Abstract
During the coronavirus disease 2019 (COVID-19) pandemic, a wave of rapid and collaborative drug discovery efforts took place in academia and industry, culminating in several therapeutics being discovered, approved and deployed in a 2-year time frame. This article summarizes the collective experience of several pharmaceutical companies and academic collaborations that were active in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antiviral discovery. We outline our opinions and experiences on key stages in the small-molecule drug discovery process: target selection, medicinal chemistry, antiviral assays, animal efficacy and attempts to pre-empt resistance. We propose strategies that could accelerate future efforts and argue that a key bottleneck is the lack of quality chemical probes around understudied viral targets, which would serve as a starting point for drug discovery. Considering the small size of the viral proteome, comprehensively building an arsenal of probes for proteins in viruses of pandemic concern is a worthwhile and tractable challenge for the community.
Subject terms: Drug discovery and development, Viral infection
The COVID-19 pandemic spurred a wave of rapid and collaborative drug discovery efforts. This Perspective article summarizes scientific drivers and considerations behind such antiviral small-molecule discovery programmes and proposes strategies to accelerate future efforts.
Introduction
Viral outbreaks are one of the gravest public health risks of our times, exemplified by the ongoing coronavirus disease 2019 (COVID-19) pandemic that has claimed more than 6 million lives. Several global trends make pandemics more likely in the future. Climate change coupled with wildlife destruction serve to increase human–animal interactions and the risk of zoonotic spillover1,2. Warming temperatures also increase the geographical areas that are hospitable to viral vectors such as mosquitoes and ticks, potentially increasing the spread of arboviruses3. The prevalence of global travel can rapidly turn a local epidemic into a global pandemic4. As such, developing effective therapeutics against current and future pandemics should be a global public health priority.
Before the COVID-19 pandemic, the focus of antiviral development was on human immunodeficiency virus (HIV) and hepatitis C virus (HCV), accounting for more than 67% of approved antivirals5. The routine drug discovery and development timescale could be of the order of decades, especially for first-generation therapeutics against a virus. COVID-19 combined the attributes of an acute, severe and rapidly transmissible viral disease. For the first time, the translational science sector has successfully executed rapid drug discovery campaigns and developed novel antivirals amid a fast-moving pandemic. Within 2 years there were two oral therapeutics with emergency use authorization (EUA): nirmatrelvir (Pfizer) and molnupiravir (Merck; developed originally for Venezuelan equine encephalitis virus, VEEV). There were also several clinical stage investigational oral therapeutics, such as ensitrelvir (S-217622; Shionogi), pomotrelvir (PBI-0451; Pardes Biosciences), bemnifosbuvir (AT-527; ATEA) and EDP-235 (Enanta). In addition, remdesivir (Gilead Sciences; developed originally for Ebola), a small-molecule therapeutic delivered intravenously, was approved early on in the pandemic.
This Perspective draws from a round table discussion between biopharmaceutical companies and public sector organizations that have substantial research and development efforts in COVID-19. We outline key lessons from rapid antiviral drug discovery efforts, or ‘sprints’, and articulate remaining open questions, specifically focusing on target selection, medicinal chemistry strategies, in vitro and in vivo models, and methods to pre-empt resistance.
Target selection and validation
Antiviral therapeutics can be directed at the host or at the virus itself. Host-directed antivirals target human proteins that are essential in the viral life cycle. Significant effort has been expended in finding host-directed therapeutics against COVID-19 (refs. 6,7), most notably through numerous drug repurposing screens. Some of these agents went into clinical trials, through platform trials such as Accelerating COVID-19 Therapeutic Interventions and Vaccines (ACTIV) and Randomized Evaluation of COVID-19 Therapies (RECOVERY)8 as well as company-sponsored trials. Nonetheless, to date there is no approved host-directed antiviral therapeutic against COVID-19. It is argued that the advantages of a host-directed approach are a higher barrier to antiviral resistance and broad-spectrum activity if the target is used by multiple viruses9. Nonetheless, downsides include possible host pathway-mediated (on-target) toxicity, lower efficacy if the viral life cycle leverages multiple redundant targets and poor translation of in vivo models. Historically, the only successful host-directed antivirals have been interferon for HCV and hepatitis B virus (HBV), and CCR5 antagonists for HIV10, as well as cyclophilin inhibitors such as alisporivir (Debio-025) in late-stage clinical development for HCV11. Most approved antivirals therefore directly target viral proteins, and we focus on targets associated with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Before embarking on a drug discovery effort, target selection is crucial. The ideal antiviral target is essential for the viral life cycle (Fig. 1), has a tractable mechanism of action, can be inhibited by small molecules with drug-like pharmaceutical properties and has a high fitness barrier to mutation12. We identify several key concepts that aid SARS-CoV-2 antiviral target selection (Table 1).
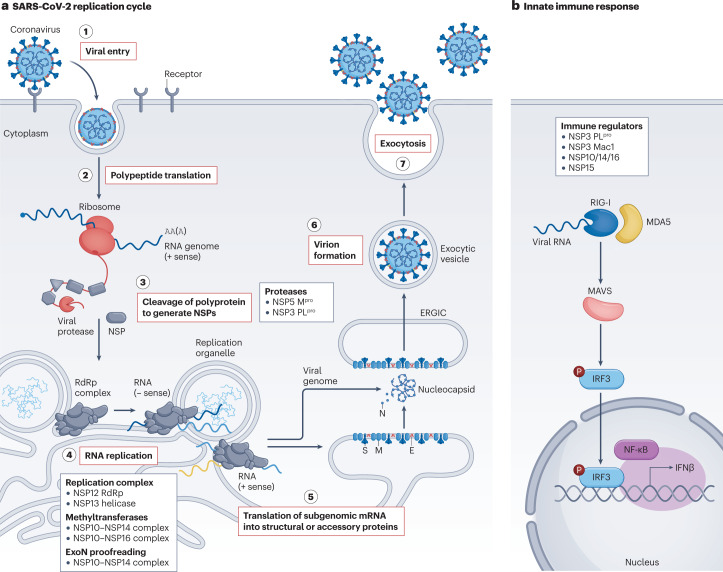
Fig. 1. Key targets in the SARS-CoV-2 replication cycle.
a, Stage 1: severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) enters the host cell upon binding to the extracellular receptors angiotensin-converting enzyme 2 (ACE2) and transmembrane protease serine 2 (TMPRSS2)119. Stage 2: following viral uncoating, the viral RNA is released and two large open reading frames (ORF1a and ORF1b) are translated into polyproteins. Stage 3: these polyproteins are co- and post-translationally processed by viral proteases into non-structural proteins (NSPs) that form the viral replication complex. Continuous cleavage of the polyprotein is required for sustained RNA synthesis, suggesting that formation of the replication complex is dynamic and occurs continually. Stage 4: the central enzyme of the replication complex is the RNA-dependent RNA polymerase (RdRp), which synthesizes viral RNA. Other enzymes such as the NSP13 helicase and the NSP14 N-methyltransferase contribute to initiation of replication, RNA unwinding, proofreading and sustaining RNA synthesis. Stage 5: genomic viral RNA is encapsulated by nucleocapsid protein (N), and viral structural proteins translocate to the endoplasmic reticulum. Stage 6: structural proteins transit through the endoplasmic reticulum-to-Golgi intermediate compartment (ERGIC) to the Golgi for glycosylation and progression into exocytic vesicles. Encapsidated genomic RNA buds into the final virion, acquiring a lipid bilayer that contains structural proteins spike (S), membrane (M) and envelope (E). Stage 7: the virion is released from the infected cell by exocytosis. Key viral targets are listed in the boxes. b, As part of the innate immune response towards SARS-CoV-2 infection, the host’s pattern recognition receptors such as proteins retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated protein 5 (MDA5) recognize viral RNA and trigger downstream signalling cascades involving the mitochondrial antiviral signalling protein (MAVS), leading to activation of the IRF3 and nuclear factor-κB (NF-κB) transcription factors that induce interferon-β (IFNβ) transcription. Viral proteins such as those listed in the box interfere with components of the pathway. Mpro, main protease; P, phosphorylation; PLpro, papain-like protease.
Table 1.
Mechanism of action validation for selected SARS-CoV-2 targets
| Target | NSP5 Mpro | NSP12 polymerase | NSP3 PLpro | NSP3 Mac1 | NSP13 helicase | NSP14 methyltransferase | NSP15 endoribonuclease | NSP16 methyltransferase | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Site | Catalytic site | Ectopic site | Catalytic site | NIRAN ADP site | Catalytic site | Catalytic site | ATPase site | Central channel | Catalytic site | ectopic site | Catalytic site | Catalytic site |
| Sequence conservation at ‘pocketome’ site (%)a | 52 | 50 | 94 | 87 | 29 | 62 | 71 | 92 | 70 | 61 | 78 | 61 |
| Chemical probe validation | ||||||||||||
| Structural/fragments/HTS data | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||
| Availability of chemical probes | ✓ | ✓ | ✓ | ✓ | ✓ | |||||||
| MoA validation | ||||||||||||
| Biological evidence (other viral families), MoA confirmed in vitro/in vivo | ✓ | ✓ | ✓ | ✓ | ||||||||
| Biological evidence (coronaviruses), MoA confirmed in vitro/in vivo | ✓ | ✓ | ✓ | |||||||||
| Clinical evidence (other viral families) | ✓ | ✓ | ✓ | ✓ | ||||||||
| Clinical evidence (coronaviruses) | ✓ | ✓ | ||||||||||
HTS, high-throughput sequencing; MoA, mechanism of action; Mpro, main protease; PLpro, papain-like protease. aSequence conservation across 27 alphacoronaviruses and betacoronaviruses for selected severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) targets (data taken from ref. 38, which considered sequences up to 31 July 2020).
Validated mechanism of action
Antiviral targets with previous clinical validation demonstrating that target inhibition leads to antiviral effects have a lower translational risk. This is a high bar to meet, and these targets might not be rapidly available in an emerging pandemic setting. However, evidence from other viruses can help to provide confidence in the target when certain viral replication mechanisms are shared. We can establish ‘target-class confidence’ if there are multiple approved therapeutics against the same target class in multiple viruses. Ongoing efforts by the National Institute for Allergy and Infectious Diseases aim to define prototype and priority pathogens — key viruses in viral families of pandemic concern — and to develop therapeutics for them13.
In the absence of clinical validation, a target is more credible if its mechanism of action is understood and it has been demonstrated that ablating protein function directly impacts viral replication in vitro or in vivo. This can be achieved via chemical probes or a reverse genetics approach.
The indirect approach of inhibiting viral proteins that are responsible for evading host immune response is less common, due to the complex interplay between multiple viral proteins and the host immune response. Likewise, care must be taken in targeting steps in the life cycle such as viral entry, where multiple pathways for infections and cell-to-cell spread exist for some viruses.
Chemical probe validation
The goal of an antiviral therapy is the chemical inhibition of the target. As such, key questions are: whether there are sites on the protein that can be engaged by a small molecule (and therefore the target is ‘druggable’); whether engaging these sites modulates protein function; and whether a chemical probe that modulates protein function also leads to viral inhibition in cellular assays.
Identifying small molecules that potently inhibit the target and have corresponding cellular antiviral activity, with changes in potency of inhibition translating to changes in antiviral activity (the structure–activity relationship), helps to build confidence that a target is validated and tractable. The availability of meaningful functional assays and linked structural data can greatly facilitate the development of chemical probes for previously untargeted viral proteins. In particular, advances in cryo-electron microscopy now allow elucidation of complex protein structures14–16. Although many chemical probes have been developed for human targets, fewer probes and structural data are available for viral targets.
Sequence conservation
In addition to direct clinical evidence or strong mechanistic evidence, the saliency of a target can be evaluated by assessing its conservation across the virus family or within circulating variants. Sequence and structural conservation can be evaluated across the entire protein or within the active binding site. Conserved targets are perceived as more robust because a target that is essential to the viral life cycle should accumulate fewer mutations. Even if a mutationally flexible target is essential, it is harder to drug because the molecule would need to potently inhibit all variants. Targeting a conserved viral protein also increases the likelihood of developing a broad-spectrum antiviral, an important priority for pandemic preparedness.
Clinically validated targets
The SARS-CoV-2 genome encodes four structural proteins and two open-reading frame polyproteins. The polyproteins are cleaved by two cysteine proteases: the non-structural protein 5 (NSP5) main protease (Mpro), responsible for cleavage at 11 positions, and the NSP3 papain-like protease (PLpro) with three cleavage positions (Fig. 1a). These cleavage steps liberate shorter viral proteins, such as the RNA-dependent RNA polymerase (RdRp) NSP12, that are crucial for viral replication and evasion of the host immune response. For example, Mpro directly cleaves NLRP12 and TAB1, two modulators of inflammatory pathways, which might point to a molecular mechanism for the enhanced production of cytokines and inflammatory response observed in patients with COVID-19 (ref. 17).
To date, the only oral SARS-CoV-2 therapeutics in clinical use or late-stage clinical trials target either the NSP5 Mpro or the NSP12 RdRp. Surveying the clinical and preclinical antiviral pipeline, we anticipate Mpro and RdRp to remain the most prevalent targeted proteins for the next 3–5 years.
The NSP5 Mpro
Numerous companies initiated programmes targeting Mpro early in the pandemic, and several SARS-CoV-2 Mpro inhibitors are now under EUA or in the clinical pipeline (Table 2). Mpro is an attractive target based on a rare confluence of factors. First, mechanistic understanding, because protease function in polyprotein processing is well-characterized and assayable given that inhibition of Mpro directly suppresses viral replication (as observed for Mpro in other viruses). Also, the rapid availability of Mpro structures facilitated structure-based drug design efforts18,19 (Fig. 2). Second, cysteine proteases are a well-characterized class of enzymes known to be druggable and to have amino acid sequence and cleavage specificity distinct from that of human cysteine proteases. Third, clinical precedent, given that multiple HIV and HCV protease inhibitors are in clinical use, and a human rhinovirus (HRV) protease inhibitor reduced viral titres in a phase II clinical trial20. Fourth, chemical probe validation, with multiple SARS-CoV chemical probes targeting Mpro reported after the 2003 SARS-CoV epidemic in Asia21,22. SARS-CoV and SARS-CoV-2 Mpro share 96% sequence similarity, therefore, some chemical probes were rapidly redeployed to SARS-CoV-2. For example, PF-00835231 was developed preclinically in response to the 2003 SARS-CoV pandemic, then shown to have activity against SARS-CoV-2 and rapidly entered clinical phase I trials in 2020 as an intravenous treatment (NCT04535167)23,24. Upon the development of the orally bioavailable follow-up compound nirmatrelvir25, no further clinical investigation of PF-00835231 was undertaken. In addition, studies on the norovirus 3C protease led to the development of GC376, a compound with good activity across norovirus, picornavirus and coronavirus families and efficacious in a cat coronavirus model26,27.
Table 2.
