Abstract
Background
A. baumannii and P. aeruginosa are important nosocomial pathogens in health-care settings. Both are intrinsically resistant to many drugs and are able to become resistant to the virtually most antimicrobial agents. An increasing prevalence of infections caused by multidrug-resistant isolates has been reported in many countries.
Methods
An institutional-based cross-sectional five-year retrospective study was conducted to assess the antimicrobial resistance trend of P. aeruginosa and A. baumani. 893 A. baumani and 729 P. aeruginosa isolates were included in the study. Conventional method was used for identification and antimicrobial susceptibility was determined by Kirby-Bauer disc-diffusion method. The isolates were from suspected bloodstream infections, wound infections, urinary tract, or surgical site nosocomial infections. Socio-demographic and other variables of interest were collected using a structured check list from a patient record data. Data were analyzed using SPSS version 26 software. P value <0.05 was considered statistically significant.
Results
A total of 1622 A. baumanii and P. aeruginosa were isolated from various clinical specimens recorded from the year 2017–2021. Out of which A. baumanni was 893 (60.6%) and P. aeruginosa was 729 (39.4%). Blood was the major source of the isolates (18.3%), followed by urine (16%), and tracheal aspirate (10.6%). Antimicrobial resistance among A. baumanni over the five years were; ampicillin (86% to 92%), ceftriaxone (66.7% to 82.2%), and ciprofloxacin (58.5% to 66.7%). In P. aeruginosa a significant increase in resistance was seen from 2017 to 2021 to Amoxicillin-clavulanate (74.1% to 84.2%), chloramphenicol (62% to 81.9%), and gentamicin (40% to 44.8%).
Conclusion
A five-year antimicrobial resistance trend analysis of A. baumanni and P. aeruginosa showed increasing multi drug resistance and resistance to highly potent antimicrobial agents in Ethiopia. It should be addressed with infection control measures, surveillance, and alternative new therapeutic strategies to circumvent the spread of multi-drug resistance.
Keywords: A. baumanii, antibiotic resistance, Ethiopia, MDR, P. aeruginosa
Introduction
Antimicrobial resistance is quickly gaining global attention, particularly as the number of microbes resistant to currently available antimicrobials rises.1 It includes both gram-positive and gram-negative bacteria and has a global prevalence rate of 60% or higher.2,3
Infections caused by drug-resistant Acinetobacter baumanii and Pseudomonas aeruginosa species are a growing source of Hospital acquired infection and a major public health concern.4 Pneumonia, bacteremia, meningitis, urinary tract infection, and wound infection are only a few of the illnesses caused by these bacteria.5,6 Infections caused by drug-resistant Acinetobacter spp. and Pseudomonas spp are linked to prolonged hospital stays and mortality.7,8
Due to its extraordinary ability to withstand medicines, eradication of P. aeruginosa has become increasingly difficult. Most antibiotics are known to be resistant to P. aeruginosa strains due to their high levels of inherent and acquired resistance mechanisms, biofilm formation.9–11 Alternative therapeutic techniques that present unique pathways against P. aeruginosa infections are becoming increasingly desired and receiving.11
Pseudomonas aeruginosa and Acinetobacter baumanni have been a leading cause of nosocomial infections, causing significant morbidity and mortality over the entire world including Ethiopia.12 Multidrug-resistant A. baumannii and P. aeruginosa thrive in hospitals, where they can easily spread from patient to patient via health-care personnel’s hands.12 The most important features of these bacteria species are their ability to persist in the hospital environment and rapidly develop resistance to a wide variety of antibiotics due to different mechanisms such as, reduced outer membrane permeability, efflux pump systems, enzymatic inactivation, and biofilm formation.13–15 As a result, practically all -lactams, aminoglycosides, and quinolones are often resistant to them. Antimicrobial drug usage, drug prescriptions without susceptibility testing, self-medication, and prolonged hospitalization have all been linked to the development of MDR.11,13 However, data on the prevalence of nosocomial multi drug resistance (MDR) A. baumannii and P. aeruginosa infections in Ethiopia, in general, and the study area, in particular, is scarce. Therefore, the purpose of this study was to assess the magnitude and AMR pattern of A. baumanni and P. aeruginosa from clinical samples in retrospective approach.
