Abstract
Strong transactivation of the β-globin genes is conferred by the β-globin locus control region (LCR), which consists of four erythroid-specific DNase I hypersensitive sites (HS1–HS4). HS2 has a powerful enhancer activity dependent upon tandem binding sites for the erythroid cell- and megakaryocyte-specific transcription factor NF-E2. An important co-activator-mediating transactivation by HS2 is the histone acetyltransferase (HAT) CREB binding protein (CBP). We showed previously that recruitment of a GAL4-CBP fusion protein to HS2 largely bypassed the requirement of the NF-E2 sites for transactivation. To determine whether GAL4-CBP recruitment is sufficient for transactivation, we assessed the importance of cis-elements within HS2. Docking of GAL4-CBP upstream of an Aγ-globin promoter lacking HS2 only weakly activated the promoter, indicating that HS2 components are required for GAL4-CBP-mediated transactivation. Sequences upstream and downstream of the NF-E2 sites were required for maximal GAL4-CBP-mediated transactivation, and HAT catalytic activity of GAL4-CBP was critical. No single factor-binding site was required for GAL4-CBP-mediated transactivation. However, deletion of two sites, a CACC site and an E-box, abolished transactivation in transient and stable transfection assays. These results suggest that NF-E2 recruits CBP as a critical step in transactivation, but additional components of HS2 are required to achieve maximal enhancer activity.
INTRODUCTION
Long-range transcriptional activation of the β-globin genes is conferred by four erythroid-specific DNase I hypersensitive sites (HS1–HS4) referred to as the β-globin locus control region (LCR) (1,2). These hypersensitive sites (HSs), located 8–50 kb upstream of the β-globin genes (3), contain many binding sites for diverse transcription factors (4). Stable transfection (5,6) and transgenic studies (7–9) provided evidence that the HSs synergize to confer long-range activation in a heterologous chromosomal environment. One model to explain the synergism is that multiple protein–protein and protein–DNA interactions involving components at all HSs are required to assemble a large ordered nucleoprotein complex or a holo-LCR (5,10). Given the fact that certain HSs such as HS2 have strong enhancer activity (11–19), subcomplexes forming on the individual HSs may have to interact specifically to form a holo-LCR. In contrast to the studies noted above, knockouts of individual murine HSs via homologous recombination, and analysis of mutant alleles in heterozygotes showed an additive effect of the HSs (20), inconsistent with the holo-LCR model. Thus, there is still much to be learned about the structure and function of the β-globin LCR.
Identifying factors responsible for conferring strong enhancer activity is important for elucidating the mechanism of long-range transactivation by the LCR. Multiple factors have been implicated in HS2 enhancer activity including NF-E2 (21–23), GATA-1 (21,24–26), TAL-1 (27,28), CBF1 (29,30), USF (17,31) and YY-1 (17,32). Intriguingly, in K562 erythroleukemia cells that express these hematopoietic and ubiquitous transcription factors, there is an absolute requirement for the tandem NF-E2 sites in conferring strong transactivation in transient and stable transfection assays (13,29,33). Although mutations of other sites have partial inhibitory effects in K562 cells (27,29), disruption of the NF-E2 sites abolishes enhancer activity. Thus, NF-E2 may be required to assemble the HS2 complex, and in its absence, other factors would not associate stably with HS2.
Despite this critical role of the NF-E2 sites for strong enhancer activity of the LCR, mice lacking the hematopoietic-specific subunit of NF-E2, p45/NF-E2, have nearly normal erythropoiesis (34,35). p45/NF-E2 null mice die of bleeding shortly after birth due to impaired megakaryopoiesis. As p45/NF-E2 null mice have an impaired oxidative-stress response in red blood cells, they appear to have an intrinsic defect in erythroid cell function (36). In contrast to in vivo analyses, disruption of the p45/NF-E2 gene in erythroid cell lines, such as CB3 murine erythroleukemia cells, results in silencing of the endogenous β-globin genes. β-Globin transcription is rescued upon expression of p45/NF-E2 (37–41), thereby confirming a causal relationship between p45/NF-E2 loss and β-globin gene silencing in cell culture. Thus, p45/NF-E2 is critical for β-globin expression in cell culture systems, but functional redundancy appears to exist in vivo.
The p45/NF-E2-mediated rescue of β-globin expression in erythroleukemia cell lines could be direct or indirect, and considering that multiple factors bind NF-E2 sites (42), it was unclear whether rescue involved HS2 binding. However, ChIP analysis showed that p45/NF-E2 could be crosslinked to HS2 in K562 and mouse erythroleukemia (MEL) cells, and in d.p.c. 14.5 murine fetal liver enriched in adult erythroid cells (41,43,44). Moreover, stable expression of p45/NF-E2 in CB3 cells results in occupancy of HS2 (41). These results strongly suggest that p45/NF-E2 functions through HS2 and that rescue involves a direct action through HS2. It thus becomes essential to understand why p45/NF-E2 has such a unique requirement relative to other HS2 binding proteins.
An increasing number of studies have implicated the histone acetyltransferases (HATs) CREB binding protein (CBP) and p300 as important coactivators for trans-acting factors functioning through promoters and enhancers (reviewed in 45–47). CBP/p300 has been implicated recently in enhancer-mediated transactivation over distances greater than several hundred base pairs (48,49). We demonstrated that CBP/p300 was required for HS2- and LCR-mediated transactivation in transient and stable transfection assays, but not for basal activity of the γ- and β-globin promoters (48). As the CBP/p300 inhibitor Adenovirus E1A repressed expression of the endogenous γ-globin genes, it is attractive to propose a model in which CBP/p300 mediates the enhancer activity of the LCR over distances up to ~50 kb, the distance between the LCR and the β-globin promoter (Fig. 1). To test this model, it is important to define the LCR components required for CBP/p300 recruitment. In this regard, p45/NF-E2 interacts physically and functionally with CBP/p300 (48,50), and the NF-E2 binding sites of HS2 were required for CBP function through HS2 (48). Thus, the importance of the NF-E2 sites may be related to the recruitment of CBP/p300.
Figure 1.