Main protease and polymerase inhibitors in the clinic
| Compound name (company) | Hit-finding strategy | Starting point | Clinical candidate structure | HLM CLint (μl min−1 mg−1) | Rat CLp (ml min−1 kg−1) | Rat oral F (%) |
SARS-CoV-2 antiviral activity (EC50 (nM)) | Refs. |
|---|---|---|---|---|---|---|---|---|
| Main protease inhibitors | ||||||||
| Nirmatrelvir (Pfizer) | Lead from historic SARS-CoV campaign | 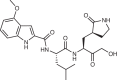 |
 |
24.5 | 27.2 | 50 | 62 (human airway epithelial cell) | 87 |
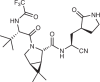
| Ensitrelvir (Shionogi) | Structure-based virtual screening and pharmacophore filtering | 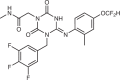 |
 |
97% after 30 min of incubation | 7.3 | 111 | 370 (Vero E6/TMPRSS2, CPE) | 18 |
|
Pomotrelvir (Pardes Biosciences) |
Historic data from patents and docking across Mpro structures | Undisclosed | 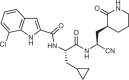 |
Undisclosed | Undisclosed | Undisclosed | 32 (WA1 strain, iPS-AT2, PFU) | 205 |
| Polymerase inhibitors | ||||||||
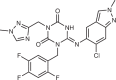
| Remdesivir (Gilead Sciences) | An Ebola antiviral obtained from screening a nucleoside library | 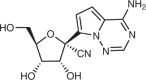 |
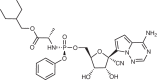 |
Undisclosed | Undisclosed | i.v. drug | 9.9 (human airway epithelial cell) | 251 |
| Molnupiravir (Merck) | Designed as a VEEV antiviral | 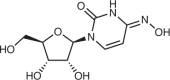 |
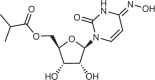 |
Undisclosed | Undisclosed | 52 | 670–2,660 (A549), 320–2,030 (Vero E6) | 252,253 |
CLp, plasma clearance; CPE, cytopathic effect; EC50, half-maximal effective concentration; F, bioavailability; HLM CLint, human liver microsome intrinsic clearance; iPS-AT2, induced pluripotent stem alveolar epithelial type II cell; i.v., intravenous; Mpro, main protease; PFU, plaque-forming unit; SARS-CoV, severe acute respiratory syndrome coronavirus; TMPRSS2, transmembrane protease serine 2; VEEV, Venezuelan equine encephalitis virus.
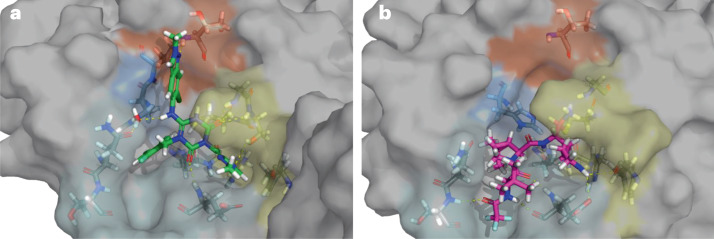
Fig. 2. Co-crystal structures of the SARS-CoV-2 main protease active site.
a, The inhibitor ensitrelvir (PDB: 7VU6) is bound18. b, The inhibitor nirmatrelvir (PDB: 7RFS) is bound25. The colours indicate the various binding pockets following the standard Schechter and Berger nomenclature for proteases254: P1′ (orange), P1 (yellow), P2 (blue) and P3–5 (cyan). SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
The NSP12 RdRp
The replication of RNA viruses requires a mechanism to synthesize viral RNAs. The RdRp catalyses the replication of RNA from a RNA template, synthesizing a complementary RNA strand. Inhibiting the RdRp therefore inhibits viral replication. Similarly to Mpro, the catalytic subunit of the SARS-CoV-2 RdRp has been validated as a target for several reasons. First, mechanistic understanding, as the structure and function of RdRp are well understood across RNA viruses. Second, there is clinical precedent, as multiple RdRp inhibitors are in clinical use or development for HIV, HCV, respiratory syncytial virus (RSV), influenza A and influenza B28, increasing confidence in the coronavirus RdRp as a relevant target. Third, a wide range of RdRp inhibitors have been developed, allowing chemical probe validation. Some have been successfully deployed against SARS-CoV-2 infection, for example, remdesivir29 and subsequently molnupiravir30. In particular, remdesivir’s broad-spectrum antiviral activity against coronaviruses was published before the SARS-CoV-2 pandemic31,32, as well as its biochemical mechanism of action33. Although both molecules engage the same target and require metabolic activation by the endogenous cellular machinery, once activated, the mechanisms of remdesivir and molnupiravir are different: remdesivir leads to delayed termination of RNA replication34, whereas molnupiravir leads to mutated RNA products35. Notably, other repositioned RdRp inhibitors such as favipiravir or sofosbuvir, albeit showing some antiviral activity in selected cellular assays, did not impact mortality and hospital admissions in SARS-CoV-2 clinical trials36,37. Fourth, the sequence of the catalytic site of RdRp is broadly conserved across coronaviruses and variants of SARS-CoV-2 (ref. 38) (Table 1). Additionally, RdRps are not encoded by human cells, although inhibition of human RNA polymerases is a source of off-target toxicity for RdRp inhibitors39.
Targets with chemical probe validation
The NSP3 PLpro
The NSP3 protease has a papain-like fold, and processes three cleavage sites in the N-terminal part of the polyproteins to produce mature NSP1, NSP2 and NSP3 (Fig. 1). The active site contains a typical protease catalytic triad, composed of Cys112–His273–Asp287. Apart from its essential role in viral replication, PLpro cleaves ubiquitin, ISG15 and IRF3, which are known regulators of host innate immune pathways17,40. Non-covalent inhibitors have been found against SARS-CoV41 and SARS-CoV-2 PLpro (refs. 42–45), with PLpro inhibition correlateing with cellular antiviral activity.
Targets with genetic evidence
Viral replication
The NSP13 helicase
NSP13 is part of the RdRp replication complex and catalyses the unwinding of RNA in a 5′ to 3′ direction46,47. It is also required for the proofreading and template switching functions of the replication complex48,49. There is no reported chemical probe against any coronavirus helicase, although a crystallographic fragment screen suggested that it is a tractable target50. Viral helicase inhibitors have been pursued for other infections; for example, amenamevir (ASP2151) is approved in Japan for treating the reactivation of varicella zoster virus (shingles)51, and pritelivir (BAY 57-1293) is in phase III clinical trials for herpes simplex virus (HSV; NCT03073967)52,53.
Evading host immunity
Upon viral infection, viral components with conserved molecular motifs termed pathogen-associated molecular patterns (PAMPs) are recognized by the host’s pattern recognition receptors (PRRs) to induce an antiviral innate immune response. Viruses subvert this innate immune response by targeting a range of signalling cascades such as Toll-like receptor signalling and intracellular signalling pathways via retinoic acid-inducible gene I (RIG-I) or melanoma differentiation-associated protein 5 (MDA5) (Fig. 1b).
The NSP3 macrodomain
As part of the innate immune response, host ADP-ribosyltransferases transfer ADP-ribose onto viral proteins, ultimately contributing to the suppression of viral replication. The viral macrodomain Mac1 from NSP3 counteracts this innate immune response by cleaving ADP-ribose already transferred onto viral proteins. Viral macrodomains are found in corona, alpha, rubi and herpes viruses54,55, and macrodomain mutations disrupt catalytic activity and decrease virulence56. To date, there is no reported Mac1 chemical probe with potent cellular antiviral activity, nor clinical evidence for inhibiting viral macrodomains. However, a crystallographic fragment screen revealed starting points for small-molecule inhibitor synthesis, therefore suggesting that Mac1 is tractable for small-molecule development54,57–59.
The NSP14 and NSP16 methyltransferases
Methyltransferases catalyse the transfer of a methyl group from S-adenosylmethionine to RNA substrates. In complex with NSP10, NSP14 catalyses the N-methylation of guanosine, and NSP16 completes the formation of the RNA cap by 2′-O-methylation of viral mRNA ribose60. The formation of the RNA cap prevents the recognition of viral RNA by host PRRs such as RIG-I, thereby subverting innate immune responses61. Recent structural biology has elucidated the mechanism of methyl transfer62,63, and chemical probes targeting NSP14 have been reported64,65. However, to date there is no evidence that chemical inhibition of NSP14 translates to an effect on cellular antiviral activity, nor clinical precedent for inhibition of viral methyltransferases. NSP14 also encodes an exoribonuclease activity that performs a proofreading function and antagonizes the innate immune response66.
The NSP15 endonuclease
The NSP15 endonuclease is an RNA uridylate-specific endoribonuclease that is part of the EndoU family67. Members of this enzyme family act on viral RNA that would activate the host innate immune response by cleaving 3′ of uridylate and thereby generating a 2′, 3′ cyclic phosphate and 5′-hydroxyl termini68–71. Nidoviral uridylate-specific endoribonucleases are highly conserved amongst the nidoviral families of Coronaviridae, Areriviridae and Roniviridae72. There are no chemical probes or approved therapeutics against coronavirus endonucleases, although a crystallographic fragment screen suggested that starting points for small-molecule inhibitor synthesis exist73. An endonuclease from a different viral family has been targeted for influenza, in which an inhibitor of the cap-dependent endonuclease, baloxavir marboxil (Xofluza), has been in clinical use for several years74. However, resistant viruses emerged in baloxavir-treated subjects at a frequency ranging from 3–11% in adults to more than 23% in children74,75.
Medicinal chemistry
Once a target has been selected, medicinal chemistry is an iterative process of designing, making and testing molecules to progress chemical starting points into development candidates. Setting a realistic goal for the profile of a therapeutic is important when designing an appropriate medicinal chemistry strategy so that the final product has sufficient therapeutic value. These goals are typically divided into target product profile (TPP) or target candidate profile (TCP)76. The TPP describes clinical attributes of a therapeutic, whereas the TCP describes the molecular attributes (such as target engagement, cellular antiviral response, safety pharmacology) that the molecule must fulfil. Here, we discuss the TPP as it informs the development of target-specific TCPs.
Target product profile
In an ideal antiviral drug discovery setting, the TPP for a directly acting antiviral aims for an orally available drug that is administered once daily or, for acute infection such as SARS-CoV-2, even once only (Table 3). In addition, a wide treatment window is desirable, to be relevant to patient populations with limited access to rapid diagnostics.
Table 3.
Example of target product profile for an acute viral infection
| Target product profile | Pandemic response | Ideal therapeutic |
|---|---|---|
| Route | Orally bioavailable | Orally bioavailable |
| Dosing | Three times a day | Once, or once a day |
| Dose | Up to 1 g three times daily | <250 mg |
| PK/PD | Cmin >1 × EC90, unbound | Higher coverage, with high barriers to resistance |
| Drug–drug interactions | Acceptable | No |
| Co-dosing with PK enhancers | Acceptable | No |
| Patient cohorts | Narrow cohort acceptable | Available to all (e.g., women of child-bearing age) |
| Therapeutic window | Within 48 h of infection | Extended up to 5 days |
Cmin, trough concentration; EC90, 90% effective concentration; PD, pharmacodynamics; PK, pharmacokinetics.
Yet, for an immediate pandemic response, many of these specifications are a luxury, and less stringent TPP requirements could be tolerated (Table 3). Considering the urgency of the situation and analysing the most vulnerable population, the following limitations alone or in combination might be acceptable for first-generation antiviral therapies. First, suboptimal dosing regimens of up to three or four times a day. Second, suboptimal delivery routes including intravenous formulations for selected high-risk patients. Albeit impractical for widespread treatment of early infection, the efficacious intravenous compound remdesivir (Veklury; Gilead Sciences) received EUA and then full FDA approval during the SARS-CoV-2 pandemic, and the first SARS-CoV-2 Mpro inhibitor entered clinical trials as an intravenous formulation23. Third, a suboptimal distribution, metabolism and pharmacokinetic (DMPK) profile while ensuring that the trough concentration (Cmin) of the free drug determined in vivo remains above the protein-adjusted 90% effective concentration (EC90) defined in cellular assays (Fig. 3). Continuous in vivo drug concentrations exceeding the EC90 are generally considered to be the minimum to achieve antiviral efficacy77, and higher trough concentrations might be desirable for second-generation antivirals once better understanding of resistance is established. Fourth, consideration of narrower patient cohorts and a willingness to monitor potential drug–drug interactions. Clinically manageable drug–drug interactions, co-dosing with pharmacokinetic enhancers or mechanisms of action precluding use in selected patient cohorts, such as in women of child-bearing potential, might be acceptable for selected high-risk patient cohorts in a pandemic. Fifth, acceptance of short therapeutic windows. For some viral therapeutics, such as oseltamivir for influenza, rapid treatment is required for antiviral efficacy78. However, it can be challenging to prescribe and distribute a drug to a patient within 48 h of symptom onset, and it is preferable if antiviral therapy remains efficient if treatment is delayed for up to 5 days after symptom onset.
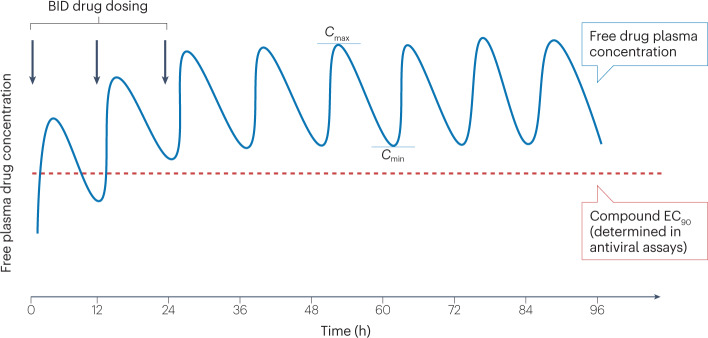
Fig. 3. The pharmacokinetic profile of a hypothetical antiviral compound administered twice daily.
The free (unbound) drug plasma concentration (blue) is depicted, including the maximum drug concentration (Cmax) and minimum drug concentration (Cmin) at steady state. The 90% effective concentration (EC90; red dotted line), the concentration at which 90% inhibition of viral replication is observed in cellular antiviral assays, is corrected for plasma protein binding. BID, twice daily.
For SARS-CoV-2, there are several clinical observations that might require additional amendments to the TPP, such as biphasic viral kinetics, or ‘rebounds’ that have been observed in treated and untreated patients79,80, and various chronic neurological, cardiovascular and gastrointestinal symptoms, known as post-acute COVID-19 syndrome (PACS) or colloquially ‘long COVID’81,82. The aetiology of these observed symptoms is as yet unknown; several factors might be at play and have implications on the TPP. First, the free concentration of the drug may be insufficient to adequately suppress viral replication, therefore, aiming for a Cmin that covers multiples of the EC90 might be required. Second, the viral kinetics might require longer treatment duration. Third, untargeted viral reservoirs might persist. In particular, the impact of SARS-CoV-2 infection on the central and peripheral nervous systems in the acute and chronic phases remains unclear83,84, so a different tissue distribution profile might be desired in the TPP. Finally, for PACS, immune triggers might be clinically relevant and potentially require a host-directed approach beyond direct-acting antivirals.
Finally, the TPP might be refined to accommodate features relevant to compounds for pandemic preparedness and to increase the barrier to resistance. Compounds with an increased antiviral treatment spectrum, aiming to cover several viral strains from a viral family — or even across viral families — might be desirable. However, increasing the spectrum comes with medicinal chemistry challenges: a compound needs to achieve the (often conflicting) goals of simultaneously inhibiting proteins from related viruses, yet avoiding related host proteins and causing off-target effects.
Accelerating medicinal chemistry
The main drivers of an accelerated drug discovery effort during a pandemic are the upfront availability of high-quality chemical matter, the willingness to move at risk, a reconsideration of the essential attributes versus the ideal therapeutic profile to address the immediate unmet medical need, and the funding to do the work. More broadly, a sense of organizational commitment and alignment with management is required to release significant financial investment at risk and execute fast, safe and rigorous campaigns.
