Abstract
The 90-kDa heat shock protein (Hsp90) is a homodimeric molecular chaperone with ATPase activity, which has become an intensely studied target for the development of drugs for the treatment of cancer, neurodegenerative and infectious diseases. The equilibrium between Hsp90 dimers and oligomers is important for modulating its function. In the absence of ATP, the passive chaperone activity of Hsp90 dimers and oligomers has been shown to stabilize client proteins as a holdase, which enhances substrate binding and prevents irreversible aggregation and precipitation of the substrate proteins. In the presence of ATP and its associated cochaperones, Hsp90 homodimers act as foldases with the binding and hydrolysis of ATP driving conformational changes that mediate client folding. Crystal structures of both wild type and W320A mutant Hsp90αMC (middle/C-terminal domain) have been determined, which displayed a preference for hexameric and dimeric states, respectively. Structural analysis showed that W320 is a key residue for Hsp90 oligomerization by forming intermolecular interactions at the Hsp90 hexameric interface through cation-π interactions with R367. W320A substitution results in the formation of a more open conformation of Hsp90, which has not previously been reported, and the induction of a conformational change in the catalytic loop. The structures provide new insights into the mechanism by which W320 functions as a key switch for conformational changes in Hsp90 self-oligomerization, and binding cochaperones and client proteins.
Keywords: Hsp90, crystal structure, conformational change, chaperone, oligomerization
1. Introduction
The 90-kD heat shock protein (Hsp90) is a molecular chaperone machine with ATPase activity that is highly conserved in bacteria and eukaryotes. The Hsp90 functions as a dimer to stabilize, regulate and activate hundreds of important client proteins under both normal and stress conditions (1–3). As a key mediator of cellular homeostasis, Hsp90 plays an important role in preventing protein misfolding and aggregation, and Hsp90 overexpression is a factor in tumorigenesis (4,5). As a host chaperone, Hsp90 is also required for viral replication (6). Therefore, Hsp90 is a promising drug target for treatment of human cancer, infectious diseases and neurodegenerative diseases. Hsp90 family members include nucleocytoplasmic Hsp90α and Hsp90β, endoplasmic reticulum GRP94/HSP90B1 and mitochondrial TRAP1 (7,8). All Hsp90 homologues exist as flexible homodimers, with each protomer composed of three domains, an N-terminal domain (NTD), a middle domain (MD) and a C-terminal domain (CTD). A dynamic highly charged linker domain connects the NTD and MD in eukaryotic Hsp90α, Hsp90β and GRP94/HSP90B1 (9).
Conformational changes of the Hsp90 dimer with open, semi-closed or closed states accompanying ATP binding, hydrolysis and release of ADP, are modulated by the binding or release of co-chaperones, post-translational modifications, and its interaction with client proteins(7,10,11). Hsp90 homodimers can also spontaneously self-associate to form oligomers under heat shock(12,13), in the presence of divalent cations(14) or even under unstressed conditions (15,16), with the Hsp90 oligomers having been demonstrated to be fully capable of binding and preventing the irreversible aggregation and precipitation of denatured substrate proteins. In vitro, the Hsp90 co-chaperone Aha1, which stimulates the ATPase activity of Hsp90 has been reported to interact more favorably with Hsp90 hexamers(17), while p23, which stabilizes the closed conformation of ATP-bound dimers, shifts the Hsp90 dimer/oligomer equilibrium towards the formation of the dimer (18).
Client protein activation by Hsp90 is dependent on binding and hydrolysis of ATP, which drives Hsp90 through a conformational cycle that is highly allosteric, propelling conformational changes in the MD and CTD. ATP binding induces the dimerization of the NTDs with a rearrangement of the interaction interface between the NTD and MD, which involves the rotation of the NTD, local structural rearrangements and the repositioning of specific elements (19). The MD participates in the ATP hydrolysis by stabilizing the ATP in the catalytic pocket, and through its interactions with co-chaperones and client proteins (20). In the MD, a key residue, W320 (W300 in yeast Hsp82) acts as a switch point that senses the interaction of Hsp90 with client proteins. The W300A mutation of yeast Hsp90 resulted in a defect in client protein activation by Hsp90 in vivo, but stimulated its ATPase activity in vitro (20,21) W320 (W300 in yeast) functions as a switch point in Hsp90 MD, important for specific conformational changes to form a semi-closed structure associated during client processing (22). While W300 mutations in yeast have shown that this residue plays an important role in conformational switching, the actual structural consequence of its mutation has not been demonstrated.
Since Hsp90s ATP binding site was discovered as the target of the potent anti-cancer natural product compounds geldanamycin and radicicol, the development of inhibitors that bind to this site (N-terminal inhibitors) has been intensely investigated(23). While a number of these drugs have made it to phase II clinical trials, in most cases their effectiveness as mono-therapy drugs has been disappointing. One of the primary deficiencies of this class of drugs is their induction of the pro-survival heat shock response (HSR). The induction of the HSR required dose escalation leading to dose-limiting toxicities(24–27). As such, new druggable sites in Hsp90 are now being sought.
Druggable sites in Hsp90s MD and CTDs are now being pursued as possible superior alternatives to N-terminal inhibitors(28–31). Docking simulations have identified several sites in the MD and CTDs for allosteric inhibitors, but these studies have been hampered because the closed conformation of Hsp90 was the only structure used for these simulation studies, and these structures lacked critical structural information on amino acid residues in critical functional regions of Hsp90. To date, there is no high resolution structural information regarding open state conformation of eukaryotic Hsp90 and it is possible that allosteric inhibitors favor the open state conformation in the MD and CTD of Hsp90. In our attempts to acquire structural information on the binding of potential inhibitors that interact with Hsp90s MD and/or CTD, the W320A mutant of Hsp90α middle and C-terminal domains (Hsp90αMC_W320A) was expressed. Our hypothesis was that the free energy of drug binding and any structural changes required for its stabilization were insufficient to overcome the free energy driving the formation of the Hsp90αMC hexamer. This mutation in Hsp90αMC domain was previously found to form a dimer, in contrast to the hexameric state favored by the WT Hsp90αMC protein in vitro (32). However, the structural consequences of this mutation were not investigated.
Here, we report the crystal structures of both the wild type (WT) Hsp90αMC hexamer and Hsp90αMC_W320A mutant dimer. Compared to the previously reported human WT_Hsp90αMC hexameric structures (PDB ID: PDB 3Q6M and PDB 3Q6N) (32), the main chain of the catalytic loop (394-LPLNISREMLQQSKIL-408) in Hsp90αMD is clearly elucidated, and the loop 616-AQALRDNSTMGYMA-629 in Hsp90αCTD (we termed as CT loop) can also be partially traced in the WT Hsp90αMC hexamer, both of which play significant roles in Hsp90 conformational changes during its functional cycle. The W320A mutant of Hsp90αMC crystallized as a dimer in a unique open conformation not previously observed. The structure of the catalytic loop was found to be altered from that present in the hexamer. The W320A caused a large rotation in the MD which disrupts the interactions required for the assembly of Hsp90αMC to be hexamer and may represent an open conformation of Hsp90 that favors HOP-mediated client binding. Structural comparison of the two structures provides atomic details behind the allosteric mechanism by which W320 acts as a key switch point in Hsp90 conformational cycle through its effects on the conformation of the catalytic loop, and gives two new structures containing new structural information for pursuing in silico drug discovery.
2. Materials and Methods
2.1. Gene cloning, protein expression and purification
Human Hsp90α_293–699 was cloned into a modified pET28b vector using NdeI and BamHI cloning sites, with an N-terminal 6xHis-tag, including a Tobacco Etch Virus (TEV) protease cleavage site between the tag and the protein sequence. The proteins were expressed in Escherichia coli strain BL21(DE3) system and first purified through a Ni-NTA affinity fast flow column. The eluted proteins were subsequently subjected to TEV protease cleavage and collected as flow through of a second subtracting Ni-NTA column, followed by a size exclusion column Superdex200 (GE Healthcare). The proteins were concentrated to 12 mg/ml in buffer (20 mM Hepes pH 7.2, 150 mM sodium chloride (NaCl)) for crystallization.
