciences), anti–CD25-FITC (clone M-A251, BD Biosciences), and anti–CD134- PE (clone L106, BD Biosciences) antibodies in PBS for 30 minutes at 4°C. Surface marker expressions of CD4+ cells were assessed by flow cytometry (LSRFortessa flow cytometer, Becton Dickinson) and analyzed by FlowJo software version 10.8.0.
Statistical analysis.
Raw, individual level data are presented in data file S1. Statistical analyses were performed using Prism v9.0.2 (GraphPad Software) and R software environment v4.0.4. P values < 0.05 were considered significant. Data were analyzed by one- or two-way analyses of variance (ANOVAs) followed by post-hoc Tukey’s test or Dunnett’s test for multiple comparisons. Non-normally distributed data were log-transformed. In the animal experiments, time to event were analyzed using Kaplan-Meier estimates and compared across groups using the Log-rank test. For the human in vitro PBMC assay, unpaired Mann-Whitney tests were applied at each concentration. We conducted gene expression analyses with R 4.0.4 using packages ‘ggplot2’, ‘dplyr’, and ‘MASS’ for the transcript abundance determination of gene arrays in each group. We log-transformed data before performing principal component analysis (PCA) and unsupervised hierarchical clustering using R package ‘stats’ with prcomp function and ‘pheatmap’ respectively. We analyzed the differential gene expression using generalized linear models (GLMs) with treatment and age as fixed effects. We then performed an over-representation analysis of the differentially expressed genes after Benjamini-Hochberg correction using the blood transcriptional module method based on an existing protocol (89): (a) we identified significant genes based on P adjusted value of ≤ 0.05 after Benjamini-Hochberg correction; (b) activity scores were computed as the mean value of member genes; and (c) over-representation analysis was performed on the significant gene list and each blood transcriptional modules using the Fisher exact test.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the members of the BCH Precision Vaccines Program (PVP) for helpful discussions as well as Kevin Churchwell, Gary Fleisher, David Williams, and August Cervini for their support of the PVP. Work within the PVP on this project was funded in part by philanthropic support from Amy and Michael Barry, Stop & Shop, and the Boston Investment Conference. We thank Barney S. Graham (NIH Vaccine Research Center) for generously providing the plasmid for pre-fusion stabilized SARS-CoV-2 spike trimer. We thank Ralph Baric for providing the SARS-CoV-2/MA10 virus. We thank Suzanne White and Lay-Hong Ang for performing lymph node sectioning and immunofluorescence staining. We thank the pharmacists of Boston Children’s Hospital for their efforts to maximize the use of SARS-CoV-2 vaccines by saving leftover or overfill of otherwise to be discarded vaccine vials. P.J.H. is affiliated to Baylor University, Hagler Institute for Advanced Study, Scowcroft Institute of International Affairs, Texas A&M University, Rice University, and University of Texas. D.J.D. would like to thank Siobhan McHugh, Geneva Boyer, Lucy Conetta and the staff of Lucy’s Daycare, the staff the YMCA of Greater Boston, Bridging Independent Living Together (BILT), Inc., and the Boston Public Schools for childcare and educational support during the COVID-19 pandemic.
FUNDING
The current study was supported in part by US National Institutes of Health (NIH)/National Institutes of Allergy and Infectious Diseases (NIAID) awards, including Adjuvant Discovery (HHSN272201400052C and 75N93019C00044) and Development (HHSN272201800047C) Program Contracts, a Massachusetts Consortium on Pathogen Readiness (Mass-CPR) award as well as philanthropic support from Amy and Michael Barry to OL; NIH grant (1R21AI137932-01A1) and Adjuvant Discovery Program contract (75N93019C00044) to DJD; the U.S. Army’s Long Term Health and Education Training Program to REH; BARDA# ASPR-20-01495, DARPA# ASPR-20-01495, NIH R01 AI148166, and NIH HHSN272201400007C to MBF; NIH R01 AI146779 and a MassCPR grant to AGS; NIGMS T32 GM007753 to BMH and TMC; T32 AI007245 to JF; NIH grants 1R01AI121066, 1R01DK115217, and 1R01AI165505, Lloyd J. Old STAR Program CRI3888, and an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund to IZ. The Precision Vaccines Program is supported in part by the BCH Department of Pediatrics and the Chief Scientific Office. EN is supported by the Daiichi Sankyo Foundation of Life Science and Uehara Memorial Foundation and is a joint Society for Pediatric Research and Japanese Pediatric Society Scholar.
Footnotes
COMPETING INTERESTS STATEMENT
EN, FB, TRO, YS, SVH, IZ, OL and DJD are named inventors on vaccine adjuvant patents assigned to Boston Children’s Hospital (“Adjuvants For Severe Acute Respiratory Syndrome-Related Coronavirus (SARS-CoV) Vaccines”, PCT/US21/34919). FB has signed consulting agreements with Merck Sharp & Dohme Corp. (a subsidiary of Merck & Co., Inc.), Sana Biotechnology, Inc., and F. Hoffmann-La Roche Ltd. IZ reports compensation for consulting services with Implicit Biosciences. MF is on the advisory board of Aikido Pharma. The Baylor College of Medicine (BCM) authors declare they are developers of a recombinant RBD technology. BCM recently licensed the technology to Biological E, an Indian manufacturer, for advancement and licensure. OL served as a paid consultant to Moody’s analytics. These commercial or financial relationships are unrelated to the current study.
DATA AND MATERIALS AVAILABILITY
All data associated with this study are in the paper or supplementary materials.
