Abstract
Vibrio harveyi, a significant opportunistic marine pathogen, has been a challenge to the aquaculture industry, leading to severe economical and production losses. Phage therapy has been an auspicious approach in controlling such bacterial infections in the era of antimicrobial resistance. In this study, we isolated and fully characterized a novel strain-specific phage, vB_VhaS_MAG7, which infects V. harveyi MM46, and tested its efficacy as a therapeutic agent in challenged gilthead seabream larvae. vB_VhaS_MAG7 is a tailed bacteriophage with a double-stranded DNA of 49,315 bp. No genes linked with virulence or antibiotic resistance were harbored in the genome. The phage had a remarkably large burst size of 1393 PFU cell−1 and showed strong lytic ability in in vitro assays. When applied in phage therapy trials in challenged gilthead seabream larvae, vB_VhaS_MAG7 was capable of improving the survival of the larvae up to 20%. Due to its distinct features and safety, vB_VhaS_MAG7 is considered a suitable candidate for applied phage therapy.
Keywords: Vibrio harveyi, aquaculture, phage therapy, gilthead seabream
1. Introduction
Vibrio harveyi is one of the most significant bacterial pathogens of aquatic animals in the marine environment [1]. It affects both fish [2] and crustaceans [3] and has been related to catastrophic losses in aquaculture in different parts of the world. In shrimp aquaculture, V. harveyi is the causative agent of luminous virbiosis [4] but it is also considered one of the causative agents of acute hepatopancreatic necrosis disease (AHPND) [5]. In finfish aquaculture, V. harveyi affects many different commercially important fish species, such as groupers (many different species), gilthead seabream (Sparus aurata), sole (Solea spp.) and European seabass (Dicentrarchus labrax). In Mediterranean aquaculture, V. harveyi has become a major issue, especially in European seabass aquaculture, in which it causes significant mortality, especially in young individuals when the water temperature is above 20 °C [6]. Currently, there are no commercially available vaccines for vibriosis caused by V. harveyi, and the disease is treated with antibiotics such as flumequine, ampicillin and oxytetracycline [7]. Several strains have shown increased resistance to antibiotics and recurrent cycles of different antibiotic treatments are not uncommon. The administration of antibiotics is problematic for the aquaculture industry, as it poses significant threats to its sustainability [8]. Antimicrobial resistance is regarded as the most important threat to humanity, and agriculture and livestock farming including aquaculture are among the major contributors to the problem [9]. Phage therapy has been considered a very promising alternative to antibiotics and is currently being investigated both in human and veterinary applications [10]. Bacteriophages, broadly referred to as phages, are highly specific bacterial viruses that exclusively infect and lyse bacterial hosts. They are the most numerous “life entity” on the planet, occurring naturally in the environment together with their hosts. Lytic phages propagate inside the bacterial cells, which lyse at the end of the propagation cycle, releasing new virions that will continue the infection cycle in the presence of suitable hosts. The targeted killing efficacy of the phages is taken advantage of in phage therapy. Phages against V. harveyi have been isolated in the past for strains infecting both shrimps and fish [11,12,13]. Because of their high host specificity, which is usually translated into a narrow spectrum of activity in only few strains of the pathogen, but also the fast emergence of bacterial resistance against phage infection, the efficacy of phage therapy is usually based on the combination of different phages in the form of a cocktail [14]. Therefore, there is an ongoing effort to isolate and characterize novel phages against V. harveyi. In this regard, herein, we present the isolation and characterization of a novel phage infecting a Vibrio harveyi, which caused vibriosis in a commercial fish farm of European seabass in Greece.
2. Results
2.1. vB_VhaS_MAG7 Morphology and Characteristics
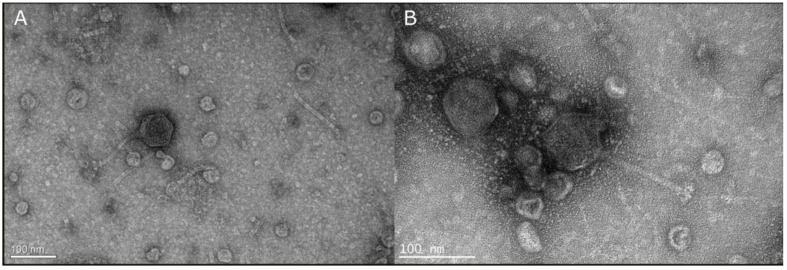
A novel phage named vB_VhaS_MAG7 was isolated from water samples obtained from a commercial fish farm at the Saronic Gulf in Greece. The novel phage formed clear pinhole plaques in the host loan. Several single plaques were collected and purified through subsequent infections for a total of six times, ensuring each time that plaque morphology was consistent. Observations in TEM revealed that vB_VhaS_MAG7 has a long, no contractile tail with an icosahedral capsid; morphological characteristics classified the novel phage to the siphovirus-like morphotype (Figure 1). The diameter of the capsid was 75 ± 04 nm, the tail was 93 ± 08 nm long and the base plate was 14 ± 04 nm wide.
Figure 1.
(A) Transmission electron micrograph of vB_VhaS_MAG7 showing the typical morphology of the Siphovirus morphology group. (B) Higher magnification of the phage showing details of the tail.
2.2. Host Range
According to host range analysis, vB_VhaS_MAG7 was found to be a strain-specific phage. No lysis was observed in the bacterial strains that were used in this study except for V. harveyi MM46 (Table 1).
Table 1.
Host range of vB_VhaS_MAG7 against Vibrio spp.
| Strain | Species | Country | Host | Lysis |
|---|---|---|---|---|
| MM46 * | V. harveyi | Greece | Sparus aurata | + |
| DSM 19623 | V. harveyi | USA | Talochestria capensis | - |
| SA 2.1 | V. harveyi | Saudi Arabia | Sparus aurata | - |
| DSM 2171 | V. alginolyticus | Japan | Trachurus trachurus | - |
| Gal 90 | V. harveyi | Greece | Sparus aurata | - |
| Vh No22 | V. harveyi | Greece | Dicentrarchus labrax | - |
| Kef 62 | V. harveyi | Greece | Dicentrarchus labrax | - |
| Kef 75 | V. harveyi | Greece | Dicentrarchus labrax | - |
| Gal 56 | V. harveyi | Greece | Dicentrarchus labrax | - |
| Gal 77 | V. harveyi | Greece | Sparus aurata | - |
| Gal 72 | V. harveyi | Greece | Dicentrarchus labrax | - |
| Gal 94 | V. harveyi | Greece | Sparus aurata | - |
| L. SUSI | V. parahaemolyticus | Philippines | Shrimp | - |
| V1 | V. alginolyticus | Greece | Sparus aurata | - |
| LAR194 | V. mediterranei | Greece | Artemia nauplii | - |
| SM1 | V. harveyi | Greece | Seriola dumerili | - |
| MAN113 | V. splendidus | Greece | Seriola dumerili | - |
| VarvA4 1.1 | V. harveyi | Greece | Sparus aurata | - |
| VH2 | V. harveyi | Greece | Seriola dumerili | - |
| VhP1 Liv | V. harveyi | Greece | Seriola dumerili | - |
| VhP1 Spl | V. harveyi | Greece | Dicentrarchus labrax | - |
| DY05 | V. owensii | Greece | Dicentrarchus labrax | - |
| SA 6.2 | V. owensii | Saudi Arabia | Oreochromis niloticus | - |
| VIB391 | V. campbellii | Thailand | Shrimp | - |
| Kef 56 | V. rotiferianus | Greece | Dicentrarchus labrax | - |
| VhSerFre | V. harveyi | Greece | Seriola dumerili | - |
| sngr | V. harveyi | Greece | Dentex dentex | - |
| ks6 | V. owensii | Greece | Dicentrarchus labrax | - |
| VH5 | V. harveyi | Greece | Seriola dumerili | - |
| RG1 | V. harveyi | Greece | Dicentrarchus labrax | - |
| Serkid | V. harveyi | Greece | Seriola dumerili | - |
| SERKID2 | V. harveyi | Greece | Seriola dumerili | - |
| SERSD | V. harveyi | Greece | Seriola dumerili | - |
| SA 5.1 | V. harveyi | Saudi Arabia | Sparus aurata | - |
| SA 6.1 | V. harveyi | Saudi Arabia | Sparus aurata | - |
| SA 9.2 | V. harveyi | Saudi Arabia | Sparus aurata | - |
| SA 1.2 | V. harveyi | Saudi Arabia | Sparus aurata | - |
| SA 7.1 | V. harveyi | Saudi Arabia | Sparus aurata | - |
| SA 3.1 | V. harveyi | Saudi Arabia | Sparus aurata | - |
| SA 4.1 | V. harveyi | Saudi Arabia | Sparus aurata | - |
| SA 2.1 | V. harveyi | Saudi Arabia | Sparus aurata | - |
| VH6 | V. harveyi | Greece | Dicentrarchus labrax | - |
| V2 | V. alginolyticus | Greece | Dentex dentex | - |
| SA 1.1 | V. owensii | Saudi Arabia | Sparus aurata | - |
| SA 9.1 | V. owensii | Saudi Arabia | Sparus aurata | - |
*: host strain, +: lysis, -: no lysis
2.3. Thermal and pH Stability of vB_VhaS_MAG7
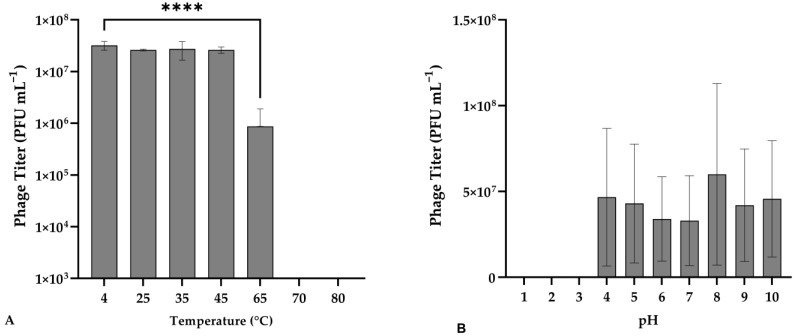
Phage vB_VhaS_MAG7 was exposed to different temperatures to assess its thermal stability. The phage titer remained stable up to 45 °C (F(3, 14) = 1.498, p = 0.6392), while a significant decrease was observed at 65 °C compared to the control (F(3, 14) = 7.774, p < 0.0001) (Figure 2).