Availability of high-quality chemical matter
Sir James Black, winner of the 1988 Nobel Prize in Physiology and Medicine, famously stated that “the most fruitful basis for the discovery of a new drug is to start with an old drug”85. One of the most pressing problems with antiviral drug discovery against a novel target is the availability of high-quality chemical matter. The only two approved RdRp-targeting antivirals, remdesivir and molnupiravir, were developed before the pandemic. Exceptionally rapid drug discovery efforts were executed against Mpro: nirmatrelvir, an example discussed above, was based on a peptidomimetic scaffold optimized for SARS-CoV in 2003, with low oral bioavailability supporting only intravenous dosing. The scaffold in turn shares structural similarity with rupintrivir, a HRV antiviral developed in the 1990s86. Significant medicinal chemistry was required to optimize oral bioavailability, leading to the novel SARS-CoV-2 Mpro inhibitor nirmatrelvir25. Another example is ensitrelvir, which is structurally different from nirmatrelvir18, but related to a P2X3 antagonist in the Shionogi clinical pipeline87. These pockets of exceptional drug discovery appear to validate Sir James’s adage, but also reflect the reality of preclinical drug development timelines. By leveraging pre-existing validated chemical matter, the timeline to first-in-human studies is dramatically shortened.
Looking ahead, for pandemic preparedness, we argue that a campaign to develop quality chemical probes and leads against targets in the viral proteome is a key opportunity for investment. Similar to efforts in systematically finding probes against human targets88, it will be worthwhile for the scientific community to find chemical probes against every viral protein for the virus families most likely to produce the next pandemic. The viral proteome is orders of magnitude smaller than the human proteome, therefore, the level of investment required to execute such an effort is likely to be much less than the human and financial toll of a future pandemic.
Hit-finding technologies
A prerequisite of running a medicinal chemistry campaign is quality biochemical assays. This is often a chicken-and-egg problem — good-quality chemical probes help to validate an assay by generating the confluence of biophysical, biochemical, structural biology and antiviral efficacy data. Establishing a suite of high-throughput, orthogonal biochemical and cell-based assays around a target is one of the challenges of rapid drug discovery against novel viral targets. As viral targets are typically conserved, pre-emptively developing assays and making them available open source to the field should aid future drug discovery and pandemic preparedness efforts.
Several enabling technologies have been deployed successfully during the pandemic to accelerate finding hit compounds against numerous targets. Experimentally, crystallographic fragment screens have seen prolific successes against multiple targets. The confluence of high-throughput crystallography, an automated processing pipeline and expanded fragment libraries has delivered dense fragment hits against Mpro (ref. 89), Mac1 (ref. 54), the NSP13 helicase50 and the NSP15 endonuclease73. The latter three are novel targets with no pre-existing inhibitors. However, going from fragment to lead compound remains a challenge. Creative approaches such as the COVID Moonshot, which used crowdsourcing to generate ideas at the fragment-to-lead stage via fragment merging90, have been attempted during the pandemic. The resulting lead is under preclinical development, although still some distance away from clinical evaluation19. Organizationally, several pharma companies in this round table have established shared hit-finding efforts, where blinded libraries of compounds were screened for Mpro biochemical activity.
Computationally, structure-based virtual screening yielded multiple successes. The discovery of S-217622 used structure-based virtual screening followed by pharmacophore filtering to generate novel non-covalent hits against Mpro (ref. 18). An unrelated effort similarly used virtual screening to discover novel chemotypes that led to potent Mpro inhibitors with broad-spectrum antiviral activity91. Beyond hit finding, free energy perturbation (FEP)-guided optimization was used to redesign the weak hit perampanel into a potent inhibitor92. However, successes in computational chemistry have so far been focused on Mpro, a target with well-developed inhibitors predating SARS-CoV-2; computational hit-finding campaigns against novel viral targets performed by members of this round table were less successful.
Late-stage drug development
Beyond the discovery stage, developing and executing process-scale chemistry for late-stage drug development, clinical trials and eventual market distribution require significant resources. A holistic view on chemistry timelines needs to be considered. For example, process-scale chemistry should be involved as soon as lead scaffolds emerge. This allows process development towards key building blocks or intermediates to commence before candidates are declared, thus reducing the lead time. Concomitantly, the commercial availability of building blocks, and the scalability of synthetic routes, should be factored into the medicinal chemistry campaign.
Further, we argue that a crucial juncture for a drug discovery sprint campaign is when to commit to scaling up synthesis, and to what scale and quality. Conventional drug discovery usually takes a stage gate approach, with separate scale-up campaigns initiated once positive results from various preclinical toxicity experiments are available. This mitigates risk, as the compound can fail at each stage of the development process. However, to accelerate drug development during a pandemic, stage gates and milestones can lead to delays by preventing time-consuming activities being performed in parallel. The most successful sprints during the pandemic benefited from a large upfront investment to enable all chemical manufacturing stages in parallel, when the evidence for the drug candidate was still at the level of a cellular antiviral assay and early pharmacokinetics. In this case, resources for process chemistry and pilot plants also need to be reserved at an early stage. However, delays in the drug discovery campaign, or a negative read-out during the development process, can mean these expensive downstream resources become underused.
Ultimately, speed comes at a cost, in both cash expenditure and increased risk. If the aim is to impact an ongoing pandemic via a drug discovery sprint, we argue that the investment has to be provided upfront in its entirety. Piecemeal investments can lead to significant delays later in the process. This point is particularly poignant for public sector investments and grants for drug discovery, as excessive risk cannot be directly economically compensated.
Cell culture models
Cellular antiviral infection models are paramount to the identification of effective small-molecule inhibitors. A crucial part of the medicinal chemistry effort is to understand the variation in biological activity through a cellular assay as a function of chemical structures, which is only feasible with a low-variance system. Especially in the later phases of drug discovery campaigns, the throughput of a cellular assay is crucial and ideally matches chemical synthesis. Assay data are often used to strategically rank compounds, so comparing potencies of different compounds across assays should be avoided.
Many assays use pathogenic infectious viral strains and therefore need to be run in laboratories with higher containment capabilities (that is, biosafety level (BSL) 3, or BSL4). This complicates assay logistics and accessibility of relevant antiviral assays and ultimately impacts assay throughput. With these restrictions in mind, the round table agrees that reproducible antiviral cellular assays that can be run in a high-throughput set-up are much preferred over noisy and/or low-throughput assays, regardless of purported biological relevance. A crucial component of the ideal assay is tracking variance, robustness and reproducibility with statistical measures such as Z scores.
In antiviral drug discovery campaigns, cellular assays are generally grouped into high-throughput tier 1 and lower-throughput tier 2 assays (Fig. 4). Tier 1 cellular assays are reliable, straightforward and scalable 2D cell cultures infected with a SARS-CoV-2 strain. Efficacy of antiviral inhibitors is generally assessed by adding different concentrations of compound to the culture (either before or after infection), and viral replication is subsequently measured using various methods. In contrast, tier 2 cellular assays are often lower throughput and might use primary cells with a higher disease relevance.
Fig. 4. A typical cascade approach for antiviral screening.
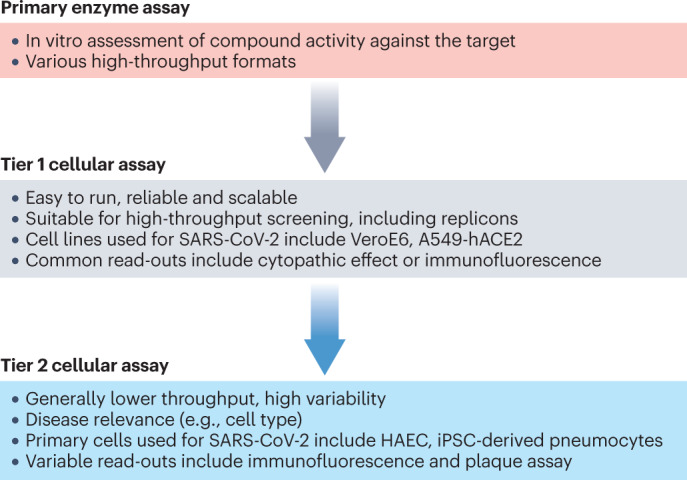
After the initial primary enzymatic assays, a high-throughput tier 1 cellular assay is used to drive medicinal chemistry, and a lower-throughput tier 2 assay is used to predict human dose. A549-hACE2, A549 cell line overexpressing human angiotensin-converting enzyme 2; HAEC, human airway epithelial cell; iPSC, induced pluripotent stem cell; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
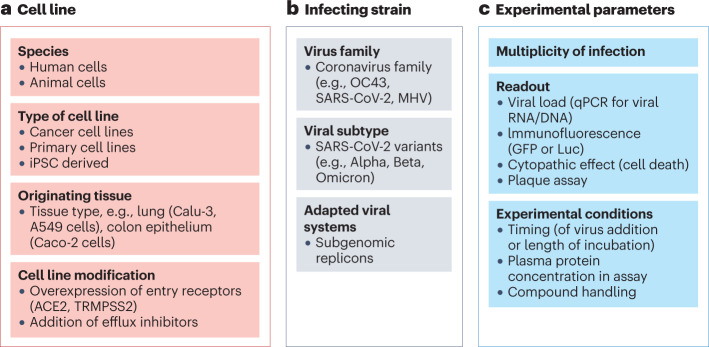
A wide range of experimental parameters influence the use and scalability of cellular assays (Fig. 5), including the type of cell lines chosen, the infecting viral strain and the experimental read-out used. Modifications to these parameters can impact efficacy measurements and could contribute to the significant lab-to-lab variability reported in antiviral efficacy measurements. As drug discovery efforts rely on comparability of results over time, early assay optimization and consistent use of a fixed protocol within an established facility are essential to medicinal chemistry efforts and understanding of structure–activity relationships.
Fig. 5. Common variables in cellular antiviral assays that impact the final read-out.
a, The cell lines used. b, The choice of infecting virus strain. c, The various experimental parameters. ACE2, angiotensin-converting enzyme 2; GFP, green fluorescent protein; iPSC, induced pluripotent stem cell; Luc, firefly luciferase; MHV, mouse hepatitis virus; qPCR, quantitative polymerase chain reaction; TMPRSS2, transmembrane protease serine 2.
Common tier 1 cellular models
Several cell lines are routinely used to assess antiviral activity against SARS-CoV-2. Initially, many assays focused on the African green monkey cell line Vero E6, a cell line commonly used for antiviral assays and previously used for SARS-CoV replication93. Vero E6 cells are very susceptible to SARS-CoV-2 viral infection and grow easily. However, nucleos(t)ide analogues often show decreased activity in these cells owing to inefficient metabolic activation compared with human cells94. In addition, SARS-CoV-2 mutates rapidly in Vero E6 cell lines95,96, often accumulating changes in the furin cleavage site of the spike protein, amongst others95,97,98. Further, the high expression of functionally active P-glycoprotein (p-gp) efflux pumps can require the additional use of p-gp inhibitors to assess antiviral activity25.
The non-small-cell lung cancer cell line Calu-3 is another commonly used line that supports SARS-CoV-2 replication99, albeit at significantly lower levels than Vero E6 cells94. Additionally, low growth rates and irregular growth patterns of Calu-3 cells have led to difficulties in scaling up and automating high-throughput assays. Other human cell lines in which SARS-CoV-2 replicates efficiently include the intestinal cell line Caco-2, in line with clinical manifestation of SARS-CoV-2 symptoms, and the liver cell line Huh7 (refs. 100,101).
Cell lines with overexpressed entry receptors
SARS-CoV-2 cellular entry occurs upon binding of its spike protein to angiotensin-converting enzyme 2 (ACE2) and transmembrane protease serine 2 (TMPRSS2) receptors7 (Fig. 1a). These entry receptors can be overexpressed to enable the use of cell lines with low physiological levels of ACE2 and TMPRSS2 that SARS-CoV-2 does not infect efficiently. For example, overexpression of human ACE2 (hACE2) or TMPRSS2 in the lung adenocarcinoma cell line A549 permits infection with SARS-CoV-2 (ref. 102). Additionally, culturing A549 cells in an air–liquid interface culture increases the endogenous expression levels of ACE2 and TMPRSS2 (ref. 103). Similarly, the overexpression of ACE2 on HeLa cervical cancer cells enables efficient SARS-CoV-2 entry104, as previously shown for SARS-CoV105. However, for the assessment of entry inhibitors, we recommend that cell lines with physiological levels of receptor expression are used, as overexpression can significantly alter antiviral activity and complicate the interpretation of results, as shown for SARS-CoV-2 monoclonal antibodies106.
Common tier 2 primary cellular models
After a small set of optimized leads or a lead candidate are identified, compounds can be evaluated in more physiologically relevant tier 2 cellular assays. These cellular models include human airway epithelial cells, normal human bronchial epithelial cells or pneumocytes derived from induced pluripotent stem cells. Primary cells offer a more physiological immune system than cancer cell lines commonly used for tier 1 assays, which can be particularly relevant for assessing molecules that interact with the host immune response107–109. In addition, primary cell models allow for a more translationally relevant understanding of drug uptake and cellular metabolism. In SARS-CoV-2 drug discovery efforts to date, data from both tier 1 and tier 2 cellular assays have been used for human dose predictions depending on availability18,110.
However, primary cells are generally not used for screening compounds, as they are expensive, more difficult to culture and scale, and assays often show high variability. For example, human airway epithelial cells and normal human bronchial epithelial cells need to grow in an air–liquid interface with differential treatment on the basal and apical cell sides111,112. In addition, the phenotype of primary cells can be maintained only for a short number of passages, which limits their utility for high-throughput assays113, as they can rapidly de-differentiate and adopt senescence phenotypes114. These limitations also apply for more complex cellular models such as bioprinted and 3D organoid models, which are not used for routine antiviral drug discovery115.
Choice of infecting strains
The choice of the infecting virus is another variable in setting up in vitro antiviral assays for drug discovery. Some viruses such as SARS-CoV-2 replicate readily in numerous human and animal cell lines, whereas replication of others such as HCV is a significant challenge116.
Based on previous knowledge from SARS-CoV, it was rapidly established that various clinical SARS-CoV-2 isolates readily infect a wide range of human and animal cell lines101. Significant variability in replication efficacy occurs for different SARS-CoV-2 strains, with Omicron demonstrating significantly longer replication cycles compared with wild-type SARS-CoV-2 and earlier variants of concern117,118. In addition, differential dependency on entry receptors has been suggested, with SARS-CoV-2 Omicron BA.1 showing higher relative affinity for ACE2 whereas SARS-CoV-2 Delta depends on high levels of TMPRSS2 expression119. Therefore, it can be challenging to compare antiviral efficacy in cellular assays against multiple SARS-CoV-2 variants.
In addition to the variable pathogenicity of variants, SARS-CoV-2 can adapt to different cell lines by accumulating mutations upon viral passaging. These viral adaptations can render additional cell lines susceptible to the virus, such as liver cell lines (Huh7 and Huh7.5) and lung cancer lines (unmodified Calu-1 and A549)120. In addition to naturally circulating viral strains, synthetically engineered viruses such as infectious cDNA clones and reporter viruses can be used in cell culture and optimized to express engineered molecular markers, facilitating high-throughput screening121–123.
