Abstract
Focused ultrasound (FUS)-enabled liquid biopsy (sonobiopsy) is an emerging technique for the noninvasive and spatiotemporally controlled diagnosis of brain cancer by inducing blood-brain barrier (BBB) disruption to release brain tumor-specific biomarkers into the blood circulation. The feasibility, safety, and efficacy of sonobiopsy were demonstrated in both small and large animal models using magnetic resonance-guided FUS devices. However, the high cost and complex operation of magnetic resonance-guided FUS devices limit the future broad application of sonobiopsy in the clinic. In this study, a neuronavigation-guided sonobiopsy device is developed and its targeting accuracy is characterized in vitro, in vivo, and in silico. The sonobiopsy device integrated a commercially available neuronavigation system (BrainSight) with a nimble, lightweight FUS transducer. Its targeting accuracy was characterized in vitro in a water tank using a hydrophone. The performance of the device in BBB disruption was verified in vivo using a pig model, and the targeting accuracy was quantified by measuring the offset between the target and the actual locations of BBB opening. The feasibility of the FUS device in targeting glioblastoma (GBM) tumors was evaluated in silico using numerical simulation by the k-Wave toolbox in glioblastoma patients. It was found that the targeting accuracy of the neuronavigation-guided sonobiopsy device was 1.7 ± 0.8 mm as measured in the water tank. The neuronavigation-guided FUS device successfully induced BBB disruption in pigs with a targeting accuracy of 3.3 ± 1.4 mm. The targeting accuracy of the FUS transducer at the GBM tumor was 5.5 ± 4.9 mm. Age, sex, and incident locations were found to be not correlated with the targeting accuracy in glioblastoma patients. This study demonstrated that the developed neuronavigation-guided FUS device could target the brain with a high spatial targeting accuracy, paving the foundation for its application in the clinic.
Keywords: Focused ultrasound, glioblastoma, blood-brain barrier, neuronavigation, targeting accuracy
I. INTRODUCTION
Glioblastoma (GBM) is the most common primary brain tumor in adults, with a median survival of 14 months from the time of diagnosis and a 5-year survival rate of less than 5% [1]. Current diagnosis of GBM typically relies on magnetic resonance imaging (MRI) and computed tomography (CT) to identify suspicious tumor lesions, followed by surgical resection or stereotactic biopsy for histological confirmation and genetic characterization. Neuroimaging cannot provide the genetic diagnosis of GBM. Surgical biopsy carries a significant risk of complications due to bleeding and inflammation. Blood-based liquid biopsy is a rapid and inexpensive way of obtaining clinically relevant genetic information about the tumor without surgery [2], [3]. However, the blood-brain barrier (BBB) limits the release of brain tumor biomarkers into the blood circulation [2], [4]. In our previous work, we developed the "sonobiopsy" technique to advance GBM diagnosis. Sonobiopsy uses focused ultrasound (FUS) in combination with microbubbles to disrupt the BBB and enhance the release of tumor-derived biomarkers from the FUS-targeted brain location into the blood circulation [5]-[8]. Sonobiopsy has the potential to enrich the circulating biomarker level, provide spatially precise genetic information about the tumor, and enable temporally controlled blood collection to minimize clearance of the released biomarkers [7].
Our previous studies have demonstrated the feasibility, safety, and efficacy of sonobiopsy in both small and large animal models [5]-[8]. It was shown that sonobiopsy increased the concentration of circulating mRNA of enhanced green fluorescent protein (eGFP) (a model tumor-specific biomarker) by 1,500–4,800 fold in GBM mouse models developed by ortho topic implantation of eGFP-transfected GBM cells [8]. It was also reported that sonobiopsy led to significantly higher plasma abundance of brain-specific protein biomarkers in wild-type pigs [6]. Recently, we found that sonobiopsy significantly enriched circulating-tumor DNA (ctDNA) in the plasma of GBM mouse and pig models and increased the detection sensitivity of GBM-specific mutations without evidence of tissue damage [7]. These animal studies are supported by preliminary clinical evidence obtained from a retrospective clinical study by Meng et al. They found that FUS-induced BBB disruption increased the concentration of circulating biomarkers, which supported the feasibility of sonobiopsy in the clinic [9]. However, all previous studies performed sonobiopsy using MR-guided FUS devices [5]-[9]. The high cost and complex operation of magnetic resonance-guided, FUS devices limit the future broad application of sonobiopsy in the clinic as a diagnostic technique.
Neuronavigation-guided FUS devices were developed for drug delivery by BBB opening [10]. A neuronavigation-guided FUS system often uses a "virtual probe" to represent a FUS transducer, with the tip of the "virtual probe" representing the transducer's geometrical focus. Wei et al. were the first to report the use of a neuronavigation-guided FUS system to perform BBB opening in a swine model [10]. Later, their team developed a clinical neuronavigation-guided FUS system (NaviFUS®) and demonstrated its feasibility and safety for BBB opening in GBM patients [11]. The NaviFUS system coupled a 256-element phased array with a clinical available neuronavigation system (Stealth S7, Medtronic plc, Dublin, Ireland) [11]. Pouliopoulos et al. developed a clinical system by integrating a veterinary neuronavigation system (BrainSight, Rogue Research Inc., Quebec, Canada) with a single-element FUS transducer [12] and clinical trials using this system are currently ongoing. An articulating or robotic arm was required for positioning the transducer for these neuronavigation-guided FUS systems [11], [12]. It is worth to mention that ultrasound-guided transcranial FUS systems are also under development, although they have not been used in clinical studies yet [13]-[15].
FUS devices for sonobiopsy have different requirements compared with those for brain drug delivery. Large-aperture phased arrays are needed for therapeutic drug delivery because drug delivery requires both a high spatial precision and a large treatment volume to efficiently deliver drugs to cover the whole diseased brain region. FUS sonication for sonobiopsy does not involve the delivery of therapeutic drugs and does not need to cover the entire tumor. Instead, sonobiopsy can be used to target specific regions inside the tumor for spatially targeted biomarker release [7]. Therefore, a small-aperture FUS transducer that is nimble and easy to integrate with existing neuronavigation systems without the need for a robotic arm is desired for sonobiopsy.
The objective of this study was to develop a neuronavigation-guided sonobiopsy device and characterize its targeting accuracy. This device integrated a commercially available neuronavigation system with a nimble, lightweight FUS transducer. The capability of the same FUS device for sonobiopsy under the guidance of MRI was demonstrated in our previous publications, which showed enhanced release of brain-specific biomarkers (GFAP and Iba1) in wild-type pigs and tumor-specific biomarkers (EGFRvIII and TERT C228T) in a pig model of GBM [7]. This study focused on developing the neuronavigation-guided FUS device for sonobiopsy. We first established a procedure for measuring the target registration error (TRE) of the neuronavigation-guided FUS device by performing hydrophone measurements in a water tank. TRE was calculated as the the difference between the projected location of a tracked point in image space and its actual location in the physical space in reference to Chaplin et. al. [16]. This procedure allowed reliable estimation of the TRE of the system in the free field and enabled future cross-platform comparisons of the TRE of different neuronavigation-guided FUS systems. We then performed in vivo experiments in pigs to verify that the developed device can induce BBB disruption and measured the targeting accuracy based on contrast-enhanced MRI images of BBB opening. At last, to evaluate the potential of the developed system in GBM patients, we performed numerical simulations of transcranial ultrasound wave propagation based on patient MRI and CT images acquired from the clinic. The distance between the simulated FUS transducer's free-field focus and the intracranial focus was calculated to evaluate the targeting accuracy of the FUS transducer.
