Abstract
Porphyromonas gingivalis, a black-pigmented, gram-negative anaerobe, is found in periodontitis lesions, and its presence in subgingival plaque significantly increases the risk for periodontitis. In contrast to many bacterial pathogens, P. gingivalis strains display considerable variability, which is likely due to genetic exchange and intragenomic changes. To explore the latter possibility, we have studied the occurrence of insertion sequence (IS)-like elements in P. gingivalis W83 by utilizing a convenient and rapid method of capturing IS-like sequences and through analysis of the genome sequence of P. gingivalis strain W83. We adapted the method of Matsutani et al. (S. Matsutani, H. Ohtsubo, Y. Maeda, and E. Ohtsubo, J. Mol. Biol. 196:445–455, 1987) to isolate and clone rapidly annealing DNA sequences characteristic of repetitive regions within a genome. We show that in P. gingivalis strain W83, such sequences include (i) nucleotide sequence with homology to tRNA genes, (ii) a previously described IS element, and (iii) a novel IS-like element. Analysis of the P. gingivalis genome sequence for the distribution of the least used tetranucleotide, CTAG, identified regions in many of the initial 218 contigs which contained CTAG clusters. Examination of these CTAG clusters led to the discovery of 11 copies of the same novel IS-like element identified by the repeated sequence capture method of Matsutani et al. This new 1,512-bp IS-like element, designated ISPg5, has features of the IS3 family of IS elements. When a recombinant plasmid containing much of ISPg5 was used in Southern analysis of several P. gingivalis strains, including clinical isolates, diversity among strains was apparent. This suggests that ISPg5 and other IS elements may contribute to strain diversity and can be used for strain fingerprinting.
Porphyromonas gingivalis, a black-pigmented, gram-negative anaerobe, is frequently found in periodontitis lesions (8, 12, 17, 30) and is a risk factor for periodontitis (13). Many bacterial pathogens display genotypic clonality; that is, clinical isolates from numerous sources fall into one or only a few groups (clonal types) based on electrophoretic patterns of their genomic DNA. However, this is rarely true for P. gingivalis. As many as 100 different clonal types have been found in periodontitis patients (2, 22, 23, 29, 43, 45). Pathologic changes associated with chronic infection in the progressing periodontitis lesion may accentuate differences between clonal types. Such differences could be caused by a variety of genetic mechanisms including genetic exchange and recombination, insertion sequence (IS) movement and related activities, and recombination events involving nonmobile repeated DNA sequences.
IS elements are transposable elements that are usually 800 to 2,500 bp in length, contain an open reading frame (ORF) encoding a transposase, and have terminal inverted repeats (usually 10 to 40 bp) (10). IS elements are usually flanked by short direct repeats (2 to 13 bp) which represent duplication of the target insertion site; the size of each repeat is specific for the particular IS (10). Thus far, three IS-like elements have been found in the genome of P. gingivalis W83. Maley and Roberts (26) identified and described the first IS element in P. gingivalis, which they designated IS1126. An analysis of laboratory isolates by Dong et al. (7) revealed that strains contained between 25 and 35 copies of IS1126. The restriction fragment length polymorphism (RFLP) pattern that they observed suggested considerable genetic heterogeneity among these strains. A second IS element in P. gingivalis, PGIS2, was described by Wang et al. (44). As with IS1126, when PGIS2 was used as a probe in Southern analysis of several P. gingivalis strains, considerable diversity in the pattern of hybridizing fragments was noted. These findings illustrate the potential utility of IS elements in studies of relatedness of individual strains and bacterial evolution and phylogeny. Lewis and Macrina (21) isolated and described a third IS-like element of P. gingivalis, IS195, which they suggested might play a role in regulation of protease genes by insertional inactivation.
Adaptation through natural genetic engineering involving the action of transposable elements and other genetic activities has been reviewed by Shapiro (39, 40). A variety of intragenomic sequence modifications are now known to be a regularly occurring, adaptively important component of genetic change. Such genetic flexibility has been postulated as a means for bacteria to adapt to different environments and ecosystems. The prominent role of IS elements in these adaptive strategies has led us to systematically extend our study of such elements in P. gingivalis. We anticipate that such work will help illuminate the dramatic nonclonality observed in clinical isolates of this pathogen.
Isolating and cloning rapidly annealing DNA from P. gingivalis allowed us to capture DNA sequences that were repeated in the genome. This approach was complemented by a search of the genomic sequence for rare oligomers. These parallel approaches complemented one another and led to the identification of the novel IS-like element described in this report.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. gingivalis strains W83, W50, ATCC 33277, V2299, V2300, V2302, V2305, V2306, V2307, and V2308 were used in this study. Strains with the prefix V are fresh clinical isolates from periodontal lesions. Bacteria were grown in brain heart infusion broth (Sigma Chemical Co., St. Louis, Mo.) supplemented with hemin (5 μg/liter; Sigma) and vitamin K1 (1 μg/liter; Sigma) under 10% H2–10% CO2–80% N2. Escherichia coli strains were grown in Luria-Bertani broth (35). Carbenicillin (50 μg/ml) was used to select for E. coli transformants.
Isolation and characterization of DNA.
