Abstract
Whole genome sequencing data of 874 Escherichia coli isolates carrying bla NDM-5 from 13 European Union/European Economic Area countries between 2012 and June 2022 showed the predominance of sequence types ST167, ST405, ST410, ST361 and ST648, and an increasing frequency of detection. Nearly a third (30.6%) of these isolates were associated with infections and more than half (58.2%) were predicted to be multidrug-resistant. Further spread of E. coli carrying bla NDM-5 would leave limited treatment options for serious E. coli infections.
Keywords: Carbapenem-resistant Enterobacterales, carbapenemase, NDM-5, Escherichia coli, surveillance, whole genome sequencing, cross-border import
Preliminary unpublished results of a survey of carbapenem- and/or colistin-resistant Enterobacterales (CCRE survey) [1] in 36 European countries showed that New Delhi metallo-β-lactamase (NDM)-5 was the most frequently reported carbapenemase in Escherichia coli with detection of bla NDM-5 in 62 (30.8%) of 201 carbapenemase-producing E. coli isolates collected in 2019. These 62 E. coli isolates were detected in 15 countries and involved various E. coli sequence types (STs), some of which are described as high-risk STs for extraintestinal infections [2]. These findings were of concern and warranted further investigation with the aim to confirm the extent of spread and describe the epidemiological and microbiological characteristics of the detected isolates.
Data collection and analysis
The European Centre for Disease Prevention and Control requested, via its EpiPulse platform, whole genome sequencing (WGS) and epidemiological data on E. coli carrying bla NDM-5 from European Union (EU)/European Economic Area (EEA) countries. In reply, data for 905 E. coli isolates were received from 13 countries. Twenty-nine isolates did not fulfil the quality criteria (assembled genome 3.7–6.4 Mbp with ≥ 90% core genome loci covered using the EnteroBase core genome multilocus sequence typing scheme (cgMLST [3]), and two isolates did not carry bla NDM-5. Therefore, data from 874 isolates from the period 2012 to June 2022 were included in the analysis (Table); these isolates are hereafter referred to as being from ‘national collections’. The data for this study were deposited in the European Nucleotide Archive under accession numbers PRJEB27363, PRJEB36710, PRJEB45009, PRJEB56146, PRJEB61153 and PRJNA845120.
Table. Dominant sequence types of Escherichia coli isolates carrying bla NDM-5 submitted from national collections, by country and period covered, EU/EEA, 2012–June 2022 (n = 874).
| E. coli sequence type | Number of isolates by country (period covered) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AT (2019–2022) |
DE (2019) |
DK (2015–2022) |
ES (2014–2021) |
FI (2016–2022) |
FR (2017–2022) |
HU (2020) |
IE (2017–2022) |
IT (2017–2018) |
NL (2012–2022) |
NO (2021–2022) |
PT (2017–2020) |
SE (2021–2022) |
Total (2012–2022) |
|
| ST167 | 0 | 2 | 23 | 1 | 5 | 95 | 0 | 8 | 4 | 42 | 3 | 6 | 11 | 200 |
| ST405 | 0 | 2 | 8 | 2 | 8 | 49 | 0 | 15 | 0 | 24 | 0 | 0 | 7 | 115 |
| ST410 | 0 | 2 | 7 | 0 | 3 | 54 | 1 | 8 | 2 | 13 | 0 | 1 | 5 | 96 |
| ST361 | 0 | 1 | 9 | 0 | 3 | 42 | 0 | 5 | 0 | 6 | 2 | 0 | 2 | 70 |
| ST648 | 0 | 1 | 5 | 0 | 6 | 18 | 0 | 11 | 0 | 12 | 1 | 0 | 11 | 65 |
| Other STs | 4 | 2 | 30 | 2 | 20 | 145 | 1 | 33 | 0 | 61 | 6 | 6 | 18 | 328 |
| Total | 4 | 10 | 82 | 5 | 45 | 403 | 2 | 80 | 6 | 158 | 12 | 13 | 54 | 874 |
AT: Austria; DE: Germany; DK: Denmark; EEA: European Economic Area: ES: Spain; EU: European Union; FI: Finland; FR: France; HU: Hungary; IE: Ireland; IT: Italy; NL: the Netherlands; NO: Norway; PT: Portugal; SE: Sweden ST: sequence type.