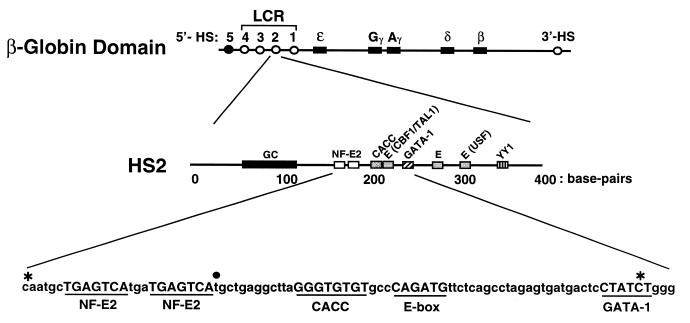
Organization of the human β-globin locus. The human β-globin gene cluster consists of one embryonic gene (ɛ), two fetal genes (Gγ and Aγ), one minor adult gene (δ) and one major adult gene (β). Four erythroid-specific HSs at the 5′ end of the locus constitute the LCR, which is critical for high-level expression of the β-globin genes. A 3′ erythroid-specific HS and a ubiquitous 5′-HS (HS5) are also shown. The organization of transcription factor binding sites within HS2 is shown in the middle. The sequence of a portion of HS2 is shown at the bottom. The asterisks denote the limits of an internal deletion of HS2 in HS2(Δ5′,GAL4,C-G)γluc. The solid circle denotes the base that was substituted with 5- and 10-bp sequences to generate TWIST5 and TWIST10 plasmids. GC, GC-rich sequences; NF-E2, tandem binding sites for the erythroid cell- and megakaryocytic-specific transcription factor NF-E2; CACC, conserved sequence with no established endogenous interactor; CBF1/TAL1, overlapping binding sites for C-promoter binding factor 1 (29) and TAL1 (27); GATA-1, binding site for the erythroid cell- and megakaryocyte-specific transcription factor GATA-1; E, E-box; USF, binding site for the upstream stimulatory factor; YY1, binding site for ying-yang 1.
In MEL cells, p45/NF-E2 binding to the LCR establishes histone H3, and to a lesser extent H4, hyperacetylation and induces RNA polymerase II recruitment at the adult β-globin promoters (41). As CBP/p300 acetylates histones and transcription factors, including the MafG subunit of NF-E2 (51) and GATA-1 (52,53), the mechanism of the CBP/p300 requirement for LCR-mediated transactivation might have multiple components. Here we describe how recruitment of CBP to the NF-E2 binding region of HS2 requires interactions with other components of HS2 to confer strong transactivation in transient transfection assays and in a chromosomal context. These results are discussed vis-à-vis a multi-step model of coactivator usage by HS2.
MATERIALS AND METHODS
Plasmid construction
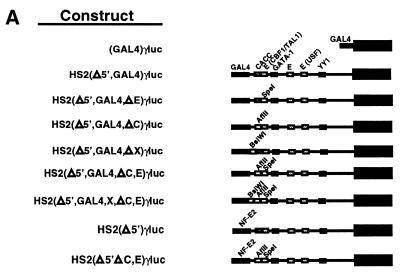
The reporter plasmid HS2γluc was described previously (5). HS2 mutations were constructed from pHS2γluc by standard PCR-based mutagenesis or by double-stranded oligo replacement and confirmed by sequencing. For all plasmids containing a GAL4 site in HS2, an adjacent XhoI (CTCGAG) and a GAL4 binding site (CGGAAGACTCTCCTCCG) were substituted for the tandem NF-E2 sites (CAATGCTGAGTCATGATGAGTCA). All Δ5′ plasmids were truncated at the XhoI site and lacked HS2 sequences upstream of the Gal4 binding site. These plasmids contained additional pBluescript sequence (GGGCCCCCC) between the KpnI site of pγluc (5) and the XhoI site. Mutations of individual binding sites were accomplished by conservative 6 bp substitutions; replacing the E-box (CAGATG) with an SpeI site (ACTAGT), CACC site (GTGTGT) with an AflII site (CTTAAG), the consensus GATA-1 site (CTATCT) with a MluI site (ACGCGT) and conserved site X (GCTGAG) with a BsiWI site (CGTACG). To generate the HS2(Δ5′,Gal4,ΔC-G)γluc plasmid, 80 bp between, and including, the NF-E2 and GATA-1 sites were replaced with an adjacent XhoI and GAL4 binding site. The endpoints of the deletion are denoted by asterisks in Figure 1. HS2 core plasmids were derived from corresponding Δ5′ plasmids and therefore lack HS2 sequences upstream of the GAL4 site. Sequences downstream and including the consensus GATA-1 site were removed by replacement of the consensus GATA-1 site with a MluI site. Lastly, the HS2 sequence between this MluI site and the MluI site of pγluc was deleted. For the plasmids TWIST5 and TWIST10, the spacing between the GAL4 binding site and CACC site was increased by addition of the sequences TGATCT and TCGAGATCTGT, respectively; these sequences were substituted for the nucleotide marked by the solid circle in Figure 1. For the plasmid (GAL4)γluc, a Gal4 binding site was introduced between the KpnI and MluI sites of the pγluc plasmid. A 5.1 kb MluI fragment from phage λ was introduced into the MluI site of (GAL4)γluc to yield (GAL4)5.1γluc.
Expression plasmids encoding the GAL4 DNA binding domain (amino acids 1–147), pRc/RSV-GAL4, and a GAL4 DNA binding domain fusion to human CBP, pGAL4-CBP(FLAG), were gifts from Dr John Chrivia (54). The expression plasmid pGAL4-CBPHAT– is identical to pGAL-CBP(FLAG) except at positions Tyr1540Tyr1541 within the coenzyme A binding domain, which were changed to alanines to disrupt acetyltransferase activity as described previously (55). The integrity of the constructs was confirmed by restriction enzyme digestion analysis and DNA sequencing.
Cell culture
The human erythroleukemia cell line K562 was propagated in Iscove’s Modified Eagle’s Medium (Biofluids, Rockville, MD) containing 25 µg/ml gentamycin, 1% antibiotic/antimycotic and 10% fetal calf serum (Gibco-BRL, Rockville, MD) (complete IMEM). Cells were grown in a humidified incubator at 37°C in the presence of 5% carbon dioxide.
Transient transfection analysis
K562 cells (5 × 105) were collected by centrifugation at 240 g for 6 min at 4°C and were resuspended in complete IMEM. For each transfection, 3 µg total DNA was suspended in 150 µl of complete IMEM, incubated with 4 µl of Superfect (Qiagen, Valencia, CA) for 15 min at room temperature and then added to cells. Except where noted, each transfection contained 1 µg of reporter plasmid and 1 µg of expression plasmid. The constitutively active β-galactosidase expression vector pCMVβ-Gal (0.1 µg) (Pharmacia, Piscataway, NJ) was included in each transfection reaction to assess differences in the transfection efficiency. After incubating for 40 h, cells were harvested and assayed for luciferase activity. β-Galactosidase activity was assayed with a luminescent substrate (Galacton Plus) according to the manufacturer’s instructions (TROPIX, Inc., Bedford, MA). The luciferase activity (relative light units = RLU) was normalized by the protein content (determined by Bradford assay using γ-globulin as a standard) and the β-galactosidase activity of the lysates. Similar results were obtained when the luciferase values were normalized by the protein content of lysates alone versus normalizing to both protein content and β-galactosidase activity.
