Abstract
Adhesin-mediated binding to extracellular matrix (ECM) proteins is thought to be a crucial step in the pathogenic process of many bacterial infections. We have previously reported conditional adherence of most Enterococcus faecalis isolates, after growth at 46°C, to ECM proteins collagen types I and IV and laminin; identified an E. faecalis-specific gene, ace, whose encoded protein has characteristics of a bacterial adhesin; and implicated Ace in binding to collagen type I. In this study, we constructed an ace disruption mutant from E. faecalis strain OG1RF that showed marked reduction in adherence to collagen types I and IV and laminin when compared to the parental OG1RF strain after growth at 46°C. Polyclonal immune serum raised against the OG1RF-derived recombinant Ace A domain reacted with a single ∼105-kDa band of mutanolysin extracts from OG1RF grown at 46°C, while no band was detected in extracts from OG1RF grown at 37°C, nor from the OG1RF ace mutant grown at 37 or 46°C. IgGs purified from the anti-Ace A immune serum inhibited adherence of 46°C-grown E. faecalis OG1RF to immobilized collagen type IV and laminin as well as collagen type I, at a concentration as low as 1 μg/ml, and also inhibited the 46°C-evoked adherence of two clinical isolates tested. We also showed in vitro interaction of collagen type IV with Ace from OG1RF mutanolysin extracts on a far-Western blot. Binding of recombinant Ace A to immobilized collagen types I and IV and laminin was demonstrated in an enzyme-linked immunosorbent assay and was shown to be concentration dependent. These results indicate that Ace A mediates the conditional binding of E. faecalis OG1RF to collagen type IV and laminin in addition to collagen type I.
Collagens, proteoglycans, and structural glycoproteins such as fibronectin and laminin (LN) are found in the extracellular matrix (ECM) of all eukaryotic tissues and are frequently exploited for colonization by microbes and initiation of infections (5, 13, 38). Collagen contains a characteristic Gly-X-Y repeating tripeptide sequence, where X and Y often are proline and hydroxyproline, respectively. Segments of the collagen polypeptides containing this repeat sequence form characteristic triple helix structures with a rope-like appearance. In mammals, collagen occurs in close to 20 genetically different types, some of which show a tissue-specific distribution. For example, collagen type IV (CIV) is found exclusively in basement membranes, whereas collagen type I (CI) has a relatively broad distribution (16). LNs, which also occur in several genetically distinct forms, are composed of three polypeptides that are partly associated to form a characteristic cross, as revealed by electron microscopy. In the long arm of the cross, the three polypeptides form a rope-like structure resembling that seen in collagen. The LNs are components of the basement membrane, where they contribute to the structural integrity of the tissue and to cell signaling (1, 2, 6, 40).
In normal tissues, most ECMs are covered by epithelial or endothelial cells and hence are not available for binding. However, any type of trauma that damages host tissues may expose the ECM and allow microbial colonization and infection. During the past decade, several microorganisms, including streptococci and staphylococci, have been shown to express surface components that recognize ECM molecules, including collagen and LN (5, 12–15, 20, 30, 31, 33–35).
Our earlier investigations on adherence of clinical isolates of Enterococcus faecalis, regardless of their source, showed that most isolates displayed conditional binding to CI, CIV, and mouse LN. The adherence phenotype was termed conditional because it was observed after growth at 46°C, but not, for most isolates, after growth at 37°C (39); in these experiments, we defined adherence as being present if >5% of total labeled cells were bound to the ECM-coated wells. We then identified a putative collagen binding gene, ace, in the E. faecalis strain V583 partial database (24), and based on structural similarities to Cna of Staphylococcus aureus, followed by some biochemical and biophysical characterization, we assigned a CI binding function to Ace (24).
In the present investigation, we constructed an E. faecalis strain OG1RF ace mutant and showed that it is deficient in adherence to CI, CIV, and LN. We also found that polyclonal anti-Ace A antibodies raised against recombinant OG1RF-derived Ace A protein inhibited adherence of wild-type OG1RF to these three ECM proteins. Using far-Western blots and solid-phase enzyme-linked immunosorbent assays (ELISAs), we confirmed in vitro Ace A binding to CI, CIV, and LN.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains and plasmids used in this study are listed in Table 1. E. faecalis strain OG1RF, a derivative of E. faecalis OG1, and E. faecalis strain V583 have been described previously (18, 25). E. faecalis strain MC02152, isolated from a patient with endocarditis, was kindly provided by J. M. Steckelberg, Mayo Clinic, Minneapolis, Minn. Escherichia coli cells were grown in Luria-Bertani (LB) broth or on LB agar with appropriate antibiotics overnight at 37°C. Enterococci were grown either in brain heart infusion (BHI) broth or agar or in Todd-Hewitt broth or agar (Difco Laboratories, Detroit, Mich.) overnight at 37°C for routine purposes and at 46°C for adherence assays. Antibiotics were used at the following concentrations: for E. coli, kanamycin, 50 μg/ml; ampicillin, 50 to 100 μg/ml; for the E. faecalis mutant, kanamycin, 2,000 μg/ml. All constructs were given TX numbers as shown in Table 1. Plasmids from these constructs were assigned respective pTEX numbers.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E. faecalis | ||
| OG1RF | Adh+ Fusr Rifr | 18, 39 |
| TX5256 | OG1RF ace::pTEX5253; ace insertion disruption mutant of OG1RF; Adh− Fusr Kanr Rifr | This study |
| E. coli | ||
| DH5α | E. coli host strain used for routine cloning | Stratagene |
| INVαF′ | E. coli host strain for cloning of PCR products | Invitrogen |
| LMG194 | E. coli strain for expression of recombinant proteins | Invitrogen |
| TX5252 | INVαF′ (pTEX5252); Ampr Kanr | This study |
| TX5253 | DH5α (pTEX5253); Kanr | This study |
| TX5254 | LMG194 (pTEX5254); Ampr | This study |
| Plasmids | ||
| pTEX4577 | Derived from pBluescript SK−, used for insertion disruption mutagenesis in enterococci | 28 |
| pBAD/HisA | Expression vector | Invitrogen |
| pTEX5252 | 1,003-bp intragenic ace PCR product cloned into pCR2.1 (TA cloning vector) | This study |
| pTEX5253 | 1,100-bp XhoI-KpnI intragenic ace fragment from pTEX5252 cloned into pTEX4577 | This study |
| pTEX5254 | 1,008-bp OG1RF ace (coding for complete A domain) cloned into pBAD/HisA expression vector | This study |
Adh+, adherence to CI, CIV, and LN after growth at 46°C; Adh−, markedly reduced adherence to CI, CIV, and LN; Ampr, ampicillin resistant; Fusr, fusidic acid resistant; Kanr, kanamycin resistant; Rifr, rifampin resistant.
