Abstract
Burkholderia pseudomallei, a facultative intracellular bacterium, is the causative agent of a broad spectrum of diseases collectively known as melioidosis. Its ability to survive inside phagocytic and nonphagocytic cells and to induce multinucleated giant cell (MNGC) formation has been demonstrated. This study was designed to assess a possible mechanism(s) leading to this cellular change, using virulent and nonvirulent strains of B. pseudomallei to infect both phagocytic and nonphagocytic cell lines. We demonstrated that when the cells were labeled with two different cell markers (CMFDA or CMTMR), mixed, and then infected with B. pseudomallei, direct cell-to-cell fusion could be observed, leading to MNGC formation. Staining of the infected cells with rhodamine-conjugated phalloidin indicated that immediately after the infection, actin rearrangement into a comet tail appearance occurred, similar to that described earlier for other bacteria. The latter rearrangement led to the formation of bacterium-containing, actin-associated membrane protrusions which could lead to a direct cell-to-cell spreading of B. pseudomallei in the infected hosts. Results from 4′,6′-diamidine-2-phenylindole dihydrochloride (DAPI) nuclear staining, poly-ADP ribose polymerase cleavage, staining of infected cells for phosphatidylserine exposure with annexin V, and electrophoresis of the DNA extracted from these infected cells showed that B. pseudomallei could kill the host cells by inducing apoptosis in both phagocytic and nonphagocytic cells.
Burkholderia pseudomallei is the causative agent of a broad spectrum of clinical manifestations collectively known as melioidosis (3, 11). Melioidosis affects humans and animals in tropical and subtropical areas and is particularly prevalent in Southeast Asian countries and northern Australia. It is a potentially fatal disease which may account for up to 40% of deaths from community-acquired septicemia in Thailand (4). One striking characteristic of the infection caused by B. pseudomallei is its dormancy state following initial subclinical infection or relapse after recovery from clinical disease (3). Infective organisms that are in the dormant state in hosts can be triggered, leading to acute and fatal disease, particularly when the immune response is depressed (3).
Different lines of evidence currently available suggest that B. pseudomallei behaves as a facultative intracellular organism. The bacilli can readily attach and multiply in the cells from infected humans and a number of naturally and experimentally infected animals, as well as (in vitro) in different phagocytic and nonphagocytic cell lines (1, 8, 9, 15, 16, 20). Several groups of investigators have reported that, after internalization, the organisms can escape from a membrane-bound phagosome into the cytoplasm (8, 9, 15). The presence of multinucleated giant cells (MNGCs) has been observed in the tissues of patients with melioidosis (25). We first demonstrated the presence of foci of MNGCs in different cultured cell lines including macrophage, epithelial, and fibroblast cell lines (W. Kespichayawattana, T. Wanun, and S. Sirisinha, Int. Congr. Melioidosis, abstr. P412, 1998). This phenomenon was subsequently confirmed and extended recently by Harley and associates (8). These investigators presented indirect evidence suggesting that the MNGC formation might be associated with cell fusion. In the present study, we provide direct evidence showing that B. pseudomallei can in fact induce cell fusion leading to MNGC formation and actin-associated membrane protrusion in both phagocytic and nonphagocytic cell lines, both of which could contribute to cell-to-cell spreading in infected hosts.
MATERIALS AND METHODS
Bacterial strains.
B. pseudomallei strains 844, UE12, UE16, UE30, and 824a were originally isolated from melioidosis patients and were of an arabinose-negative (Ara−) biotype (17, 18). Strains UE16, UE30, and 824a, however, exhibited atypical lipopolysaccharide patterns in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis analysis (2). Another strain of the Ara− biotype used (UE3) was originally isolated from a soil sample in Thailand. B. pseudomallei strains UE5, UE8, and UE11 were also isolated from soil samples in Thailand, but all were classified as a nonvirulent Ara+ biotype (18). Salmonella enterica serovar Typhi, a prototype of intracellular bacteria used for comparison, was isolated from a patient admitted at Ramathibodi Hospital (Mahidol University, Bangkok, Thailand). Escherichia coli HB101, representing noninvasive extracellular bacteria, was also used for comparative study in some experiments. All bacterial strains were routinely subcultured from stocks kept at −70°C in 20% glycerol. For use in the experiments, they were cultured in Trypticase soy broth at 37°C with shaking at 120 rpm. The overnight cultures were washed twice in phosphate-buffered saline (PBS) and adjusted to a desired concentration by measurement of the optical density at 650 nm.
Cell lines.
Murine macrophage (J774A.1 [ATCC TIB-67]), human epithelium (HeLa [ATCC CCL-2]), and mouse fibroblast (L929) cell lines were used in this study. Unless indicated otherwise, the cells were cultured and maintained in appropriate cell culture media supplemented with 10% heat-inactivated fetal bovine serum (HyClone, Logan, Utah) and 2 mM l-glutamine (Sigma Chemical Co., St. Louis, Mo.). Dulbecco's modified Eagle medium (DMEM; Gibco BRL, Grand Island, N.Y.) was used for the J774A.1 and HeLa cell lines, while RPMI 1640 (Gibco BRL) was used for the L929 cell line. Throughout the study, the cells were incubated at 37°C in a humidified incubator in the presence of 5% CO2.
Internalization of B. pseudomallei by cultured cell lines.
Infection of the cells by B. pseudomallei, E. coli, and S. enterica serovar Typhi was performed essentially as described previously (9). In brief, cells of the J774A.1 and HeLa lines (seeded at 5 × 105 cells per well) in 24-well plates were incubated overnight at 37°C with 5% CO2. On the day of infection, the cultured medium was removed and replaced with fresh medium, and the bacteria were added to each well to give a multiplicity of infection (MOI) of approximately two bacteria per cell. After 2 h at 37°C, the cells were washed with prewarmed PBS, the cultured medium containing 250 μg of kanamycin (Gibco BRL) per ml was added, and the cell culture was incubated for another 2 h to completely eliminate residual extracellular bacteria. Thereafter, the cell monolayer was washed and lysed with 0.1% Triton X-100 (Sigma Chemical Co.). Intracellular bacteria that were liberated were quantitated by dilution and plating on Trypticase soy agar. The numbers of bacterial colonies were counted after 36 to 48 h of incubation.
Intracellular survival and multiplication of B. pseudomallei in cultured cell lines.