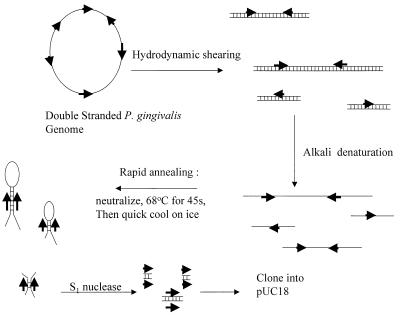
Genomic DNA was prepared from P. gingivalis by the method of Marmur (27). To isolate repeated nucleotide sequence DNA in P. gingivalis W83, we used the technique of Matsutani et al. (28) as summarized in Fig. 1. Three milligrams of this DNA suspended in 10 ml of 0.1× Tris-EDTA buffer (1 mM Tris-HCl, 0.1 mM EDTA [pH 8.0]) was sheared by 15 passes through a 26-gauge needle, denatured with alkali (pH adjusted to 12.3 with 2 M NaOH), neutralized with acid (2 M HCl), and incubated for 30 to 45 s at 68°C. This was followed by rapid cooling on ice. The DNA was then divided into 20 1.8-ml Eppendorf tubes, precipitated with ethanol, and suspended in 100 μl of S1 nuclease buffer containing 150 U of S1 nuclease to digest single-stranded DNA. This was followed by extraction with phenol and precipitation with ethanol. The resulting DNA pellet was suspended in 20 μl of Tris-EDTA buffer, treated with 2 μg of RNase/tube, and fractionated on a 1.4% agarose gel. The region of the gel from 0.8 to 1.6 kb was cut out of multiple lanes of a preparative gel and electroeluted. This DNA was treated with the Klenow fragment to ensure that the ends were blunt and then ligated into SmaI-digested, dephosphorylated pUC18 vector. After ligation, E. coli strain TB1 was transformed and spread onto Luria-Bertani plates containing carbenicillin and the indicator substrate X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Plasmid DNA preparations were made from white colonies, and EcoRI/BamHI double digests of the DNA were analyzed on 0.8% agarose gels.
FIG. 1.
The Matsutani et al. (28) method for isolating repeated DNA. Genomic DNA was subjected to hydrodynamic shearing, alkali denaturation, neutralization, and a brief period of reannealing followed by quick cooling on ice to favor intra- and intermolecular hybridization of repetitive DNA. Single-stranded DNA was digested with S1 nuclease, and remaining double-stranded DNA was cloned into pUC18. Arrows in bold indicate repetitive DNA.
Recombinant plasmid DNA was purified using a Qiagen plasmid midi kit as instructed by the manufacturer (Qiagen, Inc., Chatsworth, Calif.). Clones hybridizing with multiple bands in Southern analyses were submitted for nucleotide sequence determination. The dideoxy-chain termination nucleotide sequencing method using an ABI Prism DNA sequencing kit (Perkin-Elmer Corp.), the standard primers for pUC18 [M13F(−47) and M13R(−24)], and an Applied Biosystems (Foster City, Calif.) model 377 sequencing system at the DNA core facilities of Virginia Commonwealth University were employed. Nucleotide sequence data were then used in BLASTN and TBLASTN searches of GenBank (3). These data were also used to search the current P. gingivalis genome database (http://www.tigr.org/tdb/mdb/mdb.html).
Southern hybridization.
P. gingivalis genomic DNA was digested with BamHI/EcoRI. Electrophoretically separated DNA was transferred to positively charged nylon membranes (Boehringer Mannheim Corp., Indianapolis, Ind.), and hybridization was performed using a probe labeled with positively charged peroxidase as instructed by the manufacturer (ECL kit; Amersham Corp., Arlington Heights, Ill.). Hybridizing bands were visualized by autoradiography using intensifying screens on X-Omat LS films (Eastman Kodak Company, Rochester, N.Y.).
Whole-genome nucleotide sequence analysis.
The nucleotide sequence for the genome of P. gingivalis strain W83 was determined by the whole-genome shotgun sequencing method described for Haemophilus influenzae (9). At the time this study was initiated, the random nucleotide sequences had been assembled into 218 contigs. Simple programs were written in QuickBASIC to determine the frequency of each base and each of the possible di-, tri-, tetra-, penta-, and hexamers in the initial contigs. The distribution of the most underrepresented tetra- and hexamer nucleotide sequences in the genome was examined visually by highlighting the target nucleotide sequence in a Microsoft Word file. Contigs that contained unusual clusters of rare oligomers were searched against GenBank using BLASTN and BLASTX (3) and against the P. gingivalis genome nucleotide sequence in the current National Center for Biotechnology Information (NCBI) genome database.
Nucleotide sequence accession number.
The nucleotide sequence of ISPg5 was deposited in the GenBank nucleotide sequence database under accession no. AF224744.
RESULTS
Isolation, cloning, and characterization of rapidly annealing DNA.
