Abstract
Introduction: Models of structural connectivity in the human brain are typically simulated using tractographic approaches. However, the nonlinear fitting of anatomical pathway atlases to de novo subject brains represents a simpler alternative that is hypothesized to provide more anatomically realistic results. Therefore, the goal of this study was to perform a side-by-side comparison of the streamline estimates generated by either pathway atlas fits or tractographic reconstructions in the same subjects.
Methods: Our analyses focused on reconstruction of the corticospinal tract (CST), cerebellothalamic (CBT), and pallidothalamic (PT) pathways using example datasets from the Human Connectome Project (HCP). We used MRtrix3 to explore whole brain, as well as manual seed-to-target, tractography approaches. In parallel, we performed nonlinear fits of an axonal pathway atlas to each HCP dataset using Advanced Normalization Tools (ANTs).
Results: The different methods produced notably different estimates for each pathway in each subject. The fitted atlas pathways were highly stereotyped and exhibited low variability in their streamline trajectories. Manual tractography resulted in pathway estimates that generally corresponded with the fitted atlas pathways, but with a higher degree of variability in the individual streamlines. Pathway reconstructions derived from whole-brain tractography exhibited the highest degree of variability and struggled to create anatomically realistic representations for either the CBT or PT pathways.
Conclusion: The speed, simplicity, reproducibility, and realism of anatomical pathway model fits makes them an appealing option for some forms of structural connectivity modeling in the human brain.
Impact statement
Axonal pathway modeling is an important component of deep brain stimulation (DBS) research studies that seek to identify the brain connections that are directly activated by stimulation. The corticospinal tract, cerebellothalamic (CBT), and pallidothalamic (PT) pathways are specifically relevant to the study of subthalamic DBS for the treatment of Parkinson's disease. Our results suggest that anatomical pathway model fits of the CBT and PT pathways to de novo subject brains represent a more anatomically realistic option than tractographic approaches when studying subthalamic DBS.
Keywords: ansa lenticularis, lenticular fasciculus, dentatorubrothalamic tract, internal capsule, pyramidal tract
Introduction
Modeling techniques to estimate the structural connectivity of the human brain are a major focus of imaging research. Structural connectivity models have utility in characterizing the circuitry of brain networks (Elam et al., 2021), estimating targets for neuromodulation interventions (Horn and Fox, 2020), and prospective surgical planning for clinical brain stimulation therapies (Sheth et al., 2022). Structural connectivity estimates are typically derived from diffusion-weighted imaging (DWI) data using tractography algorithms (Wakana et al., 2007). One key advantage of using tractography approaches is that many different pathways in the brain can theoretically be reconstructed from the DWI data, given a priori knowledge of the seed and target regions for each pathway of interest. However, while tractography results are ubiquitous in human brain imaging research, their reliability, robustness, and accuracy are commonly called into question (Schilling et al., 2021). In addition, novel approaches to validate human structural connectivity estimates with intracranial electrophysiology measurements have exhibited relatively weak correlations (Adkinson et al., 2022; Howell et al., 2021).
In attempts to improve the reliability, robustness, and accuracy of structural connectivity models, several groups have used tractography data to create anatomically annotated large-scale pathway atlases for the human brain (e.g., Hansen et al., 2021; Oishi et al., 2008; Radwan et al., 2022; van Baarsen et al., 2015; Yeh et al., 2018; Zhang et al., 2018). Histology-based anatomical pathway atlases have also been created for focal brain regions consisting of small-scale pathways that exhibit a high degree of bending, crossing, and intermingling (Adil et al., 2021; Petersen et al., 2019).
Irrespective of how they were originally created, pathway atlases can then be nonlinearly warped to fit the MRI of a de novo subject, thereby representing an alternative to tractography for estimating the location of axonal streamlines in the brain. The atlas fitting process does not require the collection of DWI data, and is analogous to the coregistration of 3D nuclei atlas volumes to the subject brain (Yelnik et al., 2007). In addition, all of the available pathways in the atlas are defined in subject space simultaneously by the fitting process, which is attractive from an efficiency perspective. However, the pathway estimates derived from atlases are stereotyped results and are limited by the errors associated with the coregistration of the atlas to the subject, which can be especially inaccurate in subjects with abnormal brain anatomy (e.g., large tumors or substantial atrophy).
The goal of this study was to provide a side-by-side comparison of axonal pathway estimates generated by pathway atlas fits or tractographic reconstructions in the same subjects. We selected three different pathways for analysis, where each pathway represented a different anatomical challenge for estimating the axonal trajectories through the brain. The corticospinal tract (CST) is a large long-range pathway. The cerebellothalamic (CBT) pathway is a mid-sized mid-range tract that crosses hemispheres. The pallidothalamic (PT) pathway is a small short-range tract that exhibits a high degree of tortuosity.
We created the pathway estimates in the brains of different subjects using: (1) anatomically constrained segregation of whole-brain tractography, (2) manual seed-to-target tractography, or (3) nonlinear fitting of a pathway atlas. The analyses were performed with six example datasets randomly selected from the Human Connectome Project (HCP) (Elam et al., 2021) (Supplementary Table S1), and employed advanced processing steps to maximize the accuracy of the tractographic reconstructions (Tournier et al., 2019). The results allowed us to compare the streamline trajectories of the different pathway reconstruction methods and identify some of the caveats associated with using these various approaches to model axonal pathways in the human brain.
