Abstract
Despite extensive research in the field of thrombotic diseases, the prevention of blood clots remains an important area of study. Therefore, the development of new anticoagulant drugs with better therapeutic profiles and fewer side effects to combat thrombus formation is still needed. Herein, we report the synthesis and evaluation of novel pyrroloquinolinedione-based rhodanine derivatives, which were chosen from 24 developed derivatives by docking as potential molecules to inhibit the clotting factors Xa and XIa. For the synthesis of new hybrid derivatives of pyrrolo[3,2,1-ij]quinoline-2-one, we used a convenient structural modification of the tetrahydroquinoline fragment by varying the substituents in positions 2, 4, and 6. In addition, the design of target molecules was achieved by alkylating the amino group of the rhodanine fragment with propargyl bromide or by replacing the rhodanine fragment with 2-thioxoimidazolidin-4-one. The in vitro testing showed that eight derivatives are capable of inhibiting both coagulation factors, two compounds are selective inhibitors of factor Xa, and two compounds are selective inhibitors of factor XIa. Overall, these data indicate the potential anticoagulant activity of these molecules through the inhibition of the coagulation factors Xa and XIa.
Keywords: 5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-1,2-dione; rhodanines; anticoagulant activity; docking studies
1. Introduction
In the current trend in drug design, hybrid or multimodal structures containing covalently linked pharmacophore groups are prevalent. This combination leads to the possibility of interaction with several protein targets, due to which a synergistic effect is achieved, and it becomes possible to conduct combination therapy using a single multimodal agent and to reduce unwanted side effects (for recent reviews on the chemistry of low-molecular-weight heterocyclic hybrids and their application in medicine, see [1,2,3,4,5,6,7,8,9,10,11]).
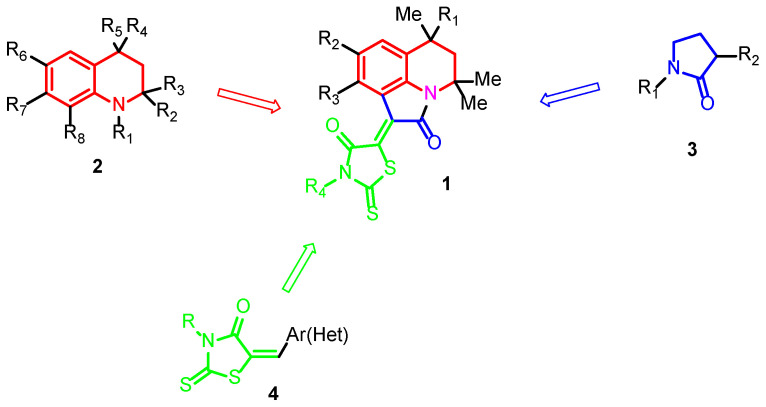
The goal of molecular hybridization set in this work was to obtain compounds 1 (Figure 1), which, according to our preliminary data [12], are effective inhibitors of the blood-coagulation factors Xa and XIa. The hope that sufficiently strong anticoagulants can be obtained in this way is based on the fact that for each of the building blocks, derivatives are known that have pronounced inhibitory activities against blood-coagulation factors.
Figure 1.
Design of target rhodanine-pyrroloquinoline-2-ones.
Tetrahydroquinoline 2 derivatives inhibit factors Xa [13,14] and XIa [15,16], pyrrolidin-2-one 3, factor Xa [17], and rhodanine 4, factor VIII [18]. Previously, as a result of the hybridization of 2 and 3, we obtained derivatives of 5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-1,2-diones, which exhibited inhibitory activity against factors Xa [19] and XIIa [20]. The introduction of the rhodanine fragment into the composition of the closest nonhydrogenated analog of the hybridization products 2 and 3 makes it possible to obtain factor Xa inhibitors (IC50~1 μM) [21].
The introduction of rhodanine into the structure of potential physiologically active compounds is associated with the risk of false-positive results in screening studies [22,23,24]. However, epalrestat containing a rhodanine fragment was successfully used to treat diabetes mellitus [25]. For compounds containing fragments of rhodanine, antibacterial activity [26,27,28,29,30,31], fungicidal [26,32], antiviral [30], anti-tuberculosis [33], anti-inflammatory [26,27,34], and anti-cancer [26,27,30,35,36,37] activities were noted. These compounds can be used in the treatment of diabetes mellitus [25,27], Alzheimer’s disease [26,27], and a number of other diseases [38,39]. Rhodanine and its derivatives are widely used in molecular hybridization. Methods for obtaining conjugates of rhodanine with benzothiophene [34,40], benzofuran [40], pyrazoles [41], quinoline [42], carbazole [43], thienopyrimidine [44], carbohydrates [37], benzyloxyarylidene [45], tetrazolquinoline [46], and biaryloxazolidinone [47] have been described.
Bioisosteric substitution is one of the most important strategies in the structural optimization of lead compounds for the development of new drugs. In particular, the introduction of halogen atoms into the molecule can significantly improve the pharmacokinetics, lipophilicity, and biological activity in general. On the other hand, halogen atoms are also the preferred structural moieties of anticoagulants due to their electron-withdrawing properties [48,49,50,51].
Thus, in this work, we continue the study of the anticoagulant activity of new hybrid molecules based on 5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-1,2-diones and rhodanines, further developing our findings published in [19,20,21]. A virtual library was constructed and screened by docking, the best selected hybrid molecules were synthesized, and their anticoagulant activities were evaluated against factors Xa and XIa.
2. Results
Molecular-docking prediction.
In the search for new inhibitors of the coagulation factors Xa and XIa, a virtual library was created, containing 543 new hybrid molecules of compounds that can be synthesized. For this virtual library, we developed a convenient synthesis of new hybrid tetrahydropyrrolo[3,2,1-ij]quinolin-1-ylidene-2-thioxothiazolidin-4-ones through the condensation of polyfunctional pyrrolo[3,2,1-ij]quinolindiones with rhodanine and 2-thioxoimidazolidin-4-one. All the molecules from this library were docked into the coagulation factors Xa and XIa using the SOL docking program, and twenty-four new hybrid molecules based on 5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-1,2-diones and rhodanines were selected, using the docking results, as the best inhibitor candidates.
The selection of the best inhibitors was made using the threshold values of the SOL score and the reliability of the docking. The threshold scores separating the good inhibitors from the inactive compounds were determined by docking native ligands and known inhibitors of the Xa and XIa factors. The atomistic models of the factors Xa and XIa used in the docking were constructed from high-quality PDB structures, 3CEN and 4CRC, respectively. The docking reliability was determined by clustering the solutions of the genetic algorithm used in the SOL program. The docking with 50 independent runs of the algorithm was considered successful if the population of the first cluster containing the most energetically favorable positions of the ligand was greater than 10. Otherwise, the docking was considered unsuccessful, and the ligands were not included in the pool of perspective inhibitors. The docking of native ligands from the 3CEN and 4CRC complexes into the corresponding proteins resulted in RMSD < 1.4 Å between the crystallized- and docked-ligand positions.
2.1. Synthesis
The 6-Methyl-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-1,2-dione [52,53] with various substituents in positions 4, 6, and 8 was used as the initial template. Therefore, in position 4, there were two methyl groups or an alicyclic fragment, while in position 6, along with the methyl group, there was another methyl group or a benzene ring with or without a substituent; alkyl, alkoxy, and acyloxy substituents or halogen atoms were in position 8.
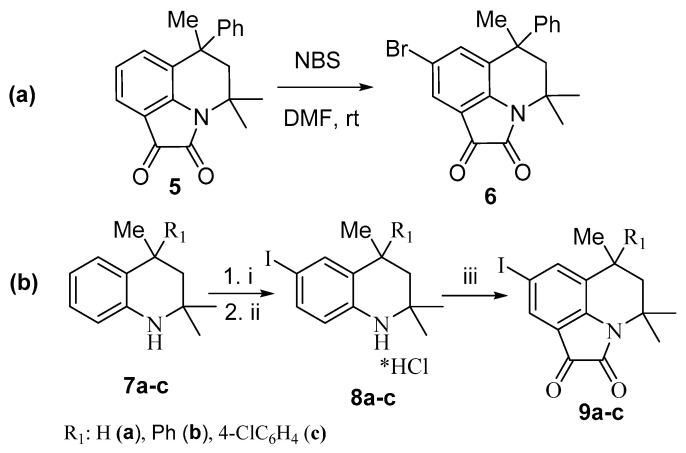
The synthesis of 8-halogen-substituted pyrroloquinolinediones is shown in Scheme 1. The bromination of 6-methyl-6-phenyl-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-1,2-dione 5 was carried out according to the procedure described in [54], using N-bromosuccinimide as a brominating agent and DMF as a solvent. To obtain 8-iodo-substituted pyrrolo[3,2,1-ij]quinoline-1,2-diones, the initial substrates, 2,2,4-trimethyl-1,2,3,4-tetrahydroquinolines, were subjected to iodination, in accordance with the procedure presented in [55]. The subsequent reaction of 6-iodo-2,2,4-trimethyl-4-R-1,2,3,4-tetrahydroquinoline hydrochlorides 8a–c with oxalyl chloride gave the corresponding diones 9a–c (Scheme 1).
Scheme 1.
Preparation of 8-bromo-pyrrolo[3,2,1-ij]quinoline-1,2-dione (a) and 8-iodo-substituted pyrrolo[3,2,1-ij]quinoline-1,2-diones (b). Reagents and conditions: (i) I2, dioxane:pyridine (1:1); (ii) HCl; (iii) (COCl)2, PhCH3, reflux, 2 h.
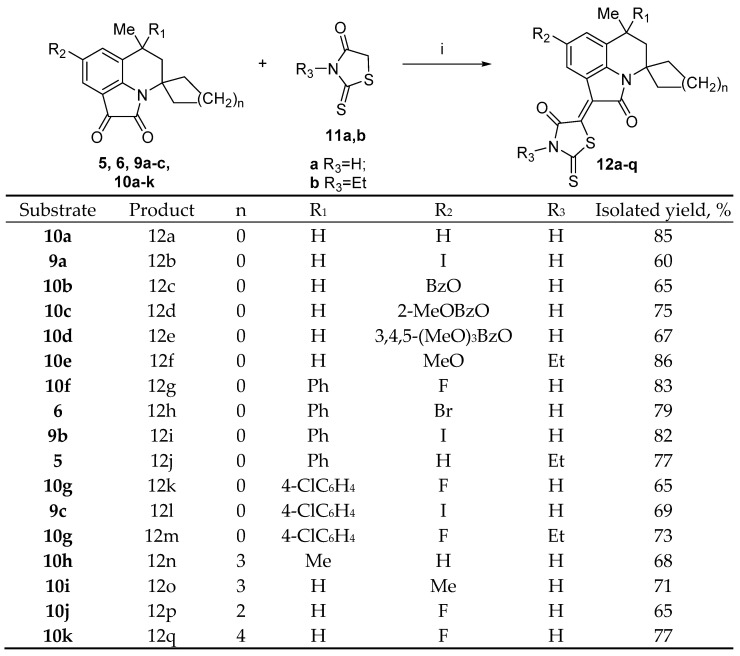
To obtain new hybrid molecules based on 6-methyl-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-1,2-diones, they were introduced into the condensation reaction with rhodanines 11a,b at reflux in an acetic-acid medium in the presence of freshly melted sodium acetate (Scheme 2).
Scheme 2.
Synthesis of the compounds 12a–q. Reagents and conditions: (i) AcOH, AcONa, and reflux, 1–6 h.
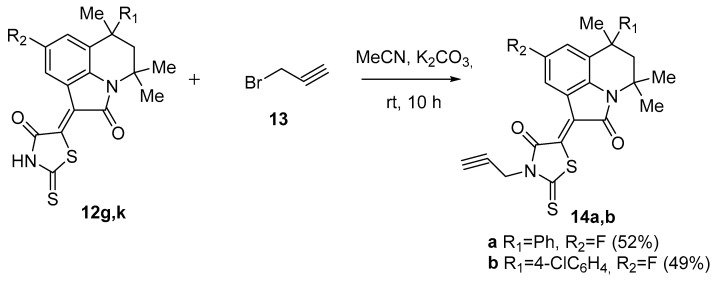
The compounds 12g and 12k containing a free NH group in the rhodanine moiety were further involved in alkylation reactions with propargyl bromide 13 (Scheme 3).
Scheme 3.
Synthesis of compounds 14a,b.
As a structural analog of rhodanine, 2-thioxoimidazolidin-4-one 15 was used in reaction with 5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-1,2-diones 5, 10 (Scheme 4).
Scheme 4.
Synthesis of compounds 16a–e. Reagents and conditions: (i) AcOH, AcONa, and reflux, 4–10 h.
2.2. Anticoagulant Studies
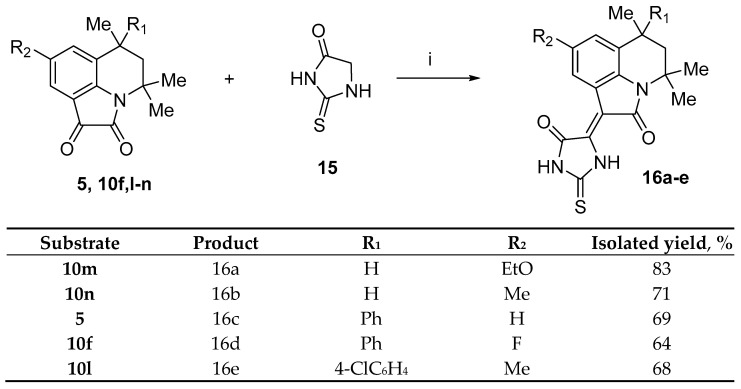
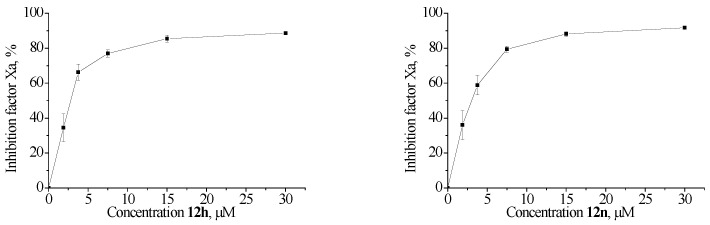
Compounds 12, 14 and 16 were evaluated for their anticoagulant activity against factors Xa and Xia, and the results are presented in Table 1. For the best Xa and XIa inhibitors, additional measurements were performed to determine the IC50 values. We measured the kinetics of the hydrolysis of the specific substrates S2765 or S2366 by factors Xa and XIa, respectively, in the buffer solution and in the presence of various concentrations of compounds. The results are presented in Figure 2 and Figure 3.
Table 1.
Results of docking and the experimental measurements of anticoagulant activity of compounds 12a–q, 14a,b, and 16a–e against factors Xa and XIa. The SOL score is the estimation of the protein–ligand-binding free energy calculated by the SOL docking program.
| N° | Factor Xa, SOL Score, kcal/mol |
Factor XIa, SOL Score, kcal/mol |
Percent Inhibition at 30 μM | IC50, μM | ||
|---|---|---|---|---|---|---|
| Xa | XIa | Xa | XIa | |||
| 12a | −5.28 | −4.54 | 94 | 22 | 9.41 ± 0.35 | - |
| 12b | −5.69 | −5.09 | 81 | 12 | 3.53 ± 0.08 | - |
| 12c | −6.04 | −5.73 | 58 | 72 | - | - |
| 12d | −6.15 | −5.84 | 63 | 98 | 2.35 ± 0.22 | 3.79 ± 0.03 |
| 12e | −5.59 | −6.13 | 49 | 94 | - | 3.74 ± 0.33 |
| 12f | −5.15 | −4.61 | 18 | 5 | - | - |
| 12g | −5.78 | −5.41 | 99 | 98 | 3.70 ± 0.03 | 5.03 ± 0.18 |
| 12h | −5.96 | −5.63 | 73 | 98 | 2.28 ± 0.23 | 3.73 ± 0.03 |
| 12i | −6.22 | −5.43 | 75 | 96 | 3.44 ± 0.06 | 5.04 ± 0.33 |
| 12j | −5.42 | −5.27 | 89 | 77 | - | - |
| 12k | −6.08 | −5.65 | 99 | 99 | 2.85 ± 0.23 | 5.24 ± 0.29 |
| 12l | −6.43 | −5.41 | 78 | 95 | 2.87 ± 0.21 | 4.30 ± 0.06 |
| 12m | −5.58 | −5.69 | 18 | 0 | - | - |
| 12n | −6.15 | −5.14 | 94 | 91 | 2.60 ± 0.50 | 9.41 ± 0.51 |
| 12o | −6.40 | −5.12 | 48 | 15 | - | - |
| 12p | −5.96 | −4.87 | 67 | 77 | 3.03 ± 0.18 | 12.22 ± 0.50 |
| 12q | −6.52 | −4.96 | 47 | 91 | - | 4.32 ± 0.06 |
| 14a | −4.84 | −5.26 | 36 | 3 | - | - |
| 14b | −5.23 | −5.30 | 49 | 17 | - | - |
| 16a | −5.14 | −4.34 | 2 | 6 | - | - |
| 16b | −5.45 | −4.66 | 31 | 5 | ||
| 16c | −5.82 | −5.30 | 45 | 57 | ||
| 16d | −5.80 | −5.19 | 17 | 2 | ||
| 16e | −6.19 | −4.88 | 59 | 86 | ||
| Rivaroxaban | −6.89 | - | 94 | 8 | ||
Figure 2.
