Abstract
The recent advances in omics and computational analysis have enabled the capacity to identify the exclusive strain-specific metabolites and novel biosynthetic gene clusters. This study analyzed eight strains of P. aurantiaca including GS1, GS3, GS4, GS6, GS7, FS2, ARS38, PBSt2, one strain of P. chlororaphis RP4, one strain of P. aeruginosa (At1RP4), and one strain of P. fluorescens (RS1) for the production of rhamnolipids, quorum-sensing signals, and osmolytes. Seven rhamnolipid derivatives were variably detected in fluorescent pseudomonads. These rhamnolipids included Rha-C10-C8, Rha-Rha-C10-C10, Rha-C10-C12db, Rha-C10-C10, Rha-Rha-C10-C12, Rha-C10-C12, and Rha-Rha-C10-C12db. Pseudomonas spp. also showed the variable production of osmoprotectants including N-acetyl glutaminyl glutamine amide (NAGGN), betaine, ectoine, and trehalose. Betaine and ectoine were produced by all pseudomonads, however, NAGGN and trehalose were observed by five and three strains, respectively. Four strains including P. chlororaphis (RP4), P. aeruginosa (At1RP4), P. fluorescens (RS1), and P. aurantiaca (PBSt2) were exposed to 1– 4% NaCl concentrations and evaluated for the changes in phenazine production profile which were negligible. AntiSMASH 5.0 platform showed 50 biosynthetic gene clusters in PB-St2, of which 23 (45%) were classified as putative gene clusters with ClusterFinder algorithm, five (10%) were classified as non-ribosomal peptides synthetases (NRPS), five (10%) as saccharides, and four (8%) were classified as putative fatty acids. The genomic attributes and comprehensive insights into the metabolomic profile of these Pseudomonas spp. strains showcase their phytostimulatory, phyto-protective, and osmoprotective effects of diverse crops grown in normal and saline soils.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-023-03607-x.
Keywords: Biosynthetic gene clusters, Genome mining, Osmolytes, Pyrrolnitrin, Pseudomonas aurantiaca, Rhamnolipids
Introduction
Secondary metabolites are low molecular weight compounds which facilitate the interaction of microbes with their environment and are usually produced at late-exponential phase of microbial growth. Microbes deploy and assemble these metabolites to mediate cross-species competitive and symbiotic interactions, as community stabilizers, regulatory signals, weapons, and to acquire diverse resources (Chevrette et al. 2019; Dar et al. 2020). Particularly, advances in the discovery of bacterial metabolites and their derivatives have unveiled several valuable bioactive metabolites with diverse applications in agriculture, medicine, and industry. Among the predominant secondary metabolites-producing genera, Pseudomonas has been extensively explored for the synthesis of antimicrobial metabolites including 2,4-diacetylphloroglucinol (DAPG), phenazines, pyrrolnitrin, pyoluteorin, pyocyanin, rhizoxin, pyoverdines, volatile hydrogen cyanide (HCN), alkylresorcinol, cyclic lipopeptides (CLPs), and rhamnolipids (Oni et al. 2015; Favre et al. 2017). However, mysteries in the synthesis of plethora of secondary metabolites are still unsolved and many cryptic or silent biosynthetic gene clusters (BGCs) have not yet elucidated.
Owing to the technological advances and convergence platforms (nano-biotechnology, computational biotechnology, etc.), there are now further avenues to be explored in locating and analyzing second metabolites of interest. Next-generation sequencing techniques and advanced computational tools have been instrumental in uncovering the silent bacterial BGCs, highlighting their prolific potential to synthesize novel products of bacterial secondary metabolism. Activation and inactivation strategies of certain BGCs including MS/MS networking, RT-qPCR, and high-throughput elicitor screening have addressed the divergent nature of bacterial secondary metabolites (Seyedsayamdost 2014; Mullins et al. 2021). These strategies allow to elucidate the function of several elicitor molecules to determine the mechanisms of induction and the functional products of silent BGCs. Substrate specificities and putative functions of conserved genes also help to determine the presence of new gene clusters in bacterial genomes. Besides this, functional core enzymes also provide the information about the products synthesized which can further be triggered through several elicitor compounds (Masschelein et al. 2017; Cesa-Luna et al. 2023; Khatri et al. 2023). Using these tools, several unique and exclusive BGCs have been identified in Pseudomonas spp. strains. For instance, in silico analysis of Pseudomonas sp. 11K1 showed the presence of two novel CLPs including brasmycin and braspeptin (Zhao et al. 2019). Similarly, BGCs coding polyhydroxyalkanoate (PHAs) were identified from Pseudomonas spp. isolated from Antarctica (Tan et al. 2020). Likewise, marker-less gene knockouts have been created to analyze the expression of heterologous biosynthetic gene clusters in Pseudomonas putida (Choi et al. 2018).
However, comparative genomic analyses have shown the substantial diversity among different species of Pseudomonas indicating the differential expression and production of secondary metabolites. Among pseudomonads, very few strains of P. aurantiaca and P. chlororaphis have been sequenced, and hence, less is known about their BGCs and unique secondary metabolites. Moreover, some secondary metabolites are exclusive to P. aurantiaca and P. chlororaphis group and need further illustration. In the present study, eleven strains of fluorescent pseudomonads including P. fluorescens, P. aeruginosa, P. chlororaphis subsp. aurantiaca, and P. chlororaphis subsp. chlororaphis were analyzed through UPLC–MS/MS to show the differences and commonalities in secondary metabolites production and synthesis. P. chlororaphis subsp. aurantiaca PB-St2 was used as a reference strain to analyze the BGCs present in its genome to compare and integrate the metabolic studies with genomic analysis. These strains have been previously characterized for the antifungal activities and plant growth-promoting (PGP) potential (Shahid et al. 2017, 2021). Detailed metabolomic analysis of these strains has demonstrated the presence of major classes of biologically active secondary metabolites. However, extended metabolomic analysis demonstrated the potential to produce rhamnolipids, osmolytes, and quorum-sensing signals.
Materials and methods
Bacterial strains
Eight strains of Pseudomonas chlororaphis subsp. aurantiaca including GS1, GS3, GS4, GS6, GS7, FS2, ARS38, PBSt2, one strain of P. chlororaphis subsp. chlororaphis (RP4), one strain of P. aeruginosa (At1RP4), and one strain of P. fluorescens (RS1) were used in this study. All bacterial strains are previously characterized and their host plants, and accession numbers are mentioned in Table S1. Strains were revived from glycerol stocks and streaked on King’s B agar plates to check the culture purity (King et al. 1954).
Identification of secondary metabolites by liquid chromatography–tandem mass spectrometry (LC–MS/MS)
Secondary metabolites for individual strains were extracted from 100 mL of 96-h-old bacterial cultures as previously described by Shahid et al. (2021). Briefly, individually grown bacterial cultures in glycerol–DMB (Davis 1949) were centrifuged at 3900 × g for 30 min (Allegra TM X-22R Centrifuge, Beckman Coulter). Supernatants were acidified to pH 2 with 6N HCl and extracted twice with equal volume (100 mL) of ethyl acetate. Organic layers were dehydrated with anhydrous Na2SO4 and evaporated to dryness in a rotary evaporator. Residual extracts were re-suspended in 4 mL of methanol–chloroform [1:1 (v/v)], and sonicated for 60 s for complete dissolution. From each extract, 200 µL aliquot was mixed with 200 µL of methanol and 100 µL of H2O. Ten microliters (10 µL) of each extract preparation were separately injected onto an Eclipse Plus C18 RRHD column (2.1 × 100 mm, 1.8 μm; Agilent, Santa Clara, CA, USA) and separated by reverse-phase liquid chromatography using a Waters Acquity UPLC system (Milford, MA, USA) at a flow rate of 0.4 mL/min. The UPLC system was coupled to an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) equipped with an EASY-Max NG electrospray ion source. The mobile phases were water/0.01% formic acid (A) and 100% acetonitrile/0.01% formic acid (B). Secondary metabolites were separated by the column with a 20-min binary solvent elution gradient (0 min, 5% B; 0–15 min, 5–100% B; 15–17 min, 100% B) followed by a 3-min column equilibration at 5% B, between injections. The MS parameters were as follows: positive-ion electrospray spray voltage, 3.9 kV; negative-ion electrospray spray voltage, 2.5 kV; capillary temperature, 325 °C; S-lens Level, 60%; sheath gas, 50; sweep gas, 1; and auxiliary gas 10. The MS survey scan was carried out over the mass range m/z 80–1800, with the data recorded in the centroid mode and at a mass resolution of 120 K FWHM (m/z 200), AGC target 4E5, and one micro-scan with a maximum injection time of 50 ms in the Quadrupole isolation mode. A lock mass at m/z 391.28426 from di-(2-ethylhexyl) phthalate (a ubiquitous plasticizer) was used for real-time internal mass calibration during the LC–MS runs with the Fourier-transform (FT) MS detection. In the LC–MS/MS runs, the top five most intense ions, with their charge state of 1 and ion counts of > 5000 in each survey scan were selected for MS/MS by collision-induced dissociation (CID) in the linear ion trap or by “higher-energy” collisional dissociation (HCD) in the collision cell in the upfront of the C-trap. Other parameters included: activation isolation window, 2 Da; AGC target for MS/MS, 5E4; maximum injection time, 30 ms; activation time, 10 ms; activation Q, 0.25; and normalized collision energy at 30% for the CID operations. Activation isolation window, 2 Da; AGC target for MS/MS, 5E4; maximum inject time, 30 ms; activation time, 10 ms; activation Q, 0.25; and normalized collision energy at 25% were used for the HCD operations.
