Abstract
DNA dodecamers have been designed with two cytosines on each end and intervening A and T stretches, such that the oligomers have fully complementary A:T base pairs when aligned in the parallel orientation. Spectroscopic (UV, CD and IR), NMR and molecular dynamics studies have shown that oligomers having the sequences d(CCATAATTTACC) and d(CCTATTAAATCC) form a parallel-stranded duplex when dissolved at 1:1 stoichiometry in aqueous solution. This is due to the C:C+ clamps on either end and extensive mismatches in the antiparallel orientation. The structure is stable at neutral and acidic pH. At higher temperatures, the duplex melts into single strands in a highly cooperative fashion. All adenine, cytosine and thymine nucleotides adopt the anti conformation with respect to the glycosidic bond. The A:T base pairs form reverse Watson–Crick base pairs. The duplex shows base stacking and NOEs between the base protons T(H6)/A(H8) and the sugar protons (H1′/H2′/H2″) of the preceding nucleotide, as has been observed in antiparallel duplexes. However, no NOEs are observed between base protons H2/H6/H8 of sequential nucleotides, though such NOEs are observed between T(CH3) and A(H8). A three-dimensional structure of the parallel-stranded duplex at atomic resolution has been obtained using molecular dynamics simulations under NMR constraints. The simulated structures have torsional angles very similar to those found in B-DNA duplexes, but the base stacking and helicoid parameters are significantly different.
INTRODUCTION
The right-handed antiparallel double-stranded DNA structure (1), commonly known as B-DNA, has played a central role in explaining several biological processes. However, both DNA and RNA are known to adopt a wide range of unusual conformations. Some examples of well-characterized conformations are single-stranded hairpins (2), triplexes (3–5), tetraplexes (6,7) and the i-motif (8). These structures arise from the fact that nucleic acid bases have multiple sites for hydrogen bonding. Within the stereochemically allowed backbone and side chain torsion angles and sugar puckers (9), alternative structures can be stabilized by favorable stacking interactions and hydrogen bonding schemes other than those involved in Watson–Crick base pairing (10,11) (hydrogen bonded base pairing schemes discussed in this paper are summarized in Fig. 1; we shall refer to these as necessary). DNA polymorphism is governed by several factors. Most important are the base sequence, concentration, temperature, pH and other solvent conditions, which can drive these systems from one conformation to another.
Figure 1.
The hydrogen bonding schemes for antiparallel and parallel DNA duplexes. The antiparallel DNA duplexes are stabilized by Watson–Crick base pairing, while other schemes are possible for parallel duplexes. (A) Watson–Crick G:C base pair, (B) Hoogsteen G:C+ base pair, (C) Donohue G:C base pair, (D) Watson–Crick A:T base pair, (E) Hoogsteen A:T base pair, (F) Donohue A:T base pair and (G) C:C+ base pair.
By nature, nucleic acids usually adopt an antiparallel-stranded (aps) duplex conformation, with one strand in the 5′→3′ and the other in the 3′→5′ orientation. However, they also have the capacity to adopt a parallel-stranded (ps) duplex conformation (12,13). Such ps DNA might play an important role in regulation of replication and transcription, genetic recombination, chromosome folding, mutational processes and RNA splicing (11). At neutral pH, parallel duplexes can be stabilized by reverse Watson–Crick G·C and A·T hydrogen bonded base pairs (14) (also called Donohue base pairs; Fig. 1). Alternatively, ps duplexes can be formed using Hoogsteen A:T base pairing (15). In mildly acidic conditions cytosine N3 is protonated (the pKa of free cytosine is 4.3, but may be different in ordered DNA) and, hence, a Hoogsteen G:C+ base pair can provide additional stabilization to a parallel duplex. Parallel-stranded purine-pyrimidine stretches form part of DNA and RNA triplexes, tetraplexes and H-type structures (3–7,16,17). There have been several theoretical and experimental reports on the formation of ps DNA duplexes (12,13,18,19). These duplexes can be divided into two categories depending on the nature of base pairing: (i) homobase pairs; (ii) heterobase pairs. The former have been extensively studied by UV, CD, IR and gel chromatography and contain A:A, T:T, G:G and C:C+ base pairs (20,21). Heterobase pairs can have complementary or mismatched base pairing (22,23). Parallel-stranded structures have been obtained in vitro by: (i) designing hairpins with polarity reversal using a 3′/3′ or a 5′/5′ linkage (24,25); (ii) use of modified oligonucleotides with a bulky group substitution on the base (26); (iii) chemical modification of backbone or glycosidic linkages, e.g. a duplex between α-anomeric oligodeoxynuleotides and complementary β-oligodeoxynucleotides (27). These structures require conditions such as low pH (where the bases are protonated) and low temperature, which are different from those existing in vivo. Ps duplex formation in such cases is sequence dependent and at high concentrations oligonucleotides may adopt higher ordered structures.
Interestingly, there have been several reports of parallel complementary stretches of DNA in different genomes. For example, Tchurikov and co-workers reported a parallel complementary region between two Drosphila DNA sequences, fragments of the suffix and a 5′-non-coding sequence of the alcohol dehrogenase gene (28,29).
One detailed structural study on ps duplexes has been on d(TCGA), which has C:C+, G:G and A:A+ homobase pairs (30). In d(CGACGAC) the repeat sequence d(CGA) helps in aligning the strands in the parallel orientation (31). A ps structure containing a C:C+ base pair has been observed in single crystals of the complex of ribo(CpA) and proflavin (32). Molecular mechanics calculations (18) on d(A)6:d(T)6, show that the aps form is favored over the ps duplex by only 0.5 kcal/mol. However, details of the three-dimensional (3D) structures of ps duplexes with complementary bases at atomic resolution are lacking.
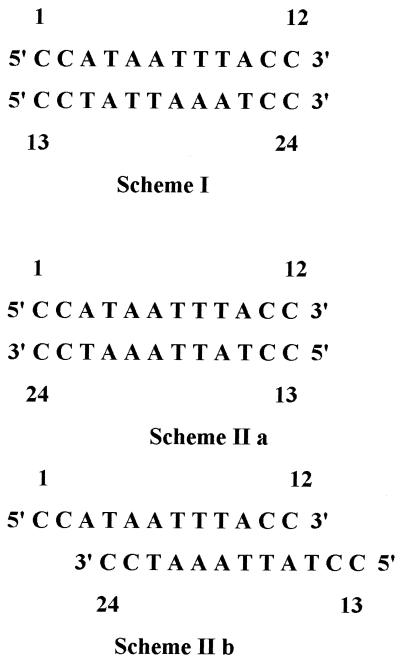
We have used NMR to look at the formation of a ps duplex with complementary bases and to unravel its intricate 3D structural details. For this study, two sequence-constrained linear DNA oligomers [d(CCATAATTTACC and d(CCTATTAAATCC)] have been synthesized such that they match perfectly with parallel polarity in formation of a duplex but are extensively mispaired in the antiparallel orientation. We have used two pairs of C:C+ residues at both ends of the duplex combining it with favorable intermediate sequences to ensure parallel polarity (Scheme 1). As observed earlier, even constrained structures may lead to formation of an aps duplex with mismatched base pairs (33) (Scheme 1) or alternative structures such as triplexes (5) or tetraplexes (6–8).
Scheme 1.
At acidic pH, C:C+ base pairing can occur (34) when the two strands containing C and C+ base stretches are in the parallel orientation and individual cytosine bases have the anti conformation (Fig. 1G). Due to the presence of three hydrogen bonds in each C:C+ base pair, such stretches are expected to constrain the intervening sequences to also be in the parallel orientation. The constraints can be switched off by raising the pH. These constructs can also be used to characterize the structures of duplexes having the same sequence but without terminal C:C+ pairs. In this paper, studies on a 1:1 molar mixture of two oligomers are described. The results support the conclusion that the resultant mixture adopts a ps duplex with reverse Watson–Crick base pairing.
