Abstract
Objective
This article describes a scalable, performant, sustainable global network of electronic health record data for biomedical and clinical research.
Materials and Methods
TriNetX has created a technology platform characterized by a conservative security and governance model that facilitates collaboration and cooperation between industry participants, such as pharmaceutical companies and contract research organizations, and academic and community-based healthcare organizations (HCOs). HCOs participate on the network in return for access to a suite of analytics capabilities, large networks of de-identified data, and more sponsored trial opportunities. Industry participants provide the financial resources to support, expand, and improve the technology platform in return for access to network data, which provides increased efficiencies in clinical trial design and deployment.
Results
TriNetX is a growing global network, expanding from 55 HCOs and 7 countries in 2017 to over 220 HCOs and 30 countries in 2022. Over 19 000 sponsored clinical trial opportunities have been initiated through the TriNetX network. There have been over 350 peer-reviewed scientific publications based on the network’s data.
Conclusions
The continued growth of the TriNetX network and its yield of clinical trial collaborations and published studies indicates that this academic-industry structure is a safe, proven, sustainable path for building and maintaining research-centric data networks.
Keywords: data warehouse, data management, real-world data, clinical trial protocols, electronic health records
INTRODUCTION
The need for both intra- and inter-institutional patient data for research has led to many efforts over the years to create institutional clinical data repositories (CDRs) and network them among organizations.1–7 Creating a network of CDRs requires the development of data models and software tools for data acquisition, transformation, and storage. Networking these repositories requires data harmonization, security and governance policies, and machine communication protocols. These projects also require funding and oversight. Typically, funding has been provided by the government, either at the federal or state level. Oversight and operational management have been either government-based or institutional, or a combination of both. While these CDR networks have successfully met their objectives to varying degrees, a common challenge to their sustainability has been their vulnerability to loss of funding. A different paradigm for the funding and operational management of CDR development and networking has recently emerged: commercial (or industry) partnerships with healthcare organizations (HCOs). This is the approach taken by TriNetX, which has, over the past 8 years, demonstrated a sustainable path to federated CDRs.
When TriNetX was launched as an industry–academic collaboration in 2014, the primary objective was the creation of a network of CDRs focused on improving the efficiency of clinical trial research in all therapeutic areas.8 This efficiency is gained via the creation of a federated network into which HCOs have agreed to share their data, enabling the evaluation of trial eligibility to maximize patient enrollment and the identification of institutions with sufficient numbers of eligible patients. Two types of organizations would be TriNetX members: HCOs that would agree to share de-identified patient data for research-purposes only (under strict security and governance rules and with a combination of technical, operational, and contractual protections) and industry organizations that would design and conduct clinical trials. The HCOs are typically academic medical center-based healthcare systems or research-focused non-academic HCOs. Industry participants include pharmaceutical companies and contract research organizations (clinical research organizations).
After 8 years, TriNetX has grown to be a global network of over 220 HCO members and 40 industry sponsors. These HCO and industry network members are geographically present in North and South America, Europe, the Middle East, Africa, and the Asia-Pacific regions. The global growth of TriNetX has produced new challenges regarding legal, cultural, and technical heterogeneities across these regions, which necessitated innovative solutions of providing a homogenous interface to different data models, terminologies, and standards used to share real-world data (RWD). Over this time frame, growing traffic in clinical trial collaborations has flowed between the academic and industry participants, and as shown by publications and platform capabilities, research activities have expanded from the initial focus on clinical trials. Currently, the platform provides a Research sub-network of 70 HCOs with data on over 101 million patients. Participant organizations in this sub-network agree to contribute their data to this combined pool for various research projects not necessarily involving clinical trial research. Other sub-networks, such as one for studying the effects of the COVID-19 pandemic,9 as well as inter-institutional collaborative projects, have been formed from the overall TriNetX global network.
The scope of RWD acquired by this network has expanded from primarily the demographics, diagnoses, procedures, medications, and laboratory results found in the HCO’s electronic medical record to include oncology-specific data such as cancer stage and tumor morphology and molecular diagnostic genomic assay results indicating the presence of a patient’s genetic variants. These data may be acquired as discrete data elements or from natural language processing of medical notes and reports. The platform also offers additional capabilities not present at its inception. There is now the ability to access external data sets, such as US medical insurance claims and mortality data. Patient encounters may now be linked across organizations while maintaining the privacy of individuals. A full suite of analytical tools is integrated with the cohort query functionality. These tools include outcomes analysis, comparison of cohorts and outcomes, competing risk, incidence and prevalence, and treatment pathways. Existing analytics are being constantly improved, and new analytics, such as patient clustering and burden of illness, are under development. A new trusted research environment has been developed and will allow users to generate their own analytics using popular languages such as Python, R, SQL, and Scala.
This article describes the global breadth of the TriNetX network and what has been accomplished through its use, as evidenced by clinical trial research activities and published research studies. Also discussed are the solutions developed by TriNetX to the various challenges which abound in creating a worldwide network of CDRs. These challenges include addressing concerns about data sharing and conforming to the various national and regional patient data privacy regulations, harmonizing the disparate data models encountered among HCOs, achieving seamless integration and linking of data from disparate sources, ensuring high utilization of the platform by member institutions, and operationally ingesting, presenting, querying, and analyzing large quantities of data in an efficient manner.