II. METHODS
A. Description of the sonobiopsy device
The neuronavigation-guided sonobiopsy device consisted of a FUS system and a neuronavigation system. The FUS system was described in our previous publications [6], [7]. In brief, the FUS system consisted of a 15-element annular-ring transducer (Imasonic, Voray sur l'Ognon, France) with a center frequency of 0.65 MHz, a focal length of 65 mm, an aperture of 65 mm, and a weight of 0.24 kg. It was driven by a multi-channel function generator coupled with a linear amplifier (Image Guided Therapy, Pessac, France). The neuronavigation system (BrainSight, Rogue Research Inc., Quebec, Canada) consisted of an optical tracking camera, two sets of rigid-body optical trackers, and a computer.
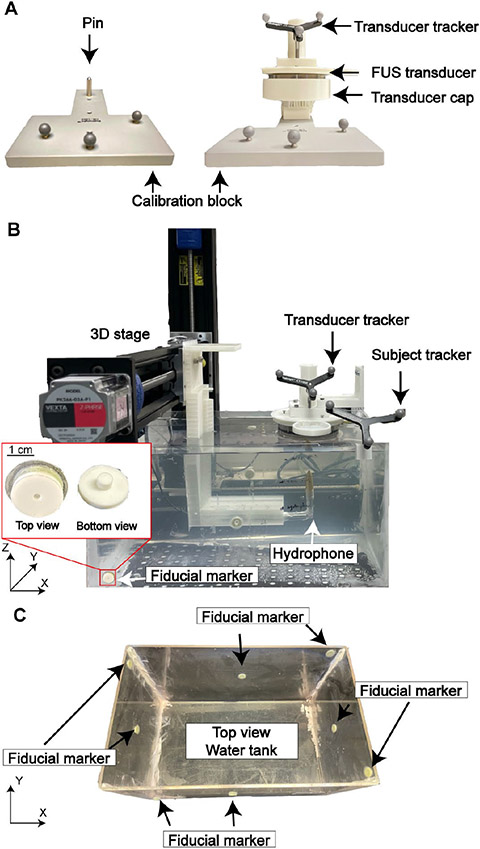
The key step in integrating the BrainSight system with the FUS transducer was to register the FUS geometrical focus location by the BrainSight software. BrainSight provided a calibration block with a pin on it (Fig. 1A). The tip location of the pin relative to the optical tracker on the calibration block was predefined in BrainSight. We 3D printed a transducer cap that tightly fits the pin. The cap was designed so that when it was positioned on the calibration block, the tip of the pin was co-axially aligned with the FUS transducer and was at a known distance from the geometrical focus of the FUS transducer as shown in Fig. 1A. During registration, the transducer with the cap was placed on the calibration block, and an optical tracker was attached to the FUS transducer. The BrainSight system then registered the tip of the pin with the optical tracker on the FUS transducer. Thereby, the location of the FUS geometrical focus was determined based on the position of the optical tracker. Once the FUS focus was registered, the BrainSight software presented a virtual probe to represent the FUS transducer with the probe's tip indicating its geometrical focus location. In this way, the FUS transducer's geometrical focus location was registered by the BrainSight system and used to guide the positioning of the FUS transducer for targeting specific locations.
Fig.1.
Experimental method for measuring the targeting accuracy of the neuronavigation-guided FUS in a water tank. (A) Setup used to calibrate the location of FUS transducer geometrical focus to the BrainSight system using a calibration block provided by BrainSight (left). The FUS transducer was inserted tightly into a 3D-printed cap (right). The cap was tightly inserted to the pin on the calibration block. (B) Experimental setup for measuring the targeting accuracy in a water tank. Position of the FUS transducer relative to the tank was tracked with BrainSight based on CT images of the water tank. The acoustic pressure fields generated by the FUS transducer was measured with a hydrophone mounted on a 3D motor stage. (C) Fiducial marker placement on the water tank.
B. Targeting accuracy quantification in a water tank
The TRE of the neuronavigation-guided sonobiopsy device was evaluated in vitro in a water tank (TREwater tank). Eight fiducial markers were sparsely placed around the water tank [17] (Fig. 1C). Two fiducials were placed on each side wall (total of 4) with one fiducial on the center of each side wall and the other on the corner. The chosen target location was relatively close to the water tank center, close towards the subject tracker. CT scans of the water tank with the fiducial markers were obtained at a resolution of 0.4 × 0.4 × 0.6 mm using a clinical CT scanner (Brilliant CT Big Bore, Philips, Amsterdam, Netherlands). The CT images were loaded to the BrainSight system to generate 3D reconstructed images of the tank with the fiducials.
Subject registration was then performed by co-registering the locations of each landmark on the tank in the physical space to the one in the image space of the BrainSight system by inserting the BrainSight pointer into the center of each fiducial marker. The subject tracker was placed on the water tank, facing the tracking camera (Fig. 1B). The fiducial markers were donut-shaped rings filled with soft multi-modality contrast agent with an opening of 4.5 mm in diameter, which was larger than the diameter of the BrainSight pointer tip (2.7 mm). This posed a challenge in precisely and accurately positioning the BrainSight pointer tip at the center of the fiducial marker, which is critical to minimizing the fiducial localization error (FLE), defined as the distance between the true position of a fiducial and its measured position [18]. To overcome this challenge, we designed and 3D-printed circular caps with a central opening of 2.7 mm to guide the pointer tip to the center of the fiducial markers (Fig. 1B). The transducer was tracked via the optical tracker placed on top of the transducer housing (Fig. 1B).
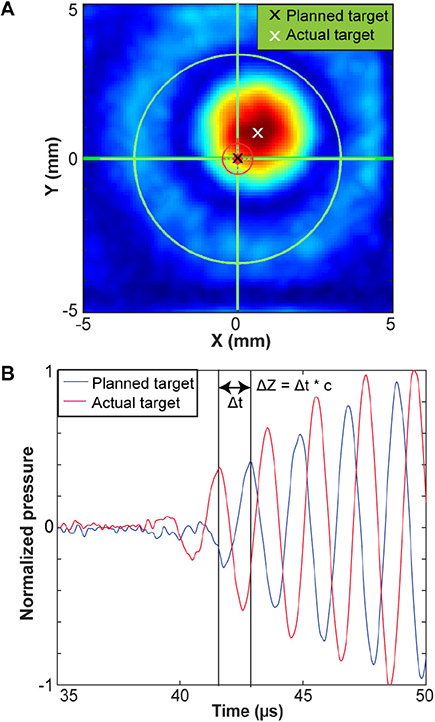
A total of three rounds of experiments were performed to characterize the TRE of the system (TREwater tank). Deionized, degassed pure water was used in all water tank experiments. Before each round of experiments, the hardware component of BrainSight system (computer and tracking camera) was turned off, and then turned on to reset the neuronavigation system. Within each round of scanning, total of four repeated scans were performed, each with an independent subject registration to evaluate the precision and repeatability of the targeting accuracy. The image space was first co-registered to the water tank by inserting optically tracked pointer into each fiducial marker on the water tank. The FUS transducer focus was registered to the BrainSight system. A hydrophone (HGL-0200, Onda Corp., Sunnyvale, CA, USA) was connected with a pre-amplifier (AG-20X0, Onda Corp., Sunnyvale, CA, USA) for measuring the acoustic pressures generated by the FUS transducer. The raw signals were acquired with a digital oscilloscope (Picoscope 5443D, St. Neots, United Kingdom) and saved in a computer. 3D volumetric scanning was automated using a 3D stage (BiSlide, Velmex Inc., Bloomfield, NY, USA) controlled by a customized MATLAB program. The MATLAB program controlled the motor to return the hydrophone to its initial position after each scan. An initial volumetric acoustic pressure scanning was performed to align the position of the hydrophone and transducer. This aligned location was then entered into the BrainSight system as the planned target location. Afterwards, the FUS transducer was removed from the water tank and the tank was registered to the BrainSight. The FUS transducer was then re-positioned back to the target location guided by BrainSight. Lastly, acoustic pressure maps centered at the BrainSight-guided target location were acquired to measure the TREwater tank. The acquired raw data were interpolated to a finer spatial resolution (0.1 mm) and then band-pass filtered to decrease noise. The targeting accuracy in the water tank, TREwater tank, along X and Y axes were calculated as the difference between the target focus (black cross in Fig. 2A) and the actual focus location (white cross in Fig. 2A) in the X–Y plane. The actual FUS focus location was determined by two methods: 1) identifying the centroid of the pressure maps; 2) identifying the location of the spatial-peak pressure. Room temperature was controlled at 23–24°C in the lab space. TREwater tank along the Z axis was calculated by calculating the difference in time of arrival between the hydrophone signal at the target and at the beam center measured in the initial round of calibration (Fig. 2B) using the sound of speed of 1496 m/s [19]. We used time-of-arrival because it has been widely used in various ultrasound localization systems for accurate location measurements [20], [21].