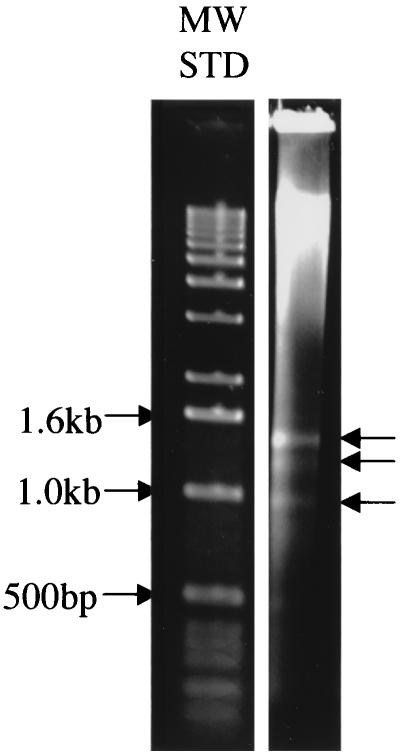
Rapidly annealing DNA, which is expected to contain repetitive nucleotide sequences, was isolated and cloned from genomic DNA of P. gingivalis strain W83 by the method of Matsutani et al. (28) (Fig. 1). The resulting double-stranded DNA was electrophoretically separated on an agarose gel (Fig. 2), and DNA in the region from 0.8 to 1.6 kb was excised, isolated, and cloned in E. coli. Plasmid DNA was prepared from transformants, and inserts were released by restriction endonuclease digestion (Fig. 3A). Clones 12, 14, 15, and 18 hybridized with multiple bands, suggesting that they were repeated in the genome of strain W83 (data for clone 14 are shown in Fig. 3B). The nucleotide sequence from insert DNA of clones 15 and 18 overlapped and together contained much of the DNA sequence of the previously reported IS1126 element (data not shown). Comparison of IS-flanking DNA found in the two clones to the P. gingivalis genomic nucleotide sequence database indicated that clones 15 and 18 contained distinct copies of IS1126 inserted at different genomic loci. Partial nucleotide sequence analysis of clone 12 indicated that it was a chimera of two nucleotide sequences not found on the same contig in the P. gingivalis database. One end of the insert displayed similarity to the peptidase T gene of Bacillus subtilis; the other showed similarity to tRNA genes of several bacterial species. These fragments were likely joined during the ligation step of the cloning procedure.
FIG. 2.
Agarose gel electrophoresis of DNA prepared by the method of Matsutani et al. (28). Arrows on the right indicate distinct bands visible by ethidium bromide staining. MW STD, molecular weight standards.
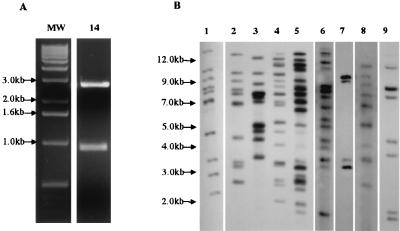
FIG. 3.
(A) Agarose gel of BamHI/EcoRI digest of clone 14, containing IS5. Upper band is pUC18 vector; lower band is insert DNA. MW, molecular weight standards. (B) Southern blot of a BamHI/EcoRI digest of nine P. gingivalis strains probed with the ISPg5 clone. Strains in lanes 1 to 9: W83, W50, V2299, V2300, V2302, V2305, V2306, V2307, and V2308, respectively. The figure is a composite photograph of different exposures of the same blot. The positions of the bands are in register to the original blot.
The DNA sequence of the insert from clone 14 was subjected to a BLASTX search of GenBank. Forty-five percent identity was found between the translated nucleotide sequence and the sequence of a transposase gene from Bacteroides thetaiotaomicron (accession no. U67062). BLASTN search of clone 14 nucleotide sequence against the P. gingivalis genome database identified regions in multiple contigs with 98 to 100% sequence identity. The region flanking the transposase-like gene in one of the P. gingivalis contigs was analyzed further. The surrounding DNA sequences were found to possess additional features of an IS, including terminal inverted repeats and a second ORF associated with the IS3 family of IS elements, as discussed below (see “Structure of ISPg5”). This suggested that clone 14 was derived from an IS-like element.
Whole-genome sequence analysis.
An element identical to that captured by the rapid annealing technique of Matsutani et al. (28) was simultaneously identified by direct analysis of the P. gingivalis W83 genomic nucleotide sequence. The discovery of this element in the genome of the W83 strain was facilitated by searching the genome for short, underrepresented nucleotide sequences. As has been found in other bacterial genomes (4), CTAG is the most underrepresented tetranucleotide sequence in the P. gingivalis genome, occurring at only 1/15 the expected frequency. After highlighting CTAG in the 218 contigs, visual inspection revealed that CTAG tetramers occurred in nonrandom clusters. Based on 605 CTAG tetranucleotide sequences in the 2.4-Mb P. gingivalis genome, the expected frequency was one occurrence in approximately 4,000 bases. However, certain 4,000-base regions had clusters of up to 20 CTAGs. BLASTX searches of GenBank with several of these regions showed that they contained homology to a transposase from B. thetaiotaomicron (accession no. U67062). When the ∼1,500-bp region homologous to U67062 was used in a BLASTN search of the P. gingivalis database, 14 contigs which contained whole or partial copies of the 1,500-base element were identified. Subsequently, as the contigs were assembled, it was determined that 11 complete copies were present. This is consistent with Southern blot analysis of strain W83 (Fig. 3B, lane 1).
Thus, the same novel IS-like sequence was identified both by direct capture using the technique of Matsutani et al. (28) and by computer analysis of available W83 genomic DNA sequence. In keeping with our proposal described in Discussion, we designated this element ISPg5 to simplify the naming of such sequences in P. gingivalis.
Structure of ISPg5.
