Abstract
Background: Studies have shown that ferroptosis- and oxidative stress-related genes (FORGs) perform crosstalk in ovarian cancer (OC). The specific role of FORGs in OC, however, remains unclear. We aimed to develop a molecular subtype and prognostic model associated with FORGs that could predict OC prognosis and evaluate the infiltration of tumor-associated immune cells. Methods: Gene expression samples were collected from the GEO (GSE53963) and Cancer Genome Atlas (TCGA) databases. Kaplan-Meier analysis was used to evaluate prognostic efficacy. Unsupervised clustering was applied to identify molecular subtypes, which was followed by tumor immune cell infiltration and functional enrichment analyses. Subtype-related differentially expressed genes (DEGs) were identified and used to establish prognostic models. Associations between the model and immune checkpoint expression, stromal scores, and chemotherapy were investigated. Results: OC patients were categorized into two FORG subtypes based on the expression characteristics of 19 FORGs. Molecular subtypes associated with patient prognosis, immune activity, and energy metabolism pathways were identified. Subsequently, DEGs in the two FORG subtypes were identified and used in prognostic models. We identified six signature genes (MEGF8, ECE1, SASH1, ARHGEF16, PLXNA1, and FCGBP) with LASSO analysis to assess the risk of OC. Patients in the high-risk group had poor prognoses and immunosuppression, while the risk scores were significantly associated with immune checkpoint expression, stromal scores, and chemotherapy sensitivity. Conclusions: Our novel clustering algorithm was used to create distinct clusters of OC patients and a prognostic model was developed that accurately predicted patient outcomes and chemotherapy responses. This approach offers effective precision medicine for OC patients.
Keywords: Ferroptosis, oxidative stress, ovarian cancer, prognosis
Introduction
Ovarian cancer (OC) is a prevalent and lethal malignant tumor in women, and it has an increasing incidence [1,2]. Although its incidence rate is lower than that of cervical cancer and endometrial cancer, which ranks third in gynecologic malignancies, its mortality rate is higher. Due to its heterogeneous clinical manifestations and molecular mechanisms, more than 70% of patients are diagnosed at a late stage [2]. Despite the application of new target drugs, including poly (ADP-ribose) polymerase (PARP) inhibitors, the survival rate of OC remains low, with a five-year survival rate of only 47.8% [3]. In clinical settings, serum carbohydrate antigen 125 (CA125) and other tumor markers are not good at predicting poor prognoses, recurrence, and chemotherapy benefits in OC patients. Moreover, single gene prognosis prediction models are often ineffective in the face of complex causative molecular mechanisms. Consequently, there is an urgent need for novel prognostic biomarkers to improve OC outcome.
Ferroptosis is a newly identified form of programmed cell death, characterized by iron-dependent accumulation of lipid peroxidation and an increase in reactive oxygen species (ROS) [4]. This form of cell death is distinct from autophagy, necrosis, cuproptosis, and apoptosis. Research has shown that ferroptosis plays a key role in OC cell proliferation and metastasis, as well as in the maintenance of tumor stemness, through abnormal expression of ferroptosis-related genes (FRGs) [5]. Bioinformatic analysis identified 57 FRGs that were abnormally expressed in tissues from OC patients compared to normal tissues, and they were associated with OC prognosis and immune cell infiltration [6]. Therefore, targeting FRGs to induce iron-dependent cell death has been proposed as a novel approach to control OC progression, even in drug-tolerant tumors. Additionally, increased levels of oxidative stress are a hallmark of tumor cells and the primary source of ROS, which can promote lipid peroxidation and ferroptosis, including NADPH oxidase (NOX) and mitochondrial-induced ROS [7]. In a moderate oxidative stress microenvironment, tumor cells can survive and proliferate, as they have evolved to maintain redox homeostasis [8]. This makes them resistant to chemotherapy. Therefore, targeting ferroptosis- and oxidative stress-related genes (FORGs) to simultaneously inhibit oxidative stress and ferroptosis signaling pathways is gaining considerable attention.
In this study, the frequency of mutations, changes in copy number, and the prognostic value of the FORGs were analyzed in OC patients. A total of 596 sample data were collected from the Gene Expression Omnibus (GEO; GSE53963) and Cancer Genome Atlas (TCGA; https://portal.gdc.cancer.gov) databases and patients were divided into two clusters. Prognostic values and immune infiltration were then analyzed within these two clusters. Differentially expressed genes (DEGs) between the two clusters were identified, and a risk prediction model was established. This model was found to be effective in predicting a patient’s overall survival, immune infiltration, and sensitivity to chemotherapy and immune checkpoint blockade therapy, indicating that our model’s signature may be a biomarker for predicting the prognosis of OC patients and guiding precise treatment strategies.