Methods
A cross-sectional retrospective analysis was conducted to assess the magnitude and AMR pattern of A. baumanni and P. aeruginosa from clinical samples that were tested for bacterial presence and subsequent susceptibility testing dated from January 2017 to December 2021, from two large teaching hospital in Ethiopia (Black lion specialized hospital and Yekatit 12 hospital medical college). All hospitals are situated in the city of Addis Ababa, Ethiopia. The clinical information extracted from the microbiology laboratories included type of sample analyzed, name of pathogens isolated and the names of antibiotics used for susceptibility testing and the susceptibility results as recorded in the laboratory report. All information were recorded by the assigned laboratory personnel/data collector and reviewed by senior microbiologist.
Laboratory Analysis, Clinical Samples and Collection of Bacterial Isolates
Several types of clinical specimens, including urine, blood, sputum, pus, tracheal aspirate, wound swab, ear discharge, pleural fluid, and cerebrospinal fluid (CSF) were cultured. Both laboratories sampled for the current study employed similar standard microbiological culturing techniques. All the laboratories performed similar microscopic identification and biochemical identification using the Clinical Laboratory Standard Institute guidelines (CLSI guideline).16 Antimicrobial susceptibility test was performed by both laboratories using the Kirby-Bauer disk diffusion method in Muller Hinton agar and interpreted using CLSI.16
Data Analysis
Collected data was entered into Microsoft Excel and loaded into SPSS, version 26 for analysis. Proportions of predominant isolates, sociodemographic characteristics, and antibiotic resistance profiles were presented using tables and figures. P < 0.05 was considered significantly associated among variables.
Results
A total of 1622 patient’s data that were isolated from various clinical specimens was recorded from the year 2017–2021 for antimicrobial susceptibility test of A. baumanni and p. aeruginosa. Out of which 881 (54.3%) were males and 741 (45.7%) were females, whereas the age ranged from 1 to 90 with the mean and median ages of 39.3 years and 37 years, respectively. A.baumanni was the predominant isolate 893 (55.05%) than P. aeruginosa which was 729 (45%) (Table 1).
Table 1.
Socio-Demographic and Clinical Characteristics of Study Participants in TASH and Yekatit 12 Hospital Medical College, 2017–2021
| Variables | Frequency | Percent | |
|---|---|---|---|
| Sex N=1622 | Male | 881 | 54.3 |
| Female | 741 | 45.7 | |
| Age N=1622 | <1 | 236 | 14.5 |
| 1–14 | 290 | 17.8 | |
| 15–24 | 254 | 15.65 | |
| 25–44 | 343 | 21.1 | |
| 45–65 | 304 | 18.7 | |
| >65 | 195 | 12 | |
| Type of specimen N=1622 | Blood | 297 | 18.3 |
| Urine | 261 | 16 | |
| CSF | 161 | 9.9 | |
| Tracheal aspirate | 173 | 10.6 | |
| Pus | 168 | 10.3 | |
| Sputum | 157 | 9.6 | |
| Wound | 154 | 9.5 | |
| Pleural fluid | 125 | 7.7 | |
| Ear discharge | 122 | 7.5 | |
| Isolated Bacteria N=1622 | A. baumanii | 893 | 55 |
| P. aeruginosa | 729 | 45 |
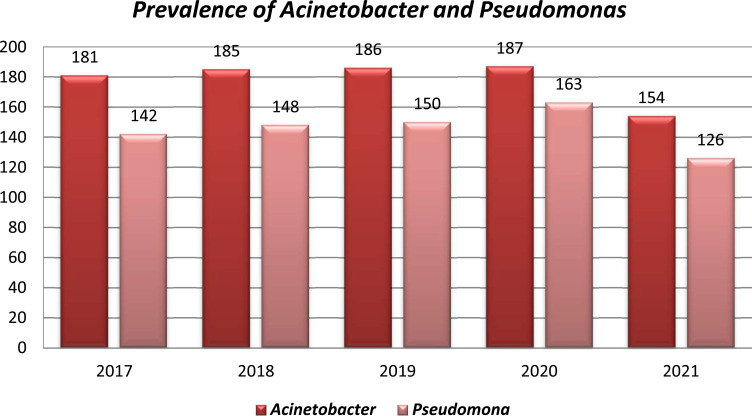
In our study, 893 A. baumanni and 729 P. aeruginosa were analyzed to investigate the trends of antimicrobial resistance. In the analysis, we found that A. baumanni had the most prevalence in all the five years ranging from 60.6% to 62.7%, while P. aeruginosa ranges was 38.2%, in 2020 A. baumanni was 56.4%, and Pseudomonas aeruginosa was 43.6% and in 2021 Acinetobacter was 62.7%, while Pseudomonas was 37.3% (Figure 1).
Figure 1.