The percentage of inhibition for the hydrolysis rate of factor-Xa-specific chromogenic substrate in buffer solution in the presence of different concentrations of inhibitors.
Figure 3.
The percentage of inhibition for the hydrolysis rate of factor-XIa-specific chromogenic substrate in buffer solution in the presence of different concentrations of inhibitors.
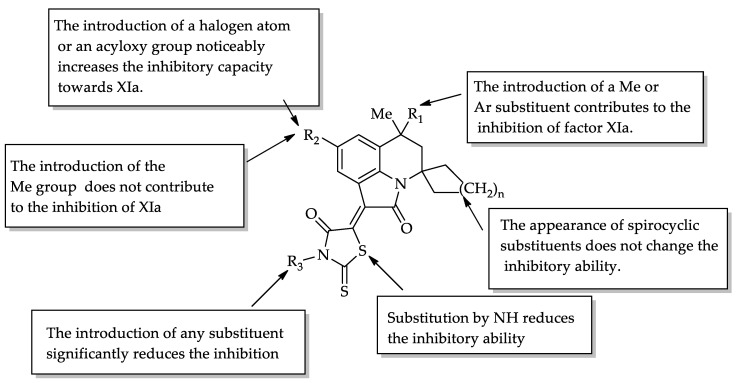
It was found that the compounds exhibited anticoagulant activity against factor Xa and XIa with a percentage inhibition in the range of 2–99% with inhibitors of 30 μM. It should be mentioned that of the 24 tested compounds, compounds 12d, 12g, 12h, 12k, 12l, and 12n showed good activity against factor Xa, with the IC50 ranging from 2.28 to 3.70 μΜ; compound 12h showed the best activity. On the other hand, compounds 12d, 12e, 12h, 12l, and 12q displayed good activity against factor Xia, with the IC50 in the range of 3.73–4.32 μΜ. Comparing the data in Table 1, we can conclude that factor XIa is much more sensitive to structural changes than factor Xa. A comparison of the data on the inhibition at a concentration of 30 μM and at IC50 allowed us to trace the following patterns (Figure 4):
-
−
The introduction of a methyl or aryl substituent at position 6, in addition to the methyl substituent, contributes to the inhibition of factor XIa (compare 12n with 12o and 12a);
-
−
The appearance of a methyl substituent in position 8 reduces the inhibitory ability of factor XIa (12o and 12p);
-
−
The appearance of a halogen atom (especially fluorine) at position 8 significantly increases the inhibitory ability (compounds 12g, 12h, 12i, 12k, and 12l), but this effect is eliminated by the appearance of substituents in the nitrogen atom of rhodanine (compounds 12m, 14a, and 14b);
-
−
The introduction of acyloxy substituents at position 8 supports the manifestation of its inhibition of factor XIa (compounds 12c, 12d, and 12e);
-
−
The appearance of substituents in the nitrogen atom of rhodanine significantly reduces the inhibitory ability (compounds 12f, 12m, 14a, and 14b);
-
−
The replacement of rhodanine by 2-thioxoimidazolidin-4-one leads to a decrease in the inhibitory ability (compounds 16a, 16b, and 16d).
Figure 4.
Structure-activity relationship.
2.3. Molecular Docking
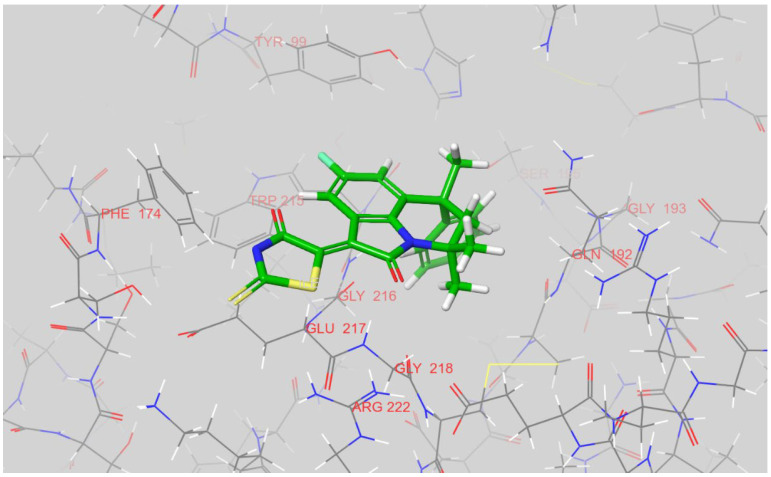
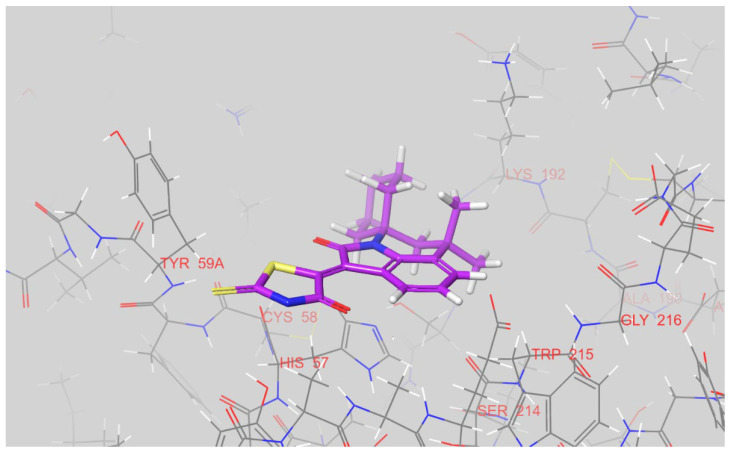
According to the predicted binding modes, all three compounds, 12k, 12g, and 12n, block the access to the catalytic triad of factor Xa. In its docked pose, the 12k places its chloro-phenyl fragment inside a S1 pocket of the enzyme while the rhodanine moiety points toward the solvent and is located near a positively charged side chain of Arg-222 (see Figure 5). A hydrophobic S4 pocket is left unoccupied.
Figure 5.
The docked pose of 12k inside the active site of factor Xa. The figure was prepared in Maestro.
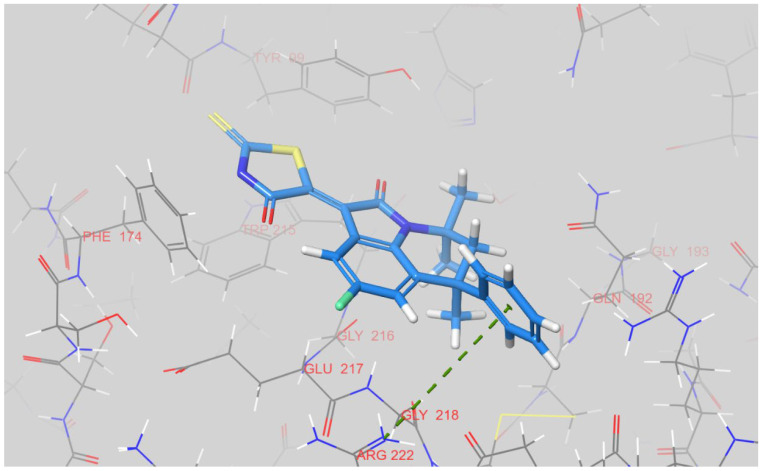
Despite high level of similarity between 12k and 12g, the latter binds to factor Xa in a different manner (Figure 6). Its rhodanine ring is placed between two aromatic residues, Tyr-99 and Phe-174, occupying the S4 pocket. A S1 pocket is almost unoccupied, with only a methyl group placed at its top. A phenyl ring is located outside the active site and it is engaged into pi-cation interaction with Arg-222. Taking this high degree of similarity into account, we assume that although they are different geometrically, both energy minima observed for 12k and 12g are very similar energetically. Both the conformations depicted on Figure 4 and Figure 5 can represent the relevant binding modes for both 12k and 12g.
Figure 6.
The docked pose of 12g inside an active site of factor Xa. A green dashed line indicates pi-cation interaction between phenyl ring of a ligand and guanidine group of Arg-222. The figure was prepared in Maestro.
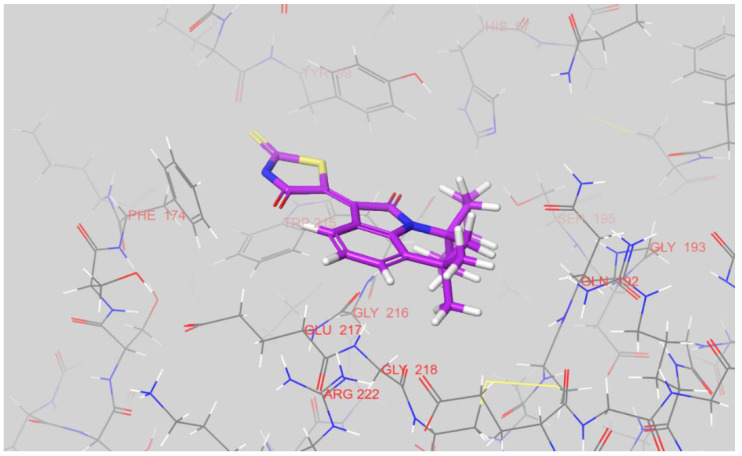
The predicted conformation of 12n is very similar to the docked pose of 12g (Figure 7). The rhodanine moiety of 12n is similarly positioned between Tyr-99 and Phe-174, inside S4 pocket. The cyclohexene ring of the spiro-system is located at the top of the S1 pocket, and does not extend inside. The S1 pocket is, therefore, left almost unoccupied. In summary, the rhodanine-containing factor-Xa inhibitors studied in the present work possess two distinct conformations in terms of the position of the rhodanine moiety. This moiety can be found either inside the S4 pocket, sandwiched between the aromatic rings of Tyr-99 and Phe-174, or pointing toward the solvent and located near a guanidine fragment of Arg-222. To profile the residues involved in binding for the rest of compounds, we calculated all the intermolecular interactions for each ligand with residues of the active site using the MMFF94 force field. An amino-acid residue was not included in the table of interactions for a given ligand if there was not a single ligand interaction with its atoms equal to or greater than 0.5 kcal/mol. The identified interaction residues of factor Xa for all the tested compounds can be found in Table S1.
Figure 7.
The docked pose of 12n inside an active site of factor Xa. The figure was prepared in Maestro.
Comparing the binding modes of 12g, 12k, and 12n with the bound geometry of rivaroxaban, a subnanomolar factor-Xa inhibitor, the three identified inhibitors were found to lack the two hydrogen bonds observed in the acyclicamide and cyclic-carbamate parts of the rivaroxaban, which interact with the backbone oxygen and backbone nitrogen of Gly-219, respectively. Similar to 12k, rivaroxaban occupies the S1 pocket through a hydrophobic aromatic ring, but it is buried more deeply in the protein. Unlike 12g and 12n, the rivaroxaban tail group lying in the S4 pocket is placed orthogonally, not in the same plane as the central part of the molecule, and it is not charged. The absence of the interactions described above can be exploited to obtain additional potency for inhibitors that are found during optimization studies in the future.
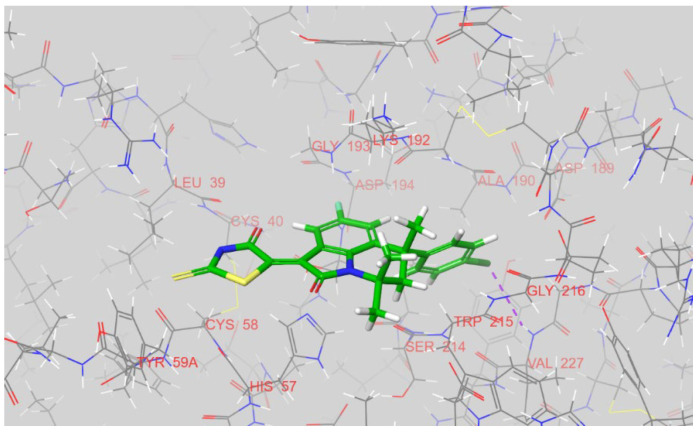
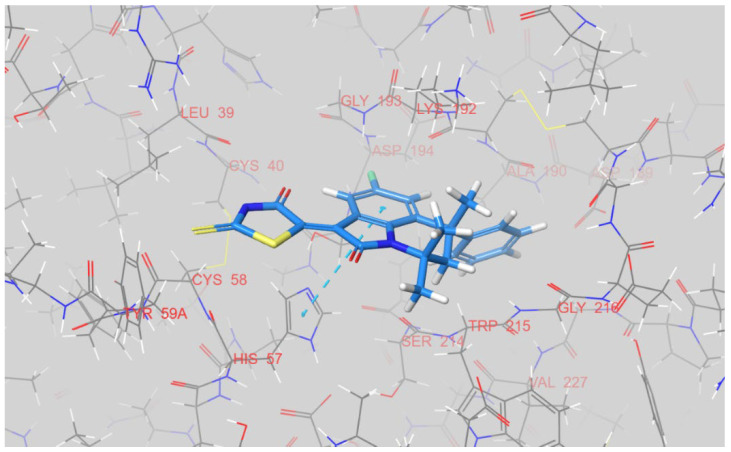
As in the case of factor Xa, 12k, 12g, and 12n were observed to have two distinct binding conformations after docking into factor Xia, with respect to the location of the rhodanine ring. The first was found for 12k, where the rhodanine cycle is positioned inside a S1’ pocket near His-57 (see Figure 8). The chloro-phenyl fragment of 12k is buried in a S1 pocket, with a halogen bond between a chlorine atom and Val-227NH observed. A S2’ pocket that is able to engage basic moieties is left unoccupied.
Figure 8.
The docked pose of 12k inside an active site of factor XIa. The dashed purple line indicates a halogen atom between a chlorine atom of the ligand and a backbone nitrogen of Val-227. The figure was prepared in Maestro.
The 12g binds to factor XIa in a similar manner to 12k (Figure 9). Again, the rhodanine ring is found in a S1’ pocket, and the phenyl ring is deeply inserted in a S1 pocket. However, since 12g lacks a chlorine atom, no halogen bonds are observed for a S1 pocket. The favourable position of the central scaffold provides pi-stacking between a fluoro-phenyl ring and His-57 instead. Similar to 12k, 12g does not occupy a S2’ pocket.
Figure 9.
Predicted binding mode of 12g with factor XIa. The dashed blue line indicates pi-stacking between the ligand central core and His-57. The figure was prepared in Maestro.
The docked pose of 12n differs from the poses of the aforementioned compounds and is depicted in Figure 10. The rhodanine moiety is predicted to be near the edge of the factor-XIa active site inside a S2 pocket. In general, it is not common for existing factor-XIa inhibitors to occupy this pocket. A cyclohexene ring of 12n partly fills a S1’ pocket. A S1 pocket is almost unoccupied, with only a methyl group placed at its top. No specific interactions are visually observable. The residues that participate in interactions with ligands for the remainder of the tested compounds are outlined in Supplementary Materials Table S1.
Figure 10.
The docked pose of 12n. The figure was prepared in Maestro.
We can conclude that, as in the case of binding to factor Xa, the rhodanine moiety does not tend to be buried in deep pockets of factor XIa. For both coagulation factors, a hydrophilic and charged rhodanine ring can instead be found in open solvent-exposed pockets and surfaces, decreasing the likelihood of a desolvation penalty.
ADME Properties
The ADME properties of all the compounds are presented in Table 2. One of the most significant issues is how well a drug is absorbed from an oral solution. In this case, all the compounds showed high levels of gastro-internal absorption.
Table 2.
