Abstract
Tree diseases constitute a significant threat to biodiversity worldwide. Pathogen discovery in natural habitats is of vital importance to understanding current and future threats and prioritising efforts towards developing disease management strategies. Ash dieback is a fungal disease of major conservational concern that is infecting common ash trees, Fraxinus excelsior, in Europe. The disease is caused by a non-native fungal pathogen, Hymenoscyphus fraxineus. Other dieback causing-species have not previously been identified in the genus Hymenoscyphus. Here, we discover the pathogenicity potential of two newly identified related species of Asian origin, H. koreanus and H. occultus, and one Europe-native related species, H. albidus. We sequence the genomes of all three Hymenoscyphus species and compare them to that of H. fraxineus. Phylogenetic analysis of core eukaryotic genes identified H. albidus and H. koreanus as sister species, whilst H. occultus diverged prior to these and H. fraxineus. All four Hymenoscyphus genomes are of comparable size (55–62 Mbp) and GC contents (42–44%) and encode for polymorphic secretomes. Surprisingly, 1133 predicted secreted proteins are shared between the ash dieback pathogen H. fraxineus and the three related Hymenoscyphus endophytes. Amongst shared secreted proteins are cell death-inducing effector candidates, such as necrosis, and ethylene-inducing peptide 1-like proteins, Nep1-like proteins, that are upregulated during in planta growth of all Hymenoscyphus species. Indeed, pathogenicity tests showed that all four related Hymenoscyphus species develop pathogenic growth on European ash stems, with native H. albidus being the least virulent. Our results identify the threat Hymenoscypohus species pose to the survival of European ash trees, and highlight the importance of promoting pathogen surveillance in environmental landscapes. Identifying new pathogens and including them in the screening for durable immunity of common ash trees is key to the long-term survival of ash in Europe.
Supplementary Information
The online version contains supplementary material available at 10.1186/s43008-023-00115-8.
Keywords: Ash dieback, Emerging fungal pathogens, Pathogenicity, Hymenoscyphus fraxineus, Forest pathology, Non-crop fungal pathogens
INTRODUCTION
Fungal diseases have huge economic and environmental impacts worldwide. Fungal diseases infecting crops are of high research priority because they directly affect food security and the global economy. In contrast, fungal diseases of non-crop plants in natural habitats and urban areas are far less studied and, consequently, less well understood. Several plant diseases affecting environmental landscapes have caused huge permanent damage to biodiversity whilst their pathogenesis was still unknown (Fisher et al. 2012; Rafiqi 2018). One such disease is ash dieback.
The ash dieback pandemic was first observed in 2002 in Poland but has quickly spread to much of Europe, invading ash trees (Fraxinus excelsior) in natural and urban habitats (Drenkhan et al. 2017). The ash dieback pandemic is expected to have huge economic and environmental consequences. For example, in the UK alone, the disease is expected to cost around £15 billion (Hill et al. 2019). The substantial loss of F. excelsior is likely to negatively affect carbon sequestration and threaten around 1000 other species that are associated with ash, including birds, mammals, invertebrates, vascular plants, lichens, and fungi (Mitchell 2014). It is anticipated that ash dieback will change the European landscape forever.
The ash dieback disease, also called Chalara dieback of ash, is caused by the ascomycete pathogen Hymenoscyphus fraxineus (formerly known under asexual, Chalara fraxinea, and sexual, Hymenoscyphus pseudoalbidus), a commensal endophyte of Asian origin that causes no or minor symptoms when colonising leaves of Asian ash host species (Zheng and Zhuang 2013; Zhao et al. 2013a; Drenkhan et al. 2017), yet is highly pathogenic on European ash. The genome of H. fraxineus is about 62 Mbp and encodes a large and diversified set of host-interacting genes (McMullan et al. 2018). The European H. fraxineus population was founded by two divergent haploid individuals and has a bottlenecked genetic diversity when compared to the native Asian population of the endophyte (McMullan et al. 2018; Gross et al. 2014). Subsequent introduction of new individuals is expected to increase the pathogen’s adaptive potential in Europe, which in turn would further jeopardise the survival of European ash trees. What triggers H. fraxineus to switch from a commensal endophyte on Asian ash to a virulent pathogen on European ash is, as yet, unknown.
It is widely accepted that all plants, including lichens, algae, and nonvascular plants, harbour microorganisms, called endophytes, living within them (Bacon and White 2000). Typically, endophytes live with no apparent symptoms on plants. However, depending on environmental conditions and interactions with the host, some endophytes have visible effects on their host’s immunity and physiology. Therefore, endophytes can be pathogenic or nonpathogenic (Hardoim et al. 2015; Brader et al. 2017; Bacon and Hinton 1996). Research in this area is scarce, although analysis of the endospheric microbiome of many plant species is increasingly detecting, through amplicon sequencing, potential pathogens that did not cause noticeable symptoms in the host at the sampling time (Pereira et al. 2019; Manzotti et al. 2020). Whether the pathogenicity of these endophytes could be triggered and how is currently unresolved. Specific ecological drivers, competition for nutrients, as well as host factors and interactions with other endophytes could all underlie the suppression of disease symptoms despite pathogen occurrence. Pathogenic fungi infecting plants in agricultural or environmental landscapes are not clustered in a particular taxonomic category that separates them from harmless endophytes. Instead, some endophytic species possess pathogenic and nonpathogenic trophic states. For example, Sydowia polyspora is both a foliar commensal endophyte and a preemergent seed pathogen in Pinus ponderosa (Ridout and Newcombe 2017). Many fungal species include both pathogenic and non-pathogenic members. For example, Fusarium oxysporum, a ubiquitous soil-borne fungus, is classified in more than 150 forms or formae speciales of pathogenic and non-pathogenic strains, each one with the ability to infect one or a group of plant species (Nirmaladevi et al. 2016). Pathogenic F. oxysporum species often carry dispensable chromosomes that are not required for non-pathogenic growth but are virulence-associated and carry genes coding for specific host-interacting proteins and toxins, also referred to as effectors. Horizontal transfer of these chromosomes from pathogenic to a non-pathogenic recipient lineage of F. oxysporum renders the latter pathogenic on the respective host (van Dam et al. 2017). Such dispensable chromosomes have, however, not yet been identified in H. fraxineus (McMullan et al. 2018).
Forecasting fungal pathogen invasions in environmental landscapes is challenging and requires cross-disciplinary experimental approaches because of a myriad of variables and the complexity of interactions between the invading species and biological and physical characteristics of the recipient ecosystem (Santini et al. 2013; Alpert et al. 2000; Hayes and Barry 2008). The main factors include pathogen virulence and residence time, host susceptibility and abundance, as well as climatic factors (Hayes and Barry 2008).
Hymenoscyphus is a large genus of the Helotiaceae family and includes over 150 species (Kirk et al. 2008), with new species increasingly being discovered (Zheng and Zhuang 2013; Çetinkaya and Uzun 2020; Gross and Han 2015). Hymenoscyphus species were long classified as saprotrophic decomposers and non-pathogenic endophytes up until the emergence of H. fraxineus as a successful invasive pathogen of ash trees in Europe. Here, we interrogate if other species of the genus Hymenoscyphus could be pathogenic and cause disease on European ash. We address this question by studying three Hymenoscyphus species closely related to H. fraxineus: H. albidus, isolated from European ash growing in Wales (UK), H. koreanus, and H. occultus; two newly-discovered species of Asian origin and not (yet) introduced to Europe; they have been isolated by Gross and Han (2015) from petioles of Korean ash (F. chinensis subsp. rhynchophylla) (Gross and Han 2015; Kosawang et al. 2020).
