Abstract
The gene encoding a nitric oxide reductase has been identified in Neisseria gonorrhoeae. The norB gene product shares significant identity with the nitric oxide reductases in Ralstonia eutropha and Synechocystis sp. and, like those organisms, the gonococcus lacks a norC homolog. The gonococcal norB gene was found to be required for anaerobic growth, but the absence of norB did not dramatically decrease anaerobic survival. In a wild-type background, induction of norB expression was seen anaerobically in the presence of nitrite but not anaerobically without nitrite or aerobically. norB expression is not regulated by FNR or NarP, but a functional aniA gene (which encodes an anaerobically induced outer membrane nitrite reductase) is necessary for expression. When aniA is constitutively expressed, norB expression can be induced both anaerobically and aerobically, but only in the presence of nitrite, suggesting that nitric oxide, which is likely to be produced by AniA as a product of nitrite reduction, is the inducing agent. This was confirmed with the use of the nitric oxide donor, spermine-nitric oxide complex, in an aniA null background both anaerobically and aerobically. NorB is important for gonococcal adaptation to an anaerobic environment, a physiologically relevant state during gonococcal infection. The presence of this enzyme, which is induced by nitric oxide, may also have implications in immune evasion and immunomodulation in the human host.
Denitrification is the process whereby microorganisms form dinitrogen (N2) from nitrate (NO3−) via nitrite (NO2−), nitric oxide (NO), and nitrous oxide (N2O) intermediates. Denitrifiers possess separate reductases which catalyze each intermediate step. Denitrification is usually seen in facultative anaerobes or microaerophilic organisms, but aerobic denitrification has been documented. The process and regulation of denitrification has been extensively studied in pseudomonads, Paracoccus spp., and Rhodobacter spp.; however, many other organisms, including fungi, denitrify. (See the recent comprehensive review by Zumft [33] for details of this process and the properties of its enzymes.)
Neisseria gonorrhoeae, the etiologic agent of the sexually transmitted disease gonorrhea, is now known to be a facultative anaerobe (17) which possesses a copper-containing nitrite reductase, AniA, in its outer membrane (5). aniA is very tightly regulated by oxygen and is only expressed under anaerobic or microaerobic conditions (13, 15). Antibodies to AniA have been found in sera from patients with gonococcal disease, indicating that AniA is expressed within the host and that anaerobiosis is a physiologically relevant environment in gonococcal infections (7). We were interested in denitrifying enzymes because copper-containing nitrite reductases are found in some of the dentrifying bacteria such as Pseudomonas aureofaciens and Rhodobacter sphaeroides, and these enzymes reduce NO2− to NO (33).
NO is known to be toxic to bacterial cells, often through its reaction with dioxygen, superoxide, or the metal centers in various enzymes. Bacteria which produce NO therefore also possess an NO reductase to eliminate the molecule. This two-subunit enzyme reduces NO to N2O and is encoded by the norC and norB genes (33).
Since it is likely that AniA produces NO and since it has been reported that gonococci produce nitrous oxide as a result of nitrite reduction (19), it is logical that the gonococcus would also possess an NO reductase. It is known that murine macrophages produce NO, and there is evidence that this is also true in human macrophages (reviewed in references 11 and 20). Therefore, an NO assault could be part of the innate immune system response to a bacterial invader. It was of interest to us to determine if the gonococcus, a strictly human pathogen, possesses this denitrifying enzyme, which could also have a role in the organism's survival in the face of an immune response. This study describes the identification and the basic regulation of the gonococcal norB gene and discusses the implications of the presence of the NorB enzyme in the pathogenesis of the organism.
MATERIALS AND METHODS
Growth of gonococcal strains.
All gonococcal strains (Table 1) were derived from strain F62 and were grown on GC medium base (Difco Laboratories, Detroit, Mich.) plates with 1% Kellogg's supplement (GCK) (16). When necessary, chloramphenicol or erythromycin was added at 1 or 2 μg ml−1, respectively. Aerobic plate cultures were grown at 37°C in a 5% CO2 incubator. Aerobic broth cultures were grown in GCK broth plus 0.042% (wt/vol) NaHCO3 in baffled flasks which were incubated with shaking at 240 rpm in a Gyrotory water bath shaker (New Brunswick Scientific Co., Edison, N.J.) at 37°C. Anaerobic cultures were incubated in a Coy anaerobic chamber (Coy Laboratory Products, Grass Lake, Mich.) at 37°C for 20 h. Nitrite was provided for anaerobically grown cultures by placing 40 μl of a 20% (wt/vol) NaNO2 solution on a sterile cellulose disk in the center of the plate. Cultures provided with nitrite grow in a characteristic halo around the nitrite disk, while cultures without nitrite remain viable but do not grow (24).
TABLE 1.