Materials and methods
Collection of OC data
RNA-seq data and patient clinical information data were acquired from TCGA and GEO (GSE53963) databases. Patients with overall survival (OS) times and status were included in the study. FORGs were acquired from a previous study [9]. Copy number variation (CNV) data and somatic mutations were downloaded from TCGA. Prognostic values of FORGs were evaluated using Kaplan-Meier (KM) methods.
Consensus clustering to identify FORG clusters
For consensus unsupervised analysis, the “ConsensusCluster-Plus” R package was used to categorize all patients into two FORG subtypes [9,10]. The number of classifications depended on the increase in intra-group correlations and the decrease in inter-group correlations. Principal component analysis (PCA) was performed to distinguish FORG clusters with the ggplot2 R package. DEGs were defined as |logFC| > 0.585 with an adjusted P-value < 0.05.
Construction of the FORG-related prognostic risk score
LASSO analysis was applied to the subtype-related DEGs to determine the optimal value of λ. The prognostic risk score was based on gene expression and correlation coefficients. Based on the median risk score, patients were separated into two risk groups, and KM survival analysis was used to assess the predictive value of this signature.
Evaluation of immune cell infiltration in OC
Single-sample gene set enrichment analysis (ssGSEA) was performed to explore immune infiltration in different FORG subtypes. The marker genes for 23 types of immune cells were descripted in the Supplementary Table 1. The immune infiltration in high- and low-risk groups was quantified with the CIBERSORT algorithm [11], and the correlation between risk scores and immune cell infiltration was analyzed with Spearman’s method [12].
Assessment of immune checkpoint expression, stromal scores, and chemotherapy effects
The immune checkpoint gene expression and stromal scores were compared between the two risk groups with the Wilcoxon test. Using the pRRophetic R package in R (version 4.1.0), the effect of molecular therapy and chemotherapy was calculated as the IC50, and then the values were compared between the high- and low-risk groups. Drug sensitivity analysis R codes were described in the Supplementary Material.
Statistical analysis
Statistical analyses were analyzed with RStudio and R (version 4.1.0). Differences between the two groups were compared with the Wilcoxon test. All analyses were two-sided, and P < 0.05 was considered significant.
Results
Genetic alterations of FORGs in OC
It has been reported that 34 FORGs were considered to be associated with ferroptosis and oxidative stress [4]. We conducted a comprehensive analysis of 596 samples from the GEO (GSE53963) and TCGA databases. Our results from a KM analysis revealed that 19 of the 34 FORGs were closely related to the prognosis of OC (Figure 1A, 1B). Moreover, CNV analysis showed that 19 genes had higher CNV increases or deletions (Figure 1C, 1D). Furthermore, somatic mutations among the FORGs in OC were analyzed and 40 (9.17%) of the 436 samples were found to be mutated (Figure 1E). Finally, a network among FORGs was constructed to reveal the correlations between the FORGs and their prognostic significance, which indicated a high correlation among these genes (Figure 1F).
Figure 1.
Genetic alterations and prognostic role of ferroptosis- and oxidative stress-related genes (FORGs) in ovarian cancer (OC). A, B. Kaplan-Meier analysis revealed that SNCA and IFNG were closely related to the prognosis of OC. C. Frequencies of copy number variation (CNV) increases and deletions and non-CNV among the FORGs. D. Locations of FORG CNV alterations on 23 chromosomes. E. Mutation frequencies of FORGs. F. Interactions among FORGs in OC. The lines represent interactions.
Identification of FORG subtypes
Patients with OC were categorized into two FORG subtypes using consensus clustering based on the expression characteristics of 19 FORGs (Figure 2A). PCA revealed significantly distinct separation between the two FORG subtypes (Supplementary Figure 1). KM analysis indicated that the prognosis of cluster A was significantly better than that of cluster B (Figure 2B). The expression pattern of FORGs and clinicopathologic factors in the two FORG subtypes are shown in a heatmap (Figure 2C), which demonstrated that most of the 19 FORGs were significantly upregulated in cluster B.
Figure 2.