The fungal secretome refers to the set of functionally diverse families of secreted proteins, many of which are involved in a range of diverse biological processes that directly affect the outcome of plant-fungi interactions. Examples include cell wall degrading enzymes, extracellular proteinases, toxins, and effector proteins that are upregulated during in planta growth of the pathogen and implicated in the suppression of host immune defences (Vincent et al. 2019). Whilst identifying and analysing predicted secretory proteins involved in disease development cannot help in accurately predicting pathogen invasions, it increasingly contributes to our understanding of pathogenicity and host responses. Here, we sequence and compare the genomes of three Hymenoscyphus species related to H. fraxineus. Since these fungal species can vary in their secretome, predominantly by gene gains and losses or by the rapid evolution of secreted proteins, we compare the full set of proteins predicted to be secreted by these four fungal species, analyse putative effectors that could be correlated with pathogenicity and, ultimately, we test the pathogenicity of these Hymecoscyphus species on European ash and discuss the threat they pose to the survival of European ash trees. The results show that in addition to the primary causative agent, H. fraxineus, the ash dieback disease can be caused by other member species in the Hymenoscyphus genus, which highlights the importance of promoting pathogen surveillance in environmental landscapes, identifying current and future threats to biodiversity, locally and at a global level.
MATERIAL AND METHODS
Fungal isolates, maintenance, and sequencing
Asian isolates: Hymenoscyphus koreanus isolate F52847-04 and H. occultus isolate F52847-22, originally known as KUS-F52847_04 and KUS-F52847_22, respectively, were collected in Korea from petioles of Korean ash and kindly provided by Dr. Andrin Gross (Swiss Federal Institute for Forest Snow and Landscape Research WSL, Birmensdorfm Switzerland). Fungal isolates were cultured in potato dextrose broth (VWR chemicals, USA) at room temperature and fungal mycelial mats were collected for DNA extraction 21 days after inoculation. Fungal DNA was extracted using E.Z.N.A HP plant DNA Mini kit (Omega Biotek, USA). The DNA was sent for library construction with TruSeq DNA PCR-free library prep kit with 350-bp insert (Illumina, USA) prior to sequencing with Illumina HiSeq 2500 platform on rapid run mode generating 2× 250 bp (Macrogen Inc, Korea).
European isolates: H. albidus was collected near Aberystwyth in Wales, UK, and kindly provided by Dr. Fiona Corke (IBERS, Plas Gogerddan, Aberystwyth University, United Kingdom) and Dr. Anne Edwards (John Innes Centre, Norwich Research Park, United Kingdom). Single spore isolates were grown in liquid culture, harvested, freeze-dried, and the genomic DNA extracted using the MagAttract HMW DNA Kit (QIAGEN, Germany). Library preparation and sequencing were done at the Earlham Institute, Norwich. Another isolate of H. albidus (ID 5/37/E5) kindly provided by Dr. Halvor Solheim, Norwegian Institute of Bioeconomy Research, Ås (NIBIO) was used for inoculation tests as described below.
Genome assembly and annotation
Paired-end raw reads were adapter and quality trimmed (Q ≤ 23) using Bbduk in the BBTools suite version 37.25 (https://jgi.doe.gov/data-and-tools/bbtools/). The draft genomes of the three Hymenoscyphus species were de novo assembled using SPAdes version 3.10.1(Bankevich et al. 2012) with the following parameters: k-mers 55, 77, 99 and 127, -- careful and –cov-cutoff auto. We used Blobtools version 1.1.1 (https://github.com/DRL/blobtools) and nucmer (Kurtz et al. 2004) to identify and dislodge scaffolds of bacterial contaminants, repetitive scaffolds and scaffolds < 1000 ≤ bp, respectively. We used the k-mer Analysis Toolkit (KAT) (Mapleson et al. 2017) and BUSCO version 3 (Simão et al. 2015) against the near-universal single-copy orthologues of Sodariomycetes odb9.
Annotation of the draft genomes of H. koreanus F52847-04 and H. occultus F52847-22 was achieved using the FunGAP pipeline version 1.0 (Min et al. 2017), while the annotation of the draft genome of H. albidus was carried out with the funannotate eukaryotic genome annotation pipeline version 1.1.1 (https://github.com/nextgenusfs/funannotate). Both FunGAP and funannotate incorporate genome masking using RepeatModeler/Repeatmasker, ab initio gene prediction using either Augustus and Maker for FunGAP or Augustus and GeneMark for funannotate and evidence-based evaluation of the predicted gene models. Functional annotation of the predicted coding sequences (CDSs) from the H. albidus, H. koreanus, and H. occultus genome assemblies depended on InterproScan version 5.25.64.0 (Jones et al. 2014) using PFAM database version 31.0 (Mistry et al. 2021) for H. koreanus and H. occultus and PFAM and CDD database version 3.16 (Mistry et al. 2021) for H. albidus. We used OrthoFinder v2.1.2 for protein clustering (Emms and Kelly 2019). The genome assemblies of H. koreanus F52847-04 and H. occultus F52847-22 and H. albidus were deposited at the Electronic Research Data Archive at the University of Copenhagen and can be accessed via https://sid.erda.dk/sharelink/C4VcS7pMHV.
Phylogenetic analysis
To generate a phylogenetic tree, core eukaryotic genes (CEGs) were identified using the Ascomycota lineage in BUSCO v5.2.1 (Simão et al. 2015). From five Hymenoscyphus species, 1560 CEG single-copy orthogroups were identified. The species investigated were H. fraxineus, H. albidus, H. koreanus, and H. occultus, with H. varicosporoides used as an outgroup. While occasionally found as root endophytes, H. varicosporoides primarily inhabits aquatic environments and is clearly placed outside our clade of interest. Orthologous genes were aligned using MAFFT v7.271 (Katoh et al. 2002). A maximum-likelihood tree was generated using RaxML v8.2.12 (Stamatakis 2014) with the PROTGAMMAGTR model and H. varicosporoides as the root. The tree was visualised using FigTree v1.4.3 (Rambaut 2012).
Secretome prediction
We used a previously described pipeline (Rafiqi et al. 2022) to predict fungal secretomes of H. albidus, H. occultus, and H. koreanus. The pipeline is similar to the one previously used to mine the secretome of H. fraxineus (McMullan et al. 2018), and uses SignalP 4. 1f (Nielsen 2017) to filter proteins that contain predicted signal peptides. The set of predicted secreted proteins was further used in the pipeline to predict transmembrane helices with TMHMM 2.0c (Krogh et al. 2001) and cellular localization signals with TargetP 1.1b (Emanuelsson et al. 2000). Protein sequences containing predicted transmembrane helices or mitochondrial targeting signal were removed from the list. The remaining set of proteins was annotated with Hmmer (Zhang et al. 2018) against PfamScan (Finn et al. 2014a, 2014b) for domain information, TargetP (Almagro Armenteros et al. 2019) for subcellular localisation, Predictnls (Cokol et al. 2000) for prediction of nuclear localization signals, T-Reks (Jorda and Kajava 2009) for detection of repeats, Disulfinder (database: uniprotkb/swiss-prot) for prediction of the disulfide bond, and MOTIF search for the search of known motifs. The positions of the motifs RxLR, [LI]xAR, [RK]CxxCx12}H, [YFW]xC, YxSL[RK], G[IFY][ALST]R, DELD, and [SG]PC[KR]P were identified with a script based on regular expressions.
Genetic diversity
To investigate genetic diversity within coding sequences, OrthoFinder v.2.2.6 (Emms and Kelly 2019) was used to identify orthogroups from four Hymenoscyphus species: H. fraxineus, H. albidus, H. koreanus, and H. occultus. Single-copy orthogroups were selected and orthologous genes from the four species were each combined into a single file. Genes were aligned using GUIDANCE2 in codons with the MAFFT algorithm (Sela et al. 2015). Due to erroneous sequence lengths, 37 genes were removed, leaving 7308 genes.
Synonymous (dS) and non-synonymous (dN) mutations were quantified using PAML v4.9 with the yn00 method on the single-copy orthogroups (Yang 2007). The dN/dS ratio was calculated. DnaSP (Rozas et al. 2003) was run on the orthologous genes to investigate single nucleotide variant density per gene, which was calculated by dividing the number of SNPs by the net number of sites. DnaSP failed to run on two genes, leaving 7306 genes.