Human Hsp90α_293–714_W320A mutant was cloned using PCR-based site-directed mutagenesis method, which is expressed and purified by the same procedure as the wild type (WT) protein.
2.2. Crystallization and Data collection
10mg/ml of the WT hHsp90α_293–699 protein was crystallized in a solution containing 0.1 M sodium acetate pH 6.0, 12 % PEG3350 by using sitting drop vapor diffusion method at 20 °C. The hHsp90α_293–714_W320A mutant protein was crystallized in a solution containing 0.1 M Hepes PH 7.5, 20% PEG 400. 20% glycerol was added to each of the mother liquids as cryoprotectant. All data were collected at the Advanced Photon Source, Argonne National Laboratory.
2.3. Data Collection and Structural determination
All data were collected from the beamline 19-ID at the Advanced Photon Source, Argonne National Laboratory. Diffraction data were collected at processed and scaled using HKL-3000 (33). The structures of hHsp90α_293–699 and hHsp90α_293–714_W320A were both solved by molecular replacement method using phaser (34), with PDB 3Q6M as the template (32). The PHENIX program was used for the refinement (35). Translation, libration, and screw-rotation displacement groups used in the refinement were defined by the TLMSD server (36). COOT was used for the iterative manual structural building (37). The current models are of good geometry and refinement statistics (Table I). Although the mutant construct contains 15 additional residues at its C-terminal end, nearly all are disordered and not visible in the structure. All molecular graphic figures were generated with PYMOL (38). The structures were deposited into the protein data bank, PDB ID: 7RXZ for hHsp90α_293–699, and PDB ID: 7RY0 for hHsp90α_293–714_W320A.
Table 1.
Crystallographic data collection and refinement statistics
| Data collection | Hsp90αMC | Hsp90αMC_W320A |
| Beamline | 19-ID, APS | 19-ID, APS |
| Wavelength, Å | 0.97922 | 0.97918 |
| Space group | P21 | P212121 |
| Cell parameters a, b, c, Å | 153.8, 88.0, 166.2 β=114.36° |
38.9, 154.3, 156.7 |
| Resolution, Å | 50.00–3.15 (3.26–3.15) |
50.00– 2.20 (2.28–2.20) |
| Total reflections | 231,203 | 317,128 |
| Unique reflections | 70,092 (6,937) | 49,628 (4,888) |
| Redundancy | 3.3 (3.2) | 6.4 (5.9) |
| Completeness, % | 99.7 (99.7) | 99.9 (99.7) |
| I/σ | 11.6 (1.7) | 35.4 (1.0) |
| Rsym, % | 13.2 (81.1) | 11.1 (101.5) |
| Rpim, % | 8.6 (53.1) | 4.7 (43.5) |
| CC1/2 | 0.620 (0.732) | 0.754 (0.885) |
| Refinement statistics | ||
| Resolution range used, Å | 48.1–3.15 | 49.49–2.20 |
| No. reflections used | 64,549 | 49,018 |
| Rwork/Rfree, % | 19.2/24.8 | 20.5/25.4 |
| Rmsd bond lengths, Å | 0.012 | 0.010 |
| Rmsd bond angles, ° | 1.405 | 1.302 |
| Wilson B value, Å2 | 76.9 | 41.2 |
| Number of atoms (average B, Å2) | ||
| Protein | 19,425 (88.8) | 6,312 (89.9) |
| Ligand | 0 | 24 (83.8) |
| Water | 0 | 138 (51.0) |
| Ramachandran values | ||
| Preferred regions, % | 95.1 | 97.5 |
| Allowed regions, % | 4.9 | 2.5 |
Values in parentheses are for the highest-resolution shell. . Rfree was calculated using 5% of data.
2.4. Dynamic light scattering
The particle sizes and estimated molecular weights of the proteins were analyzed using the Dynamic Light Scattering (DLS) instrument from Malvern zetasizer. 5μl of each protein at 3mg/ml in buffer (20mM Hepes pH 7.2, 150mM NaCl) was loaded in the cuvette and the DLS measurements were performed at 25 °C. The estimated MW by DLS was used to analyze the oligomeric states of each protein.
3. Results
3.1. Crystal packing forms of WT Hsp90αMC and Hsp90αMC_W320A mutant
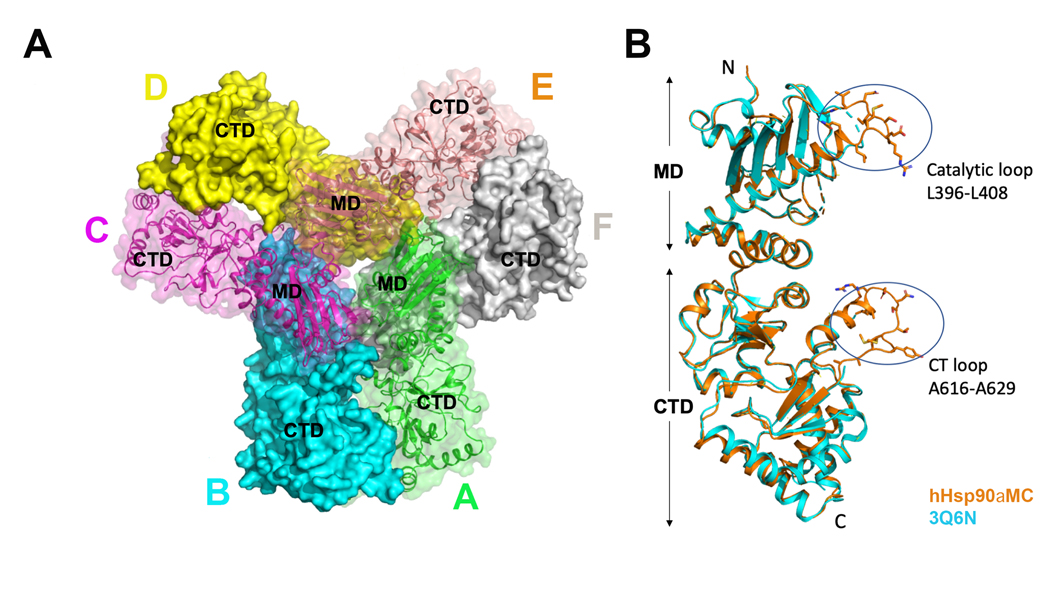
Both WT Hsp90αMC and the Hsp90αMC_W320A mutant proteins could be crystallized although under different conditions, and the structures have been determined at the resolutions of 3.15 Å and 2.20 Å respectively. They were crystallized in different crystal packing forms, with different oligomeric states in the asymmetric unit (ASU). The WT Hsp90αMC protein was crystalized in space group P21, which is the same as one of the crystal forms reported before, although the crystallization conditions are totally different (32). There are three Hsp90αMC homodimers forming a hexamer in each ASU, with 2 asymmetric units packing in the unit cell (Figures 1A, S1A). The Hsp90αMC_W320A mutant was crystalized in space group P212121, which is totally different from the crystal forms of WT Hsp90αMC. There is only one Hsp90αMC homodimer in each ASU, with four asymmetric units packing in the unit cell (Figure S1B). The interaction interfaces involved in the hexamer and dimers were analyzed with PISA server (https://www.ebi.ac.uk/pdbe/pisa/). In the hexamer, the average interface areas between molecules A-B, C-D and E-F is 1,674 Å2, that for C-B, E-D is 1,420 Å2, and for A-F is 991 Å2. In the W320A mutant dimer structure, the interface of molecules A-B is 1,288 Å2. The average accessible surface area (ASA) of residue W320 in the six molecules of WT Hsp90 hexamer structure is 190 Å2. However, in the mutant structure, the average ASA of the residue A320 is reduced to 100 Å2. When W320 was replaced by a phenylalanine, the calculated ASA of residue F320 should be 190 Å2, the same as that from W320.
Figure 1.
(A) Crystal structure of WT hHsp90αMC hexamer (chain A, green; chain B, cyan; chain C, magenta; chain D, yellow; chain E, orange; chain F, grey. (B) Structural alignment of the monomer chain E in WT hHsp90αMC (orange) with that in the previous solved structure (PDB: 3Q6N) (cyan).