Chemicals.
CI and CIV were purchased from Sigma Chemical Co. (St. Louis, Mo.). Mouse LN (isolated from the Engelbreth-Holm-Swarm sarcoma) was purchased from Life Technologies (Grand Island, N.Y.). Tran35S-label and bovine serum albumin (BSA) were purchased from ICN Biomedicals, Inc. (Costa Mesa, Calif.). Oligonucleotide primers were purchased from Life Technologies. PCR buffers were purchased from Invitrogen Corporation (Carlsbad, Calif.). All other chemicals used in the investigation were of molecular biology grade.
General DNA techniques.
DNA preparation, purification, restriction digestion, agarose gel electrophoresis, and ligation were performed by standard methods (26). Chromosomal DNA from E. faecalis was prepared according to the method described by Murray and colleagues (18). PCR amplification of DNA was performed on a DNA thermal cycler (Perkin-Elmer Corp., Norwalk, Conn.). Preparation of agarose plugs, pulsed-field gel electrophoresis (PFGE), and Southern blot analysis were carried out according to previously described methods (17, 22). Radioactive DNA probes were prepared by random-primed labeling according to the protocol supplied (Life Technologies). Electroporation of E. coli and E. faecalis was carried out with a Bio-Rad Gene Pulser as described previously (11). Isopropylthio-β-d-galactoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-galactoside (X-Gal) were used at 0.5 mM and 80 μg/ml, respectively. DNA sequencing reactions were performed by the Taq dye-deoxy terminator method on an automated ABI Prism sequencer (Applied Biosystems, Foster City, Calif.).
Construction of a mutation in the ace gene of E. faecalis OG1RF.
The E. faecalis OG1RF ace gene was disrupted by using a suicide vector pTEX4577 (28) containing an internal fragment of the ace gene. A 1,003-bp internal fragment (coding for the A domain of Ace of E. faecalis strain OG1RF) was amplified by PCR with AceF2 (5′-GAGCAAAAGTTCAATCGTTGAC-3′) and AceR3 (5′-GTCTGTCTTTTCACTTGTTTCT-3′) primers and cloned into the TA cloning vector pCR2.1 (Invitrogen Corp., Carlsbad, Calif.) resulting in TX5252. A 1,100-bp XhoI-KpnI DNA fragment from pTEX5252 was recloned into a pBluescript derivative, pTEX4577, and the resulting recombinant plasmid was designated as pTEX5253. Competent cells of E. faecalis OG1RF were electroporated with 5 μg of purified pTEX5253 in 2 μl of sterile water (22). Transformants showing growth on Todd-Hewitt agar supplemented with 2,000 μg of kanamycin per ml were selected, and one was designated as TX5256. Chromosomal DNA from agarose plugs was analyzed by PFGE after NotI or SmaI restriction digestion and hybridization to confirm the disruption. To further confirm the location of pTEX4577 within ace, chromosomal DNA from TX5256 was PCR amplified with the AceF1 (5′-CTATTGTCAACTTCTGAAAAAG-3′) primer and T7 or T3 primers from pTEX4577, and the resulting PCR products were sequenced. To test for the stability of this disruption mutation, OG1RF ace::pTEX5253 (TX5256) was grown overnight at 37 or 46°C in BHI broth without kanamycin and then reinoculated into BHI broth, grown again overnight two times, and then plated on BHI agar. Approximately 3,000 colonies grown on BHI agar were subpatched on BHI agar supplemented with 2,000 μg of kanamycin per ml to screen for colonies that had lost resistance to kanamycin.
Adherence assay.
Adherence to CI, CIV, and LN was tested by a previously described assay with some modifications (39). Bacteria were streaked from freezer vials onto BHI agar and incubated at 37°C overnight. A few colonies were picked and resuspended in BHI, and 108 CFU were inoculated into 5 ml of BHI broth with 10 μCi of Tran35S-label per ml. The cultures were grown at 46°C for 16 h and then harvested by centrifugation at 2,800 × g for 15 min. The cell pellets were washed three times in phosphate-buffered saline (PBS) buffer and resuspended in 0.1% Tween 80–0.1% BSA in PBS. The cell density was adjusted to an optical density at 600 nm (OD600) of 0.2. One microgram of ECM proteins in a total volume of 50 μl of PBS was used to coat Immulon 1 Removawells (Dynatech Laboratories, Chantilly, Va.) and incubated at 4°C overnight. After decanting, the wells were blocked with 200 μl of 0.2% BSA in PBS at 4°C for 2 h and then washed with PBS three times. A total volume of 50 μl of labeled bacteria was added to each well and incubated at room temperature for 2 h with gentle shaking at 70 rpm. The wells were washed with 0.1% Tween 80–0.1% BSA in PBS three times. Each detachable well was separated and placed into a vial with 2 ml of scintillation liquid and counted in a liquid scintillation counter (LKB Wallace, San Francisco, Calif.). Fifty microliters of labeled bacteria (adjusted to an OD600 of 0.2) was counted to determine the total amount of radioactivity added to each well. The percentage of adherence was calculated with the formula (radioactivity of bound cells/radioactivity of total cells added) × 100. The assays were performed in duplicate. Isolates were considered to adhere to ECM proteins if >5% of total labeled cells bound to the well.
Cloning, expression, and purification of Ace A from OG1RF.
A 1,008-bp DNA fragment coding for the complete A domain was amplified from pTEX5252 (derived from OG1RF) by using AceFc (5′-CAGAACTCGAGTTGAGCAAAAGTTCAATC-3′) and AceRc (5′-TGGAGGTACCCTAGTCTGTCTTTTCACTTG-3′) primers (introduced restriction sites are underlined) and cloned into pBAD/HisA expression vector (Invitrogen) followed by electroporation into the E. coli host LMG194, and one of the resulting colonies (designated as TX5254) was verified for the fidelity of the sequence and confirmed as correct by sequencing. Following electrophoresis of lysates on 10% NuPAGE Bis-Tris gels (NOVEX, San Diego, Calif.), Western transfer was carried out according to the protocol supplied by NOVEX, and His-tagged recombinant protein was detected with anti-His (penta) antibodies (Qiagen, Inc., Valencia, Calif.).