Stem inoculation
An inoculation experiment was performed on 4/7/2018 to determine if inoculation with the fungi (H. koreanus F52847-04, H. occultus F52847-22, and H. albidus 5/37/E5) could cause the development of necrotic lesions on Fraxinus excelsior. Two-year-old open-pollinated offspring from clone #35 from the clonal seed orchard Tuse Næs (N55° 45 57.99 E11° 42 47.48) that had been kept under disease-free conditions were used for the experiment. The seedlings were inoculated with infected wood plugs. Before inoculation, sterile wood plugs (3 × 4 × 10 mm3 in size) had been placed on five week old cultures allowing the mycelium to colonize the wood plugs. An incision was cut into the stem of the plant and the infected wood plug was placed in the incision. Parafilm was wrapped around the stem. A culture of H. fraxineus strain 18.3 (Kosawang et al. 2020) was used as a positive control and sterile wood plugs as negative controls. Three biological replicates were performed for each inoculation type (i.e. the three species and the positive and negative controls). During the experiment, the plants were kept in a closed chamber at room temperature and 16 h of light. The experiment was terminated after 5 weeks (8/8/2018). Parafilm was removed and photos of the lesions were taken for documentation.
qRT-PCR of selected Hymenoschyphus effector candidates
Six, two-week-old mycelia plugs of the three Hymenoscyphus species and H. fraxineus strain 18.3 were grown on detached leaves of common ash (Fraxinus excelsior) clone # 27× rinsed with running tap water for 30 min. The plugs were placed face-down allowing the fungal mycelia to interact with the leaves. Three of the plugs were harvested after 3 and 6 days after inoculation (DAI) for RNA extraction. For the control treatment, the plugs were grown in malt extract broth and were collected accordingly after 3 and 6 DAI. Total RNA was isolated from the plugs using an E.Z.N.A Plant RNA kit (Omega Bio-tek, USA) with on-column DNase I digestion, and cDNA was prepared from 500 ng total RNA using a qScript cDNA synthesis kit (Quanta Biosciences, USA) following the manufacturer’s instruction. Three biological replicates of control and treatment were performed for each species of Hymenoscyphus.
Gene expression analysis was performed in two technical replicates for each biological replicate on a Mx3005P qPCR system (Stratagene, USA) and FIREPol EvaGreen qPCR Mix Plus (Solis Biodyne, Estonia). Analysis of melting curves was carried out at the end of each run to detect nonspecific amplifications. Relative expression of the effector-encoding genes was calculated in relation to the expression of the beta-tubulin gene according to the 2−ΔΔCT method (Livak and Schmittgen 2001). Gene expression data were analysed statistically using student t-test with a confidence level of 95% implemented in Microsoft Excel version 16.30 (Microsoft Corp., USA).
RESULTS AND DISCUSSION
Whole genome sequencing of three endophytic fungal species related to H. fraxineus
Genome sequences of three Hymenoscyphus species, H. albidus (76× coverage), H. koreanus (114× coverage) and H. occultus (102× coverage), were generated and compared to the genome of related and previously published H. fraxineus (McMullan et al. 2018). We assessed the completeness of the three Hymenoscyphus genome assemblies, using KAT and BUSCO. The k-mer distribution of such assemblies is compatible with the distribution of a complete one, and the estimated genome size is close to the actual size of the genome assembly. Approximately 96% of single copy ortholog genes from the Sodariomycetes odb9 were completely assembled in all three genomes (Additional file 1: Table S1). Among a total of 3725 tested BUSCO genes, only 61, 73, and 78 single-copy orthologs were missing in the H. albidus, H. koreanus, and H. occultus genomes, respectively. These results suggest that draft genome sequences generated in this study were relatively complete in terms of both assembly completeness and gene content. Genome features of H. albidus, H. koreanus, and H. occultus are detailed in Table 1.
Table 1.
Genome assembly statistics for Hymenoscyphus albidus, H. koreanus F52847-04 and H. occultus F52847-22 isolates
| Characteristic | H. albidus | H. koreanus F52847-04 | H. occultus F52847-22 |
|---|---|---|---|
| Genome size (Mb) | 55.7 | 57.4 | 61.6 |
| No. of scaffolds | 2002 | 1517 | 728 |
| N50/L50 scaffold length (Kb) | 265/53.5 | 225/76.2 | 115/154.7 |
| Coverage | 76× | 114× | 102× |
| %GC | 44.3 | 44.1 | 42.1 |
| No. of CDSs | 13,657 | 15,042 | 14,686 |
| Average gene length | 1510 | 1540 | 1573 |
All Hymenoscyphus genomes assembled here are of similar size (55–62 Mbp) and GC content (42–44%) and are comparable to H. fraxineus. The total number of coding sequences (CDSs) varied among the three genomes: 13,657 in H. albidus, 15,042 in H. koreanus and 14,686 in H. occultus (Table 1). Of these CDSs, 7791 (57.0%) in H. albidus, 8313 (55.2%) in H. koreanus, and 8180 (55.6%) in H. occultus harboured recognised protein signatures in Pfam/CDD databases. When compared to other Hymenoscyphus species, the predicted number of CDSs in the three genome assemblies were slightly greater than that of H. fraxineus (11,097 CDSs) (McMullan et al. 2018), but were in the same range as that of H. varicosporoides PMI_453 (14,929 CDSs; https://mycocosm.jgi.doe.gov/Hymvar1/Hymvar1.home.html).
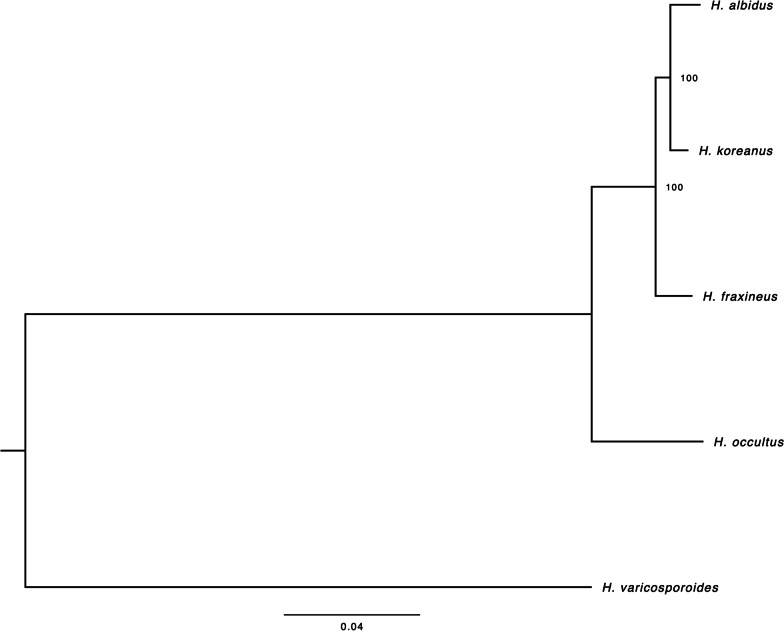
OrthoFinder clustering of proteins of all four Hymenoscyphus species (H. fraxineus and the three other related species addressed here) assigned 49,045 proteins (90% of total proteins) to 12,426 orthogroups (OGs), 8665 (69.7%) of which contained proteins from all four Hymenoscyphus species. Transporter, synthase, and cytochrome 450 coding proteins were among the largest OGs. We found a total of 71 proteins, representing 9 OGs specific to H. fraxineus (2 proteins, 2 OGs), H. albidus (4 proteins, 1 OG), and H. occultus (63 proteins, 6 OGs), respectively. No proteins specific to H. koreanus were found. The fact that only a small fraction, less than 1%, of predicted CDSs in all four Hymenoscyphus species are species-specific emphasises that these closely-related species likely share many common adaptations to similar environmental conditions. Phylogenetic analysis of core eukaryotic genes (CEGs) showed H. albidus and H. koreanus as sister species, whilst H. occultus was a more distant relative in the clade (Fig. 1). In previous studies, H. fraxineus and H. albidus were initially thought to be sister species, mostly due to their similar morphologies and high level of synteny (Elfstrand et al. 2021). In the present study we gain additional resolution with the inclusion of the newly discovered related species H. koreanus and H. occultus. Our expectation is, that with an increase in attention to this genus, additional species will be discovered and further our understanding of the biology of this group. Our finding supports the phylogenetic relatedness of Hymenoscyphus species previously described by (Gross and Han 2015).
Fig. 1.