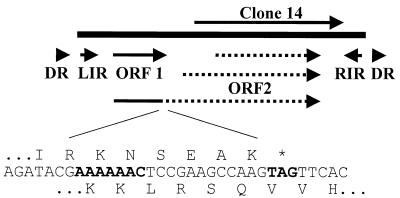
The structure of ISPg5 is illustrated in Fig. 4. The 1,512-bp element is bounded on either side by 30-bp perfect inverted repeats that share homology with the terminal inverted repeats of the IS3 family of IS elements (25). Four base pairs downstream from the end of the left inverted repeat is the start codon for a 126-amino-acid ORF with limited homology to an ORF present in IS elements of the IS3 family termed ORF1 or ORFA (25). As noted previously for a variety of IS elements, the distance between ORF1 and the left inverted repeat could accommodate a −10 promoter hexamer but not a −35 hexamer. Several other IS elements with this structure possess −35 hexamers within their right inverted repeats that can reconstitute an intact ORF1 promoter upon IS circularization or promote expression of adjacent genes following IS insertion (25).
FIG. 4.
Organization of ISPg5. The position of the insert DNA in clone 14 is shown, along with the following features of ISPg5: DR (3-bp direct repeat), LIR (left inverted repeat), RIR (right inverted repeat), ORF1, and three possible versions of ORF2, including a fusion with ORF1 caused by frameshifting. The nucleotide and amino acid sequences of the fusion region are shown, with the frameshift-promoting region and the stop codon for ORF1 indicated in bold.
A second ORF with homology to the ORF2 or ORFB of IS3 elements (25) occurs downstream from ORF1 in ISPg5. A potential ATG start site for ORF2 occurs 203 bp downstream from ORF1, the use of which would produce a 262-amino-acid protein. Alternatively, the use of a CTG start site 53 bp downstream from ORF1 would produce a 312-amino-acid protein. Finally, common to transposases of many IS elements in the IS1 and IS3 families (5, 25, 37), it appears that ribosomal frameshifting may produce a fusion of ORF1 to ORF2 in ISPg5 (Fig. 4). The nucleotide sequence A6C, which occurs at the site of the −1 frameshift in IS1 (five codons from the stop codon of the first ORF and overlapping the second ORF), occurs in a similar location in ISPg5 (four codons from the stop codon of ORF1 and overlapping ORF2, which is in the −1 frame). A similar nucleotide sequence, A6G, has been shown to promote ribosomal frameshifting in the IS3 family of elements, IS150 and IS911 (5). Regardless of the start site utilized, ORF2 contains a perfectly conserved DD(35)E motif, which is common to transposases and retroviral integrases (25). Thus, the size and organization of ISPg5, the size and nucleotide sequence of its terminal inverted repeats, and the nucleotide sequence homology and features of its ORFs all suggest that it belongs to the IS3 family of bacterial IS elements. Analysis of the P. gingivalis genome indicates that 5 of the 11 ISPg5 elements have perfect three-base duplicated target sites flanking the element. The remaining six elements have mismatched repeats indicating recombination events mediated by these IS elements. The G+C content of the element is 47.7%, compared to ∼48.2% for the unfinished genomic nucleotide sequence, suggesting that ISPg5 was not acquired recently by horizontal transfer.
None of the eleven ISPg5 insertions interrupted an identifiable gene. The flanking genes for each insertion are shown in Table 1. Six of the flanking regions contained conserved hypothetical genes. Elements 4 and 7 inserted into iterons (1) with the consensus sequence (CCAAAACCACTAGTAGATTTTC)4–5. Note that this iteron contains the rare tetramer CTAG (in boldface). Element 4 is split by insertion of an ISPg1 and is adjacent to an ISPg7. Element 6 is between two copies of ISPg1. Element 10 has a 246-base insert of an unknown type that is found at four other locations in the P. gingivalis genome.
TABLE 1.
ISPg5 insertion sites
| Site no. | Positiona | Targetb | Upstream genec | Downstream gene | Commentd |
|---|---|---|---|---|---|
| 1 | 7,070–8,582 | CCGAGATTGT--GTGGTTCGTA | AAD06682 | AA19233 | |
| Conserved hypothetical | ATP-dependent C1p protease | ||||
| E = 10−46 | E = 0 | ||||
| 2 | 48,048–49,560 | GCGGAAAGCT--CTTCTTGGTC | AAC07737 | AAC06734 | |
| 50S ribosomal protein L19 | Serine hydroxymethyl transferase | ||||
| E = 10−27 | E = 10−133 | ||||
| 3 | 464,145–465,657 | AGAAGAAAGT--AGAAGAGAAT | CAA97754 | AL132824 | |
| Conserved hypothetical (esterase) E = 10−14 | Oxidoreductase E = 10−164 | ||||
| 4 | 500,511–503,365 | TTCAGAAAAA--AAATCTACTA | No match <10−10 | AAF50582 | Split by ISPg1 |
| Conserved hypothetical 679-amino-acid protein | Conserved hypothetical E = 10−36 | Adjacent to ISPg7 In iteron | |||
| 5 | 638,157–649,667 | CTAAAAACGT--CGTGGCGTGA | AAB18619 | CAB42780 | |
| GMP synthetase | Ribosomal protein L31 | ||||
| E = 10−156 | E = 10−21 | ||||
| 6 | 1,002,186–1,003,697 | ACGACAGGGG--GGGGTTATTC | CAB79748 | AAC62424 | Between 2 copies of ISPg1 |
| Ca2+-transporting ATPase | NA+ transporter | ||||
| E = 10−130 | E = 10−41 | ||||
| 7 | 1,505,121–1,506,632 | TTCGGAAACT--ACTACCGGTG | CAA05255 | FEPE | Adjacent to iteron |
| DNA polymerase III | Ferredoxin | ||||
| E = 10−72 | E = 10−17 | ||||
| 8 | 1,724,699–1,726,181 | TTTCGGTTTT--CTCACGCCGC | AAB40238 | AAC05444 | |
| Cu+-transporting ATPase | Cardiolipin synthetase | ||||
| E = 10−142 | E = 10−93 | ||||
| 9 | 1,795,031–1,796,543 | CATGAAAAGC--AGCACAATTG | CAB13019 | AAD51075 | |
| Conserved hypothetical | Immunoreactive 87-kDa protein | ||||
| E = 10−18 | E = 10−79 | ||||
| 10 | 2,154,750–2,156,508 | GAAGTCGTTT--AACTTTTCGG | BAA10771 | AAF12166 | Adjacent to IsPg1 |
| Conserved hypothetical | Thymidylate synthetase | Has insert | |||
| E = 10−46 | E = 10−112 | ||||
| 11 | 2,233,854–2,235,366 | AAAAAAACAT--CATGGCTCCG | AAC06592 | BBA33066 | |
| Conserved hypothetical | Immunoreactive 60-kDa protein | ||||
| E = 10−40 | E = 0 |
Position in genome, release dated 6 March 2000.