MATERIALS AND METHODS
Challenge: data sharing and privacy protection
HCOs that participate in the TriNetX network provide healthcare data as a de-identified, pseudo-anonymized, or limited data set (depending on local privacy regulations), and HCOs grant the use of that data, for research purposes, on the TriNetX platform. This participation typically requires approval by an institutional review board or independent ethics committee, information technology department security teams, and institutional counsel. Contractual agreements between the HCO and TriNetX are not exclusionary, nor do they have any penalty for termination or lack of involvement. In exchange for contributing their data, HCOs incur no financial costs and receive data query, analytic, and visualization capabilities, as well as the hardware needed to execute the software. TriNetX is a federated ecosystem, so the data resides on hardware located within the HCO’s data center. A cloud-hosted option is also available. TriNetX is compliant with the data privacy regulations applicable to the contributing HCO, such as the Health Insurance Portability and Accountability Act (HIPAA) in the United States, the General Data Protection Regulation (GDPR) in Europe, Lei Geral de Proteção de Dados (LGPD) in Brazil, and other regional laws depending on the legislative landscape. The process by which data are de-identified has been attested to through a formal determination by a qualified expert as defined in Section §164.514(b)(1) of the HIPAA Privacy Rule.10 TriNetX also provides technical engineering support for onboarding the HCO’s patient data.
Challenge: data harmonization
New data are first harmonized syntactically by ingestion into the TriNetX common data model. Initially, TriNetX used existing i2b2 instances as the sole upstream source of data. However, this approach limited which HCOs could join TriNetX. To expand the network, it was essential to develop new custom connectors and toolkits for ingesting data from a full spectrum of common data models and healthcare information exchange standards, including OMOP, FHIR, HL7, and custom direct-to-EHR. TriNetX has also incorporated APIs into its data pipelines.
The data are further harmonized semantically by mapping it to a set of standards that comprise the TriNetX interface terminology. The standards are selected to be as close as possible to the way most of the data in any given domain are captured (eg, ICD for diagnoses) to minimize the need for data mapping. The mappings are made available to data providers upon request and the original codes are maintained but are not made available for querying on the platform. Standards are refreshed quarterly for medication terminology and yearly for other terminologies. All unmapped concepts (terms that have no match in the standards) are tracked, periodically re-evaluated, and compared with the refreshed standards. Table 1 provides an overview of the salient standards and associated mapping activities with the core clinical concepts captured in TriNetX.
Table 1.
An overview of the salient standards used in TriNetX, the associated mapping activities, and challenges introduced by the global heterogeneity
| Data type | Source vocabulary | Target terminology | Method |
|---|---|---|---|
| Demographics | EHR and ADT | HL7 v3 vocabulary for sex, race, ethnicity, vital status; ISO 639 for language | Manual mapping |
| Encounters | EHR and ADT, or derived by TriNetX | HL7 v3 vocabulary for visit type (eg, inpatient, outpatient, ER) | Manual mapping |
| Diagnoses |
|
ICD-10-CM |
|
| Procedures |
|
|
|
| Medications and Vaccinations |
|
RxNorm, OMOP extension of RxNorm, CVX Group codes | Semi-automated methods involving the use of external sources such as RxNorm “ApproximateTerm” API are utilized. For national catalogues of medications, TriNetX maps medications to RxNorm Ingredients + Route + Brand + Strength. |
| Lab results, clinical findings, and vital signs | Local lab coding or LOINC | LOINC |
|
| Genomics | Structured data from: molecular diagnostic labs (XML, JSON, CSV files), annotated VCF files, cancer registry data from NAACCR records | HGNC (gene symbols), HGVS (SNVs), ISCN (SVs, cytogenomic), Genomic Coordinates, rsID, LOINC (eg, IHC, MSI) |
|
| Oncology |
|
ICD-O | NAACCR-based data sources (United States) are almost always linked to ICD-O, but other regions (eg, EMEA or Australia) frequently do not provide ICD-O data. However, when oncology data are provided using ICD-10-CM codes, additional mappings from ICD-10-CM to ICD-O topographies are applied. Additionally, when morphologies are not provided, some ICD-10-CM codes provide morphology information, enabling the derivation of ICD-O morphologies. |
| Cross-domain mappings | selected HCPCS, SNOMED, and ICD-10-PCS codes | RxNorm | Data types are not homogeneous across regions, and some medications are frequently reported within procedures data sources (eg, CPT or OPS), so cross-domain mappings are also required to maximize the data coverage of TriNetX at a global scale. |
ADT: Admission Discharge Transfer; AEMPS: Agencia Española de Medicamentos y Productos Sanitarios; ATC: Anatomical Therapeutic Chemical; CPT: Current Procedural Terminology; CSV: Comma Separated Variable; DM + D: Dictionary of Medicines and Devices; EAN: European Article Numbering; HCPCS: Healthcare Common Procedure Coding System; HGNC: HUGO Gene Nomenclature Committee; HGVS: Human Genome Variation Society; HL-7: Health Level Seven; ICD-9: International Classification of Diseases, Ninth Revision; ICD-10-CM: International Classification of Diseases, Tenth Revision, Clinical Modification; ICD-10-GM: International Classification of Diseases, Tenth Revision, German Modification; ICD-10-PCS: International Classification of Diseases, Tenth Revision, Procedure Coding System; ICD-O-3: International Classification of Diseases for Oncology, third edition; IHC: Immunohistochemistry; ISCN: International System for Human Cytogenomic Nomenclature; JSON: JavaScript Object Notation; LOINC: Logical Observation Identifiers Names and Codes; MSI: Microsatellite instability; NAACCR: North American Association of Central Cancer Registries; NLP: natural language processing; NDC: National Drug Code; OPCS-4: OPCS Classification of Surgical Operations and Procedures (4th revision); OPS: Operationen- und Prozedurenschlüssel; rsID: Reference SNP cluster ID; SNOMED: Systematized Nomenclature of Medicine; SNV: single-nucleotide variants; SV: structural variant; VCF: Variant Call Format; XML: Extensible Markup Language.
The following examples illustrate some specific challenges with the semantic harmonization of data. For instance, TriNetX uses RxNorm as the standard for representing medication data. However, RxNorm only covers drugs on the US market. For ex-US coverage, the medication terminology was extended with OMOP’s RxNorm Extensions.16 Procedures are another challenging data domain because, unlike diagnosis, there is no globally adopted standard to represent procedures, and many countries use their national standards. For the US data, TriNetX uses ICD-10-PCS (as well as HCPCS and CPT) terminologies. Our typical approach would be to map local terminologies to one of these standards; however, mapping to ICD-10-PCS is particularly challenging.12 Instead, SNOMED Procedures are used as an additional target standard in TriNetX Terminology to accommodate the countries that do not utilize ICD-10-PCS.
