Abstract
Moraxella catarrhalis is a respiratory pathogen responsible for acute bacterial otitis media in children and exacerbation of chronic bronchitis in adults. M. catarrhalis strains are frequently resistant to the bactericidal activity of normal human serum. In order to determine if the lipooligosaccharide (LOS) of M. catarrhalis has a role in serum resistance, the UDP-glucose-4-epimerase (galE) gene was identified, cloned, and sequenced and a deletion/insertion mutation was introduced into M. catarrhalis strain 2951. GalE enzymatic activity, measured in whole-cell lysates, was ablated in M. catarrhalis 2951 galE. Mass spectrometric analysis of LOS isolated with hot phenol-water confirmed that strain 2951 produced a type A LOS. These studies showed that the LOS from 2951 galE had lost two hexose residues due to the galE mutation and that the resultant LOS structure lacked the (Galα1-4Galβ1-4Glc) Pk epitope found on M. catarrhalis 2951. Wild-type M. catarrhalis 2951 is resistant to complement-mediated serum bactericidal activity. In contrast, a greater than 2-log10-unit reduction in CFU occurred after incubation of 2951 galE in either 50 or 25% pooled human serum (PNHS), and CFU in 10% PNHS decreased by about 1 log10 unit. These studies suggest that the Pk epitope of the LOS may be an important factor in the resistance of M. catarrhalis to the complement-mediated bactericidal effect of normal human serum.
Moraxella catarrhalis is a human respiratory pathogen that is currently the third leading cause of otitis media along with Streptococcus pneumoniae and Haemophilus influenzae (10). Studies from various centers in the United States, Europe, and Asia used tympanocentesis to demonstrate that 15 to 20% of the middle-ear infections occurring in young children were caused by M. catarrhalis (10, 15, 16, 18, 46). M. catarrhalis has also been implicated as an important cause of respiratory disease in adults with predisposing conditions (41). Studies from several centers have reported clusters of nosocomial outbreaks of M. catarrhalis, most of which occurred in pulmonary care units (43, 45).
Although multiple studies have described specific bacterial components considered potential virulence factors, the steps involved in the pathogenesis of M. catarrhalis colonization and infection remain elusive (28, 41). One feature of this organism which has stimulated the interest of a number of investigators is its resistance to killing by normal human serum. Recent studies have focused on components of the bacterial outer membrane, as these structures would most likely be available for interaction with the host immune response. One prominent bacterial surface component, implicated as a potential virulence factor, is the lipooligosaccharide (LOS).
The LOS is similar to those of other airway pathogens such as H. influenzae, Neisseria meningitidis, and Bordetella pertussis in lacking O antigens typical of the enteric gram-negative bacilli. There are three M. catarrhalis serotypes (A, B, and C) based on chemically defined differences in the LOS antigen structures (12, 13, 38, 52). The LOSs of all three serotypes consist of a multiantenneray carbohydrate structure, but in all three serotypes, one of the oligosaccharide chains terminates in Galα1-4Galβ1-4Glc. Mandrell and Apicella showed that M. catarrhalis LOS reacted with monoclonal antibody (MAb) Gal 1-3 specific for the Pk (Galα1-4Galβ1-4Glc) epitope (36). The role of the Moraxella LOS in human infection has not been clearly defined. Most Moraxella strains have been shown to be highly resistant to complement-mediated killing in normal human serum (41, 53). In this paper, we present studies that investigate the role that the terminal Galα1-4Galβ1-4Glc structure of Moraxella LOS plays in resistance to complement-mediated killing by normal human serum. To perform these investigations, we created a mutation in the UDP-glucose 4-epimerase gene, resulting in a truncated LOS structure lacking terminal galactose residues. This change resulted in the loss of the Pk epitope from the LOS. These studies indicate that the Pk epitope may be a factor responsible for the resistance of M. catarrhalis to the complement-mediated bactericidal effect of normal human serum.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacteria and plasmids used in this study are described in Table 1. All M. catarrhalis clinical isolates were kindly provided by Timothy Murphy (Veterans Administration Medical Center, Buffalo, N.Y.) and Howard Faden (Children's Hospital, Buffalo, N.Y.). Neisseria gonorrhoeae strain 1291 and the 1291a-e pyocin mutant were described elsewhere (11, 26).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype, relevant phenotype, or selection marker | Source or reference |
|---|---|---|
| Strains | ||
| E. coli XL1-Blue MRF′ | Δ(mcrA) 183 Δ(mcrCB-hsdSMR-mrr) 173 endA1 sup E44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| M. catarrhalis | Wild-type serotype A | ATCC 25238 |
| M. catarrhalis 2951 | Wild-type clinical isolate, serotype A | This study |
| M. catarrhalis 2951 galE | UDP-glucose 4-epimerase deficient | This study |
| Plasmids | ||
| pMCSau2 | Ampicillin | This study |
| pMCApoA | Ampicillin | This study |
| pMCPvu1a | Ampicillin | This study |
| pMCgalE | Ampicillin | This study |
| pMCgalEBBR | Ampicillin, spectinomycin | This study |
| pUC18 | Ampicillin | Pharmacia Biotech |
| pUC19 | Ampicillin | Pharmacia Biotech |
| pTAV1 | Ampicillin | 6 |
| pMC2951galETA | Ampicillin | This study |
| pABR3 | Ampicillin, spectinomycin | 2 |
| pD2galE | Kanamycin | 31 |
Development of MAb 4G5.
MAb 4G5 was isolated from a previously described fusion (34). The antibody was defined as an immunoglobulin G2a using mouse MonoAb-ID (Zymed Laboratories). MAb 9E9, which is specific for the high-molecular-mass (HMW) protein of M. catarrhalis, was a gift from Timothy Murphy (Veterans Administration Medical Center, Buffalo, N.Y.) (30).
Bacterial growth.
Escherichia coli was grown at 37°C in Luria-Bertani medium with or without agar (1.5%) and supplemented with antibiotics as needed. Wild-type M. catarrhalis was grown either on gonococcal agar (GCA) supplemented with 1% IsoVitaleX (BBL Laboratories, Cockeysville, Md.) or brain heart infusion (BHI) agar (Difco Laboratories, Detroit, Mich.) supplemented with 2.5% heat-inactivated fetal calf serum (FCS) at 37°C in 5% CO2 with 85% relative humidity. Spectinomycin-resistant M. catarrhalis was grown on supplemented BHI agar with 15 μg of spectinomycin/ml or in supplemented BHI broth containing 5.0 μg of spectinomycin/ml. Selection was carried out without CO2.
Recombinant DNA and transformation methods.
All recombinant DNA techniques were performed as outlined previously (47). Transformations of Moraxella were performed as previously described by Catlin (8) and modified by Stephens et al. (48).
Cloning and mutagenesis of the UDP-glucose 4-epimerase gene (galE).
The cloning of M. catarrhalis strain 25238 galE was accomplished in three steps (Fig. 1A to D). A 32P-labeled probe made from bp 1 to 931 of the N. meningitidis galE (31) was used to probe a Southern blot of a Sau3AI partial digest of M. catarrhalis genomic DNA. This probe hybridized to a 2.8-kb DNA genomic fragment which was ligated into pUC18 and used to transform DH5α. Transformants were screened with the same probe. Plasmids isolated from colonies to which this probe hybridized contained an M. catarrhalis DNA fragment that contained the 3′ 425 bp of the putative M. catarrhalis galE (pMCSau2) (Fig. 1A).