Stable transfection analysis
Pools of stably transfected K562 cells were generated as described previously (5). Cell lysates were assayed for luciferase activity, and the luciferase activity was normalized by the protein content of the lysates as described above.
RESULTS
CBP recruitment upstream of an Aγ-globin promoter is insufficient for strong transactivation
Studies on the mechanism of the erythroid-specific enhancer activity of the LCR have been carried out in both in vitro and in vivo systems. Importantly, in numerous reports, the cis-acting and trans-acting requirements for LCR-mediated transactivation in transient and stable transfection assays were similar if not identical. For example, as noted above, the tandem binding sites for NF-E2 within HS2 are critical for strong enhancer activity in transient and stable transfection assays and in transgenic mice. A conserved E-box within HS2 contributes to the strong transactivation in transient and stable transfection assays, but is considerably less important than the NF-E2 sites (27,29). Moreover, a recently characterized negative element of HS2 was active in both transient and stable transfection assays (56). Based on these common requirements, and considering that multiple co-activators have been identified and characterized through transient transfection analyses (57–61), transient assays were used herein to assess how CBP/p300 is utilized by HS2.
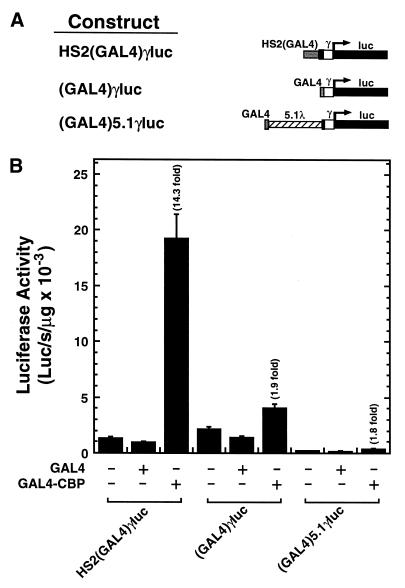
Previously, we generated a reporter, HS2(GAL4)γluc, in which the tandem NF-E2 sites of HS2 were replaced with a single GAL4 binding site, while maintaining the spacing between neighboring cis-elements of HS2 (48). Expression of GAL4-CBP strongly activated HS2(GAL4)γluc (14.3-fold) (Fig. 2). A GAL4 fusion to the unrelated HAT PCAF did not activate the reporter in transfected K562 cells (K.D.Johnson, J.E.Norton and E.H.Bresnick, unpublished data), even though the identical GAL4-PCAF construct conferred activation through a GAL4 site placed within the SV40 enhancer, positioned 3.1 kb downstream of a synthetic promoter in a transient transfection assay in B78 melanoma cells (49). Thus, not all HATs can function through the NF-E2 binding region of HS2.
Figure 2.
CBP recruitment is insufficient for strong transactivation of an Aγ-globin promoter. (A) Structure of test constructs. (B) K562 cells were transiently co-transfected with test constructs and either the blank vector pcDNA3 or expression vectors encoding the DNA binding domain of GAL4 alone or the GAL4 DNA binding domain fused to CBP. The graph depicts the luciferase activities normalized by the protein content of the lysates (mean ± SE, n ≥ 6). Fold activation values are also shown. Note that the activity of HS2(GAL4)γluc is similar to the promoter-only construct (GAL4)γluc, as the NF-E2 sites are critical for the enhancer activity of HS2 in this assay.
To assess whether CBP recruitment is sufficient for activation of the Aγ-globin promoter, we tested whether GAL4-CBP can activate reporters containing a GAL4 site immediately upstream [(GAL4)γluc] or 5.1 kb upstream [(GAL4)5.1γluc] of an Aγ-globin promoter. In contrast to HS2(GAL4)γluc, GAL4-CBP only weakly influenced the activity of the promoter-only constructs (1.9- and 1.8-fold, respectively) (Fig. 2). This result suggests that recruitment of GAL4-CBP near the promoter is insufficient for strong activation, and that additional components of HS2 are required. As the GAL4 site replaced the tandem NF-E2 sites of HS2(GAL4)γluc, and GAL4-CBP strongly activates this reporter, one can rule out NF-E2 as being essential for GAL4-CBP-mediated activation, once recruitment has taken place.
CBP cooperates with multiple components of HS2 to confer strong transactivation
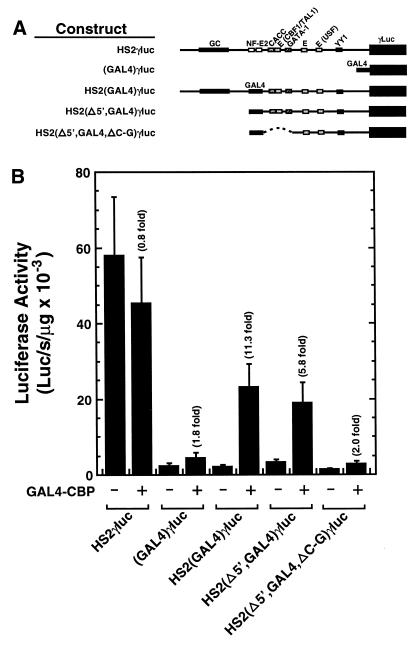
A simple model to explain the results in Figure 2 is that NF-E2 engages in a protein–protein interaction with CBP/p300, allowing it to dock on HS2. The strong enhancer activity of HS2 would then require additional components of HS2 to functionally interact with CBP/p300. To define cis-elements of HS2 required for GAL4-CBP-mediated activation, we generated HS2 mutants lacking sequences at the 5′-side of the GAL4 site with [HS2(Δ5′,GAL4,ΔC-G)γluc] or without [HS2(Δ5′,GAL4)γluc] an internal deletion on the 3′-side of the GAL4 site, which removed three conserved factor binding sites, a CACC site, an E-box and a GATA-1 site. As in Figure 2, GAL4-CBP weakly (1.8-fold) and strongly (11.3-fold) activated (GAL4)γluc and HS2(GAL4)γluc reporters, respectively (Fig. 3). Deletion of the 5′ sequences lowered GAL4-CBP-mediated activation to 5.8-fold, while mutation of additional CACC, E-box and GATA-1 sites strongly reduced activation to 2-fold, comparable with (GAL4)γluc. Thus, sequences at the 5′- and 3′-ends of HS2 are important for GAL4-CBP-mediated activation of HS2. Considering that multiple additional factor binding sites reside on the 3′-side of the internal deletion (4), these remaining sites are insufficient to confer strong activation by GAL4-CBP.