The J774A.1 and HeLa cells were infected and treated as described above for the internalization experiment. After killing of the extracellular bacteria with kanamycin (250 μg/ml), the cells were washed and incubated in the culture medium containing 20 μg of kanamycin per ml to inhibit the growth of residual extracellular bacteria. The incubation periods were 6 and 8 h for J774A.1 cells and 12 and 24 h for HeLa cells before the numbers of intracellular bacteria were determined as described above.
Giemsa staining of B. pseudomallei-infected cell lines.
Cells were seeded and grown overnight on glass coverslips. At different intervals after infection with B. pseudomallei, the coverslips were washed with PBS, fixed for 15 min with 1% paraformaldehyde, and then washed with 50 and 90% ethanol for 5 min each. The coverslips were air dried before staining with the Giemsa stain. For evaluation of MNGC formation, at least 1,000 nuclei per coverslip were counted, and the percent MNGC formation was calculated as follows: (number of nuclei within multinucleated cells/total number of nuclei counted) × 100.
Plaque assay.
Burkholderia-induced plaque formation and assay were performed essentially as described earlier for Shigella (14) with the exception that the process was carried out in the presence of kanamycin instead of gentamicin. Briefly, the HeLa cell monolayers were infected with either B. pseudomallei strain 844 or UE5 at an MOI of 1:10 for 2 h in the absence of any antibiotics. The infected cell monolayers were washed 3 times with PBS before a 0.5% agarose overlay consisting of DMEM, 250 μg of kanamycin per ml, and 4.5 mg of d-glucose per ml was added. The plates were incubated at 37°C in a humidified 5% CO2 atmosphere for 24 h. To enhance visualization of the plaques, another similar agarose overlay containing in addition 0.01% neutral red was added, and the plaques were observed 4 h later.
Cell fusion assay.
Confluent cell monolayers were harvested, washed with PBS, and separated into two tubes for staining with CellTracker Green CMFDA (Molecular Probes, Eugene, Oreg.) or CellTracker Orange CMTMR (Molecular Probes). Labeling of the cells was performed as described by the manufacturer. Briefly, the cell suspensions were incubated with the dyes at a concentration of 5 μM for J774A.1 cells or 25 μM for HeLa and L929 cells. After 15 min of incubation at 37°C in a water bath, the cells were pelleted, and the remaining dyes were discarded. The culture medium, supplemented with 10% fetal bovine serum and 2 mM l-glutamine, was added to the cells, and the mixture was incubated at 37°C for an additional 30 min to complete the labeling process. The labeled cells were washed twice with large volumes of PBS, counted, and adjusted to the desired concentration. Equal numbers of CMFDA-labeled and CMTMR-labeled cells were mixed and plated in a six-well plate containing a 22- by 22-mm glass coverslip. After an overnight incubation, the mixed-cell cocultures were infected with B. pseudomallei at an MOI of approximately 50:1 as described above. After different intervals of incubation, the cells were washed and fixed with 3.7% formaldehyde in PBS and observed with a fluorescence microscope equipped with a dual-wavelength filter.
Fluorescence staining of actin and bacteria.
Cells were cultured on 22- by 22-mm glass coverslips seeded in the six-well plates. After an overnight incubation, the cells were infected with B. pseudomallei at an MOI of approximately 50:1. After 2 h of incubation, the extracellular bacteria were washed away, and fresh culture medium containing kanamycin (250 μg/ml) was added. This mixture was incubated further until the experiment was performed. At that time, the cells were washed with PBS, fixed with 3.7% formaldehyde in PBS for 15 min, and then permeabilized by a 5-min treatment with 0.1 or 1% Triton X-100 in PBS. To minimize nonspecific binding, the cells were blocked for 30 min with 1% bovine serum albumin (Sigma Chemical Co.) before proceeding further. To stain intracellular bacteria, the permeabilized infected cells were allowed to react with a precalibrated dilution of either mouse monoclonal antibodies (17) (for staining the Ara− biotype) or rabbit polyclonal antibodies raised against B. pseudomallei (for staining both biotypes). Rhodamine-conjugated phalloidin (Molecular Probes) was simultaneously added to these cells to stain actin fibers (1 U per microscopic slide as recommended by the manufacturer). The slides were washed with PBS (containing 1% bovine serum albumin) before adding fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin (Ig) (DAKO, Glostrup, Denmark) or fluorescein isothiocyanate-conjugated goat anti-rabbit IgG (Zymed Laboratories, Inc., San Francisco, Calif.) at the dilutions recommended by the manufacturers. The coverslips were finally washed three times before they were examined for the presence of actin-associated bacteria under a fluorescence microscope equipped with a dual-wavelength filter.
Assessment of apoptosis.
Cells were seeded at 1 × 106 cells per well for HeLa and L929 cells or 2 × 106 for J774A.1 cells in six-well plates and incubated for 18 to 20 h prior to infection. After 2 h of infection with B. pseudomallei at an MOI of approximately 50:1, the cells were washed and further incubated in the presence of 250 μg of kanamycin per ml. At different time intervals, the cells were removed and lysed in buffer (10 mM Tris-HCl, pH 8.0–100 mM NaCl–0.5% SDS–35 mM EDTA) and then treated with proteinase K (0.1 mg/ml) at 50°C overnight. Protein was removed by extraction with phenol-chloroform-isoamyl alcohol (25:24:1). Nucleic acids were then precipitated by the addition of ethanol and centrifuged at 9,500 × g for 30 min. The pellets were air dried and resuspended in TE buffer (10 mM Tris-HCl, pH 8.0–1 mM EDTA). The DNA solution was incubated at 37°C for 1 h in the presence of RNase (0.1 mg/ml) before it was subjected to electrophoresis in 1.8% agarose gel (10). The gel was then stained with ethidium bromide, and the DNA ladders were viewed under a UV light.
In some experiments, the cells were seeded and infected as described earlier, on glass coverslips in a six-well plate, and at different intervals, the cells were washed, fixed with 3.7% formaldehyde, and stained with 4′,6′-diamidine-2′-phenylindole dihydrochloride (DAPI) at 1 μg/ml, for the observation of nuclear morphology. The proportions of condensed and fragmented apoptotic nuclei were calculated from counting a total of 1,000 nuclei. In other experiments, the translocation of phosphatidylserine (PS) from the inner side to the external surface of B. pseudomallei-infected cells was detected by staining the cells with fluorescein-labeled annexin V and analyzing FITC-positive viable cells by flow cytometry (22). Viability of the cells was determined by the exclusion of propidium iodide (i.e., only PI− cells were counted).
