Abstract
Background: Alzheimer’s disease (AD) is a widespread neurodegenerative disease that primarily affects the elderly. Unfortunately, the lack of convenient early diagnostic tools makes it difficult to intervene and treat the disease during its initial stages. Methods: We obtained four bulk and single-cell RNA-sequencing peripheral blood samples related to AD from public databases. Using Boruta and LASSO machine learning algorithms, we screened the signature genes and constructed a diagnostic model using lightGBM. The model was further validated in a test cohort. Additionally, we extracted hub biomarkers using the protein-protein interactions method and validated them in a single-cell RNA-seq dataset. Results: Our analysis revealed the identification of 37 AD-related peripheral blood signature genes, with their main enrichment in ribosome-related biological functions. Four core biomarkers, RPL24, RPL5, RPS27A, and RPS4X, were identified and exhibited good diagnostic power in the testing cohort. Immune infiltration analysis revealed a higher proportion of CD4+ T cells in AD patients’ peripheral blood compared to healthy controls, with a negative correlation with the four ribosome-associated core genes. Validation in a single-cell RNA-seq dataset confirmed these findings. Conclusions: Ribosomal family proteins have the potential to serve as biomarkers for the diagnosis and treatment of AD, and are associated with CD4+ T cell activation.
Keywords: Neurodegenerative diseases, machine learning algorithms, bioinformatics, serum biomarker, single-cell sequencing
Introduction
Alzheimer’s disease (AD) is a neurodegenerative disorder that is characterized by progressive cognitive decline and memory impairment. The typical pathological hallmarks of AD include extracellular plaques of amyloid β (Aβ) depositions and neurofibrillary tangles, which are formed due to hyperphosphorylation of microtubule-associated tau protein. These pathological changes are accompanied by neuronal loss, synaptic degeneration, glial cell activation, and neuroinflammation [1]. The burden of AD on patients and their families is significant, and it has become a significant social problem. Unfortunately, there is currently no cure for AD, and available drugs can only delay the progression of the disease. Therefore, early diagnosis and intervention are critical. Imaging techniques, such as positron emission tomography (PET) and magnetic resonance imaging (MRI), are commonly used for the early diagnosis of AD. However, the high cost of PET and the low specificity of MRI present significant challenges to their practical application [2].
Recently, molecular biomarkers that reflect changes in the structure and function of neurons have been discovered, which has greatly improved the efficiency of AD diagnosis. For example, cerebrospinal fluid biomarkers, such as Aβ40, Aβ42, and tau, can directly reflect AD neuropathology [3]. However, the invasive nature of cerebrospinal fluid collection and the associated risks limit their use for screening healthy individuals. As a result, there is a growing need to identify biomarkers that are specific, sensitive, and minimally invasive.
Recent research has examined the use of peripheral blood biomarkers for AD diagnosis [4]. However, the abundance of biomarkers in the blood that reflect neuropathological damage to brain tissue is extremely low due to the blood-brain barrier. Traditional techniques, such as enzyme-linked immunosorbent assays and electrochemiluminescence, are not sensitive enough for quantitative and efficient detection. Therefore, the value of blood indicators in AD diagnosis is limited. However, the development of advanced detection techniques, such as genomic, proteomic, and metabolomic analysis, is anticipated to lead to the identification of reliable and widely applicable peripheral blood biomarkers for early screening of AD.
AD is a complex disease that involves interactions between multiple factors, and it is challenging to understand using simplified models. Machine learning has emerged as a powerful tool for identifying patterns in complex data and can be used to diagnose and determine prognoses in AD. Previous studies have demonstrated the application of machine learning in identifying potential biomarkers for AD by analyzing gene expression profiles and constructing differential co-expression networks [5]. Other studies have explored the role of long non-coding RNA expression in AD pathogenesis [6]. Despite these promising findings, the application of machine learning in AD diagnosis and prognosis remains in its early stages.
In this study, we used transcriptome and single-cell transcriptome data from public databases to identify serum biomarkers for AD using a range of bioinformatics methods and machine learning algorithms. We also investigated immune infiltration in AD. Figure 1 provides an overview of our research methodology.
Figure 1.
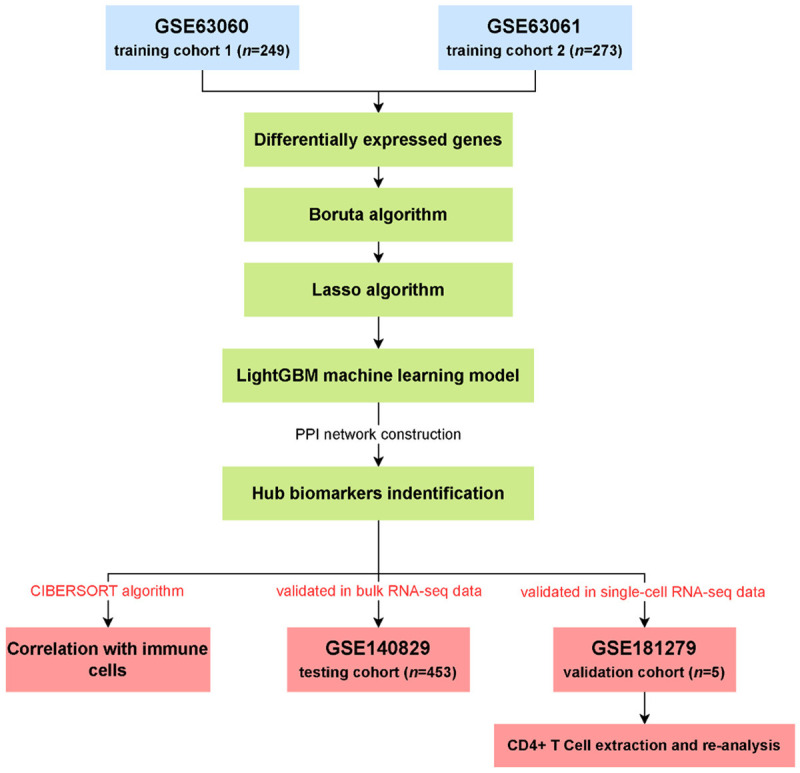
The flow chart of this study.
Methods
Datasets preprocessing in training, testing, and validation cohorts
Four datasets related to Alzheimer’s disease were acquired through the Gene Expression Omnibus (GEO). All datasets were collected from blood samples taken from patients with AD or healthy controls. Two of these datasets, GSE63060 and GSE63061, were selected for use as the training cohort. Both datasets were based on the GPL6947 chip and were derived from peripheral blood samples. GSE63060 includes 145 AD samples and 104 control samples, while GSE63061 has 139 AD samples and 134 control samples [7]. The RNA-Seq dataset GSE140829 was used for the testing cohort, consisting of 204 AD blood samples and 249 healthy control samples [8]. Additionally, we used a single-cell RNA-Seq dataset GSE181279, which contains 36,849 peripheral blood mononuclear cells from three AD and two healthy control samples, as the validation cohort [9]. Detailed information about the datasets used in this study is shown in Table 1. The clinical baseline characteristics of the AD and control groups from the training and testing cohorts are displayed in Table 2.
Table 1.
The detailed information for gene datasets used in this study
| GEO accession | Platform; Data type | Sample (number) | Tissue | Attribute |
|---|---|---|---|---|
| GSE63060 | GPL6947; bulk RNA-seq | AD (145); Control (104) | Blood | Training cohort |
| GSE63061 | GPL10558; bulk RNA-seq | AD (139); Control (134) | Blood | Training cohort |
| GSE140829 | GPL15988; bulk RNA-seq | AD (204); Control (249) | Blood | Testing cohort |
| GSE181279 | GPL24676; scRNA-seq | AD (3); Control (2) | Blood | Validation cohort |
Table 2.