Challenge: data integration and linking
Integrating data from a single institution, even if it originates from different sources (EHR, unstructured text of documents, cancer registry, other departmental systems), is predicated on having the same patient identifier across all the sources. The next frontier is to link patient records across de-identified HCO datasets. This is possible using privacy-preserving record linking. Cryptographic tokens are generated and managed by a third party and represent a distinct person without revealing their identity. Using such tokens, multiple de-identified datasets and data types can be brought together to form a composite network with capabilities exceeding the original sources.
TriNetX has taken advantage of privacy-preserving record linkage to build a new Linked Network among HCOs who opt-in to the linking process. In the Linked Network, clinical data drawn primarily from an EHR has been linked to, and therefore enriched with, closed claims data. In addition, the hospital mortality data has been augmented with data coming from government and private sources. In this Linked Network, EHR data provides details about individual episodes of care, while the claims data contributes the longitudinal view across many providers.
It is difficult to understate the utility of bringing together disparate data sources to arrive at an overall picture of health and illness. This linking technology presents opportunities to incorporate data representing variables such as social determinants of health, lifestyle factors, and quality-of-life metrics into more comprehensive disease analyses. However, new data types must be added cautiously, thoughtfully, and in compliance with all regulatory guidelines, prioritizing patient privacy and minimizing the risk of re-identification.
Challenge: platform utilization
Once the data are ingested and harmonized, investigators at TriNetX HCOs can use TriNetX to access their institution’s de-identified patient data to design institutional investigator-initiated trials and assess their feasibility, as well as collaborative trials with other HCOs, cooperative groups, government agencies, or industry. Researchers also use TriNetX to design and conduct non-trial-related clinical and biomedical research.
Industry participants obtain the same clinical trial design tools as HCOs and can run queries against the entire network to capture aggregate patient counts at each HCO of patients who match the trial’s eligibility criteria. Industry participants cannot run queries against individual HCOs; only HCOs can run individual queries against their own data. The performance of the TriNetX network allows industry participants to gain rapid feedback about the impact of individual inclusion and exclusion criteria on the size of potential cohorts at the trial design phase, enabling real-time iteration and the optimization of the protocol before its release to HCOs. Knowing potential aggregate patient populations at the trial design phase can result in better recruitment expectations (both patient- and time-wise) and potentially less need for time-consuming and costly trial amendments. In this way, pharmaceutical companies and contract research organizations can address some of the issues cited in a study by the Tufts Center for the Study of Drug Development that prevent the efficient execution of clinical trial feasibility and site selection and often waste the time of HCOs in the process, with protocols with little to no chance of success.17
This model for networked CDRs relies on both of the member organizational types satisfying their objectives. HCOs must find value in the enterprise analytics capabilities, the ability to more easily collaborate with other HCOs, and the increased visibility to sponsored trial opportunities. In addition, industry members must realize increased trial efficiencies and accelerated clinical research.
Challenge: networks as means of organizing and segmenting the data
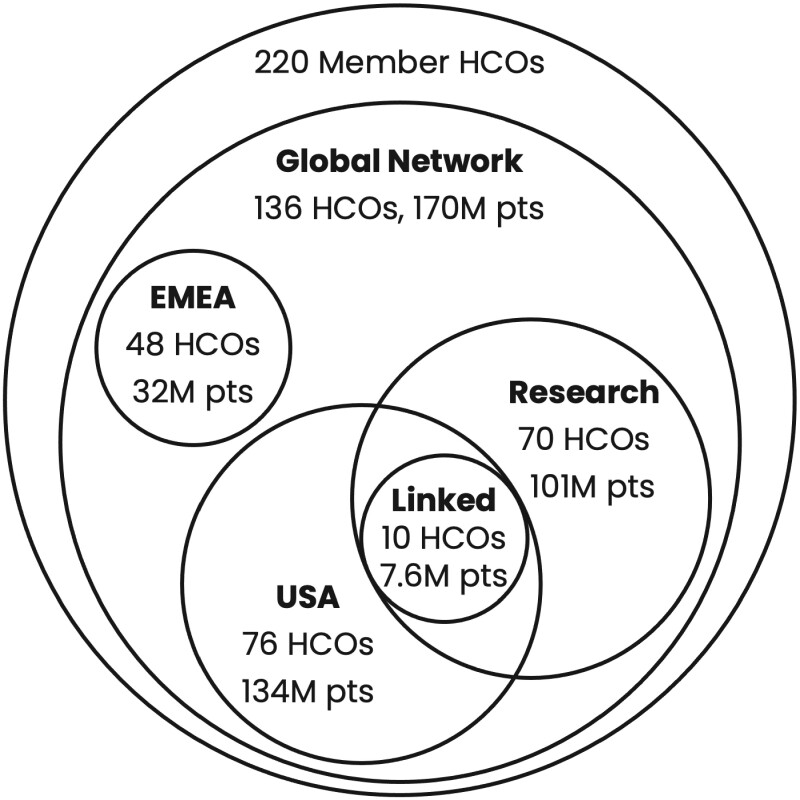
The TriNetX ecosystem currently has over 220 member HCOs in various stages of implementation, of whom 136 have data currently accessible on the platform for clinical trial optimization, site selection, and research via the Global Network (Figure 1). These 136 HCOs are then segmented into different specialty networks that vary by regional coverage, data sources, and capabilities. For example, the EMEA Network, which includes data from 48 HCOs located in Europe and the Middle East, provides query capabilities for clinical trial optimization but does not allow for quantitative analytics or data downloads. By comparison, the Research Network, which includes data from 70 HCOs located predominantly in the United States but inclusive of some HCOs from the Latin America and Asia Pacific regions, allows for on-platform advanced cohort analyses with exact counts (for variables with results greater than 10), as well as data downloads for further off-platform analyses. Figure 1 only shows a small portion of the specialty networks available to users depending on institutional contractual agreements.
