SUMMARY
DNA N6-methyldeoxyadenosine (6mA) modification was first discovered in Bacterium coli in the 1950s. Over the next several decades, 6mA was recognized as a critical DNA modification in the genomes of prokaryotes and protists. While important in prokaryotes, less is known about the presence and functional roles of DNA 6mA in eukaryotes, particularly in mammals. Taking advantage of recent technology advances that made 6mA detection and sequencing possible, studies over the past several years have brought new insights into 6mA biology in mammals. In this perspective, we present recent progress, discuss challenges, and pose four questions for future research regarding mammalian DNA 6mA.
A BRIEF HISTORY OF MAMMALIAN 6mA OR LACK THEREOF
5-Methyldeoxycytidine (5mC) has dominated studies of covalent DNA modification for several decades.1 It is the most abundant DNA modification (~3%–8% of all cytosine) in the genomes of most high eukaryotes.1,2 Robust sequencing methods such as bisulfite sequencing allow site-specific and genome-wide detection of 5mC.3–6 In prokaryotes, it is a different story altogether. N6-methyldeoxyadenosine (6mA), first discovered in Bacterium coli as early as 1955, is the most abundant DNA modification in most bacterial genomes, while 5mC is much less abundant or undetectable in many bacterial species.7–9 6mA plays important roles ranging from protecting bacterial genome against restriction enzymes to regulating DNA mismatch repair, chromosome replication, and transcription.10–13 The interests of 6mA extended to other species with studies showing its presence in fungi, protists, plants, invertebrates, and non-mammalian vertebrates, with varied abundances.13
Interests in DNA 6mA in eukaryotes re-emerged in 2015 and 2016, with several groups showing the presence and potential regulatory roles in different eukaryotic species.14–19 Subsequent studies have shown roles of 6mA in the genomes of eukaryotic species, including fungi, Chlamydomonas (green algae), Tetrahymena, and Drosophila.14–16,20–23 However, its presence and functional roles in high eukaryotes including mammals have been challenged because of (1) the bacterial contamination concerns, (2) the potential misincorporation of ribo-N6-methyladenosine (m6A), and (3) the lack of truly accurate sequencing approaches to reveal the presence and exact genomic locations of 6mA.2,3,24–26 The low abundance, prevalent contamination from bacterial DNA, and ambiguous function have made the presence of 6mA in invertebrates and vertebrates a big question mark.
Ever since the report of 6mA detection in human tissues using HPLC-MS/MS,27 methods to map DNA 6mA in different eukaryotic organisms, in particular mammals, have been developed and reported.2,17,18,24,26–45 Different reports using different methods have led to conflicting conclusions, with the most recent report observing a significantly lower 6mA abundance in many eukaryotic samples than those reported previously and called for the reassessment of 6mA across eukaryotes, especially mammals.26 In this perspective, we provide an overview of these previous studies, focusing on the presence and functional roles of DNA 6mA in mammalian systems, highlight four “enigmas” that will need to be addressed, and also provide our views on future research directions.
ENIGMA #1: TRULY EXIST OR ARTIFACTS?
Is 6mA truly present in mammalian genomes? Excluding probable artifacts, are there true 6mA sites in the genome accumulating to reasonable levels? The answers to these questions largely depend on the methods for detection. So far, LC-MS/ MS and dot blot have been used for identifying mammalian 6mA levels, while 6mA DIP-seq, ChIP-exo/6mACE-seq, single-molecule real-time (SMRT)/nanopore sequencing, and 6mA-RE-seq/DA-6mA-seq have been developed for whole-genome profiling of 6mA (Table 1).17,26,27,30–35,38,40,42,43,45–47 These methods, while useful in different applications, all have limitations.
Table 1.
A summary of mammalian 6mA mapping methods and the representative applications
| Method | Strengths/limitations | Year/reference | Tissues/cell lines | Abundance | Motif | Enriched regions | Functions |
|---|---|---|---|---|---|---|---|
| 6mA-DIP-seq | efficient and easy-to-use/large DNA input amount; low resolution; low sensitivity; not quantitative; high false-positive rate because of the non-specific binding | 2017; Yao et al.30 | mouse prefrontal cortex | 37,937 gain-of-6mA regions and 21,974 loss-of-6mA regions upon stress | ‐ | intergenic and intronic regions | negatively correlates with neuronal gene expression |
| 2018; Xie et al.45 | normal human astrocytes and human patient-derived glioma stem cells | 7,282–17,263 peaks | GGAAT | heterochromatin | transcriptional silencing of oncogenic pathways | ||
| 2018; Xiao et al.32 | human blood | 21,129 high-confidence peaks | ‐ | exon-coding regions; mtDNA | ‐ | ||
| 2019; Kweon et al.34 | mouse ES cells | 4,922 peaks | AGAAGAGGA | Intergenic regions | triggering proteolysis of its cognate sensor proteins | ||
| 2020; Li et al.38 | mouse ES cells to trophoblast cells | 20,318 differentially increased peaks | ‐ | intergenic regions, such as LINE-1s | repressing SIDD-SATB1 interactions and regulating gene expression during trophoblast development | ||
| 2022; Chen et al.43 | human breast cancer cells (MDA-MB-453) | 17,294 high-confidence peaks | [G/C]AGG | introns, intergenic regions, enhancers, and upstream promoter regions | repressing the expression of cell cycle inhibitor genes | ||
| SMRT-seq/Nanopore sequencing/6mASCOPE | base resolution; quantitative/large DNA input amount; high false positive rate when the abundance of modification is low | 2016; Wu et al.17 | mouse TT2 ES cells | 37,581 sites with Alkbh1 knockout | GAA; AATA | intergenic regions, young full-length LINE-1 transposon | repressing the expression of nearby genes |
| 2018; Zhu et al.31 | Human lymphoblastoid cells (hLCLs) | ‐ | AG | promotors of young full-length LINE-1 transposon | ‐ | ||
| 2018; Xiao et al.32 | Human lymphocytes (HX1) | 881,240 sites | [G/C]AGG[C/T] | exon-coding regions; mtDNA | repressing tumorigenesis | ||
