Abstract
The purpose of this study was to identify the functional significance of the binding of soluble CD14 (sCD14) to bacterial peptidoglycan (PGN) and to compare the structural requirements of sCD14 for the binding to PGN and lipopolysaccharide (LPS) and for sCD14-mediated enhancement of PGN- and LPS-induced cell responses. sCD14 did not facilitate the responses of membrane CD14 (mCD14)-negative pre-B 70Z/3 cells to PGN, although it facilitated the responses of these cells to LPS and although mCD14 facilitated the responses of 70Z/3 cells to PGN. sCD14 enhanced mCD14-mediated cell activation by both PGN and LPS, but only the responses to LPS, and not to PGN, were enhanced by LPS-binding protein. Four 4- or 5-amino-acid-long sequences within the 65-amino-acid N-terminal region of sCD14 were needed for binding to both PGN and LPS and for enhancement of cell activation by both PGN and LPS. However, deletions of individual sequences had different effects on the ability of sCD14 to bind to PGN and to LPS and on the ability to enhance the responses to PGN and to LPS. Thus, there are different structural requirements of sCD14 for binding to PGN and to LPS and for the enhancement of PGN- and LPS-induced cell activation.
Peptidoglycan (PGN), the major constituent of the cell walls of gram-positive bacteria, can reproduce major clinical manifestations of bacterial infections, including fever, inflammation, hypotension, leukocytosis, sleepiness, decreased appetite, malaise, and arthritis (5, 6), through the activation of macrophages and stimulation of secretion of mediators of inflammation, primarily cytokines and chemokines (5, 6, 21). The first step in macrophage activation by PGN is the binding of PGN to its specific receptor, membrane CD14 (mCD14) (4–7, 22, 23). mCD14 is a pattern recognition receptor; i.e., it also serves as the receptor for other bacterial macrophage activators, primarily lipopolysaccharide (LPS) or endotoxin from gram-negative bacteria, and also lipoteichoic acid from gram-positive bacteria, lipoproteins from spirochetes, lipoarabinomannan from mycobacteria, and others (5, 6). However, mCD14 is a glycosylphosphatidylinositol-linked rather than a transmembrane molecule, and mCD14 by itself probably cannot transmit the activating signal into the cell. Therefore, the second step in macrophage activation by PGN is transmission of the activating signal from the receptor into the cell, which most likely occurs through the Toll-like receptor-2 (13, 15, 25).
Soluble CD14 (sCD14) lacks the glycosylphosphatidylinositol anchor but has the same amino acid sequence as mCD14 and is present in normal human serum at 4 to 6 μg/ml (5, 6). Serum also contains an acute-phase protein, LPS-binding protein (LBP), which facilitates the binding of LPS to CD14 by catalytically transferring monomeric LPS from LPS aggregates onto mCD14 or sCD14 molecules. Therefore, LBP greatly enhances CD14-mediated responses to LPS (9, 17, 18, 24, 26).
Although both PGN and LPS bind to CD14 and activate cells through CD14, there are several differences in the function of CD14 as the PGN and LPS receptor. First, LBP enhances CD14-mediated cell activation by LPS but not by PGN (7, 22). Second, LBP increases the affinity of binding of LPS, but not of PGN, to CD14 (4). Third, the binding sites for PGN and LPS on CD14 and the sites needed for cell activation are partially different (4, 7). Fourth, PGN and LPS induce differential activation of mitogen-activated protein kinases (3) and induce different patterns of gene activation (21). And fifth, sCD14-LPS complexes activate mCD14-negative endothelial and epithelial cells, whereas sCD14-PGN complexes do not (10).
The reason for the unresponsiveness of these mCD14-negative cells to sCD14-PGN complexes is unknown (10), although sCD14 binds to PGN with high affinity (Kd = 25 nM) and forms stable complexes with PGN at an approximately 1:1 molar ratio (4, 23). Nonmyeloid cells are also unresponsive to PGN when they are transfected with and express mCD14, even though these cells are fully responsive to LPS through mCD14 or sCD14 (10). By contrast, lymphoid and monocytic cells expressing mCD14 are responsive to PGN (7, 10).
Therefore, the first aim of our current study was to determine if sCD14-PGN complexes activate mCD14-negative lymphoid cells. Because we found here that sCD14-PGN complexes do not activate CD14-negative lymphoid cells, the functional significance of sCD14-PGN binding still remained unknown. However, in addition to facilitating activation of mCD14-negative cells by LPS, sCD14 has another function; i.e., sCD14 enhances mCD14-mediated cell activation by LPS by binding LPS and then by transferring LPS to mCD14 (9). Therefore, the second aim of this study was to determine if sCD14 enhances mCD14-mediated cell activation by PGN. Because different regions of sCD14 and of mCD14 are important for LPS binding and cell activation (4, 7, 19, 20), and also because different regions of mCD14 are important for PGN- and LPS-induced cell activation (7), the third aim of this study was to compare the regions of sCD14 involved in sCD14-mediated enhancement of cell activation by PGN and LPS and binding to PGN and LPS. Because LBP increases low-affinity binding of PGN to CD14 (4), our fourth aim was to determine if LBP had any effect on mCD14- and sCD14-mediated cell activation by PGN.
MATERIALS AND METHODS
Materials.