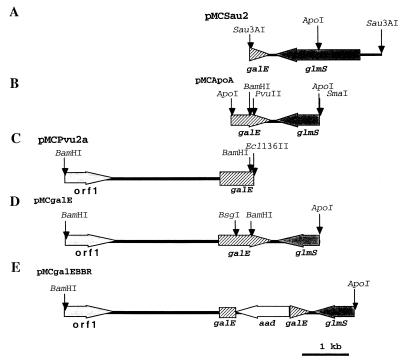
FIG. 1.
M. catarrhalis DNA inserts from the five plasmids used to create the mutant and wild-type constructs. (A) M. catarrhalis DNA fragment from pMCSau2 cloned into the BamHI site of pUC18. (B) M. catarrhalis DNA fragment from pMCApoA cloned into the EcoRI site in pUC19. (C) M. catarrhalis DNA fragment from pMCPvu2a cloned into the SmaI site in pUC18. (D) M. catarrhalis DNA fragment constructed from those shown in panels B and C cloned into pUC18. (E) M. catarrhalis DNA fragment from pMCgalE with the aad gene inserted into the BsgI and BamHI gap of galE in pUC18. Multiple restriction enzyme sites have been omitted for clarity.
The M. catarrhalis genomic DNA fragment from pMCSau2 was used to probe an ApoI digest of M. catarrhalis chromosomal DNA. A 1.9-kb fragment was identified and cloned into pUC19. Plasmids were isolated from colonies which hybridized to this probe. Sequence analysis revealed another 414 bp of M. catarrhalis galE 5′ to that found in pMCSau2. This plasmid was designated pMCApoA (Fig. 1B). DNA sequencing reactions were performed by using dye terminator cycle sequencing chemistry with AmpliTaq DNA polymerase and FS enzyme (PE Applied Biosystems, Foster City, Calif.). The reactions were run on and analyzed with an Applied Biosystems model 373A stretch fluorescence automated sequencer at the University of Iowa DNA Facility.
A third Southern blot was produced using a randomly primed 32P probe constructed from a 482-bp ApoI/PvuII fragment of the M. catarrhalis DNA in pMCApoA. This probed hybridized to a 4,000-bp fragment. A sublibrary was constructed in pUC18 with this DNA fragment. Clones containing portions of the M. catarrhalis putative galE open reading frame (ORF) were identified with the same randomly primed probe. The plasmid within these clones contained 710 bp of the 5′ end of M. catarrhalis galE in addition to the region upstream of this gene. This plasmid was designated pMCPvu1a (Fig. 1C).
Based on information gained from the above transformations and sequence analysis, a plasmid was constructed by ligating the BamHI/SmaI 1,506-bp fragment from pMCApoA and the BamHI/Ecl136II fragment (6,676 bp) from pMCPvu1a. This plasmid was designated pMCgalE. The M. catarrhalis DNA in the plasmid was sequenced (6,471 bp) and was demonstrated to contain the entire M. catarrhalis galE (Fig. 1D). galE from M. catarrhalis strain 2591 was amplified by PCR from genomic DNA using the forward primer 5′TATGACAAACACAGGGACAAC′3 and the reverse primer 5′ATCAATGCCCACAACCAG. The resulting 1,140-bp product was cloned into the pTAV1 cloning vector.
UDP-glucose 4-epimerase enzyme assays.
M. catarrhalis strains were grown in BHI broth supplemented with 2% FCS and spectinomycin as required. To prepare extracts for enzymatic assay, 100-ml cultures were inoculated with 0.01 volume of fresh overnight culture grown in supplemented BHI broth and incubated at 37°C with shaking. Separate cultures were grown to either exponential phase or stationary phase and washed twice in 1× phosphate-buffered saline. The washed pellets were resuspended in 5 ml of assay buffer (125 mM potassium bicinate [pH 8.5], 1 mM phenylmethylsulfonyl fluoride) and lysed in a French press using 16,000 lb/in2. The bacterial debris was sedimented by centrifugation at 15,800 × g for 30 min at 4°C. The supernatants were transferred to prechilled microcentrifuge tubes and kept on ice. The total protein content of the crude cell extracts was determined using the Bio-Rad protein assay reagent by following the microassay protocol with bovine serum albumin as the standard. Extracts containing equal amounts of total protein were added to the two-step UDP-glucose 4-epimerase assay mixture as described below.
The two-step UDP-glucose 4-epimerase assay described here is a modification of a procedure described elsewhere (57, 58). We modified the assay slightly to optimize it for Moraxella extracts. The first step of the two-step assay was carried out in a 500-μl reaction volume (125 mM bicinate [pH 8.5], 0.44 mM UDP-galactose) at 37°C for 15 min. The reaction mixture was then placed in a boiling water bath for 90 s, chilled on ice for 5 min, and then centrifuged at 15,800 × g for 10 min at 4°C. A 400-μl aliquot of the supernatant was added to the mixture from the second step of the assay in a 600-μl total volume (0.125 mM bicinate, [pH 8.5], 1.25 mM NAD+, 0.02 U of UDP-glucose dehydrogenase). The reaction was observed in a quartz cuvette, and the increase in absorbance was measured every 15 s at 340 nm. All extracts including appropriate controls were assayed in triplicate.
Determination of UDP-glucose 4-epimerase activity levels.
The net absorbance was determined after adjusting for endogenous UDP-galactose and UDP-glucose and UDP-glucose contamination of exogenous UDP-galactose preparations. The initial velocities (Vi) of the second reaction (UDP-glucose to UDP-glucuronic acid) were determined over the first 30 s. Vi, which is indicative of the starting concentration of UDP-glucose, was converted to nanomoles of NADH generated per minute per nanogram of total protein using the Beer-Lambert law and ɛNADH = 6.2 × 103 M−1 cm−1.
SDS-PAGE and Western blotting of isolated LOS.
LOS was isolated from 6 liters of supplemented BHI broth for strain 2951 and 6 liters of supplemented BHI broth cultures with 5 μg of spectinomycin/ml for the 2951 galE mutant by a modified Westphal hot phenol-water preparation (31). Whole-bacterial-cell proteinase K lysates were made from bacteria grown on supplemented BHI agar. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed as described by Lesse et al. (32). Western blotting was performed by the method of Towbin et al. (51).
Mass spectrometric analysis.
LOS structures from M. catarrhalis 2951 and 2951 galE were analyzed by mass spectrometry (MS). In each case, approximately 0.5 mg of LOS was treated with mild hydrazine for 30 min at 37°C (21) for conversion into the corresponding water-soluble O-deacylated LOSs, which are more amenable to mass spectrometric analysis (19).
O-deacylated samples were taken up in water and desalted by drop dialysis using a 0.025-μm-pore-size nitrocellulose membrane (Millipore, Bedford, Mass.). The dialyzed sample was mixed in a 1:1 ratio with 320 mM 2,5-dihydroxybenzoic acid solution in 4:1 (vol/vol) acetone-water containing 175 mM 1-hydroxyisoquinoline (40), desalted with cation-exchange resin beads (DOWEX, 50X; NH4+) (42), and then air dried on a stainless steel target.