Raw data files were recorded and processed using the XCalibur 4.1.31.9 (Thermo Scientific) software suite. For relative quantification of secondary metabolites produced by Pseudomonas strains, each sample solution was injected in triplicate and the average of peak areas were taken. Bar graphs were plotted with error bars to show the analytical standard deviations.
Detection of pyoverdines
Second method was specific for the extraction of pyoverdines as described by Deveau et al. (2016). One candidate strain from each species including P. aeruginosa (At1RP4), P. fluorescens (RS1), P. chlororaphis subsp. aurantiaca (PBSt2), and P. chlororaphis subsp. chlororaphis (RP4) was grown in 100 mL of King’s B broth for 96 h at 28 °C and 150 rpm. Cells were removed by centrifugation at 3900 × g and 4 °C for 20 min. The pH of the supernatants was adjusted to 6 with 6 N HCl and 5 g of Amberlite XAD-4 adsorbent resin (Sigma Aldrich, USA) were added in each supernatant. The mixtures were agitated at 28 °C and 100 rpm for 4 h and passed through fritted funnel. The bound metabolites were eluted with methanol/H2O (50:50) under agitation for 4 h and dried in rotary evaporator. Extracts were re-suspended in 2 mL of methanol/H2O (20:80) and stored at 4 °C for further analysis.
Effect of salt stress on the differential expression of secondary metabolites
Four candidate strains including P. chlororaphis subsp. aurantiaca (PBSt2), P. chlororaphis subsp. chlororaphis (RP4), P. aeruginosa (At1RP4), and P. fluorescens (RS1) were selected for this experiment. The selection was made to compare the effect of salt stress on metabolites production by four different bacterial species under study. Isolates were individually grown in 100 mL of DMB medium (Davis 1949) containing 1, 2, 3, and 4% NaCl and harvested after 96 h. Supernatants were extracted as described above and processed for LCMS/MS analysis using the same method as mentioned earlier. The experiment was repeated in triplicate and each sample was subjected to LCMS/MS analysis three times. Average of peak areas of each detected compound was used for construction of Heatmap. Each value was assigned a color through the selection of the color scale, and data was visualized through the changing color pattern to see the increase or decrease of a particular metabolite on the given salt stress.
Genomic properties and secondary metabolites prediction by antiSMASH in reference strain P. chlororaphis subsp. aurantiaca PB-St2
Genome sequencing of reference strain P. chlororaphis subsp. aurantiaca (PBSt2) has shown the presence of characteristic secondary metabolites in the strain (Mehnaz et al. 2014). The complete number of genes associated with the general Cluster of Orthologous Genes (COG) functional categories and the genomic properties of PB-St2 were re-evaluated. Biosynthetic gene clusters (BGCs) of PB-St2 were analyzed using antiSMASH 5.0 platform following the guidelines (Blin et al. 2019). PB-St2 GenBank FASTA files (Accession no. AYUD00000000) were uploaded on antiSMASH bacterial version 5.0 for the analysis of NRPS/PKS BGCs. Basic analysis features including active-site finders, Cluster Blast, and Cluster Pfam-analysis were selected for analysis and output data was analyzed.
Results and discussion
Secondary metabolites detected using liquid chromatography–mass spectrometry (LC–MS/MS)
Detection of secondary metabolites of Pseudomonas spp. was based on LC/ESI-FTMS in the full-mass scan mode. The most abundant peaks observed were of phenazines which have been reported previously from these strains (Shahid et al. 2021). In addition to phenazines, pyrrolnitrin, lahorenoic acids (A, B, & C), a range of quorum-sensing signals, rhamnolipids, and osmolytes were produced by these strains. Chemical formulas, monoisotopic neutral masses, observed m/z values, and retention times of these compounds are shown in Table 1. Rhamnolipid derivatives were detected in all fluorescent pseudomonads. However, MS spectra for rhamnolipids showed trace amounts. Supplementary Fig. S1 shows the retention times of seven rhamnolipid derivatives variably detected in fluorescent pseudomonads. Rhamnolipid Rha-C10-C8 (molecular formula = C24H44O9, monoisotopic m/z = 477.3063) was detected at RT = 9.65 in all Pseudomonas strains except for P. fluorescens RS-1, and P. aurantiaca GS-3 and GS-7. Rha-Rha-C10-C10 (molecular formula = C32H58O13), monoisotopic m/z = 651.3955 was detected at RT = 10.18, and Rha-C10-C12db (molecular formula = C28H50O9), monoisotopic m/z = 531.3532 was detected at RT = 10.94 in three P. aurantiaca strains including PB-St2, ARS-38, and FS-2, P. chlororaphis RP-4, P. fluorescens RS-1, and P. aeruginosa At1RP4. Five Pseudomonas spp. strains including P. aurantiaca ARS-38, PB-St2, and GS-3, P. fluorescens RS-1, and P. chlororaphis RP-4 showed rhamnolipid Rha-C10-C10 (molecular formula = C26H48O9), monoisotopic m/z = 505.3376, at RT = 10.96. Rha-Rha-C10-C12 (molecular formula = C34H62O13), monoisotopic m/z = 679.4268 was eluted at RT = 11.42, in strains P. aurantiaca PB-St2, FS2, and P. aeruginosa At1RP4. Likewise, Rha-C10-C12 (molecular formula = C28H52O9), monoisotopic m/z = 533.3689 showed elution at RT = 11.43 in strains P. aurantiaca ARS-38, PB-St2, and GS-3, P. fluorescens RS-1, and P. aeruginosa At1RP4. Rha-Rha-C10-C12db with (molecular formula = C34H50O9) monoisotopic m/z = 677.4111, was detected in P. aurantiaca PB-St2, and P. aeruginosa At1RP4 at RT = 12.94. MS/MS plots of all the strains were uploaded on Global Natural Product Social molecular networking (GNPS) to check the similarities with previously detected rhamnolipids present in database. The GNPS mirror plots with MS/MS chromatograms showing GOLD category and cosine similarity (< 90%) were extracted and have been shown in Supplementary Figs. S2–S8. MS/MS chromatograms were uploaded on Mass Search Tool (MASST) and searched through entire GNPS libraries (Wang et al. 2016). The search parameters including parent mass tolerance, score threshold, and minimum matched peaks were used as default settings. Minimum library class to consider in search was adjusted as GOLD, and maximum mass shift between library and putative analogs was 100 Da. Default filters were used to search exact matches and mirror plots were analyzed without considering analogs for accurate identification of metabolites produced by these strains.
Table 1.
