Abstract
Gut microbiota dysbiosis with increased pathogenic bacteria and decreased beneficial bacteria is associated with colorectal cancer (CRC) development. This study examined the effect of a newly developed probiotic formula in modulating CRC-related bacteria. We developed a probiotic formula containing three bifidobacteria (B. adolescentis, B. longum, and B. bifidum) based on the identification of bacterial species that showed significant correlations with CRC-related bacteria including Fusobacterium nucleatum (Fn), Lachnoclostridium sp. m3, Clostridium hathewayi (Ch), and Bacteroides clarus (Bc). We co-cultured Fn with each bifidobacterium or the combined formula and examined the growth of Fn by qPCR. The three individual bifidobacteria significantly inhibited the growth of Fn compared to the control treatment (24~65% inhibition; all p < 0.001). The combination of the three bifidobacteria showed a greater inhibitory effect on Fn growth (70% inhibition) than the individual bifidobacteria (all p < 0.05). We further examined the effect of the probiotic formula in a pilot study of 72 subjects (40 on probiotics; 32 with no intervention) for 4 weeks and followed them up for 12 weeks. The relative fecal abundances of the bifidobacteria in the formula and the CRC-related markers (Fn, m3, Ch, and Bc) were quantitated by qPCR before and after the intervention, and the combined CRC risk score (4Bac; Fn, m3, Ch, and Bc) was evaluated. Subjects with probiotics intervention showed significantly increased abundances of the bifidobacteria from week 2 to week 5 compared to baseline (p < 0.05), and the abundances dropped to baseline levels after the cessation of the intervention. There were significant decreases in the levels of CRC-related markers (Fn and m3) and the CRC risk score (4Bac) from week 2 to week 12 compared to baseline levels (p < 0.05) in the intervention group but not in the control group. A novel probiotic formula containing B. adolescentis, B. longum, and B. bifidum was effective in inhibiting the growth of F. nucleatum in vitro and improving the gut microbial environment against CRC development.
Keywords: colorectal cancer, probiotics, Fusobacterium nucleatum, Bifidobacterium
1. Introduction
Gut microbiota dysbiosis has been associated with colorectal tumorigenesis [1,2,3,4]. Specifically, some pathogenic bacteria, such as Fusobacterium nucleatum, have been shown to play important roles in CRC development [5,6]. Therefore, it is anticipated that reducing or eliminating pathogenic bacteria associated with CRC can reduce the risk of CRC development. Probiotics can exert beneficial effects on gut microbiota and help maintain homeostasis [7]. Probiotics can directly influence colonization of microbes via the production of inhibitory compounds (bacteriocins, short-chain fatty acids, etc.) and substrates that might nourish other microbes (secreted exopolysaccharides, vitamins, etc.). Probiotics can also indirectly modulate microbiota by affecting the host immune system and intestinal barrier integrity [7]. The anti-tumor effects of probiotics against CRC have been reported. In vitro studies have demonstrated that the co-culturing of certain probiotic strains inhibit the proliferation of and induce apoptosis in colon cancer cells [8,9]. In vivo studies have shown the effectiveness of probiotics in reducing cancer incidence or suppressing tumor growth in carcinogen-treated animals [10,11]. In human studies, probiotics have been mainly used as an adjuvant treatment during chemotherapy, but fewer have been used in prevention due to study difficulties. They have also shown to be associated with a reduced risk of post-operative complications. An earlier prospective study from Italy showed that subjects with an increased consumption of yogurt for 12 years had lower CRC incidence, suggesting that microbiota modulation via diet may play a role in CRC prevention, although the mechanisms supporting this benefit have not been elucidated [12].
However, it is unknown whether probiotic bacteria inhibit CRC development via the amelioration of gut microbiota dysbiosis. Whether the pathogenic bacteria involved in CRC development can be inhibited by specific probiotic bacteria has not been investigated, and nor have their mechanisms of action. We hypothesize that bacteria associated with CRC development can be altered by microbiota modulation via a targeted probiotic formula. By using metagenomics analysis to compare the fecal microbiome of CRC patients and healthy subjects, we identified 20 bacterial gene markers for the non-invasive diagnosis of CRC, 8 of which were enriched in CRC patients (8Up), while the other 12 were decreased in CRC patients (12Down) [1]. Using targeted quantification by quantitative PCR (qPCR), we further demonstrated that a panel of four bacterial markers (4Bac), composed of Fusobacterium nucleatum (Fn), Bacteroides clarus (Bc), Clostridium hathewayi (Ch), and Lachnoclostridium sp. (m3), showed good diagnostic performance for colorectal adenoma (including in the non-advanced stage) and CRC [13]. These findings pave the way for assessing CRC risk based on bacterial markers using qPCR or metagenome sequencing. In this study, we first developed a probiotic formula based on the metagenome sequencing data of a CRC cohort to identify probiotic species that inversely correlate with our previously identified bacterial markers for CRC risk. We then investigated the effects of a formula containing the identified probiotic species in reducing pathogenic bacteria of CRC in human subjects.
2. Materials and Methods
2.1. Metagenomics Dataset
We analyzed fecal metagenomic sequencing data from 589 Hong Kong Chinese subjects (184 CRC, 185 adenoma, and 220 control subjects) consisting of a discovery cohort of 74 subjects with CRC and 54 controls for the identification of the 20 CRC-related bacterial gene markers [1,14]. This study has been approved by The Joint Chinese University of Hong Kong, New Territories East Cluster Clinical Research Ethics Committee (The Joint CUHK-NTEC CREC, CREC Ref. No: 2021.126). Written informed consent was obtained from all subjects. Abundances of the 20 gene markers were analyzed as described in our previous study [13]. Relative abundances of species were analyzed by MetaPhlAn3 [15].