Polymeric non-cross-linked soluble PGN (PGN) was purified by vancomycin affinity chromatography and analyzed as before (12). PGN contained <24 pg of endotoxin/mg, as determined by the Limulus lysate assay (12). LPS from Salmonella enterica serovar Minnesota Re 595 (a minimal, naturally occurring endotoxic structure of LPS), obtained by phenol-chloroform-petroleum ether extraction, was purchased from Sigma, and its purity was analyzed as described before (2).
Recombinant human full-length wild-type (wt) sCD14 (residues 1 to 323), containing at the C terminus an affinity tag of six histidines, was expressed in a baculovirus system, affinity purified on nickel-agarose, and analyzed as described previously (20). sCD14 deletion mutants, lacking one (DDED, PQPD, DPRQY, AVEVE), two (DDED + PQPD), or all four (QUAD) indicated 4- to 5-amino-acid-long regions within the N-terminal 65 amino acids (see Fig. 1), were constructed, expressed, and purified as described (20). Recombinant human LBP was the same as before (4). The endotoxin content of sCD14 and LBP preparations was <1 pg/μg, determined by the Limulus lysate assay.
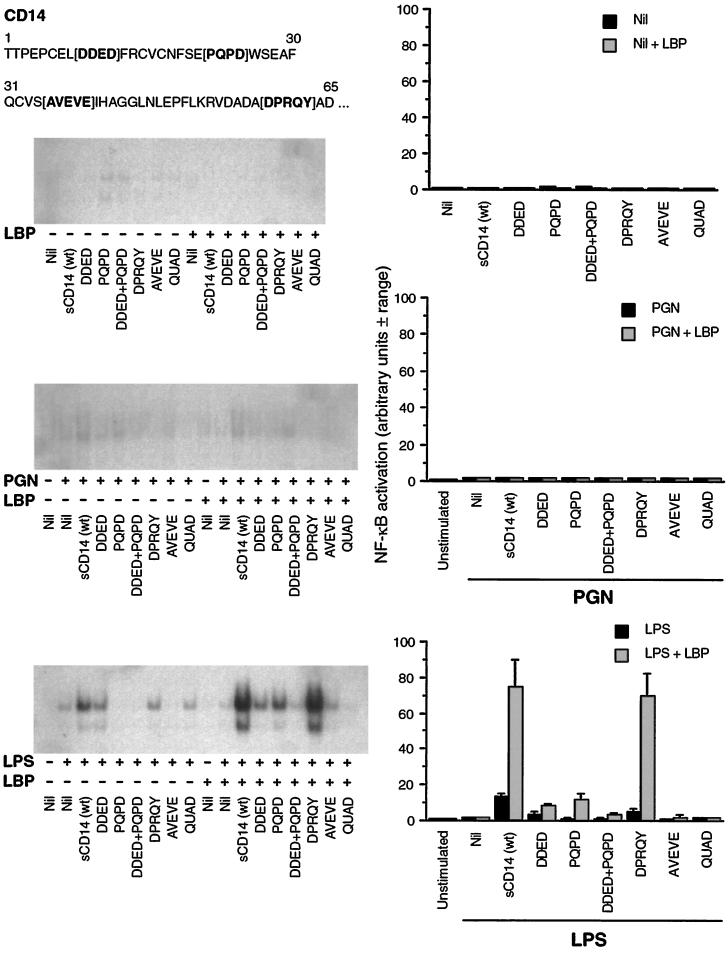
FIG. 1.
sCD14-PGN and sCD14 deletion mutant-PGN complexes, in contrast to sCD14-LPS complexes, do not activate 70Z/3 cells. 70Z/3 cells were incubated in serum-free medium alone or with 10 μg of wt sCD14/ml or 10 μg of sCD14/ml from which the amino acids in the N-terminal region indicated in bold type in brackets were deleted, with or without 0.2 μg of LBP/ml and with or without 15 μg of PGN/ml or 100 ng of LPS/ml, as indicated. Activation of NF-κB in nuclear extracts was determined by the electrophoretic mobility shift assay. Autoradiograms from one out of two similar experiments are shown on the left, and the quantified results (average of two experiments ± range) are shown as bar graphs on the right. Nil, no addition.
Cells and activation of NF-κB.
Untransfected mouse 70Z/3 immature B cells and stable transfectants of 70Z/3 cells, expressing glycosylphosphatidylinositol-linked full-length human mCD14 obtained as described before (7), were cultured in RPMI 1640 with 10 mM HEPES and 7.5% fetal calf serum and, for 70Z/3-CD14 transfectants, with the addition of 1 mg of G418/ml (7). The expression of CD14 was verified by flow cytometry (7). Before stimulation, cells were washed three times with RPMI 1640 (without serum) at 37°C, suspended in RPMI 1640 with 10 mM HEPES at 3 × 106 cells/ml, dispensed in 0.3-ml aliquots into wells of 48-well tissue culture plates, and incubated for 30 min at 37°C in 5% CO2. Stimulants (PGN or LPS) were mixed with sCD14 (wt or mutants) with or without LBP in RPMI 1640 (as indicated in Results), incubated at 37°C in 5% CO2 for 1 h, and added to cell cultures to yield the following final concentrations: PGN, 15 μg/ml for 70Z/3 cells or 4 μg/ml for 70Z/3-CD14 transfectants; LPS, 100 ng/ml for 70Z/3 cells or 10 ng/ml for 70Z/3-CD14 transfectants; sCD14 (wt or mutants), 10 μg/ml; and LBP, 0.2 μg/ml. Cells were left unstimulated (control) or were stimulated with PGN for 90 min (70Z/3) or 60 min (70Z/3-CD14), or with LPS for 45 min (70Z/3) or 30 min (70Z/3-CD14) (optimum times) and were harvested, and nuclear extracts were prepared as previously described (7). Activation of NF-κB in the nuclear extracts was determined by the electrophoretic mobility shift assay as described (7). The NF-κB bands on autoradiograms were quantified using Kodak Digital Science Image Station 440CF and the Image Analysis Software 3.0. The results were then normalized to unstimulated groups (which equaled 1 arbitrary unit) and expressed as means ± ranges or standard errors (SE).