The different crystal packing forms of WT Hsp90αMC and Hsp90αMC_W320A suggested that residue W320 plays a key role in Hsp90 oligomerization. Sequence alignment of Hsp90s showed that W320 is conserved in all eukaryotic nucleocytoplasmic Hsp90s although it is a phenylalanine (F) in prokaryotic Hsp90 (HTPG_E. Coli, Figure S2).
3.2. Crystal structure of WT Hsp90αMC hexamer
The hexameric structure of the WT hHsp90αMC is similar to the structures of hHsp90αMC reported before (PDB 3Q6M and PDB 3Q6N) (32). The superimposition of each protomer molecule A in WT Hsp90αMC structure with that in PDB 3Q6N structure shows a root mean square deviation (r.m.s.d.) of 0.86 Å over 393 aligned Cα atoms (Figure 1B) (39). However, the current structure differs from the previous ones mainly in two loop regions, including the catalytic loop in the MD containing Arg400 that stabilizes the closed conformation of Hsp90 required for ATP hydrolysis, and the CT loop following the C-terminal amphipathic helix. Both loops are disordered in the earlier structures while the polypeptide main chain atoms and some of the side chains in the current structure are clearly visible, with the catalytic loop traced in all of the six molecules and the CT loop completely traced in molecules B, C and E (Figure 1B, Figures S3 & S4). However, parts of the central region of the Src-loop region (349-FDLFENRKKK-358) of Hsp90 MD were disordered in human Hsp90αMC hexameric structures reported here and previously (32).
3.3. Hexameric interface of WT Hsp90αMC structure
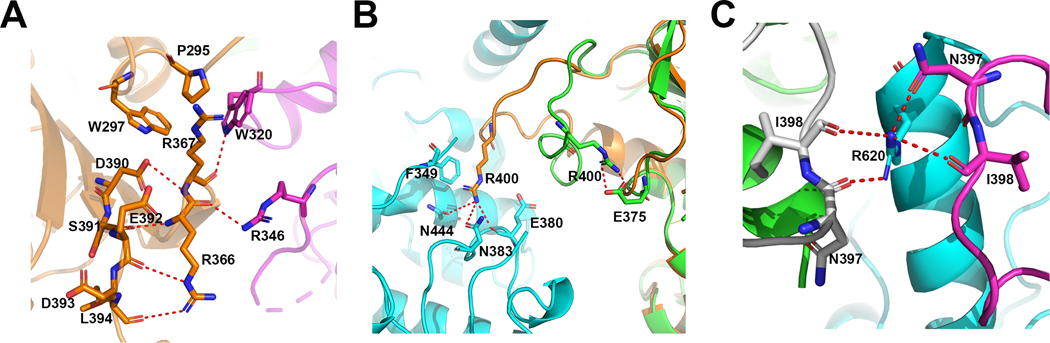
We found W320 and R346 are involved in important intermolecular interactions at the hexameric interface of WT hHsp90αMC. The side chain of W320 is in involved in strong a hydrophobic interaction with P295 in a third molecule from a neighboring dimer, which is stabilized by a π-π interaction with W297 and cation-π interaction with R367 in the third molecule, and the NE1 atom on the indole side chain of W320 forms a hydrogen bond interaction with the main chain carbonyl of R367 in the third molecule (Figure 2A). The NH2 atom on the side chain of R346 forms a hydrogen bond with the main chain carbonyl of R366 in the third molecule (Figure 2A). These molecular interactions are similar to those observed before (32). However, additional intermolecular interactions of the catalytic loop with the residues on the MD and CTD at the hexameric interface were identified in the newly solved WT hHsp90αMC structure (Table S1), which were consistent with the MDCC binding hHsp90αMC hexameric structure(40) but not observed in the previous apo WT hHsp90αMC structures (PDB ID: 3Q6M, 3Q6N). These regions are known to be important in the modulation of Hsp90 function.
Figure 2.
(A) Interactions of W320 and R346 with P295, R366 and R367 in a third molecule at the hexameric interface. (B)Conformational differences of the essential catalytic residue R400 (shown in stick) in the structures of WT hHsp90αMC (orange) and hHsp90αMC_W320A (green). (C) Interactions of R620 with residues N397 and I398 on the catalytic loops in WT hHsp90αMC hexamer structure.
The catalytic loop interacts with the MD of a third molecule in an inactive closed conformational state and R400 forms the hydrogen bonds with the main chain of E380, and the side chains of N383 and N444 on the third molecule. In addition, R400 forms cation-π interaction with F349 on the third molecule (Figure 2B). The interaction of the catalytic loop region with a third molecule further contributes to the stabilization of the hexamer interface.
Besides the contributions of W320 and R400 in the formation of the hexamer interface interactions, another key residue in the WT Hsp90αMC hexameric structure is R620, which is located in the CTD loop region and interacts with residues on the catalytic loops from two other molecules. In the WT hHsp90αMC hexamer structure, although the R620 can only be traced in one of the molecules in each homodimer block (B, C, E), it forms intermolecular hydrogen bonds to stabilize the hexamer interface, i.e. R620 in molecule B is hydrogen bonded with N397 and I398 on the catalytic loops of molecules C and F, respectively (Figure 2C).
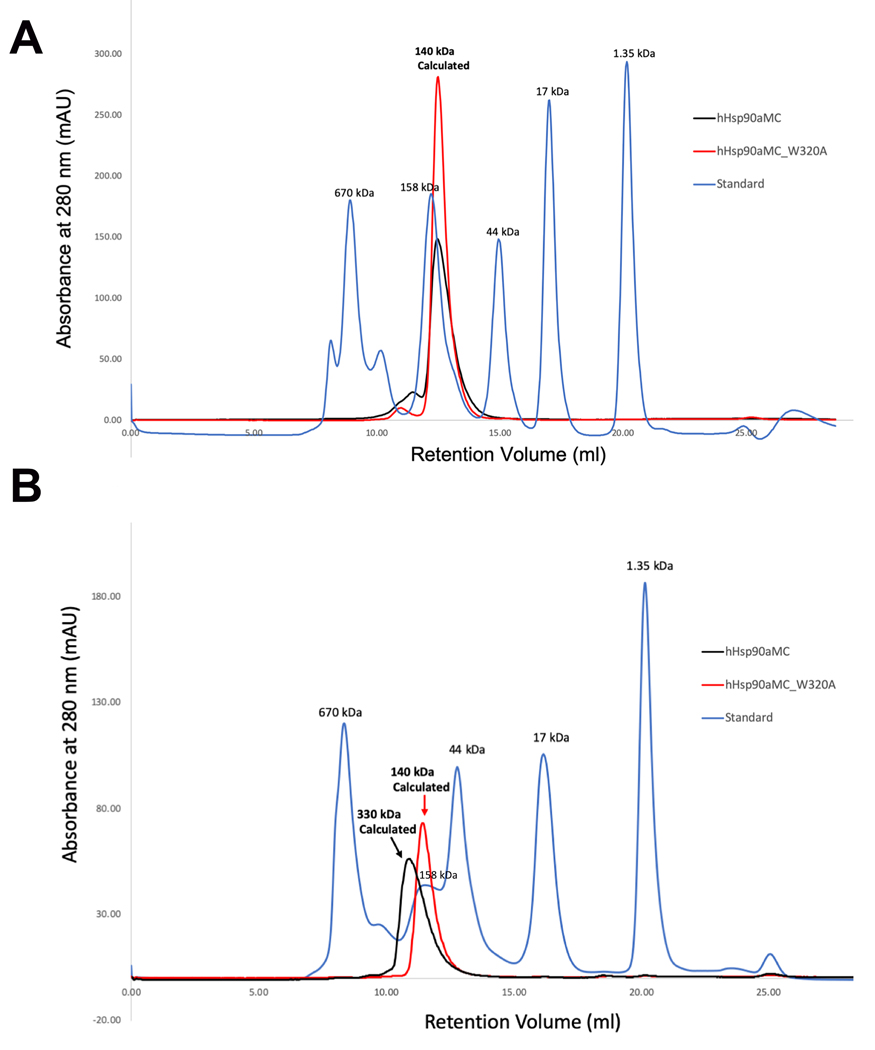
3.4. W320A mutant disrupts hexamer association in solution
W320 was mutated to alanine to evaluate the role of W320 in the hexameric association of hHsp90α. Both WT hHsp90αMC and hHsp90αMC/W320A proteins were purified initially as dimers in solution by SEC at 4°C (Figure 3A). However, the purified WT hHsp90αMC dimeric protein was observed to oligomerize over time at room temperature, at low concentrations of salt or after a freeze thaw cycle following storage at −80 °C, which is consistent to that described before (32). The thawed frozen hHsp90αMC sample from −80 °C displayed a heterogeneous/polydisperse oligomer from SEC (hexamer or tetramer) (Figure 3B). In contrast, hHsp90αMC/W320A protein continued to elute as a dimer over time by SEC (Figure 3B). The oligomeric states of protein samples by SEC results are consistent with the results of DLS experiments (Table S2).