Samples were then analyzed by matrix-assisted laser desorption ionization (MALDI)-MS using a PE Biosystems (Framingham, Mass.) Voyager DE time-of-flight mass spectrometer operated with a nitrogen laser (337 nm) in the negative-ion mode under delayed-extraction conditions (55). The delay time was 175 ns, and the grid voltage was 93.5% of full acceleration voltage (20 to 30 kV). Spectra were acquired, averaged, and mass calibrated with an external calibrator consisting of an equimolar mixture of angiotensin II, bradykinin, luteinizing hormone-releasing factor, bombesin, α-MSH (CZE mixture; Bio-Rad), and adrenocorticotropin 1-24 (Sigma).
Electrospray mass spectra were obtained using a quadrupole ion trap mass analyzer fitted with an electrospray ionization source (Finnigan LCQ; Finnigan MAT, San Jose, Calif.). For sample delivery, direct infusion with a syringe pump at a flow rate of 0.5 to 2 μl/min was used. The mobile phase was 70% acetonitrile in water. Ions were produced with a spray voltage of 2.9 keV with the heated capillary set at 200°C. Spectra were collected in the negative-ion mode by averaging 20 individual scans, consisting of three “microscans.” Collision-induced dissociation was carried out in the mass analyzer on an ion selected from the mass spectrum by using He as the collision gas in the ion trap.
Bactericidal assay.
Bacteria were grown to early log phase, A600 = 0.2, in supplemented BHI broth. A 0.5-ml aliquot of each strain was centrifuged for 1 min at 2,000 × g in a Beckman microcentrifuge at room temperature. The pellet was resuspended in 1.0 ml of phosphate-buffered salt solution (PBSS) consisting of 10 mM K2HPO4, 10 mM KH2PO4, 136 mM NaCl, 5 mM KCl, 1 mM CaCl2, 0.3 mM MgCl2 · 6H2O, 1 mM MgSO4 · 7H2O, and 0.01% bovine serum albumin, pH 7.0.
The bactericidal assay, modified from that reported by Andreoni et al. (4), was carried out in a 96-well plate in a 200-μl final volume. Pooled normal human serum (PNHS; a 20-donor pool of serum from human volunteers who had no previous history of neisserial infections) was diluted to 10, 25, or 50% in PBSS. A control containing PNHS heat-inactivated for 30 min at 56°C was included in each experiment. Ten microliters (106 cells) of the resuspended bacteria was diluted into 190 μl of PBSS, and serial 1/10 dilutions were made in PBSS. Twenty microliters of each dilution was spread on GCA and grown overnight at 37°C in 5% CO2. The colonies in these reaction mixtures were counted and used as the initial CFU. Ten microliters of the bacterial stock was incubated in the diluted serum for 30 min with shaking at 200 rpm in a 37°C incubator (Inova 4080; New Brunswick Scientific, Edison, N.J.). Serial 1/10 dilutions of the reaction mixtures were diluted into PBSS and were spread on GCA plates. These were grown overnight at 37°C in 5% CO2, and emerging colonies were counted the next day. The resulting CFU value was the 30-min value.
Killing was assessed by comparing the number of CFU from the 30-min serum incubation with the number of initial CFU. Results were expressed as the log10 change in CFU at 30 min compared to the initial CFU.
Statistical analysis and DNA sequence construction and analysis.
Statistical analysis of the data from bactericidal and UDP-glucose 4-epimerase activity assays was carried out using the paired t test and analysis of variance functions found in Statview, version 4.0 (Abacus Concepts, Inc., Berkeley, Calif.).
DNA sequence construction and analysis were performed using the Wisconsin Package, version 10.0 (Genetics Computer Group, Madison, Wis.), AssemblyLIGN, version 1.0, (Oxford Molecular Group Inc., Oxford, United Kingdom), Gene Works, version 2.5.1 (Oxford Molecular Group Inc.), and Sharedraw, version 2.0 (Pierce Software Inc., San Jose, Calif.).
Nucleotide sequence accession numbers.
The nucleotide sequence of M. catarrhalis galE is available from the GenBank database under accession no. AF248583 and AF248584.
RESULTS
Homology analysis.
The Blast search protocol at the National Center for Biotechnology Information website was used to compare the M. catarrhalis DNA sequence with the nonredundant database (3). Three large ORFs were identified in the 6,841 bp of M. catarrhalis DNA that had been cloned and sequenced. The galE sequence of M. catarrhalis contained 1,094 bp (Fig. 1D). Figure 2 shows the degree of similarity among two M. catarrhalis strains and four other bacterial species. The predicted amino acid sequence had 59% identity and 70% similarity over 334 amino acid residues to GalE of Bacillus subtilis. There was 53% identity and 68% similarity to GalE of N. meningitidis, 55% identity and 67% similarity to H. influenzae GalE, and 52% identity and 66% similarity to N. gonorrhoeae GalE.
FIG. 2.
Predicted amino acid alignment of GalE proteins from various gram-negative bacteria. The consensus sequence is shown at the bottom. Both strains of M. catarrhalis have an additional 23 residues at the N-terminal end of the protein. Boldface, amino acids that are in the binding pocket of the folded protein and that have been shown to interact with the substrate (UDP-sugar) or the cofactor (NAD+). Residues that interact with NAD+ are in italics, and residues that interact with UDP-sugar are underlined (15, 33, 49, 50). M. cat. 25238, M. catarrhalis ATCC strain 25238 (GenBank accession no. AF248583); M. cat. 2951, M. catarrhalis strain 2951 (GenBank accession no. AF248584); N. men. B, N. meningitidis serotype B strain FAM20 (SwissProt accession no. Q59624); N. gon., N. gonorrhoeae (SwissProt accession no. Q05026); B. sub., B. subtilis (SwissProt accession no. P55180); H. inf., H. influenzae (SwissProt accession no. P24325). E. coli SwissProt accession no., P09147.
A large ORF (1,836 bp) reading in the opposite orientation and ending 82 bp immediately 3′ to galE had homology to the gene encoding glucosamine fructose-6-phosphate aminotransferase, glmS (Fig. 1D). The predicted amino acid sequence had 57% identity and 70% similarity over 611 amino acids residues to the product of glmS from Thiobacillus ferrooxidans.
The product of a third 1,104-bp partial ORF at the 5′ end of the M. catarrhalis sequence showed homology to UDP-glucose dehydrogenase (Fig. 1D). The homology of the predicted amino acid sequence was 30% identity and 49% similarity to a putative enzyme described in Burkholderia pseudomallei.
Mutagenesis of the UDP-glucose 4-epimerase gene (galE).
Strain 2951 galE mutant was constructed by excision of a 429-bp DNA fragment from the galE gene in pMCgalE by partial digestion with BamHI followed by complete digestion using BsgI (Fig. 1E). Blunt ends were produced by using T4 DNA polymerase as previously described (47). A spectinomycin resistance gene (aad) was ligated into the blunt-ended pMCgalE. Colonies were selected by growth on Luria-Bertani agar containing ampicillin and spectinomycin. The location and orientation of the spectinomycin resistance gene within galE were confirmed by diagnostic restriction endonuclease digestions and direct DNA sequencing. This plasmid was designated pMCgalEBBR.