Figure 1.
HCO members of TriNetX and a sample of various networks they comprise. For each network, the number of HCOs and patients is shown. Circle sizes are approximate. EMEA: Europe, Middle East and Africa.
Through TriNetX, HCO can more easily achieve the objective of sharing data with another TriNetX HCO member through the creation of a highly performant, highly available, federated “virtual data mart.” As other data network projects show, harmonizing data models and managing queries between organizations can be a daunting task, but one which is made easier by technical assistance provided by TriNetX. But once an HCO is onboarded—its data model harmonized with the TriNetX model—data can easily be shared among network members after the execution of Data Use Agreements between HCOs choosing to form a Collaborative Network.
RESULTS
HCOs that have evaluated the details around data sharing and privacy protection, data harmonization, and data integration have continued to opt into the TriNetX network, resulting in a variety of benefits to their research enterprises.
Network growth
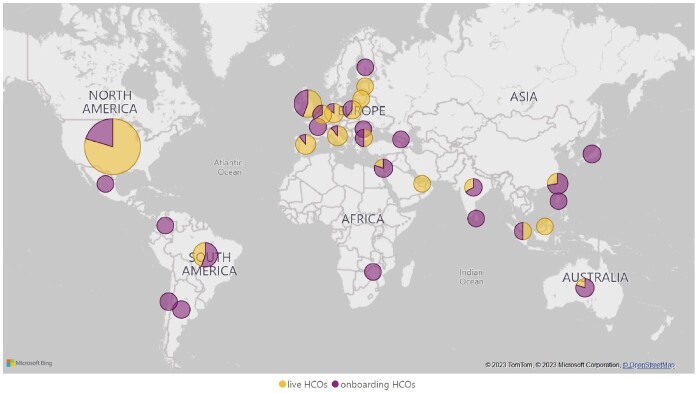
One important characteristic of any network is its dimensions. The first TriNetX HCO agreements were signed in late 2014, and today there are approximately 220 organizations across 30 countries in total under contract and in different stages of onboarding (Figure 2). Eight years later, TriNetX is a global company with a presence in both the Americas, Europe, the Middle East, Africa, and Asia-Pacific regions. Figure 2 represents the current geographical presence. Life science industry membership, composed of pharmaceutical companies and clinical research organizations, now has 40 participants, including most of the leading organizations in each market segment.
Figure 2.
Map of HCOs currently under contract. Microsoft Bing Maps screenshot reprinted with permission from Microsoft Corporation.
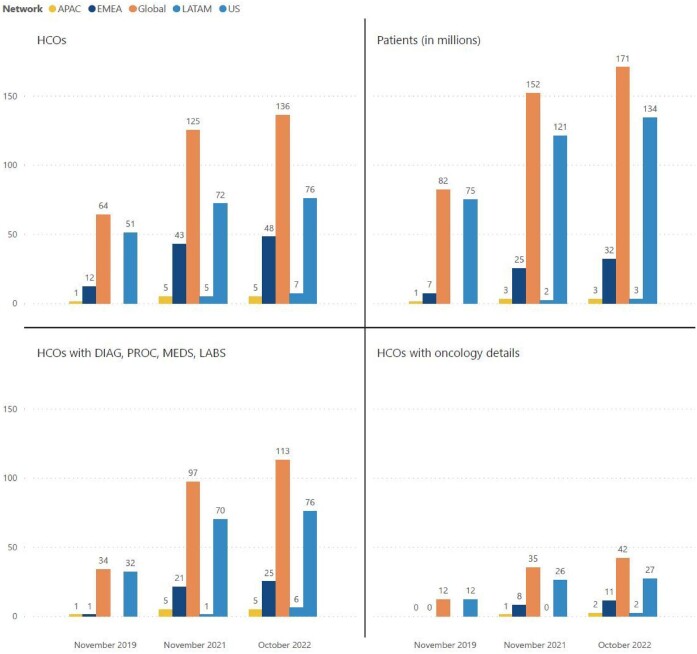
The volume of addressable data grows as new sites join the network (see Figure 3); there are currently roughly 56 billion facts representing various observations about patients drawn primarily, but not exclusively, from electronic health record systems. Growing the breadth of the data asset is achieved by expanding the data domains covered in the platform. We began with easily accessible structured data such as demographics, encounters, diagnoses, procedures, meds, and labs.18 From there, the data were expanded with small additions like vital signs to major ones like genomics (genes and specific variants plus additional attributes), and additional data domains continue to be evaluated for inclusion. Growing the depth of the data is achieved by targeting a more detailed and nuanced representation of the data. For example, medication details such as brand, strength, and route of administration have been added to the ingredient-level representation of medications. We leveraged additional sources (eg, cancer registries), methods (eg, mining unstructured data from notes to generate structured facts), curation (eg, simple calculation and more complex algorithmic derivation of facts based on existing data such as chemotherapy lines of treatment), and technologies (eg, privacy-preserving patient record linkage) to improve the breadth and fidelity of the data. Focusing on both breadth and depth of the data leads to an ever-more representative picture of the patient and their longitudinal history.
Figure 3.
Growth of data on the TriNetX platform between 2019 and 2022. Counts for 2020 are omitted.
Network scalability is fundamental to positive user experience. In other words, the high level of performance and availability must be maintained as more organizations join and more nodes are added. On average, over the last 90 days, response to basic queries returning the number of patients with an arbitrary set of criteria across over 100 million patients in a federated environment across a global roster of HCOs took under 0.5 s. This performance is made possible by the network architecture, hardware characteristics, and the choice of database technologies. The average overall response time on the platform is under 1 s for basic queries such as patient counts, and approximately 20 s for all queries (including advanced analytics). By instilling an expectation that an answer is only seconds away, this level of performance can enable rapid iteration on query criteria. TriNetX also aims for high availability of the platform, with uptime maintained in excess of 99.5%.