MATERIALS AND METHODS
DNA synthesis and purification
The two DNA dodecamers 5′-d(CCATAATTTACC)-3′ and 5′-d(CCTATTAAATCC)-3′ were synthesized with an Applied Biosystems Model 380B DNA synthesizer using solid state phosphoramidate chemistry (20 µM preparation). After deprotection by heating in concentrated ammonium hydroxide at 55°C for 16 h, oligomers were purified on 15% denaturing polyacrylamide/bis-acrylamide (19:1) gels containing 7.6 M urea, 0.09 M Tris–borate buffer pH 8.3, and 2 mM EDTA. Molar extinction coefficients were measured by phosphate analysis as described previously (35).
NMR
The samples for NMR study were prepared by mixing the oligomers in a 1:1 molar ratio. The final solution contained ∼5 mM concentration of each strand, 0.05 M deuterated sodium acetate buffer and 0.1 M NaCl. For experiments in 2H2O, the sample was lyophilized three times from 2H2O to deuterate all exchangeable protons, prior to dissolution in 0.6 ml of 99.9% 2H2O. For experiments in H2O, a mixture of 90% H2O and 10% 2H2O was used. 1H NMR experiments were carried out in Varian Unity+ 600 and Bruker AMX 500 spectrometers. The spectra in a mixed solvent of 90% H2O + 10% 2H2O included one-dimensional (1D) 1H NMR spectra recorded with a P1 1 pulse sequence (36) and two-dimensional (2D) nuclear Overhauser enhancement spectroscopy (NOESY) (37) with P1 1 detection pulse sequence and a mixing time of 200 ms. The 2D experiments in 2H2O include exclusive correlation spectroscopy (E-COSY) (38), clean total correlation spectroscopy (clean TOCSY) (39) with a mixing time of 80 ms and a set of NOESY spectra with different mixing times (50, 100, 150, 200, 250, 300, 350 and 450 ms). Most NMR experiments were recorded at four temperatures (275, 298, 303 and 308 K) and two pH values (5.5 and 6.8), although 1D 1H experiments were carried out in the range 275–328 K and the pH range 5.2–7.0. In all experiments the 1H carrier frequency was kept at the water resonance. In 2D experiments the time domain data points were 512 and 4096 in the t1 and t2 dimensions, respectively. The data were multiplied by sine bell window functions shifted by π/4 and π/8 along the t1 and t2 axes, respectively, and zero filled to 1024 data points in the t1 dimension prior to 2D-FT. 1H chemical shift calibrations were carried out with respect to the methyl signal (at 0.0 p.p.m.) of 3-(trimethylsilyl)[3,3,2,2-2H] propionate-d4, which was used as an external reference.
Distance restraints
The inter-proton distances have been estimated from a set of NOESY spectra recorded with mixing times of 50, 100, 150, 200, 250, 300, 350 and 450 ms. As a prelude to distance estimation, we monitored the build-up of the NOE volumes of each resolved cross-peak (40–42). Distances were estimated from the initial build-up rates of the NOE curves by the two-spin approximation formula rij = rref(Rij/Rref)1/6, where rij is the distance between protons i and j, rref is a reference distance and Rij and Rref are the initial build-up rates, respectively. The inter-proton distances were estimated using the volume integral of the intranucleotide C(H5)–C(H6) and T(CH3)–T(H6) cross-peaks as reference, except in the estimation of intra-sugar distances. For intra-sugar inter-proton distances, the volume integral of the H2″–H2′ cross-peak, which is independent of the pseudorotation phase angle (P) (43), was used as a reference. Estimated inter-proton distances have been used as constraints with the upper and lower bounds ±0.5 Å in the energy minimization and molecular dynamics calculations. No distance estimation was carried out from NOESY cross-peaks, which either partially overlapped or were weak at short mixing times (50 and 100 ms). However, this information was used to restrain the corresponding proton pairs with lower and upper bounds of 2.0 and 5.0 Å, respectively.
The 1H NMR spectrum shows that four C:C+ and eight A:T base pairs stabilize the formation of ps DNA in the system under study. Based on these data, the hydrogen bond distances C(O2)–C(H42), C(H42)–C(O2) and C(N3)–C(H3) (for C:C+), and A(H61)–T(02) and A(N1)–T(H3) (for A:T) within such base pairs were restrained in the range 1.9–2.2 Å. The heavy atoms in these hydrogen bonds were restrained within the range 2.9–3.1 Å. Corresponding to the strong NOE peaks observed between A(H2) and T(H3) belonging to A:T base pairs, these distances were restrained in the range 3.3–4.0 Å. On the other hand, the distances corresponding to the NOE peaks observed between A(H2) and T(H3) belonging to sequential A:T base pairs were restrained in the range 3.4–4.4 Å. For all these distance constraints a force constant of 100 kcal mol–1 Å–2 was used.
Torsion angle restraints
The information about the range of pseudorotational phase angles (P) obtained from the E-COSY spectrum has been used to define two of the five sugar ring torsion angles (-C2′-C3′-C4′-O4′- and -C1′-C2′-C3′-C4′-). This information was also used to define the lower and upper bounds for one of the backbone torsion angles, δ. Further, the information about glycosidic torsional angles (χ), derived from the intra-nucleotide H6/H8–H1′/H2′/H2″ NOE connectivities, has been used to constrain χ. In addition to the experimental restraints, the rest of the backbone torsion angle constraints have been used according to their preferred ranges of values reported in the literature (9). The ranges of the various torsion angles used are given in Table 1. For all these torsion constraints, a force constant of 100 kcal mol–1 rad–2 was used.
Table 1. Ranges of torsion angles used in structure calculation of the ps duplex.
| Torsion angle | (°) |
|---|---|
| α (O3′-P-O5′-C5′) | 260–310 |
| β (P-O5′-C5′-C4′) | 150–210 |
| γ (O5′-C5′-C4′-C3′) | 30–90 |
| δ (C5′-C4′-C3′-O3′) | 130–160 |
| ɛ (-C4′-C3′-O3′-P-) | 180–240 |
| ζ (-C3′-O3′-P-O5′-) | 260–310 |
| χPy (-O4′-C1′-N1-C2-) | 40–80 |
| χPu (-O4′-C1′-N9-C4-) | 40–80 |
Starting structure
As a first step in modeling the structure, the two individual strands shown in Scheme 1 were generated using the molecular modeling package INSIGHT II (Molecular Simulations Inc., San Diego, CA) on a SGI (Indigo II) workstation. Other steps involved in generating the structure were as follows: (i) parallel orientation of both strands; (ii) initial values of torsion angles chosen to be similar to that of B-DNA; (iii) all glycosidic torsion angles kept in the anti conformation; (iv) the two strands brought close to each other with a C1′–C1′ distance of 12 Å; (v) the structure energy minimized to achieve good inter-strand inter-base hydrogen bonds to arrive at the starting structure.