The baseline characteristics of AD and control groups from training and test cohorts
| Training cohort | Testing cohort | ||||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| AD | Control | P value | AD | Control | P value | ||
| n | 284 | 238 | 204 | 249 | |||
| Gender (%) | Female | 184 (64.8) | 143 (60.1) | 0.31 | 104 (51.0) | 139 (55.8) | 0.35 |
| Male | 100 (35.2) | 95 (39.9) | 100 (49.0) | 110 (44.2) | |||
| Age (Year, mean (SD)) | 76.62 (6.73) | 74.02 (6.32) | 0.112 | 73.00 (7.09) | 73.66 (6.25) | 0.288 | |
AD: Alzheimer’s disease; SD: standard deviation.
To begin the analysis, the microarray gene datasets GSE63060 and GSE63061 were downloaded, normalized, and log2-transformed using the “limma” package in R software (version 4.2). The raw RNA-Seq dataset GSE140829 was annotated using the human genome to build 38 references and formatted to transcripts per million (TPM) data for subsequent analysis. Finally, samples in the single-cell RNA-Seq (scRNA-Seq) dataset GSE181279 were normalized, and batch effects were eliminated using the canonical correlation analysis in the “Seurat” R package (version 4.0) [10].
Filtering for differentially expressed genes
To identify differentially expressed genes (DEGs) from the GSE63060 and GSE63061 datasets, we applied the “sva” package [11] for data integration and batch correction. The “limma” package [12] was then utilized to analyze the expression data for DEG identification. Our criteria for DEG screening were set at |log2Foldchange| >1 and adjusted P < 0.05.
Features selection using Boruta and LASSO
In this section, we describe the methods used to select relevant features for further analysis. Firstly, we applied the Boruta algorithm to exclude genes that have low correlation with AD. To accomplish this, we reshuffled the original features to obtain new features and trained a random forest model based on the input features. The importance of each feature was then calculated, and relevant features were selected from the original features using the feature importance in the new features as a reference. The iterations were stopped after all features were judged to be important or useless. This process helped to identify signature genes that may be associated with the onset and development of AD.
Next, we continued the screening of signature genes using the LASSO algorithm, which builds a regularized linear model and filters important features for classification by eliminating features that are worthless or redundant. To assess the discriminative ability of the hub genes, we applied principal component analysis (PCA).
To implement these methods, we utilized the “Boruta” (v7.0) [13] and “glmnet” (v4.1) [14] R packages for filtering and analyzing genetic data. By conducting this analysis, we aimed to select genes that are most relevant to AD, and that may be used as potential biomarkers or therapeutic targets.
Construction and validation of the diagnostic model in the testing cohort
After selecting signature genes using the Boruta and LASSO algorithms, we constructed an Alzheimer’s disease diagnosis model using the lightGBM algorithm. To evaluate the model’s performance, we utilized the testing cohort GSE140829 and conducted PCA to estimate the model’s ability to distinguish Alzheimer’s disease samples from healthy controls. We used the “lightGBM” R package (v.3.3.2) [15] for model construction and receiver operating characteristic (ROC) analysis to evaluate the model’s performance.
Hub biomarker identification
In order to identify the core biomarkers associated with AD, we conducted a protein-protein interaction (PPI) network analysis using genes from the diagnostic model. The String online tool (https://www.string-db.org/, v11.5) was employed with a high confidence score of 0.7 to construct the PPI network. Subsequently, we performed enrichment analysis to identify genes that play pivotal roles in the network. To identify the hub biomarkers, we conducted molecular complex detection (MCODE) analysis on the PPI network. The predictive power of these hub biomarkers was then analyzed and demonstrated in the training and testing cohorts. Additionally, we presented the expression profile of each core gene in different cohorts for both AD and control data using box plots.
Immune cell infiltration and association with hub biomarkers
To further explore the relationship between hub biomarkers and immune cell infiltration in AD, we utilized the CIBERSORT algorithm [16]. To analyze immune cell fraction in the merged gene expression matrix of GSE63060 and GSE63061. The gene expression matrix of the training dataset was uploaded to the official online platform (https://cibersortx.stanford.edu/) to estimate the immune cell infiltration in each sample. Immune cells with P-values less than 0.05 were selected for further analysis. We then calculated and demonstrated the correlation between immune cell infiltration and the expression of hub biomarkers.
Validation in sing-cell RNA seq dataset
To validate the results obtained from bulk RNA-seq data, we further analyzed scRNA-Seq data. To begin with, we used the “Seurat” package to normalize the scRNA-Seq data and identify highly variable features for each sample using the “FindVariableFeatures” function. We then employed the “FindIntegrationAnchors” method to find anchors based on variable features, and used the “IntegrateData” function to integrate the five scRNA-Seq samples. The data were scaled, and the cells were clustered and analyzed using the uniform manifold approximation and projection for dimension reduction (UMAP) algorithm. Cell types for cell clustering were identified using the CellMarker database [17] and the “singleR” package (v1.10) [18].
In addition, we calculated the proportions of various cell types in different samples and demonstrated expression differences of hub biomarkers across samples and different cell types.
Biological enrichment investigation in CD4+ T cells
To explore the potential mechanisms in CD4+ T cells, we extracted this subset cell type from the scRNA-Seq dataset and performed gene set enrichment analysis (GSEA). Unlike focusing on the enrichment of a few significantly different genes, GSEA sorts all the different genes between the two groups of samples by logFC value and examines the overall trends of enrichment in the gene set. This approach helps to prevent important but non-significant biological traits from being overlooked. Terms with adjusted P-values < 0.05 were considered significant in our analysis.
Results
Important feature genes associated with AD
We initially identified 267 differentially expressed genes (DEGs) from the gene expression matrix in the two combined training cohorts, which consisted of 131 upregulated and 136 downregulated DEGs (Table S1; Figure 2A).
Figure 2.
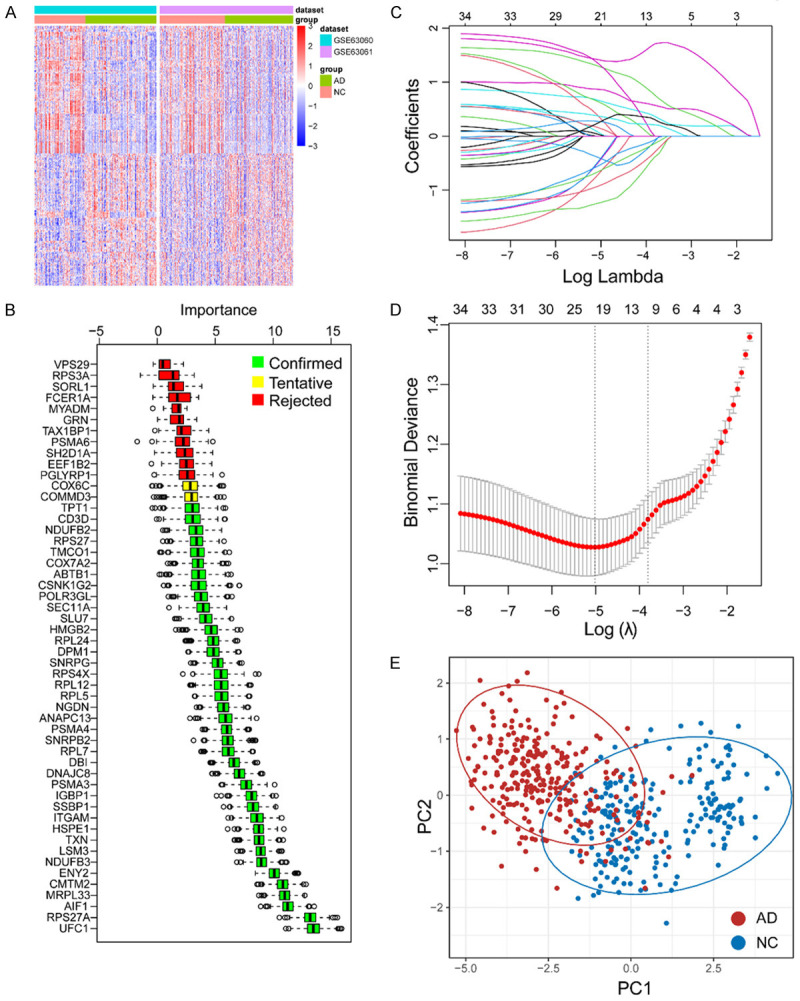
Identification of important genes associated with AD. A. The heatmap of differentially expressed genes in combined training cohort (GSE63060 and GSE63061). B. Confirmed important genes associated with AD by using Boruta algorithm. C, D. LASSO algorithm was applied for further narrow down the important genes related to AD. E. PCA plot has shown the ability of the hub genes to distinguish between AD and normal groups. Abbreviation: AD: Alzheimer’s disease; NC: normal control.