The phylogenetic relationship between Hymenoscyphus species: H. fraxineus, H. koreanus (HK), H. occultus, H. albidus. H. varicosporoides was used as an outgroup. The tree was generated using 1560 orthologous core eukaryotic genes
Comparative analysis of the in silico secretomes of H. albidus, H. koreanus, H. occultus and H. fraxineus
Common gene ontologies
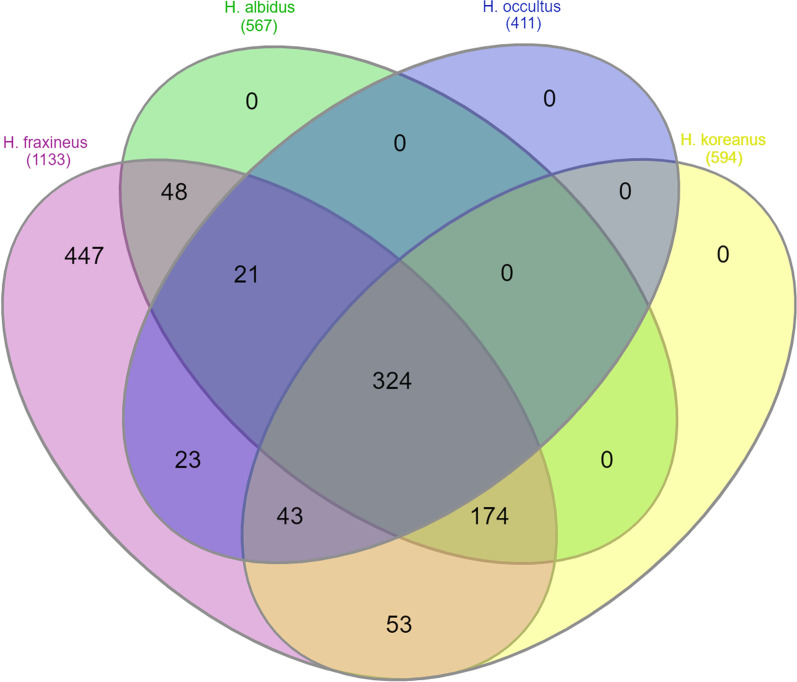
We mined the genomes of H. albidus, H. koreanus and H. occultus for predicted secreted proteins, using a pipeline described in materials and methods. All three fungi code for secretomes of similar sizes: 1446, 1579 and 1674 predicted secreted proteins, respectively (Additional file 2: Table S2). Secretomes of these three species were compared to that of related pathogenic H. fraxineus. Whilst the four predicted secretomes are highly heterogenous, strikingly a high number of proteins (1133), were shared between the ash dieback pathogen H. fraxineus and the three related Hymenoscyphus endophytes. 594 proteins were shared with H. koreanus, 567 proteins with H. albidus and 411 with H. occultus, with an overlap of 324 proteins shared amongst all four secretomes (Fig. 2). Genes in common between pathogenic species that are involved in plant interaction shed light on a foundation of pathogenicity whereas novel genes may play a role in niche adaptation or disease development. Regardless of their phylogenetic distance, the fact that all four Hymenoscyphus species share a significant number of predicted secreted proteins suggests possible common infection strategies among species, which is in agreement with the observed similarity between H. fraxineus and H. albidus in developing necrotrophic growth on ash leaves (Hietala et al. 2022).
Fig. 2.
Venn diagram of the number of proteins shared between the computed secretomes of the three Hymenoscyphus species: H. koreanus, H. occultus and H. albidus, and the computed secretome of the previously published genome of the ash dieback pathogen H. fraxineus
Polymorphism and positive selection
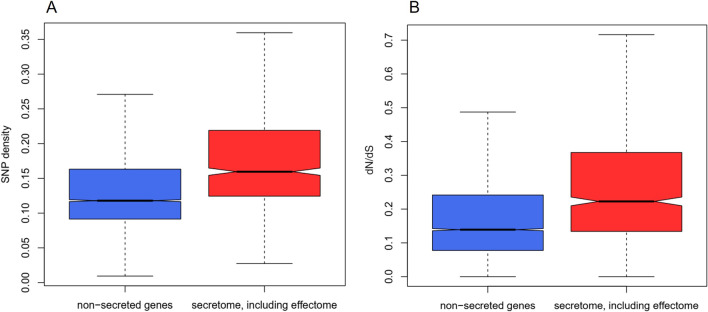
We analysed genetic diversity within coding sequences in all four Hymenoscyphus species. Genes coding for secreted proteins were associated with a SNP density that is significantly higher on average than that observed in other genes (Fig. 3A). In addition, SNPs observed in secretome genes were associated with an increase in the rate of non-synonymous nucleic acid changes to the rate of synonymous nucleic acid changes (dN/dS) (Fig. 3B). The polymorphism observed here in conjunction with high adaptive diversity in secretome genes among all four Hymenoscyphus species suggest genes coding for host-interacting proteins, including but not limited to effectors, undergo a faster rate of adaptive evolution than non-secreted genes and may play a key role in the infection process and pathogenesis on ash trees.
Fig. 3.
Polymorphism and positive selection in the secretomes of Hymenoscyphus species. A The proportion of SNPs is higher in secreted protein (red) compared to non-secreted protein-coding (blue) genes of Hymenoscyphus species. In secretome genes, the mean proportion of SNPs was 0.182 (standard deviation = 0.079; standard error mean = 0.004). In non-effector genes, the mean proportion of SNPs was 0.140 (standard deviation = 0.078; standard error mean = 0.001). B dN/dS ratio is higher in secretome genes (red) compared to non-secreted protein-coding genes (blue). In secretome genes, the mean dN/dS ratio was 0.288 (standard deviation = 0.210; standard error mean = 0.011). In non-secreted protein-coding genes, the mean dN/dS ratio was 0.189 (standard deviation = 0.166; S.E.M. = 0.002). Wilcox test gave a p value of 2.2 × 10−16 suggesting a significant difference
Large repertoires of CAZymes
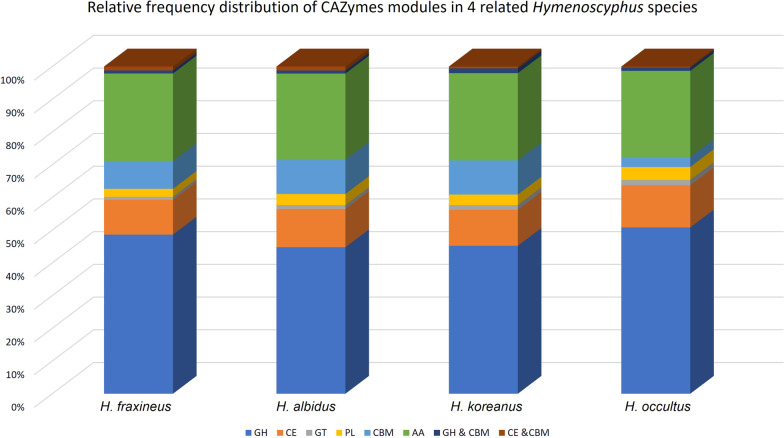
To identify carbohydrate-active enzyme (CAZyme) gene content, protein sequences from the three Hymenoscyphus species and pathogenic H. fraxineus were submitted for CAZymes prediction, using at least two integrated automated dbCAN2 tools (Zhang et al. 2018). Secretomes of all four species encode comparably high numbers (292–307) of CAZymes (Fig. 4). Glycoside hydrolases (GH) appear to be the most highly represented (around 40–50%) of all CAZymes modules, followed by auxiliary activities (AA), carbohydrate esterases (CE) and carbohydrate-binding modules (CBM), respectively. CAZymes are plant cell wall degrading enzymes (PCWDs) that play a critical role in disintegrating components of the host plant cell wall, which provides pathogens with a carbon source and access to other nutrients in the host apoplast. Plant pathogenic fungi tend to secrete more CAZymes than saprophytes and symbionts. Within plant pathogens, necrotrophic and hemibiotrophic fungi contain more CAZymes than biotrophic fungi (Zhao et al. 2013c). The high number of CAZymes in the predicted secretomes of Hymenoscyphus species could, therefore, be attributed to their trophic mode (Stenlid et al. 2017). Cytological details of how these fungi grow in planta are yet unavailable, although some evidence for the formation of biotrophic fungal structures was observed in ash leaf cells inoculated with the ash dieback pathogen, H. fraxineus, (Mansfield et al. 2018), suggesting a hemibiotrophic lifestyle, though details of how long the initial biotrophic phase takes before the pathogen switches into a necrotrophic growth phase has not yet been identified (Mansfield et al. 2019). Large sets of CAZymes are more frequent in necrotrophic pathogens (Zhao et al. 2013b). In agreement with this, a new classification of fungi based exclusively on genome-derived analysis of CAZymes has grouped H. fraxineus with polymertroph pathogens with a narrow host range, corresponding to necrotrophs (Hane et al. 2020). Indeed a recent comparative study by (Hietala et al. 2022) showed that both H. fraxineus and H. albidus develop similar necrotrophic growth on leaves of European ash. Detailed cytological studies are needed to understand the nature of the life style of Hymenoscyphus species on living ash trees and how CAZymes are involved.