M. catarrhalis strain 25238 was resistant to transformation by homologous recombination with XbaI-restricted DNA from pMCgalEBBR. We tested three other M. catarrhalis strains, and only one (M. catarrhalis 2951) was transformable with this restricted plasmid DNA. To insure that the galE genes were comparable, the galE gene from strain 2951 was first amplified by PCR using primers made according to the sequences of the 5′ and 3′ ends of the strain 25238 galE gene. A PCR product of the expected size was obtained and cloned into pTAV1 (Table 1) and transformed into E. coli XL1-Blue MRF′. Colonies were screened by diagnostic restriction endonuclease digestions of plasmid preparations. A plasmid having the correct diagnostic restriction endonuclease fragments was sequenced. It was found that there was divergence at 16 nucleotide residues, which translated to four amino acid differences between the products of galE of strain 25238 and strain 2951. A comparison of the predicted amino acid sequences of GalE from strain 25238 and strain 2951 with a consensus GalE sequence showed that none of the amino acid changes involved active sites in the enzyme (Fig. 2).
Comparison of GalE activity in M. catarrhalis strain 2951 and 2951 galE mutant.
The levels of UDP-glucose 4-epimerase activity were measured by a two-step assay of whole-cell extracts obtained from bacteria in both the exponential and stationary growth phases (Table 2). GalE activity from lysates of galE mutant bacteria harvested during either the exponential or stationary growth phase was at the lower detectable limits of the assay. Lysates from strain 2951 harvested during the exponential growth phase demonstrated high levels of GalE activity (average of 13.640 nmol of NADH generated per min per μg of total protein). Lysates from strain 2951 obtained at stationary growth phase had activity that was not detectable and that was no different from the activity in the lysate of strain 2951 galE at either exponential or stationary growth. Increases in A340 that occurred upon addition of UDP-glucose (0.45 mM final concentration) to these lysates showed that they did not contain inhibitors of the UDP-glucose dehydrogenase used in the second step of the assay.
TABLE 2.
UDP-glucose 4-epimerase activity
| Bacterial strain | UDP-glucose 4-epimerase activity (nmol of NADH/min/μg of total protein) ± SD in:
|
|
|---|---|---|
| Log phase | Stationary phase | |
| 2951 (wild type) | 13.640a ± 3.104 | 0.0073b ± 0.0180 |
| 2951 galE | 0.079c ± 0.176 | 0.0006 ± 0.0010 |
P < 0.0001 versus log-phase 2951 galE.
P < 0.0001 versus log-phase wild type.
P = 0.9490 versus stationary-phase 2951 galE.
Characterization of the LOS epitope recognized by MAb 4G5 and reactivity with strain 2951 and 2951 galE.
Western blot analysis showed that MAb 4G5 reacted to the LOS of M. catarrhalis strain 7169, the strain used for immunization of the donor mice. Flow cytometry confirmed that the LOS 4G5 epitope was expressed on the surface of strain 7169. Subsequent blots confirmed that the LOS from 22 other M. catarrhalis strains also expressed the MAb 4G5 epitope. These data demonstrated that the LOS epitope defined by MAb 4G5 is conserved on a diversity of clinical isolates from different geographic regions, from both adults and children and from various body sites and fluids. In addition, the M. catarrhalis isolates that represent the strains used to define the three major LOS serotypes also reacted with MAb 4G5. MAb 4G5 bound to the LOS of strain 2951 but did not react with LOS from strain 2951 galE.
We performed Western blot assays with MAb 4G5 using proteinase K lysates from 8 strains of H. influenzae, 11 strains of N. meningitidis, 12 strains of N. gonorrhoeae, 2 strains of Neisseria lactamica, and 2 strains of Neisseria cinerea. The only strain recognized by MAb 4G5 that was not a Moraxella species was the gonococcal pyocin-derived mutant, 1291b. The LOS of the 1291b mutant terminates in a structure that is immunochemically identical to the Pk antigen (Galα1-4Galβ1-4Glc) found on human cells (26, 37). These data indicate that MAb 4G5 reacted with the Galα1-4Galβ1-4Glc structure that is found as a terminal structure on all three M. catarrhalis LOS serotypes. It also indicated that this structure was no longer present on the LOS of strain 2951 galE. The mass-spectrometric analysis presented below confirmed the results of these MAb studies.
Mass-spectrometric analysis of M. catarrhalis strain 2951 and 2951 galE LOSs.
The structures of the major LOSs for the three serotypes A (Fig. 3), B, and C have been previously reported (12–14). To assign the serotype of strain 2951 LOS and define the structure of the galE mutant LOS, several MS experiments were carried out on the LOSs from both strains.
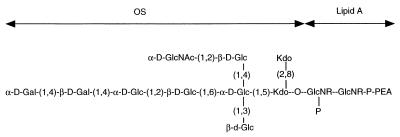
FIG. 3.
Structure of the LOS from serotype A. OS, oligosaccharide.
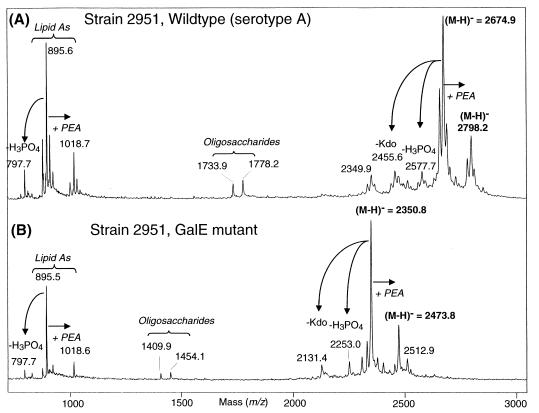
To determine the precise molecular masses of the LOS glycoforms, LOSs from both the parental strain 2951 and the 2951 galE were converted into the water-soluble O-deacylated form and analyzed in the negative-ion mode by MALDI-time of flight MS. MALDI-MS spectra of the O-deacylated LOS preparations are shown in Fig. 4. In both spectra, a single dominant deprotonated molecular ion peak, (M-H)−, was observed at m/z 2,674.9 and m/z 2,350.5 for the wild-type and mutant strains, respectively. A less abundant second peak 123 Da higher in mass, corresponding to the presence of an additional phosphoethanolamine (PEA) moiety (i.e., m/z 2,798.2 and m/z 2,473.8 for the wild-type and mutant strains, respectively) was also detected. These masses are consistent with a composition of Hex7HexNAcKdo2-lipid A for the wild type and Hex5HexNAcKdo2-lipid A for the galE mutant, with elements of LOSs from both strains being partially replaced with PEA. These compositions are consistent with the previously published structure of a serotype A LOS (38) for the parental strain and with the expected loss of two galactose residues (Galα1 4Galβ1) on the nonreducing terminus of the largest oligosaccharide branch to form the truncated galE mutant LOS. The presence of oligosaccharide and lipid A “prompt fragments” (i.e., fragments generated from facile decomposition of the intact LOS species prior to acceleration) in these spectra adds further support to these assignments (Fig. 5). For example, lipid A peaks present in both spectra at m/z 895.6 and 1,018.7 are those expected based on the previously published lipid A structure (38) with the exception of a partial substitution of PEA. Likewise, the oligosaccharide fragments at m/z 1,778.2 (wild type) and m/z 1,453.1 (galE mutant) are consistent with the monosaccharide compositions as stated above.
FIG. 4.