Figure 2.
(A) Effect of different temperatures on the stability of vB_VhaS_MAG7. Incubation at 4 °C for 1 h was used as control. (B) Effect of pH on the stability of vB_VhaS_MAG7. Incubation with pH = 7 for 2 h was used as control. Phage titer was measured against V. harveyi MM46. Error bars were shown for the mean of n = 3. Statistical significance indicated by **** at p < 0.0001.
Regarding the behavior of vB_VhaS_MAG7 to various pH, no significant reduction in the phage titer was observed from 4 to 10 pH [F (9, 20) = 1426, p = 0.2425]. However, the phage was completely inactivated at pH values of 1, 2 and 3 (Figure 3).
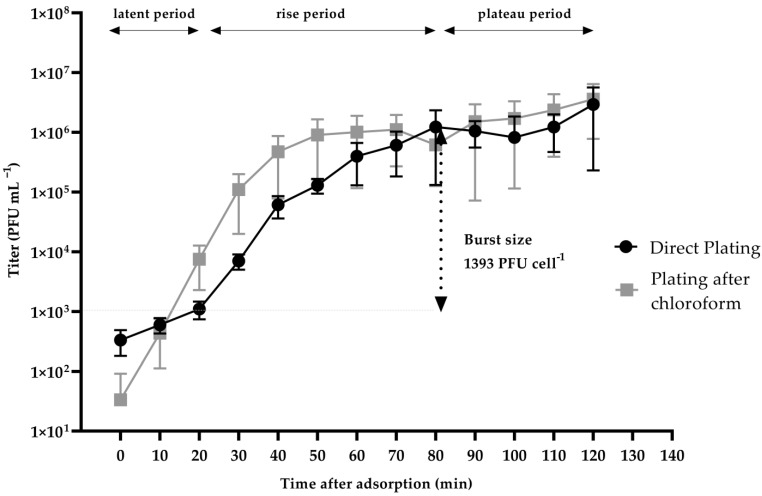
Figure 3.
One-step growth of vB_VhaS_MAG7 measured against V. harveyi MM46 at multiplicity of infection (MOI) 0.01. Error bars were shown for the mean of n = 3.
2.4. One-Step Growth of vB_VhaS_MAG7
A one-step growth assay revealed that the latent phase for the vB_VhaS_MAG7 was 20 min. The eclipse phase (the time required for the phage to produce the first mature viral particle inside the bacterium) was estimated to be ~10 min, while the burst size was found to be 1393 PFU cell−1. vB_VhaS_MAG7 reached the plateau phase at approximately 80 min after the infection (Figure 3).
2.5. In Vitro Cell Lysis
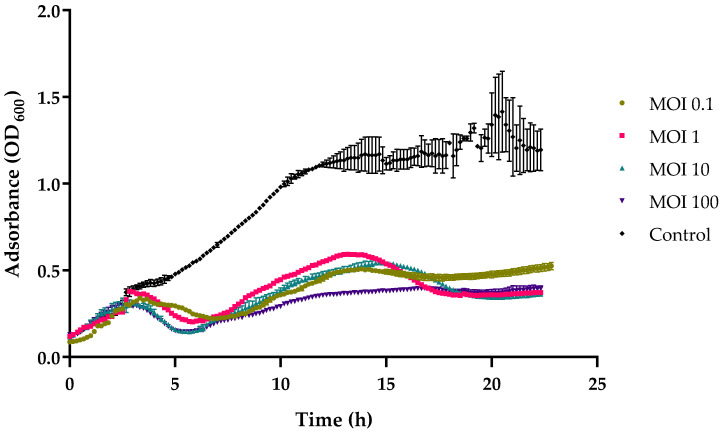
vB_VhaS_MAG7 was able to significantly inhibit the bacterial growth of MM46 up to 33% in MOI 0.1, 1 and 100 compared to the uninfected population (F (4, 670) = 193.3, p < 0.0001) after 24 h of incubation (Figure 4) as shown via in vitro analysis. A major decrease in the bacterial population was observed at about 5 h after infection at a multiplicity of infection of 100.
Figure 4.
In vitro lysis of vB_VhaS_MAG7 against V. harveyi MM46 at MOIs 0.1, 1, 10, and 100 for 24 h. Bacterial growth indicated by the absorbance (OD600) read. Error bars were shown for the mean of n = 3.
2.6. Genomic Analysis of vB_VhaS_MAG7
Genome sequencing of vB_VhaS_MAG7 produced 14,574,756 raw reads with an average reading length of 150 bp. The phage genome was assembled into a single contig with a minimum coverage of 5× The genome size was 49,315 bp, and the gene distribution was rather dense, as indicated by 1.55 genes per kbp. A total of 76 open reading frames (ORFs) were identified (Table 2), in which 70 (92%) ORFs used ATG as a start codon, 3 ORFs (4%) used GTG and 3 ORFs (4%) used TTG. The vast majority of ORFs were annotated as hypothetical proteins, while 33 ORFs were assigned protein functions based on the similarity with homolog proteins in NCBI and Swissprot databases. No tRNAs were found. Moreover, there were no homologs of integrase, virulence-associated genes or genes encoding antibiotic resistance (Figure 5). Bacphlip analysis showed a 90% probability that vB_VhaS_MAG7 follows a lytic lifestyle.
Table 2.
Summary table of vB_VhaS_MAG7 genes annotated with relevant information based on important amino acid sequences and structural protein homologations (E-value ≤ 10−3).