Methods
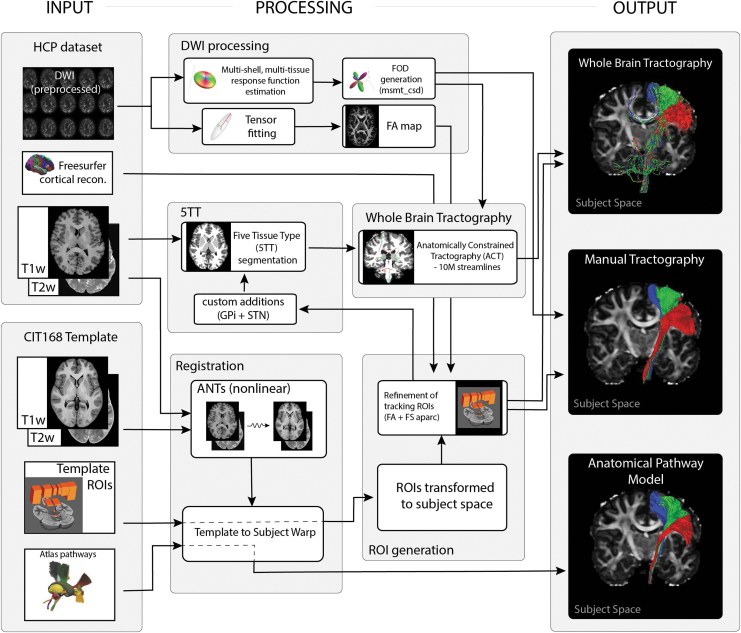
We compared the nonlinear fits of atlas pathway models with prominent methods for pathway reconstruction with tractography (Fig. 1). Our goal was to generate the best possible representation of three example axonal pathways using tractographic algorithms. The three pathways of interest were the CST, CBT, and PT pathways. We relied on HCP preprocessed datasets to ensure that the quality of the DWI data was sufficient for detailed tractographic analyses (Glasser et al., 2013). Six HCP subjects (three males, three females) were randomly selected from the “HCP 1200 Subject Release” for our analyses (Supplementary Table S1).
FIG. 1.
Flowchart illustrating the key processing steps for each of the three pathway reconstruction methods. FA, fractional anisotropy; FOD, fiber orientation distribution; GPi, globus pallidus internus; STN, subthalamic nucleus.
We employed one reconstruction strategy based on whole-brain tractography, and another strategy based on manual seed-to-target tractography. Both strategies were implemented using MRtrix3 (Tournier et al., 2019) and the default (iFOD2) probabilistic tractography algorithm. In parallel, we performed nonlinear fits of the Petersen et al. (2019) axonal pathway atlas to each HCP dataset using Advanced Normalization Tools (ANTs) (Avants et al., 2014). Table 1 provides estimates of the processing time associated with the estimating the pathways in brain imaging data.
Table 1.
Estimated Processing Time
| |
Requirements |
Reproducible |
Processing time |
||||
|---|---|---|---|---|---|---|---|
| Imaging data | Other | Template registration | DWI preprocessinga | Tractographyb | Total | ||
| Whole-brain ACT | T1w DWI |
— | Yes | 39 min | 5 h 45 min | 1 h 12 min | 7 h 36 min |
| Manual tractography | T1w DWI |
Neuroanatomical knowledge | No | 39 min | 5 h 45 min | Manual tracking + filtering | 6 h 24 min + manual filtering |
| Anatomical pathway model | T1w | — | Yes | 39 min | — | — | 39 min |
Time estimates for a single subject for the three processing pipelines using the minimum data necessary. If a T2w dataset is available then multimodal registration may improve registration accuracy but will increase processing time by ∼20 min. (T1w data for template registration and DWI for streamline generation). To provide more realistic estimates for this table we used a non-HCP unprocessed DWI dataset (2 mm isotropic resolution, 180 directions, multishell [0, 700, 1200, 2800 sec/mm2], reverse-phase-encoded b0 volumes only, acquired on a Siemens 3T Skyra Magnetom). All processing were done using a workstation equipped with a Xeon Gold 5120 CPU (14c/28t @ 2.20GHz).
The DWI preprocessing includes: denoising and Gibbs unringing (MRtrix), as well as correction for EPI distortions, subject motion, and Eddy currents (FSL).
The tractography stage includes generation of functions and for whole-brain ACT only, generation of tissue segmentation map (5TT), and 10 million streamlines (ACT).
ACT, anatomically constrained tractography; DWI, diffusion-weighted imaging; FOD, fiber orientation distribution.
Tractographic reconstruction of the CST is a relatively straightforward process that generally yields good results (e.g., Archer et al., 2018) (Fig. 2). Reconstructing the CBT pathway with tractography is more challenging, especially when considering the decussation in the midbrain (e.g., Nowacki et al., 2018) (Fig. 3). The PT pathway is very difficult to reconstruct with tractography because of the small fibers, high tortuosity of the trajectory, and crossing of the internal capsule (IC) (e.g., Kwon et al., 2021) (Fig. 4).
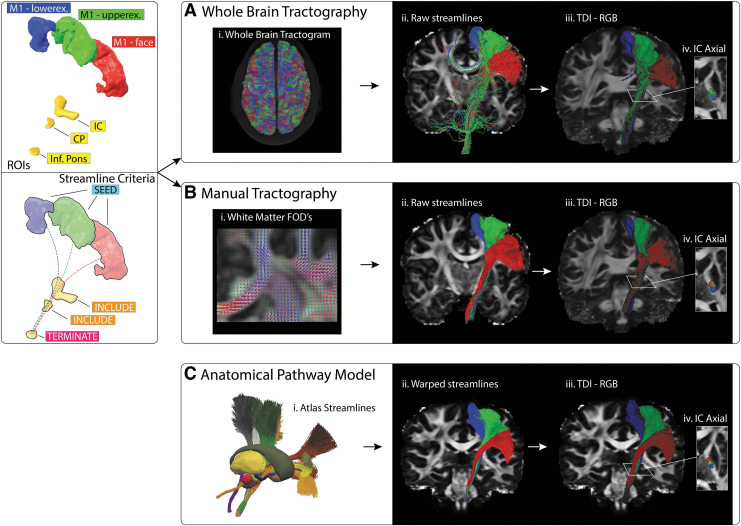
FIG. 2.