Non-infectious cellular models with replicons
Non-infectious subgenomic replicons can be used to connect enzymatic assays and cellular systems. Subgenomic replicons are artificially constructed RNA molecules that contain all the viral genome except genes that encode the structural proteins. They can replicate in cells, but are unable to infect other cells, thus are safer to operate and can often be handled in a BSL2 laboratory environment. Replicons enable a cell-based assay system to interrogate the fitness of different protein mutants, as well as to screen for potential antivirals. Historically, replicon systems have been crucial in drug discovery for viruses where cellular replication was difficult to achieve, such as HCV, or where laboratory handling is associated with significant health risks124–126.
For SARS-CoV-2, several replicon systems have been reported. In principle, these are based on the deletion of selected viral proteins (S, E and/or M proteins) and the addition of genes encoding firefly luciferase (Luc), green fluorescent protein (GFP) or other reporters127–129. SARS-CoV-2 replicons suitable for BSL2 environments rely on a variety of technologies, including transient reporter replicons130, trans-complementation systems131 and attenuated viruses with deletions of viral accessory genes132.
However, replicons have limitations. They cannot be used to assess targets that are not included in the construct; interdependency of host and viral targets and immunological responses might not be modelled; and compounds optimized against a replicon might not show activity against wild-type virus, so should always be cross-checked during development. In addition, with increasing availability of high-throughput SARS-CoV-2 BSL3 screening facilities133,134, the future importance of SARS-CoV-2 replicon systems as a screening option remains an open question.
Animal models
Various animal models for SARS-CoV-2 infection have been described in the NIH COVID-19 Open Data Portal and reviewed135,136. Based on animal models for SARS-CoV137,138, the ferret and139,140 hamster models141,142 that are susceptible to SARS-CoV-2 infection were rapidly deployed early in 2020. However, both models have significant logistical and financial overheads. More recent studies use mouse models, which are easier to deploy owing to their high accessibility, low cost, rapid breeding speed and ease of manipulation. Mouse models expressing specific SARS-CoV-2 entry receptors (for example K18-hACE2) and infected with wild-type virus143, or wild-type mice infected with mouse-adapted viral strains144 have been frequently used. In addition, the acquisition of the 501Y mutation in variants of concern enables the infection of wild-type mice and other rodents, particularly aged animals, albeit at overall lower viral loads145. Preclinical studies in non-human primates have been predictive of COVID vaccine outcomes in clinical efficacy studies146, but are not routinely used in small-molecule drug discovery.
Similarly to human infections, animals infected with SARS-CoV-2 show major differences in viral load and pathology dependent on the infecting viral strain147. Specifically, animal susceptibility appears to be linked to the affinity of the SARS-CoV-2 spike protein for the ACE2 receptors, with variant-specific spike substitutions such as N501Y, D614G and V367F affecting transmission in animals146. Significant variant-specific differences in viral load distribution and disease pathology have been described148–151, with lower viral loads and little weight loss observed for the Omicron variant compared with Delta in wild-type and hACE2-transgenic Syrian hamsters152. These results are in line with clinical data suggesting that infectious viral load was lower in Omicron-infected individuals than in Delta-infected individuals153,154. Overall, these differences in how variants infect both animals and humans are expected to delay defining effective animal models for every new variant of concern. Therefore, firmly establishing the translational relevance of an animal model might not be significantly less arduous than performing clinical studies in patients.
We assert that animal efficacy models should not be crucial for small-molecule antiviral discovery against SARS-CoV-2. Indeed, the first directly acting antivirals against SARS-CoV-2 were advanced solely on the basis of in vitro cellular data and projections for human drug exposure that exceeded the level sufficient for a pharmacodynamic effect. We therefore suggest that it is sufficient to define a cellular EC90 in a primary cell model and determine the human dose and dosing frequency that is required to remain above the cellular benchmark at all times (for example, the free fraction of the drug at Cmin over EC90 for treatment duration). Although efficacy in animal models is a generally recommended (but not absolute) requirement for regulatory approval of human therapeutics, HIV and HCV antivirals have set a precedent for approval in the absence of animal models. However, animal models can offer reassurance on the mechanism of action during preclinical development, especially if investigated inhibitors can be shown to impact significantly on SARS-CoV-2 viral load and histopathological end points, and dose-related effects can be linked to unbound drug exposure25,94,155,156.
Animal models should also have an important role in pandemic preparedness, particularly for diseases such as Ebola where a human phase IIa study is not possible if disease is not circulating at the time or human challenge models are not available157–159. In these cases, animal efficacy in a relevant model, in combination with human pharmacokinetics and safety studies, might be sufficient for drug approval160.
Despite potential differences in pathology between animal and human models, studies of infected animals might also provide insights into issues such as disease transmission and the impact of viral load on transmission112, the effect of age and comorbidities on disease progression161, organ and brain involvement, vascular symptoms, secondary infections, ‘long COVID’162 and immunological sequelae163. However, for most of these presentations, the human pathogenicity remains unclear, and relevant animal models require further validation146.
Human viral challenge models
Ultimately, the most relevant model to assess natural infection, viral load distribution and efficacy of antiviral inhibitors is a human viral challenge (HVC) model. This model enables viral load kinetics and viral shedding to be assessed in human healthy volunteers, with certainty of the time of infection and prospective assessment of symptoms164.
HVCs have been used to assess the natural disease course of acute respiratory viruses, including RSV, influenza, HRV and most recently SARS-CoV-2 (refs. 165,166). Generally, the availability of ‘rescue’ treatments and upfront knowledge on potential human disease pathology and complications are paramount to the conduct of HVCs. Nevertheless, for novel human pathogens such as SARS-CoV-2 with incomplete understanding of long-term disease implications, complex ethical issues have to be carefully assessed before embarking on HVCs.
Potential confounding factors in HVCs are the typically short time frame between infection and the start of treatment, which can be precisely controlled in a HVC but is less controllable in natural infection, potentially leading to an overestimation of compound efficacy. The careful selection of healthy volunteers and regular sampling might also explain some of the differences noted between natural infection and HVC trials for SARS-CoV-2 and other respiratory viruses166. For example, higher peak viral loads were measured for SARS-CoV-2 HVCs than for natural infection166,167, and more common upper respiratory tract infections were noted in RSV HVCs, compared with more lower respiratory tract infections in natural infection168. Overall, data generated in HVC studies can be highly variable dependent on the inoculation dose, the viral strain used and the immune profile and age of the healthy volunteers.
A key point at which HVCs can contribute to clinical decision-making is in determining treatment duration based on the evolution of viral load. This is enabled by assessing viral circulation clearance using regular viral load measurements after a standardized infection dose. The TPP, including the treatment duration, can then be specified, as exemplified for influenza infection169. However, translation from efficacy in an HVC to natural infection is not certain: for rupintrivir, an investigational protease inhibitor against HRV, efficacy in HVCs overestimated its utility in natural infection20,170.
Pre-empting resistance
Resistance mutations can render an antiviral therapy inefficient. Viral mutations can occur spontaneously and are selected to preferentially replicate under immunological pressure or selection pressure exerted by drug therapy. Nonetheless, only mutants that are transmissible and cause adverse pathologies are of concern.
The likelihood of a virus developing mutations depends first on viral factors, such as how readily the virus mutates and whether it has a polymerase proofreading function. Second, it depends on host factors, including the patient’s HLA type, immunosuppression and other disease or co-infection. Third, it depends on environmental factors such as whether antiviral treatment is given, the drug target, whether treatment is single or combination therapy or whether infection is acute or chronic. Drug-induced resistance has been observed across the spectrum of antiviral therapeutics and is largely independent of treatment duration; it has been reported after short courses for acute infection, such as with the influenza drugs oseltamivir171 and baloxavir172, as well as after longer treatment courses used for chronic viral infections173.
Although the lower likelihood of viral drug resistance is commonly used as an argument for host-targeting antivirals, this is not necessarily a solution. For example, targeting cyclophilin with small-molecule inhibitors resulted in amino acid substitutions in the HCV NS5a protein174, and targeting CCR5 led to substitutions in the HIV gp120 protein175,176.
We suggest that the essential strategies to circumvent drug-induced resistance are to design compounds that sit tightly in the substrate binding site, to drive the free drug concentration as high as safely possible and to consider combination therapies. At the discovery stage for SARS-CoV-2 drugs, several approaches can pre-empt the development of resistance.
Selecting the target
A commonly used approach to identify viral targets that carry a low tolerance to resistance mutations is based on sequence conservation, with the assumption that highly conserved proteins are more likely to be essential and their alteration would be detrimental to viral replication38,177–179. Various levels of sequence conservation can be considered, for example, across the Coronaviridae family, such as across alphacoronaviruses (including 229E and NL63), betacoronaviruses (SARS, SARS-CoV-2 and OC43), or within circulating variants of SARS-CoV-2. For certain conserved targets such as the polymerase, it is feasible to even consider targeting strategies across other viral families; for example, molnupiravir is active against Filoviridae (Ebola), Togaviridae (VEEV) and Coronaviridae180.
Another approach entails the incorporation of data on resistance mutations identified through sequence surveillance in patients infected with SARS-CoV-2. This is particularly relevant if there is target-specific evidence of drug-induced resistance, as described for the early protease inhibitors developed for HIV and HCV. Of note, coronaviruses as a family accumulate fewer mutations than other RNA viruses such as HIV and HCV181, for which closely related mutant spectra, termed viral quasispecies, can be detected in each infected individual, due to the lack of RdRp proofreading function and the individual’s immune response182–185. In contrast, coronaviruses have a unique error-correcting mechanism that was unknown among RNA viruses before its discovery in SARS-CoV186. The NSP14 exoribonuclease excises nucleotides misincorporated by the low-fidelity RdRp and thereby lowers the replication error rate compared with other RNA viruses187–189.
When interpreting phylogenetic data and resistance mutations in treated patients, several caveats have to be taken into account. First, natural mutations occur without drug selection pressure and do not imply pre-existing drug resistance for a certain target. For example, baseline resistance-associated mutations do not predict treatment failure190. Second, variants need to persist and remain transmissible to become relevant resistance-associated variants (RAVs). Viruses with resistance-associated mutations in their genome often have a fitness cost or growth disadvantage compared with the wild-type virus in the absence of selective pressure. Evolutionary competition between wild-type virus and mutants has been observed in patients (for example with HCV), where variants rapidly expand in the presence of selection pressure. However, once the drug is removed, the wild-type virus again outcompetes the RAVs190. Nevertheless, several known drug-induced variants are fit and transmissible, such as those in patients with influenza who are treated with neuraminidase inhibitors191,192 or cap-dependent endonuclease inhibitors193–195. Finally, mutations observed only under treatment do not per se imply drug-induced escape. This is particularly important when interpreting data from clinical case reports196–198. Large-scale clinical studies that sequence the whole viral genome and monitor viral load longitudinally across the treatment duration are required to identify causative drug-induced mutations (for example, the PANORAMIC trial; EudraCT number: 2021-005748-31)199. Even if the mutant is replication competent and persists, the impact on disease presentation and progression has to be assessed on a large scale.
Drugging the target
Strategic approaches to drugging the target can mitigate resistance development at the discovery stage. Members of our round table deploy structural biology to define regions of the target that should be robust to resistance, biochemical enzymatic assays to screen for antivirals that avoid resistance and cell culture systems to provide insight into resistance development.
Structural biology
A structure-based discovery approach allows the medicinal chemistry campaign to focus not only on maximizing protein–ligand interaction, but also on defining the region of the protein to target with chemical inhibitors. To circumvent viral resistance mutations from the start, the first consideration is to select the relevant sequence to crystallize. This is a pragmatic decision, as even single mutations can affect the propensity to form crystals, but it is also important for the analysis of resistance. Common approaches include selecting a wild-type sequence, a clinically relevant strain or a consensus sequence that comprises the most frequently occurring residues across variants.
The next consideration is whether to target the active site or an allosteric site. Our consensus is that it is preferable to directly target the enzyme active site, as allosteric inhibitors can have a lower barrier to resistance. For example, HCV NS5B polymerase has highly polymorphic allosteric sites, and non-nucleoside inhibitors targeting these sites were associated with a lower barrier to resistance compared with nucleoside inhibitors targeting the active site200,201. In addition to targeting allosteric sites, targeting distal pockets of the active site can rapidly lead to resistance mutations during therapy, as shown for baloxavir, an inhibitor of the influenza cap-dependent endonuclease172,202.
Another consideration is the concept that within the active site, a high barrier to resistance can be achieved by designing compact inhibitors that limit their binding to the substrate envelope. The substrate envelope is defined as the space spanned by a key set of residues that interact with diverse native substrate sequences203,204. Any resistance mutations that occur within the substrate envelope are likely to cause a significant disruption to enzyme function and loss of viral fitness, whereas mutations outside the envelope incur significantly less fitness cost. The substrate envelope approach can be further refined by designing the inhibitor to interact predominantly with consensus residues within the substrate envelope that are shared across a viral family205.
This concept has been applied to HIV and HCV protease inhibitors206,207 and recently deployed in SARS-CoV-2 (ref. 208). For HIV, analysing structures of protein–ligand complexes for FDA-approved protease inhibitors revealed that contacts outside the substrate envelope correspond to the primary drug resistance mutation sites206. For SARS-CoV-2, the substrate envelope of Mpro was defined by crystallizing a library of substrate peptides bound to the native cleavage site208. Then, analysis of several protease inhibitors revealed that contacts mostly lie within the substrate binding envelope, although some parts of the molecules interact with residues outside the envelope. However, without comparative in vitro studies using Mpro inhibitors engaging different regions of the substrate envelope, or comprehensive sequencing data on patients treated with Mpro inhibitors, it is too early to tell whether these interactions will manifest as clinically observed resistance mutations.
Enzymology
The impact of selected resistance mutations on enzyme function can be defined experimentally, with saturation mutagenesis providing a more comprehensive assessment. For example, every amino acid in SARS-CoV-2 Mpro was mutated to reveal which mutations still resulted in a functional enzyme209. Reassuringly, the saturation mutagenesis approach showed that mutationally intolerant residues are also conserved across homologues. Mutationally intolerant sites were revealed at both the active site and the dimer interface, and at the allosteric communication network between these regions. However, further work is needed to interrogate the fitness of these mutant enzymes in authentic viral systems using reverse genetics. A similar mutagenesis approach has been successfully applied to predict clinically relevant patterns of resistance in bacterial β-lactamase210,211.
In addition to assessing variants within SARS-CoV-2, further robustness to resistance can be engineered by designing inhibitors that are active against enzymes from different viruses within the same family. However, we have found that routinely screening compounds against a broad panel of enzymes as primary screens is laborious and expensive. An alternative approach could be to use enzyme engineering to design a ‘consensus enzyme’212,213, namely a single model enzyme that contains the most frequent residues across a viral family.
Ideally, as understanding of viral evolution increases, the screening cascade should include enzymatic assays of circulating variants or clinically observed drug-resistant mutations. However, in the context of a drug design campaign that races against an emerging and evolving pandemic, constantly moving goalposts by including new variants in the assay cascade is not feasible.
Beyond direct enzyme inhibition, we identify the degrader technology as another potentially viable way to overcome resistance. This approach exploits intracellular proteolysis to degrade the target protein. The paradigmatic example is proteolysis-targeting chimeras (PROTACs), which are heterobifunctional molecules in which one ligand binds to the target protein and the other ligand engages with an ubiquitin ligase, resulting in ubiquitylation of the target and subsequent degradation by the proteasome. Potential advantages of PROTACs are that lower-affinity binders can have a biological effect, as the target protein is irreversibly removed; degradation by PROTACs is catalytic, as the ligand is not consumed; and the pharmacodynamic efficacy is driven by the viral protein production rate and can extend beyond the detectable pharmacokinetic presence of the PROTAC molecule. The feasibility of a PROTAC approach has been explored for the HCV protease214. The degrader paradigm has been extended to RNA degradation by using ligands that recruit a ribonuclease, and this approach has been applied to target SARS-CoV-2 RNA215.