Fig.2.
Methods for measuring the targeting registration error (TRE) in the water tank. (A) TRE in the lateral X–Y plane was measured by the distance between the planned target (black) and the actual measured target, which was the beam centroid (white). (B) TRE in the Z direction was measured by the difference between the time-of-arrival of signals acquired at the planned target (blue) and reference signal acquired at the actual target (red).
C. Targeting accuracy quantification in vivo using a pig model
1). Experimental procedure
All animal procedures were reviewed and approved by the Institutional Animal Care and Use Committee at Washington University in St. Louis in accordance with the Guide for the Care and Use of Laboratory Animals and Animal Welfare Act (Washington University in St. Louis, 19-1062, 02/14/2020). A total of 5 piglets (age: ~ 4 weeks old; sex: male; body weight: 6.8–9.1 kg) were used in this study. These pigs were kept alive for 3 weeks. One or two separate FUS sonication sessions were performed on week 2 and/or week 3 for each pig. The pigs were sedated, intubated, and maintained under general anesthesia during the whole procedure. The blood oxygen level and pulse rate were monitored. The hair on the pig head was removed. A catheter was placed in the ear vein for microbubble and MRI contrast agent injections.
For treatment planning, the animal was first transported to the MRI scanner room. MRI images of the pig with fiducial markers placed on the head were obtained. Eight fiducial markers were sparsely placed on the pig head and jaw joints while avoid soft muscle and fat tissue to minimize movement of the fiducial markers when BrainSight pointer was inserted in each marker during subject registration. Additional fiducial markers were placed on the side bar and the water chamber as backup in case the fiducial markers on the side of the pig slipped off during study (which did not happen in all the studies). T1-weighted and T2-weighted MRI images of the pig brain were acquired using a 3 T Siemens scanner (MAGNETOM Prisma, Siemens, Munich, Germany). Images were then sent to the BrainSight computer. One targeted brain location was selected in the BrainSight software for each pig subject during each procedure.
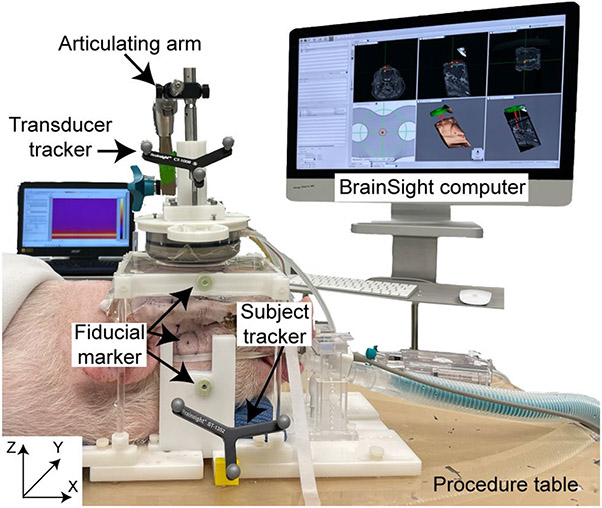
After the pre-procedure MRI images were acquired, pigs were transported to the procedural room and positioned on a custom-made plastic frame with two sidebars clamping the pig's head firmly to the frame (Fig.3). FUS parameters (frequency: 0.65 MHz, pressure 3 MPa (free field), pulse repetition frequency 1 Hz, duty cycles: 1%, pulse length: 10 ms, treatment duration 3 min) and Definity® microbubble solution (dose: 100 μL/kg) were the same as our previous pig studies [7].
Fig.3.
Experimental setup for in vivo pig study. Both the animal and transducer were tracked by BrainSight neuronavigation system with an individual optical tracker. The pig head was immobilized by a bite bar and two side-supports. The transducer was coupled to the pig head through a water chamber. The location of one fiducial marker on the side of the pig head was marked by a circle. Fiducial markers were also attached to the side support and water chamber as backups.
The optical tracking camera was positioned facing toward both the transducer and subject optical trackers. The BrainSight pointer was then inserted into the center of each fiducial marker using the 3D printed inserts to register the physical space with the image space in the BrainSight system. The transducer focus was co-registered to the transducer tracker using the calibration block as mentioned above. The transducer was moved to each target with a free-moving, multi-joint, articulating arm (Medtronic plc., Dublin, Ireland). Once the setup was complete, movement of the pig was avoided to minimize FLE [18], since locations of the fiducial markers on the pig surface may move from the locations when they were registered and contribute to a higher FLE.
2). Targeting accuracy measurements in vivo
After the FUS sonication finished, gadolinium contrast was injected at a 0.2 mL/kg dose. Then the pig was transported back to the MRI scanning room. T1-weighted 3D gradient-echo MRI scans (TR/TE: 8.16/3.76 ms; slice thickness: 1 mm; in-plane resolution: 1 mm ×1 mm; matrix size: 152 × 150; flip angle 10°) were performed to evaluate extravasation of the MRI contrast agent, gadolinium dimeglumine (Gd-BOPTA; MultiHance, Bracco Diagnostics Inc., Monroe Township, NJ, US), as an indication of FUS-induced BBB opening outcome. Locations of BBB opening were identified by comparing the treated with untreated brain hemispheres using a customized MATLAB script. Finally, the targeting accuracy in vivo in pigs (TApig) was measured by subtracting the coordinates of the localized BBB opening region with the BrainSight defined target locations. The coordinates for the tracking accuracy measurement were defined along Anterior/Posterior (X+/X−), Left/Right (Y+/Y−), and Superior/Inferior (Z+/Z−) directions.
D. Accuracy of the FUS transducer in targeting GBM tumors
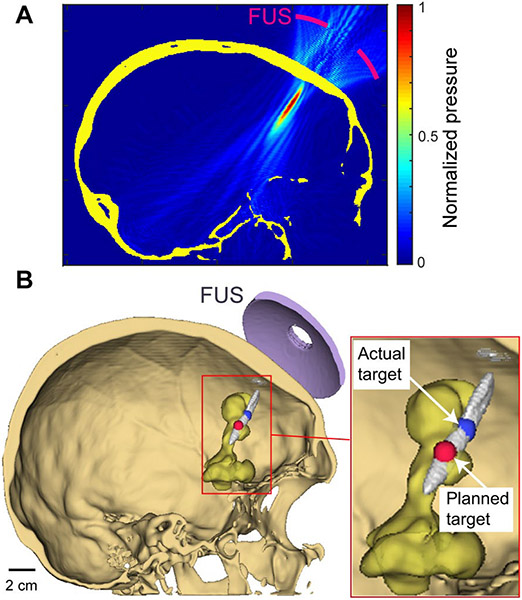
The study protocol was approved by the Institutional Review Board at the Washington University School of Medicine (Washington University in St. Louis, 201312136, 02/18/2019). All patient-identifying information was removed. Patient anonymity was preserved, and the principles of the Declaration of Helsinki were followed. All procedures followed the guidelines set forth by the Health Insurance Portability and Accountability Act Privacy Rule. This study selected 68 patients with newly diagnosed, supratentorial, histologic, or molecular GBM diagnosed between November 2014 and September 2019. Demographic parameters, including patients' age, gender, and tumor incident locations, were recorded. Fourteen patients were excluded from this study since their target locations were behind the ear and impractical to be targeted by FUS. For each patient, MR images were acquired using an MRI scanner (Philips 1.5 Tesla Ingenia MRI, Philips, Amsterdam, Netherlands) and CT images were acquired using a CT scanner (SOMATOM Confidence, Siemens, Munich, Germany). The incident locations of each patient were identified by the physicians. Figure 4B shows an example of the patient's CT image with the tumor segmented based on MRI. The MATLAB k-Wave acoustic modeling toolbox was used to simulate the intracranial acoustic pressure field distribution for each patient. k-Wave is a linear wave propagation simulator using a pseudo-spectral method to achieve both accurate and fast simulations [22], [23].