CST estimation. Left panel: ROIs used to reconstruct face, upper extremity, and lower extremity CST bundles. Right panel: (A) (i) The whole-brain tractogram was combined with the ROIs to extract; (ii) streamlines passing through all regions; (iii) Streamlines resampled into a track density image and each voxel color coded red (face), green (upper extremity), or blue (lower extremity) to help visualize any distinct separation between bundles. (B) Streamlines were generated manually using the ROIs as seed and inclusion masks. (C) The CST anatomical pathway model was nonlinearly transformed into subject space. CST, corticospinal tract; ROIs, regions of interests.
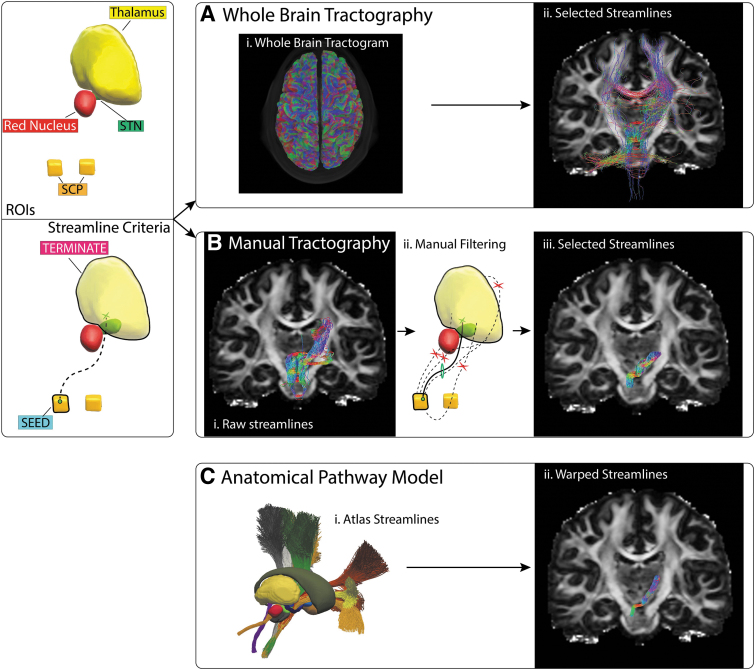
FIG. 3.
CBT pathway estimation. Left panel: Tracking ROIs and streamline criteria for a valid trajectory. Right panel: (A) (i–ii) Streamlines extracted from the whole-brain tractogram. (B) (i–ii) Manual tractography was seeded from the SCP with the contralateral thalamus used as termination mask. (iii) The resulting streamlines were manually filtered to select (iv) a bundle of plausible streamlines. (C) The CBT anatomical pathway model was nonlinearly transformed into subject space. CBT, cerebellothalamic; SCP, superior cerebellar peduncle.
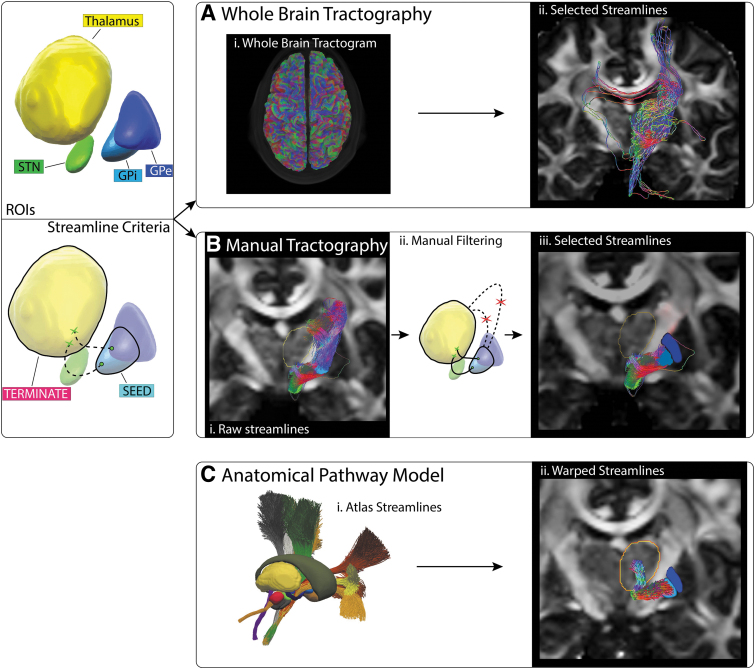
FIG. 4.
PT pathway estimation. Left panel: Tracking ROIs and streamline criteria for a valid trajectory following the AL sweeping around the IC or the LF crossing through the IC. Right panel: (A) (i–ii) Streamlines extracted from whole-brain tractogram. (B) (i–ii) Manual tractography seeded from the GPi with the contralateral thalamus used as termination mask. (iii) The resulting streamlines were manually filtered to select (iv) a bundle of plausible streamlines. (C) The anatomical pathway model of the PT pathway was nonlinearly transformed into subject space. AL, ansa lenticularis; LF, lenticular fasciculus; PT, pallidothalamic; IC, internal capsule.