Cell culture systems
In addition to evaluating resistance mutations at the structural and enzymatic levels, viruses or replicons harbouring mutations can be propagated in cell culture to determine the effect of compounds. Modification of the previously described non-infectious replicon systems so that they contain resistance mutations is a nascent method in the SARS-CoV-2 field. However, for HCV, a large body of literature has elucidated mechanisms of resistant replicon formation against protease inhibitors216,217, polymerase inhibitors218,219 and combinations220, as well as comparing the barrier of resistance of nucleoside and non-nucleoside inhibitors221. Inhibitors can be designed to have a favourable profile against a panel of replicons harbouring mutations from various viral genotypes, or against drug-resistant mutations observed after treatment with other inhibitors from the same class, as reported for HCV222,223. Further, the replication efficiency of a replicon system can quantify the fitness cost of a mutation216. Of note, mutations with efficient growth in a replicon may not be fit in vivo224, thus careful validation of in vitro–in vivo correlation and comparison with clinically observed mutations225 are required to elucidate the fitness cost of resistance.
Beyond replicons, cell culture systems with live virus can be used to model resistance development under drug pressure. A typical approach is serial passaging — subjecting the virus to low concentrations of the inhibitor over long periods of time, isolating and growing the surviving mutants, then re-exposing these to another cycle of inhibitor treatment. The number of cycles required for resistant mutants to emerge is a metric that assesses the barrier to resistance. The replicative fitness of the mutants can be quantified by measuring the growth rate of the mutant, as well as by cellular competition studies in which mutants are co-infected with wild-type virus. Further, sequencing of viral mutants can reveal residues that are susceptible to mutations in cell culture. In cases where the chemical compound is discovered phenotypically, serial passaging studies can help to elucidate the viral target226,227. Viral passaging can be used during lead optimization, where the barrier to resistance can be used as an additional factor to select the lead series228. Overall, significant series-specific differences in the barrier to resistance are common, as different chemical series typically contact different sets of residues in the binding site. Passaging experiments using remdesivir in the SARS-CoV-2-related virus mouse hepatitis virus (MHV)229, and later for SARS-CoV-2 (refs. 230,231), revealed potential point mutations that could confer resistance. Viral passaging on MHV with the Mpro inhibitor nirmatrelvir was used to investigate the potential mutation sites accompanied by reduced nirmatrelvir sensitivity110. Additional mutations were reported for serial passaging using probe compound ALG-097161, leading to a combination of mutations that also confer resistance to nirmatrelvir in vitro232. However, the relevance of these mutations detected in serial passaging experiments has to be independently verified by clinical trials. Furthermore, serial passaging studies of clinically approved inhibitors are contentious owing to the potential to select for resistant variants, which could impact biosecurity and public health. As such, funding agencies have put restrictions in place for such studies233.
Deploying the therapeutic
Strategic approaches to pre-empt resistance development can also be used at the clinical stage. For example, combination therapies that target multiple viral proteins have a lower likelihood of selecting for escape mutants234,235 and are commonly used in clinical settings for several viruses, including HIV and HCV. Only influenza and HSV have monotherapies in routine clinical use, and optimal treatment regimens for SARS-CoV-2 are still being explored. Further, it is likely that certain patient populations, such as immunosuppressed patients, might require combination therapy as they are more likely to harbour resistant viruses.
As a second approach, maintaining high drug concentrations (with Cmin of the free drug as high multiples of the EC90) might lower the likelihood of inducing viral resistance. For SARS-CoV-2, the appropriate level of Cmin above the EC90 is not yet clear, with early clinical data still emerging. The clinical strategy for nirmatrelvir and pomotrelvir 236,237 is to select dosing that enables a Cmin greater than EC90 for 90% of the patient population. For ensitrelvir, less stringent criteria of a Cmin greater than the half-maximal effective concentration (EC50) were reported for animal efficacy18. We argue that during a drug discovery sprint, a pragmatic approach is required, driving the medicinal chemistry campaigns to optimize EC90 and drug exposure, while recognizing that the pharmacokinetic–pharmacodynamic (PK–PD) relationship might be a point of product differentiation between the ideal and pandemic TPP (Table 3).
Concluding remarks
The exigencies of the COVID-19 pandemic have spurred a wave of rapid drug discovery campaigns. In less than 2 years, several antivirals were discovered, clinically evaluated and approved. We have summarized key scientific drivers and considerations behind these sprint discovery campaigns and how knowledge from previous antiviral drug discovery campaigns was fruitfully deployed. Beyond scientific approaches, the broader organizational context has enabled rapid discovery, outlining future directions for the community.
We find that a common strand underlying many campaigns is the optimism among the project team and support from all levels. Planning and executing future steps assuming that previous experiments will be successful, while also having a mitigating plan in place in case of failure, is crucial to moving quickly. Further, upfront investment is needed to move rapidly. In the fast-moving research environment of a pandemic, it remains an open question how public sector funding, traditionally more risk-averse, can rapidly enable drug discovery.
Another fruitful experiment during the pandemic was the unprecedentedly collaborative philosophy across biopharmaceutical companies, nucleating public–private partnerships such as IMI CARE, close-knit consortia of companies, and even completely open science consortia with company participation19. Further, the rapid implementation of clinical trial networks, such as ACTIV8, RECOVERY (NCT04381936)238 and PANORAMIC (EudraCT number: 2021-005748-31), has greatly contributed to the accelerated assessment of potential COVID-19 therapies. The unique severity of the pandemic created the strong impetus for these collaborations. The successes of these projects can inspire more openness and collaboration in the biopharmaceutical industry, and we believe commercial organizations should be more willing to work quickly and collaboratively during global emergencies.
Furthermore, the pandemic has been exceptional for fast-moving and rigorous regulatory emergency review and approval, with agencies establishing rapid response mechanisms as early as May 2020 (refs. 239,240). Regulatory agencies have outlined more efficient processes to give rapid feedback on supporting data, with the aim to enable clinical trials and provide upfront definitions of clinical end points241. We argue that in view of the rapidly changing clinical picture — with increasing natural immunity and vaccination rates, as well as variable pathogenicity of circulating strains — a regular review of acceptable clinical trial end points should be conducted. End points initially used for small-molecule clinical trials in COVID-19, such as mortality and hospitalization rate, are now considered too low in frequency to support reasonably sized clinical trials. Instead, additional primary end points such as viral replication could be considered. These are easier to standardize than symptom-related end points and are considered acceptable for other viral diseases such as HIV and HCV infections242–244. In particular, viral kinetic data from human challenge trials, not immediately available at the start of the pandemic, could feed into defining alternative end points.
Ultimately, a successful drug discovery campaign is contingent upon selecting and drugging the right protein target. We note that mechanism-free repurposing has been widely attempted during the COVID-19 pandemic, but has failed to deliver therapeutics245, with early positive results partly due to confounding factors such as phospholipidosis246. This is perhaps not surprising, as most therapeutics and chemical compound libraries are optimized for human targets, which are generally dissimilar to viral targets.
Therefore, we conclude with a call to arms on pandemic preparedness. A responsive mode to antiviral drug discovery, even armed with modern technology, is too slow to prevent the significant human and economic catastrophe of a fast-moving pandemic — it took a decade to offer a therapeutic for HIV and about 2 years for COVID. With significant funding in place, for example, the decision by the NIH to allocate $577 million to fund nine Antiviral Drug Discovery (AViDD) Centers for Pathogens of Pandemic Concern247, the question becomes how do we best nucleate a concerted effort for pandemic preparedness?
One strategy is to systematically target the viral proteome. Of the 16 non-structural proteins and four structural proteins in SARS-CoV-2, only three have validated chemical probes. Once these existing targets are drugged and resistance inevitably emerges, we will need to uncover new viable viral targets, new mechanisms of action, or judicious combinations of therapeutics with no cross-resistance. Another related strategy is open dissemination of tools such as assay protocols, building on successful precedents for human targets such as the RAS initiative248 and the Structural Genomics Consortium249. Focused efforts should be invested in ensuring that the myriad of assays developed, and resulting data, are disseminated with good data management practices, to ensure ‘Findability, Accessibility, Interoperability and Reusability’250.
Considering the broad toll of a pandemic, we argue that public sector investment should be put into systematically developing chemical probes and focused libraries against proteins in viruses of pandemic concern. Concerted funding should be in place to push chemical matter against promising targets into early clinical development, so that it can enter phase II clinical trials when a pandemic of the same viral family strikes. This is even more pressing for other viruses of pandemic concern, such as Dengue and Zika, which are endemic in the Global South and currently lacking effective therapeutics. Therefore, a global health angle to drug discovery and development is acutely needed to alleviate significant unmet medical need.
Acknowledgements
A.v.D. and A.A.L. are members of the The COVID Moonshot Consortium. This work was supported by the NIH and NIAID Antiviral Drug Discovery (AViDD) grant (U19AI171399) and Wellcome Trust grant (224021/Z/21/Z) to A.A.L. and A.v.D., by a Royal Society University Research Fellowship to A.A.L. (URF\R1\201543), support of NIHR Biomedical Research Centre Oxford to A.v.D. and support of the intramural research programme of the NCATS, NIH to M.D.H. The authors acknowledge M. Boby for preparation of Fig. 2.
Glossary
- Cmin
The minimum or trough concentration of a drug in plasma over the treatment duration, with the free or unbound Cmin defined as the minimum concentration multiplied by the percentage of compound unbound in plasma.
- Crystallographic fragment screen
A sensitive biophysical technique for finding chemical motifs that can be expanded into leads and eventually drugs; protein crystals are soaked or grown at high concentrations with libraries of very small molecules (fragments, <250 Da) carefully selected to contain signatures present in drugs and ligands.
- EC90
The drug concentration at which 90% inhibition of viral replication is observed in cellular assays. Cellular assay medium can contain protein components such as fetal calf serum. To calculate the protein-adjusted EC90, the EC90 is multiplied by the percentage of unbound compound measured for the cellular assay medium.
- Free drug
The concentration of the drug that is not bound to plasma, also termed unbound drug, as assessed by measuring the plasma protein binding in vitro. Typically only the free drug can act on its target in tissues and cause pharmacology-relevant effects.
- Pharmacokinetic–pharmacodynamic
(PK–PD). The relationship between pharmacokinetics (the concentration of drug in plasma or a tissue of interest) and the drug’s therapeutic effect. In antiviral therapeutics, the PK–PD relationship required for an efficacious therapeutic is typically a free Cmin greater than (at least 1×) the protein-adjusted EC90.
- Structure–activity relationship
The relationship between the chemical structure of a molecule and its biological activity.
- Virtual screening
A computational technique used in drug discovery to identify chemical structures that are most likely to bind to a specific target.
Author contributions
All authors researched data for the article and contributed substantially to discussion of the content. A.v.D. and A.A.L. wrote the article. All authors reviewed and edited the manuscript before submission.
Peer review
Peer review information
Nature Reviews Drug Discovery thanks Timothy Sheahan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Competing interests
All authors and/or their employers are involved in COVID drug discovery or development programmes, and may continue to be involved in future programmes. These programmes may lead to the generation of intellectual property for their respective employers, and may directly or indirectly contribute to past, present or future revenue of their respective employers. M.D.H. is an employee of NCATS, A.K. is an employee of Pardes Biosciences, L.A.P. is an employee of Vir Biotechnology, K.S.S. is an employee of Takeda California, Inc., U.S. is an employee of Gilead Sciences, J.A.T. is an employee of Novartis Institutes for Biomedical Research, A.v.D. is an employee of Oxford University, and consults for PostEra, Inc. A.A.L. is an employee of Cambridge University and CSO for PostEra, Inc.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
ACTIV trial: https://www.nih.gov/research-training/medical-research-initiatives/activ
Consortia of companies: https://cen.acs.org/biological-chemistry/infectious-disease/How-big-pharma-firms-quietly-collaborating-on-new-coronavirus-antivirals/98/i18
IMI CARE: https://www.imi-care.eu/
NIH COVID-19 Open Data Portal: https://opendata.ncats.nih.gov/covid19/animal
PANORAMIC trial: https://www.panoramictrial.org/
RECOVERY trial: https://www.recoverytrial.net/
These authors contributed equally: Annette von Delft, Alpha A. Lee.
Contributor Information
Annette von Delft, Email: Annette.vondelft@cmd.ox.ac.uk.
Alpha A. Lee, Email: alpha.lee@postera.ai
References
- 1.Gibb R, Franklinos LHV, Redding DW, Jones KE. Ecosystem perspectives are needed to manage zoonotic risks in a changing climate. Br. Med. J. 2020;371:m3389. doi: 10.1136/bmj.m3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vidal, J. Destroyed habitat creates the perfect conditions for coronavirus to emerge. [Reprinted from Ensia] Scientific Americanhttps://www.scientificamerican.com/article/destroyed-habitat-creates-the-perfect-conditions-for-coronavirus-to-emerge/ (18 March 2020).
- 3.Messina JP, et al. The current and future global distribution and population at risk of dengue. Nat. Microbiol. 2019;4:1508–1515. doi: 10.1038/s41564-019-0476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldin, I. & Mariathasan, M. The Butterfly Defect: How Globalization Creates Systemic Risks, and What to Do about It (Princeton Univ. Press, 2015).
- 5.Chaudhuri S, Symons JA, Deval J. Innovation and trends in the development and approval of antiviral medicines: 1987–2017 and beyond. Antiviral Res. 2018;155:76–88. doi: 10.1016/j.antiviral.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon DE, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann M, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins FS, Stoffels P. Accelerating COVID-19 therapeutic interventions and vaccines (ACTIV): an unprecedented partnership for unprecedented times. JAMA. 2020;323:2455–2457. doi: 10.1001/jama.2020.8920. [DOI] [PubMed] [Google Scholar]
- 9.Kumar N, et al. Host-directed antiviral therapy. Clin. Microbiol. Rev. 2020;33:e00168-19. doi: 10.1128/CMR.00168-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wood, A. & Armour, D. In Progress in Medicinal Chemistry (eds King, F. D. & Lawton, G.) 239–271 (Elsevier, 2005).
- 11.Paeshuyse J, et al. The non-immunosuppressive cyclosporin DEBIO-025 is a potent inhibitor of hepatitis C virus replication in vitro. Hepatology. 2006;43:761–770. doi: 10.1002/hep.21102. [DOI] [PubMed] [Google Scholar]
- 12.Hall MD, et al. Report of the National Institutes of Health SARS-CoV-2 antiviral therapeutics summit. J. Infect. Dis. 2021;224:S1–S21. doi: 10.1093/infdis/jiab305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.NIAID pandemic preparedness plan targets “prototype” and priority pathogens. NIAIDhttps://www.niaid.nih.gov/news-events/niaid-pandemic-preparedness-plan-targets-prototype-and-priority-pathogens (2022).
