Abstract
Background:
We compared plasma metabolites of amino acid (AA) oxidation and the tricarboxylic acid (TCA) cycle in youth with and without type 1 diabetes mellitus (T1DM) and related the metabolites to glomerular filtration rate (GFR), renal plasma flow (RPF), and albuminuria. Metabolites associated with impaired kidney function may warrant future study as potential biomarkers or even future interventions to improve kidney bioenergetics.
Methods:
Metabolomic profiling of fasting plasma samples using a targeted panel of 644 metabolites and an untargeted panel of 19,777 metabolites was performed in 50 youth with T1DM≤10 years and 20 controls. GFR and RPF were ascertained by iohexol and p-aminohippurate clearance, and albuminuria calculated as urine albumin to creatinine ratio. Sparse partial least squares discriminant analysis and moderated t tests were used to identify metabolites associated with GFR and RPF.
Results:
Adolescents with and without T1DM were similar in age (16.1±3.0 vs. 16.1±2.9 years) and BMI (23.4±5.1 vs. 22.7±3.7 kg/m2), but those with T1DM had higher GFR (189±40 vs. 136±22 ml/min) and RPF (820±125 vs. 615±65 ml/min). Metabolites of AA oxidation and the TCA cycle were significantly lower in adolescents with T1DM vs. controls, and the measured metabolites were able to discriminate diabetes status with an AUC of 0.82 (95% CI: 0.71, 0.93) and error rate of 0.21. Lower glycine (r:−0.33, q=0.01) histidine (r:−0.45, q<0.001), methionine (r:−0.29, q=0.02), phenylalanine (r:−0.29, q=0.01), serine (r:−0.42, q<0.001), threonine (r:−0.28, q=0.02), citrate (r:−0.35, q=0.003), fumarate (r:−0.24, q=0.04), and malate (r:−0.29, q=0.02) correlated with higher GFR. Lower glycine (r:−0.28, q=0.04), phenylalanine (r:−0.3, q=0.03), fumarate (r:−0.29, q=0.04), and malate (r:−0.5, q<0.001) correlated with higher RPF. Lower histidine (r:−0.28, q=0.02) was correlated with higher mean ACR.
Conclusion:
In conclusion, adolescents with relatively short T1DM duration exhibited lower plasma levels of carboxylic acids that associated with hyperfiltration and hyperperfusion.
Keywords: type 1 diabetes, plasma metabolomics, elevated albumin excretion, kidney oxygenation, hyperfiltration, blood oxygen level dependent (BOLD) magnetic resonance imaging (MRI)
Graphical Abstract
Introduction
Diabetic kidney disease (DKD) is one of the leading causes of morbidity and mortality and accelerates early cardiovascular disease (CVD) in people with type 1 diabetes mellitus (T1DM) [1, 2]. Emerging data suggest that in addition to β-cell injury and hyperglycemia, the pathologic features of DKD in T1DM also include insulin resistance and mitochondrial dysfunction [3].
The kidneys are one of the most energy-demanding organs and are second only to the heart with respect to oxygen consumption per tissue mass [4, 5]. The high oxygen demand is necessary to maintain adequate adenosine triphosphate (ATP) production, especially in the proximal tubules [4]. The principal determinants of kidney ATP consumption are tubular sodium reabsorption and glomerular filtration rate (GFR)[4, 6]. To maintain their energy-intensive active transport mechanisms, the proximal tubules contain more mitochondria than any other structure of the kidney [7], and mitochondrial dysfunction has been implicated in playing a role in the early stages of DKD [8]. Indeed, emerging animal data suggest that the kidneys are unable to sufficiently compensate for the increased rate of ATP consumption in diabetes due to the effects of mitochondrial dysfunction on substrate utilization, which can predispose to kidney hypoxia [9, 10]. However, little is known about mitochondrial dysfunction in T1DM of relatively short duration and how it might relate to kidney function.
The objectives of this study were to identify plasma signatures of mitochondrial dysfunction in adolescents with recently diagnosed T1DM (≤10 years duration) and correlate these metabolites with measures of GFR, renal plasma flow (RPF), and albuminuria. We hypothesized that adolescents with T1DM would exhibit lower plasma metabolites of amino acid oxidation compared with healthy controls, and that reduced concentrations of these metabolites would associate with intraglomerular hemodynamic dysfunction. Additionally, we used untargeted metabolomics to identify novel potential biomarkers of early kidney dysfunction in an unbiased manner, for future study.
Methods:
Study Design and Participants
Fifty adolescents with T1DM (12-21 years of age, diabetes duration 1-10 years and HbA1c <11%) from the Copeptin in Adolescent Participants with Type 1 Diabetes and Early Renal Hemodynamic Function study (CASPER, NCT03618420) and 20 healthy adolescent controls (12-21 years of age) from the Renal Hemodynamics, Energetics and Insulin Resistance in Youth Onset Type 2 Diabetes Study (Renal-HEIR, NCT03584217) were included in this analysis. Participants with T1DM were recruited from the pediatric clinics at the Barbara Davis Center for Diabetes at the Anschutz Medical Campus in Aurora, Colorado. T1DM was defined by American Diabetes Association criteria plus the presence of glutamic acid decarboxylase, islet cell, zinc transporter 8 and/or insulin autoantibodies at diagnosis, and the need for exogenous insulin. Exclusion criteria are detailed in Supplementary Table 1. All participants were examined at our clinical research center, where tests were performed in the morning after a 12-h fast, preceded by 3 days of restricted physical activity and a fixed-macronutrient, sodium, and protein replete, weight-maintenance diet. Participants remained fasting until all study procedures were completed. The CASPER and Renal-HEIR cohorts were intentionally harmonized (see below) and were both approved by the Colorado Multiple Institutional Review Board (COMIRB). Participants or parents provided written informed assent and/or consent as appropriate for age. Puberty was assessed by a pediatric endocrinologist using the standards of Tanner and Marshall for breast development in girls (inspection and palpation) and genital development in boys. Vital signs including height, weight, systolic and diastolic blood pressure were measured according to standard procedures.