The recombinant Ace A domain was overexpressed by inoculating 1 liter of LB broth with 10 ml of an overnight culture of TX5254. Following 2.5 h of growth at 37°C, arabinose was added to a final concentration of 0.2% to induce protein expression, and incubation was continued for an additional 6 h. The bacterial cell pellet was lysed by sonication in denaturing lysis buffer (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris-HCl [pH 8.0]) containing 5 mM imidazole, and the supernatant was purified by metal chelation (Ni2+) chromatography. The bound proteins were washed with a mixture of 8 M urea, 0.1 M NaH2PO4, and 0.01 M Tris-HCl (pH 6.3) and neutralized with renaturing buffer (50 mM Tris-HCl, 50 mM NaCl, 50 mM NaH2PO4 [pH 8.0]); after washing with renaturing buffer containing 20 to 40 mM imidazole, the recombinant protein was eluted with a linear gradient of 50 to 400 mM imidazole in renaturing buffer, and the fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Fractions containing eluted recombinant Ace A were pooled, dialyzed against 25 mM Tris-HCl (pH 8.0), concentrated by lyophilization, and repurified by metal chelation (Ni2+) chromatography. Purified recombinant Ace A protein showed a single band on SDS-PAGE.
Production of rabbit polyclonal serum.
After verifying a single reacting band of His-tagged recombinant Ace A on a Western blot probed with anti-His (penta) antibodies (Qiagen, Inc.), this protein was used to raise polyclonal antibodies by immunization of rabbits at Bethyl Laboratories, Inc. (Montgomery, Tex.) and stored at −70°C. Antibody titers of sera were determined by enzyme-linked immunosorbent assay (ELISA) with preimmune serum as a control.
Protein extraction and Western blotting.
Protein extracts from E. faecalis OG1RF were prepared by using mutanolysin (Sigma Chemical Co.). E. faecalis OG1RF cells grown at 37 and 46°C were washed and resuspended in 1/10 volume of 0.02 M Tris-HCl (pH 7.0)–0.01 M MgSO4 buffer containing 100 mM phenylmethylsulfonyl fluoride (PMSF). Mutanolysin was added to a final concentration of 5 U/1 OD600 of cells and incubated at 37°C for 1 h in a rotating shaker. The supernatant collected after centrifugation at 11,750 × g for 15 min was concentrated by lyophilization. Protein concentrations were estimated by bicinchoninic acid assay (Pierce, Rockford, Ill.). Mutanolysin extracts from the E. faecalis OG1RF wild type and Ace insertion mutant (TX5256) were electrophoresed on 4 to 12% NuPAGE Bis-Tris gels (NOVEX) under reducing conditions in MOPS (3-[N-morpholino]propanesulfonic acid) buffer, and transferred to a polyvinylidene difluoride (PVDF) membrane. Electrophoresis and transfer were carried out according to the protocol supplied by NOVEX. Membranes were then incubated with either anti-Ace A polyclonal antiserum or preimmune serum (antibody I) followed by protein A-horseradish peroxidase conjugate (antibody II), and developed with 4-chloronaphthol in the presence of H2O2.
Far-Western blot assay.
Mutanolysin-PMSF extracts from the parental E. faecalis OG1RF and its ace insertion mutant (TX5256) were electrophoresed on 4 to 12% NuPAGE Bis-Tris gel (NOVEX) under nonreducing conditions in MOPS buffer and transferred to a PVDF membrane. After overnight renaturation in blocking buffer, the membrane was further incubated with 10 μg of CIV per ml for about 16 h at 4°C with gentle shaking. Bound CIV on Western blots was detected with anti-CIV monoclonal antibodies (Sigma Chemical Co.) followed by horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) antibodies (Life Technologies, Inc.), and developed with 4-chloronaphthol in the presence of H2O2.
Elution of antibodies specific to Ace.
Because the Ace immune serum previously described (24) was found to react with several bands, we eluted Ace A-specific antibodies from anti-Ace (OG1RF) rabbit immune serum by the method described below and used in the adherence inhibition assay. Recombinant Ace A protein was electrophoresed on 10% NuPAGE Bis-Tris gels (NOVEX) and transferred to a PVDF membrane. Membranes were blocked with 5% skim milk and incubated with polyclonal serum raised against recombinant Ace A of OG1RF. Following visualization of the antibodies bound to Ace on a cut strip by the procedure described in the previous section, the area containing the antigen-anti-Ace antibody complex was excised and incubated with 10 ml of 100 mM glycine (pH 2.5) for 15 min at room temperature to elute Ace-specific antibodies. After neutralization with 1 ml of 1 M Tris (pH 8.0), the solution was transferred to a clean tube and stored at −20°C until use (7).
IgG purification and inhibition of adherence.
IgGs were purified from both preimmune rabbit serum and polyclonal immune rabbit serum raised against recombinant Ace by affinity column chromatography with the Immunopure (G) IgG purification kit as per the supplied protocol (Pierce). Labeled bacteria were incubated with various concentrations of either preimmune rabbit IgGs or anti-Ace A IgGs for 1 h at 37°C and then centrifuged at 2,800 × g followed by resuspension in PBS with 0.1% Tween 80 and 0.1% BSA to remove excess unbound IgGs, prior to addition of labeled cells to the ECM-coated wells in adherence assay described earlier. Eluted Ace-specific antibodies were also used in the inhibition assay.
Binding of recombinant Ace to collagens and LN.
Microtiter plates were coated with 10 μg of ECM proteins or BSA in 100 μl of PBS and allowed to incubate overnight at 4°C. Wells were washed five times with PBST (PBS with 0.01% Tween 20). After blocking wells with 5% BSA, wells were again washed. Various concentrations of recombinant Ace A (1 to 200 μg/100 μl) in PBS with 0.1% BSA were added to the wells and incubated at 37°C. After 4 h, unbound protein was removed by washing with PBST. Bound proteins were detected by penta-His monoclonal antibodies (Qiagen, Inc.) that recognize the His tag of the recombinant Ace A protein, followed by horseradish peroxidase-conjugated goat anti-mouse IgG antibodies (Life Technologies, Inc.). Relative binding was measured by monitoring A450 following the addition of 3, 3′, 5, 5′-tetramethylbenzidine and H2O2. In a parallel set of experiments, CI-, CIV-, and LN-precoated wells were digested with collagenase VII (Sigma Chemical Co.) before incubation with recombinant Ace A.