Shown are 10 bases on either side of ISPg5. All sequences are oriented in the forward direction relative to ISPg5. Bases in bold are matching direct repeats.
Closest gene with BLAST score E < 10−10 other than another IS element. The top line is the GenBank accession number.
Other information on insert site.
Distribution of ISPg5 among laboratory strains and clinical isolates.
Given the clonal diversity of P. gingivalis (2, 22, 23, 29, 43, 45), we examined the prevalence and distribution of ISPg5 on the genomes of several laboratory strains and clinical isolates. Southern blots were prepared from BamHI/EcoRI-digested genomic DNA from these strains and probed with labeled clone 14 DNA, which contains a large segment of ISPg5 including much of the transposase. Note that ISPg5 lacks both BamHI and EcoRI restriction sites. Hybridizing bands were apparent for 9 of the 10 P. gingivalis strains listed above (Fig. 3B). Consistent with the presence of ISPg5 in multiple copies in the P. gingivalis W83 database, the blot of strain W83 had 11 hybridizing bands, 2 of which comigrate (Fig. 3B, lane 1). Strain W50 had several bands in common with W83. The remaining P. gingivalis strains had remarkably diverse RFLP patterns indicating that these strains had a wide variation in copy number and insertion sites for ISPg5. It is also interesting that one strain, ATCC 33277 lacked this IS element (data not shown).
DISCUSSION
Genetic events associated with IS activity in bacteria are believed to dynamically influence genome architecture (39, 40). Such natural genetic engineering may result in gene activation or inactivation, as well as sequence duplication, deletion, or transposition (39, 40). Reports of IS-like elements isolated individually (21, 26, 44) or identified by genomic sequence inspection (H. Dong, M. Duncan, T. Chen, F. Dewhirst, J. L. Galvin, R. Fleischmann, and C. Fraser, J. Dent. Res. 78:508, abstr. 3127, 1999) suggest that P. gingivalis contains a rich assortment of these mobile genetic elements. The preponderance of nonclonal P. gingivalis strains (2, 22, 23, 29, 43, 45) suggests that IS activity may contribute to the variability of this organism's genotype as reflected in its multiple RFLPs.
Initial analysis of the genomic sequence of P. gingivalis W83 revealed the presence of multiple copies of four different IS-like elements (Dong, et al., J. Dent. Res., 1999), but this number recently has been revised to eight (H. Dong, M. Duncan, T. Chen, F. Dewhirst, J. L. Galvin, R. Fleischmann, and C. Fraser, unpublished data). As postgenomic analysis of the W83 strain proceeds, study of the occurrence and location of these elements is likely to provide information useful in our understanding of this pathogen's life-style. However, we may not be able to extrapolate such findings to all clinical isolates, due to the large number of clonotypes associated with human periodontal disease. The availability of a collection of IS-like elements from different P. gingivalis clinical isolates would help us in comparative strain analysis, however. This report demonstrates that building such a collection of novel IS-like elements from P. gingivalis strains will be possible using the technique of Matsutani et al. (28).
ISPg5, the novel element described here, showed nucleotide sequence homology with and had features characteristic of the IS3 family of bacterial IS elements. Our designation of this element follows a scheme that we propose be adapted for the logical naming of IS and IS-like elements in P. gingivalis. Specifically, such elements are given the designation ISPgn. In this scheme, the previously described P. gingivalis IS-like elements would be given synonyms: IS1126 = ISPg1, PGIS2 = ISPg2, and IS195 = ISPg3. Newly discovered IS elements are designated with the suggested ISPg prefix followed by a number (n) reflecting their order of discovery. Using this scheme, ISPg4 is a novel 10-copy member of the IS4 family. ISPg5 is the novel 11-copy member of the IS3 family described here. ISPg6 is a single-copy member related to ISPg2, and ISPg8 is a single-copy element related to ISPg3 (Dong et al., J. Dent. Res., 1999); Dong et al., unpublished data); both ISPg2 and ISPg6 appear to be members of the ISAs1 family (where As is Aeromonas salmonicida), as defined by Mahillon and Chandler (25). ISPg7 is a single-copy member related to ISRm102-F34-1 (38) (where Rm is Sinorhizobium meliloti).