| Type | Predicted Functions | Start | End | Length | Strand |
|---|---|---|---|---|---|
| ORF 1 | Hypothetical protein | 401 | 629 | 72 | Forward |
| ORF 2 | Hypothetical protein | 633 | 868 | 74 | Forward |
| ORF 3 | Bifunctional DNA primase/polymerase | 871 | 3183 | 767 | Forward |
| ORF 4 | Coil-containing protein | 3237 | 3762 | 170 | Forward |
| ORF 5 | Putative helicase subunit | 3762 | 4551 | 259 | Forward |
| ORF 6 | Hypothetical protein | 4542 | 5095 | 180 | Forward |
| ORF 7 | PD-(D/E)XK nuclease superfamily protein | 5151 | 6229 | 355 | Forward |
| ORF 8 | P-loop-containing nucleoside triphosphate hydrolase | 6212 | 7641 | 471 | Forward |
| ORF 9 | Helicase superfamily 1/2 ATP-binding domain protein | 7611 | 9317 | 563 | Forward |
| ORF 10 | Hypothetical protein | 9307 | 9739 | 138 | Forward |
| ORF 11 | Hypothetical protein | 9870 | 10,209 | 108 | Forward |
| ORF 12 | Hypothetical protein | 10,308 | 10,496 | 59 | Forward |
| ORF 13 | Hypothetical protein | 10,481 | 10,903 | 136 | Forward |
| ORF 14 | Hypothetical protein | 10,889 | 11,070 | 56 | Forward |
| ORF 15 | Hypothetical protein | 11,055 | 11,462 | 131 | Forward |
| ORF 16 | Hypothetical protein | 11,449 | 11,992 | 176 | Forward |
| ORF 17 | Hypothetical protein | 11,959 | 12,164 | 65 | Forward |
| ORF 18 | NTP-ppase-like protein | 12,158 | 12,601 | 144 | Forward |
| ORF 19 | Yopx protein | 12,699 | 13,096 | 128 | Forward |
| ORF 20 | Hypothetical protein | 13,112 | 13,233 | 36 | Forward |
| ORF 21 | Hypothetical protein | 13,220 | 13,370 | 45 | Forward |
| ORF 22 | Tmhelix-containing protein | 13,358 | 13,627 | 86 | Forward |
| ORF 23 | Endodeoxyribonuclease I | 13,614 | 14,082 | 152 | Forward |
| ORF 24 | Hypothetical protein | 14,070 | 14,594 | 169 | Forward |
| ORF 25 | Hypothetical Protein | 14,582 | 14,737 | 47 | Forward |
| ORF 26 | Membrane lipoprotein | 14,795 | 15,477 | 223 | Forward |
| ORF 27 | Hypothetical Protein | 15,565 | 15,765 | 63 | Reverse |
| ORF 28 | Intramolecular chaperone auto-processing domain protein | 15,769 | 18,027 | 750 | Reverse |
| ORF 29 | Hypothetical protein | 18,117 | 18,535 | 135 | Reverse |
| ORF 30 | Tail fiber protein | 18,526 | 19,525 | 329 | Reverse |
| ORF 31 | Hypothetical protein | 19,566 | 19,908 | 110 | Reverse |
| ORF 32 | Tmhelix-containing protein | 19,910 | 20,263 | 113 | Reverse |
| ORF 33 | Hypothetical protein | 20,311 | 20,720 | 130 | Reverse |
| ORF 34 | Hypothetical protein | 20,700 | 20,813 | 34 | Reverse |
| ORF 35 | Hypothetical protein | 20,804 | 21,118 | 104 | Reverse |
| ORF 36 | Hypothetical protein | 21,129 | 21,385 | 80 | Forward |
| ORF 37 | Hypothetical protein | 21,542 | 21,658 | 34 | Forward |
| ORF 38 | Hypothetical protein | 21,688 | 22,548 | 282 | Reverse |
| ORF 39 | Hypothetical protein | 22,567 | 23,346 | 255 | Reverse |
| ORF 40 | Baseplate J-like protein | 23,334 | 24,534 | 397 | Reverse |
| ORF 41 | Hypothetical protein | 24,520 | 24,943 | 137 | Reverse |
| ORF 42 | Hypothetical protein | 25,020 | 25,217 | 62 | Reverse |
| ORF 43 | Hypothetical protein | 25,205 | 25,360 | 51 | Reverse |
| ORF 44 | Tmhelix-containing protein | 25,390 | 25,630 | 75 | Reverse |
| ORF 45 | Hypothetical protein | 25,617 | 26,541 | 303 | Reverse |
| ORF 46 | Hypothetical protein | 26,612 | 27,280 | 218 | Reverse |
| ORF 47 | Hypothetical Protein | 27,243 | 27,408 | 50 | Reverse |
| ORF 48 | Cytosine specific methyltransferase | 27,395 | 28,586 | 392 | Reverse |
| ORF 49 | Putative tail protein | 28,637 | 29,857 | 403 | Reverse |
| ORF 50 | DNA circularization protein | 30,519 | 31,778 | 415 | Reverse |
| ORF 51 | Phosphoesterase | 31,837 | 32,371 | 175 | Reverse |
| ORF 52 | Tmhelix-containing protein | 32,364 | 32,485 | 36 | Reverse |
| ORF 53 | Tmhelix-containing protein | 32,550 | 32,842 | 90 | Reverse |
| ORF 54 | Hypothetical Protein | 32,876 | 33,085 | 65 | Reverse |
| ORF 55 | Tail tape measure protein | 33,191 | 34,803 | 533 | Reverse |
| ORF 56 | Hypothetical protein | 34,796 | 34,994 | 60 | Reverse |
| ORF 57 | Tail assembly chaperone protein | 35,029 | 35,467 | 141 | Reverse |
| ORF 58 | DNA binding HTH domain protein | 35,609 | 35,835 | 71 | Forward |
| ORF 59 | Hypothetical protein | 35,826 | 36,023 | 61 | Forward |
| ORF 60 | Hypothetical protein | 36,005 | 36,180 | 54 | Forward |
| ORF 61 | Hypothetical protein | 36,153 | 36,356 | 64 | Forward |
| ORF 62 | Hypothetical protein | 36,340 | 36,634 | 93 | Forward |
| ORF 63 | Major tail protein | 36,673 | 37,064 | 125 | Reverse |
| ORF 64 | Tail protein | 37,052 | 38,522 | 486 | Reverse |
| ORF 65 | Hypothetical protein | 38,524 | 39,202 | 223 | Reverse |
| ORF 66 | Hypothetical protein | 39,185 | 39,580 | 127 | Reverse |
| ORF 67 | Hypothetical protein | 39,568 | 39,932 | 117 | Reverse |
| ORF 68 | Major capsid protein | 39,934 | 41,028 | 360 | Reverse |
| ORF 69 | Hypothetical protein | 41,031 | 41,455 | 136 | Reverse |
| ORF 70 | Putative ATP dependent Clp protease | 41,441 | 42,692 | 412 | Reverse |
| ORF 71 | Portal protein | 42,679 | 44,325 | 543 | Reverse |
| ORF 72 | Hypothetical protein | 44,386 | 44,657 | 85 | Reverse |
| ORF 73 | Terminase large subunit | 44,708 | 46,677 | 652 | Reverse |
| ORF 74 | Hypothetical protein | 46,666 | 47,374 | 232 | Reverse |
| ORF 75 | Putative HNH endonuclease | 47,435 | 47,948 | 167 | Reverse |
| ORF 76 | Coil-containing protein | 48,283 | 49,029 | 245 | Reverse |
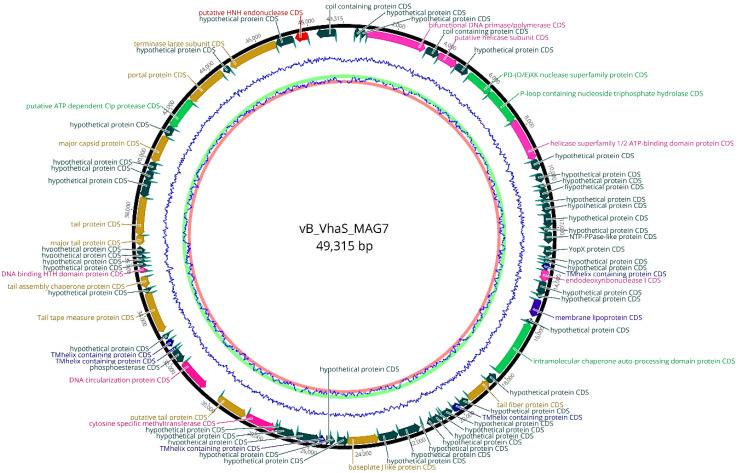
Figure 5.
Visual representation of vB_VhaS_MAG7 genome. The GC content of the genome is shown by the inner blue line. Predicted ORFs are shown in the outer circle. ORF color refers to annotated biochemical function: phage assembly proteins (brown), DNA replication proteins, repair and recombination (purple), miscellaneous proteins (light green), transmembrane proteins (blue), hypothetical proteins (dark green), Shine Dalgarno sequences (cyan).
Several genes linked to phage packaging and structural assembly were identified, as well as genes that are associated with DNA replication and nucleotide metabolism. No gene clusters or subclusters were observed, indicating that there is no specific genome arrangement.
Genes related to tail structure and assembly such as tail fiber protein (ORF 30), baseplate protein (ORF 40), putative tail protein (ORF 49), tail assembly chaperone protein (ORF 57), major tail protein (ORF 63), tail protein (ORF 64) and tape measure protein (ORF 55) were identified, as well as genes that encode major structural components such as major capsid protein (ORF 68). Large terminase subunit (ORF 73) and portal protein (ORF 71), which are involved in the packaging process, were also present in the genome.