To refine the selection of important feature genes, we applied the Boruta algorithm and identified 39 genes as significant feature genes in the blood of AD (Table S2; Figure 2B). To further narrow down the selection, we employed the LASSO algorithm and selected 21 hub genes, which were then used to construct a gene model (Table S3; Figure 2C, 2D). The PCA plots demonstrated that these hub genes effectively discriminated between samples from AD patients and healthy controls (Figure 2E).
Construction of gene model and validation in testing cohort
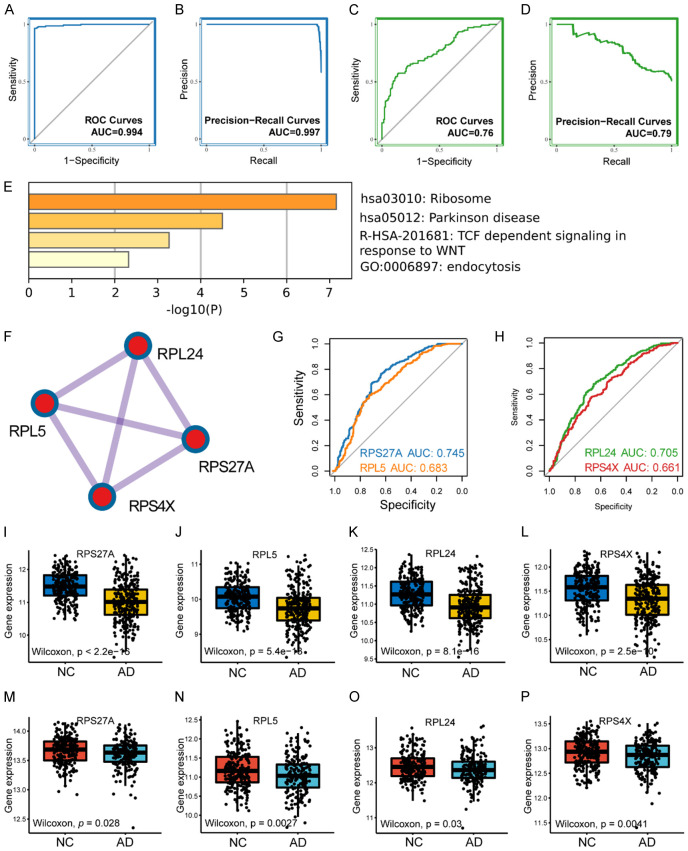
The hub genes identified from the LASSO analysis were utilized to construct a diagnostic model for AD using the lightGBM machine learning algorithm (Table S4). Subsequently, we evaluated the model’s diagnostic performance. In the training cohort, the area under the ROC curve (AUC) and precision-recall curves were 0.994 and 0.997, respectively (Figure 3A, 3B). In the testing cohort, the AUC and precision-recall curves were 0.76 and 0.79, respectively (Figure 3C, 3D). These results indicate that the diagnostic model possessed satisfactory diagnostic capability.
Figure 3.
Gene model validation and hub biomarkers identification. Receiver operator characteristic curve of gene model in training cohort (A) and testing cohort (C). Precision-recall curve of gene model in training cohort (B) and testing cohort (D). (E) The bar plot has displayed the results of functional enrichment analysis of the gene model. (F) Hub biomarkers identified by molecular complex detection algorithm. (G) ROC analysis of RPS27A and RPL5 in testing cohort. (H) ROC analysis of RPL24 and RPS4X in testing cohort. The gene expression profile of RPS27A (I), RPL5 (J), RPL24 (K) and RPS4X (L) in training cohort. The boxplots have demonstrated the expression of RPS27A (M), RPL5 (N), RPL24 (O) and RPS4X (P) in testing cohort. Abbreviation: ROC: receiver operator characteristic; AUC: area under curve; AD: Alzheimer’s disease; NC: normal control.
Hub biomarkers identification
The gene module was input into the STRING platform to construct a protein-protein interaction (PPI) network. The biological pathways mainly enriched in the PPI network were ribosomal pathways (Figure S1; Figure 3E). Subsequently, the MCODE algorithm identified four hub biomarkers (RPL24, RPL5, RPS27A, and RPS4X) (Figure 3F). The ROC curves showed that these ribosomal biomarkers had good diagnostic abilities for AD, with AUC values of 0.745 for RPS27A, 0.683 for RPL5, 0.705 for RPL24, and 0.661 for RPS4X (Figure 3G, 3H).
To further investigate the role of these ribosomal biomarkers in AD, we compared the expression levels of these biomarkers between healthy controls and patients with AD in the training cohort. The results showed that the expression levels of these ribosomal biomarkers were significantly higher in healthy controls than in patients with AD (P < 2.2e-16 for RPS27A, P = 5.4e-13 for RPL5, P = 8.1e-16 for RPL24, and P = 2.5e-10 for RPS4X) (Figure 3I-L). A similar trend was observed in the testing cohort (P = 0.028 for RPS27A, P = 0.0027 for RPL5, P = 0.03 for RPL24, and P = 0.0041 for RPS4X) (Figure 3M-P).
Immune cell infiltration in the training cohort
Using the CIBERSORT analysis, we examined the proportion of seven different immune cells and found statistically significant differences between the control and AD groups. Our results revealed that dendritic cells, M2 macrophages, and CD8+ T cells had higher proportions in control samples than in AD samples, while mast cells, natural killer cells, and CD4+ T cells had a higher proportion in the AD samples (Figure 4A).
Figure 4.