2.2. Design of a Probiotic Formula against CRC-Associated Bacteria
Firstly, Spearman correlation was analyzed between abundances of all detected probiotics (22 species detected in our cohort as listed in Figure 1A) and disease development from normal to adenoma and further to CRC to identify probiotic species that significantly decreased with disease development. To identify probiotic species that potentially modulate CRC-related bacteria, we analyzed correlations in the abundances between the probiotic species and 20 CRC-related bacterial gene markers previously identified by our team [1]. The combined scores of the 8 markers increased in CRC (8Up) and the 12 markers decreased in CRC (12Down) were included in correlation analysis. We also included the four individual gene markers that were targeted by our qPCR test for non-invasive diagnosis of CRC and adenoma, mapping to Fusobacterium nucleatum (Fn), Lachnoclostridium sp. m3, Clostridium hathewayi (Ch), and Bacteroides clarus (Bc) [13]. Probiotic species inversely correlated with markers enriched in CRC or positively correlated with markers decreased in CRC were selected. Finally, three Bifidobacterium species that significantly decreased with CRC development and significantly correlated with CRC-related markers were included in the probiotic formula, including B. adolescentis, B. longum, and B. bifidum. For clinical trial, we also included three specific prebiotics (Galactooligosaccharides, xylooligosaccharide, and resistant dextrin) in the formula to stimulate the favorable growth and/or enhance activities of the probiotic bacteria [16,17,18,19]. Capsules containing the three bifidobacteria (25 billion CFU per capsule) and prebiotics were prepared as described in our previous study [20].
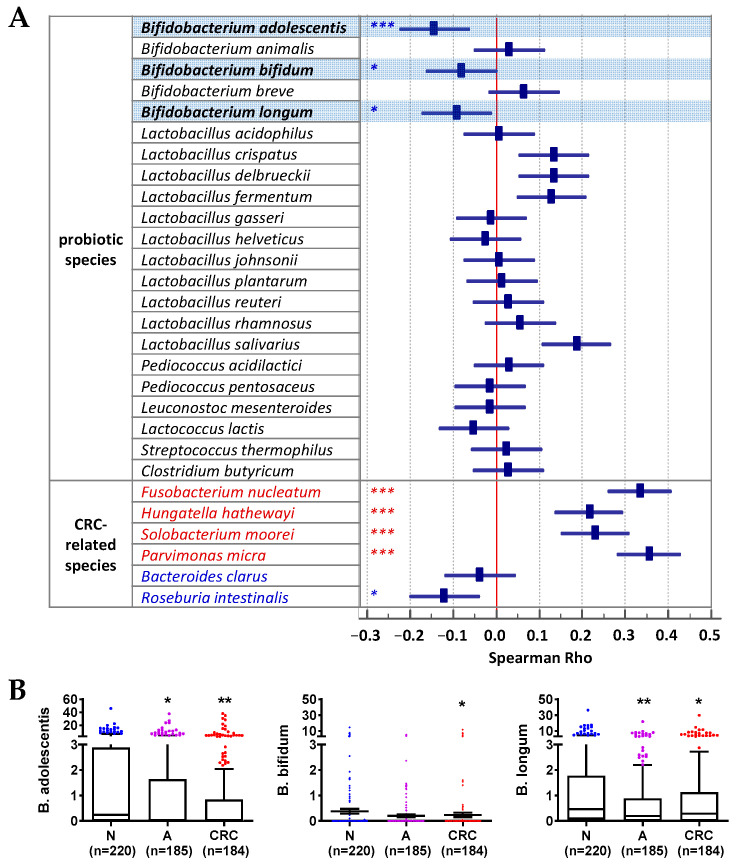
Figure 1.
Correlations of probiotics with colorectal neoplasm development. (A) Spearman correlation of detected probiotic species and CRC-related species with colorectal neoplasm development. * p < 0.05, *** p < 0.0001; blue denotes decrease, and red denotes increase. (B) Relative abundances of species of interest in fecal samples from normal subjects (N), patients with adenoma (A), and CRC. * p < 0.05, ** p < 0.001 vs. N.
2.3. Bacterial Strains and Culture Conditions
The bifidobacterium strains (B. adolescentis DSM 18351, B. longum DSM 16603, and B. bifidum DSM 22892) were obtained from Probiotical (Novara, Italy). Fn (ATCC 25586) and Enterocloster aldenensis (ATCC BAA 1318) were obtained from American Type Culture Collection (Manassas, VA, USA). All the bacteria were cultured in RCM medium (Sigma-Aldrich, St. Louis, MO, USA) under anaerobic condition at 37 °C.
2.4. Bacterial Co-Culture Assay
Co-culture experiments were performed to investigate whether the selected bifidobacteria inhibited the growth of Fn. B. adolescentis, B. longum, and B. bifidum, either individually or in combination, were co-cultured with Fn, with E. aldenensis (an anaerobic species that has shown no correlation with Fn) used as control treatment. Specifically, five co-culture groups in final volumes of 20 mL were analyzed: (1) B. adolescentis (109 CFU) plus Fn (109 CFU), (2) B. longum (109 CFU) plus Fn (109 CFU), (3) B. bifidum (109 CFU) plus Fn (109 CFU), (4) probiotics combination (109 CFU) plus Fn (109 CFU), and (5) E. aldenensis (109 CFU) plus Fn (109 CFU). The ratio of B. adolescentis to B. longum to B. bifidum is 4.6:2.7:2.7. Each treatment was repeated in triplicate. All treatment/control groups started with the same copies of Fn in the same volumes (20 mL) and were cultured under anaerobic conditions at 37 °C for 12 h. After that, the culture media containing bacteria were mixed fully, and 10 μL of each were diluted with 90 μL ultrapure water (total 100 μL) and boiled at 100 °C for 30 min for bacterial DNA extraction. Then, 2 μL of each of the 100 μL DNA extracts were used as templates in each qPCR reaction. A set of primers and a probe carrying a 5′ reporter dye FAM (6-carboxy fluorescein) specifically targeting Fn, as listed in Table S1, was used. qPCR amplifications were performed in a 20 µL reaction system of TaqMan Universal Master Mix II (Applied Biosystems) containing 0.3 µM of each primer and 0.2 µM of the probe in MicroAmp fast optical 96-well reaction plates (Applied Biosystems, Waltham, MA, USA) with adhesive sealing. Thermal cycler parameters were 95 °C for 10 min and (95 °C 15 s, 60 °C 1 min) × 45 cycles on an ABI QuantStudio sequence detection system. The growth of Fn was then assessed as the relative abundance of Fn in each sample (equivalent to equal volumes of cultures) using ∆Cq method compared to the control group (Power (2, −(Cqtreatment/control − Cqcontrol))) and shown as percentages in reference to control treatment.