Identification of ferroptosis- and oxidative stress-related genes (FORGs) subtypes. A. Two FORG clusters were identified using consensus clustering analyses. B. Kaplan-Meier analysis indicated that the prognosis of cluster A was significantly better than that of cluster B. C. The expression pattern of FORGs and clinicopathologic factors in the two FORG subtypes are shown in the heatmap.
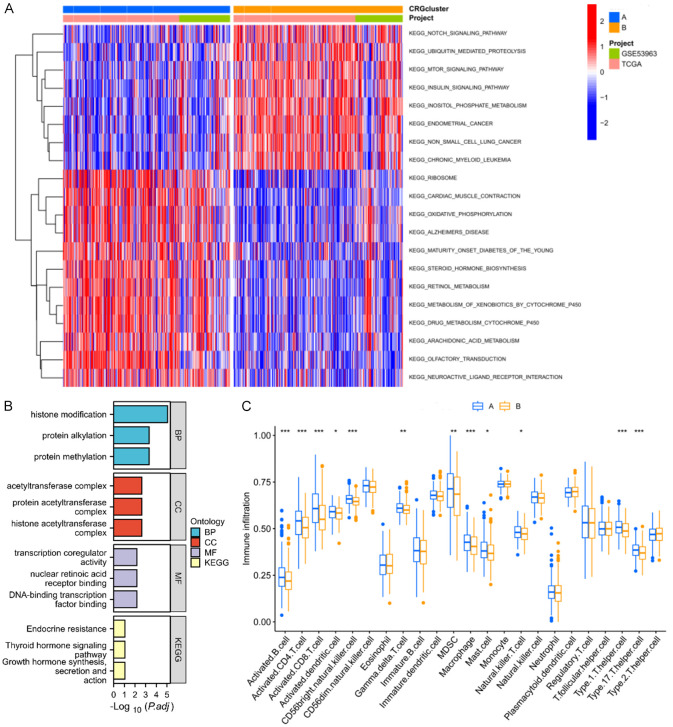
Characteristics of biological function and immune infiltration in different FORG subtypes
To further investigate the biological functions and pathways associated with different FORG subtypes, the DEGs between gene set variation analysis (GSVA), Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were identified. The GSVA results showed that subtype A was mainly related to oxidative phosphorylation, metabolism of xenobiotics by cytochrome P450, and drug metabolism cytochrome P450. Subtype B was mainly enriched in the NOTCH signaling pathway, mTOR signaling pathway, and insulin signaling pathway (Figure 3A). To identify the biologic function of each of the FORG subtypes in OC, 308 DEGs were analyzed with the “limma” R package. GO terms were closely associated with histone modification, protein alkylation, and protein methylation, while KEGG terms were enriched in endocrine resistance (Figure 3B). Finally, a ssGSEA analysis was performed to assess the immune infiltration scores of the FORG subtypes (Figure 3C). In subtype A, the highest scores were observed for activated B cells, activated CD4 T cells, activated CD8 T cells, activated dendritic cells, activated CD56bright natural killer (NK) cells, eosinophils, myeloid-derived suppressor cells, macrophages, mast cells, NK cells, Type 1 T helper cells, and Type 17 T helper cells.
Figure 3.
Functional analyses and tumor microenvironment in ferroptosis- and oxidative stress-related genes (FORGs) clusters. A. GSVA analysis showed the enriched pathways in two FORG clusters. B. GO and KEGG analyses. C. ssGSEA analysis was performed to assess the immune infiltration scores of the FORG subtypes.
Construction of the FORG prognostic signature
LASSO analysis was applied to the 308 subtype-related DEGs (Supplementary Table 2) to determine the optimal value of λ (Figure 4A). The results indicated that MEGF8, ECE1, SASH1, ARHGEF16, PLXNA1, and FCGBP were identified as signature genes for the construction of a prognostic model according to expression levels. All OC patients were divided into two groups according to the median of the risk scores. KM survival analysis demonstrated that low-risk patients had a better prognosis than high-risk patients (Figure 4B). To further investigate the differences in the composition of immune infiltration among the two groups, the percentages of the 22 immune cells were calculated with the CIBERSORT algorithm analysis. The results showed that resting CD4 memory T cells, resting NK cells, and M2 macrophages were positively correlated with risk scores, while activated CD4 memory T cells, activated NK cells, and M1 macrophages were negatively correlated with risk scores (Figures 4C, 5).
Figure 4.