| 2019; Pacini et al.46 | Human lymphoblastoid cell line (AK1); human hydatidiform cell lin (CHM1) | 74,345 sites in CHM1 and 80,561 sites in AK1 | AG/GA | introns, exons, in the 5’ UTR and near transcriptional start sites (TSSs) | ‐ | ||
| 2022; Cui et al.42 | hepatocellular carcinoma (HCC) | 500 ppm | AGG | intergenic and intronic regions | positively correlated with gene expression | ||
| 2022; Kong et al.26 | HEK293T; human glioblastoma brain tissues; human peripheral blood mononuclear cells (PBMCs) | 1; 2–3; 17 ppm | no reliable motif | not supporting the enrichment of 6mA in young L1 elements or mtDNA | ‐ | ||
| ChIP-exo/6mACE-seq | base resolution/large DNA input amount; low sensitivity; high false positive rate because of the non-specific binding | 2018; Koh et al.33 | HEK293T | 14,000 sites | AATGG | young and active LINE and SINE subfamilies; mtDNA | destabilizing dsDNA and regulating mitochondrial function |
| 2020; Hao et al.40 | HepG2 | 23 sites under normoxia; 34 sites under hypoxia (mtDNA only) | CTTATC | ‐ | repressing the transcription of mitochondrial genes | ||
| 6mA-RE-seq/DA-6mA-seq | base resolution/limited motifs | 2019; Li et al.35 | mouse primary cortical neuron | 2,033,704 + 306,207 GATC sites for extinction training group; 2,033,704 + 212,326 GATC sites for retention control group | ‐ | LINE1 elements | positively correlated with gene expression and the formation of fear extinction memory |
Dot blot is not quantitative and can be affected by the prokaryotic DNA contamination in samples and the specificity of the antibodies used when detecting mammalian 6mA. This method is not recommended for future quantification of 6mA or other DNA or RNA modifications in general. Note that a previous report showed that antibodies were able to detect 6mA as low as 0.003% of 6mA/A ratio but unmodified adenine also showed signals.25 We do not recommend quantifying 6mA below ~0.01% of 6mA/A using dot blot. We also suggest verifying the specificity and sensitivity of the selected antibody before applying them for 6mA detection. It is well known that different antibodies from different companies or different clones of antibodies from the same company exhibit varied properties. For example, antibodies from synaptic systems may exhibit higher sensitivity, while the monoclonal antibody from Abcam (cat. no. 151230) tends to be more selective.18,33 In addition, the isotype-matched control IgG should be used side by side when applying the selected antibody for immunoprecipitation-based studies.
LC-MS/MS, especially UHPLC-QQQ-MS/MS, is capable to quantify modified DNA or RNA bases with high specificity and sensitivity, reporting the exact mass and reaching the detection limit of around 1 ppm (parts per million). It has been utilized for detecting the presence and abundance of 6mA in various human, mouse, rat, and pig tissues or cell lines in numerous studies, with the results varying from less than 1 ppm to thousands of ppm.2,18,27,28,30,45 The main limitation of LC-MS/MS is the fact that it measures the sum of 6mA regardless of the source. It is now clear that many of the previous studies were conducted on samples contaminated by prokaryotic-origin 6mA from either mycoplasma contamination, plasmid transfection, or in some cases 6mA contamination from bacterial systems used to prepare reagents for DNA digestion.2,25 Proper controls are required to ensure the elimination of possibilities for contamination when measuring the absolute level of DNA 6mA in the genomes of mammals as well as other high eukaryotes.
Despite the limitation, we and others have applied LC-MS/MS to various samples and have attempted to eliminate all possible contaminations. In many mammalian cells and tissues, we have observed 6mA levels close to the background, suggesting extremely low levels of 6mA in genomic DNA.26,40,48 However, in mouse testis and glioblastoma cells, we and others consistently observed DNA 6mA with measurable abundances higher than 1 ppm, suggesting the presence of noticeable levels of DNA 6mA in the genomes of these cells.17,18,45,48 In addition, after careful purification of mitochondrial DNA (mtDNA), we observed that the level of 6mA in mtDNA is at least 1,300-fold higher than that from gDNA, indicating the presence of relatively abundant 6mA modification in mtDNA.40 Another study made similar observations.33 We thus suspect mtDNA 6mA contributes to baseline level DNA 6mA for many cells and tissues examined using LC-MS/MS; however, certain tissue or cell lines do contain elevated gDNA 6mA.
While LC-MS/MS cannot trace the origin of 6mA, sequencing-based approaches, if accurate, offer perhaps the best options to measure the presence and relative abundance of 6mA in the whole genome and at specific loci. The antibody-dependent methods such as 6mA DIP-seq and ChIP-exo (6mACE-seq), including the dot blot, have been challenged for antibody non-specificity, particularly when applied to study low abundant 6mA.25,49 In one experiment, ~137,557 antibody-enriched regions, most of which were located at short tandem repeats, were detected by DIP-seq in mouse embryonic stem cells (mESCs) when using just a non-specific mouse IgG antibody.49 This number already exceeds the 6mA frequency estimated by LC-MS/MS in most mammalian gDNA,2 suggesting a source of non-specific immunoprecipitation using some of the anti-6mA antibodies. Another study also suggested the potential RNA m6A origin of some of the 6mA peaks detected using DIP-seq.25 Clearly, much more accurate methods with high specificity and sensitivity are required to clearly dissect the presence and distribution of 6mA in mammalian genomes.
Third-generation sequencing (TGS), including PacBio SMRT sequencing, and nanopore sequencing, are other technologies used for mapping 6mA, especially in bacterial genomes, with the distinct advantage of single-base resolution and high sensitivity.17,26,31,32,42,50 However, since the level of 6mA in mammalian genomes is much lower than that in bacterial genomes, the accuracy of TGS methods is usually non-ideal when working on mammalian 6mA.25,31 The low abundance and indirect readout pose challenges when applying TGS for 6mA detection.25,26
ENIGMA #2: GENOMIC LOCATIONS?