Gonococcal strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| RUG7001a | Wild type | 25 |
| RUG7002 | ΔaniA | This study |
| RUG7021 | Promoterless lacZ | 25 |
| RUG7058 | norB::ermC | This study |
| RUG7060 | norB::ermC ΔaniA | This study |
| RUG7500 | norB′-′lacZ | This study |
| RUG7501 | norB′-′lacZ aniA::ermC | This study |
| RUG7502 | norB′-′lacZ tacP::aniA | This study |
| RUG7503 | norB′-′lacZ narP::ermC | This study |
RUG7001 is an F62 strain which contains aniA′-′lacZ.
Gonococcal transformation.
Type 1 gonococcal cells were transformed as previously described (15) and plated on GCK plates containing the appropriate antibiotic.
Construction of norB′-′lacZ fusion.
The region upstream of the norB open reading frame (ORF) was amplified by PCR, using primers which created BamHI sites on both ends. The amplified fragment contained 177 bp upstream of the putative start codon and the first three codons of the ORF. This DNA fragment was digested with BamHI and ligated to pLES94 (25), which had also been digested with BamHI, to create a translational lacZ fusion. This ligation mixture was used to transform Escherichia coli MC1061. Transformants were selected by plating on Luria-Bertani medium plates containing ampicillin at 100 μg ml−1 and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) at 40 μg ml−1. After overnight incubation at 37°C, blue colonies were screened for plasmids containing the insert in the correct orientation. When an appropriate plasmid was identified (pEF1), it was used to transform gonococcal strain F62. Transformants were plated on GCK plates containing chloramphenicol. Chromosomal DNA was isolated from chloramphenicol-resistant colonies (15), and PCR was used to confirm the presence of the reporter construct. The resulting PCR product was sequenced to confirm the desired nucleotide sequence. The resulting strain was designated RUG7500.
Construction of a norB null mutant.
norB was amplified in two fragments by PCR with primers which created a ClaI site within the coding region. The two DNA fragments were purified and digested with ClaI. These digested fragments were ligated to an erythromycin resistance cassette (ermC) which had been amplified from pHSS23 (30) and digested with HpaII. This ligation mixture was transformed into N. gonorrhoeae RUG7001. This resulted in interruption of the norB gene with the erythromycin resistance cassette and the creation of strain RUG7058. The interruption of norB was confirmed by PCR.
The norB null mutant was also combined with an aniA null mutation. The insertionally inactivated norB was amplified by PCR from strain RUG7058. The resulting PCR product was used to transform RUG7002 which contains a 493-bp deletion in the aniA gene and the aniA′-′lacZ reporter construct. This norB aniA double mutant was designated RUG7060. Both mutations were confirmed by PCR.
Construction of the aniA null and constitutive strains.
To combine the norB′-′lacZ reporter with an aniA null background, the aniA gene insertionally inactivated with the ermC gene from strain RUG7011 (15) was amplified by PCR. This PCR product was used to transform RUG7500, which created strain RUG 7501. The presence of both the reporter construct and the ermC cassette was confirmed by PCR. To create the combination with the aniA constitutive mutant, pEF1 (norB′-′lacZ) was used to transform the F62 strain containing the previously described constitutive aniA (15), creating strain RUG7502. PCR was used to confirm the presence of both the reporter construct and the constitutive construct.
Construction of the narP null strain.
The norB′-′lacZ reporter was combined with a narP (encodes the response regulator of a two component system [15, 19]) null background by amplifying the insertionally inactivated narP from strain RUG7036 (15) by PCR. This PCR product was used to transform RUG7500, creating RUG7503. The presence of both the reporter construct and the ermC gene within narP was confirmed by PCR.
Anaerobic survival plate counts.
Nonpiliated colonies of desired strains were suspended in GCK broth at an optical density at 600 nm of approximately 0.1. Suspensions were serially diluted, and the appropriate dilutions were plated onto GCK plates in duplicate. Each strain was plated onto GCK with or without 2 mM NaNO2 and incubated both aerobically and anaerobically. Aerobic cultures were incubated for 48 h before enumeration. Anaerobic cultures were incubated for 24 h anaerobically and then 48 h aerobically to allow for visible colony formation. The number of CFU from the anaerobic cultures was compared to the CFU counts in the corresponding aerobic culture. The results are reported as the percentage of aerobic CFU and are the means of three independent assays for each strain.
Induction of norB by NO.
For each plate tested, 10 mg of the NO generator, spermine-nitric oxide complex (SPER-NO; Sigma RBI, St. Louis, Mo.) was dissolved in 50 μl of 0.1 N NaOH and used to saturate a cellulose disk. This disk was placed in the center of a plate which had been inoculated with a heavy suspension of organism, and the plate was incubated for 20 h either in the CO2 incubator or the anaerobic chamber. Cells were harvested with a sterile swab from around the disk and tested for β-galactosidase (β-Gal) activity. Disks saturated with 50 μl of 0.1 N NaOH were used for the no SPER-NO controls.
β-Gal assays.
Gonococcal cultures were assayed for β-Gal by the method of Miller (22) as previously described (15). Activity is reported in Miller units. The results reported are the averages of at least three independent assays for each strain.
Oligonucleotides and DNA sequencing.
All oligonucleotide synthesis and DNA sequencing was performed by the University of Rochester Core Nucleic Acid Laboratory.
