Abstract
We report a case of prolonged shedding of the infective SARS-CoV-2 omicron variant BA.1.1.2 in a 79-year-old male patient with diffuse large B-cell lymphoma, after receiving chemotherapy with rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisolone (R-CHOP). The patient was admitted to our hospital in late March 2022 for the sixth course of R-CHOP chemotherapy. Initially, the patient tested negative for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) using an in-hospital loop-mediated amplification assay with a nasopharyngeal swab, both on the day of admission and three days later. However, the patient developed fever and was diagnosed with coronavirus disease (COVID-19) six days after admission and was suspected to have contracted the infection in the ward. Viral shedding continued for more than three months, with confirmed viral infectivity. As compared to the original Wuhan-Hu-1/2019 strain, amino acid substitutions including S36 N in non-structural protein (NSP)2, S148P, S1265del and L1266I in NSP3, G105D in NSP4, G496S, A831V, or V987F in spike protein, and I45T in open-reading frame (ORF)9b were randomly detected in isolated viruses. Although the patient had received two doses of the BNT162b2 vaccine approximately six months earlier and the third dose on day 127 after the infection, both serum anti-spike and anti-nuclear protein IgG and IgM tests were negative at day 92, 114, and 149 after the infection. The patient finally cleared the virus after the third course of remdesivir and did not have further recurrence.
Keywords: Diffuse large B cell lymphoma, Rituximab, BA.1.1.2, Prolonged viral shedding, Remdesivir
1. Introduction
B-cell depletion therapy with anti-cluster of differentiation (CD) 20 monoclonal antibodies, such as rituximab or obinutuzumab, are used to treat B-cell associated hematologic malignancies and autoimmune diseases such as rheumatoid arthritis and systemic lupus erythematosus [1]. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is associated with an increased risk of severe disease, mortality, and prolonged viral shedding in patients undergoing B-cell depletion therapy [2,3]. In such patients, administration of monoclonal antibodies, convalescent plasma, nirmatrelvir/ritonavir, and repetitive or prolonged remdesivir, in combination or a single agent, have been reported [[4], [5], [6], [7]]. Here, we report a case of diffuse large B-cell lymphoma (DLBCL) with prolonged infective SARS-CoV-2 omicron variant shedding in a patient treated with rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisolone (R-CHOP).
2. Case report
A 79-year-old man was diagnosed with DLBCL in October 2021. R-CHOP therapy was initiated from early November at three-to four-week intervals. The following were administered on the first day of the R-CHOP course: rituximab 375 mg/m2/day (500 mg/day), cyclophosphamide 750 mg/m2/day (1000 mg/day), doxorubicin 50 mg/m2/day (70 mg/day), and vincristine 1.4 mg/m2/day (1.9 mg/day), in addition to prednisolone 30 mg/m2/day (50 mg/day) for the first to fifth day. Vincristine was discontinued after the third course of therapy due to peripheral neuritis.
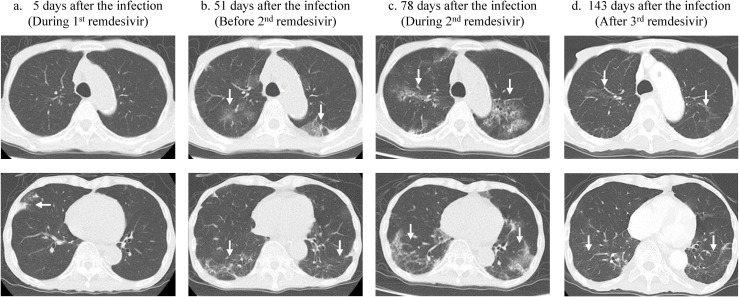
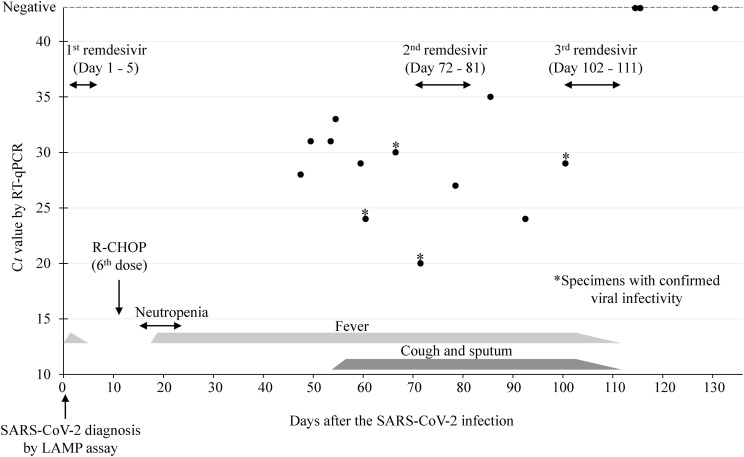
The patient was admitted to our hospital in late March 2022 for his sixth course of R-CHOP therapy. Laboratory tests on admission showed a hemoglobin level of 9.2 g/dL and hematocrit count of 28.6% with normocytic and normochromic blood picture. The white blood cell count was 3700 cells/μL with 73.8% neutrophils, 17.9% lymphocytes, 6.7% monocytes, 0.3% eosinophils, and 1.3% basophils. The patient's serum C-reactive protein level was 0.06 mg/dL. The total protein and albumin levels were 6.5 g/dL and 4.0 g/dL, respectively. Antibodies against hepatitis B and C viruses and human immunodeficiency virus were negative. On admission, the patient was tested for SARS-CoV-2 using an in-hospital rapid nicking enzyme amplification