UHPLC-QQQ-MS/MS has revealed the presence of 6mA in not only mammalian mtDNA but also genomes of mouse trophoblast stem cells and glioblastoma cells.38,45,48 These observations hint at the potential regulatory roles of 6mA. If 6mA does exist in the genomes of certain mammalian cells, is it randomly distributed or enriched to specific motifs at distinct loci? A few studies, although supporting the presence of 6mA in mammalian genomes, argued against 6mA acting as a functional DNA mark in mammalian cells, but rather non-directed, random incorporation.3,24 In one of the published studies ribo-m6A was fed to cells and was shown to convert to 6mATP and incorporated into DNA.24 Despite this possibility several reports have suggested consistent genomic features for the distribution of mammalian 6mA (Table 1), arguing against the random incorporation model. Taking mESCs as an example, 6mA was shown to exist at intergenic regions and LINE-1 retrotransposon elements in multiple studies using independent methods.17,34,38 Similar genomic distribution patterns of 6mA were also suggested in mouse cortex and human lymphoblastoid cells (hLCLs), human hepatocellular carcinoma (HCC), and HEK293T cell line,30,31,33,42 despite very low total gDNA 6mA levels observed in some of these systems. One study directly compared 6mA profiles based on published SMRT-seq datasets and found that 6mA signals do occur consistently at the same genomic location within a given human cell type.46 Although reports are claiming that no reliable 6mA motif was identified,26 we noticed that other studies did report the same consensus motif of AG(G).31,32,34,42,43,46
Sequencing methods that give base-resolution information with modification level at each site are required to confirm the presence of 6mA in mammalian gDNA. Progress has already been made. We have recently introduced DR-6mA-seq,48 and uncovered the presence and genome-wide distribution of 6mA in the genomes of specific mouse tissues and a transformed mouse glioblastoma model cell line. Most genomic 6mA sites appear to localize at non-coding regions. The genetic features of 6mA also appear to differ among different cell types.
In HepG2 mtDNA, 159 high-confidence 6mA sites were detected by DR-6mA-seq and overlapped very well with the 29 6mA sites previously detected by ChIP-exo, confirming the presence of 6mA in the mammalian mitochondrial genome.40,48 Using an SMRT-based method, 6mASCOPE, the presence of 6mA was also detected but at a lower level (29 ppm) in HEK293T mtDNA.26 We cannot exclude random exclusion of 6mATP derived from ribo-m6A into mammalian gDNA; however, reasonable levels of 6mA could be detected in certain mouse tissues and glioblastoma model cells using MS. Our recent base-resolution sequencing also uncovered an accumulation of 6mA to certain motifs and specific genomic locations. In fact, we have validated at least two 6mA sites accumulating over 50% fraction in mouse glioblastoma cells when applying amplicon sequencing to measure the modification stoichiometry.48 We have also confirmed this observation using an orthogonal method of silver-ion-mediated base-paring affinity assay, which could detect 6mA at specific sites even at low modification fraction (<20%).51,52 Although gDNA 6mA is scarce and likely non-existing in most mammalian tissues and cells, these recent observations did suggest the presence and accumulation of 6mA in specific genomic loci in certain mammalian genomes.
ENIGMA #3: AN ENZYMATICALLY REGULATED MODIFICATION IN MAMMALS?
Although non-enzymatic covalent modifications (NECMs) on DNA have been shown to potentially affect gene expression regulation,53 an enzymatically regulated DNA modification would add new pathways to the network of gene expression regulation. The conserved motif and high stoichiometry of 6mA at specific sites revealed by sequencing in certain mammalian genomes may suggest enzymatic installation through methyltransferase(s). The identity of the methyltransferase requires further research. The presence of demethylases that mediate the demethylation of DNA 6mA, as has been known for RNA m6A,54–56 as well as the potential 6mA-binding proteins that bind preferentially to 6mA-modified DNA and mediate downstream regulation also require further research. Considering the low modification level and the fact that no homolog of the bacterial 6mA methyltransferase (Dam) has been found in mammals, it has been challenging to determine 6mA effector proteins in mammals.57 Many of the 6mA effector proteins proposed in the past several years either lack experimental evidence or are inconsistent in different studies (Figure 1).58,59
Figure 1. Proposed functions and effector proteins for 6mA on mammalian genomic DNA and mtDNA.