All of our DWI-based pathway reconstructions used custom anatomical regions of interests (ROIs). The ROIs were initially defined in the high-resolution CIT168 brain atlas template (Pauli et al., 2018). These ROIs were then nonlinearly warped to each HCP dataset. For the CST reconstruction, the ROIs were further refined using the subject-specific Freesurfer cortical parcellation map provided with the HCP data (primary motor cortex ROIs) and the DWI-derived FA map (IC ROI) (Supplementary Fig. S1). All nonlinear registrations and warps were carried out with the ANTs toolbox (three stage, rigid and affine and b-spline symmetric normalization, optional arguments left at default values) and used both T1w and T2w data for multimodal registration between CIT168 template and HCP structural data (0.7 mm isotropic). ROIs used with the DWI data for streamline generation were resliced to match the lower data resolution (1.25 mm isotropic). All registrations were manually inspected to ensure adequate results from the nonlinear warping.
Whole-brain tractography was carried out using multishell multitissue constrained spherical deconvolution (MSMT-CSD) (Jeurissen et al., 2014) and the Anatomically Constrained Tractography (ACT) framework (Smith et al., 2012), as implemented in the MRtrix3 toolbox (Fig. 1). In brief, multitissue orientation distribution functions (ODFs) were generated using MSMT-CSD. Tissue segmentation masks (cortical gray matter [GM], subcortical GM, white matter [WM], and cerebrospinal fluid) were generated from the T1w data. The masks provided biologically realistic constraints for streamline propagation and reduced false positives in the whole-brain tractogram. We used rules that the streamlines cannot terminate in WM or CSF, and if a streamline entered a GM structure, then it had to terminate within and could not exit to propagate further. Ten million streamlines were generated for each HCP dataset. Additional parameters used in ACT generation were—backtracking: on, crop_at_gmwmi: on, seed_dynamic using wm-ODFs, maxlength: 250, cutoff: 0.06. Subsequent streamline selection was done by filtering the full ten million using specified inclusion masks.
The two DWI-based CST reconstructions were carried out using three sets of CST-ROIs. Each CST-ROI was associated with either the face, upper extremity, or lower extremity regions of primary motor cortex (Fig. 2). For manual tractography, each cortical region was used as a seed mask along with three subsequent inclusion masks at the level of the IC, the cerebral peduncle, and the inferior pons. Once a streamline passed through all inclusion regions, it was automatically terminated. For each bundle (i.e., face, arm, leg), 5000 streamlines were selected, with a max length threshold of 150 mm. No further manual filtering of the streamlines was applied. For the whole-brain approach, the same three sets of four ROIs were used to extract streamlines that passed through the specified regions.
For manual tractography of the CBT pathway, two ROIs were used (Fig. 3). A seed mask was placed in the superior cerebellar peduncle and a target mask covered the contralateral thalamus. The thalamus mask was generated using FSL First (Patenaude et al., 2011). Five thousand streamlines were generated using unidirectional seeding and a max length threshold of 100 mm. The resulting raw streamlines were manually filtered to remove obvious erroneous streamlines (e.g., crossing the midline outside the cerebellar decussation, or following the IC before turning sharply and entering the superior aspect of the thalamus). For the whole-brain approach, the same two ROIs were used to extract streamlines that passed through the specified regions.
For manual tractography of the PT pathway, globus pallidus internus (GPi) volume was used as the seed mask, and the thalamus mask was used as the termination mask (Fig. 4). Five thousand streamlines were generated using unidirectional seeding and a max length threshold of 50 mm. The resulting streamlines were manually filtered to remove implausible trajectories such as those exiting the superior aspect of the GPi or streamlines entering the thalamus through regions clearly not consistent with the thalamic fasciculus. For the whole-brain approach, the same two ROIs were used to extract streamlines that passed through the specified regions.
Results
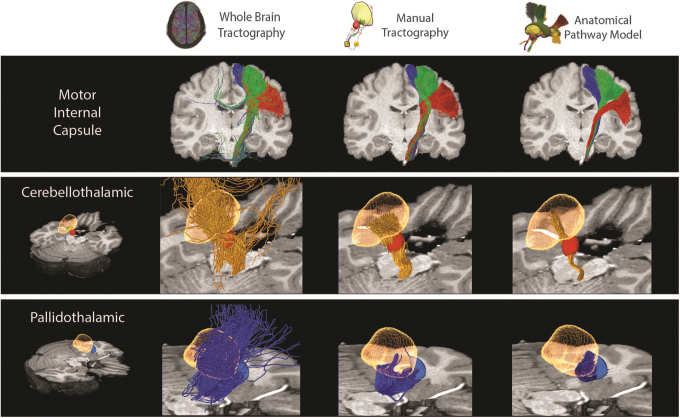
This study compared streamline estimates of axonal trajectories in the CST, CBT, and PT pathways that were defined within the context of HCP datasets. Each pathway modeling method provided a collection of streamlines for each pathway in each brain we analyzed. However, the different methods produced noticeably different estimates for each pathway (Fig. 5) in each subject (Supplementary Figs. S2–S4). The fitted atlas pathways were highly stereotyped and exhibited low variability in their streamline trajectories. Manual tractography resulted in pathway estimates that generally corresponded with the fitted atlas pathways, but with a higher degree of variability in the individual streamlines. Pathway reconstructions derived from whole-brain tractography exhibited the highest degree of variability and struggled to create anatomically realistic representations for either the CBT or PT pathways (Fig. 5).
FIG. 5.