ISPg5 is most similar to a transposon nucleotide sequence from B. thetaiotaomicron (accession no. U67062). There appears to be a frameshift in the reported nucleotide sequence that can be corrected by insertion of a G at position 2400, which would extend the region of high homology to 144 bases or 48 amino acids. The corrected B. thetaiotaomicron nucleotide sequence now contains the full DDE motif region. The aligned nucleotide sequences of these Bacteroides phylum organisms appear to define a new subfamily of the IS3 family, with the following DDE consensus in the style of Mahillon and Chandler (25):
iiVADITYV(63)IHHSDRGvQY(35) ISMTQ-GDPLHNALAERM-NTiKN-W
The probable inverted repeat for B. thetaiotaomicron was found by similarity to ISPg5 and distance from the stop codon. The left inverted repeats are aligned as follows:
TGATGTGTACTGGTTTTAGTTGACAACTTCG B. thetaiotaomicron TGA GTGTACTG AGTTGAC A TT G ConsensusTGACGTGTACTGAAAAAAGTTGAC-AGTTGG P. gingivalis
ISPg5 appears to generate a 3-bp direct repeat upon insertion. As shown in Table 1, 6 of 11 copies contained matching direct repeats. We propose that the remaining five copies have lost matching direct repeats due to recombination events. In other P. gingivalis IS families, we have inferred large genomic inversions by recombinations between IS elements which result in exchanged direct repeats (unpublished observation). However, for the five pairs of unpaired ISPg5 direct repeats, there are no candidate exchanges—all 10 repeats are unique.
Southern analysis of several clinical isolates using ISPg5 as a probe indicated considerable diversity among P. gingivalis strains with regard to number of copies and insertion sites of ISPg5. A similar analysis in which genomic digests of laboratory strains were probed with IS1126 (ISPg1 in P. gingivalis) showed that RFLP patterns were similar in separately maintained descendants of a single strain, but different strains showed different patterns (7). Such differences in IS loci could account for clonal diversity as determined by DNA-based methods. Loss of the duplicated target sites from several copies of IS1126, as was also observed with ISPg5 in this study, was indicative of past homologous recombination events between elements. These recombinations are likely to be responsible for organizational differences, such as deletions and inversions, between genomes.
The importance of IS elements in the life-style of procaryotes, especially pathogens, is well established. IS elements also have been shown to activate cryptic genes by appropriate juxtaposition of promoter sequences (20, 32, 33) and the inactivation of gene expression by insertional mutagenesis (6, 19, 21, 31, 41, 42). Large- and small-scale genomic rearrangements promoted by IS elements also are known to result in altered phenotypic expression (10, 11, 14, 24, 36). For example, Hacker et al. reported that spontaneous E. coli mutants with deletions in hemolysin gene occurred with high frequency and that this event was linked to recombination between two IS-like elements (16). IS elements are often found associated with so-called pathogenicity islands, linkage groups of virulence genes believed to be horizontally transferred among strains (15, 34).
There is growing circumstantial evidence linking IS-like elements with important phenotypic characteristics of P. gingivalis. Lewis and Macrina (21) identified IS-like sequences associated with the Lys-gingipain virulence gene in three different P. gingivalis strains. Their data suggested one such element, IS195, might have a role in virulence gene regulation by an insertional mutagenesis mechanism. The ragAB locus of P. gingivalis W50, which encodes an immunodominant antigen, is flanked by a copy of IS1126, and contains a G+C content lower than that of the rest of the genome, an indication that it was acquired by horizontal gene transfer with the potential involvement of the IS (18). Finally, at the genotypic level, Dong et al. have examined insertion sites and flanking DNA regions for copies of IS1126 contained in the genomes of P. gingivalis strains ATCC 33277 and W83 (7). Their studies revealed that gene order was conserved over large regions of both genomes; however, the loci of the IS elements were different in the two strains. These authors suggest that the separate lineages of these two strains may have derived from IS transposition as well as from homologous recombination between similar IS elements.
We hypothesize that the genotypic diversity displayed by P. gingivalis is related to genomic plasticity that creates appropriate phenotypic changes in response to ecological pressures encountered in the periodontal pocket. It is reasonable to link the nonclonality of P. gingivalis isolates with genetic recombination promoted by IS elements. This study was performed in strain W83 in part because data from its genomic nucleotide sequence were available to allow for assessment of the utility of the reannealing capture technique. Although most of the clones recovered from the reannealing reactions have yet to be characterized, it is clear from our initial results that this technique allows the ready physical isolation of IS-like elements in P. gingivalis. Thus, the application of this technique to genotypically distinct strains of P. gingivalis should allow for isolation of other IS-like elements in these strains (28). The capture of new elements in addition to those found in W83 will provide a panel of genetic probes that will facilitate mapping studies and virulence gene localization in nonclonal isolates for which complete genomic sequence data are not available. Results from such molecular genetic approaches will likely contribute to our understanding of the taxonomy, evolution, and pathogenicity of P. gingivalis.
ACKNOWLEDGMENTS
This research was supported in part by NIH grants DE 12082 (to R.D.F., C.M.F., M.J.D., and F.E.D.) and DE04224 (to F.L.M.) from the National Institute for Dental and Craniofacial Research.