There has also been growth in the utilization of the network. The volume of queries by researchers from HCOs and industry averages ∼45 000 per month. These are executed by approximately 1300 active users per month. Participation in TriNetX Research—a more recent offering—opens up access to a pool of shared de-identified data, with no patient re-identification capabilities and no HCO attribution, that can be interrogated with TriNetX’s advanced analytics suite. Participation in TriNetX Research allows researchers to access a dataset of EHR-derived clinical data that is magnitudes larger than what is typically available at a single institution. Today the Research network stands at over 101M patients across 70 contributing HCOs.
Clinical trials opportunities
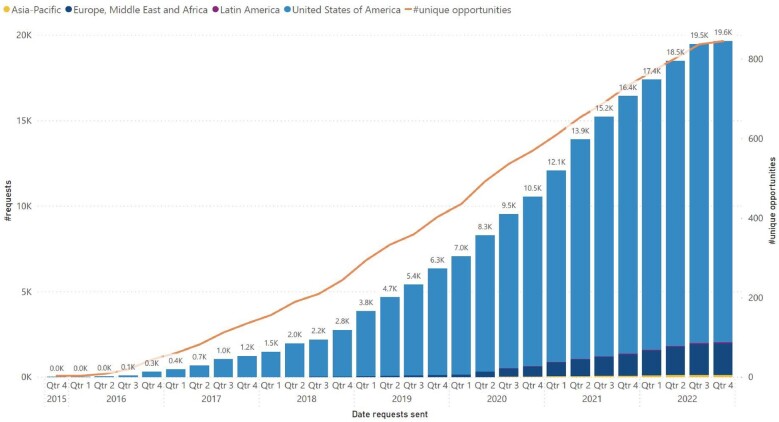
The initial use case for TriNetX was to accelerate clinical research by providing tools for optimizing clinical trial design and site selection.8,19 Since the network began commercial operations in 2016, the success of the network in facilitating clinical trial collaborations can be seen in the increase in requests from pharmaceutical companies and contract research organizations to member HCOs for industry-sponsored clinical trial participation (see Figure 4).
Figure 4.
Cumulative requests to HCOs from pharmaceutical companies and contract research organizations for participation in clinical trials. The bar graph shows the cumulative number of requests, broken down by geographic regions, and the orange line—unique opportunities available to HCOs.
Scientific publications
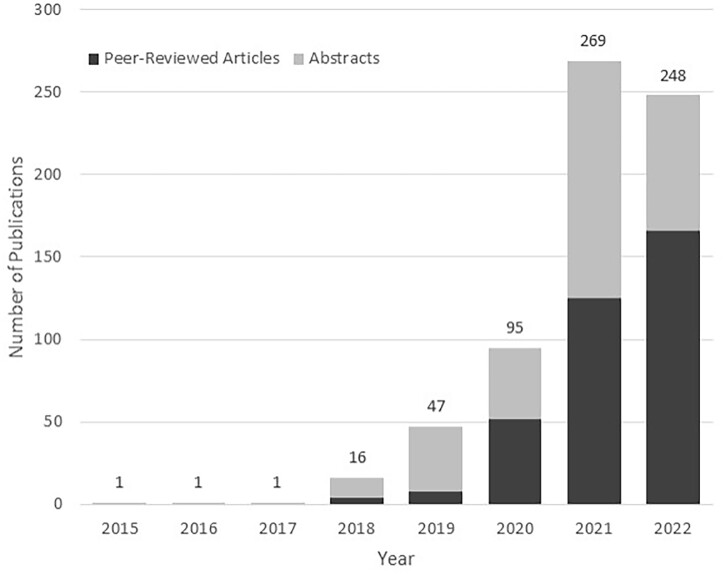
While clinical trial design optimization and site selection remain a core functionality that is further strengthened by continued regional diversification of the network, the utility of global data aggregation and harmonization extends far beyond this initial use case. Peer-reviewed publications and abstracts utilizing data from the TriNetX network have grown since 2015, more than doubling each year from 2018 to 2021 (see Figure 5). Nowhere has this application of the TriNetX network data been more evident than during the recent COVID-19 pandemic. By creating a pressing need for global real-time RWD, the pandemic accelerated interest in and utilization of global CDRs and intuitive analytic platforms. Researchers have used TriNetX to explore a broad range of COVID-19-related topics, including understanding the characteristics of and outcomes among COVID-19 patients,20–22 assessing vaccine efficacy and breakthrough infection incidence,23,24 examining outcomes among patient subgroups,25–32 exploring the incidence and outcomes of cardiovascular events,33–36 and identifying a connection between COVID-19 and neuropsychiatric conditions.37–39 In addition to clinical outcomes, researchers have also used TriNetX to explore the pandemic’s impact on healthcare access and healthcare-seeking behavior.9,40–43 While the use of TriNetX for research purposes is relatively new, the rapid adoption during the pandemic demonstrates the value of TriNetX to the research community.
Figure 5.
Number of conference abstracts and peer-reviewed journal articles from studies utilizing TriNetX. The counts for 2022 are partial and cover publications from January to September.
DISCUSSION
There have been several data-sharing network projects that have become unsustainable in the past, as illustrated by the caBIG and other examples in the United States, or projects like EHR4CR and INTEGRATE in Europe.44,45 The long-term financial stability of such initiatives is of paramount importance for an HCO as it represents a significant investment in time, resources, and intellectual capital. HCOs are faced with the challenge of finding resources to do the work of creating a common data model, platform, and network infrastructure. Past solutions have predominantly been government-funded initiatives. One notable exception is the InSite platform in Europe—a commercial implementation of the EHR4CR project—whose objectives included determining how to ensure the CDR solution was sustainable. InSite was acquired by TriNetX in 2018.