reaction (NEAR) assay with a nasopharyngeal swab, which was negative. The patient was afebrile and exhibited no respiratory symptoms. Three days after admission, another patient in the same room developed respiratory symptoms and was diagnosed with coronavirus disease (COVID-19). Therefore, our patient was tested again for SARS-CoV-2 using an in-hospital loop-mediated isothermal amplification (LAMP) assay, which, too, was negative. However, six days after admission, the patient developed fever, and a repeated LAMP assay yielded positive results. At that time, the patient's body temperature increased to 38.6 °C. The peripheral blood SpO2 was approximately 97–99% while breathing ambient air. Chest computed tomography (CT) after five days revealed small areas of ground-glass opacity only in the right middle lobe (Fig. 1 -a). The patient was diagnosed as having symptomatic COVID-19 Moderate I according to the criteria of the Japanese Ministry of Health and Welfare, which corresponds to a patient who has a shortness of breath or pneumonia without respiratory failure. Remdesivir was administered for five days (200 mg on the first day, then 100 mg/day from days two to five). The patient became afebrile two days after symptom onset. As the patient's physical condition seemed to improve, no nucleic acid detection test was performed to confirm the disappearance of viral shedding. Eleven days after the infection, the sixth course of R-CHOP was administered. Eighteen days after the infection, the patient developed fever (Fig. 2 ). Initially, since the fever was considered to be due to R-CHOP neutropenia, piperacillin and tazobactam were administered; but the fever persisted, although the neutrophils recovered to normal levels. A nasopharyngeal RT-qPCR swab for SARS-CoV-2 on day 47 post-infection yielded a positive cycle threshold value of 28. As the patient gradually developed cough and sputum production chest CT was performed on day 51, which revealed bilateral ground-glass opacities in both the upper and lower lobes (Fig. 1-b). The time-course of the patient's infection based on the results of the RT-qPCR tests for SARS-CoV-2 during the hospitalization is shown in Fig. 2. The genotype of the viruses detected in the patient after admission were uniformly omicron variant BA.1.1.2. To determine the infectivity of the virus detected in the nasopharyngeal swabs, the specimens were sent to the Fukushima Prefectural Institute of Public Health. Using VeroE6/TMPRSS2 cells [8], infectious virus was isolated from specimens obtained at 60, 66, and 71 days after infection (Fig. 2). A second course of remdesivir was administered for 10 days, from day 72–81 after infection. However, repeat chest CT showed progression of bilateral infiltration in both lungs (Fig. 1-c). Low-grade fever and cough persisted even after administration of the second dose of remdesivir. As the patient's nasopharyngeal swab specimen taken 100 days after the infection still had infectious virus, remdesivir treatment was administered again for 10 days from days 102–111 after the infection. After the third course of remdesivir treatment, the patient became afebrile, and his cough gradually improved. The nasopharyngeal specimen obtained on day 114 showed no evidence of infection. Chest CT on day 143 after infection showed improvement in bilateral infiltration of SARS-CoV-2 (Fig. 1-d). Although the patient had received the BNT162b2 vaccine twice about six months before the infection and a third dose on day 120 after the infection, serum anti-spike and anti-nuclear IgG and IgM antibody levels were below the detection limit on day 92, 114, and 149 after the infection. After successful viral clearance, the patient was discharged to another long-term care facility 183 days after the infection. A rapid antigen test performed at the facility on day 200 after the infection was negative, and he did not experience recurrence of COVID-19 after the third remdesivir treatment.