Moreover, genes responsible for DNA manipulation and nucleotide metabolism, namely helicase (ORF 5), DNA primase/polymerase (ORF 3), a homing endonuclease (ORF 75), endodeoxyribonuclease (ORF 23), cytosine specific methyltransferase (ORF 48) and DNA binding HTH domain protein (ORF 58), were found. Interestingly, miscellaneous genes such as PD-(D/E)XK nuclease superfamily protein (ORF 7) were also included in the phage genome.
2.7. Genomic Synteny and Phylogenetic Analysis
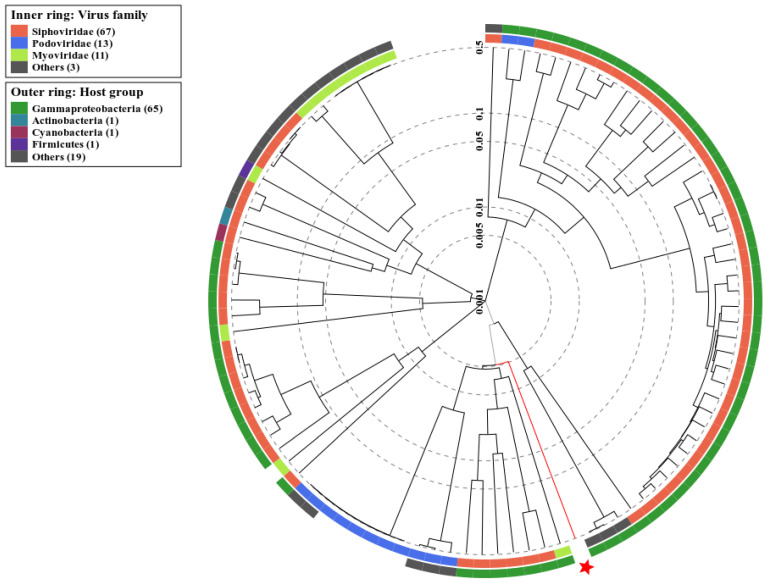
Phage vB_VhaS_MAG7 belongs to the class of Caudoviricetes and is part of a Siphoviridae morphology cluster, as shown using proteomic tree analysis (Figure 6) that confirmed the TEM morphological observations. Furthermore, the potential host of MAG7 was found to belong to the taxa of Gammaproteobacteria, which includes the Vibrionaceae family. Genome comparative analysis among vB_VhaS_MAG7 and other similar phages revealed that vB_VhaS_MAG7 is a novel phage, since the genetic distance between vB_VhaS_MAG7 and the closest phage was 53.9%, which is below the 95% species threshold (Supplementary Data, Figure S1).
Figure 6.
Identification of classes and host group for Vb_VhaS_MAG7 according to the proteomic tree produced by VIPTree. Vb_VhaS_MAG7 belongs to a Siphoviridae cluster and infects hosts from the Gammaproteobacteria group (red star and line). The branch length scale was calculated as log values. The inner and outer rings indicate the virus taxonomic family and the host group, respectively.
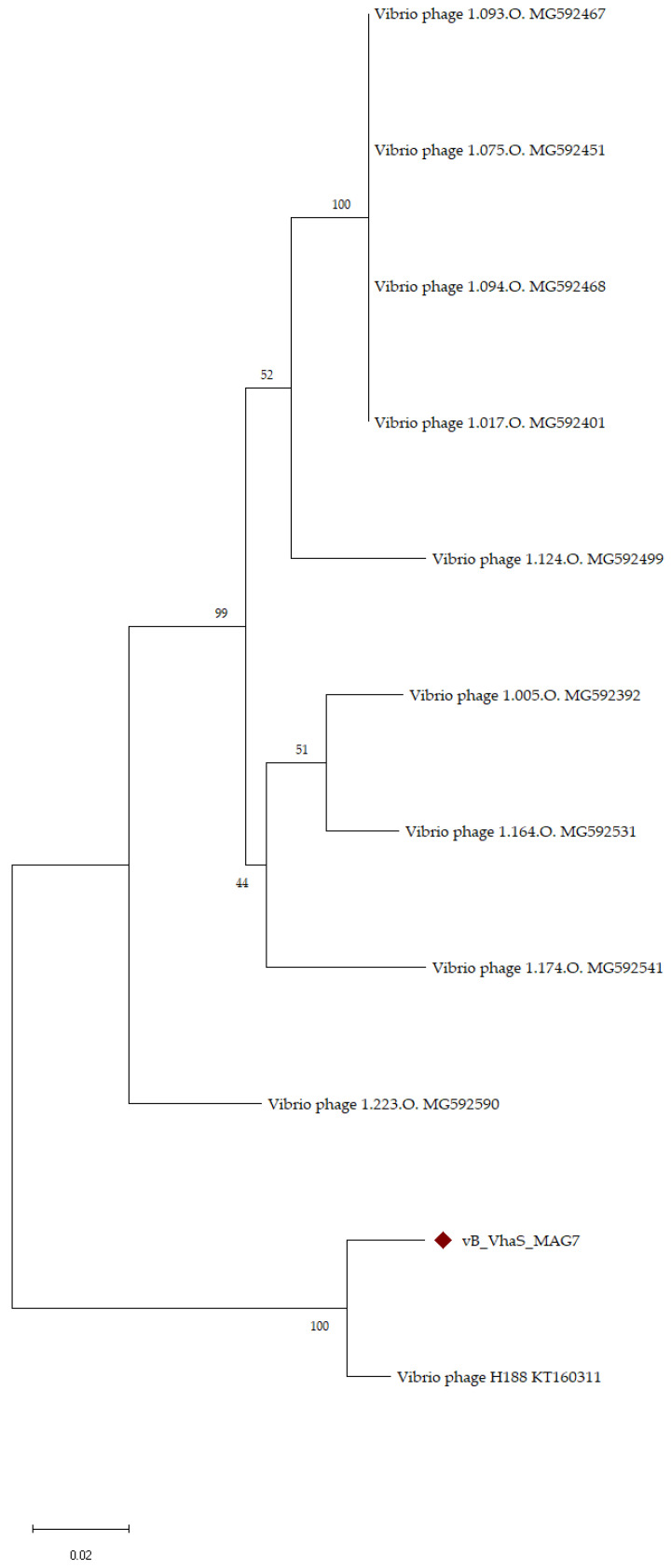
Phylogenetic analysis using large terminase subunits of the closest phages in Vb_VhaS_MAG7 (Figure 7) showed that MAG7 has a recent common ancestor with Vibrio phage H188, with a bootstrap value of 100%. In addition, the length of the branch indicates that MAG7 has a greater number of amino acid substitutions in its large subunit, as it deviates from its common ancestor with H188.
Figure 7.
Phylogenetic tree of vB_VhaS_MAG7 and closest phages based on the large terminase subunits. Large phage terminase subunits were downloaded from the NCBI database and aligned using MUSCLE. A rooted phylogenetic tree of maximum probability (bootstrap = 1000) was constructed using MEGA X. The phages with the largest genomic distance from vB_VhaS_MAG7 were arbitrarily used as an outgroup. The “diamond” symbol represents vB_VhaS_MAG7.
2.8. In Vivo Phage Therapy in Gilthead Seabream Larvae
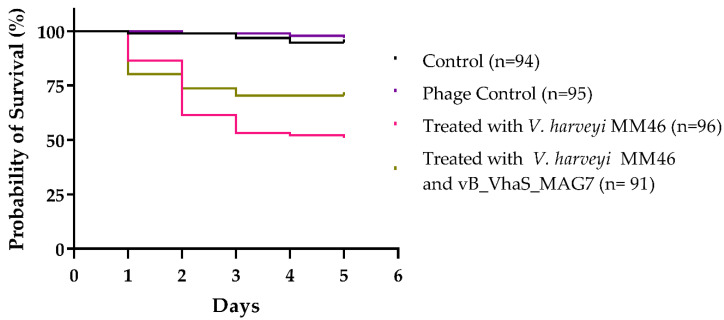
In vivo phage treatment trials were carried out in gilthead sea bream larvae to evaluate the effectiveness of vB_VhaS_MAG7 in the control of Vibrio harveyi MM46. The bacterial strain V. harveyi MM46 was found to be pathogenic, significantly reducing the survival of the larvae to up to 51% compared to the control group, in which 94.6% of the larvae survived during the 5-day trial (X2(1, 190) = 46.77, p < 0.0001). The survival of gilthead sea bream larvae increased significantly (significance level p < 0.05) when treated with vB_VhaS_MAG7 at MOI 10 compared to the group infected with Vibrio harveyi MM46 (X2(1, 187) = 5.295, p = 0.0214). The control group of the phage (without the addition of bacteria) also did not have a significant difference compared to the control group (X2(1, 189) = 0.5529, p = 0.4571), indicating the safety of the phage suspension and possibly the absence of endotoxins (Figure 8).