Figure 3.
Strong GAL4-CBP-mediated transactivation of an Aγ-globin promoter requires sequences upstream and downstream of the NF-E2 sites of HS2. (A) Structure of test constructs. (B) K562 cells were transiently co-transfected with test constructs and either the blank vector pcDNA3 or an expression vector encoding GAL4 fused to CBP. Expression of the GAL4 DNA binding domain alone yielded activities comparable with that of the pcDNA3 control. The graph depicts the absolute luciferase activities normalized by the protein content of the lysates (mean ± SE, n = 6). Fold activation values are also shown.
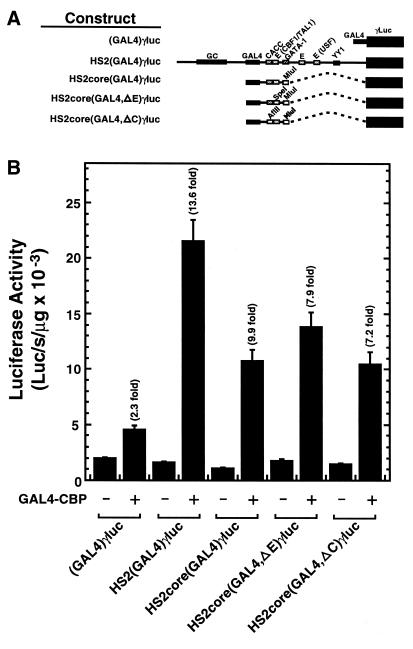
Since HS2(Δ5′,GAL4,ΔC-G)γluc, lacking HS2 sequences on the 5′-side, and CACC, E-box and GATA-1 sites on the 3′-side of the GAL4 site, were not strongly activated by GAL4-CBP, and HS2(Δ5′,GAL4)γluc was activated 5.8-fold, we asked whether an HS2 fragment containing the GAL4 site, the 3′ CACC site and the neighboring E-box [HS2core(GAL4)γluc] was sufficient for strong activation. GAL4-CBP activated HS2core(GAL4)γluc 9.9-fold compared with 13.6-fold for HS2(GAL4)γluc (Fig. 4). Single mutations of the E-box or CACC site, in the context of the minimal core construct, only caused a small reduction in GAL4-CBP-mediated activation (7.9- and 7.2-fold, respectively). Thus, sequences at the 3′-side of the NF-E2 sites containing the E-box and CACC site mediate strong activation, but neither site is essential.
Figure 4.
A minimal core sequence of HS2 is sufficient for strong GAL4-CBP-mediated transactivation of a Aγ-globin promoter. (A) Structure of test constructs. (B) K562 cells were transiently co-transfected with the test constructs and either the blank vector pcDNA3 or an expression vector encoding the GAL4 DNA binding domain fused to CBP. Expression of the GAL4 DNA binding domain alone yielded activities comparable with that of the pcDNA3 control. The graph depicts the luciferase activities normalized by the protein content of the lysates (mean ± SE, n ≥ 6). Fold activation values are also shown.
The erythroid cell- and megakaryocyte-specific and HS2 binding factor GATA-1 functionally interacts with CBP in the context of a synthetic promoter containing GATA-1 binding sites (62). However, the consensus GATA-1 site (24,25) of HS2 was not required for CBP/p300 recruitment in K562 cells (48). The activity of an HS2 mutant lacking this site was abolished by adenovirus E1A, an inhibitor of CBP/p300, inconsistent with its requirement for CBP/p300 recruitment in this system. Nevertheless, it is possible that GATA-1 or a GATA-1-associated co-activator (63) cooperates with CBP/p300 once CBP/p300 docks on HS2. However, as HS2core(GAL4)γluc lacks a consensus GATA-1 site and was strongly activated by GAL4-CBP, this site is not required for GAL4-CBP-mediated activation of HS2. Furthermore, in the context of HS2(GAL4)γluc, the consensus GATA-1 site is also not important for strong GAL4-CBP-mediated transactivation (K.D.Johnson, J.E.Norton and E.H.Bresnick, unpublished data). Thus, the binding of GATA-1 to the consensus site is dispensable for strong GAL4-CBP-mediated activation of HS2 once GAL4-CBP has docked on HS2.
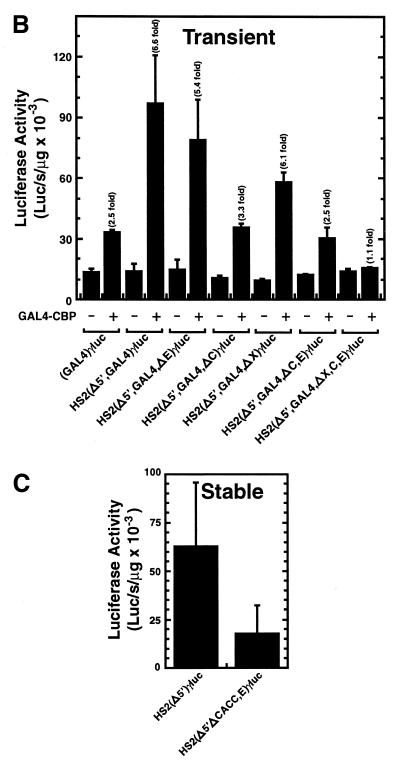
To define whether specific combinations of sites are necessary for strong GAL4-CBP-mediated activation of HS2, additional site mutations were generated in the context of HS2(Δ5′,GAL4)γluc. GAL4-CBP activated HS2(Δ5′,GAL4)γluc 6.6-fold, while single site mutations of the E-box and CACC site decreased activation to 5.4- and 3.3-fold, respectively (Fig. 5). As a conserved 6 bp sequence lies just to the 3′-side of the NF-E2 sites (site X) and is present in the GAL4 site-substituted constructs, a mutant lacking this site [HS2(Δ5′,GAL4,ΔX)γluc] was also generated. GAL4-CBP activated HS2(Δ5′,GAL4,ΔX)γluc 6.1-fold, similar to the activation seen with HS2(Δ5′,GAL4)γluc. Constructs containing multiple site mutations were also generated. Mutation of both the E-box and CACC site strongly reduced the level of activation to 2.5-fold, equivalent to that of (GAL4)γluc. Similarly, mutation of the E-box, the CACC site and site X abolished activation (1.1-fold). Insertion of 5 or 10 bp spacers between the CACC site and the GAL4 site (TWIST5 and TWIST10 plasmids, respectively) only had minor effects on GAL4-CBP-mediated activation (data not shown), suggesting that the helical orientation of the CACC site with respect to the docking site of GAL4-CBP is not critical. Thus, strong GAL4-CBP-mediated activation of HS2 requires the E-box and CACC site and, as shown in Figures 3B and 4B, requires sequences at the 5′-side of the GAL4 site.