Molecular dynamics and energy minimization methods
Molecular dynamics (MD) simulations were performed with DISCOVER software (Molecular Simulations Inc.). The AMBER force field was used to calculate the energy of the system. Electrostatic interactions were calculated using Coulomb’s law with point charges (44) and a distance-dependent dielectric constant. van der Waals contributions were calculated with a 6–12 Lennard–Jones potential. A time step of 1 fs was used. To obtain the starting structure, an initial steepest descent minimization of 100 steps was performed on the initial structure, followed by conjugate gradient minimization of 1000 steps. The best-fit structure thus obtained was used for restrained MD simulations. Initial random velocities were assigned with a Maxwell–Boltzmann distribution for a temperature of 600 K. Two hundred structures were collected at 1 ps intervals along the restrained MD trajectory. These structures were significantly different from each other, as is evident from their pairwise root mean square deviations (RMSDs). Each of these structures was cooled to 300 K in steps of 50 K. After each temperature step, the system was allowed to equilibrate for 10 ps. This was followed by 500 steps of steepest descent minimization and 1000 steps of conjugate gradient minimization to monitor convergence and for structure analysis. In the event of any constraint violation, another round of dynamics was performed by varying the initial temperature as well as the weight of the restraints. The molecule was then cooled to 300 K and energy minimized as mentioned. This procedure was repeated until well-converged structures were obtained with zero violations. In these calculations the NMR derived restraints were applied with force constants of 25 kcal mol–1 Å–2 for all NOEs involving non-exchangeable protons, and 100 kcal mol–1 Å–2 for all NOEs involving exchangeable protons and the atoms involved in hydrogen bonds. The structures thus simulated have been analyzed using INSIGHT II and CURVES (45,46).
RESULTS AND DISCUSSION
UV, CD and IR temperature and concentration profiles
UV melting curves were measured with a Cary Model 210 spectrophotometer interfaced to an IBM AT computer. Data were transferred from the AT computer, using the program PC-NFS (Sun Microsystems), to a SUN SPARC Station 2 computer for analysis using the program LAP, written by J. Powell, J. Robinson and J. T. Morris (DCRT, NIH, Bethesda, MD). Cooperative thermal dissociation curves (Fig. 2A) were observed for mixtures containing 25 µmol concentrations of each strand with UV, which indicate that the DNA adopts a distinct ordered conformation below the Tm. The UV melting profiles are consistent with the formation of a pH-, concentration- and temperature-dependent ordered structure when the two strands in Scheme 1 are mixed in aqueous solution (Fig. 2A). These thermal transition profiles vividly show the influence of pH on stability of the structure. At a pH of 5.0 the complex melts cooperatively over the range 43–51°C with a Tm of 47°C. The Tm decreases linearly with pH over the range 5–6.5 as a result of deprotonation of the stabilizing C:C+ terminal groups, but evidence for ordered structure is seen even at a pH of 7.4.
Figure 2.
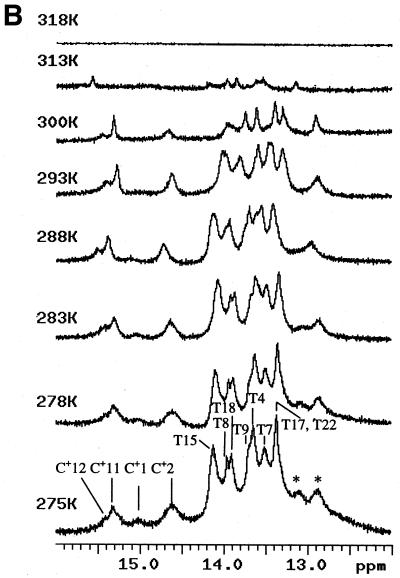
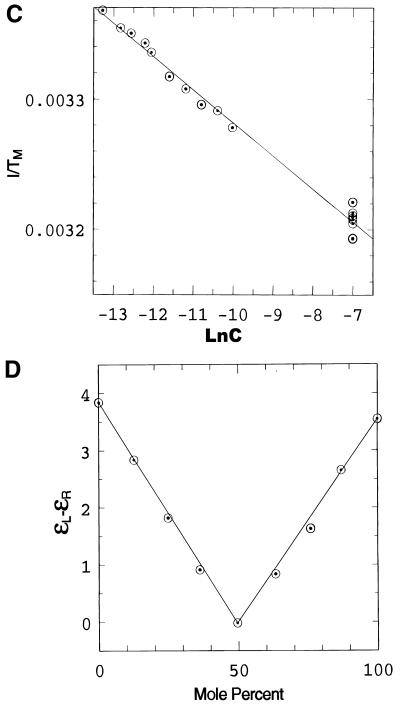
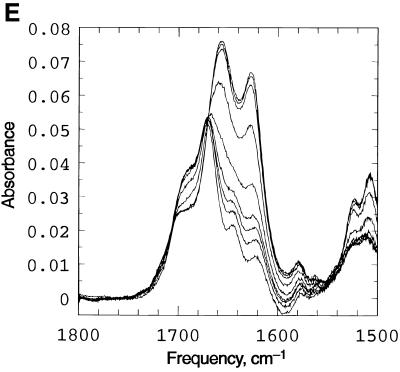
Spectroscopic studies showing the effect of temperature, pH and concentration on the the complex formed when d(CCATAATTTACC) (C1) and d(CCTATTAAATTC) (C2) are mixed in 1:1 stoichiometry. (A) UV temperature profiles as a function of pH. The numerals identify the pH values as follows: 1, pH 5.04; 2, pH 5.57: 3, pH 5.85; 4, pH 6.19; 5, pH 6.18; 6, pH 7.0; 7, pH 7.4. (Inset) Melting temperature of the duplex as a function of pH. (B) Temperature dependence of the imino (3NH) proton region in the 1D 1H NMR spectrum of the same system recorded in a mixed solvent of 90% H2O and 10% 2H2O at pH 5.5. The numbering of the cytosine and thymine nucleotides is as given in Scheme 1. The assignments of C+/T(3NH) protons are as described in the text. (C) Dependence of 1/Tm on the natural logarithm of the concentration (C) of the oligomers at pH 5. The concentration is here expressed as mol phosphate. Points at lnC < –10 are experimental data from the UV melting curve, while points at lnC = –7 correspond to experimental data from the IR melting curve (D) CD mixing curve as a function of C2 mol fraction (0.01 M Na cacodylate pH 5, 1.0 M NaCl, 8°C). Identical curves were obtained using UV. (E) IR spectra of C1:C2 in D2O as a function of temperature (0.2 M Na cacodylate pH 5.0, 1.0 M NaCl, 0.02 M total oligomer concentration). The lowest absorbance curve corresponds to the fully helical complex at 20.1°C and the temperature steps reflected by increasing absorbance are 27.5, 32.0, 34.3, 37.2, 40.0, 42.8, 46.1 and, at the maximum absorbance, 50.9°C.
We have also measured the change in transition temperature Tm as a function of concentration (C) of the two oligomers mixed in a 1:1 ratio. These have been measured over a wide range of concentrations (between 0.1 and 5 µmol strand concentrations by UV and CD, at 1 mM by IR and at 5 mM by NMR) (Fig. 2B). Over these ranges of concentrations the value of Tm varies between 295 and 314 K. The dependence of 1/Tm as a function of lnC (Fig. 2C) is strictly linear over this wide concentration range. This indicates that below the Tm only one species is present over the entire range of concentrations that could be studied by different techniques.
Titrations carried out using UV and CD (mixing curves) show a sharp transition at a 1:1 ratio of the two oligomers, thus establishing the stoichiometry of the complex (Fig. 2D). The IR spectra observed as a function of temperature (Fig. 2E) exhibit an isosbestic point at 1665 cm–1, consistent with the presence of only two states during the process of melting, an ordered duplex and the two single random coiled strands. These observations rule out formation of self-paired duplexes or any other ordered structures.
The melting temperature (Tm) is related to the thermodynamic parameters for the association of single strands into a multi-stranded helix. For the association of n non-self-complementary sequences forming a single nmer structure, the plot of 1/Tm against lnC is expected to be a straight line (47). The slope of such a linear relation is given by (n – 1)R/ΔH0, where R is the gas constant and ΔH0 is the enthalpy of association. If one assumes that the complex is a duplex (n = 2), then one estimates a value of –6.5 kcal mol–1 for the enthalpy of association for each base pair from the slope. This value is similar to the enthalpy values of A:T base pairs (7–8 kcal mol–1) observed for other ordered DNA systems. If, however, the complex was a quadruplex (n = 4) formed from two strands of each oligomer (which will also have a 1:1 stoichiometry), the value of enthalpy is estimated as –2.17 kcal mol–1 for each base pair. This value is too low for the formation of an ordered structure. Further, gel electrophoresis studies carried out on the complex formed when the two strands are mixed in 1:1 proportions (not shown here) show a single band corresponding to the molecular weight of a duplex. Thus, this evidence suggests that the complex present below the Tm is a duplex.