2.5. Pilot Clinical Study
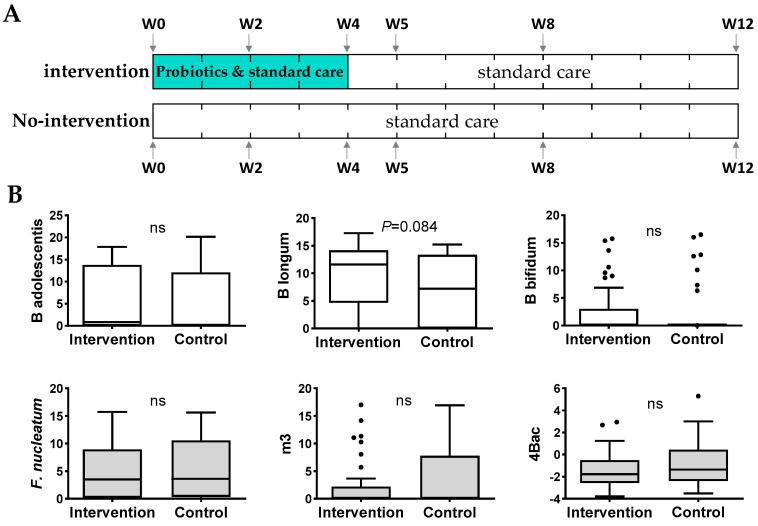
We have previously conducted a pilot clinical trial in which intervention using the probiotics formula containing B. adolescentis, B. longum, and B. bifidum was included [20]. Here, we made use of these samples to test the effects of this probiotics formula on modulating the abundances of CRC-related bacteria. This was an open-label study of consecutive hospitalized patients with COVID-19 in a tertiary referral center in Hong Kong. Patients were excluded if they were below the age of 18, on mechanical ventilation, admitted to intensive care unit, on peritoneal dialysis or haemodialysis, immunocompromised, had an active or known history of infective endocarditis, pregnant, or had a history of suspected intolerance to the probiotic formula or its components. The latter was defined as any condition such as allergic reaction or any discomfort that rendered the subject not suitable to participate in this study. A designated pharmacist was responsible for dispensing the study capsules. Forty patients (age: 50.4 ± 12.3y (mean ± SD); 24 males) received two doses of the probiotic formula (100 billion CFU) per day, while 32 patients (age: 50.3 ± 15.1y; 15 males) in the control group received no probiotic intervention. All the subjects were recruited consecutively, and the baseline stool samples were collected before probiotic administration. There were no significant differences in the age (p = 0.99) and sex (p = 0.24 by Fisher’s exact test) distributions between the two groups. All patients took study capsules together with standard meals for 4 weeks. Both intervention group and no-intervention group received the same treatment protocol for COVID-19 endorsed by the local health authority. This study was approved by the Clinical Research Ethics Committees (2020.407) and was registered in the Clinical Trials Registry (NCT04581018). Written informed consent was obtained from all patients. Stool samples were collected at baseline (week 0), week 2, week 4, week 5, week 8, and week 12 from all participants in both intervention group and no-intervention group. Stools were collected in Norgen’s Stool Preservative (Norgen Biotek Corp, Thorold, ON, Canada) and stored in a −80 °C freezer within 24 h until further analysis.
2.6. DNA Extraction and Quantification of Microbial Markers by Duplex qPCR
Fecal DNA was extracted using Maxwell RSC PureFood GMO and Authentication Kit (Promega, Madison, WI, USA) following manufacturer’s instructions. Fecal levels of four microbial DNA markers for CRC (Fn, m3, Bc, and Ch) and the three biofidobacteria (B. adolescentis, B. longum, and B. bifidum) were quantified by qPCR. Primer and probe sequences targeting the markers and 16s rDNA internal control are listed in Table S1. Each probe carried a 5′ reporter dye FAM (6-carboxy fluorescein) or VIC (4,7,2′-trichloro-7′-phenyl-6-carboxyfluorescein) and a 3′ quencher dye TAMRA (6-carboxytetramethyl-rhodamine). Primers and hydrolysis probes were synthesized by Invitrogen (Carlsbad, CA, USA). qPCR amplifications were performed on an ABI QuantStudio sequence detection system as previously described, with thermal cycler parameters of 95 °C for 10 min and (95 °C 15 s, 60 °C 1 min) × 45 cycles [21]. Positive controls of the markers and a negative control (H2O as template) were included within every experiment. Measurements were performed in triplicates for each sample. Relative level of each marker was calculated using ∆Cq method, compared to internal control (Power (2, −(Cqtarget − Cqcontrol))), and shown as log value of ‘*10e6+1′.
2.7. Scoring Algorithms
Among the 20 CRC-associated gene markers, 8 were increased and 12 were decreased in CRC patients compared to control subjects. The 8Up score was calculated as Log10 [Sum(8 increased markers) +*1e-20]. The 12Down score was calculated as Log10 [Sum(12 decreased markers) +*1e-20]. The combined score of four microbial markers (4Bac) using a logistic regression model (4Bac score = 0.23162 × Fn + 0.13451 × m3 − 0.10075 × Bc + 0.32841 × Ch − 2.73836) was determined in our previous study [13].
2.8. Statistical Analysis
Values were all expressed as mean ± SD or median (interquartile range (IQR)) as appropriate. The differences in bacterial levels between two groups were determined by Mann–Whitney U test or paired t-test. Correlations between bacterial species, abundances/gene marker, and abundances/scores were conducted by Pearson correlation analysis. Correlations between bacterial species abundances and colorectal neoplasm stage were analyzed by Spearman’s correlation analysis. The changes in markers across different time points were analyzed by one-way ANOVA multiple comparison for linear trend where appropriate. All tests were conducted by GraphPad Prism 9.4.1 (GraphPad Software Inc., San Diego, CA, USA) or MedCalc® Statistical Software version 19.6 (MedCalc Software Ltd., Ostend, Belgium; https://www.medcalc.org; accessed on 20 December 2020). p < 0.05 was considered statistical significance.
3. Results
3.1. Identification of Bacterial Species Inversely Correlated with CRC Development from Human Metagenomic Datasets
Based on the Spearman correlation analysis of the probiotic species detected in our cohort (22 species detected), we identified three bifidobacterial species, including B. adolescentis (rho = −0.144; p < 0.001), B. longum (rho = −0.092; p < 0.05), and B. bifidum (rho = −0.081; p < 0.05), that inversely correlated with disease development from the control samples to the CRC samples (Figure 1A). On the other hand, other probiotics showed no significant trends of change, and Lactobacillus salivarius showed an increasing trend from the normal subjects to the CRC patients (p < 0.0001) (Figure 1A). Bacteria known to be associated with CRC development, such as F. nucleatum (Fn) and Clostridium hathewayi (Ch), were significantly increased in stool samples from the normal group to the CRC group, while beneficial species, such as Roseburia intestinalis and Bacteroides clarus (Bc), showed decreasing trends (Figure 1A). Importantly, the abundances of B. adolescentis, B. longum, and B. bifidum in stool were significantly decreased in subjects with CRC compared to those of the controls, and B. adolescentis and B. longum were also significantly decreased in subjects with adenoma compared to the controls (all p < 0.05; Figure 1B).