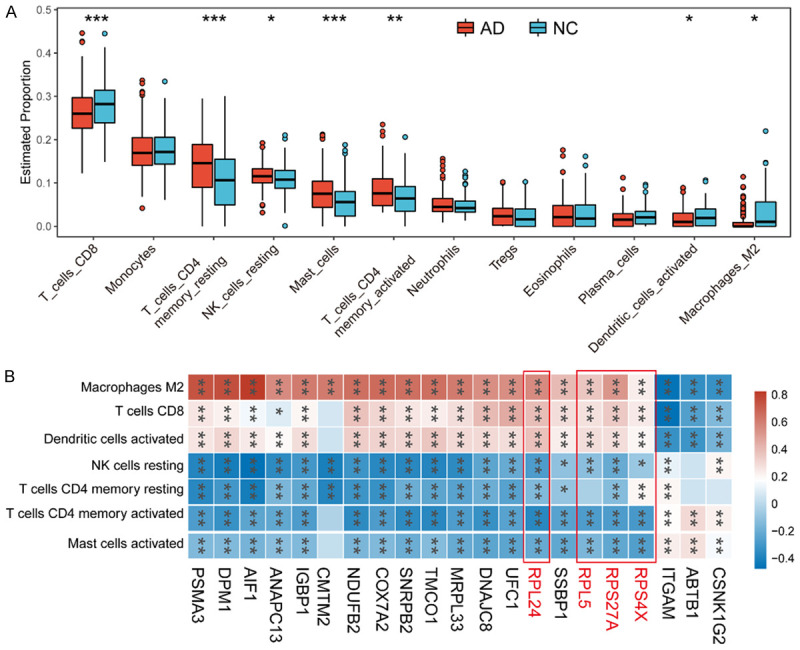
Immune cell infiltration in training cohort. A. The boxplot has shown the proportions of different types of immune cells in AD and normal control samples. B. Correlation heatmap has displayed the association between the expression of hub biomarkers and immune cell infiltration. Abbreviation: AD: Alzheimer’s disease; NC: normal control.
Furthermore, we evaluated the relationships between the hub biomarkers and immune cells using correlation heatmaps. Our analysis showed that RPL24, RPL5, RPS27A, and RPS4X were positively correlated with dendritic cells, M2 macrophages, and CD8+ T cells, whereas they all showed a negative correlation with mast cells, natural killer cells, and CD4+ T cells (Figure 4B).
Validation using scRNA-Seq data
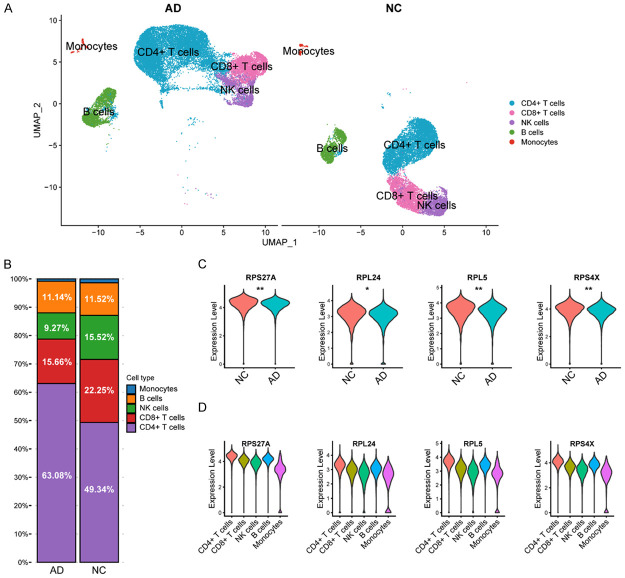
Following pre-processing and application of the UMAP algorithm, we identified 19 cell clusters (Figure S2), and ultimately identified five different cell types, including monocytes, B cells, natural killer cells, CD8+ T cells, and CD4+ T cells (Figure 5A). Notably, CD4+ T cells were the most abundant cell type and had a higher proportion in AD samples (63.08%) compared to healthy controls (49.34%), suggesting a potentially significant role in AD progression (Figure 5B).
Figure 5.
Validation in single-cell RNA-Seq dataset. A. Identification of varied cell types in the single-cell RNA-Seq dataset by using UMAP algorithm. B. Proportions of different cell types in AD and normal control samples. C. The violin diagram shows the differences in the expression of RPS27A, RPL24, RPL5 and RPS4X in AD and normal samples (Wilcoxon t-test, *P < 0.05, **P < 0.01). D. The expression profile of RPS27A, RPL24, RPL5 and RPS4X in different types of cells in scRNA-Seq dataset. Abbreviation: AD: Alzheimer’s disease; NC: normal control.
Subsequent analysis revealed that the expression levels of RPL24, RPL5, RPS27A, and RPS4X were higher in healthy controls compared to patients with AD (Figure 5C). Furthermore, violin plots demonstrated that these hub biomarkers were primarily expressed in CD4+ T cells, indicating the important role of ribosomal genes in CD4+ T cells (Figure 5D).
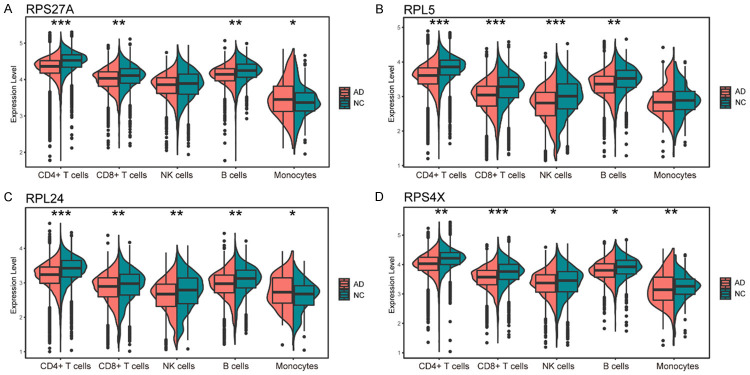
Specifically, all four biomarkers showed higher expression levels in the control group in CD4+ and CD8+ T cells and B cells, with RPL5, RPL24, and RPS4X significantly more highly expressed in natural killer cells of the control group. The expression pattern of these four gene markers in monocytes varied, with RPS27A and RPL24 being highly expressed in the AD group and RPS4X showing lower expression (Figure 6A-D).
Figure 6.
Violin diagrams show the differential expression of RPS27A (A), RPL5 (B), RPL24 (C) and RPS4X (D) in various cell types in AD patients and normal controls, respectively. (Wilcoxon t-test, *P < 0.05, **P < 0.01, ***P < 0.001). Abbreviation: AD: Alzheimer’s disease; NC: normal control.
Biological functions and pathway enrichment in CD4+ T cells
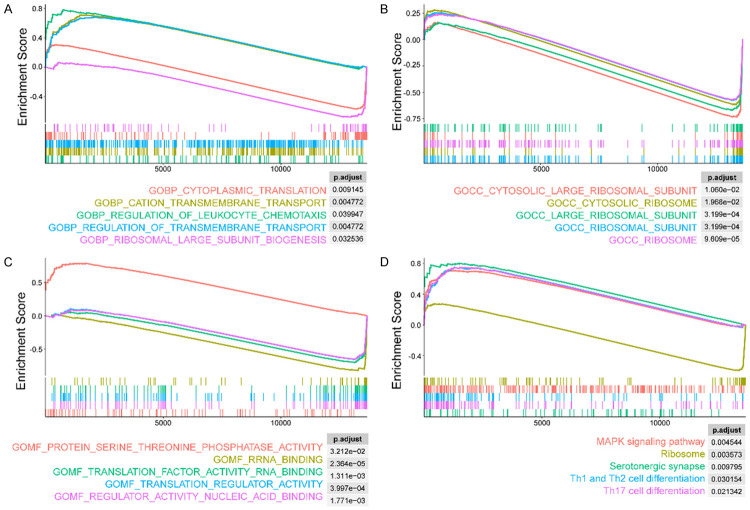
Based on the results of GSEA the top five GO and KEGG terms were selected and presented. As shown in Figure 7A, in patients with AD, several upregulated biological processes (BP) were identified, including “regulation of leukocyte chemotaxis”, “regulation of cation transmembrane transport”, and “regulation of transmembrane transport”. In contrast, “cytoplasmic translation” and “ribosomal large subunit biogenesis” were significantly downregulated. In terms of cellular components (CC), terms with significant statistical differences in the AD group were mainly related to ribosome function and included “cytosolic large ribosomal subunit” and “cytosolic ribosome” (Figure 7B). Patients with AD showed upregulated “protein serine threonine phosphatase activity” in molecular functions (MF), whereas RNA synthesis functions were downregulated, including “translation regulator activity nucleic acid binding” and “translation regulator activity” (Figure 7C). Additionally, pathway analysis revealed that the MAPK signaling and T-cell differentiation pathways were upregulated in the AD group, while the “ribosome” pathway was downregulated, consistent with the results of GO analysis (Figure 7D).