Chemical formulas, monoisotopic m/z, and observed peaks of detected metabolites in Pseudomonas spp.
| Sr. no | Metabolites | Chemical formula | Monoisotopic m/z | Retention time | Mass error (ppm) |
|---|---|---|---|---|---|
| [M + H]+ | |||||
| AHLs | |||||
| 1 | Homoserine Lactone | C4H7NO2 | 102.0549 | 1.63 | 0.5 |
| 2 | 3-OH-C6-HSL | C10H17NO4 | 216.1235 | 2.13 | 1.7 |
| 3 | 3-oxo-C6-HSL | C10H16NO4 | 214.1079 | 1.89 | 2.8 |
| 4 | C6-HSL | C10H17NO3 | 200.1286 | 3.04 | 2.4 |
| 5 | 3-OH-C8-HSL | C12H21NO4 | 244.1548 | 4.05 | 3.6 |
| 6 | 3-oxo-C16-HSL | C20H35NO4 | 354.2638 | 6.13 | 1.5 |
| 7 | 3-OH-C10-HSL | C14H25NO4 | 272.1861 | 6.28 | 1.2 |
| 8 | HHQ | C16H21NO | 244.1701 | 6.91 | 0.6 |
| 9 | 3-OH-C12-HSL | C16H29NO4 | 300.2174 | 7.34 | 0.3 |
| 10 | 3-oxo-C12-HSL | C16H27NO4 | 298.2012 | 8.48 | 1.5 |
| 11 | 2-nonyl-4-quinolone | C18H25NO | 272.2008 | 8.63 | 1.5 |
| Rhamnolipids | |||||
| 12 | Rha-C10-C8 | C24H44O9 | 477.3063 | 9.65 | 0.0 |
| 13 | Rha-Rha-C10-C10 | C32H58O13 | 651.3955 | 10.18 | 0.3 |
| 14 | Rha-C10-C12db | C28H50O9 | 531.3532 | 10.94 | 0.3 |
| 15 | Rha-C10-C10 | C26H48O9 | 505.3376 | 10.96 | 1.7 |
| 16 | Rha-Rha-C10-C12 | C34H62O13 | 679.4268 | 11.42 | 1.0 |
| 17 | Rha-C10-C12 | C28H52O9 | 533.3689 | 11.43 | 0.7 |
| 18 | Rha-Rha-C10-C12db | C34H50O9 | 677.4111 | 12.94 | 0.3 |
| Osmolytes | |||||
| 19 | Ectoine | C6H10N2O2 | 225.0658 | 0.84 | 0.7 |
| Betaine | C5H11NO2 | 181.0759 | 1.85 | 0.6 | |
| NAGGN | C12H21N5O5 | 213.0658 | 2.24 | 0.6 | |
| Trehalose | C12H22O11 | 241.0607 | 1.14 | 0.5 |
Rhamnolipids are eco-friendly biosurfactants with the potential applications in agriculture, petroleum industry, and environmental remediation (Shi et al. 2021). They have been widely reported from P. aeruginosa strains exhibiting oil displacement efficiencies and emulsifying activities (Zhao et al. 2020). Robineau et al. (2020) demonstrated the antifungal activities of mono-rhamnolipids against Botrytis cinerea and showed their high emulsification activities against crude oils. Likewise, rhamnolipids isolated from P. aeruginosa strain MR01 showed considerable anticancer potential against MCF-7 human breast cancer cells (Rahimi et al. 2019). Similarly, rhamnolipids from P. aeruginosa PA14 showed effective dispersal of biofilms of clinical bacterial isolates and sulfate-reducing bacteria which lead to iron corrosion (Wood et al. 2018). However, most of the studies report the rhamnolipids from P. aeruginosa and P. fluorescens. However, there may be only few reports of rhamnolipids from P. chlororaphis group. This study showed the production of diverse mono- and di-rhamnolipid derivatives from all strains of fluorescent pseudomonads indicating the potential of these strains in developing ecofriendly value-added products for large-scale industrial processes. Besides this, the Pseudomonas spp. strains used in this study are biofungicides and biofertilizers, and the rhamnolipid production by these strains is an additional feature for their use in bioremediation of oil-contaminated agricultural soils.
Among osmolytes, four metabolites including ectoine, betaine, and N-acetylglutaminyl glutamine amide (NAGGN) were differentially detected in Pseudomonas spp. (Supplementary Fig. S9). Ectoine with (molecular formula = C6H10N2O2) monoisotopic m/z = 225.0658 and betaine with (molecular formula = C5H11NO2) monoisotopic m/z = 181.0759 were detected in all strains. However, NAGGN (molecular formula = C12H21N5O5), monoisotopic m/z = 213.0658 was detected in P. aurantiaca PB-St2, ARS38, and P. chlororaphis RP4. Trehalose was detected at RT = 1.14 in P. aurantiaca PB-St2, ARS38, and FS2, P. chlororaphis RP4, and P. fluorescens RS1 (Supplementary Figs. S10–S17). Osmolytes are produced by rhizospheric bacteria as protective molecules in response to increased salt concentrations in the soil. These osmoprotectants destroy 1-aminocyclopropane-1-carboxylic acid and subsequently reduce the induction of ethylene. However, osmolytes have been mainly reported from halophilic bacteria and their studies are limited in non-halophiles and particularly in P. chlororaphis group. Pseudomonas spp. have the potential to utilize osmoprotectants as the sole carbon source and induce systemic resistance in plants increasing their stability under saline conditions. For example, trehalose-producing Pseudomonas sp. UW4 has been shown to protect tomato plants from negative effects of salinity (Orozco-Mosqueda et al. 2019). Similarly, a recent study has shown that disruption of trehalose challenges the survival of P. aeruginosa under stressed conditions (Woodcock et al. 2021). Another research study reported the potential of NAGGN in protecting plants against Ralstonia solanacearum in addition to its role as osmoprotectant (Clough et al. 2022). This study also reports the differential production of osmoprotectants including ectoine, betaine, trehalose, and NAGGN in different strains of Pseudomonas spp. Among these strains, P. chlororaphis RP4, P. aurantiaca strains GS1, GS3, GS4, GS6, GS7, and FS2, and P. aeruginosa At1RP4 were isolated from extreme habitats and showed the presence of these osmoprotectants.
Fluorescent pseudomonads have been shown to synthesize several quorum-sensing (QS) signaling molecules, which also play additional roles as cytotoxic, immune-modulatory, and iron-acquiring agents. Strong signals corresponding to 2-nonyl-4-quinolone; m/z [M + H]+ = 272.2001 at RT 8.63, and 4-hydroxy-2-H-quinolone; m/z [M + H]+ = 244.1692 at 6.91 were detected. Among other quorum-sensing signals, homoserine lactone, 3-OH-C6-HSL, C6-HSL, 3-OH-C8-HSL, 3-OH-C10-HSL, 3-OH-C12-HSL, 3-oxo-C12-HSL, and 3-oxo-C16-HSL were produced by all eleven strains of pseudomonads. However, 3-oxo-C6-HSL was detected in P. aurantiaca PB-St2, ARS38, and FS2, P. chlororaphis RP4, and P. fluorescens RS1 (Fig. S18). Strong [M + H]+ signals corresponding to L-homoserine lactone (HSL), monoisotopic m/z = 102.0549 at RT = 1.63, C6-HSL, monoisotopic m/z = 200.1286 at RT = 3.04, 3-OH-C6-HSL, monoisotopic m/z = 216.1235 at RT = 2.13, 3-OH-C8-HSL, monoisotopic m/z = 244.1548 at RT = 4.05, and 3-oxo-C6-HSL, monoisotopic m/z = 214.1079 at RT = 1.89 were noted in positive-ion mode analysis. Signals for other AHLs including 3-OH-C10-HSL, 3-OH-C12-HSL, 3-oxo-C12-HSL, and 3-oxo-C16-HSL were noted in MS spectrum. Nonetheless, their MS/MS spectrum could not be retrieved because of the low abundance of these compounds. UPLC–MS and MS/MS chromatograms for homoserine lactones are shown in Supplementary Figs. S19–S32. These quorum-sensing signals not only activate the complex QS networks in bacterial cells but also mediate iron-acquisition, and cytotoxicity, and modulate host immune responses (Freund et al. 2018). A research study has demonstrated role of AHLs in regulation of social behavior and virulence of Pseudomonas syringae pv. actinidiae (Cellini et al. 2020). Another study described the phyto-stimulatory potential of Pseudomonas spp. AHLs in enhancing germination and growth of Solanum lycopersicum (Ferreira et al. 2020). Likewise, AHLs and quinolones from P. fluorescens DR397 have been reported to promote growth of legume cultivars through optimization of transcriptional responses (Nishu et al. 2022). However, most of these AHLs have not been pitched in P. chlororaphis group. The results of this study also demonstrate the presence of several AHLs and quinolones in P. aurantiaca and P. chlororaphis strains which offer the tremendous potential of these strains to be used as phytostimulants of several crops. Pyoverdine C55H85N17O22; m/z [M + H]+ = 1336.6127 was detected at RT 10.24 (Supplementary Fig. S33). It was most noticeable in P. aeruginosa At1RP4, followed by P. fluorescens RS1, whereas its concentration was less in other Pseudomonas strains. Pyoverdines are fluorescent dihydroxyquinoline fluorophore-based siderophores, which are integral to the growth of pseudomonads in iron-limited conditions. Production of these molecules allows the survival of Pseudomonas spp. across diverse hosts ranging from plants to humans. Particularly, the role of pyoverdines becomes significant for plants under phytopathogenic attack, where they induce systemic resistance in plants increasing their survivability (Ringel and Brüser 2018).