Figure 3.
Size-exclusion Chromatographs for WT Hsp90⍺MC and Hsp90αMC_W320A mutant proteins. (A). Chromatographs from fresh protein samples. The MWs are indicated for the protein peaks. The theoretic MW of hHsp90αMC is about 50kDa. Due to its elongated shape, the apparent MW is calculated as 67kDa in solution (www.fluidic.com) based on its hydrodynamic radius of 35 Å calculated from the current crystal structure(58). The estimated MW of the protein from the retention volume is about 140 kDa, representing a dimer in solution. (B).Chromatographs from the thawed protein samples after frozen at −80 oC. The retention volumes as well as the estimated MWs are indicated for the protein peaks. Note WT protein oligomerized while W320A mutant protein retained as a dimer in solution.
3.5. Crystal structure of Hsp90αMC_W320A dimer
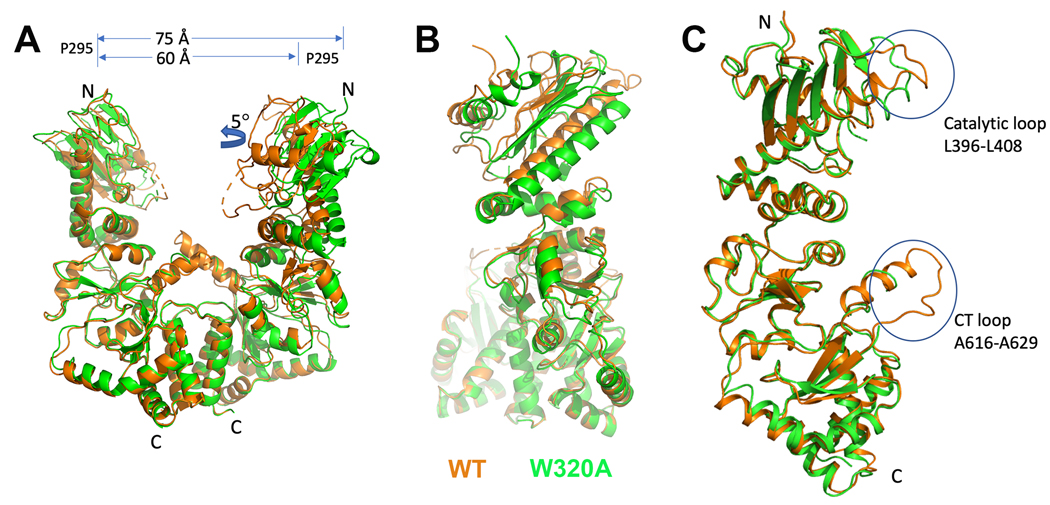
The Hsp90αMC_W320A crystallized as a dimer rather than a hexamer, displaying a large conformational change compared to the WT hHsp90αMC hexameric structures (Figures 4 & S1B). When the hHsp90αMC_W320A mutant dimeric structure is compared with the dimeric blocks from the WT hHsp90αMC hexameric structure, it displayed a r.m.s.d. of 0.75 Å over 768 aligned Cα atoms (Figure 4A) (39). However, the W320A mutant structure displayed a more open conformation when compared with the block dimers in the hexamer. The distance between two P295 residues in each homodimer is about 60 Å in the hexameric structure but 75 Å in the dimeric W320A mutant structure (Figure 4). In addition, the MD of one protomer of the mutant dimer displayed a twist of about 5°. This new structure has not been previously observed, and precludes the interactions required for the formation of the hexamer.
Figure 4.
(A) Structural comparison of the homodimers of W320A vs WT hHsp90αMC (orange: WT hHsp90αMC; green, W320A mutant dimer). (B) Structural alignment of the monomer in WT hHsp90αMC (orange) and that in the W320A mutant (green) structures.
Similar to the WT hexamer structure, the catalytic loop region can also be traced in the mutant dimeric structure, albeit displaying different conformations (Figures 4C, S5, S6). However, the CT loop region is disordered in the mutant dimeric structure, while the main chain can be traced in molecules B, C and E of the WT Hsp90αMC hexameric structure (Figures 4C, S4).
3.6. W320A mutant results in conformational changes of the catalytic loop
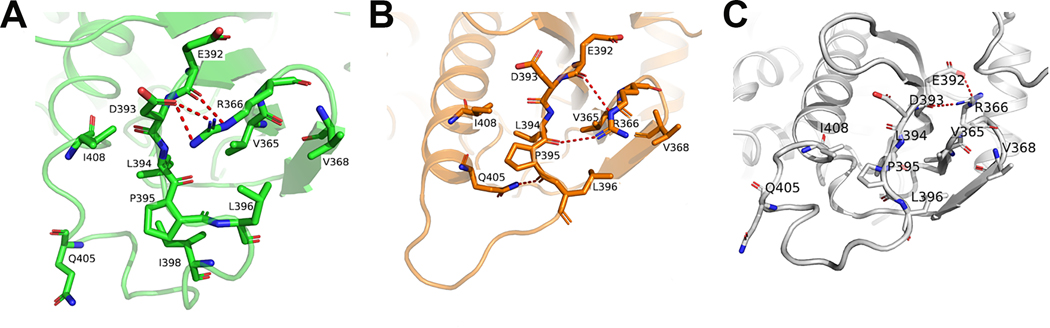
When comparing each monomer in the W320A mutant dimeric structure with that in the WT hexameric structure, we found that the W320A mutation lead to the loss of its hydrophobic interaction with P295, and the hydrogen bond and cation-π interactions with R367 as in the WT hexameric structure. In addition, the hydrogen bond interaction of R366 with R346 (Figure 2A) is also lost. The loss of these intermolecular interactions led to the disruption of hexameric interface and caused further conformational changes of the MD, in particular the catalytic loop region (Figures 2B, 5A, S3, S5). Specifically, R366 forms a salt bridge with the side chain of D393 within the same molecule (Figure 5A), while in the WT hexameric structure it forms a hydrogen bond with the main chain carbonyl group of L394 located in the catalytic loop (Figure 5B). The salt bridge between D393-R366 in the W320A mutant dimeric structure induces conformational changes of D393 and L394, which further leads to the conformational changes of P395 and L396 in the catalytic loop. L396 forms hydrophobic interactions with V365 and V368, which do not exist in the hexametric structure (Figure 5A). In four of the main chains (chains B, C, D & E) of the WT Hsp90αMC hexamer structure, P395 forms a hydrogen bond with the side chain of Q405, while its side chain forms hydrophobic interactions with the side chain of I408 (Figures 5B, S6A). In the other two main chains (chains A & F) of WT Hsp90αMC hexamer structure, P395 forms hydrophobic interactions with the side chain of I408 but does not hydrogen bond with the side chain of Q405 (Figures 5C, S6A). On the other hand, in the Hsp90αMC_W320 mutant, the H-bond with Q405 is not present and Pro395 is sandwiched by hydrophobic interactions with L394, L396, and I398 (Figures 5A, S6B). The structural comparisons indicate that Q405 plays an important role for a transition from coil to helix in the catalytic loop, which is stabilized by hydrogen bonding via its side chain (Figures 5, S6).