Inhibition of binding of 32P-sCD14 to PGN-agarose and LPS-agarose.
Full-length human sCD14 (wt), containing at the C terminus a phosphorylation site for protein kinase A (16), was labeled with 32P (4), and the binding of 32P-sCD14 to PGN-agarose or LPS (from S. enterica serovar Minnesota Re 595)-agarose in the absence or presence of increasing concentrations (0.2 to 200 μg/ml) of sCD14 (wt or deletion mutants) and with or without 1 μg of LBP/ml was determined as previously described (4). The 50% inhibitory concentrations (IC50) were calculated using a logarithmic curve fit generated by SigmaPlot software (Jandel Scientific, San Rafael, Calif.).
RESULTS
70Z/3 cells are unresponsive to sCD14-PGN complexes.
To test whether sCD14-PGN complexes activate mCD14-negative cells of lymphoid origin, we selected 70Z/3 immature mouse B cells, because when transfected with mCD14, 70Z/3 cells (similarly to human monocytes) become fully responsive to PGN in a CD14-dependent manner (7). The experiments were done in serum-free medium on cells washed before the assay to avoid interference from sCD14 and LBP contained in the serum or CD14 released from CD14 transfectants during culture.
sCD14-PGN complexes did not activate NF-κB in 70Z/3 cells, regardless of the presence of LBP (Fig. 1). Increasing the concentration of PGN (up to 30 μg/ml) and the time of incubation with cells (up to 3 h) still did not activate NF-κB in 70Z/3 cells (data not shown). By contrast, as expected, sCD14-LPS complexes activated NF-κB in 70Z/3 cells, and this activation was greatly enhanced by LBP (Fig. 1).
We have previously found that various CD14 deletion mutants were either fully responsive, partially responsive, or unresponsive to PGN and LPS, and these deletion mutants also showed unchanged, decreased, increased, or abolished binding to PGN and LPS (7, 19, 20, and this paper below). Moreover, the responsiveness and binding of different deletion mutants were different for PGN and LPS (7, and this paper below), and the behavior of sCD14 and mCD14 mutants was also different (20).
One possibility for the unresponsiveness of 70Z/3 cells to sCD14-PGN complexes might have been a too high or a too low affinity of binding of PGN to sCD14. Therefore, we wanted to test if sCD14 deletion mutants (which had different affinities for PGN, see below) facilitated the responses of 70Z/3 cells to PGN. However, none of the deletion mutants (despite their various abilities to bind to PGN, see below) facilitated the response of 70Z/3 cells to PGN, regardless of the presence of LBP (Fig. 1). By contrast, in the absence of LBP, two deletion mutants (DDED and DPRQY) facilitated the responses of 70Z/3 cells to LPS, although they were much less effective than wt sCD14 (Fig. 1). LBP enhanced LPS-induced activation by DDED, PQPD, and DPRQY deletion mutants, and in the presence of LBP, the DPRQY deletion mutant was as active as wt sCD14 (Fig. 1). LBP by itself (without sCD14) did not facilitate the response of 70Z/3 cells to LPS (Fig. 1). Neither LBP alone nor any of the sCD14 preparations alone without or with LBP activated NF-κB in 70Z/3 cells (Fig. 1).
The responsiveness of 70Z/3-CD14 transfectants to PGN is enhanced by sCD14.
To test the hypothesis that sCD14 and/or LBP enhance mCD14-mediated cell activation by PGN, we selected 70Z/3-CD14 transfectants, because their responsiveness to PGN, similar to the responsiveness of human monocytes (22), is almost completely CD14 dependent (7).
PGN activated NF-κB in 70Z/3-CD14 transfectants in serum-free medium, and LBP had no effect on mCD14-mediated cell activation by PGN (Fig. 2), confirming our previous results (7) that in 70Z/3 cells expression of mCD14 alone is sufficient for NF-κB activation. By contrast, as expected, LPS alone (at a low concentration of 10 ng/ml) induced very weak activation of NF-κB in 70Z/3-CD14 transfectants in serum-free medium, but this activation was greatly enhanced by LBP (Fig. 2).