- 14.Ke Z, et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wrapp D, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dolan KA, et al. Structure of SARS-CoV-2 M protein in lipid nanodiscs. eLife. 2022;11:e81702. doi: 10.7554/eLife.81702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moustaqil M, et al. SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): implications for disease presentation across species. Emerg. Microbes Infect. 2021;10:178–195. doi: 10.1080/22221751.2020.1870414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Unoh Y, et al. Discovery of S-217622, a noncovalent oral SARS-CoV-2 3CL protease inhibitor clinical candidate for treating COVID-19. J. Med. Chem. 2022;65:6499–6512. doi: 10.1021/acs.jmedchem.2c00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The COVID Moonshot Consortium Open science discovery of oral non-covalent SARS-CoV-2 main protease inhibitor therapeutics. bioRxiv. 2023 doi: 10.1101/2020.10.29.339317. [DOI] [Google Scholar]
- 20.Hayden FG, et al. Phase II, randomized, double-blind, placebo-controlled studies of ruprintrivir nasal spray 2-percent suspension for prevention and treatment of experimentally induced rhinovirus colds in healthy volunteers. Antimicrob. Agents Chemother. 2003;47:3907–3916. doi: 10.1128/AAC.47.12.3907-3916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang H, Yang J. A review of the latest research on Mpro targeting SARS-COV inhibitors. RSC Med. Chem. 2021;12:1026–1036. doi: 10.1039/D1MD00066G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pillaiyar T, Manickam M, Namasivayam V, Hayashi Y, Jung S-H. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boras B, et al. Preclinical characterization of an intravenous coronavirus 3CL protease inhibitor for the potential treatment of COVID19. Nat. Commun. 2021;12:6055. doi: 10.1038/s41467-021-26239-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffman RL, et al. Discovery of ketone-based covalent inhibitors of Coronavirus 3CL proteases for the potential therapeutic treatment of COVID-19. J. Med. Chem. 2020;63:12725–12747. doi: 10.1021/acs.jmedchem.0c01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owen DR, et al. An oral SARS-CoV-2 M inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374:1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 26.Kim Y, et al. Reversal of the progression of fatal coronavirus infection in cats by a broad-spectrum Coronavirus protease inhibitor. PLoS Pathog. 2016;12:e1005531. doi: 10.1371/journal.ppat.1005531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim Y, et al. Broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, noroviruses, and coronaviruses. J. Virol. 2012;86:11754–11762. doi: 10.1128/JVI.01348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian L, et al. RNA-dependent RNA polymerase (RdRp) inhibitors: the current landscape and repurposing for the COVID-19 pandemic. Eur. J. Med. Chem. 2021;213:113201. doi: 10.1016/j.ejmech.2021.113201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beigel JH, et al. Remdesivir for the treatment of Covid-19 - final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jayk Bernal A, et al. Molnupiravir for oral treatment of Covid-19 in nonhospitalized patients. N. Engl. J. Med. 2022;386:509–520. doi: 10.1056/NEJMoa2116044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheahan TP, et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9:eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheahan TP, et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11:222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tchesnokov EP, Feng JY, Porter DP, Götte M. Mechanism of inhibition of Ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019;11:326. doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yin W, et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabinger F, et al. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat. Struct. Mol. Biol. 2021;28:740–746. doi: 10.1038/s41594-021-00651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vegivinti CTR, et al. Efficacy of antiviral therapies for COVID-19: a systematic review of randomized controlled trials. BMC Infect. Dis. 2022;22:107. doi: 10.1186/s12879-022-07068-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brimacombe KR, et al. An OpenData portal to share COVID-19 drug repurposing data in real time. bioRxiv. 2020 doi: 10.1101/2020.06.04.135046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yazdani S, et al. Genetic variability of the SARS-CoV-2 pocketome. J. Proteome Res. 2021;20:4212–4215. doi: 10.1021/acs.jproteome.1c00206. [DOI] [PubMed] [Google Scholar]
- 39.Arnold JJ, et al. Sensitivity of mitochondrial transcription and resistance of RNA polymerase II dependent nuclear transcription to antiviral ribonucleosides. PLoS Pathog. 2012;8:e1003030. doi: 10.1371/journal.ppat.1003030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin D, et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ratia K, et al. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl Acad. Sci. USA. 2008;105:16119–16124. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osipiuk J, et al. Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat. Commun. 2021;12:743. doi: 10.1038/s41467-021-21060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shan H, et al. Development of potent and selective inhibitors targeting the papain-like protease of SARS-CoV-2. Cell. Chem. Biol. 2021;28:855–865.e9. doi: 10.1016/j.chembiol.2021.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen Z, et al. Design of SARS-CoV-2 PLpro inhibitors for COVID-19 antiviral therapy leveraging binding cooperativity. J. Med. Chem. 2022;65:2940–2955. doi: 10.1021/acs.jmedchem.1c01307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma C, et al. Discovery of SARS-CoV-2 Papain-like protease Inhibitors through a combination of high-throughput screening and a FlipGFP-based reporter assay. ACS Cent. Sci. 2021;7:1245–1260. doi: 10.1021/acscentsci.1c00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanner JA, et al. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5’ to 3’ viral helicases. J. Biol. Chem. 2003;278:39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malone B, Urakova N, Snijder EJ, Campbell EA. Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design. Nat. Rev. Mol. Cell. Biol. 2022;23:21–39. doi: 10.1038/s41580-021-00432-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen J, et al. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell. 2020;182:1560–1573.e13. doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malone B, et al. Structural basis for backtracking by the SARS-CoV-2 replication–transcription complex. Proc. Natl Acad. Sci. USA. 2021;118:e2102516118. doi: 10.1073/pnas.2102516118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Newman JA, et al. Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase. Nat. Commun. 2021;12:4848. doi: 10.1038/s41467-021-25166-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chono K, et al. ASP2151, a novel helicase-primase inhibitor, possesses antiviral activity against varicella-zoster virus and herpes simplex virus types 1 and 2. J. Antimicrob. Chemother. 2010;65:1733–1741. doi: 10.1093/jac/dkq198. [DOI] [PubMed] [Google Scholar]
- 52.Kleymann G, et al. New helicase-primase inhibitors as drug candidates for the treatment of herpes simplex disease. Nat. Med. 2002;8:392–398. doi: 10.1038/nm0402-392. [DOI] [PubMed] [Google Scholar]
- 53.Crute JJ, et al. Herpes simplex virus helicase-primase inhibitors are active in animal models of human disease. Nat. Med. 2002;8:386–391. doi: 10.1038/nm0402-386. [DOI] [PubMed] [Google Scholar]
- 54.Schuller M, et al. Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking. Sci. Adv. 2021;7:eabf8711. doi: 10.1126/sciadv.abf8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fehr AR, Jankevicius G, Ahel I, Perlman S. Viral macrodomains: unique mediators of viral replication and pathogenesis. Trends Microbiol. 2018;26:598–610. doi: 10.1016/j.tim.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fehr AR, et al. The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. mBio. 2016;7:e01721-16. doi: 10.1128/mBio.01721-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sherrill LM, et al. Design, synthesis and evaluation of inhibitors of the SARS-CoV-2 nsp3 macrodomain. Bioorg. Med. Chem. 2022;67:116788. doi: 10.1016/j.bmc.2022.116788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gahbauer S, et al. Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2. Proc. Natl Acad. Sci. USA. 2023;120:e2212931120. doi: 10.1073/pnas.2212931120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roy A, et al. Discovery of compounds that inhibit SARS-CoV-2 Mac1-ADP-ribose binding by high-throughput screening. Antiviral Res. 2022;203:105344. doi: 10.1016/j.antiviral.2022.105344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krafcikova P, Silhan J, Nencka R, Boura E. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat. Commun. 2020;11:3717. doi: 10.1038/s41467-020-17495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Daffis S, et al. 2’-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosas-Lemus M, et al. High-resolution structures of the SARS-CoV-2 2’-O-methyltransferase reveal strategies for structure-based inhibitor design. Sci. Signal. 2020;13:eabe1202. doi: 10.1126/scisignal.abe1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilamowski M, et al. 2’-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography. Proc. Natl Acad. Sci. USA. 2021;118:e2100170118. doi: 10.1073/pnas.2100170118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ahmed-Belkacem R, et al. Potent Inhibition of SARS-CoV-2 nsp14 N7-methyltransferase by sulfonamide-based bisubstrate analogues. J. Med. Chem. 2022;65:6231–6249. doi: 10.1021/acs.jmedchem.2c00120. [DOI] [PubMed] [Google Scholar]
- 65.Otava T, et al. The structure-based design of SARS-CoV-2 nsp14 methyltransferase ligands yields nanomolar inhibitors. ACS Infect. Dis. 2021;7:2214–2220. doi: 10.1021/acsinfecdis.1c00131. [DOI] [PubMed] [Google Scholar]
- 66.Case JB, et al. Murine hepatitis virus nsp14 exoribonuclease activity is required for resistance to innate immunity. J. Virol. 2017;92:e01531-17. doi: 10.1128/JVI.01531-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim Y, et al. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020;29:1596–1605. doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ulferts R, Ziebuhr J. Nidovirus ribonucleases: structures and functions in viral replication. RNA Biol. 2011;8:295–304. doi: 10.4161/rna.8.2.15196. [DOI] [PubMed] [Google Scholar]
- 69.Kindler E, et al. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog. 2017;13:e1006195. doi: 10.1371/journal.ppat.1006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deng X, et al. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc. Natl Acad. Sci. USA. 2017;114:E4251–E4260. doi: 10.1073/pnas.1618310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hackbart M, Deng X, Baker SC. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc. Natl Acad. Sci. USA. 2020;117:8094–8103. doi: 10.1073/pnas.1921485117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ivanov KA, et al. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl Acad. Sci. USA. 2004;101:12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Godoy, A. S. et al. SARS-CoV-2 nidoviral RNA uridylate‐specific endoribonuclease (NSP15): a target enabling package 10.5281/zenodo.4452975 (2020).
- 74.Hayden FG, et al. Baloxavir marboxil for uncomplicated influenza in adults and adolescents. N. Engl. J. Med. 2018;379:913–923. doi: 10.1056/NEJMoa1716197. [DOI] [PubMed] [Google Scholar]
- 75.Hirotsu N, et al. Baloxavir marboxil in Japanese pediatric patients with Influenza: safety and clinical and virologic outcomes. Clin. Infect. Dis. 2020;71:971–981. doi: 10.1093/cid/ciz908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Katsuno K, et al. Hit and lead criteria in drug discovery for infectious diseases of the developing world. Nat. Rev. Drug Discov. 2015;14:751–758. doi: 10.1038/nrd4683. [DOI] [PubMed] [Google Scholar]
- 77.Zhang X, et al. Anti-SARS-CoV-2 repurposing drug database: clinical pharmacology considerations. CPT Pharmacomet. Syst. Pharm. 2021;10:973–982. doi: 10.1002/psp4.12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Highlights of prescribing information: TAMIFLU (oseltamivir phosphate). FDAhttps://www.accessdata.fda.gov/drugsatfda_docs/label/2012/021087s062lbl.pdf (2012).
- 79.Ranganath N, et al. Rebound phenomenon after nirmatrelvir/ritonavir treatment of coronavirus disease-2019 in high-risk persons. Clin. Infect. Dis. 2023;76:e537–e539. doi: 10.1093/cid/ciac481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dai EY, et al. Viral kinetics of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron infection in mRNA-vaccinated individuals treated and not treated with nirmatrelvir–ritonavir. medRxiv. 2022 doi: 10.1101/2022.08.04.22278378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Crook H, Raza S, Nowell J, Young M, Edison P. Long covid — mechanisms, risk factors, and management. Br. Med. J. 2021;374:n1648. doi: 10.1136/bmj.n1648. [DOI] [PubMed] [Google Scholar]
- 82.Choutka J, Jansari V, Hornig M, Iwasaki A. Unexplained post-acute infection syndromes. Nat. Med. 2022;28:911–923. doi: 10.1038/s41591-022-01810-6. [DOI] [PubMed] [Google Scholar]
- 83.Douaud G, et al. SARS-CoV-2 is associated with changes in brain structure in UK Biobank. Nature. 2022;604:697–707. doi: 10.1038/s41586-022-04569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Serrano GE, et al. Mapping of SARS-CoV-2 brain invasion and histopathology in COVID-19 disease. medRxiv. 2021 doi: 10.1101/2021.02.15.21251511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Raju TN. The Nobel chronicles. 1988: James Whyte Black (b 1924), Gertrude Elion (1918–99), and George H Hitchings (1905–98) Lancet. 2000;355:1022. doi: 10.1016/S0140-6736(05)74775-9. [DOI] [PubMed] [Google Scholar]
- 86.Patick AK, et al. In vitro antiviral activity of AG7088, a potent inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 1999;43:2444–2450. doi: 10.1128/AAC.43.10.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kai H, et al. Discovery of clinical candidate Sivopixant (S-600918): lead optimization of dioxotriazine derivatives as selective P2X3 receptor antagonists. Bioorg. Med. Chem. Lett. 2021;52:128384. doi: 10.1016/j.bmcl.2021.128384. [DOI] [PubMed] [Google Scholar]
- 88.Müller S, et al. Target 2035 - update on the quest for a probe for every protein. RSC Med. Chem. 2022;13:13–21. doi: 10.1039/D1MD00228G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Douangamath A, et al. Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat. Commun. 2020;11:5047. doi: 10.1038/s41467-020-18709-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chodera J, Lee AA, London N, von Delft F. Crowdsourcing drug discovery for pandemics. Nat. Chem. 2020;12:581. doi: 10.1038/s41557-020-0496-2. [DOI] [PubMed] [Google Scholar]
- 91.Luttens A, et al. Ultralarge virtual screening identifies SARS-CoV-2 main protease inhibitors with broad-spectrum activity against coronaviruses. J. Am. Chem. Soc. 2022;144:2905–2920. doi: 10.1021/jacs.1c08402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang C-H, et al. Potent noncovalent inhibitors of the main protease of SARS-CoV-2 from molecular sculpting of the drug perampanel guided by free energy perturbation calculations. ACS Cent. Sci. 2021;7:467–475. doi: 10.1021/acscentsci.1c00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Simmons G, et al. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl Acad. Sci. USA. 2004;101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pruijssers AJ, et al. Remdesivir inhibits SARS-CoV-2 in human lung cells and chimeric SARS-CoV expressing the SARS-CoV-2 RNA polymerase in mice. Cell Rep. 2020;32:107940. doi: 10.1016/j.celrep.2020.107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ogando NS, et al. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020;101:925–940. doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sasaki M, et al. SARS-CoV-2 variants with mutations at the S1/S2 cleavage site are generated in vitro during propagation in TMPRSS2-deficient cells. PLoS Pathog. 2021;17:e1009233. doi: 10.1371/journal.ppat.1009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Davidson AD, et al. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020;12:68. doi: 10.1186/s13073-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sonnleitner ST, et al. The mutational dynamics of the SARS-CoV-2 virus in serial passages in vitro. Virol. Sin. 2022;37:198–207. doi: 10.1016/j.virs.2022.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schultz DC, et al. Pyrimidine inhibitors synergize with nucleoside analogues to block SARS-CoV-2. Nature. 2022;604:134–140. doi: 10.1038/s41586-022-04482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chu H, et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020;1:e14–e23. doi: 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhu N, et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chang C-W, et al. A newly engineered A549 cell line expressing ACE2 and TMPRSS2 is highly permissive to SARS-CoV-2, including the Delta and Omicron variants. Viruses. 2022;14:1369. doi: 10.3390/v14071369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sasaki M, et al. Air-liquid interphase culture confers SARS-CoV-2 susceptibility to A549 alveolar epithelial cells. Biochem. Biophys. Res. Commun. 2021;577:146–151. doi: 10.1016/j.bbrc.2021.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shang J, et al. Cell entry mechanisms of SARS-CoV-2. Proc. Natl Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nie Y, et al. Highly infectious SARS-CoV pseudotyped virus reveals the cell tropism and its correlation with receptor expression. Biochem. Biophys. Res. Commun. 2004;321:994–1000. doi: 10.1016/j.bbrc.2004.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lempp FA, et al. Lectins enhance SARS-CoV-2 infection and influence neutralizing antibodies. Nature. 2021;598:342–347. doi: 10.1038/s41586-021-03925-1. [DOI] [PubMed] [Google Scholar]
- 107.Wang R, et al. Human airway lineages derived from pluripotent stem cells reveal the epithelial responses to SARS-CoV-2 infection. Am. J. Physiol. Lung Cell Mol. Physiol. 2022;322:L462–L478. doi: 10.1152/ajplung.00397.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Davis JD, Wypych TP. Cellular and functional heterogeneity of the airway epithelium. Mucosal Immunol. 2021;14:978–990. doi: 10.1038/s41385-020-00370-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sims AC, Burkett SE, Yount B, Pickles RJ. SARS-CoV replication and pathogenesis in an in vitro model of the human conducting airway epithelium. Virus Res. 2008;133:33–44. doi: 10.1016/j.virusres.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.FDA. Emergency use authorization for paxlovid: CDER Review, https://www.fda.gov/media/155194/download (2021).