Linear MALDI-time of flight spectra of the parental wild-type M. catarrhalis strain 2951 (A) and the isogenic galE mutant (B). The observed molecular ions for the individual LOS species appear as unresolved isotope clusters whose centroids correspond to the average mass. In addition to the peaks described in the text, peaks due to prompt ion fragmentations are also present due to β-elimination of phosphoric acid from the parent (M-H)− peaks (m/z 2,578.3, m/z 2,253.4) and lipid A fragments (m/z 797.7) and loss of a Kdo (m/z 2,456.3, m/z 2,131.8), as are peaks resulting from facile loss of CO2 from a Kdo residue of the oligosaccharides (m/z 1,734.2 and 1410.1). Fragmentations of these types have been discussed in detail elsewhere (19). Masses shown in Fig. 5 are the theoretically predicted masses based on the structures shown. The experimental masses listed in the two spectra above may differ but are within the expected experimental mass accuracy (± 0.1%).
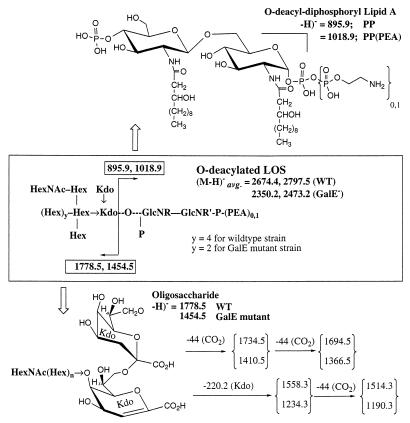
FIG. 5.
Schematic of prompt fragmentation processes observed for O-deacylated LOS by negative-ion MALDI-MS. All masses shown are average masses based on those calculated for both the wild-type and galE mutant LOSs. Hex, hexose; HexNAc, N-acetylhexosamine; Kdo, 2-deoxy-3-keto-octulosonic acid. See the legend for Fig. 4 and text for further discussion.
Since the oligosaccharide structures are known to be highly branched, we examined these in more detail using a successive series of tandem MS (MSn) experiments. The MSn experiments were all carried out using an ion trap instrument operating under negative ion electrospray ionization conditions (data not shown). The original full-scan mass spectrum of the O-deacylated LOS from the galE mutant strain showed four major peaks with monoisotopic masses at m/z 1,452.3 (singly charged oligosaccharide fragment), m/z 1,174.0 (doubly charged parent mass), m/z 895.4 (singly charged lipid A fragment), and m/z 508.2 (doubly charged lipid A-PEA fragment; m = 1,016.4). The peak corresponding to the oligosaccharide at m/z 1,452.3 was selected, and a tandem experiment (MS2) was performed yielding a peak at m/z 1,232.3 that indicated the loss of Kdo (220 Da). Further fragmentation of this peak (MS3) yielded several peaks corresponding to loss of CO2 (m/z 1,188.4) followed by the further loss of hexose (m/z 1,026.3), hexose and HexNAc (m/z 823.3), and two hexoses and HexNAc (m/z 660.8). These data are all consistent with the serotype A structure. Moreover, since the mass difference observed for the LOS and the oligosaccharide fragments between the wild-type and the mutant strains correspond to two hexose residues (ΔM, 324 Da), the galE mutant strain would be predicted to produce a structure identical to that of a previously reported heptasaccharide containing a minor glycoform of serotype C (12). This structure differs from the LOS of serotype A only by the absence of a galactose disaccharide linked α1 α4 on the nonreducing terminus of the largest branch.
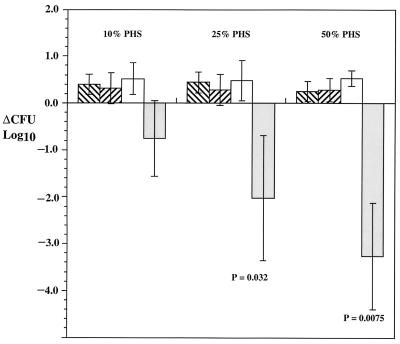
Comparison of bactericidal activity of PNHS against M. catarrhalis strain 2951 and the 2951 galE mutant.
Figure 6 shows the change in serum bactericidal activity of PNHS against strain 2951 and 2951 galE. M. catarrhalis strain 2951 was resistant to serum-mediated killing at all serum concentrations tested (Fig. 6). In contrast, incubation of strain 2951 galE in 50 or 25% PNHS resulted in a greater-than-2-log10-unit reduction in CFU (P = 0.0075 and P = 0.0352, respectively). Survival of the mutant strain was also impaired in 10% PHS, but substantial variability (range in log reduction, 0.42 to 1.30) decreased the statistical significance (P = 0.0910) of the results.
FIG. 6.
Sensitivity to serum complement-mediated bactericidal activity for M. catarrhalis strain 2951 and 2591 galE mutant after 30 min. ▧, results obtained with M. catarrhalis 2951 and heat-inactivated PNHS; ▨, results obtained with M. catarrhalis 2951 galE and inactivated PNHS; □, results obtained with M. catarrhalis 2951 and PNHS;  , results obtained with M. catarrhalis 2951 galE and PNHS. P values, comparisons between survival of strain 2951 and that of 2951 galE in PNHS. Each result is the mean from four separate experiments ± 1 standard deviation.
, results obtained with M. catarrhalis 2951 galE and PNHS. P values, comparisons between survival of strain 2951 and that of 2951 galE in PNHS. Each result is the mean from four separate experiments ± 1 standard deviation.
DISCUSSION
The presence of antibody and complement in various body environments constantly poses a challenge for the colonization and spread of bacteria. Resistance to these factors is frequently a requirement for pathogenesis. Multiple studies have demonstrated that resistance to complement-mediated killing is an important virulence factor for M. catarrhalis infections. Hol et al. reported in two studies that 156 of 179 (87%) and 124 of 200 (62%) clinical isolates from adult patients with pulmonary infections were serum resistant (23, 24). Recently, Verduin et al. have reported that 89% of M. catarrhalis strains isolated from adults with lower respiratory disease are completely or partially resistant to the bactericidal action of serum (54). In contrast, it has been reported that 58% of M. catarrhalis isolates carried in the pharynges of asymptomatic, healthy children are serum sensitive and do not appear to cause disease (23, 24, 54).
There are multiple reports that implicate various bacterial factors potentially involved in serum resistance. Verduin and coworkers have reported that resistant M. catarrhalis strains either bind or bind and inactivate a terminal complement component or intermediate involved in the formation of the membrane attack complex (53). Other investigators have demonstrated that complement resistance may involve an HMW outer membrane protein (OMP) or ubiquitous surface proteins (UspA1 and UspA2) (22, 30; C. M. Verduin, H. J. Bootsma, C. Hol, A. Fleer, M. Jansze, K. L. Klingman, T. F. Murphy, and H. van Dijk, Abstr. 95th Gen. Meet. Am. Soc. Microbiol. 1995, abstr. B137, p. 189, 1995; C. M. Verduin, M. Jansze, J. Verhoef, A. Fleer, and H. van Dijk, Clin. Exp. Immunol., abstr. 143, p. 50, 1994). Further studies have suggested that this HMW OMP binds human vitronectin, which subsequently inhibits complement activity (Verduin et al., Abstr. 95 Gen. Meet. Am. Soc. Microbiol. 1995; Verduin et al., Clin. Exp. Immunol.).