We downloaded WGS data from the National Center for Biotechnology Information (NCBI) Pathogen Detection on 20 July 2022 and included an additional 2,561 E. coli isolates carrying bla NDM-5 after quality control using the same criteria as described above. We determined the STs with the MLST 2.0 tool from the Danish Technical University (DTU) (version 2.0.9, database version 26 July 2022, using the Achtman MLST scheme for E. coli) [4]. In line with previously published definitions [5], major STs were defined as representing > 10% and minor STs as 5–10% of the isolates from national collections. Resistance genes were determined using E. coli analysis plugin of BioNumerics 7.6.3 (Applied Maths NV/bioMérieux, Sint-Martens-Latem, Belgium) with thresholds of ≥ 90% sequence identity and ≥ 60% sequence length for gene coverage. Fluoroquinolone resistance point mutations were detected using the PointFinder tool from DTU (reference sequences collected on 29 September 2022) [6]. Clusters of related isolates were determined with a threshold of 10 allelic differences based on previously published studies [7] and using the EnteroBase cgMLST scheme [3].
The epidemiological variables collected for isolates from national collections included year and month of sample collection, the location of the healthcare institution submitting the sample (city and National Territorial Units for Statistics level 2 region), the sample type (screening or clinical sample) and site of sampling for clinical samples, the clinical relevance (infection or carriage), the status of the patient (inpatient or outpatient), the type of acquisition (community- or healthcare-associated), age and sex of the patient, travel/hospitalisation and country of travel/hospitalisation in the 6 months before sampling and an epidemiological link to another patient.
Distribution of sequence types
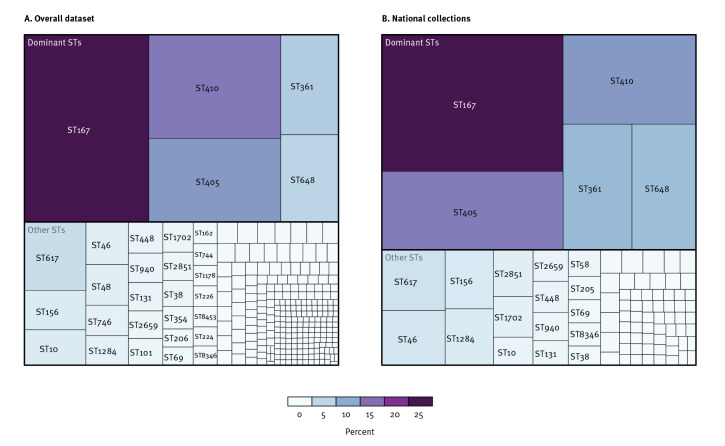
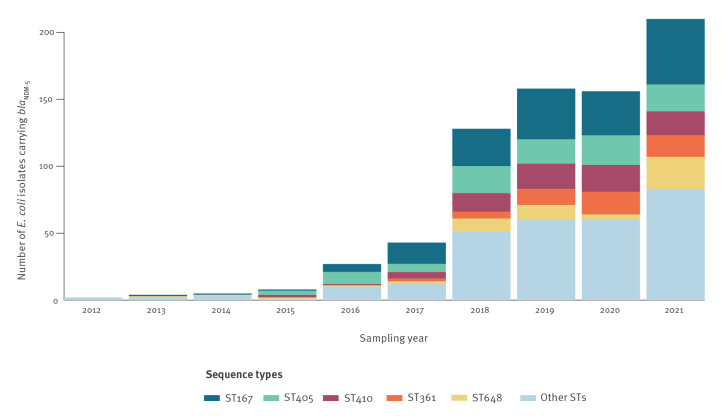
The overall dataset included 3,435 E. coli isolates carrying bla NDM-5 belonging to 267 STs. Of these, 83 STs were present in the 874 isolates from national collections, with five predominant STs: ST167 (n = 200; 22.9%), ST405 (n = 115; 13.2%), ST410 (n = 96; 11.0%), ST361 (n = 70; 8.0%) and ST648 (n = 65; 7.4%) (Figure 1). Based on the national collections and the definitions outlined above, ST167, ST405 and ST410 were classified as major STs and ST361 and ST648 as minor STs, and they were grouped together as dominant STs for further analyses. The five dominant STs were detected in all countries that submitted more than 20 isolates, i.e. Denmark, Finland, France, Ireland, the Netherlands and Sweden (Table). The number of detected E. coli isolates carrying bla NDM-5 increased over time (Figure 2). Isolates from national collections (n = 500) were involved in 162 clusters of which 114 were multi-country clusters.