Metabolomics
Fasting plasma samples were collected to measure metabolites of mitochondrial function by Q Exactive UHPLC-MS Analysis at the University of Colorado School of Medicine Metabolomics Core. Samples were stored between 1 and 12 months after collection at −80 C. Plasma samples (20 µl) were extracted using 480 ul of ice cold methanol:acetonitrile:water (5:3:2) supplemented with the following stable isotope labeled standards: amino acids (2.5 uM, Cambridge Isotope Laboratories cat no MSK-A2-1.2), D4-citrate (1 uM), 13C5-α-ketoglutarate (1 uM), 13C4-succinate (1 uM), 13C2-fumarate (1 uM). All standards were purchased from Cambridge Isotope Laboratories. Extractions were performed for 30 min at 4 C as described [11]. Extracts were analyzed on a Thermo Vanquish ultra-high pressure liquid chromatograph (UHPLC) coupled online to a Thermo Q Exactive mass spectrometer using a 5 min C18 gradient method in positive and negative modes exactly as described previously [12]. An untargeted data acquisition was utilized alongside a targeted data analysis (i.e., using an established metabolite panel) to assess mitochondrial energy metabolism. Peak integration and metabolite assignment were performed using Maven (Princeton University) against the KEGG database, confirmed against chemical formula determination from isotopic patterns and accurate mass, and validated against experimental retention times for 644 standard compounds for the targeted panel and 19,777 for the untargeted panel (Sigma Aldrich; MLSMS, IROATech, Bolton, MA, USA) [13]. Instrument stability was assessed using replicate injections of a quality control mix injected every 10 runs. Peak areas were drift-corrected using the MetaboDrift Excel macro. We performed absolute quantification of metabolites already known to be altered in later stages of DKD, including metabolites of the tricarboxylic acid (TCA) cycle and amino acid (AA) metabolism [8].
GFR and RPF by iohexol and p-aminohippurate (PAH) clearance techniques, UACR and blood pressure
An intravenous (IV) line was placed, and participants were asked to empty their bladders. Spot plasma and urine samples were collected prior to iohexol and PAH infusion, as previously described [14, 15]. We report absolute GFR (mL/min) and RPF (mL/min) in the main analyses because the practice of indexing GFR and RPF for body surface area (BSA) underestimates hyperfiltration and hyperperfusion [16], and BSA calculations introduce noise into the clearance measurements. In adolescents with T1DM, GFR and RPF were measured during mild hyperglycemia (goal blood glucose 170-190 mg/dL [9.4-10.6 mmol/l]) achieved by a modified hyperglycemic clamp with paired 20% dextrose and insulin IV infusions. This approach was chosen to mimic the typical glycemic milieu of an adolescent with T1DM (equivalent to HbA1c ~7.6-8.2%) and to maintain steady-state glycemic and insulin concentrations during kidney measures [17, 18]. In controls, GFR and RPF were measured fasting without glucose control to represent non-diabetic physiology. UACR was measured in spot urine samples obtained while fasting before and after renal clearance assessment and averaged.
Blood pressure was assessed according to American Heart Association guidelines[19]. Participants were seated quietly with their back supported, feet flat on the floor and arm at heart level. BP was be measured with an automated oscillometric sphygmomanometer. Measurements were performed in triplicate over the brachial artery of the non-dominant arm after 5 minutes of quiet rest, with one minute between measures. Systolic blood pressure (SBP) and diastolic blood pressure (DBP) were be defined as the average of the 3 measures.
Laboratory Assessments
All laboratory assays for the CASPER and Renal-HEIR cohorts were performed by the University of Colorado CTRC Core Laboratories or by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) laboratory in Phoenix, Arizona. Insulin was measured via Clinical Laboratory Improvement Amendments (CLIA)-certified chemiluminescent immunoassay (Beckman Coulter). Iohexol and PAH concentrations were measured in Phoenix by high-performance liquid chromatography (Waters, Milford, MA). Other fasting laboratory evaluations included: total cholesterol, low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein (HDL-C) cholesterol, triglycerides, glucose and HbA1c (DCCT-calibrated); assays were performed by standard methods in the CTRC laboratory.
Statistical Analysis
Data were assessed for normality and potential outliers. Continuous demographic variables were compared using t-tests or Kruskal-Wallis Rank Sum tests, and chi-squared or Fisher’s exact tests were used for categorical variables.
Sparse partial least squares discriminant analysis (sPLS-DA) was used to select metabolites that best differentiated between controls and participants with T1DM [20]. Performance was evaluated for 10 sPLS-DA components using “leave one out” cross validation, and the use of two components resulted in the lowest overall classification error rate [21]. Permutation tests indicated that the sPLS-DA results are not due to overfitting, a known limitation of sPLS-DA [21]. Confidence intervals for sPLS-DA area under the curve (AUC) were based on 1000 bootstrap samples.
Metabolites were compared between groups using moderated t-tests [22]. Correlations between metabolites and other measures were examined using Spearman’s correlation coefficient in the full cohort, as this study was not powered to detect an interaction effect. Correlations were examined for all targeted metabolites and for 13 identifiable metabolites from the untargeted metabolomics that were selected by sPLS-DA and were also in the list of the top 20 most significant metabolites by moderated t-test. All statistical analyses were performed using R version 4.0.3 (R Core Team, Vienna), with unadjusted p-values and false discovery rate (FDR)-adjusted q-values presented for all tests. sPLS-DA was performed using the mixOmics package (https://CRAN.R-project.org/package=mixOmics) [23].
Data and Resource Availability
The datasets generated and analyzed during this study, in addition to GFR, and RPF are available from the corresponding author upon reasonable request.
Results
Participants’ characteristics stratified by T1DM status
Adolescents with and without T1DM were similar in age, pubertal stage, ethnicity, body mass index (BMI), fat mass, blood pressure and lipids (Table 1). Participants with T1DM had higher HbA1c, as well as higher GFR (189±40 vs. 136±22 ml/min and 183±26 vs. 139±8 ml/min/1.73m2, respectively; p<0.0001 for both) and RPF (820±125 vs. 615±65 ml/min and 824±120 vs. 634±85 ml/min/1.73m2, respectively; p<0.0001 for both), consistent with hyperfiltration and hyperperfusion.
Table 1.