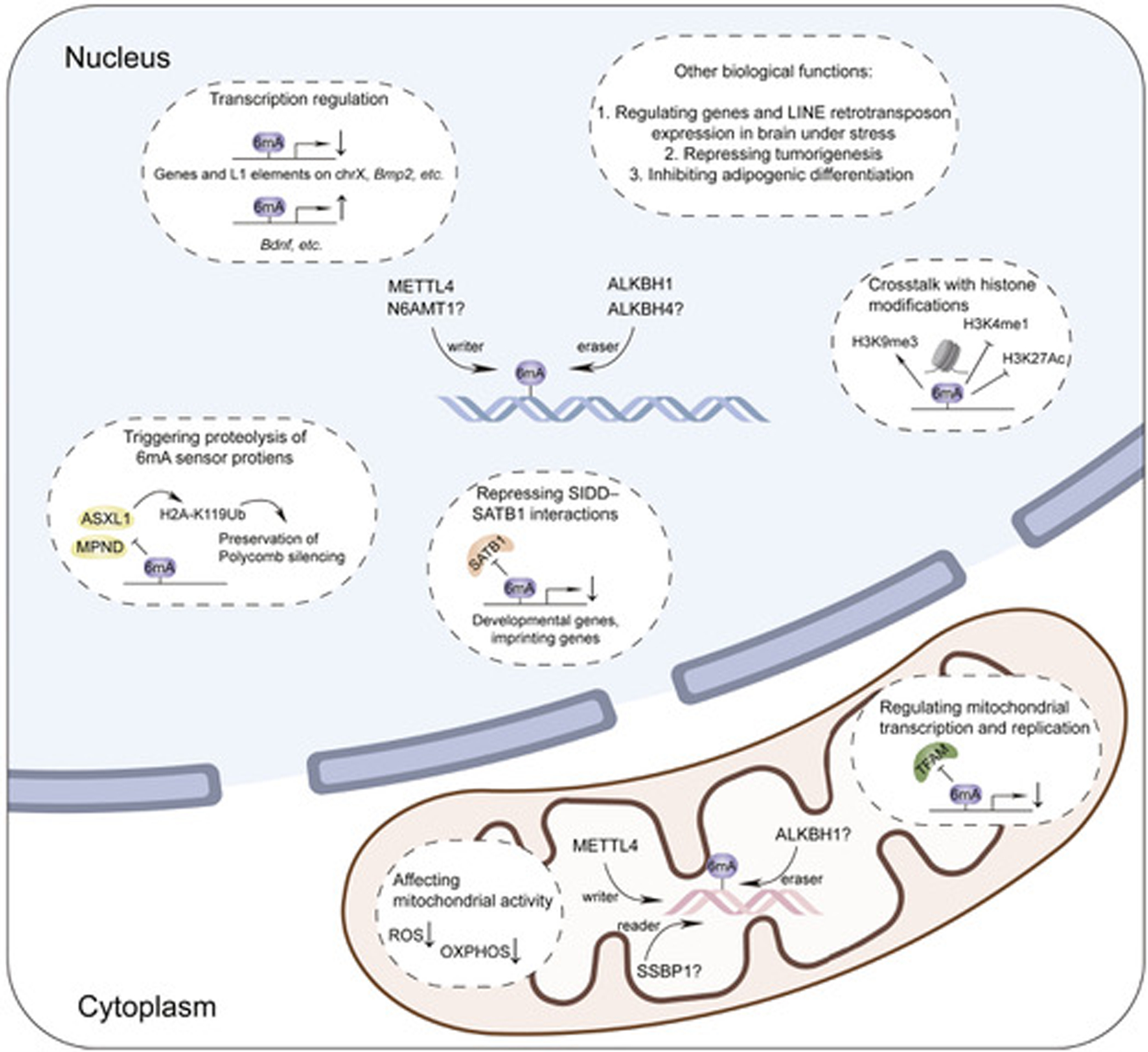
The downward and upward arrows refer to the repression or upregulation of transcription, respectively. The question marks refer to the putative 6mA methylases, demethylases, and binding proteins but with conflicting data or a lack of strong evidence.
METTL4, a homolog of DAMT-1, and ALKBH1, a homolog of the AlkB family demethylase, are the only two promising 6mA effector proteins identified in mammals so far.58 The methyltransferase activity of METTL4 on 6mA has been validated both in vitro and inside cells in human mtDNA and mouse gDNA and has been shown to impact mitochondrial replication, transcription, mitochondrial activities, and adipogenesis40,60; its homolog DAMT-1 has been suggested as a 6mA methyltransferase in C. elegans and mediates the crosstalk between methylations of histone H3K4 and adenines, whereas NMAD-1, the homolog of a putative mammalian 6mA demethylase in vitro, was also reported to demethylate 6mA in C. elegans and regulate DNA replication and repair.15,61
ALKBH1 was reported to erase 6mA both in vitro and in mESCs, patient-derived human glioblastoma models, human mesenchymal stem cells, and human cancer cell lines.17,32,41,45 It has also been proposed as a 6mA demethylase in human mtDNA, affecting oxidative phosphorylation.33 A recent complex structural study has demonstrated its distinct substrate recognition mode toward 6mA sites on bulged DNA.62 However, ALKBH1 also mediates tRNA oxidation and demethylation in mammalian cytosol and mitochondria, and it only catalyzes 6mA demethylation in ssDNA but not dsDNA, raising concerns about whether its biological effects mostly arise from tRNA oxidation or DNA 6mA demethylation.62–65
Other proteins potentially involved in mammalian 6mA deposition, recognition, and removal so far lack supporting experimental evidence. N6AMT1, or N6-adenine-specific DNA methyltransferase, was reported to regulate 6mA in multiple mammalian cells but has been challenged for the lack of methyltransferase activity even in vitro.32,35,36,66 ALKBH4 displayed demethylation activity on dsDNA in vitro,34 but the activity is very weak and requires further in vitro and cell-based supports. Two potential mammalian 6mA binding proteins were also reported. SSBP1, a housekeeping protein involved in mitochondrial biogenesis, was shown to bind at 6mA-modified regions of human mtDNA.33 However, SSBP1 is an ssDNA-binding protein and 6mA peaks show overlap with ssDNA regions. Direct evidence from biochemical binding assays and functional studies is required to verify SSBP1 as a 6mA-binding protein.38,67 Another candidate binding protein, SATB1, is a DNA-binding protein antagonized by DNA 6mA.38 In a different indirect reading mechanism, the presence of 6mA makes dsDNA stiff, which affects DNA bending when bound by certain DNA-binding proteins (TFAM, etc.) to impact downstream regulation.40 Similarly, DNA 6mA antagonizes the binding of SATB1 to SIDD sequences and regulates chromatin structure during early development, although the mechanism of how 6mA actively repels the binding of SATB1 remains to be determined.58 It should be noted that RNA m6A methyltransferases and demethylases, including the METTL3–14 complex, PCIF1, FTO, and ALKBH5, were reported to exert enzymic activity in vitro on DNA dA or 6mA, preferentially on ssDNA, although the biological significance in vivo remains to be elucidated.34,55,68–70
In summary, testing the robust biochemical activities on dsDNA or ssDNA is the first requirement to assign the potential methylases or demethylases for DNA 6mA. However, the biochemical evidence is insufficient to confirm an enzyme as a true 6mA effector protein, considering the enzyme could be forced to act on the substrates in vitro without having real biological functions on them in vivo. It is possible that 6mA is deposited and removed when dsDNA is melted to ssDNA during replication or other processes. This may explain the low levels of 6mA in gDNA in general. A 6mA-binding protein will need to bind preferentially to the 6mA-modified dsDNA, either through direct recognition of 6mA or through indirect mechanisms such as changing the physical properties of the modified regions of gDNA. Now, with several 6mA sites defined in specific mammalian cells, the community can study these questions regarding these sites. Eventually, the effector proteins are the ones that lead to functional outcomes if 6mA is functionally relevant in mammals. It is also possible that these effector proteins may have other cellular functions and are “hijacked” to install, read, or erase 6mA during specific biological processes.
ENIGMA #4: FUNCTIONAL RELEVANCE?