3.2. Identification of Probiotic Species with Inverse Correlation with CRC-Associated Microbial Risk
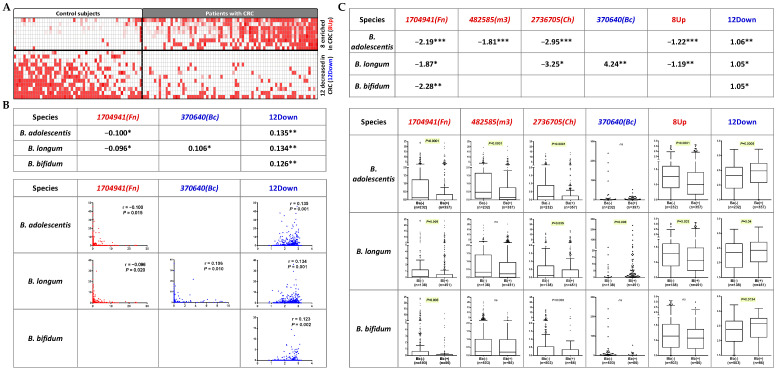
The correlations with CRC markers also identified the same three bifidobacterial species (B. adolescentis, B. longum, and B. bifidum) as being significantly associated with one or more of the “pathogenic” CRC-related bacterial gene markers (Fn, Ch, m3, and 8Up) and of the “beneficial” markers (Bc and 12Down) (Figure 2A). B. adolescentis and B. longum were inversely correlated with Fn (Pearson’s r of −0.1 and −0.096, respectively, both p < 0.05). B. longum was positively correlated with Bc (r = 0.106, p = 0.01). B. adolescentis, B. longum, and B. bifidum were positively correlated with 12Down (Pearson’s r of 0.135, 0.134 and 0.126, respectively, all p ≤ 0.002) (Figure 2B). We further checked the fold changes of the CRC gene markers with the presence of the bifidobacteria compared to the samples without the corresponding bifidobacteria. The presence of each of the three bifidobacterial species was significantly associated with a decrease in at least one of the “pathogenic” markers (Fn, Ch, m3, and 8Up) and an increase in at least one of the “beneficial” markers (Bc and 12Down) (Figure 2C). These data support the potential effect of B. adolescentis, B. longum, and B. bifidum in CRC prevention based on the inverse correlation with CRC-associated microbial risk markers.
Figure 2.
Correlations of probiotics species and CRC-related bacterial gene markers. (A) Heatmap illustration of the 20 bacterial gene markers identified by our previous study, with 8 significantly enriched and 12 significantly decreased in CRC patients compared to in healthy subjects. White color denotes relative abundance of 0. (B) Three bifidobacterial species (B. adolescentis, B. longum, and B. bifidum) inversely correlate with the pathogenic markers (shown in red) or positively correlate with the beneficial markers (shown in blue). (C) Fold changes of the CRC-associated gene makers in samples negative in B. adolescentis, B. longum, or B. bifidum compared to those positive in the corresponding bifidobacteria. * p < 0.05; ** p < 0.001; *** p < 0.0001; ns, not significant.
3.3. B. adolescentis, B. longum, and B. bifidum Suppressed the Growth of F. nucleatum In Vitro
We next evaluated whether B. adolescentis, B. longum, and B. bifidum can inhibit the growth of Fn. The co-culturing of the individual bifidobacteria and the combined three bifidobacteria with Fn led to an inhibition of the growth of Fn compared to the control treatment (26~70% inhibition; all p < 0.0001). The combination of the three bifidobacteria showed a significantly greater inhibitory effect on Fn growth (70% inhibition) than the individual bifidobacteria (B. adolescentis 65%, B. longum 26%, and B. bifidum 62%; all p < 0.05) (Figure 3), which is also greater than the sum of the proportions of the individual effects (53.7%). These data support the synergistic effect of the combination of three bifidobacterial species on suppressing the growth of Fn.
Figure 3.
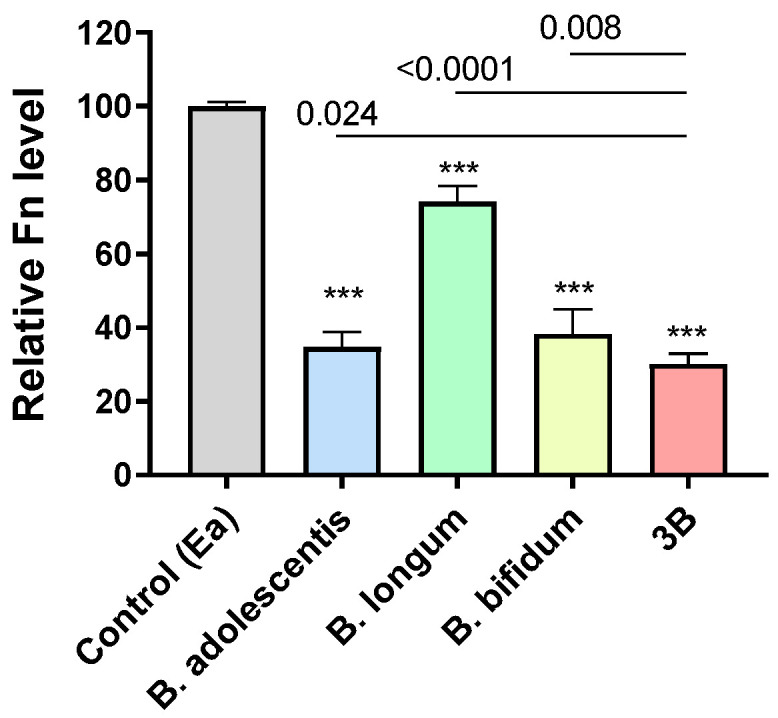
In vitro co-culturing with bifidobacteria significantly inhibited the growth of F. nucleatum (Fn) compared to the control treatment by E. aldenensis (Ea). 3B, combination of B. adolescentis, B. longum, and B. bifidum. Dosages of all treatments: Fn = 1:1. *** p < 0.0001 vs. control.