From its inception, a major objective of TriNetX was to create a unique, alternative sustainability model that reduced the barriers to cooperation between industry and academic/research institutions. With a conservative security and governance model as the core architectural principle, industry participants would pay for access to the network and its benefits, enabling them to utilize the technology for trial design, feasibility, site selection, and optimization of clinical trial operations, as well as for outcomes, health economics, and epidemiological research. As a result, grant funding is not required to expand and improve the capabilities of the network. HCOs get to use the same platform and its analytics capabilities to work with their own data along with data on Collaborative Networks and TriNetX networks (such as TriNetX Research) to which they contribute. Additionally, the HCOs receive increased opportunities to participate in industry-sponsored clinical trials and to use the platform to support their endeavors to publish their research in scientific journals.
Building a global network
To accomplish our mission, the network of clinical data for research must be as representative of the various patient populations as possible. Therefore, the data must be global, and the network must support the global nature of clinical and basic research. Working across geographic boundaries presents a number of challenges, including working with multiple languages, regional terminologies, different legal frameworks, and heterogeneous data sources. With staff in the United States, Europe, Asia-Pacific, and Latin America, and HCOs in 30 countries under contract, the goal of building a global network has been realized within the TriNetX platform.
Making clinical data more accessible
EHR data have only recently become more accessible, and researchers are still gaining familiarity with its strengths, weaknesses, and capabilities. For those familiar with the potential uses of EHR data in research applications, the need for advanced data analytics skills to interrogate the data often remains a barrier to discovery. The TriNetX platform is designed to be usable by clinical researchers who are not necessarily familiar with complex data analytics tools. The integration of data and analytics is one of the central tenets of the TriNetX approach—users should be able to formulate their questions and quickly obtain answers in an intuitive environment. To facilitate this goal, TriNetX continuously curates its interface terminology (such as the custom rollup of lab tests for SARS-CoV-2 PCR and serology testing), maintains a Google-like semantic search capability to ensure that users can easily locate their preferred search terms, implements push-button advanced analytics such as propensity score matching and Kaplan–Meyer survival curves, and just introduced a trusted research environment where users can program their own analytics.
CONCLUSIONS
CDRs are essential tools in the armamentarium of informaticians and data scientists. They underpin cohort identification tools—often the first class of data analytics tools encountered by researchers—that not only give them crucial insight into patient populations but inform them about the availability of clinical data for research. Today, there are a wide variety of available common data models, corresponding CDRs, and networks that can connect them to each other. For instance, in the United States, there are i2b2/SHRINE,1 the ACT network,46 OMOP,2 PCORnet,3 and All of Us Research,47 while in Europe, there is eHealth,48 and EHDEN.49 The COVID-19 pandemic led to the rapid development of new ones such as 4CE and N3C.50,51 TriNetX has become a viable CDR as a variety of developments demonstrate—the number of queries being run, the number of papers being published, and the participation in important initiatives such as N3C.
Importantly, this model depends on gaining and holding the trust of participating HCOs, ensuring them that their data are protected technically, and that the use of their data is for research only, and conservatively governed based on well-understood contractual obligations agreed upon by all parties that choose to become members of the network. With that trust in mind, HCOs gain access to a variety of advantages for their organizations including:
An increase in the number of sponsored trial opportunities they have historically received from pharmaceutical companies and CROs.
Access to resources that will shoulder most of the burden of creating a data asset consisting of both phenotypic and genotypic data, along with data from unstructured data sources and third-party data assets (mortality, claims), for the benefit of researchers across the HCO enterprise.
Quality data through the review of the conformance, completeness, and plausibility52 of each HCO’s data against the network as a whole.
A cohort identification, analysis, feasibility tool for their researchers to use against their own HCO data.
The elimination of most obstacles historically encountered in the formation of HCO-only collaborative networks and the participation in external networks such as N3C.
Access to a large network of de-identified patient data for use in outcomes and epidemiological research use-cases in combination with a suite of on-line analytics tools.
Publication and grant application support via the data and analytics assets available in the platform.
An analytics and data environment that provides access to medical school students, residents, and fellows, enabling them to develop their research skills as part of their professional development.
Industry participants join TriNetX for 2 primary reasons:
To use a technology platform that will help them address obstacles they have historically faced in the clinical trial protocol design, feasibility, and site selection process
To monitor the safety and efficacy of their products after they have been introduced to the market
TriNetX has endeavored to create a global data and analytics platform that is characterized by technical and contractual patient privacy protections, a business model that requires no fees from HCOs, and a way to deliver clinical trial opportunities from industry participants to HCOs that have joined the network. The ongoing growth of this platform of electronic health record data continues to be guided by the original goals of liberating clinical data and enabling biomedical and clinical research in a trusted ecosystem.
Contributor Information
Matvey B Palchuk, TriNetX, LLC, Cambridge, Massachusetts, USA; Harvard Medical School, Boston, Massachusetts, USA.
Jack W London, Thomas Jefferson University, Philadelphia, Pennsylvania, USA.
David Perez-Rey, Biomedical Informatics Group, Artificial Intelligence Department, Universidad Politécnica de Madrid, Madrid, Spain.
Zuzanna J Drebert, TriNetX, LLC, Cambridge, Massachusetts, USA.
Jessamine P Winer-Jones, TriNetX, LLC, Cambridge, Massachusetts, USA.
Courtney N Thompson, TriNetX, LLC, Cambridge, Massachusetts, USA.
John Esposito, TriNetX, LLC, Cambridge, Massachusetts, USA.
Brecht Claerhout, TriNetX, LLC, Cambridge, Massachusetts, USA.
FUNDING
This work was funded by TriNetX, LLC.
AUTHOR CONTRIBUTIONS
MBP: conception and analysis, drafting and revising, final approval, accountable for the work. JWL: conception and analysis, drafting and revising, final approval, accountable for the work. D.P-R: conception and analysis, drafting and revising, final approval, accountable for the work. ZJD: conception and analysis, drafting and revising, accountable for the work. JPW-J: conception and analysis, drafting and revising, accountable for the work. CNT: drafting and revising, accountable for the work. JE: conception and analysis, drafting and revising, final approval, accountable for the work. BC: conception and analysis, final approval, accountable for the work.