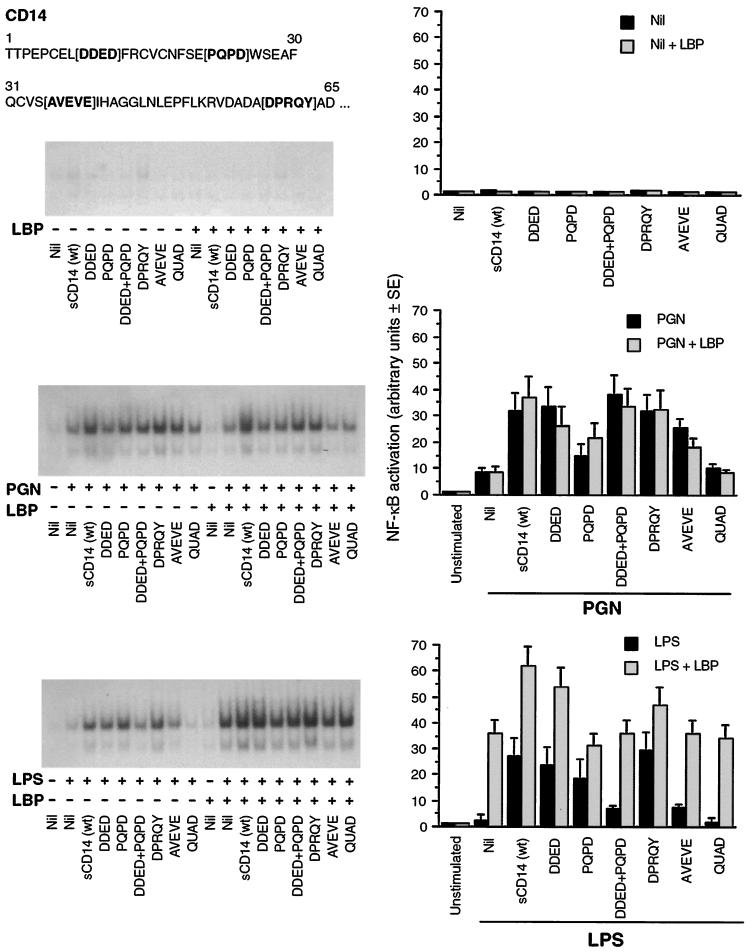
FIG. 2.
Responsiveness of 70Z/3-CD14 transfectants to PGN is enhanced by sCD14 and sCD14 deletion mutants. 70Z/3-CD14 transfectants were incubated in serum-free medium alone or with 10 μg of wt sCD14/ml or 10 μg of sCD14/ml from which the amino acids in the N-terminal region indicated in bold type in brackets were deleted, with or without 0.2 μg of LBP/ml and with or without 4 μg of PGN/ml or 10 ng of LPS/ml, as indicated. Activation of NF-κB in nuclear extracts was determined by the electrophoretic mobility shift assay. Autoradiograms from one out of four similar experiments are shown on the left, and the quantified results (average of four experiments ± SE) are shown as bar graphs on the right. Nil, no addition.
sCD14 (wt) enhanced mCD14-mediated activation of NF-κB by PGN, and LBP had no effect on this enhancement (Fig. 2). sCD14 (wt), as expected, also enhanced mCD14-mediated activation of NF-κB by LPS, but in contrast to PGN, this sCD14 enhancement of LPS response was further potentiated by LBP (Fig. 2).
Because in our previous studies deletions of four different regions in the N-terminal sequence of mCD14 had different effects on PGN- and LPS-induced cell activation (7), and because structural requirements of sCD14 and mCD14 for LPS-induced cell activation are not identical (20), we next tested the effects of the same four deletions on the capacity of sCD14 to enhance mCD14-mediated responses.
All single-deletion sCD14 mutants and the double-deletion mutant could still enhance mCD14-mediated cell activation by PGN, and some mutants (DDED, DPRQY, and DDED + PQPD) had activity similar to that of the wt sCD14. However, two other mutants (PQPD and AVEVE) had their enhancing activity diminished by 70% and 25%, respectively (Fig. 2). LPS-induced activation was enhanced by the DDED, DPRQY, and PQPD sCD14 deletion mutants to the same extent as by the wt sCD14. However, the enhancing activities on LPS responses of the double DDED + PQPD deletion mutant and single AVEVE deletion mutant were diminished by more than 80% (Fig. 2). The quadruple sCD14 deletion mutant had no effect on PGN- and LPS-induced responses.
LBP had no effect on the activity of sCD14 mutants (similar to that on wt sCD14) in PGN-induced responses (Fig. 2). However, LBP further potentiated the enhancing effect of the DDED and DPRQY deletion mutants on the LPS-induced responses and had no effect on the responses induced by other deletion mutants (or this effect was not greater than the enhancing effect of LBP alone on mCD14-mediated responses) (Fig. 2). Neither LBP alone nor any of the sCD14 preparations alone without or with LBP activated NF-κB in 70Z/3-CD14 transfectants (Fig. 2).
These results demonstrate that sCD14 enhances mCD14-mediated responses to both PGN and LPS and that individual deletions have different effects on the enhancing activity of sCD14 for PGN- and LPS-induced responses.
Differential binding of sCD14 deletion mutants to PGN and LPS.
Because the requirements for specific sequences needed for LPS binding and cell activation through mCD14 were different (7, 19), we then wanted to determine whether the changes in the enhancing activity of the sCD14 mutants were due to the changes in their binding capacity for PGN and LPS. To answer this question, the binding of sCD14 and all deletion mutants to PGN and LPS was compared by measuring their IC50 for the binding of 32P-sCD14 (wt) to PGN and LPS in the absence and presence of LBP.