Fig. 4.
Relative frequency distribution of CAZymes modules in the four related Hymenoschyphus species. GH glycoside hydrolases, AA auxiliary activities, CE carbohydrate esterases, CBM carbohydrate-binding modules
Shared Cell-death inducing effectors
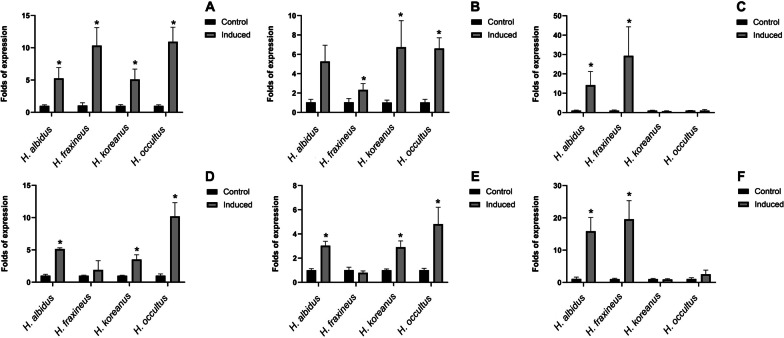
Amongst predicted secreted proteins shared between pathogenic H. fraxineus and the three additional Hymenoscyphus species studied here are plant cell death inducing proteins. These include proteins with predicted nuclease activity. Typically, each Hymenoscyphus species is predicted to secrete three to four S1/P1 nucleases, one Ribonuclease 2-5A and three close homologues of T2 RNAse family protein. Secreted nuclease effectors are increasingly identified in the secretomes of fungal pathogens (Kumakura et al. 2021; Rafiqi et al. 2021, 2022; Pennington et al. 2019b). They might either act intracellularly as a cytotoxin by scavenging host nucleic acids (Balabanova et al. 2012; Pennington et al. 2019a) or extracellularly to interact with the damage-associated molecular pattern extracellular RNA and DNA (Kumakura et al. 2021; Park et al. 2019; Widmer 2018). Other cell-death inducing effector candidates in the secretomes of this Hymenoscyphus group include homologues of necrosis-inducing toxins, called Nep1-like proteins (NLPs) (Fellbrich et al. 2002; Bailey 1995). NLPs cause necrosis in dicotyledonous plants and are suggested to function as virulence factors that accelerate disease development and pathogen growth in the host (Feng et al. 2014). Reverse transcription quantitative PCR (RT-qPCR) of a selected set of genes coding for cell death inducing effector candidates showed upregulation of these genes during in planta growth of all Hymenoscyphus species (Fig. 5). Predicted S1/P1 nuclease genes were induced 3dpi and 6dpi in the three Hymenoscyphus species as well as in pathogenic H. fraxineus (Fig. 5A). NLP homologues are 15- and 30-fold induced in both H. albidus and H. fraxineus, respectively (Fig. 5C, F), suggesting a potential involvement in virulence. It is particularly surprising to find that H. albidus, which has long been regarded as a nonpathogenic endophyte in European ash trees, is expressing PCD-inducing toxins. This is in line with a recent increasing number of studies showing that H. albidus and H. fraxineus, the ash dieback primary causal agent, develop similar necrotic growth on leaves of common ash trees. Although it causes shorter lesions in ash stems than those caused by H. fraxineus, H. albidus has been consistently shown to induce comparable lesions on leaves of European ash (F. excelsior), American green ash (F. pennsylvanica) as well as Manchurian ash (F. mandshurica) (Kowalski et al. 2015; Gross and Sieber 2016; Hietala et al. 2022). Both expressing PCD-inducing effector candidates and causing necrosis on the host leaf tissue suggest that H. albidus is a mild leaf pathogen of ash trees rather than a nonpathogenic endophyte, as it has long been considered. The fact that H. albidus has not been associated with the recent continental breakout of the ash dieback epidemic in Europe may be attributed to its relatively low abundance, perhaps due to its low fecundity, in addition to potential environmental selection pressures that may have acted disadvantageously on its spread and virulence within its native range (Drenkhan et al. 2016; Hietala et al. 2022) as compared with invasive H. fraxineus. Furthermore, European ash has coexisted with native H. albidus for a much longer time and may, therefore, have evolved to minimise the cost of infection by this pathogen, but was in fact previously unexposed and immunologically naïve to invasive H. fraxineus.
Fig. 5.
Real-time qPCR analysis of selected in planta-inducible effector candidate genes in H. fraxineus, H. koreanus, H. occultus and H. albidus during growth on detached F. excelsior leaves 3dpi (a–c) and 6dpi (d–f). a and d S1/P1 nuclease, b and e Ribonuclease T2 family, c and f NLP. Relative expression of the effector-encoding genes was calculated in relation to the expression of beta tubulin gene
Pathogenicity on European ash
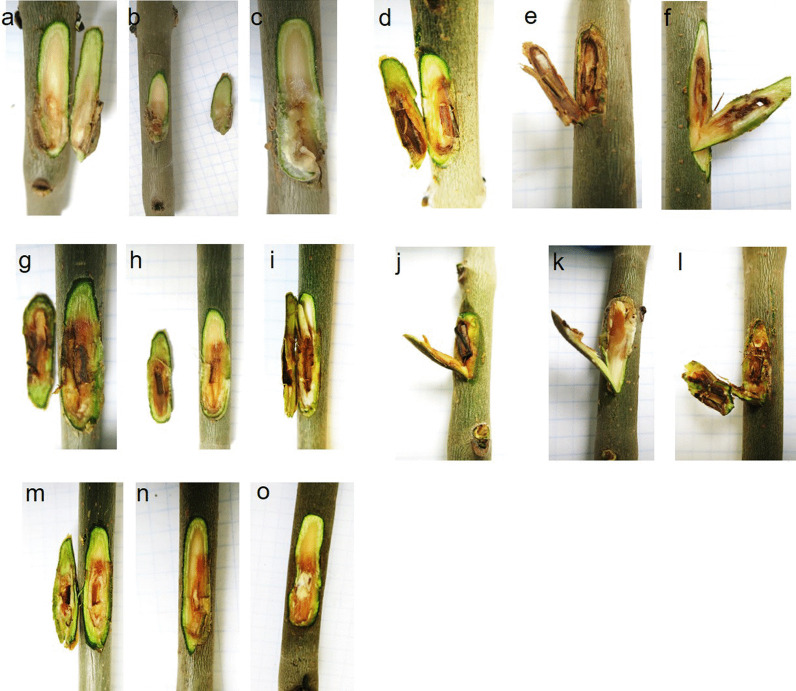
Even with the availability of large genome data of numerous groups of plant-interacting fungi, no study has, to date, addressed a clear correlation between genomic features or secretome composition of fungi and their pathogenicity potential. This is mainly due to the complexity of interactions between fungi and their host plants. As such, many fungal species include pathogenic as well as non-pathogenic members (Nirmaladevi et al. 2016), and others possess pathogenic and nonpathogenic states (Ridout and Newcombe 2017). Therefore, predicting the pathogenicity of fungal microorganisms is, as yet, not accurate. Nevertheless, we set out to investigate the pathogenicity of all three Hymenoscyphus species, studied here. Stems of European ash seedlings were inoculated with mycelia of H. koreanus, H. occultus and H. albidus. H. fraxineus was used as a positive control and sterile wood plugs as negative controls. These latter did not induce the formation of any necrotic lesions on ash stems (Fig. 6a–c). However, inoculations with each of the four Hymenoscyphus species resulted in necrotic lesions manifested by discoloration of the stem tissue to light or dark brown (Fig. 6d–o). In our pathogenicity test, lesions developed by H. fraxineus, H. koreanus and H. occultus were relatively deeper than those formed by H. albidus (Fig. 6m–o), suggesting a slight difference in virulence. Our results demonstrate that all four related Hymenoscyphus species develop pathogenic growth on European ash stems.