CONFLICT OF INTEREST STATEMENT
MBP, ZJD, JPW-J, CNT, JE, and BC are employees of TriNetX, LLC. JWL and DP-R have a Consulting Role with TriNetX, LLC.
DATA AVAILABILITY
There are no new data associated with this article.
REFERENCES
- 1. Murphy SN, Mendis M, Hackett K, et al. Architecture of the open-source clinical research chart from Informatics for Integrating Biology and the Bedside. AMIA Annu Symp Proc 2007; 2007: 548–52. [PMC free article] [PubMed] [Google Scholar]
- 2. Overhage JM, Ryan PB, Reich CG, et al. Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc 2012; 19 (1): 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Califf RM. The Patient-Centered Outcomes Research Network: a national infrastructure for comparative effectiveness research. N C Med J 2014; 75 (3): 204–10. [DOI] [PubMed] [Google Scholar]
- 4. Rahm AK, Ladd I, Burnett-Hartman AN, et al. The Healthcare Systems Research Network (HCSRN) as an environment for dissemination and implementation research: a case study of developing a multi-site research study in precision medicine. EGEMS (Wash DC) 2019; 7 (1): 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jacobson RS, Becich MJ, Bollag RJ, et al. A federated network for translational cancer research using clinical data and biospecimens. Cancer Res 2015; 75 (24): 5194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Magid DJ, Gurwitz JH, Rumsfeld JS, Go AS.. Creating a research data network for cardiovascular disease: the CVRN. Expert Rev Cardiovasc Ther 2008; 6 (8): 1043–5. [DOI] [PubMed] [Google Scholar]
- 7. McNeil MM, Gee J, Weintraub ES, et al. The Vaccine Safety Datalink: successes and challenges monitoring vaccine safety. Vaccine 2014; 32 (42): 5390–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Topaloglu U, Palchuk MB.. Using a federated network of real-world data to optimize clinical trials operations. JCO Clin Cancer Inform 2018; 2: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. London JW, Fazio-Eynullayeva E, Palchuk MB, et al. Effects of the COVID-19 pandemic on cancer-related patient encounters. JCO Clin Cancer Inform 2020; 4: 657–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ama-assn.org. 2023. HIPAA Privacy Standards. http://www.ama-assn.org/ama/pub/physician-resources/solutions-managing-your-practice/coding-billing-insurance/hipaahealth-insurance-portability-accountability-act/hipaa-privacy-standards.page. Accessed December 9, 2021.
- 11. National Library of Medicine. SNOMED CT to ICD-10-CM Map. 2021. https://www.nlm.nih.gov/research/umls/mapping_projects/snomedct_to_icd10cm.html. Accessed December 9, 2021.
- 12. Fung KW, Xu J, Ameye F, et al. Map-assisted generation of procedure and intervention encoding (Magpie): an innovative approach for ICD-10-PCS coding. Stud Health Technol Inform 2019; 264: 428–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schulz S, Steffel J, Polster P, et al. Aligning an administrative procedure coding system with SNOMED CT. CEUR Workshop Proc 2019; 2518: ODLS8. http://ceur-ws.org/Vol-2518/paper-ODLS8.pdf. Accessed June 3, 2021. [Google Scholar]
- 14. Millan-Fernandez-Montes A, Perez-Rey D, Hernandez-Ibarburu G, et al. Mapping clinical procedures to the ICD-10-PCS: the German operation and procedure classification system use case. J Biomed Inform 2020; 109: 103519. [DOI] [PubMed] [Google Scholar]
- 15. Regenstrief Institute. RELMA. https://loinc.org/relma/. Accessed December 9, 2021.
- 16. Dymshyts D, Ostropolets A, Bolshutkin V, et al. International RxNorm Extension to support the expansion of the OHDSI research network beyond the US. 2016. https://www.ohdsi.org/web/wiki/lib/exe/fetch.php?media=resources:ohdsi-submission-rxe_2016.docx. Accessed November 29, 2021.
- 17. Getz KA, Stergiopoulos S, Short M, et al. The impact of protocol amendments on clinical trial performance and cost. Ther Innov Regul Sci 2016; 50 (4): 436–41. [DOI] [PubMed] [Google Scholar]
- 18. Murphy SN, Morgan MM, Barnett GO, et al. Optimizing healthcare research data warehouse design through past COSTAR query analysis. Proc AMIA Symp 1999: 892–6. [PMC free article] [PubMed] [Google Scholar]
- 19. Stacey J, Mehta M.. Using EHR data extraction to streamline the clinical trial process. Clin Researcher 2017; April: 2–7. www.acrpnet.org/2017/04/01/using-ehr-data-extraction-streamline-clinical-trial-process/. accessed December 9, 2021 [Google Scholar]
- 20. Harrison SL, Fazio-Eynullayeva E, Lane DA, et al. Comorbidities associated with mortality in 31,461 adults with COVID-19 in the United States: a federated electronic medical record analysis. PLoS Med 2020; 17 (9): e1003321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Parcha V, Booker KS, Kalra R, et al. A retrospective cohort study of 12,306 pediatric COVID-19 patients in the United States. Sci Rep 2021; 11 (1): 10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taquet M, Dercon Q, Luciano S, et al. Incidence, co-occurrence, and evolution of long-COVID features: a 6-month retrospective cohort study of 273,618 survivors of COVID-19. PLoS Med 2021; 18 (9): e1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hadi YB, Thakkar S, Shah-Khan SM, et al. COVID-19 vaccination is safe and effective in patients with inflammatory bowel disease: analysis of a large multi-institutional research network in United States. Gastroenterology 2021; 161 (4): 1336–9.