Wild-type sCD14 and the DDED, PQPD, and DDED + PQPD deletion mutants showed essentially similar binding to PGN, consistent with the high-affinity sCD14 (wt) binding that was demonstrated before (4), whereas DPRQY and AVEVE deletion mutants showed 3 to 4 times lower binding, as measured by their IC50 for 32P-sCD14 (wt) binding (Table 1). This binding of wt sCD14 or sCD14 deletion mutants to PGN was not significantly enhanced by LBP. The quadruple mutant showed no binding to PGN (IC50 > 1,000 μg/ml).
TABLE 1.
Inhibition of 32P-sCD14 binding to PGN-agarose and LPS-agarose by sCD14 deletion mutantsa
| Inhibitor | IC50 (μg/ml) of:
|
|||
|---|---|---|---|---|
| PGN-agarose
|
LPS-agarose
|
|||
| No LBP | + LBP | No LBP | + LBP | |
| sCD14 (wt) | 8.2 ± 1.9 | 5.5 ± 0.9 | 56.1 ± 12 | 0.66 ± 0.2 |
| DDED | 6.3 ± 0.2 | 4.9 ± 0.9 | 48.3 ± 8.6 | 11.2 ± 3.7 |
| PQPD | 14.9 ± 3.2 | 15.8 ± 0.7 | >300 | 34.6 ± 5.7 |
| DDED + PQPD | 9.0 ± 2.6 | 17.4 ± 4.0 | >300 | 188 ± 55 |
| DPRQY | 33.4 ± 7.2 | 32.9 ± 5.0 | enhanced 4.7×b | enhanced 1.5×b |
| AVEVE | 43.6 ± 13 | 70.3 ± 21 | enhanced 1.9×b | >1,000 |
| QUAD | >1,000 | >1,000 | >1,000 | >1,000 |
Inhibition of binding of 32P-sCD14 to PGN-agarose and LPS-agarose by various concentrations of the indicated unlabeled sCD14 wt or deletion mutants, with or without 1 μg of LBP/ml, was measured, and the IC50 was calculated based on a logarithmic curve fit. The results are means of three experiments ± SE.
Binding of 32P-sCD14 to LPS-agarose was increased with the addition of some unlabeled sCD14 deletion mutants. The maximum fold enhancement, which was seen at 20 μg/ml, is shown.
The IC50 of sCD14 (wt) for 32P-sCD14 binding to LPS (without LBP) was seven times higher than for PGN, confirming the lower affinity of sCD14 (wt) for LPS than for PGN in the absence of LBP (4). However, in the presence of LBP, the IC50 of sCD14 (wt) binding to LPS was 85 times enhanced by LBP, confirming higher affinity of binding of sCD14 (wt) to LPS than to PGN in the presence of LBP (Table 1) (4). The IC50 of binding of the sCD14 DDED deletion mutant to LPS (without LBP) was similar to the IC50 of sCD14 (wt), but the binding of the DDED mutant was much less enhanced by LBP than that of sCD14 (wt). The IC50 of the PQPD, DDED + PQPD, and QUAD mutants (without LBP) were very high, indicating very poor binding or no binding to LPS (Table 1). The binding of the PQPD and DDED + PQPD mutants (but not of the QUAD mutant) to LPS was enhanced by LBP. The DPRQY and AVEVE mutants not only did not inhibit the binding of 32P-sCD14 (wt) to LPS (without LBP) but enhanced this binding, suggesting that they either form aggregates with 32P-sCD14 (wt) and LPS or enhance transfer of 32P-sCD14 (wt) onto LPS. This enhancing effect was reduced by LBP.
DISCUSSION
Our results demonstrate that (i) sCD14 does not facilitate the responses of mCD14-negative lymphoid cells to PGN, (ii) sCD14 enhances mCD14-mediated cell activation by PGN, (iii) this enhancement is not influenced by LBP, and (iv) four 4- to 5-amino-acid-long sequences within the 65-amino-acid N-terminal region of sCD14 are needed for this enhancing activity. By contrast, sCD14 both facilitates the responses of the same mCD14-negative cells to LPS and enhances mCD14-mediated cell activation by LPS, and both of these activities of LPS and sCD14 are greatly enhanced by LBP. Our results also demonstrate different structural requirements of sCD14 for the binding to PGN and LPS and also for the enhancement of cell activation by PGN and LPS.
These results extend previous findings (10) by showing that lymphoid cells, which are responsive to PGN when transfected with mCD14 (7), are still unresponsive to sCD14-PGN complexes. Although the reason for this unresponsiveness is not clear, the main function of sCD14-PGN complexes in vivo could be enhancement of the responses of mCD14-positive cells to PGN.
The structural requirements of sCD14 and mCD14 for the enhancement (through sCD14) or induction (through mCD14) of NF-κB responses by PGN and for the binding of sCD14 to PGN are similar: all four deletions (DDED, PQPD, DPRQY, and AVEVE) are needed to totally abolish both responses and the binding, and the same single or double deletions (DDED, DPRQY, and DDED + PQPD) have little effect on the activities of either sCD14 or mCD14, whereas other deletions (AVEVE and PQPD) have only a partial effect (Table 2).
TABLE 2.