Baseline Participant Characteristics Stratified by Study
| T1DM (n=50) | Controls (n=20) | p-value | |
|---|---|---|---|
| Age (years) | 16.0 ± 3.0 | 16.1 ± 2.9 | 0.94 |
| Tanner stage | 5 (4-5) | 5 (4-5) | 0.99 |
| T1DM duration (years) | 5.7 ± 2.6 | -- | -- |
| Sex (female, %) | 50% | 70% | 0.13 |
| Race/ethnicity (%) | |||
| Black non-Hispanic | 2 | 5 | 0.41 |
| Hispanic | 6 | 0 | |
| White non-Hispanic | 92 | 95 | |
| BMI (kg/m2) | 23.4 ± 5.1 | 22.7 ± 3.7 | 0.58 |
| Weight (kg) | 67.5 ± 17.6 | 62.7 ± 14.5 | 0.28 |
| Fat mass (kg) | 20.3 ± 9.4 | 19.2 ± 6.7 | 0.62 |
| Blood pressure (mm Hg) | |||
| Systolic blood pressure | 119 ± 9 | 117 ± 8 | 0.30 |
| Diastolic blood pressure | 74 ± 11 | 73 ± 11 | 0.89 |
| Mean arterial pressure | 89 ± 9 | 88 ± 9 | 0.66 |
| UACR (mg/g) | 6 (5–14) | 7 (4–10) | 0.87 |
| GFR (ml/min) | 189 ± 40 | 136 ± 22 | <0.0001 |
| GFR (ml/min/1.73m2) | 183 ± 26 | 139 ± 8 | <0.0001 |
| RPF (ml/min) | 820 ± 125 | 615 ± 65 | <0.0001 |
| RPF (ml/min/1.73m2) | 824 ± 120 | 634 ± 85 | <0.0001 |
| HbA1c (%) | 8.7 ± 1.3 | 5.2 ± 0.2 | <0.0001 |
| LDL-C (mg/dL) | 89 ± 24 | 92 ± 24 | 0.65 |
| HDL-C (mg/dL) | 51 ± 10 | 46 ± 9 | 0.08 |
| Triglycerides (mg/dL) | 78 ± 35 | 88 ± 36 | 0.27 |
Data are presented as mean ± SD, median (IQR), or percent.
Abbreviations: BMI, body mass index; GFR, glomerular filtration rate; GIR, glucose infusion rate; HDL-C, high-density lipoprotein cholesterol; HbA1c, hemoglobin A1c; LDL-C, low-density lipoprotein cholesterol; RPF, renal plasma flow; T1DM, type 1 diabetes mellitus; UACR, urinary albumin/creatinine ratio.
Of note similar table was published in Diabetes for a separate analysis
Plasma metabolites in adolescents with T1DM and controls
Table 2 summarizes the 21 plasma metabolites that were absolutely quantified stratified by T1DM status. Compared to controls, all metabolites except alanine, cystine, proline, serine, and α-ketoglutarate were statistically significantly attenuated in T1DM. After adjusting for GFR, arginine, glycine, methionine, and α-ketoglutarate were no longer statistically significant. By using sPLS-DA, the 21 plasma metabolites that were absolutely quantified discriminated T1DM status with an AUC of 0.82 (95% CI: 0.71, 0.93; permutation p<0.001) and error rate of 0.21 (Figure 1). The sPLS-DA discrimination performance was further improved with untargeted metabolomics with an AUC of 0.95 (95% CI: 0.94, 0.99; permutation p<0.001) and error rate of 0.16.
Table 2.
Absolutely quantified metabolites stratified by diabetes status.
| T1DM (n=50) | Controls (n=20) | p-value | q-value | p-value* | q-value* | |
|---|---|---|---|---|---|---|
| Alanine (μM) | 301.19±94.73 | 354.42±112.14 | 0.048 | 0.173 | 0.057 | 0.075 |
| Arginine (μM) | 66.77±19.54 | 83.28±21.15 | 0.003 | 0.016 | 0.047 | 0.065 |
| Cystine (μM) | 37.49±13.98 | 42.55±27.06 | 0.307 | 0.307 | 0.204 | 0.214 |
| Glutamine (μM) | 27.11±11.72 | 38.45±12.81 | 0.001 | 0.008 | <0.001 | <0.001 |
| Glycine (μM) | 383.64±83.69 | 467.79±132.43 | 0.002 | 0.016 | 0.064 | 0.079 |
| Histidine (μM) | 80.41±26.62 | 117.61±46.57 | <0.001 | 0.001 | 0.025 | 0.043 |
| Isoleucine (μM) | 59.33±19.14 | 80.55±24.48 | <0.001 | 0.004 | 0.007 | 0.015 |
| Leucine (μM) | 108.53±29.05 | 146.13±42.79 | <0.001 | 0.001 | 0.001 | 0.006 |
| Lysine (μM) | 142.25±37.45 | 180.59±46.06 | 0.001 | 0.008 | 0.002 | 0.006 |
| Methionine (μM) | 22.74±6.95 | 29.24±8.23 | 0.001 | 0.015 | 0.038 | 0.057 |
| Phenylalanine (μM) | 52.00±12.11 | 71.67±19.80 | <0.001 | <0.001 | <0.001 | 0.002 |
| Proline (μM) | 127.63±45.93 | 150.05±50.93 | 0.078 | 0.173 | 0.102 | 0.119 |
| Serine (μM) | 125.60±71.32 | 218.99±240.43 | 0.015 | 0.073 | 0.276 | 0.276 |
| Threonine (μM) | 89.52±32.67 | 128.87±53.79 | <0.001 | 0.006 | 0.011 | 0.024 |
| Tyrosine (μM) | 61.01±19.10 | 78.88±24.68 | 0.002 | 0.016 | 0.024 | 0.043 |
| Valine (μM) | 149.85±40.31 | 195.26±53.18 | <0.001 | 0.004 | 0.002 | 0.006 |
| Citrate (μM) | 209.20±49.37 | 261.34±84.23 | 0.002 | 0.016 | 0.163 | 0.18 |
| α-Ketoglutarate (μM) | 11.81±2.62 | 13.27±2.83 | 0.043 | 0.173 | 0.036 | 0.057 |
| Succinate (μM) | 5.75±1.30 | 6.89±1.26 | 0.001 | 0.015 | 0.002 | 0.006 |
| Fumarate (μM) | 1.71±0.52 | 2.19±0.46 | 0.001 | 0.008 | 0.006 | 0.015 |
| Malate (μM) | 29.97±15.36 | 53.85±17.69 | <0.001 | <0.001 | <0.001 | <0.001 |
Data are presented as mean ± SD. All metabolite concentrations are presented in μM. q-values represent FDR-adjusted p-values.
Indicates a p-value or q-value from models adjusted for GFR
Figure 1. Sample plots from the sPLS-DA of targeted (left panel) and untargeted (right panel) metabolomics.