REFERENCES
- 1.Koff WC, Schenkelberg T, Williams T, Baric RS, McDermott A, Cameron CM, Cameron MJ, Friemann MB, Neumann G, Kawaoka Y, Kelvin AA, Ross TM, Schultz-Cherry S, Mastro TD, Priddy FH, Moore KA, Ostrowsky JT, Osterholm MT, Goudsmit J, Development and deployment of COVID-19 vaccines for those most vulnerable. Science Translational Medicine 13, eabd1525 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, McGettigan J, Kehtan S, Segall N, Solis J, Brosz A, Fierro C, Schwartz H, Neuzil K, Corey L, Gilbert P, Janes H, Follmann D, Marovich M, Mascola J, Polakowski L, Ledgerwood J, Graham BS, Bennett H, Pajon R, Knightly C, Leav B, Deng W, Zhou H, Han S, Ivarsson M, Miller J, Zaks T, Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. The New England journal of medicine, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, Frenck RW Jr., Hammitt LL, Türeci Ö, Nell H, Schaefer A, Ünal S, Tresnan DB, Mather S, Dormitzer PR, Şahin U, Jansen KU, Gruber WC, Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. The New England journal of medicine 383, 2603–2615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sadoff J, Le Gars M, Shukarev G, Heerwegh D, Truyers C, de Groot AM, Stoop J, Tete S, Van Damme W, Leroux-Roels I, Berghmans PJ, Kimmel M, Van Damme P, de Hoon J, Smith W, Stephenson KE, De Rosa SC, Cohen KW, McElrath MJ, Cormier E, Scheper G, Barouch DH, Hendriks J, Struyf F, Douoguih M, Van Hoof J, Schuitemaker H, Interim Results of a Phase 1–2a Trial of Ad26.COV2.S Covid-19 Vaccine. The New England journal of medicine, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corbett KS, Edwards DK, Leist SR, Abiona OM, Boyoglu-Barnum S, Gillespie RA, Himansu S, Schäfer A, Ziwawo CT, DiPiazza AT, Dinnon KH, Elbashir SM, Shaw CA, Woods A, Fritch EJ, Martinez DR, Bock KW, Minai M, Nagata BM, Hutchinson GB, Wu K, Henry C, Bahl K, Garcia-Dominguez D, Ma L, Renzi I, Kong WP, Schmidt SD, Wang L, Zhang Y, Phung E, Chang LA, Loomis RJ, Altaras NE, Narayanan E, Metkar M, Presnyak V, Liu C, Louder MK, Shi W, Leung K, Yang ES, West A, Gully KL, Stevens LJ, Wang N, Wrapp D, Doria-Rose NA, Stewart-Jones G, Bennett H, Alvarado GS, Nason MC, Ruckwardt TJ, McLellan JS, Denison MR, Chappell JD, Moore IN, Morabito KM, Mascola JR, Baric RS, Carfi A, Graham BS, SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 586, 567–571 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corbett KS, Flynn B, Foulds KE, Francica JR, Boyoglu-Barnum S, Werner AP, Flach B, O’Connell S, Bock KW, Minai M, Nagata BM, Andersen H, Martinez DR, Noe AT, Douek N, Donaldson MM, Nji NN, Alvarado GS, Edwards DK, Flebbe DR, Lamb E, Doria-Rose NA, Lin BC, Louder MK, O’Dell S, Schmidt SD, Phung E, Chang LA, Yap C, Todd JM, Pessaint L, Van Ry A, Browne S, Greenhouse J, Putman-Taylor T, Strasbaugh A, Campbell TA, Cook A, Dodson A, Steingrebe K, Shi W, Zhang Y, Abiona OM, Wang L, Pegu A, Yang ES, Leung K, Zhou T, Teng IT, Widge A, Gordon I, Novik L, Gillespie RA, Loomis RJ, Moliva JI, Stewart-Jones G, Himansu S, Kong WP, Nason MC, Morabito KM, Ruckwardt TJ, Ledgerwood JE, Gaudinski MR, Kwong PD, Mascola JR, Carfi A, Lewis MG, Baric RS, McDermott A, Moore IN, Sullivan NJ, Roederer M, Seder RA, Graham BS, Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. The New England journal of medicine 383, 1544–1555 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogel AB, Kanevsky I, Che Y, Swanson KA, Muik A, Vormehr M, Kranz LM, Walzer KC, Hein S, Güler A, Loschko J, Maddur MS, Ota-Setlik A, Tompkins K, Cole J, Lui BG, Ziegenhals T, Plaschke A, Eisel D, Dany SC, Fesser S, Erbar S, Bates F, Schneider D, Jesionek B, Sänger B, Wallisch AK, Feuchter Y, Junginger H, Krumm SA, Heinen AP, Adams-Quack P, Schlereth J, Schille S, Kröner C, de la Caridad Güimil Garcia R, Hiller T, Fischer L, Sellers RS, Choudhary S, Gonzalez O, Vascotto F, Gutman MR, Fontenot JA, Hall-Ursone S, Brasky K, Griffor MC, Han S, Su AAH, Lees JA, Nedoma NL, Mashalidis EH, Sahasrabudhe PV, Tan CY, Pavliakova D, Singh G, Fontes-Garfias C, Pride M, Scully IL, Ciolino T, Obregon J, Gazi M, Carrion R Jr., Alfson KJ, Kalina WV, Kaushal D, Shi PY, Klamp T, Rosenbaum C, Kuhn AN, Türeci Ö, Dormitzer PR, Jansen KU, Sahin U, BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature 592, 283–289 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Mercado NB, Zahn R, Wegmann F, Loos C, Chandrashekar A, Yu J, Liu J, Peter L, McMahan K, Tostanoski LH, He X, Martinez DR, Rutten L, Bos R, van Manen D, Vellinga J, Custers J, Langedijk JP, Kwaks T, Bakkers MJG, Zuijdgeest D, Rosendahl Huber SK, Atyeo C, Fischinger S, Burke JS, Feldman J, Hauser BM, Caradonna TM, Bondzie EA, Dagotto G, Gebre MS, Hoffman E, Jacob-Dolan C, Kirilova M, Li Z, Lin Z, Mahrokhian