Molecular biology techniques.
General techniques were performed according to standard protocols (2, 23). Plasmid preparations were obtained with the Wizard Plus Minipreps kit (Promega Corp., Madison, Wis.) or the QIAprep Spin Miniprep kit (Qiagen, Inc., Valencia, Calif.). DNA fragments were purified with Wizard PCR Preps kit (Promega), or with GENECLEAN Spin or MERmaid Spin kits (Bio 101, Vista, Calif.).
RESULTS
Identification of the norB gene in N. gonorrhoeae.
The work in our laboratory has focused on the aniA gene and its gene product in N. gonorrhoeae F62. Since AniA is a copper-containing nitrite reductase (5), it is likely that the product of nitrite reduction is NO. The next enzyme in the denitrifying pathway is the NO reductase (i.e., Nor). The sequence of what appears to be the entire coding region of a norB homolog was recently completed in the gonococcal genome (Gonococcal Genome Sequencing Project [http://dna1.chem.ou.edu/gono.html]). Surprisingly, N. gonorrhoeae does not encode the expected NorCB NO reductase which can be found in denitrifying organisms such as Pseudomonas aeruginosa (1), Pseudomonas stutzeri (4, 34), Paracoccus denitrificans (9), R. sphaeroides (3), and others. Instead, the gonococcus seems to encode a single subunit NO reductase, consisting only of NorB.
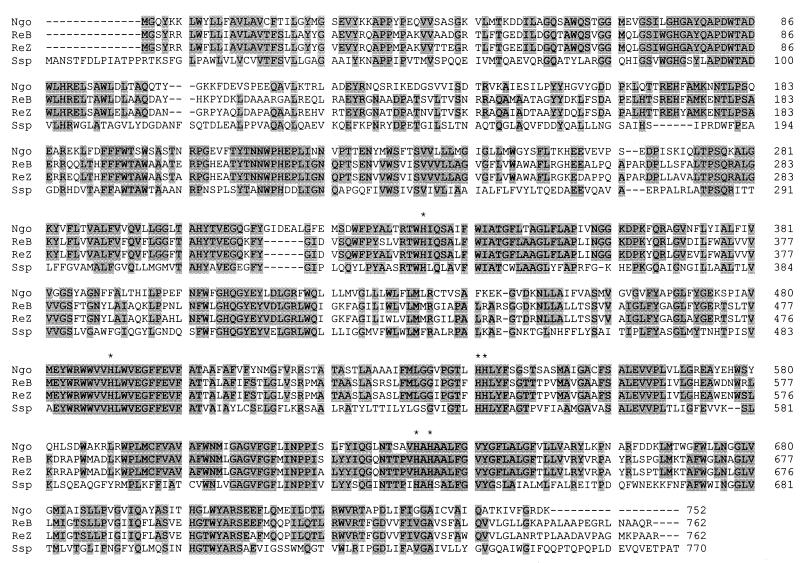
The predicted NorB protein has a calculated molecular mass of 84.3 kDa and an amino-terminal extension of approximately 260 to 270 amino acids. A BLAST search revealed significant identity to the Ralstonia eutropha (Alcaligenes eutrophus) and Synechocystis NO reductases along the entire length of the protein (61 and 42%, respectively) (Fig. 1). As in the gonococcus, the R. eutropha norB and norZ (genes encoding two isofunctional NO reductases) and Synechocystis norB predicted gene products have amino-terminal extensions, and the organisms lack norC genes (8). Like the R. eutropha NorB, the gonococcal NorB contains 14 potential transmembrane domains (data not shown) and all six conserved histidine residues (Fig. 1) and lacks a heme c binding motif (C-X-X-C-H).
FIG. 1.
Amino acid sequence alignment of NorB. The N. gonorrhoeae NorB (Ngo) predicted amino acid sequence is compared to the R. eutropha NorB (ReB; nucleotide sequence accession no. AF002217), the R. eutropha NorZ (ReZ; nucleotide sequence accession no. AF002661), and the Synechocystis sp. strain PCC6803 NorB (Ssp; nucleotide sequence accession no. D90917). Identical residues are shaded in gray, and conserved His residues are indicated by asterisks. Sequence alignment was performed using CLUSTAL W (http://www2.ebi.ac.uk/clustalw/).
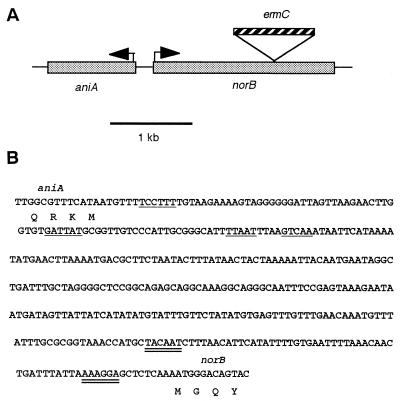
Upstream of the norB ORF is a gonococcal ribosome-binding site, AAAGGA, at position −11 from the putative translational start, and a potential ς70 −10 sequence, TACAAT, at position −61 (Fig. 2B). In contrast to other NO reductase genes, we did not find a complete FNR binding site, only a potential half site, TTGAA, at position −96.