REFERENCES
- 1.Abeles A, Reaves L, Youngren-Grimes B, Austin S. Control of P1 plasmid replication by iterons. Mol Microbiol. 1995;18:903–912. doi: 10.1111/j.1365-2958.1995.18050903.x. [DOI] [PubMed] [Google Scholar]
- 2.Ali R W, Martin L, Haffajee A D, Socransky S S. Detection of identical ribotypes of Porphyromonas gingivalis in patients residing in the United States, Sudan, Romania, and Norway. Oral Microbiol Immunol. 1997;12:106–111. doi: 10.1111/j.1399-302x.1997.tb00625.x. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burge C, Campbell A M, Karlin S. Over- and under-representation of short oligonucleotides in DNA sequences. Proc Natl Acad Sci USA. 1992;89:1358–1362. doi: 10.1073/pnas.89.4.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandler M, Fayet O. Translational frame shifting in the control of transposition in bacteria. Mol Microbiol. 1993;7:497–503. doi: 10.1111/j.1365-2958.1993.tb01140.x. [DOI] [PubMed] [Google Scholar]
- 6.Collins C M, Gutman D M. Insertional inactivation of Escherichia coli urease gene by IS3411. J Bacteriol. 1992;174:883–888. doi: 10.1128/jb.174.3.883-888.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong H, Chen T, Dewhirst F, Fleischmann R, Fraser C, Duncan M. Genomic loci of the Porphyromonas gingivalis insertion element IS1126. Infect Immun. 1999;67:3416–3423. doi: 10.1128/iai.67.7.3416-3423.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dzink J L, Tanner A C R, Haffajee A D, Socransky S S. Gram negative species associated with active destructive periodontal lesions. J Clin Periodontol. 1985;12:648–659. doi: 10.1111/j.1600-051x.1985.tb00936.x. [DOI] [PubMed] [Google Scholar]
- 9.Fleischmann R, Adams M, White O, Clayton R, Kirkness E, Kerlavage A, Bult C, Tomb J, Dougherty B, Merrick J. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 10.Galas D, Chandler M. Bacterial insertion sequences. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 109–162. [Google Scholar]
- 11.Garcia M I, Labigne A, Le Bouguenec C. Nucleotide sequence of the afimbrial-adhesin-encoding afa-3 gene cluster and its translocation via flanking IS1 insertion sequences. J Bacteriol. 1994;176:7601–7613. doi: 10.1128/jb.176.24.7601-7613.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Genco R J, Zambon J J, Christersson L A. The origin of periodontal infections. Adv Dent Res. 1988;2:245–259. doi: 10.1177/08959374880020020901. [DOI] [PubMed] [Google Scholar]
- 13.Grossi S, Zambon J, Ho A, Koch G, Dunford R, Machtei E, Norderyd O, Genco R. Assessment of risk for periodontal disease. I. Risk indicators for attachment loss. J Periodontol. 1994;65:260–267. doi: 10.1902/jop.1994.65.3.260. [DOI] [PubMed] [Google Scholar]
- 14.Haack K R, Roth J R. Recombination between chromosomal IS200 elements supports frequent duplication formation in Salmonella typhimurium. Genetics. 1995;141:1245–1252. doi: 10.1093/genetics/141.4.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 16.Hacker J, Knapp S, Goebel W. Spontaneous deletions and flanking regions of the chromosomally inherited hemolysin determinant of an Escherichia coli O6 strain. J Bacteriol. 1983;154:1145–1152. doi: 10.1128/jb.154.3.1145-1152.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haffajee A D, Socransky S S. Microbial etiological agents of destructive periodontal diseases. Periodontol 2000. 1994;5:78–111. doi: 10.1111/j.1600-0757.1994.tb00020.x. [DOI] [PubMed] [Google Scholar]
- 18.Hanley S A, Aduse-Opoku J, Curtis M A. A 55-kilodalton immunodominant antigen of Porphyromonas gingivalis W50 has arisen via horizontal gene transfer. Infect Immun. 1999;67:1157–1171. doi: 10.1128/iai.67.3.1157-1171.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones B D, Kockatell C V, Johnson D E, Warren J W, Mobley H L. Construction of a urease-negative mutant of Proteus mirabilis: analysis of virulence in a mouse model of ascending urinary tract infection. Infect Immun. 1990;58:1120–1123. doi: 10.1128/iai.58.4.1120-1123.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee K Y, Hopkins J D, Syvanen M. Direct involvement of IS26 in an antibiotic resistance operon. J Bacteriol. 1990;172:3229–3236. doi: 10.1128/jb.172.6.3229-3236.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis J P, Macrina F L. IS195, an insertion sequence-like element associated with protease genes in Porphyromonas gingivalis. Infect Immun. 1998;66:3035–3042. doi: 10.1128/iai.66.7.3035-3042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loos B G, Dyer D W. Restriction fragment length polymorphism analysis of the fimbrillin locus, fimA, of Porphyromonas gingivalis. J Dent Res. 1992;71:1173–1181. doi: 10.1177/00220345920710050901. [DOI] [PubMed] [Google Scholar]
- 23.Loos B G, Mayrand D, Genco R J, Dickinson D P. Genetic heterogeneity of Porphyromonas gingivalis by genomic DNA fingerprinting. J Dent Res. 1990;69:1488–1493. doi: 10.1177/00220345900690080801. [DOI] [PubMed] [Google Scholar]
- 24.Louarn J M, Bouche J P, Legendre F, Louarn J, Patte J. Characterization and properties of very large inversions of the E. coli chromosome along the origin-to-terminus axis. Mol Gen Genet. 1985;250:467–476. doi: 10.1007/BF00331341. [DOI] [PubMed] [Google Scholar]