SH, Maxfield LF, Nampanya F, Nityanandam R, Nkolola JP, Patel S, Ventura JD, Verrington K, Wan H, Pessaint L, Van Ry A, Blade K, Strasbaugh A, Cabus M, Brown R, Cook A, Zouantchangadou S, Teow E, Andersen H, Lewis MG, Cai Y, Chen B, Schmidt AG, Reeves RK, Baric RS, Lauffenburger DA, Alter G, Stoffels P, Mammen M, Van Hoof J, Schuitemaker H, Barouch DH, Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature 586, 583–588 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gebre MS, Brito LA, Tostanoski LH, Edwards DK, Carfi A, Barouch DH, Novel approaches for vaccine development. Cell 184, 1589–1603 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katz IT, Weintraub R, Bekker LG, Brandt AM, From Vaccine Nationalism to Vaccine Equity - Finding a Path Forward. The New England journal of medicine 384, 1281–1283 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Keech C, Albert G, Cho I, Robertson A, Reed P, Neal S, Plested JS, Zhu M, Cloney-Clark S, Zhou H, Smith G, Patel N, Frieman MB, Haupt RE, Logue J, McGrath M, Weston S, Piedra PA, Desai C, Callahan K, Lewis M, Price-Abbott P, Formica N, Shinde V, Fries L, Lickliter JD, Griffin P, Wilkinson B, Glenn GM, Phase 1–2 Trial of a SARS-CoV-2 Recombinant Spike Protein Nanoparticle Vaccine. The New England journal of medicine 383, 2320–2332 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richmond P, Hatchuel L, Dong M, Ma B, Hu B, Smolenov I, Li P, Liang P, Han HH, Liang J, Clemens R, Safety and immunogenicity of S-Trimer (SCB-2019), a protein subunit vaccine candidate for COVID-19 in healthy adults: a phase 1, randomised, double-blind, placebo-controlled trial. Lancet (London, England), (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia S, Zhang Y, Wang Y, Wang H, Yang Y, Gao GF, Tan W, Wu G, Xu M, Lou Z, Huang W, Xu W, Huang B, Wang H, Wang W, Zhang W, Li N, Xie Z, Ding L, You W, Zhao Y, Yang X, Liu Y, Wang Q, Huang L, Yang Y, Xu G, Luo B, Wang W, Liu P, Guo W, Yang X, Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: a randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect Dis 21, 39–51 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Zeng G, Pan H, Li C, Hu Y, Chu K, Han W, Chen Z, Tang R, Yin W, Chen X, Hu Y, Liu X, Jiang C, Li J, Yang M, Song Y, Wang X, Gao Q, Zhu F, Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18–59 years: a randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect Dis 21, 181–192 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kyriakidis NC, López-Cortés A, González EV, Grimaldos AB, Prado EO, SARS-CoV-2 vaccines strategies: a comprehensive review of phase 3 candidates. NPJ vaccines 6, 28 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.CTRI. Biological E’s novel Covid-19 vaccine of SARS-CoV-2 for protection against Covid-19 disease., <http://ctri.nic.in/Clinicaltrials/pmaindet2.php?trialid=48329&EncHid=&userName=covid-19%20vaccine> Accessed May 1, 2021
- 17.The New York Times. Coronavirus Vaccine Tracker, <https://www.nytimes.com/interactive/2020/science/coronavirus-vaccine-tracker.html>, Accessed May 3, 2021
- 18.Yang J, Wang W, Chen Z, Lu S, Yang F, Bi Z, Bao L, Mo F, Li X, Huang Y, Hong W, Yang Y, Zhao Y, Ye F, Lin S, Deng W, Chen H, Lei H, Zhang Z, Luo M, Gao H, Zheng Y, Gong Y, Jiang X, Xu Y, Lv Q, Li D, Wang M, Li F, Wang S, Wang G, Yu P, Qu Y, Yang L, Deng H, Tong A, Li J, Wang Z, Yang J, Shen G, Zhao Z, Li Y, Luo J, Liu H, Yu W, Yang M, Xu J, Wang J, Li H, Wang H, Kuang D, Lin P, Hu Z, Guo W, Cheng W, He Y, Song X, Chen C, Xue Z, Yao S, Chen L, Ma X, Chen S, Gou M, Huang W, Wang Y, Fan C, Tian Z, Shi M, Wang FS, Dai L, Wu M, Li G, Wang G, Peng Y, Qian Z, Huang C, Lau JY, Yang Z, Wei Y, Cen X, Peng X, Qin C, Zhang K, Lu G, Wei X, A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature 586, 572–577 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Piccoli L, Park YJ, Tortorici MA, Czudnochowski N, Walls AC, Beltramello M, Silacci-Fregni C, Pinto D, Rosen LE, Bowen JE, Acton OJ, Jaconi S, Guarino B, Minola A, Zatta F, Sprugasci N, Bassi J, Peter A, De Marco A, Nix JC, Mele F, Jovic S, Rodriguez BF, Gupta SV, Jin F, Piumatti G, Lo Presti G, Pellanda AF, Biggiogero M, Tarkowski M, Pizzuto MS, Cameroni E, Havenar-Daughton C, Smithey M, Hong D, Lepori V, Albanese E, Ceschi A, Bernasconi E, Elzi L, Ferrari P, Garzoni C, Riva A, Snell G, Sallusto F, Fink K, Virgin HW, Lanzavecchia A, Corti D, Veesler D, Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell 183, 1024–1042.e1021 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Premkumar L, Segovia-Chumbez B, Jadi R, Martinez DR, Raut R, Markmann A, Cornaby C, Bartelt L, Weiss S, Park Y, Edwards CE, Weimer E, Scherer EM, Rouphael N, Edupuganti S, Weiskopf D, Tse LV, Hou YJ, Margolis