Fig.4.
Method for quantifying the targeting accuracy of the FUS transducer in GBM patients. (A) Representative acoustic pressure field simulation for a GBM patient using k-Wave at the actual focus sagittal plan. (B) 3-D visualization of the acoustic beam (>50% peak pressure) relative to the tumor (yellow). Actual target location and planned target location were identified and 3-D visualized in 3D Sheer [28]. TAGBM was calculated as the distance between the planned target and actual target location.
The FUS transducer (fc = 0.65 MHz, diameter = 65 mm, radius = 65 mm) [6] was modeled as a single-element transducer with the same frequency, aperture, and focal length as the transducer in in our experimental studies. The sound speed, density, and absorption coefficient at each pixel on the computation grid were calculated based on CT scans of the GBM patients. CT scans were performed using the clinical CT scanner with a peak voltage of 120 kV. Resolution of the CT images was not controlled in the retrospective study. Therefore, a linear interpolation was performed to achieve an isotropic pixel resolution of 0.44 mm for CT images across all patients. The density and sound speed of the non-skull region, including intra-cranial and outer-cranial space, were defined as 1000 kg/m3 and 1540 m/s. The density and sound speed profile for the skull was calculated using methods reported by Marsac et al. [24] as follows,
| (1) |
| (2) |
| (3) |
where is density, HU is Hounsfield Unit, is the speed of sound, abs is absorption, is absorption coefficient, is a power index for porosity.
The relationships described in equations (1) and (2) show that density and sound speed can be linearly mapped to the distribution of the Hounsfield Unit (HU) profile if the slope, is given. As shown by Webb et al., this slope can be different for different scan parameters, including effective tube energy and post-processing kernel size used in the scan [25]. To control the scan parameters, all CT images used in this study were acquired using the same scanner with consistent tube energy and post-processing kernel. The relationship between density and HU was measured to be using an established calibration process [26]. Then the maps of density and sound speed for each patient were translated from HU of their respective CT images using the results of the scanner calibrations. Absorption of the skull was calculated based on the absorption coefficient , where is absorption coefficient and f is the center frequency, multiplied with the porosity term in eqn. (3). We used , b = 1.1, β = 0.5 in reference to Constans et al. [27].
The size of the simulation grid was varied based on the size of the CT scan of each patient. Typically, the simulation was set up with 600 × 600 × 500 pixels. The simulations were performed in parallel on 4 GPUs, each with 32 GB of dedicated memory (Tesla V100-SMX2, NVIDIA, Santa Clara, CA). Each simulation took about one hour to finish with GPU acceleration. The timespan in the simulation was 150 μs, with a timestep of 18 ns. A steady state was achieved with the given simulated pulse duration and timestep.
After each simulation was completed, the simulation results were visualized first as raw pressure field (Fig. 4A) and then in 3D Slicer, an open-source 3D data visualization tool [28] (Fig.4B). The targeting accuracy and skull attenuation were then analyzed in MATLAB. In the MATLAB analysis script, the intracranial space was first separated from the extracranial space using active contouring in MATLAB. Then the full width half maximum (FWHM) volume was segmented, and the location of the local pressure maximum was identified within the FWHM as the actual target location. Lastly, targeting accuracy in silico (TAGBM) was calculated and recorded as the distance between the planned target and actual target (Fig. 4B). The distance was calculated along axial and lateral directions of the FUS transducer, similar to that reported by Jiang et al. and Deffieux et al [29], [30]. The attenuation was calculated as the ratio between trans-skull peak pressure and the free field peak pressure. The dependencies of targeting accuracy and attenuation to biological variables including age, gender, and tumor incident locations were evaluated. In our patient cohort, tumors were located in the temporal lobe (N = 22), frontal lobe (N = 13), parietal lobe (N = 13), occipital lobe (N = 6). Patients with a tumor in the parietal or occipital lobe were combined due to their limited patient numbers. Patients (N = 14) with a tumor location deeper than the focal depth of the FUS transducer and inaccessible to FUS transducer such as behind ears were excluded.
E. Statistical analysis
The measured data are presented as mean ± standard deviation. Statistical analyses were performed with GraphPad Prism (Version 8.3, GraphPad Software Inc., La Jolla, CA, USA). Comparisons among two groups were conducted using an unpaired t-test. Comparisons among more than two groups with a single variable (gender and age) were conducted using a one-way analysis of variance (ANOVA). Comparisons with multiple variables (tumor incident locations) were conducted using a two-way ANOVA. A P-value < 0.05 was used to determine statistical significance.
III. RESULTS
A. Target registration error quantification in the water tank
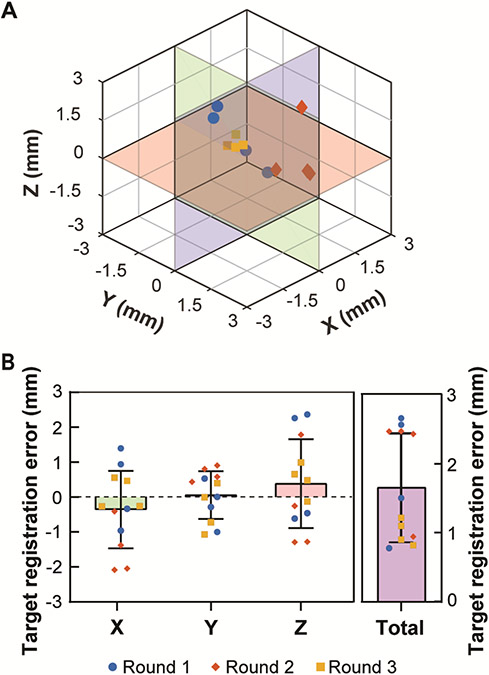
Figure 5A shows the actual target locations measured based on the centroid of the pressure maps relative to the planned target location ([0, 0, 0]) as measured in the water tank experiment. The TREwater tank was on average 1.7 ± 0.8 mm with [X, Y, Z] = [−0.4 ± 1.1, 0.1 ± 0.7, 0.4 ± 1.3] mm (Fig. 5B). The maximum errors were [X, Y, Z] = [−2.1, −1.1, 2.4] mm. The target accuracy in the water tank in Euclidean distance was 1.7 ± 0.6 mm. The targeting accuracy measured based on the spatial-peak pressure was [lateral, axial] = [−0.4 ± 1.2, 0.1 ± 0.7, 0.4 ± 1.3] mm, approximately the same as those measured based on the centroid of the pressure maps.
Fig.5.
Results of target registration error measured in the water tank. (A) 3D-visualization of actual focus locations relative to the planned target position (0, 0, 0). (B) Summary of the targeting accuracy in water tank along X, Y, Z directions and total Euclidean distances. Repeated measurements (N = 4) were performed during each of the 3 rounds of measurements.