FIG. 2.
Orientation of norB and aniA. (A) Schematic representation of the orientation of the norB and aniA genes in the gonococcal chromosome. Direction of transcription is indicated by arrows, and the location of the ermC insertion in strain RUG7058 is also indicated. (B) Nucleotide sequence of the aniA-norB intergenic region from gonococcal strain FA1090 obtained from the Genome Sequencing Project. The previously reported ribosome-binding site, ς70 −10, and FNR binding site for aniA (GenBank accession no. AF082184) are marked by a single underline. The putative ribosome-binding site and ς70 −10 for norB are indicated by double underlines. The first four amino acids for each gene product are also indicated below the nucleotide sequence.
aniA is located upstream of norB.
After locating the norB gene in the gonococcal genome (contig 17 on 03/03/00), we investigated the other ORFs surrounding norB. Considering our work with aniA, it was interesting to find aniA positioned about 375 bp upstream of norB, transcribed in the opposite direction (Fig. 2). This is similar to other organisms in which the nor genes are found in proximity to nitrite reductase (nir) genes. The two ORFs immediately downstream of norB are not homologous to other nir or nor genes.
norB is required for the anaerobic growth, but not the survival, of N. gonorrhoeae.
It has been reported previously that aniA is required for anaerobic growth in the gonococcus (15, 21). We were interested in determining if norB was also required. The norB gene was insertionally inactivated with the ermC gene. The resulting strain, RUG7058, was not attenuated in aerobic growth either in shaken broth cultures or on plates (data not shown) but, like the aniA mutant, was unable to grow anaerobically. However, RUG7058 survived anaerobic incubation and grew into visible colonies when transferred to a CO2 incubator (data not shown).
To quantify anaerobic survival, strains RUG7001 (wild type) and RUG7058 (norB) were plated and counted as described in Materials and Methods. Plate counts from the anaerobic plates were compared to the plate counts of the corresponding aerobic cultures. As seen in Table 2, in the absence of nitrite, only approximately 20% of both RUG7001 and RUG7058 (norB) survived anaerobic incubation. In contrast, in the presence of nitrite, virtually 100% of RUG7001 survived, while about 65% of RUG7058 (norB) survived. The morphology and size of the colonies of the two anaerobically incubated strains were identical when the cultures were subsequently incubated aerobically. Interestingly, this was not the case with the cultures that were grown aerobically. In the absence of nitrite, both strains grew with normal colony morphology and to normal size. In the presence of nitrite, however, the norB mutant exhibited a small colony phenotype (0.5 versus 3 mm) only when grown aerobically, while the colonies of the wild-type strain were normal. RUG7058 (norB) plates counts did not differ in the presence or absence of nitrite (data not shown), but the colony morphology was distinct and consistent.
TABLE 2.
Anaerobic survival of N. gonorrhoeae norB and aniA mutants
| Strain | Relevant genotype | Anaerobic survivala (% CFU ± SEM)
|
|
|---|---|---|---|
| +NO2 | −NO2 | ||
| RUG7001 | Wild type | 97 ± 14 | 16.7 ± 0.9 |
| RUG7058 | norB::ermC | 62.7 ± 8.7 | 18 ± 3.6 |
| RUG7002 | ΔaniA | 6.7 ± 2.3 | 26 ± 9.3 |
| RUG7060 | ΔaniA norB::ermC | 6.3 ± 1.5 | 13.3 ± 2.4 |
Expressed as a percentage of aerobic CFU ml−1 ± the SEM; + NO2, in the presence of 2 mM sodium nitrite; −NO2, in the absence of sodium nitrite.
We have previously observed low levels of aniA expression in gonococci grown on a plate overnight in the CO2 incubator (25). In the absence of NorB, the NO produced by AniA in the presence of nitrite could react with O2, forming toxic compounds such as N2O3, which has been found to cause mutations in bacteria (see references 26 and 32 and references therein). This accumulation of toxic molecules could account for the small-colony phenotype of RUG7058 (norB) observed only in the presence of nitrite on aerobically grown plates.
We reasoned that, similar to the results in P. stutzeri (4), if we eliminated the source of NO, AniA, this would rescue the small-colony phenotype observed aerobically in the presence of nitrite. RUG7002 (aniA) and RUG7060 (aniA norB) were plated for colony counts aerobically and anaerobically as described above. As we predicted, the colonies of the aniA norB double mutant were of normal size both in the presence and in the absence of nitrite when grown aerobically. This suggests that the production of NO in the absence of NorB was the initial cause for the small-colony phenotype. The anaerobic survival results for strains RUG7002 (aniA) and RUG7060 (norB aniA) were surprising (Table 2). While their survival in the absence of nitrite was comparable to RUG7001 and RUG7058 (norB), the anaerobic survival rate of both of the aniA mutants in the presence of nitrite was only 6%. This seems to indicate that, in anaerobic conditions, 2 mM nitrite is more toxic than NO to the gonococcus.
norB is induced anaerobically in the presence of nitrite.