Effect of salt stress on the differential expression of secondary metabolites
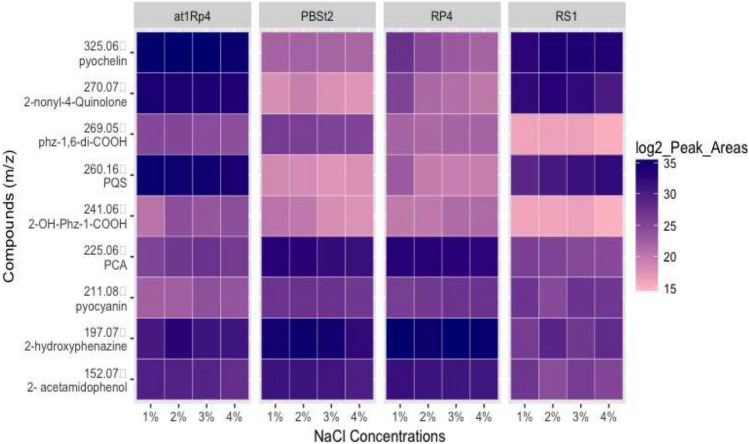
Four candidate strains from four different species of Pseudomonas spp. were subjected to salt stress and the difference was observed. Average peak areas from triplicate experiments were used to prepare the heat map. It was noticed that production of pyochelin [C14H16N2O3S2, monoisotopic m/z [M + H]+ = 325.06751] was not affected in P. aeruginosa At1RP4, and P. fluorescens RS1, when subjected to 1–4% NaCl stress in the culture medium. As widely reported in literature, 1% of NaCl was considered as control as it is regularly added in the different bacterial culture media. However, pyochelin production remained unchanged in At1RP4, and RS1 even in the stressed conditions. Nonetheless, its production got affected with increasing NaCl concentration in P. aurantiaca PB-St2, and P. chlororaphis RP-4. Similar trend was observed in the production of 2-nonyl-4-quinole, m/z [M + H]+ = 272.2001, and Pseudomonas quinolone signal [PQS; C16H21NO2 = m/z [M + H]+ = 260.1645] which did not show any change under salt-stress in P. aeruginosa At1RP4, and P. fluorescens RS1. However, a significant decline in production was observed in P. aurantiaca PB-St2, and P. chlororaphis RP-4 with increasing salinity concentration. Phenazines showed variable trend when exposed to different concentrations of NaCl. Phenazine carboxylic acid [PCA = C13H8N2O2, m/z [M + H]+ = 225.0658] did not show any considerable change in production with respect to changing NaCl concentrations in all four strains. Likewise, 2-acetamidophenol [C8H9NO2 = m/z [M + H]+ = 152.0706] showed negligible change under salinity stress except a small variation in production was observed in P. fluorescens RS1 at 2% NaCl level. On the other hand, phenazine-1-carboxylic acid [C14H8N2O4 = m/z [M + H]+ = 269.0553] showed variable trend. In P. aeruginosa At1RP4, its production remained unaffected with respect to changing salinity levels, whereas, it showed gradual decrease in production with increasing concentration of NaCl. Similarly, 2-OH-Phz-1-COOH [C13H8N2O3 = m/z [M + H]+ = 241.0604] showed increased levels at 2–4% of NaCl concentration in P. aeruginosa At1RP4, and P. chlororaphis RP-4. However, a gradual decline in its levels was observed in P. aurantiaca PB-St2, and P. fluorescens RS1. The amount of 2-hydroxyphenazine [C12H8N2O = m/z [M + H]+ = 197.0703] slightly decreased in all other strains as compared to P. chlororaphis RP-4, where it remained same throughout different salinity levels. Pyocyanin production increased in 3% and 4% of NaCl concentration in P. aeruginosa At1RP4, and P. fluorescens RS1, whereas, it did not change in P. chlororaphis RP-4, and P. aurantiaca PB-St2 (Fig. 1). Several studies have reported the differential production and regulation of osmolytes under salinity. Nonetheless, there is hardly any report about the differential regulation of phenazines under varying salinity levels. In this study, exposing Pseudomonas spp. strains to different NaCl concentrations did not show significant change in the production of phenazines indicating the stability of these compounds under natural saline conditions. P. chlororaphis and P. aeruginosa strains were isolated from para grass and Atriplex rhizoplanes, hence, the stability of these compounds can be attributed to their niches, whereas the stability of P. aurantiaca and P. fluorescens can be because of the differential regulation by AHLs and QS signals under challenged conditions. This fact has been previously reported in P. aeruginosa PA1201 where QS signals have differentially regulated the production of phenazine-1-carboxylic acid (Sun et al. 2016). In another study, phenazine producing halotolerant P. aeruginosa GS‐33 suppressed charcoal rot of soybean in saline conditions, showing that phenazine production and QS networks did not get bother by salinization (Patil et al. 2016). Stability and activity profiles in saline conditions of the strains under study indicate their broad potential applications across diverse agricultural conditions. Agricultural landscapes show changes in soil architecture, salinity levels, moisture and mineral contents. However, the biggest threat to agricultural productivity is salinity which is attributed to decreased crop yield and leaves the soil unfit for future use. Many of the biofertilizers and biofungicides fail to perform in salinized soils because of the compromised metabolic activities of the bioinocula in response to salinity stress. Nonetheless, the strains used in this study did not demonstrate marked fluctuations or decreased production of secondary metabolites despite being exposed to high saline conditions indicating their potential for the use in salinized soils. The stable production of antifungal phenazines and active metabolic pathways of these strains indicate toward the underlying differentially regulated genetic mechanisms which can adjust themselves in response to external stimuli and salinity conditions across diverse agricultural systems. One of the reasons for the stability and consistent production of metabolites by these strains can be attributed to the simultaneous production of osmoprotectants which keep metabolic pathways intact. As discussed above, osmoprotectants scavenge the damaging radicals and metabolites and can be pivotal for the activity of these strains under salinized soils.
Fig. 1.
Heat map for the LC–MS/MS profiles of selected specialized metabolites produced by Pseudomonas spp. strains PB-St2, RP-4, At1RP4, and RS1, treated with increasing concentrations of NaCl grown on DMB medium for 96 h. Blue color indicates increased metabolite production with respect to salinity concentration, whereas pink color indicates decreased production. Metabolites production remained relatively consistent for all the strains regardless of NaCl concentration
Genomic properties of reference strain P. chlororaphis subsp. aurantiaca PB-St2
Detailed analysis of P. chlororaphis subsp. aurantiaca PB-St2 to explore the biosynthetic gene clusters (BGCs) involved in the synthesis of secondary metabolites indicated unique features of the strain. The genome comprised 23 contigs with a total size of 6,569,731 bp and a calculated GC content of 63.25%, and showed the protein coding density of 89.13% with an average intergenic length of 136.17 bp. In addition, genome encoded, further, 60 tRNA genes and 4 rRNA genes (5S:2, 16S:1 and 23S:1 rRNA) as shown in Table 2 and Supplementary Fig. S34. The genome of P. chlororaphis subsp. aurantiaca strain PB-St2 contained 6,248 predicted protein-encoding genes, of which 4,737 (76%) have been assigned tentative functions. The remaining 1,511 ORFs were hypothetical/unknown proteins and 4950 (app. 79%) of all predicted protein-encoding genes could be allocated to the 23 functional COGs. Analysis of COGs revealed that ~ 21% of all protein-encoding genes fell into four main categories: energy metabolism (5.1%), amino acid transport and metabolism (10.4%), coenzyme transport and metabolism (3.0%); replication, recombination, and repair (2.8%). Diverse genomic attributes of this strain hint toward the presence of unique and exclusive secondary metabolism pathways adding to the ecological fitness of this strain. For instance, the strain showed the production of distinctive antimicrobial metabolites Lahorenoic acids A, B, and C, previously (Mehnaz et al. 2014). Similarly, the strain has also been evaluated for an array of metabolic compounds including phenazines, aromatic acids, siderophores, diketopiperazines, phenols, and pyrroles and the genomic features of PB-St2 complement these reports. Furthermore, in-depth analysis of hypothetical or unknown proteins can also be pitched in future to elucidate their metabolic potential.
Table 2.