- 111.Do TND, et al. A robust SARS-CoV-2 replication model in primary human epithelial cells at the air liquid interface to assess antiviral agents. Antiviral Res. 2021;192:105122. doi: 10.1016/j.antiviral.2021.105122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Abdelnabi R, et al. The oral protease inhibitor (PF-07321332) protects Syrian hamsters against infection with SARS-CoV-2 variants of concern. Nat. Commun. 2022;13:719. doi: 10.1038/s41467-022-28354-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rayner RE, Makena P, Prasad GL, Cormet-Boyaka E. Optimization of normal human bronchial epithelial (NHBE) cell 3D cultures for in vitro lung model studies. Sci. Rep. 2019;9:500. doi: 10.1038/s41598-018-36735-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hiemstra PS, Grootaers G, van der Does AM, Krul CAM, Kooter IM. Human lung epithelial cell cultures for analysis of inhaled toxicants: Lessons learned and future directions. Toxicol. Vitr. 2018;47:137–146. doi: 10.1016/j.tiv.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 115.Jansen J, et al. SARS-CoV-2 infects the human kidney and drives fibrosis in kidney organoids. Cell Stem Cell. 2022;29:217–231.e8. doi: 10.1016/j.stem.2021.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lohmann V, Bartenschlager R. On the history of hepatitis C virus cell culture systems. J. Med. Chem. 2014;57:1627–1642. doi: 10.1021/jm401401n. [DOI] [PubMed] [Google Scholar]
- 117.Hui KPY, et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature. 2022;603:715–720. doi: 10.1038/s41586-022-04479-6. [DOI] [PubMed] [Google Scholar]
- 118.Mautner L, et al. Replication kinetics and infectivity of SARS-CoV-2 variants of concern in common cell culture models. Virol. J. 2022;19:76. doi: 10.1186/s12985-022-01802-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Meng B, et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature. 2022;603:706–714. doi: 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ramirez S, et al. Overcoming culture restriction for SARS-CoV-2 in human cells facilitates the screening of compounds inhibiting viral replication. Antimicrob. Agents Chemother. 2021;65:e0009721. doi: 10.1128/AAC.00097-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xie X, et al. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe. 2020;27:841–848.e3. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xie X, et al. A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. Nat. Commun. 2020;11:5214. doi: 10.1038/s41467-020-19055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Muruato AE, et al. A high-throughput neutralizing antibody assay for COVID-19 diagnosis and vaccine evaluation. Nat. Commun. 2020;11:4059. doi: 10.1038/s41467-020-17892-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lohmann V, et al. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 125.Hannemann H. Viral replicons as valuable tools for drug discovery. Drug. Discov. Today. 2020;25:1026–1033. doi: 10.1016/j.drudis.2020.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tao W, Gan T, Guo M, Xu Y, Zhong J. Novel stable Ebola virus minigenome replicon reveals remarkable stability of the viral genome. J. Virol. 2017;91:e01316-17. doi: 10.1128/JVI.01316-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.He X, et al. Generation of SARS-CoV-2 reporter replicon for high-throughput antiviral screening and testing. Proc. Natl Acad. Sci. USA. 2021;118:e2025866118. doi: 10.1073/pnas.2025866118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kotaki T, Xie X, Shi P-Y, Kameoka MA. PCR amplicon-based SARS-CoV-2 replicon for antiviral evaluation. Sci. Rep. 2021;11:2229. doi: 10.1038/s41598-021-82055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ricardo-Lax I, et al. Replication and single-cycle delivery of SARS-CoV-2 replicons. Science. 2021;374:1099–1106. doi: 10.1126/science.abj8430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Xia H, et al. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33:108234. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang X, et al. A trans-complementation system for SARS-CoV-2 recapitulates authentic viral replication without virulence. Cell. 2021;184:2229–2238.e13. doi: 10.1016/j.cell.2021.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liu Y, et al. A live-attenuated SARS-CoV-2 vaccine candidate with accessory protein deletions. Nat. Commun. 2022;13:4337. doi: 10.1038/s41467-022-31930-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chiu W, et al. Development and optimization of a high-throughput screening assay for in vitro anti-SARS-CoV-2 activity: evaluation of 5676 phase 1 passed structures. J. Med. Virol. 2022;94:3101–3111. doi: 10.1002/jmv.27683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bakowski MA, et al. Drug repurposing screens identify chemical entities for the development of COVID-19 interventions. Nat. Commun. 2021;12:3309. doi: 10.1038/s41467-021-23328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chu H, Chan JF-W, Yuen K-Y. Animal models in SARS-CoV-2 research. Nat. Methods. 2022;19:392–394. doi: 10.1038/s41592-022-01447-w. [DOI] [PubMed] [Google Scholar]
- 136.de Vries RD, et al. Animal models of SARS-CoV-2 transmission. Curr. Opin. Virol. 2021;50:8–16. doi: 10.1016/j.coviro.2021.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chu Y-K, et al. The SARS-CoV ferret model in an infection-challenge study. Virology. 2008;374:151–163. doi: 10.1016/j.virol.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Czub M, Weingartl H, Czub S, He R, Cao J. Evaluation of modified vaccinia virus Ankara based recombinant SARS vaccine in ferrets. Vaccine. 2005;23:2273–2279. doi: 10.1016/j.vaccine.2005.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shi J, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ryan KA, et al. Dose-dependent response to infection with SARS-CoV-2 in the ferret model and evidence of protective immunity. Nat. Commun. 2021;12:81. doi: 10.1038/s41467-020-20439-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Imai M, et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl Acad. Sci. USA. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chan JF-W, et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020;71:2428–2446. doi: 10.1093/cid/ciaa644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bao L, et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 144.Dinnon KH, 3rd, et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shuai H, et al. Emerging SARS-CoV-2 variants expand species tropism to murines. eBioMedicine. 2021;73:103643. doi: 10.1016/j.ebiom.2021.103643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Muñoz-Fontela C, et al. Advances and gaps in SARS-CoV-2 infection models. PLoS Pathog. 2022;18:e1010161. doi: 10.1371/journal.ppat.1010161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Munster VJ, et al. Subtle differences in the pathogenicity of SARS-CoV-2 variants of concern B.1.1.7 and B.1.351 in rhesus macaques. Sci. Adv. 2021;7:eabj3627. doi: 10.1126/sciadv.abj3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.McMahan K, et al. Reduced pathogenicity of the SARS-CoV-2 Omicron variant in hamsters. Med. 2022;3:262–268.e4. doi: 10.1016/j.medj.2022.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang Y-N, et al. Different pathogenesis of SARS-CoV-2 Omicron variant in wild-type laboratory mice and hamsters. Signal Transduct. Target. Ther. 2022;7:62. doi: 10.1038/s41392-022-00930-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rissmann M, et al. Pulmonary lesions following inoculation with the SARS-CoV-2 Omicron BA.1 (B.1.1.529) variant in Syrian golden hamsters. Emerg. Microbes Infect. 2022;11:1778–1786. doi: 10.1080/22221751.2022.2095932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Natekar JP, et al. Differential pathogenesis of SARS-CoV-2 variants of concern in human ACE2-expressing mice. Viruses. 2022;14:1139. doi: 10.3390/v14061139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Halfmann PJ, et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603:687–692. doi: 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Laitman AM, et al. The SARS-CoV-2 Omicron variant does not have higher nasal viral loads compared to the Delta variant in symptomatic and asymptomatic individuals. J. Clin. Microbiol. 2022;60:e0013922. doi: 10.1128/jcm.00139-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Puhach O, et al. Infectious viral load in unvaccinated and vaccinated individuals infected with ancestral, Delta or Omicron SARS-CoV-2. Nat. Med. 2022;28:1491–1500. doi: 10.1038/s41591-022-01816-0. [DOI] [PubMed] [Google Scholar]
- 155.Sasaki M, et al. S-217622, a SARS-CoV-2 main protease inhibitor, decreases viral load and ameliorates COVID-19 severity in hamsters. Sci. Transl. Med. 2022;15:eabq40. doi: 10.1126/scitranslmed.abq4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dinnon KH, 3rd, et al. SARS-CoV-2 infection produces chronic pulmonary epithelial and immune cell dysfunction with fibrosis in mice. Sci. Transl. Med. 2022;14:eabo5070. doi: 10.1126/scitranslmed.abo5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sullivan NJ, Martin JE, Graham BS, Nabel GJ. Correlates of protective immunity for Ebola vaccines: implications for regulatory approval by the animal rule. Nat. Rev. Microbiol. 2009;7:393–400. doi: 10.1038/nrmicro2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Beasley DWC, Brasel TL, Comer JE. First vaccine approval under the FDA animal rule. NPJ Vaccines. 2016;1:16013. doi: 10.1038/npjvaccines.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.FDA. Product Development under the Animal Rule: Guidance for Industry (Center for Drug Evaluation and Research, 2015).
- 160.Animal rule approvals. FDAhttps://www.fda.gov/drugs/nda-and-bla-approvals/animal-rule-approvals (2021).
- 161.Osterrieder N, et al. Age-dependent progression of SARS-CoV-2 infection in Syrian hamsters. Viruses. 2020;12:779. doi: 10.3390/v12070779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sefik E, et al. A humanized mouse model of chronic COVID-19. Nat. Biotechnol. 2022;40:906–920. doi: 10.1038/s41587-021-01155-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Brodin P, Arditi M. Severe acute hepatitis in children: investigate SARS-CoV-2 superantigens. Lancet Gastroenterol. Hepatol. 2022;7:594–596. doi: 10.1016/S2468-1253(22)00166-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lambkin-Williams R, Noulin N, Mann A, Catchpole A, Gilbert AS. The human viral challenge model: accelerating the evaluation of respiratory antivirals, vaccines and novel diagnostics. Respir. Res. 2018;19:123. doi: 10.1186/s12931-018-0784-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Rapeport G, et al. SARS-CoV-2 human challenge studies - establishing the model during an evolving pandemic. N. Engl. J. Med. 2021;385:961–964. doi: 10.1056/NEJMp2106970. [DOI] [PubMed] [Google Scholar]
- 166.Killingley B, et al. Safety, tolerability and viral kinetics during SARS-CoV-2 human challenge in young adults. Nat. Med. 2022;28:1031–1041. doi: 10.1038/s41591-022-01780-9. [DOI] [PubMed] [Google Scholar]
- 167.He X, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 168.DeVincenzo J, et al. A randomized, placebo-controlled, respiratory syncytial virus human challenge study of the antiviral efficacy, safety, and pharmacokinetics of RV521, an inhibitor of the RSV-F protein. Antimicrob. Agents Chemother. 2020;64:e01884-19. doi: 10.1128/AAC.01884-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Canini L, Woolhouse MEJ, Maines TR, Carrat F. Heterogeneous shedding of influenza by human subjects and its implications for epidemiology and control. Sci. Rep. 2016;6:38749. doi: 10.1038/srep38749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Binford SL, et al. In vitro resistance study of rupintrivir, a novel inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 2007;51:4366–4373. doi: 10.1128/AAC.00905-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Moscona A. Oseltamivir resistance–disabling our influenza defenses. N. Engl. J. Med. 2005;353:2633–2636. doi: 10.1056/NEJMp058291. [DOI] [PubMed] [Google Scholar]
- 172.Gubareva LV, Fry AM. Baloxavir and treatment-emergent resistance: public health insights and next steps. J. Infect. Dis. 2020;221:337–339. doi: 10.1093/infdis/jiz245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Strasfeld L, Chou S. Antiviral drug resistance: mechanisms and clinical implications. Infect. Dis. Clin. North Am. 2010;24:413–437. doi: 10.1016/j.idc.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Chatterji U, et al. HCV resistance to cyclosporin A does not correlate with a resistance of the NS5A-cyclophilin A interaction to cyclophilin inhibitors. J. Hepatol. 2010;53:50–56. doi: 10.1016/j.jhep.2010.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ratcliff AN, Shi W, Arts EJ. HIV-1 resistance to maraviroc conferred by a CD4 binding site mutation in the envelope glycoprotein gp120. J. Virol. 2013;87:923–934. doi: 10.1128/JVI.01863-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Roche M, et al. A common mechanism of clinical HIV-1 resistance to the CCR5 antagonist maraviroc despite divergent resistance levels and lack of common gp120 resistance mutations. Retrovirology. 2013;10:43. doi: 10.1186/1742-4690-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Cagliani R, Forni D, Clerici M, Sironi M. Coding potential and sequence conservation of SARS-CoV-2 and related animal viruses. Infect. Genet. Evol. 2020;83:104353. doi: 10.1016/j.meegid.2020.104353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ryder SP, Morgan BR, Coskun P, Antkowiak K, Massi F. Analysis of emerging variants in structured regions of the SARS-CoV-2 genome. Evol. Bioinform. Online. 2021;17:11769343211014167. doi: 10.1177/11769343211014167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Chan AP, Choi Y, Schork NJ. Conserved genomic terminals of SARS-CoV-2 as coevolving functional elements and potential therapeutic targets. mSphere. 2020;5:e00754-20. doi: 10.1128/mSphere.00754-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Painter GR, et al. The prophylactic and therapeutic activity of a broadly active ribonucleoside analog in a murine model of intranasal venezuelan equine encephalitis virus infection. Antiviral Res. 2019;171:104597. doi: 10.1016/j.antiviral.2019.104597. [DOI] [PubMed] [Google Scholar]
- 181.Lythgoe KA, et al. SARS-CoV-2 within-host diversity and transmission. Science. 2021;372:eabg0821. doi: 10.1126/science.abg0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Forns X, Purcell RH, Bukh J. Quasispecies in viral persistence and pathogenesis of hepatitis C virus. Trends Microbiol. 1999;7:402–410. doi: 10.1016/S0966-842X(99)01590-5. [DOI] [PubMed] [Google Scholar]