β-Gal assays were performed on RUG7500 (norB′-′lacZ) grown both aerobically and anaerobically measuring norB expression (Table 3). In highly aerated broth cultures, the level of norB expression was about 20 Miller units both with and without 2 mM NO2−. In contrast, when RUG7500 was incubated anaerobically in the presence of nitrite, expression increased to about 1800 Miller units, while levels in cultures without nitrite remained low at 34 Miller units. In all cases the promoterless lacZ control (RUG7021) expression was virtually nil.
TABLE 3.
β-Gal activity of the gonococcal norB′-′lacZ fusion
| Strain | Relevant genotype | β-Gal activity (Miller units ± SEM)a
|
|||
|---|---|---|---|---|---|
| Aerobic cultures
|
Anaerobic cultures
|
||||
| +NO2 | −NO2 | +NO2 | −NO2 | ||
| RUG7021 | Promoterless lacZ | 0.1 ± 0.01 | 0.1 ± 0.01 | 0.4 ± 0.11 | 0.1 ± 0.03 |
| RUG7500 | norB′-′lacZ | 23 ± 1.5 | 20 ± 1.6 | 1,798 ± 18.4 | 34 ± 1.9 |
| RUG7501 | norB′-′lacZ aniA::ermC | 20 ± 1.2 | 21 ± 0.5 | 42 ± 6.4 | 29 ± 0.5 |
| RUG7502 | norB′-′lacZ tacP::aniA | 164 ± 11 | 19 ± 0.1 | 1,094 ± 33 | 41 ± 9.5 |
| RUG7503 | norB′-′lacZ narP::ermC | 21 ± 0.5 | 21 ± 0.4 | 1,081 ± 36 | 22 ± 2 |
Aerobic cultures were grown in GCK broth with shaking in baffled flasks in the presence (+) or absence (−) of 2 mM sodium nitrite (NO2). Anaerobic cultures were grown on plates with (+) or without (−) disks saturated with 20% (wt/vol) sodium nitrite (NO2).
The levels of norB expression exhibited a different pattern than what was observed with aniA (15). In the case of aniA, we observed significant induction due to anaerobiosis alone, with an additional increase in β-Gal activity when nitrite was added to the cultures, while norB remained at a basal level of expression during anaerobiosis with significant induction seen only in the presence of nitrite (Table 3). This suggests a different method of regulation.
norB is not regulated by NarP.
Since induction of norB was seen in the presence of nitrite, we investigated whether norB is activated by NarP. The narP gene was insertionally inactivated in the norB′-′lacZ reporter strain. The β-Gal activity obtained from this strain, RUG7503, was not greatly decreased compared to the wild-type background (Table 3), indicating that NarP plays no role in norB regulation.
AniA is required for the induction of norB.
To determine the effects of an aniA null mutation on norB expression, we created strain RUG7501, as described in Materials and Methods. Interestingly, when aniA was inactivated, we no longer observed the induction of norB in the presence of nitrite. Levels of β-Gal activity were 42 and 29 Miller units with and without NO2−, respectively (Table 3). There was no effect aerobically. Since it is likely that AniA produces NO as a product of NO2− reduction, these results suggest that NO may be the inducer for norB.
Induction of norB is seen aerobically when AniA is constitutively expressed.
When gonococcal cultures are grown in a highly aerated culture, no induction of the aniA gene is detectable due to the requirement for FNR (15). To produce AniA, and thus NO, aerobically and to determine the effects on norB expression, we utilized the previously described aniA constitutive mutant in which aniA is under the control of the tac promoter (15). RUG7502 contains the norB′-′lacZ fusion in an aniA constitutive background. Anaerobically, RUG7502 produced over 1,000 Miller units of β-Gal activity in the presence of nitrite. In highly aerated cultures, norB induction was seen at levels of over 150 Miller units only in the presence of nitrite, while levels in cultures without nitrite remained low. This suggested that AniA was indeed producing NO as a result of nitrite reduction, which in turn induced norB.
An NO generator induces norB expression in the absence of AniA.
To confirm that NO was responsible for norB induction, we utilized the NO generator SPER-NO. To eliminate the potential role of AniA in norB induction, we tested the NO generator on strain RUG7501, in which aniA is insertionally inactivated. For each plate tested, 10 mg of SPER-NO was solubilized in 0.1 N NaOH and was used to saturate a paper disk, a technique similar to the method used for adding nitrite to gonococcal plated cultures. While the actual levels of activity in the anaerobic cultures were somewhat variable, we consistently obtained significant induction of norB in the presence of SPER-NO both aerobically and anaerobically. Aerobic β-Gal levels (Miller units ± the standard error of the mean [SEM]) were 464 ± 91 in the presence of SPER-NO and 24 ± 3.3 in the absence of SPER-NO. Anaerobic levels were 884 ± 199 in the presence of SPER-NO and 28 ± 3.7 in the absence of SPER-NO. This provided convincing data that the gonococcal norB gene is induced by NO.