B. Targeting accuracy in vivo in pigs
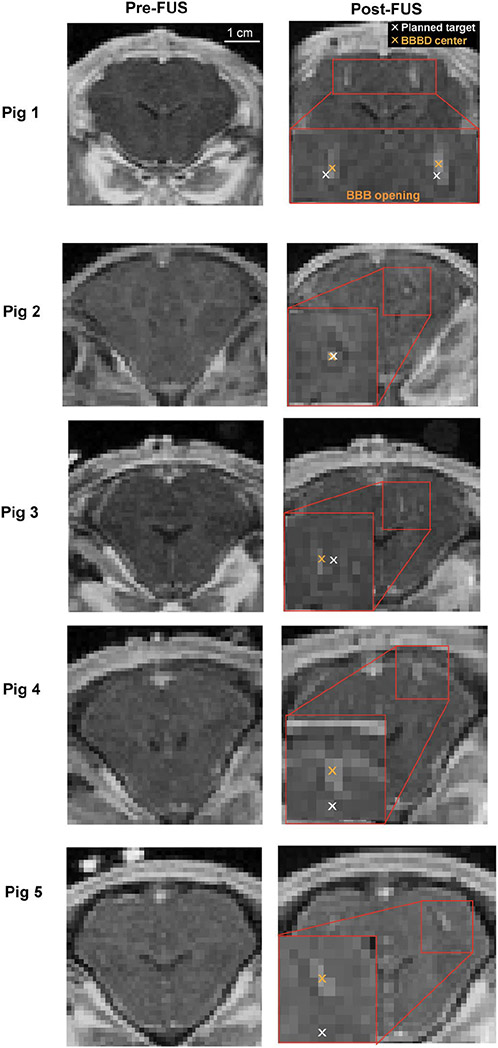
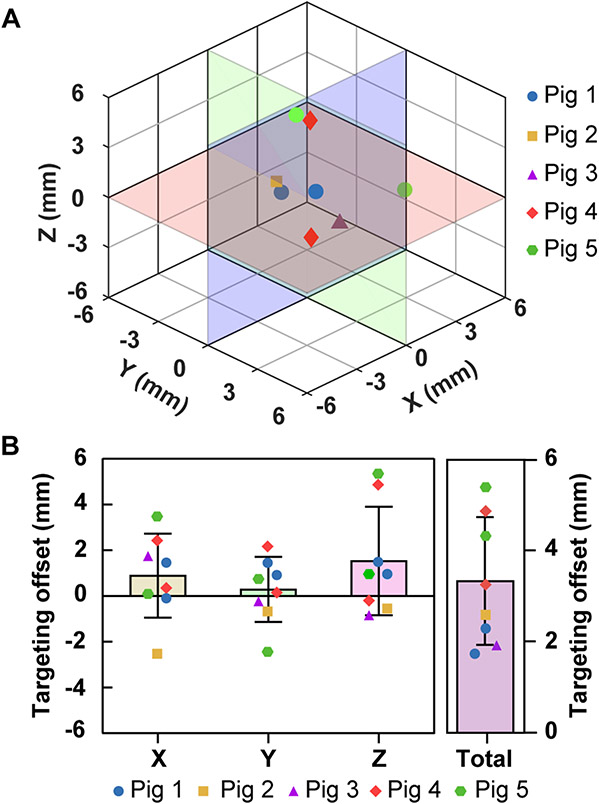
The T1-weighted contrast-enhanced MRI images post-FUS treatments show localized contrast enhancement, indicating successful BBB opening. Representative contrast-enhanced MRI images of the pigs’ pre-FUS and post-FUS are shown in Fig. 6. Figure 7A presents the 3D locations of the actual targets relative to the planned target in the pig experiment. The TApig were on average 3.3 ± 1.4 mm with [X, Y, Z] = [0.9 ± 1.8, 0.3 ± 1.4, 1.5 ± 2.4] mm (Fig. 7B). The maximum errors were [X, Y, Z] = [3.5, −2.4, 5.4] mm. The Euclidean distance was 3.3 ± 1.4 mm.
Fig.6.
In vivo targeting accuracy measured by contrast-enhanced MRI in pigs. Representative pre- and post-FUS T1W MRI images in the coronal view are presented for each pig. Targeting accuracy was quantified as the difference between the target (white cross) and the centroid of the BBB opening location (orange cross).
Fig.7.
Summary of targeting accuracy measured in vivo. (A) 3D visualization of the BBB opening centroid locations. (B) Targeting accuracy in pigs along X, Y, Z directions and total Euclidean distances.
C. Accuracy of the FUS transducer in targeting GBM tumors
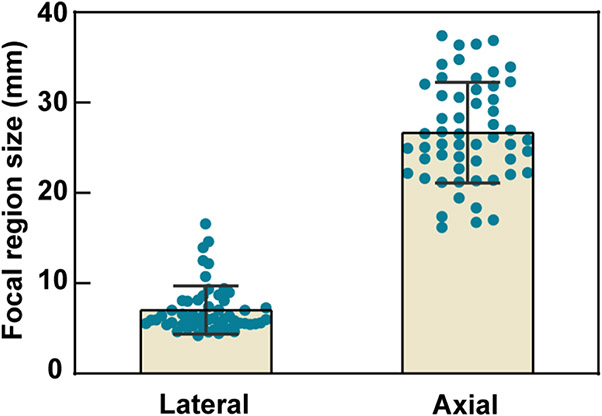
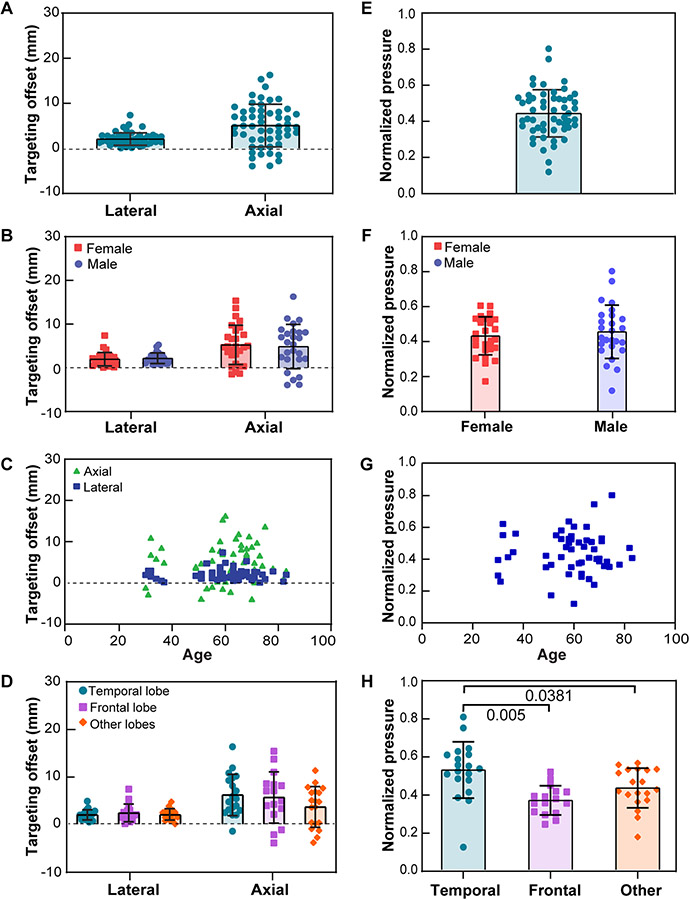
The averaged intracranial beam dimension is [lateral, axial] = [7.0 ± 2.7, 26.7 ± 5.6] mm as shown in Fig. 8. Numerical simulations revealed that the beam offsets caused by skull aberration were higher in the axial direction than in the lateral direction in Fig. 9A. The simulation showed the TAGBM between free field focus locations and intracranial focus locations were on average 6.4 ± 3.8 mm with [lateral, axial] = [2.1 ± 1.4, 5.1 ± 4.7] mm (N = 54). The TAGBM in male patients were on average [lateral, axial] = [2.2 ± 1.2, 4.9 ± 5.1] mm (N = 26). The TAGBM in female patients were on average [lateral, axial] = [2.0 ± 1.5, 5.3 ± 4.5] mm (N = 28). An unpaired t-test showed no significant differences between the TAGBM of male and female groups (Fig. 9B). Age was not found to be correlated with TAGBM (Fig. 9C). The TAGBM associated with different tumor incident locations were on average [lateral, axial] = [2.0 ± 1.1, 6.2 ± 4.4] mm (N = 19) in temporal lobe, [2.4 ± 1.9, 5.6 ± 5.4] mm (N = 16) in frontal lobe, [2.0 ± 1.1, 3.7 ± 4.3] mm (N = 19) in occipital and parietal lobes (Fig. 9D). A significantly lower skull attenuation was found when transmitting through temporal bone than through frontal bone (p = 0.0050, Fig. 9H) and other skull bone regions (p = 0.0381, Fig. 9H). No significant differences were found in the targeting accuracy when transmitting through different bone regions. The correlation analysis of the beam offset against age shows no significant trend between age and skull-induced beam offset.