Figure 7.
The GSEA plots have shown the biological function and pathway enrichment of DEGs in CD4+ T cells in the peripheral blood of AD patients and normal control. By using gene ontology gene sets, we conducted enrichment analysis of BP (A), CC (B) and MF (C) for DEGs and only the top 5 terms were displayed. For explore the molecular pathway enrichment for DEGs, the KEGG analysis was introduced, and the top 5 enriched pathway were demonstrated (D). Abbreviation: BP: biological process; CC: cellular component; MF: molecular function; DEGs: differentially expressed genes.
Discussion
Alzheimer’s disease (AD) is a multifaceted and intricate process that involves multiple systems, making it challenging for a single biomarker to capture the entire pathological process. Several studies have highlighted the importance of combining the detection of multiple blood biomarkers to significantly enhance diagnostic efficiency for AD [3,9,19]. High-throughput sequencing technologies offer tremendous opportunities for investigating AD. Transcriptome sequencing can be employed to identify differences in mRNA expression at the transcriptome level, and follow-up studies can subsequently validate the findings using protein analyses. However, given the high-dimensional gene expression profile data, an increasing number of researchers are turning to machine learning rather than traditional statistical techniques for data analysis to effectively reveal the biological properties of AD [20].
In this study, we employed the Boruta algorithm to identify characteristic genes associated with AD. Compared to traditional feature-selection algorithms, the Boruta algorithm is known to provide superior results in identifying the importance of variables. In the field of AD research, the Boruta algorithm has been widely used for feature selection in multi-omics data. For example, it can be used to screen biomarkers of various neurodegenerative diseases at the microRNA level to determine the characteristics of these diseases [21]. Additionally, the Boruta feature filtering method has been employed in a study to analyze the characteristics of each brain region according to the systematic methylation map in patients with AD [22]. The study revealed important methylation signatures that contribute to the development of the disease. Moreover, the Boruta algorithm has also been applied to single nucleotide polymorphism data to aid in the early detection of AD [23]. The above studies demonstrate the versatility and wide applicability of the Boruta algorithm in the context of AD research.
Next, we utilized the LASSO algorithm to further refine characteristic genes and employed the lightGBM algorithm to construct a gene model with potential relevance to AD. The performance of this model was assessed using ROC analysis, which demonstrated favorable predictive ability in both the training and testing sets, suggesting that it may be a valuable tool for identifying patients with AD. Furthermore, we utilized the MCODE algorithm to identify the core genes of the model, and found that these genes were primarily involved in the regulation of ribosomal activity. In cells, ribosomes play a central role in the translation of proteins from mRNA, but with age, ribosomal function deteriorates, leading to an increase in defective proteins [24]. Synthesis of new proteins is a critical process for neuronal activity-dependent learning and memory, with ribosome biosynthesis being the key rate-limiting step in intracellular protein translation [25]. Multiple studies have reported a reduction in ribosome numbers and increased oxidation of ribosomal RNA in the brains of AD patients, resulting in reduced protein synthesis viability due to ribosomal dysfunction, which is a key feature of metabolic disorders in the AD brain [26]. A decrease in total ribosomal RNA and total RNA can be observed in the cerebrum of AD patients [27]. Oxidative stress in the cerebrum of AD patients has also been found to significantly impair the synthesis of 5S ribosomal RNA, which is responsible for ribosome stability, and specifically impairs ribosome function [28]. Animal studies have shown that Aβ injections lead to protein synthesis impairment by heavy polyribosomes formation in the hippocampus of the AD rat model [29]. Similarly, pathological tau proteins can bind to ribosomes and damage RNA translation [30]. Impaired protein synthesis due to ribosome dysfunction has been observed not only in patients with AD, but also in various other neurodegenerative pathologies [31].
Furthermore, we identified four core genes, namely RPL24, RPS27A, RPL5, and RPS4X, that exhibited lower expression levels in AD. These genes showed a strong ability for the diagnosis of AD as suggested by the ROC analysis. RPL24 is located in the cytoplasm and is an essential constituent of the 60S subunit of ribosomes. Its primary biological functions include RNA binding and forming the structure of the ribosomal protein complex. Previous studies have demonstrated that RPL24 can inhibit translation elongation and improve protein synthesis homeostasis, thereby inhibiting protein synthesis in tumor cells [32,33]. Nevertheless, the understanding of the role of RPL24 in AD is still limited. RPS27A, a member of the 40S subunit of the ribosome, is one of the core genes identified in multiple bioinformatics studies investigating mild cognitive impairment, and its levels are significantly altered in the blood of AD patients [34,35]. Recent studies have shown that RPS27A, as part of a triplet with interleukin (IL)-18 and CX3CL1, acts as a potential upstream regulator in microglia cells to reduce IL-18 and alleviate neurodegenerative diseases [36]. As one of the components of the 60S subunit of the ribosome, RPL5 has been shown to inhibit tumorigenesis by activating downstream tumor suppressor factors and downregulating oncoprotein expression [37]. RPL5 can also suppress breast cancer cell growth by regulating E2F transcription factor 1 and endoplasmic reticulum stress of tumor cells [38]. However, the role of RPL5 in AD remains unclear. Previous research has shown that numerous ribosomal genes, including RPS4X, are highly assembled and translated in axons far from the cellular body of neurons to maintain local ribosomal function [39]. Interestingly, proteomic analysis of high-purity cerebral capillaries isolated from the gray and white matter of four donors with AD and three controls showed that RPS4X was upregulated in AD brain vessels, but no significant difference was observed in brain parenchyma [40]. This finding may explain the low expression of RPS4X in the peripheral blood of AD patients.
Further immune infiltration analysis was conducted on the training set and demonstrated that the level of CD4+ T cells was significantly higher in patients with AD as compared to the control group, and this showed a negative correlation with the previously identified ribosomal biomarkers. These findings suggest that the low expression of key ribosomal genes in AD is associated with the accumulation of CD4+ T cells. To validate this observation, peripheral blood scRNA-Seq data from the AD and control groups were utilized as a validation set, which also showed significant enrichment of CD4+ T cells and reduction of ribosomal key genes in AD, thereby corroborating the results obtained from the bulk RNA-Seq data. Previous studies have reported that the increased levels of activated CD4+ T cells and CD8+ T cells in the peripheral blood of AD patients are closely linked to cognitive deficits and magnetic resonance imaging changes in specific brain regions [41]. CD4+ T cells can aggravate or alleviate AD symptoms based on their infiltrating subgroups and constitute a major source of pro-inflammatory cytokines that decrease endothelial integrity and stimulate astrocytes, leading to Aβ production [42]. Furthermore, in vitro studies have demonstrated that β-secretase 1 levels in 5xFAD-transgenic mice were higher in CD4+ T cells, and its activation was enhanced [43,44]. Recent studies have demonstrated the significance of ribosomal proteins in the regulation of the immune system. Immune cells, such as T cells and B cells, require high levels of protein synthesis to support their proliferation and activation during immune responses, and ribosomal proteins play a vital role in this process. Dysregulation of ribosomal proteins can affect the immune response, and certain ribosomal proteins, such as RPL5 and RPL11, can interact directly with the p53 tumor suppressor protein to regulate the expression of genes involved in cell growth and proliferation [45]. Moreover, ribosomal stress caused by ribosomal protein dysfunction can activate the p53 pathway and induce cell cycle arrest or apoptosis, resulting in the elimination of potentially harmful cells.