Figure 8.
Survival of gilthead seabream larvae infected with V. harveyi MM46 in an experimental phage treatment trial for 5 days. Gilthead seabream larvae infected with MM46 (~107 CFU mL−1) were treated with vB_VhaS_MAG7 at MOI 10, two hours after infection. Statistical significance at p < 0.0001.
3. Discussion
The number of outbreaks caused by Vibrio harveyi is on the rise as the effects of climate change become more prominent. This bacterium poses a threat to various marine organisms such as abalones, shrimps, corals, and fishes [15,16,17,18] and can result in major economic losses for the aquaculture industry. Moreover, many V. harveyi strains are considered multi-drug resistant, and therefore pose a great challenge. To address this issue, phage therapy has been proposed to control vibriosis. In this context, there has been an increasing number of studies regarding the isolation of phages against V. harveyi [13,19,20,21]. In this study, we isolated a new lytic bacteriophage, vB_VhaS_MAG7, which targets V. harveyi MM46 and we evaluated its potential as a therapeutic agent.
As evidenced by its comparison to other phages in the NCBI nr database using a BLAST search and VIRIDIC, vB_VhaS_MAG7 is considered a novel phage. The closest related phage, Vibrio phage H188, which infects Vibrio kanaloae [22], was found to have 95% similarity over 48% query cover with MAG7. Although analyzing the evolutionary relationships of phages can be difficult and uncertain due to their wide variety and the mosaic nature of their genomes, phylogenetic analysis based on the signature gene, large terminase subunits, was well supported, further enforcing our findings. Phage vB_VhaS_MAG7 is a siphovirus, as shown in TEM. Viral proteomic analysis also showed that the phage was part of a cluster consisting of siphoviruses. vB_VhaS_MAG7 is a tailed phage with a double-stranded DNA genome and therefore is a part of the Caudoviricetes class, as of the new taxonomic system [23].
Vibrio phages have been linked to inducing virulence in their hosts [24,25]. However, no evidence of virulence factors such as toxins or AMR genes was found in vB_VhaS_MAG7 genome, pointing out its safety as a therapeutic agent. Moreover, no integrases or genes that are associated with genome integration or recombination were present, indicating a vB_VhaS_MAG7 lytic lifestyle. This was further supported by the BACPHLIP analysis. The genome of vB_VhaS_MAG7 contained genes encoding potential structural proteins including capsid protein, major tail protein, and baseplate protein. Genes linked to phage assembly such as large terminase subunit, portal protein and tail assembly chaperone protein were also identified. Interestingly, a gene encoding a cytosine-specific methyltransferase was present. Mono-specific orphan methyltransferases provide protection against digestion by several restriction host’s endonucleases and are considered an important defense mechanism of the phage against the R m modification systems of the bacterial host [26]. Therefore, a more in-depth study is necessary to gain a clearer comprehension of the function of this gene, thereby improving our understanding of phage–host interactions.
Temperature and acidity are crucial factors that play a major role in phage adsorption to the host and phage neutralization, determining the efficacy of phage treatment [27]. Phage vB_VhaS_MAG7 remained consistent in a broad range of temperatures and pH values, which makes it a highly practical candidate for phage therapy.
As shown in the host range analysis, vB_VhaS_MAG7 appears to be a strain-specific phage, solely infecting V. harveyi MM46. Usually, phages with a wide host range are considered ideal for phage therapy, particularly in the field of aquaculture, where pathogenic strains and species exhibit a high degree of diversity. However, in this case, we consider vB_VhaS_MAG7 an excellent candidate due to its efficient lytic ability both in vitro and in vivo. Remarkably, vB_VhaS_MAG7 burst size was unusually large. A few phages have been previously reported [13,28,29,30] to produce a substantial number of virions, including Vibrio phage Virtus, which also targets V. harveyi. Burst size is a vital characteristic for a phage and it is affected by a number of elements, such as the metabolic activity of the host bacteria, the surrounding environment, and the host’s protein synthesis system [28,31,32]. That said, further investigation is required to comprehend the molecular mechanism associated with its remarkable burst size.
Multiple studies have shown that phages are effective at treating vibriosis in various animal models [11,21,33]. For instance, the survival of abalone was improved by 70% in 7 days as shown by Wang et al. [33], and Vinod et al. showed a significant increase in the survival of giant tiger prawn (Penaeus monodon) treated with phages [11]. Moreover, we have previously demonstrated a successful phage therapy scheme whereby Vibrio phage Virtus [13] was able to increase the survival of gilthead seabream larvae by 35% compared to the untreated population. Here, gilthead seabream larvae that were treated with vB_VhaS_MAG7 had 19% higher probability of survival in comparison with the untreated group. In addition, the phage control group showed no difference in terms of survival compared to the control group, corroborating vB_VhaS_MAG7 safety as a therapeutic agent.
In conclusion, Vb_VhaS_MAG7 is a novel strain-specific phage against V. harveyi. The distinct biological and genetic features of Vb_VhaS_MAG7, as well as its efficacy as a therapeutic agent, make it a suitable candidate for phage therapy as a safe and effective option.
4. Materials and Methods
4.1. Bacterial Strains
Bacterial strains that were used in this study (Table 1) belong to the bacterial collection of Laboratory of Aquaculture Microbiology, Institute of Marine Biology, Biotechnology and Aquaculture (IMBBC), Hellenic Center for Marine Research (HCMR) in Heraklion, Crete. They were identified at the species level either through whole-genome sequencing (WGS) and/or through sequencing of 16s rRNA and mreB genes and PCR detection of toxR, as described in Droubogiannis et al. [34]. Vibrio harveyi strain MM46 which had been isolated from diseased seabass from the same fish farm was used as a host for phage isolation. Identification of the host was performed following whole-genome sequencing (NZ_JAPPTO000000000.1). All bacterial strains were maintained in microbeads (MicroBank, Pro-Lab Diagnostics, Ontario, Canada) at −80 °C and were grown in lysogeny broth (10 gL−1 tryptone, 5 gL−1 NaCl, 1 L deionized water, 0.75 gL−1 MgSO4, 1.5 gL−1 KCl, 0.73 gL−1 CaCl2) at 25 °C prior to the experiments.
4.2. Antibiotic Susceptibility Testing
The bacterial strains used in this study were tested for antibiotic susceptibility using a standard disk diffusion test [35]. To carry out this test, bacterial suspensions of the V. harveyi MM46 were diluted to obtain a specific absorbance reading at OD600. The diluted bacterial suspensions were then plated on Mueller–Hinton agar (a type of nutrient agar) with 2% NaCl. Antimicrobial susceptibility disks (provided by ThermoFisher Scientific, Waltham, Massachusetts, United States) were placed on the agar plates, and the plates were incubated at 25 °C (which is the optimal temperature for the bacteria used) for 24 h. The diameters of the zones of inhibition around the disks were measured and recorded, and these measurements were interpreted as susceptible, medium, or resistant according to the Clinical Laboratory Standards Institute (CLSI) guidelines (specifically, CLSI M45-A2 [36] and CLSI M100-S25 [37]). Table 3 shows the details of the antimicrobial susceptibility disks used in the study and the corresponding interpretations of the recorded diameters.
Table 3.
Antibiotic Susceptibility Testing of V. harveyi MM46.
| Antimicrobial Agent | Zone Diameter (mm) | Interpretation | |
|---|---|---|---|
| Flumequine | UB | 30 | S |
| Tetracycline | TE | 25 | S |
| Florfenicol | FFC | 25 | S |
| Oxytetracycline | OT | 26 | I |
| Oxolinic acid | OA | 22 | S |
| Trimethoprim/sulfamethoxazole | SXT | 15 | I |
| Ampicillin | AMP | - | R |
| Piperacillin | PRL | - | R |
Abbreviations: S, sensitive; I, medium; R, resistant.