Is mammalian 6mA too scarce to be functionally relevant in mammals? Functional relevance is intimately linked to genomic location and effector proteins of 6mA. Two criteria will need to be met to assign true functions of 6mA: (1) the function needs to be related to specific 6mA sites on DNA; (2) the function needs to depend on effector proteins that either deposit, recognize, or potentially erase specific 6mA on DNA. Diverse functions have been proposed for mammalian DNA 6mA in recent years (Figure 1).17,30,34,35,38,41,45,71 Few of these studies met these criteria.
6mA is abundant in mammalian mtDNA. METTL4 is one enzyme that could install mtDNA 6mA. The knockdown of METTL4 has led to reduced mtDNA 6mA levels, upregulated transcription, increased mitochondrial copy number, and elevated mitochondrial respiration activity.40 However, METTL4 also has nuclear roles, and one cannot completely exclude the possibility that these mitochondrial effects are actually a consequence of the nuclear function of METTL4.72 Similarly, ALKBH1, a potential 6mA demethylase, mediates tRNA oxidation in both cytosol and mitochondria.63,64 Its functional impact through either DNA 6mA demethylation or tRNA oxidation needs to be clearly defined.
To satisfy the two criteria, we need to develop and apply quantitative methods that detect 6mA sites. Targeted DNA demethylation systems using dCas9 fused with the catalytic domains of 6mA demethylase will also need to be developed and used to manipulate discrete 6mA sites at specific DNA loci to demonstrate the direct functional effects. A dioxygenase from fungus, CcTet, has recently been shown to mediate preferential 6mA oxidation and demethylation over 5mC in dsDNA.73 This demethylase, if fused with dCas9, could be very useful in reversing 6mA to A at specific sites to investigate subsequent functional consequences. In principle, dCas9-fused methyltransferase could be useful as well. We currently do not know any mammalian dsDNA 6mA methyltransferase, although METTL4 can mediate 6mA methylation of ssDNA.34,40 dsDNA 6mA methyltransferases from low eukaryotes could be employed for functional interrogations. The glioblastoma model cell line reported recently, with discrete 6mA sites possessing high modification fraction, offers an exemplary system for such studies.45,48
Aside from mtDNA, the chance for gDNA 6mA to be functional in most mammalian adult tissues is very low, because of the exceedingly low levels of gDNA 6mA. It is more likely that 6mA on mammalian gDNA functions in certain biological processes such as tumorigenesis and early development. Particularly, accumulating pieces of evidence have suggested functional roles of 6mA during early development, with at least one potential 6mA binding protein identified.17,18,38 With the base-resolution sequencing method available, the functional impacts of these 6mA sites can be more thoroughly investigated.
CONCLUDING REMARKS
After an initial rush of eukaryotic DNA 6mA research, the community has come back to a more realistic picture. This DNA modification may play notable roles in gene expression regulation in low eukaryotic species, but its role in high eukaryotes, in particular mammals, is limited. Mammalian mtDNA is frequently modified with 6mA but not 5mC, presenting an intriguing system for more thorough mechanistic interrogations. The 6mA-modified dsDNA is more resistant to bending and could have functional consequences during mitochondrial transcription and replication. The levels of 6mA on mammalian genomic DNA are low across most tissues and cell types. However, during early development and in certain cancer cells, 6mA appears to accumulate at specific sites or loci, suggesting functional relevance.
Moving forward, the four questions we list are also opportunities for the community to clearly define DNA 6mA function in mammals. We will need to (1) detect the presence of 6mA on gDNA; (2) apply reliable methods to map the exact locations of 6mA. Some of these sites will need to accumulate to reasonable modification stoichiometry to be functionally relevant; (3) effector proteins such as methylases, demethylases, or binding proteins will need to be identified and perturbed for functional characterizations; (4) CRISPR-based systems to reverse 6mA at specific sites will need to be established to assign direct function. Functional characterizations should focus on biological processes in which 6mA accumulates to measurable levels in gDNA, such as during early development and tumorigenesis, and we may view 6mA as a DNA mark that plays primarily localized roles at specific gDNA loci and in mtDNA.
ACKNOWLEDGMENTS
We acknowledge the funding by the National Institutes of Health (HG006827 and HG008935). C.H. is a Howard Hughes Medical Institute Investigator.
Footnotes
DECLARATION OF INTERESTS
C.H. is a scientific founder; a member of the scientific advisory board and equity holder of Aferna Green, Inc. and AccuaDX Inc.; and a scientific co-founder and equity holder of Accent Therapeutics, Inc.