Evidence suggests that the RPS4X gene is associated with immune cell infiltration. For instance, the RPS4X gene is upregulated in peripheral blood mononuclear cells (PBMCs) from patients with systemic lupus erythematosus (SLE), a disease characterized by chronic immune cell infiltration and inflammation [46]. Additionally, RPS4X expression is positively correlated with disease activity and the number of infiltrating immune cells. Another study found that RPS4X expression was significantly down-regulated in CD4+ T cells from multiple sclerosis (MS) patients compared with healthy controls. MS is a chronic inflammatory disease of the central nervous system characterized by immune cell infiltration, and RPS4X expression was inversely correlated with disease severity and the frequency of infiltrating T cells in the brain [47]. Furthermore, RPS27A has been shown to modulate immune responses in a variety of disease settings by regulating the recruitment and activity of tumor-associated macrophages, which play a crucial role in shaping the tumor microenvironment and promoting cancer progression [48]. Studies have shown that RPL5 gene expression is related to immune cell infiltration in different types of cancer. For example, in colorectal cancer, high RPL5 expression is associated with increased infiltration of CD8+ T cells and natural killer cells, as well as improved patient survival [49]. However, the role played by the interaction between ribosomes and CD4 cells in the pathogenesis of AD remains unclear. A possible explanation is that as the degree of ageing and inflammation increases in AD patients, abnormalities in the translation of proteins by ribosomes occur, leading to the activation of CD4 cells [50].
In this study, we utilized the GSEA algorithm to examine the functional enrichment of CD4+ T cells from patients with Alzheimer’s disease (AD). Our analysis revealed that ribosomes and their synthesis-related functions and pathways were inhibited in these cells. Previous studies have highlighted the critical role of ribosomes in the protein production of effector CD4+ T cells following TCR stimulation [51]. Moreover, ribosomal proteins (RPs) have been linked to a variety of physiological and pathological processes, including the regulation of T-cell development and immune-related diseases [52]. For instance, Noc4L-mediated ribosome biogenesis controls the activation of regulatory T cells (Tregs) and maintains immune homeostasis [53]. Additionally, post-transcriptional mechanisms have been shown to regulate ribosomes in murine CD4+ T cells after 24 hours of activation [54]. Nonetheless, further studies are needed to comprehensively elucidate the role of RPs in AD and their potential as a therapeutic target.
Furthermore, we observed that leukocyte chemoattraction-related pathways were highly expressed in CD4+ T cells from AD patients. Leukocyte chemotaxis refers to the process by which white blood cells migrate to specific sites of inflammation or infection in response to chemical signals. In AD, leukocyte chemotaxis enhances the recruitment of activated immune cells into the brain via chemotaxis [55]. Aβ has been found to be chemotactic for monocytes and it induces the secretion of proinflammatory cytokines and chemokines in the periphery as well as the brain [56]. Additionally, microglia and monocyte-derived cells have been shown to play an important role in promoting proinflammatory and neurotoxic pathways [57]. Notably, blood samples from AD patients with dementia have revealed neutrophil hyperactivation associated with increased reactive oxygen species production [58].
Overall, ribosome family proteins have been shown to play a role in Alzheimer’s disease (AD) pathogenesis. Studies have reported that alterations in ribosomal function and biogenesis are associated with AD [59,60]. Ribosomal proteins have been identified as potential biomarkers for AD, and changes in the expression levels of certain ribosomal proteins have been observed in AD patients [26]. Additionally, several studies have suggested that ribosomal dysfunction can lead to the accumulation of misfolded proteins, including amyloid-beta (Aβ) and tau, which are hallmarks of AD pathology [29,61]. Therefore, ribosome family proteins may play an important role in AD pathogenesis, and further research is needed to fully elucidate the mechanisms involved.
However, the present study has certain limitations that must be considered. Firstly, although bioinformatics methods are useful for generating hypotheses, it is essential to verify the findings using in vivo or in vitro experiments. Secondly, a larger sample size and more extensive clinical data collection could enhance the validity and reliability of the conclusions. Therefore, future studies are warranted to confirm the results of the present investigation and to expand upon the findings reported here.
Conclusions
In conclusion, our study utilized machine learning and bioinformatics approaches to analyze multiple datasets of bulk RNA-Seq and scRNA data, identifying four under expressed ribosome family protein biomarkers (RPL24, RPS27A, RPL5, RPS4X) in AD peripheral blood. Further, our findings suggest that these biomarkers are negatively correlated with CD4+ T cells activation in AD patients.
Acknowledgements
This work was supported by Hangzhou Medical Health Science and Technology Projects (B20210660, B20200287). Additionally, we wish to acknowledge to the patients and researchers who contributed to the Gene Expression Omnibus project.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Scheltens P, De Strooper B, Kivipelto M, Holstege H, Chetelat G, Teunissen CE, Cummings J, van der Flier WM. Alzheimer’s disease. Lancet. 2021;397:1577–1590. doi: 10.1016/S0140-6736(20)32205-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dubois B, Villain N, Frisoni GB, Rabinovici GD, Sabbagh M, Cappa S, Bejanin A, Bombois S, Epelbaum S, Teichmann M, Habert MO, Nordberg A, Blennow K, Galasko D, Stern Y, Rowe CC, Salloway S, Schneider LS, Cummings JL, Feldman HH. Clinical diagnosis of Alzheimer’s disease: recommendations of the International Working Group. Lancet Neurol. 2021;20:484–496. doi: 10.1016/S1474-4422(21)00066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ossenkoppele R, van der Kant R, Hansson O. Tau biomarkers in Alzheimer’s disease: towards implementation in clinical practice and trials. Lancet Neurol. 2022;21:726–734. doi: 10.1016/S1474-4422(22)00168-5. [DOI] [PubMed] [Google Scholar]
- 4.Hansson O. Biomarkers for neurodegenerative diseases. Nat Med. 2021;27:954–963. doi: 10.1038/s41591-021-01382-x. [DOI] [PubMed] [Google Scholar]
- 5.Wang L, Liu ZP. Detecting diagnostic biomarkers of Alzheimer’s disease by integrating gene expression data in six brain regions. Front Genet. 2019;10:157. doi: 10.3389/fgene.2019.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou M, Zhao H, Wang X, Sun J, Su J. Analysis of long noncoding RNAs highlights region-specific altered expression patterns and diagnostic roles in Alzheimer’s disease. Brief Bioinform. 2019;20:598–608. doi: 10.1093/bib/bby021. [DOI] [PubMed] [Google Scholar]