Pathway estimate comparison. Examples of the streamlines associated with different axonal pathways, as simulated by different modeling methods from a single representative subject. All of the methods generated reasonable approximations of the CST (red: face, green: arm, blue: leg streamlines). Whole-brain tractography was not effective at reconstructing the CBT (orange streamlines) or PT (blue streamlines) pathways. Manual tractography generated reasonable approximations of the CBT pathway, but failed to capture the anatomical trajectory of the PT pathway. Yellow volume: thalamus, red volume: red nucleus, blue volume: pallidum.
Each method provided reasonable estimates for the CST, which were in general agreement with each other (Fig. 5; Supplementary Figs. S2 and S5). However, at the level of the thalamus, the tractographic reconstructions exhibited a posterior bias in the position of the streamlines within the IC, relative to the fitted atlas pathways (Fig. 2; Supplementary Fig. S5). In addition, the somatotopic organization of the CST streamlines were generally maintained with manual tractography, but not with whole-brain tractography (Fig. 2; Supplementary Fig. S5).
The manual tractography and fitted atlas pathways both provided reasonable estimates of the CBT pathway, which were in general agreement with each other (Fig. 5; Supplementary Figs. S3 and S6). The manual tractography CBT streamlines spanned a larger volume of tissue than the fitted atlas pathways (Fig. 3; Supplementary Fig. S6). However, this discrepancy could conceivably be adjusted by defining a more focused termination mask that was specifically focused on the cerebellar receiving area of the thalamus. Alternatively, the CBT pathway streamlines estimated from whole-brain tractography were not consistent with the expected anatomical trajectory (Fig. 5; Supplementary Fig. S6).
Each method provided differing estimates for the trajectory of the PT pathway (Fig. 5; Supplementary Fig. S4 and S7). The anatomically defined trajectories of the ansa lenticularis and lenticular fasciculus project from the pallium, cross the IC, enter the fields of Forel above the subthalamic nucleus (STN), and then make a relatively dramatic dorsal turn into the pallidal receiving area of thalamus (Gallay et al., 2008; Parent and Parent, 2004). However, the streamlines from manual tractography got pulled ventrally in the IC as they exited the pallidum. Once they crossed the IC, they generally emerged several millimeters below and anterior to the fields of Forel, and then wrapped around the medial side of the STN, before eventually reaching the thalamus (Figs. 4 and 5). Some of the PT streamlines from whole-brain tractography followed a similar trajectory as described for manual tractography, but most of them got pulled into the IC and exhibited spurious paths before eventually reaching the thalamus (Figs. 4 and 5). Alternatively, the PT streamlines of the fitted atlas pathways followed the expected anatomical trajectory.
Overall, whole-brain tractography generated the most extreme examples of erroneous streamlines for the pathways we simulated (Fig. 5). All of the streamlines displayed in the figures passed through all of the inclusion masks. However, in some instances the whole brain streamlines could loop back on themselves and continue until they hit a gray matter region as determined by the tractography algorithm. Erroneous streamlines typically occur because of the imperfect nature of streamline propagation guided by local diffusion orientations that are gross approximations of the underlying tissue microstructure. In addition, some erroneous streamlines appear in the results due to slight mismatches between the interpolated partial volume tissue map (used for anatomical constraints in the ACT generation pipeline) and the binary cortical ROIs (used for pathway extraction).
Discussion
The creation of axonal pathway models within subject-specific MRI datasets has utility in both scientific and clinical analyses (Elam et al., 2021; Horn and Fox, 2020). However, there are many different approaches to estimate structural connectivity in the human brain, and the pathway estimates generated by any given method are dependent upon the quality of the imaging data that is available for the analysis (Maier-Hein et al., 2017). As such, different tractography approaches are commonly compared with each other (Schilling et al., 2019). Unfortunately, the general lack of consistency in the anatomical accounts of WM pathways in the human brain (Bullock et al., 2022), makes it difficult to define one tractography method as superior to all others (Schilling et al., 2021). Alternatively, our recent work has focused on transitioning away from pathway estimation based on DWI data, and pivoted toward the creation of human axonal pathway priors derived from histological results and anatomical landmarks (Petersen et al., 2019).
We hypothesize that a detailed pathway atlas can be quickly and easily fit to a de novo subject brain to provide a higher degree of anatomical accuracy, reproducibility, and robustness than is possible with traditional tractographic approaches (Table 1 and Fig. 5). Along this line, we propose that atlas pathway fits are especially relevant when attempting to study focal brain regions with smaller scale pathways that cross each other and/or follow tortuous trajectories. One such example is the subthalamic region (Emmi et al., 2020; Jeon et al., 2022), which is pertinent to the analysis of clinical deep brain stimulation (DBS) for the treatment of Parkinson's disease.
Recent trends in subthalamic DBS research have gravitated toward estimating the activation of axonal pathways from the stimulation and subsequently correlating those predictions with behavioral outcome metrics (e.g., Chaturvedi et al., 2010; Horn et al., 2019; Vanegas-Arroyave et al., 2016). The basic goal of these kinds of analyses is to identify specific axonal pathways whose activation is linked to the generation of a specific therapeutic effect, or side effect, of the stimulation. Along this line, the CST, CBT, and PT pathways are directly relevant to DBS research. DBS of the CST is known to generate motor contraction side effects (e.g., Tommasi et al., 2008). Strong evidence implicates activation of the CBT pathway with the control of tremor (e.g., Coenen et al., 2014). Evolving results suggest that stimulating the PT pathway might improve rigidity (e.g., Butson et al., 2011). Continued examination of these hypotheses on a patient-specific level requires the creation of the axonal pathway models within the context of their individual MRI datasets (Noecker et al., 2021). However, there is ongoing debate on the best method for estimating the pathway trajectories in patient-specific brain models (Wang et al., 2021).