Construction of the ferroptosis- and oxidative stress-related genes (FORGs) prognostic signature. A. Cross-validation for tuning parameter selection in the LASSO model. B. Kaplan-Meier survival analysis demonstrated that low-risk patients had a better prognosis than high-risk patients. C. Correlation between the immune cells, prognostic signature genes, and risk scores.
Figure 5.

Evaluation of immune cell infiltration in high- and low-risk groups. A, C, E. Resting CD4 memory T cells, resting natural killer (NK) cells, and M2 macrophages were positively correlated with risk scores. B, D, F. Activated CD4 memory T cells, activated NK cells, and M1 macrophages were negatively correlated with risk scores.
Analysis of immune checkpoint expression, stromal scores, and chemotherapy effects between the two groups
Expression levels of immune checkpoint genes in the two groups were compared, and the results showed that the high-risk group had higher immune checkpoint expression levels, including TNFRSF8, TNFRSF25, and CD276, indicating that immunotherapy may be more effective in high-risk patients (Figure 6A, 6B). Additionally, we also found that stromal scores were increased in the high-risk group compared with the low-risk group (Figure 6C). Subsequently, we evaluated the therapeutic effects of cisplatin and pazopanib using IC50 values. We found that the IC50 values of cisplatin were significantly increased in the high-risk group, while the IC50 values of pazopanib and axitinib (Supplementary Figure 2) were decreased in the same group, suggesting that the prognostic signature is closely associated with the efficacy of chemotherapy drugs (Figure 6D, 6E).
Figure 6.

Comparison analysis of immune checkpoint expression, stromal scores, and chemotherapy effects between the two groups. A, B. The immune checkpoint gene expressions were different in the high-risk and low-risk groups. C. Stromal scores and risk groups. D. IC50 values of cisplatin were significantly increased in the high-risk group. E. IC50 values of pazopanib were decreased in the high-risk group.
Discussion
Recent studies have highlighted the use of combining ferroptosis and oxidative stress pathways as prognostic signatures to predict OS of OC patients, immune infiltration, and the effects of immunotherapy and chemotherapy. To date, only a few risk models have been constructed that integrate the two pathways. We propose a novel risk model that combines ferroptosis and oxidative stress pathways to improve the prognostic accuracy of OC patient outcomes. Nineteen FORGs were closely investigated in relation to the prognosis of ovarian cancer (OC). Among them, MAPK1 has been previously demonstrated to be involved in OC proliferation, migration, invasion, and chemosensitivity [13,14]. Additionally, GSTM1 has been suggested as a biomarker to predict the efficacy of paclitaxel-based chemotherapy in OC [15]. Furthermore, P62 has been found to inhibit vK3-induced oxidative damage through the KEAP1/NRF2 pathway in OC cells. Genetic alterations, including CNVs and somatic mutations of the 19 FORGs, were also analyzed, and most of them showed increased or decreased CNV.
Based on the expression characteristics of 19 FORGs in OC, patients were categorized into two molecular subtypes, with cluster A exhibiting a better prognosis than cluster B. To investigate the underlying mechanisms, GSVA results revealed that both clusters were mainly enriched in energy metabolism pathways, suggesting that targeting these pathways may be beneficial for OC prognoses. Furthermore, immune infiltration levels were examined between the two clusters, and cluster A was found to be enriched in activated immune cells, including activated B cells, activated CD4 T cells, activated CD8 T cells, activated dendritic cells, activated CD56bright NK cells, and eosinophils. These findings suggest that the higher infiltration levels of activated immune cells in cluster A may partially explain the better prognoses of OC patients. Moreover, OC with distinct prognoses and immune microenvironments were better classified by the two FORG subtypes, indicating that these FORGs may be associated with the formation of a complex immune microenvironment and an influence on OC progression. This evidence facilitates our understanding of the crosstalk between FORGs, immune cell infiltration, and OC progression.