In order to elucidate the possible mechanism of apoptotic cell death induced by B. pseudomallei, a cleavage of poly-ADP ribose polymerase (PARP) was determined as described previously (21). Briefly, the J774A.1 cell monolayers were infected with B. pseudomallei at an MOI of 100:1. After 30 min, extracellular bacteria were removed by washing 3 times with PBS. The infected cells were then reincubated in DMEM containing 250 μg of kanamycin per ml and then harvested 1, 2, and 3 h later by lysing in lysis buffer (containing 62.5 mM Tris [pH 6.8], 6 M urea, 10% glycerol, 2% SDS, 0.003% bromphenol blue, and 5% 2-mercaptoethanol). Twenty microliters of the lysates was electrophoresed on a 0.1% SDS–10% polyacrylamide gel and electrotransferred to a polyvinylidene difluoride membrane. The membrane was blocked in 5% skimmed milk for 1 h before reacting with antibody to PARP (anti-cII-10; Centre Hospitalier De l'Universite, Laval, Quebec, Canada). The reaction was detected with horseradish peroxidase-conjugated rabbit anti-mouse IgG using the enhanced chemiluminescence method as recommended by the manufacturer (Pierce, Rockford, Ill.).
RESULTS
Bacterial internalization and intracellular multiplication.
In this experiment, one phagocytic cell line (J774A.1) and one nonphagocytic cell line (HeLa) were exposed to several strains of virulent Ara− and nonvirulent Ara+ B. pseudomallei at an MOI of 2:1, and the number of intracellular bacteria was determined 4 h after exposure. Similar tests were conducted with S. enterica Typhi serving as a virulent, invasive control and E. coli as a noninvasive control. The results presented in Table 1 show that all six isolates of B. pseudomallei tested could be readily phagocytosed by the macrophage cells (J774A.1). At 4 h, the percent internalization of B. pseudomallei in J774A.1 cells infected with different isolates was slightly below that of the S. enterica serovar Typhi (Table 1), both of which were, however, noticeably higher than that of E. coli. Similar to S. enterica serovar Typhi, B. pseudomallei could also invade cells of the nonphagocytic HeLa line. However, on average, the number of intracellular B. pseudomallei recovered after 4 h was 3 to 4 orders of magnitude lower than that of S. enterica serovar Typhi. At the same time, the number of E. coli organisms found inside the HeLa cells was 1 to 2 orders of magnitude below that of B. pseudomallei. Data in Table 2 show that these B. pseudomallei isolates could not only survive but also multiply inside phagocytic and nonphagocytic cells, at a rate roughly comparable to that in the broth culture (unpublished observations). It should also be mentioned that B. pseudomallei could also invade and multiply in the L929 cells at a rate similar to that of the HeLa cells.
TABLE 1.
Internalization of bacteria into cultured cell linesa
| Bacterium | Host cell line | Inoculum size (CFU) | No. of intracellular bacteria (CFU)b | % Internalization |
|---|---|---|---|---|
| B. pseudomallei | ||||
| 844 | J774A.1 | 0.78 × 106 | (3.85 ± 0.26) × 105 | 49.36 |
| HeLa | 0.49 × 106 | (0.23 ± 0.04) × 102 | 0.0047 | |
| UE3 | J774A.1 | 1.27 × 106 | (4.57 ± 0.40) × 105 | 35.98 |
| HeLa | 2.12 × 106 | (3.95 ± 0.77) × 102 | 0.0186 | |
| UE5 | J774A.1 | 1.11 × 106 | (2.32 ± 0.14) × 105 | 20.92 |
| HeLa | 1.02 × 106 | (0.28 ± 0.09) × 102 | 0.0027 | |
| UE8 | J774A.1 | 1.02 × 106 | (3.73 ± 0.27) × 105 | 36.57 |
| HeLa | 1.62 × 106 | (1.83 ± 0.46) × 102 | 0.0113 | |
| UE16 | J774A.1 | 0.82 × 106 | (1.68 ± 0.47) × 105 | 20.49 |
| HeLa | 1.28 × 106 | (1.73 ± 0.38) × 102 | 0.0135 | |
| UE30 | J774A.1 | 1.01 × 106 | (4.25 ± 0.23) × 105 | 42.08 |
| HeLa | 1.53 × 106 | (1.35 ± 0.17) × 102 | 0.0088 | |
| S. enterica serovar Typhi | J774A.1 | 1.23 × 106 | (6.45 ± 0.96) × 105 | 52.44 |
| HeLa | 2.22 × 106 | (1.79 ± 0.06) × 105 | 8.06 | |
| E. coli HB101 | J774A.1 | 2.03 × 106 | (0.12 ± 0.01) × 105 | 0.59 |
| HeLa | 2.60 × 106 | (0.20 ± 0.20) × 101 | 0.0001 |
Inocula were added, and the mixtures were incubated for 4 h.
No. of CFU of liberated intracellular bacteria ± standard error of the mean from triplicate wells.
TABLE 2.
Intracellular survival and multiplication of B. pseudomallei strains
| Time (h)b | Survival in cell linea
|
|||||
|---|---|---|---|---|---|---|
| J774A.1
|
HeLa
|
|||||
| 844 | UE16 | UE5 | 844 | UE16 | UE5 | |
| 4 | (5.82 ± 1.76) × 104 | (3.82 ± 0.18) × 105 | (1.30 ± 0.30) × 104 | (0.02 ± 0.02) × 102 | (0.08 ± 0.02) × 102 | (0.02 ± 0.02) × 102 |
| 6 | (7.28 ± 2.22) × 105 | (5.62 ± 0.52) × 105 | (2.86 ± 1.20) × 105 | ND | ND | ND |
| 8 | (4.68 ± 1.08) × 106 | (1.24 ± 0.12) × 106 | (1.86 ± 0.02) × 106 | ND | ND | ND |
| 12 | ND | ND | ND | (1.20 ± 0.95) × 102 | (1.70 ± 1.00) × 102 | (0.85 ± 0.20) × 102 |
| 24 | ND | ND | ND | (5.11 ± 2.39) × 104 | (2.51 ± 2.34) × 105 | (3.97 ± 1.92) × 104 |
Results are mean CFU of intracellular bacteria ± SEM from duplicate wells. ND, not done.
Hours after inoculation of bacteria into host cells.