M. catarrhalis mutants defective in expression of UspA1 or UspA2 have been constructed and analyzed for sensitivity to human serum (1, 39). Whereas UspA1 is primarily involved in attachment, the putative function of UspA2 appears to be associated with the resistance of M. catarrhalis to the bactericidal activity of normal human sera. In comparative studies, the M. catarrhalis mutant defective in UspA2 expression was shown to be extremely sensitive to killing by normal human serum, whereas the wild type and the uspA1 mutant were completely resistant (1). Analysis using HMW OMP-specific MAb 9E9 (30) of strain 2951 and strain 2951 galE indicates that the HMW OMP reacts with both strains in a Western blot (data not shown).
More recently, a study by Verduin and coworkers analyzed 75 strains of M. catarrhalis with various degrees of complement susceptibility by pulsed-field gel electrophoresis and automated ribotyping (54). These studies divided the complement-sensitive and complement-resistant strains into two groups. Therefore, these investigators conclude that M. catarrhalis complement resistance represents a separate lineage in the species.
The role of the LOS in the serum resistance of M. catarrhalis is less clear. Ninety-five percent of the M. catarrhalis clinical isolates can be grouped into three serotypes based on the reaction of their LOS (52) with a polyclonal rabbit antibody raised against whole bacteria. The chemical structures corresponding to these three serotypes have been defined (25). Sixty-one percent of clinical isolates belong to serogroup A. M. catarrhalis strain 2951 LOS is type A based on mass spectrometric analysis.
In contrast to normal nonimmune serum, convalescent serum from patients with Moraxella infection was found to be bactericidal to the patient's own isolate (9) and to contain anti-LOS antibodies (20). However, antibodies to LOS in a patient's convalescent serum do not appear to be serotype specific (44). In addition, there does not appear to be any linkage between the serotype and the site of infection or severity of disease (44).
The 2951 galE mutant is sensitive to complement-mediated killing in normal human serum, whereas the parent strain is resistant. These data suggest that resistance to serum complement-mediated killing may be related to the presence of the two terminal galactose residues on the LOS. These residues form part of a structure that has been shown to be immunochemically identical to the Pk (Galα1-4Galβ1-4Glc) antigen found on a number of human cells including erythrocytes, as well as gastrointestinal, ureteral, and bladder epithelial cells (7, 29, 35). It would appear that the Moraxella Galα1-4Galβ1-4Glc structure may act as a human self-antigen and that antibodies to it are not present in normal human serum. The increased sensitivity to the bactericidal effects of normal human serum that occurs in the galE mutant suggests that removal of this epitope exposes a LOS antigen with which naturally occurring human antibodies can react resulting in complement-mediated lysis. Mutations in galE genes of other bacteria have been shown to alter the pathogenic potential of these strains. N. meningitidis group B strain B1940 was made serum sensitive by the introduction of this mutation (56). A galE mutation introduced into N. meningitidis strain NMB by Kahler et al. produced a serum-sensitive mutant in a capsule-positive bacterium (27). Pasteurella multocida with a galE mutation showed reduced virulence when tested by intraperitoneal inoculation in mice (17).
Resistance to killing by normal human serum has been shown to be important in the pathogenesis of the closely related species N. gonorrhoeae and N. meningitidis. Strains of N. gonorrhoeae isolated from disseminated gonococcal infections are typically serum resistant. In contrast, mucosal gonococcal isolates are serum sensitive, as typically studied in vitro. Previous studies have indicated that the LOS structure is an important factor in both of these phenotypes (4, 5).
It is of interest that the LOS of serum-sensitive gonococci undergoes sialylation in vivo, a modification that allows the isolate to become serum resistant and that seems critical in the pathogenesis of mucosal infection. Unlike the LOS from these gonococci, M. catarrhalis serotype A LOS lacks the lactosamine (Galβ1-4 GlcNAc) sialylation site. Only serotype C LOS contains this potential sialylation site (12). Studies in our laboratory confirm that serotype A LOS of M. catarrhalis does not undergo sialylation at an alternative site (data not shown). Hence, the intact LOS carbohydrate chain appears to confer protection against complement-mediated killing.
The resistance of bacteria to killing by normal human serum is the result of a complex array of bacterial surface factors. These studies suggest that in M. catarrhalis as with other pathogens LOS is one of these factors (4, 5).
ACKNOWLEDGMENTS
This research was supported by Public Health Service Research grants AI45728, AI44642 (M.A.A.), AI46469 (A.A.C.), and AI31254 (B.W.G.). The MALDIMS was performed with instrumentation kindly provided by PE Biosystems, and the ESI MS was performed at the mass spectrometry facility at the University of California, Berkeley (Department of Chemistry). The University of Iowa DNA facility is supported in part by the Diabetes Endocrinology Research Center with National Institutes of Health grant DK25295 and by the College of Medicine.
REFERENCES
- 1.Aebi C, Lafontaine E R, Cope L D, Latimer J L, Lumbley S L, McCracken G H, Jr, Hansen E J. Phenotypic effect of isogenic uspA1 and uspA2 mutations on Moraxella catarrhalis 035E. Infect Immun. 1998;66:3113–3119. doi: 10.1128/iai.66.7.3113-3119.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexeyev M F. Three kanamycin resistance gene cassettes with different polylinkers. BioTechniques. 1995;18:52–56. [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreoni J, Kayhty H, Densen P. Vaccination and the role of capsular polysaccharide antibody in prevention of recurrent meningococcal disease in late complement component-deficient individuals. J Infect Dis. 1993;168:227–231. doi: 10.1093/infdis/168.1.227. [DOI] [PubMed] [Google Scholar]
- 5.Apicella M A, Westerink M A, Morse S A, Schneider H, Rice P A, Griffiss J M. Bactericidal antibody response of normal human serum to the lipooligosaccharide of Neisseria gonorrhoeae. J Infect Dis. 1986;153:520–526. doi: 10.1093/infdis/153.3.520. [DOI] [PubMed] [Google Scholar]
- 6.Borovkov A Y, Rivkin M I. XcmI-containing vector for direct cloning of PCR products. BioTechniques. 1997;22:812–814. doi: 10.2144/97225bm04. [DOI] [PubMed] [Google Scholar]