Distribution of A. bauumanni and P. aeruginosa from 2017 to 2021.
Among the two labs, 9 different types of clinical specimen were processed, but the highest level of the isolates was found in the blood 203 (26.3%), followed by urine 167 (21.6%), tracheal aspirate 79 (10.2%), and pus 74 (9.6%) specimen. P. aeruginosa was the predominant isolate in urinary tract infection (25%), sepsis (16.4%), and respiratory infections (13.5%) (Figure 2).
Figure 2.
Distribution of P. aeruginosa and A. baumanii across clinical specimens from 2017 to 2021.
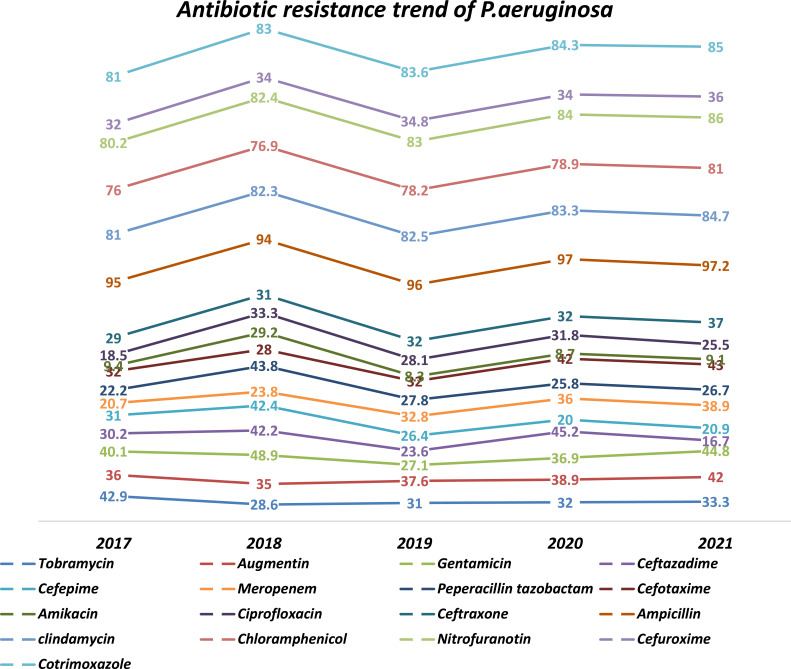
P. aeruginosa resistance to antibiotics over the five years showed that higher resistance to Ampicillin (98%), followed by Amoxicillin-clavulanate (91.3%), and Nitrofurantoin (86%), while it was most sensitive to Tobramycin 68 (39.1%), Cefoxitin, Azithromycin 33.3%, and Amikacin 135 (24.5%).
Most P. aeruginosa in these study sites were recovered from urine, followed by blood, sputum, pus and tracheal aspirate. A significant increase in resistance was seen in Amoxicillin-clavulanic acid (94.1% to 100%), Chloramphenicol (88.9% to 100%), Gentamicin (40% to 44.8%), Cefepime (31% to 90.9%), P. tazobactam (22.2% to 26.7%), Meropenem (20.7% to 38.9%), Cefotaxime (71.4% to 100%), Ciprofloxacin (18.5% to 25.5%), Tobramycin (42.9% to 33.3%), Ceftazidime (30.2% to 16.7%), Ceftriaxone (76.5% to 66.7%), Amikacin (9.4% to 9.2%) showed decrease in resistance pattern (Figure 3).
Figure 3.
Trend of antimicrobial resistance pattern for Pseudomonas aeruginosa.
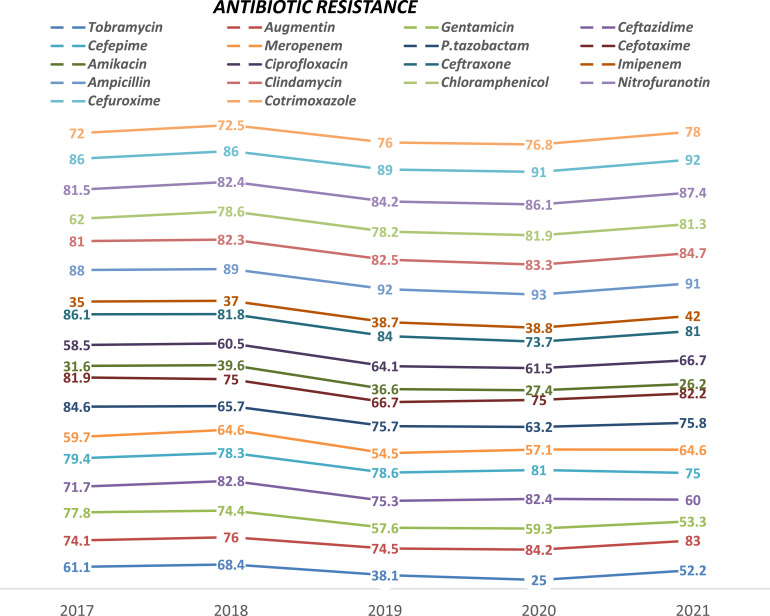
An increasing trend of antimicrobial resistance for A. baumanni was observed in the five years ampicillin (86% to 92%), followed by Amoxicillin-clavulanate (81% to 84.7%), chloramphenicol (76% to 82.3%), and Cefotaxime (66.7% to 82.2%). However, a decreasing trend of resistance was observed for tobramycin (61.1% to 25.2%), followed by Tazobactam (84.6% to 63.2%), and Amikacin (31.6% to 26.2%) (Figure 4).