B. pseudomallei-induced plaque formation.
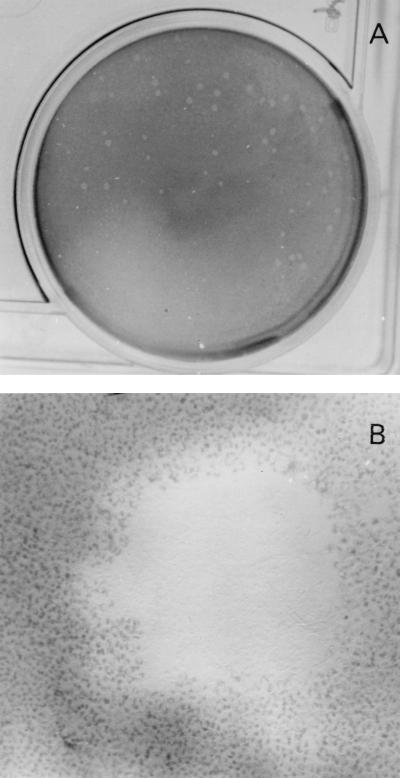
To determine if B. pseudomallei could spread directly from cell to cell, a bacterial plaque assay was performed using cells of the nonphagocytic HeLa line. The results, presented in Fig. 1, included B. pseudomallei plaques with an average diameter of about 1.0 mm at 24 h after the infection. At a higher magnification, cells at the periphery were found to harbor large numbers of bacteria. There was no visible plaque formation when the plates were shifted from 37 to 4°C after the initial absorption period. From a limited number of isolates tested, it appeared that the virulent B. pseudomallei were more efficient than the nonvirulent strains in plaque induction.
FIG. 1.
Plaque formation in HeLa cells by B. pseudomallei. Burkholderia plaques occurred in the presence of kanamycin at 24 h after infection. The cell monolayer was stained with neutral red to enhance visibility. Lysis of cells in the center of the plaque could be readily observed (B [magnification, ×40]), leaving at times a considerable amount of visible debris. Cells in the periphery contained a large number of intracellular bacteria.
B. pseudomallei-induced cell fusion and MNGC formation.
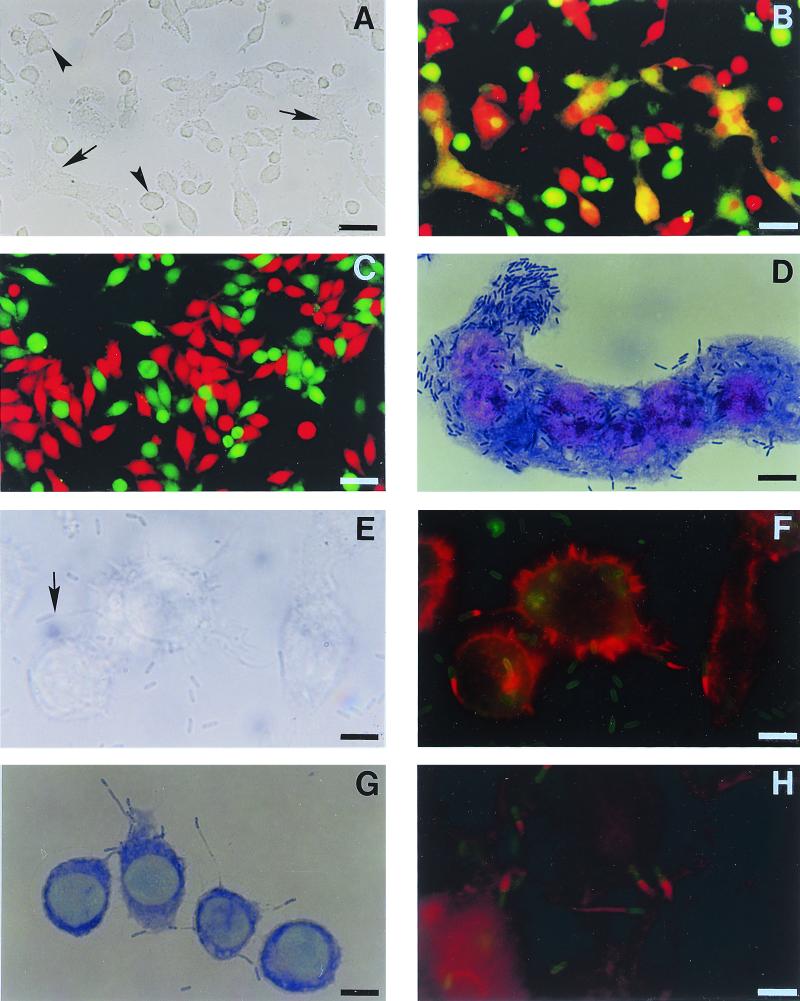
We demonstrate here for the first time (Fig. 2 A through C) that the MNGCs were formed as a result of direct cell-to-cell fusion. As shown in Fig. 2, when the J774A.1 cells were labeled separately with CMTMR (red) and CMFDA (green), mixed together, and then cocultured before the addition of B. pseudomallei, the orange MNGC (Fig. 2B) could be readily observed within 4 to 6 h after the infection. This indicated that a fusion between CMTMR-labeled cells and CMFDA-labeled cells had occurred. In the same field, MNGCs (Fig. 2A and B) resulting from fusion of the like labeling cells could be readily observed. As is to be expected, no MNGC formation could be found in the labeled cell coculture in the absence of B. pseudomallei (Fig. 2C). Similar results were obtained when the experiment was carried out with HeLa and L929 cells. The difference between the phagocytic and nonphagocytic cells was the rate of MNGC formation, which was considerably lower in the HeLa and L929 cells (data not presented). The latter finding is consistent with the presence of the lower number of intracellular bacteria in these two cell types compared with the J774A.1 cells.
FIG. 2.
Morphological changes of cells J774A.1 (A through G) and HeLa (H) cells infected with B. pseudomallei. The J774A.1 cells were separately labeled with CMTMR (red) and CMFDA (green) cell markers, mixed, and then cocultured in the presence (A and B) or absence (C) of B. pseudomallei. Cell fusion was observed 6 h later under phase-contrast (A) or fluorescence (B and C) microscopes. Fusion of the differently labeled cells, appearing as orange-staining cells (arrow), could be readily observed (B) in the presence of B. pseudomallei. In the same field (A and B), fusion of the same colored labeling cells can also be seen (arrowhead). In the absence of B. pseudomallei (C), no fusion occurred. An MNGC loaded with numerous bacilli (as indicated by Giemsa stain) could be readily observed at 6 h (D). Phase-contrast (E) and fluorescence (F) photomicrographs demonstrate the presence of actin-based peripheral membrane protrusions (arrow) that occurred 4 h after the infection. The actin tail (red) attached to one pole of the bacterium (green) can be readily observed (F). Contact of the bacterium located at the tip of each protrusion with adjacent cells (as shown by Giemsa stain) is shown in panel G. Similar membrane protrusions with typical actin tails were also noted in nonphagocytic cells (H). Bars = 50 μm (A through C) and 10 μm (D through H).