Number of genes of P. aurantiaca PBSt2 associated with the general COG functional categories
| Attribute | Value | % of Total | |
|---|---|---|---|
| Genome size (bp) | 6,572,501 | 100.00 | |
| DNA Coding (bp) | 5,849,525 | 89.00 | |
| DNA G + C (bp) | 4,157,107 | 63.25 | |
| DNA scaffolds | 23 | – | |
| Total genes | 6260 | 100.00 | |
| Protein coding genes | 6248 | 99.80 | |
| RNA genes | 60 | 0.96 | |
| Pseudo-gene | 12 | 0.19 | |
| Genes in internal clusters | 2874 | 46.00 | |
| Genes with function prediction | 4271 | 68.35 | |
| Genes assigned to COGs | 4950 | 79.07 | |
| Genes with Pfam domains | 4311 | 60.01 | |
| Genes with signal peptides | 689 | 11.02 | |
| Genes with transmembrane helices | 1235 | 19.76 | |
| CRISPR repeats | 00 | 00 | |
Secondary metabolites prediction by antiSMASH in reference strain P. chlororaphis subsp. aurantiaca PB-St2
Putative biosynthetic gene clusters (BGCs) predicted in the genome of P. aurantiaca PB-St2
Using antiSMASH 5.0 platform, 50 biosynthetic gene clusters were identified in Pseudomonas aurantiaca PB-St2 of which 23 clusters (45%) were classified as putative gene clusters with ClusterFinder algorithm. Out of these 23 putative biosynthetic gene clusters, significant ClusterBlast Hits were not found for clusters 2, 3, 5, 7, 8, 11, 12, 13, 20, 34, 35, 37, 38, 39, 41, 42, 43, 44, and 50. Biosynthetic gene clusters are the physical groups of different gene assemblages coding for pathway-specific enzymes and secondary metabolites in microorganisms. Two predominant BGCs in Pseudomonas spp. are polyketide synthases (PKS) and non-ribosomal peptide synthases (NRPS) encoding metabolites for diverse applications in medicine, phyto-stimulants, biofungicides, and immune-suppressants (Palazzotto and Weber 2018). The following gene clusters identified in the genome of PB-St2 are indicative of its biocontrol and plant growth-promoting potential and can be engineered to over-produce the desired metabolites.
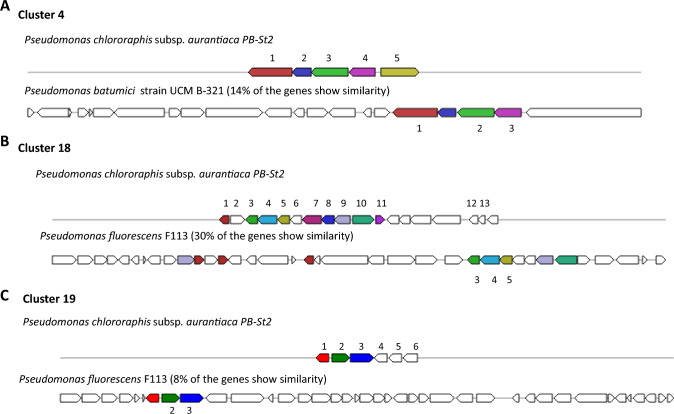
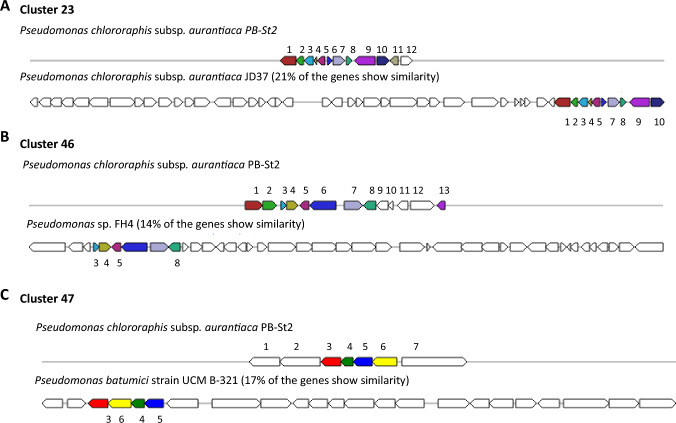
In PB-St2 genome, biosynthetic gene cluster 4, classified as putative, was assigned to fengycin and 14% of its genes showed similarity with Pseudomonas batumici strain UCM B-321 (Fig. 2A). Fengycin is antimicrobial cyclic lipopeptide acting against pathogenic fungi, bacteria, and nematodes and is synthesized at modular multi-enzymatic templates (Chen et al. 2007). Putative biosynthetic gene cluster 18 was classified as syringolin A and 30% of its genes showed similarity with Pseudomonas fluorescens F113 (Fig. 2B). Syringolin A is a tripeptide derivative which was isolated from pathogenic gamma-proteobacterium Pseudomonas syringae and is synthesized by a mixed non-ribosomal peptide synthetase/polyketide synthetase (Amrein et al. 2004). Similarly, putative cluster 19 was assigned to diffusible hemolytic cyclic lipopeptide entolysin showing 8% similarity with Pseudomonas fluorescens F113 (Fig. 2C). Entolysin is synthesized by three non-ribosomal peptide synthetases (NRPS) and essential for the swarming motility of P. entomophila (Vallet-Gely et al. 2010). Putative burkholderic acid biosynthetic gene cluster showed 21% similarity with P. chlororaphis subsp. aurantiaca JD37 strain (Fig. 3A). Burkholderic acid is non-canonical cryptic polyketide (PK) from pathogenic Burkholderia species which is natively silent metabolite but affects the virulence of these bacteria (Biggins et al. 2012). Fourteen percent genes of cluster 46 of P. aurantiaca PB-St2 showed the homology with Grincamycin Biosynthetic gene cluster of Pseudomonas sp. strain FH4 (Fig. 3B). Grincamycin is a polyketide angucycline antibiotic of Streptomyces sp. and was reported to be active against Sarcoma 180 solid tumors in mice (Basnet et al. 2006). Putative gene cluster 47 was assigned to sphingan polysaccharide gene cluster and 17% of its genes showed similarity with Pseudomonas batumici strain UCM B-321 (Fig. 3C). Sphingans are diverse, structurally related polysaccharides secreted by Sphingomonas spp. and are well-known rheological control agents in food and industrial processes (Schmid et al. 2014). Similarities and dissimilarities in BGCs may be indicative of their evolution over the period of time and can enable these strains to associate with exceptional hosts, for which pseudomonads are particularly known.
Fig. 2.
A Cluster 4: Genetic map of putative Fengycin Biosynthetic gene cluster 4 detected by antiSMASH 5.0. The genes were designated by colors. Same colors represent equal genes in reference and query strains while un-colored genes indicate other genes. 1. methylcrotonyl CoA carboxylase subunit alpha; 2. gamma-carboxygeranoyl CoA hydratase; 3. methylcrotonyl CoA carboxylase; 4. isovaleryl CoA dehydrogenase; 5. AMP-binding protein; B Cluster 18: Genetic map of Syringolin A Biosynthetic gene cluster 18 detected by antiSMASH 5.0. The genes were designated by colors. Same colors represent equal genes in reference and query strains while un-colored genes indicate other genes. 1. 3-oxo-acyl ACP reductase; 2. aldose-1 epimerase; 3. arabinose ABC transporter permease; 4. arabinose ABC transporter ATP-binding protein; 5. arabinose ABC transporter ATP–substrate-binding protein; 6. senescence marker 30-family protein; 7. 2,5-dioxovalerate dehydrogenase; 8. GguC protein; 9. MFS transporter; 10. dihydroxy-acid dehydratase; 11. GntR family transcriptional regulator; 12. 7-cyano-7-deazaguanine synthase; 13. 7-carboxy-7-deazaguanine synthase; C Cluster 19: Genetic map of putative Entolysin Biosynthetic gene cluster 19 detected by antiSMASH 5.0. The genes were designated by colors. Same colors represent equal genes in reference and query strains while un-colored genes indicate other genes. 1. AraC family transcriptional regulator; 2. 4-hydroxybenzoate monooxygenase; 3. DSBA oxidoreductase; 4. Cache domain containing protein; 5. ATPase; 6. 3-beta hydroxysteroid dehydrogenase
Fig. 3.