Fig. 1.
Chest computed tomography of the patient after SARS-CoV-2 infection. Upper and lower images show upper and lower lobes of the lungs, respectively, on the indicated day. 1-a. Five days after the infection, a small infiltration was observed only at the right middle lobe (white arrow). 1-b. After 51 days of infection, bilateral infiltrations appeared before the second dose of remdesivir therapy (white arrows). 1-c. After 78 days of infection, during the second remdesivir dose, the infiltrations progressed (white arrows). 1-d. After the third remdesivir dose, and 143 days after the infection, the infiltrations had almost disappeared (white arrows).
Fig. 2.
Time-course of the patient's infection and treatment based on the Ct values of SARS-CoV-2 in nasopharyngeal specimens. The patient's peripheral white blood cells were below 500 cells/mm3 during the period of neutropenia. *Specimens with confirmed viral infectivity. LAMP: loop-mediated isothermal amplification; R-CHOP: rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisolone; Ct: cycle threshold.
The results of the whole-genome sequencing analysis for viral RNA isolated from the patient's swab specimens are shown in Table 1 . In total, eight non-synonymous mutation sites, two synonymous mutation sites, and a deletion, were randomly observed among the isolated viruses, compared to the original Wuhan strain isolated in 2019 (Wuhan-Hu-1/2019, GenBank accession number MN908947). Base substitutions T911A and C912A lead to amino acid substitution S36 N in non-structural protein (NSP)2, T3161C and 6513-6515Del lead to S36N, S1265del and L1266I in NSP3, G8868A leads to G105D in NSP4, G23048A, C24054T, or G24521T lead to G496S, A831V, or V987F in spike protein, and T28417C leads to I45T in open-reading frame (ORF)9b, respectively, according to data from Nextclade v2.10.0 [13]. The number of mutations tended to increase after the second course of remdesivir treatment (Table 1).
Table 1.
Base and amino acid substitutions/deletions in isolated SARS-CoV-2 after COVID-19 symptoms began.
| Base substitution and deletion* |
G910A |
T911A, C912A |
C2197T |
T3161C |
6513-6515 Del |
G8868A |
G23048A |
C24054T |
G24521T |
T28417C |
|---|---|---|---|---|---|---|---|---|---|---|
| Amino acid substitution | - | S36 N in NSP2 | - | S148P in NSP3 | S1265del and L1266I in NSP3 | G105D in NSP4 | G496S in S | A831V in S | V987F in S | I45T in ORF9b |
| Day 47 | - | - | - | - | - | - | ● | - | - | ● |
| Day 60 | - | - | - | - | ● | - | ● | - | - | ● |
| Day 66 | - | - | - | - | ● | - | ● | ● | - | ● |
| Day 71 | - | - | - | - | ● | - | ● | ● | - | ● |
| Day 78 | - | - | - | - | ● | - | - | - | - | - |
| Day 92 | ○ | ● | - | - | ● | - | ● | ● | - | ● |
| Day 100 | - | - | ○ | ● | ● | ● | - | - | ● | - |
Footnote: Remdesivir administration: first, between day 1–5 (five days); second, from day 72–81 (10 days); third, from day 102–111 (10 days). *These base substitutions and a deletion are relative to the reference strain Wuhan-Hu-1/2019 (GenBank accession number MN908947).
Abbreviations: Del, deletion; NSP, non-structural protein; S, spike protein; ORF, open-reading frame; -, no substitution detected; ○, synonymous mutation; ●, non-synonymous mutation.
Written informed consent was obtained from the patient regarding publication of this case report.
3. Discussion
In the present case, we describe a patient being treated with R-CHOP for DLBCL who developed prolonged infective SARS-CoV-2 omicron variant. Viral shedding continued for more than three months with confirmed viral infectivity, and multiple mutations were detected in the infectious virus samples. The third course of remdesivir administered finally cleared the viral secretion, and this may be a treatment option for COVID-19 patients with prolonged infective viral shedding who have undergone B cell-depleting therapy.