ADMET values of Rivaroxaban and tested compounds.
| No | Model Name—Predicted Value | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Water Solubility | Caco2 Permeability | Intestinal Absorption (Human) | Skin Permeability | P-Glycoprotein Substrate | P-Glycoprotein I Inhibitor | P-Glycoprotein II Inhibitor | VDss (Human) | Fraction Unbound (Human) | BBB Permeability | CNS Permeability | CYP2D6 Substrate | CYP3A4 Substrate | CYP2D6 Inhibitor | CYP3A4 Inhibitor | Total Clearance | Renal OCT2 Substrate | AMES Toxicity | Max. Tolerated Dose (Human) | Oral Rat Acute Toxicity (LD50) | Hepatotoxicity | Skin Sensitization | |
| 12a | −5.119 | 1.112 | 92.065 | −3.099 | No | Yes | No | 0.477 | 0.164 | 0.173 | −1.834 | No | Yes | No | No | −0.094 | No | No | −1.104 | 2.913 | No | No |
| 12b | −5.833 | 1.099 | 91.664 | −3.123 | No | Yes | No | 0.422 | 0.121 | 0.085 | −1.728 | No | Yes | No | Yes | −0.537 | No | No | −1.041 | 2.913 | No | No |
| 12c | −5.570 | 1.002 | 91.414 | −2.732 | Yes | Yes | Yes | 0.403 | 0 | −0.250 | −1.672 | No | Yes | No | Yes | −0.277 | No | No | −0.470 | 3.116 | Yes | No |
| 12d | −5.196 | 1.195 | 93.192 | −2.788 | Yes | Yes | Yes | 0.175 | 0.004 | −0.670 | −1.844 | No | Yes | No | Yes | −0.141 | No | Yes | −0.340 | 3.384 | Yes | No |
| 12e | −5.339 | 1.325 | 93.529 | −2.770 | Yes | Yes | Yes | −0.095 | 0 | −1.117 | −2.847 | No | Yes | No | Yes | 0.271 | No | No | −0.144 | 3.137 | No | No |
| 12f | −5.250 | 1.187 | 93.334 | −3.599 | Yes | Yes | No | 0.259 | 0.140 | −0.178 | −1.996 | No | Yes | No | No | −0.059 | No | No | −1.082 | 3.007 | Yes | No |
| 12g | −5.565 | 1.008 | 94.135 | −2.745 | Yes | Yes | Yes | 0.461 | 0.016 | −0.199 | −1.667 | No | Yes | No | Yes | −0.246 | No | Yes | −0.669 | 3.126 | Yes | No |
| 12h | −5.785 | 0.959 | 92.948 | −2.769 | Yes | Yes | Yes | 0.744 | 0.007 | −0.074 | −1.462 | No | Yes | No | Yes | −0.251 | No | Yes | −0.605 | 2.971 | Yes | No |
| 12i | −5.759 | 0.957 | 93.583 | −2.771 | Yes | Yes | Yes | 0.746 | 0.009 | −0.079 | −1.495 | No | Yes | No | Yes | −0.540 | No | Yes | −0.619 | 0.953 | Yes | No |
| 12j | −5.553 | 0.977 | 94.048 | −2.754 | Yes | Yes | Yes | 0.834 | 0.020 | 0.121 | −1.463 | No | Yes | No | Yes | 0.092 | No | No | −0.541 | 2.910 | No | No |
| 12k | −5.985 | 0.995 | 92.660 | −2.754 | Yes | Yes | Yes | 0.455 | 0.008 | −0.225 | −1.548 | No | Yes | No | Yes | −0.378 | No | No | −0.638 | 3.233 | No | No |
| 12l | −6.128 | 0.944 | 92.108 | −2.774 | Yes | Yes | Yes | 0.730 | 0 | −0.105 | −1.376 | No | Yes | No | Yes | −0.672 | No | No | −0.594 | 2.998 | No | No |
| 12m | −6.188 | 1.001 | 92.450 | −2.742 | Yes | Yes | Yes | 0.515 | 0 | −0.117 | −1.489 | No | Yes | No | Yes | −0.187 | No | No | −0.572 | 3.271 | No | No |
| 12n | −6.079 | 1.108 | 91.003 | −3.101 | Yes | Yes | Yes | 0.570 | 0.063 | 0.223 | −1.514 | No | Yes | No | Yes | −0.003 | No | No | −1.274 | 2.912 | No | No |
| 12o | −6.194 | 1.115 | 90.689 | −3.054 | Yes | Yes | Yes | 0.606 | 0.057 | 0.320 | −2.537 | No | Yes | No | Yes | 0.196 | No | No | −1.247 | 2.918 | No | No |
| 12p | −5.785 | 1.294 | 91.302 | −3.416 | Yes | Yes | Yes | 0.290 | 0.088 | −0.009 | −1.690 | No | Yes | No | Yes | −0.152 | No | No | −1.176 | 2.985 | No | No |
| 12q | −6.27 | 1.290 | 90.525 | −3.365 | Yes | Yes | Yes | 0.344 | 0.043 | 0.017 | −1.507 | No | Yes | No | Yes | −0.127 | No | No | −1.243 | 3.008 | No | No |
| 14a | −6.077 | 1.27 | 97.261 | −2.789 | Yes | Yes | Yes | 0.553 | 0 | −0.132 | −1.537 | No | Yes | No | Yes | 0.037 | No | No | −0.321 | 2.954 | No | No |
| 14b | −6.530 | 1.261 | 96.471 | −2.790 | Yes | Yes | Yes | 0.602 | 0 | −0.133 | −1.537 | No | Yes | No | Yes | −0.094 | No | No | −0.279 | 3.070 | No | No |
| 16a | −4.326 | 0.956 | 94.396 | −3.340 | Yes | No | No | 0.000 | 0.233 | −0.581 | −1.985 | No | Yes | No | No | −0.317 | No | No | −0.248 | 2.712 | No | No |
| 16b | −4.539 | 1.269 | 94.806 | −3.441 | Yes | No | No | 0.110 | 0.252 | −0.119 | −2.116 | No | No | No | No | −0.422 | No | No | −0.262 | 2.866 | No | No |
| 16c | −4.521 | 0.969 | 96.467 | −2.811 | Yes | Yes | Yes | 0.545 | 0.024 | −0.106 | −2.016 | No | Yes | No | Yes | −0.313 | No | No | −0.118 | 2.511 | Yes | No |
| 16d | −4.721 | 1.418 | 96.112 | −2.876 | Yes | Yes | Yes | 0.310 | 0.017 | −0.240 | −2.049 | No | Yes | No | Yes | −0.457 | No | No | −0.215 | 2.855 | No | No |
| 16e | −5.087 | 0.951 | 95.655 | −2.814 | Yes | Yes | Yes | 0.634 | 0.018 | −0.111 | −1.817 | No | Yes | No | Yes | −0.557 | No | No | −0.139 | 2.766 | No | No |
| Rivaroxaban | −4.382 | 1.2876 | 92.803 | −2.967 | No | Yes | No | −0.687 | 0 | −1.022 | −2.660 | No | Yes | No | Yes | 0.296 | No | Yes | −0.232 | 2.707 | Yes | No |
| Unit | Numeric (log mol/L) | Numeric (log Papp in 10−6 cm/s) | Numeric (% Absorbed) | Numeric (log Kp) | Categorical (Yes/No) | Categorical (Yes/No) | Categorical (Yes/No) | Numeric (log L/kg) | Numeric (Fu) | Numeric (log BB) | Numeric (log PS) | Categorical (Yes/No) | Categorical (Yes/No) | Categorical (Yes/No) | Categorical (Yes/No) | Numeric (log mL/min/kg) | Categorical (Yes/No) | Categorical (Yes/No) | Numeric (log mg/kg/day) | Numeric (mol/kg) | Categorical (Yes/No) | Categorical (Yes/No) |
Moreover, the positive values of all the compounds, except for the compounds 12a and 12b and the reference drug, Rivaroxaban, showed that they can be transported across the cell membrane by the ATP-binding cassette (ABC) transporter, which is a component of P-glycoprotein. By contrast, the compounds 12a–12q and 14a–14b and the reference drug, Rivaroxaban, were also predicted to be P-glycoprotein I and II inhibitors, which can inhibit their transport. Regarding skin permeability, if the logKp is greater than −2.5, a drug cannot be skin-permeable. In this framework, all the compounds demonstrated improved skin permeability.
The volume of distribution (VDss) suggests the total amount of drug that needs to be homogeneously distributed in the blood. The VDss values are considered low if they are below 0.15 log VDss, and high VDss values are >0.45 log VDss. Therefore, only compounds 12e, 16a, and 16b and Rivaroxaban can be regarded as low-VDss compounds. The permeability of the blood–brain barrier (BBB) indicates whether a substance can enter the brain. Since it is assumed that compounds with logBB > 0.3 can pass through the BBB, all the compounds’ logBB values indicate a low ability to pass through the BBB. Similarly, the majority of the compounds have low central nervous system (CNS) permeability, except for compounds 12e and 12o and Rivaroxaban, whose logPS values are lower than −2.0.
The metabolism prediction suggested that most of the compounds are both substrates and inhibitors of CYP3A4. The maximum tolerated dose (MRTD) is a crucial criterion in toxicity evaluation; a value of less than 0.44 log(mg/kg/day) is considered low, while a value of more than 0.477 log(mg/kg/day) is considered high. Thus, none of the compounds showed an optimal MRTD. Moreover, most of the compounds showed no AMES toxicity and no hepatotoxicity. They ae therefore considered non-toxic, except for compounds 12d and 12g–i, as well as the reference drug, Rivaroxaban.
3. Materials and Methods
The 1H- and 13C-NMR spectra were recorded on Bruker DRX-500 (500.13 and 125.76 MHz, respectively), Bruker AV 600, and Bruker AM 300 (600.13, 300.13 and 150.90 MHz, respectively) in DMSO-d6, with TMS as internal standard. The IR spectra of solid samples were recorded on a Vertex 70 FT-IR spectrometer using a Platinum ATR (Bruker) attachment equipped with a diamond prism in a frequency range from 4000 to 400 cm−1, with a resolution of 2 cm−1. The results were obtained by averaging 16 scans. High-resolution mass spectra were recorded on an Agilent Technologies LCMS 6230B spectrometer (electrospray ionization). Melting points were determined on a Stuart SMP30 apparatus. Assaying of the purity of the starting materials and the synthesized compounds, as well as the analysis of reaction mixtures, was conducted by TLC on Merck TLC silica-gel 60 F254 plates. Eluents: CHCl3, petroleum ether, ethyl acetate, and their mixtures, in different ratios. Visualization of TLC plates was prepared under UV light or in iodine vapor. Commercially available solvents and reagents (Sigma-Aldrich, Merck, Acros Organics) were used in this work.
3.1. Synthesis
Procedure for the synthesis of 8-bromo-4,4,6-trimethyl-6-phenyl-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-1,2-dione 6.
The following procedure was adapted from previously published research [54]. A mixture of 4,4,6-trimethyl-6-phenyl-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-1,2-dione (1 g, 3.28 mmol) and N-bromosuccinimide (0.64 g, 3.61 mmol) in DMF (20 mL) was stirred at room temperature for 12 h. The mixture was diluted with water (40 mL). The resulting precipitate was filtered, washed in water, dried, and recrystallized from 2-PrOH.
Red powder, 1.18 g; yield 94 %; Rf = 0.27 (chloroform); m.p. 168–170 °C; IR (KBr, νmax/cm−1): 1729 (C=O), 1295, 775, 707 (C-Br), 468; 1H NMR (500.13 MHz, DMSO-d6), δ (ppm): 0.75 (3H, s, C6-CH3); 1.59 (3H, s, C4-CH3); 1.71 (3H, s, C4-CH3); 2.09 (1H, d, J = 14.4 Hz, C5-H); 2.45 (1H, d, J = 14.4 Hz, C5-H); 7.14 (2H, d, J = 7.8 Hz, Ph); 7.20 (1H, t, J = 7.2 Hz, Ph); 7.29 (2H, t, J = 7.6 Hz, Ph); 7.65 (1H, s, H-7(9)); 7.77 (1H, s, H-7(9)); 13C NMR (125.76 MHz, DMSO-d6), δ (ppm): 24.9, 27.7, 30.2, 39.7, 50.3, 53.9, 114.9, 118.2, 125.2, 126.4, 126.6, 128.3, 130.0, 138.0, 146.5, 147.1, 156.4, 182.8. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C20H18BrNO2 + H+ 384.0595, found 384.0598.
General procedure for the synthesis of 6-iodo-2,2,4-trimethyl-4-R1-1,2,3,4-tetrahydroquinoline hydrochloride 8a–c.
The following procedure was adapted from previously published research [55]. To a solution of corresponding 2,2,4-trimethyl-1,2,3,4-tetrahydroquinoline 7 (0.01 mol) in mixture of dioxane:pyridine (1:1, 500 mL) at 0–5 °C was added molecular iodine (0.03 mol) in small portions, and mixture was subjected to constant stirring for 20–30 min. After 30 min, the ice bath was removed and the mixture was allowed to reach room temperature after 1 h. Progress of the reaction was monitored by thin-layer chromatography (TLC) on SiO2 with 10:1 petroleum ether–ethyl acetate as the eluent and visualization by iodine vapor and UV radiation. After the completion of the conversion, the reaction mixture was quenched by addition of saturated aqueous solution of Na2S2O3 until complete discoloration of the solution. The resulting solution was extracted with CH2Cl2 (3 × 10 mL); the combined organic layers were washed with saturated brine, dried over anhydrous Na2SO4, and concentrated under vacuo at 50–60 °C. The oily residue was dissolved in minimal amounts of acetone, and concentrated HCl (0.01 mol, 1 mL) was added dropwise under stirring. The precipitate formed was filtered off and ether was washed.
6-Iodo-2,2,4-trimethyl-1,2,3,4-tetrahydroquinoline hydrochloride (8a). White powder, 1.78 g; yield 59%; Rf = 0.48 (5:1 petroleum ether/ethyl acetate); m.p. 125–127 °C; 1H NMR (600.13 MHz, DMSO-d6), δ (ppm): 1.02–1.22 (9H, set of singlets, C4-CH3 +2 C2-CH3); 1.63 (1H, dd, J = 12.5 Hz, J = 5.5 Hz, C3-H); 1.69 (1H, dd, J = 12.5 Hz, J = 5.5 Hz, C3-H); 2.72–2.82 (1H, m, C4-H); 6.27–7.39 (3H, CHarom); 13C NMR (150.90 MHz, DMSO-d6), δ (ppm): 20.5, 27.4, 27.8, 30.9, 43.9, 48.7, 84.9, 116.7, 126.9, 134.9, 135.1, 144.4. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C12H16IN + H+ 302.0414, found 302.0412.
6-Iodo-2,2,4-trimethyl-4-phenyl-1,2,3,4-tetrahydroquinoline hydrochloride (8b). White powder, 2.75 g; yield 73%; Rf = 0.49 (5:1 petroleum ether/ethyl acetate); m.p. 140–142 °C; IR (KBr, νmax/cm−1): 3324, 3251 (NH), 702, 517; 1H NMR (600.13 MHz, DMSO-d6), δ (ppm): 0.86 (3H, s, C4-CH3); 1.22 (3H, s, C2-CH3); 1.64 (3H, s, C2-CH3); 1.96 (1H, d, J = 14.3 Hz, C3-H); 2.27 (1H, d, J = 14.3 Hz, C5-H); 6.80 (1H, d, J = 8.5 Hz, H-8); 7.12–7.15 (3H, m, Ph); 7.22–7.25 (2H, m, Ph); 7.32 (1H, s, H-5(7)); 7.47 (1H, d, J = 8.5 Hz, H-5(7)); 13C-NMR (150.90 MHz, DMSO-d6), δ (ppm): 28.2, 28.7, 31.5, 40.7, 49.9, 52.1, 84.7, 121.2, 126.4, 127.0, 127.1, 128.6, 134.1, 136.4, 138.1, 150.0. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C18H20IN + H+ 378.0713, found 378.0715.
4-(4-Chlorophenyl)-6-iodo-2,2,4-trimethyl-1,2,3,4-tetrahydroquinoline hydrochloride (8c). White powder, 2.51 g; yield 61%; Rf = 0.55 (5:1 petroleum ether/ethyl acetate); m.p. 146–148 °C; IR (KBr, νmax/cm−1): 2457, 809, 819, 500; 1H NMR (600.13 MHz, DMSO-d6), δ (ppm): 0.90 (3H, s, C4-CH3); 1.22 (3H, s, C2-CH3); 1.64 (3H, s, C2-CH3); 1.97 (1H, d, J = 14.4 Hz, C3-H); 2.24 (1H, d, J = 14.4 Hz, C3-H); 6.83 (1H, d, J = 8.5 Hz, H-8); 7.15 (2H, d, J = 8.4 Hz, C6H4); 7.28 (2H, d, J = 8.3 Hz, C6H4); 7.34 (1H, s, H-5(7)); 7.49 (1H, d, J = 8.5 Hz, H-7(5)); 13C-NMR (150.90 MHz, DMSO-d6), δ (ppm): 28.0, 28.4, 31.4, 40.5, 49.6, 52.4, 85.7, 121.7, 128.4, 128.6, 129.2, 129.2, 131.1, 131.2, 136.6, 138.0, 149.0. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C18H19ClIN + H+ 412.0323, found 412.0327.
8-R2-4,4,6-trimethyl-6-R1-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-1,2-diones 5, 9a–c, 10a–o were obtained according to the published method [52,53]. General procedure: oxalyl chloride (4.7 mL, 5.5 mmol) was added to corresponding tetrahydroquinoline hydrochloride (5 mmol) in anhydrous toluene (50 mL). The reaction mixture was refluxed for 1.5–2 h until complete dissolution of the salt. Excess solvent was removed under vacuum, and the orange–red precipitate that formed after cooling was filtered off, washed with EtOH, and recrystallized from 2-PrOH.