4.3. Isolation and Purification of Bacteriophages
The bacteriophage vB_Vh_MAG7 was isolated from water samples taken from a commercial fish farm in Greece. Grab sampling was used to collect water using a sterile container which was then transported at low temperature—to prevent the growth of any phages present in the sample—to the laboratory for analysis. The standard isolation and phage purification procedures were followed according to Clokie et al. [38], as described by Droubogiannis et al. [13]. Specifically, the enrichments underwent centrifugation at a speed of 13,000 rpm for a duration of 3 min, and the resulting supernatants were then passed through a sterile filter with a pore size of 0.22 µm (manufactured by GVS Life Sciences located in Sanford, ME, USA). Next, 10 µL of each sample was placed on bacterial lawns of the host strain, and the clearest plaques were collected after a 24 h incubation period. These plaques were subsequently propagated against the host strain using the double agar layer method, as originally described by Clokie et al [38] and reported in the work of Droubogiannis et al. [13]. This entire procedure was repeated five times before the phage was considered to be purified. The purified phage was maintained either in lysogeny broth (10 gL−1 tryptone, 5 gL−1 NaCl, 1 L deionized water, 0.75 gL−1 MgSO4, 1.5 gL−1 KCl, 0.73 gL−1 CaCl2) or in SM buffer (5.8 gL−1 NaCl, 2 gL−1 MgSO4, 50 mL Tris-Cl (1 M, pH 7.5)) at 4 °C. Prior to the experiments, we determined the titer of the phage using a double agar assay, as described in Clokie et al. [38].
4.4. Transmission Electron Microscopy
Transmission Electron Microscopy was conducted at the Electron Microscopy Lab of the University of Crete using a JEOL JEM-2100 (JEOL Ltd., Akishima, Tokyo, Japan) transmission electron microscope in order to observe phage morphology. Phage sample preparation included negative staining with 4% w/v uranyl acetate, 7.2 pH as previously described in Misol et al. [21]. Morphological characteristics of the phage were analyzed and measured using ImageJ (software version 1.53p). A total of n = 30 measurements was performed.
4.5. Host Range Assay
Several bacterial strains were used to evaluate the host range of the phage (Table 1). Briefly, 1 mL of a fresh broth bacterial culture (~107 CFU mL−1) was added to 3 mL of LB soft agar (0.75% agar) following the solidification of the mixture. A spot test was then performed where 10 μL of the phage dilutions (initial titer ~109 PFU mL−1) were spotted in the bacterial lawn of each individual bacterial strain in order to evaluate the efficacy of the phage. Phage plaques were counted the following day and the titer was calculated as PFU mL−1.
4.6. Stability of Phage in Different Temperatures and pH Values
The thermal stability of the novel phage was examined as described in Droubogiannis et al. [13] by exposing aliquots of the phage (titer: 107 PFU mL−1) to various temperatures (4, 25, 35, 45, 65, 70 and 80 °C). The aliquots were incubated at each temperature for 1 h and then rested at room temperature (RT) for 10 min. Each aliquot was then serially diluted and spotted (10 µL/spot) on a host bacterial lawn. After a 24 h incubation of the agar plates, the phage titer was determined for each temperature and 4 °C was used as the control.
The stability of phage titer at various pH levels was assessed as described by Pan et al. [28]. The desired pH values, ranging from 1 to 10, were achieved by adding NaOH or HCl to LB broth following the suspension of phages with a final titer of ~10−7 PFU mL−1. The suspensions were stored at 4 °C for 2 h. After a 10 min rest to allow the suspension to adjust to room temperature (RT), aliquots were taken and serially diluted and spotted onto the host bacterial lawn. After a 24 h incubation of the agar plates, the phage titer was determined for each pH value and pH = 7 was used as the control. Both the thermal and pH stability was tested in triplicate.
4.7. One-Step Growth Assay
A one-step growth assay was conducted as described in Droubogiannis et al. [34]. In brief, 1 mL of a fresh culture (107 CFU mL−1) was centrifuged at 13,000 rpm for 3 min. The supernatant was discarded and the pellet was saline washed. The procedure was repeated twice, and the pellet was resuspended in LB, infected with the phage at a MOI of 0.01 and rested for 15 min at room temperature. The supernatant was removed after 3 min of centrifugation at 13,000 rpm, and the pellet was dissolved in 1 mL saline. The infected culture was then transferred to a fresh tube containing 25 mL LB, and aliquots were taken and deposited in empty Eppendorf vials. The aliquots were then centrifuged at 13,000 rpm for 3 min and the supernatant was transferred to a new 2 mL Eppendorf vial, serially diluted, and spotted onto the host bacterial lawn on LB ½ agar plates. This procedure was repeated every 10 min for 120 min. The phage titer was determined after 24 h of incubation of the agar plates at 25 °C. The burst size was calculated as the ratio of the final count of released virions at the end of the burst period to the initial count of infected bacterial cells at the beginning of the latent period.
4.8. In Vitro Cell Lysis
To assess the effectiveness of vB_VhaS_MAG7 in vitro against V. harveyi MM46, 180 µL of fresh host bacterial culture (~106 CFU mL−1) was loaded into each well of sterile 96-well plates. The plates were then read at OD600 using a TECAN microplate reader (Infinite PRO 200) with orbital shaking at 25 °C. When the host culture was in the exponential phase (≈107 CFU mL−1), vB_VhaS_MAG7 was added at MOIs of 0.1, 1, 10, and 100. Two control treatments were also used in triplicate; one negative control with only LB and one positive control where V. harveyi MM46 population was added. The growth curves of the cultures were measured every 10 min for 24 h, and all assays were performed in triplicate.
4.9. Genomic Analysis
The genetic material of vB_Vh_MAG7 was extracted using the phenol–chloroform method, as described by Higuera et al. [39]. At least 5 µg of high-purity bacterial DNA was used to generate a paired-end 300 PE genomic library using Nextera library preparation kit (Illumina, San Diego, CA, USA). The quality of DNA was tested using a BioAnalyzer (Bio-Rad, Chicago, IL, USA), as described previously [14]. Sequencing was performed using an Illumina NovaSeq 6000 sequencing platform (Illumina, San Diego, CA, USA) according to the manufacturer’s protocol. Possibly contaminated, primer, N-terminus and 30- or 50-low quality reads were trimmed off (threshold: 0.05).
The Geneious assembler was used for the assembly of the paired reads using Geneious Prime software. The genome annotation pipeline, previously described by Droubogiannis et al. [34], was followed. Both structural and functional annotation were conducted in the Galaxy webserver [40] environment. Predicted ORFs were manually checked for the presence of a valid start and stop codon and a Shine-Dalgarno sequence. Several databases, including NCBI Basic Local Alignment Search Tool (BLAST) adjusted at a non-redundant (nr) protein database [41], Swissprot, Gene Ontology [42], Interpro [43] and TMHMM 2.0 were used to assess protein homology for functional annotation. Genes associated with integration, virulence and antibiotic resistance in the phage genome were searched for using the INTEGRALL Database webserver [44] and Virulence Factor Database (VFDB) [45], as well as the VirulenceFinder and ResFinder webservers [46]. The phage lifestyle was predicted using BACPHLIP through Galaxy Webserver [47]. The genome of vB_VhaS_MAG7 with annotated predicted ORFs was then visualized as a circular representation with Geneious software (Geneious v9.1, Biomatters, Auckland, New Zealand). Comparative phage genome analysis was conducted using VIRIDIC, which calculates intergenomic similarities between viral genomes [48].
4.10. In Vivo Phage Therapy Trial in Gilthead Seabream Larvae
In vivo phage therapy trials were conducted according to Droubogiannis et al. [13]. Gilthead seabream eggs at the same developmental stage were obtained from HCMR hatchery, washed three times with sterile sea water, and placed individually in a 96-well microplate (1 egg/well) containing 200 μL sterile sea water. After one day of incubation, the quality of eggs was evaluated according to Panini et al. [49]. The challenge test started when larvae were hatched.
Bacteria used in the challenge test were cultivated in LB overnight and diluted 1:100 in fresh LB. After a 2 h incubation at 25 °C, cells were centrifuged and washed twice with buffer A (saline 0.9%, MgCl2 10 mM). The bacterial suspensions were adjusted to ~107 CFU mL−1 with buffer A. No treatment was performed in the first group of larvae. The second group was treated with vB_VhaS_MAG7 alone (without addition of bacteria) at an approximate concentration of 108 PFU mL−1 and served as a negative phage control. The third group was treated with 106 CFU mL−1 Vibrio harveyi MM46. Finally, the fourth group was treated with 106 CFU mL−1 Vibrio harveyi MM46 and vB_VhaS_MAG7 at 10 ΜOΙ. Phage suspensions were treated with 10% (w/v) PEG overnight at 4 °C to remove possible endotoxins in the phage lysate and diluted in SM buffer (NaCl 100 mM, MgSO47H2O 8 mM, Tris-Cl 1 M; pH 7.5). The phage titer was also determined prior to the experiment with a double agar assay. Phage suspensions were added to the corresponding treatments two hours after infection. In addition, all controls were treated the same way, but instead of phage lysate, saline 0.9% was added to each well. The survival of gilthead seabream larvae was monitored daily for the following five days.