D, Sette A, Collins MH, Schmitz J, Baric RS, de Silva AM, The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci Immunol 5, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esposito D, Mehalko J, Drew M, Snead K, Wall V, Taylor T, Frank P, Denson JP, Hong M, Gulten G, Sadtler K, Messing S, Gillette W, Optimizing high-yield production of SARS-CoV-2 soluble spike trimers for serology assays. Protein Expr Purif 174, 105686 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dalvie NC, Rodriguez-Aponte SA, Hartwell BL, Tostanoski LH, Biedermann AM, Crowell LE, Kaur K, Kumru OS, Carter L, Yu J, Chang A, McMahan K, Courant T, Lebas C, Lemnios AA, Rodrigues KA, Silva M, Johnston RS, Naranjo CA, Tracey MK, Brady JR, Whittaker CA, Yun D, Brunette N, Wang JY, Walkey C, Fiala B, Kar S, Porto M, Lok M, Andersen H, Lewis MG, Love KR, Camp DL, Silverman JM, Kleanthous H, Joshi SB, Volkin DB, Dubois PM, Collin N, King NP, Barouch DH, Irvine DJ, Love JC, Engineered SARS-CoV-2 receptor binding domain improves manufacturability in yeast and immunogenicity in mice. Proceedings of the National Academy of Sciences of the United States of America 118, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang S, Li Y, Dai L, Wang J, He P, Li C, Fang X, Wang C, Zhao X, Huang E, Wu C, Zhong Z, Wang F, Duan X, Tian S, Wu L, Liu Y, Luo Y, Chen Z, Li F, Li J, Yu X, Ren H, Liu L, Meng S, Yan J, Hu Z, Gao L, Gao GF, Safety and immunogenicity of a recombinant tandem-repeat dimeric RBD-based protein subunit vaccine (ZF2001) against COVID-19 in adults: two randomised, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Infect Dis, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai L, Zheng T, Xu K, Han Y, Xu L, Huang E, An Y, Cheng Y, Li S, Liu M, Yang M, Li Y, Cheng H, Yuan Y, Zhang W, Ke C, Wong G, Qi J, Qin C, Yan J, Gao GF, A Universal Design of Betacoronavirus Vaccines against COVID-19, MERS, and SARS. Cell 182, 722–733.e711 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hauser BM, Sangesland M, Lam EC, Feldman J, Yousif AS, Caradonna TM, Balazs AB, Lingwood D, Schmidt AG, Engineered receptor binding domain immunogens elicit pan-coronavirus neutralizing antibodies. bioRxiv, (2020). [Google Scholar]
- 26.Tan TK, Rijal P, Rahikainen R, Keeble AH, Schimanski L, Hussain S, Harvey R, Hayes JWP, Edwards JC, McLean RK, Martini V, Pedrera M, Thakur N, Conceicao C, Dietrich I, Shelton H, Ludi A, Wilsden G, Browning C, Zagrajek AK, Bialy D, Bhat S, Stevenson-Leggett P, Hollinghurst P, Tully M, Moffat K, Chiu C, Waters R, Gray A, Azhar M, Mioulet V, Newman J, Asfor AS, Burman A, Crossley S, Hammond JA, Tchilian E, Charleston B, Bailey D, Tuthill TJ, Graham SP, Duyvesteyn HME, Malinauskas T, Huo J, Tree JA, Buttigieg KR, Owens RJ, Carroll MW, Daniels RS, McCauley JW, Stuart DI, Huang KA, Howarth M, Townsend AR, A COVID-19 vaccine candidate using SpyCatcher multimerization of the SARS-CoV-2 spike protein receptor-binding domain induces potent neutralising antibody responses. Nature communications 12, 542 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walls AC, Fiala B, Schäfer A, Wrenn S, Pham MN, Murphy M, Tse LV, Shehata L, O’Connor MA, Chen C, Navarro MJ, Miranda MC, Pettie D, Ravichandran R, Kraft JC, Ogohara C, Palser A, Chalk S, Lee EC, Guerriero K, Kepl E, Chow CM, Sydeman C, Hodge EA, Brown B, Fuller JT, Dinnon KH 3rd, Gralinski LE, Leist SR, Gully KL, Lewis TB, Guttman M, Chu HY, Lee KK, Fuller DH, Baric RS, Kellam P, Carter L, Pepper M, Sheahan TP, Veesler D, King NP, Elicitation of Potent Neutralizing Antibody Responses by Designed Protein Nanoparticle Vaccines for SARS-CoV-2. Cell 183, 1367–1382.e1317 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang L, Tian D, Han JB, Fan W, Zhang Y, Li Y, Sun W, Wei Y, Tian X, Yu DD, Feng XL, Cheng G, Bi Y, Zheng YT, Liu W, A recombinant receptor-binding domain in trimeric form generates protective immunity against SARS-CoV-2 infection in nonhuman primates. Innovation (N Y) 2, 100140 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He L, Lin X, Wang Y, Abraham C, Sou C, Ngo T, Zhang Y, Wilson IA, Zhu J, Single-component, self-assembling, protein nanoparticles presenting the receptor binding domain and stabilized spike as SARS-CoV-2 vaccine candidates. Sci Adv 7, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borriello F, Nanishi E, Seo H-S, O’Meara TR, McGrath ME, Saito Y, Haupt RE, Chen J, Diray-Arce J, Song K, Xu AZ, Caradonna TM, Feldman J, Hauser BM, Schmidt AG, Baden LR, Ernst RK, Dillen C, Weston SM, Johnson RM, Hammond HL, Yu J, Chang A, Hilgers L, Platenburg PP, Dhe-Paganon S, Barouch DH, Ozonoff A, Zanoni I, Frieman MB, Dowling DJ, Levy O, An adjuvanted SARS-CoV-2 RBD nanoparticle elicits neutralizing antibodies and fully protective immunity in aged mice. bioRxiv, 2021.2009.2009.459664 (2021). [Google Scholar]