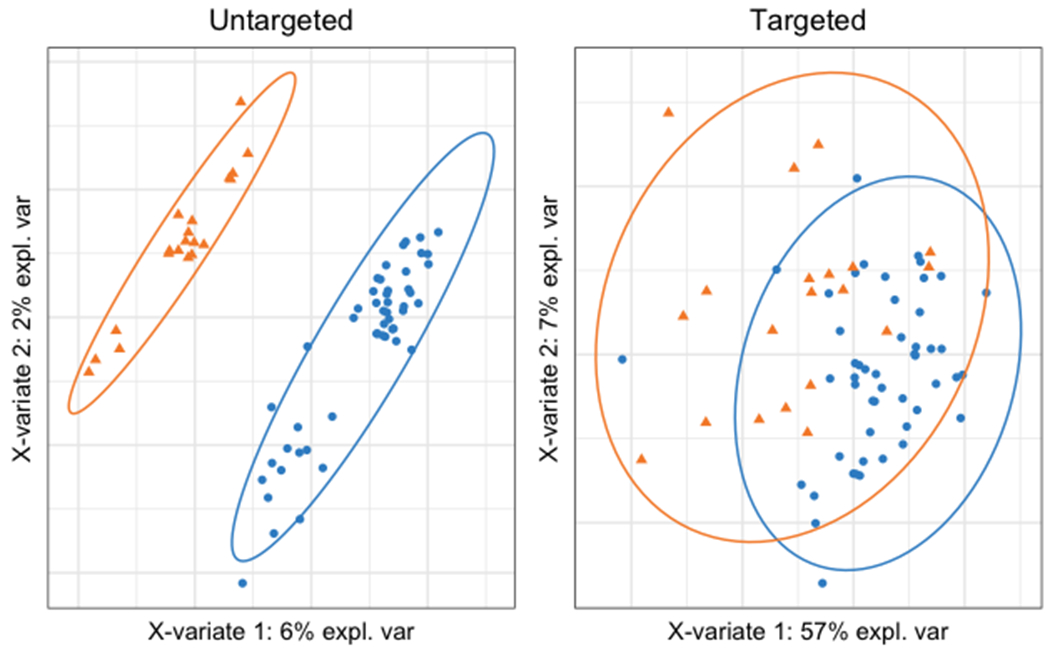
Orange triangles represent controls and blue circles represent participants with T1DM. X-variate 1 is the first component of PLS-DA and X-variate 2 is the second component. PLS-DA performed using the untargeted metabolites resulted in better separation between groups than PLS-DA using targeted metabolites.
Relationships among plasma metabolites, kidney energetics and function
Figure 2 shows the relationships among GFR, RPF, albuminuria, and plasma metabolites from targeted and untargeted metabolomics, respectively. Lower glycine (r:−0.33, q=0.01) histidine (r:−0.45, q<0.001), methionine (r:−0.29, q=0.02), phenylalanine (r:−0.29, q=0.01), serine (r:−0.42, q<0.001), threonine (r:−0.28, q=0.02), citrate (r:−0.35, q=0.003), fumarate (r:−0.24, q=0.04), and malate (r:−0.29, q=0.02) correlated with higher GFR. Lower glycine (r:−0.28, q=0.04), phenylalanine (r:−0.30, q=0.03), fumarate (r:−0.29, q=0.04), and malate (r:−0.5, q<0.001) correlated with higher RPF. Lower histidine (r:−0.28, q=0.02) was correlated with higher mean ACR (Supplemental Table 2).
Figure 2.
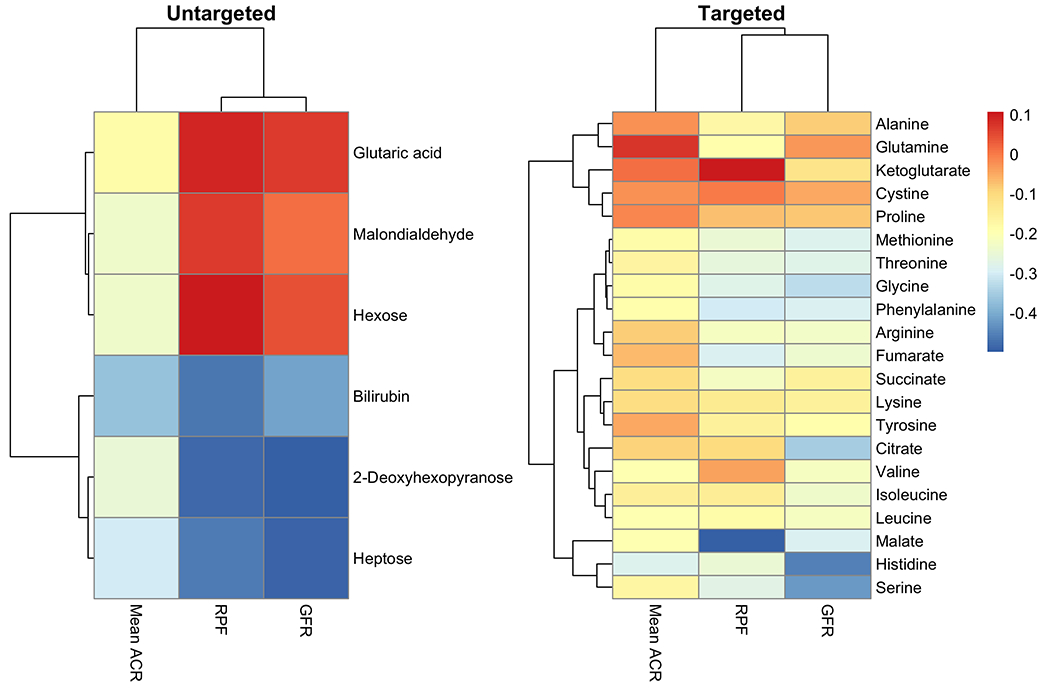
Heatmap of the correlations between the untargeted (left panel) and targeted (right panel) metabolomics and clinical variables.
Of the clinically important untargeted metabolites selected based on sPLS-DA and moderated t-tests, 2-Deoxyhexopyranose correlated negatively with RPF (r=−0.49, q<0.001) and GFR (r=−0.51, q<0.001). Likewise, higher heptose associated with lower GFR (r=−0.5, q<0.001) and lower RPF (r=−0.49, q=0.001). Higher concentrations of glutaric acid, malondialdehyde, hexose associated with higher RPF and GFR. Finally, higher concentrations of bilirubin associated with lower RPF (r:−0.45, q=0.001), UACR (r:−0.27, q=0.02), and GFR (r:−0.35, q=0.003) (Supplemental Table 3).
When stratified by T1DM status, these patterns of association between plasma metabolites and kidney function were similar across groups. However, this study was not adequately powered for detection of interaction effects across targeted and untargeted metabolomics while accounting for multiple comparisons.