Figure 5.
Various conformations of the catalytic loops. (A) W320A mutant (green), (B) chain E of WT hHsp90αMC (orange), (C) chain F of WT hHsp90αMC (grey).
The conformation of the catalytic loop region in hHsp90αMD is flexible, which allows it to adopt multiple conformations, and as mentioned previously, the electron densities for this region often could not be assigned. With ATP binding, the catalytic residue R400 (R380 in yeast Hsp82) displays the open active state and forms the hydrogen bond with the γ-phosphate ATP in the NT domain, and without ATP binding it is held in an inactive state (10,41). Structural alignment of each protomer in the wildtype hexamer and W320A mutant dimer showed that they both displayed the inactive closed state with different conformations (Figure 4C). In the hHsp90αMC_W320A mutant dimer structure, the conformation of the catalytic loop is semi-closed, and R400 forms the hydrogen bond with E375 in the same molecule (Figure 2B). However, in the WT hexamer structure, the catalytic loop displays an inactive closed conformation by interacting with the MD of a third molecule (Figure 2B), with loops showing two somewhat different conformations due altered intramolecular H-bonding patterns. Therefore, W320 is important for protein oligomerization by both direct interaction with a third molecule and inducing the conformational change of the catalytic loop responsible for hexameric association.
4. Discussion
In this study, we report two structures of hHsp90αMC, the WT and the W320A mutant. We found W320A mutation disrupted the hexameric association of Hsp90 and displayed a dimeric structure instead, due to the loss of molecular interactions from the side chain of W320 when replaced with alanine. The mutant dimer structure displays a large conformational change in the MD and its catalytic loop region, and adopts a more open conformation when compared to the hexameric WT structure. Our structural comparison provides insights into how conformational changes of Hsp90 associated with W320 in the MD could play a critical role in the allosteric regulation of its biological function.
We found that residues W320 of Hsp90α is important for MC domain to oligomerize and form a hexamer, which is consistent with a previous report (Figure 2A) (32). The structural data show that W320 plays an important role in tethering the hexamer interface, by forming exquisite intermolecular interactions through cation-π and hydrogen bond interactions (Figure 2A). The W320A mutation abolished these intermolecular interactions at the hexamer interface, resulting in the disruption of the hexamer via inducing a more opened conformation and a large conformational change of the catalytic loop.
Our structural observation shows that W320 as well as other residues including P295, R366 and R367 are involved in hexameric association of WT Hsp90. These residues were also observed previously in various conformations of Hsp90 and when in complex with either client protein or cochaperones. Collectively, our data and those from others suggest that these residues could play important roles in the regulation of Hsp90 conformational changes associated with its oligomerization, client and cochaperone binding and therefore its cellular function. W320 is highly conserved in both eukaryotic nucleocytoplasmic Hsp90s, although it is a F257 in E. coli HtpG (Figure S2). P295 and R366 are conserved in eukaryotic nucleocytoplasmic Hsp90s and R367 is conserved in both eukaryotic nucleocytoplasmic Hsp90s and E. Coli HptG. W320 plays an important role in client protein interaction-induced conformational changes via a cation-π interaction (22). W320 is located on a loop that is sandwiched by an α-helix and another loop, each containing a conserved basic amino acid, K314 and R346 respectively in the eukaryotic nucleocytoplasmic Hsp90s (Figure S7). In the closed state structure of homodimeric Hsp90 full length protein bound with AMPPNP and complexed with p23 (PDB ID: 7L7J), W320 is tethered to the MD through intramolecular hydrogen bond interaction with K314 and cation-π interaction with R346 (42).
Apo-Hsp90 dimers exist in an equilibrium predominated by two open states, with one conformation being more extended that the other (19,43,44). Hsp90 adopts multiple conformations when transitioning between open, semi-closed and closed states during its functional chaperone cycle driven by its interactions with co-chaperones, client proteins and the binding and hydrolysis of ATP. Molecular dynamic studies of new cryo-EM structures implicate the catalytic loop region, W320, F395 in the src-loop in the MD, and the amphipathic helix and its following loop in CTD as critical regions involved in the allosteric communications that occur between all three domains during Hsp90s reaction cycle(45).
Interaction interfaces and the effect of co-chaperones and clients on Hsp90 structure have been studied using a variety of biophysical techniques including SAXS, NMR, FRET, PET, X-ray crystallography and cryo-EM (reviewed in (46,47)). When analyzing the complex structures of Hsp90 with co-chaperones reported by Agard’s group, W320 was found to be essential for binding Cdc37 and Aha1 (48,49). In the Hsp90-Cdc37-Cdk4 structure (PDB ID: 5FWL), Hsp90β W312 (W320 in Hsp90α) forms the cation-π interaction and hydrogen bond with residue R231 in the middle domain of Cdc37 (48) (Figure S8A). It is noteworthy that the W300 mutation in Hsp82 in yeast decreases its ability to fold a stabilize v-src (20). In the apo structure of yeast Hsc82 bound to Aha1 (PDB ID: 6XLB), the NT-domain of Hsc82 is undocked from the middle domain and the CTD of Aha1 is bound to F328 (F352 of Hsp90α) in a semi-closed conformation. In the yeast Hsc82-Aha1CTD-AMPPNP structure (PDB ID: 6XLE), both W296 (W300 in yeast Hsp82, W320 in human Hsp90α) and R322 (R326 in yeast Hsp82, R346 in human Hsp90α) are involved in association with the C-terminal domain of Aha1(49) (Figure S8B). Specifically, W296 forms π-π interactions with Y335 and W285, and hydrogen bonds with N331 and N267, while R322 forms a cation-π interaction with Y335 on the CT domain of Aha1. In contrast, both W320 and R346 are involved in intermolecular interactions at the hexamer interface of WT Hsp90αMC structure. Overlay of the cryo-EM structures and the hHsp90αMC hexamer indicates that these interaction interfaces are buried in the hexameric structure, and structure is incompatible with the binding of either of these co-chaperones.
In the closed homodimer structure of the full length (FL) Hsp90 bound with p23 (PDB ID: 7L7J) R400 coordinates the γ-phosphate of AMPNP, while P295, R366 and R367 are located at the interface between the MD and the NTD (Figure S9A) (42), indicating that the hexameric structure of hHsp90αMC is incompatible with the binding of p23. A recent study using FRET measurements and molecular dynamic simulations suggests that upon ATP hydrolysis, R380 (R400 in human) becomes completely detached from the nucleotide. This converts Hsp90 from the closed state A to the closed state B, with structural conformational changes transferred via the central long MD α-helix (50), suggesting that changes in the conformation of the catalytic loop play an important role in the allosteric communication between the three domains of Hsp90.
When overlaying the dimeric unit of WT Hsp90αMC structure with the newly available cryo-EM structures of Hsp90 in complex with co-chaperones and client, the best matching structure was that of the Hsp90-Hsp70-HOP-GR loading intermediate before dimerization of Hsp90s NTD (7KW7, r.m.s.d= 1.58Å over 688 amino acids). While the catalytic loops of the two structures have somewhat similar conformations, it should be noted that the Hsp90-Hsp70-HOP-GR loading intermediate is in a more compact semi-closed conformation(51). Overlay of the two structures indicates that only the interface of Hsp90 that interacts with TRP2A of HOP is accessible and does not clash with the hexameric structure of Hsp90αMC. However, the overlay indicates that the structure of the TPR2B domain that is adopted in the Hsp90-Hsp70-HOP-GR loading intermediate has major steric clashes, and the DP2 binding interface is buried in the hexameric structure of the Hsp90αMC (not shown). As WT Hsp90αMC was initially purified as a dimer, these observations suggest that the current structure represents a more opened structural intermediate prior to client loading or a precursor to an inactive hexamer in the absence of an abundance of Hsp90 clientele (32,52). In regards to the latter, the work from both Kamal and coworkers (53) and Rodina and coworkers (54) suggests that in non-transformed cells the bulk of Hsp90 exists in state with a paucity of interactions with its co-chaperones.