DISCUSSION
As a copper-containing nitrite reductase, it is highly probable that AniA produces NO as a result of its nitrite reduction, and it has been demonstrated that gonococci produce nitrous oxide as the product of nitrite reduction (19). We therefore set out to identify the NO reductase gene and to investigate the role of this enzyme in the anaerobic growth of the organism. It is also intriguing to speculate about the potential role of this enzyme in host immune evasion by the gonococcus.
The gene for the gonococcal NO reductase was identified by BLAST analysis of the gonococcal genome project. Surprisingly, we found only a norB homolog, while most denitrifiers possess a two-subunit enzyme encoded by norC and norB. The gonococcal NorB has significant identity (61%) to R. eutropha NorB and NorZ along the entire length of the predicted protein. In contrast, when gonococcal NorB is compared to the NorB found in P. denitrificans, for example, there is only 28% identity, and the gonococcal NorB has an amino-terminal extension of 271 amino acids (data not shown). Cramm et al. speculated that the amino-terminal extension may serve the same role as the NorC subunit and proposed further studies to explore this possibility (8).
When the norB gene is inactivated in N. gonorrhoeae, the organism can no longer grow anaerobically. However, a norB mutant survives anaerobic incubation in the presence of nitrite better than an aniA mutant (Table 2). This indicates that NO is not extremely toxic to the gonococcus under anaerobic conditions. In other organisms the inability to grow anaerobically in the absence of an NO reductase was attributed to the toxic effects of NO accumulation (4, 18). This does not seem to be the case in N. gonorrhoeae under these culture conditions, as shown by the anaerobic survival data.
Our studies on AniA indicate that it is anchored in the outer membrane of the gonococcus (5, 6, 14). This external location makes it unlikely that AniA is linked to anaerobic respiration and energy production. However, the gonococcal norB gene product appears to have multiple transmembrane domains indicative of a protein integral to the cytoplasmic membrane. From this location in the cytoplasmic membrane, it is quite feasible that NorB is involved in anaerobic respiration. Thus, the lack of NorB would result in the inability to respire under anaerobic conditions, preventing the growth of the organism until it is switched to environmental conditions which are permissive for aerobic respiration.
Our initial experiments looking at norB regulation revealed significant induction of the gene in the presence of nitrite but none due to anaerobiosis alone (Table 3). This is in contrast to aniA, which is induced anaerobically by FNR. Our previous work indicated that aniA was the only gene induced by FNR that was required for anaerobic growth (15). It was not surprising, therefore, that we could not locate an FNR binding site upstream of norB and that norB was not induced due to anaerobiosis alone. The data indicating that norB expression can be induced aerobically also rule out involvement of FNR. Since aniA requires NarP for the induction due to nitrite (15), we investigated whether or not norB also required NarP. Insertionally inactivating narP (RUG7503) had only a small effect on norB expression (Table 3). This decrease from 1,800 to 1,100 U is most likely due to the decrease in aniA expression due to the lack of NarP (15), resulting in diminished NO production and norB induction.
The results obtained with the aniA null and constitutive strains (Table 3) suggested that norB was induced by NO. Confirmation of this was achieved by providing exogenous NO through the use of a NO generator in the absence of AniA. When SPER-NO was added to either aerobically or anaerobically grown plated cultures of RUG7501, significant induction of norB was observed (see Results) compared to the low β-Gal activity observed both without an inducer and in the presence of nitrite (Table 3). This observation is not without precedence, since the NO reductases from both R. sphaeroides and P. denitrificans have been shown to be induced by NO (18, 27, 29).
The question that remains is which activating protein is responding to NO and inducing the gonococcal norB? The R. sphaeroides and P. denitrificans NO reductases are activated by members of the FNR family of proteins (NnrR [27] and NNR [28], respectively). These proteins bind to sequences which are very similar to the FNR consensus in E. coli (summarized in reference 33). In N. gonorrhoeae, it seems unlikely that a member of the FNR family is involved in norB induction since we are unable to locate a full FNR binding site upstream of norB, although we cannot rule out a different binding consensus in the gonococcus. We have also searched the gonococcal genome with the protein sequence of several members of the FNR family of proteins and have only found the previously reported FNR (15, 19). Two other well-characterized NO-responsive regulators are SoxR and SoxS in E. coli (reviewed in reference 31). Again, BLAST analysis of the gonococcal genome found no homologs to these regulators. Further experimentation will be required, including pinpointing a binding site upstream of norB, in order to identify the norB activator.
We are intrigued by the possibility that the gonococcal NorB may function in immune evasion. Flavohemoglobin (Hmp), SoxRS, Cu,Zn-superoxide dismutase, and homocysteine have all been found to provide some pathogenic bacteria protection against NO (reviewed in reference 31). Specifically, E. coli soxRS mutants are more sensitive to macrophage killing, and the Salmonella enterica serovar Typhimurium Cu,Zn-superoxide dismutase protects against phagocyte-derived NO. In addition to SoxRS, we have also been unable to locate an Hmp homolog in the gonococcal genome, and although there is a partial coding region in GenBank for a gonococcal sod gene (accession no. U11277), the sod ORF identified in the genome appears to contain a frameshift, and no one has reported a functional superoxide dismutase in N. gonorrhoeae. In the absence of any of these known proteins involved in the defense against NO, it is possible that the gonococcal NorB is the means by which the organism protects itself against NO assault from macrophages.