Both virulent (Ara−) and nonvirulent (Ara+) biotypes could induce cell fusion and MNGC formation in all three cell lines. However, the data presented in Table 3 show that the MNGC formation in cells of the J774A.1 line could be induced at a faster rate by the virulent strain (strain 844). Four hours after infection with the virulent Ara− isolate, the MNGCs could be readily observed, and the number gradually increased to reach a peak at around 7 to 8 h, when the experiment was terminated. With the nonvirulent strain (strain UE5), a negligible number of the infected cells participated in the MNGC formation at 4 h. The results shown by phase-contrast photomicrographs (Fig. 3) also gave the impression that the cellular damage caused by the virulent strain was more extensive 6 h after the infection was initiated. The quantitative data on the number of MNGC (Table 4) and on plaque formation are consistent with this conclusion. However, at the end of the experiment, both biotypes gave essentially similar degrees of MNGC formation.
TABLE 3.
MNGC formation in J774A.1 cells infected with B. pseudomallei
| Time (h) after infection | % MNGC formationa with B. pseudomallei strains
|
|
|---|---|---|
| 844 | UE5 | |
| 4 | 1.79 | 0.66 |
| 5 | 8.86 | 2.95 |
| 6 | 32.23 | 9.19 |
| 7 | 74.58 | 27.07 |
| 8 | 73.61 | 68.61 |
Calculated as follows (number of nuclei within multinucleated giant cells/total number counted) × 100.
FIG. 3.
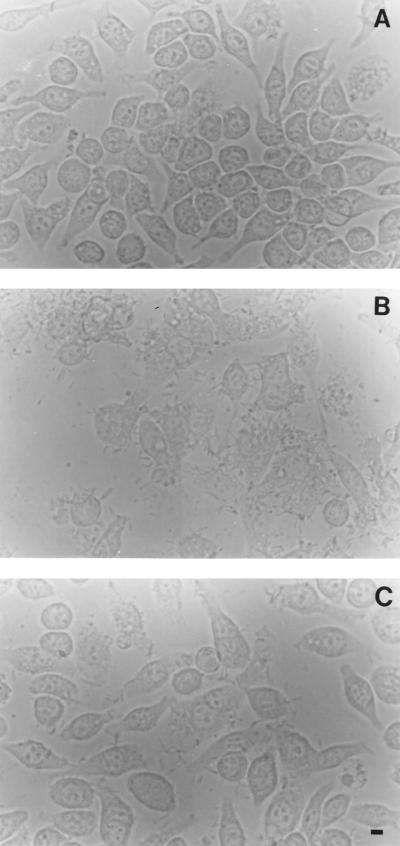
Destruction of J774A.1 cells infected with Ara− and Ara+ B. pseudomallei. The cell monolayers (A) were infected with virulent Ara− (B) or nonvirulent Ara+ (C) B. pseudomallei for 6 h and then observed under a phase-contrast microscope for MNGC formation and cell destruction. A more extensive morphological change can be readily observed with the virulent organisms (compare panel B with panel C). Bar = 10 μm.
TABLE 4.
MNGC formationa
| B. pseudomallei group (strain) | % MNGC formationb |
|---|---|
| Virulent (844) | 43 ± 6.15 |
| Virulent with atypical LPSc (UE16) | 62 ± 5.07 |
| Nonvirulent (UE5) | 11.69 ± 2.06 |
| Nonvirulent (UE8) | 12.01 ± 0.97 |
| Nonvirulent (UE11) | 20.86 ± 5.71 |
Percentages were determined 6 h after infection of cells of the J774A.1 line with indicated strains of B. pseudomallei.
Data are means ± SEMs from at least two independent experiments.
LPS, lipopolysaccharide.
B. pseudomallei-induced cellular actin rearrangement and membrane protrusion.
Photomicrographs presented in Fig. 2E through H clearly demonstrate that B. pseudomallei could induce the formation of peripheral membrane protrusions in both phagocytic (J774A.1) and nonphagocytic (HeLa) cells. Many of these bacterium-containing protrusions extended to and some of them eventually touched the neighboring cells (Fig. 2G) and, at times, appeared to be pushing the latter, like the protrusions described previously for other bacteria, e.g., Listeria monocytogenes and Shigella flexneri (5–7, 19, 26, 27). When these B. pseudomallei-infected cells were stained with rhodamine-conjugated phalloidin for actin fibers, actin rearrangement in a “comet” tail formation (12) could be readily observed (Fig. 2F and H). Actin rearrangement occurring at only one polar end of the bacilli could be noted at 4 h of infection when the observation was made. Because our laboratory is not equipped to take video pictures to observe intracellular motility of B. pseudomallei, we could not state with certainty whether such an association existed. However, with careful observation at different time points, a movement of bacterium-containing protrusions could be noted occasionally.
Induction of apoptosis.
Although at the early stage of infection, when a majority of the infected cells including the MNGCs were still viable as shown by exclusion in the trypan blue dye test, evidence suggesting that these cells were undergoing apoptotic death could already be noted. For example, 4 h after the infection of J774A.1 cells with B. pseudomallei, DAPI nuclear staining showed the presence of many cells with condensed and fragmented nuclei typical of apoptotic cells (Fig. 4). Depending on the experimental conditions, the proportion of cells with apoptotic nuclei gradually increased from an average of 3% at 2 h to 43% at 6 h when the experiment was carried out with the virulent strain. For the nonvirulent strain, these proportions were 1 and 23%, respectively. These nuclear changes could be readily observed in both single and unfused nucleated cells (Fig. 4A) and MNGCs (Fig. 4B). At times, both normal and abnormal appearing nuclei could also be seen in the same MNGCs. As is to be expected from the previous experiments, this phenomenon could also be observed in nonphagocytic cells, although at a rate lower than in the phagocytic cells. Consistent with these nuclear changes, the plasma membrane of these B. pseudomallei-infected cells was also altered after the infection. The limited data presented in Table 5 show that the percentage of J774A.1 cells that stained positively with annexin V, a marker for PS, gradually increased with the time of infection. The data presented again showed that virulent strain 844 induced a more drastic change than nonvirulent strain UE5.