Figure 5.
Strong GAL4-CBP-mediated transactivation of an Aγ-globin promoter is abrogated by mutation of multiple cis-elements of HS2 in transient and stable transfection assays. (A) Structure of test constructs. (B) K562 cells were transiently co-transfected with the test constructs and either the blank vector pcDNA3 or an expression vector encoding the GAL4 DNA binding domain fused to CBP. Expression of the GAL4 DNA binding domain alone yielded activities comparable with that of the pcDNA3 control. The graph depicts the luciferase activities normalized by the protein content of the lysates (mean ± SE, n ≥ 8). Fold activation values are also shown. (C) K562 cells were stably transfected with the indicated constructs. The graph depicts the luciferase activities normalized by the protein content of the lysates (mean ± SE, n = 4).
As the CACC site and E-box were essential for strong GAL4-CBP-mediated activation, we asked whether these sites were important for HS2 enhancer activity upon integration into chromosomal DNA. Stably transfected pools of K562 cells were generated with constructs containing the tandem NF-E2 binding sites with [HS2(Δ5′)γluc] or without the CACC site and E-box [HS2(Δ5′,ΔC,E)γluc]. Mutation of the CACC site and E-box resulted in considerably lower activity in the stable transfection assay (Fig. 5C). Thus, under conditions in which NF-E2 normally mediates CBP/p300 usage by HS2, there is an important requirement for the CACC site and E-box for strong enhancer activity.
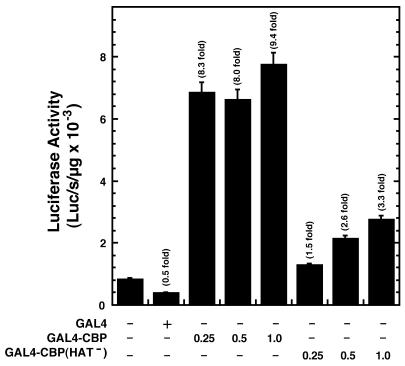
Given that CBP and p300 are histone and protein acetylases and can physically interact with diverse proteins, it was instructive to ask whether strong GAL4-CBP-mediated transactivation requires the acetylase function of CBP. A HAT-defective mutant of CBP (GAL4-CBPHAT–) was generated, based on residues known to be important for CBP/p300 catalytic activity, and its ability to activate HS2(GAL4)γluc was assessed. As shown in Figure 6, GAL4-CBPHAT– only weakly activated the reporters, confirming that strong activation requires the acetylase function of CBP.
Figure 6.
Requirement of HAT activity for strong GAL4-CBP-mediated transactivation of an Aγ-globin promoter. K562 cells were transiently co-transfected with HS2(GAL4)γluc and either the blank vector pcDNA3 or expression vectors encoding the GAL4 DNA binding domain, the GAL4 DNA binding domain fused to CBP or the GAL4 DNA binding domain fused to a HAT-defective mutant of CBP. The graph depicts the luciferase activities normalized by the protein content of the lysates (mean ± SE, n = 6). Fold activation values are also shown.
DISCUSSION
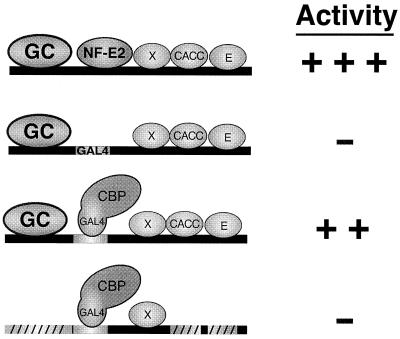
The results described herein are summarized in Figure 7. NF-E2 physically recruits a CBP/p300-containing coactivator complex to HS2. In addition to CBP/p300 recruitment, other components of HS2 are required for strong transactivation. These components require cis-elements upstream of the NF-E2 sites and a CACC site and E-box downstream of the NF-E2 sites. The requirement for these components may result from (i) acetylation of the cognate binding proteins by GAL4-CBP, (ii) direct stimulation of GAL4-CBP HAT activity or (iii) recruitment of additional factors to HS2 by the cognate binding proteins, which synergize with GAL4-CBP.
Figure 7.
Summary of cis-element requirements for GAL4-CBP-mediated activation of HS2. Schematic representations of protein–DNA interactions occuring on test constructs are shown on the left. Relative luciferase activities are indicated on the right. +++, high activity; ++, intermediate activity; –, low activity. GC, factors binding to GC-rich sequences; X, factor binding to conserved sequence; CACC, CACC site binding factor; E, E-box binding factor; diagonal lines depict mutated factor binding site.
Multiple targets for the acetylase function of CBP have been implicated, including non-histone proteins (64,65), the acetylase ACTR (66) and histones (66). An increasing number of transcription factors have been shown to be acetylated by CBP/p300 (reviewed in 46,67). GATA-1 is acetylated by CBP/p300 in vitro and in vivo, and acetylation has been reported to stimulate transactivation and DNA binding functions of GATA-1 (52,53). Although the consensus GATA-1 site was not required for GAL4-CBP-mediated activation of HS2, the presence of an imperfect GATA-1 site immediately upstream of the consensus site leaves open the possibility that GATA-1 acetylation may still be relevant. However, as demonstrated in Figure 5, mutations of the CACC site and E-box abolish activation despite the presence of the consensus GATA-1 site. Thus, GATA-1 binding to HS2 is insufficient for GAL4-CBP-mediated activation. Interestingly, acetylation of the erythroid-specific transcription factor EKLF increases EKLF-dependent transactivation, and EKLF binds CACC sites (68). As acetylated EKLF has enhanced affinity for the Swi/Snf chromatin remodeling complex (69), acetylation might facilitate Swi/Snf recruitment. The requirement of the CACC site for strong GAL4-CBP-mediated transactivation may be explained by acetylation of EKLF, and it would be intriguing if NF-E2-recruited CBP/p300 interacts functionally with EKLF-recruited Swi/Snf. However, such a mechanism remains speculative. Lastly, the association of the hematopoietic-specific transcription factor TAL1 with the Sin3A co-repressor is destabilized by PCAF-mediated acetylation of TAL1, and acetylation enhances TAL1 DNA binding (70). As TAL1 can bind to the E-box of HS2 in vitro (27), cooperation between GAL4-CBP and TAL1 might be important for strong GAL4-CBP-mediated activation of HS2.