Discussion
In the current study, we are the first to show that adolescents with ≤10 years of T1DM exhibit lower plasma levels of amino acids and carboxylic acid compared to controls of similar age and BMI. Additionally, in untargeted metabolomics we found differences in numerous metabolites, including branched chain amino acids, between adolescents with and without T1DM. We also demonstrated novel associations among plasma levels of carboxylic acids, branched chain amino acids, and kidney function. Together, these novel observations support an innovative theory that metabolic changes in critical energy-intensive kidney functions are an early feature of T1DM. The extent to which these changes reflect perturbed mitochondrial bioenergetics involved in the pathogenesis of DKD is the focus of ongoing investigations.
T1DM is a complex multifaceted metabolic disorder with pathologic features beyond β-cell injury, insulinopenia and hyperglycemia, including insulin resistance and mitochondrial dysfunction. Adolescents and adults with T1DM exhibit insulin resistance compared with controls without diabetes, independent of body habitus and hyperglycemia [24–27]. Additionally, our group has previously shown that normal-weight adolescents with T1DM demonstrate directly measured slowed post-exercise ATP re-synthesis in muscle, evidence of mitochondrial dysfunction, which also relate to insulin resistance [28]. Both insulin resistance and mitochondrial dysfunction have been related to the development of DKD in adults with T1DM and T2D [3, 8, 29–31], and in youth with T2D with very short duration of T2D [32]. The kidneys are highly metabolically active and the high oxygen demand is necessary to maintain adequate ATP production, as 95% of the ATP produced in the kidney is through aerobic metabolism [4, 6]. Indeed, the kidneys require a large number of mitochondria to sustain filtration, fluid and electrolyte balance [7]. Proximal tubules contain more mitochondria than any other kidney structure due to their high energy expenditure; ~80% of filtrate that passes through the glomerulus is reabsorbed by the proximal tubule [7].
Kidney tissue hypoxia, i.e., a metabolic mismatch between kidney energy expenditure and substrate metabolism, has been proposed as a unifying pathway in the development of DKD. Emerging animal data suggest that in diabetes the kidneys are unable to sufficiently compensate for the increased ATP consumption due to the effects of insulin resistance and mitochondrial dysfunction on substrate utilization (impaired ATP generation, Figure 1) [9, 10, 33, 34]. The resultant ATP deficit is followed by increased perfusion or kidney blood flow to deliver more O2 to the kidneys. However, in the kidneys increased kidney blood flow leads to increased GFR, and thus a higher Na+ load which further increases kidney O2 consumption. Similarly, hypoxia-induced incapacity to sustain mitochondrial synthesis of ATP results in breakdown and deamination of purines (e.g., hypoxanthine, xanthine) that are rapidly catabolized to urate with the concomitant generation of deleterious hydrogen peroxide following reperfusion, a metabolic phenomenon underlying the ischemia/reperfusion injury damage in acute kidney ischemia and, at least in part, recapitulated in chronic kidney disease [35–38].
Amino acid metabolism accounts for 10-15% of total energy production and ATP synthesis, and relies on interplay among small intestines, liver, kidneys, and muscle (Supplemental Figure 1). Perturbations in serum metabolites, particularly amino acids, have been associated with kidney dysfunction in adults with both T1DM and T2D [39–41]. Studies suggest increased substrate utilization of amino acids in diabetes, which could explain their lower circulating concentrations [42–44]. For example, lower concentrations of amino acids isoleucine, leucine, and valine have been associated with greater risk of kidney failure in individuals with T2D [41]. However, investigations in youth with T1DM and use of gold standard measurements of kidney function have previously been missing from the literature. Our study revealed that essential amino acids and branched amino acids were depleted in young persons with T1DM compared to healthy controls. Branched amino acids play critical roles in signaling pathways; leucine is a key regulator of mammalian target of Rapamycin (mTOR), a central regulator of cellular growth and insulin signaling [45]. Additionally, amino acids and carboxylic acids, including histidine, serine and malate related to kidney function using gold-standard methods. The mechanisms by which alterations in amino acids contribute to early kidney dysfunction are poorly understood. Possible links include mitochondrial dysfunction and impaired tubular reabsorption resulting in amino acid depletion [8, 46–48]. Additionally, malate has been ascribed to influence nitric oxide concentrations, which may explain its relationship with RPF [49].
Furthermore, the untargeted analyses revealed interesting associations with non-amino acid metabolites, including plasma malondialdehyde, a recognized marker of oxidative stress. Indeed, higher concentrations of plasma malondialdehyde associated with lower kidney oxygen availability [50]. Higher concentrations of monosaccharides heptose and hexose also related to lower kidney oxygen availability. 2-Deoxyhexopyranose, which has been implicated in the sodium-independent transport of sugar in kidney tubular cells, associated with kidney function and oxygen availability [51]. Finally, higher concentrations of bilirubin associated with higher kidney oxygen availability and lower UACR. This finding is consistent with the literature as circulating unconjugated bilirubin is thought to be a strong antioxidant and has shown to be protective against kidney and cardiovascular disease [52].
Strengths of this study include directly measured GFR and RPF by gold-standard methods, GFR and RPF assessed in both T1DM and non-T1DM cohorts, and comprehensive metabolic phenotyping including targeted and untargeted plasma metabolomics. We also provided careful control of glycemia during the kidney measures, as well as control of dietary intake and physical activity prior to study measurements. Methods to quantify mitochondrial function more directly, including determining acetate-turnover in the TCA cycle by 11C-acetate positron emission tomography (PET), tissue-specific mitochondrial bioenergetics by positional isotopomer NMR tracer analysis (PINTA) and spatial metabolomics from kidney tissue, are likely superior but less suitable in pediatrics due to their invasiveness and radiation exposure. Additionally, urine metabolomics would have been informative and potentially more reflective of kidney metabolic perturbations than plasma metabolites. By design, our study enrolled adolescents with T1DM ≤10 years to examine perturbed kidney energetics in the earliest stages of kidney dysfunction (i.e., hyperfiltration). Consequently, our study findings may not be generalizable to adolescents with longstanding T1DM or more advanced stages of kidney dysfunction. Hyperfiltration may also not necessarily imply or predict progressive kidney disease in T1DM. Moreover, the effects of age and T1DM duration warrant further study, as previous work by Mathew et al. showed conflicting results for certain amino acids in a significantly older cohort, which might be ascribed to differences in GFR [53]. Other limitations include the cross-sectional study design limiting causal inferences, and the relatively small control group.
In conclusion, adolescents with T1DM for ≤10 years demonstrate attenuated metabolites of mitochondrial function that are associated with gold-standard measures of kidney function. These observations may implicate perturbed mitochondrial bioenergetics in the pathogenesis of early DKD. Future directions include interrogating the quantification and distribution of mitochondrial metabolites in kidney tissue using matrix-assisted laser desorption/ionization mass spectrometry imaging. Furthermore, improving mitochondrial bioenergetics has the potential to restore kidney function and offer promise as a future intervention to mitigate DKD risk.