- 31.Reed SG, Tomai M, Gale MJ Jr., New horizons in adjuvants for vaccine development. Curr Opin Immunol 65, 97–101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pulendran B, S. A. P, D. T. O’Hagan, Emerging concepts in the science of vaccine adjuvants. Nature reviews. Drug discovery, 1–22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Hagan DT, Lodaya RN, Lofano G, The continued advance of vaccine adjuvants - ‘we can work it out’. Seminars in immunology 50, 101426 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Lal H, Cunningham AL, Godeaux O, Chlibek R, Diez-Domingo J, Hwang SJ, Levin MJ, McElhaney JE, Poder A, Puig-Barberà J, Vesikari T, Watanabe D, Weckx L, Zahaf T, Heineman TC, Efficacy of an adjuvanted herpes zoster subunit vaccine in older adults. The New England journal of medicine 372, 2087–2096 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Cunningham AL, Lal H, Kovac M, Chlibek R, Hwang SJ, Díez-Domingo J, Godeaux O, Levin MJ, McElhaney JE, Puig-Barberà J, Vanden Abeele C, Vesikari T, Watanabe D, Zahaf T, Ahonen A, Athan E, Barba-Gomez JF, Campora L, de Looze F, Downey HJ, Ghesquiere W, Gorfinkel I, Korhonen T, Leung E, McNeil SA, Oostvogels L, Rombo L, Smetana J, Weckx L, Yeo W, Heineman TC, Efficacy of the Herpes Zoster Subunit Vaccine in Adults 70 Years of Age or Older. The New England journal of medicine 375, 1019–1032 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Vinuesa CG, Linterman MA, Yu D, MacLennan IC, Follicular Helper T Cells. Annual review of immunology 34, 335–368 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Crotty S, T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity 50, 1132–1148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sage PT, Tan CL, Freeman GJ, Haigis M, Sharpe AH, Defective TFH Cell Function and Increased TFR Cells Contribute to Defective Antibody Production in Aging. Cell Rep 12, 163–171 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leist SR, Dinnon KH 3rd, Schäfer A, Tse LV, Okuda K, Hou YJ, West A, Edwards CE, Sanders W, Fritch EJ, Gully KL, Scobey T, Brown AJ, Sheahan TP, Moorman NJ, Boucher RC, Gralinski LE, Montgomery SA, Baric RS, A Mouse-Adapted SARS-CoV-2 Induces Acute Lung Injury and Mortality in Standard Laboratory Mice. Cell 183, 1070–1085.e1012 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lederer K, Castaño D, Gómez Atria D, Oguin TH 3rd, Wang S, Manzoni TB, Muramatsu H, Hogan MJ, Amanat F, Cherubin P, Lundgreen KA, Tam YK, Fan SHY, Eisenlohr LC, Maillard I, Weissman D, Bates P, Krammer F, Sempowski GD, Pardi N, Locci M, SARS-CoV-2 mRNA Vaccines Foster Potent Antigen-Specific Germinal Center Responses Associated with Neutralizing Antibody Generation. Immunity 53, 1281–1295.e1285 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang JG, Su D, Song TZ, Zeng Y, Huang W, Wu J, Xu R, Luo P, Yang X, Zhang X, Luo S, Liang Y, Li X, Huang J, Wang Q, Huang X, Xu Q, Luo M, Huang A, Luo D, Zhao C, Yang F, Han JB, Zheng YT, Liang P, S-Trimer, a COVID-19 subunit vaccine candidate, induces protective immunity in nonhuman primates. Nature communications 12, 1346 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia-Beltran WF, Lam EC, St Denis K, Nitido AD, Garcia ZH, Hauser BM, Feldman J, Pavlovic MN, Gregory DJ, Poznansky MC, Sigal A, Schmidt AG, Iafrate AJ, Naranbhai V, Balazs AB, Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuzmina A, Khalaila Y, Voloshin O, Keren-Naus A, Boehm-Cohen L, Raviv Y, Shemer-Avni Y, Rosenberg E, Taube R, SARS-CoV-2 spike variants exhibit differential infectivity and neutralization resistance to convalescent or post-vaccination sera. Cell host & microbe, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen X, Tang H, Pajon R, Smith G, Glenn GM, Shi W, Korber B, Montefiori DC, Neutralization of SARS-CoV-2 Variants B.1.429 and B.1.351. The New England journal of medicine, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z, Schmidt F, Weisblum Y, Muecksch F, Barnes CO, Finkin S, Schaefer-Babajew D, Cipolla M, Gaebler C, Lieberman JA, Oliveira TY, Yang Z, Abernathy ME, Huey-Tubman KE, Hurley A, Turroja M, West KA, Gordon K, Millard KG, Ramos V, Da Silva J, Xu J, Colbert RA, Patel R, Dizon J, Unson-O’Brien C, Shimeliovich I, Gazumyan A, Caskey M, Bjorkman PJ, Casellas R, Hatziioannou T, Bieniasz PD, Nussenzweig MC, mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 592, 616–622 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abu-Raddad LJ, Chemaitelly H, Butt AA, Effectiveness of the BNT162b2 Covid-19 Vaccine against the B.1.1.7 and B.1.351 Variants. The New England journal of