Comparison of binding and activity of mCD14 and sCD14 deletion mutants towards PGN and LPSa
| CD14 | PGN
|
LPS
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inhibition of 32P-sCD14 binding when LBP was:
|
Activation through mCD14 (7) | Enhancement by sCD14 through mCD14 when LBP was:
|
Activation through sCD14 when LBP was:
|
Inhibition of 32P-sCD14 binding when LBP was:
|
Activation through mCD14 (7) | Enhancement by sCD14 through mCD14 when LBP was:
|
Activation through sCD14 when LBP was:
|
|||||||
| Absent | Present | Absent | Present | Absent | Present | Absent | Present | Absent | Present | Absent | Present | |||
| Wild type | +++ | +++ | ++++ | ++ | ++ | − | − | + | ++++ | ++++ | ++ | ++++ | ++ | ++++ |
| DDED | +++ | +++ | ++++ | ++ | ++ | − | − | + | ++ | +++ | ++ | ++++ | + | ++ |
| PQPD | ++ | ++ | + | + | + | − | − | − | + | − | ++ | ++ | − | ++ |
| DDED + PQPD | +++ | ++ | +++ | ++ | ++ | − | − | − | +/− | − | +/− | +/− | − | − |
| DPRQY | + | + | +++ | ++ | ++ | − | − | ↑↑b | ↑ | +++ | ++ | +++ | + | ++++ |
| AVEVE | + | + | + | + | + | − | − | ↑ | − | − | +/− | +/− | − | − |
| QUAD | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
Data are from this study and from reference 7, where the activity and binding of PGN and LPS were compared simultaneously in the same assays. The binding, activation, or enhancement of activation were graded from the greatest (++++) to the lowest (+), borderline (+/−), or none (−); for quantitative results see Fig. 1 and 2, Table 1, and reference 7. Data on the binding to LPS and activation by LPS from other studies, which used different assay systems and in which PGN was not tested together with LPS, are described in Discussion.
↑, enhancement of 32P-sCD14 binding (see Table 1).
The same four sequences are also important for sCD14-mediated or mCD14-mediated enhancement or induction of NF-κB responses by LPS and for the binding of sCD14 to LPS, because deletion of all four sequences totally abolished all LPS responses and binding (Table 2). However, the roles of specific sequences in the responsiveness to PGN and LPS are different, because, in contrast to PGN, the single AVEVE deletion or double DDED + PQPD deletion completely or almost completely abolished the binding of sCD14 to LPS and the activity of mCD14 or sCD14 for both LPS responses, whereas they had almost no effect (double DDED + PQPD deletion) or much less effect (AVEVE deletion) on the activity and binding to PGN. Another difference between PGN and LPS is the effect of LBP: all LPS, but not PGN, responses and high-affinity binding are enhanced by LBP (Table 2).
Whereas our current and previous (7) results demonstrate similar effects of deletions on the binding and activity of sCD14 and mCD14 towards PGN (as described above), our current and previous (7, 19, 20) results demonstrate different behavior of mCD14 and sCD14 deletion mutants towards LPS (Table 2). All mCD14 single- or double-deletion mutants expressed in CHO cells lost their serum-dependent high-affinity binding of 3H-LPS (19), whereas most sCD14 mutants (except DPRQY) retained binding of soluble LPS (20) but had impaired binding or lost their binding to solid-phase-bound LPS (Table 1). Moreover, in the enzyme-linked immunosorbent assay (ELISA) and fluorescence assays (20), the sCD14 DDED + PQPD double-deletion mutant bound LPS several times better than the wt sCD14, whereas in this study (inhibition of wt sCD14 binding to solid-phase LPS), the same double-deletion mutant showed much poorer binding than the wt sCD14 (Table 1). Some other mutants (DPRQY and AVEVE), which showed good LPS binding in the ELISA and fluorescence assays (20), not only did not inhibit binding of labeled wt sCD14 to LPS but enhanced its binding (Table 1), suggesting that they form aggregates with LPS and sCD14. LPS seemed to have been required for this aggregation, because the same mutants showed good inhibition of wt sCD14 binding to solid-phase PGN (rather than the enhancement seen with LPS [Table 1]).
The reasons for these differences in binding are not clear, but they may be related to several factors, such as the ability of these different assays to detect different affinities of binding, the requirement for LBP in the cell-binding assay (19), the absence of LBP in the assays for the binding of soluble LPS to sCD14 (20), or different structural requirements of sCD14 for the binding to different forms of LPS (monomeric, aggregated, or solid-phase bound) under different conditions (in solution, solid-phase-bound sCD14, membrane-bound CD14, or sCD14 mutants competing for the binding of wt sCD14). The last possibility seems to be supported by results showing that in the ELISA with biotinylated PGN, similar to the assay in which the DDED + PQPD double-deletion mutant showed better binding to LPS than wt sCD14 (20), the DDED + PQPD double-deletion mutant and DDED single-deletion mutant also showed higher binding to PGN than the wt sCD14 (S. Viriyakosol, T. N. Kirkland, and R. Dziarski, unpublished data). These differences in the behavior of various sCD14 mutants towards LPS are less apparent in the functional assays than in the binding assays discussed above, because similar sCD14 mutants are active in the assays for activation of U373 (20) and 70Z/3 (this study) cells, although the 70Z/3 cells seem to be more responsive than the U373 cells.
