Abstract
We used an expressed sequence tag approach to analyze genes expressed by the infective larvae of the rodent filarial parasite Litomosoides sigmodontis. One hundred fifty two new genes were identified, including several proposed as vaccine candidates in studies with human filarial parasites. Our findings have important implications for the use of L. sigmodontis as a model for filarial infection.
The rodent filarial parasite Litomosoides sigmodontis has recently been proposed as an important model of filariasis because it is the only filarial species in which the full development cycle can take place in inbred laboratory mice (1, 19). The power of murine genetics and immunology can now be brought to bear on fundamental questions in filarial biology that have previously been intractable. Perhaps the most immediate application of this model is the rigorous testing of vaccine strategies. The main objective of a filarial vaccine would be to target the incoming vector-derived larvae without inducing an immune response to later stages that might damage the host. We and others have been studying genes that are specific to the third larval (L3) stage of human filarial parasites (12, 13, 27). This work has identified several possible vaccine targets, with the abundant larval transcript 1 (alt-1) gene emerging as a particularly promising candidate (11).
The aim of this study was to sample genes expressed by the L3 stage of L. sigmodontis for comparison with gene expression in onchocerciasis and lymphatic filariasis parasites and thus to assess the utility of this rodent filaria as a model for vaccine studies. With this information, it will be possible to assess the validity of using the Litomosoides model to evaluate specific vaccine candidates. Further, the data will provide an important resource for comparative analysis of gene expression between species both within and outside the filariae.
For this study, we used an L. sigmodontis L3 cDNA library (in UniZap XR; Stratagene, La Jolla, Calif.). Randomly chosen recombinants were picked and cDNA inserts (with an average size of ∼640 bp) were PCR amplified using vector primers. Two hundred thirty PCR products were prepared using shrimp alkaline phosphatase and exonuclease I (Amersham Pharmacia Biotech Ltd., Uppsala, Sweden) (6) and sequenced using the 5′ vector primer SAC (GGGAACAAAAGCTGGAG) and ABI Big DYE terminators (the Perkin-Elmer Corporation, Norwalk, Conn.). Sequencing reactions were analyzed using an ABI 377 automated sequencer. There were 197 successful sequences with an average read length of 421 bp. The clones are archived and are freely available to the research community.
Sequences were edited to remove vector and poor 3′ sequences and then compared to the public databases using the BLAST family of algorithms (2). Expressed sequence tags (ESTs) with no significant similarity to any protein sequences in the databases using a minimum BLASTX score of 80, with a probability of <1 × e−8, were designated as novel. Sequences were clustered using AssemblyLIGN (Oxford Molecular, Oxford, United Kingdom), and where clusters contained more than one EST, an overlapping nucleotide sequence was used to generate an improved consensus sequence. Sixteen clusters with more than one EST and 136 clusters containing only one EST were defined. ESTs within a single cluster are assumed to be derived from the same gene. Each cluster is designated with an L. sigmodontis cluster (LSC) number. Nucleotide sequences were translated into putative peptide sequences and aligned to homologues from other species using ClustalW as implemented in MacVector.
Although small, the EST data set gave us both the information we were looking for and significant new information on the biology of this organism. None of these 152 genes have been previously identified from L. sigmodontis. Table 1 gives an overview of the 16 clusters containing more than one EST and their relationship to other filarial genes. A more comprehensive analysis of all the genes sequenced can be found at http://www.ed.ac.uk/∼mbx/LitoWeb/LitoESTs.html.
TABLE 1.