Supplementary Material
Supplemental Figure 1. Amino acid metabolism and their role in the cellular energetic metabolism. At the top of the figure are the main classification criteria of amino acids. On the right are the organs where amino acid metabolism takes place with the respective physiological pathways (from top to bottom: kidney, skeletal muscle, liver, and gut). The bottom of the figure summarizes the urea and Krebs cycles and their interactions (green arrows). The Krebs cycle is the most important source of cellular energy and amino acids are involved at various stages. The urea cycle is the biochemical pathway to dispose of nitrogen, which derives from the catabolism of amino acids. Both cycles play a central role in cellular energetic metabolism.
Acknowledgements:
The authors are grateful to Dr. Fabia Gamboni for assistance with metabolomics analysis. Drs. Pyle and Bjornstad, as well as Mr. Vigers, are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Support:
Financial support for this work provided by the NIDDK Diabetic Complications Consortium (RRID:SCR_001415, www.diacomp.org), grants DK076169 and DK115255 (18AU3871 [P.B.]), JDRF grant number 2-SRA-2018-627-M-B (P.B.), and NIH/NIDDK grant number K23-DK116720 (P.B.), K24-HL145076 (K.J.N.), UL1-RR025780 (University of Colorado Denver), support from Center for Women’s Health Research at University of Colorado, the Department of Pediatrics, Section of Endocrinology and Barbara Davis Center for Diabetes at University of Colorado School of Medicine, by Interagency Agreement 16FED1604631 with the Centers for Disease Control and Prevention (R.G.N.), and by the Intramural Research Program of the NIDDK.
Disclaimer:
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
Trial Registration: ClinicalTrials.gov NCT03618420 and NCT03584217
Financial Disclosure/Conflict of Interest: TV: none. CV: none. LL: none. PP: none. HH: none. AD: none. JAR: none. FP: none. DZC: none. DHV: none. KJN: none. MEP: none. RGN: none. LP: none. PB: has acted as a consultant for AstraZeneca, Bayer, Bristol-Myers Squibb, Boehringer Ingelheim, Eli-Lilly, Sanofi, Novo Nordisk, and Horizon Pharma. PB serves on the advisory boards of AstraZeneca, Boehringer Ingelheim, and XORTX.
Ethics Approval and Patient Consent Statement: The research protocols of CASPER and Renal-HEIR were approved by COMIRB, and all participants gave written informed consent.
References (Vancouver):
- 1.Groop PH, Thomas MC, Moran JL, Wadèn J, Thorn LM, Mäkinen VP, Rosengård-Bärlund M, Saraheimo M, Hietala K, Heikkilä O, Forsblom C, Group FS (2009) The presence and severity of chronic kidney disease predicts all-cause mortality in type 1 diabetes. Diabetes 58:1651–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orchard TJ, Secrest AM, Miller RG, Costacou T (2010) In the absence of renal disease, 20 year mortality risk in type 1 diabetes is comparable to that of the general population: a report from the Pittsburgh Epidemiology of Diabetes Complications Study. Diabetologia 53:2312–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjornstad P, Snell-Bergeon JK, Rewers M, Jalal D, Chonchol MB, Johnson RJ, Maahs DM (2013) Early diabetic nephropathy: a complication of reduced insulin sensitivity in type 1 diabetes. Diabetes Care 36:3678–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soltoff SP (1986) ATP and the regulation of renal cell function. Annu Rev Physiol 48:9–31. [DOI] [PubMed] [Google Scholar]
- 5.Hesp AC, Schaub JA, Prasad PV, Vallon V, Laverman GD, Bjornstad P, van Raalte DH (2020) The role of renal hypoxia in the pathogenesis of diabetic kidney disease: a promising target for newer renoprotective agents including SGLT2 inhibitors? Kidney Int 98:579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen JJ (1979) Is the function of the renal papilla coupled exclusively to an anaerobic pattern of metabolism? Am J Physiol 236:F423–433. [DOI] [PubMed] [Google Scholar]
- 7.Bhargava P, Schnellmann RG (2017) Mitochondrial energetics in the kidney. Nat Rev Nephrol 13:629–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma K, Karl B, Mathew AV, Gangoiti JA, Wassel CL, Saito R, Pu M, Sharma S, You YH, Wang L, Diamond-Stanic M, Lindenmeyer MT, Forsblom C, Wu W, Ix JH, Ideker T, Kopp JB, Nigam SK, Cohen CD, Groop PH, Barshop BA, Natarajan L, Nyhan WL, Naviaux RK (2013) Metabolomics reveals signature of mitochondrial dysfunction in diabetic kidney disease. J Am Soc Nephrol 24:1901–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nangaku M (2006) Chronic hypoxia and tubulointerstitial injury: a final common pathway to end-stage renal failure. J Am Soc Nephrol 17:17–25. [DOI] [PubMed] [Google Scholar]
- 10.Singh DK, Winocour P, Farrington K (2008) Mechanisms of disease: the hypoxic tubular hypothesis of diabetic nephropathy. Nat Clin Pract Nephrol 4:216–226. [DOI] [PubMed] [Google Scholar]
- 11.Nemkov T, D’Alessandro A, Hansen KC (2015) Three-minute method for amino acid analysis by UHPLC and high-resolution quadrupole orbitrap mass spectrometry. Amino Acids 47:2345–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nemkov T, Reisz JA, Gehrke S, Hansen KC, D’Alessandro A (2019) High-Throughput Metabolomics: Isocratic and Gradient Mass Spectrometry-Based Methods. Methods Mol Biol 1978:13–26. [DOI] [PubMed] [Google Scholar]
- 13.Nemkov T, Hansen KC, D’Alessandro A (2017) A three-minute method for high-throughput quantitative metabolomics and quantitative tracing experiments of central carbon and nitrogen pathways. Rapid Commun Mass Spectrom 31:663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seegmiller JC, Wolf BJ, Albtoush N, Melena I, Gross SP, Vinovskis C, Ix JH, Bjornstad P (2020) Tubular Secretion Markers, Glomerular Filtration Rate, Effective Renal Plasma Flow, and Filtration Fraction in Healthy Adolescents. Kidney Medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vinovskis C, Li LP, Prasad P, Tommerdahl K, Pyle L, Nelson RG, Pavkov ME, van Raalte D, Rewers M, Pragnell M, Mahmud FH, Cherney DZ, Johnson RJ, Nadeau KJ, Bjornstad P (2020) Relative Hypoxia and Early Diabetic Kidney Disease in Type 1 Diabetes. Diabetes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delanaye P, Radermecker RP, Rorive M, Depas G, Krzesinski JM (2005) Indexing glomerular filtration rate for body surface area in obese patients is misleading: concept and example. Nephrol Dial Transplant 20:2024–2028. [DOI] [PubMed] [Google Scholar]