- 7.Brodin N T, Dahmen J, Nilsson B, Messeter L, Martensson S, Heldrup J, Sjogren H O, Lundblad A. Monoclonal antibodies produced by immunization with neoglycoproteins containing Gal alpha 1-4Gal beta 1-4Glc beta-O and Gal alpha 1-4Gal beta 1-4GlcNAc beta-O residues: useful immunochemical and cytochemical reagents for blood group P antigens and a differentiation marker in Burkitt lymphoma and other B-cell malignancies. Int J Cancer. 1988;42:185–194. doi: 10.1002/ijc.2910420208. [DOI] [PubMed] [Google Scholar]
- 8.Catlin B W. Transformation of Neisseria meningitidis by deoxyribonucleates from cells and from culture slime. J Bacteriol. 1960;79:579–590. doi: 10.1128/jb.79.4.579-590.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman A J J, Musher D M, Jonsson S, Clarridge J E, Wallace R J J. Development of bactericidal antibody during Branhamella catarrhalis infection. J Infect Dis. 1985;151:878–882. doi: 10.1093/infdis/151.5.878. [DOI] [PubMed] [Google Scholar]
- 10.Del Beccaro M A, Mendelman P M, Inglis A F, Richardson M A, Duncan N O, Clausen C R, Stull T L. Bacteriology of acute otitis media: a new perspective. J Pediatr. 1992;120:81–84. doi: 10.1016/s0022-3476(05)80605-5. [DOI] [PubMed] [Google Scholar]
- 11.Dudas K C, Apicella M A. Selection and immunochemical analysis of lipooligosaccharide mutants of Neisseria gonorrhoeae. Infect Immun. 1988;56:499–504. doi: 10.1128/iai.56.2.499-504.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edebrink P, Jansson P-E, Rahman M M, Widmalm G, Holme T, Rahmam M. Structural studies of the O-antigen oligosaccharides from two strains of Moraxella catarrhalis serotype C. Carbohydr Res. 1995;266:237–261. doi: 10.1016/0008-6215(94)00276-l. [DOI] [PubMed] [Google Scholar]
- 13.Edebrink P, Jansson P-E, Widmalm G, Holme T, Rahman M. The structure of oligosaccharide isolated from the lipopolysaccharide of Moraxella catarrhalis serotype B, strain CCUG 3292. Carbohydr Res. 1996;295:127–146. doi: 10.1016/s0008-6215(96)90132-9. [DOI] [PubMed] [Google Scholar]
- 14.Edebrink P, Jansson P E, Rahman M M, Widmalm G, Holme T, Rahman M, Weintraub A. Structural studies of the O-polysaccharide from the lipopolysaccharide of Moraxella (Branhamella) catarrhalis serotype A (strain ATCC 25238) Carbohydr Res. 1994;257:269–284. doi: 10.1016/0008-6215(94)80040-5. [DOI] [PubMed] [Google Scholar]
- 15.Faden H, Bernstein J, Brodsky L, Stanievich J, Ogra P L. Effect of prior antibiotic treatment on middle ear disease in children. Ann Otol Rhinol Laryngol. 1992;101:87–91. doi: 10.1177/000348949210100119. [DOI] [PubMed] [Google Scholar]
- 16.Faden H, Hong J, Murphy T F. Immune response to outer membrane antigens of Moraxella catarrhalis in children with otitis media. Infect Immun. 1992;60:3824–3829. doi: 10.1128/iai.60.9.3824-3829.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandez de Henestrosa A R, Badiloa I, Saco M, Perez de Rosa A M, Campoy S, Barbe J. Importance of the galE gene on the virulence of Pasteurella multocida. FEMS Microbiol Lett. 1997;154:311–316. doi: 10.1111/j.1574-6968.1997.tb12661.x. [DOI] [PubMed] [Google Scholar]
- 18.Gan V N, Kusmiesz H, Shelton S, Nelson J D. Comparative evaluation of loracarbef and amoxicillin-clavulanate for acute otitis media. Antimicrob Agents Chemother. 1991;35:967–971. doi: 10.1128/aac.35.5.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson B W, Engstrom J J, John C M, Hines W, Falick A M. Characterization of bacterial lipooligosaccharides by delayed extraction matrix-assisted laser desorption ionization time-of-flight mass spectrometry. J Am Soc Mass Spectrom. 1997;8:645–658. [Google Scholar]
- 20.Gu X X, Chen J, Barenkamp S J, Robbins J B, Tsai C M, Lim D J, Battey J. Synthesis and characterization of lipooligosaccharide-based conjugates as vaccine candidates for Moraxella (Branhamella) catarrhalis. Infect Immun. 1998;66:1891–1897. doi: 10.1128/iai.66.5.1891-1897.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helander O M, Nummila K, Kilpelaien I, Vaara M. Increased substitution of phosphate groups in lipopolysaccharides and lipid A of polymyxin-resistant mutants of Salmonella typhimurium and Escherichia coli. Prog Clin Biol Res. 1995;392:15–23. [PubMed] [Google Scholar]
- 22.Helminen M E, Maciver I, Paris M, Latimer J L, Lumbley S L, Cope L D, McCracken G H, Jr, Hansen E J. A mutation affecting expression of a major outer membrane protein of Moraxella catarrhalis alters serum resistance and survival in vivo. J Infect Dis. 1993;168:1194–1201. doi: 10.1093/infdis/168.5.1194. [DOI] [PubMed] [Google Scholar]
- 23.Hol C, Verduin C M, van Dijke E, Verhoef J, van Dijk H. Conplement resistance in Branhamella (Moraxella) catarrhalis. Lancet. 1993;341:1281. doi: 10.1016/0140-6736(93)91185-o. [DOI] [PubMed] [Google Scholar]
- 24.Hol C, Verduin C M, van Dijke E E, Verhoef J, Fleer A, van Dijk H. Complement resistance is a virulence factor of Branhamella (Moraxella) catarrhalis. FEMS Immunol Med Microbiol. 1995;11:207–211. doi: 10.1111/j.1574-695X.1995.tb00118.x. [DOI] [PubMed] [Google Scholar]
- 25.Holme T, Rahman M, Jansson P E, Widmalm G. The lipopolysaccharide of Moraxella catarrhalis structural relationships and antigenic properties. Eur J Biochem. 1999;265:524–529. doi: 10.1046/j.1432-1327.1999.00731.x. [DOI] [PubMed] [Google Scholar]
- 26.John C M, Griffiss J M, Apicella M A, Mandrell R E, Gibson B W. The structural basis for pyocin resistance in Neisseria gonorrhoeae lipooligosaccharides. J Biol Chem. 1991;266:19303–19311. [PubMed] [Google Scholar]
- 27.Kahler C M, Martin L E, Shih G C, Rahman M M, Carlson R W, Stephens D S. The (α2→8)-linked polysialic acid capsule and lipooligosaccharide structure both contribute to the ability of serogroup B Neisseria meningitidis to resist the bactericidal activity of normal human serum. Infect Immun. 1998;66:5939–5947. doi: 10.1128/iai.66.12.5939-5947.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karalus R, Campagnari A A. Moraxella catarrhalis: a review of an important human mucosal pathogen. Microbes Infect. 2000;5:1–13. doi: 10.1016/s1286-4579(00)00314-2. [DOI] [PubMed] [Google Scholar]
- 29.Kasai K, Galton J, Terasaki P I, Wakisaka A, Kawahara M, Root T, Hakomori S I. Tissue distribution of the Pk antigen as determined by a monoclonal antibody. J Immunogenet. 1985;12:213–220. doi: 10.1111/j.1744-313x.1985.tb00848.x. [DOI] [PubMed] [Google Scholar]