Formation of C:C+ base pairs provide evidence for ps DNA
Figure 2B shows the imino proton region of the 1H NMR spectra of the complex formed when the two DNA oligomers are mixed in a 1:1 stoichiometry. There are a total of four cytosines, four thymines and four adenosines in each DNA strand. It is known that the hydrogen bonded imino protons of T and C+ nucleotides give rise to resonances in the range 11–16 p.p.m. Non-hydrogen bonded protons appear at higher fields and are often too broad to be observed. From the observed resonances of imino protons in the range 13.0–15.5 p.p.m. it is obvious that the molecular system acquires a highly ordered structure under the experimental conditions used and that the imino protons of several pyrimidine and purine bases are involved in inter-base hydrogen bonding. For the sake of clarity, we have marked the positions of various proton resonances in Figure 2B, even though this information was available only after the complete sequential assignments.
Some of the possible ordered structures for the complex formed when the two DNA oligomers are mixed in a 1:1 stoichiometry are: (i) a ps DNA duplex with either reverse Watson–Crick or Hoogsteen base pairing for the eight A:T base pairs, which can be further stabilized by the four C:C+ clamps at slightly acidic pH (Scheme 1); (ii) antiparallel DNA duplexes with Watson–Crick and mismatched base pairs (Scheme 1IIa). For maximum Watson–Crick base pairs, the two strands may be aligned in several different ways. Structure II(a) can be stabilized by C:C base pairs provided one of the two cytosines has a syn conformation. Structure II(b) does not have C:C base pairs but has a relatively more favorable A:T base pairing. Other structures can be proposed, such as self-paired duplexes, a quadruplex, etc., but are ruled out by the IR, CD and UV data presented above.
The stability and dissociation of the DNA complex were monitored by temperature-dependent 1D 1H NMR spectra (Fig. 2B). The broadening and ultimate disappearance of imino protons is indicative of the breaking of hydrogen bonded base pairs. As the temperature increases, the rapidly exchanging C+ imino proton signals broaden considerably, followed by the imino proton resonances belonging to T(3NH) of the complex. Between 313 and 318 K there is a sudden disappearance of all the exchangeable imino proton resonances. Most of the resonances are visible even in the 313 K spectrum and vanish collectively thereafter. Thus, the ordered structure melts in a highly cooperative fashion, with most of the imino protons broadening to noise level in this narrow temperature range. This also shows that all resonances belong to a single conformer and that all 12 bases in one strand are hydrogen bonded to the corresponding bases in the second strand, as indicated in Scheme 1.
We have also recorded 1D 1H NMR spectra of the complex as a function of pH (not shown). Although the pKa of free cytosine is 4.2 at 298 K, the duplex structure is intact even at a pH of 6.8. This shows the relatively higher stability of the C:C+ clamps in the ordered state (Scheme 1), because of which C(N3) remains protonated even at neutral pH. As expected, the melting temperature of the ordered structure is lower at higher pH.
Below the Tm, the basic NMR spectral features do not change with pH, temperature or concentration. In particular, we looked carefully in the T(CH3) region for any spectral changes or splitting of lines as a function of experimental conditions, but did not find any evidence which may suggest multiple conformations. Barring a few broad and weak resonances, all cross-peaks in the 2D spectra could be assigned.
Sequential assignments of non-exchangeable base and sugar protons
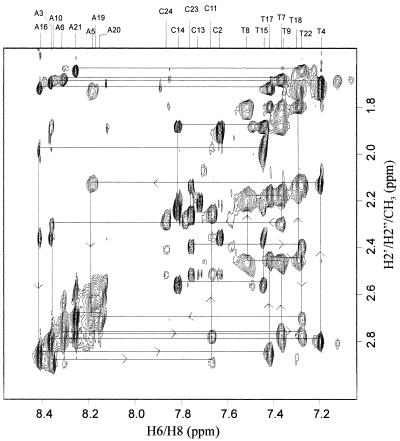
Sequence-specific resonance assignments of non-exchangeable protons in the duplex have been carried out following established procedures (48), with the combined use of 2D TOCSY and 2D NOESY spectra in ∼99.9% 2H2O. Figure 3 shows a selected region of the pure absorption NOESY spectrum of the complex with self and sequential NOE connectivities between H2′/H2″/CH3 and H2/H6/H8. Figure S1 shows self and sequential NOE connectivities between H1′/CH5 and H2/H6/H8. These NOEs allow almost complete sequence-specific resonance assignment of the non-exchangeable protons. In these spectral regions we observe that intra-nucleotide NOEs between the base (H6/H8) and the sugar (H1′, H2′ and H2″) protons mostly follow a general behavior, where the (H6/H8)→H2′ NOE cross-peak is stronger than (H6/H8)→H2″, which in turn is stronger than (H6/H8)→H1′. In fact, most of the H6/H8→H1′ NOE cross-peaks are very weak. This pattern shows that the glycosyl bond torsion angles of the individual A and T nucleotide units adopt the anti conformation. In addition, of the eight cytosines present in the complex at least seven (C2, C11, C12, C13, C14, C23 and C24) show such signatures (Fig. 3A) and reveal that the respective glycosyl bond torsion angles adopt the anti conformation with respect to their individual sugar moieties. In the case of the 3′-end C12 and C24 neucleotides, although the H2′ and H2″ protons were found to be equivalent, their respective H6–H1′ NOEs were found to be relatively very weak. For C1, no intra-nucleotide NOEs between the base (H6) and the sugar (H1′, H2′ and H2″) protons could be seen. The stereospecific assignment of individual H2′ and H2″ protons could be achieved by intensity comparison of the H1′–H2′ and H1′–H2″ cross-peaks in the NOESY spectrum, where the latter is found to be stronger than the former. These results rule out the existence of aps duplex structures II(a) and II(b).
Figure 3.
Selected region of the pure absorption NOESY spectrum of the non-exchangeable protons in the duplex formed between d(CCATAATTTACC) and d(CCTATTAAATCC), recorded in 99.9% 2H2O at pH 5.5. This sub-spectrum shows self and sequential NOE cross-peaks between the sugar H2′, H2″ and CH3 protons and the base H2/H6/H8 protons. The assignment of various peaks was completed following the standard procedure and the walk steps are indicated in the figure. Experimental parameters were as follows: τm = 200 ms, recycle delay 1 s, 64 scans/t1 increment, time domain data points of 600 and 4096 in the t1 and t2 dimensions, respectively. The 1H carrier frequency was kept at the water resonance. The data were multiplied by sine bell window functions shifted by π/4 and π/8 along the t1 and t2 axes, respectively, and zero filled to 1024 data points in the t1 dimension prior to 2D-FT. The digital resolution along the ω1 and ω2 axes correspond to 5.8 and 1.5 Hz/point, respectively.
Sequence-specific resonance assignment of exchangeable protons
Information on the hydrogen bond schematics in DNA duplexes can be derived from exchangeable imino and amino proton resonances and their NOE interactions with other intra- and inter-strand base protons. Each base pair in an ordered DNA structure has one or two imino protons, which are involved in hydrogen bonding. In addition, amino protons belonging to A, C and C+ may be involved in hydrogen bonding. Thus, the number of imino and amino proton signals and their positions in a 1H NMR spectrum throw light upon the type of hydrogen bonded base pairs present in the system.