Of interest is that a similar overlay of the Hsp90-Hsp70-HOP-GR loading intermediate with the Hsp90αMC_W320A dimer structure indicates that the mutant appears to be fully capable of binding the TPR2A-TPR2B_DP2 domain construct of HOP (Figure S10). In addition, amino acid residues previously implicated in client binding are all solvent exposed and accessible(46,47). These observations suggest that the conformation of the W320A mutant dimer may represent that which precedes HOP mediated client binding.
Lopez and coworkers have recently demonstrated that the shift from the open to closed state are allosterically transmitted across all three domains of Hsp90s, with the conserved R380 and R32 (R400 and R46 in human) residues playing a central role in the rotation of Hsp90s NTD that is required in the open state allowing its dimerization upon nucleotide binding (55). Relevant to these observations, in the open state structures of FL and an NM-domain construct of apo-HtpG (PDB 2IOQ, 1Y4U), R303 (R367 in human Hsp90) is totally exposed (Figure S9B) (56). In the hexameric WT hHsp90αMC structure, R367 forms intermolecular cation-π interaction with W320 at the hexamer interface, while in the W320A mutant structure, it is solvent exposed. In addition, in the open state of the Hsp90 homologue HtpG, the NTD and MD are tethered together by an important intramolecular interaction between R33 (R32 in yeast Hsp82 and R46 in hHsp90α) and F257 via cation-π interactions (Figure S11A). It was previously shown that R32 in yeast Hsp82 plays a critical role in stabilizing the catalytic center for ATP hydrolysis in the closed state (57), suggesting this R33/F257 cation-π interaction should be removed to allow conformational changes of NTD in order to carry out catalysis. Superimposition of the Hsp90αMC_W320A structure with the 2IOQ and 1Y4U HtpG structures indicates that R33 is in position to form a cation-π interaction as A320 and F257 overlap (Figure S7B). Mutation of A320 back to tryptophan in the Hsp90αMC_W320A structure using PyMol indicates that the orientation of the W320 residue fills the same pocket as F257 and can form a similar cation-π interaction with R33 (Figure S11B). In addition, the position of R336 in the two HtpG structures and R400 in the W320 mutant are similarly placed, with amino acids substitutions in the catalytic loop possibly accounting for the somewhat different structures of the loops (Figure S11C). However, in a similar overlay of the WT hHsp90αMC dimeric unit with the apo-HtpG structure, all rotamers of W320 showed substantial steric clashes with amino acid residues in the pocket that accommodates F257, and the structure of the catalytic loops show no similarity (not shown). These observations suggest that the conformational change displayed by the hHsp90αMC_W320A mutant is representing the unique open state of human Hsp90MC and is likely physiologically relevant. It is expected in the FL hHsp90 that the W320A mutation would abolish its cation-π interaction with R46 and offer greater flexibility of NTD to change its conformation to the closed state for ATP binding and catalysis. Therefore, our study suggests a mechanism of conformational switch by W320 in regulating Hsp90 intramolecular and intermolecular interactions for its chaperone functions. In support of this conjecture, previous work on characterizing the effect of mutating W300 to Ala in full length yeast Hsp82 demonstrated that the W300A mutant had a more closed compact conformation in the presence of ATP (21), a higher ATPase activity than wildtype Hsp82 (20,22), and a decrease in Aha1 stimulated ATP hydrolysis (48). The Hsp82 W300 mutation also exhibited a differential ability to affect the maturation of two of its major clients: the mutant demonstrated a marked decreased ability to chaperone the maturation of v-src at 30°C, but not the glucocorticoid receptor(20,21). While these experiments have been found to be experimentally tractable using the power of the yeast genetic system, they have yet to be carried out with the human Hsp90α or -β isoforms.
In general, conformational changes of the catalytic loop is essential for Hsp90 activity (10). Our study found that in addition to performing the function of ATP hydrolysis, the catalytic loop is also important for Hsp90 assembly to form a hexamer by interacting with residues in MD and CTD domain in other Hsp90 molecules. Mutation of W320 will disrupt the hexamerization by further regulating the conformational change of the catalytic loop in dimeric Hsp90, which is a previous unobserved phenomenon. W320 is important for intramolecular interactions with NTD, intermolecular interactions with cochaperone and client proteins, as well as forming the hexmeric interface in Hsp90 hexamer. Therefore, our research result provided the basic structural information on W320 of Hsp90 to act as a key switch point in allosterically regulating its intramolecular and intermolecular interactions of key structural regions required for chaperone functions.
Supplementary Material
Acknowledgements
We gratefully acknowledge the staff of beam-line 19ID at the Advanced Photon Source for their support. This work is supported by NIH grant R15CA219907 (R.L.M., J.D.) and Oklahoma Agricultural Experiment Station at Oklahoma State University under project OKL03060 (J.D.) and OKL03159 (R.L.M.). J.D. is additionally supported by NIH AI149295. The research is also supported in part by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health (Award P20GM103640).
Footnotes
The authors declare no competing interest.
References
- 1.Schopf FH, Biebl MM, and Buchner J. 2017. The HSP90 chaperone machinery. Nat Rev Mol Cell Biol. 18(6):345–360, doi: 10.1038/nrm.2017.20, https://www.ncbi.nlm.nih.gov/pubmed/28429788. [DOI] [PubMed] [Google Scholar]
- 2.Sima S, and Richter K. 2018. Regulation of the Hsp90 system. Biochim Biophys Acta Mol Cell Res. 1865(6):889–897, doi: 10.1016/j.bbamcr.2018.03.008, https://www.ncbi.nlm.nih.gov/pubmed/29563055. [DOI] [PubMed] [Google Scholar]
- 3.Pearl LH 2016. Review: The HSP90 molecular chaperone-an enigmatic ATPase. Biopolymers. 105(8):594–607, doi: 10.1002/bip.22835, https://www.ncbi.nlm.nih.gov/pubmed/26991466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ou JR, Tan MS, Xie AM, Yu JT, and Tan L. 2014. Heat shock protein 90 in Alzheimer’s disease. Biomed ResInt. 2014:796869, doi: 10.1155/2014/796869, https://www.ncbi.nlm.nih.gov/pubmed/25374890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birbo B, Madu EE, Madu CO, Jain A, and Lu Y. 2021. Role of HSP90 in Cancer. Int J Mol Sci. 22(19), doi: 10.3390/ijms221910317, https://www.ncbi.nlm.nih.gov/pubmed/34638658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wan Q, Song D, Li H, and He ML. 2020. Stress proteins: the biological functions in virus infection, present and challenges for target-based antiviral drug development. Signal Transduct Target Ther. 5(1):125, doi: 10.1038/s41392-020-00233-4, https://www.ncbi.nlm.nih.gov/pubmed/32661235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biebl MM, and Buchner J. 2019. Structure, Function, and Regulation of the Hsp90 Machinery. Cold Spring Harb Perspect Biol. 11(9), doi: 10.1101/cshperspect.a034017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoter A, El-Sabban ME, and Naim HY. 2018. The HSP90 Family: Structure, Regulation, Function, and Implications in Health and Disease. Int J Mol Sci. 19(9), doi: 10.3390/ijms19092560, https://www.ncbi.nlm.nih.gov/pubmed/30158430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Soroka J, and Buchner J. 2012. The Hsp90 chaperone machinery: conformational dynamics and regulation by co-chaperones. Biochim Biophys Acta. 1823(3):624–635, doi: 10.1016/j.bbamcr.2011.09.003, https://www.ncbi.nlm.nih.gov/pubmed/21951723. [DOI] [PubMed] [Google Scholar]