Interestingly, the acetylation activity of CBP towards nucleosomal but not free histones increases upon interaction with certain transcription factors, including p45/NF-E2 (71). As certain CBP-interacting proteins lacked this activity, CBP binding was insufficient to stimulate acetylation. Upon recruitment of GAL4-CBP to HS2, factors binding the E-box and CACC site might stimulate the HAT activity of GAL4-CBP. p45/NF-E2 regulates histone acetylation based on our recent analysis of the histone acetylation state of the endogenous β-globin locus in MEL and CB3 cells (41). Binding of p45/NF-E2 to the LCR established hyperacetylation at and near the adult β-globin genes, which reside 40–50 kb downstream of the LCR. Considering that CBP/p300 is an important p45/NF-E2 coactivator, CBP/p300 is a good candidate for a mediator of the p45/NF-E2-dependent acetylation changes. As CBP and p300 are complex proteins with multiple interaction surfaces, it would not be surprising if multiple functions of CBP/p300 contributed to HS2-mediated activation. Kraus et al. (72) described four distinct transactivation functions of p300, which were necessary for estrogen receptor-mediated activation of transcription from chromatin templates in vitro. The relative importance of the individual activation functions differed for two activators, the estrogen receptor and GAL4-VP16, suggesting that they may function in a context-dependent manner.
Since our experiments used transient transfection assays, can one infer that GAL4-CBP-mediated activation of HS2 does not involve chromatin modification? Transiently transfected DNA can exist in a heterogeneous state with regard to chromatin structure (73) but can adopt a nucleosomal structure (74). Even if only a fraction of the transfected DNA molecules assemble into chromatin, a coactivator function requiring chromatin modification would probably be apparent with a mixed population of templates. Nearly all studies on transactivation mediated by CBP/p300 have used transient transfections, and transient assays have been used to analyze the mechanism of enhancer blocking activity by a chromatin-insulating element (75). As the critical requirement for the CACC site and the E-box is apparent upon integration of constructs into chromosomal DNA (Fig. 5C), clearly these elements are not only required in transient assays.
As summarized in Figure 7, a single cis-element, the tandem NF-E2 sites, is critical for initiating activation, while additional cis-elements, including the CACC site and E-box contribute in a combinatorial fashion, potentially at a subsequent step(s). We propose that this mechanism involving coactivator recruitment, followed by functional interactions between the co-activator and regulatory factors not mediating recruitment may be common in long-range transactivation. However, additional studies are required to define whether the functional interactions occur concomitant with recruitment or post-recruitment. To further analyze the mechanism, it will be important to identify physiological components of HS2 cooperating with CBP/p300 and to determine if related proteins are used by other long-range regulatory elements, which may share a common mechanism of regulated coactivator usage.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Tony Kouzarides, Yoshihiro Nakatani, Edward Korzus and Mark Chrivia for plasmids. The authors acknowledge support from the Milwaukee Foundation, the Leukemia Society of America and the National Institutes of Health (grant DK55700). E.H.B. is a Leukemia Society of America Scholar and a Shaw Scientist.
REFERENCES
- 1.Grosveld F., van Assendelft,G.B., Greaves,D.R. and Kollias,G. (1987) Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell, 51, 975–985. [DOI] [PubMed] [Google Scholar]
- 2.Forrester W.C., Takegawa,S., Papayannopoulou,T., Stamatoyannopoulos,G. and Groudine,M. (1987) Evidence for a locus activation region: the formation of developmentally stable hypersensitive sites in globin-expressing hybrids. Nucleic Acids Res., 15, 10159–10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuan D. and London,I.M. (1984) Mapping of DNase I-hypersensitive sites in the upstream DNA of human embryonic epsilon-globin gene in K562 leukemia cells. Proc. Natl Acad. Sci. USA, 81, 2718–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hardison R., Slightom,J.L., Gumucio,D.L., Goodman,M., Stojanovic,N. and Miller,W. (1997) Locus control regions of mammalian beta-globin gene clusters: combining phylogenetic analyses and experimental results to gain functional insights. Gene, 205, 73–94. [DOI] [PubMed] [Google Scholar]
- 5.Bresnick E.H. and Tze,L. (1997) Synergism between hypersensitive sites confers long-range gene activation by the beta-globin locus control region. Proc. Natl Acad. Sci. USA, 94, 4566–4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Molete J.M., Petrykowska,H., Bouhassira,E.E., Feng,Y.Q., Miller,W. and Hardison,R.C. (2001) Sequences flanking hypersensitive sites of the beta-globin locus control region are required for synergistic enhancement. Mol. Cell. Biol., 21, 2969–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peterson K.R., Clegg,C.H., Navas,P.H., Norton,E.J., Kimbrough,T.G. and Stamatoyannopoulos,G. (1996) Effect of deletion of 5′HS3 or 5′HS2 of the human beta-globin locus control region on the developmental regulation of globin gene expression in beta-globin locus yeast artificial chromosome transgenic mice. Proc. Natl Acad. Sci. USA, 93, 6605–6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bungert J., Dave,U., Lim,K.C., Lieuw,K.H., Shavit,J.A., Liu,Q. and Engel,J.D. (1995) Synergistic regulation of human beta-globin gene switching by locus control region elements HS3 and HS4. Genes Dev., 9, 3083–3096. [DOI] [PubMed] [Google Scholar]
- 9.Navas P.A., Peterson,K.R., Li,Q., McArthur,M. and Stamatoyannopoulos,G. (2001) The 5′HS4 core element of the human beta-globin locus control region is required for high-level globin gene expression in definitive but not in primitive erythropoiesis. J. Mol. Biol., 312, 17–26. [DOI] [PubMed] [Google Scholar]