Fig. 8.
Simulated transcranial FUS beam dimensions in GBM patients. The lateral and axial full-width half-maximum (FWHM) dimensions of the focal region are plotted for all 54 patients.
Fig.9.
FUS transducer targeting accuracy in GBM patients and intracranial peak acoustic pressure normalized by that simulated in free field. (A) TAGBM in lateral and axial directions. (B). TAGBM for male and female (C). TAGBM for different ages. (D). TAGBM for different tumor incident brain lobes. (E) Overall beam peak pressure in GBM patients normalized to peak pressure in free field simulation. (F). Summary of normalized pressure versus gender. (G). Summary of normalized pressure vs. age. (H) Normalized pressure in different tumor incident brain lobes. Only groups with statistically significant differences were labeled with P-values.
IV. Discussion
This study developed a neuronavigation-guided sonobiopsy device by integrating a commercial neuronavigation system with a FUS system. The targeting accuracy of the device was evaluated in vitro by hydrophone measurement in a water tank, in vivo using a pig model. The targeting accuracy of the FUS transducer in GBM patients was evaluated in silico using numerical simulations based on MRI and CT images of GBM patients.
The water tank measurements provided a straightforward approach for characterizing the targeting accuracy of the neuronavigation-guided FUS system. The targeting accuracy of previous systems was characterized either in vitro using MR thermometry in a phantom or in vivo by measuring the offset between the assigned and actual BBB opening location in swine and non-human primate models. Chaplin et al. measured the accuracy in guiding the FUS focus on a phantom by the offset between locations of the planned target and centroid of a hot spot in MRI thermometry [16]. Wei et al. measured the accuracy in guiding FUS-induced BBB opening in a swine model by the offset between the assigned and actual BBB opening location on the focal plane based on contrast-enhanced MRI images after FUS sonication [10]. Similarly, Wu et al. measured the targeting accuracy of their neuronavigation-guided system for BBB opening in non-human primates using contrast-enhanced MRI [31]. These approaches successfully quantified the targeting accuracies of their respective neuronavigation-guided FUS systems, but there is a lack of procedures for the straightforward measurement of the targeting accuracy that can be easily adopted for cross-platform comparison. The water tank experiment can be performed straightforwardly using a water tank and hydrophone and can be used to benchmark and compare the targeting accuracy of different neuronavigation-guided FUS systems. The TRE could be affected by many factors. One critical factor is the attachment pattern of fiducial markers. Steinmeier et al. showed that clustered placed fiducial markers led to 1.5 mm worse targeting error than the sparsely placed scenarios [17]. The number of fiducial markers was also found to be critical. Steinmeier et al. showed that eight fiducial markers improved targeting accuracy by ~1 mm versus four fiducial scenarios [17]. The targeting accuracy of our system as measured in the water tank was 1.7 ± 0.8 mm, which was lower than the in vitro thermometry-measured results (3.1–3.3 mm) using a made-in-house neuronavigation system [16], [17], that may be due, in part, to the higher number of fiducials we used (N = 8) versus Chaplin et al. (N = 6) [16]. To minimize the FLE, we designed 3D-printed caps for both the fiducial marker and the transducer to minimize the amount of ambiguity when registering the locations of each fiducial and the focus. We were able to suppress the error for each fiducial marker after registration transform to < 1 mm, which is slightly lower than the previously reported value (1.5 mm) for the BrainSight system [31]. The root mean square error (RMSE) of fiducial registration error for all the fiducial markers used in the tank test was 0.9 mm. It is worth noting that the 3D printer used to print the caps had a printing resolution of 0.2 mm, which could contribute to the FLE.
Our neuronavigation-guided FUS device achieved successful BBB opening with high spatial targeting accuracy. The in vivo targeting accuracy (Euclidean distance) of our study was 3.3 ± 1.4 mm, which was comparable to those reported by Wei et al. (2.3 ± 0.9 mm, swine model) and Wu et al. (3.1 mm on average, non-human primate) [10], [31]. It is worth to note that the in vivo targeting accuracy of current clinical MRI-guided transcranial FUS device [32]-[34] is better than existing neuronavigation-guided device. One key challenge in accurate targeting using a neuronavigation system for animal studies is ensuring minimal change in animal positioning after it has been co-registered. It was also important to place the fiducial markers on the bony structure. One of the key aspects of precise subject registration is concordance between locations of the fiducial markers during the MRI scan and subject registration during the neuronavigation-guided procedure. Placing fiducial markers on bony structures can reduce the potential movement of fiducial markers when inserting the BrainSight pointer into each fiducial marker during the subject registration and reduce the FLE. Error injected in this step may partially contribute to the higher targeting offset in neuronavigation-guided FUS systems compared with MRI-guided transcranial FUS studies [32]-[34].
The pig study demonstrated the feasibility of the neuronavigation-guided FUS system in BBB opening and showed high targeting accuracy. However, to further support the clinical application of our device for sonobiopsy, there is a need to evaluate the feasibility of performing FUS sonication in GBM patients using our FUS transducer. Although numerical simulations of transcranial ultrasound wave propagation using CT scans of small cohort of skulls specimens [29], [35] patients with medical conditions (intracranial clots, cancer) [12], [36] were reported in previous studies, our study was the first to perform the simulation in a large cohort of GBM patients. Our patient population covered a wide range of ages (30–83 years), both sexes, and various GBM incident locations (temporal, frontal, parietal, and occipital lobes).
Simulation results showed that the beam offset from the skull aberration effect in the lateral direction (focal plane) is much less than that of the axial direction ([lateral, axial] = [2.1 ± 1.4, 5.1 ± 4.7] mm at 0.65 MHz), and the beam was consistently shifted towards the transducer in the axial direction. This finding aligns with reports from Jiang et al. ([lateral, axial] = [1.5 mm, 9.5 mm] at 0.8 MHz), Deffieux et al. ([lateral, axial] = [1.8 mm, 8.3 mm] at 0.3 MHz; [1.10 mm, 13.00 mm] at 0.5 MHz and [3.8 mm, 28 mm] at 0.7 MHz), and Pouliopoulos et al. ([lateral, axial] = [0.1 mm, 6.1 mm] at 0.25 MHz) [12], [29], [30].
The beam offset was largely along the axial direction, which may be compensated through axial beam steering using our annular-ring FUS transducer. Age, sex, and tumor incident locations were not correlated with the targeting accuracy in GBM patients. Similarly, age and sex were not correlated with the simulated peak acoustic pressure. The simulation results indicated a significantly lower beam attenuation effect in the temporal bone than other skull bones because the temporal bone had the thinnest thickness [37]. The sonobiopsy device does not currently include aberration correction capabilities to target through thicker bones, but acoustic lenses could be added in the future to achieve aberration correction [38], [39].