RESULTS
Construction of an ace disruption mutation and stability.
Following electroporation of OG1RF with the suicide vector pTEX5253 and selection on kanamycin, 14 recombinant OG1RF colonies were recovered. DNA from three kanamycin-resistant OG1RF derivative colonies was digested with NotI or SmaI, followed by PFGE; hybridization with an ace probe, prepared by amplification with the AceF2 and AceR3 primers, showed two ace hybridizing bands (as expected for insertion duplication mutants, since there are single NotI and SmaI restriction sites in pTEX5253). OG1RF processed the same way showed a single hybridizing fragment. One of these colonies was designated as TX5256. The correct insertion, resulting from integration of pTEX5253, was also verified by sequencing of the PCR product amplified from TX5256 genomic DNA by using AceF1 and T7 primers and was found to have occurred at nucleotide 1101.
All colonies of TX5256 tested after passing through multiple generations without antibiotic selection retained the ability to grow on BHI agar supplemented with 2,000 μg of kanamycin per ml, indicating the stability of this mutation.
Adherence of the OG1RF ace mutant (OG1RF ace::pTEX5253).
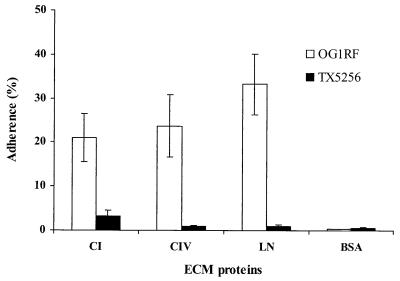
Adherence of OG1RF and the mutant TX5256 to ECM proteins (CI, CIV, and LN) was tested. The ace mutant grown at 46°C showed a 6.4-fold decrease in percentage of binding to CI (from 21.1% to 3.3%) when compared to OG1RF. Similarly, the ace mutant grown at 46°C showed a substantial decrease in adherence to CIV (27.4-fold decrease relative to OG1RF) and LN (32.9-fold decrease relative to OG1RF) (Fig. 1). This reduced adherence was also found in two other ace-disrupted kanamycin-resistant colonies tested (data not shown).
FIG. 1.
Adherence of E. faecalis OG1RF and TX5256 (OG1RF ace::pTEX5253) to immobilized CI, CIV, LN, and BSA. Adherence was tested in wells coated with 1 μg of ECM proteins (see text). Bars represent the mean percentages of cells bound ± standard deviations for six wells. Results are representative of three independent experiments. BSA was used as a negative control.
Western analysis of mutanolysin preparations of E. faecalis OG1RF and the OG1RF ace mutant.
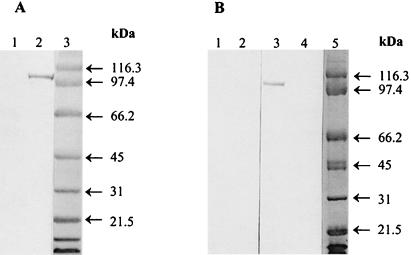
Anti-Ace A polyclonal immune rabbit serum reacted with a single ∼105-kDa band of mutanolysin-PMSF extracts prepared from 46°C-grown OG1RF, whereas no bands were detected from mutanolysin-PMSF extracts of 37°C-grown OG1RF (Fig. 2A). The apparent observed molecular mass is higher than predicted (calculated based on the sequence described in the companion paper [19]), perhaps due to the acidic nature of the Ace protein (24), which has a pI of 4.2 as calculated from the amino acid sequence. The OG1RF ace mutant (TX5256) grown at 46°C demonstrated loss of the ∼105-kDa immunoreactive protein band seen in OG1RF grown at 46°C (Fig. 2B).
FIG. 2.
Western blots of E. faecalis OG1RF and its ace mutant, TX5256 (OG1RF ace::pTEX5253). (A) Mutanolysin surface preparations probed with anti-Ace A polyclonal immune serum. Lanes: 1 and 2, protein extracts from 37°C- and 46°C-grown OG1RF, respectively; 3, molecular mass standards. (B) Mutanolysin surface preparations of 46°C-grown E. faecalis OG1RF and TX5256. Lanes: 1 and 2, OG1RF and TX5256 protein extracts, respectively, probed with rabbit preimmune serum; 3 and 4, OG1RF and TX5256 protein extracts, respectively, probed with anti-Ace A polyclonal immune serum; and 5, molecular mass standards.
Influence of anti-Ace IgGs on adherence of E. faecalis OG1RF to ECM proteins.
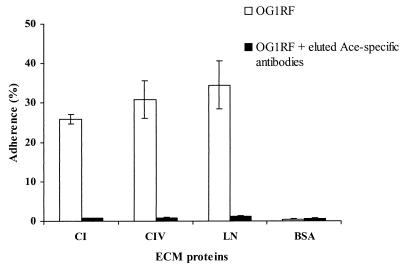
We have previously reported inhibition of adherence of a 46°C-grown E. faecalis OG1RF gelE mutant (28) to CI by IgGs purified from E. faecalis EF1 anti-Ace A antibodies (24). However, that serum reacted with several bands on Western blots. Using IgGs that were purified from E. faecalis OG1RF anti-Ace A polyclonal immune serum, we tested the influence of anti-Ace IgGs on adherence of OG1RF to CIV and LN as well as to CI. Using 0.001 to 100 μg of either purified preimmune IgGs or purified anti-Ace IgGs per ml, inhibition of adherence was tested. Preincubation with as little as 1 μg of anti-Ace IgGs per ml considerably inhibited adherence to CIV and LN, in addition to CI, whereas preimmune serum had no effect on adherence over the range of concentrations tested (Table 2). Antibodies eluted from recombinant Ace A were also tested in the adherence inhibition assay. As shown in Fig. 3, eluted antibodies at a 1-μg/ml concentration eliminated 46°C-grown OG1RF adherence to the three ECM proteins, CI, CIV, and LN (28- to 37-fold decrease relative to OG1RF).
TABLE 2.