Genes identified as abundant ESTs in the L. sigmodontis dataset
| Cluster identifier | No. of ESTs | Length of consensus (bp) | Gene name (description) | Species containing homologues
|
|
|---|---|---|---|---|---|
| Filariae | Other nematodes | ||||
| LSC00018 | 16 | 611 | alt-1 (abundant larval transcript) | B. malayi, Bp, O. volvulus, W. bancrofti, A. viteae, D. immitis | None |
| LSC00017 | 8 | 1,311 | cpl-1 (cathepsin L-like cysteine protease) | B. malayi, O. volvulus, B. pahangi, D. immitis | C. elegans, Pristionchus pacificus |
| LSC00030 | 5 | 890 | cox-1 (mitochondrial cytochrome oxidase subunit 1) | B. malayi, O. volvulus | C. elegans, P. pacificus, Haemonchus contartus |
| LSC00036 | 3 | 588 | tpx-1 (thioredoxin peroxidase) | B. malayi, O. volvulus, D. immitis | C. elegans |
| LSC00047 | 3 | 818 | pbp-1 (similar to phosphatidyl-ethanolamine binding protein) | B. malayi, Onchocerca ochengi | C. elegans, T. canis |
| LSC00080 | 3 | 557 | col (similar to cuticular collagen) | B. malayi, O. volvulus | C. elegans, Caenorhabditis briggsae, P. pacificus, Necator americanus |
| LSC00108 | 3 | 502 | rbp-1 (putative RNA binding protein) | B. malayi, O. volvulus, W. bancroth | C. elegans |
| LSC00014 | 2 | 561 | Novel | None | None |
| LSC00025 | 2 | 535 | ndh-5 (mitochondrial NADH dehydrogenase subunit 5) | B. malayi, O. volvulus | C. elegans, Ascaris suum |
| LSC00040 | 2 | 404 | nap-1 (nucleosome assembly protein) | B. malayi | C. elegans, P. pacificus |
| LSC00042 | 2 | 672 | ral-2 (Ascaris and Onchocerca antigen homologue) | B. malayi, O. volvulus, W. bancrofti | C. elegans, P. pacificus, A. suum |
| LSC00055 | 2 | 689 | tph-1 (translationally controlled tumor protein homologue) | B. malayi, O. volvulus | C. elegans |
| LSC00066 | 2 | 605 | Novel (similar to C. elegans hypothetical protein) | B. malayi, O. volvulus | C. elegans |
| LSC00139 | 2 | 592 | Novel (similar to C. elegans hypothetical protein) | B. malayi, O. volvulus | C. elegans, P. pacificus, T. canis |
| LSC00152 | 2 | 275 | col-1 (collagen) | B. malayi, B. pahangi | C. elegans |
| LSC00194 | 2 | 602 | asp-1 (Ancylostoma secreted protein homologue) | B. malayi, O. volvulus | C. elegans, Ancylostoma caninum, N. americanus, H. contortus |
Because extensive EST data sets are available for the human pathogens Brugia malayi (3) and Onchocerca volvulus (25), it is possible to directly compare the information from our study with gene expression data from these human pathogens. The results of EST analysis of the most abundantly expressed genes from the L3 stage of B. malayi and O. volvulus are shown in Table 2. Of the twenty genes most abundantly expressed in B. malayi or O. volvulus L3, eight were identified in the L. sigmodontis data set. A more extensive EST analysis will almost certainly identify more shared genes between L. sigmodontis and these human pathogens.
TABLE 2.