In the case of a ps duplex, four C:C+ and eight A:T base pairs are expected, which should give rise to four C+(3NH) and eight T(3NH) proton resonances, respectively. Generally, C+(3NH) protons resonate downfield in the chemical shift range 14.5–16.0 p.p.m. and, hence, can be easily identified. We indeed see broad imino proton resonances downfield of 14.5 p.p.m. (between 14.5 and 15.5 p.p.m.) and assign them as those arising from C+(3NH) protons. Integration of these broad lines accounts for four imino protons with their individual chemical shifts at 14.62, 15.05, 15.30 and 15.40 p.p.m. (Fig. 2B). Among them, resonances at 15.05 and 15.30 p.p.m. are broader compared with the ones at 14.62 and 15.40 p.p.m. This difference can be attributed to the exchange rates of these imino protons with the solvent. Imino protons belonging to the terminal C:C+ base pairs C1:C13+ and C12:C24+ (Scheme 1) would exchange faster compared with the interior C2:C14+ and C11:C23+ pairs. This interpretation is supported by the 1D 1H NMR temperature dependence study, wherein the resonances assigned to the terminal C:C+ base pairs broaden and disappear first as the temperature is increased, followed by those belonging to the interior ones. The observation of these imino proton resonances confirms the formation of C:C+ base pairs (Fig. 1), particularly in the parallel orientation, as seven of eight cytosines were found with their respective glycosyl bond torsion angles in the anti conformation with respect to their individual sugar moieties. If they had paired in the antiparallel orientation, one of the cytosines in the C:C+ base pairs should have shown the signature of the syn orientation. This interpretation could have been further supported by the direct observation of intra C:C+ base pair NOEs between 3NH and 4NH2 protons. However, 4NH2 proton reosnances were too broad to show any NOEs, even at 275 K.
The formation of C:C+ base pairs with both individual bases in the anti conformation rules out the possibility of an antiparallel DNA duplex as well as self-paired structures. For example, if one of the strands self-paired, the other strand would also have the capacity to self-pair and this would have given rise to a complex NMR spectrum, wherein one would have seen resonances arising from four different 12mer strands. As we could account for only two 12mer strands, such self-pairing is ruled out. On the other hand, an intercalated stucture of any kind is ruled out because we did not observe H11–H11 cross-peaks in the NOESY spectrum recorded in 2H2O.
We observed six broad imino proton resonances in the range 13.0–14.5 p.p.m. The degeneracy of these individual resonances due to small chemical shift differences could be removed in the 2D NOESY spectrum in most cases, because of the better dispersion of chemical shift of the second proton, giving rise to a resolved cross-peak (Fig. 4). We can indeed account for eight resonances in this region (13–14.5 p.p.m.). This allows fairly unambiguous sequential assignment of resonances for exchangeable and for most of the non-exchangeable protons belonging to the eight A:T base pairs present in Scheme 1. The discussions below are based on the final assignments, which were cross-checked for consistencies among the various NOE cross-peaks. Imino protons of all eight thymines present in the ps duplex can be unambiguously assigned following the sequential NOE connectivities seen between various imino proton resonances participating in the individual base pairings. Although we could walk along the eight base pairs, we could not resolve the directionality of the connectivity pathway, i.e. whether it is from T15(3NH) to T22(3NH) [i.e. T15(3NH) (belonging to base pair A3:T15)→T4(3NH) (T4:A16)→T17(3NH) (A5:T17)→T18(3NH) (A6:T18)→T7(3NH) (T7:A19)→T8(3NH) (T8:A20)→T9(3NH) (T9:A21)→T22(3NH) (A10:T22)] or vice versa, i.e. from T22(3NH) to T15(3NH). These connectivities are illustrated in Figure 4A. Such assignments of imino protons are substantiated by other NOE connectivities to these protons with H2 protons of individual adenosines (Fig. 4B). For example, in the A3:T15 base pair, a strong NOE is seen between T15(3NH) and A3(H2). One also expects NOEs to be present diagonally across the strands [such as T17(3NH)–A6(H2)] or within the same strand [such as T15(3NH)–A16(H2)], between two neighboring base pairs. Hence, the walk through T(3NH)–A(H2) NOEs throughout the stretch of eight A:T base pairs provides further support for the assignment of all T(3NH) resonances. As expected, the imino protons belonging to the terminal A:T base pairs, A3:T15 and A10:T22, showed one intra-base pair and one inter-base pair T(3NH)–A(H2) NOE, while the rest showed one intra-base pair and two inter base-pair T(3NH)–A(H2) NOEs. The C(4-NH2) and A(6-NH2) resonances, which could have resolved the directionality problem of this assignment of exchangeable and A(H2) protons, are broad and hence could not be characterized in detail. As far as the assignment of other non-exchangeable protons are concerned, there was no such directionality problem. At the end, there were only two unassigned broad resonances at 12.85 and 13.12 p.p.m. (indicated by asterisks in Fig. 2B). Except for these two resonances, no other resonance was left unassigned in the 1D 1H NMR spectrum.
Figure 4.
Selected regions of the pure absorption NOESY spectrum involving exchangeable protons in the duplex formed between d(CCATAATTTACC) and d(CCTATTAAATCC), recorded in a mixed solvent of 90% H2O and 10% 2H2O at pH 5.5. (A) Sequential NOEs between thymine (3NH) imino protons; (B) self and sequential NOEs between T(3NH) and A(H2) protons showing that all thymines are involved in hydrogen bonding with adenines. The assignment of various peaks was completed following the standard procedure and the walk steps are indicated in the figure. Experimental parameters were as follows: τm = 200 ms, recycle delay 1 s, 128 scans/t1 increment, time domain data points of 600 and 4096 in the t1 and t2 dimensions, respectively. The 1H carrier frequency was kept at the water resonance. The data were multiplied by sine bell window functions shifted by π/4 and π/8 along the t1 and t2 axes, respectively, and zero filled to 1024 data points in the t1 dimension prior to 2D-FT. During the assignment, although we could walk along the 8 bp, we could not resolve the directionality of the connectivity pathway, whether it is from T15(3NH) to T22(3NH) or vice versa, i.e. from T22(3NH) to T15(3NH) (see text). This means that the chemical shift of T15(3NH) may take the value of T22(3NH) and T22(3NH) that of T15(3NH). Likewise, T4(3NH) may take the value of T9(3NH) and T9(3NH) that of T4(3NH), and so on. This ambiguity in the assignment is highlighted in the chemical shift table.
The chemical shifts of all exchangeable and non-exchangeable protons thus obtained are deposited in the Protein Data Bank (PDB) (RCSB ID code RCSB014208; PDB ID accession code 1JUU). (See also Supplementary Material, Table S1.)
Nature of hydrogen bonding in the ps duplex
One of the strong pieces of evidence for the formation of a parallel duplex is the observation of C:C+ hydrogen bonded base pairs involving the C+(3NH) imino protons. The aps duplex cannot form C:C+ base pairs when both bases are in the anti conformation. The A:T base pairing in the parallel-stranded duplex can be either reverse Watson–Crick or Hoogsteen (Fig. 1). A Hoogsteen base pair is expected to show a T(3NH)–A(H8) NOE, while the reverse Watson–Crick base pair is expected to show a T(3NH)–A(H2) interaction. In the present study we observed only the T(3NH)–A(H2) NOE cross-peaks. This supports the formation of reverse Watson–Crick A:T base pairs.