Our results further support the notion that the binding sites for LPS (1, 11, 14) and PGN (4–6) on CD14 are conformational. The sequences that are common and most critical for CD14 binding to both LPS and PGN and for cell activation are amino acids 35 to 39 (AVEVE deletion) and amino acids 51 to 64 (the site of deletion of amino acids 59 to 63 [DPRQY] and the binding site of anti-CD14 monoclonal antibody [MAb] MEM-18), because AVEVE is the single deletion that has the greatest effect on the binding and activation by both PGN and LPS (7, 19) and because anti-CD14 MAb MEM-18 is most efficient in inhibiting CD14 binding both to LPS (by over 95%) and to PGN (by over 80%) (4) and in inhibiting cell activation by both LPS and PGN (5, 6). Similarly, alanine substitutions revealed that the LPS-binding region is located between amino acid 39 and 44, whereas [A9–13]mCD14 (the site of our DDED deletion), as well as [A57, A59, A61–63]mCD14 (the site of our DPRQY deletion) are still able to bind LPS and to activate transfected CHO cells (14), which is consistent with our results (Table 2).
These two regions, however, may not be sufficient for binding of LPS and PGN, because MAbs specific to other regions of CD14 or deletions in other regions of CD14 also partially or almost totally inhibit binding to LPS, and to a lesser extent to PGN, and cell activation by LPS and PGN (4, 7, 14). However, these other sequences that contribute to LPS and PGN binding and cell activation are at least partially different, because there are other anti-CD14 MAbs (directed to more N-terminal regions of CD14) (4) or other deletions (Table 2) that inhibit LPS binding but not PGN binding, and one MAb (directed to a more C-terminal region of CD14) that inhibits PGN binding but not LPS binding (4).
These results also show that wt CD14 is the most versatile molecule, whereas various deletion mutants are not as versatile as the wt CD14 and function well only in some assays with some ligands. Specific mutations affect different aspects of CD14 functions in various ways, causing more or less severe changes in the function depending on the assay system and the ligand or activator, and this effect is especially evident with LPS but not as much with PGN. Moreover, the differences between the functions of sCD14 and mCD14 may not be due entirely to different behavior of the soluble and membrane forms of CD14 but may also depend on the cell type; i.e., in lymphoid cells stimulated with LPS, sCD14 mutants behave more similarly to the mCD14 mutants (7 and Fig. 2). Therefore, the behavior of sCD14 and mCD14 depends on the ligand (e.g., LPS versus PGN), the form of the ligand (soluble versus solid-phase bound), the presence of soluble accessory molecules (LBP), the cell type, and the form of CD14 (soluble versus membrane).
ACKNOWLEDGMENTS
We are grateful to Peter Tobias and Richard Tapping for providing sCD14 with the phosphorylation site and LBP.
This work was supported by United States Public Health Service Grants AI2879 (to R.D.) and GM37696 (to T.N.K.) from NIH and by a grant from the Medical Research Service of the Department of Veterans Affairs (to T.N.K.).
REFERENCES
- 1.Cunningham M D, Shapiro R A, Seachord C, Ratcliffe K, Cassiano L, Darveau R P. CD14 employs hydrophilic regions to “capture” lipopolysaccharide. J Immunol. 2000;164:3255–3263. doi: 10.4049/jimmunol.164.6.3255. [DOI] [PubMed] [Google Scholar]
- 2.Dziarski R, Gupta D. Heparin, sulfated heparinoids, and lipoteichoic acid bind to the 70 kDa peptidoglycan/lipopolysaccharide receptor protein on lymphocytes. J Biol Chem. 1994;269:2100–2110. [PubMed] [Google Scholar]
- 3.Dziarski R, Jin Y, Gupta D. Differential activation of extracellular signal-regulated kinase (ERK) 1, ERK2, p38, and c-Jun NH2-terminal kinase mitogen-activated protein kinases by bacterial peptidoglycan. J Infect Dis. 1996;174:777–785. doi: 10.1093/infdis/174.4.777. [DOI] [PubMed] [Google Scholar]
- 4.Dziarski R, Tapping R I, Tobias P. Binding of bacterial peptidoglycan to CD14. J Biol Chem. 1998;273:8680–8690. doi: 10.1074/jbc.273.15.8680. [DOI] [PubMed] [Google Scholar]
- 5.Dziarski R, Ulmer A, Gupta D. Interactions of bacterial lipopolysaccharide and peptidoglycan with mammalian CD14. In: Doyle R J, editor. Glycomicrobiology. New York, N.Y: Kluwer Academic/Plenum Publishers; 2000. pp. 145–186. [Google Scholar]
- 6.Dziarski R, Ulmer A, Gupta D. Interactions of CD14 with components of Gram-positive bacteria. Chem Immunol. 2000;74:83–107. doi: 10.1159/000058761. [DOI] [PubMed] [Google Scholar]
- 7.Gupta D, Kirkland T N, Viriyakosol S, Dziarski R. CD14 is a cell-activating receptor for bacterial peptidoglycan. J Biol Chem. 1996;271:23310–23316. doi: 10.1074/jbc.271.38.23310. [DOI] [PubMed] [Google Scholar]