3.4. Intervention with B. adolescentis, B. longum, and B. bifidum Significantly Reduced Microbial Risk for CRC
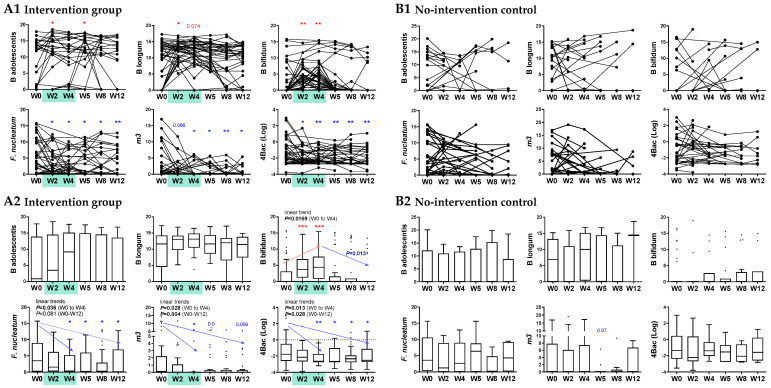
We further evaluated the changes of the CRC-associated microbial markers (Fn, m3, Ch, and Bc, as well as their combined score 4Bac) in human subjects after taking a high dose of the probiotic formula, supplemented with prebiotics, for four weeks compared to the no-intervention control subjects (Figure 4A). At baseline, there was no significant difference in the relative abundances of the three Bifidobacterium species or CRC-markers (Fn, m3, and the combined 4Bac score) in fecal samples between the intervention and control groups (Figure 4B). In the intervention group, the relative abundances of B. adolescentis, B. longum, and B. bifidum significantly increased at week 2 and week 4/5 by a pairwise comparison to baseline levels (p < 0.05), and they decreased after stopping probiotics intake and showed no significant difference compared to baseline at weeks 8 and 12 (Figure 5A1). The abundances of Fn and m3 and the 4Bac score were significantly decreased compared to baseline at almost all time points (except m3 at week 2) from week 2 to week 12 (2 months after stopping probiotics intake) (all p < 0.05 by paired t-test; Figure 5A1). By observing the trends of change, we found increasing trends in the three Bifidobacterium species from baseline to week 4, although only the increase in B. bifidum was significant (linear trend p < 0.05). The abundances of the three bifidobacteria dropped after stopping the probiotics intake, and the decrease in B. bifidum from week 4 to week 12 was significant (p < 0.05) (Figure 5A2). Fn, m3, and 4Bac dropped significantly from baseline to week 4 or from baseline to week 12 (linear trends all p < 0.05 except for Fn (week 0–12); Figure 5A2). In the no-intervention controls, no significant changes in the three bifidobacterial species or the CRC-markers were observed from week 2 to week 12 compared to baseline (Figure 5B1,B2).
Figure 4.
(A) Flow diagram of the pilot clinical study. The recruited patients in the intervention group took probiotic capsules daily from baseline (week 0, W0) to W4. Stools were collected from both groups of patients at W0, W2, W4, W5, W8, and W12. All patients in both intervention and no-intervention groups received the same treatment protocol for COVID-19 endorsed by the local health authority. (B) Baseline levels of B. adolescentis, B. longum, B. bifidum, and the CRC-related microbial markers showed no significant difference between the intervention and no-intervention groups. ns, not significant.
Figure 5.
Intervention with the probiotic formula containing B. adolescentis, B. longum, and B. bifidum reduced microbial risk for CRC. (A) Changes in B. adolescentis, B. longum, and B. bifidum and the CRC-related microbial markers were detected in the intervention group by paired t-test vs. W0 (A1) and Mann–Whitney U test vs. W0 (A2). (B) No significant changes in B. adolescentis, B. longum, B. bifidum, and the CRC-related microbial markers were found in the no-intervention group by paired t-test vs. W0 (B1) or Mann–Whitney U test vs. W0 (B2). * p < 0.05, ** p < 0.001, *** p < 0.0001; red denotes increase and blue denotes decrease. Changes are not significant if not indicated.
4. Discussion
This is the first study providing evidence for a probiotic formula in reducing CRC-related microbial risk, thus implying the potential for CRC prevention. This probiotic formula, involving three bifidobacteria (B. adolescentis, B. longum, and B. bifidum), was established based on the identification of probiotic species that decreased significantly with colorectal neoplasia progression and was inversely correlated with CRC-enriched markers including Fn. We showed the effects of the selected bifidobacteria on suppressing the growth of Fn in vitro. Moreover, our clinical intervention trial further demonstrated the usefulness of the probiotic formula in reducing the microbial risk for CRC development as indicated by our previously devised test for CRC risk assessment.
The anti-tumor effects of B. adolescentis, B. longum, and B. bifidum have been reported previously but only individually and mainly in in vitro and animal studies. Whole cells, cell extracts and cell-free supernatants of B. adolescentis have been shown to inhibit the proliferation of colon cancer cells in vitro [22,23]. B. bifidum has been shown to exert a cytotoxicity effect on colon cancer cells [24]. The administration of B. bifidum attenuated tumorigenesis in azoxymethane (AOM)/dextran sulphate sodium (DSS)-induced colitis-associated colon cancer in mice via modulating gut microbiota and metabolome [25]. B. longum has also been shown to reduce inflammation and tumor incidence in AOM/DSS-induced colon cancer in mice [26]. The combination of five bifidobacterium strains (1 B. longum strain, 2 B. bifidum, and 2 B. breve strains) has been shown to induce the apoptosis of colon cancer cells and significantly reduce the incidence and inhibit the progression of tumors in CRC mouse models [27]. However, these findings mainly focused on the effects of the probiotics on suppressing the growth of cancer cells in vitro or tumors in carcinogen-treated mice, which seems more for therapeutics.
