SUMMARY
OBJECTIVE:
In this study, we aimed to identify a microRNA expression signature that could be used to distinguish methamphetamine from control samples. We also utilized the existing bioinformatics tools in order to predict the candidate microRNAs that could play potential key roles in regulating drug addiction-related genes.
METHODS:
Methamphetamine samples from 21 ventral tegmental area and 21 nucleus accumbens samples and their control regions were obtained from the Council of Forensic Medicine (Istanbul). Quantitative analysis of let-7b-3p was studied using quantitative reverse transcription PCR. Statistical analysis was carried out using Student’s t-test. The receiver operating characteristic curves were plotted with Statistical Package for the Social Sciences (SPSS 20.0).
RESULTS:
Our quantitative reverse transcription PCR results revealed that let-7b-3p was significantly overexpressed in brain tissues of the methamphetamine-user group. Let-7b-3p had significant power to discriminate methamphetamine from control samples in the ventral tegmental area (AUC; 0.922) and nucleus accumbens (AUC; 0.899) regions.
CONCLUSION:
We have shown for the first time in the literature the differential expression of let-7b-3p in samples from methamphetamine-addicted individuals. We suggest that let-7b-3p could be a powerful marker for the diagnosis of methamphetamine addiction. Our results showed that differentially expressed let-7b-3p in methamphetamine users could be used as a diagnostic and therapeutic marker.
KEYWORDS: Methamphetamine, MicroRNAs, Autopsy, Biomarkers, Brain
INTRODUCTION
Methamphetamine, usually known as “crystal meth”, is the most widely used psychoactive component of illicit drugs 1 . Similar to other amphetamines, methamphetamine increases the levels of neurotransmitters such as dopamine and norepinephrine and shows a notably strong serotonergic effect 2 . Methamphetamine is one of the most widely used illegal synthetic drugs, especially in Europe (nearly 15 million users), Oceania, and North America 3 . The European Drug Report has estimated that the availability and use of methamphetamine have increased and the trend will continue to rise. The report also highlighted that the number of methamphetamine deaths has dramatically increased among adolescents and young adults in recent years 4 .
Drug addiction is a serious psychiatric disorder that is characterized by loss of control over drug consumption 5 . Addictive drugs converge on the mesocorticolimbic dopaminergic [DA] circuitry, which contains the nucleus accumbens (NAc) and ventral tegmental area (VTA) in the brain’s limbic system 6 . Therefore, VTA and NAc regions are the major components of the brain reward system and also play a highly important role in response to drug addiction 7 . Drug addiction induces adjustment in neuroplasticity, which is regulated by permanent alterations in gene expression and protein function in the VTA and NAc 8 . As potent regulators of post-transcriptional gene expression, microRNAs (miRNAs) are poised to play key roles in the addiction-relevant reprogramming of neuronal gene expression in the corticostriatal system 9–11 . miRNAs are a class of non-coding 18–25 bp long nucleotide RNAs that can regulate the expression of hundreds of genes, either by translational suppression or by degrading mRNAs to bind their complementary sequences in the 3´ UTR 12,13 . Recent studies demonstrated that miRNA expression plays a key role in drug addiction in the NAc and VTA as well as in other regions of the mesocorticolimbic DA system 14,15 . Furthermore, the impact of drugs such as heroin, morphine, and alcohol on differentially expressed miRNAs has been shown both in vivo and in vitro. However, the role of miRNAs in methamphetamine-seeking behaviors and the specific targets of key regulatory miRNAs need to be identified. In our previous study, we have worked on unraveling the molecular mechanisms underlying 3,4-methylenedioxymethamphetamine (MDMA)-seeking behavior to develop specific biomarkers of therapeutic approaches in postmortem human brain tissues of MDMA users 16 .
In this study, we have utilized diverse bioinformatics resources to predict potential miRNA regulators of drug addiction. Based on our literature search and bioinformatics analysis, we selected let-7b-3p as a top candidate biomarker for methamphetamine-seeking behavior. The family members of let-7 are extremely evolutionarily conserved across various animal species, including flies and mammals. Some known biological roles for let-7b, which was the first defined human miRNA, include the regulation of stem-cell differentiation, cell differentiation, and neuromuscular development. Many studies have shown that let-7b has putative target sites on several addiction-related genes and causes neurodegeneration diseases 17 . The findings of our study highlight a new role of let-7b-3p in methamphetamine-seeking behavior.