Inhibition of adherence of 46°C-grown E. faecalis OG1RF to ECM proteins by IgGs purified from anti-Ace A (OG1RF-derived) rabbit immune serum
| ECM | % of cells bound at IgG concn ofa:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 μg/ml | 0.001 μg/ml
|
0.01 μg/ml
|
0.1 μg/ml
|
1 μg/ml
|
10 μg/ml
|
100 μg/ml
|
|||||||
| PI | Anti-Ace | PI | Anti-Ace | PI | Anti-Ace | PI | Anti-Ace | PI | Anti-Ace | PI | Anti-Ace | ||
| CI | 24.6 ± 3.88 | 25.3 ± 6.78 | 20.1 ± 4.04 | 27.2 ± 4.95 | 18.0 ± 4.09 | 24.5 ± 5.35 | 6.3 ± 1.04 | 21.6 ± 2.86 | 2.2 ± 0.38 | 22.0 ± 3.12 | 2.3 ± 0.44 | 23.8 ± 3.66 | 1.5 ± 0.37 |
| CIV | 29.6 ± 5.31 | 30.3 ± 6.15 | 26.2 ± 8.35 | 28.8 ± 6.12 | 20.3 ± 3.05 | 28.9 ± 6.61 | 7.9 ± 3.49 | 25.9 ± 8.98 | 3.2 ± 1.89 | 26.6 ± 5.12 | 2.8 ± 0.81 | 26.0 ± 4.73 | 2.2 ± 1.25 |
| LN | 32.0 ± 5.91 | 30.2 ± 1.94 | 26.2 ± 5.15 | 31.4 ± 7.89 | 21.8 ± 1.97 | 29.1 ± 4.67 | 8.6 ± 2.04 | 26.4 ± 7.51 | 3.5 ± 1.25 | 28.3 ± 4.25 | 2.8 ± 0.56 | 32.0 ± 3.03 | 2.1 ± 0.49 |
| BSA | 0.9 ± 0.12 | 0.7 ± 0.05 | 0.5 ± 0.09 | 0.1 ± 0.05 | 0.7 ± 0.09 | 0.6 ± 0.08 | 0.5 ± 0.03 | 0.8 ± 0.16 | 0.6 ± 0.26 | 0.6 ± 0.26 | 0.7 ± 0.12 | 0.6 ± 0.26 | 0.4 ± 0.06 |
Values are means ± standard deviations for six wells. Results are representative of three independent experiments. PI, IgGs purified from preimmune rabbit serum; Anti-Ace, IgGs purified from polyclonal anti-Ace A rabbit immune serum.
FIG. 3.
Inhibition of adherence of E. faecalis OG1RF to ECM proteins by eluted Ace A-specific antibodies. These antibodies were eluted with recombinant Ace A on the Western blot and were from anti-Ace A polyclonal immune serum. Labeled bacteria were incubated with 1 μg of eluted Ace-specific antibodies per ml for 1 h at 37°C. Adherence was tested in wells coated with 1 μg of ECM proteins (see text). Bars represent the mean percentages of cells bound ± standard deviations for four wells. Results are representative of two independent experiments.
We also examined the ability of these anti-Ace A IgGs (purified from rabbit polyclonal immune serum raised against OG1RF-derived recombinant Ace A) to inhibit adherence of two clinical E. faecalis strains, V583 and MC02152, to CI, CIV, and LN that showed conditional binding at 46°C. Preincubation of these strains with anti-Ace A IgGs at a 20-μg/ml concentration inhibited adherence to CI, CIV, and LN; relative to preimmune serum, adherence decreased by about 8.5- to 13.4-fold as shown in Table 3. Purified IgGs from preimmune serum had no effect on adherence at this concentration.
TABLE 3.
Inhibition of adherence of 46°C-grown E. faecalis clinical isolates V583 and MC02152 to ECM proteins by IgGs (20 μg/ml) purified from anti-Ace A (OG1RF-derived) rabbit immune serum
| ECM | % of cells bounda
|
|||||
|---|---|---|---|---|---|---|
| V583
|
MC02152
|
|||||
| No IgG | PI | Anti-Ace | No IgG | PI | Anti-Ace | |
| CI | 29.2 ± 1.87 | 28.1 ± 2.84 | 3.2 ± 0.17 | 26.0 ± 1.61 | 24.4 ± 1.31 | 2.9 ± 0.46 |
| CIV | 38.1 ± 8.66 | 33.6 ± 6.81 | 2.5 ± 0.17 | 27.7 ± 0.39 | 26.5 ± 1.84 | 2.3 ± 0.13 |
| LN | 29.4 ± 1.96 | 27.6 ± 3.53 | 2.7 ± 0.78 | 26.7 ± 1.22 | 23.5 ± 0.76 | 1.9 ± 0.52 |
| BSA | 0.7 ± 0.10 | 0.6 ± 0.07 | 0.6 ± 0.01 | 0.6 ± 0.12 | 0.5 ± 0.19 | 0.6 ± 0.17 |
Values are mean percentages of cells bound ± standard deviations for six wells. Results are representative of three independent experiments. PI, IgGs purified from preimmune rabbit serum; Anti-Ace, IgGs purified from polyclonal anti-Ace A rabbit immune serum.
CIV interaction with Ace by far-Western blotting.
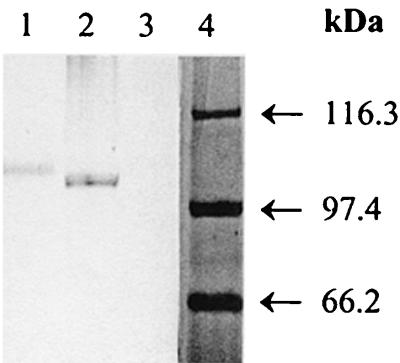
We used far-Western analysis and examined CIV's interaction with the ∼105-kDa Ace protein to determine the direct association of CIV with Ace. Probing of mutanolysin-PMSF extracts (prepared from 46°C-grown OG1RF) on a Western blot with CIV, followed by detection with anti-CIV monoclonal antibodies, identified a single ∼105-kDa protein band, whereas no band was detected from mutanolysin extracts of the OG1RF ace mutant (Fig. 4).
FIG. 4.
Far-Western blot assay of extracts of 46°C-grown E. faecalis OG1RF and its ace mutant TX5256. OG1RF and TX5256 extracts on PVDF membrane were probed with 10 μg of CIV per ml, and bound CIV was detected with anti-CIV monoclonal antibodies. Lanes: 1, CIV (positive control); 2 and 3, OG1RF and TX5256 mutanolysin extracts; and 4, molecular mass standards.