The physicochemical and spectral data of 4,4,6-trimethyl-6-phenyl-5,6-dihydro-1H-pyrrolo[3,2,1-ij]quinoline-1,2(4H)-dione (5), 4,4,6-trimethyl-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-1,2-dione (10a), 4,4,6-trimethyl-1,2-dioxo-1,2,5,6-tetrahydro-4H-pyrrolo[3,2,1-ij]quinolin-8-yl benzoate (10b), 8-methoxy-4,4,6-trimethyl-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-1,2-dione (10e) and 4,4,6,8-tetramethyl-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-1,2-dione (10n) are described in [52]. The data of 6′,6′-dimethyl-5′,6′-dihydrospiro-[cyclohexane-1,4′-pyrrolo[3,2,1-ij]quinoline]-1′,2′-dione (10h), 6′,8′-dimethyl-5′,6′-dihydrospiro-[cyclohexane-1,4′-pyrrolo[3,2,1-ij]quinoline]-1′,2′-dione (10i), 8′-fluoro-6′-methyl-5′,6′ -dihydrospiro[cyclopentane-1,4′-pyrrolo[3,2,1-ij]quinoline]-1′,2′-dione (10j), and 8′-fluoro-6′-methyl-5′,6′-dihydrospiro[cycloheptane-1,4′-pyrrolo[3,2,1-ij]quinoline]-1′,2′-dione (10k) are described in [53].
8-Iodo-4,4,6-trimethyl-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-1,2-dione (9a). Red powder, 1.28 g; yield 72%; Rf = 0.33 (5:1 petroleum ether/ethyl acetate); m.p. 168–170 °C; IR (KBr, νmax/cm−1): 3427, 1720 (C=O), 1296, 873, 443; 1H NMR (300.13 MHz, DMSO-d6), δ (ppm): 1.30–1.33 (6H, m, C6-CH3 + C4-CH3); 1.52 (1H, t, J = 12 Hz, C5-H); 1.56 (3H, s, C4-CH3); 1.84 (1H, dd, J = 12 Hz, J = 3 Hz, C5-H); 2.89 (1H, m, C6-H); 7.63 (1H, s, H-7(9)); 7.83 (1H, s, H-7(9)); 13C NMR (150.90 MHz, DMSO-d6), δ (ppm): 18.4, 24.8, 25.7, 26.9, 44.6, 54.3, 86.5, 118.2, 129.9, 130.2, 142.5, 147.5, 156.6, 183.1. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C14H14NIO2 + H+ 356.0142, found 356.0139.
8-Iodo-4,4,6-trimethyl-6-phenyl-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-1,2-dione (9b). Orange powder, 1.81 g; 84%; Rf = 0.25 (5:1 petroleum ether/ethyl acetate); m.p. 231–233 °C; IR (KBr, νmax/cm−1): 1736 (C=O), 1298, 700. 1H NMR (600.13 MHz, DMSO-d6), δ (ppm): 0.73 (3H, s, C6-CH3); 1.58 (3H, s, C4-CH3); 1.69 (3H, s, C4-CH3); 2.06 (1H, d, J = 14.4 Hz, C5-H); 2.42 (1H, d, J = 14.4 Hz, C5-H); 7.13 (2H, d, J = 7.8 Hz, Ph); 7.20 (1H, t, J = 7.3 Hz Ph); 7.29 (2H, t, J = 7.6 Hz Ph); 7.76 (1H, s, H-7(9)); 7.89 (1H, s, H-7(9)); 13C-NMR (150.90 MHz, DMSO-d6), δ (ppm): 24.8, 27.6, 30.0, 39.3, 50.1, 53.7, 85.9, 118.4, 126.2, 126.4, 128.2, 130.0, 130.5, 143.4, 146.7, 147.0, 156.0, 182.5. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C20H18NIO2 + H+ 432.0455, found 432.0457.
6-(4-Chlorophenyl)-8-iodo-4,4,6-trimethyl-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-1,2-dione (9c). Orange powder, 1.70 g; yield 73%; Rf = 0.41 (5:1 petroleum ether/ethyl acetate); m.p. 220–222 °C; 1H NMR (600.13 MHz, DMSO-d6), δ (ppm): 0.76 (3H, s, C6-CH3); 1.58 (3H, s, C4-CH3); 1.69 (3H, s, C4-CH3); 2.07 (1H, d, J = 14.5 Hz, C5-H); 2.41 (1H, d, J = 14.5 Hz, C5-H); 7.16 (2H, d, J = 8.4 Hz, CHarom); 7.35 (2H, d, J = 8.4 Hz, CHarom); 7.77 (1H, s, H-7(9)); 7.90 (1H, s, H-7(9)); 13C-NMR (150.90 MHz, DMSO-d6), δ (ppm): 24.9, 27.5, 29.9, 39.1, 49.9, 53.6, 86.1, 118.5, 128.1, 128.5, 129.5, 130.7, 130.9, 143.3, 146.1, 146.6, 156.1, 182.5. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C20H17ClINO2 + H+ 466.0065, found 466.0063.
4,4,6-Trimethyl-1,2-dioxo-1,2,5,6-tetrahydro-4H-pyrrolo[3,2,1-ij]quinolin-8-yl-2-methoxybenzoate (10c). Red solid, 1.53 g; yield 81%; Rf = 0.43 (5:1 petroleum ether/ethyl acetate); m.p. 165–167 °C; 1H NMR (600.13 MHz, DMSO-d6), δ (ppm): 1.32 (3H, d, J = 6.7 Hz, C6-CH3); 1.37 (3H, s, C4-CH3); 1.58 (1H, t, J = 12.9 Hz, C5-H); 1.70 (3H, s, C4-CH3); 1.89 (1H, dd, J = 13.7 Hz, J = 4.5 Hz, C5-H); 2.90–2.96 (1H, m, C6-H); 3.88 (3H, s, MeO); 7.10 (1H, t, J = 7.5 Hz, CHarom); 7.23 (1H, d, J = 8.4 Hz, CHarom); 7.28 (1H, s, CHarom); 7.46 (1H, s, CHarom); 7.64–7.66 (1H, m, CHarom); 7.94 (1H, d, J = 7.4 Hz, CHarom); 13C-NMR (150.90 MHz, DMSO-d6), δ (ppm): 17.9, 24.2, 25.3, 26.5, 44.2, 53.7, 55.8, 112.6, 115.7, 116.0, 118.2, 120.0, 127.7, 127.9, 131.5, 134.6, 145.1, 146.1, 156.8, 158.9, 163.8, 183.3. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C22H21NO5 + H+ 380.1494, found 380.1490.
4,4,6-Trimethyl-1,2-dioxo-1,2,5,6-tetrahydro-4H-pyrrolo[3,2,1-ij]quinolin-8-yl 3,4,5-trimethoxybenzoate (10d). Orange solid, 1.69 g; yield 77%; Rf = 0.50 (5:1 petroleum ether/ethyl acetate); m.p. 213–215 °C; m.p. 165–167 °C; 1H NMR (600.13 MHz, DMSO-d6), δ (ppm): 1.32 (3H, d, J = 6.7 Hz, C6-CH3); 1.37 (3H, s, C4-CH3); 1.58 (1H, t, J = 13.0 Hz, C5-H); 1.71 (3H, s, C4-CH3); 1.89 (1H, dd, J = 13.7 Hz, J = 4.5 Hz, C5-H); 2.91–2.95 (1H, m, C6-H); 3.78 (3H, s, MeO); 3.87 (6H, s, 2MeO); 7.31 (1H, d, J = 1.0 Hz, CHarom); 7.39 (2H, s, CHarom); 7.51 (1H, s, CHarom); 13C-NMR (150.90 MHz, DMSO-d6), δ (ppm): 18.0, 24.3, 25.4, 26.5, 44.3, 53.8, 56.1, 60.2, 107.2, 115.7, 116.0, 123.7, 127.8, 128.0, 142.4, 145.2, 146.3, 152.8, 156.9, 164.4, 183.4. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C24H25NO7 + H+ 440.1705, found 440.1703.
8-Fluoro-4,4,6-trimethyl-6-phenyl-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-1,2-dione (10f). Orange solid, 1.11 g; yield 69%; Rf = 0.55 (5:1 petroleum ether/ethyl acetate); m.p. 160–162 °C; IR (KBr, νmax/cm−1): 1724 (C=O), 704, 427; 1H NMR (600.13 MHz, DMSO-d6), δ (ppm): 0.72 (3H, s, C6-CH3); 1.59 (3H, s, C4-CH3); 1.69 (3H, s, C4-CH3); 2.09 (1H, d, J = 14.4 Hz, C5-H); 2.48 (1H, d, J = 14.4 Hz, C5-H, 7.15 (2H, d, J = 8.1 Hz, Ph); 7.20 (1H, t, J = 7.3 Hz, Ph); 7.29 (2H, t, J = 7.6 Hz, Ph); 7.42 (1H, dd, J = 6.8 Hz, J = 2.2 Hz, H-7(9)); 7.58 (1H, dd, J = 10.3 Hz, J = 2.2 Hz, H-7(9)); 13C-NMR (150.90 MHz, DMSO-d6), δ (ppm): 24.6 and 24.7 (stereoisomers), 27.6, 30.2, 39.5, 50.1, 53.6, 109.8, 109.9, 117.06, 117.11, 122.5, 122.7, 126.2, 126.4, 128.17, 129.21, 143.6, 146.9, 156.6 (C-F) and 157.7 (C-F), 159.3, 183.2. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C20H18FNO2+ H+ 324.1395, found 324.1401.
6-(4-Chlorophenyl)-8-fluoro-4,4,6-trimethyl-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-1,2-dione (10g). Orange solid, 1.28 g; yield 72%; Rf = 0.48 (5:1 petroleum ether/ethyl acetate); m.p. 143–145 °C; IR (KBr, νmax/cm−1): 1727 (C=O), 1302, 818, 453; 1H NMR (600.13 MHz, DMSO-d6), δ (ppm): 0.75 (3H, s, C6-CH3); 1.60 (3H, s, C4-CH3); 1.69 (3H, s, C4-CH3); 2.10 (1H, d, J = 14.5 Hz, C5-H); 2.47 (1H, d, J = 14.5 Hz, C5-H); 7.18 (2H, d, J = 8.5 Hz, C6H4); 7.35 (2H, d, J = 8.5 Hz, C6H4); 7.42 (1H, dd, J = 6.8 Hz, J = 2.3 Hz, H-7(9)); 7.58 (1H, dd, J = 10.3 Hz, J = 2.4 Hz, H-7(9))13C-NMR (150.90 MHz, DMSO-d6), δ (ppm): 24.8, 27.5, 30.2, 39.3, 49.9, 53.5, 110.0, 110.1, 117.2, 117.3, 122.4, 122.5, 128.1, 128.5, 128.59, 128.63, 130.9, 143.6, 146.0, 156.6, 157.7, 159.3, 183.2. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C20H17ClFNO2+ H+ 358.1006, found 358.1008.
6-(4-Chlorophenyl)-4,4,6,8-tetramethyl-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-1,2-dione (10l). Orange solid, 1.11 g; yield 63%; Rf = 0.44 (5:1 petroleum ether/ethyl acetate); m.p. 169–171 °C; 1H NMR (600.13 MHz, DMSO-d6), δ (ppm): 0.76 (3H, s, C6-CH3); 1.59 (3H, s, C4-CH3); 1.68 (3H, s, C4-CH3); 2.08 (1H, d, J = 14.5 Hz, C5-H); 2.33 (3H, s, C8-CH3); 2.44 (1H, d, J = 14.5 Hz, C5-H); 7.17 (2H, d, J = 8.5 Hz, CHarom); 7.32–7.35 (3H, m, CHarom); 7.48 (1H, s, CHarom); 13C-NMR (150.90 MHz, DMSO-d6), δ (ppm): 20.3, 24.9, 27.6, 30.2, 38.9, 50.1, 53.5, 116.3, 123.3, 126.6, 128.0, 128.5, 130.8, 132.3, 136.6, 145.0, 146.5, 156.7, 184.1. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C21H20ClNO2+ H+ 354.1256, found 354.1255.
8-Ethoxy-4,4,6-trimethyl-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-1,2-dione (10m). Orange solid, 1.16 g; yield 85%; Rf = 0.32 (chloroform); m.p. 169–171 °C; 1H NMR (300.13 MHz, DMSO-d6), δ (ppm): 1.28–1.32 (9H, m, C6-CH3 +C4-CH3 +, OCH2CH3); 1.57 (1H, t, J = 15 Hz, C5-H); 1.67 (3H, s, C4-CH3); 1.84 (1H, dd, J = 15 Hz, J = 6 Hz, C5-H); 2.86 (1H, m, C6-H); 4.02 (2H, q, J = 6 Hz, OCH2CH3); 6.93 (1H, s, H-7); 7.13 (1H, s, H-9); 13C-NMR (150.90 MHz, DMSO-d6), δ (ppm): 15.1, 18.6, 24.5, 25.9, 27.1, 45.0, 54.0, 64.4, 107.4, 116.4, 122.6, 128.5, 142.1, 155.4, 157.2, 184.7. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C16H19NO3+ H+ 274.1439, found 274.1436.
6-(4-Chlorophenyl)-4,4,6-trimethyl-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinoline-1,2-dione (10o). Orange solid, 0.97 g; yield 57%; Rf = 0.26 (chloroform); m.p. 113–115 °C; 1H NMR (600.13 MHz, DMSO-d6), δ (ppm): 0.77 (3H, s, C6-CH3); 1.61 (3H, s, C4-CH3); 1.69 (3H, s, C4-CH3); 2.10 (1H, d, J = 14.5 Hz, C5-H); 2.46 (1H, d, J = 14.5 Hz, C5-H); 7.15–7.20 (3H, m, CHarom); 7.33 (2H, d, J = 8.5 Hz, CHarom); 7.51 (1H, d, J = 7.3 Hz, CHarom); 7.64 (1H, d, J = 7.8 Hz, CHarom); 13C-NMR (150.90 MHz, DMSO-d6), δ (ppm): 25.0, 27.7, 30.4, 39.1, 50.0, 53.6, 116.5, 123.0, 123.1, 126.8, 128.1, 128.6, 130.9, 136.5, 146.6, 147.2, 156.8, 183.9. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C20H18ClNO2+ H+ 340.1099, found 340.1097.
General procedure for the synthesis of (2,4,5,6-tetrahydro-1H-pyrrolo[3,2,1-ij]quinolin-1-ylidene)-2-thioxothiazolidin-4-ones 12a–q. A mixture of the corresponding pyrrolo[3,2,1-ij]quinoline-1,2-dione (5, 6, 9a–c, 10a–l) (1 mmol) and 2-thioxothiazolidin-4-one (11a,b) (1 mmol) in glacial acetic acid (25 mL) and freshly fused sodium acetate (2 mmol) was heated under reflux for 1–6 h. The resulting precipitate was filtered off, washed with water, and recrystallized from 2-PrOH/acetic acid.
(Z)-2-Thioxo-5-(4,4,6-trimethyl-2-oxo-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-1(2H)-ylidene)thiazolidin-4-one (12a). Dark-brown solid, 0.29 g; yield 85%; Rf = 0.50 (10:1 chloroform/ethyl acetate); m.p. 261–263 °C; 1H NMR (600.13 MHz, DMSO-d6), δ (ppm): 1.32 (3H, d, J = 6.3 Hz, C6-CH3); 1.35 (3H, s, C4-CH3); 1.56 (2H, t, J = 12.8 Hz, C5-H); 1.72 (3H, s, C4-CH3); 1.86–1.90 (1H, m, C6-H); 7.01 (1H, t, J = 12.8 Hz, H-8); 7.33 (1H, d, J = 7.7 Hz, H-7(9)); 8.52 (1H, d, J = 7.7 Hz, H-7(9)); 13.88 (1H, br.s, NH); 13C NMR (150.90 MHz, DMSO-d6), δ (ppm): 17.9, 24.4, 25.4, 26.7, 44.9, 54.1, 117.63, 122.2, 124.7, 125.2, 125.3, 129.0, 133.4, 141.1, 165.36, 169.1, 199.7. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C17H16N2O2S2+ H+ 345.0727, found 345.0732.
(Z)-5-(8-Iodo-4,4,6-trimethyl-2-oxo-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-1(2H)-ylidene)-2-thioxothiazolidin-4-one (12b). Dark-red solid, 0.28 g; yield 60%; Rf = 0.65 (10:1 chloroform/ethyl acetate); m.p. 294–296 °C; 1H NMR (600.13 MHz, DMSO-d6), δ (ppm): 1.31 (3H, d, J = 6.6 Hz, C6-CH3); 1.33 (3H, s, C4-CH3); 1.56 (1H, m, C5-H); 1.71 (3H, s, C4-CH3); 1.86–1.88 (1H, m, C5-H); 2.92 (1H, s, C6-H); 7.64–7.68 (1H, m, H-7(9)); 8.87–8.91 (1H, m, H-7(9)); 14.12 (1H, br.s, NH); 13C NMR (150.90 MHz, DMSO-d6), δ (ppm): 17.8, 24.4, 25.5, 26.5, 44.5, 54.3, 85.7, 119.7, 122.9, 128.1, 133.1, 136.8, 140.6, 164.9, 199.6. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C17H15IN2O2S2+ H+ 470.9692, found 470.9698.