Figure 1.
Distribution of sequence types of Escherichia coli isolates carrying bla NDM-5 from two datasets: overall dataset (n = 267)a, and national collections (n = 83), EU/EEA, 2012–June 2022
EEA: European Economic Area; EU: European Union; ST: sequence type.
Isolates without a specified ST were excluded (n = 53 for overall dataset and n = 35 for national collections).
a The overall dataset included isolates from the following countries: Argentina, Australia, Austria, Bangladesh, Cambodia, Canada, Chile, China, Czechia, Denmark, Egypt, Finland, France, Germany, Hungary, India, Ireland, Israel, Italy, Japan, Kenya, Laos, Lebanon, Malawi, Myanmar/Burma, Nepal, the Netherlands, Niger, Nigeria, Norway, Pakistan, Portugal, Qatar, Singapore, South Korea, Spain, Sweden, Switzerland, Taiwan, Tanzania, Thailand, United Kingdom, United States and Vietnam, collected or uploaded to NCBI between 2014 and 2022.
Figure 2.
Frequency of sequence types of Escherichia coli isolates carrying bla NDM-5 over time by year of sampling in the dataset from national collections, EU/EEA, 2012–2021a (n = 741)
EEA: European Economic Area; EU: European Union; ST: sequence type.
a The time distribution illustrated above should not be interpreted as an epidemic curve as year-to-year variation is probably affected by detection and reporting biases and may not reflect true temporal trends in incidence. Isolates from 2022 (n = 133) are not displayed because data for that year were incomplete at the time of analysis. Seventy STs other than the dominant ones are grouped as “Other STs”.
Resistance determinants
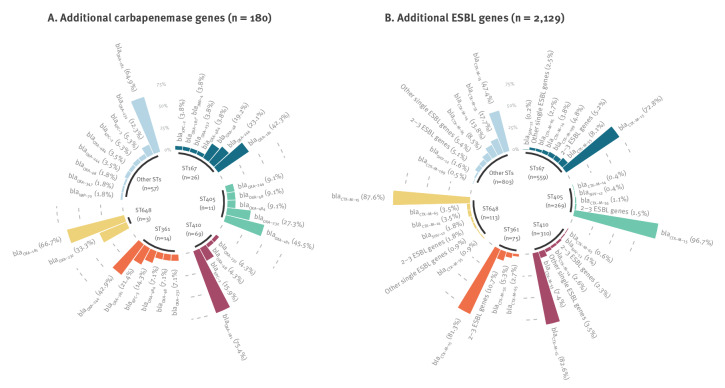
In the overall dataset, a small percentage (n = 180; 5.2%) of E. coli isolates carrying bla NDM-5 harboured additional carbapenemase genes, with 68.3% of these genes detected in the dominant STs and bla OXA-181 the most commonly co-carried carbapenemase gene (n = 110; 61.1%) (Figure 3A). Among the 446 ST410 isolates, 69 (15.5%) carried two carbapenemase genes, which was the highest proportion compared with other dominant STs (1.8–7.4%).
Figure 3.
Distribution of Escherichia coli isolates with bla NDM-5 co-carrying additional carbapenemase (n = 180) or ESBL (n = 2,129) genes, by dominant sequence types from the overall dataseta
ESBL: extended-spectrum β-lactamase; ST: sequence type.
Single ESBL genes found in less than 20 isolates are categorised as “Other single ESBL genes”. Two or three ESBL genes are grouped as “2–3 ESBL genes”.
a The overall dataset included isolates from the following countries: Argentina, Australia, Austria, Bangladesh, Cambodia, Canada, Chile, China, Czechia, Denmark, Egypt, Finland, France, Germany, Hungary, India, Ireland, Israel, Italy, Japan, Kenya, Laos, Lebanon, Malawi, Myanmar/Burma, Nepal, the Netherlands, Niger, Nigeria, Norway, Pakistan, Portugal, Qatar, Singapore, South Korea, Spain, Sweden, Switzerland, Taiwan, Tanzania, Thailand, United Kingdom, United States and Vietnam, collected or uploaded to NCBI between 2014 and 2022.