- 10.Ellis J., Tan-Un,K.C., Harper,A., Michalovich,D., Yannoutsos,N., Philipsen,S. and Grosveld,F. (1996) A dominant chromatin-opening activity in 5′ hypersensitive site 3 of the human beta-globin locus control region. EMBO J., 15, 562–568. [PMC free article] [PubMed] [Google Scholar]
- 11.Tuan D.Y., Solomon,W.B., London,I.M. and Lee,D.P. (1989) An erythroid-specific, developmental stage-independent enhancer far upstream of the human β-like globin genes. Proc. Natl Acad. Sci. USA, 86, 2554–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryan T.M., Behringer,R.R., Martin,N.C., Townes,T.M., Palmiter,R.D. and Brinster,R.L. (1989) A single erythroid-specific DNase I super-hypersensitive site activates high levels of human beta-globin gene expression in transgenic mice. Genes Dev., 3, 314–323. [DOI] [PubMed] [Google Scholar]
- 13.Ney P.A., Sorrentino,B.P., Lowrey,C.H. and Nienhuis,A.W. (1990) Inducibility of the HS II enhancer depends on binding of an erythroid specific nuclear protein. Nucleic Acids Res., 18, 6011–6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talbot D., Philipsen,S., Fraser,P. and Grosveld,F. (1990) Detailed analysis of the site 3 region of the human beta-globin dominant control region. EMBO J., 9, 2169–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lloyd J.A., Krakowsky,J.M., Crable,S.C. and Lingrel,J.B. (1992) Human gamma- to beta-globin gene switching using a mini construct in transgenic mice. Mol. Cell. Biol., 12, 1561–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardison R., Xu,J., Jackson,J., Mansberger,J., Selifonova,O., Grotch,B., Biesecker,J., Petrykowska,H. and Miller,W. (1993) Comparative analysis of the locus control region of the rabbit beta-like gene cluster: HS3 increases transient expression of an embryonic epsilon-globin gene. Nucleic Acids Res., 21, 1265–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellis J., Talbot,D., Dillon,N. and Grosveld,F. (1993) Synthetic human beta-globin 5′HS2 constructs function as locus control regions only in multicopy transgene concatamers. EMBO J., 12, 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu D., Chang,J.C., Moi,P., Liu,W., Kan,Y.W. and Curtin,P.T. (1992) Dissection of the enhancer activity of beta-globin 5′ DNase I-hypersensitive site 2 in transgenic mice. Proc. Natl Acad. Sci. USA, 89, 3899–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraser P., Pruzina,S., Antoniou,M. and Grosveld,F. (1993) Each hypersensitive site of the human beta-globin locus control region confers a different developmental pattern of expression on the globin genes. Genes Dev., 7, 106–113. [DOI] [PubMed] [Google Scholar]
- 20.Bender M.A., Roach,J.N., Halow,J., Close,J., Alami,R., Bouhassira,E.E., Groudine,M. and Fiering,S.N. (2001) Targeted deletion of 5′HS1 and 5′HS4 of the beta-globin locus control region reveals additive activity of the DNase I hypersensitive sites. Blood, 98, 2022–2027. [DOI] [PubMed] [Google Scholar]
- 21.Talbot D. and Grosveld,F. (1991) The 5′HS2 of the globin locus control region enhances transcription through the interaction of a multimeric complex binding at two functionally distinct NF-E2 binding sites. EMBO J., 10, 1391–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrews N.C., Erdjument-Bromage,H., Davidson,M.B., Tempst,P. and Orkin,S.H. (1993) Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature, 362, 722–728. [DOI] [PubMed] [Google Scholar]
- 23.Ney P.A., Andrews,N.C., Jane,S.M., Safer,B., Purucker,M.E., Weremowicz,S., Morton,C.C., Goff,S.C., Orkin,S.H. and Nienhuis,A.W. (1993) Purification of the human NF-E2 complex: cDNA cloning of the hematopoietic cell-specific subunit and evidence for an associated partner. Mol. Cell. Biol., 13, 5604–5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evans T. and Felsenfeld,G. (1989) The erythroid-specific transcription factor Eryf1: a new finger protein. Cell, 58, 877–885. [DOI] [PubMed] [Google Scholar]
- 25.Tsai S.F., Martin,D.I., Zon,L.I., D’Andrea,A.D., Wong,G.G. and Orkin,S.H. (1989) Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature, 339, 446–451. [DOI] [PubMed] [Google Scholar]
- 26.Caterina J.J., Ciavatta,D.J., Donze,D., Behringer,R.R. and Townes,T.M. (1994) Multiple elements in human beta-globin locus control region 5′ HS 2 are involved in enhancer activity and position-independent, transgene expression. Nucleic Acids Res., 22, 1006–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elnitski L., Miller,W. and Hardison,R. (1997) Conserved E boxes function as part of the enhancer in hypersensitive site 2 of the beta-globin locus control region. Role of basic helix–loop–helix proteins. J. Biol. Chem., 272, 369–378. [DOI] [PubMed] [Google Scholar]
- 28.Gould K.A. and Bresnick,E.H. (1998) Sequence determinants of DNA binding by the hematopoietic helix–loop–helix transcription factor TAL1: importance of sequences flanking the E-box core. Gene Expr., 7, 87–101. [PMC free article] [PubMed] [Google Scholar]
- 29.Lam L.T. and Bresnick,E.H. (1996) A novel DNA-binding protein, HS2NF5, interacts with a functionally important sequence of the human beta-globin locus control region. J. Biol. Chem., 271, 32421–32429. [DOI] [PubMed] [Google Scholar]
- 30.Lam L.T. and Bresnick,E.H. (1998) Identity of the beta-globin locus control region binding protein HS2NF5 as the mammalian homolog of the notch-regulated transcription factor suppressor of hairless. J. Biol. Chem., 273, 24223–24231. [DOI] [PubMed] [Google Scholar]
- 31.Bresnick E.H. and Felsenfeld,G. (1993) Evidence that the transcription factor USF is a component of the human beta-globin locus control region heteromeric protein complex. J. Biol. Chem., 268, 18824–18834. [PubMed] [Google Scholar]