EST analysis of abundantly expressed genes in the infective L3 stage of B. malayi and O. volvulus
| Genea |
B. malayi
|
O. volvulus
|
|||||
|---|---|---|---|---|---|---|---|
| Clusterb | Abundance rankc | % of EST data set | Clusterb | Abundance rankc | % of EST data set | ||
| alt-2d | BMC00213* | 1 | 3.15 | OVC00048* | 1 | 12.59 | |
| asp-1 | BMC00351 | 2 | 2.07 | OVC00237* | 4 | 1.98 | |
| alt-1d | BMC00123* | 3 | 1.46 | OVC00109* | 9 | 1.07 | |
| tin-2 | BMC00030 | 4 | 0.89 | OVC00071 | 6 | 1.43 | |
| tpx-2 | BMC00211 | 5 | 0.84 | OVC00018 | 5 | 1.55 | |
| cpl-1 | BMC04934* | 7 | 0.80 | OVC00916* | 0.63 | ||
| tin-1 | BMC00135 | 8 | 0.65 | OVC00099* | 15 | 0.63 | |
| mlc-1 | BMC00126 | 9 | 0.65 | OVC00888 | 0.11 | ||
| ant-2 | BMC00133* | 10 | 0.65 | OVC00092* | 10 | 0.99 | |
| nlt-1 | BMC03432* | 11 | 0.65 | OVC00579 | |||
| rpp-1 | BMC00166 | 12 | 0.61 | OVC00288 | 0.14 | ||
| cpi-2 | BMC01649 | 13 | 0.61 | OVC00142 | 3 | 2.34 | |
| alt-3 | BMC00136* | 14 | 0.56 | OVC00025* | 7 | 1.27 | |
| spn-1 | BMC04832* | 15 | 0.47 | OVC00784* | 0.11 | ||
| col | BMC06828 | OVC00039 | 2 | 3.53 | |||
| col | BMC03905 | 0.28 | OVC00036 | 8 | 1.15 | ||
| thi-1 | BMC00153 | 0.28 | OVC00287 | 11 | 0.79 | ||
| fba-1 | BMC00771 | 0.09 | OVC00662 | 12 | 0.75 | ||
| col-2 | BMC02934 | 0.18 | OVC00762 | 13 | 0.67 | ||
| act-1 | BMC00540 | 0.09 | OVC00082 | 14 | 0.63 | ||
The 15 most abundantly expressed genes in the infective L3 stage of B. malayi and O. volvulus were analyzed. Genes in bold have homologues in the L. sigmodontis EST data set. Gene products (4): act, actin (28); alt, abundant larval transcript (12, 14); ant, abundant novel transcript; asp, ancylostoma secreted protein; col, collagen; cpi, cysteine protease inhibitor (12, 18); cpl, cathepsin l; fba, fructose bisphosphate aldolase; mlc, myosin light chain; nlt, novel larval transcript (8); rpp, ribosomal phosphoprotein; spn, serpin (27); thi, thioredoxin; tin, troponin; tpx, thioredoxin peroxidase (10, 17, 22).
*, all ESTs from this cluster are from L3 stages. The EST data sets were clustered as described by Blaxter et al. (3).
Rank in abundance in B. malayi or O. volvulus L3 EST data set.
The O. volvulus gene alt-1 is the orthologue of B. malayi alt-2, and O. volvulus alt-2 is the orthologue of B. malayi alt-1.
Of particular interest was the identification of a gene similar to alt-1 of B. malayi as the most abundantly expressed gene in the L. sigmodontis dataset (EST cluster LSC00018) (Table 1). alt-1 was originally identified as one of the most abundant spliced leader-trans-spliced mRNAs expressed by B. malayi L3 (12) and has subsequently been shown to be specific to the infective L3 stage (11). Vaccine studies with gerbils have demonstrated the highest level of protection elicited by a recombinant filarial antigen to date (11). A related gene, alt-2, has also been identified in B. malayi (12). Additional alt-like genes have been sequenced from Dirofilaria immitis (20/22 antigen) (7), Acanthocheilonema viteae (20), Brugia pahangi (GenBank accession no. AJ275489), Wuchereria bancrofti (GenBank accession no. AF084553), and O. volvulus (14). Recognition of the ALT antigen has also been associated with protective immunity in experimental models of D. immitis (7) and O. volvulus infection (14).