Anti-CD20 monoclonal antibodies, which are widely used to treat hematological and autoimmune diseases [1], target specific Fab domains of CD20+ or CD19+ B lymphocytes, causing selective depletion of circulating B cells through natural killer cell-mediated, antibody-dependent cellular cytotoxicity, complement-dependent cytotoxicity, and antibody-triggered apoptosis [2,3]. Prolonged shedding of infectious SARS-CoV-2 has been reported in patients who are immunocompromised, particularly those receiving B-cell depletion therapy [[1], [2], [3]]. In the present case, the patient had impaired humoral immunity, which was indicated by the absence of anti-SARS-CoV-2 spike or anti-nuclear proteins IgM and IgG after SARS-CoV-2 infection. We administered the second and third courses of remdesivir after confirming detection of infective virus. A number of case reports describe prolonged viral shedding in patients with COVID-19 receiving B cell-depletion therapy and re-administration of remdesivir [4,5]. Most of the retreated patients received two courses of remdesivir [4,5]. A patient with non-Hodgkin lymphoma in remission received three courses of remdesivir each lasting five, three, and five days, respectively, and was discharged without oxygen supplementation [4]. Another report described an immunocompromised patient with secondary hypogammaglobulinemia caused by rituximab administration for two years who secreted viable SARS-CoV-2 for five months and finally cleared the virus after three courses of remdesivir (first, five days; second, 10 days; third, 30 days) in addition to two administrations of convalescent plasma. The patient experienced no remarkable side effects or laboratory abnormalities due to the extended course of remdesivir [5]. Our patient, too, had no remarkable side effects from the three courses of remdesivir. However, the risk of de novo emergence of a remdesivir resistance due to E802D in nsp12 mutation in an immunocompromised patient after remdesivir treatment has been reported [6]. The benefits and drawbacks of remdesivir administration for more than 10 days or three or more courses of remdesivir treatment need further investigation.
As our patient's serum did not show any detectable level of anti-spike or anti-nuclear protein antibodies, monoclonal antibody administration might be another treatment option [7,9]. Currently, three monoclonal antibodies, namely casirivimab and imdevimab, sotrovimab, and tixagevimab co-packaged with cilgavimab, are approved in Japan. Casirivimab and imdevimab have reduced activity against omicron variants [10]. Sotrovimab administration was a treatment option because it retains binding ability to the spike protein of BA.1 and BA.1.1, although slightly reduced against the latter [10]. In this case, the third administration of remdesivir finally cleared the virus; however, if it was not successful, sotrovimab could have been administered. Another treatment option includes combination antiviral therapy with remdesivir and nirmatrelvir/ritonavir in addition to sotrovimab. This therapy has been administered as a salvage therapy for highly immunocompromised patients with leukemia receiving B-cell depletion treatment [11,12]. Tixagevimab co-packaged with cilgavimab was approved in August 2022 in Japan for prophylactic use only. In this case, considering that the patient's serum anti-spike and -nuclear protein IgG and IgM were negative during the hospital stay, administration of tixagevimab co-packaged with cilgavimab before transferring to the long-term care facility could have been considered.
In the whole-genome analysis, viruses isolated from the patient were found to have developed both synonymous and non-synonymous mutations, one deletion, compared to the original viral strain isolated in Wuhan-Hu-1/2019. These point mutations occur in immunocompromised patients with prolonged viral infection such as this case and may alter the replication ability of the virus and the monoclonal antibody-binding ability to viral spike proteins [14].
Further studies are required to understand the best clinical and therapeutic management strategies for immunocompromised patients with COVID-19 receiving B cell depletion therapy.
Disclosure statement
The authors state that they have no conflicts of interest.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author contributions
All authors met the ICMJE authorship criteria. Nakamura K, Sugiyama M, Ishizuka H, Kitakawa K: study design, and investigation; Sasajima S, Minakawa Y, Sato H, Miyazawa M, Fujita S, Saito N, Kashiwabara N, Kohata H: data acquisition and interpretation; Hara Y, Kanari Y, Shinka T, Kanemitsu K: analysis; Nakamura K, Kitakawa K, Sugiyama M: writing-original draft; all authors: writing-review and editing of the final manuscript.