Sugar puckers derived from three-bond coupling constants (3J)
An E-COSY experiment has been used to resolve conformation-dependent characteristic multiplet structures of H2′–H1′ and H2″–H1′ cross-peaks. The ω2 axis in these cross-peaks contains information about 3J(H1′–H2′) and 3J(H1′–H2″). Though the estimated J values are not very precise, one can certainly conclude which of them is larger and thus fix a range for the sugar puckers. In the present study, even though there are 24 furanose rings in the ps duplex, the H2′/H2″–H1′ region of the E-COSY spectrum shows fairly well resolved cross-peaks (Supplementary Material, Fig. S1). Wherever the cross-peaks are well resolved, the 3J(H1′–H2′) and 3J(H1′–H2″) values were estimated. In all cases, 3J(H1′–H2′) was found to be larger than 3J(H1′–H2″). These J values qualitatively indicate that the corresponding sugar rings adopt conformations in the S domain of the pseudorotational map with P ranging from C1′-exo to C3′-exo (P = 90–198). The 3J(H2″–H3′) and 3J(H3′–H4′) values which could have helped in further narrowing the domain of the sugar puckers could not be estimated from the E-COSY spectrum because of the low intensities of the corresponding peaks.
NMR structure determination of the ps duplex
Restrained MD simulation and energy minimization calculations were performed on the ps duplex following the procedure described in Materials and Methods. A total of 78 inter-proton distance constraints measured by NOE, 56 hydrogen bond constraints and 162 torsion constraints were used with the force constants described earlier. Among the NOE constraints, 14 were inter-strand, 27 intra-residue and the remaining were intra-strand inter-residue constraints. All these constraints have been deposited in the PDB (RCSB ID code RCSB014208; PDB ID accession code 1JUU). Of the 200 calculated structures, there were 19 structures which lay within 4.0 kcal mol–1 above the minimum energy structure. These 20 structures are characterized by low all-atom pairwise RMSDs, which range from 0.18 to 1.26 (Supplementary Material, Table S2). The stereochemistry of all 20 structures was critically examined for acceptable hydrogen bond lengths and angles and for stereochemical feasibility of the torsion angles. Non-bonded inter-atomic distances were also required to be in allowable ranges. All 20 structures satisfied these criteria.
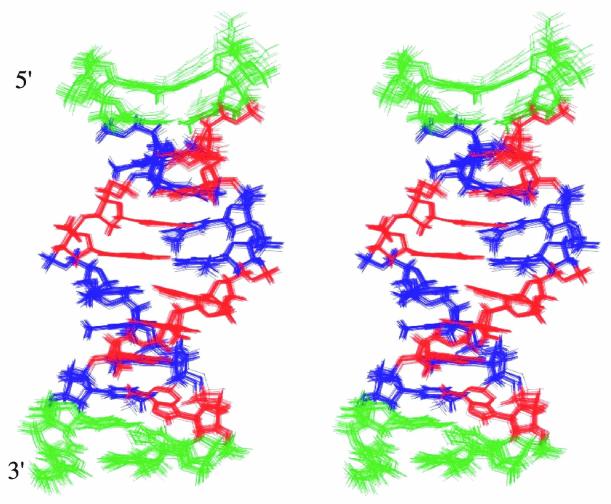
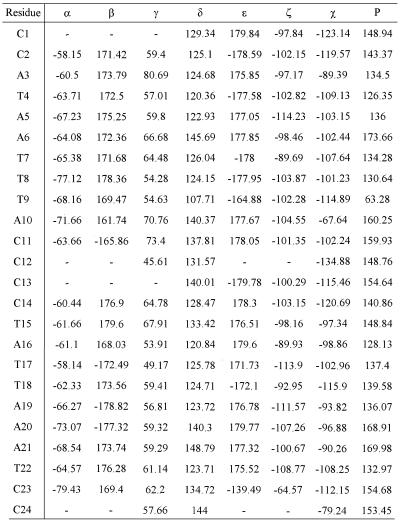
Figure 5 shows the best fit superimposition of these 20 structures. The corresponding structure files in PDB format have been deposited in the PDB (RCSB ID code RCSB014208; PDB ID accession code 1JUU). The backbone torsion angles, glycosidic torsion angle and pseudorotation angle of each residue in the lowest energy conformation of the ps duplex are listed in Table 2. The range of backbone torsion angles, glycosidic torsion angles (χ) and P values for the selected 20 structures are shown in the Supplementary Material (Fig. S2). Even though considerable flexibility has been allowed in the sugar and backbone torsion angles, the final structures converged into a narrow range of torsion angles at the end of the MD simulations. The deviations were found to be larger only near the ends of these structures. Sugar conformations are in the S domain and vary from C1′-exo to C2′-endo for all individual nucleotides, except for T9, which has a C4′-exo pucker. For T9, A10, C14, T15 and T22, P values varied over a wider range. Glycosidic torsion (χ) angles for all individual nucleotide units adopt the anti conformation. Nucleotides A3, T15 and T22 show a somewhat larger spread of torsional angles among the 20 selected structures. Further, the glycosidic bonds for all the base pairs are in the trans orientation with respect to each other.
Figure 5.
Stereo view of 20 lowest energy structures simulated by molecular dynamics under NMR constraints. As expected, the structures show wider differences at the two ends of the parallel duplex while the middle parts converge fairly close to each other.
Table 2. Torsion angles of each residue in the lowest energy conformation of ps duplex.
The lowest energy structure of the ps duplex has been selected for further discussion. For this structure the backbone torsion angles lie in a restricted region as compared with the glycosidic torsion and pseudorotation angles (Table 2). Angles α (-O3′-P-O5′-C5′-), β (-P-O5′-C5′-C4′-), γ (-O5′-C5′-C4′-C3′-) and ɛ (-C4′-C3′-O3′-P-) for individual nucleotides in the ps DNA are mostly locked into the gauche– (g–), trans (t), gauche+ (g+) and trans (t) conformations, respectively, similar to the angles observed in B-DNA. The average values for α, β, γ and ɛ are –65°, –172°, 60° and 175°. The ζ (-C3′-O3′-P-O5′-) values are –105° on average and range from –64° to –114°. The values of δ (-C5′-C4′-C3′-O3′-) show more flexibility and vary through the length of the duplex. The δ values are 130° on average and range from 107° to 148°. It should be pointed out that the value of δ is linked to the sugar pucker.
The interstrand C1′–C1′ distances for hydrogen bonded base pairs in the lowest energy structure of the ps duplex are close to 11.4 Å, which is greater by ∼2 Å compared with the value observed in aps duplexes for the A:T base pair region. Inter-atomic distances between non-exchangeable base protons (H2/H6/H8) of the successive nucleotides obtained from the structure are >4 Å, explaining the absence of sequential NOEs. The helix diameter is ∼19 Å for the A:T stretch and 17 Å for the C:C+ region. The major and minor grooves are of similar width (8–9 Å), which is a consequence of reverse Watson–Crick base pairing, with T(CH3) and A(H2) on the same groove. The groove widths for an aps duplex with a similar sequence are 6 and 12 Å, respectively, and the diameter is ∼19 Å. It has been suggested that larger groove widths allow higher flexibility of the phosphate backbone (49) and this may be the case with the ps duplex.
Inter-strand hydrogen bonding and base stacking
As discussed earlier, the C:C+ base pairs at the two ends of the ps duplex with three hydrogen bonds each provide major stability to parallel duplexes. Each of the eight A:T base pairs present in the ps duplex has two hydrogen bonds. Base pair stacking provides an additional source of stability in DNA duplexes. There are nine pyrimidine–pyrimidine, ten pyrimidine–purine and three purine–purine stacking interactions in the ps duplex. The base stacking interactions and hydrogen bonding for the ps duplex are shown in Supplementary Material, Figure S3. We observe that polar substituents do not stack well on the aromatic ring of the adjacent base. It is well known that protonated pyrimidines do not stack well (10). Thus, the stacking interactions in ps duplexes may be somewhat weaker than in aps duplexes.