Many bla NDM-5-positive E. coli (n = 2,129; 62.0%) also harboured at least one extended-spectrum β-lactamase (ESBL) gene, with 62.3% of isolates being of the dominant STs (Figure 3B). The most frequently identified ESBL genes were bla CTX-M family genes (n = 2,096; 98.4%) with most isolates positive for bla CTX-M-15 (n = 1,464; 68.8%). Plasmid-mediated AmpC β-lactamase determinants were also prevalent (n = 1,027; 29.9%), with 67.7% of them identified in the dominant STs. The most often identified genes encoding for acquired AmpC β-lactamases were bla CMY-42 (n = 490; 47.7%) and bla CMY-2 (n = 406; 39.5%). Of note, the highest proportion of E. coli isolates harbouring bla CMY-2 (n = 266; 65.5%) belonged to ST410 compared with other dominant STs (< 11% each).
High predicted resistance to aminoglycosides (n = 3,236; 94.2%) was observed among E. coli isolates carrying bla NDM-5, the most frequent genes being aadA2 (74.9%), aac(6’)-Ib-cr (39.4%), aadA5 (34.8%) and rmtB (24.6%). Predicted resistance to fluoroquinolones was also common based on one or more mutations in type II topoisomerase genes (n = 3,195; 93.0%). Combining the resistance markers, more than half (n = 2,000; 58.2%) of the E. coli isolates carrying bla NDM-5 in the overall dataset were predicted to be multidrug-resistant with resistance to all β-lactams (including aztreonam), aminoglycosides and fluoroquinolones for treatment of serious E. coli infections.
Similar trends were observed for predicted trimethoprim (n = 3,185; 92.7%) and sulfamethoxazole (n = 3,223; 93.8%) resistance, the most frequent genes being dfrA12 and dfrA17 as well as sul1 and sul2, respectively. Predicted resistance to colistin (plasmid-mediated mcr-1.1 carriage: n = 176; 5.1%) and tigecycline (tet(X) carriage: n = 10; 0.3%) was low.
Epidemiological findings
Among the 874 isolates from national collections, 618 (70.7%) had available information on infection/carriage status and 189 (30.6%) of those were associated with infection. Of 766 (87.6%) isolates with available information on sample type, 234 (30.5%) originated from clinical samples. The most frequent specimen type was urine (n = 178; 76.1%), while isolation from blood (n = 23; 9.8%) and the respiratory tract (n = 6; 2.6%) was infrequent. Of 341 E. coli isolates carrying bla NDM-5 with available information on prior travel and/or hospitalisation within the past 6 months before sampling, a link to a country outside of the EU/EEA was reported for 287 (84.2%) isolates, mostly countries in Asia (158/341; 46.3% isolates) or in Africa (125/341; 36.7% isolates).
Discussion
Our results confirmed the emergence of E. coli isolates carrying bla NDM-5 in the EU/EEA initially detected in the CCRE survey and showed the predominance of five STs (ST167, ST410, ST405, ST361 and ST648) known as high-risk clones based on their rapid global spread and their association with virulence and multidrug resistance [8-16]. While these STs carrying bla NDM-5 have previously been described in the EU/EEA, reports were limited to a single ST in one [8,9] or two [10] institutions, or to the description of the national epidemiology of one single ST [11] or several STs [12,13]. Only one study including 33 isolates of E. coli carrying bla NDM-5 has previously reported cross-border emergence in Germany and Switzerland [14]. Our study included WGS data on 874 isolates from national reference laboratories in 13 countries and thus provides a more comprehensive analysis of the spread of these high-risk STs in Europe. Of the isolates with available information on prior travel and/or hospitalisation, 84.2% had a link with a country outside of the EU/EEA, mainly on the African and Asian continents, suggesting potential acquisition outside of the EU/EEA.