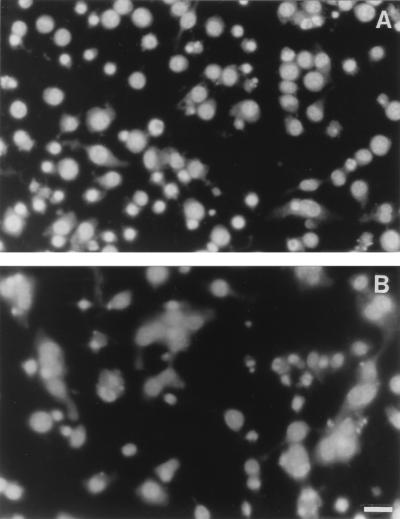
FIG. 4.
Apoptosis of J774A.1 cells infected with B. pseudomallei. The cell monolayer was fixed, and the nuclei were stained with DAPI 4 h (A) and 6 h (B) after the infection. Condensed and fragmented nuclei typical of apoptotic cell death could be readily observed as early as 4 h, when most of the cells were still viable and only a small number of MNGCs had formed. Six hours after the infection, a large number of MNGCs could be readily observed; normal and apoptotic nuclei can appear together within the same MNGC (B). Bar = 50 μm.
TABLE 5.
Percentages of annexin V-positive J774A.1 cells infected with B. pseudomallei
| Time (h) after infection | Uninfected | % of J774A.1 cells infected with B. pseudomallei straina:
|
|
|---|---|---|---|
| 844 | UE5 | ||
| 2 | ND | 1.42 | 2.57 |
| 4 | 2.25 | 4.01 | 2.07 |
| 6 | 0.60 | 18.39 | 5.05 |
Percentage of FITC+ PI− cells by flow cytometric analysis. Only PI− cells (viable cells) were counted. ND, not done.
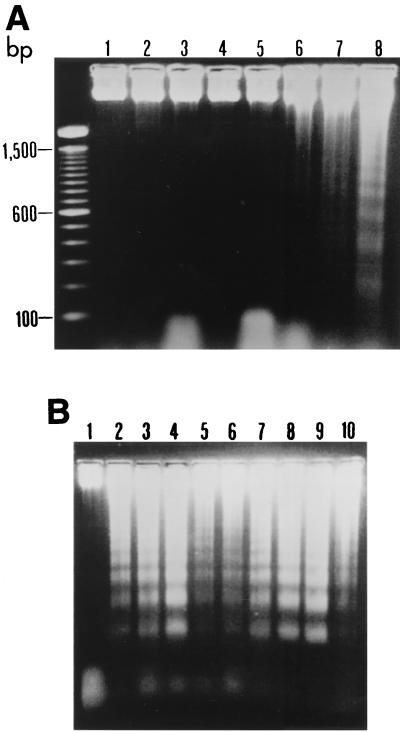
Results presented in Fig. 5 clearly demonstrate that B. pseudomallei could readily induce DNA breakage, as shown by a DNA ladder formation from 18 h of infection onward. All nine strains of B. pseudomallei tested (six virulent and three nonvirulent) could readily induce this change. However, one of the six virulent strains (strain 824a) and two of the three nonvirulent strains (UE5 and UE8) appeared to cause less extensive damage. The difference could not be explained based on the lower number of B. pseudomallei used, as the experiment was carried out using the same number of bacteria.
FIG. 5.
DNA fragmentation of HeLa cells infected with B. pseudomallei. (A) The cells were infected with a virulent strain of B. pseudomallei (strain 844) and, at the indicated intervals (lanes: 3, 12 h; 4, 14 h; 5, 16 h; 6, 18 h; 7, 20 h; and 8, 24 h), the cells were removed, and the DNA was extracted, subjected to electrophoresis in 1.8% agarose, and stained with ethidium bromide. DNA ladders typical for apoptotic cells could be observed from 18 h of infection onward. Lanes 1 and 2 represent the DNA of uninfected cells from the HeLa line taken at 12 h and 24 h of incubation, respectively. The left lane is the base pair markers. (B) The DNA ladders observed when the cells were infected for 24 h with different strains of B. pseudomallei. Lanes: 2, 3, and 4, virulent strains 844, UE3 and UE12, respectively; 5, 6, and 7, nonvirulent strains UE5, UE8, and UE11, respectively; and 8, 9, and 10, virulent strains, with atypical lipopolysaccharide pattern, UE16, UE30, and 824a, respectively. Lane 1 represents uninfected HeLa cells at 24 h of incubation.
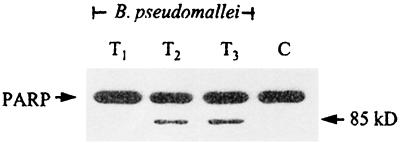
In order to determine the possible mechanism leading to the apoptotic cell death caused by B. pseudomallei, biochemical changes occurring at earlier stages of infection were analyzed. This was carried out by determining the degree of PARP cleavage in J774A.1 cells heavily infected with B. pseudomallei (MOI of 100 bacteria per cell). It is clearly demonstrated in Fig. 6 that the PARP cleavage could be detected within 2 h of infection, judging from the appearance of a faster-moving protein band as early as 2 h after the infection. This is indicative of caspase pathway involvement.
FIG. 6.
PARP cleavage. J774A.1 cells were heavily infected with B. pseudomallei at an MOI of 100:1 for 30 min. After a washing, the infected cells were incubated further for different intervals in the presence of kanamycin, and the cells were then harvested as described in Materials and Methods. Samples removed at 1, 2, and 3 h (lanes T1, T2, and T3, respectively) were lysed and then subjected to immunoblotting. Cleaved PARP (85 kDa) could be readily detected from 2 h (T2) onward. Lane C, Uninfected cell control.
DISCUSSION
The results presented in this study demonstrate that, after exposure to B. pseudomallei in vitro, both phagocytic and nonphagocytic cells exhibited certain morphological changes including (i) cell fusion leading to MNGC formation, (ii) cellular actin rearrangement initiated at one pole of the bacterium, typical of actin-based motility noted for some other bacteria (12), (iii) finger-like actin-associated peripheral membrane protrusion, and (iv) morphological and biochemical changes typical of apoptotic cell death. Although some of these characteristics have previously been reported to occur in several other bacterial infections (5–7, 26, 27), none of these infections have been reported to cause cell fusion. Thus, this phenomenon appears to be unique for B. pseudomallei infection. It is most likely that the cell fusion noted here is directly responsible for an MNGC formation previously noted by us (Int. Congr. Melioidosis) and subsequently by Harley and associates (8). Both the cell fusion and MNGC formation are not uncommon in, for example, viral infections. B. pseudomallei must in some way alter the external surfaces of the infected cells, which causes the surfaces to fuse with the membranes of neighboring cells. In the present study, we noted a translocation of membrane PS from the cytosol side to the external surface of the infected cells, but whether this change is associated with cell fusion remains to be investigated.