Subtype-related DEGs in two FORG clusters were used to establish prognostic models for OC. Through LASSO analyses, a signature gene set, including MEGF8, ECE1, SASH1, ARHGEF16, PLXNA1, and FCGBP, was identified for OC risk assessment. Of these genes, ECE1 was significantly upregulated and is associated with OC development [16]. SASH1 has been reported to be associated with OC lymph node metastasis [17]. OC patients were stratified into two risk groups according to risk scores, and patients in the low-risk group had a better prognosis. It has been suggested that the immune microenvironment is essential for tumor development and chemotherapy resistance [18]. Some immune cells were significantly correlated with risk scores. Specifically, resting CD4 memory T cells, resting NK cells, and M2 macrophages were positively correlated with risk scores, whereas activated CD4 memory T cells, activated NK cells, and M1 macrophages were negatively correlated with risk scores, suggesting that higher risk scores were associated with immunosuppression. Previous studies have demonstrated that macrophages play a diverse role in the tumor microenvironment [19]. M2 macrophages may contribute to the immune escape of tumor cells by suppressing inflammation while simultaneously promoting tumor proliferation, and they are associated with a poor prognosis in OC and other cancers [20-22]. In contrast, M1 macrophages are generally thought to promote inflammation and are associated with a good prognosis in patients with OC [20,23]. Our results are consistent with previous studies, but the precise nature of the interactions between the model genes, immune microenvironment, and tumor cells remains to be elucidated. We also examined immune checkpoint expression, stromal scores, and chemotherapy effects between the two risk groups our results indicated that prognostic models may facilitate effective treatment with chemotherapy and targeted drugs.
In summary, we constructed a novel FORG cluster and prognostic model that showed efficiency in predicting patient prognoses, immune cell infiltration, and chemotherapy responses in OC. Our findings provide new perspectives on crosstalk between FORGs, immune cell infiltration, and OC progression.
Acknowledgements
This work was supported by the Suzhou Municipal Hospital Gynecological Clinical Trial and Improvement Project (grant number SLT201955), the Maternal and Child Health Research Project of Jiangsu Province (grant number F201904), the Top Talent Support Program for Young and Middle-aged People of Wuxi Health Committee (grant number BJ2020047), and the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (grant number 2019PT310002).
Disclosure of conflict of interest
None.
Supplementary Material, Supplementary Figures 1 and 2
Supplementary Table 1
Supplementary Table 2
References
- 1.Sun X, Xu P, Zhang F, Sun T, Jiang H, Lu X, Zhang M, Li P. The cuproptosis-related gene signature serves as a potential prognostic predictor for ovarian cancer using bioinformatics analysis. Ann Transl Med. 2022;10:1021. doi: 10.21037/atm-22-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Webb PM, Jordan SJ. Epidemiology of epithelial ovarian cancer. Best Pract Res Clin Obstet Gynaecol. 2017;41:3–14. doi: 10.1016/j.bpobgyn.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Block MS, Dietz AB, Gustafson MP, Kalli KR, Erskine CL, Youssef B, Vijay GV, Allred JB, Pavelko KD, Strausbauch MA, Lin Y, Grudem ME, Jatoi A, Klampe CM, Wahner-Hendrickson AE, Weroha SJ, Glaser GE, Kumar A, Langstraat CL, Solseth ML, Deeds MC, Knutson KL, Cannon MJ. Th17-inducing autologous dendritic cell vaccination promotes antigen-specific cellular and humoral immunity in ovarian cancer patients. Nat Commun. 2020;11:5173. doi: 10.1038/s41467-020-18962-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, Xu Y, Dai L, Yu Z, Wang M, Chan S, Sun R, Han Q, Chen J, Zuo X, Wang Z, Hu X, Yang Y, Zhao H, Hu K, Zhang H, Chen W. A novel oxidative stress- and ferroptosis-related gene prognostic signature for distinguishing cold and hot tumors in colorectal cancer. Front Immunol. 2022;13:1043738. doi: 10.3389/fimmu.2022.1043738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Zhao G, Condello S, Huang H, Cardenas H, Tanner EJ, Wei J, Ji Y, Li J, Tan Y, Davuluri RV, Peter ME, Cheng JX, Matei D. Frizzled-7 identifies platinum-tolerant ovarian cancer cells susceptible to ferroptosis. Cancer Res. 2021;81:384–399. doi: 10.1158/0008-5472.CAN-20-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ye Y, Dai Q, Li S, He J, Qi H. A novel defined risk signature of the ferroptosis-related genes for predicting the prognosis of ovarian cancer. Front Mol Biosci. 