The goal of this study was to provide a side-by-side comparison of different techniques to represent the CST, CBT, and PT pathways in individual subject brains. Our results demonstrate that the specific methods used to estimate the pathways are likely to influence the pathway activation predictions from DBS simulations. This is because the trajectory of the streamline dictates its proximity to the implanted DBS electrode contacts. Then, the neural response to DBS is highly dependent upon the extracellular voltage distribution along the trajectory of the axon model (Gunalan et al., 2018). Therefore, attention should be paid to not only the accuracy of electrode localization in the brain of each subject, but also the anatomical validity of the streamline trajectories used to estimate the pathways (Noecker et al., 2021).
Our results suggest that anatomical pathway models, nonlinearly transformed into subject space, are likely to be the most accurate approach currently available to estimate axonal trajectories in the subthalamic region. This is because even when great care is taken when simulating tractography streamlines to represent subthalamic axonal pathways, the results are highly variable and not necessarily consistent with current anatomical understanding (Fig. 5).
Given the widespread use of Lead-DBS (Horn et al., 2019), the most common approach for estimating structural connectivity in clinical DBS research studies has been to use a whole-brain tractogram. Whole-brain tractography has the theoretical advantage of generally representing many of the structural connections in the brain, all within a single dataset. However, constructing a whole-brain tractogram is a time-consuming process (Table 1), and the anatomical accuracy of the simulated axonal trajectories are questionable (Fig. 5). Alternatively, when an a priori definition of the axonal pathway of interest is available, then seed-to-target tractography approaches can be employed to define anatomically inspired pathway models (Petersen et al., 2017). These methods can generate results with a higher degree of anatomical fidelity (Schilling et al., 2021), but they often require substantial levels of manual intervention to define the specific streamlines that constitute the pathway model (Figs. 2–4). Unfortunately, these general issues associated with tractography-based approaches are difficult to overcome because of the reliance on relatively low-resolution DWI data that are describing a water diffusion displacement profile in an underdetermined inverse problem (Thomas et al., 2014).
Recent trends in human structural connectivity modeling have begun to embrace the concept of using anatomically defined axonal pathway priors in parallel with tractographic algorithms to define the trajectory of specific connections in the brain (Bullock et al., 2022). Such strategies represent a welcome advance to the field of human brain connectomics. However, they also introduce the question, if detailed anatomical pathway priors exist, what additional value is provided by clinical-grade DWI data when attempting to define subject-specific structural connectivity models? Therefore, we attempted to begin to address this question in the subthalamic region with side-by-side comparisons of tractography-based pathway estimates and anatomical pathway priors fitted to the same subjects (Fig. 5).
The nonlinear fitting of an anatomical pathway atlas to a de novo subject brain can bypass the use of subject-specific DWI data in structural connectivity modeling (Fig. 1). This is advantageous when high-quality DWI data are not available for a subject, or when trying to reconstruct pathways that are known to be difficult for tractography algorithms (e.g., CBT or PT pathways). However, fitted pathway atlases generate stereotyped results that may fail to capture the real intersubject variability in the pathways (Fig. 5). This is a major limitation in applications where the goal is to compare the density or integrity of a given pathway across subjects in the study. Alternatively, when considering DBS pathway activation studies, stereotyped pathways could be considered an advantage. From the perspective of population-based statistics, if every streamline in the analysis (i.e., activated or not activated by the stimulus) is consistent across every subject in the study, this can help to standardize the pathway activation calculations. Therefore, the most appropriate method for subject-specific structural connectivity modeling is likely to be highly dependent upon the scientific question that is being explored.
Our results provide a side-by-side comparison of the streamline trajectories generated by different structural connectivity modeling methods, but they do not answer the question of streamline accuracy. This is because no real “gold standard” exists for these pathways in the human brain (Petersen et al., 2019). There are countless different approaches to simulating streamlines and brain connectivity with tractography algorithms (Schilling et al., 2021). We choose two example methods that we propose generate representative streamline estimates for either whole-brain or manual tractography analyses. However, a key limitation of this study was the possibility that another tractography method could have generated better results than the examples we presented.
Tractographic pathway modeling efforts are dependent upon the quality of DWI data used for the analyses (Maier-Hein et al., 2017). In this study, we used HCP datasets that are measurably superior to typical clinical brain imaging data (Elam et al., 2021). Nonetheless, an important limitation of HCP datasets is that they were acquired on young, healthy, normal subjects. Given that the anatomical pathway models were also derived from normative datasets, they are likely to be best suited to simulate the anatomy of other normal subjects. Alternatively, a powerful feature of tractographic approaches is that they can be directly applied to patient-specific datasets to evaluate brain connectivity in subjects with abnormal anatomy. Therefore, it remains unclear how well anatomical pathway model fits perform within the context of brains of subjects suffering from different disease states.
Conclusion
Different methods to simulate axonal pathways near the subthalamic region of the brain generate noticeably different estimates for the streamline trajectories. Our results demonstrate that anatomical pathway models can be applied to de novo subjects to estimate the location of pathways, and when considering pathways that are difficult to reconstruct with tractography, likely provide estimates that are more anatomically realistic. Therefore, we currently rely on nonlinear fits of anatomical pathway models to patient-specific imaging data as our preferred strategy for connectomic DBS modeling in the subthalamic region because of the speed, simplicity, reproducibility, and realism of the results relative to tractographic approaches.
Supplementary Material
Authors' Contributions
Conceptualization: M.V.P. and C.C.M. Formal analysis: M.V.P. Methodology: M.V.P. Writing: M.V.P. and C.C.M. Funding: C.C.M.