This study laid the foundation for translating the neuronavigation-guided sonobiopsy device to the clinic. Neuronavigation guidance allows the sonobiopsy procedure to be performed in an outpatient facility outside the MRI and makes sonobiopsy a potentially accessible and affordable procedure. Findings from the numerical simulation suggest that this system can be broadly used in a wide range of GBM patients of different ages, sex, and tumor locations. The performance of the FUS device can be improved in the future by integrating with cavitation monitoring and feedback control to ensure consistent and safe FUS sonication [40]. Future studies are needed to evaluate the performance of this system in the clinic.
V. CONCLUSION
In this study, we developed a neuronavigation-guided sonobiopsy device and characterized its performance in vitro, in vivo, and in silico. This device integrated a nimble, lightweight FUS transducer with a neuronavigation system. We established a straightforward procedure for calibrating the targeting accuracy of the system in a water tank by performing hydrophone measurements. We also demonstrated that the developed system could successfully induce BBB opening in pigs in vivo with a high targeting accuracy. The feasibility of the FUS device in transcranial targeting of GBM tumors in patients was demonstrated by numerical simulation using the k-Wave toolbox. The beam offsets induced by the skull aberration effect were mostly in the axial direction, and the offset was independent of sex, age, and incident bone location. In summary, a neuronavigation-guided sonobiopsy device was developed and fully characterized for future clinical application.
Acknowledgment
The authors would like to thank Matthew Bledsoe for his assistance in the large animal studies. The authors also would like to thank Yi Huang for his help in the statistical analysis. The authors also want to thank Professor Jean-Francois Aubry for his guidance in selecting the human skull simulation model.
This work was supported in part by the the National Institutes of Health grants (R01EB027223, R01EB030102, R01MH116981, and UG3MH126861). It was also partially supported by the Office of Naval Research (grant number # N00014-19-1-2335).
Contributor Information
Lu Xu, Department of Biomedical Engineering, Washington University in St. Louis, St. Louis, MO 63105 USA..
Christopher P. Pacia, Department of Biomedical Engineering, Washington University in St. Louis, St. Louis, MO 63105 USA..
Yan Gong, Department of Biomedical Engineering, Washington University in St. Louis, St. Louis, MO 63105 USA..
Zhongtao Hu, Department of Biomedical Engineering, Washington University in St. Louis, St. Louis, MO 63105 USA..
Chih-Yen Chien, Department of Biomedical Engineering, Washington University in St. Louis, St. Louis, MO 63105 USA..
Leqi Yang, Department of Biomedical Engineering, Washington University in St. Louis, St. Louis, MO 63105 USA..
H. Michael Gach, Department of Radiation Oncology, Department of Radiology, Department of Biomedical Engineering, Washington University in St. Louis, MO 63110 USA..
Yao Hao, Department of Radiation Oncology, Washington University School of Medicine, St. Louis, MO 63110 USA..
Hassanzadeh Comron, Department of Radiation Oncology, Washington University School of Medicine, St. Louis, MO 63110 USA..
Jiayi Huang, Department of Radiation Oncology, Washington University School of Medicine, St. Louis, MO 63110 USA..
Eric C. Leuthardt, Department of Neurosurgery, Department of Biomedical Engineering, Department of Neuroscience, Center for Innovation in Neuroscience and Technology, Washington University in St. Louis, MO 63110 USA.
Hong Chen, Department of Biomedical Engineering and the Department of Radiation Oncology, Washington University in St. Louis, MO 63110 USA..
REFERENCES
- [1].Hanif F, Muzaffar K, Perveen K, Malhi SM, and Simjee SU, “Glioblastoma multiforme: A review of its epidemiology and pathogenesis through clinical presentation and treatment,” Asian Pacific Journal of Cancer Prevention, vol. 18, no. 1, pp. 3–9, 2017, doi: 10.22034/APJCP.2017.18.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Connolly ID, Li Y, Gephart MH, and Nagpal S, “The ‘Liquid Biopsy’: the Role of Circulating DNA and RNA in Central Nervous System Tumors,” Curr Neurol Neurosci Rep, vol. 16, no. 3, pp. 1–8, 2016, doi: 10.1007/s11910-016-0629-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Alix-Panabières C and Pantel K, “Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy,” Cancer Discov, vol. 6, no. 5, pp. 479–491, May 2016, doi: 10.1158/2159-8290.CD-15-1483. [DOI] [PubMed] [Google Scholar]
- [4].Bettegowda C et al. , “Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies,” Sci Transl Med, vol. 6, no. 224, pp. 1–12, Feb. 2014, doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhu L, Id AN, Pham C, Id P, Yue Y, and Id HC, “Focused ultrasound for safe and effective release of brain tumor biomarkers into the peripheral circulation,” PLoS One, vol. 15, no. 6, p. e0234182, 2020, doi: 10.1371/journal.pone.0234182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pacia CP et al. , “Feasibility and safety of focused ultrasound-enabled liquid biopsy in the brain of a porcine model,” Sci Rep, vol. 10, no. 1, p. 7449, Dec. 2020, doi: 10.1038/s41598-020-64440-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pacia CP et al. , “Sonobiopsy for minimally invasive, spatiotemporally-controlled, and sensitive detection of glioblastoma-derived circulating tumor DNA,” Theranostics, vol. 27, no. 1, pp. 362–378, 2022, doi: 10.7150/THNO.65597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhu L et al. , “Focused Ultrasound-enabled Brain Tumor Liquid Biopsy,” Sci Rep, vol. 8, no. 1, p. 6553, Dec. 2018, doi: 10.1038/s41598-018-24516-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Meng Y et al. , “MR-guided focused ultrasound liquid biopsy enriches circulating biomarkers in patients with brain tumors,” Neuro Oncol, vol. 23, no. 10, pp. 1789–1797, Oct. 2021, doi: 10.1093/neuonc/noab057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wei K-C et al. , “Neuronavigation-Guided Focused Ultrasound-Induced Blood-Brain Barrier Opening: A Preliminary Study in Swine,” American Journal of Neuroradiology, vol. 34, no. 1, pp. 115–120, Jan. 2013, doi: 10.3174/ajnr.A3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chen K-T et al. , “Neuronavigation-guided focused ultrasound for transcranial blood-brain barrier opening and immunostimulation in brain tumors,” Sci Adv, vol. 7, no. 6, Feb. 2021, doi: 10.1126/sciadv.abd0772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pouliopoulos AN, Wu SY, Burgess MT, Karakatsani ME, Kamimura HAS, and Konofagou EE, “A Clinical System for Non-invasive Blood–Brain Barrier Opening Using a Neuronavigation-Guided Single-Element Focused Ultrasound Transducer,” Ultrasound Med Biol, vol. 46, no. 1, pp. 73–89, 2020, doi: 10.1016/j.ultrasmedbio.2019.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Deng L, Hughes A, and Hynynen K, “A Noninvasive Ultrasound Resonance Method for Detecting Skull Induced Phase Shifts May Provide a Signal for Adaptive Focusing,” IEEE Trans Biomed Eng, vol. 67, no. 9, pp. 2628–2637, 2020, doi: 10.1109/TBME.2020.2967033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Adams C et al. , “Implementation of a Skull-Conformal Phased Array for Transcranial Focused Ultrasound Therapy,” IEEE Trans Biomed Eng, vol. 68, no. 11, pp. 3457–3468, 2021, doi: 10.1109/TBME.2021.3077802. [DOI] [PubMed] [Google Scholar]