METHODS
Postmortem human brain tissue acquisition
The NAc and VTA regions of postmortem human brain tissues were collected from the Morgue Department, Council of Forensic Medicine, Istanbul, Turkey, and local ethical approval was obtained (approval number: 2020/38). The study consisted of 21 subjects (13 males and 8 females) whose deaths were ruled methamphetamine intoxication based on toxicology findings and 21 drug-free control subjects (13 males and 8 females) matched pairwise with methamphetamine users for age and gender. Post-hoc analysis elicited no meaningful differentiation between the study of methamphetamine users and the control subjects for any demographic parameter. The brain samples were collected under full ethical clearance. Cases with a history of poly-drug abuse or other complicating conditions such as HIV/AIDS were excluded. Brain specimens of VTA and NAc were collected as part of the routine autopsy process, and the tissues were flash-frozen in isopentane in liquid nitrogen. The frozen samples were stored at -80°C until further use.
Total RNA extraction
An amount of 50–100 mg of NAc and VTA brain tissues were homogenized in 1 mL TRIzol reagent (Invitrogen, USA). Total RNA was extracted following the manufacturer’s instructions. Total RNA concentrations and purities of the samples were determined by spectrophotometry using a NanoDrop ND-2000 system (Thermo Fisher Scientific Inc., Wilmington, DE).
Validation by real-time RT-PCR analysis
Validation of let-7b-3p was studied from the VTA and NAc regions of 21 methamphetamine users and 21 matched controls. Let-7b-3p and RNU43 assays were purchased from Applied Biosystems, Foster City, CA. Single-stranded complementary DNA (cDNA) was synthesized from total RNA using the TaqMan® MicroRNA Reverse Transcription Kit according to the manufacturer’s protocol (Applied Biosystems, Foster City, CA, USA). Following reverse transcription, quantitative reverse transcription PCR (qRT-PCR) reactions were carried out in duplicate using TaqMan® MicroRNA Assays (Applied Biosystems) in a Light Cycler 480-II real-time thermal cycler (Roche, Switzerland, Basel). RNU43 was used for the normalization of miRNA expression analyses. Relative quantification analysis was performed by delta-delta-Ct (2-ΔΔCT) calculation as described previously 18 .
Statistical analysis
Data presented as means±standard error and Student’s t-test (unpaired, two-tailed) were used for statistical analysis of the qRT-PCR. A two-tailed p-value of 0.05 or below was considered statistically significant. The receiver operating characteristic (ROC) curves were plotted using SPSS 21.0 (Statistical Package for the Social Sciences) to determine the power of control and validated let-7b-3p to differentiate between the samples.
Target analysis and miRNA target prediction
We used miRDB (http://www.mirdb.org/), TargetScan 6.0 (www.targetscan.org), and mirTarBase (http://mirtarbase.mbc.nctu.edu.tw/) to identify the predicted let-7b-3p targeting mRNAs with p-value smaller than 0.01 involved in addiction. Using STRING (http://string-db.org/) to demonstrate the interrelationships between genes and their interactive functional networks, we demonstrated the common let-7b-3p targets in protein-protein interaction (PPI) networks.
RESULTS
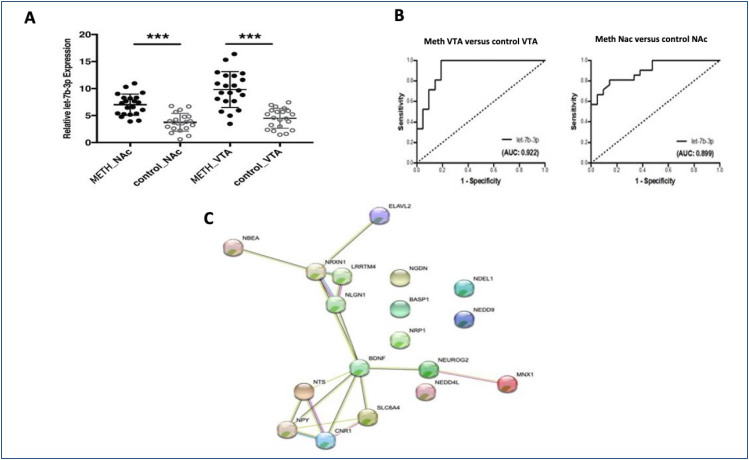
A total of 21 methamphetamine tissues (21 VTA and 21 NAc) and 21 normal tissues (21 VTA and 21 NAc) were obtained from the Council of Forensic Medicine, Istanbul, Turkey. We performed qRT-PCR to investigate the expression levels of let-7b-3p in order to determine the differences between methamphetamine and control subjects in VTA and NAc parts of human postmortem brain tissues. The mean age of methamphetamine subjects was 27.61±9.08, whereas controls had an average age of 30.09±10.43 years (Table 1). The methamphetamine group was found to have methamphetamine in the blood ranging from 569.3 to 1025.6 ng/mL with a median of 798 ng/mL (Table 1). When the two groups were compared in terms of the postmortem interval (PMI) samples, the difference between the mean PMI levels of the two groups was not significant. We investigated the expression profiles of let-7b-3p in methamphetamine samples and controls in VTA and NAc parts of brain tissues using qRT-PCR analysis. The results showed that the expression levels of let-7b-3p in VTA samples and NAc samples in Figure 1A (p<0.002 and p<0.004, respectively) were significantly upregulated in methamphetamine-addicted brain tissue samples compared to normal tissues.