Data Availability Statement
The HCP datasets used in our analyses are publicly available at the Connectome Coordination Facility website. The processed streamline data presented in the figures are available on request from author M.V.P.
Author Disclosure Statement
C.C.M. is a paid consultant for Boston Scientific Neuromodulation, receives royalties from Hologram Consultants, Neuros Medical, Qr8 Health, and is a shareholder in the following companies: Hologram Consultants, Surgical Information Sciences, BrainDynamics, CereGate, Autonomic Technologies, Cardionomic, and Enspire DBS.
Funding Information
This work was supported by a grant from the National Institutes of Health (R01 NS105690).
Supplementary Material
References
- Adil SM, Calabrese E, Charalambous LT, et al. A high-resolution interactive atlas of the human brainstem using magnetic resonance imaging. Neuroimage 2021;237:118135; doi: 10.1016/j.neuroimage.2021.118135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adkinson JA, Tsolaki E, Sheth SA, et al. Imaging versus electrographic connectivity in human mood-related fronto-temporal networks. Brain Stimul 2022;15(3):554–565; doi: 10.1016/j.brs.2022.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer DB, Vaillancourt DE, Coombes SA. A Template and Probabilistic Atlas of the human sensorimotor tracts using diffusion MRI. Cereb Cortex 2018;28(5):1685–1699; doi: 10.1093/cercor/bhx066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Stauffer M, et al. The Insight ToolKit image registration framework. Front Neuroinform 2014;8:44; doi: 10.3389/fninf.2014.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock DN, Hayday EA, Grier MD, et al. A taxonomy of the Brain's white matter: Twenty-one major tracts for the 21st century. Cereb Cortex 2022;32(20):4524–4548; doi: 10.1093/cercor/bhab500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butson CR, Cooper SE, Henderson JM, et al. Probabilistic analysis of activation volumes generated during deep brain stimulation. Neuroimage 2011;54(3):2096–2104; doi: 10.1016/j.neuroimage.2010.10.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi A, Butson CR, Lempka SF, et al. Patient-specific models of deep brain stimulation: Influence of field model complexity on neural activation predictions. Brain Stimul 2010;3(2):65–67; doi: 10.1016/j.brs.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenen VA, Allert N, Paus S, et al. Modulation of the cerebello-thalamo-cortical network in thalamic deep brain stimulation for tremor: A diffusion tensor imaging study. Neurosurgery 2014;75(6):657–669; doi: 10.1227/NEU.0000000000000540 [DOI] [PubMed] [Google Scholar]
- Elam JS, Glasser MF, Harms MP, et al. The Human Connectome Project: A retrospective. Neuroimage 2021;244:118543; doi: 10.1016/j.neuroimage.2021.118543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmi A, Antonini A, Macchi V, et al. Anatomy and connectivity of the subthalamic nucleus in humans and non-human primates. Front Neuroanat 2020;14:13; doi: 10.3389/fnana.2020.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallay MN, Jeanmonod D, Liu J, et al. Human pallidothalamic and cerebellothalamic tracts: Anatomical basis for functional stereotactic neurosurgery. Brain Struct Funct 2008;212(6):443–463; doi: 10.1007/s00429-007-0170-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, et al. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage 2013;80:105–124; doi: 10.1016/j.neuroimage.2013.04.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunalan K, Howell B, McIntyre CC. Quantifying axonal responses in patient-specific models of subthalamic deep brain stimulation. Neuroimage 2018;172:263–277; doi: 10.1016/j.neuroimage.2018.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen CB, Yang Q, Lyu I, et al. Pandora: 4-D white matter bundle population-based atlases derived from diffusion MRI fiber tractography. Neuroinformatics 2021;19(3):447–460; doi: 10.1007/s12021-020-09497-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn A, Fox MD. Opportunities of connectomic neuromodulation. Neuroimage 2020;221:117180; doi: 10.1016/j.neuroimage.2020.117180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn A, Li N, Dembek TA, et al. Lead-DBS v2: Towards a comprehensive pipeline for deep brain stimulation imaging. Neuroimage 2019;184:293–316; doi: 10.1016/j.neuroimage.2018.08.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B, Isbaine F, Willie JT, et al. Image-based biophysical modeling predicts cortical potentials evoked with subthalamic deep brain stimulation. Brain Stimul 2021;14(3):549–563;doi: 10.1016/j.brs.2021.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon H, Lee H, Kwon DH, et al. Topographic connectivity and cellular profiling reveal detailed input pathways and functionally distinct cell types in the subthalamic nucleus. Cell Rep 2022;38(9):110439; doi: 10.1016/j.celrep.2022.110439 [DOI] [PubMed] [Google Scholar]
- Jeurissen B, Tournier JD, Dhollander T, et al. Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion MRI data. Neuroimage 2014;103:411–426; doi: 10.1016/j.neuroimage.2014.07.061 [DOI] [PubMed] [Google Scholar]
- Kwon DH, Paek SH, Kim YB, et al. In vivo 3D reconstruction of the human pallidothalamic and nigrothalamic pathways with super-resolution 7T MR Track Density Imaging and Fiber Tractography. Front Neuroanat 2021;15:739576; doi: 10.3389/fnana.2021.739576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier-Hein KH, Neher PF, Houde JC, et al. The challenge of mapping the human connectome based on diffusion tractography. Nat Commun 2017;8(1):1349; doi: 10.1038/s41467-017-01285-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noecker AM, Frankemolle-Gilbert AM, Howell B, et al. StimVision v2: Examples and applications in subthalamic deep brain stimulation for Parkinson's Disease. Neuromodulation 2021;24(2):248–258; doi: 10.1111/ner.13350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki A, Schlaier J, Debove I, et al. Validation of diffusion tensor imaging tractography to visualize the dentatorubrothalamic tract for surgical planning. J Neurosurg 2018;130(1):99–108; doi: 10.3171/2017.9.JNS171321 [DOI] [PubMed] [Google Scholar]