- 25.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maley J, Roberts I S. Characterisation of IS1126 from Porphyromonas gingivalis W83: a new member of the IS4 family of insertion sequence elements. FEMS Microbiol Lett. 1994;123:219–224. doi: 10.1111/j.1574-6968.1994.tb07225.x. [DOI] [PubMed] [Google Scholar]
- 27.Marmur J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 28.Matsutani S, Ohtsubo H, Maeda Y, Ohtsubo E. Isolation and characterization of IS elements repeated in the bacterial chromosome. J Mol Biol. 1987;196:445–455. doi: 10.1016/0022-2836(87)90023-4. [DOI] [PubMed] [Google Scholar]
- 29.Menard C, Mouton C. Clonal diversity of the taxon Porphyromonas gingivalis assessed by random amplified polymorphic DNA fingerprinting. Infect Immun. 1995;63:2522–2531. doi: 10.1128/iai.63.7.2522-2531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore W E, Moore L V. The bacteria of periodontal diseases. Periodontol 2000. 1994;5:66–77. doi: 10.1111/j.1600-0757.1994.tb00019.x. [DOI] [PubMed] [Google Scholar]
- 31.Ou J T, Baron L S, Rubin F A, Kopecko D J. Specific insertion and deletion of insertion sequence 1-like DNA element causes the reversible expression of the virulence capsular antigen Vi of Citrobacter freundii in Escherichia coli. Proc Natl Acad Sci USA. 1988;85:4402–4405. doi: 10.1073/pnas.85.12.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Podglajen I, Breuil J, Casin I, Collatz E. Genotypic identification of two groups within the species Bacteroides fragilis by ribotyping and by analysis of PCR-generated fragment patterns and insertion sequence content. J Bacteriol. 1995;177:5270–5275. doi: 10.1128/jb.177.18.5270-5275.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Podglajen I, Breuil J, Collatz E. Insertion of a novel DNA sequence, IS1186, upstream of the silent carbapenemase gene cfiA, promotes expression of carbapenem resistance in clinical isolates of Bacteroides fragilis. Mol Microbiol. 1995;12:105–114. doi: 10.1111/j.1365-2958.1994.tb00999.x. [DOI] [PubMed] [Google Scholar]
- 34.Ritter A, Blum G, Emody L, Kerenyi M, Bock A, Neuhierl B, Rabsch W, Scheutz F, Hacker J. tRNA genes and pathogenicity islands: influence on virulence and metabolic properties of uropathogenic Escherichia coli. Mol Microbiol. 1995;17:109–121. doi: 10.1111/j.1365-2958.1995.mmi_17010109.x. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Savic D J, Romac S P, Ehrlich S D. Inversion of the lactose region of Escherichia coli K-12: inversion termini map within IS3 elements alpha 3 beta 3 and beta 5 alpha 5. J Bacteriol. 1983;155:943–946. doi: 10.1128/jb.155.2.943-946.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sekine Y, Eisaki N, Ohtsubo E. Translational control in production of transposase and in transposition of insertion sequence IS3. J Mol Biol. 1994;235:1406–1420. doi: 10.1006/jmbi.1994.1097. [DOI] [PubMed] [Google Scholar]
- 38.Selbitschka W, Zekri S, Schroeder G, Puhler A, Toro N. The Sinorhizobium meliloti insertion sequence (IS) elements ISRm102F34-1/ISRm7 and ISRm220-13-5 belong to a new family of insertion sequence elements. FEMS Microbiol Lett. 1999;172:1–7. doi: 10.1111/j.1574-6968.1999.tb13441.x. [DOI] [PubMed] [Google Scholar]
- 39.Shapiro J. Genome system architecture and natural genetic engineering in evolution. Ann N Y Acad Sci. 1999;870:23–35. doi: 10.1111/j.1749-6632.1999.tb08862.x. [DOI] [PubMed] [Google Scholar]
- 40.Shapiro J A. Natural genetic engineering, adaptive mutation, and bacterial evolution. In: Rosenberg E, editor. Microbial ecology and infectious disease. Washington, D.C.: ASM Press; 1999. pp. 259–275. [Google Scholar]
- 41.Simonet M, Riot B, Fortineau N, Berche P. Invasion production by Yersinia pestis is abolished by insertion of an IS200-like element within the inv gene. Infect Immun. 1996;64:375–379. doi: 10.1128/iai.64.1.375-379.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simpson W, Wang C Y, Mikolajczyk-Pawlinska J, Potempa J, Travis J, Bond V C, Genco C A. Transposition of the endogenous insertion sequence element IS1126 modulates gingipain expression in Porphyromonas gingivalis. Infect Immun. 1999;67:5012–5020. doi: 10.1128/iai.67.10.5012-5020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teanpaisan R, Douglas C W I, Eley A R, Walsh T F. Clonality of Porphyromonas gingivalis, Prevotella intermedia, Prevotella nigrescens isolated from periodontally diseased and healthy sites. J Periodontal Res. 1996;31:423–432. doi: 10.1111/j.1600-0765.1996.tb00511.x. [DOI] [PubMed] [Google Scholar]
- 44.Wang C Y, Bond V C, Genco C A. Identification of a second endogenous Porphyromonas gingivalis insertion element. J Bacteriol. 1997;179:3808–3812. doi: 10.1128/jb.179.11.3808-3812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y J, Yasui S, Yoshimura F, Ishikawa I. Multiple restriction fragment length polymorphism genotypes of Porphyromonas gingivalis in single periodontal pockets. Oral Microbiol Immunol. 1995;10:125–128. doi: 10.1111/j.1399-302x.1995.tb00132.x. [DOI] [PubMed] [Google Scholar]