About 30% of the E. coli isolates carrying bla NDM-5 were documented as associated with infections, emphasising the clinical relevance and the need for early detection. Sixty-two per cent of isolates carried ESBL genes conferring resistance to aztreonam which is not inactivated by metallo-β-lactamases. Together with predicted resistance to aminoglycosides and fluoroquinolones, this leaves very limited options for the treatment of serious E. coli infections. With respect to aminoglycosides, the rmtB-encoded 16S rRNA methylase conferring resistance to novel aminoglycosides is a particular concern [12,17]. Moreover, studies have shown that isolates (over)-producing NDM-5 had increased minimum inhibitory concentrations to cefiderocol, one of the last treatment options for metallo-β-lactamase-producing Enterobacterales [18,19].
A limitation of this study is that it is not based on a standardised sampling protocol but on regular national surveillance with differences in sample collection protocols, national coverage and data completeness and that it is subject to detection and reporting bias. However, the frequent detection in 13 countries indicates that E. coli carrying bla NDM-5 is now established in the EU/EEA. The proportion of carbapenem resistance in E. coli in the EU/EEA has so far been very low, i.e. only 0.2% of invasive isolates [20]. Further spread of E. coli isolates carrying bla NDM-5 in community and healthcare settings, and within and between countries, is likely to increase dissemination with resulting higher levels of carbapenem and multidrug resistance in E. coli infections.
Conclusion
The spread of E. coli carrying bla NDM-5 is occurring rapidly and on a large geographical scale with a considerable risk for increasing carbapenem resistance in E. coli in the EU/EEA within a few years. Early detection and control are required to mitigate adverse consequences for patients and healthcare systems.
Ethical statement
All data were anonymised and collected in accordance with the European Parliament and Council decisions on the epidemiological surveillance and control of communicable disease in the European Community. Ethical approval and informed consent were thus not required.
Data availability
The whole genome sequencing data for this study were deposited in the European Nucleotide Archive under accession numbers PRJEB27363, PRJEB36710, PRJEB45009, PRJEB56146, PRJEB61153 and PRJNA845120.
Conflict of interest: None declared.
Authors’ contributions: ML: design of study, bioinformatic analysis and drafting of the manuscript; RAB: interpretation of results and drafting of manuscript; EA, OS: bioinformatic analysis; PA, RH, HH, LR, KR, LD, NP, JBH, AT, LB, MC, ND, MM, MG, APAH, ØS, AKP, MC, VM, JOI, MPV, KW, BM: compilation and analysis of national data, interpretation of results; DP, DLM: design of study and interpretation of results; AK: design and coordination of study, epidemiological analysis, and drafting of manuscript. All authors: review of manuscript.
References
- 1.European Centre for Disease Prevention and Control (ECDC). ECDC study protocol for genomic-based surveillance of carbapenem-resistant and/or colistin-resistant Enterobacteriaceae at the EU level. Version 2.0. Stockholm: ECDC; 2018. Available from: https://ecdc.europa.eu/en/publications-data/ecdc-study-protocol-genomic-based-surveillance-carbapenem-resistant-andor
- 2. Manges AR, Geum HM, Guo A, Edens TJ, Fibke CD, Pitout JDD. Global extraintestinal pathogenic Escherichia coli (ExPEC) lineages. Clin Microbiol Rev. 2019;32(3):e00135-18. 10.1128/CMR.00135-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou Z, Alikhan NF, Mohamed K, Fan Y, Agama Study Group. Achtman M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020;30(1):138-52. 10.1101/gr.251678.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50(4):1355-61. 10.1128/JCM.06094-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peirano G, Chen L, Nobrega D, Finn TJ, Kreiswirth BN, DeVinney R, et al. Genomic epidemiology of global carbapenemase-producing Escherichia coli, 2015-2017. Emerg Infect Dis. 2022;28(5):924-31. 10.3201/eid2805.212535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zankari E, Allesøe R, Joensen KG, Cavaco LM, Lund O, Aarestrup FM. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother. 2017;72(10):2764-8. 10.1093/jac/dkx217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schürch AC, Arredondo-Alonso S, Willems RJL, Goering RV. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene-based approaches. Clin Microbiol Infect. 2018;24(4):350-4. 10.1016/j.cmi.2017.12.016 [DOI] [PubMed] [Google Scholar]