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang L, Wang Q, Davis PB, et al. Increased risk for COVID-19 breakthrough infection in fully vaccinated patients with substance use disorders in the United States between December 2020 and August 2021. World Psychiatry 2022; 21 (1): 124–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alkhouli M, Nanjundappa A, Annie F, et al. Sex differences in case fatality rate of COVID-19: insights from a multinational registry. Mayo Clin Proc 2020; 95 (8): 1613–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. D'Silva KM, Jorge A, Cohen A, et al. COVID-19 outcomes in patients with systemic autoimmune rheumatic diseases compared to the general population: a US multicenter, comparative cohort study. Arthritis Rheumatol 2021; 73 (6): 914–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hadi YB, Naqvi SFZ, Kupec JT, et al. Characteristics and outcomes of COVID-19 in patients with HIV: a multicentre research network study. Aids 2020; 34 (13): F3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jorge A, D'Silva KM, Cohen A, et al. Temporal trends in severe COVID-19 outcomes in patients with rheumatic disease: a cohort study. Lancet Rheumatol 2021; 3 (2): e131–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nyland JE, Raja-Khan NT, Bettermann K, et al. Diabetes, drug treatment and mortality in COVID-19: a multinational retrospective cohort study. Diabetes 2021; 70 (12): 2903–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang Q, Schultz JL, Aldridge GM, et al. COVID-19 case fatality and Alzheimer’s disease. J Alzheimers Dis 2021; 84 (4): 1447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Singh S, Khan A.. Clinical characteristics and outcomes of coronavirus disease 2019 among patients with preexisting liver disease in the United States: a multicenter research network study. Gastroenterology 2020; 159 (2): 768–71.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Turk MA, Landes SD, Formica MK, et al. Intellectual and developmental disability and COVID-19 case-fatality trends: TriNetX analysis. Disabil Health J 2020; 13 (3): 100942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Annie F, Bates MC, Nanjundappa A, et al. Prevalence and outcomes of acute ischemic stroke among patients≤ 50 years of age with laboratory confirmed COVID-19 infection. Am J Cardiol 2020; 130: 169–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chima M, Williams D, Thomas NJ, et al. COVID-19-associated pulmonary embolism in pediatric patients. Hosp Pediatr 2021; 11 (6): e90–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Harrison SL, Buckley BJR, Lane DA, et al. Associations between COVID-19 and 30-day thromboembolic events and mortality in people with dementia receiving antipsychotic medications. Pharmacol Res 2021; 167: 105534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Harrison SL, Fazio-Eynullayeva E, Lane DA, et al. Higher mortality of ischaemic stroke patients hospitalized with COVID-19 compared to historical controls. Cerebrovasc Dis 2021; 50 (3): 326–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nalleballe K, Reddy Onteddu S, Sharma R, et al. Spectrum of neuropsychiatric manifestations in COVID-19. Brain Behav Immun 2020; 88: 71–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Taquet M, Geddes JR, Husain M, et al. 6-month neurological and psychiatric outcomes in 236 379 survivors of COVID-19: a retrospective cohort study using electronic health records. Lancet Psychiatry 2021; 8 (5): 416–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Taquet M, Luciano S, Geddes JR, et al. Bidirectional associations between COVID-19 and psychiatric disorder: retrospective cohort studies of 62 354 COVID-19 cases in the USA. Lancet Psychiatry 2021; 8 (2): 130–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Khan A, Bilal M, Morrow V, et al. Impact of the coronavirus disease 2019 pandemic on gastrointestinal procedures and cancers in the United States: a multicenter research network study. Gastroenterology 2021; 160 (7): 2602–4.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Parcha V, Kalra R, Glenn AM, et al. Coronary artery bypass graft surgery outcomes in the United States: Impact of the coronavirus disease 2019 (COVID-19) pandemic. JTCVS Open 2021; 6: 132–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Onteddu SR, Nalleballe K, Sharma R, et al. Underutilization of health care for strokes during the COVID-19 outbreak. Int J Stroke 2020; 15 (5): NP9–NP10. [DOI] [PubMed] [Google Scholar]
- 43. Sheng S, Wang X, Gil Tommee C, et al. Continued underutilization of stroke care during the COVID-19 pandemic. Brain Behav Immun Health 2021; 15: 100274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. De Moor G, Sundgren M, Kalra D, et al. Using electronic health records for clinical research: the case of the EHR4CR project. J Biomed Inform 2015; 53: 162–73. [DOI] [PubMed] [Google Scholar]
- 45. Kondylakis H, Claerhout B, Keyur M, et al. The INTEGRATE project: delivering solutions for efficient multi-centric clinical research and trials. J Biomed Inform 2016; 62: 32–47. [DOI] [PubMed] [Google Scholar]
- 46. Visweswaran S, Becich MJ, D'Itri VS, et al. Accrual to Clinical Trials (ACT): a Clinical and Translational Science Award Consortium Network. JAMIA Open 2018; 1 (2): 147–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Denny JC, Rutter JL, Goldstein DB. et al. ; All of Us Research Program Investigators. The “All of Us” Research Program. N Engl J Med 2019; 381: 668–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. European Commission. Electronic cross-border health services. https://ec.europa.eu/health/ehealth/electronic_crossborder_healthservices_en. Accessed November 29, 2021.
- 49. European Health Data & Evidence Network. EHDEN. https://www.ehden.eu/. Accessed November 29, 2021.
- 50. Brat GA, Weber GM, Gehlenborg N, et al. International electronic health record-derived COVID-19 clinical course profiles: the 4CE consortium. NPJ Digit Med 2020; 3: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Haendel MA, Chute CG, Bennett TD, et al. ; N3C Consortium. The National COVID Cohort Collaborative (N3C): rationale, design, infrastructure, and deployment. J Am Med Inform Assoc 2021; 28 (3): 427–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kahn MG, Callahan TJ, Barnard J, et al. A harmonized data quality assessment terminology and framework for the secondary use of electronic health record data. eGEMs 2016; 4 (1): 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There are no new data associated with this article.