(Z)-4,4,6-Trimethyl-2-oxo-1-(4-oxo-2-thioxothiazolidin-5-ylidene)-1,2,5,6-tetrahydro-4H-pyrrolo[3,2,1-ij]quinolin-8-yl benzoate (12c). Brown–purple-needle crystals, 0.30 g; yield 65%; Rf = 0.68 (10:1 chloroform/ethyl acetate); m.p. 276–278 °C; 1H NMR (600.13 MHz, DMSO-d6), δ (ppm): 1.32 (3H, d, J = 6.7 Hz, C6-CH3); 1.37 (3H, s, C4-CH3); 1.60 (1H, t, J = 12.9 Hz, C5-H); 1.74 (3H, s, C4-CH3); 1.93 (1H, dd, J = 13.7 Hz, J = 4.5 Hz, C5-H); 2.93–2.98 (1H, m, C6-H); 7.34 (1H, s, H-7(9)); 7.61 (2H, t, J = 7.7 Hz, CHarom); 7.76 (1H, t, J = 7.5 Hz, CHarom); 8.15 (2H, d, J = 7.7 Hz, CHarom); 8.42 (1H, s, H-7(9)); 14.01 (1H, br.s, NH); 13C NMR (150.90 MHz, DMSO-d6), δ (ppm): 17.8, 24.4, 25.7, 26.7, 44.7, 54.2, 117.9, 118.8, 122.7, 123.9, 126.3, 128.7, 128.9, 129.7, 134.0, 135.0, 138.9, 145.6, 164.9, 165.4, 169.3, 199.7. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C24H20N2O4S2+ H+ 465.0938, found 465.0944.
(Z)-4,4,6-Trimethyl-2-oxo-1-(4-oxo-2-thioxothiazolidin-5-ylidene)-1,2,5,6-tetrahydro-4H-pyrrolo[3,2,1-ij]quinolin-8-yl 2-methoxybenzoate (12d). Dark-blue-needle crystals, 0.37 g; yield 75%; Rf = 0.45 (10:1 chloroform/ethyl acetate); m.p. 287–289 °C; 1H NMR (600.13 MHz, DMSO-d6), δ (ppm): 1.33 (3H, d, J = 5.9 Hz, C6-CH3); 1.36 (3H, s, C4-CH3); 1.60 (1H, t, J = 13.0 Hz, C5-H); 1.74 (3H, s, C4-CH3); 1.92 (1H, dd, J = 13.7 Hz, J = 4.5 Hz, C5-H); 2.94–2.98 (1H, m, C6-H); 3.90 (3H, s, O-CH3); 7.10 (1H, t, J = 7.5 Hz, CHarom); 7.24 (1H, d, J = 8.5 Hz, CHarom); 7.29 (1H, s, H-7(9)); 7.66 (1H, t, J = 7.6 Hz, CHarom); 7.93 (1H, d, J = 7.3 Hz, CHarom); 8.41 (1H, d, J = 1.2 Hz H-7(9)); 14.01 (1H, br.s, NH); 13C NMR (150.90 MHz, DMSO-d6), δ (ppm): 17.9, 24.4, 25.7, 26.6, 44.7, 54.2, 55.8, 112.6, 118.0, 118.5, 118.9, 120.1, 122.7, 124.0, 126.2, 131.4, 134.5, 135.0, 138.8, 145.7, 158.9, 164.3, 165.4, 169.4, 199.7. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C25H22N2O5S2+ H+ 495.1044, found 495.1045.
(Z)-4,4,6-Trimethyl-2-oxo-1-(4-oxo-2-thioxothiazolidin-5-ylidene)-1,2,5,6-tetrahydro-4H-pyrrolo[3,2,1-ij]quinolin-8-yl 3,4,5-trimethoxybenzoate (12e). Brown–purple-needle crystals, 0.37 g; yield 67%; Rf = 0.58 (10:1 chloroform/ethyl acetate); m.p. 314–316 °C; IR (KBr, νmax/cm−1): 3221, 3106 (NH), 1687 (N-C=O), 1321, 1241 (C=S), 1129 (C-S), 736; 1H NMR (600.13 MHz, DMSO-d6), δ (ppm): 1.32 (3H, d, J = 6.6 Hz, C6-CH3); 1.37 (3H, s, C4-CH3); 1.61 (1H, t, J = 12.9 Hz, C5-H); 1.74 (3H, s, C4-CH3); 1.93 (1H, dd, J = 13.7 Hz, J = 4.3 Hz, C5-H); 2.94–2.98 (1H, m, C6-H); 3.34 (3H, s, O-CH3); 3.78 (3H, s, O-CH3); 3.88 (3H, s, O-CH3); 7.31 (1H, s, H-7); 7.42 (2H, s, CHarom); 8.40 (1H, s, H-9); 14.01 (1H, br.s, NH); 13C NMR (150.90 MHz, DMSO-d6), δ (ppm): 17.9, 24.4, 25.6, 26.6, 44.7, 54.2, 56.0, 60.1, 107.0, 117.9, 118.0, 118.9, 122.6, 123.6, 123.9, 126.3, 138.9, 142.3, 145.7, 152.8, 164.6, 165.4, 199.8. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C27H26N2O7S2+ H+ 555.1256, found 555.1259
(Z)-3-Ethyl-5-(8-methoxy-4,4,6-trimethyl-2-oxo-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-1(2H)-ylidene)-2-thioxothiazolidin-4-one (12f). Dark-blue-needle crystals, 0.35 g; yield 86%; Rf = 0.59 (10:1 chloroform/ethyl acetate); m.p. 230–232 °C; 1H NMR (500.13 MHz, DMSO-d6), δ (ppm): 1.23 (3H, t, J = 7.1 Hz, CH2CH3); 1.33 (3H, d, J = 6.7 Hz, C6-CH3); 1.35 (3H, s, C4-CH3); 1.55–1.66 (1H, m, C5-H); 1.71 (3H, s, C4-CH3); 1.89 (1H, dd, J = 13.7 Hz, J = 4.7 Hz, C5-H); 2.89–2.94 (1H, m, C6-H); 3.80 (3H, s, O-CH3); 4.10 (1H, q, J = 7.1 Hz, CH2CH3); 6.96 (1H, s, H-7(9)); 8.28 (1H, s, H-7(9)); 13C NMR (125.76 MHz, DMSO-d6), δ (ppm): 11.8, 18.1, 24.3, 25.9, 26.7, 39.1, 45.4, 54.1, 55.9, 111.6, 116.1, 118.3, 126.2, 126.3, 135.7, 155.5, 165.3, 165.4, 197.4. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C20H22N2O3S2+ H+ 403.1146, found 403.1141.
(Z)-5-(8-Fluoro-4,4,6-trimethyl-2-oxo-6-phenyl-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-1(2H)-ylidene)-2-thioxothiazolidin-4-one (12g). Dark-blue-needle crystals, 0.36 g; yield 83%; Rf = 0.47 (10:1 chloroform/ethyl acetate); m.p. 308–310 °C; 1H NMR (600.13 MHz, DMSO-d6), δ (ppm): 0.70 (3H, s, C6-CH3); 1.62 (3H, s, C4-CH3); 1.70 (3H, s, C4-CH3); 2.13 (1H, d, J = 14.1 Hz, C5-H); 2.53 (1H, d, J = 14.3 Hz, C5-H); 7.08 (2H, d, J = 7.7 Hz, CHarom); 7.18 (1H, t, J = 7.2 Hz, CHarom); 7.26 (2H, t, J = 6.6 Hz, CHarom); 7.37 (1H, d, J = 9.8 Hz, H-7(9)); 8.51 (1H, d, J = 7.7 Hz, H-7(9)); 14.10 (1H, br.s, NH); 13C NMR 150.90 MHz, DMSO-d6), δ (ppm): 24.8, 27.8, 30.2, 39.9, 50.5, 54.2, 112.9, 113.1, 117.3, 117.5, 118.8, 118.9, 123.5, 126.3, 126.4, 127.5, 127.6 (C-F), 128.3, 136.0, 137.5, 147.3, 157.2, 158.8, 165.2, 169.4, 199.5. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C23H19FN2O2S2+ H+ 439.0946, found 439.0948.
(Z)-5-(8-Bromo-4,4,6-trimethyl-2-oxo-6-phenyl-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-1(2H)-ylidene)-2-thioxothiazolidin-4-one (12h). Dark-brown solid, 0.39 g; yield 79%; Rf = 0.47 (10:1 chloroform/ethyl acetate); m.p. 291–293 °C; 1H NMR (500.13 MHz, DMSO-d6), δ (ppm): 0.74 (3H, s, C6-CH3); 1.62 (3H, s, C4-CH3); 1.72 (3H, s, C4-CH3); 2.14 (1H, d, J = 14.4 Hz, C5-H); 2.50 (1H, d, J = 14.1 Hz, C5-H); 7.07 (2H, d, J = 7.7 Hz, CHarom); 7.18 (1H, t, J = 7.2 Hz, CHarom); 7.27 (2H, t, J = 7.5 Hz, CHarom); 7.60 (1H, s, H-7(9)); 8.90 (1H, s, H-7(9)); 14.10 (1H, br.s, NH); 13C NMR (125.76 MHz, DMSO-d6), δ (ppm): 24.9, 27.9, 39.8, 40.0, 50.6, 54.2, 114.5, 120.1, 122.9, 126.4, 126.5, 128.4, 128.6, 132.7, 136.5, 140.3, 147.4, 165.2, 169.6, 199.6. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C23H19BrN2O2S2+ H+ 499.0145, found 499.0151.
(Z)-5-(8-Iodo-4,4,6-trimethyl-2-oxo-6-phenyl-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-1(2H)-ylidene)-2-thioxothiazolidin-4-one (12i). Brown solid, 0.44 g; yield 82%; Rf = 0.51 (10:1 chloroform/ethyl acetate); m.p. 269–271 °C; 1H NMR (600.13 MHz, DMSO-d6), δ (ppm): 0.71 (3H, s, C6-CH3); 1.61 (3H, s, C4-CH3); 1.70 (3H, s, C4-CH3); 2.13 (1H, d, J = 14.3 Hz, C5-H); 2.48 (1H, d, J = 14.4 Hz, C5-H); 7.06 (2H, d, J = 7.7 Hz, CHarom); 7.18 (1H, t, J = 7.2 Hz, CHarom); 7.27 (2H, t, J = 7.6 Hz, CHarom); 7.75 (1H, s, H-7(9)); 9.07 (1H, s, H-7(9)); 14.10 (1H, br.s, NH); 13C NMR (150.90 MHz, DMSO-d6), δ (ppm): 24.8, 27.8, 30.0, 50.4, 54.2, 85.8, 120.3, 122.7, 126.2, 126.4, 128.2, 128.7, 133.9, 136.0, 138.3, 140.6, 147.3, 164.8, 169.4, 199.4. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C23H19IN2O2S2+ H+ 547.0005, found 547.0001.
(Z)-3-Ethyl-2-thioxo-5-(4,4,6-trimethyl-2-oxo-6-phenyl-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-1(2H)-ylidene)thiazolidin-4-one (12j). Dark-red solid, 0.35 g; yield 77%; Rf = 0.76 (10:1 chloroform/ethyl acetate); m.p. 254–256 °C; 1H NMR (500.13 MHz, DMSO-d6), δ (ppm): 0.75–0.77 (3H, m, C6-CH3); 1.23 (3H, tt, J = 7.0 Hz, J = 2.9 Hz, CH2CH3); 1.64 (3H, s, C4-CH3); 1.71 (3H, s, C4-CH3); 2.15 (1H, d, J = 14.3 Hz, C5-H); 2.52 (1H, d, J = 14.3 Hz, C5-H); 4.10–4.13 (2H, m, CH2CH3); 7.07 (2H, d, J = 7.7 Hz, CHarom); 7.14–7.26 (4H, m, H-8 +CHarom); 7.46 (1H, d, J = 7.7 Hz, H-7(9)); 8.77 (1H, d, J = 7.8 Hz, H-7(9)); 13C NMR (125.76 MHz, DMSO-d6), δ (ppm): 11.9, 24.8, 27.9, 30.3, 50.6, 54.1, 118.3, 122.5, 125.3, 126.1, 126.3, 126.4, 128.1, 131.1, 131.3, 141.3, 147.7, 165.3, 166.3, 197.1. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C25H24N2O2S2+ H+ 449.1353, found 449.1352.
(Z)-5-(6-(4-Chlorophenyl)-8-fluoro-4,4,6-trimethyl-2-oxo-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-1(2H)-ylidene)-2-thioxothiazolidin-4-one (12k). Dark-blue-needle crystals, 0.31 g; yield 65%; Rf = 0.48 (10:1 chloroform/ethyl acetate); m.p. 274–276 °C; 1H NMR (600.13 MHz, DMSO-d6), δ (ppm): 0.74 (3H, s, C6-CH3); 1.63 (3H, s, C4-CH3); 1.69 (3H, s, C4-CH3); 2.13 (1H, d, J = 14.2 Hz, C5-H); 2.50 (1H, d, J = 14.2 Hz, C5-H); 7.11 (2H, d, J = 7.6 Hz, CHarom); 7.32 (2H, d, J = 8.1 Hz, CHarom); 7.36 (1H, d, J = 9.9 Hz, H-7(9)); 8.51 (1H, d, J = 9.4 Hz, H-7(9)); 14.10 (1H, br.s, NH); 13C NMR (150.90 MHz, DMSO-d6), δ (ppm): 24.9, 27.7, 30.1, 39.6, 50.2, 54.1, 113.0, 113.2, 117.2, 117.3, 118.9, 119.0, 123.4, 126.9, 127.0, 128.1, 128.4 (C-F), 130.1, 136.1, 137.4, 146.3, 157.2, 158.8, 165.1, 169.4, 199.4. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C23H18ClFN2O2S2+ H+ 473.0556, found 473.0559.
(Z)-5-(6-(4-Chlorophenyl)-8-iodo-4,4,6-trimethyl-2-oxo-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-1(2H)-ylidene)-2-thioxothiazolidin-4-one (12l). Dark-purple solid, 0.40 g; yield 69%; Rf = 0.59 (10:1 chloroform/ethyl acetate); m.p. 285–287 °C; 1H NMR (600.13 MHz, DMSO-d6), δ (ppm): 0.76 (3H, s, C6-CH3); 1.64 (3H, s, C4-CH3); 1.69 (3H, s, C4-CH3); 2.15 (1H, d, J = 14.1 Hz, C5-H); 2.47 (1H, d, J = 14.3 Hz, C5-H); 7.09 (2H, d, J = 8.1 Hz, CHarom); 7.19 (1H, t, J = 7.8 Hz, CHarom); 7.31 (2H, d, J = 8.3 Hz, CHarom); 7.46 (1H, d, J = 7.7 Hz, H-7(9)); 8.74 (1H, d, J = 7.8 Hz, H-7(9)); 13.99 (1H, br.s, NH); 13C NMR (150.90 MHz, DMSO-d6), δ (ppm): 25.0, 27.8, 30.3, 33.1, 50.5, 54.1, 85.8, 118.5, 120.5, 122.6, 124.5, 125.7, 126.4, 128.1, 128.5, 130.9, 134.3, 138.3, 141.0, 146.9, 165.4, 169.3, 199.7. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C23H18ClIN2IO2S2+ H+ 580.9616, found 580.9615.
(Z)-5-(6-(4-Chlorophenyl)-8-fluoro-4,4,6-trimethyl-2-oxo-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-1(2H)-ylidene)-3-ethyl-2-thioxothiazolidin-4-one (12m). Brown solid, 0.37 g; yield 73%; Rf = 0.43 (10:1 chloroform/ethyl acetate); m.p. 235–237 °C; IR (KBr, νmax/cm−1): 3104, 2909 (NH), 1687(N-C=O), 1321, 1241 (C=S), 1129 (C-S), 736; 1H NMR (500.13 MHz, DMSO-d6), δ (ppm): 0.85, (3H, s, C6-CH3); 1.26 (3H, t, J = 6.4 Hz, CH2CH3), 1.64 (3H, s, C4-CH3); 1.71 (3H, s, C4-CH3); 2.15 (1H, d, J = 14.3 Hz, C5-H); 2.50 (1H, d, J = 12.3 Hz, C5-H; 4.15 (2H, q, J = 6.6 Hz, CH2CH3); 7.11 (2H, d, J = 8.1 Hz, CHarom); 7.32 (2H, d, J = 7.9 Hz, CHarom); 7.41 (1H, s, H-7); 8.53 (1H, s, H-9); 13C NMR (125.76 MHz, DMSO-d6), δ (ppm): 11.7, 25.1, 27.7, 30.1, 39.2, 39.8, 50.9, 54.3, 112.9, 113.2, 117.3, 117.4, 124.6, 127.4, 127.5, 128.1, 128.4, 131.1, 133.0, 137.8, 146.3, 157.2, 159.1, 165.3, 166.5, 196.9. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C25H22ClFN2O2S2+ H+ 501.0869, found 501.0871
(Z)-5-(6′,6′-Dimethyl-2′-oxo-5′,6′-dihydrospiro[cyclohexane-1,4′-pyrrolo[3,2,1-ij]quinolin]-1′(2′H)-ylidene)-2-thioxothiazolidin-4-one (12n). Brown solid, 0.27 g; yield 68%; Rf = 0.58 (10:1 chloroform/ethyl acetate); m.p. 254–256 °C; IR (KBr, νmax/cm−1): 3385, 3225 (NH), 1686 (N-C=O), 1434, 1320, 1243 (C=S), 1131 (C-S), 747; 1H NMR (500.13 MHz, DMSO-d6), δ (ppm): 1.20–1.32 (2H, m, (CH2)5); 1.33 (6H, s, C6-(CH3)2); 1.55–1.70 (6H, m, (CH2)5 + C5-H); 2.05 (2H, s, (CH2)5); 2.53–2.60 (1H, m, C5-H + (CH2)5); 7.04 (1H, t, J = 7.3 Hz, H-8); 7.43 (1H, d, J = 7.7 Hz, H-7(9)); 8.58 (1H, d, J = 7.6 Hz, H-7(9)); 13.85 (1H, br.s, NH); 13C NMR (125.76 MHz, DMSO-d6), δ (ppm): 21.8, 25.0, 30.7, 31.5, 33.8, 42.3, 57.9, 118.2, 122.5, 125.1, 125.5, 129.0, 129.7, 133.8, 140.3, 166.0, 169.3, 200.1. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C21H22N2O2S2+ H+ 399.1197, found 399.1196.