Figure 4.
Trend of antimicrobial resistance of A. baumanni from 2017 to 2021, Ethiopia.
Overall, 73.7% of A. baumanni and 58.9% of P. aeruginosa were multidrug resistant. 9.7% of A. baumanni isolates showed resistance to greater than nine antibiotics while 6.7% P. aeruginosa were resistant to more than nine antibiotics (Table 2).
Table 2.
MDR Pattern of P. Aeruginosa and A. Baumanni Isolates from Clinical Isolates 2017–2021, Ethiopia
| Bacteria Species | Antibiogram | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| R1 | R2 | R3 | R4 | R5 | R6 | R7 | R8 | >R9 | |
| A. baumanii | 135 | 97 | 107 | 98 | 91 | 102 | 84 | 80 | 87 |
| P. aeruginosa | 208 | 89 | 91 | 66 | 63 | 59 | 53 | 49 | 49 |
Abbreviations: R1, resistance to one class of antibiotic; R1, resistance to two antibiotics; R3, resistance to three class of antibiotics; >R9, Resistance to more than nine class of antibiotics.
Discussion
The nightmare of the rising numbers of multidrug-resistant organisms (MDROs) requires the implementation of effective stewardship programs.11,17 However, this should be preceded by making available evidence-based knowledge regarding the local antimicrobial resistance pattern, which is fundamental. The excessive use of antibiotics has led to a vast widespread prevalence of antimicrobial resistance.2 As time passes, bacterial pathogens will defy every antibacterial option, thus, becoming extremely hard to control. Hence, the WHO identified it as an international health prime concern.13,18
In our study, 893 A. baumanni and 729 P. aeruginosa were analyzed to investigate the trends of antimicrobial resistance. In the analysis we found that A. baumanni had the most prevalence in all the five years. The prevalence of A. baumanni in 2017 was 60.6%, while P. aeruginosa was 39.4%, and in 2021 A. baumanni was 62.7%, while P. aeruginosa was 37.3%. The difference regarding type and frequency of pathogens could be linked to several factors like environmental conditions, health practices, patient conditions, personal hygiene, number of patients involved in each study, and laboratory procedures.19
In our study, the highest prevalence of infections due to P. aeruginosa was observed in the Urinary tract (25%) specimens followed by blood (16.4%), and sputum (13.5%). De Francesco et al, and Yayn et al reported that most of the isolates of P. aeruginosa were obtained from the respiratory tract followed by urinary tract, wound and blood3,20 While the highest prevalence infection due to A. baumanni was observed from blood (32.7%), urinary tract (19.4%) followed by CSF (12.6%) and different studies also reported the isolation of A. baumanni from these clinical specimens. However, the current finding was lower than the study conducted in Dessie which isolated A. baumanni from wound specimen at a rate of 51.4% and in Iran at the rate of 56.7% which is higher than this study.10,12
In this study, the multidrug resistance prevalence showed that 73.7% of A. baumanni and 58.9% of P. aeruginosa were multidrug resistant. The highest percentage of resistance among A. baumanni were exhibited towards Ampicillin that showed the most resistant 147 (98%) followed by Amoxicillin-clavulanic acid 146 (91.3%), Nitrofurantoin 33 (86%), and Ceftriaxone 160 (82.5%), while Amikacin being the most sensitive 135 (24.5%). Resistance in P. aeruginosa and A. baumannii to broad-spectrum antimicrobial agents in hospital settings is now an emerging issue worldwide.21 Surveillance studies indicate that the rates of resistance to antimicrobial agents among A. baumannii are increasing in Mexico,22 the Arabian Peninsula and in many parts of the globe.23,24 While in P. aeruginosa, out of the drugs tested for the resistance pattern Ampicillin, Amoxicillin-clavulanate, chloramphenicol, Cefotaxime and Ceftriaxone had shown higher resistance pattern. However, these isolates showed the maximum sensitivity pattern for tobramycin (61.1% to 25.2%), followed by tazobactam (84.6% to 63.2%), and amikacin (31.6% to 26.2%). Compared to a cross-sectional study conducted at Ethiopian public health institute (EPHI) in 2021 the Pseudomonas species were highly resistant to amikacin in our study (9.1%), Ceftazidime was lower in our study (16.7%) compared to the study by Tesfa et al which was 35%. Most of the findings in different countries confirmed that pseudomonas was becoming increasingly resistant