4.11. Statistical Analysis
One-way ANOVA was performed for the thermal, pH stability and in vitro assays along with Dunnett’s multiple comparison test [50]. Tukey’s HSD post hoc test [51] was used as a multiple comparison tool after ANOVA was performed. Kaplan–Meier survival analysis [52] was performed for the in vivo phage therapy trial in gilthead seabream larvae. All statistical analyses were carried out using GraphPad Prism version 9.0.0 for Windows, GraphPad Software, San Diego, CA, USA).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24098200/s1.
Author Contributions
Writing and formal analysis, S.D., L.P. and P.K.; genome sequencing D.S. and E.F.; drafting the paper D.S. and E.F.; writing—original draft preparation, S.D.; funding acquisition, P.K.; writing—review and editing, S.D. and P.K. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The genome sequence of phage vB_VhaS_MAG7 is available in GenBank under accession number OK381870. The genome sequence of V. harveyi MM46 is available in GenBank under accession number NZ_JAPPTO000000000.1.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the Greek General Secretariat of Research and Technology under the Operational Program Competitiveness, Entrepreneurship and Innovation 2014–2020 and the grant funded “ROBUST-Prevention of Vibriosis caused by Vibrio harveyi with innovative tools, MIS5045915”.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Austin B., Zhang X. Vibrio harveyi: A Significant Pathogen of Marine Vertebrates and Invertebrates. Lett. Appl. Microbiol. 2006;43:119–124. doi: 10.1111/j.1472-765X.2006.01989.x. [DOI] [PubMed] [Google Scholar]
- 2.Saeed M.O. Association of Vibrio harveyi with Mortalities in Cultured Marine Fish in Kuwait. Aquaculture. 1995;136:21–29. doi: 10.1016/0044-8486(95)01045-9. [DOI] [Google Scholar]
- 3.Montero A.B., Austin B. Characterization of Extracellular Products from an Isolate of Vibrio harveyi Recovered from Diseased Post-Larval Penaeus Vannamei (Bonne) J. Fish Dis. 1999;22:377–386. doi: 10.1046/j.1365-2761.1999.00189.x. [DOI] [Google Scholar]
- 4.Moriarty D.J.W. Control of Luminous Vibrio Species in Penaeid Aquaculture Ponds. Aquaculture. 1998;164:351–358. doi: 10.1016/S0044-8486(98)00199-9. [DOI] [Google Scholar]
- 5.Mai H.N., Caro L.F.A., Cruz-Flores R., White B.N., Dhar A.K. Differentially Expressed Genes in Hepatopancreas of Acute Hepatopancreatic Necrosis Disease Tolerant and Susceptible Shrimp (Penaeus vannamei) Front. Immunol. 2021;12:634152. doi: 10.3389/fimmu.2021.634152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cascarano M.C., Stavrakidis-Zachou O., Mladineo I., Thompson K.D., Papandroulakis N., Katharios P. Mediterranean Aquaculture in a Changing Climate: Temperature Effects on Pathogens and Diseases of Three Farmed Fish Species. Pathogens. 2021;10:1205. doi: 10.3390/pathogens10091205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rigos G., Kogiannou D., Padrós F., Cristòfol C., Florio D., Fioravanti M., Zarza C. Best Therapeutic Practices for the Use of Antibacterial Agents in Finfish Aquaculture: A Particular View on European Seabass (Dicentrarchus labrax) and Gilthead Seabream (Sparus aurata) in Mediterranean Aquaculture. Rev. Aquac. 2021;13:1285–1323. doi: 10.1111/raq.12523. [DOI] [Google Scholar]
- 8.Cabello F.C., Godfrey H.P., Tomova A., Ivanova L., Dölz H., Millanao A., Buschmann A.H. Antimicrobial Use in Aquaculture Re-Examined: Its Relevance to Antimicrobial Resistance and to Animal and Human Health. Environ. Microbiol. 2013;15:1917–1942. doi: 10.1111/1462-2920.12134. [DOI] [PubMed] [Google Scholar]
- 9.O’Neill J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. The Review on Antimicrobial Resistance. Government of the United Kingdom; London, UK: 2016. [Google Scholar]
- 10.Thiel K. Old Dogma, New Tricks[Mdash]21st Century Phage Therapy. Nat. Biotech. 2004;22:31–36. doi: 10.1038/nbt0104-31. [DOI] [PubMed] [Google Scholar]
- 11.Vinod M.G., Shivu M.M., Umesha K.R., Rajeeva B.C., Krohne G., Karunasagar I., Karunasagar I. Isolation of Vibrio harveyi Bacteriophage with a Potential for Biocontrol of Luminous Vibriosis in Hatchery Environments. Aquaculture. 2006;255:117–124. doi: 10.1016/j.aquaculture.2005.12.003. [DOI] [Google Scholar]
- 12.Wu L., Tian Y., Pang M., Yang Z., Bao H., Zhou Y., Sun L., Wang R., Zhang H. A Novel Vibriophage VB_VhaS_PcB-1G Capable of Inhibiting Virulent Vibrio harveyi Pathogen. Aquaculture. 2021;542:736854. doi: 10.1016/j.aquaculture.2021.736854. [DOI] [Google Scholar]
- 13.Droubogiannis S., Katharios P. Genomic and Biological Profile of a Novel Bacteriophage, Vibrio Phage Virtus, Which Improves Survival of Sparus Aurata Larvae Challenged with Vibrio harveyi. Pathogens. 2022;11:630. doi: 10.3390/pathogens11060630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hyman P. Phages for Phage Therapy: Isolation, Characterization, and Host Range Breadth. Pharmaceuticals. 2019;12:35. doi: 10.3390/ph12010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cardinaud M., Barbou A., Capitaine C., Bidault A., Dujon A.M., Moraga D., Paillard C. Vibrio harveyi Adheres to and Penetrates Tissues of the European Abalone Haliotis Tuberculata within the First Hours of Contact. Appl. Environ. Microbiol. 2014;80:6328–6333. doi: 10.1128/AEM.01036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin G.G., Rubin N., Swanson E. Vibrio Parahaemolyticus and V. Harveyi Cause Detachment of the Epithelium from the Midgut Trunk of the Penaeid Shrimp Sicyonia Ingentis. Dis. Aquat. Organ. 2004;60:21–29. doi: 10.3354/dao060021. [DOI] [PubMed] [Google Scholar]
- 17.Haldar S., Maharajan A., Chatterjee S., Hunter S.A., Chowdhury N., Hinenoya A., Asakura M., Yamasaki S. Identification of Vibrio harveyi as a Causative Bacterium for a Tail Rot Disease of Sea Bream Sparus Aurata from Research Hatchery in Malta. Microbiol. Res. 2010;165:639–648. doi: 10.1016/j.micres.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Luna G.M., Bongiorni L., Gili C., Biavasco F., Danovaro R. Vibrio harveyi as a Causative Agent of the White Syndrome in Tropical Stony Corals. Environ. Microbiol. Rep. 2010;2:120–127. doi: 10.1111/j.1758-2229.2009.00114.x. [DOI] [PubMed] [Google Scholar]
- 19.Kang S., Zhang L., Liao J., Zhang D., Wu S., Zhang X., Qin Q., Wei J. Isolation and Characterization of a Newly Discovered Phage, V-YDF132, for Lysing Vibrio harveyi. Viruses. 2022;14:1802. doi: 10.3390/v14081802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lelin C., Thirumalaikumar E., Uma G., Babu M.M., Ajan C., Vimal S., Citarasu T. Isolation and Partial Characterization of Bacteriophages Infecting Vibrio harveyi from Shrimp Farm Effluent Water. Aquac. Int. 2022;30:2081–2094. doi: 10.1007/s10499-022-00891-x. [DOI] [Google Scholar]
- 21.Misol G.N., Kokkari C., Katharios P. Biological and Genomic Characterization of a Novel Jumbo Bacteriophage, Vb_VhaM_pir03 with Broad Host Lytic Activity against Vibrio harveyi. Pathogens. 2020;9:1–38. doi: 10.3390/pathogens9121051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y., Wang M., Liu Q., Song X., Wang D., Ma Y., Shao H., Jiang Y. Complete Genomic Sequence of Bacteriophage H188: A Novel Vibrio Kanaloae Phage Isolated from Yellow Sea. Curr. Microbiol. 2016;72:628–633. doi: 10.1007/s00284-015-0984-6. [DOI] [PubMed] [Google Scholar]