- 183.Raghwani J, et al. Evolution of HIV-1 within untreated individuals and at the population scale in Uganda. PLoS Pathog. 2018;14:e1007167. doi: 10.1371/journal.ppat.1007167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Barton JP, et al. Relative rate and location of intra-host HIV evolution to evade cellular immunity are predictable. Nat. Commun. 2016;7:11660. doi: 10.1038/ncomms11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Powdrill MH, et al. Contribution of a mutational bias in hepatitis C virus replication to the genetic barrier in the development of drug resistance. Proc. Natl Acad. Sci. USA. 2011;108:20509–20513. doi: 10.1073/pnas.1105797108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Rausch JW, Capoferri AA, Katusiime MG, Patro SC, Kearney MF. Low genetic diversity may be an Achilles heel of SARS-CoV-2. Proc. Natl Acad. Sci. USA. 2020;117:24614–24616. doi: 10.1073/pnas.2017726117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Takada K, Ueda MT, Watanabe T, Nakagawa S. Genomic diversity of SARS-CoV-2 can be accelerated by a mutation in the nsp14 gene. iScience. 2023;26:106210. doi: 10.1016/j.isci.2023.106210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Eckerle LD, et al. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 2010;6:e1000896. doi: 10.1371/journal.ppat.1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Smith EC, Blanc H, Surdel MC, Vignuzzi M, Denison MR. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 2013;9:e1003565. doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bonsall D, et al. Characterization of hepatitis C virus resistance to grazoprevir reveals complex patterns of mutations following on-treatment breakthrough that are not observed at relapse. Infect. Drug Resist. 2018;11:1119–1135. doi: 10.2147/IDR.S156581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Dharan NJ, et al. Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States. J. Am. Med. Assoc. 2009;301:1034–1041. doi: 10.1001/jama.2009.294. [DOI] [PubMed] [Google Scholar]
- 192.Hauge SH, Dudman S, Borgen K, Lackenby A, Hungnes O. Oseltamivir-resistant influenza viruses A (H1N1), Norway, 2007-08. Emerg. Infect. Dis. 2009;15:155–162. doi: 10.3201/eid1502.081031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Imai M, et al. Influenza A variants with reduced susceptibility to baloxavir isolated from Japanese patients are fit and transmit through respiratory droplets. Nat. Microbiol. 2020;5:27–33. doi: 10.1038/s41564-019-0609-0. [DOI] [PubMed] [Google Scholar]
- 194.Lee LY, et al. Evaluating the fitness of PA/I38T-substituted influenza A viruses with reduced baloxavir susceptibility in a competitive mixtures ferret model. PLoS Pathog. 2021;17:e1009527. doi: 10.1371/journal.ppat.1009527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ikematsu H, et al. Duration of fever and PA/I38X-substituted virus emergence in patients treated with baloxavir in the 2018-2019 influenza season. J. Infect. Chemother. 2020;26:400–402. doi: 10.1016/j.jiac.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 196.Gandhi S, et al. De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS-CoV-2 infection in an immunocompromised patient: a case report. Nat. Commun. 2022;13:1547. doi: 10.1038/s41467-022-29104-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Malsy J, et al. Sustained response after remdesivir and convalescent plasma therapy in a B-cell-depleted patient with protracted coronavirus disease 2019 (COVID-19) Clin. Infect. Dis. 2021;73:e4020–e4024. doi: 10.1093/cid/ciaa1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Helleberg M, et al. Persistent COVID-19 in an immunocompromised patient temporarily responsive to two courses of remdesivir therapy. J. Infect. Dis. 2020;222:1103–1107. doi: 10.1093/infdis/jiaa446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Butler CC, et al. Molnupiravir plus usual care versus usual care alone as early treatment for adults with COVID-19 at increased risk of adverse outcomes (PANORAMIC): an open-label, platform-adaptive randomised controlled trial. Lancet. 2023;401:281–293. doi: 10.1016/S0140-6736(22)02597-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wyles DL. Antiviral resistance and the future landscape of hepatitis C virus infection therapy. J. Infect. Dis. 2013;207:S33–S39. doi: 10.1093/infdis/jis761. [DOI] [PubMed] [Google Scholar]
- 201.Shi ST, et al. In vitro resistance study of AG-021541, a novel nonnucleoside inhibitor of the hepatitis C virus RNA-dependent RNA polymerase. Antimicrob. Agents Chemother. 2008;52:675–683. doi: 10.1128/AAC.00834-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Omoto S, et al. Characterization of influenza virus variants induced by treatment with the endonuclease inhibitor baloxavir marboxil. Sci. Rep. 2018;8:9633. doi: 10.1038/s41598-018-27890-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Matthew AN, et al. Drug design strategies to avoid resistance in direct-acting antivirals and beyond. Chem. Rev. 2021;121:3238–3270. doi: 10.1021/acs.chemrev.0c00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Prabu-Jeyabalan M, Nalivaika E, Schiffer CA. Substrate shape determines specificity of recognition for HIV-1 protease: analysis of crystal structures of six substrate complexes. Structure. 2002;10:369–381. doi: 10.1016/S0969-2126(02)00720-7. [DOI] [PubMed] [Google Scholar]
- 205.Discovery and Development of PBI-0451. A novel oral protease inhibitor for the potential treatment of SARS-CoV-2. Pardes Bioscienceshttps://ir.pardesbio.com/static-files/fc7c4f8c-e0bd-4b97-8c9c-eff09bafd4db (2022).
- 206.King NM, Prabu-Jeyabalan M, Nalivaika EA, Schiffer CA. Combating susceptibility to drug resistance: lessons from HIV-1 protease. Chem. Biol. 2004;11:1333–1338. doi: 10.1016/j.chembiol.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 207.Romano KP, Ali A, Royer WE, Schiffer CA. Drug resistance against HCV NS3/4A inhibitors is defined by the balance of substrate recognition versus inhibitor binding. Proc. Natl Acad. Sci. USA. 2010;107:20986–20991. doi: 10.1073/pnas.1006370107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Shaqra AM, et al. Defining the substrate envelope of SARS-CoV-2 main protease to predict and avoid drug resistance. Nat. Commun. 2022;13:3556. doi: 10.1038/s41467-022-31210-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Flynn JM, et al. Comprehensive fitness landscape of SARS-CoV-2 Mpro reveals insights into viral resistance mechanisms. eLife. 2022;11:e77433. doi: 10.7554/eLife.77433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Deng Z, et al. Deep sequencing of systematic combinatorial libraries reveals β-lactamase sequence constraints at high resolution. J. Mol. Biol. 2012;424:150–167. doi: 10.1016/j.jmb.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Firnberg E, Labonte JW, Gray JJ, Ostermeier M. A comprehensive, high-resolution map of a gene’s fitness landscape. Mol. Biol. Evol. 2014;31:1581–1592. doi: 10.1093/molbev/msu081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Porebski BT, Buckle AM. Consensus protein design. Protein Eng. Des. Sel. 2016;29:245–251. doi: 10.1093/protein/gzw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sternke M, Tripp KW, Barrick D. Consensus sequence design as a general strategy to create hyperstable, biologically active proteins. Proc. Natl Acad. Sci. USA. 2019;116:11275–11284. doi: 10.1073/pnas.1816707116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.de Wispelaere M, et al. Small molecule degraders of the hepatitis C virus protease reduce susceptibility to resistance mutations. Nat. Commun. 2019;10:3468. doi: 10.1038/s41467-019-11429-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Haniff HS, et al. Targeting the SARS-CoV-2 RNA genome with small molecule binders and ribonuclease targeting chimera (RIBOTAC) degraders. ACS Cent. Sci. 2020;6:1713–1721. doi: 10.1021/acscentsci.0c00984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Trozzi C, et al. In vitro selection and characterization of hepatitis C virus serine protease variants resistant to an active-site peptide inhibitor. J. Virol. 2003;77:3669–3679. doi: 10.1128/JVI.77.6.3669-3679.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Lu L, et al. Mutations conferring resistance to a potent hepatitis C virus serine protease inhibitor in vitro. Antimicrob. Agents Chemother. 2004;48:2260–2266. doi: 10.1128/AAC.48.6.2260-2266.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Le Pogam S, et al. In vitro selected Con1 subgenomic replicons resistant to 2’-C-methyl-cytidine or to R1479 show lack of cross resistance. Virology. 2006;351:349–359. doi: 10.1016/j.virol.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 219.Nguyen TT, et al. Resistance profile of a hepatitis C virus RNA-dependent RNA polymerase benzothiadiazine inhibitor. Antimicrob. Agents Chemother. 2003;47:3525–3530. doi: 10.1128/AAC.47.11.3525-3530.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Pelosi LA, Voss S, Liu M, Gao M, Lemm JA. Effect on hepatitis C virus replication of combinations of direct-acting antivirals, including NS5A inhibitor daclatasvir. Antimicrob. Agents Chemother. 2012;56:5230–5239. doi: 10.1128/AAC.01209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.McCown MF, et al. The hepatitis C virus replicon presents a higher barrier to resistance to nucleoside analogs than to nonnucleoside polymerase or protease inhibitors. Antimicrob. Agents Chemother. 2008;52:1604–1612. doi: 10.1128/AAC.01317-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ng TI, et al. In vitro antiviral activity and resistance profile of the next-generation hepatitis C virus NS5A inhibitor pibrentasvir. Antimicrob. Agents Chemother. 2017;61:e02558-16. doi: 10.1128/AAC.02558-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Han B, et al. Sofosbuvir susceptibility of genotype 1 to 6 HCV from DAA-naïve subjects. Antiviral Res. 2019;170:104574. doi: 10.1016/j.antiviral.2019.104574. [DOI] [PubMed] [Google Scholar]
- 224.Bukh J, et al. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc. Natl Acad. Sci. USA. 2002;99:14416–14421. doi: 10.1073/pnas.212532699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Sarrazin C, et al. Clinical significance of in vitro replication-enhancing mutations of the hepatitis C virus (HCV) replicon in patients with chronic HCV infection. J. Infect. Dis. 2005;192:1710–1719. doi: 10.1086/497142. [DOI] [PubMed] [Google Scholar]
- 226.Belema M, et al. Discovery and development of hepatitis C virus NS5A replication complex inhibitors. J. Med. Chem. 2014;57:1643–1672. doi: 10.1021/jm401793m. [DOI] [PubMed] [Google Scholar]
- 227.Kaptein SJF, et al. A pan-serotype dengue virus inhibitor targeting the NS3–NS4B interaction. Nature. 2021;598:504–509. doi: 10.1038/s41586-021-03990-6. [DOI] [PubMed] [Google Scholar]
- 228.Cheng K-C, Korfmacher WA, White RE, Njoroge FG. Lead optimization in discovery drug metabolism and pharmacokinetics/case study: the hepatitis C virus (HCV) protease inhibitor SCH 503034. Perspect. Med. Chem. 2007;1:1–9. [PMC free article] [PubMed] [Google Scholar]
- 229.Agostini ML, et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9:e00221-18. doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Szemiel AM, et al. In vitro selection of Remdesivir resistance suggests evolutionary predictability of SARS-CoV-2. PLoS Pathog. 2021;17:e1009929. doi: 10.1371/journal.ppat.1009929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Stevens LJ, et al. Mutations in the SARS-CoV-2 RNA-dependent RNA polymerase confer resistance to remdesivir by distinct mechanisms. Sci. Transl. Med. 2022;14:eabo0718. doi: 10.1126/scitranslmed.abo0718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Jochmans D, et al. The substitutions L50F, E166A and L167F in SARS-CoV-2 3CLpro are selected by a protease inhibitor in vitro and confer resistance to nirmatrelvir. mBio. 2022;14:e02815–e02822. doi: 10.1128/mbio.02815-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Research involving enhanced potential pandemic pathogen. NIHhttps://www.nih.gov/news-events/research-involving-potential-pandemic-pathogens (2021).
- 234.Manns MP, et al. The way forward in HCV treatment–finding the right path. Nat. Rev. Drug Discov. 2007;6:991–1000. doi: 10.1038/nrd2411. [DOI] [PubMed] [Google Scholar]
- 235.Baumert TF, Berg T, Lim JK, Nelson DR. Status of direct-acting antiviral therapy for hepatitis C virus infection and remaining challenges. Gastroenterology. 2019;156:431–445. doi: 10.1053/j.gastro.2018.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Singh RSP, et al. Innovative randomized phase I study and dosing regimen selection to accelerate and inform pivotal COVID-19 trial of nirmatrelvir. Clin. Pharmacol. Ther. 2022;112:101–111. doi: 10.1002/cpt.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Spengler JR, et al. Meeting report: 35th international conference on antiviral research in Seattle, Washington, USA – March 21–25, 2022. Antiviral Res. 2023;211:105521. doi: 10.1016/j.antiviral.2022.105521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.RECOVERY Collaborative Group. Dexamethasone in hospitalized patients with Covid-19. N. Engl. J. Med. 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.EMA initiatives for acceleration of development support and evaluation procedures for COVID-19 treatments and vaccines. EMAhttps://www.ema.europa.eu/en/documents/other/ema-initiatives-acceleration-development-support-evaluation-procedures-covid-19-treatments-vaccines_en.pdf (2022).
- 240.COVID-19 Public Health Emergency: general considerations for pre-IND meeting requests for COVID-19 related drugs and biological products. FDAhttps://www.fda.gov/regulatory-information/search-fda-guidance-documents/covid-19-public-health-emergency-general-considerations-pre-ind-meeting-requests-covid-19-related (2022).
- 241.COVID-19: developing drugs and biological products for treatment or prevention: guidance for industry. FDAhttps://www.fda.gov/media/137926/download (2021).
- 242.Summary of product characteristics: Maviret. EMAhttps://www.ema.europa.eu/en/documents/product-information/maviret-epar-product-information_en.pdf (2022).
- 243.Human immunodeficiency virus-1 infection: developing antiretroviral drugs for treatment: guidance for industry. FDAhttps://www.fda.gov/files/drugs/published/Human-Immunodeficiency-Virus-1-Infection--Developing-Antiretroviral-Drugs-for-Treatment.pdf (2015).
- 244.Chronic hepatitis C virus infection: developing direct-acting antiviral drugs for treatment: guidance for industry. FDAhttps://www.fda.gov/media/79486/download (2017).
- 245.Edwards A, Hartung IV. No shortcuts to SARS-CoV-2 antivirals. Science. 2021;373:488–489. doi: 10.1126/science.abj9488. [DOI] [PubMed] [Google Scholar]
- 246.Tummino TA, et al. Drug-induced phospholipidosis confounds drug repurposing for SARS-CoV-2. Science. 2021;373:541–547. doi: 10.1126/science.abi4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.NIAID announces antiviral drug development awards. NIAIDhttps://www.niaid.nih.gov/news-events/niaid-announces-antiviral-drug-development-awards (2022).
- 248.Nissley DV, McCormick F. RAS at 40: update from the RAS initiative. Cancer Discov. 2022;12:895–898. doi: 10.1158/2159-8290.CD-21-1554. [DOI] [PubMed] [Google Scholar]
- 249.Williamson AR. Creating a structural genomics consortium. Nat. Struct. Biol. 2000;7:953. doi: 10.1038/80726. [DOI] [PubMed] [Google Scholar]
- 250.Wilkinson MD, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Highlights of Prescribing Information for VEKLURY. FDAhttps://www.accessdata.fda.gov/drugsatfda_docs/label/2020/214787Orig1s000lbl.pdf (2020).
- 252.Fact sheet for healthcare providers: emergency use authorization for LAGEVRIO (molnupiravir) capsules. FDAhttps://www.fda.gov/media/155054/download (2021).
- 253.Use of molnupiravir for the treatment of COVID-19. EMAhttps://www.ema.europa.eu/en/documents/referral/lagevrio-also-known-molnupiravir-mk-4482-covid-19-article-53-procedure-assessment-report_en.pdf (2022).
- 254.Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/S0006-291X(67)80055-X. [DOI] [PubMed] [Google Scholar]