On the other hand, here, we showed the effect of our probiotic formula on reducing levels of CRC-associated bacteria, which is indicative of the preventative potential against CRC development. Clinical results (Figure 1 and Figure 2) showed opposite trends of change from normal controls via adenoma to CRC or inverse correlations between the bifidobacteria and Fn, and B. adolescentis and B. longum were significantly decreased in adenoma patients compared to the control subjects. These data demonstrate that the bifidobacteria decreased at an early stage of CRC development, and their decrease may provide a more suitable intestinal environment for the growth of Fn. These data are in line with the in vitro (Figure 3) data, which showed the effects of the selected bifidobacteria on suppressing the growth of Fn in vitro. Abnormality in the composition of the gut microbiota has been implicated as a potentially important etiological factor in the initiation and progression of CRC [28]. With the widespread application of metagenomic analyses in the investigation of gut microbiota, an increasing number of bacteria have been identified to be positively associated with CRC [1,2,3,4]. Recent basic research has established a critical function for the dysbiotic microbiota [29] and specific bacterial species, such as Fn and Ch in promoting colorectal tumorigenesis. It has been well demonstrated that Fn induces inflammation and modulates the host immune response to promote tumor development [5,6]. Ch has been shown to promote colonic epithelial cell proliferation in mouse models [30]. The Lachnoclostridium sp. m3 is a novel pathogenic species that potentially contributes to colorectal adenoma and cancer development [13]. Bc was found to be depleted in CRC patients in our previous study [21]. Although the role of Bc remains largely unexplored, some species of Bacteroides are considered to be the next generation of probiotics [31]. Therefore, the decreased levels of Fn, Ch, and m3 and the increased level of Bc, contributing to a decreased combined score of 4Bac, reflect a decreased microbial risk for developing CRC. With the significantly reduced levels of 4Bac, the effect of our probiotic formula on reducing microbial risk for CRC was well demonstrated in this study. Probiotics suppress the growth of pathogens by the production of inhibitory compounds, such as bacteriocins and organic acids [32], and the competition of colonization sites with pathogens [33]. The mechanisms by which the three bifidobacteria suppressed the growth of Fn, as well as other CRC-associated bacteria, warrant further investigation.
The probiotic formula used in the clinical trial was supplemented with prebiotics, which may also contribute to the modulation of gut microbiota. However, as the dosage of prebiotics was much smaller than the daily intake of dietary fiber of the study subjects, its influence on the gut microbiota would have been limited. The bifidobacteria in the intervention group detected by qPCR at week 2 to 4/5 were probably the transient passages and the probiotics failed to colonize the colon. While most probiotics are transient in nature, many other factors influence the probiotic viability and mucoadhesive properties [34]. Therefore, strategies to improve the colonization of probiotics, or determining the appropriate time and dosage of probiotic supplementation, are important for future clinical application. Regardless, according to the data from our clinical trial, although the bifidobacteria cannot colonize in the subjects’ colons and resume to baseline level within one month of the discontinuance of probiotics intake, the effect on the reduced CRC markers can last for at least another eight weeks after the daily intake of the probiotic formula, supplemented with prebiotics, for four weeks. As the clinical trial involved patients who already had gut dysbiosis, the effectiveness of the probiotic formula on reducing CRC-associated bacteria and modulating CRC-associated microbiome function needs to be further verified or investigated by future clinical trials involving subjects representative of the general population.
Different strains of the same probiotic species may show differential properties, such as B. adolescentis of various origins [35]. B. adolescentis isolated from the feces of a new-born increased the body weight of mice, while those isolated from elderly humans significantly decreased body weight and increased serum leptin concentrations and the relative abundances of potentially beneficial genera (e.g., Bacteroides, Parabacteroides, and Faecalibaculum). The strains of the B. adolescentis, B. longum, and B. bifidum that were present in the subjects of our metagenome sequencing cohort might be different from the probiotic strains used in the in vitro experiments and the pilot clinical trial, which were originally isolated in non-Chinese populations. In addition to characterizing strain-specific functions, whether the strains isolated from a certain population would be more efficient at colonizing this population deserves further investigation.
Future studies are warranted to further verify the effects of the identified bifidobacterial species and the probiotic formula. In vitro assays are needed to test their inhibiting effects on the growth of other pathogenic bacteria for CRC. The suppressive effects and mechanisms of the identified bifidobacteria against the growth-promoting effects of Fn, Ch, and m3 on colon cancer cells should be investigated. In vivo studies are also warranted to assess the formula in preventing/suppressing CRC development. Most importantly, a long-term prospective clinical study is warranted to demonstrate the CRC prevention effect of the formula.
In summary, a novel probiotic formula consisting of B. adolescentis, B. longum, and B. bifidum was effective in inhibiting the growth of F. nucleatum and improved the gut microbial environment against CRC development. The new probiotic formula composed of food grade components will be suitable to be taken by average risk adults to reduce microbial risk for CRC development.
Acknowledgments
The authors acknowledge the technical support of Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cells12091244/s1. Table S1: Nucleotide sequences of primers and probes used in this study.
Author Contributions
Conceptualization, J.Q.L.; methodology, J.Q.L.; validation, Y.Z., E.Y.T.L., Y.S., Y.H. and T.Z.; formal analysis, J.Q.L. and Y.Z.; investigation, J.Q.L., Y.Z., E.Y.T.L., Y.S., T.Z. and Z.X.; resources, J.Y., S.C.N. and F.K.L.C.; data curation, J.Q.L. and Y.Z.; writing—original draft preparation, J.Q.L.; writing—review and editing, S.C.N. and F.K.L.C.; visualization, J.Q.L. and Y.Z.; supervision, J.Q.L., S.C.N. and F.K.L.C.; project administration, J.Q.L.; funding acquisition, J.Q.L. All authors have read and agreed to the published version of the manuscript.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. This study was approved by The Joint Chinese University of Hong Kong, New Territories East Cluster Clinical Research Ethics Committee (The Joint CUHK-NTEC CREC, CREC Ref. Nos: 2020.407 and 2021.126) and was registered in the Clinical Trials Registry (NCT04581018).
Conflicts of Interest
I have read the journal’s policy and the authors of this manuscript have the following competing interest: F.K.L.C. and S.C.N. are the scientific co-founders and sit on the board of Directors of GenieBiome Ltd. S.C.N. has served as an advisory board member for Pfizer, Ferring, Janssen, and Abbvie and as a speaker for Ferring, Tillotts, Menarini, Janssen, Abbvie, and Takeda. She has received research grants from Olympus, Ferring, and Abbvie. F.K.L.C. has served as an advisor and lecture speaker for Eisai Co., Ltd., AstraZeneca, Pfizer Inc., Takeda Pharmaceutical Co. and Takeda (China) Holdings Co., Ltd., J.Q.L. and Z.X. are part-time employees of GenieBiome Ltd. J.Q.L., S.C.N. and F.K.L.C. are named inventors of patent applications held by the CUHK in relation to this work. All other co-authors have no conflict of interest.