REFERENCES
- 1.Vanyushin BF, Tkacheva SG, and Belozersky AN (1970). Rare bases in animal DNA. Nature 225, 948–949. 10.1038/225948a0. [DOI] [PubMed] [Google Scholar]
- 2.O’Brown ZK, Boulias K, Wang J, Wang SY, O’Brown NM, Hao Z, Shibuya H, Fady P-E, Shi Y, He C, et al. (2019). Sources of artifact in measurements of 6mA and 4mC abundance in eukaryotic genomic DNA. BMC Genomics 20, 445. 10.1186/s12864-019-5754-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bochtler M, and Fernandes H (2021). DNA adenine methylation in eukaryotes: enzymatic mark or a form of DNA damage? BioEssays 43, e2000243. 10.1002/bies.202000243. [DOI] [PubMed] [Google Scholar]
- 4.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, and Paul CL (1992). A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA 89, 1827–1831. 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, Siejka-Zielińska P, Velikova G, Bi Y, Yuan F, Tomkova M, Bai C, Chen L, Schuster-Böckler B, and Song C-X (2019). Bisulfite-free direct detection of 5-methylcytosine and 5-hydroxymethylcytosine at base resolution. Nat. Biotechnol 37, 424–429. 10.1038/s41587-019-0041-2. [DOI] [PubMed] [Google Scholar]
- 6.Liu C, Cui X, Zhao BS, Narkhede P, Gao Y, Liu J, Dou X, Dai Q, Zhang LS, and He C (2020). DNA 5-methylcytosine-specific amplification and sequencing. J. Am. Chem. Soc 142, 4539–4543. 10.1021/jacs.9b12707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vanyushin BF, Belozersky AN, Kokurina NA, and Kadirova DX (1968). 5-methylcytosine and 6-Methylaminopurine in bacterial DNA. Nature 218, 1066–1067. 10.1038/2181066a0. [DOI] [PubMed] [Google Scholar]
- 8.Dunn DB, and Smith JD (1955). Occurrence of a new base in the deoxyribonucleic acid of a strain of bacterium coli. Nature 175, 336–337. 10.1038/175336a0. [DOI] [PubMed] [Google Scholar]
- 9.O’Brown ZK, and Greer EL (2016). N6-methyladenine: a conserved and dynamic DNA mark. Adv. Exp. Med. Biol 945, 213–246. 10.1007/978-3-319-43624-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naito T, Kusano K, and Kobayashi I (1995). Selfish behavior of restriction-modification systems. Science 267, 897–899. 10.1126/science.7846533. [DOI] [PubMed] [Google Scholar]
- 11.Messer W, and Noyer-Weidner M (1988). Timing and targeting: the biological functions of Dam methylation in E. coli. Cell 54, 735–737. 10.1016/S0092-8674(88)90911-7. [DOI] [PubMed] [Google Scholar]
- 12.Lu M (1994). SeqA: A negative modulator of replication initiation in E. coli. Cell 77, 413–426. 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 13.Low DA, Weyand NJ, and Mahan MJ (2001). Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect. Immun 69, 7197–7204. 10.1128/IAI.69.12.7197-7204.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang G, Huang H, Liu D, Cheng Y, Liu X, Zhang W, Yin R, Zhang D, Zhang P, Liu J, et al. (2015). N6-methyladenine DNA modification in Drosophila. Cell 161, 893–906. 10.1016/j.cell.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 15.Greer EL, Blanco MA, Gu L, Sendinc E, Liu J, Aristizábal-Corrales D, Hsu C-H, Aravind L, He C, and Shi Y (2015). DNA methylation on N6-adenine in C. elegans. Cell 161, 868–878. 10.1016/j.cell.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu Y, Luo GZ, Chen K, Deng X, Yu M, Han D, Hao Z, Liu J, Lu X, Doré LC, et al. (2015). N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas. Cell 161, 879–892. 10.1016/j.cell.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K, Liu Y, Byrum SD, Mackintosh SG, Zhong M, et al. (2016). DNA methylation on N6-adenine in mammalian embryonic stem cells. Nature 532, 329–333. 10.1038/nature17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J, Zhu Y, Luo G-Z, Wang X, Yue Y, Wang X, Zong X, Chen K, Yin H, Fu Y, et al. (2016). Abundant DNA 6mA methylation during early embryogenesis of zebrafish and pig. Nat. Commun 7, 13052. 10.1038/ncomms13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koziol MJ, Bradshaw CR, Allen GE, Costa ASH, Frezza C, and Gurdon JB (2016). Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications. Nat. Struct. Mol. Biol 23, 24–30. 10.1038/nsmb.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He S, Zhang G, Wang J, Gao Y, Sun R, Cao Z, Chen Z, Zheng X, Yuan J, Luo Y, et al. (2019). 6mA-DNA-binding factor Jumu controls maternal-to-zygotic transition upstream of Zelda. Nat. Commun 10, 2219. 10.1038/s41467-019-10202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mondo SJ, Dannebaum RO, Kuo RC, Louie KB, Bewick AJ, LaButti K, Haridas S, Kuo A, Salamov A, Ahrendt SR, et al. (2017). Widespread adenine N6-methylation of active genes in fungi. Nat. Genet 49, 964–968. 10.1038/ng.3859. [DOI] [PubMed] [Google Scholar]
- 22.Luo G-Z, Hao Z, Luo L, Shen M, Sparvoli D, Zheng Y, Zhang Z, Weng X, Chen K, Cui Q, et al. (2018). N6-methyldeoxyadenosine directs nucleosome positioning in Tetrahymena DNA. Genome Biol 19, 200. 10.1186/s13059-018-1573-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shah K, Cao W, and Ellison CE (2019). Adenine methylation in Drosophila is associated with the tissue-specific expression of developmental and regulatory genes. G3 (Bethesda) 9, 1893–1900. 10.1534/g3.119.400023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Musheev MU, Baumgärtner A, Krebs L, and Niehrs C (2020). The origin of genomic N6-methyl-deoxyadenosine in mammalian cells. Nat. Chem. Biol 16, 630–634. 10.1038/s41589-020-0504-2. [DOI] [PubMed] [Google Scholar]
- 25.Douvlataniotis K, Bensberg M, Lentini A, Gylemo B, and Nestor CE (2020). No evidence for DNA N6-methyladenine in mammals. Sci. Adv 6, eaay3335. 10.1126/sciadv.aay3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong Y, Cao L, Deikus G, Fan Y, Mead EA, Lai W, Zhang Y, Yong R, Sebra R, Wang H, et al. (2022). Critical assessment of DNA adenine methylation in eukaryotes using quantitative deconvolution. Science 375, 515–522. 10.1126/science.abe7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang D, Wang H, Song W, Xiong X, Zhang X, Hu Z, Guo H, Yang Z, Zhai S, Zhang L-H, et al. (2016). The decreased N6-methyladenine DNA modification in cancer cells. Biochem. Biophys. Res. Commun 480, 120–125. 10.1016/j.bbrc.2016.09.136. [DOI] [PubMed] [Google Scholar]