medicine, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kustin T, Harel N, Finkel U, Perchik S, Harari S, Tahor M, Caspi I, Levy R, Leshchinsky M, Ken Dror S, Bergerzon G, Gadban H, Gadban F, Eliassian E, Shimron O, Saleh L, Ben-Zvi H, Keren Taraday E, Amichay D, Ben-Dor A, Sagas D, Strauss M, Shemer Avni Y, Huppert A, Kepten E, Balicer RD, Netzer D, Ben-Shachar S, Stern A, Evidence for increased breakthrough rates of SARS-CoV-2 variants of concern in BNT162b2-mRNA-vaccinated individuals. Nature medicine 27, 1379–1384 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bates TA, Leier HC, Lyski ZL, McBride SK, Coulter FJ, Weinstein JB, Goodman JR, Lu Z, Siegel SAR, Sullivan P, Strnad M, Brunton AE, Lee DX, Adey AC, Bimber BN, O’Roak BJ, Curlin ME, Messer WB, Tafesse FG, Neutralization of SARS-CoV-2 variants by convalescent and BNT162b2 vaccinated serum. Nature communications 12, 5135 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iwasaki A, Medzhitov R, Control of adaptive immunity by the innate immune system. Nat Immunol 16, 343–353 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pollard AJ, Bijker EM, A guide to vaccinology: from basic principles to new developments. Nature reviews. Immunology 21, 83–100 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grant SM, Lou M, Yao L, Germain RN, Radtke AJ, The lymph node at a glance - how spatial organization optimizes the immune response. J Cell Sci 133, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Acton SE, Reis e Sousa C, Dendritic cells in remodeling of lymph nodes during immune responses. Immunol Rev 271, 221–229 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Williamson EJ, Walker AJ, Bhaskaran K, Bacon S, Bates C, Morton CE, Curtis HJ, Mehrkar A, Evans D, Inglesby P, Cockburn J, McDonald HI, MacKenna B, Tomlinson L, Douglas IJ, Rentsch CT, Mathur R, Wong AYS, Grieve R, Harrison D, Forbes H, Schultze A, Croker R, Parry J, Hester F, Harper S, Perera R, Evans SJW, Smeeth L, Goldacre B, Factors associated with COVID-19-related death using OpenSAFELY. Nature 584, 430–436 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Centers for Disease Control and Prevention (CDC). COVID-19 Hospitalization and Death by Age, Accessed Apr 3rd, 2021, 2021
- 55.Hotez PJ, Bottazzi ME, Developing a low-cost and accessible COVID-19 vaccine for global health. PLoS Negl Trop Dis 14, e0008548 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McMahan K, Yu J, Mercado NB, Loos C, Tostanoski LH, Chandrashekar A, Liu J, Peter L, Atyeo C, Zhu A, Bondzie EA, Dagotto G, Gebre MS, Jacob-Dolan C, Li Z, Nampanya F, Patel S, Pessaint L, Van Ry A, Blade K, Yalley-Ogunro J, Cabus M, Brown R, Cook A, Teow E, Andersen H, Lewis MG, Lauffenburger DA, Alter G, Barouch DH, Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arunachalam PS, Walls AC, Golden N, Atyeo C, Fischinger S, Li C, Aye P, Navarro MJ, Lai L, Edara VV, Röltgen K, Rogers K, Shirreff L, Ferrell DE, Wrenn S, Pettie D, Kraft JC, Miranda MC, Kepl E, Sydeman C, Brunette N, Murphy M, Fiala B, Carter L, White AG, Trisal M, Hsieh CL, Russell-Lodrigue K, Monjure C, Dufour J, Spencer S, Doyle-Meyers L, Bohm RP, Maness NJ, Roy C, Plante JA, Plante KS, Zhu A, Gorman MJ, Shin S, Shen X, Fontenot J, Gupta S, O’Hagan DT, Van Der Most R, Rappuoli R, Coffman RL, Novack D, McLellan JS, Subramaniam S, Montefiori D, Boyd SD, Flynn JL, Alter G, Villinger F, Kleanthous H, Rappaport J, Suthar MS, King NP, Veesler D, Pulendran B, Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nature 594, 253–258 (2021). [DOI] [PubMed] [Google Scholar]
- 58.Chen WH, Wei J, Kundu RT, Adhikari R, Liu Z, Lee J, Versteeg L, Poveda C, Keegan B, Villar MJ, de Araujo Leao AC, Rivera JA, Gillespie PM, Pollet J, Strych U, Zhan B, Hotez PJ, Bottazzi ME, Genetic modification to design a stable yeast-expressed recombinant SARS-CoV-2 receptor binding domain as a COVID-19 vaccine candidate. Biochim Biophys Acta Gen Subj 1865, 129893 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen WH, Tao X, Agrawal AS, Algaissi A, Peng BH, Pollet J, Strych U, Bottazzi ME, Hotez PJ, Lustigman S, Du L, Jiang S, Tseng CK, Yeast-expressed SARS-CoV recombinant receptor-binding domain (RBD219-N1) formulated with aluminum hydroxide induces protective immunity and reduces immune enhancement. Vaccine 38, 7533–7541 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pollet J, Chen WH, Versteeg L, Keegan B, Zhan B, Wei J, Liu Z, Lee J, Kundu R, Adhikari R, Poveda C, Villar MJ, de Araujo Leao AC, Altieri Rivera J, Momin Z, Gillespie PM, Kimata JT, Strych U, Hotez PJ, Bottazzi ME, SARS-CoV-2 RBD219-N1C1: A yeast-expressed SARS-CoV-2 recombinant receptor-binding domain candidate vaccine stimulates virus neutralizing antibodies and T-cell immunity in mice. Human vaccines & immunotherapeutics, 1–11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paavonen J, Naud P, Salmerón J, Wheeler CM, Chow SN, Apter D, Kitchener H, Castellsague X, Teixeira JC, Skinner SR, Hedrick J, Jaisamrarn U, Limson G, Garland S, Szarewski A, Romanowski B, Aoki FY, Schwarz TF, Poppe WA, Bosch FX, Jenkins D, Hardt K, Zahaf T, Descamps D, Struyf F, Lehtinen M, Dubin G, Efficacy of human papillomavirus (HPV)-16/18 AS04-adjuvanted vaccine against cervical infection and precancer caused by oncogenic HPV types (PATRICIA): final analysis of a double-blind, randomised study in young women. Lancet (London, England) 374, 301–314 (2009). [DOI] [PubMed] [Google Scholar]