Table 1. Age, brain pH, postmortem interval, and methamphetamine in the blood level of methamphetamine and control postmortem samples that were involved in the study.
| Methamphetamine (n=21) |
Control (n=21) |
p-value | |
|---|---|---|---|
| Age | 27.61±9.08 | 30.09±10.43 | 0.42 |
| Brain pH | 6.61±0.25 | 6.48±0.23 | 0.09 |
| PMI | 17.38±4.55 | 16.9±4.47 | 0.74 |
| Blood level of methamphetamine | 798 ng/mL (569.3–1025.6 ng/mL min–max) |
– | – |
PMI: postmortem interval.
Figure 1. (A) Relative expression levels of let-7b-3p in control versus methamphetamine-addicted brain tissues. (B) ROC analysis of let-7b-3p in methamphetamine versus control samples (NAc and VTA regions). Let-7b-3p cooperative power to discriminate two sets of samples composed of 21 methamphetamine and 21 control samples. (C) PPI network of commonly deregulated let-7b-3p targets. Pink: experimentally determined (known interactions); blue: from curated databases (known interactions); yellow: text mining; green: gene neighborhood (predicted interactions); black: co-expression. The interaction score was set to high confidence of 0.49. ***p<0.01.
To test the power of let-7b-3p for distinguishing the methamphetamine group from controls, we plotted ROC curves and the results showed that let-7b-3p had a higher area under the curve for NAc (AUC; 0.899) and VTA regions (AUC; 0.922) (Figure 1B).
The biological process, molecular function, cellular component, and KEGG pathways analysis of the potential targets of let-7b-3p with functional enrichments in the PPI network showed that the genes shown in Table 2 play an important role in the mechanisms of neurexins and neuroligins and in the neuronal system (Figure 1C).
Table 2. Biological, molecular, cellular functions gene ontology, and Kyoto Encyclopedia of Genes and Genomes pathway analysis of let-7p-3p potential targets for functional enhancements in the protein-protein interaction network.
| Pathway ID | Biological process (GO) | Count in network | False discovery rate |
|---|---|---|---|
| Pathway description | |||
| GO:0048699 | Generation of neurons | 11 | 8.43e-06 |
| GO:0031175 | Neuron projection development | 8 | 1.54e-05 |
| GO:0030182 | Neuron differentiation | 9 | 1.60e-05 |
| GO:0010976 | Positive regulation of neuron projection development | 6 | 2.16e-05 |
| GO:0051962 | Positive regulation of nervous system development | 7 | 3.63e-05 |
| Molecular function (GO) | |||
| Pathway description | |||
| GO:0099106 | Ion channel regulator activity | 3 | 0.0177 |
| GO:0005184 | Neuropeptide hormone activity | 2 | 0.022 |
| GO:0098772 | Molecular function regulator | 7 | 0.0248 |
| GO:0005246 | Calcium channel regulator activity | 2 | 0.0248 |
| Cellular component (GO) | |||
| Pathway description | |||
| GO:0150034 | Distalaxon | 6 | 1.43e-05 |
| GO:0098793 | Presynapse | 6 | 2.28e-05 |
| GO:0043005 | Neuron projection | 9 | 2.28e-05 |
| GO:0030426 | Growth cone | 5 | 2.28e-05 |
| GO:0030424 | Axon | 7 | 2.28e-05 |
| KEGG pathways | |||
| Pathway description | |||
| GO:6794361 | Neurexins and neuroligins | 3 | 0.0016 |
| GO:112316 | Neuronal system | 4 | 0.0078 |
GO: gene ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes.