to different types of antimicrobial agents.25 In Ethiopia, a high prevalence of carbapenem-resistant Acinetobacter spp. was reported from one previous phenotypic study by Ayenew et al,26 which is comparable to the prevalence of carbapenem resistance in this study. However, a systematic review in Africa indicates P. aeruginosa and A. baumannii showed that the lowest prevalence of carbapenemase-producing A. baumannii (4.7%).27 However, most available studies including a study for hospital environment28 reported higher prevalence of the MDR P. aeruginosa and A. baumannii in Ethiopia, calling for the application of genotypic methods to studies on mechanisms of resistance and spread.
The trend analysis in antimicrobial resistant from 2017 to 2021 showed that an increasing pattern from year to year for both A. baumanni and P. aeruginosa. Antibiotic policies and infection control measures are considered of immense value in fighting the mounting trends of nosocomial incidents. In addition, genuine efforts are needed to develop new antimicrobial agents against these pathogens and to monitor the efficacy of the presently accessible drugs.29,30 For instance, hand hygiene has been found to have impact in controlling infection. Infections due to drug-resistant gram-negative rods are an emerging risk factor for increased mortality in ICU. A number of studies have indicated the rising trends of pathogens from other body systems and hospital wards.19,29–31
Compared to the previous years, carbapenem resistance in both P. aeruginosa and A.baumanni has increased significantly in 2021 and this is comparable with previous studies conducted in Africa and Europe.25,32–34 The trend analysis in antimicrobial resistance showed that in the year 2021 there was high resistant trait in Ampicillin, Chloramphenicol, Cefepime, Gentamycin, which is comparable with similar study in Saudi Arabia and this could be due to pan drug abuse, intrinsic resistance, acquired resistance and other environmental factors.17,23,35
A study conducted at Kathmandu university hospital in Nepal in 2020, Pseudomonas spp was resistant to ciprofloxacin and Ceftazidime while ours was 31.8% for Ciprofloxacin and 45.2% for Ceftazidime. In the same study 94.4% of Acinetobacter were resistant to Ciprofloxacin, 88.89% were resistant to Piperacillin-tazobactam while our result was 61.5% for Ciprofloxacin and 63.2% for Piperacillin-tazobactam.36 The difference between our study and other studies could be linked to several factor-like environmental conditions, health practices, patient conditions, personal hygiene, drug abuse and number of patients involved.31,37,38
This study carries some limitations as the retrospective nature of it cannot eliminate the risk of selection bias and did not allow to make a better correlation between timing, type, and duration of empirical antimicrobial therapy as reported risk factors and the exact correlation between resistance and clinical outcomes. For the same reason, we could not verify the exact mechanism of resistance of A. baumannii and P.aeruginosa.
Conclusion
The majority of the pathogens in the studied hospital have evolved resistance to most of the antibiotics. In conclusion, there has been an increasing trend of antimicrobial resistance in A. baumanni and P. aeruginosa. Both species remain a therapeutic challenge in hospitals and health-care setting due to the increasing rate of with traits of MDR and resistance to high potent antimicrobial agents. Continuous surveillance and appropriate infection prevention and control program needs to be strengthened to circumvent the spread of these pathogens in health-care facilities.
Data Sharing Statement
The data sets used or analyzed during the current study are available from the corresponding author on reasonable request.
Ethical Declaration: Ethics Approval and Consent to Participate
Ethical clearance for this study was provided by Addis Ababa University, College of health science, department of medical Laboratory ethical review committee DRERC/479/21/MLS. Participants’ study data were recorded in codes and were kept private and confidentially. As this was a retrospective study informed consent was not collected from study participants but a permission was obtained from the hospitals to collect and access the laboratory logbook and it was also in accordance to Helsinki II declaration.