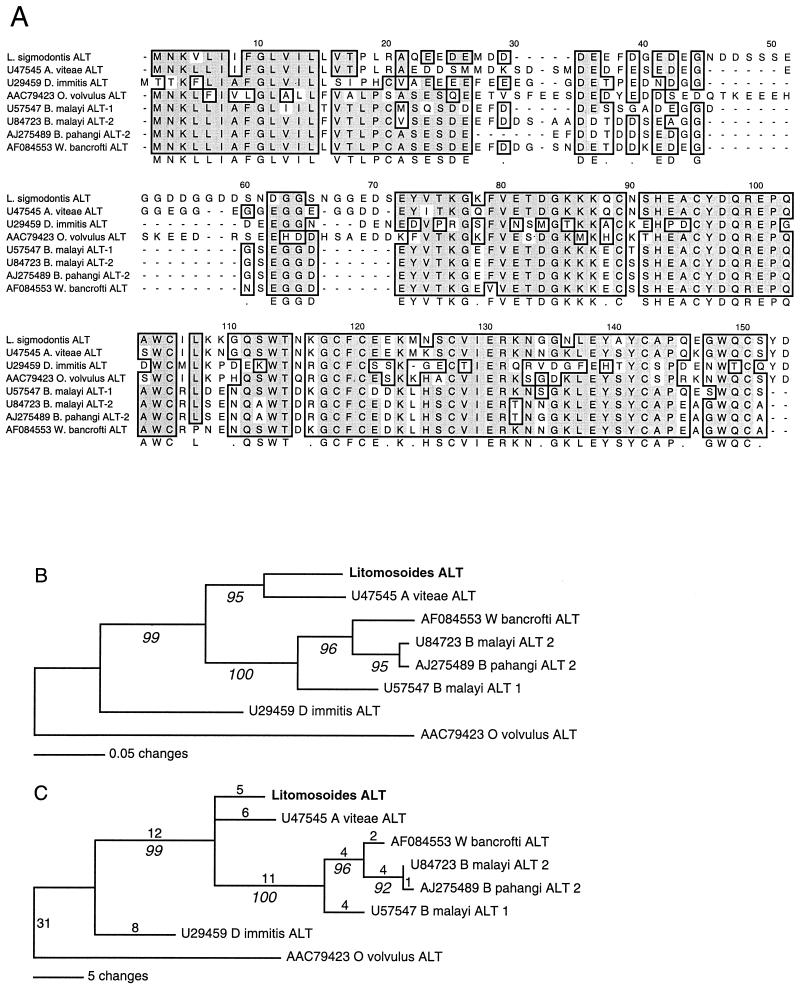
The EST data sets from B. malayi and O. volvulus confirm the stage specificity of expression of the alt genes identified and also reveal additional members of the alt gene family with distinct sequence and expression patterns (D. Guiliano, B. Gregory, and M. Blaxter, unpublished data). Among the L. sigmodontis ESTs in this study, only a single alt gene was identified. Comparison to published sequences shows that the L. sigmodontis ALT is most similar to that from the rodent parasite A. viteae (Fig. 1). Phylogenetic analysis shows that L. sigmodontis and A. viteae lie basal to the human infective filaria.This is consistent with analyses published previously (26) as well as our own data from small-subunit rRNA genes (not shown). The biological function of the ALT proteins is not known, but their highly regulated expression, abundance, and presence in excreted-secreted products of mammalian stage nematodes (7, 12) suggest that they may play an important role in establishing and maintaining infection.
FIG. 1.
(A) Alignment of ALT homologues from filarial nematodes. The alignment was constructed using ClustalW and adjusted by eye. Residues identical in more than four of eight aligned sequences are shaded grey, while similar residues are not shaded but boxed. A five-of-eight majority rule consensus is given below the alignment. (B and C) Analysis of similarity of ALTs. Phylogenetic trees were constructed using only the conserved C-terminal portion (residues 72 to 153 of the alignment in panel A). (B) Minimum evolution, neighbor joining; (C) maximum parsimony (gaps included). Italicized numerals give bootstrap support (1,000 replicates). Trees were arbitrarily rooted by using O. volvulus ALT.