A Cluster 23: Genetic map of putative burkholderic acid Biosynthetic gene cluster 23 detected by antiSMASH 5.0. The genes were designated by colors. Same colors represent equal genes in reference and query strains while un-colored genes indicate other genes. 1. D-alanine-D-alanine ligase; 2. GNAT family acetyltransferase; 3. lysine transporter LysE; 4. Cro/Cl family transcriptional regulator; 5. membrane protein; 6. hypothetical protein; 7. AraC family transcriptional regulator; 8. cupin; 9. MFS transporter; 10. LysR family transcriptional regulator; 11. flavin reductase; 12. short-chain dehydrogenase; B Cluster 46: Genetic map of Grincamycin Biosynthetic gene cluster 46 detected by antiSMASH 5.0. The genes were designated by colors. Same colors represent equal genes in reference and query strains while un-colored genes indicate other genes. 1. adenine permease; 2. tRNA methyltransferase; 3. 3-dehydroquinate dehydratase; 4. shikimate 5-dehydrogenase; 5. TetR family transcriptional regulator; 6. 4-hydroxyphenylpyruvate dioxygenase; 7. membrane protein; 8. membrane protein; 9. xylose isomerase; 10. hypothetical protein; 11. IclR family transcriptional regulator; 12. FAD-binding dehydrogenase; 13. 3,4 dioxygenase subunit alpha; C Cluster 47: Genetic map of Sphingan polysaccharide Biosynthetic gene cluster 47 detected by antiSMASH 5.0. The genes were designated by colors. Same colors represent equal genes in reference and query strains while un-colored genes indicate other genes. 1. fis family transcriptional regulator; 2. ATPase; 3. dTDP-4 dehydrorhamnose reductase; 4. dTDP-4 dehydrorhamnose 3,5 epimerase; 5. glucose-1-phosphate thymidylyltransferase; 6. dTDP-glucose 4,6 dehydratase
Nonribosomal peptide synthetase (NRPS) biosynthetic gene clusters predicted in the genome of P. aurantiaca PB-St2
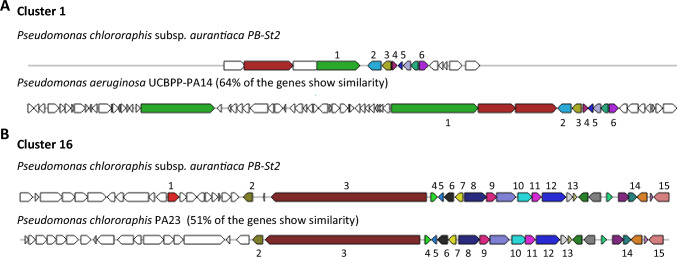
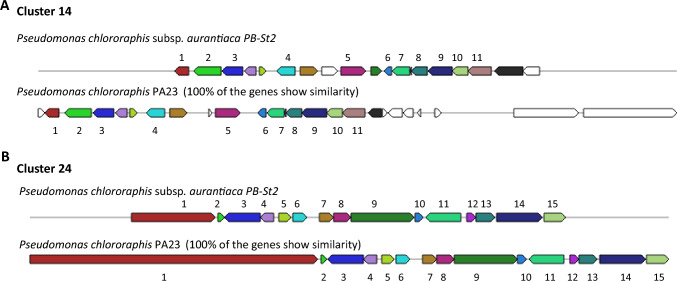
Five gene clusters (10%) were classified as NRPS gene clusters in the genome of P. aurantiaca PB-St2 strain. Two NRPS biosynthetic gene clusters including cluster 9 and 26 did not show similarity with known genes in ClusterBlast using ClusterFinder algorithm. NRPS clusters 1 and 16 were related to pyoverdines production (Fig. 4). Pyoverdines are characterized as high-affinity ferric iron chelator siderophores synthesized by fluorescent pseudomonads under iron-limited conditions. Pyoverdines biosynthesis is a complex process and to date, 60 different pyoverdines derivates have been characterized from different strains of Pseudomonas spp. (Trapet et al. 2016). Two clusters, 14 and 24 were identified as Pyrrolnitrin BGC and Mangotoxin BGC, respectively (Fig. 5). NRPS biosynthetic gene cluster 25 was classified as ralsolamycin and 40% of the genes showed similarity with Pseudomonas amygdali strain 107 (Supplementary Fig. S35). Ralsolamycin is a recently characterized secondary metabolite of phytopathogenic bacterium Ralstonia solanacearum and induces chlamydospore formation in fungi (Baldeweg et al. 2017). NRPS BGCs are the most essential BGCs and confer broad-spectrum antimicrobial and survival properties to the producing strains adding to their ecological fitness and existence in unprecedented circumstances. Pyoverdines producing gene clusters 1 and 16 of PB-St2 are fluorescent siderophores and can scavenge iron and other metals from the environment, making them unavailable for competing microbes. The reason for wide antifungal and plant growth-promoting abilities of PB-St2 can be additionally because of the diverse derivatives of siderophores which add to their survival in extraordinary niches.
Fig. 4.
Genetic map of Pyoverdine Biosynthetic gene clusters1 and 16 detected by AntiSMASH 5.0. The genes were designated by colors. Same colors represent equal genes in reference and query strains while un-colored genes indicate other genes. A Cluster 1: 1. pyoverdine biosynthesis protein; 2. ferripyoverdine receptor; 3. pyoverdine biosynthesis protein PvdE; 4. pyoverdine synthetase PvdF; 5. chromophore maturation protein PvdO; 6. chromophore maturation protein PvdP, B Cluster 16: 1. transcriptional regulator; 2. 2-succinyl-6-hydroxy 2,4 carboxylate synthase; 3. peptide synthase; 4. RNA polymerase sigma 70; 5. pyoverdine biosynthesis protein; 6. amino acid ABC transporter substrate-binding protein; 7. TetR family transcriptional regulator; 8. terpene utilization protein AtuA; 9. 2,4-dienoyal-CoA reductase; 10. acyl-CoA dehydrogenase; 11. enoyl-CoA hydratase; 12. 3-methylcrotonyl-CoA carboxylase; 13. exonuclease; 14. LuxR family transcriptional regulator; 15. cofactor biosynthesis protein MoaA
Fig. 5.
Genetic map of Pyrrlonitrin biosynthetic gene cluster 14, and Mangotoxin biosynthetic gene cluster 24 detected by AntiSMASH 5.0. The genes were designated by colors. Same colors represent equal genes in reference and query strains while un-colored genes indicate other genes. A Cluster 14: Genetic map of Pyrrolnitrin gene cluster 14 detected by AntiSMASH 5.0. The genes were designated by colors. Same colors represent equal genes in reference and query strains while un-colored genes indicate other genes. 1. AraC family transcriptional regulator; 2. feruloyl CoA synthase; 3. salicylaldehyde dehydrogenase; 4. porin; 5. dipeptidase; 6. flavin reductase; 7. potassium transporter; 8. 2Fe-2S ferredoxin (PrnD); 9. FAD-dependent oxidoreductase (PrnC); 10. PrnB; 11. tryptophan halogenase (PrnA). B Cluster 24: 1. Large adhesive protein; 2. tryptophan synthase subunit beta; 3. DeoR-family transcriptional regulator; 4. hypothetical protein; 5. GntR family transcriptional regulator; 6. thioesterase; 7. hypothetical protein; 8. P-aminobenzoate N-oxygenase AurF; 9. peptide transporter; 10. polyketide cyclase; 11. chemotaxis protein; 12. RNA polymerase sigma factor; 13. peptide ABC transporter substrate-binding; 14. ferrioxamine B receptor; 15. Peptidase
Saccharide biosynthetic gene clusters predicted in the genome of P. aurantiaca PB-St2
Five (10%) of the total BGCs were classified as saccharides and related to polysaccharide B, O-antigen, S-layer glycan, and lipopolysaccharide. Putative saccharide gene cluster 31 did not show similarity with known gene clusters. Cluster 6 was characterized as polysaccharide B and 34% of its genes showed similarity with P. fluorescens strain NT0133 (Supplementary Fig. S36). Cluster 17 of this category was classified as O-antigen and showed 28% similarity with Pseudomonas chlororaphis strain UFB2 (Supplementary Fig. S37). Cluster 27 was assigned as S-layer glycan BGC and indicated 29% similarity with P. chlororaphis strain UFB2 (Supplementary Fig. S38). Lipopolysaccharide BGC 49 showed 27% similarity with Pseudoalteromonas sp. strain S2471 (Supplementary Fig. S39). Lipopolysaccharides (LPS)-associated proteins are expressed as a mechanism of resistance to detergents and hydrophobic antibiotics and are secreted to the bacterial outer surface. O-antigen lipopolysaccharide is associated to adhesion and considered as the principal surface-associated virulence factor of P. aeruginosa (Allison and Castric 2016). S-layer glycoproteins (SLP) are the outermost para-crystalline bi-dimensional protein arrays involved in bacterial protection against detrimental environmental conditions, adherence to various substrates, and aggregation. Polysaccharide B is also considered as adhesion and plays a vital role during biofilm formation (Gerbino et al. 2015). The presence of these extraordinary active BGCs is not common to all bacteria and prove the supremacy of PB-St2 to mitigate the competitors for survival, shelter, and food at its habitat.