- 7.Sood S, Gallagher IJ, Lunnon K, Rullman E, Keohane A, Crossland H, Phillips BE, Cederholm T, Jensen T, van Loon LJ, Lannfelt L, Kraus WE, Atherton PJ, Howard R, Gustafsson T, Hodges A, Timmons JA. A novel multi-tissue RNA diagnostic of healthy ageing relates to cognitive health status. Genome Biol. 2015;16:185. doi: 10.1186/s13059-015-0750-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nachun D, Ramos E, Karydas A, Dokuru D, Gao F, Yang Z, Van Berlo V, Sears R, Kramer J, Boxer A, Rosen H, Miller B, Coppola G. Systems-level analysis of peripheral blood gene expression in dementia patients reveals an innate immune response shared across multiple disorders. bioRxiv. 2019:2019.2012.2013.875112. [Google Scholar]
- 9.Xu H, Jia J. Single-Cell RNA sequencing of peripheral blood reveals immune cell signatures in Alzheimer’s disease. Front Immunol. 2021;12:645666. doi: 10.3389/fimmu.2021.645666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902. e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kursa MB, Rudnicki WR. Feature selection with the boruta package. Journal of Statistical Software. 2010;36:1–13. [Google Scholar]
- 14.Simon N, Friedman J, Hastie T, Tibshirani R. Regularization paths for Cox’s proportional hazards model via coordinate descent. J Stat Softw. 2011;39:1–13. doi: 10.18637/jss.v039.i05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan J, Xu Y, Cheng Q, Jiang S, Wang Q, Xiao Y, Ma C, Yan J, Wang X. LightGBM: accelerated genomically designed crop breeding through ensemble learning. Genome Biol. 2021;22:271. doi: 10.1186/s13059-021-02492-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Lan Y, Xu J, Quan F, Zhao E, Deng C, Luo T, Xu L, Liao G, Yan M, Ping Y, Li F, Shi A, Bai J, Zhao T, Li X, Xiao Y. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019;47:D721–D728. doi: 10.1093/nar/gky900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, Chak S, Naikawadi RP, Wolters PJ, Abate AR, Butte AJ, Bhattacharya M. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Z, Li M, Zhang L, Liu Y. Electrochemical immunosensor based on superwettable microdroplet array for detecting multiple Alzheimer’s disease biomarkers. Front Bioeng Biotechnol. 2022;10:1029428. doi: 10.3389/fbioe.2022.1029428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie C, Zhuang XX, Niu Z, Ai R, Lautrup S, Zheng S, Jiang Y, Han R, Gupta TS, Cao S, Lagartos-Donate MJ, Cai CZ, Xie LM, Caponio D, Wang WW, Schmauck-Medina T, Zhang J, Wang HL, Lou G, Xiao X, Zheng W, Palikaras K, Yang G, Caldwell KA, Caldwell GA, Shen HM, Nilsen H, Lu JH, Fang EF. Amelioration of Alzheimer’s disease pathology by mitophagy inducers identified via machine learning and a cross-species workflow. Nat Biomed Eng. 2022;6:76–93. doi: 10.1038/s41551-021-00819-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z, Guo W, Ding S, Chen L, Feng K, Huang T, Cai YD. Identifying key MicroRNA signatures for neurodegenerative diseases with machine learning methods. Front Genet. 2022;13:880997. doi: 10.3389/fgene.2022.880997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Guo W, Zeng T, Yin J, Feng K, Huang T, Cai YD. Detecting brain structure-specific methylation signatures and rules for Alzheimer’s disease. Front Neurosci. 2022;16:895181. doi: 10.3389/fnins.2022.895181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmed H, Soliman H, Elmogy M. Early detection of Alzheimer’s disease using single nucleotide polymorphisms analysis based on gradient boosting tree. Comput Biol Med. 2022;146:105622. doi: 10.1016/j.compbiomed.2022.105622. [DOI] [PubMed] [Google Scholar]
- 24.Stein KC, Morales-Polanco F, van der Lienden J, Rainbolt TK, Frydman J. Ageing exacerbates ribosome pausing to disrupt cotranslational proteostasis. Nature. 2022;601:637–642. doi: 10.1038/s41586-021-04295-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yordanova MM, Loughran G, Zhdanov AV, Mariotti M, Kiniry SJ, O’Connor PBF, Andreev DE, Tzani I, Saffert P, Michel AM, Gladyshev VN, Papkovsky DB, Atkins JF, Baranov PV. AMD1 mRNA employs ribosome stalling as a mechanism for molecular memory formation. Nature. 2018;553:356–360. doi: 10.1038/nature25174. [DOI] [PubMed] [Google Scholar]
- 26.Ding Q, Markesbery WR, Chen Q, Li F, Keller JN. Ribosome dysfunction is an early event in Alzheimer’s disease. J Neurosci. 2005;25:9171–9175. doi: 10.1523/JNEUROSCI.3040-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding Q, Markesbery WR, Cecarini V, Keller JN. Decreased RNA, and increased RNA oxidation, in ribosomes from early Alzheimer’s disease. Neurochem Res. 2006;31:705–710. doi: 10.1007/s11064-006-9071-5. [DOI] [PubMed] [Google Scholar]
- 28.Ding Q, Zhu H, Zhang B, Soriano A, Burns R, Markesbery WR. Increased 5S rRNA oxidation in Alzheimer’s disease. J Alzheimers Dis. 2012;29:201–209. doi: 10.3233/JAD-2012-111058. [DOI] [PubMed] [Google Scholar]
- 29.Maina MB, Bailey LJ, Doherty AJ, Serpell LC. The involvement of Abeta42 and Tau in nucleolar and protein synthesis machinery dysfunction. Front Cell Neurosci. 2018;12:220. doi: 10.3389/fncel.2018.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meier S, Bell M, Lyons DN, Rodriguez-Rivera J, Ingram A, Fontaine SN, Mechas E, Chen J, Wolozin B, LeVine H 3rd, Zhu H, Abisambra JF. Pathological Tau promotes neuronal damage by impairing ribosomal function and decreasing protein synthesis. J Neurosci. 2016;36:1001–1007. doi: 10.1523/JNEUROSCI.3029-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turi Z, Lacey M, Mistrik M, Moudry P. Impaired ribosome biogenesis: mechanisms and relevance to cancer and aging. Aging (Albany NY) 2019;11:2512–2540. doi: 10.18632/aging.101922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Challa S, Khulpateea BR, Nandu T, Camacho CV, Ryu KW, Chen H, Peng Y, Lea JS, Kraus WL. Ribosome ADP-ribosylation inhibits translation and maintains proteostasis in cancers. Cell. 2021;184:4531–4546. e4526. doi: 10.1016/j.cell.2021.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knight JR, Vlahov N, Gay DM, Ridgway RA, Faller WJ, Proud C, Mallucci GR, von der Haar T, Smales CM, Willis AE, Sansom OJ. Rpl24(Bst) mutation suppresses colorectal cancer by promoting eEF2 phosphorylation via eEF2K. Elife. 2021;10:e69729. doi: 10.7554/eLife.69729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ji W, An K, Wang C, Wang S. Bioinformatics analysis of diagnostic biomarkers for Alzheimer’s disease in peripheral blood based on sex differences and support vector machine algorithm. Hereditas. 2022;159:38. doi: 10.1186/s41065-022-00252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tao Y, Han Y, Yu L, Wang Q, Leng SX, Zhang H. The predicted key molecules, functions, and pathways that bridge Mild Cognitive Impairment (MCI) and Alzheimer’s Disease (AD) Front Neurol. 2020;11:233. doi: 10.3389/fneur.2020.