- 10.Prodromou C 2016. Mechanisms of Hsp90 regulation. Biochem J. 473(16):2439–2452, doi: 10.1042/BCJ20160005, https://www.ncbi.nlm.nih.gov/pubmed/27515256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Backe SJ, Sager RA, Woodford MR, Makedon AM, and Mollapour M. 2020. Post-translational modifications of Hsp90 and translating the chaperone code. J Biol Chem. 295(32):11099–11117, doi: 10.1074/jbc.REV120.011833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nemoto TK, Ono T, and Tanaka K. 2001. Substrate-binding characteristics of proteins in the 90 kDa heat shock protein family. Biochem J. 354(Pt 3):663–670, doi: 10.1042/0264-6021:3540663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yonehara M, Minami Y, Kawata Y, Nagai J, and Yahara I. 1996. Heat-induced chaperone activity of HSP90. J Biol Chem. 271(5):2641–2645, doi: 10.1074/jbc.271.5.2641, https://www.ncbi.nlm.nih.gov/pubmed/8576234. [DOI] [PubMed] [Google Scholar]
- 14.Moullintraffort L, Bruneaux M, Nazabal A, Allegro D, Giudice E, Zal F, Peyrot V, Barbier P, Thomas D, and Garnier C. 2010. Biochemical and biophysical characterization of the Mg2+-induced 90-kDa heat shock protein oligomers. J Biol Chem. 285(20):15100–15110, doi: 10.1074/jbc.M109.094698, https://www.ncbi.nlm.nih.gov/pubmed/20228408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nemoto T, and Sato N. 1998. Oligomeric forms of the 90-kDa heat shock protein. Biochem J. 330 ( Pt 2):989–995, doi: 10.1042/bj3300989, https://www.ncbi.nlm.nih.gov/pubmed/9480920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jakob U, Meyer I, Bugl H, Andre S, Bardwell JC, and Buchner J. 1995. Structural organization of procaryotic and eucaryotic Hsp90. Influence of divalent cations on structure and function. J Biol Chem. 270(24):14412–14419, doi: 10.1074/jbc.270.24.14412, https://www.ncbi.nlm.nih.gov/pubmed/7782303. [DOI] [PubMed] [Google Scholar]
- 17.Lepvrier E, Moullintraffort L, Nigen M, Goude R, Allegro D, Barbier P, Peyrot V, Thomas D, Nazabal A, and Garnier C. 2015. Hsp90 Oligomers Interacting with the Aha1 Cochaperone: An Outlook for the Hsp90 Chaperone Machineries. Anal Chem. 87(14):7043–7051, doi: 10.1021/acs.analchem.5b00051, https://www.ncbi.nlm.nih.gov/pubmed/26076190. [DOI] [PubMed] [Google Scholar]
- 18.Lepvrier E, Nigen M, Moullintraffort L, Chat S, Allegro D, Barbier P, Thomas D, Nazabal A, and Garnier C. 2015. Hsp90 oligomerization process: How can p23 drive the chaperone machineries? Biochim Biophys Acta. 1854(10 Pt A):1412–1424, doi: 10.1016/j.bbapap.2015.07.003, https://www.ncbi.nlm.nih.gov/pubmed/26151834. [DOI] [PubMed] [Google Scholar]
- 19.Krukenberg KA, Förster F, Rice LM, Sali A, and Agard DA. 2008. Multiple conformations of E. coli Hsp90 in solution: insights into the conformational dynamics of Hsp90. Structure. 16(5):755–765, doi: 10.1016/j.str.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer P, Prodromou C, Hu B, Vaughan C, Roe SM, Panaretou B, Piper PW, and Pearl LH. 2003. Structural and functional analysis of the middle segment of hsp90: implications for ATP hydrolysis and client protein and cochaperone interactions. Mol Cell. 11(3):647–658, doi: 10.1016/s1097-2765(03)00065-0, https://www.ncbi.nlm.nih.gov/pubmed/12667448. [DOI] [PubMed] [Google Scholar]
- 21.Hawle P, Siepmann M, Harst A, Siderius M, Reusch HP, and Obermann WM. 2006. The middle domain of Hsp90 acts as a discriminator between different types of client proteins. Mol Cell Biol. 26(22):8385–8395, doi: 10.1128/MCB.02188-05, https://www.ncbi.nlm.nih.gov/pubmed/16982694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rutz DA, Luo Q, Freiburger L, Madl T, Kaila VRI, Sattler M, and Buchner J. 2018. A switch point in the molecular chaperone Hsp90 responding to client interaction. Nat Commun. 9(1):1472, doi: 10.1038/s41467-018-03946-x, https://www.ncbi.nlm.nih.gov/pubmed/29662162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khandelwal A, Crowley VM, and Blagg BSJ. 2016. Natural Product Inspired N-Terminal Hsp90 Inhibitors: From Bench to Bedside? Med Res Rev. 36(1):92–118, doi: 10.1002/med.21351, https://www.ncbi.nlm.nih.gov/pubmed/26010985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trepel J, Mollapour M, Giaccone G, and Neckers L. 2010. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 10(8):537–549, doi: 10.1038/nrc2887, https://www.ncbi.nlm.nih.gov/pubmed/20651736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neckers L, Blagg B, Haystead T, Trepel JB, Whitesell L, and Picard D. 2018. Methods to validate Hsp90 inhibitor specificity, to identify off-target effects, and to rethink approaches for further clinical development. Cell Stress Chaperones. 23(4):467–482, doi: 10.1007/s12192-018-0877-2, https://www.ncbi.nlm.nih.gov/pubmed/29392504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong DS, Banerji U, Tavana B, George GC, Aaron J, and Kurzrock R. 2013. Targeting the molecular chaperone heat shock protein 90 (HSP90): lessons learned and future directions. Cancer Treat Rev. 39(4):375–387, doi: 10.1016/j.ctrv.2012.10.001, https://www.ncbi.nlm.nih.gov/pubmed/23199899. [DOI] [PubMed] [Google Scholar]
- 27.Mielczarek-Lewandowska A, Hartman ML, and Czyz M. 2020. Inhibitors of HSP90 in melanoma. Apoptosis. 25(1–2):12–28, doi: 10.1007/s10495-019-01577-1, https://www.ncbi.nlm.nih.gov/pubmed/31659567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goode KM, Petrov DP, Vickman RE, Crist SA, Pascuzzi PE, Ratliff TL, Davisson VJ, and Hazbun TR. 2017. Targeting the Hsp90 C-terminal domain to induce allosteric inhibition and selective client downregulation. Biochim Biophys Acta Gen Subj. 1861(8):1992–2006, doi: 10.1016/j.bbagen.2017.05.006, https://www.ncbi.nlm.nih.gov/pubmed/28495207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eskew JD, Sadikot T, Morales P, Duren A, Dunwiddie I, Swink M, Zhang X, Hembruff S, Donnelly A, Rajewski RA, Blagg BS, Manjarrez JR, Matts RL, Holzbeierlein JM, and Vielhauer GA. 2011. Development and characterization of a novel C-terminal inhibitor of Hsp90 in androgen dependent and independent prostate cancer cells. BMC Cancer. 11:468, doi: 10.1186/1471-2407-11-468, https://www.ncbi.nlm.nih.gov/pubmed/22039910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burlison JA, Avila C, Vielhauer G, Lubbers DJ, Holzbeierlein J, and Blagg BS. 2008. Development of novobiocin analogues that manifest anti-proliferative activity against several cancer cell lines. J Org Chem. 73(6):2130–2137, doi: 10.1021/jo702191a, https://www.ncbi.nlm.nih.gov/pubmed/18293999. [DOI] [PubMed] [Google Scholar]
- 31.Armstrong HK, Koay YC, Irani S, Das R, Nassar ZD, Australian Prostate Cancer B, Selth LA, Centenera MM, McAlpine SR, and Butler LM. 2016. A Novel Class of Hsp90 C-Terminal Modulators Have Pre-Clinical Efficacy in Prostate Tumor Cells Without Induction of a Heat Shock Response. Prostate. 76(16):1546–1559, doi: 10.1002/pros.23239, https://www.ncbi.nlm.nih.gov/pubmed/27526951. [DOI] [PubMed] [Google Scholar]