References
- 1.D'Abramo A., Vita S., Maffongelli G., Mariano A., Agrati C., Castilletti C., et al. Spallanzani COVID-19 Case Investigation Team. Prolonged and severe SARS-CoV-2 infection in patients under B-cell-depleting drug successfully treated: a tailored approach. Int J Infect Dis. 2021;107:247–250. doi: 10.1016/j.ijid.2021.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Camprubí D., Gaya A., Marcos M.A., Martí-Soler H., Soriano A., Mosquera M.D.M., et al. Persistent replication of SARS-CoV-2 in a severely immunocompromised patient treated with several courses of remdesivir. Int J Infect Dis. 2021;104:379–381. doi: 10.1016/j.ijid.2020.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corey L., Beyrer C., Cohen M.S., Michael N.L., Bedford T., Rolland M. SARS-CoV-2 variants in patients with immunosuppression. N Engl J Med. 2021;385:562–566. doi: 10.1056/nejmsb2104756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Heeti O., Kumar R.N., Kling K., Angarone M., Achenbach C., Taiwo B. Remdesivir retreatment: another unproven intervention for COVID-19. J Antimicrob Chemother. 2022;77:854–856. doi: 10.1093/jac/dkab472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez M.A., Chen T.Y., Choi H., Hwang M., Navarathna D., Hao L., et al. Extended remdesivir infusion for persistent coronavirus disease 2019 infection. Open Forum Infect Dis. 2022;9:ofac382. doi: 10.1093/ofid/ofac382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gandhi S., Klein J., Robertson A.J., Peña-Hernández M.A., Lin M.J., Roychoudhury P., et al. De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS-CoV-2 infection in an immunocompromised patient: a case report. Nat Commun. 2022;13:1547. doi: 10.1038/s41467-022-29104-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin–Blondel G., Marcelin A.G., Soulié C., Kaisaridi S., Lusivika-Nzinga C., Zafilaza K., et al. Time to negative PCR conversion amongst high-risk patients with mild-to-moderate Omicron BA.1 and BA.2 COVID-19 treated with sotrovimab or nirmatrelvir. Clin Microbiol Infect. 2023;29:543. doi: 10.1016/j.cmi.2022.12.016. e5–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendoza E.J., Manguiat K., Wood H., Drebot M. Two detailed plaque assay protocols for the quantification of infectious SARS-CoV-2. Curr Protoc Microbiol. 2020;57 doi: 10.1002/cpmc.105. 10.1002/cpmc.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng M.M., Reyes C., Satram S., Birch H., Gibbons D.C., Drysdale M., et al. Real-world effectiveness of sotrovimab for the early treatment of COVID-19 during SARS-CoV-2 Delta and Omicron waves in the USA. Infect Dis Ther. 2023;11:1–15. doi: 10.1007/s40121-022-00755-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takashita E., Kinoshita N., Yamayoshi S., Sakai-Tagawa Y., Fujisaki S., Ito M., et al. Efficacy of antibodies and antiviral drugs against Covid-19 Omicron variant. N Engl J Med. 2022;386:995–998. doi: 10.1056/NEJMc2119407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ford E.S., Simmons W., Karmarkar E.N., Yoke L.H., Braimah A.B., Orozco J.J., et al. Successful treatment of prolonged, severe coronavirus disease 2019 lower respiratory tract disease in a B cell acute lymphoblastic leukemia patient with an extended course of remdesivir and nirmatrelvir/ritonavir. Clin Infect Dis. 2023;76:926–929. doi: 10.1093/cid/ciac868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baldi F., Dentone C., Mikulska M., Fenoglio D., Mirabella M., Magnè F., et al. Case report: sotrovimab, remdesivir and nirmatrelvir/ritonavir combination as salvage treatment option in two immunocompromised patients hospitalized for COVID-19. Front Med. 2023;9 doi: 10.3389/fmed.2022.1062450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nextcladev2.11.0 Clade assignment, mutation calling, and sequence quality Checks. https://clades.nextstrain.org/
- 14.Nussenblatt V., Roder A.E., Das S., de Wit E., Youn J.H., Banakis S., et al. Year-long COVID-19 infection reveals within-host evolution of SARS-CoV-2 in a patient with B cell depletion. medRxiv. 2021;225(7):1118–1123. doi: 10.1101/2021.10.02.21264267. [DOI] [PMC free article] [PubMed] [Google Scholar]