2021;8:645845. doi: 10.3389/fmolb.2021.645845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu J, Xiong Y, Zhang Y, Wen J, Cai N, Cheng K, Liang H, Zhang W. The molecular mechanisms of regulating oxidative stress-induced ferroptosis and therapeutic strategy in tumors. Oxid Med Cell Longev. 2020;2020:8810785. doi: 10.1155/2020/8810785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aboelella NS, Brandle C, Kim T, Ding ZC, Zhou G. Oxidative stress in the tumor microenvironment and its relevance to cancer immunotherapy. Cancers (Basel) 2021;13:986. doi: 10.3390/cancers13050986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Q, Lin W, Liu T, Hu J, Zhu Y. Immunological classification of glioblastoma and its prognostic implications. Am J Transl Res. 2022;14:8009–8022. [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Pang ZQ, Han YS, Shen LK, Yao WH, Zhang L, He JX, Shan YJ, Ren MH. The predictive value of pyroptosis for the prognosis and immune escape of bladder cancer. Am J Transl Res. 2022;14:7744–7757. [PMC free article] [PubMed] [Google Scholar]
- 11.Yin S, Li W, Wang J, Wu H, Hu J, Feng Y. Screening of key genes associated with m6A methylation in diabetic nephropathy patients by CIBERSORT and weighted gene coexpression network analysis. Am J Transl Res. 2022;14:2280–2290. [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, Li Y, Zhang C, Chen X, Zhu L, Luo T. Role of ferroptosis-related molecular patterns in hepatocellular carcinoma microenvironment. Am J Transl Res. 2022;14:86–102. [PMC free article] [PubMed] [Google Scholar]
- 13.Yiwei T, Hua H, Hui G, Mao M, Xiang L. HOTAIR interacting with MAPK1 regulates ovarian cancer skov3 cell proliferation, migration, and invasion. Med Sci Monit. 2015;21:1856–1863. doi: 10.12659/MSM.893528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu ZH, Yao TZ, Liu W. miR-378a-3p sensitizes ovarian cancer cells to cisplatin through targeting MAPK1/GRB2. Biomed Pharmacother. 2018;107:1410–1417. doi: 10.1016/j.biopha.2018.08.132. [DOI] [PubMed] [Google Scholar]
- 15.Ferracini AC, Lopes-Aguiar L, Lourenço GJ, Yoshida A, Lima CSP, Sarian LO, Derchain S, Kroetz DL, Mazzola PG. GSTP1 and ABCB1 polymorphisms predicting toxicities and clinical management on carboplatin and paclitaxel-based chemotherapy in ovarian cancer. Clin Transl Sci. 2021;14:720–728. doi: 10.1111/cts.12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rayhman O, Klipper E, Muller L, Davidson B, Reich R, Meidan R. Small interfering RNA molecules targeting endothelin-converting enzyme-1 inhibit endothelin-1 synthesis and the invasive phenotype of ovarian carcinoma cells. Cancer Res. 2008;68:9265–9273. doi: 10.1158/0008-5472.CAN-08-2093. [DOI] [PubMed] [Google Scholar]
- 17.Ren X, Liu Y, Tao Y, Zhu G, Pei M, Zhang J, Liu J. Downregulation of SASH1 correlates with tumor progression and poor prognosis in ovarian carcinoma. Oncol Lett. 2016;11:3123–3130. doi: 10.3892/ol.2016.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019;79:4557–4566. doi: 10.1158/0008-5472.CAN-18-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dehne N, Mora J, Namgaladze D, Weigert A, Brüne B. Cancer cell and macrophage cross-talk in the tumor microenvironment. Curr Opin Pharmacol. 2017;35:12–19. doi: 10.1016/j.coph.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 20.Bruni D, Angell HK, Galon J. The immune contexture and immunoscore in cancer prognosis and therapeutic efficacy. Nat Rev Cancer. 2020;20:662–680. doi: 10.1038/s41568-020-0285-7. [DOI] [PubMed] [Google Scholar]
- 21.Vitale I, Manic G, Coussens LM, Kroemer G, Galluzzi L. Macrophages and metabolism in the tumor microenvironment. Cell Metab. 2019;30:36–50. doi: 10.1016/j.cmet.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Zou J, Wu B, Lin C, Ding Q, Li J. MiR-216b targets CPEB4 to suppress colorectal cancer progression through inhibiting IL-10-mediated M2 polarization of tumor-associated macrophages. Am J Transl Res. 2022;14:8129–8145. [PMC free article] [PubMed] [Google Scholar]
- 23.Min L, Wang H, Qi H. Astragaloside IV inhibits the progression of liver cancer by modulating macrophage polarization through the TLR4/NF-κB/STAT3 signaling pathway. Am J Transl Res. 2022;14:1551–1566. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.