Fatty acid biosynthetic gene clusters predicted in the genome of P. aurantiaca PB-St2
Four gene clusters (8%) including cluster 30, 32, 33, and 36 were classified as putative fatty acid BGCs in P. aurantiaca PB-St2 genome. BGCs 30 and 33 did not show similarity with any of the known gene clusters, whereas 23% genes of the cluster 32 showed similarity with lipopolysaccharide BGC (Supplementary Fig. S40). Cluster 36 of this category was identified as orfamide BGC and 61% of its genes showed similarity with Pseudomonas protegens strain Pf-5 (Supplementary Fig. S41). Orfamide cyclic lipopeptides (CLPs) are biosurfactant metabolites which are important for bacterial surface motility and demonstrate surface tension reduction activity (D'aes et al. 2014). For cluster 45, 45% of the genes showed similarity with Pseudomonas sp. strain GM18 and classified as Pseudomonas quinolone signal (PQS) biosynthetic gene cluster (Supplementary Fig. S42). The presence of these gene clusters augments the overall fitness and survival of P. aurantiaca PB-St2 in diverse ecological niches. Besides, the gene clusters which could not be assigned any designated function could offer extended features to the strain and can be further explored for their similarities and dissimilarities with the previously reported gene clusters.
Conclusions
Pseudomonas spp. possess prolific metabolomic diversity which enables them to survive in unusual habitats. This study has categorically analyzed the genomic properties and BGCs of P. aurantiaca PB-St2 strain and the under-pitched secondary metabolites, AHLs and QS signals, produced by the group of Pseudomonas spp., which capacitate them to be the determinants of plant growth promotion and biocontrol of certain diseases. The BGCs share high similarities with some of the predicted gene clusters in Pseudomonas spp. group. BGCs showing less similarities are the subject of further investigation as their biological functions are yet to be determined. Furthermore, the unchanged profiles of phenazines production of Pseudomonas strains in response to salinity stress indicate their successful colonization under external environmental biotic stressors.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Authors are grateful to Muhmmad Adeel (English Language Expert) for editing the manuscript.
Author contributions
IS wrote the manuscript and performed practical work. CHB and JH helped in LCMS data analysis. Heatmap and statistical analysis was performed by SH. SM conceived the study, edited the manuscript and HAE proofread the manuscript.
Data availability
No specific data of this manuscript is identified for data availability.
Declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
References
- Allison TM, Castric P. Selective distribution of Pseudomonas aeruginosa O-antigen among strains producing group I pilin. FEMS Pathogens Dis. 2016 doi: 10.1093/femspd/ftv102. [DOI] [PubMed] [Google Scholar]
- Amrein H, Makart S, Granado J, Shakya R, Schneider-Pokorny J, Dudler R. Functional analysis of genes involved in the synthesis of syringolin A by Pseudomonas syringae pv. syringae B301 D-R. Mol Plant Microbe Interact. 2004;17:90–97. doi: 10.1094/MPMI.2004.17.1.90. [DOI] [PubMed] [Google Scholar]
- Baldeweg F, Kage H, Sebastian S, Allen C, Hoffmeister D, Nett M. Structure of ralsolamycin the interkingdom morphogen from the crop plant pathogen Ralstonia solanacearum. Org Lett. 2017;19:4868–4871. doi: 10.1021/acs.orglett.7b02329. [DOI] [PubMed] [Google Scholar]
- Basnet DB, Oh TJ, Vu TT, Sthapit B, Liou K, Lee HC, Yoo JC, Sohng JK. Angucyclines Sch 47554 and Sch 47555 from Streptomyces sp. SCC-2136: cloning, sequencing, and characterization. Mol Cells. 2006;22:154–162. [PubMed] [Google Scholar]
- Biggins JB, Ternei MA, Brady SF. Malleilactone, a polyketide synthase-derived virulence factor encoded by the cryptic secondary metabolome of Burkholderia pseudomallei group pathogens. J Am Chem Soc. 2012;134:13192–13195. doi: 10.1021/ja3052156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, et al. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019;47:W81–W87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cellini A, Donati I, Fiorentini L, Vandelle E, Polverari A, Venturi V, et al. N-Acyl homoserine lactones and Lux solos regulate social behaviour and virulence of Pseudomonas syringae pv. actinidiae. Microbial Ecol. 2020;79:383–396. doi: 10.1007/s00248-019-01416-5. [DOI] [PubMed] [Google Scholar]
- Cesa-Luna C, Geudens N, Girard L, De Roo V, Maklad HR, Martins JC, Höfte M, De Mot R. Charting the ipopeptidome of nonpathogenic Pseudomonas. Msystems. 2023;8:e00988–e1022. doi: 10.1128/msystems.00988-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XH, Koumoutsi A, Scholz R, Eisenreich A, Schneider K, Heinemeyer I, et al. Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat Biotechnol. 2007;25:1007–1014. doi: 10.1038/nbt1325. [DOI] [PubMed] [Google Scholar]
- Chevrette MG, Carlson CM, Ortega HE, et al. The antimicrobial potential of Streptomyces from insect microbiomes. Nat Commun. 2019 doi: 10.1038/s41467-019-08438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KR, Cho JS, Cho IJ, Park D, Lee SY. Markerless gene knockout and integration to express heterologous biosynthetic gene clusters in Pseudomonas putida. Metabol Eng. 2018;47:463–474. doi: 10.1016/j.ymben.2018.05.003. [DOI] [PubMed] [Google Scholar]
- Clough SE, Jousset A, Elphinstone JG, Friman VP. Combining in vitro and in vivo screening to identify efficient Pseudomonas biocontrol strains against the phytopathogenic bacterium Ralstonia solanacearum. Microbiol Open. 2022;11:e1283. doi: 10.1002/mbo3.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'aes J, Kieu NP, Léclère V, Tokarski C, Olorunleke FE, De Maeyer K, et al. To settle or to move? The interplay between two classes of cyclic lipopeptides in the biocontrol strain Pseudomonas CMR12a. Environ Microbiol. 2014;16:2282–2300. doi: 10.1111/1462-2920.12462. [DOI] [PubMed] [Google Scholar]
- Dar D, Thomashow LS, Weller DM, Newman DK. Global landscape of phenazine biosynthesis and biodegradation reveals species-specific colonization patterns in agricultural soils and crop microbiomes. Elife. 2020;9:e59726. doi: 10.7554/eLife.59726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BD. The isolation of biochemically deficient mutants of bacteria by means of penicillin. Proc Natl Acad Sci. 1949;35:1–10. doi: 10.1073/pnas.35.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau A, Gross H, Palin B, Mehnaz S, Schnepf M, Leblond P, et al. Role of secondary metabolites in the interaction between Pseudomonas fluorescens and soil microorganisms under iron-limited conditions. FEMS Microbiol Ecol. 2016;92:fiw107. doi: 10.1093/femsec/fiw107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favre L, Ortalo-Magné A, Greff S, Pérez T, Thomas OP, Martin JC, Culioli G. Discrimination of four marine biofilm-forming bacteria by LC-MS metabolomics and influence of culture parameters. J Proteome Res. 2017;16:1962–1975. doi: 10.1021/acs.jproteome.6b01027. [DOI] [PubMed] [Google Scholar]
- Ferreira NP, Ximenez GR, Chiavelli LU, Lucca DL, Santin SM, Zuluaga MY, et al. Acyl-homoserine lactone from plant-associated Pseudomonas sp. influences Solanum lycopersicum germination and root growth. J Chem Ecol. 2020;46:699–706. doi: 10.1007/s10886-020-01186-2. [DOI] [PubMed] [Google Scholar]
- Freund JR, Mansfield CJ, Doghramji LJ, Adappa ND, Palmer JN, Kennedy DW, et al. Activation of airway epithelial bitter taste receptors by Pseudomonas aeruginosa quinolones modulates calcium, cyclic-AMP, and nitric oxide signaling. J Biol Chem. 2018;293:9824–9840. doi: 10.1074/jbc.RA117.001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbino E, Carasi P, Mobili P, Serradell M, Gómez-Zavaglia A. Role of S-layer proteins in bacteria. World J Microbiol Biotechnol. 2015 doi: 10.1007/s11274-015-1952-9. [DOI] [PubMed] [Google Scholar]