- 23.Turner D., Kropinski A.M., Adriaenssens E.M. A Roadmap for Genome-Based Phage Taxonomy. Viruses. 2021;13:506. doi: 10.3390/v13030506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oakey H.J., Owens L. A New Bacteriophage, VHML, Isolated from a Toxin-Producing Strain of Vibrio harveyi in Tropical Australia. J. Appl. Microbiol. 2000;89:702–709. doi: 10.1046/j.1365-2672.2000.01169.x. [DOI] [PubMed] [Google Scholar]
- 25.Khemayan K., Prachumwat A., Sonthayanon B., Intaraprasong A., Sriurairatana S., Flegel T.W. Complete Genome Sequence of Virulence-Enhancing Siphophage VHS1 from Vibrio harveyi. Appl. Environ. Microbiol. 2012;78:2790–2796. doi: 10.1128/AEM.05929-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy J., Mahony J., Ainsworth S., Nauta A., van Sinderen D. Bacteriophage Orphan DNA Methyltransferases: Insights from Their Bacterial Origin, Function, and Occurrence. Appl. Environ. Microbiol. 2013;79:7547–7555. doi: 10.1128/AEM.02229-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pradeep A.N., Ramasamy S., Veniemilda J.K., Kumar C.S.V. Effect of PH & Temperature Variations on Phage Stability-A Crucial Prerequisite for Successful Phage Therapy. Int. J. Pharm. Sci. Res. 2022;13:5178–5182. doi: 10.13040/IJPSR.0975-8232.13(12).5178-82. [DOI] [Google Scholar]
- 28.Pan L., Li D., Sun Z., Lin W., Hong B., Qin W., Xu L., Liu W., Zhou Q., Wang F., et al. First Characterization of a Hafnia Phage Reveals Extraordinarily Large Burst Size and Unusual Plaque Polymorphism. Front. Microbiol. 2022;12:4264. doi: 10.3389/fmicb.2021.754331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan H., Huang Y., Mi Z., Yin X., Wang L., Fan H., Zhang Z., An X., Chen J., Tong Y. Complete Genome Sequence of IME13, a Stenotrophomonas Maltophilia Bacteriophage with Large Burst Size and Unique Plaque Polymorphism. J. Virol. 2012;86:11392–11393. doi: 10.1128/JVI.01908-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng H., Li C., Luo D., Zhang J., Ding Y., Chen M., Yang X., Lei T., Wu S., Ye Q., et al. Novel Phage VB_CtuP_B1 for Controlling Cronobacter Malonaticus and Cronobacter Turicensis in Ready-to-Eat Lettuce and Powered Infant Formula. Food Res. Int. 2021;143:110255. doi: 10.1016/j.foodres.2021.110255. [DOI] [PubMed] [Google Scholar]
- 31.Sui B., Qi X., Wang X., Ren H., Liu W., Zhang C. Characterization of a Novel Bacteriophage Swi2 Harboring Two Lysins Can Naturally Lyse Escherichia coli. Front. Microbiol. 2021;12:670799. doi: 10.3389/fmicb.2021.670799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hyman P., Abedon S.T. In: Practical Methods for Determining Phage Growth Parameters BT-Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions. Clokie M.R.J., Kropinski A.M., editors. Humana Press; Totowa, NJ, USA: 2009. pp. 175–202. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y., Barton M., Elliott L., Li X., Abraham S., O’Dea M., Munro J. Bacteriophage Therapy for the Control of Vibrio harveyi in Greenlip Abalone (Haliotis laevigata) Aquaculture. 2017;473:251–258. doi: 10.1016/j.aquaculture.2017.01.003. [DOI] [Google Scholar]
- 34.Droubogiannis S., Pavlidi L., Tsertou M.I., Kokkari C., Skliros D., Flemetakis E., Katharios P. Vibrio Phage Artemius, a Novel Phage Infecting Vibrio Alginolyticus. Pathogens. 2022;11:848. doi: 10.3390/pathogens11080848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bauer A.W., Kirby W.M., Sherris J.C., Turck M. Antibiotic Susceptibility Testing by a Standardized Single Disk Method. Am. J. Clin. Pathol. 1966;45:493–496. doi: 10.1093/ajcp/45.4_ts.493. [DOI] [PubMed] [Google Scholar]
- 36.Hindler J.A., Richter S.S. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria: M45. Clinical and Laboratory Standards Institute; Malvern, PA, USA: 2016. [Google Scholar]
- 37.Patel J.B., Cockerill F.R., Bradford P.A. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fifth Informational Supplement. Clinical and Laboratory Standards Institute; Malvern, PA, USA: 2015. [Google Scholar]
- 38.Clokie M.R.J., Kropinski A.M. Bacteriophages. Volume 1. Springer; Berlin/Heidelberg, Germany: 2009. [Google Scholar]
- 39.Higuera G., Bastías R., Tsertsvadze G., Romero J., Espejo R.T. Recently Discovered Vibrio Anguillarum Phages Can Protect against Experimentally Induced Vibriosis in Atlantic Salmon, Salmo Salar. Aquaculture. 2013;392–395:128–133. doi: 10.1016/j.aquaculture.2013.02.013. [DOI] [Google Scholar]
- 40.Ramsey J., Rasche H., Maughmer C., Criscione A., Mijalis E., Liu M., Hu J.C., Young R., Gill J.J. Galaxy and Apollo as a Biologist-Friendly Interface for High-Quality Cooperative Phage Genome Annotation. PLoS Comput. Biol. 2020;16:1–19. doi: 10.1371/journal.pcbi.1008214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 42.Harris M.A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., et al. The Gene Oncology (GO) Database and Informatics Resource. Nucleic Acids Res. 2004;32:258–261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitchell A.L., Attwood T.K., Babbitt P.C., Blum M., Bork P., Bridge A., Brown S.D., Chang H.Y., El-Gebali S., Fraser M.I., et al. InterPro in 2019: Improving Coverage, Classification and Access to Protein Sequence Annotations. Nucleic Acids Res. 2019;47:D351–D360. doi: 10.1093/nar/gky1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moura A., Soares M., Pereira C., Leitão N., Henriques I., Correia A. INTEGRALL: A Database and Search Engine for Integrons, Integrases and Gene Cassettes. Bioinformatics. 2009;25:1096–1098. doi: 10.1093/bioinformatics/btp105. [DOI] [PubMed] [Google Scholar]
- 45.Chen L., Yang J., Yu J., Yao Z., Sun L., Shen Y., Jin Q. VFDB: A Reference Database for Bacterial Virulence Factors. Nucleic Acids Res. 2005;33:325–328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kleinheinz K.A., Joensen K.G., Larsen M.V. Applying the ResFinder and VirulenceFinder Web-Services for Easy Identification of Acquired Antibiotic Resistance and E. Coli Virulence Genes in Bacteriophage and Prophage Nucleotide Sequences. Bacteriophage. 2014;4:e27943. doi: 10.4161/bact.27943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hockenberry A.J., Wilke C.O. BACPHLIP: Predicting Bacteriophage Lifestyle from Conserved Protein Domains. PeerJ. 2021;9:e11396. doi: 10.7717/peerj.11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moraru C., Varsani A., Kropinski A.M. VIRIDIC—A Novel Tool to Calculate the Intergenomic Similarities of Prokaryote-Infecting Viruses. Viruses. 2020;12:1268. doi: 10.3390/v12111268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Panini E.B., Mylonas C.C., Zanuy S., Carrillo M., Ramos J., Bruce M.P. Incubation of embryos and larvae of marine fish using microtiter plates. Aquac. Int. . 2001;9:189–196. doi: 10.1023/A:1014261830098. [DOI] [Google Scholar]
- 50.Dunnett C.W. A Multiple Comparison Procedure for Comparing Several Treatments with a Control. J. Am. Stat. Assoc. 1955;50:1096–1121. doi: 10.1080/01621459.1955.10501294. [DOI] [Google Scholar]
- 51.Haynes W. Tukey’s Test. In: Dubitzky W., Wolkenhauer O., Cho K.-H., Yokota H., editors. Encyclopedia of Systems Biology. Springer; New York, NY, USA: 2013. pp. 2303–2304. [Google Scholar]
- 52.Kishore J., Goel M., Khanna P. Understanding Survival Analysis: Kaplan-Meier Estimate. Int. J. Ayurveda Res. 2010;1:274. doi: 10.4103/0974-7788.76794. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequence of phage vB_VhaS_MAG7 is available in GenBank under accession number OK381870. The genome sequence of V. harveyi MM46 is available in GenBank under accession number NZ_JAPPTO000000000.1.