It is estimated that approximately 50% of all primary gonococcal infections in women are asymptomatic, which leads to higher rates of complicated gonococcal disease, such as pelvic inflammatory disease and disseminated gonococcal infection (10). The effects of NO on the inflammatory response are dependent on the concentration. Low levels of NO appear to be anti-inflammatory, while higher levels such as those produced by iNOS (NOS-2) in response to bacterial lipopolysaccharide, can be proinflammatory (reviewed in reference 12). Gonococci, with the ability to both produce and reduce NO, may exert a modulatory effect on NO levels and thus on the host inflammatory response. This may result in the development of symptomatic or asymptomatic infections, depending on the relative activities of gonococcal nitrite reductase and NO reductase.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant RO1 AI11709 from the National Institutes of Health to V.L.C. T.C.H. was supported by National Institutes of Health training grant T32 AI07363.
We thank Lin Silver and Lori Wright for their expert technical assistance.
REFERENCES
- 1.Arai H, Igarashi Y, Kodama T. The structural genes for nitric oxide reductase from Pseudomonas aeruginosa. Biochim Biophys Acta. 1995;1261:279–284. doi: 10.1016/0167-4781(95)00018-c. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 3.Bartnikas T B, Tosques I E, Laratta W P, Shi J, Shapleigh J P. Characterization of the nitric oxide reductase-encoding region in Rhodobacter sphaeroides 2.4.3. J Bacteriol. 1997;179:3534–3540. doi: 10.1128/jb.179.11.3534-3540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun C, Zumft W G. The structural genes of the nitric oxide reductase complex from Pseudomonas stutzeri are part of a 30-kilobase gene cluster for dentrification. J Bacteriol. 1992;174:2394–2397. doi: 10.1128/jb.174.7.2394-2397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardinale J A. Ph.D. thesis. Rochester, N.Y: University of Rochester; 1999. [Google Scholar]
- 6.Clark V L, Campbell L A, Palermo D A, Evans T M, Klimpel K W. Induction and repression of outer membrane proteins by anaerobic growth of Neisseria gonorrhoeae. Infect Immun. 1987;55:1359–1364. doi: 10.1128/iai.55.6.1359-1364.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark V L, Knapp J S, Thompson S, Klimpel K W. Presence of antibodies to the major anaerobically induced gonococcal outer membrane protein in sera from patients with gonococcal infections. Microb Pathog. 1988;5:381–390. doi: 10.1016/0882-4010(88)90038-1. [DOI] [PubMed] [Google Scholar]
- 8.Cramm R, Siddiqui R A, Friedrich B. Two isofunctional nitric oxide reductases in Alcaligenes eutrophus H16. J Bacteriol. 1997;179:6769–6777. doi: 10.1128/jb.179.21.6769-6777.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Boer A P N, van der Oost J, Reijnders W N M, Westerhoff H V, Stouthamer A H, van Spanning R J M. Mutational analysis of the nor gene cluster which encodes nitric-oxide reductase from Paracoccus denitrificans. Eur J Biochem. 1996;242:592–600. doi: 10.1111/j.1432-1033.1996.0592r.x. [DOI] [PubMed] [Google Scholar]
- 10.Division of STD Prevention. Sexually transmitted disease surveillance, 1998. Atlanta, Ga: Department of Health and Human Services, Atlanta. Centers for Disease Control and Prevention; 1999. [Google Scholar]
- 11.Fang F C. Mechanisms of nitric oxide-related antimicrobial activity. J Clin Investig. 1997;99:2818–2825. doi: 10.1172/JCI119473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grisham M B, Jourd'heuil D, Wink D A. Nitric oxide I. Physiological chemistry of nitric oxide and its metabolites: implications in inflammation. Am J Physiol. 1999;276:G315–G321. doi: 10.1152/ajpgi.1999.276.2.G315. [DOI] [PubMed] [Google Scholar]
- 13.Hoehn G T, Clark V L. Isolation and nucleotide sequence of the gene (aniA) encoding the major anaerobically induced outer membrane protein of Neisseria gonorrhoeae. Infect Immun. 1992;60:4695–4703. doi: 10.1128/iai.60.11.4695-4703.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoehn G T, Clark V L. The major anaerobically induced outer membrane protein of Neisseria gonorrhoeae, Pan 1, is a lipoprotein. Infect Immun. 1992;60:4704–4708. doi: 10.1128/iai.60.11.4704-4708.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Householder T C, Belli W A, Lissenden S, Cole J A, Clark V L. cis- and trans-acting elements involved in regulation of aniA, the gene encoding the major anaerobically induced outer membrane protein in Neisseria gonorrhoeae. J Bacteriol. 