Funding Statement
J.Q.L. received funding for this work, grant number MRP/058/20, by the Midstream Research Programme for Universities, ITF, Hong Kong. https://www.itf.gov.hk/en/itcfas/ (accessed on 1 April 2021). SCN is supported by the Croucher Senior Medical Research Fellowship. https://croucher.org.hk/funding/recognising-outstanding-hong-kong-scientists-mid-career/croucher-senior-medical-research-fellowships (accessed on 1 January 2023). YS received additional support from the Hong Kong Ph.D. Fellowship Scheme (HKPFS). https://cerg1.ugc.edu.hk/hkpfs/index.html (accessed on 1 August 2021). The funders had no role in the study design, the data collection and analysis, the decision to publish, or the preparation of the manuscript.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Yu J., Feng Q., Wong S.H., Zhang D., Liang Q.Y., Qin Y., Tang L., Zhao H., Stenvang J., Li Y., et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2015;66:70–78. doi: 10.1136/gutjnl-2015-309800. [DOI] [PubMed] [Google Scholar]
- 2.Nakatsu G., Li X., Zhou H., Sheng J., Wong S.H., Wu W.K.K., Ng S.C., Tsoi H., Dong Y., Zhang N., et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat. Commun. 2015;6:8727. doi: 10.1038/ncomms9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dai Z., Coker O.O., Nakatsu G., Wu W.K.K., Zhao L., Chen Z., Chan F.K.L., Kristiansen K., Sung J.J.Y., Wong S.H., et al. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome. 2018;6:70. doi: 10.1186/s40168-018-0451-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tilg H., Adolph T.E., Gerner R.R., Moschen A.R. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell. 2018;33:954–964. doi: 10.1016/j.ccell.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Kostic A.D., Chun E., Robertson L., Glickman J.N., Gallini C.A., Michaud M., Clancy T.E., Chung D.C., Lochhead P., Hold G.L., et al. Fusobacterium nucleatum Potentiates Intestinal Tumorigenesis and Modulates the Tumor-Immune Microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rubinstein M.R., Wang X., Liu W., Hao Y., Cai G., Han Y.W. Fusobacterium nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/β-Catenin Signaling via its FadA Adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slizewska K., Markowiak-Kopec P., Slizewska W. The Role of Probiotics in Cancer Prevention. Cancers. 2020;13:20. doi: 10.3390/cancers13010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han K.J., Lee N.-K., Park H., Paik H.-D. Anticancer and Anti-Inflammatory Activity of Probiotic Lactococcus lactis NK34. J. Microbiol. Biotechnol. 2015;25:1697–1701. doi: 10.4014/jmb.1503.03033. [DOI] [PubMed] [Google Scholar]
- 9.Tiptiri-Kourpeti A., Spyridopoulou K., Santarmaki V., Aindelis G., Tompoulidou E., Lamprianidou E.E., Saxami G., Ypsilantis P., Lampri E.S., Simopoulos C., et al. Lactobacillus casei Exerts Anti-Proliferative Effects Accompanied by Apoptotic Cell Death and Up-Regulation of TRAIL in Colon Carcinoma Cells. PLoS ONE. 2016;11:e0147960. doi: 10.1371/journal.pone.0147960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gosai V., Ambalam P., Raman M., Kothari C., Kothari R., Vyas B., Sheth N.R. Protective effect of Lactobacillus rhamnosus 231 against N-Methyl-N′-nitro-N-nitrosoguanidine in animal model. Gut Microbes. 2011;2:319–325. doi: 10.4161/gmic.18755. [DOI] [PubMed] [Google Scholar]
- 11.Zhang M., Fan X., Fang B., Zhu C., Zhu J., Ren F. Effects of Lactobacillus salivarius Ren on cancer prevention and intestinal microbiota in 1, 2-dimethylhydrazine-induced rat model. J. Microbiol. 2015;53:398–405. doi: 10.1007/s12275-015-5046-z. [DOI] [PubMed] [Google Scholar]
- 12.Pala V., Sieri S., Berrino F., Vineis P., Sacerdote C., Palli D., Masala G., Panico S., Mattiello A., Tumino R., et al. Yogurt consumption and risk of colorectal cancer in the Italian European prospective investigation into cancer and nutrition cohort. Int. J. Cancer. 2011;129:2712–2719. doi: 10.1002/ijc.26193. [DOI] [PubMed] [Google Scholar]
- 13.Liang J.Q., Li T., Nakatsu G., Chen Y.-X., Yau T.O., Chu E., Wong S., Szeto C.H., Ng S.C., Chan F.K.L., et al. A novel faecal Lachnoclostridium marker for the non-invasive diagnosis of colorectal adenoma and cancer. Gut. 2019;69:1248–1257. doi: 10.1136/gutjnl-2019-318532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakatsu G., Zhou H., Wu W.K., Wong S.H., Coker O.O., Dai Z., Li X., Szeto C.H., Sugimura N., Lam T.Y.-T., et al. Alterations in Enteric Virome Are Associated With Colorectal Cancer and Survival Outcomes. Gastroenterology. 2018;155:529–541.e5. doi: 10.1053/j.gastro.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 15.Beghini F., McIver L.J., Blanco-Míguez A., Dubois L., Asnicar F., Maharjan S., Mailyan A., Manghi P., Scholz M., Thomas A.M., et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. eLife. 2021;10 doi: 10.7554/eLife.65088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amaretti A., Bernardi T., Leonardi A., Raimondi S., Zanoni S., Rossi M. Fermentation of xylo-oligosaccharides by Bifidobacterium adolescentis DSMZ 18350: Kinetics, metabolism, and β-xylosidase activities. Appl. Microbiol. Biotechnol. 2012;97:3109–3117. doi: 10.1007/s00253-012-4509-y. [DOI] [PubMed] [Google Scholar]
- 17.Van Laere K.M.J., Hartemink R., Bosveld M., Schols H.A., Voragen A.G.J. Fermentation of Plant Cell Wall Derived Polysaccharides and Their Corresponding Oligosaccharides by Intestinal Bacteria. J. Agric. Food Chem. 2000;48:1644–1652. doi: 10.1021/jf990519i. [DOI] [PubMed] [Google Scholar]