Binding of recombinant Ace A domain to ECM proteins.
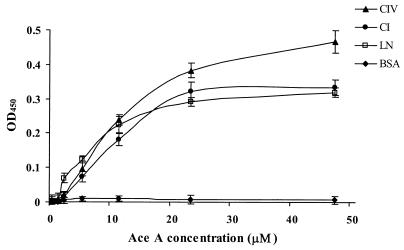
The results from an ELISA demonstrating the binding of recombinant Ace A to immobilized CI, CIV, and LN are shown in Fig. 5. Binding of recombinant Ace protein to the collagens and laminin was found to be concentration dependent and exhibited saturation kinetics. Fibrinogen, to which none of the E. faecalis isolates bound in our assay (39), was used as a control. The percentage of Ace bound to fibrinogen was the same as to BSA at all of the concentrations tested (data not shown). As evident from Fig. 5, binding of Ace A to CIV was slightly greater than its binding to CI and LN. The results from collagenase VII-treated wells showed that the binding of recombinant Ace A to CI and CIV was markedly reduced after collagenase treatment, whereas LN-coated wells were unaffected by collagenase treatment, indicating that the LN was not substantially contaminated by collagen (data not shown).
FIG. 5.
Binding of recombinant Ace A to immobilized ECM proteins CI, CIV, and LN (10 μg) as a function of concentration of Ace A. BSA was used as a negative control. All OD450 values were corrected for the response of penta-His monoclonal antibodies with CI, CIV, and LN, respectively. Data points represent the mean OD450 values ± standard deviations for six wells representing three independent experiments.
DISCUSSION
We previously showed that the majority of E. faecalis isolates adhered, after growth at 46°C, to CI, CIV, and LN (39). An examination of the adherence process revealed that (i) E. faecalis strain OG1RF adherence was dependent on the amounts of CI, CIV, and LN in the substrates; (ii) OG1RF adherence to ECM proteins was inhibited after preincubation of the bacteria with soluble ECM proteins; (iii) trypsin treatment of the bacteria rendered the cells nonadhesive; (iv) digestion of the CI and CIV substrates with collagenase destroyed their ability to support adhesion of strain OG1RF, whereas bacteria still adhered to collagenase-digested LN substrate; and (v) scanning electron microscopy of E. faecalis OG1RF adhered to LN-coated wells showed single cells evenly distributed over the substrate (39). We recently identified a gene, ace, in the E. faecalis strain V583 partial genome database that encodes a protein with a structural organization similar to the collagen binding microbial surface component recognizing adhesive matrix molecules (MSCRAMM), Cna, from S. aureus. Both proteins contain features characteristic of cell wall-anchored proteins at the C terminus preceded by a region composed of B repeats and an N-terminal A region. The A region of Cna contains the collagen binding domain, which has a β-barrel structure as revealed by X-ray analysis of protein crystals (36). One of the β-sheets contains a “trench” that was identified as a putative collagen binding site. Computational docking experiments showed that the binding trench could accommodate the rope-like collagen structure (23, 36). The A region of Ace has significant sequence similarity to the corresponding domain of Cna. We therefore expressed a recombinant form of the Ace A region and showed by circular dichroism spectroscopy that the secondary structures of the Ace and Cna A regions are very similar (24). In fact, computational analysis suggested that the putative ligand binding domain of Ace adopts a structure very similar to that determined for the corresponding domain of Cna with a predicted binding trench. Furthermore, the recombinant Ace A region bound CI, and antibodies raised against the recombinant protein inhibited adherence of E. faecalis to CI substrate (24).
In the current study, we have characterized the E. faecalis-specific (R. W. Duh, K. V. Singh, K. Malathum, and B. E. Murray, submitted for publication) ace gene from strain OG1RF and found that an OG1RF ace mutant showed markedly reduced binding not only to immobilized CI, but also to CIV and LN, raising the possibility that the same MSCRAMM is responsible for adherence to the three ECM proteins. However, since this mutant was generated by a chromosomal insertion, the possibility remains that the inserted plasmid may have had a polar effect on downstream genes that are responsible for some of the observed effect.
To detect the Ace protein in E. faecalis OG1RF, we raised polyclonal antibodies against recombinant Ace A of OG1RF that has been expressed in E. coli. These anti-Ace A antibodies detected an ∼105-kDa protein in 46°C-grown OG1RF, but not in 37°C-grown OG1RF; the OG1RF ace mutant was found to lack the ∼105-kDa protein. Detection of this anti-Ace reactive band in 46°C-grown OG1RF mutanolysin extracts, but not in 37°C-grown OG1RF, correlates with the previously reported conditional (growth at 46°C) binding (39). We have also sequenced the complete ace gene from OG1RF (19). The deduced amino acid sequence of OG1RF Ace adhesin predicts a 75.6-kDa protein, which is ∼30 kDa smaller than the observed molecular size on the Western blot. Similar results were found for Ace proteins of the other E. faecalis strains studied in the companion paper (19) as well as for E. faecalis EF1 and EF2 (24). This difference may be due to the highly acidic nature of the Ace protein (24). Another possibility is that the difference in migration might be due to association of Ace with peptidoglycan, although mutanolysin treatment is known to free at least some proteins from peptidoglycan (8). However, several lines of evidence strongly indicate that the protein identified in mutanolysin-PMSF extracts is indeed the ace gene product. The evidence includes the following: (i) anti-Ace A polyclonal antibodies reacted with a single ∼105-kDa band in mutanolysin extracts prepared from 46°C-grown E. faecalis OG1RF; (ii) preimmune serum did not react with any band in these extracts; and (iii) there was loss of the ∼105-kDa protein band in the ace insertion mutant. In a companion paper, we also report that protein size variation among various E. faecalis strains corresponds to the number of B repeats (19).