Helicoid parameters
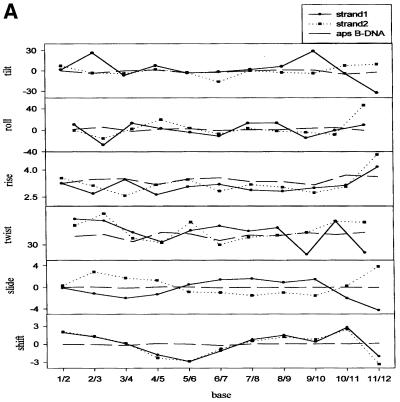
Unlike X-ray crystallography, NMR data does not provide direct evidence on helicoid parameters. However, it is of interest to compare the parameters of the simulated structures of the ps duplex with a model aps duplex having A:T stretches. The inter-base parameters (50) for the ps duplex are given in Figure 6A–C. For comparison, we generated a model B-DNA structure with A:T stretches using INSIGHT II and CURVES (45,46). The values for the aps B-DNA duplex are shown for comparison as dotted lines.
Figure 6.
Comparison of various helicoid parameters of the minimum energy structure for the ps duplex with that of a model aps duplex (B-DNA) (see text) having A:T stretches.
Rise (Dz) varies over the length of the duplex. Dz is lower for the ps duplex compared with the aps duplex. However, Dz is relatively larger at the 3′-end. The deviations from aps duplex conformation are larger at the 3′-end for the ps duplex. Average twist was found to be 38.4°. On the other hand, propeller twist and tip show the maximum deviations from that of the B-DNA conformation. The average propeller twist is 120° and 121° for the ps duplex, while it is –16° for B-DNA. Tip for the second strand (residues 13–24) also differs significantly from that of B-DNA. As expected, the values indicate that the two strands in the ps duplex do not have equivalent helicoid parameters.
CONCLUSION
Though it is widely accepted that the basic structure of DNA corresponds to an antiparallel double helix as proposed by Watson and Crick, the possibility of a parallel duplex has not been ruled out. Parallel-stranded DNA stretches in biological systems have recently been proposed and, therefore, this is a new and emerging area of research. In this paper we have demonstrated the structural feasibility of a parallel-stranded DNA. We have obtained its 3D structure by NMR spectroscopy. The duplex is thermodynamically stable under physiological conditions. The structure has backbone, glycosidic torsion and pseudorotation angles similar to those observed in an aps B-DNA. However, the A:T base pairs adopt reverse Watson–Crick pairing. The helicoid parameters of the ps and aps duplexes are significantly different and so also are the patterns of base pair stacking.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The facilities provided by the National Facility for High Field NMR, supported by the Department of Science and Technology, Department of Biotechnology, Council of Scientific and Industrial Research and Tata Institute of Fundamental Research, Mumbai, India, are gratefully acknowledged.
PDB no. 1JUU
REFERENCES
- 1.Watson J.D. and Crick,F.H. (1953) Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature, 171, 737–738. [DOI] [PubMed] [Google Scholar]
- 2.Orbons L.P.M., van der Marcel,G.A., van Boom,A.H. and Altona,C. (1986) Hairpin and duplex formation of the DNA octamer d(m5C-G-m5C-G-T-G-m5C-G) in solution. An NMR study. Nucleic Acids Res., 14, 4187–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Felsenfield G., Davies,D.R. and Rich,A. (1957) Formation of a three-stranded polynucleotide molecule. J. Am. Chem. Soc., 79, 2023–2024. [Google Scholar]
- 4.Lee J.S., Johnson,D.A. and Morgan,A.R. (1979) Complexes formed by (pyrimidine)n·(purine)n DNAs on lowering the pH are three-stranded. Nucleic Acids Res., 6, 6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhaumik S.R., Chary,K.V.R., Govil,G., Liu,K. and Miles,H.T. (1995) NMR characterisation of a triple stranded complex formed by homo-purine and homo-pyrimidine DNA strands at 1:1 molar ratio and acidic pH. Nucleic Acids Res., 23, 4116–4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aboul-Ela F., Murchie,A.I.H., Norman,D.G. and Lilley,D.M. (1994) Solution structure of a parallel-stranded tetraplex formed by d(TG4T) in the presence of sodium ions by nuclear magnetic resonance spectroscopy. J. Mol. Biol., 243, 458–471. [DOI] [PubMed] [Google Scholar]
- 7.Phillips K., Dauter,Z., Murchie,A.I., Lilley,D.M. and Luisi,B. (1997) The crystal structure of a parallel-stranded guanine tetraplex at 0.95 Å resolution. J. Mol. Biol., 273, 171–182. [DOI] [PubMed] [Google Scholar]
- 8.Gehring K., Leroy,J.L. and Guéron,M. (1993) A tetrameric DNA structure with protonated cytosine.cytosine base pairs. Nature, 363, 561–565. [DOI] [PubMed] [Google Scholar]
- 9.Govil G. and Hosur,R.V. (1982) Conformation of Biological Molecules. NMR, Vol. 20, Springer-Verlag, New York, NY.
- 10.Saenger W. (1984) Principles of Nucleic Acid Structure. Springer-Verlag, New York, NY, p. 556.
- 11.Soyfer V.N. and Potaman,V.N. (1996) Triple-helical Nucleic Acids, Springer-Verlag, New York, NY.
- 12.Ramsing N.B., Rippe,K. and Jovin,T.M. (1989) Helix-coil transition of parallel-stranded DNA. Thermodynamics of hairpin and linear duplex oligonucleotides. Biochemistry, 28, 9528–9535. [DOI] [PubMed] [Google Scholar]
- 13.van de Sande J.H., Ramsing,N.B., Germann,M.W., Elhorst,W., Kalisch,B.W., Kitzing,V.E., Pon,R.T., Clegg,R.C. and Jovin,T.M. (1988) Parallel stranded DNA. Science, 241, 551–557. [DOI] [PubMed] [Google Scholar]
- 14.Otto C., Rippe,K., Thomas,K., Ramsing,N.B. and Jovin,T.M. (1991) The hydrogen-bonding structure in parallel-stranded duplex DNA is reverse Watson-Crick. Biochemistry, 30, 3062–3069. [DOI] [PubMed] [Google Scholar]
- 15.Raghunathan G., Miles,H.T. and Sasisekharan,V. (1994) Parallel nucleic acid helices with Hoogsteen base pairing: symmetry and structure. Biopolymers, 34, 1573–1581. [DOI] [PubMed] [Google Scholar]
- 16.Smith F.W. and Feigon,J. (1992) Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature, 356, 126–131. [DOI] [PubMed] [Google Scholar]
- 17.Mirkin S.M. and Frank-Kamenetskii,M.D. (1994) H-DNA and related structures. Annu. Rev. Biophys. Biomol. Struct., 23, 541–576. [DOI] [PubMed] [Google Scholar]
- 18.Pattabiraman N. (1986) Can the double helix be parallel? Biopolymers, 25, 1603–1606. [DOI] [PubMed] [Google Scholar]
- 19.Rippe K. and Jovin,T.M. (1992) Parallel-stranded duplex DNA. Methods Enzymol., 211, 199–220. [DOI] [PubMed] [Google Scholar]
- 20.Suda T., Mishima,Y., Asakura,H. and Kominami,R. (1995) Formation of a parallel-stranded DNA homoduplex by d(GGA) repeat oligonucleotides. Nucleic Acids Res., 23, 3771–3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westof E. and Sundaralingam,M. (1980) X-ray-structure of a cytidylyl-3′,5′-adenosine-proflavine complex: a self-paired parallel-chain double helical dimer with an intercalated acridine dye. Proc. Natl Acad. Sci. USA, 77, 1852–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Förtsch I., Fritzsche,H., Birch-Hirschfeld,E., Evertsz,E., Klement,R., Jovin,T.M. and Zimmer,C. (1996) Parallel-stranded duplex DNA containing dA.dU base pairs. Biopolymers, 38, 209–220. [DOI] [PubMed] [Google Scholar]