- 30.Klingman K L, Murphy T F. Purification and characterization of a high-molecular-weight outer membrane protein of Moraxella (Branhamella) catarrhalis. Infect Immun. 1994;62:1150–1155. doi: 10.1128/iai.62.4.1150-1155.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee F K, Gibson B W, Melaugh W, Zaleski A, Apicella M A. Relationship between UDP-glucose 4-epimerase activity and oligoglucose glycoforms in two strains of Neisseria meningitidis. Infect Immun. 1999;67:1405–1414. doi: 10.1128/iai.67.3.1405-1414.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lesse A J, Campagnari A A, Bittner W E, Apicella M A. Increased resolution of lipopolysaccharides and lipooligosaccharides utilizing tricine sodium dodecyl sulfate polyacrylamide gel electrophoresis. J Immunol Methods. 1990;126:109–117. doi: 10.1016/0022-1759(90)90018-q. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Thoden J B, Kim J, Berger E, Gulick A M, Ruzicka F J, Holden H M, Frey P A. Mechanistic roles of tyrosine 149 and serine 124 in UDP-galactose 4-epimerase from Escherichia coli. Biochemistry. 1997;36:10675–10684. doi: 10.1021/bi970430a. [DOI] [PubMed] [Google Scholar]
- 34.Luke N R, Russo T A, Luther N, Campagnari A A. Use of an isogenic mutant constructed in Moraxella catarrhalis to identify a protective epitope of outer membrane protein B1 defined by monoclonal antibody 11C6. Infect Immun. 1999;67:681–687. doi: 10.1128/iai.67.2.681-687.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mandrell R E. Further antigenic similarities of Neisseria gonorrhoeae lipooligosaccharides and human glycosphingolipids. Infect Immun. 1992;60:3017–3020. doi: 10.1128/iai.60.7.3017-3020.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mandrell R E, Apicella M A. Lipo-oligosaccharides (LOS) of mucosal pathogens: molecular mimicry and host-modification of LOS. Immunobiology. 1993;187:382–402. doi: 10.1016/S0171-2985(11)80352-9. [DOI] [PubMed] [Google Scholar]
- 37.Mandrell R E, McLaughlin R, Abu Kwaik Y, Lesse A, Yamasaki R, Gibson B, Spinola S M, Apicella M A. Lipooligosaccharides (LOS) of some Haemophilus species mimic human glycosphingolipids, and some LOS are sialylated. Infect Immun. 1992;60:1322–1328. doi: 10.1128/iai.60.4.1322-1328.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masoud H, Perry M B, Richards J C. Characterization of the lipopolysaccharide of Moraxella catarrhalis: structural analysis of the lipid A from M. catarrhalis serotype A lipopolysaccharide. Eur J Biochem. 1994;220:209–216. doi: 10.1111/j.1432-1033.1994.tb18616.x. [DOI] [PubMed] [Google Scholar]
- 39.McMichael J C, Fiske M J, Fredenburg R A, Chakravarti D N, VanDerMeid K R, Barniak V, Caplan J, Bortell E, Baker S, Arumugham R, Chen D. Isolation and characterization of two proteins from Moraxella catarrhalis that bear a common epitope. Infect Immun. 1998;66:4374–4381. doi: 10.1128/iai.66.9.4374-4381.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohr M D, Bornsen K O, Widmer H M. Matrix-assisted laser desorption/ionization mass spectrometry: improved matrix for oligosaccharides. Rapid Commun Mass Spectrom. 1995;9:809–814. doi: 10.1002/rcm.1290090919. [DOI] [PubMed] [Google Scholar]
- 41.Murphy T F. Branhamella catarrhalis: epidemiology, surface antigenic structure, and immune response. Microbiol Rev. 1996;60:267–269. doi: 10.1128/mr.60.2.267-279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nordhoff E, Ingendoh A, Cramer R, Overberg A, Stahl B, Karas M, Hillenkamp F, Crain P F. Matrix-assisted laser desorption/ionization mass spectrometry of nucleic acids with wavelengths in the ultraviolet and infrared. Rapid Commun Mass Spectrom. 1992;6:771–776. doi: 10.1002/rcm.1290061212. [DOI] [PubMed] [Google Scholar]
- 43.Picard B, Goullet P, Denamur E, Suermondt G. Esterase electrophoresis: a molecular tool for studying the epidemiology of Branhamella catarrhalis nosocomial infection. Epidemiol Infect. 1989;103:547–554. doi: 10.1017/s0950268800030946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rahman M, Holme T, Jonsson I, Krook A. Lack of serotype-specific antibody response to lipopolysaccharide antigens of Moraxella catarrhalis during lower respiratory tract infection. Eur J Clin Microbiol Infect Dis. 1995;14:297–304. doi: 10.1007/BF02116522. [DOI] [PubMed] [Google Scholar]
- 45.Richards S J, Greening A P, Enright M C, Morgan M G, McKenzie H. Outbreak of Moraxella catarrhalis in a respiratory unit. Thorax. 1993;48:91–92. doi: 10.1136/thx.48.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruuskanen O, Heikkinen T. Otitis media: etiology and diagnosis. Pediatr Infect Dis J. 1994;13:S23–S26. [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 48.Stephens D S, McAllister C F, Zhou D, Lee F K, Apicella M A. Tn916-generated, lipooligosaccharide mutants of Neisseria meningitidis and Neisseria gonorrhoeae. Infect Immun. 1994;62:2947–2952. doi: 10.1128/iai.62.7.2947-2952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swanson B A, Frey P A. Identification of lysine 153 as a functionally important residue in UDP-galactose 4-epimerase from Escherichia coli. Biochemistry. 1993;32:13231–13236. doi: 10.1021/bi00211a035. [DOI] [PubMed] [Google Scholar]
- 50.Thoden J B, Frey P A, Holden H M. Molecular structure of the NADH/UDP-glucose abortive complex of UDP-galactose 4-epimerase from Escherichia coli: implications for the catalytic mechanism. Biochemistry. 1996;35:5137–5144. doi: 10.1021/bi9601114. [DOI] [PubMed] [Google Scholar]
- 51.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vaneechoutte M, Verschraegen G, Claeys G, van den Abelle A-M. Serological typing of Branhamella catarrhalis strains on the basis of lipopolysaccharide antigens. J Clin Microbiol. 1990;28:182–187. doi: 10.1128/jcm.28.2.182-187.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verduin C M. Differences in complement activation between complement-resistant and complement-sensitive Moraxella (Branhamella) catarrhalis strains occur at the level of membrane attack complex formation. Infect Immun. 1994;62:589–595. doi: 10.1128/iai.62.2.589-595.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verduin C M, Kools-Sijmons M, van der Plas J, Vlooswijk J, Tromp M, van Dijk H, Banks J, Verbrugh H, van Belkum A. Complement-resistant Moraxella catarrhalis forms a genetically distinct lineage within the species. FEMS Microbiol Lett. 2000;184:1–8. doi: 10.1111/j.1574-6968.2000.tb08981.x. [DOI] [PubMed] [Google Scholar]
- 55.Vestal M L, Juhasz P, Martin S A. Delayed extraction matrix-assisted laser desorption time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 1995;9:1044–1050. [Google Scholar]
- 56.Vogel U, Weinberger A, Frank R, Muller A, Kohl J, Atkinson J P, Frosch M. Complement factor C3 deposition and serum resistance in isogenic capsule and lipooligosaccharide sialic acid mutants of serogroup B Neisseria meningitidis. Infect Immun. 1997;65:4022–4029. doi: 10.1128/iai.65.10.4022-4029.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilson D B, Hogness D S. The enzymes of the galactose operon in Escherichia coli. 3. The size and composition of galactokinase. J Biol Chem. 1969;244:2137–2142. [PubMed] [Google Scholar]
- 58.Wilson D B, Hogness D S. The enzymes of the galactose operon in Escherichia coli. IV. The frequencies of translation of the terminal cistrons in the operon. J Biol Chem. 1969;244:2143–2148. [PubMed] [Google Scholar]