- 62.Campbell JD, Development of the CpG Adjuvant 1018: A Case Study. Methods in molecular biology (Clifton, N.J.) 1494, 15–27 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Cooper CL, Davis HL, Morris ML, Efler SM, Adhami MA, Krieg AM, Cameron DW, Heathcote J, CPG 7909, an immunostimulatory TLR9 agonist oligodeoxynucleotide, as adjuvant to Engerix-B HBV vaccine in healthy adults: a double-blind phase I/II study. J Clin Immunol 24, 693–701 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Maletto B, Rópolo A, Morón V, Pistoresi-Palencia MC, CpG-DNA stimulates cellular and humoral immunity and promotes Th1 differentiation in aged BALB/c mice. Journal of leukocyte biology 72, 447–454 (2002). [PubMed] [Google Scholar]
- 65.Maletto BA, Rópolo AS, Liscovsky MV, Alignani DO, Glocker M, Pistoresi-Palencia MC, CpG oligodeoxinucleotides functions as an effective adjuvant in aged BALB/c mice. Clinical immunology (Orlando, Fla.) 117, 251–261 (2005). [DOI] [PubMed] [Google Scholar]
- 66.Manning BM, Enioutina EY, Visic DM, Knudson AD, Daynes RA, CpG DNA functions as an effective adjuvant for the induction of immune responses in aged mice. Exp Gerontol 37, 107–126 (2001). [DOI] [PubMed] [Google Scholar]
- 67.Ming F, Yang J, Chu P, Ma M, Shi J, Cai H, Huang C, Li H, Jiang Z, Wang H, Wang W, Zhang S, Zhang L, Immunization of aged pigs with attenuated pseudorabies virus vaccine combined with CpG oligodeoxynucleotide restores defective Th1 immune responses. PloS one 8, e65536 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qin W, Jiang J, Chen Q, Yang N, Wang Y, Wei X, Ou R, CpG ODN enhances immunization effects of hepatitis B vaccine in aged mice. Cell Mol Immunol 1, 148–152 (2004). [PubMed] [Google Scholar]
- 69.Sen G, Chen Q, Snapper CM, Immunization of aged mice with a pneumococcal conjugate vaccine combined with an unmethylated CpG-containing oligodeoxynucleotide restores defective immunoglobulin G antipolysaccharide responses and specific CD4+-T-cell priming to young adult levels. Infection and immunity 74, 2177–2186 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sablan BP, Kim DJ, Barzaga NG, Chow WC, Cho M, Ahn SH, Hwang SG, Lee JH, Namini H, Heyward WL, Demonstration of safety and enhanced seroprotection against hepatitis B with investigational HBsAg-1018 ISS vaccine compared to a licensed hepatitis B vaccine. Vaccine 30, 2689–2696 (2012). [DOI] [PubMed] [Google Scholar]
- 71.Janssen JM, Jackson S, Heyward WL, Janssen RS, Immunogenicity of an investigational hepatitis B vaccine with a toll-like receptor 9 agonist adjuvant (HBsAg-1018) compared with a licensed hepatitis B vaccine in subpopulations of healthy adults 18–70 years of age. Vaccine 33, 3614–3618 (2015). [DOI] [PubMed] [Google Scholar]
- 72.Kuo TY, Lin MY, Coffman RL, Campbell JD, Traquina P, Lin YJ, Liu LT, Cheng J, Wu YC, Wu CC, Tang WH, Huang CG, Tsao KC, Chen C, Development of CpG-adjuvanted stable prefusion SARS-CoV-2 spike antigen as a subunit vaccine against COVID-19. Sci Rep 10, 20085 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lien CE, Lin YJ, Chen C, Lian WC, Kuo TY, Campbell JD, Traquina P, Lin MY, Liu LT, Chuang YS, Ko HY, Liao CC, Chen YH, Jan JT, Ma HH, Sun CP, Lin YS, Wu PY, Wang YC, Tao MH, Lin YL, CpG-adjuvanted stable prefusion SARS-CoV-2 spike protein protected hamsters from SARS-CoV-2 challenge. Sci Rep 11, 8761 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Press Information Bureau Government of India. Department of Biotechnology Mission COVID Suraksha Supported Biological E Limited Novel Covid-19 Vaccine candidate – CORBEVAX receives DCGI approval for Two Clinical Trials. https://pib.gov.in/PressReleasePage.aspx?PRID=1751664, Accessed Sep 28, 2021,
- 75.Kayraklioglu N, Horuluoglu B, Klinman DM, CpG Oligonucleotides as Vaccine Adjuvants. Methods in molecular biology (Clifton, N.J.) 2197, 51–85 (2021). [DOI] [PubMed] [Google Scholar]