1999;181:541–551. doi: 10.1128/jb.181.2.541-551.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kellogg D S, Jr, Peacock W L, Jr, Deacon W E, Brown L, Pirkle C I. Neisseria gonorrhoeae I. Virulence genetically linked to clonal variation. J Bacteriol. 1963;85:1274–1279. doi: 10.1128/jb.85.6.1274-1279.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knapp J S, Clark V L. Anaerobic growth of Neisseria gonorrhoeae coupled to nitrite reduction. Infect Immun. 1984;46:176–181. doi: 10.1128/iai.46.1.176-181.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwiatkowski A V, Shapleigh J P. Requirement of nitric oxide for induction of genes whose products are involved in nitric oxide metabolism in Rhodobacter spheroides 2.4.3. J Biol Chem. 1996;271:24382–24388. doi: 10.1074/jbc.271.40.24382. [DOI] [PubMed] [Google Scholar]
- 19.Lissenden, S., S. Mohan, T. Overton, T. Regan, H. Crooke, J. A. Cardinale, T. C. Householder, D. O'Conner, V. L. Clark, H. Smith, and J. A. Cole. Identification of transcription activators which regulate gonococcal adaptation from aerobic to anaerobic or oxygen-limited growth. Mol. Microbiol., in press. [DOI] [PubMed]
- 20.MacMicking J, Xie Q-W, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 21.Mellies J, Jose J, Meyer T F. The Neisseria gonorrhoeae gene aniA encodes an inducible nitrite reductase. Mol Gen Genet. 1997;256:525–532. doi: 10.1007/s004380050597. [DOI] [PubMed] [Google Scholar]
- 22.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. Assay of β-galactosidase; pp. 352–355. [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Short H B, Clark V L, Kellogg D S, Jr, Young F E. Anaerobic survival of clinical isolates and laboratory strains of Neisseria gonorrhoeae: use in transfer and storage. J Clin Microbiol. 1982;15:915–919. doi: 10.1128/jcm.15.5.915-919.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silver L E, Clark V L. Construction of a translational lacZ fusion system to study gene regulation in Neisseria gonorrhoeae. Gene. 1995;166:101–104. doi: 10.1016/0378-1119(95)00605-6. [DOI] [PubMed] [Google Scholar]
- 26.Squadrito G L, Pryor W A. Oxidative chemistry of nitric oxide: the roles of superoxide, peroxynitrite, and carbon dioxide. Free Radic Biol Med. 1998;25:392–403. doi: 10.1016/s0891-5849(98)00095-1. [DOI] [PubMed] [Google Scholar]
- 27.Tosques I E, Shi J, Shapleigh J P. Cloning and characterization of nnrR, whose product is required for the expression of proteins involved in nitric oxide metabolism in Rhodobacter sphaeroides 2.4.3. J Bacteriol. 1996;178:4958–4964. doi: 10.1128/jb.178.16.4958-4964.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Spanning R J M, De Boer A P N, Reijnders W N M, Spiro S, Westerhoff H V, Stouthamer A H, Van der Oost J. Nitrite and nitric oxide reduction in Paracoccus denitrificans is under the control of NNR, a regulatory protein that belongs to the FNR family of transcriptional activators. FEBS Lett. 1995;360:151–154. doi: 10.1016/0014-5793(95)00091-m. [DOI] [PubMed] [Google Scholar]
- 29.Van Spanning R J M, Houben E, Reijnders W N M, Spiro S, Westerhoff H V, Saunders N. Nitric oxide is a signal for NNR-mediated transcription activation in Paracoccus denitrificans. J Bacteriol. 1999;181:4129–4132. doi: 10.1128/jb.181.13.4129-4132.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wainwright L A, Pritchard K H, Seifert H S. A conserved DNA sequence is required for efficient gonococcal pilin antigenic variation. Mol Microbiol. 1994;13:75–87. doi: 10.1111/j.1365-2958.1994.tb00403.x. [DOI] [PubMed] [Google Scholar]
- 31.Watmough N J, Butland G, Cheesman M R, Moir J W B, Richardson D J, Spiro S. Nitric oxide in bacteria: synthesis and consumption. Biochim Biophys Acta. 1999;1411:456–474. doi: 10.1016/s0005-2728(99)00032-8. [DOI] [PubMed] [Google Scholar]
- 32.Wink D A, Mitchell J B. Chemical biology of nitric oxide: insights into regulatory, cytotoxic, and cytoprotective mechanisms of nitric oxide. Free Radic Biol Med. 1998;25:434–456. doi: 10.1016/s0891-5849(98)00092-6. [DOI] [PubMed] [Google Scholar]
- 33.Zumft W G. Cell biology and molecular basis of dentrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zumft W G, Braun C, Cuypers H. Nitric oxide reductase from Pseudomonas stutzeri. Eur J Biochem. 1994;219:481–490. doi: 10.1111/j.1432-1033.1994.tb19962.x. [DOI] [PubMed] [Google Scholar]