- Oishi K, Zilles K, Amunts K, et al. Human brain white matter atlas: Identification and assignment of common anatomical structures in superficial white matter. Neuroimage 2008;43(3):447–457; doi: 10.1016/j.neuroimage.2008.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent M, Parent A. The pallidofugal motor fiber system in primates. Parkinsonism Relat Disord 2004;10(4):203–211; doi: 10.1016/j.parkreldis.2004.02.007 [DOI] [PubMed] [Google Scholar]
- Patenaude B, Smith SM, Kennedy DN, et al. A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage 2011;56(3):907–922; doi: 10.1016/j.neuroimage.2011.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli WM, Nili AN, Tyszka JM. A high-resolution probabilistic in vivo atlas of human subcortical brain nuclei. Sci Data 2018;5:180063; doi: 10.1038/sdata.2018.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen MV, Lund TE, Sunde N, et al. Probabilistic versus deterministic tractography for delineation of the cortico-subthalamic hyperdirect pathway in patients with Parkinson disease selected for deep brain stimulation. J Neurosurg 2017;126(5):1657–1668; doi: 10.3171/2016.4.JNS1624 [DOI] [PubMed] [Google Scholar]
- Petersen MV, Mlakar J, Haber SN, et al. Holographic reconstruction of axonal pathways in the human brain. Neuron 2019;104(6):1056–1064; doi: 10.1016/j.neuron.2019.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radwan AM, Sunaert S, Schilling K, et al. An atlas of white matter anatomy, its variability, and reproducibility based on constrained spherical deconvolution of diffusion MRI. Neuroimage 2022;254:119029; doi: 10.1016/j.neuroimage.2022.119029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling KG, Nath V, Hansen C, et al. Limits to anatomical accuracy of diffusion tractography using modern approaches. Neuroimage 2019;185:1–11; doi: 10.1016/j.neuroimage.2018.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling KG, Rheault F, Petit L, et al. Tractography dissection variability: What happens when 42 groups dissect 14 white matter bundles on the same dataset? Neuroimage 2021;243:118502; doi: 10.1016/j.neuroimage.2021.118502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth SA, Bijanki KR, Metzger B, et al. Deep brain stimulation for depression informed by intracranial recordings. Biol Psychiatry 2022;92(3):246–251; doi: 10.1016/j.biopsych.2021.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RE, Tournier JD, Calamante F, et al. Anatomically-constrained tractography: Improved diffusion MRI streamlines tractography through effective use of anatomical information. Neuroimage 2012;62(3):1924–1938; doi: 10.1016/j.neuroimage.2012.06.005 [DOI] [PubMed] [Google Scholar]
- Thomas C, Ye FQ, Irfanoglu MO, et al. Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. Proc Natl Acad Sci U S A 2014;111(46):16574–16579; doi: 10.1073/pnas.1405672111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasi G, Krack P, Fraix V, et al. Pyramidal tract side effects induced by deep brain stimulation of the subthalamic nucleus. J Neurol Neurosurg Psychiatry 2008;79(7):813–819; doi: 10.1136/jnnp.2007.117507 [DOI] [PubMed] [Google Scholar]
- Tournier JD, Smith R, Raffelt D, et al. MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. Neuroimage 2019;202:116137; doi: 10.1016/j.neuroimage.2019.116137 [DOI] [PubMed] [Google Scholar]
- van Baarsen KM, Kleinnijenhuis M, Jbabdi S, et al. A probabilistic atlas of the cerebellar white matter. Neuroimage 2016;124(Pt A):724–732; doi: 10.1016/j.neuroimage.2015.09.014 [DOI] [PubMed] [Google Scholar]
- Vanegas-Arroyave N, Lauro PM, Huang L, et al. Tractography patterns of subthalamic nucleus deep brain stimulation. Brain 2016;139(Pt 4):1200–1210; doi: 10.1093/brain/aww020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakana S, Caprihan A, Panzenboeck MM, et al. Reproducibility of quantitative tractography methods applied to cerebral white matter. Neuroimage 2007;36(3):630–644; doi: 10.1016/j.neuroimage.2007.02.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Akram H, Muthuraman M, et al. Normative vs. patient-specific brain connectivity in deep brain stimulation. Neuroimage 2021;224:117307; doi: 10.1016/j.neuroimage.2020.117307 [DOI] [PubMed] [Google Scholar]
- Yeh FC, Panesar S, Fernandes D, et al. Population-averaged atlas of the macroscale human structural connectome and its network topology. Neuroimage 2018;178:57–68; doi: 10.1016/j.neuroimage.2018.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelnik J, Bardinet E, Dormont D, et al. A three-dimensional, histological and deformable atlas of the human basal ganglia. I. Atlas construction based on immunohistochemical and MRI data. Neuroimage 2007;34(2):618–638; doi: 10.1016/j.neuroimage.2006.09.026 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Descoteaux M, Zhang J, et al. Mapping population-based structural connectomes. Neuroimage 2018;172:130–145; doi: 10.1016/j.neuroimage.2017.12.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The HCP datasets used in our analyses are publicly available at the Connectome Coordination Facility website. The processed streamline data presented in the figures are available on request from author M.V.P.