In addition to alt-1, a number of other genes that have been proposed as vaccine candidates were identified. These included a member of the Ancylostoma secreted protein family of antigens (LSC00194) (23) and a RAL-2-related protein (LSC00042) (21, 24). In addition, LSC00047 encodes a phosphatidylethanolamine binding protein-related gene; homologues have been identified as an important diagnostic antigen in onchocerciasis (16) and as a surface molecule on Toxocara canis larvae (9). Also in the data set are enzymes and antienzymes under study in other filarial systems for drug or vaccine potential. The thioredoxin peroxidases of filarial nematodes have received significant attention in recent years because they may play a key role in detoxifying host oxyradicals deposited on the parasite (5, 10, 15, 17). The L. sigmodontis ESTs include both a thioredoxin peroxidase (LSC00036) and a thioredoxin (LSC00102). Proteases may mediate important processes in larval invasion of the host and establishment of the parasite, while protease inhibitors secreted by filarial L3 may play roles in the life cycle (in particular molting) (18) and in interfering with the host immune response (27). The L. sigmodontis EST data set includes clusters encoding cathepsin L-like (LSC00017 and LSC00209) and aspartyl (LSC00143) proteases and a cystatin-like protease inhibitor (LSC00180). The cystatin is most like B. malayi cystatin-2, an inhibitor synthesized throughout the B. malayi lifecycle (B. Gregory and R. Maizels, personal communication).
Several clusters encode apparent structural and housekeeping proteins and mitochondrially encoded genes (see http://www.ed.ac.u/∼mbx/LitoWeb/LitoESTs.html). For other clusters, a domain similarity can be discerned that is suggestive of function. For example, LSC00029 and LSC00181 have immunoglobulin-like domains, most similar to immunoglobulin domain-containing proteins from Caenorhabditis elegans. For many clusters (23 are described at http://www.ed.ac.u/∼mbx /LitoWeb/LitoESTs.html), the closest or only homologue in the public databases is a protein of unknown function predicted from the C. elegans genome project. For two such clusters, LSC00066 and LSC00139, more than one EST was sequenced, suggesting that these genes may be expressed at high levels in the vector-derived L3. Homologues of these clusters are also expressed at high levels in B. malayi. Whatever functions they perform, their abundance in the L3 data set suggests that they might be of importance to the larvae.
If L. sigmodontis is to be a model of real value in discovery and testing of vaccine candidates, it will be necessary to understand the relationship of this organism to the filarial pathogens which cause human disease. Importantly, in this study we were able to verify that, as in the human filarial nematodes, alt-1 is a highly abundant larval transcript in L. sigmodontis. However, it is important to recognize that 34% of the genes identified were unique to L. sigmodontis. This is likely to reflect the different migration patterns and host specificities of the parasites. Studies of novel genes unique to a particular parasite may further our understanding of the biology of filarial nematodes and their relationship to the mammalian host. Although it is small, the EST data set provided a substantial amount of basic information about this organism on which to build further studies and will greatly enhance our ability to use this important model system.
Nucleotide sequence accession numbers.
The sequences of L. sigmodontis genes found in this study were submitted to the EST database section of GenBank (accession numbers AW152683 to AW152860 and BE140074 to BE140093).
Acknowledgments
This work was supported by the Edna McConnell Clark Foundation and the Medical Research Council (UK). J. Allen is an MRC Senior Fellow.