- 17.Cherney DZ, Kanbay M, Lovshin JA (2020) Renal physiology of glucose handling and therapeutic implications. Nephrol Dial Transplant 35:i3–i12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cherney DZ, Perkins BA, Soleymanlou N, Maione M, Lai V, Lee A, Fagan NM, Woerle HJ, Johansen OE, Broedl UC, von Eynatten M (2014) Renal hemodynamic effect of sodium-glucose cotransporter 2 inhibition in patients with type 1 diabetes mellitus. Circulation 129:587–597. [DOI] [PubMed] [Google Scholar]
- 19.Whelton PK, Carey RM, Aronow WS, Casey DE Jr, Collins KJ, Dennison Himmelfarb C, DePalma SM, Gidding S, Jamerson KA, Jones DW, MacLaughlin EJ, Muntner P, Ovbiagele B, Smith SC Jr, Spencer CC, Stafford RS, Taler SJ, Thomas RJ, Williams KA Sr, Williamson JD, Wright JT Jr. (2018) 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Hypertension 71:e13–e115. [DOI] [PubMed] [Google Scholar]
- 20.Le Cao KA, Rossouw D, Robert-Granie C, Besse P (2008) A sparse PLS for variable selection when integrating omics data. Stat Appl Genet Mol Biol 7:Article 35. [DOI] [PubMed] [Google Scholar]
- 21.Westerhuis JA, Hoefsloot HCJ, Smit S, Vis DJ, Smilde AK, van Velzen EJJ, van Duijnhoven JPM, van Dorsten FA (2008) Assessment of PLSDA cross validation. Metabolomics 4:81–89. [Google Scholar]
- 22.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research 43:e47–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneidman D, Rohart F, Gautier B, Singh A, Lê Cao K- A (2017) mixOmics: An R package for ‘omics feature selection and multiple data integration. PLOS Computational Biology 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cree-Green M, Stuppy JJ, Thurston J, Bergman BC, Coe GV, Baumgartner AD, Bacon S, Scherzinger A, Pyle L, Nadeau KJ (2018) Youth With Type 1 Diabetes Have Adipose, Hepatic, and Peripheral Insulin Resistance. J Clin Endocrinol Metab 103:3647–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Millstein RJ, Pyle LL, Bergman BC, Eckel RH, Maahs DM, Rewers MJ, Schauer IE, Snell-Bergeon JK (2018) Sex-specific differences in insulin resistance in type 1 diabetes: The CACTI cohort. J Diabetes Complications 32:418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nadeau KJ, Regensteiner JG, Bauer TA, Brown MS, Dorosz JL, Hull A, Zeitler P, Draznin B, Reusch JE (2010) Insulin resistance in adolescents with type 1 diabetes and its relationship to cardiovascular function. J Clin Endocrinol Metab 95:513–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schauer IE, Snell-Bergeon JK, Bergman BC, Maahs DM, Kretowski A, Eckel RH, Rewers M (2011) Insulin resistance, defective insulin-mediated fatty acid suppression, and coronary artery calcification in subjects with and without type 1 diabetes: The CACTI study. Diabetes 60:306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cree-Green M, Newcomer BR, Brown MS, Baumgartner AD, Bergman B, Drew B, Regensteiner JG, Pyle L, Reusch JE, Nadeau KJ (2015) Delayed skeletal muscle mitochondrial ADP recovery in youth with type 1 diabetes relates to muscle insulin resistance. Diabetes 64:383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sas KM, Kayampilly P, Byun J, Nair V, Hinder LM, Hur J, Zhang H, Lin C, Qi NR, Michailidis G, Groop PH, Nelson RG, Darshi M, Sharma K, Schelling JR, Sedor JR, Pop-Busui R, Weinberg JM, Soleimanpour SA, Abcouwer SF, Gardner TW, Burant CF, Feldman EL, Kretzler M, Brosius FC 3rd, Pennathur S (2016) Tissue-specific metabolic reprogramming drives nutrient flux in diabetic complications. JCI Insight 1:e86976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saulnier PJ, Darshi M, Wheelock KM, Looker HC, Fufaa GD, Knowler WC, Weil EJ, Tanamas SK, Lemley KV, Saito R, Natarajan L, Nelson RG, Sharma K (2018) Urine metabolites are associated with glomerular lesions in type 2 diabetes. Metabolomics 14:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tofte N, Vogelzangs N, Mook-Kanamori D, Brahimaj A, Nano J, Ahmadizar F, Willems van Dijk K, Frimodt-Moller M, Arts I, Beulens JWJ, Rutters F, van der Heijden AA, Kavousi M, Stehouwer CDA, Nijpels G, van Greevenbroek MMJ, van der Kallen CJH, Rossing P, Ahluwalia TS, t Hart LM (2020) Plasma metabolomics identifies markers of impaired renal function: A meta-analysis of 3,089 persons with type 2 diabetes. J Clin Endocrinol Metab. [DOI] [PubMed] [Google Scholar]
- 32.Cree-Green M, Gupta A, Coe GV, Baumgartner AD, Pyle L, Reusch JE, Brown MS, Newcomer BR, Nadeau KJ (2017) Insulin resistance in type 2 diabetes youth relates to serum free fatty acids and muscle mitochondrial dysfunction. J Diabetes Complications 31:141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haase VH (2006) The VHL/HIF oxygen-sensing pathway and its relevance to kidney disease. Kidney Int 69:1302–1307. [DOI] [PubMed] [Google Scholar]
- 34.Mudaliar S, Alloju S, Henry RR (2016) Can a Shift in Fuel Energetics Explain the Beneficial Cardiorenal Outcomes in the EMPA-REG OUTCOME Study? A Unifying Hypothesis. Diabetes Care 39:1115–1122. [DOI] [PubMed] [Google Scholar]