It is logical to postulate that cell fusion is one of the mechanisms that B. pseudomallei uses for direct cell-to-cell spreading, thus allowing them to survive any detrimental effect of extracellular environment and serum and to evade host defense. The ability of B. pseudomallei to induce plaque formation in the presence of kanamycin indicates direct cell-to-cell spreading. This, together with its ability to invade and to multiply in a number of nonphagocytic cells, may be partly responsible for the dormant state of B. pseudomallei in vivo in infected hosts. A possible molecular mechanism of cell-to-cell spreading in melioidosis is the ability of B. pseudomallei to initially induce actin-associated peripheral membrane protrusions like the ones most commonly reported for L. monocytogenes and S. flexneri (5–7). For these organisms, the bacterium-containing protrusions have been shown to reach nearby cells and to be phagocytosed by these cells. However, neither cell fusion nor MNGC formation has been observed in these bacterial infections. In the case of B. pseudomallei, the morphological changes did not stop at the stage of cytoplasmic protrusions, but our data indicated that following this stage, there was an intermediate process of cell fusion which eventually led to the formation of MNGC. The remnant of bacterium-containing cytoplasmic protrusions on the MNGC could thereafter infect neighboring cells, resulting in additional cell fusion and followed by a new cycle of infection and multiplication. The continuous process, initiated by the ability of Burkholderia to induce actin rearrangement, could give rise to a giant cell containing as many as 50 to 60 nuclei (unpublished observations). However, with the data available, we could not be certain if this process also depends on the microtubule function as has been shown for Actinobacillus actinomycetemcomitans (13).
Under laboratory conditions of these experiments, both biotypes of B. pseudomallei could infect and kill both phagocytic and nonphagocytic cells within 12 to 48 h of infection. Vorachit and associates (23) suggested that, in the presence of biofilms reported to be produced by some B. pseudomallei isolates, these bacteria could remain quiescent for quite some time. It is possible that disease-producing B. pseudomallei may have the ability to synthesize biofilms, thus allowing it to survive inside these and some other types of cells (which are yet to be determined) without killing them, and this occurrence might explain the dormancy state and relapse which are so common in melioidosis (3). Different lines of evidence presented in this study showed that both the extent and rate of cellular damages observed with the nonvirulent Ara+ biotype were less than those of the virulent Ara− biotype.
Very recently, a type III secretion-associated gene cluster has been identified in B. pseudomallei (24). It is logical therefore to speculate that B. pseudomallei also possesses the type III secretion system similar to systems described earlier for some other gram-negative bacilli, e.g., Shigella, Salmonella, and Yersinia (5, 7, 26, 27). However, these three genera of gram-negative bacilli are not known to induce cell fusion and MNGC formation, and, among the three, only Shigella can induce actin-associated peripheral membrane protrusions. In general, this secretion system is known to involve the host cell protein tyrosine kinase. Kanai and Kondo (11) presented evidence suggesting the involvement of protein tyrosine kinase in pathogenicity of B. pseudomallei. Their observation is consistent with the recent report of Harley and associates (8) showing that in some cell lines, the MNGC formation is partially inhibited by genistein, a chemical known to also inhibit the activity of protein kinase. Our data taken together with data from other groups of investigators make it tempting to suggest that the internalization of B. pseudomallei by phagocytosis in the case of macrophages or induced phagocytosis in the case of nonphagocytic cells, peripheral membrane protrusions, and direct cell-to-cell fusion induced by this bacterium can partially explain the involvement of B. pseudomallei in different tissues and organs. Its ability to directly spread from cell to cell and to produce biofilms (23) enables it to survive inside hosts with high antibody levels, and such a situation may be associated with the relapse which is frequently noted in areas of both endemicity and nonendemicity of B. pseudomallei infection.
Finally, very little is currently known about the molecular mechanism of host cell killing by B. pseudomallei. In the present study, we have presented different lines of evidence consistent with the induction of programmed cell death, including (i) condensed and fragmented nuclei, (ii) DNA ladder formation, (iii) cleavage of one of the DNA-repairing enzymes, PARP, and (iv) translocation of membrane PS from the cytoplasmic side to the external surface, which is typical for cells undergoing apoptotic change. Moreover, a typical peripheral chromatin condensation of cells infected with B. pseudomallei could be visualized by a transmission electron microscopic analysis (unpublished observations). Altogether, the data presented in our study clearly demonstrate that once inside either phagocytic or nonphagocytic cells, B. pseudomallei induces membrane-bound cytoplasmic protrusion and cell fusion, thus leading to direct cell-to-cell spreading and multinucleated cell formation, and that these changes are followed by apoptotic cell death. However, these observations cannot readily explain the viruence and pathogenicity of the disease-producing Ara− biotype, because the nonvirulent Ara+ biotype can also induce these changes, although at a lower rate and to a much lesser extent. It is clear therefore that this point needs further investigation.
ACKNOWLEDGMENTS
The work was supported by a research grant from Chulabhorn Research Institute (Thailand).
We are grateful to W. Prachyabrued (Faculty of Dentistry, Mahidol University) for valuable suggestions and to Maurice Broughton (Faculty of Science, Mahidol University) for editing the manuscript.