REFERENCES
- 1.Al-Qaoud K M, Taubert A, Zahner H, Fleischer B, Hoerauf A. Infection of BALB/c mice with the filarial nematode Litomosoides sigmodontis: role of CD4+ T cells in controlling larval development. Infect Immun. 1997;65:2457–2461. doi: 10.1128/iai.65.6.2457-2461.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Blaxter M, Aslett M, Guiliano D, Daub J the Filarial Genome Project. Parasitic helminth genomics. Parasitology. 1999;188:S39–S51. doi: 10.1017/s0031182099004060. [DOI] [PubMed] [Google Scholar]
- 4.Blaxter M L, Guiliano D B, Scott A L, Williams S A. A unified nomenclature of filarial genes. Parasitol Today. 1997;13:416–417. doi: 10.1016/s0169-4758(97)01140-x. [DOI] [PubMed] [Google Scholar]
- 5.Chandrashekar R, Curtis K C, Lu W, Weil G J. Molecular cloning of an enzymatically active thioredoxin peroxidase from Onchocerca volvulus. Mol Biochem Parasitol. 1998;93:309–312. doi: 10.1016/s0166-6851(98)00041-3. [DOI] [PubMed] [Google Scholar]
- 6.Daub J, Loukas A, Pritchard D I, Blaxter M. A survey of genes expressed in adults of the human hookworm, Necator americanus. Parasitology. 1999;120:171–184. doi: 10.1017/s0031182099005375. [DOI] [PubMed] [Google Scholar]
- 7.Frank G R, Grieve R B. Purification and partial characterization of three larval excretory-secretory proteins of Dirofilaria immitis. Mol Biochem Parasitol. 1996;75:221–229. doi: 10.1016/0166-6851(95)02533-2. [DOI] [PubMed] [Google Scholar]
- 8.Frank G R, Wisnewski N, Brandt K S, Carter C R D, Jennings N S, Selkirk M E. Molecular cloning of the 22-24 kDa excretory-secretory 22U protein of Dirofilaria immitis and other filarial nematode parasites. Mol Biochem Parasitol. 1999;98:297–302. doi: 10.1016/s0166-6851(98)00173-x. [DOI] [PubMed] [Google Scholar]
- 9.Gems D, Ferguson C J, Robertson B D, Nieves R, Page A P, Blaxter M L, Maizels R M. An abundant, trans-spliced mRNA from Toxocara canis infective larvae encodes a 26 kDa protein with homology to phosphatidylethanolamine-binding proteins. J Biol Chem. 1995;31:18517–18522. doi: 10.1074/jbc.270.31.18517. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh I, Eisinger S W, Raghavan N, Scott A L. Thioredoxin peroxidases from Brugia malayi. Mol Biochem Parasitol. 1998;91:207–220. doi: 10.1016/s0166-6851(97)00213-2. [DOI] [PubMed] [Google Scholar]
- 11.Gregory W F, Atmadja A K, Allen J E, Maizels R M. The alt-1 and alt-2 genes of Brugia malayi encode stage-specific candidate vaccine antigens for filariasis. Infect Immun. 2000;68:4174–4179. doi: 10.1128/iai.68.7.4174-4179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gregory W F, Blaxter M L, Maizels R M. Differentially expressed, abundant trans-spliced cDNAs from larval Brugia malayi. Mol Biochem Parasitol. 1997;87:85–95. doi: 10.1016/s0166-6851(97)00050-9. [DOI] [PubMed] [Google Scholar]
- 13.Hunter S J, Martin S A M, Thompson F J, Tetley L, Devaney E. The isolation of differentially expressed cDNA clones from the filarial nematode Brugia pahangi. Parasitology. 1999;119:189–198. doi: 10.1017/s0031182099004576. [DOI] [PubMed] [Google Scholar]
- 14.Joseph G T, Huima T, Lustigman S. Characterization of an Onchocerca volvulus L3-specific larval antigen, Ov-ALT-1. Mol Biochem Parasitol. 1998;96:177–183. doi: 10.1016/s0166-6851(98)00094-2. [DOI] [PubMed] [Google Scholar]
- 15.Klimowski L, Chandrashekar R, Tripp C A. Molecular cloning, expression and enzymatic activity of a thioredoxin peroxidase from Dirofilaria immitis. Mol Biochem Parasitol. 1997;90:297–306. doi: 10.1016/s0166-6851(97)00167-9. [DOI] [PubMed] [Google Scholar]