- Khatri S, Sazinas P, Strube ML, Ding L, Dubey S, Shivay YS, Sharma S, Jelsbak L. Pseudomonas is a key player in conferring disease suppressiveness in organic farming. Plant Soil. 2023;18:1–20. doi: 10.1007/s11104-023-05927-6. [DOI] [Google Scholar]
- King EO, Ward MK, Raney DE. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clinical Med. 1954;44:301–307. [PubMed] [Google Scholar]
- MacLean BX, Pratt BS, Egertson JD, MacCoss MJ, Smith RD, Baker ES. Using skyline to analyze data-containing liquid chromatography, ion mobility spectrometry, and mass spectrometry dimensions. J Am Soc Mass Spectrom. 2018;29:2182–2188. doi: 10.1007/s13361-018-2028-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masschelein J, Jenner M, Challis GL. Antibiotics from Gram-negative bacteria: a comprehensive overview and selected biosynthetic highlights. Nat Prod Rep. 2017;34:712–783. doi: 10.1039/c7np00010c. [DOI] [PubMed] [Google Scholar]
- Mehnaz S, Bauer JS, Gross H. Complete genome sequence of the sugar cane endophyte Pseudomonas aurantiaca PB-St2, a disease-suppressive bacterium with antifungal activity toward the plant pathogen Colletotrichum falcatum. Genome Announc. 2014;2:e01108–e01113. doi: 10.1128/genomeA.01108-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins AJ, Webster G, Kim HJ, Zhao J, Petrova YD, Ramming CE, Jenner M, Murray JA, Connor TR, Hertweck C, Challis GL. Discovery of the Pseudomonas polyyne protegencin by a phylogeny-guided study of polyyne biosynthetic gene cluster diversity. Mbio. 2021;12:e00715–e721. doi: 10.1128/mBio.00715-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishu SD, No JH, Lee TK. Transcriptional response and plant growth promoting activity of Pseudomonas fluorescens DR397 under drought stress conditions. Microbiol Spectr. 2022;10:e00979–e1022. doi: 10.1128/spectrum.00979-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oni FE, Kieu Phuong N, Höfte M. Recent advances in Pseudomonas biocontrol. In: Murillo J, Vinatzer BA, Jackson RW, Arnold DL, editors. Bacterial-plant interactions: advance research and future trends. Poole: Caister Academic Press; 2015. pp. 167–198. [Google Scholar]
- Orozco-Mosqueda MDC, Duan J, DiBernardo M, Zetter E, Campos-García J, Glick BR, Santoyo G. The production of ACC deaminase and trehalose by the plant growth promoting bacterium Pseudomonas sp. UW4 synergistically protect tomato plants against salt stress. Front Microbiol. 2019;10:1392. doi: 10.3389/fmicb.2019.01392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzotto E, Weber T. Omics and multi-omics approaches to study the biosynthesis of secondary metabolites in microorganisms. Curr Opin Microbiol. 2018;45:109–116. doi: 10.1016/j.mib.2018.03.004. [DOI] [PubMed] [Google Scholar]
- Patil S, Paradeshi J, Chaudhari B. Suppression of charcoal rot in soybean by moderately halotolerant Pseudomonas aeruginosa GS-33 under saline conditions. J Microbiol. 2016;56:889–899. doi: 10.1002/jobm.201600008. [DOI] [PubMed] [Google Scholar]
- Rahimi K, Lotfabad TB, Jabeen F, Ganji SM. Cytotoxic effects of mono-and di-rhamnolipids from Pseudomonas aeruginosa MR01 on MCF-7 human breast cancer cells. Colloids Surf B. 2019;181:943–952. doi: 10.1016/j.colsurfb.2019.06.058. [DOI] [PubMed] [Google Scholar]
- Ringel MT, Brüser T. The biosynthesis of pyoverdines. Microbial Cell. 2018;5:424. doi: 10.15698/mic2018.10.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robineau M, Le Guenic S, Sanchez L, Chaveriat L, Lequart V, Joly N, Calonne M, Jacquard C, Declerck S, Martin P, Dorey S (2020) Synthetic mono-rhamnolipids display direct antifungal effects and trigger an innate immune response in tomato against Botrytis cinerea. Molecules 25:3108 [DOI] [PMC free article] [PubMed]
- Schmid J, Sperl N, Sieber V. A comparison of genes involved in sphingan biosynthesis brought up to date. Appl Microbiol Biotechnol. 2014;98:7719–7733. doi: 10.1007/s00253-014-5940-z. [DOI] [PubMed] [Google Scholar]
- Seyedsayamdost MR. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. PNAS. 2014;111:7266–7271. doi: 10.1073/pnas.1400019111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid I, Rizwan M, Baig DN, Saleem RS, Malik KA, Mehnaz S. Secondary metabolites production and plant growth promotion by Pseudomonas chlororaphis subsp. aurantiaca strains isolated from cotton, cactus, and para grass. J Microbiol Biotechnol. 2017;27:480–491. doi: 10.4014/jmb.1601.01021. [DOI] [PubMed] [Google Scholar]
- Shahid I, Han J, Hardie D, Baig DN, Malik KA, Borchers CH, Mehnaz S. Profiling of antimicrobial metabolites of plant growth promoting Pseudomonas spp. isolated from different plant hosts. 3 Biotech. 2021;11:1–14. doi: 10.1007/s13205-020-02585-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Chen Y, Liu X, Li D. Rhamnolipid production from waste cooking oil using newly isolated halotolerant Pseudomonas aeruginosa M4. J Clean Product. 2021;278:123879. doi: 10.1016/j.jclepro.2020.123879. [DOI] [Google Scholar]
- Sun S, Zhou L, Jin K, Jiang H, He YW. Quorum sensing systems differentially regulate the production of phenazine-1-carboxylic acid in the rhizobacterium Pseudomonas aeruginosa PA1201. Sci Rep. 2016;6:1–14. doi: 10.1038/srep30352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan IKP, Foong CP, Tan HT, Lim H, Zain NAA, Tan YC, et al. Polyhydroxyalkanoate (PHA) synthase genes and PHA-associated gene clusters in Pseudomonas spp. and Janthinobacterium spp. isolated from Antarctica. J Biotechnol. 2020;313:18–28. doi: 10.1016/j.jbiotec.2020.03.006. [DOI] [PubMed] [Google Scholar]
- Trapet P, Avoscan L, Klinguer A, Pateyron S, Citerne S, Chervin C, Mazurier S, Lemanceau P, Wendehenne D, Besson-Bard A. The Pseudomonas fluorescens siderophore pyoverdine weakens Arabidopsis thaliana defense in favor of growth in iron-deficient conditions. Plant Physiol. 2016;171:675–693. doi: 10.1104/pp.15.01537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet-Gely I, Novikov A, Augusto L, Liehl P, Bolbach G, Péchy-Tarr M, et al. Association of hemolytic activity of Pseudomonas entomophila, a versatile soil bacterium, with cyclic lipopeptide production. Appl Environ Microbiol. 2010;76:910–921. doi: 10.1128/AEM.02112-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TL, Gong T, Zhu L, Miller J, Miller DS, Yin B, Wood TK. Rhamnolipids from Pseudomonas aeruginosa disperse the biofilms of sulfate-reducing bacteria. NPJ Biofilms Microbiol. 2018;4:1–8. doi: 10.1038/s41522-018-0066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcock SD, Syson K, Little RH, Ward D, Sifouna D, Brown JKM, et al. Trehalose and α-glucan mediate distinct abiotic stress responses in Pseudomonas aeruginosa. PLoS Genet. 2021;17:e1009524. doi: 10.1371/journal.pgen.1009524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Liu YP, Zhang LQ. In silico and genetic analyses of cyclic lipopeptide synthetic gene clusters in Pseudomonas sp. 11K1. Front Microbiol. 2019;10:544. doi: 10.3389/fmicb.2019.00544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Han S, Zhang Y. Comparative studies on the structural composition, surface/interface activity and application potential of rhamnolipids produced by Pseudomonas aeruginosa using hydrophobic or hydrophilic substrates. Bioresour Technol. 2020;295:122269. doi: 10.1016/j.biortech.2019.122269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No specific data of this manuscript is identified for data availability.