- 35.Clendenen N, Nunns GR, Moore EE, Gonzalez E, Chapman M, Reisz JA, Peltz E, Fragoso M, Nemkov T, Wither MJ, Sauaia A, Silliman CC, Hansen K, Banerjee A, D’Alessandro A, Moore HB (2019) Selective organ ischaemia/reperfusion identifies liver as the key driver of the post-injury plasma metabolome derangements. Blood Transfus 17:347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fox BM, Gil HW, Kirkbride-Romeo L, Bagchi RA, Wennersten SA, Haefner KR, Skrypnyk NI, Brown CN, Soranno DE, Gist KM, Griffin BR, Jovanovich A, Reisz JA, Wither MJ, D’Alessandro A, Edelstein CL, Clendenen N, McKinsey TA, Altmann C, Faubel S (2019) Metabolomics assessment reveals oxidative stress and altered energy production in the heart after ischemic acute kidney injury in mice. Kidney Int 95:590–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie T, Chen C, Peng Z, Brown BC, Reisz JA, Xu P, Zhou Z, Song A, Zhang Y, Bogdanov MV, Kellems RE, D’Alessandro A, Zhang W, Xia Y (2020) Erythrocyte Metabolic Reprogramming by Sphingosine 1-Phosphate in Chronic Kidney Disease and Therapies. Circ Res 127:360–375. [DOI] [PubMed] [Google Scholar]
- 38.Chouchani ET, Pell VR, Gaude E, Aksentijevic D, Sundier SY, Robb EL, Logan A, Nadtochiy SM, Ord ENJ, Smith AC, Eyassu F, Shirley R, Hu CH, Dare AJ, James AM, Rogatti S, Hartley RC, Eaton S, Costa ASH, Brookes PS, Davidson SM, Duchen MR, Saeb-Parsy K, Shattock MJ, Robinson AJ, Work LM, Frezza C, Krieg T, Murphy MP (2014) Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature 515:431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tofte N, Suvitaival T, Trost K, Mattila IM, Theilade S, Winther SA, Ahluwalia TS, Frimodt-Moller M, Legido-Quigley C, Rossing P (2019) Metabolomic Assessment Reveals Alteration in Polyols and Branched Chain Amino Acids Associated With Present and Future Renal Impairment in a Discovery Cohort of 637 Persons With Type 1 Diabetes. Front Endocrinol (Lausanne) 10:818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Welsh P, Rankin N, Li Q, Mark PB, Wurtz P, Ala-Korpela M, Marre M, Poulter N, Hamet P, Chalmers J, Woodward M, Sattar N (2018) Circulating amino acids and the risk of macrovascular, microvascular and mortality outcomes in individuals with type 2 diabetes: results from the ADVANCE trial. Diabetologia 61:1581–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niewczas MA, Sirich TL, Mathew AV, Skupien J, Mohney RP, Warram JH, Smiles A, Huang X, Walker W, Byun J, Karoly ED, Kensicki EM, Berry GT, Bonventre JV, Pennathur S, Meyer TW, Krolewski AS (2014) Uremic solutes and risk of end-stage renal disease in type 2 diabetes: metabolomic study. Kidney Int 85:1214–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, Zheng S, Wu G (2020) Amino Acid Metabolism in the Kidneys: Nutritional and Physiological Significance. Adv Exp Med Biol 1265:71–95. [DOI] [PubMed] [Google Scholar]
- 43.Hou Y, Hu S, Li X, He W, Wu G (2020) Amino Acid Metabolism in the Liver: Nutritional and Physiological Significance. Adv Exp Med Biol 1265:21–37. [DOI] [PubMed] [Google Scholar]
- 44.Pasiakos SM, Carbone JW (2014) Assessment of skeletal muscle proteolysis and the regulatory response to nutrition and exercise. IUBMB Life 66:478–484. [DOI] [PubMed] [Google Scholar]
- 45.Lynch CJ, Adams SH (2014) Branched-chain amino acids in metabolic signalling and insulin resistance. Nat Rev Endocrinol 10:723–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Costeas PA, Chinsky JM (1996) Effects of insulin on the regulation of branched-chain alpha-keto acid dehydrogenase E1 alpha subunit gene expression. Biochem J 318 ( Pt 1):85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giesbertz P, Daniel H (2016) Branched-chain amino acids as biomarkers in diabetes. Curr Opin Clin Nutr Metab Care 19:48–54. [DOI] [PubMed] [Google Scholar]
- 48.Lotta LA, Scott RA, Sharp SJ, Burgess S, Luan J, Tillin T, Schmidt AF, Imamura F, Stewart ID, Perry JR, Marney L, Koulman A, Karoly ED, Forouhi NG, Sjogren RJ, Naslund E, Zierath JR, Krook A, Savage DB, Griffin JL, Chaturvedi N, Hingorani AD, Khaw KT, Barroso I, McCarthy MI, O’Rahilly S, Wareham NJ, Langenberg C (2016) Genetic Predisposition to an Impaired Metabolism of the Branched-Chain Amino Acids and Risk of Type 2 Diabetes: A Mendelian Randomisation Analysis. PLoS Med 13:e1002179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hou E, Sun N, Zhang F, Zhao C, Usa K, Liang M, Tian Z (2017) Malate and Aspartate Increase L-Arginine and Nitric Oxide and Attenuate Hypertension. Cell Reports 19:1631–1639. [DOI] [PubMed] [Google Scholar]
- 50.Nielsen F, Mikkelsen BB, Nielsen JB, Andersen HR, Grandjean P (1997) Plasma malondialdehyde as biomarker for oxidative stress: reference interval and effects of life-style factors. Clin Chem 43:1209–1214. [PubMed] [Google Scholar]
- 51.Kleinzeller A (1972) The Na-Iindependent Transport of Sugar in Renal Tubular Cells. Na-linked Transport of Organic Solutes, pp 109–115. [Google Scholar]
- 52.Boon AC, Bulmer AC, Coombes JS, Fassett RG (2014) Circulating bilirubin and defense against kidney disease and cardiovascular mortality: mechanisms contributing to protection in clinical investigations. Am J Physiol Renal Physiol 307:F123–136. [DOI] [PubMed] [Google Scholar]
- 53.Mathew AV, Jaiswal M, Ang L, Michailidis G, Pennathur S, Pop-Busui R (2019) Impaired Amino Acid and TCA Metabolism and Cardiovascular Autonomic Neuropathy Progression in Type 1 Diabetes. Diabetes 68:2035–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Amino acid metabolism and their role in the cellular energetic metabolism. At the top of the figure are the main classification criteria of amino acids. On the right are the organs where amino acid metabolism takes place with the respective physiological pathways (from top to bottom: kidney, skeletal muscle, liver, and gut). The bottom of the figure summarizes the urea and Krebs cycles and their interactions (green arrows). The Krebs cycle is the most important source of cellular energy and amino acids are involved at various stages. The urea cycle is the biochemical pathway to dispose of nitrogen, which derives from the catabolism of amino acids. Both cycles play a central role in cellular energetic metabolism.