Fig. 6.
Pathogenicity test of Hymenoscyphus species on European ash, F. excelsior, stems. Necrosis developed on the stems inoculated with H. fraxineus (d–f), H, koreanus (g–i), H. occultus (j–l) and H. albidus (m–o). Sterile plugs were used as control (a–c)
CONCLUSIONS
In this study, we explored the pathogenicity of three fungal species closely related to the ash dieback pathogen, H. fraxineus. We sequenced the genomes of two new Hymenoscyphus species of Asian origin; H. koreanus and H. occultus as well as a native European isolate of H. albidus, which has long been considered as a harmless native decomposer and commensal on F. excelsior (Hietala and Solheim 2011). Recently isolated from ash tree collections, the two new Asian species have not previously been reported as pathogens (Gross and Han 2015). The genomes of all three sequenced species are of similar size and GC content and are comparable to the H. fraxineus genome. Phylogenetic analysis of core eukaryotic genes identified H. albidus and H. koreanus as sister species, whilst H. occultus was a more distant relative in the clade (Fig. 1), which is in agreement with previous findings (Gross and Han 2015). The past two decades have witnessed a surge in newly-discovered phytopathogenic species, which has impacted the interpretation of phylogenetic analyses performed previously. One such example is the fungal pathogen Phytophthora ramorum that causes sudden oak death, which was first isolated in 2000 (Werres et al. 2001); by 2015, over 50 other Phytophthora species have been discovered and added to the Tree of Life. Currently, a large part of the inventory of pathogenic forest fungi remains undescribed; they are more likely to be discovered when they start causing noticeable, sometimes irreparable, damage to trees and the environmental landscape as a whole.
To identify genes that play a role in Hymenoscyphus-Fraxinus interaction, we analysed the secretomes of H. albidus, H. koreanus, and H. occultus, and we compared them to the previously described secretome of H. fraxineus. Whilst most predicted secreted proteins are highly heterogenous in all four secretomes, it is noteworthy that 1133 proteins were identified as common between the ash dieback pathogen H. fraxineus and the three other related Hymenoscyphus fungi. One particular group of shared proteins, toxins and cell-death inducing proteins, not only expressed, but was also induced upon growth of the fungal species on detached leaves of European ash. These closely related species may, thus, share common niche adaptation, infection mechanisms or disease development strategies on European ash, raising the intriguing possibility that the three Hymenoscyphus species sequenced here may be pathogenic on European ash. Indeed, pathogenicity testing using artificial infection of European ash stems with mycelia of all four Hymenoscyphus species produced necrotic lesions under controlled conditions, confirming the virulence of these fungal species on European ash trees (Fig. 6). Lesions induced by all species were comparable, except those induced by H. albidus, which were slightly smaller in size and reduced in depth. Introduction of Asian H. koreanus and H. occultus populations into Europe is likely to jeopardise the survival of European ash, hence the need for heightened biosecurity awareness to guide conservation strategies. This study is the first to discover new potential ash dieback pathogens, raising awareness and promoting plant disease surveillance and investigations in environmental landscapes. As a consequence of human activity, trade, and climate change, plant pathogens are broadening their geographical ranges and increasingly appearing in new habitats (Rafiqi et al. 2018). Forests are a particularly vulnerable ecosystem. Thus, the increasing need for pathogen surveillance and forecast for better detection and response to future outbreaks.
Supplementary Information
Additional file 1. Table S1: List of 1560 orthologous core eukaryotic genes used for phylogenetic analysis.
Additional file 2. Table S2: In silico secretomes of H. albidus, H. koreanus and H. occultus.
Acknowledgements
We thank Dr Andrin Gross (The Swiss Federal Institute for Forest, Snow and Landscape Research, Switzerland), Dr Fiona Corke (Aberystwyth University, Aberystwyth, UK), Dr Anne Edwards, Professor Allan Downie (John Innes Centre, Norwich, UK) and Dr Halvor Solheim (Norwegian Institute of Bioeconomy Research, Ås (NIBIO)) for providing fungal material.
Author contributions
MR, MM and LRN planned and designed the research. MR, CK, JAP, JL, HY, MM and LRN performed experiments and analysed data. MR, CK, MM and LRN wrote the manuscript. MR and CK contributed equally in performing experiments. All authors read and approved the final manuscript.
Funding
Work at Royal Botanic Gardens Kew (RBG Kew; MR) was supported by the Pilot Study Fund 11231-105. Work at the University of Copenhagen (UC; LRN & CK) was supported by Godfred Birkedal Hartmanns Familiefond (2017) and the Danish Council for Independent Research (Grant No. 6111-00254). Work at the Earlham Institute (EI; MM, JAP & HY) was supported by Biotechnology and Biological Sciences Research Council (BBSRC) grants (BB/CCG1720/1) and (BBS/E/T/000PR9818) WP1 Signatures of Domestication and Adaptation. Sequencing at the EI is supported by BBSRC National Capability in Genomics and Single Cell Analysis (BBS/E/T/000PR9816) by members of the Genomics Pipelines and Core Bioinformatics Groups. HY & JAP were also supported by the BBSRC funded Norwich Research Park Biosciences Doctoral Training Partnership grants BB/M011216/1 & BB/T008717/1 as well as JAP Year in Industry (BBS/E/T/000PR9811).
Availability of data and materials
The genome assemblies and annotations of H. koreanus F52847-04, H. occultus F52847-22 and H. albidus were deposited at the Electronic Research Data Archive at the University of Copenhagen and can be accessed via https://sid.erda.dk/sharelink/DljEqJhxMG.
Declarations
Ethics approval and consent to participate
Not applicable.
Adherence to national and international regulations
The authors confirm adherence to Plant Health and Quarantine regulations of the biotic materials used in the study in accordance with Danish legislation.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Maryam Rafiqi, Email: m.rafiqi@kew.org.