- 8.Hailman E, Lichenstein H S, Wurfel M M, Miller D S, Johnson D A, Kelley M, Busse L A, Zukowski M M, Wright S D. Lipopolysaccharide (LPS)-binding protein accelerates the binding of LPS to CD14. J Exp Med. 1994;179:269–277. doi: 10.1084/jem.179.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hailman E, Vasselon T, Kelley M, Busse L A, Hu M C T, Lichenstein H S, Detmers P A, Wright S D. Stimulation of macrophages and neutrophils by complexes of lipopolysaccharide and soluble CD14. J Immunol. 1996;156:4384–4390. [PubMed] [Google Scholar]
- 10.Jin Y, Gupta D, Dziarski R. Endothelial and epithelial cells do not respond to complexes of peptidoglycan with soluble CD14, but are activated indirectly by peptidoglycan-induced tumor necrosis factor-α and interleukin-1 from monocytes. J Infect Dis. 1998;177:1629–1638. doi: 10.1086/515318. [DOI] [PubMed] [Google Scholar]
- 11.Juan T S C, Hailman E, Kelley M J, Busse L A, Davy E, Empig C J, Narhi L O, Wright S D, Lichenstein H S. Identification of a lipopolysaccharide binding domain in CD14 between amino acids 57 and 64. J Biol Chem. 1995;270:5219–5224. doi: 10.1074/jbc.270.10.5219. [DOI] [PubMed] [Google Scholar]
- 12.Rosenthal R S, Dziarski R. Isolation of peptidoglycan and soluble peptidoglycan fragments. Methods Enzymol. 1994;235:253–285. doi: 10.1016/0076-6879(94)35146-5. [DOI] [PubMed] [Google Scholar]
- 13.Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning C J. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by Toll-like receptor 2. J Biol Chem. 1999;274:17406–17409. doi: 10.1074/jbc.274.25.17406. [DOI] [PubMed] [Google Scholar]
- 14.Stelter F, Bernheiden M, Menzel R, Jack R S, Witt S, Fan X, Pfister M, Schutt C. Mutation of amino acids 39–44 of human CD14 abrogates binding of lipopolysaccharide and Escherichia coli. Eur J Biochem. 1997;243:100–109. doi: 10.1111/j.1432-1033.1997.00100.x. [DOI] [PubMed] [Google Scholar]
- 15.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Differential roles of TLR2 and TLR4 in recognition of Gram-negative and Gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 16.Tapping R I, Tobias P S. Cellular binding of soluble CD14 requires lipopolysaccharide (LPS) and LPS-binding protein. J Biol Chem. 1997;272:23157–23164. doi: 10.1074/jbc.272.37.23157. [DOI] [PubMed] [Google Scholar]
- 17.Tobias P S, Soldau K, Gegner J A, Mintz D, Ulevitch R J. Lipopolysaccharide binding protein-mediated complexation of lipopolysaccharide with soluble CD14. J Biol Chem. 1995;270:10482–10488. doi: 10.1074/jbc.270.18.10482. [DOI] [PubMed] [Google Scholar]
- 18.Tobias P S, Soldau K, Kline L, Lee J D, Kato K, Martin T P, Ulevitch R J. Cross-linking of lipopolysaccharide (LPS) to CD14 on THP-1 cells mediated by LPS-binding protein. J Immunol. 1993;150:3011–3021. [PubMed] [Google Scholar]
- 19.Viriyakosol S, Kirkland T N. A region of human CD14 required for lipopolysaccharide binding. J Biol Chem. 1995;270:361–368. doi: 10.1074/jbc.270.1.361. [DOI] [PubMed] [Google Scholar]
- 20.Viriyakosol S, Mathison J C, Tobias P S, Kirkland T N. Structure-function analysis of CD14 as a soluble receptor for lipopolysaccharide. J Biol Chem. 2000;275:3144–3149. doi: 10.1074/jbc.275.5.3144. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z-M, Liu C, Dziarski R. Chemokines are the main pro-inflammatory mediators in human monocytes activated by Staphylococcus aureus, peptidoglycan, and endotoxin. J Biol Chem. 2000;275:20260–20267. doi: 10.1074/jbc.M909168199. [DOI] [PubMed] [Google Scholar]
- 22.Weidemann B, Brade H, Rietschel E T, Dziarski R, Bazil V, Kusumoto S, Flad H-D, Ulmer A J. Soluble peptidoglycan-induced monokine production can be blocked by anti-CD14 monoclonal antibodies and by lipid A partial structures. Infect Immun. 1994;62:4709–4715. doi: 10.1128/iai.62.11.4709-4715.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weidemann B, Schletter J, Dziarski R, Kusumoto S, Stelter F, Rietschel E T, Flad H-D, Ulmer A J. Specific binding of soluble peptidoglycan and muramyldipeptide to CD14 on human monocytes. Infect Immun. 1997;65:858–864. doi: 10.1128/iai.65.3.858-864.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright S D, Ramos R A, Tobias P S, Ulevitch R J, Mathison J C. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 25.Yoshimura A, Lien E, Ingalls R R, Tuomanen E, Dziarski R, Golenbock D. Recognition of Gram-positive bacterial cell wall components by the innate immune system occurs via Toll-like receptor 2. J Immunol. 1999;163:1–5. [PubMed] [Google Scholar]
- 26.Yu B, Wright S D. Catalytic properties of lipopolysaccharide (LPS) binding protein. Transfer of LPS to soluble CD14. J Biol Chem. 1996;271:4100–4105. doi: 10.1074/jbc.271.8.4100. [DOI] [PubMed] [Google Scholar]