- 32.Lee CC, Lin TW, Ko TP, and Wang AH. 2011. The hexameric structures of human heat shock protein 90. PLoS One. 6(5):e19961, doi: 10.1371/journal.pone.0019961, https://www.ncbi.nlm.nih.gov/pubmed/21647436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minor W, Cymborowski M, Otwinowski Z, and Chruszcz M. 2006. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 62(Pt 8):859–866, doi: 10.1107/S0907444906019949, https://www.ncbi.nlm.nih.gov/pubmed/16855301. [DOI] [PubMed] [Google Scholar]
- 34.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr. 40(Pt 4):658–674, doi: 10.1107/S0021889807021206, https://www.ncbi.nlm.nih.gov/pubmed/19461840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, and Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 66(Pt 2):213–221, doi: 10.1107/S0907444909052925, https://www.ncbi.nlm.nih.gov/pubmed/20124702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Painter J, and Merritt EA. 2006. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 62(Pt 4):439–450, doi: 10.1107/S0907444906005270, https://www.ncbi.nlm.nih.gov/pubmed/16552146. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, and Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 60(Pt 12 Pt 1):2126–2132, doi: 10.1107/S0907444904019158, https://www.ncbi.nlm.nih.gov/pubmed/15572765. [DOI] [PubMed] [Google Scholar]
- 38.Schrodinger L The PyMOL Molecular Graphics System, Version 2.1. [Google Scholar]
- 39.Krissinel E, and Henrick K. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 60(Pt 12 Pt 1):2256–2268, doi: 10.1107/S0907444904026460, https://www.ncbi.nlm.nih.gov/pubmed/15572779. [DOI] [PubMed] [Google Scholar]
- 40.Peng S, Woodruff J, Pathak PK, Matts RL, and Deng J. 2022. Crystal structure of the middle and C-terminal domains of Hsp90α labeled with a coumarin derivative reveals a potential allosteric binding site as a drug target. Acta Crystallogr D Struct Biol. 78(Pt 5):571–585, doi: 10.1107/s2059798322002261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, and Pearl LH. 2006. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 440(7087):1013–1017, doi: 10.1038/nature04716, https://www.ncbi.nlm.nih.gov/pubmed/16625188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee K, Thwin AC, Nadel CM, Tse E, Gates SN, Gestwicki JE, and Southworth DR. 2021. The structure of an Hsp90-immunophilin complex reveals cochaperone recognition of the client maturation state. Mol Cell. 81(17):3496–3508 e3495, doi: 10.1016/j.molcel.2021.07.023, https://www.ncbi.nlm.nih.gov/pubmed/34380015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bron P, Giudice E, Rolland JP, Buey RM, Barbier P, Díaz JF, Peyrot V, Thomas D, and Garnier C. 2008. Apo-Hsp90 coexists in two open conformational states in solution. Biol Cell. 100(7):413–425, doi: 10.1042/bc20070149. [DOI] [PubMed] [Google Scholar]
- 44.Southworth DR, and Agard DA. 2008. Species-dependent ensembles of conserved conformational states define the Hsp90 chaperone ATPase cycle. Mol Cell. 32(5):631–640, doi: 10.1016/j.molcel.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verkhivker GM 2022. Exploring Mechanisms of Allosteric Regulation and Communication Switching in the Multiprotein Regulatory Complexes of the Hsp90 Chaperone with Cochaperones and Client Proteins : Atomistic Insights from Integrative Biophysical Modeling and Network Analysis of Conformational Landscapes. J Mol Biol.167506, doi: 10.1016/j.jmb.2022.167506. [DOI] [PubMed] [Google Scholar]
- 46.Didenko T, Duarte AM, Karagöz GE, and Rüdiger SG. 2012. Hsp90 structure and function studied by NMR spectroscopy. Biochim Biophys Acta. 1823(3):636–647, doi: 10.1016/j.bbamcr.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 47.Prodromou C, and Bjorklund DM. 2022. Advances towards Understanding the Mechanism of Action of the Hsp90 Complex. Biomolecules. 12(5), doi: 10.3390/biom12050600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verba KA, Wang RY, Arakawa A, Liu Y, Shirouzu M, Yokoyama S, and Agard DA. 2016. Atomic structure of Hsp90-Cdc37-Cdk4 reveals that Hsp90 traps and stabilizes an unfolded kinase. Science. 352(6293):1542–1547, doi: 10.1126/science.aaf5023, https://www.ncbi.nlm.nih.gov/pubmed/27339980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y, Sun M, Myasnikov AG, Elnatan D, Delaeter N, Nguyenquang M, and Agard DA. 2020. Cryo-EM structures reveal a multistep mechanism of Hsp90 activation by co-chaperone Aha1. bioRxiv.2020.2006.2030.180695, doi: 10.1101/2020.06.30.180695, https://www.biorxiv.org/content/biorxiv/early/2020/07/01/2020.06.30.180695.full.pdf. [DOI] [Google Scholar]
- 50.Wolf S, Sohmen B, Hellenkamp B, Thurn J, Stock G, and Hugel T. 2021. Hierarchical dynamics in allostery following ATP hydrolysis monitored by single molecule FRET measurements and MD simulations. Chem Sci. 12(9):3350–3359, doi: 10.1039/d0sc06134d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang RY, Noddings CM, Kirschke E, Myasnikov AG, Johnson JL, and Agard DA. 2022. Structure of Hsp90-Hsp70-Hop-GR reveals the Hsp90 client-loading mechanism. Nature. 601(7893):460–464, doi: 10.1038/s41586-021-04252-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lepvrier E, Thomas D, and Garnier C. 2018. Hsp90 Quaternary Structures and the Chaperone Cycle: Highly Flexible Dimeric and Oligomeric Structures and Their Regulation by Co-Chaperones. Current Proteomics. 16(1):5–11, doi: 10.2174/1570164615666180522095147. [DOI] [Google Scholar]
- 53.Kamal A, Thao L, Sensintaffar J, Zhang L, Boehm MF, Fritz LC, and Burrows FJ. 2003. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature. 425(6956):407–410, doi: 10.1038/nature01913, https://www.ncbi.nlm.nih.gov/pubmed/14508491. [DOI] [PubMed] [Google Scholar]
- 54.Rodina A, Wang T, Yan P, Gomes ED, Dunphy MP, Pillarsetty N, Koren J, Gerecitano JF, Taldone T, Zong H, Caldas-Lopes E, Alpaugh M, Corben A, Riolo M, Beattie B, Pressl C, Peter RI, Xu C, Trondl R, Patel HJ, Shimizu F, Bolaender A, Yang C, Panchal P, Farooq MF, Kishinevsky S, Modi S, Lin O, Chu F, Patil S, Erdjument-Bromage H, Zanzonico P, Hudis C, Studer L, Roboz GJ, Cesarman E, Cerchietti L, Levine R, Melnick A, Larson SM, Lewis JS, Guzman ML, and Chiosis G. 2016. The epichaperome is an integrated chaperome network that facilitates tumour survival. Nature. 538(7625):397–401, doi: 10.1038/nature19807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lopez A, Dahiya V, Delhommel F, Freiburger L, Stehle R, Asami S, Rutz D, Blair L, Buchner J, and Sattler M. 2021. Client binding shifts the populations of dynamic Hsp90 conformations through an allosteric network. Sci Adv. 7(51):eabl7295, doi: 10.1126/sciadv.abl7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shiau AK, Harris SF, Southworth DR, and Agard DA. 2006. Structural Analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell. 127(2):329–340, doi: 10.1016/j.cell.2006.09.027, https://www.ncbi.nlm.nih.gov/pubmed/17055434. [DOI] [PubMed] [Google Scholar]
- 57.Mader SL, Lopez A, Lawatscheck J, Luo Q, Rutz DA, Gamiz-Hernandez AP, Sattler M, Buchner J, and Kaila VRI. 2020. Conformational dynamics modulate the catalytic activity of the molecular chaperone Hsp90. Nat Commun. 11(1):1410, doi: 10.1038/s41467-020-15050-0, https://www.ncbi.nlm.nih.gov/pubmed/32179743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fleming PJ, and Fleming KG. 2018. HullRad: Fast Calculations of Folded and Disordered Protein and Nucleic Acid Hydrodynamic Properties. Biophys J. 114(4):856–869, doi: 10.1016/j.bpj.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.