(Z)-5-(6′,8′-Dimethyl-2′-oxo-5′,6′-dihydrospiro[cyclohexane-1,4′-pyrrolo[3,2,1-ij]quinolin]-1′(2′H)-ylidene)-2-thioxothiazolidin-4-one (12o). Dark-purple solid, 0.28 g; yield 71%; m.p. 294–296 °C; Rf = 0.61 (10:1 chloroform/ethyl acetate); 1H NMR (500.13 MHz, DMSO-d6), δ (ppm): 1.20–1.31 (2H, m, (CH2)5); 1.33 (3H, d, J = 6.8 Hz, C6-CH3); 1.45–1.60 (4H, m, (CH2)5); 1.69–1.72 (1H, m, (CH2)5); 1.77–1.87 (3H, m, C5-H + (CH2)5); 2.30 (3H, s, C8-CH3); 2.52 (1H, dd, J = 14.0 Hz, J = 4.5 Hz, C5-H); 2.76–2.81 (1H, m, C6-H); 7.13 (1H, s, H-7(9)); 8.37 (1H, s, H-7(9)); 13.85 (1H, br.s, NH); 13C NMR (125.76 MHz, DMSO-d6), δ (ppm): 18.2, 20.9, 21.4, 21.9, 24.8, 24.9, 31.4, 33.5, 37.8, 58.0, 117.9, 125.4, 125.6, 125.8, 129.7, 131.0, 132.9, 139.7, 165.7, 168.9, 199.8. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C21H22N2O2S2+ H+ 399.1196, found 399.1198.
(Z)-5-(8′-Fluoro-6′-methyl-2′-oxo-5′,6′-dihydrospiro[cyclopentane-1,4′-pyrrolo[3,2,1-ij]quinolin]-1′(2′H)-ylidene)-2-thioxothiazolidin-4-one (12p). Dark-blue-needle crystals, 0.25 g; yield 65%; Rf = 0.55 (10:1 chloroform/ethyl acetate); m.p. 288–290 °C; 1H NMR (500.13 MHz, DMSO-d6), δ (ppm): 1.31 (3H, d, J = 6.8 Hz, C6-CH3); 1.52–1.59 (2H, m, (CH2)4); 1.61–1.65 (1H, m, (CH2)4); 1.66–1.82 (3H, m, (CH2)4); 1.95–2.03 (2H, m, (CH2)4+ C5-H); 2.85–2.95 (2H, m, (CH2)4 + C6-H); 7.16 (1H, dd, J = 8.4 Hz, J = 1.7 Hz, H-7(9)); 8.24–8.27 (1H, m, H-7(9)); 13.85 (1H, br.s, NH); 13C NMR (125.76 MHz, DMSO-d6), δ (ppm): 17.7, 24.6, 25.0, 26.6, 35.3, 37.8, 42.9, 64.1, 111.6, 111.8, 115.5, 115.7, 117.8, 117.9, 123.8, 127.0, 127.1, 135.2, 137.9, 157.0, 158.9, 164.9, 169.2, 199.48. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C19H17FN2O2S2+ H+ 389.0789, found 389.0791.
(Z)-5-(8′-Fluoro-6′-methyl-2′-oxo-5′,6′-dihydrospiro[cycloheptane-1,4′-pyrrolo[3,2,1-ij]quinolin]-1′(2′H)-ylidene)-2-thioxothiazolidin-4-one (12q). Brown solid, 0.32 g; yield 77%; Rf = 0.66 (10:1 chloroform/ethyl acetate); m.p. 343–345 °C; 1H NMR (500.13 MHz, DMSO-d6), δ (ppm): 1.30 (3H, d, J = 6.8 Hz, C6-CH3); 1.32–1.45 (2H, m, (CH2)6); 1.50–1.65 (6H, m, (CH2)6); 1.66–1.75 (2H, m, (CH2)6); 1.83–1.90 (2H, m, C5-H + (CH2)6); 2.24 (1H, dd, J = 9.5 Hz, J = 4.3 Hz, C6-H); 2.79–2.84 (1H, m, C6-H); 3.06–3.12 (1H, m, (CH2)6); 7.18 (1H, dd, J = 8.2 Hz, J = 1.8 Hz, H-7(9)); 8.27 (1H, dd, J = 8.4 Hz, J = 2.4 Hz, H-7(9)); 13.95 (1H, br.s, NH); 13C NMR (125.76 MHz, DMSO-d6), δ (ppm): 18.1, 22.5, 23.1, 25.3, 29.7, 29.9, 36.3, 37.5, 39.7, 42.4, 61.3, 111.7, 111.9, 115.7, 115.9, 118.2, 118.3, 123.9, 127.1, 127.2, 135.2, 137.7, 157.0, 158.9, 165.4, 169.2, 199.6. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C21H21FN2O2S2+ H+ 417.1102, found 417.1100.
General procedure for synthesis of substituted 5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-1(2H)-ylidene)-3-(prop-2-yn-1-yl)-2-thioxo- thiazolidin-4-one 14a,b. To a suspension of compound 12 (0.75 mmol) in acetonitrile (20 mL) were added potassium carbonate (1.1 mmol) and propargyl bromide 13 (0.9 mmol), and the mixture was stirred at room temperature for 10 h. Completion of reaction was monitored by TLC. Next, the reaction mixture was diluted with water; brown solid was precipitated out. The precipitated product was filtered, washed with water, dried, and recrystallized from 2-PrOH.
(Z)-5-(8-Fluoro-4,4,6-trimethyl-2-oxo-6-phenyl-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-1(2H)-ylidene)-3-(prop-2-yn-1-yl)-2-thioxothiazolidin-4-one (14a). Brown solid, 0.18 g; yield 52%; Rf = 0.25 (10:1 chloroform/ethyl acetate); m.p. 190–192 °C; IR (KBr, νmax/cm−1): 2361, 2342 (C≡C), 1686 (N-C=O), 1433, 1321, 1241 (C=S), 1130 (C-S), 736;1H NMR (500.13 MHz, DMSO-d6), δ (ppm): 0.71 (3H, s, C6-CH3); 1.63 (3H, s, C4-CH3); 1.71 (3H, s, C4-CH3); 2.14 (1H, d, J = 14.4 Hz, C5-H); 2.54 (1H, d, J = 14.4 Hz, C5-H); 3.47–3.48 (1H, t, J = 2.2 Hz, ≡CH); 4.35 (2H, d, J = 1.4 Hz, CH2); 7.08–7.10 (2H, m, CHarom); 7.18 (1H, t, J = 7.3 Hz, CHarom); 7.24–7.29 (2H, m, CHarom); 7.41–7.45 (1H, m, H-7(9)); 8.62 – 8.65 (1H, m, H-7(9)); 13C NMR (125.76 MHz, DMSO-d6), δ (ppm): 21.3, 24.7, 27.7, 30.2, 39.9, 50.4, 54.2, 75.8, 78.0, 113.2, 113.4, 117.9, 118.1, 118.8, 126.2, 126.4, 126.6, 127.6, 128.2, 134.6, 137.4, 147.2, 157.2, 158.7, 165.3, 178.6, 196.3. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C26H21FN2O2S2+ H+ 477.1102, found 477.1108.
(Z)-5-(6-(4-Chlorophenyl)-8-fluoro-4,4,6-trimethyl-2-oxo-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-1(2H)-ylidene)-3-(prop-2-yn-1-yl)-2-thioxothiazolidin-4-one (14b). Brown solid, 0.19 g; yield 49%; Rf = 0.20 (10:1 chloroform/ethyl acetate); m.p. 125–127 °C; 1H NMR (500.13 MHz, DMSO-d6), δ (ppm): 0.85 (3H, s, C6-CH3); 1.64 (3H, s, C4-CH3); 1.71 (3H, s, C4-CH3); 2.15 (1H, d, J = 14.5 Hz, C5-H); 2.50 (1H, d, J = 14.5 Hz, C5-H); 3.25 (1H, s, ≡CH); 4.33 (1H, s, CH2); 7.11–7.14 (2H, m, CHarom); 7.28–7.31 (3H, m, CHarom); 8.63–8.65 (1H, m, H-7(9)); 13C NMR (125.76 MHz, DMSO-d6), δ (ppm): 21.4, 25.1, 27.7, 30.1, 39.8, 50.9, and 51.0 (stereoisomers), 54.3, 75.5, 77.8, 113.4 and 113.6, 117.6 and 117.8, 119.2 and 119.3, 127.37 and 127.43, 128.1, 128.4, 128.6, 131.1, 134.7, 137.5, 146.3, 157.2 and 159.1, 165.5, 178.5, 196.2. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C26H20ClFN2O2S2 + H+ 511.0713, found 511.0717.
General procedure for synthesis of substituted 4,4,6-trimethyl-1-(5-oxo-1-R3-2-thioxoimidazolidin-4-ylidene)-6-R1-8-R2-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-2(1H)-one 16a-g. A mixture of pyrrolo[3,2,1-ij]quinoline-1,2-dione (10,11) (1 mmol) and 2-thioxoimidazolidin-4-one (16a–c) (1 mmol), in glacial acetic acid (25 mL) and freshly fused sodium acetate (2 mmol), was heated under reflux for 4–10 h. The solid obtained after cooling was filtered, washed with water, dried and recrystallized from 2-PrOH.
(Z)-8-Ethoxy-4,4,6-trimethyl-1-(5-oxo-2-thioxoimidazolidin-4-ylidene)-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-2(1H)-one (16a). Brown solid, 0.31 g; yield 83%; Rf = 0.69 (10:1 chloroform/ethyl acetate); m.p. 343–345 °C; 1H NMR (500.13 MHz, DMSO-d6), δ (ppm): 1.31 (3H, d, J = 7.4 Hz, C6-CH3); 1.32 (3H, s, C4-CH3); 1.35 (3H, t, J = 7 Hz, OCH2CH3); 1.56 (1H, t, J = 13 Hz, C5-H); 1.72 (3H, s, C4-CH3); 1.97 (1H, dd, J = 13.8 Hz, J = 4.8 Hz, C5-H); 2.88–2.92 (1H, m, C6-H); 4.03 (2H, q, J = 7 Hz, OCH2CH3); 6.89 (1H, d, J = 1.9 Hz, H-7(9)); 8.51 (1H, d, J = 2.3 Hz, H-7(9)); 9.32 (1H, s, NH); 9.55 (1H, br.s, NH); 13C NMR (125.76 MHz, DMSO-d6), δ (ppm): 14.8, 18.2, 24.3, 25.8, 26.8, 45.4, 53.8, 63.6, 112.1, 114.6, 118.8, 125.4, 125.5, 133.6, 138.5, 154.2, 166.2, 178.8, 180.2. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C19H21N3O3S + H+ 372.1377, found 372.1376.
(Z)-4,4,6,8-Tetramethyl-1-(5-oxo-2-thioxoimidazolidin-4-ylidene)-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-2(1H)-one (16b). Light-brown powder, 0.24 g; yield 71%; Rf = 0.55 (10:1 chloroform/ethyl acetate); m.p. 357–359 °C; IR (KBr, νmax/cm−1): 3287, 3095 (NH), 1662 (N-C=O), 1543, 1353, 1220 (C=S), 769, 495; 1H NMR (500.13 MHz, DMSO-d6), δ (ppm): 1.31 (3H, d, J = 6.7 Hz, C6-CH3); 1.34 (3H, s, C4-CH3); 1.55 (1H, t, J = 13 Hz, C5-H); 1.72 (3H, s, C4-CH3); 1.86 (1H, dd, J = 13.6 Hz, J = 4.6 Hz, C5-H); 2.31 (3H, s, C8-CH3); 2.86–2.91 (1H, m, C6-H); 7.11 (1H, s, H-7(9)); 8.61 (1H, s, H-7(9)); 9.23 (1H, s, NH); 9.50 (1H, br.s, NH); 13C NMR (125.76 MHz, DMSO-d6), δ (ppm): 18.1, 21.2, 24.3, 25.4, 26.8, 45.3, 53.7, 118.3, 124.3, 125.2, 125.9, 128.1, 130.5, 137.4, 137.9, 166.4, 178.7, 180.0. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C18H19N3O2S + H+ 342.1272, found 342.1271.
(Z)-4,4,6-Trimethyl-1-(5-oxo-2-thioxoimidazolidin-4-ylidene)-6-phenyl-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-2(1H)-one (16c). Orange powder, 0.28 g; yield 69%; Rf = 0.66 (10:1 chloroform/ethyl acetate); m.p. 357–359 °C; 1H NMR (500.13 MHz, DMSO-d6), δ (ppm): 0.75 (3H, s, C6-CH3); 1.65 (3H, s, C4-CH3); 1.71 (3H, s, C4-CH3); 2.15 (1H, d, J = 14.3 Hz, C5-H); 2.51 (1H, d, J = 14.0 Hz, C5-H); 7.06 (1H, d, J = 7.8 Hz, H-7(9)); 7.14–7.19 (3H, m, CHarom); 7.24 (2H, t, J = 7.6 Hz, CHarom); 7.40 (1H, d, J = 7.8 Hz, CHarom); 8.96 (1H, d, J = 7.8 Hz, H-7(9)); 9.31 (1H, s, NH); 9.55 (1H, br.s, NH); 13C NMR (125.76 MHz, DMSO-d6), δ (ppm): 24.9, 28.1, 30.5, 39.4, 51.0, 54.0, 119.0, 122.1, 124.9, 125.7, 126.1, 126.2, 126.6, 128.2, 129.6, 138.9, 139.7, 148.1, 166.5, 178.7, 180.2. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C23H21N3O2S + H+ 404.1428, found 404.1427.
(Z)-8-Fluoro-4,4,6-trimethyl-1-(5-oxo-2-thioxoimidazolidin-4-ylidene)-6-phenyl-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-2(1H)-one (16d). Orange powder, 0.27 g; yield 64%; Rf = 0.38 (10:1 chloroform/ethyl acetate); m.p. 352–354 °C; IR (KBr, νmax/cm−1): 1726 (C=O), 1434, 1303, 1270 (C=S), 817, 454; 1H NMR (500.13 MHz, DMSO-d6), δ (ppm): 0.72 (3H, s, C6-CH3); 1.64 (3H, s, C4-CH3); 1.71 (3H, s, C4-CH3); 2.15 (1H, d, J = 14.3 Hz, C5-H); 2.52 (1H, d, J = 14.3 Hz, C5-H); 7.08 (2H, d, J = 7.6 Hz, CHarom); 7.18 (1H, t, J = 7.1 Hz, CHarom); 7.24–7.32 (3H, m, CHarom); 8.78 (1H, d, J = 9.5 Hz, CHarom); 9.42 (1H, s, NH); 9.68 (1H, s, NH); 13C NMR (125.76 MHz, DMSO-d6), δ (ppm): 24.8, 27.9, 30.3, 39.9, 50.8, 54.0, 113.0, 113.2, 115.7, 115.9, 119.5, 119.6, 124.0, 126.2, 126.5, 126.89, 126.94, 128.2, 136.1, 140.6, 147.5, 157.1, 158.9, 166.2, 178.5, 180.1. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C23H20FN3O2S + H+ 422.1334, found 422.1339.