- 23.Dolinnaya N., Ulku,A. and Fresco,J.R. (1997) Parallel-stranded linear homoduplexes of d(A+-G)n > 10 and d(A-G)n > 10 manifesting the contrasting ionic strength sensitivities of poly(A+.A+) and DNA. Nucleic Acids Res., 25, 1100–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lancelot G., Guesnet,J.L. and Vovelle,F. (1989) Solution structure of the parallel-stranded duplex oligonucleotide α-d(TCTAAAC)-β-d(AGATTTG) via complete relaxation matrix analysis of the NOE effects and molecular mechanics calculations. Biochemistry, 28, 7871–7878. [DOI] [PubMed] [Google Scholar]
- 25.Thuong N.T., Asseline,U., Roig,V., Takasugi,M. and Hélène,C. (1987) Oligo(α-deoxynucleotide)s covalently linked to intercalating agents: differential binding to ribo- and deoxyribopolynucleotides and stability towards nuclease digestion. Proc. Natl Acad. Sci. USA, 84, 5129–5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishikawa F., Frazier,J., Howard,F.B. and Miles,H.T. (1972) Polyadenylate polyuridylate helices with non-Watson-Crick hydrogen bonding. J. Mol. Biol., 70, 475–490. [DOI] [PubMed] [Google Scholar]
- 27.Praseuth D., Chassignol,M., Takasugi,M., Le Doan,T., Thuong,N.T. and Hélène,C. (1987) Double helices with parallel strands are formed by nuclease-resistant oligo-[α]-deoxynucleotides and oligo-[α]-deoxynucleotides covalently linked to an intercalating agent with complementary oligo-[β]-deoxynucleotides. J. Mol. Biol., 196, 939–942. [DOI] [PubMed] [Google Scholar]
- 28.Tchurikov N.A., Ebralidze,A.K. and Georgiev,G.P. (1986) The suffix sequence is involved in processing the 3′ ends of different mRNAs in Drosophila melanogaster. EMBO J., 5, 2341–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tchurikov N.A., Chernov,B.K., Golova,Y.B. and Nechipurenko,Y.D. (1989) Parallel DNA: generation of a duplex between two Drosophila sequences in vitro. FEBS Lett., 257, 415–418. [DOI] [PubMed] [Google Scholar]
- 30.Wand Y. and Patel,D.J. (1994) Solution structure of the d(T-C-G-A) duplex at acidic pH. A parallel-stranded helix containing C+.C, G.G and A.A pairs. J. Mol. Biol., 242, 508–526. [PubMed] [Google Scholar]
- 31.Robinson H. and Wang,A.H. (1993) 5′-CGA sequence is a strong motif for homo base-paired parallel-stranded DNA duplex as revealed by NMR analysis. Proc. Natl Acad. Sci. USA, 90, 5224–5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coll M., Solans,X., Altaba,F.M. and Subirana,J.A. (1987) Crystal and molecular structure of the sodium salt of the dinucleotide duplex d(CpG). J. Biomol. Struct. Dyn., 4, 797–811. [DOI] [PubMed] [Google Scholar]
- 33.Bhaumik S.R., Chary,K.V.R., Govil,G., Liu,K. and Miles,H.T. (1997) Homopurine and homopyrimidine strands complementary in parallel orientation form an antiparallel duplex at neutral pH with A-C, G-T and T-C mismatched base pairs. Biopolymers, 41, 773–785. [DOI] [PubMed] [Google Scholar]
- 34.Langridge R. and Rich,A. (1963) Molecular structure of helical polycytidylic acid. Nature, 198, 725. [DOI] [PubMed] [Google Scholar]
- 35.Muraoka M., Miles,H.T. and Howard,F.B. (1980) Copolymers of adenylic and 2-aminoadenylic acids. Effect of progressive changes in hydrogen bonding and stacking on interaction with poly(uridylic acid). Biochemistry, 19, 2429–2439. [DOI] [PubMed] [Google Scholar]
- 36.Hore P.J. (1983) Solvent suppresion in Fourier transform nuclear magnetic resonance. J. Magn. Reson., 55, 283–300. [Google Scholar]
- 37.Kumar A., Wagner,G., Ernst,R.R. and Wuthrich,K. (1980) Studies of J-connectivities and selective 1H-1H Overhauser effects in H2O solutions of biological macromolecules by two-dimensional NMR experiments. Biochem. Biophys. Res. Commun., 96, 1156–1163. [DOI] [PubMed] [Google Scholar]
- 38.Griesinger C., Sorensen,O.W. and Ernst,R.R. (1986) Correlation of connected transitions by two-dimensional NMR spectroscopy. J. Chem. Phys., 85, 6837–6852. [Google Scholar]
- 39.Grieseinger C., Otting,G., Wuthrich,K. and Ernst,R.R. (1988) Clean-TOCSY for 1H spin system identification in macromolecules. J. Am. Chem. Soc., 110, 7870–7872. [Google Scholar]
- 40.Kumar A., Wagner,G., Ernst,R.R. and Wuthrich,K. (1981) Buildup rates of the nuclear Overhauser effect measured by two-dimensional proton magnetic resonance spectroscopy: implications for studies of protein conformation. J. Am. Chem. Soc., 103, 3654–3658. [Google Scholar]
- 41.Wagner G. and Wuthrich,K. (1979) Truncated driven nuclear Overhauser effect (TOE): a new technique for studies of selective 1H-1H Overhauser effects in the presence of spin diffusion. J. Magn. Reson., 33, 675–680. [Google Scholar]
- 42.Chary K.V.R., Hosur,R.V., Govil,G., Chen,C. and Miles,H.T. (1988) Sequence-specific solution structure of d-GGTACGCGTACC. Biochemistry, 27, 3858–3867. [DOI] [PubMed] [Google Scholar]
- 43.Chary K.V.R. and Modi,S. (1988) Analysis of intrasugar interproton NOESY cross-peaks as an aid to determine sugar geometries in DNA fragments. FEBS Lett., 233, 319–325. [DOI] [PubMed] [Google Scholar]
- 44.Cornell W.D., Cieplak,P., Bayly,C.I., Gould,I.R., Merz,K.M., Ferguson,D.M., Spellmeyer,D.C., Fox,T., Caldwell,J.W. and Kollman,P.A. (1995) J. Am. Chem. Soc., 117, 5179–5197. [Google Scholar]
- 45.Lavery R. and Sklenar,H. (1988) The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn., 6, 63–91. [DOI] [PubMed] [Google Scholar]
- 46.Lavery R. and Sklenar,H. (1989) Defining the structure of irregular nucleic acids: conventions and principles. J. Biomol. Struct. Dyn., 6, 655–667. [DOI] [PubMed] [Google Scholar]
- 47.Breslauer K.J. (1995) Extracting thermodynamic data from equilibrium melting curves for DNA. Methods Enzymol., 259, 221–242. [DOI] [PubMed] [Google Scholar]
- 48.Wüthrich K. (1986) NMR of Proteins and Nucleic Acids. Wiley, New York, NY.
- 49.Garcia A.E., Soumpasis,D.M. and Jovin,T.M. (1994) Dynamics and relative stabilities of parallel- and antiparallel-stranded DNA duplexes. Biophys. J., 66, 1742–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ravishanker G., Swaminathan,S., Beveridge,D.L., Lavery,R. and Sklenar,H. (1989) Conformational and helicoidal analysis of 30 PS of molecular dynamics on the d(CGCGAATTCGCG) double helix: “curves”, dials and windows. J. Biomol. Struct. Dyn., 6, 669–699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.