- [15].Deng L, Yang SD, Oareilly MA, Jones RM, and Hynynen K, “An Ultrasound-Guided Hemispherical Phased Array for Microbubble-Mediated Ultrasound Therapy,” IEEE Trans Biomed Eng, vol. 69, no. 5, pp. 1776–1787, 2022, doi: 10.1109/TBME.2021.3132014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chaplin V et al. , “On the accuracy of optically tracked transducers for image-guided transcranial ultrasound,” Int J Comput Assist Radiol Surg, vol. 14, no. 8, pp. 1317–1327, 2019, doi: 10.1007/s11548-019-01988-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Steinmeier R, Rachinger J, Kaus M, Ganslandt O, Huk W, and Fahlbusch R, “Factors Influencing the Application Accuracy of Neuronavigation Systems,” Stereotact Funct Neurosurg, vol. 75, no. 4, pp. 188–202, 2000, doi: 10.1159/000048404. [DOI] [PubMed] [Google Scholar]
- [18].West JB and Maurer CR, “Designing optically tracked instruments for image-guided surgery,” IEEE Trans Med Imaging, vol. 23, no. 5, pp. 533–545, 2004, doi: 10.1109/TMI.2004.825614. [DOI] [PubMed] [Google Scholar]
- [19].del Grosso VA and Mader CW, “Speed of Sound in Pure Water,” J Acoust Soc Am, vol. 52, no. 5B, pp. 1442–1446, Nov. 1972, doi: 10.1121/1.1913258. [DOI] [Google Scholar]
- [20].Gillespie D and Macaulay J, “Time of arrival difference estimation for narrow band high frequency echolocation clicks,” J Acoust Soc Am, vol. 146, no. 4, pp. EL387–EL392, Oct. 2019, doi: 10.1121/1.5129678. [DOI] [PubMed] [Google Scholar]
- [21].Hu Z et al. , “3-D Transcranial Microbubble Cavitation Localization by Four Sensors,” IEEE Trans Ultrason Ferroelectr Freq Control, vol. 68, no. 11, pp. 3336–3346, 2021, doi: 10.1109/TUFFC.2021.3091950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Treeby BE and Cox BT, “k-Wave: MATLAB toolbox for the simulation and reconstruction of photoacoustic wave fields,” J Biomed Opt, vol. 15, no. 2, p. 021314, 2010, doi: 10.1117/1.3360308. [DOI] [PubMed] [Google Scholar]
- [23].Martin E, Jaros J, and Treeby BE, “Experimental validation of k-Wave: Nonlinear wave propagation in layered, absorbing fluid media,” IEEE Trans Ultrason Ferroelectr Freq Control, vol. 67, no. 1, pp. 81–91, 2020, doi: 10.1109/TUFFC.2019.2941795. [DOI] [PubMed] [Google Scholar]
- [24].Marsac L et al. , “Ex vivo optimisation of a heterogeneous speed of sound model of the human skull for non-invasive transcranial focused ultrasound at 1 MHz,” International Journal of Hyperthermia, vol. 33, no. 6, pp. 635–645, Aug. 2017, doi: 10.1080/02656736.2017.1295322. [DOI] [PubMed] [Google Scholar]
- [25].Webb TD et al. , “Measurements of the Relationship Between CT Hounsfield Units and Acoustic Velocity and How It Changes With Photon Energy and Reconstruction Method,” IEEE Trans Ultrason Ferroelectr Freq Control, vol. 65, no. 7, pp. 1111–1124, Jul. 2018, doi: 10.1109/TUFFC.2018.2827899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Szymanowski H, Oelfke U, and Rana M, The calibration of CT Hounsfield units for radiotherapy treatment planning,” Phys Med Biol, vol. c, pp. 110–124, 1996, doi: 10.1088/0031-9155/41/1/009. [DOI] [PubMed] [Google Scholar]
- [27].Constans C, Deffieux T, Pouget P, Tanter M, and Aubry JF, “A 200-1380-kHz Quadrifrequency Focused Ultrasound Transducer for Neurostimulation in Rodents and Primates: Transcranial in Vitro Calibration and Numerical Study of the Influence of Skull Cavity,” IEEE Trans Ultrason Ferroelectr Freq Control, vol. 64, no. 4, pp. 717–724, 2017, doi: 10.1109/TUFFC.2017.2651648. [DOI] [PubMed] [Google Scholar]
- [28].Fedorov A et al. , “3D Slicer as an image computing platform for the Quantitative Imaging Network,” Magn Reson Imaging, vol. 30, no. 9, pp. 1323–1341, Nov. 2012, doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Deffieux T and Konofagou E, “Numerical study of a simple transcranial focused ultrasound system applied to blood-brain barrier opening,” IEEE Trans Ultrason Ferroelectr Freq Control, vol. 57, no. 12, pp. 2637–2653, 2010, doi: 10.1109/TUFFC.2010.1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jiang C, Li D, Xu F, Li Y, Liu C, and Ta D, “Numerical Evaluation of the Influence of Skull Heterogeneity on Transcranial Ultrasonic Focusing,” Front Neurosci, vol. 14, no. April, pp. 1–12, Apr. 2020, doi: 10.3389/fnins.2020.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wu S-Y et al. , “Efficient Blood-Brain Barrier Opening in Primates with Neuronavigation-Guided Ultrasound and Real-Time Acoustic Mapping,” Sci Rep, vol. 8, no. 1, p. 7978, Dec. 2018, doi: 10.1038/s41598-018-25904-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Moser D, Zadicario E, Schiff G, and Jeanmonod D, “Correction: MR-guided focused ultrasound technique in functional neurosurgery: targeting accuracy,” J Ther Ultrasound, vol. 1, no. 1, p. 17, 2013, doi: 10.1186/2050-5736-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chauvet D and AuBry J-F, “Targeting accuracy of transcranial magnetic resonance–guided high-intensity focused ultrasound brain therapy: a fresh cadaver model,” vol. 118, no. May, pp. 1046–1052, 2013. [DOI] [PubMed] [Google Scholar]
- [34].Gallay MN, Moser D, and Jeanmonod D, “Safety and accuracy of incisionless transcranial MR-guided focused ultrasound functional neurosurgery: Single-center experience with 253 targets in 180 treatments,” J Neurosurg, vol. 130, no. 4, pp. 1234–1243, 2019, doi: 10.3171/2017.12.JNS172054. [DOI] [PubMed] [Google Scholar]
- [35].Clement GT and Hynynen K, “A non-invasive method for focusing ultrasound through the human skull,” Phys Med Biol, vol. 47, no. 8, pp. 1219–1236, 2002, doi: 10.1088/0031-9155/47/8/301. [DOI] [PubMed] [Google Scholar]
- [36].Baron C, Aubry JF, Tanter M, Meairs S, and Fink M, “Simulation of Intracranial Acoustic Fields in Clinical Trials of Sonothrombolysis,” Ultrasound Med Biol, vol. 35, no. 7, pp. 1148–1158, 2009, doi: 10.1016/j.ultrasmedbio.2008.11.014. [DOI] [PubMed] [Google Scholar]
- [37].Mahinda HAM and Murty OP, “Variability in thickness of human skull bones and sternum - An autopsy experience,” Journal of Forensic Medicine and Toxicology, vol. 26, no. 2, pp. 26–31, 2009. [Google Scholar]
- [38].Maimbourg G, Houdouin A, Deffieux T, Tanter M, and Aubry JF, “3D-printed adaptive acoustic lens as a disruptive technology for transcranial ultrasound therapy using single-element transducers,” Phys Med Biol, vol. 63, no. 2, 2018, doi: 10.1088/1361-6560/aaa037. [DOI] [PubMed] [Google Scholar]
- [39].Maimbourg G, Houdouin A, Deffieux T, Tanter M, and Aubry JF, “Steering capabilities of an acoustic lens for transcranial therapy: Numerical and experimental studies,” IEEE Trans Biomed Eng, vol. 67, no. 1, pp. 27–37, 2020, doi: 10.1109/TBME.2019.2907556. [DOI] [PubMed] [Google Scholar]
- [40].Chien C-Y, Yang Y, Gong Y, Yue Y, and Chen H, “Blood-Brain Barrier Opening by Individualized Closed-Loop Feedback Control of Focused Ultrasound,” BME Front, vol. 2022, pp. 1–11, Apr. 2022, doi: 10.34133/2022/9867230. [DOI] [PMC free article] [PubMed] [Google Scholar]