- 18.Wronkowska M., Soral-Śmietana M., Biedrzycka E. Utilization of resistant starch of native tapioca, corn and waxy corn starches and their retrograded preparations by Bifidobacterium. Int. J. Food Sci. Nutr. 2008;59:80–87. doi: 10.1080/09637480701663862. [DOI] [PubMed] [Google Scholar]
- 19.Garrido D., Ruiz-Moyano S., Jimenez-Espinoza R., Eom H.-J., Block D.E., Mills D.A. Utilization of galactooligosaccharides by Bifidobacterium longum subsp. infantis isolates. Food Microbiol. 2013;33:262–270. doi: 10.1016/j.fm.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L., Xu Z., Mak J.W., Chow K.M., Lui G., Li T.C., Wong C.K., Chan P.K., Ching J.Y., Fujiwara Y., et al. Gut microbiota-derived synbiotic formula (SIM01) as a novel adjuvant therapy for COVID-19: An open-label pilot study. J. Gastroenterol. Hepatol. 2022;37:823–831. doi: 10.1111/jgh.15796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang Q., Chiu J., Chen Y., Huang Y., Higashimori A., Fang J., Brim H., Ashktorab H., Ng S.C., Ng S.S.M., et al. Fecal Bacteria Act as Novel Biomarkers for Noninvasive Diagnosis of Colorectal Cancer. Clin. Cancer Res. 2017;23:2061–2070. doi: 10.1158/1078-0432.CCR-16-1599. [DOI] [PubMed] [Google Scholar]
- 22.Kim Y., Lee D., Kim D., Cho J., Yang J.W., Chung M., Kim K., Ha N. Inhibition of proliferation in colon cancer cell lines and harmful enzyme activity of colon bacteria by Bifidobacterium adolescentis SPM0212. Arch. Pharmacal Res. 2008;31:468–473. doi: 10.1007/s12272-001-1180-y. [DOI] [PubMed] [Google Scholar]
- 23.Lee D.K., Jang S., Kim M.J., Kim J.H., Chung M.J., Kim K.J., Ha N.J. Anti-proliferative effects of Bifidobacterium adolescentis SPM0212 extract on human colon cancer cell lines. BMC Cancer. 2008;8:310. doi: 10.1186/1471-2407-8-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bahmani S., Azarpira N., Moazamian E. Anti-colon cancer activity of Bifidobacterium metabolites on colon cancer cell line SW742. Turk. J. Gastroenterol. 2019;30:835–842. doi: 10.5152/tjg.2019.18451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q., Wang K., Wu W., Lv L., Bian X., Yang L., Wang Q., Li Y., Ye J., Fang D., et al. Administration of Bifidobacterium bifidum CGMCC 15068 modulates gut microbiota and metabolome in azoxymethane (AOM)/dextran sulphate sodium (DSS)-induced colitis-associated colon cancer (CAC) in mice. Appl. Microbiol. Biotechnol. 2020;104:5915–5928. doi: 10.1007/s00253-020-10621-z. [DOI] [PubMed] [Google Scholar]
- 26.Valadez-Bustos N., Escamilla-Silva E.M., García-Vázquez F.J., Gallegos-Corona M.A., Amaya-Llano S.L., Ramos-Gómez M. Oral Administration of Microencapsulated B. Longum BAA-999 and Lycopene Modulates IGF-1/IGF-1R/IGFBP3 Protein Expressions in a Colorectal Murine Model. Int. J. Mol. Sci. 2019;20:4275. doi: 10.3390/ijms20174275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parisa A., Roya G., Mahdi R., Shabnam R., Maryam E., Malihe T. Anti-cancer effects of Bifidobacterium species in colon cancer cells and a mouse model of carcinogenesis. PLoS ONE. 2020;15:e0232930. doi: 10.1371/journal.pone.0232930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irrazábal T., Belcheva A., Girardin S.E., Martin A., Philpott D.J. The Multifaceted Role of the Intestinal Microbiota in Colon Cancer. Mol. Cell. 2014;54:309–320. doi: 10.1016/j.molcel.2014.03.039. [DOI] [PubMed] [Google Scholar]
- 29.Wong S.H., Zhao L., Zhang X., Nakatsu G., Han J., Xu W., Xiao X., Kwong T.N.Y., Tsoi H., Wu W.K.K., et al. Gavage of Fecal Samples From Patients With Colorectal Cancer Promotes Intestinal Carcinogenesis in Germ-Free and Conventional Mice. Gastroenterology. 2017;153:1621–1633.e6. doi: 10.1053/j.gastro.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 30.Xia X., Wu W.K.K., Wong S.H., Liu D., Kwong T.N.Y., Nakatsu G., Yan P.S., Chuang Y.-M., Chan M.W.-Y., Coker O.O., et al. Bacteria pathogens drive host colonic epithelial cell promoter hypermethylation of tumor suppressor genes in colorectal cancer. Microbiome. 2020;8:108. doi: 10.1186/s40168-020-00847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El Hage R., Hernandez-Sanabria E., Van De Wiele T. Emerging Trends in “Smart Probiotics”: Functional Consideration for the Development of Novel Health and Industrial Applications. Front. Microbiol. 2017;8:1889. doi: 10.3389/fmicb.2017.01889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prabhurajeshwar C., Chandrakanth R.K. Probiotic potential of Lactobacilli with antagonistic activity against pathogenic strains: An in vitro validation for the production of inhibitory substances. Biomed. J. 2017;40:270–283. doi: 10.1016/j.bj.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherman P.M., Ossa J.C., Johnson-Henry K. Unraveling Mechanisms of Action of Probiotics. Nutr. Clin. Pract. 2009;24:10–14. doi: 10.1177/0884533608329231. [DOI] [PubMed] [Google Scholar]
- 34.Han S., Lu Y., Xie J., Fei Y., Zheng G., Wang Z., Liu J., Lv L., Ling Z., Berglund B., et al. Probiotic Gastrointestinal Transit and Colonization After Oral Administration: A Long Journey. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.609722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang B., Kong Q., Cui S., Li X., Gu Z., Zhao J., Zhang H., Chen W., Wang G. Bifidobacterium adolescentis Isolated from Different Hosts Modifies the Intestinal Microbiota and Displays Differential Metabolic and Immunomodulatory Properties in Mice Fed a High-Fat Diet. Nutrients. 2021;13:1017. doi: 10.3390/nu13031017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.