- 16.Lobos E, Weiss N, Karam M, Taylor H R, Ottesen E A, Nutman T B. An immunogenic Onchocerca volvulus antigen: a specific and early marker of infection. Science. 1991;251:1603–1605. doi: 10.1126/science.2011741. [DOI] [PubMed] [Google Scholar]
- 17.Lu W, Egerton G L, Bianco A E, Williams S A. Thioredoxin peroxidase from Onchocerca volvulus: a major hydrogen peroxide detoxifying enzyme in filarial parasites. Mol Biochem Parasitol. 1998;91:221–235. doi: 10.1016/s0166-6851(97)00230-2. [DOI] [PubMed] [Google Scholar]
- 18.Lustigman S, Brotman B, Huima T, Prince A M, McKerrow J H. Molecular cloning and characterization of onchocystatin, a cysteine proteinase inhibitor of Onchocerca volvulus. J Biol Chem. 1992;267:17339–17346. [PubMed] [Google Scholar]
- 19.Maréchal P, Petit G, Diagne M, Taylor D W, Bain O. Use of the Litomosoides sigmodontis - mouse model in development of an Onchocerca vaccine. II. L. sigmodontis in the BALB/c mouse: vaccination experiments; preliminary immunological studies. Parasite. 1994;1:31–32. [Google Scholar]
- 20.Pogonka T, Oberländer W, Marti T, Lucius R. Acanthocheilonema viteae: characterization of a molt-associated excretory/secretory 18-kDa protein. Exp Parasitol. 1999;93:73–81. doi: 10.1006/expr.1999.4445. [DOI] [PubMed] [Google Scholar]
- 21.Rao K V N, Eswaran M, Ravi V, Gnanasekhar B, Narayanan R B, Kaliraj P, Jayaraman K, Marson A, Raghavan N, Scott A L. The Wuchereria bancrofti orthologue of Brugia malayi SXP1 and the diagnosis of bancroftian filariasis. Mol Biochem Parasitol. 2000;107:71–80. doi: 10.1016/s0166-6851(99)00231-5. [DOI] [PubMed] [Google Scholar]
- 22.Schrum S, Bialonski A, Marti T, Zipfel P F. Identification of a peroxidoxin protein (OvPXN-2) of the human parasitic nematode Onchocerca volvulus by sequential protein fractionation. Mol Biochem Parasitol. 1998;94:131–135. doi: 10.1016/s0166-6851(98)00034-6. [DOI] [PubMed] [Google Scholar]
- 23.Sen L K, Ghosh Z, Bin S, Qiang M G, Thompson J M, Hawdon R A, Koski X, Shuhua X, Hotez P J. Hookworm burden reductions in BALB/c mice vaccinated with recombinant Ancylostoma secreted proteins (ASPs) from Ancylostoma duodenale, Ancylostoma caninum and Necator americanus. Vaccine. 2000;18:1096–1102. doi: 10.1016/s0264-410x(99)00371-0. [DOI] [PubMed] [Google Scholar]
- 24.Wang S H, Zheng H J, Dissanayake S, Cheng W F, Tao Z H, Lin S Z, Piessens W F. Evaluation of recombinant chitinase and SXP1 antigens as antimicrofilarial vaccines. Am J Trop Med Hyg. 1997;56:474–481. doi: 10.4269/ajtmh.1997.56.474. [DOI] [PubMed] [Google Scholar]
- 25.Williams S A, Johnston D A. Helminth genome analysis: the current status of the filarial and schistosome genome projects. Filarial Genome Project. Schistosome Genome Project. Parasitology. 1999;118:S19–S38. doi: 10.1017/s0031182099004473. [DOI] [PubMed] [Google Scholar]
- 26.Xie H, Bain O, Williams S A. Molecular phylogenetic studies on filarial parasites based on 5s ribosomal spacer sequences. Parasite. 1994;1:141–151. doi: 10.1051/parasite/1994012141. [DOI] [PubMed] [Google Scholar]
- 27.Yenbutr P, Scott A L. Molecular cloning of a serine proteinase inhibitor from Brugia malayi. Infect Immun. 1995;63:1745–1753. doi: 10.1128/iai.63.5.1745-1753.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng W, Donelson J E. The actin genes of Onchocerca volvulus. Mol Biochem Parasitol. 1992;55:207–216. doi: 10.1016/0166-6851(92)90141-6. [DOI] [PubMed] [Google Scholar]