To confirm the direct involvement of Ace, and not possible downstream gene products, in 46°C-evoked adherence of E. faecalis to CI, CIV, and LN, we tested the ability of anti-Ace A IgGs to inhibit binding of E. faecalis OG1RF to these immobilized ECM proteins. The inhibition of the adherence of 46°C-grown OG1RF to CI, CIV, and LN by anti-Ace A IgGs as well as by eluted Ace-specific antibodies provides evidence that the ∼105-kDa protein of OG1RF is the adhesin that mediates binding to these three ECM proteins. We also tested the ability of these IgGs to block adherence of the two clinical strains V583 and MC02152 after growth at 46°C. The inhibited adherence to CI, CIV, and LN by anti-Ace IgGs in these two strains further corroborates the involvement of Ace A in strains that showed conditional adherence. We confirmed the CIV affinity to the ∼105-kDa OG1RF Ace protein by using a far-Western blot and then extended this result to test the binding ability of the recombinant Ace A domain to CI, CIV, and LN in an ELISA. In the ELISAs, OG1RF-derived recombinant Ace A protein bound to CI as well as to CIV and LN. These ELISA results implicate the A domain of Ace in binding to CI, CIV, and LN. It is tempting to speculate that the proposed trench on the Ace A domain that has been implicated in binding the triple helix collagen structure (Y. Xu, R. T. Owens and M. Höök, unpublished results) is also responsible for binding the rigid triple helix structure of the LN long arm. By analogy, the collagen binding integrins α1β1 and α2β1, both of which both contain a trench in the binding domain, have been shown to bind LN in addition to several types of collagens, including CI and CIV (3, 21, 32).
Similar to the E. faecalis Ace adhesin, other adhesins have been reported to bind to different ECM proteins. The plasmid-encoded outer membrane protein YadA of Yersinia enterocolitica has been shown to bind to several types of collagens (27), LN (4, 29), and fibronectin (37). A 150-kDa fibrinogen binding adhesin of Porphyromonas (Bacteroides) gingivalis also recognized fibronectin (9, 10). Switalski et al. (34) showed that the collagen-binding MSCRAMM from S. aureus (later identified as Cna) recognizes many types of collagens and McGavin et al. (15) identified an S. aureus surface protein that could bind with broad specificity to several ECM proteins, including fibrinogen, fibronectin, and vitronectin.
In conclusion, the results from the constructed OG1RF ace mutant and the inhibition of binding of OG1RF and of two clinical isolates to all three ECM proteins by anti-Ace A antibodies demonstrate that the A domain of Ace mediates adherence of E. faecalis to CIV and LN in addition to CI. Further supporting evidence for Ace A-mediated binding was obtained from the CIV far-Western analysis and the ELISAs showing binding of recombinant Ace A to both collagens and LN. Additional studies will be needed to determine what if any contribution ace may make to the ability of E. faecalis to colonize and/or cause infection in humans.
ACKNOWLEDGMENT
This work was supported by NIH grant AI33516 from NIAID, the Division of Microbiology and Infectious Diseases, to B. E. Murray.
REFERENCES
- 1.Beck K, Hunter I, Engel J. Structure and function of laminin: anatomy of a multidomain glycoprotein. FASEB J. 1990;4:148–160. doi: 10.1096/fasebj.4.2.2404817. [DOI] [PubMed] [Google Scholar]
- 2.Engel J. Laminins and other strange proteins. Biochemistry. 1992;31:10643–10651. doi: 10.1021/bi00159a001. [DOI] [PubMed] [Google Scholar]
- 3.Ettner N, Göhring W, Sasaki T, Mann K, Timpl R. The N-terminal globular domain of the laminin α1 chain binds to α1β1 and α2β1 integrins and to the heparan sulfate-containing domains of perlecan. FEBS Lett. 1998;430:217–221. doi: 10.1016/s0014-5793(98)00601-2. [DOI] [PubMed] [Google Scholar]
- 4.Flügel A, Schulze-Koops H, Heesemann J, Kühn K, Sorokin L, Burkhardt H, von der Mark K, Emmrich F. Interaction of enteropathogenic Yersinia enterocolitica with complex basement membranes and the extracellular matrix proteins collagen type IV, laminin-1 and -2, and nidogen/entactin. J Biol Chem. 1994;269:29732–29738. [PubMed] [Google Scholar]
- 5.Foster T J, Höök M. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 1998;6:484–488. doi: 10.1016/s0966-842x(98)01400-0. [DOI] [PubMed] [Google Scholar]
- 6.Gehlsen K R, Dillner L, Engvall E, Ruoslahti E. The human laminin receptor is a member of the integrin family of cell adhesion receptors. Science. 1988;241:1228–1229. doi: 10.1126/science.2970671. [DOI] [PubMed] [Google Scholar]
- 7.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 8.Juarez Z E, Stinson M W. An extracellular protease of Streptococcus gordonii hydrolyzes type IV collagen and collagen analogues. Infect Immun. 1999;67:271–278. doi: 10.1128/iai.67.1.271-278.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lantz M S, Allen R D, Bounelis P, Switalski L M, Höök M. Bacteroides gingivalis and Bacteroides intermedius recognize different sites on human fibrinogen. J Bacteriol. 1990;172:716–726. doi: 10.1128/jb.172.2.716-726.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lantz M S, Allen R D, Duck L W, Blume J L, Switalski L M, Höök M. Identification of Porphyromonas gingivalis components that mediate its interactions with fibronectin. J Bacteriol. 1991;173:4263–4270. doi: 10.1128/jb.173.14.4263-4270.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Weinstock G M, Murray B E. Generation of auxotrophic mutants of Enterococcus faecalis. J Bacteriol. 1995;177:6866–6873. doi: 10.1128/jb.177.23.6866-6873.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindmark H, Guss B. SFS, a novel fibronectin-binding protein from Streptococcus equi, inhibits the binding between fibronectin and collagen. Infect Immun. 1999;67:2383–2388. doi: 10.1128/iai.67.5.2383-2388.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ljungh A, Moran A P, Wadström T. Interactions of bacterial adhesins with extracellular matrix and plasma proteins: pathogenic implications and therapeutic possibilities. FEMS Immunol Med Microbiol. 1996;16:117–126. doi: 10.1111/j.1574-695X.1996.tb00128.x. [DOI] [PubMed] [Google Scholar]