DISCUSSION
Drug addiction is believed to be a neurological dysfunction because altered gene expression affects neuronal function and eventually behavior. Early detection and correct diagnosis are especially important for methamphetamine addiction in the therapy decision-making process. miRNAs have become a popular research subject among scientists in recent years. It has been shown that miRNAs can be used to explain several addiction processes, and determining the levels of miRNAs is proposed as an important approach to overcome therapeutic resistance in drug addiction. They are potential diagnostic and therapeutic biomarkers that have been shown to be present in the use of various types of drugs in recent years 19 .
MicroRNAs, which are among the non-coding RNAs, have a critical role in gene expression. The first known human miRNA, let-7, is expressed in both embryonic and adult mammalian brains. Let-7b serves as a key regulator of neural stem cell proliferation and differentiation. Overexpression of let-7 elicits neuronal cell proliferation and accelerated neural differentiation 20 .
The let-7 miRNA family was identified as a top candidate based on the number of assumed target sites. This study experimentally validated the in silico prediction that let-7b-3p, which is a member of the let-7 miRNA family, can interact with methamphetamine addiction.
Downregulation of let-7 was found to increase morphine and related μ-opioid receptor (MOR) expression in a human neuroblastoma cell line. This suggests that MOR is a target of let-7 because the expression of MOR is under constitutive suppression by let-7. Accordingly, morphine treatment causes an increase in let-7. Chronic morphine treatment notably upregulated let-7 expression in sensory neurons and brain stem nuclei 21 . The brain expression of the level of let-7 increases after morphine treatment, temporally correlating with the development of tolerance to morphine. Treatment with a let-7 inhibitor decreases brain let-7 levels and opioid tolerance. Let-7b has been previously proposed as an important factor for distinguishing morphine exposed from non-morphine-exposed brain tissues 22 . In parallel with this finding, our results showed that let-7b-3p is significantly overexpressed in brain tissues.
Recent studies have identified the role of several miRNAs in mammalian midbrain dopaminergic neurons and that they are related to addictive behaviors. Deregulation of let-7 seems to play a key role in neurological disorders 23 . Also, cocaine addiction affects the expression of let-7d, highlighting the possibility that some miRNAs are important regulators of the brain reward pathway and likely implicated in addiction 21 . Toll-like receptor (TLR) signaling is known to be a key component of neurodegeneration, and TLR7 responds to miRNAs in promoting immune responses leading to neurodegeneration 22 . Moreover, an alcoholic individual’s brain contains more ATP (adenosine triphosphate) than a non-alcoholic individual’s brain, and alcohol dependence is associated with hippocampal degeneration. Studies of postmortem human alcoholic brain hippocampal tissues have shown that increased expression of TLR7 and let-7b causes neurodegeneration 24 .
CONCLUSION
Our findings show that let-7b-3p is differentially expressed in methamphetamine users and let-7b-3p could serve as a potential biomarker for predicting methamphetamine abuse and treatment response. Let-7b-3p has been linked to mechanisms of drug abuse, and further studies would be very important in developing preventive strategies and new therapeutic interventions for methamphetamine abuse.
ACKNOWLEDGMENTS
We gratefully acknowledge the directors of the Council of Forensic Medicine for tissue acquisition.
Footnotes
Funding: none.