- 8. Garcia-Fernandez A, Villa L, Bibbolino G, Bressan A, Trancassini M, Pietropaolo V, et al. Novel insights and features of the NDM-5-producing Escherichia coli sequence type 167 high-risk clone. MSphere. 2020;5(2):e00269-20. 10.1128/mSphere.00269-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barrado L, Pérez-Vázquez M, Del Pozo JL, Martín-Salas C, Leiva J, Mazón A, et al. Clonal transmission of NDM-5-producing Escherichia coli belonging to high-risk sequence type ST405. Int J Antimicrob Agents. 2018;52(1):123-4. 10.1016/j.ijantimicag.2018.05.018 [DOI] [PubMed] [Google Scholar]
- 10. Bibbolino G, Di Lella FM, Oliva A, Lichtner M, Del Borgo C, Raponi G, et al. Molecular epidemiology of NDM-5-producing Escherichia coli high-risk clones identified in two Italian hospitals in 2017-2019. Diagn Microbiol Infect Dis. 2021;100(4):115399. 10.1016/j.diagmicrobio.2021.115399 [DOI] [PubMed] [Google Scholar]
- 11. Roer L, Overballe-Petersen S, Hansen F, Schønning K, Wang M, Røder BL, et al. Escherichia coli sequence type 410 is causing new international high-risk clones. MSphere. 2018;3(4):e00337-18. 10.1128/mSphere.00337-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Turton JF, Pike R, Perry C, Jenkins C, Turton JA, Meunier D, et al. Wide distribution of Escherichia coli carrying IncF plasmids containing blaNDM-5 and rmtB resistance genes from hospitalized patients in England. J Med Microbiol. 2022;71(8). 10.1099/jmm.0.001569 [DOI] [PubMed] [Google Scholar]
- 13. Hans JB, Pfennigwerth N, Neumann B, Pfeifer Y, Fischer MA, Eisfeld J, et al. Molecular surveillance reveals the emergence and dissemination of NDM-5-producing Escherichia coli high-risk clones in Germany, 2013 to 2019. Euro Surveill. 2023;28(10):2200509. 10.2807/1560-7917.ES.2023.28.10.2200509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chakraborty T, Sadek M, Yao Y, Imirzalioglu C, Stephan R, Poirel L, et al. Cross-border emergence of Escherichia coli producing the carbapenemase NDM-5 in Switzerland and Germany. J Clin Microbiol. 2021;59(3):e02238-20. 10.1128/JCM.02238-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cummins EA, Snaith AE, McNally A, Hall RJ. The role of potentiating mutations in the evolution of pandemic Escherichia coli clones. Eur J Clin Microbiol Infect Dis. 2021. 10.1007/s10096-021-04359-3 [DOI] [PubMed] [Google Scholar]
- 16. Schaufler K, Semmler T, Wieler LH, Trott DJ, Pitout J, Peirano G, et al. Genomic and functional analysis of emerging virulent and multidrug-resistant Escherichia coli lineage sequence type 648. Antimicrob Agents Chemother. 2019;63(6):e00243-19. 10.1128/AAC.00243-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cox G, Ejim L, Stogios PJ, Koteva K, Bordeleau E, Evdokimova E, et al. Plazomicin retains antibiotic activity against most aminoglycoside modifying enzymes. ACS Infect Dis. 2018;4(6):980-7. 10.1021/acsinfecdis.8b00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bonnin RA, Emeraud C, Jousset AB, Naas T, Dortet L. Comparison of disk diffusion, MIC test strip and broth microdilution methods for cefiderocol susceptibility testing on carbapenem-resistant Enterobacterales. Clin Microbiol Infect. 2022;28(8):1156.e1-5. 10.1016/j.cmi.2022.04.013 [DOI] [PubMed] [Google Scholar]
- 19. Simner PJ, Mostafa HH, Bergman Y, Ante M, Tekle T, Adebayo A, et al. Progressive development of cefiderocol resistance in Escherichia coli during therapy is associated with an increase in blaNDM-5 copy number and gene expression. Clin Infect Dis. 2022;75(1):47-54. 10.1093/cid/ciab888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.European Centre for Disease Prevention and Control (ECDC). Antimicrobial resistance in the EU/EEA (EARS-Net) - Annual epidemiological report for 2021. Stockholm: ECDC; 2022. Available from: https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2021