Consent for Publication
Not applicable as details like; videos or images related to study subjects were not recorded for this study.
Disclosure
The authors listed above declare that there is no competing interest.
References
- 1.Gupta V, Datta P, Chander J. Prevalence of metallo-beta lactamase (MBL) producing Pseudomonas spp. and Acinetobacter spp. in a tertiary care hospital in India. J Infect. 2006;52(5):311–314. doi: 10.1016/j.jinf.2005.08.013 [DOI] [PubMed] [Google Scholar]
- 2.Banerjee T, Mishra A, Das A, Sharma S, Barman H, Yadav G. High prevalence and endemicity of multidrug resistant Acinetobacter spp. in intensive care unit of a tertiary care hospital, Varanasi, India. J Pathog. 2018;2018:9129083. doi: 10.1155/2018/9129083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Francesco MA, Ravizzola G, Peroni L, Bonfanti C, Manca N. Prevalence of multidrug-resistant Acinetobacter baumannii and Pseudomonas aeruginosa in an Italian hospital. J Infect Public Health. 2013;6(3):179–185. doi: 10.1016/j.jiph.2012.11.006 [DOI] [PubMed] [Google Scholar]
- 4.Kaur A, Singh S. Prevalence of Extended Spectrum Betalactamase (ESBL) and Metallobetalactamase (MBL) producing Pseudomonas aeruginosa and Acinetobacter baumannii isolated from various clinical samples. J Pathog. 2018;2018:6845985. doi: 10.1155/2018/6845985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loivukene K, Sepp E, Adamson V, et al. Prevalence and antibiotic susceptibility of Acinetobacter baumannii, Pseudomonas aeruginosa and Klebsiella pneumoniae in Estonian intensive care units in comparison with European data. Scand J Infect Dis. 2006;38(11–12):1001–1008. doi: 10.1080/00365540600786507 [DOI] [PubMed] [Google Scholar]
- 6.Mirzaei B, Bazgir ZN, Goli HR, Iranpour F, Mohammadi F, Babaei R. Prevalence of multi-drug resistant (MDR) and extensively drug-resistant (XDR) phenotypes of Pseudomonas aeruginosa and Acinetobacter baumannii isolated in clinical samples from Northeast of Iran. BMC Res Notes. 2020;13(1):380. doi: 10.1186/s13104-020-05224-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohd Rani F, Ismail S, Abdullah FH, et al. Prevalence and antimicrobial susceptibilities of Acinetobacter baumannii and non-baumannii Acinetobacters from Terengganu, Malaysia and their carriage of carbapenemase genes. J Med Microbiol. 2018;67(11):1538–1543. doi: 10.1099/jmm.0.000844 [DOI] [PubMed] [Google Scholar]
- 8.Mohd Sazlly Lim S, Zainal Abidin A, Liew SM, Roberts JA, Sime FB. The global prevalence of multidrug-resistance among Acinetobacter baumannii causing hospital-acquired and ventilator-associated pneumonia and its associated mortality: a systematic review and meta-analysis. J Infect. 2019;79(6):593–600. doi: 10.1016/j.jinf.2019.09.012 [DOI] [PubMed] [Google Scholar]
- 9.Monnheimer M, Cooper P, Amegbletor HK, et al. High prevalence of carbapenemase-producing Acinetobacter baumannii in wound infections, Ghana, 2017/2018. Microorganisms. 2021;9(3):537. doi: 10.3390/microorganisms9030537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nasiri MJ, Zamani S, Fardsanei F, et al. Prevalence and mechanisms of carbapenem resistance in Acinetobacter baumannii: a comprehensive systematic review of cross-sectional studies from Iran. Microb Drug Resist. 2020;26(3):270–283. doi: 10.1089/mdr.2018.0435 [DOI] [PubMed] [Google Scholar]
- 11.Ababneh Q, Abu Laila S, Jaradat Z. Prevalence, genetic diversity, antibiotic resistance and biofilm formation of Acinetobacter baumannii isolated from urban environments. J Appl Microbiol. 2022;133(6):3617–3633. doi: 10.1111/jam.15795 [DOI] [PubMed] [Google Scholar]
- 12.Tilahun M, Gedefie A, Bisetegn H, Debash H. Emergence of high prevalence of extended-spectrum beta-lactamase and carbapenemase producing Acinetobacter species and pseudomonas aeruginosa among hospitalized patients at Dessie comprehensive specialized Hospital, North-East Ethiopia. Infect Drug Resist. 2022;15:895–911. doi: 10.2147/IDR.S358116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benmahmod AB, Said HS, Ibrahim RH. Prevalence and mechanisms of carbapenem resistance among Acinetobacter baumannii clinical isolates in Egypt. Microb Drug Resist. 2019;25(4):480–488. doi: 10.1089/mdr.2018.0141 [DOI] [PubMed] [Google Scholar]