REFERENCES
- 1.Costa G, Gołembiowska K. Neurotoxicity of MDMA: main effects and mechanisms. Exp Neurol. 2022;347:113894–113894. doi: 10.1016/j.expneurol.2021.113894. [DOI] [PubMed] [Google Scholar]
- 2.Pantoni MM, Kim JL, Alstyne KR, Anagnostaras SG. MDMA and memory, addiction, and depression: dose-effect analysis. Psychopharmacology (Berl) 2022;239(3):935–949. doi: 10.1007/s00213-022-06086-9. doi: 10.1007/s00213-022-06086-9. Erratum in: Psychopharmacology (Berl). 2022;239(7):2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.UNODC . Word drug report. Vienna: UNODC; 2014. [Google Scholar]
- 4.European Monitoring Centre for Drugs and Drug Addiction . European drug report 2017: trends and developments. Lisbon: European Monitoring Centre for Drugs and Drug Addiction; 2017. [Google Scholar]
- 5.Nestler EJ, Lüscher C. The molecular basis of drug addiction: linking epigenetic to synaptic and circuit mechanisms. Neuron. 2019;102(1):48–59. doi: 10.1016/j.neuron.2019.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan B, Hu Z, Yao W, Le Q, Xu B, Liu X, et al. MiR-218 targets MeCP2 and inhibits heroin seeking behavior. Sci Rep. 2017;7:40413–40413. doi: 10.1038/srep40413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamilton PJ, Nestler EJ. Epigenetics and addiction. Curr Opin Neurobiol. 2019;59:128–136. doi: 10.1016/j.conb.2019.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nestler EJ. Molecular basis of long-term plasticity underlying addiction. Nat Rev Neurosci. 2001;2(2):119–128. doi: 10.1038/35053570. [DOI] [PubMed] [Google Scholar]
- 9.Gowen AM, Odegaard KE, Hernandez J, Chand S, Koul S, Pendyala G, et al. Role of microRNAs in the pathophysiology of addiction. Wiley Interdiscip Rev RNA. 2021;12(3):e1637. doi: 10.1002/wrna.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith ACW, Kenny PJ. MicroRNAs regulate synaptic plasticity underlying drug addiction. Genes Brain Behav. 2018;17(3):e12424. doi: 10.1111/gbb.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sartor GC, St Laurent G, Wahlestedt C. The emerging role of non-coding RNAs in drug addiction. Front Genet. 2012;3:106–106. doi: 10.3389/fgene.2012.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pashaei E, Guzel E, Ozgurses ME, Demirel G, Aydin N, Ozen M. A meta-analysis: identification of common Mir-145 target genes that have similar behavior in different GEO datasets. PLoS One. 2016;11(9):e0161491. doi: 10.1371/journal.pone.0161491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guzel E, Karatas OF, Semercioz A, Ekici S, Aykan S, Yentur S, et al. Identification of microRNAs differentially expressed in prostatic secretions of patients with prostate cancer. Int J Cancer. 2015;136(4):875–879. doi: 10.1002/ijc.29054. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Wei T, Zhao W, Ren Z, Wang Y, Zhou Y, et al. MicroRNA-181a is involved in methamphetamine addiction through the ERAD pathway. Front Mol Neurosci. 2021;14:667725–667725. doi: 10.3389/fnmol.2021.667725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eipper-Mains JE, Kiraly DD, Palakodeti D, Mains RE, Eipper BA, Graveley BR. microRNA-seq reveals cocaine-regulated expression of striatal microRNAs. RNA. 2011;17(8):1529–1543. doi: 10.1261/rna.2775511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Demirel G, Guzel E, Creighton CJ, Ozturk YE, Kucuk C, Asliyuksek H, et al. MDMA abuse in relation to MicroRNA variation in human brain ventral tegmental area and nucleus accumbens. Iran J Pharm Res. 2019;18(4):1989–1999. doi: 10.22037/ijpr.2019.15097.12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu W, Zhao M, Lin Z, Liu H, Ma H, Hong Q, et al. Increased expression of plasma hsa-miR-181a in male patients with heroin addiction use disorder. J Clin Lab Anal. 2020;34(11):e23486. doi: 10.1002/jcla.23486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Johnson EE, Chieng B, Napier I, Connor M. Decreased mu-opioid receptor signalling and a reduction in calcium current density in sensory neurons from chronically morphine-treated mice. Br J Pharmacol. 2006;148(7):947–955. doi: 10.1038/sj.bjp.0706820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He Y, Yang C, Kirkmire CM, Wang ZJ. Regulation of opioid tolerance by let-7 family microRNA targeting the mu opioid receptor. J Neurosci. 2010;30(30):10251–8. doi: 10.1523/JNEUROSCI.2419-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandrasekar V, Dreyer JL. MicroRNAs miR-124, let-7d and miR-181a regulate cocaine-induced plasticity. Mol Cell Neurosci. 2009;42(4):350–362. doi: 10.1016/j.mcn.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 22.Crews FT, Lawrimore CJ, Walter TJ, Coleman LG. The role of neuroimmune signaling in alcoholism. Neuropharmacology. 2017;122:56–73. doi: 10.1016/j.neuropharm.2017.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crews FT, Walter TJ, Coleman LG, Jr, Vetreno RP. Toll-like receptor signaling and stages of addiction. Psychopharmacology (Berl) 2017;234(9-10):1483–1498. doi: 10.1007/s00213-017-4560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coleman LG, Jr, Zou J, Crews FT. Microglial-derived miRNA let-7 and HMGB1 contribute to ethanol-induced neurotoxicity via TLR7. J Neuroinflammation. 2017;14(1):22–22. doi: 10.1186/s12974-017-0799-4. [DOI] [PMC free article] [PubMed] [Google Scholar]