- 76.Dowling DJ, van Haren SD, Scheid A, Bergelson I, Kim D, Mancuso CJ, Foppen W, Ozonoff A, Fresh L, Theriot TB, Lackner AA, Fichorova RN, Smirnov D, Vasilakos JP, Beaurline JM, Tomai MA, Midkiff CC, Alvarez X, Blanchard JL, Gilbert MH, Aye PP, Levy O, TLR7/8 adjuvant overcomes newborn hyporesponsiveness to pneumococcal conjugate vaccine at birth. JCI insight 2, e91020 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garrido C, Curtis AD 2nd, Dennis M, Pathak SH, Gao H, Montefiori D, Tomai M, Fox CB, Kozlowski PA, Scobey T, Munt JE, Mallory ML, Saha PT, Hudgens MG, Lindesmith LC, Baric RS, Abiona OM, Graham B, Corbett KS, Edwards D, Carfi A, Fouda G, Van Rompay KKA, De Paris K, Permar SR, SARS-CoV-2 vaccines elicit durable immune responses in infant rhesus macaques. Sci Immunol 6, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Soni D, Van Haren SD, Idoko OT, Evans JT, Diray-Arce J, Dowling DJ, Levy O, Towards Precision Vaccines: Lessons From the Second International Precision Vaccines Conference. Frontiers in immunology 11, 590373 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bar-On YM, Goldberg Y, Mandel M, Bodenheimer O, Freedman L, Kalkstein N, Mizrahi B, Alroy-Preis S, Ash N, Milo R, Huppert A, Protection of BNT162b2 Vaccine Booster against Covid-19 in Israel. The New England journal of medicine, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Falsey AR, Frenck RW Jr., Walsh EE, Kitchin N, Absalon J, Gurtman A, Lockhart S, Bailey R, Swanson KA, Xu X, Koury K, Kalina W, Cooper D, Zou J, Xie X, Xia H, Türeci Ö, Lagkadinou E, Tompkins KR, Shi PY, Jansen KU, Şahin U, Dormitzer PR, Gruber WC, SARS-CoV-2 Neutralization with BNT162b2 Vaccine Dose 3. The New England journal of medicine, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu K, Choi A, Koch M, Elbashir S, Ma L, Lee D, Woods A, Henry C, Palandjian C, Hill A, Quinones J, Nunna N, O’Connell S, McDermott AB, Falcone S, Narayanan E, Colpitts T, Bennett H, Corbett K, Seder R, Graham BS, Stewart-Jones GB, Carfi A, Edwards DK, Variant SARS-CoV-2 mRNA vaccines confer broad neutralization as primary or booster series in mice. bioRxiv, 2021.2004.2013.439482 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Baylor NW, Egan W, Richman P, Aluminum salts in vaccines--US perspective. Vaccine 20 Suppl 3, S18–23 (2002). [DOI] [PubMed] [Google Scholar]
- 83.Borriello F, Pietrasanta C, Lai JCY, Walsh LM, Sharma P, O’Driscoll DN, Ramirez J, Brightman S, Pugni L, Mosca F, Burkhart DJ, Dowling DJ, Levy O, Identification and Characterization of Stimulator of Interferon Genes As a Robust Adjuvant Target for Early Life Immunization. Front Immunol 8, 1772 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tan CW, Chia WN, Qin X, Liu P, Chen MI, Tiu C, Hu Z, Chen VC, Young BE, Sia WR, Tan YJ, Foo R, Yi Y, Lye DC, Anderson DE, Wang LF, A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat Biotechnol 38, 1073–1078 (2020). [DOI] [PubMed] [Google Scholar]
- 85.Yu J, Tostanoski LH, Peter L, Mercado NB, McMahan K, Mahrokhian SH, Nkolola JP, Liu J, Li Z, Chandrashekar A, Martinez DR, Loos C, Atyeo C, Fischinger S, Burke JS, Slein MD, Chen Y, Zuiani A, Lelis FJN, Travers M, Habibi S, Pessaint L, Van Ry A, Blade K, Brown R, Cook A, Finneyfrock B, Dodson A, Teow E, Velasco J, Zahn R, Wegmann F, Bondzie EA, Dagotto G, Gebre MS, He X, Jacob-Dolan C, Kirilova M, Kordana N, Lin Z, Maxfield LF, Nampanya F, Nityanandam R, Ventura JD, Wan H, Cai Y, Chen B, Schmidt AG, Wesemann DR, Baric RS, Alter G, Andersen H, Lewis MG, Barouch DH, DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 369, 806–811 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu J, Li Z, He X, Gebre MS, Bondzie EA, Wan H, Jacob-Dolan C, Martinez DR, Nkolola JP, Baric RS, Barouch DH, Deletion of the SARS-CoV-2 Spike Cytoplasmic Tail Increases Infectivity in Pseudovirus Neutralization Assays. Journal of Virology, JVI.00044–00021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.van Haren SD, Dowling DJ, Foppen W, Christensen D, Andersen P, Reed SG, Hershberg RM, Baden LR, Levy O, Age-Specific Adjuvant Synergy: Dual TLR7/8 and Mincle Activation of Human Newborn Dendritic Cells Enables Th1 Polarization. Journal of immunology (Baltimore, Md. : 1950) 197, 4413–4424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Berenbaum MC, Correlations between methods for measurement of synergy. The Journal of infectious diseases 142, 476–480 (1980). [DOI] [PubMed] [Google Scholar]
- 89.Li S, Rouphael N, Duraisingham S, Romero-Steiner S, Presnell S, Davis C, Schmidt DS, Johnson SE, Milton A, Rajam G, Kasturi S, Carlone GM, Quinn C, Chaussabel D, Palucka AK, Mulligan MJ, Ahmed R, Stephens DS, Nakaya HI, Pulendran B, Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat Immunol 15, 195–204 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are in the paper or supplementary materials.