- 28.Ratel D, Ravanat J-L, Charles M-P, Platet N, Breuillaud L, Lunardi J, Berger F, and Wion D (2006). Undetectable levels of N6-methyl adenine in mouse DNA: cloning and analysis of PRED28, a gene coding for a putative mammalian DNA adenine methyltransferase. FEBS Lett 580, 3179–3184. 10.1016/j.febslet.2006.04.074. [DOI] [PubMed] [Google Scholar]
- 29.Schiffers S, Ebert C, Rahimoff R, Kosmatchev O, Steinbacher J, Bohne A-V, Spada F, Michalakis S, Nickelsen J, Müller M, et al. (2017). Quantitative LC-MS liefert keinen Hinweis auf m 6 dA oder m 4 dC im Genom von Mausstammzellen und -geweben. Angew. Chem 129, 11422–11425. 10.1002/ange.201700424. [DOI] [PubMed] [Google Scholar]
- 30.Yao B, Cheng Y, Wang Z, Li Y, Chen L, Huang L, Zhang W, Chen D, Wu H, Tang B, et al. (2017). DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress. Nat. Commun 8, 1122. 10.1038/s41467-017-01195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu S, Beaulaurier J, Deikus G, Wu TP, Strahl M, Hao Z, Luo G, Gregory JA, Chess A, He C, et al. (2018). Mapping and characterizing N6-methyladenine in eukaryotic genomes using single-molecule real-time sequencing. Genome Res 28, 1067–1078. 10.1101/gr.231068.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao C-L, Zhu S, He M, Chen D, Zhang Q, Chen Y, Yu G, Liu J, Xie SQ, Luo F, et al. (2018). N6-methyladenine DNA modification in the human genome. Mol. Cell 71, 306–318.e7. 10.1016/j.molcel.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 33.Koh CWQ, Goh YT, Toh JDW, Neo SP, Ng SB, Gunaratne J, Gao YG, Quake SR, Burkholder WF, and Goh WSS (2018). Single-nucleotide-resolution sequencing of human N 6-methyldeoxyadenosine reveals strand-asymmetric clusters associated with SSBP1 on the mitochondrial genome. Nucleic Acids Res 46, 11659–11670. 10.1093/nar/gky1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kweon S-M, Chen Y, Moon E, Kvederaviciutė K, Klimasauskas S, and Feldman DE (2019). Polycomb silencing. Mol. Cell 74, 1138–1147.e6. 10.1016/j.molcel.2019.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, Zhao Q, Wei W, Lin Q, Magnan C, Emami MR, Wearick-Silva LE, Viola TW, Marshall PR, Yin J, et al. (2019). The DNA modification N6-methyl-2′-deoxyadenosine (m6dA) drives activity-induced gene expression and is required for fear extinction. Nat. Neurosci 22, 534–544. 10.1038/s41593-019-0339-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li W, Shi Y, Zhang T, Ye J, and Ding J (2019). Structural insight into human N6amt1–Trm112 complex functioning as a protein methyltransferase. Cell Discov 5, 51. 10.1038/s41421-019-0121-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian L-F, Liu Y-P, Chen L, Tang Q, Wu W, Sun W, Chen Z, and Yan X-X (2020). Structural basis of nucleic acid recognition and 6mA demethylation by human ALKBH1. Cell Res 30, 272–275. 10.1038/s41422-019-0233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z, Zhao S, Nelakanti RV, Lin K, Wu TP, Alderman MH, Guo C, Wang P, Zhang M, Min W, et al. (2020). N6-methyladenine in DNA antagonizes SATB1 in early development. Nature 583, 625–630. 10.1038/s41586-020-2500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Zhang X-M, Luan M-W, Xing J-F, Chen J, and Xie S-Q (2020). Distribution patterns of DNA N6-methyladenosine modification in non-coding RNA genes. Front. Genet 11, 268. 10.3389/fgene.2020.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hao Z, Wu T, Cui X, Zhu P, Tan C, Dou X, Hsu K-W, Lin Y-T, Peng P-H, Zhang L-S, et al. (2020). N6-deoxyadenosine methylation in mammalian mitochondrial DNA. Mol. Cell 78, 382–395.e8. 10.1016/j.molcel.2020.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y, Chen Y, Wang Y, Jiang S, Lin W, Wu Y, Li Q, Guo Y, Liu W, and Yuan Q (2022). DNA demethylase ALKBH1 promotes adipogenic differentiation via regulation of HIF-1 signaling. J. Biol. Chem 298, 101499. 10.1016/j.jbc.2021.101499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cui H, Rong W, Ma J, Zhu Q, Jiang B, Zhang L, Li C, Zhuo Z, and Chen M (2022). DNA N6-adenine methylation in HBV-related hepatocellular carcinoma. Gene 822, 146353. 10.1016/j.gene.2022.146353. [DOI] [PubMed] [Google Scholar]
- 43.Chen J, Zhuang Y, Wang P, Ning J, Liu W, Huang Y, Lin X, Peng L, and Zhang D (2022). Reducing N6AMT1-mediated 6mA DNA modification promotes breast tumor progression via transcriptional repressing cell cycle inhibitors. Cell Death Dis 13, 216. 10.1038/s41419-022-04661-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schiffers S, Ebert C, Rahimoff R, Kosmatchev O, Steinbacher J, Bohne AV, Spada F, Michalakis S, Nickelsen J, Müller M, et al. (2017). Quantitative LC-MS provides no evidence for m6dA or m4dC in the genome of mouse embryonic stem cells and tissues. Angew. Chem. Int. Ed. Engl 56, 11268–11271. 10.1002/anie.201700424. [DOI] [PubMed] [Google Scholar]
- 45.Xie Q, Wu TP, Gimple RC, Li Z, Prager BC, Wu Q, Yu Y, Wang P, Wang Y, Gorkin DU, et al. (2018). N6-methyladenine DNA modification in glioblastoma. Cell 175, 1228–1243.e20. 10.1016/j.cell.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pacini CE, Bradshaw CR, Garrett NJ, and Koziol MJ (2019). Characteristics and homogeneity of N6-methylation in human genomes. Sci. Rep 9, 5185. 10.1038/s41598-019-41601-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo G-Z, Wang F, Weng X, Chen K, Hao Z, Yu M, Deng X, Liu J, and He C (2016). Characterization of eukaryotic DNA N6-methyladenine by a highly sensitive restriction enzyme-assisted sequencing. Nat. Commun 7, 11301. 10.1038/ncomms11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng X, Cui X, Zhang L-S, Ye C, Wang P, Zhong Y, Zheng Z, and He C (2023). Sequencing of N6-methyl-deoxyadenosine at single-base resolution across the mammalian genome. bioRxiv 2023.01.16.524325. 