- 14.De AS, Kumar SH, Baveja SM. Prevalence of metallo-beta-lactamase producing Pseudomonas aeruginosa and Acinetobacter species in intensive care areas in a tertiary care hospital. Indian J Crit Care Med. 2010;14(4):217–219. doi: 10.4103/0972-5229.76089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gildas Comlan Zohoun A, Moket D, El Hamzaoui S. Résistance à l’imipénème par production de métallo-β-lactamases par Acinetobacter baumannii et Pseudomonas aeruginosa à l’Hôpital militaire d’instruction Mohammed V de Rabat [Prevalence of Acinetobacter baumannii and Pseudomonas aeruginosa isolates resistant to imipenem by production of metallo-beta-lactamases in Rabat Military teaching hospital Mohammed V]. Ann Biol Clin. 2013;71(1):27–30. French doi: 10.1684/abc.2012.0778. [DOI] [PubMed] [Google Scholar]
- 16.CLSI. Performance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2017. [Google Scholar]
- 17.Al-Tawfiq JA, Mohandhas TX. Prevalence of antimicrobial resistance in Acinetobacter calcoaceticus-baumannii complex in a Saudi Arabian hospital. Infect Control Hosp Epidemiol. 2007;28(7):870–872. doi: 10.1086/518842 [DOI] [PubMed] [Google Scholar]
- 18.Cai B, Echols R, Magee G, et al. Prevalence of carbapenem-resistant gram-negative infections in the United States Predominated by Acinetobacter baumannii and Pseudomonas aeruginosa. Open Forum Infect Dis. 2017;4(3):ofx176. doi: 10.1093/ofid/ofx176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fazeli H, Rad TM, Esfahani BN, Pourdad A, Akbari M. Risk factors and prevalence of high resistant Acinetobacter spp among hospitalized patients. J Res Med Sci. 2014;19(5):480–481. [PMC free article] [PubMed] [Google Scholar]
- 20.Yayan J, Ghebremedhin B, Rasche K. Antibiotic resistance of pseudomonas aeruginosa in pneumonia at a Single University Hospital Center in Germany over a 10-Year Period. PLoS One. 2015;10(10):e0139836. doi: 10.1371/journal.pone.0139836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dehkordi SMH, Anvar SA, Rahimi E, Ahari H, Ataee M. Molecular investigation of prevalence, phenotypic and genotypic diversity, antibiotic resistance, frequency of virulence genes and genome sequencing in Pseudomonas aeruginosa strains isolated from lobster. Int J Food Microbiol. 2022;382:109901. doi: 10.1016/j.ijfoodmicro.2022.109901 [DOI] [PubMed] [Google Scholar]
- 22.Garza-Gonzalez E, Llaca-Diaz JM, Bosques-Padilla FJ, Gonzalez GM. Prevalence of multidrug-resistant bacteria at a tertiary-care teaching hospital in Mexico: special focus on Acinetobacter baumannii. Chemotherapy. 2010;56(4):275–279. doi: 10.1159/000319903 [DOI] [PubMed] [Google Scholar]
- 23.Al-Agamy MH, Shibl AM, Tawfik AF, Radwan HH. High prevalence of metallo-beta-lactamase-producing Pseudomonas aeruginosa from Saudi Arabia. J Chemother. 2009;21(4):461–462. doi: 10.1179/joc.2009.21.4.461 [DOI] [PubMed] [Google Scholar]
- 24.Aljindan R, Bukharie H, Alomar A, Abdalhamid B. Prevalence of digestive tract colonization of carbapenem-resistant Acinetobacter baumannii in hospitals in Saudi Arabia. J Med Microbiol. 2015;64(Pt 4):400–406. doi: 10.1099/jmm.0.000033 [DOI] [PubMed] [Google Scholar]