Lene R. Nielsen, Email: lron@ign.ku.dk
References
- Almagro Armenteros JJ, Salvatore M, Emanuelsson O, Winther O, von Heijne G, Elofsson A, Nielsen H (2019) Detecting sequence signals in targeting peptides using deep learning. LID—10.26508/lsa.201900429 [doi] LID—e201900429 (2575-1077 (Electronic)) [DOI] [PMC free article] [PubMed]
- Alpert P, Bone E, Holzapfel C. Invasiveness, invasibility and the role of environmental stress in the spread of non-native plants. Perspect Plant Ecol Evol Syst. 2000;3(1):52–66. doi: 10.1078/1433-8319-00004. [DOI] [Google Scholar]
- Bacon CW, Hinton DM. Symptomless endophytic colonization of maize by Fusarium moniliforme. Can J Bot. 1996;74(8):1195–1202. doi: 10.1139/b96-144. [DOI] [Google Scholar]
- Bacon CW, White J. Microbial endophytes. CRC Press; 2000. [Google Scholar]
- Bailey BA. Purification of a protein from culture filtrates of Fusarium oxysporum that induces ethylene and necrosis in leaves of Erythroxylum coca. Phytopathology. 1995;85:1250–1255. doi: 10.1094/Phyto-85-1250. [DOI] [Google Scholar]
- Balabanova LA, Gafurov YM, Pivkin MV, Terentyeva NA, Likhatskaya GN, Rasskazov VA. An extracellular S1-type nuclease of marine fungus Penicillium melinii. Mar Biotechnol. 2012;14(1):87–95. doi: 10.1007/s10126-011-9392-5. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brader G, Compant S, Vescio K, Mitter B, Trognitz F, Ma L-J, Sessitsch A. Ecology and genomic insights into plant-pathogenic and plant-nonpathogenic endophytes. Annu Rev Phytopathol. 2017;55(1):61–83. doi: 10.1146/annurev-phyto-080516-035641. [DOI] [PubMed] [Google Scholar]
- Çetinkaya AS, Uzun Y. Hymenoscyphus caudatus, a new ascomycete record for the mycobiota of Turkey. Anatol J Bot. 2020;6:66. [Google Scholar]
- Cokol M, Nair R, Rost B. Finding nuclear localization signals. EMBO Rep. 2000;1(5):411–415. doi: 10.1093/embo-reports/kvd092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drenkhan R, Riit T, Adamson K, Hanso M. The earliest samples of Hymenoscyphus albidus vs. H. fraxineus in Estonian mycological herbaria. Mycol Prog. 2016;15(8):835–844. doi: 10.1007/s11557-016-1209-5. [DOI] [Google Scholar]
- Drenkhan R, Solheim H, Bogacheva A, Riit T, Adamson K, Drenkhan T, Maaten T, Hietala AM. Hymenoscyphus fraxineus is a leaf pathogen of local Fraxinus species in the Russian Far East. Plant Pathol. 2017;66(3):490–500. doi: 10.1111/ppa.12588. [DOI] [Google Scholar]
- Elfstrand M, Chen J, Cleary M, Halecker S, Ihrmark K, Karlsson M, Davydenko K, Stenlid J, Stadler M, Durling MB. Comparative analyses of the Hymenoscyphus fraxineus and Hymenoscyphus albidus genomes reveals potentially adaptive differences in secondary metabolite and transposable element repertoires. BMC Genomics. 2021;22(1):503. doi: 10.1186/s12864-021-07837-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence (0022-2836 (Print)) [DOI] [PubMed]
- Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20(1):238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellbrich G, Romanski A, Varet A, Blume B, Brunner F, Engelhardt S, Felix G, Kemmerling B, Krzymowska M, Nurnberger T. NPP1, a Phytophthora-associated trigger of plant defense in parsley and Arabidopsis. Plant J. 2002;32(3):375–390. doi: 10.1046/j.1365-313X.2002.01454.x. [DOI] [PubMed] [Google Scholar]
- Feng BZ, Zhu XP, Fu L, Lv RF, Storey D, Tooley P, Zhang XG. Characterization of necrosis-inducing NLP proteins in Phytophthora capsici. BMC Plant Biol. 2014;14:126. doi: 10.1186/1471-2229-14-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M (2014a) Pfam: the protein families database. Nucleic Acids Res 42(Database issue):D222–D230 [DOI] [PMC free article] [PubMed]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M (2014b) Pfam: the protein families database (1362-4962 (Electronic)) [DOI] [PMC free article] [PubMed]
- Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, Gurr SJ. Emerging fungal threats to animal, plant and ecosystem health. Nature. 2012;484(7393):186–194. doi: 10.1038/nature10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross A, Han JG. Hymenoscyphus fraxineus and two new Hymenoscyphus species identified in Korea. Mycol Prog. 2015;14(4):19. doi: 10.1007/s11557-015-1035-1. [DOI] [Google Scholar]
- Gross A, Sieber TN. Virulence of Hymenoscyphus albidus and native and introduced Hymenoscyphus fraxineus on Fraxinus excelsior and Fraxinus pennsylvanica. Plant Pathol. 2016;65(4):655–663. doi: 10.1111/ppa.12450. [DOI] [Google Scholar]
- Gross A, Hosoya T, Queloz V. Population structure of the invasive forest pathogen Hymenoscyphus pseudoalbidus. Mol Ecol. 2014;23(12):2943–2960. doi: 10.1111/mec.12792. [DOI] [PubMed] [Google Scholar]
- Hane JK, Paxman J, Jones DAB, Oliver RP, de Wit P. “CATAStrophy”, a genome-informed trophic classification of filamentous plant pathogens—How many different types of filamentous plant pathogens are there? Front Microbiol. 2020;10:66. doi: 10.3389/fmicb.2019.03088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardoim PR, van Overbeek LS, Berg G, Pirttilä AM, Compant S, Campisano A, Döring M, Sessitsch A. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev. 2015;79(3):293–320. doi: 10.1128/MMBR.00050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes KR, Barry SC. Are there any consistent predictors of invasion success? Biol Invas. 2008;10(4):483–506. doi: 10.1007/s10530-007-9146-5. [DOI] [Google Scholar]
- Hietala AM, Solheim H. Hymenoscyphus species associated with European ash1. EPPO Bull. 2011;41(1):3–6. doi: 10.1111/j.1365-2338.2010.02426.x. [DOI] [Google Scholar]
- Hietala AM, Agan A, Nagy NE, Børja I, Timmermann V, Drenkhan R, Solheim H. The native Hymenoscyphus albidus and the invasive Hymenoscyphus fraxineus are similar in their necrotrophic growth phase in ash leaves. Front Microbiol. 2022;13:66. doi: 10.3389/fmicb.2022.892051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill L, Jones G, Atkinson N, Hector A, Hemery G, Brown N. The £15 billion cost of ash dieback in Britain. Curr Biol. 2019;29(9):R315–R316. doi: 10.1016/j.cub.2019.03.033. [DOI] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorda J, Kajava AV (2009) T-REKS: identification of Tandem REpeats in sequences with a K-meanS based algorithm (1367-4811 (Electronic)) [DOI] [PubMed]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk PM, Cannon PF, David JC, Stalpers JA (2008) Ainsworth and Bisby’s Dictionary of the fungi. International Mycological Institute, Wallingford
- Kosawang C, McKinney LV, Nielsen LR, Kjær ED. Variation in aggressiveness of Hymenoscyphus fraxineus genotypes amid the ash dieback epidemic. Plant Pathol. 2020;69(4):677–684. doi: 10.1111/ppa.13158. [DOI] [Google Scholar]
- Kowalski T, Bilanski P, Holdenrieder O (2015) Virulence of Hymenoscyphus albidus and H. fraxineus on Fraxinus excelsior and F. pennsylvanica. PLoS ONE 10(10):e0141592 [DOI] [PMC free article] [PubMed]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes (0022-2836 (Print)) [DOI] [PubMed]
- Kumakura N, Singkaravanit-Ogawa S, Gan P, Tsushima A, Ishihama N, Watanabe S, Seo M, Iwasaki S, Narusaka M, Narusaka Y, Takano Y, Shirasu K. Guanosine-specific single-stranded ribonuclease effectors of a phytopathogenic fungus potentiate host immune response. bioRxiv. 2021;6:66. doi: 10.1111/nph.19582. [DOI] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome Biol. 2004;5(2):R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield JW, Galambos N, Saville R. The use of ascospores of the dieback fungus Hymenoscyphus fraxineus for infection assays reveals a significant period of biotrophic interaction in penetrated ash cells. Plant Pathol. 2018;67(6):1354–1361. doi: 10.1111/ppa.12844. [DOI] [Google Scholar]
- Mansfield J, Brown I, Papp-Rupar M. Life at the edge—the cytology and physiology of the biotroph to necrotroph transition in Hymenoscyphus fraxineus during lesion formation in ash. Plant Pathol. 2019;68(5):908–920. doi: 10.1111/ppa.13014. [DOI] [Google Scholar]