10.1101/2023.01.16.524325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lentini A, Lagerwall C, Vikingsson S, Mjoseng HK, Douvlataniotis K, Vogt H, Green H, Meehan RR, Benson M, and Nestor CE (2018). A reassessment of DNA-immunoprecipitation-based genomic profiling. Nat. Methods 15, 499–504. 10.1038/s41592-018-0038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fang G, Munera D, Friedman DI, Mandlik A, Chao MC, Banerjee O, Feng Z, Losic B, Mahajan MC, Jabado OJ, et al. (2012). Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat. Biotechnol 30, 1232–1239. 10.1038/nbt.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hong T, Yuan Y, Wang T, Ma J, Yao Q, Hua X, Xia Y, and Zhou X (2017). Selective detection of N6-methyladenine in DNA via metal ion-mediated replication and rolling circle amplification. Chem. Sci 8, 200–205. 10.1039/C6SC02271E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li W, Yang H, Wang Y, Weng X, and Wang F (2021). Highly sensitive detection of 6mA at single-base resolution based on A–C mismatch. Analyst 146, 4450–4453. 10.1039/D1AN00918D. [DOI] [PubMed] [Google Scholar]
- 53.Zheng Q, Maksimovic I, Upad A, and David Y (2020). Non-enzymatic covalent modifications: a new link between metabolism and epigenetics. Protein Cell 11, 401–416. 10.1007/s13238-020-00722-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X, Wei LH, Wang Y, Xiao Y, Liu J, Zhang W, Yan N, Amu G, Tang X, Zhang L, et al. (2019). Structural insights into FTO’s catalytic mechanism for the demethylation of multiple RNA substrates. Proc. Natl. Acad. Sci. USA 116, 2919–2924. 10.1073/pnas.1820574116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol 7, 885–887. 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29. 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guarné A (2012). The functions of MutL in mismatch repair. Prog. Mol. Biol. Transl. Sci 110, 41–70. 10.1016/B978-0-12-387665-2.00003-1. [DOI] [PubMed] [Google Scholar]
- 58.Boulias K, and Greer EL (2022). Means, mechanisms and consequences of adenine methylation in DNA. Nat. Rev. Genet 23, 411–428. 10.1038/s41576-022-00456-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen C, Wang K, Deng X, and Chen J (2022). DNA N6-methyldeoxyadenosine in mammals and human disease. Trends Genet 38, 454–467. 10.1016/j.tig.2021.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Z, Hou Y, Wang Y, Gao T, Ma Z, Yang Y, Zhang P, Yi F, Zhan J, Zhang H, et al. (2020). Regulation of adipocyte differentiation by METTL4, a 6mA methylase. Sci. Rep 10, 8285. 10.1038/s41598-020-64873-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang SY, Mao H, Shibuya H, Uzawa S, O’Brown ZK, Wesenberg S, Shin N, Saito TT, Gao J, Meyer BJ, et al. (2019). The demethylase NMAD-1 regulates DNA replication and repair in the Caenorhabditis elegans germline. PLoS Genet 15, e1008252. 10.1371/journal.pgen.1008252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang M, Yang S, Nelakanti R, Zhao W, Liu G, Li Z, Liu X, Wu T, Xiao A, and Li H (2020). Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA. Cell Res 30, 197–210. 10.1038/s41422-019-0237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J, Wang X, Hao Z, Dai Q, Zheng G, et al. (2016). ALKBH1-mediated tRNA demethylation regulates translation. Cell 167, 816–828.e16. 10.1016/j.cell.2016.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kawarada L, Suzuki T, Ohira T, Hirata S, Miyauchi K, and Suzuki T (2017). ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res 45, 7401–7415. 10.1093/nar/gkx354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rashad S, Han X, Sato K, Mishima E, Abe T, Tominaga T, and Niizuma K (2020). The stress specific impact of ALKBH1 on tRNA cleavage and tiRNA generation. RNA Biol 17, 1092–1103. 10.1080/15476286.2020.1779492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Woodcock CB, Yu D, Zhang X, and Cheng X (2019). Human HemK2/KMT9/N6AMT1 is an active protein methyltransferase, but does not act on DNA in vitro, in the presence of Trm112. Cell Discov 5, 50. 10.1038/s41421-019-0119-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miralles Fusté J, Shi Y, Wanrooij S, Zhu X, Jemt E, Persson Ö, Sabouri N, Gustafsson CM, and Falkenberg M (2014). In vivo occupancy of mitochondrial single-stranded DNA binding protein supports the strand displacement mode of DNA replication. PLoS Genet 10, e1004832. 10.1371/journal.pgen.1004832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu D, Horton JR, Yang J, Hajian T, Vedadi M, Sagum CA, Bedford MT, Blumenthal RM, Zhang X, and Cheng X (2021). Human MettL3-MettL14 RNA adenine methyltransferase complex is active on double-stranded DNA containing lesions. Nucleic Acids Res 49, 11629–11642. 10.1093/nar/gkab460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu D, Zhou J, Chen Q, Wu T, Blumenthal RM, Zhang X, and Cheng X (2022). Enzymatic characterization of in vitro activity of RNA methyltransferase PCIF1 on DNA. Biochemistry 61, 1005–1013. 10.1021/acs.biochem.2c00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zou S, Toh JDW, Wong KHQ, Gao Y-G, Hong W, and Woon ECY (2016). N6-methyladenosine: a conformational marker that regulates the substrate specificity of human demethylases FTO and ALKBH5. Sci. Rep 6, 25677. 10.1038/srep25677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ouyang L, Su X, Li W, Tang L, Zhang M, Zhu Y, Xie C, Zhang P, Chen J, and Huang H (2021). ALKBH1-demethylated DNA N6-methyladenine modification triggers vascular calcification via osteogenic reprogramming in chronic kidney disease. J. Clin. Invest 131, e146985. 10.1172/JCI146985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen H, Gu L, Orellana EA, Wang Y, Guo J, Liu Q, Wang L, Shen Z, Wu H, Gregory RI, et al. (2020). METTL4 is an snRNA m6Am methyltransferase that regulates RNA splicing. Cell Res 30, 544–547. 10.1038/s41422-019-0270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mu Y, Zhang L, Hu J, Zhou J, Lin H-W, He C, Chen H-Z, and Zhang L (2022). A fungal dioxygenase CcTet serves as a eukaryotic 6mA demethylase on duplex DNA. Nat. Chem. Biol 18, 733–741. 10.1038/s41589-022-01041-3. [DOI] [PubMed] [Google Scholar]