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khayer N, Mirzaie M, Marashi SA, Jalessi M. Rps27a might act as a controller of microglia activation in triggering neurodegenerative diseases. PLoS One. 2020;15:e0239219. doi: 10.1371/journal.pone.0239219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie J, Zhang W, Liang X, Shuai C, Zhou Y, Pan H, Yang Y, Han W. RPL32 promotes lung cancer progression by facilitating p53 degradation. Mol Ther Nucleic Acids. 2020;21:75–85. doi: 10.1016/j.omtn.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma X, Li Y, Zhao B. Ribosomal protein L5 (RPL5)/E2F transcription factor 1 (E2F1) signaling suppresses breast cancer progression via regulating endoplasmic reticulum stress and autophagy. Bioengineered. 2022;13:8076–8086. doi: 10.1080/21655979.2022.2052672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shigeoka T, Koppers M, Wong HH, Lin JQ, Cagnetta R, Dwivedy A, de Freitas Nascimento J, van Tartwijk FW, Strohl F, Cioni JM, Schaeffer J, Carrington M, Kaminski CF, Jung H, Harris WA, Holt CE. On-site ribosome remodeling by locally synthesized ribosomal proteins in axons. Cell Rep. 2019;29:3605–3619. e3610. doi: 10.1016/j.celrep.2019.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki M, Tezuka K, Handa T, Sato R, Takeuchi H, Takao M, Tano M, Uchida Y. Upregulation of ribosome complexes at the blood-brain barrier in Alzheimer’s disease patients. J Cereb Blood Flow Metab. 2022;42:2134–2150. doi: 10.1177/0271678X221111602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu J, Duan J, Mo J, Xiao H, Huang Y, Chen W, Xiang S, Yang F, Chen Y, Xu S. Mild cognitive impairment patients have higher regulatory T-cell proportions compared with Alzheimer’s disease-related dementia patients. Front Aging Neurosci. 2021;12:624304. doi: 10.3389/fnagi.2020.624304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kubick N, Flournoy PCH, Enciu AM, Manda G, Mickael ME. Drugs modulating CD4+ T cells blood-brain barrier interaction in Alzheimer’s disease. Pharmaceutics. 2020;12:880. doi: 10.3390/pharmaceutics12090880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hernandez-Mir G, Raphael I, Revu S, Poholek CH, Avery L, Hawse WF, Kane LP, McGeachy MJ. The Alzheimer’s Disease-associated protein BACE1 modulates T cell activation and Th17 function. J Immunol. 2019;203:665–675. doi: 10.4049/jimmunol.1800363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dai L, Wang Q, Lv X, Gao F, Chen Z, Shen Y. Elevated beta-secretase 1 expression mediates CD4(+) T cell dysfunction via PGE2 signalling in Alzheimer’s disease. Brain Behav Immun. 2021;98:337–348. doi: 10.1016/j.bbi.2021.08.234. [DOI] [PubMed] [Google Scholar]
- 45.Lindstrom MS, Bartek J, Maya-Mendoza A. p53 at the crossroad of DNA replication and ribosome biogenesis stress pathways. Cell Death Differ. 2022;29:972–982. doi: 10.1038/s41418-022-00999-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi ZR, Han YF, Yin J, Zhang YP, Jiang ZX, Zheng L, Tan GZ, Wang L. The diagnostic benefit of antibodies against ribosomal proteins in systemic lupus erythematosus. Adv Rheumatol. 2020;60:45. doi: 10.1186/s42358-020-00148-2. [DOI] [PubMed] [Google Scholar]
- 47.Tajouri L, Mellick AS, Ashton KJ, Tannenberg AE, Nagra RM, Tourtellotte WW, Griffiths LR. Quantitative and qualitative changes in gene expression patterns characterize the activity of plaques in multiple sclerosis. Brain Res Mol Brain Res. 2003;119:170–183. doi: 10.1016/j.molbrainres.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 48.Luo J, Zhao H, Chen L, Liu M. Multifaceted functions of RPS27a: an unconventional ribosomal protein. J Cell Physiol. 2023;238:485–497. doi: 10.1002/jcp.30941. [DOI] [PubMed] [Google Scholar]
- 49.Zhang H, Liu J, Dang Q, Wang X, Chen J, Lin X, Yang N, Du J, Shi H, Liu Y, Han J. Ribosomal protein RPL5 regulates colon cancer cell proliferation and migration through MAPK/ERK signaling pathway. BMC Mol Cell Biol. 2022;23:48. doi: 10.1186/s12860-022-00448-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu B, Jadhav RR, Gustafson CE, Le Saux S, Ye Z, Li X, Tian L, Weyand CM, Goronzy JJ. Distinct age-related epigenetic signatures in CD4 and CD8 T cells. Front Immunol. 2020;11:585168. doi: 10.3389/fimmu.2020.585168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Asmal M, Colgan J, Naef F, Yu B, Lee Y, Magnasco M, Luban J. Production of ribosome components in effector CD4+ T cells is accelerated by TCR stimulation and coordinated by ERK-MAPK. Immunity. 2003;19:535–548. doi: 10.1016/s1074-7613(03)00268-1. [DOI] [PubMed] [Google Scholar]
- 52.Chen C, Peng J, Ma S, Ding Y, Huang T, Zhao S, Gao L, Liang X, Li C, Ma C. Ribosomal protein S26 serves as a checkpoint of T-cell survival and homeostasis in a p53-dependent manner. Cell Mol Immunol. 2021;18:1844–1846. doi: 10.1038/s41423-021-00699-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu X, Zhang W, Guo J, Zhang X, Li L, Wang T, Yan J, Zhang F, Hou B, Gao N, Gao GF, Zhou X. Noc4L-mediated ribosome biogenesis controls activation of regulatory and conventional T cells. Cell Rep. 2019;27:1205–1220. e1204. doi: 10.1016/j.celrep.2019.03.083. [DOI] [PubMed] [Google Scholar]
- 54.Jurgens AP, Popovic B, Wolkers MC. T cells at work: How post-transcriptional mechanisms control T cell homeostasis and activation. Eur J Immunol. 2021;51:2178–2187. doi: 10.1002/eji.202049055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goldeck D, Larbi A, Pellicano M, Alam I, Zerr I, Schmidt C, Fulop T, Pawelec G. Enhanced chemokine receptor expression on leukocytes of patients with Alzheimer’s disease. PLoS One. 2013;8:e66664. doi: 10.1371/journal.pone.0066664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pietronigro E, Zenaro E, Constantin G. Imaging of leukocyte trafficking in Alzheimer’s disease. Front Immunol. 2016;7:33. doi: 10.3389/fimmu.2016.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hohsfield LA, Humpel C. Migration of blood cells to beta-amyloid plaques in Alzheimer’s disease. Exp Gerontol. 2015;65:8–15. doi: 10.1016/j.exger.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dong Y, Lagarde J, Xicota L, Corne H, Chantran Y, Chaigneau T, Crestani B, Bottlaender M, Potier MC, Aucouturier P, Dorothee G, Sarazin M, Elbim C. Neutrophil hyperactivation correlates with Alzheimer’s disease progression. Ann Neurol. 2018;83:387–405. doi: 10.1002/ana.25159. [DOI] [PubMed] [Google Scholar]
- 59.Hernandez-Ortega K, Garcia-Esparcia P, Gil L, Lucas JJ, Ferrer I. Altered machinery of protein synthesis in Alzheimer’s: from the nucleolus to the ribosome. Brain Pathol. 2016;26:593–605. doi: 10.1111/bpa.12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Evans HT, Taylor D, Kneynsberg A, Bodea LG, Gotz J. Altered ribosomal function and protein synthesis caused by tau. Acta Neuropathol Commun. 2021;9:110. doi: 10.1186/s40478-021-01208-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koren SA, Hamm MJ, Meier SE, Weiss BE, Nation GK, Chishti EA, Arango JP, Chen J, Zhu H, Blalock EM, Abisambra JF. Tau drives translational selectivity by interacting with ribosomal proteins. Acta Neuropathol. 2019;137:571–583. doi: 10.1007/s00401-019-01970-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.