(Z)-6-(4-Chlorophenyl)-4,4,6,8-tetramethyl-1-(5-oxo-2-thioxoimidazolidin-4-ylidene)-5,6-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-2(1H)-one (16e). Light-brown powder, 0.31 g; yield 68%; Rf = 0.41 (10:1 chloroform/ethyl acetate); m.p. 258–260 °C; 1H NMR (500.13 MHz, DMSO-d6), δ (ppm): 0.75 (3H, s, C6-CH3); 1.62 (3H, s, C4-CH3); 1.69 (3H, s, C4-CH3); 2.12 (1H, d, J = 14.3 Hz, C5-H); 2.37 (3H, s, C8-CH3); 2.48 (1H, d, J = 14.4 Hz, C5-H); 7.10 (2H, d, J = 8.6 Hz, CHarom); 7.27–7.32 (3H, m, CHarom); 8.60 (1H, s, CHarom); 12.8 (1H, s, NH); 13C NMR (125.76 MHz, DMSO-d6), δ (ppm): 21.3, 25.0, 25.5, 27.9, 30.3, 50.8, 54.0, 118.6, 125.3, 126.6, 127.0, 128.2, 128.6, 130.8, 130.9, 131.3, 132.4, 138.3, 147.1, 165.7, 167.3, 170.9. HPLC-HRMS-ESI, m/z ([M + H]+), calcd for C24H22ClN3O2S + H+ 452.1195, found 452.1201.
Spectra for all compounds are in supplementary file.
3.2. In Vitro Assays
The inhibition of blood-clotting factors Xa and XIa by the synthesized compounds 12a–q, 14a,b, and 16a–e was studied by measuring the kinetics of hydrolysis of substrates specific to each of these enzymes in the presence of these compounds. A specific low-molecular-weight chromogenic substrate, S2765 (Z-D-Arg-Gly-Arg-pNA·2HCl), was used in the case of factor Xa, while the substrate S2366 (pyroGlu-Pro-ArgpNA·HCl) (both substrates from Chromogenix, USA) was used for factor XIa.
A buffer containing 140 mM NaCl, 20 mM HEPES, and 0.1% PEG 6000 (pH 8.0) was placed in the wells of a 96-well plate, followed by the addition of factor Xa (final concentration, 2.5 nmol·L–1) or XIa (final concentration 0.8 nmol·L–1), the substrate S2765 (final concentration, 200 μmol·L–1) or S2366 (final concentration, 200 μmol·L–1), and a solution of the test compound in DMSO (final concentration, 30 μmol·L–1; the DMSO content in the well was no more than 2%). The kinetics of the formation of p-nitroaniline were measured using a THERMOmax Microplate Reader (Molecular Devices Corporation, USA) using the absorption of light with a wavelength of 405 nm. The initial rate of substrate degradation was determined from the initial slope of the 4-nitroaniline-formation curve. The rate of substrate degradation by the enzyme in the presence of the inhibitor was expressed as a percentage relative to the rate of substrate degradation in the absence of the inhibitor. The results are presented in Table 1. The data obtained were processed using GraphPad Prism and OriginPro 8 software.
3.3. Molecular-Docking Studies
All simulations were performed by the SOL docking program [56,57]. This program docks flexible low-molecular-weight ligands into the active site of a rigid target protein based on the docking paradigm, which assumes that the ligand bonds with the protein near the global energy minimum of the protein–ligand complex. The search for the global energy minimum was performed using a genetic algorithm with following main parameters: the population size and the number of generations are 30,000 and 1000, respectively. First, using the SOLGRID module, a grid of potentials describing the interaction of the probe-ligand atoms with the protein was calculated. These interactions were obtained using the MMFF94 force field. The SOL scoring function estimated the free energy of protein–ligand binding. Docking solutions that were the ligand-docked positions obtained in several dozen (50 by default) independent runs of the genetic algorithm, and that corresponded to the lowest energy of the protein–ligand complex, were subjected to cluster analysis to assess the reliability of the global energy minimum. A high population of the first cluster-containing-ligand poses with lowest energy and a low number of clusters were the main clues that identified the reliability of docking results. Otherwise, re-docking was carried out with increases in parameters of the genetic algorithm.
The SOL docking program was validated and successfully used for the development of inhibitors of a number of target proteins: thrombin, urokinase (uPA), coagulation factor Xa (see references in [57]), and coagulation factors XIa [12] and XIIa [20,58]. For all these target proteins, when docking native ligands using the SOL program, a high accuracy of ligand positioning was achieved and the score cut-off separating inhibitors from inactive compounds was estimated. The high positioning accuracy meant that the root-mean-square deviation, RMSD, between all atoms in the crystallized-native-ligand pose and the docked-ligand pose, corresponding to the lowest energy of the protein–ligand complex, was less than 2.0 Å. The SOL score cut-offs were determined as explained below.
To predict the anticoagulant activity by docking, we retrieved high-quality structures of target proteins from PDB: structure of FXa from the 3CEN complex and FXIa from 4CRC. Both crystal complexes possess good resolution (<2.0 Å) and no missing residues. These structures were used to construct atomistic models of FXa and FXIa. Protein structures were manually cleaned from native ligands, water molecules, and salt ions. Their protonation was made in the Aplite program [59]. Extracted native ligands were protonated using Avogadro [60]. Validation of prepared protein structures was performed by the docking of native ligands and docking of known FXa and FXIa inhibitors. Both docking procedures for native ligands (the native ligand into the FXa model prepared from 3CEN and the native ligand into the FXIa model prepared from 4CRC) were successful, with RMSD values between the best docking pose and the native conformation of less than 1.4 Å. To study the known binders, we retrieved 10 crystal complexes of FXa and 10 complexes of FXIa with highly active inhibitors (see Table S2). The mean score for the known FXa inhibitors was −6.76 ± 0.44 kcal/mol and the mean score for the known FXIa inhibitors was −5.53 ± 0.56 kcal/mol. These values further served as score cut-offs for the selection of candidates.
ADME Properties
For the prediction of the ADMET properties of all compounds, the free web pkCSM server was used: https://biosig.lab.uq.edu.au/pkcsm/prediction (accessed 12 April 2023). For the prediction, all the molecules were first converted into SMILES (simplified molecule input line entry specification).
4. Conclusions
We applied docking in the search for new coagulation-factor-Xa and -XIa inhibitors. Eight of the twenty-four synthesized and experimentally tested quinolin-1,2-dione and rhodanine compounds exhibited selective or dual inhibitory activity against the coagulation factors Xa and Xia, with the IC50 ranging from 2.28–12.22 µM.
By varying the substituents in the central core and the rhodanine ring, we explored the primary SAR for the identified chemical series of dihydropyrrolo[3,2,1-ij]quinolinones condensed with rhodanine. Some substitutions were found to compromise activities for both factor Xa and factor XIa. For example, the N-alkylation of rhodanine is detrimental to anticoagulant activity, possibly due to its position close to active site walls in docking-predicted binding modes, or to the removal of negative charge. Several substitutions are associated with an increase in inhibition activity against only one of the two coagulation factors. For example, the introduction of acyloxy or halogen atoms at the 8th position of dihydropyrrolo[3,2,1-ij]quinolinone scaffold helps to increase activity against factor XIa.
According to the binding modes predicted by the docking, the three best factor-Xa inhibitors, 12k, 12g, and 12n block access to the catalytic triad of factor Xa. The inhibitors 12g and 12n, on one hand, and of 12k, on the other, bind to factor Xa in a different manner. While the 12k inhibitor occupies the S1 pocket and the S4 hydrophobic pocket remains unoccupied, in the case of the 12g and 12n inhibitors, the S4 pocket is occupied, while the S1 pocket is almost unoccupied. These factor-Xa inhibitors have two different conformations with respect to the position of the rhodanine fragment: either they are located inside the S4 pocket, sandwiched between the aromatic rings of Tyr-99 and Phe-174, while the S1 pocket is practically unoccupied; or the rhodanine is directed into the solvent and located next to the guanidine fragment of Arg-222.
For both coagulation factors, the hydrophilic and charged ring of rhodanine is not buried in deep pockets of the target, but can rather be found in open pockets and on solvent-exposed surfaces, reducing the likelihood of a desolvation penalty.
As can be seen from the results of this work, for a more accurate selection of compounds for experiments to determine their inhibitory activities, docking plays a key role. However, docking alone is not sufficient, and it must be supplemented with post-processing. In post-processing, for the ligands with the best docking characteristics, quantum-chemical calculations of the protein–ligand-binding enthalpy can be performed, or the relative stability of the ligands in their docked positions can be estimated using sufficiently long (about 100 ns) molecular dynamic trajectories.
In addition to exploring SAR, we highlighted some optimization points for the identified chemical series, which were inferred from the comparison of the predicted binding modes with bound conformations of known inhibitors, and which can be exploited to increase potency against factor Xa. In all the cases studied, the P1 part of the identified inhibitors binding to the S1 pocket can be extended to fit the pocket better, and to be buried more deeply. The other aromatic rings in this part also need to be explored. The di-methyl part of 12k, 12g, and 12n does not seem to contribute to activity specifically and can be substituted with other groups bearing H-bond acceptors/donors, considering its position near Gly-219 after docking to factor Xa. These and other modifications are can be exploited in the further development of the design.
Acknowledgments
All docking calculations were carried out using the equipment of the shared research facilities of the HPC computing resources at Lomonosov Moscow State University, including the Lomonosov supercomputer [68]. The authors acknowledge Anthi Petrou for her help in the prediction of toxicity.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/molecules28093851/s1. The copies of 1H- and 13C-NMR spectra, IR, and data from HPLC-MS-ESI analysis for all new synthesized compounds were submitted along with the manuscript [61,62,63,64,65,66,67].
Author Contributions
Conceptualization, K.S.S.; methodology, N.P.N., A.M.L. and S.M.M.; investigation (anticoagulant studies), N.A.P., M.A.P., I.S.I., A.V.S., A.S.T. and V.B.S.; data curation, V.B.S. and K.S.S.; writing—original draft preparation, A.S.S. and N.P.N.; writing—review and editing, N.P.N., M.Y.K. and K.S.S. and A.G.; supervision K.S.S. and A.G. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
See the file Electronic Supplementary Material.pdf containing 1H and 13C NMR, IR and data from HPLC-MS-ESI analysis for new synthesized compounds (Figures S1–S127).
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the Russian Science Foundation, grant number 18-74-10097, https://rscf.ru/project/21-74-03011/) (accessed on 20 March 2023).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Ivasiv V., Albertini C., Gonçalves A.E., Rossi M., Bolognesi M.L. Molecular Hybridization as a Tool for Designing Multitarget Drug Candidates for Complex Diseases. Curr. Top. Med. Chem. 2019;19:1694–1711. doi: 10.2174/1568026619666190619115735. [DOI] [PubMed] [Google Scholar]
- 2.Prasher P., Sharma M., Aljabali A.A.A., Gupta G., Negi P., Kapoor D.N., Singh I., Zacconi F.C., de Jesus Andreoli Pinto T., da Silva M.W., et al. Hybrid molecules based on 1,3,5-triazine as potential therapeutics: A focused review. Drug Dev. Res. 2020;81:837–858. doi: 10.1002/ddr.21704. [DOI] [PubMed] [Google Scholar]
- 3.Auti P.S., George G., Paul A.T. Recent advances in the pharmacological diversification of quinazoline/quinazolinone hybrids. RSC Adv. 2020;10:41353–41392. doi: 10.1039/D0RA06642G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Decker M. Recent advances in the development of hybrid molecules/designed multiple compounds with antiamnesic properties. Mini Rev. Med. Chem. 2007;7:221–229. doi: 10.2174/138955707780059817. [DOI] [PubMed] [Google Scholar]
- 5.Mishra S., Singh P. Hybrid molecules: The privileged scaffolds for various pharmaceuticals. Eur. J. Med. Chem. 2016;124:500–536. doi: 10.1016/j.ejmech.2016.08.039. [DOI] [PubMed] [Google Scholar]
- 6.Oliveira R., Miranda D., Magalhães J., Capela R., Perry M.J., O’Neill P.M., Moreira R., Lopes F. From hybrid compounds to targeted drug delivery in antimalarial therapy. Bioorg. Med. Chem. 2015;23:5120–5130. doi: 10.1016/j.bmc.2015.04.017. [DOI] [PubMed] [Google Scholar]
- 7.Anusionwu C.G., Aderibigbe B.A., Mbianda X.Y. Hybrid Molecules Development: A Versatile Landscape for the Control of Antifungal Drug Resistance: A Review. Mini Rev. Med. Chem. 2019;19:450–464. doi: 10.2174/1389557519666181210162003. [DOI] [PubMed] [Google Scholar]
- 8.Decker M. Design of Hybrid Molecules for Drug Development. Elsevier; Oxford, UK: 2017. [Google Scholar]
- 9.Panda P., Chakroborty S. Navigating the Synthesis of Quinoline Hybrid Molecules as Promising Anticancer Agents. ChemistrySelect. 2020;5:10187–10199. doi: 10.1002/slct.202002790. [DOI] [Google Scholar]
- 10.Shiri P. Novel Hybrid Molecules Based on triazole-β-lactam as Potential Biological Agents. Mini Rev. Med. Chem. 2021;21:536–553. doi: 10.2174/1389557520666201027160436. [DOI] [PubMed] [Google Scholar]
- 11.Upadhyay H.C. Coumarin-1,2,3-triazole Hybrid Molecules: An Emerging Scaffold for Combating Drug Resistance. Curr. Top. Med. Chem. 2021;21:737–752. doi: 10.2174/1568026621666210303145759. [DOI] [PubMed] [Google Scholar]
- 12.Novichikhina N., Ilin I., Tashchilova A., Sulimov A., Kutov D., Ledenyova I., Krysin M., Shikhaliev K., Gantseva A., Gantseva E., et al. Synthesis, Docking, and In Vitro Anticoagulant Activity Assay of Hybrid Derivatives of Pyrrolo[3,2,1-ij]Quinolin-2(1H)-one as New Inhibitors of Factor Xa and Factor XIa. Molecules. 2020;25:1889. doi: 10.3390/molecules25081889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ilin I., Lipets E., Sulimov A., Kutov D., Shikhaliev K., Potapov A., Krysin M., Zubkov F., Sapronova L., Ataullakhanov F., et al. New factor Xa inhibitors based on 1,2,3,4-tetrahydroquinoline developed by molecular modelling. J. Mol. Graph. Model. 2019;89:215–224. doi: 10.1016/j.jmgm.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 14.Santana-Romo F., Lagos C.F., Duarte Y., Castillo F., Moglie Y., Maestro M.A., Charbe N., Zacconi F.C. Innovative Three-Step Microwave-Promoted Synthesis of N-Propargyltetrahydroquinoline and 1,2,3-Triazole Derivatives as a Potential Factor Xa (FXa) Inhibitors: Drug Design, Synthesis, and Biological Evaluation. Molecules. 2020;25:491. doi: 10.3390/molecules25030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quan M.L., Wong P.C., Wang C., Woerner F., Smallheer J.M., Barbera F.A., Bozarth J.M., Brown R.L., Harpel M.R., Luettgen J.M., et al. Tetrahydroquinoline Derivatives as Potent and Selective Factor XIa Inhibitors. J. Med. Chem. 2014;57:955–969. doi: 10.1021/jm401670x. [DOI] [PubMed] [Google Scholar]