- 14.Lopes J D, dos Reis M, Brentani R R. Presence of laminin receptors in Staphylococcus aureus. Science. 1985;229:275–277. doi: 10.1126/science.3160113. [DOI] [PubMed] [Google Scholar]
- 15.McGavin M H, Krajewska-Pietrasik D, Rydén C, Höök M. Identification of a Staphylococcus aureus extracellular matrix-binding protein with broad specificity. Infect Immun. 1993;61:2479–2485. doi: 10.1128/iai.61.6.2479-2485.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller E J, Gay S. The collagens: an overview and update. Methods Enzymol. 1987;144:3–41. doi: 10.1016/0076-6879(87)44170-0. [DOI] [PubMed] [Google Scholar]
- 17.Murray B E, Singh K V, Heath J D, Sharma B R, Weinstock G M. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–2063. doi: 10.1128/jcm.28.9.2059-2063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray B E, Singh K V, Ross R P, Heath J D, Dunny G M, Weinstock G M. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. J Bacteriol. 1993;175:5216–5223. doi: 10.1128/jb.175.16.5216-5223.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nallapareddy, S. R., K. V. Singh, R.-W. Duh, G. M. Weinstock, and B. E. Murray. Diversity of ace, a gene encoding a microbial surface component recognizing adhesive matrix molecules, from different strains of Enterococcus faecalis and evidence for production of Ace during human infections. Infect. Immun. 68:5210–5217. [DOI] [PMC free article] [PubMed]
- 20.Patti J M, Jonsson H, Guss B, Switalski L M, Wiberg K, Lindberg M, Höök M. Molecular characterization and expression of a gene encoding a Staphylococcus aureus collagen adhesin. J Biol Chem. 1992;267:4766–4772. [PubMed] [Google Scholar]
- 21.Pfaff M, Göhring W, Brown J C, Timpl R. Binding of purified collagen receptors (α1β1, α2β1) and RGD-dependent integrins to laminins and laminin fragments. Eur J Biochem. 1994;225:975–984. doi: 10.1111/j.1432-1033.1994.0975b.x. [DOI] [PubMed] [Google Scholar]
- 22.Qin X, Teng F, Xu Y, Singh K V, Weinstock G M, Murray B E. Targeted mutagenesis of enterococcal genes. Methods Cell Sci. 1998;20:21–33. [Google Scholar]
- 23.Rich R L, Demeler B, Ashby K, Deivanayagam C C S, Petrich J W, Patti J M, Narayana S V L, Höök M. Domain structure of the Staphylococcus aureus collagen adhesin. Biochemistry. 1998;37:15423–15433. doi: 10.1021/bi981773r. [DOI] [PubMed] [Google Scholar]
- 24.Rich R L, Kreikemeyer B, Owens R T, LaBrenz S, Narayana S V L, Weinstock G M, Murray B E, Höök M. Ace is a collagen-binding MSCRAMM from Enterococcus faecalis. J Biol Chem. 1999;274:26939–26945. doi: 10.1074/jbc.274.38.26939. [DOI] [PubMed] [Google Scholar]
- 25.Sahm D F, Kissinger J, Gilmore M S, Murray P R, Mulder R, Solliday J, Clarke B. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother. 1989;33:1588–1591. doi: 10.1128/aac.33.9.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Schulze-Koops H, Burkhardt H, Heesemann J, von der Mark K, Emmrich F. Plasmid-encoded outer membrane protein YadA mediates specific binding of enteropathogenic yersiniae to various types of collagen. Infect Immun. 1992;60:2153–2159. doi: 10.1128/iai.60.6.2153-2159.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh K V, Qin X, Weinstock G M, Murray B E. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J Infect Dis. 1998;178:1416–1420. doi: 10.1086/314453. [DOI] [PubMed] [Google Scholar]
- 29.Skurnik M, el Tahir Y, Saarinen M, Jalkanen S, Toivanen P. YadA mediates specific binding of enteropathogenic Yersinia enterocolitica to human intestinal submucosa. Infect Immun. 1994;62:1252–1261. doi: 10.1128/iai.62.4.1252-1261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spellerberg B, Rozdzinski E, Martin S, Weber-Heynemann J, Schnitzler N, Lütticken R, Podbielski A. Lmb, a protein with similarities to the LraI adhesin family, mediates attachment of Streptococcus agalactiae to human laminin. Infect Immun. 1999;67:871–878. doi: 10.1128/iai.67.2.871-878.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Speziale P, Raucci G, Visai L, Switalski L M, Timpl R, Höök M. Binding of collagen to Staphylococcus aureus Cowan 1. J Bacteriol. 1986;167:77–81. doi: 10.1128/jb.167.1.77-81.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sung U, O'Rear J J, Yurchenco P D. Cell and heparin binding in the distal long arm of laminin: identification of active and cryptic sites with recombinant and hybrid glycoprotein. J Cell Biol. 1993;123:1255–1268. doi: 10.1083/jcb.123.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Switalski L M, Murchison H, Timpl R, Curtiss III R, Höök M. Binding of laminin to oral and endocarditis strains of viridans streptococci. J Bacteriol. 1987;169:1095–1101. doi: 10.1128/jb.169.3.1095-1101.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Switalski L M, Speziale P, Höök M. Isolation and characterization of a putative collagen receptor from Staphylococcus aureus strain Cowan 1. J Biol Chem. 1989;264:21080–21086. [PubMed] [Google Scholar]
- 35.Switalski L M, Speziale P, Höök M, Wadström T, Timpl R. Binding of Streptococcus pyogenes to laminin. J Biol Chem. 1984;259:3734–3738. [PubMed] [Google Scholar]
- 36.Symersky J, Patti J M, Carson M, House-Pompeo K, Teale M, Moore D, Jin L, Schneider A, DeLucas L J, Höök M, Narayana S V L. Structure of the collagen-binding domain from a Staphylococcus aureus adhesin. Nat Struct Biol. 1997;4:833–838. doi: 10.1038/nsb1097-833. [DOI] [PubMed] [Google Scholar]
- 37.Tertti R, Skurnik M, Vartio T, Kuusela P. Adhesion protein YadA of Yersinia species mediates binding of bacteria to fibronectin. Infect Immun. 1992;60:3021–3024. doi: 10.1128/iai.60.7.3021-3024.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Westerlund B, Korhonen T K. Bacterial proteins binding to the mammalian extracellular matrix. Mol Microbiol. 1993;9:687–694. doi: 10.1111/j.1365-2958.1993.tb01729.x. [DOI] [PubMed] [Google Scholar]
- 39.Xiao J, Höök M, Weinstock G M, Murray B E. Conditional adherence of Enterococcus faecalis to extracellular matrix proteins. FEMS Immunol Med Microbiol. 1998;21:287–295. doi: 10.1111/j.1574-695X.1998.tb01176.x. [DOI] [PubMed] [Google Scholar]
- 40.Yurchenco P D, Schittny J C. Molecular architecture of basement membranes. FASEB J. 1990;4:1577–1590. doi: 10.1096/fasebj.4.6.2180767. [DOI] [PubMed] [Google Scholar]