- Manzotti A, Bergna A, Burow M, Jørgensen HJL, Cernava T, Berg G, Collinge DB, Jensen B. Insights into the community structure and lifestyle of the fungal root endophytes of tomato by combining amplicon sequencing and isolation approaches with phytohormone profiling. FEMS Microbiol Ecol. 2020;96(5):66. doi: 10.1093/femsec/fiaa052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapleson D, Garcia Accinelli G, Kettleborough G, Wright J, Clavijo BJ. KAT: a K-mer analysis toolkit to quality control NGS datasets and genome assemblies. Bioinformatics. 2017;33(4):574–576. doi: 10.1093/bioinformatics/btw663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullan M, Rafiqi M, Kaithakottil G, Clavijo BJ, Bilham L, Orton E, Percival-Alwyn L, Ward BJ, Edwards A, Saunders DGO, Garcia Accinelli G, Wright J, Verweij W, Koutsovoulos G, Yoshida K, Hosoya T, Williamson L, Jennings P, Ioos R, Husson C, Hietala AM, Vivian-Smith A, Solheim H, MaClean D, Fosker C, Hall N, Brown JKM, Swarbreck D, Blaxter M, Downie JA, Clark MD. The ash dieback invasion of Europe was founded by two genetically divergent individuals. Nat Ecol Evol. 2018;2(6):1000–1008. doi: 10.1038/s41559-018-0548-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min B, Grigoriev IV, Choi IG. FunGAP: Fungal Genome Annotation Pipeline using evidence-based gene model evaluation. Bioinformatics. 2017;33(18):2936–2937. doi: 10.1093/bioinformatics/btx353. [DOI] [PubMed] [Google Scholar]
- Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, Finn RD, Bateman A. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021;49(D1):D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell R (2014) The potential ecological impact of ash dieback in the UK
- Nielsen H (2017) Predicting secretory proteins with SignalP (1940-6029 (Electronic)) [DOI] [PubMed]
- Nirmaladevi D, Venkataramana M, Srivastava RK, Uppalapati SR, Gupta VK, Yli-Mattila T, Clement Tsui KM, Srinivas C, Niranjana SR, Chandra NS. Molecular phylogeny, pathogenicity and toxigenicity of Fusarium oxysporum f. sp. Lycopersici. Sci Rep. 2016;6(1):21367. doi: 10.1038/srep21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H-J, Wang W, Curlango-Rivera G, Xiong Z, Lin Z, Huskey DA, Hawes MC, VanEtten HD, Turgeon BG (2019) A DNase from a fungal phytopathogen is a virulence factor likely deployed as counter defense against host-secreted extracellular DNA. mBio 10(2):e02805-18 [DOI] [PMC free article] [PubMed]
- Pennington HG, Jones R, Kwon S, Bonciani G, Thieron H, Chandler T, Luong P, Morgan SN, Przydacz M, Bozkurt T, Bowden S, Craze M, Wallington EJ, Garnett J, Kwaaitaal M, Panstruga R, Cota E, Spanu PD. The fungal ribonuclease-like effector protein CSEP0064/BEC1054 represses plant immunity and interferes with degradation of host ribosomal RNA. PLoS Pathog. 2019;15(3):e1007620. doi: 10.1371/journal.ppat.1007620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington HG, Jones R, Kwon S, Bonciani G, Thieron H, Chandler T, Luong P, Morgan SN, Przydacz M, Bozkurt T, Bowden S, Craze M, Wallington EJ, Garnett J, Kwaaitaal M, Panstruga R, Cota E, Spanu PD. The fungal ribonuclease-like effector protein CSEP0064/BEC1054 represses plant immunity and interferes with degradation of host ribosomal RNA. PLoS Pathog. 2019;15(3):e1007620–e1007620. doi: 10.1371/journal.ppat.1007620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira E, Vázquez de Aldana BR, San Emeterio L, Zabalgogeazcoa I. A survey of culturable fungal endophytes from Festuca rubra subsp pruinosa, a grass from marine cliffs, reveals a core microbiome. Front Microbiol. 2019;9:3321. doi: 10.3389/fmicb.2018.03321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiqi M (2018) Plant Killers-State of the World's Fungi 2018, pp 56–61
- Rafiqi M, Saunders D, McMullan M, Oliver R, Bone R, Fones H, Gurr SA, Vincent D, Coker TAB, Richard A (2018) Plant Killers: Royal Botanic Gardens, Kew
- Rafiqi M, Jelonek L, Diouf AM, Mbaye A, Rep M, Diarra A. Profile of the in silico secretome of the palm dieback pathogen, a fungus that puts natural oases at risk. bioRxiv. 2021;6:66. doi: 10.1371/journal.pone.0260830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiqi M, Jelonek L, Diouf AM, Mbaye A, Rep M, Diarra A (2022) Profile of the in silico secretome of the palm dieback pathogen, Fusarium oxysporum f. sp. albedinis, a fungus that puts natural oases at risk. PLoS ONE 17(5):e0260830 [DOI] [PMC free article] [PubMed]
- Rambaut A (2012) Figtree 1.4.3. http://tree.bio.ed.ac.uk/software/figtree/
- Ridout M, Newcombe G. Sydowia polyspora is both a Foliar Endophyte and a preemergent seed pathogen in Pinus ponderosa. Plant Dis. 2017;102(3):640–644. doi: 10.1094/PDIS-07-17-1074-RE. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19(18):2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Santini A, Ghelardini L, De Pace C, Desprez-Loustau ML, Capretti P, Chandelier A, Cech T, Chira D, Diamandis S, Gaitniekis T, Hantula J, Holdenrieder O, Jankovsky L, Jung T, Jurc D, Kirisits T, Kunca A, Lygis V, Malecka M, Marcais B, Schmitz S, Schumacher J, Solheim H, Solla A, Szabò I, Tsopelas P, Vannini A, Vettraino AM, Webber J, Woodward S, Stenlid J. Biogeographical patterns and determinants of invasion by forest pathogens in Europe. New Phytol. 2013;197(1):238–250. doi: 10.1111/j.1469-8137.2012.04364.x. [DOI] [PubMed] [Google Scholar]
- Sela I, Ashkenazy H, Katoh K, Pupko T. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 2015;43(W1):W7–14. doi: 10.1093/nar/gkv318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenlid J, Elfstrand M, Cleary M, Ihrmark K, Karlsson M, Davydenko K, Durling MB. Genomes of Hymenoscyphus fraxineus and Hymenoscyphus albidus encode surprisingly large cell wall degrading potential, balancing saprotrophic and necrotrophic signatures. Baltic for. 2017;23(1):41–51. [Google Scholar]
- van Dam P, Fokkens L, Ayukawa Y, van der Gragt M, Ter Horst A, Brankovics B, Houterman PM, Arie T, Rep M. A mobile pathogenicity chromosome in Fusarium oxysporum for infection of multiple cucurbit species. Sci Rep. 2017;7(1):9042. doi: 10.1038/s41598-017-07995-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent D, Rafiqi M, Job D. The multiple facets of plant-fungal interactions revealed through plant and fungal secretomics. Front Plant Sci. 2019;10:1626. doi: 10.3389/fpls.2019.01626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werres S, Marwitz R, Man In't veld WA, De Cock AWAM, Bonants PJM, De Weerdt M, Themann K, Ilieva E, Baayen RP (2001) Phytophthora ramorum sp. nov., a new pathogen on Rhododendron and Viburnum. Mycol Res 105(10):1155–1165
- Widmer H (2018) Functional characterisation of a fungal endonuclease effector and regulated host cell death
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, Busk PK, Xu Y, Yin Y. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018;46(W1):W95–W101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y-J, Hosoya T, Baral H-O, Hosaka K, Kakishima M. Hymenoscyphus pseudoalbidus, the correct name for Lambertella albida reported from Japan. Mycotaxon. 2013;122(1):25–41. doi: 10.5248/122.25. [DOI] [Google Scholar]
- Zhao Z, Liu H, Wang C, Xu J-R. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics. 2013;14(1):274. doi: 10.1186/1471-2164-14-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Liu H, Wang C, Xu JR. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics. 2013;14:274. doi: 10.1186/1471-2164-14-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Zhuang W. Four new species of the genus Hymenoscyphus (fungi) based on morphology and molecular data. Sci China Life Sci. 2013;56(1):90–100. doi: 10.1007/s11427-012-4429-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Table S1: List of 1560 orthologous core eukaryotic genes used for phylogenetic analysis.
Additional file 2. Table S2: In silico secretomes of H. albidus, H. koreanus and H. occultus.
Data Availability Statement
The genome assemblies and annotations of H. koreanus F52847-04, H. occultus F52847-22 and H. albidus were deposited at the Electronic Research Data Archive at the University of Copenhagen and can be accessed via https://sid.erda.dk/sharelink/DljEqJhxMG.