REFERENCES
- 1.Ahmed K, Enciso H D, Masaki H, Tao M, Omori A, Tharavichikul P, Nagatake T. Attachment of Burkholderia pseudomallei to pharyngeal epithelial cells. A highly pathogenic bacteria with low attachment ability. Am J Trop Med Hyg. 1999;60:90–93. doi: 10.4269/ajtmh.1999.60.90. [DOI] [PubMed] [Google Scholar]
- 2.Anuntagool N, Intachote P, Wuthiekanun V, White N J, Sirisinha S. Lipopolysaccharide from nonvirulent Ara+Burkholderia pseudomallei isolates is immunologically indistinguishable from lipopolysaccharide from virulent Ara− clinical isolates. Clin Diagn Lab Immunol. 1998;5:225–229. doi: 10.1128/cdli.5.2.225-229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaowagul W, Suputtamongkol Y, Dance D A B, Rachanuvong A, Pattaraarechachai J, White N J. Relapse in melioidosis: incidence and risk factors. J Infect Dis. 1993;168:1181–1185. [PubMed] [Google Scholar]
- 4.Chaowagul W, White N J, Dance D A B, Wattanagoon Y, Naigowit P, Davis T M E, Looareesuwan S, Pitakwatchara N. Melioidosis: a major cause of community-acquired septicemia in northeastern Thailand. J Infect Dis. 1989;159:890–899. doi: 10.1093/infdis/159.5.890. [DOI] [PubMed] [Google Scholar]
- 5.Cossart P, Boquet P, Normark S, Rappuoli R, editors. Cellular microbiology. Washington, D.C.: ASM Press; 2000. [DOI] [PubMed] [Google Scholar]
- 6.Dabiri G A, Sanger J M, Portnoy D A, Southwick F S. Listeria monocytogenes moves rapidly through the host-cell cytoplasm by inducing directional actin assembly. Proc Natl Acad Sci USA. 1990;87:6068–6072. doi: 10.1073/pnas.87.16.6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finlay B B, Cossart P. Exploitation of mammalian host cell functions by bacterial pathogens. Science. 1997;276:718–725. doi: 10.1126/science.276.5313.718. [DOI] [PubMed] [Google Scholar]
- 8.Harley V S, Dance D A B, Drasar B S, Tavey G. Effects of Burkholderia pseudomallei and other Burkholderia species on eukaryotic cells in tissue culture. Microbios. 1998;96:71–93. [PubMed] [Google Scholar]
- 9.Jones A L, Beveridge T J, Woods D E. Intracellular survival of Burkholderia pseudomallei. Infect Immun. 1996;64:782–790. doi: 10.1128/iai.64.3.782-790.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jordan M A, Wendall K, Gardiner S, Derry W B, Copp H, Wilson L. Mitotic block induced in HeLa cells by low concentrations of paclitaxel (taxol) results in abnormal mitotic exit and apoptotic cell death. Cancer Res. 1996;56:816–825. [PubMed] [Google Scholar]
- 11.Kanai K, Kondo E. Recent advances in biomedical science of Burkholderia pseudomallei (basonym: Pseudomonas pseudomallei) Jpn J Med Sci Biol. 1994;47:1–45. doi: 10.7883/yoken1952.47.1. [DOI] [PubMed] [Google Scholar]
- 12.Machesky L M. Rocket-based motility: a universal mechanism? Nat Cell Biol. 1999;1:E29–E31. doi: 10.1038/10020. [DOI] [PubMed] [Google Scholar]
- 13.Meyer D H, Rose J E, Lippmann J E, Fives-Taylor P M. Microtubules are associated with intracellular movement and spread of the periodontopathogen Actinobacillus actinomycetemcomitans. Infect Immun. 1999;67:6518–6525. doi: 10.1128/iai.67.12.6518-6525.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oaks E V, Wingfield M E, Formal S B. Plaque formation by virulent Shigella flexneri. Infect Immun. 1985;48:124–129. doi: 10.1128/iai.48.1.124-129.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pruksachartvuthi S, Aswapokee N, Thankerngool K. Survival of Pseudomonas pseudomallei in human phagocytes. J Med Microbiol. 1990;31:109–114. doi: 10.1099/00222615-31-2-109. [DOI] [PubMed] [Google Scholar]
- 16.Razak N, Ismail G. Interaction of human polymorphonuclear leukocytes with Pseudomonas pseudomallei. J Gen Appl Microbiol. 1982;28:509–518. [Google Scholar]
- 17.Sirisinha S, Anuntagool N, Intachote P, Wuthiekanun V, Puthucheary S D, Vadivelu J, White N J. Antigenic differences between clinical and environmental isolates of Burkholderia pseudomallei. Microbiol Immunol. 1998;42:731–737. doi: 10.1111/j.1348-0421.1998.tb02346.x. [DOI] [PubMed] [Google Scholar]
- 18.Smith M D, Angus B J, Wuthiekanun V, White N J. Arabinose assimilation defines a nonvirulent biotype of Burkholderia pseudomallei. Infect Immun. 1997;65:4319–4321. doi: 10.1128/iai.65.10.4319-4321.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Theriot J A. The cell biology of infection by intracellular bacterial pathogens. Annu Rev Cell Dev Biol. 1995;11:213–239. doi: 10.1146/annurev.cb.11.110195.001241. [DOI] [PubMed] [Google Scholar]
- 20.Ulett G C, Ketheesan N, Hirst R G. Macrophage-lymphocyte interactions mediate anti-Burkholderia pseudomallei activity. FEMS Immunol Med Microbiol. 1998;21:283–286. doi: 10.1111/j.1574-695X.1998.tb01175.x. [DOI] [PubMed] [Google Scholar]
- 21.Utaisincharoen P, Ubol S, Tangthawornchaikul N, Chaisuriya P, Sirisinha S. Binding of tumor necrosis factor-alpha (TNF-α) to TNF-RI induces caspase(s)-dependent apoptosis in human cholangiocarcinoma cell lines. Clin Exp Immunol. 1999;116:41–47. doi: 10.1046/j.1365-2249.1999.00856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled annexin V. J Immunol Methods. 1995;184:39–51. doi: 10.1016/0022-1759(95)00072-i. [DOI] [PubMed] [Google Scholar]
- 23.Vorachit M, Lam K, Jayanetra P, Costerton J W. Electron microscopic study of the mode of growth of Pseudomonas pseudomallei in vivo and in vitro. J Trop Med Hyg. 1995;98:379–391. [PubMed] [Google Scholar]
- 24.Winstanley C, Halles B A, Hart C A. Evidence for the presence in Burkholderia pseudomallei of a type III secretion system-associated gene cluster. J Med Microbiol. 1999;48:649–656. doi: 10.1099/00222615-48-7-649. [DOI] [PubMed] [Google Scholar]
- 25.Wong K T, Puthucheary S D, Vadivelu J. The histopathology of human melioidosis. Histopathology. 1995;26:51–55. doi: 10.1111/j.1365-2559.1995.tb00620.x. [DOI] [PubMed] [Google Scholar]
- 26.Zeile W L, Purich D L, Southwick F S. Recognition of two classes of oligoproline sequences in profilin-mediated acceleration of actin-based Shigella motility. J Cell Biol. 1996;133:49–59. doi: 10.1083/jcb.133.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou D, Mooseker M S, Galan J E. Role of the S. typhimurium actin-binding protein Sip A in bacterial internalization. Science. 1999;283:2092–2095. doi: 10.1126/science.283.5410.2092. [DOI] [PubMed] [Google Scholar]