- 16.Quan M.L., Pinto D.J.P., Smallheer J.M., Ewing W.R., Rossi K.A., Luettgen J.M., Seiffert D.A., Wexler R.R. Factor XIa Inhibitors as New Anticoagulants. J. Med. Chem. 2018;61:7425–7447. doi: 10.1021/acs.jmedchem.8b00173. [DOI] [PubMed] [Google Scholar]
- 17.Young R.J., Borthwick A.D., Brown D., Burns-Kurtis C.L., Campbell M., Chan C., Charbaut M., Chung C., Convery M.A., Kelly H.A., et al. Structure and property based design of factor Xa inhibitors: Pyrrolidin-2-ones with biaryl P4 motifs. Bioorg. Med. Chem. Lett. 2008;18:23–27. doi: 10.1016/j.bmcl.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 18.Nicolaes G.A.F., Kulharia M., Voorberg J., Kaijen P.H., Wroblewska A., Wielders S., Schrijver R., Sperandio O., Villoutreix B.O. Rational design of small molecules targeting the C2 domain of coagulation factor VIII. Blood. 2014;123:113–120. doi: 10.1182/blood-2013-05-503227. [DOI] [PubMed] [Google Scholar]
- 19.Medvedeva S.M., Potapov A.Y., Gribkova I.V., Katkova E.V., Sulimov V.B., Shikhaliev K.S. Synthesis, docking, and anticoagulant activity of new factor-Xa inhibitors in a series of pyrrolo[3,2,1-ij]quinoline-1,2-diones. Pharm. Chem. J. 2018;51:975–979. doi: 10.1007/s11094-018-1726-4. [DOI] [Google Scholar]
- 20.Tashchilova A., Podoplelova N., Sulimov A., Kutov D., Ilin I., Panteleev M., Shikhaliev K.S., Medvedeva S., Novichikhina N., Potapov A., et al. New Blood Coagulation Factor XIIa Inhibitors: Molecular Modeling, Synthesis, and Experimental Confirmation. Molecules. 2022;27:1234. doi: 10.3390/molecules27041234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sulimov V.B., Gribkova I.V., Kochugaeva M.P., Katkova E.V., Sulimov A.V., Kutov D.C., Shikhaliev K.S., Medvedeva S.M., Krysin M.Y., Sinauridze E.I., et al. Application of Molecular Modeling to Development of New Factor Xa Inhibitors. BioMed Res. Intern. 2015;2015:120802. doi: 10.1155/2015/120802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baell J.B., Holloway G.A. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem. 2010;53:2719–2740. doi: 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- 23.Baell J.B. Observations on screening-based research and some concerning trends in the literature. Future Med. Chem. 2010;2:1529–1546. doi: 10.4155/fmc.10.237. [DOI] [PubMed] [Google Scholar]
- 24.Baell J.B., Walters M.A. Chemical con artists foil drug discovery. Nature. 2014;513:481–483. doi: 10.1038/513481a. [DOI] [PubMed] [Google Scholar]
- 25.Kamar A.K.D.A., Yin L.J., Liang C.T., Fung G.T., Avupati V.R. Rhodanine scaffold: A review of antidiabetic potential and structure–activity relationships (SAR) Med. Drug Discov. 2022;15:100131–100160. doi: 10.1016/j.medidd.2022.100131. [DOI] [Google Scholar]
- 26.Tomašić T., Mašič L.P. Rhodanine as a Privileged Scaffold in Drug Discovery. Curr. Med. Chem. 2009;16:1596–1629. doi: 10.2174/092986709788186200. [DOI] [PubMed] [Google Scholar]
- 27.Kaminskyy D., Kryshchyshyn A., Lesyk R. Recent developments with rhodanine as a scaffold for drug discovery. Exp. Opinion Drug Discov. 2017;12:1233–1252. doi: 10.1080/17460441.2017.1388370. [DOI] [PubMed] [Google Scholar]
- 28.Mendgen T., Steuer C., Klein C.D. Privileged Scaffolds or Promiscuous Binders: A Comparative Study on Rhodanines and Related Heterocycles in Medicinal Chemistry. J. Med. Chem. 2012;55:743–753. doi: 10.1021/jm201243p. [DOI] [PubMed] [Google Scholar]
- 29.Maddila S., Gorle S., Jonnalagadda S.B. Drug screening of rhodanine derivatives for antibacterial activity. Exp. Opin. Drug Discov. 2020;15:203–229. doi: 10.1080/17460441.2020.1696768. [DOI] [PubMed] [Google Scholar]
- 30.Mousavi S.M., Zarei M., Hashemi S.A., Babapoor A., Amani A.M. A conceptual review of rhodanine: Current applications of antiviral drugs, anticancer and antimicrobial activities. Art. Cells Nanomed. Biotech. 2019;47:1132–1148. doi: 10.1080/21691401.2019.1573824. [DOI] [PubMed] [Google Scholar]
- 31.Verma R., Verma S.K., Rakesh K.P., Girish Y.R., Ashrafizadeh M., Kumar K.S.S., Rangappa K.S. Pyrazole-based analogs as potential antibacterial agents against methicillin-resistance staphylococcus aureus (MRSA) and its SAR elucidation. Eur. J. Med. Chem. 2021;212:113134. doi: 10.1016/j.ejmech.2020.113134. [DOI] [PubMed] [Google Scholar]
- 32.Makki M.S.T., Abdel-Rahman R.M., Alshammari N.A.H. Synthesis of Novel Fluorine Compounds Substituted-4-thiazolidinones Derived from Rhodanine Drug as Highly Bioactive Probes. Curr. Org. Synth. 2019;16:413–422. doi: 10.2174/1570179416666190312150046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trotsko N. Antitubercular properties of thiazolidin-4-ones: A review. Eur. J. Med. Chem. 2021;212:113266. doi: 10.1016/j.ejmech.2021.113266. [DOI] [PubMed] [Google Scholar]
- 34.Mohsin N.A., Ahmad M. Hybrid organic molecules as antiinflammatory agents; a review of structural features and biological activity. Turk. J. Chem. 2018;42:1–20. doi: 10.3906/kim-1706-58. [DOI] [Google Scholar]
- 35.Szczepański J., Tuszewska H., Trotsko N. Anticancer Profile of Rhodanines: Structure–Activity Relationship (SAR) and Molecular Targets—A Review. Molecules. 2022;27:3750. doi: 10.3390/molecules27123750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin L.J., Kamar A.K.D.A., Fung G.T., Liang C.T., Avupati V.R. Review of anticancer potentials and structure-activity relationships (SAR) of rhodanine derivatives. Biomed. Pharmacother. 2022;145:112406. doi: 10.1016/j.biopha.2021.112406. [DOI] [PubMed] [Google Scholar]
- 37.Khodair A.I., Awadb M.K., Gessonc J.-P., Elshaier Y.A.M.M. New N-ribosides and N-mannosides of rhodanine derivatives with anticancer activity on leukemia cell line: Design, synthesis, DFT and molecular modelling studies. Carbohydr. Res. 2020;487:107894. doi: 10.1016/j.carres.2019.107894. [DOI] [PubMed] [Google Scholar]
- 38.Liu J., Wu Y., Piao H., Zhao X., Zhang W., Wang Y., Liu M. A Comprehensive Review on the Biological and Pharmacological Activities of Rhodanine Based Compounds for Research and Development of Drugs. Mini Rev. Med. Chem. 2018;18:948–961. doi: 10.2174/1389557516666160928162724. [DOI] [PubMed] [Google Scholar]
- 39.Mermer A. The Importance of Rhodanine Scaffold in Medicinal Chemistry: A Comprehensive Overview. Mini Rev. Med. Chem. 2021;21:738–789. doi: 10.2174/1389557521666201217144954. [DOI] [PubMed] [Google Scholar]
- 40.El-Miligy M.M.M., Hazzaa A.A., El-Messmary H., Nassra R.A., El-Hawash S.A.M. New hybrid molecules combining benzothiophene or benzofuran with rhodanine as dual COX-1/2 and 5-LOX inhibitors: Synthesis, biological evaluation and docking study. Bioorg. Chem. 2017;72:102–115. doi: 10.1016/j.bioorg.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 41.Andleeb H., Tehseen Y., Shah S.J.A., Khan I., Iqbalb J., Hameeda S. Identification of novel pyrazole–rhodanine hybrid scaffolds as potent inhibitors of aldose reductase: Design, synthesis, biological evaluation and molecular docking analysis. RSC Adv. 2016;6:77688–77700. doi: 10.1039/C6RA14531K. [DOI] [Google Scholar]
- 42.Guo M., Zheng C.-J., Song M.-X., Wu Y., Sun L.-P., Li Y.-J., Liu Y., Piao H.-R. Synthesis and biological evaluation of rhodanine derivatives bearing a quinoline moiety as potent antimicrobial agents. Bioorg. Med. Chem. Lett. 2013;23:4358–4361. doi: 10.1016/j.bmcl.2013.05.082. [DOI] [PubMed] [Google Scholar]
- 43.Jiang H., Zhang W.-J., Li P.-H., Wang J., Dong S.-Z., Zhang K., Chen H.-X., Du Z.-Y. Synthesis and biological evaluation of novel carbazole-rhodanine conjugates as topoisomerase II inhibitors. Bioorg. Med. Chem. Lett. 2018;28:1320–1323. doi: 10.1016/j.bmcl.2018.03.017. [DOI] [PubMed] [Google Scholar]
- 44.Nagarajua K., Maddilaa S.N., Maddilaa S., Sobhanapuramb S., Jonnalagaddaa S.B. Synthesis and Antimicrobial Activity of Novel Thienopyrimidine Linked Rhodanine Derivatives. Can. J. Chem. 2018;97:94–99. doi: 10.1139/cjc-2018-0220. [DOI] [Google Scholar]
- 45.Kumar V., Ramu R., Shirahatti P.S., Kumari V.B.C., Sushma P., Mandal S.P., Patil S.M. α-Glucosidase, α-Amylase Inhibition, Kinetics and Docking Studies of Novel (2-Chloro-6-(trifluoromethyl)benzyloxy) arylidene) Based Rhodanine and Rhodanine Acetic Acid Derivatives. ChemistrySelect. 2021;6:9637–9644. doi: 10.1002/slct.202101954. [DOI] [Google Scholar]
- 46.Subhedar D.D., Shaikh M.H., Nawale L., Yeware A., Sarkar D., Khan F.A.K., Sangshetti J.N., Shingate B.B. Novel Tetrazoloquinoline-Rhodanine Conjugates: Highly Efficient Synthesis and Biological Evaluation. Bioorg. Med. Chem. Lett. 2016;26:2278–2283. doi: 10.1016/j.bmcl.2016.03.045. [DOI] [PubMed] [Google Scholar]
- 47.Wu Y., Ding X., Xu S., Yang Y., Zhang X., Wang C., Lei H., Zhao Y. Design and synthesis of biaryloxazolidinone derivatives containing a rhodanine or thiohydantoin moiety as novel antibacterial agents against Gram-positive bacteria. Bioorg. Med. Chem. Lett. 2019;29:496–502. doi: 10.1016/j.bmcl.2018.12.012. [DOI] [PubMed] [Google Scholar]
- 48.Yin Y., Sasaki S., Taniguchi Y. Effects of 8-halo-7-deaza-2′-deoxyguanosine triphosphate on DNA synthesis by DNA polymerases and cell proliferation. Bioorg. Med. Chem. 2016;24:3856–3861. doi: 10.1016/j.bmc.2016.06.030. [DOI] [PubMed] [Google Scholar]
- 49.Li M.L., Ren Y.J., Dong M.H., Ren W.X. Design, synthesis and structural exploration of novel fluorinated dabigatran derivatives as direct thrombin inhibitors. Eur. J. Med. Chem. 2015;96:122–138. doi: 10.1016/j.ejmech.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 50.Li X., Guo T., Feng Q., Bai T., Wu L., Liu Y., Zheng X., Jia J., Pei J., Wu S., et al. Progress of thrombus formation and research on the structure-activity relationship for antithrombotic drugs. Eur. J. Med. Chem. 2022;228:114035. doi: 10.1016/j.ejmech.2021.114035. [DOI] [PubMed] [Google Scholar]
- 51.Zimmermann M.O., Lange A., Wilcken R., Cieslik M.B., Exner T.E., Joerger A.C., Koch P., Boeckler F.M. Halogen-enriched fragment libraries as chemical probes for harnessing halogen bonding in fragment-based lead discovery. Future Med. Chem. 2014;6:617–639. doi: 10.4155/fmc.14.20. [DOI] [PubMed] [Google Scholar]
- 52.Lescheva E.V., Medvedeva S.M., Shikhaliev K.S. Synthesis of 4,4,6-trimethyl-8-R-4H-pyrrolo[3,2,1-ij]quinoline-1,2-diones. J. Org. Pharm. Chem. 2014;12:15–20. doi: 10.24959/ophcj.14.798. [DOI] [Google Scholar]
- 53.Medvedeva S.M., Krysin M.Y., Zubkov F.I., Nikitina E.V., Shikhaliev K.S. New Heterocyclic Systems Based on Substituted 3,4-Dihydro-1H-Spiro[Quinoline-2,1′-Cycloalkanes] Chem. Heterocycl. Compd. 2014;50:1280–1290. doi: 10.1007/s10593-014-1590-4. [DOI] [Google Scholar]
- 54.Kojima E., Fujimori S., Awano K. Cyclic Anthranilic Acid Derivatives and Process for Their Preparation. No. US4956372A. US Patent. 1992 September 11;
- 55.Hsu Y.P., Hall E., Booher G., Murphy B., Radkov A.D., Yablonowski J., Mulcahey C., Alvarez L., Cava F., Brun Y.V., et al. Fluorogenic D-amino acids enable real-time monitoring of peptidoglycan biosynthesis and high-throughput transpeptidation assays. Nat. Chem. 2019;11:335–341. doi: 10.1038/s41557-019-0217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sulimov A.V., Kutov D.C., Oferkin I.V., Katkova E.V., Sulimov V.B. Application of the docking program SOL for CSAR benchmark. J. Chem. Inf. Model. 2013;53:1946–1956. doi: 10.1021/ci400094h. [DOI] [PubMed] [Google Scholar]
- 57.Sulimov V.B., Ilin I.S., Kutov D.C., Sulimov A.V. Development of docking programs for Lomonosov supercomputer. J. Turkish Chem. Soc. Sect. A Chem. 2020;7:259–276. doi: 10.18596/jotcsa.634130. [DOI] [Google Scholar]
- 58.Ilin I., Podoplelova N., Sulimov A., Kutov D., Tashchilova A., Panteleev M., Shikhaliev K., Krysin M., Stolpovskaya N., Potapov A., et al. Experimentally Validated Novel Factor XIIa Inhibitors Identified by Docking and Quantum Chemical Post-Processing. Mol. Inform. 2022;41:2200205. doi: 10.1002/minf.202200205. [DOI] [PubMed] [Google Scholar]
- 59.Kutov D.C., Katkova E.V., Sulimov A.V., Kondakova O.A., Sulimov V.B. Influence of the method of hydrogen atoms incorporation into the target protein on the protein-ligand binding energy. Bull. SUSU MMCS. 2017;10:94–107. doi: 10.14529/mmp170308. [DOI] [Google Scholar]
- 60.Hanwell M.D., Curtis D.E., Lonie D.C., Vandermeersch T., Zurek E., Hutchison G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012;4:17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wong P.C., Quan M.L., Watson C.A., Crain E.J., Harpel M.R., Rendina A.R., Luettgen J.M., Wexler R.R., Schumacher W.A., Seiffert D.A. In vitro, antithrombotic and bleeding time studies of BMS-654457, a small-molecule, reversible and direct inhibitor of factor Xia. J. Thromb. Thrombolysis. 2015;40:416–423. doi: 10.1007/s11239-015-1258-7. [DOI] [PubMed] [Google Scholar]
- 62.Pinto D.J.P., Orwat M.J., Smith L.M., Quan M.L., Lam P.Y.S., Rossi K.A., Apedo A., Bozarth J.M., Wu Y., Zheng J.J., et al. Discovery of a Parenteral Small Molecule Coagulation Factor XIa Inhibitor Clinical Candidate (BMS-962212) J. Med. Chem. 2017;60:9703–9723. doi: 10.1021/acs.jmedchem.7b01171. [DOI] [PubMed] [Google Scholar]
- 63.Fjellstrom O., Akkaya S., Beisel H.-G., Eriksson P.-O., Erixon K., Gustafsson D., Jurva U., Kang D., Karis D., Knecht W., et al. Creating novel activated factor XI inhibitors through fragment based lead generation and structure aided drug design. PLoS ONE. 2015;10:e0113705. doi: 10.1371/journal.pone.0113705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu Z., Wang C., Han W., Rossi K.A., Bozarth J.M., Wu Y., Sheriff S., Myers J.E., Jr., Luettgen J.M., Seiffert D.A., et al. Pyridazine and pyridazinone derivatives as potent and selective factor XIa inhibitors. Bioorg. Med. Chem. Lett. 2018;28:987–992. doi: 10.1016/j.bmcl.2018.02.049. [DOI] [PubMed] [Google Scholar]
- 65.Corte J.R., Fang T., Hangeland J.J., Friends T.J., Rendina A.R., Luettgen J.M., Bozarth J.M., Barbera F.A., Rossi K.A., Wei A., et al. Pyridine and pyridinone-based factor XIa inhibitors. Bioorg. Med. Chem. Lett. 2015;25:925–930. doi: 10.1016/j.bmcl.2014.12.050. [DOI] [PubMed] [Google Scholar]
- 66.Corte J.R., Fang T., Pinto D.J.P., Orwat M.J., Rendina A.R., Luettgen J.M., Rossi K.A., Wei A., Ramamurthy V., Myers J.E., et al. Orally bioavailable pyridine and pyrimidine-based Factor XIa inhibitors: Discovery of the methyl N-phenyl carbamate P2 prime group. Bioorg. Med. Chem. 2016;24:2257–2272. doi: 10.1016/j.bmc.2016.03.062. [DOI] [PubMed] [Google Scholar]
- 67.Podoplelova N.A., Sulimov V.B., Tashchilova A.S., Ilin I.S., Panteleev M.A., Ledeneva I.V., Shikhaliev K.S. Blood coagulation in the 21st cetrury: Existing knowledge, current strategies for treatment and perspective. Pediatr. Hematol. Immunopathol. 2020;19:139–157. (In Russian) [Google Scholar]
- 68.Voevodin V.V., Antonov A.S., Nikitenko D.A., Shvets P.A., Sobolev S.I., Sidorov I.Y., Stefanov K.S., Voevodin V.V., Zhumatiy S.A. Supercomputer Lomonosov-2: Large scale, deep monitoring and fine analytics for the user community. Supercomput. Front. Innov. 2019;6:4–11. doi: 10.14529/jsfi190201. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
See the file Electronic Supplementary Material.pdf containing 1H and 13C NMR, IR and data from HPLC-MS-ESI analysis for new synthesized compounds (Figures S1–S127).