- 25.Addis T, Araya S, Desta K. Occurrence of multiple, extensive and pan drug-resistant Pseudomonas Aeruginosa and Carbapenemase production from presumptive isolates stored in a biobank at Ethiopian public health institute. Infect Drug Resist. 2021;14:3609–3618. doi: 10.2147/IDR.S327652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ayenew Z, Tigabu E, Syoum E, Ebrahim S, Assefa D, Tsige E. Multidrug resistance pattern of Acinetobacter species isolated from clinical specimens referred to the Ethiopian Public health institute: 2014 to 2018 trend analysis. PLoS One. 2021;16(4):e0250896. doi: 10.1371/journal.pone.0250896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kindu M, Derseh L, Gelaw B, Moges F. Carbapenemase-producing non-glucose-fermenting gram-negative Bacilli in Africa, Pseudomonas aeruginosa and Acinetobacter baumannii: a systematic review and meta-analysis. Int J Microbiol. 2020;2020:9461901. doi: 10.1155/2020/9461901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solomon FB, Wadilo F, Tufa EG, Mitiku M. Extended spectrum and metalo beta-lactamase producing airborne Pseudomonas aeruginosa and Acinetobacter baumanii in restricted settings of a referral hospital: a neglected condition. Antimicrob Resist Infect Control. 2017;6:106. doi: 10.1186/s13756-017-0266-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alvarez-Otero J, Lamas-Ferreiro JL, Gonzalez-Gonzalez L, et al. Resistencia a carbapenemas en Pseudomonas aeruginosa aisladas en urocultivos: prevalencia y factores de riesgo [Carbapenem resistance in Pseudomonas aeruginosa isolated from urine cultures: prevalence and risk factors]. Rev Esp Quimioter. 2017;30(3):183–194. Spanish. [PubMed] [Google Scholar]
- 30.Ibrahim ME. Prevalence of Acinetobacter baumannii in Saudi Arabia: risk factors, antimicrobial resistance patterns and mechanisms of carbapenem resistance. Ann Clin Microbiol Antimicrob. 2019;18(1):1. doi: 10.1186/s12941-018-0301-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mody L, Gibson KE, Horcher A, et al. Prevalence of and risk factors for multidrug-resistant Acinetobacter baumannii colonization among high-risk nursing home residents. Infect Control Hosp Epidemiol. 2015;36(10):1155–1162. doi: 10.1017/ice.2015.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anane AY, Apalata T, Vasaikar S, Okuthe GE, Songca S. Prevalence and molecular analysis of multidrug-resistant Acinetobacter baumannii in the extra-hospital environment in Mthatha, South Africa. Braz J Infect Dis. 2019;23(6):371–380. doi: 10.1016/j.bjid.2019.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ayobami O, Willrich N, Harder T, Okeke IN, Eckmanns T, Markwart R. The incidence and prevalence of hospital-acquired (carbapenem-resistant) Acinetobacter baumannii in Europe, Eastern Mediterranean and Africa: a systematic review and meta-analysis. Emerg Microbes Infect. 2019;8(1):1747–1759. doi: 10.1080/22221751.2019.1698273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowings M, Ehlers MM, Dreyer AW, Kock MM. High prevalence of oxacillinases in clinical multidrug-resistant Acinetobacter baumannii isolates from the Tshwane region, South Africa - an update. BMC Infect Dis. 2015;15:521. doi: 10.1186/s12879-015-1246-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aly MM, Abu Alsoud NM, Elrobh MS, Al Johani SM, Balkhy HH. High prevalence of the PER-1 gene among carbapenem-resistant Acinetobacter baumannii in Riyadh, Saudi Arabia. Eur J Clin Microbiol Infect Dis. 2016;35(11):1759–1766. doi: 10.1007/s10096-016-2723-8 [DOI] [PubMed] [Google Scholar]
- 36.Dawadi P, Khadka C, Shyaula M, et al. Prevalence of metallo-beta-lactamases as a correlate of multidrug resistance among clinical Pseudomonas aeruginosa isolates in Nepal. Sci Total Environ. 2022;850:157975. doi: 10.1016/j.scitotenv.2022.157975 [DOI] [PubMed] [Google Scholar]
- 37.Hu Y, Qing Y, Chen J, et al. Prevalence, risk factors, and molecular epidemiology of intestinal carbapenem-resistant Pseudomonas aeruginosa. Microbiol Spectr. 2021;9(3):e0134421. doi: 10.1128/Spectrum.01344-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi Q, Huang C, Xiao T, Wu Z, Xiao Y. A retrospective analysis of Pseudomonas aeruginosa bloodstream infections: prevalence, risk factors, and outcome in carbapenem-susceptible and -non-susceptible infections. Antimicrob Resist Infect Control. 2019;8:68. doi: 10.1186/s13756-019-0520-8 [DOI] [PMC free article] [PubMed] [Google Scholar]