- 32.Yant S.R., Zhu,W., Millinoff,D., Slightom,J.L., Goodman,M. and Gumucio,D.L. (1995) High affinity YY1 binding motifs: identification of two types (ACAT and CCAT) and distribution of potential binding sites within the human beta globin cluster. Nucleic Acids Res., 23, 4353–4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moi P. and Kan,Y.W. (1990) Synergistic enhancement of globin gene expression by activator protein-1-like proteins. Proc. Natl Acad. Sci. USA, 87, 9000–9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shivdasani R.A. and Orkin,S.H. (1995) Erythropoiesis and globin gene expression in mice lacking the transcription factor NF-E2. Proc. Natl Acad. Sci. USA, 92, 8690–8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shivdasani R.A., Rosenblatt,M.F., Zucker-Franklin,D., Jackson,C.W., Hunt,P., Saris,C.J. and Orkin,S.H. (1995) Transcription factor NF-E2 is required for platelet formation independent of the actions of thrombopoietin/MGDF in megakaryocyte development. Cell, 81, 695–704. [DOI] [PubMed] [Google Scholar]
- 36.Chan J.Y., Kwong,M., Lo,M., Emerson,R. and Kuypers,F.A. (2001) Reduced oxidative-stress response in red blood cells from p45/NF-E2-deficient mice. Blood, 97, 2151–2158. [DOI] [PubMed] [Google Scholar]
- 37.Lu S.J., Rowan,S., Bani,M.R. and Ben-David,Y. (1994) Retroviral integration within the Fli-2 locus results in inactivation of the erythroid transcription factor NF-E2 in Friend erythroleukemias: evidence that NF-E2 is essential for globin expression. Proc. Natl Acad. Sci. USA, 91, 8398–8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kotkow K.J. and Orkin,S.H. (1995) Dependence of globin gene expression in mouse erythroleukemia cells on the NF-E2 heterodimer. Mol. Cell. Biol., 15, 4640–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bean T.L. and Ney,P.A. (1997) Multiple regions of p45 NF-E2 are required for beta-globin gene expression in erythroid cells. Nucleic Acids Res., 25, 2509–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mosser E.A., Kasanov,J.D., Forsberg,E.C., Kay,B.K., Ney,P.A. and Bresnick,E.H. (1998) Physical and functional interactions between the transactivation domain of the hematopoietic transcription factor NF-E2 and WW domains. Biochemistry, 37, 13686–13695. [DOI] [PubMed] [Google Scholar]
- 41.Johnson K.D., Christensen,H.M., Zhao,B. and Bresnick,E.H. (2001) Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol. Cell, 8, 465–471. [DOI] [PubMed] [Google Scholar]
- 42.Motohashi H., Shavit,J.A., Igarashi,K., Yamamoto,M. and Engel,J.D. (1997) The world according to Maf. Nucleic Acids Res., 25, 2953–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daftari P., Gavva,N.R. and Shen,C.K. (1999) Distinction between AP1 and NF-E2 factor-binding at specific chromatin regions in mammalian cells. Oncogene, 18, 5482–5486. [DOI] [PubMed] [Google Scholar]
- 44.Forsberg E.C., Downs,K.M. and Bresnick,E.H. (2000) Direct interaction of NF-E2 with hypersensitive site 2 of the beta-globin locus control region in living cells. Blood, 96, 334–339. [PubMed] [Google Scholar]
- 45.Vo N. and Goodman,R.H. (2001) CREB-binding protein and p300 in transcriptional regulation. J. Biol. Chem., 276, 13505–13508. [DOI] [PubMed] [Google Scholar]
- 46.Blobel G.A. (2000) CREB-binding protein and p300: molecular integrators of hematopoietic transcription. Blood, 95, 745–755. [PubMed] [Google Scholar]
- 47.Mayr B. and Montminy,M. (2001) Transcriptional regulation by the phosphorylation-dependent factor CREB. Nature Rev. Mol. Cell. Biol., 2, 599–609. [DOI] [PubMed] [Google Scholar]
- 48.Forsberg E.C., Johnson,K., Zaboikina,T.N., Mosser,E.A. and Bresnick,E.H. (1999) Requirement of an E1A-sensitive coactivator for long-range transactivation by the beta-globin locus control region. J. Biol. Chem., 274, 26850–26859. [DOI] [PubMed] [Google Scholar]
- 49.Krumm A., Madisen,L., Yang,X.J., Goodman,R., Nakatani,Y. and Groudine,M. (1998) Long-distance transcriptional enhancement by the histone acetyltransferase PCAF. Proc. Natl Acad. Sci. USA, 95, 13501–13506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng X., Reginato,M.J., Andrew,N.C. and Lazar,M.A. (1997) The transcriptional integrator CREB-binding protein mediates positive cross talk between nuclear hormone receptors and the hematopoietic bZip protein p45/NF-E2. Mol. Cell. Biol., 17, 1407–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hung H.L., Kim,A.Y., Hong,W., Rakowski,C. and Blobel,G.A. (2001) Stimulation of NF-E2 DNA binding by CREB-binding protein (CBP)-mediated acetylation. J. Biol. Chem., 276, 10715–10721. [DOI] [PubMed] [Google Scholar]
- 52.Hung H.L., Lau,J., Kim,A.Y., Weiss,M.J. and Blobel,G.A. (1999) CREB-binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol. Cell. Biol., 19, 3496–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boyes J., Byfield,P., Nakatani,Y. and Ogryzko,V. (1998) Regulation of activity of the transcription factor GATA-1 by acetylation. Nature, 396, 594–598. [DOI] [PubMed] [Google Scholar]
- 54.Swope D.L., Mueller,C.L. and Chrivia,J.C. (1996) CREB-binding protein activates transcription through multiple domains. J. Biol. Chem., 271, 28138–28145. [DOI] [PubMed] [Google Scholar]
- 55.Korzus E., Torchia,J., Rose,D.W., Xu,L., Kurokawa,R., McInerney,E.M., Mullen,T.M., Glass,C.K. and Rosenfeld,M.G. (1998) Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science, 279, 703–707. [DOI] [PubMed] [Google Scholar]
- 56.Elnitski L., Li,J., Noguchi,C.T., Miller,W. and Hardison,R. (2001) A negative cis-element regulates the level of enhancement by hypersensitive site 2 of the beta-globin locus control region. J. Biol. Chem., 276, 6289–6298. [DOI] [PubMed] [Google Scholar]
- 57.Hong H., Kohli,K., Trivedi,A., Johnson,D.L. and Stallcup,M.R. (1996) GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc. Natl Acad. Sci. USA, 93, 4948–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Onate S.A., Boonyaratanakornkit,V., Spencer,T.E., Tsai,S.Y., Tsai,M.J., Edwards,D.P. and O‘Malley,B.W. (1998) The steroid receptor coactivator-1 contains multiple receptor interacting and activation domains that cooperatively enhance the activation function 1 (AF1) and AF2 domains of steroid receptors. J. Biol. Chem., 273, 101–108. [DOI] [PubMed] [Google Scholar]
- 59.Heery D.M., Kalkhoven,E., Hoare,S. and Parker,M.G. (1997) A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature, 387, 733–736. [DOI] [PubMed] [Google Scholar]
- 60.Chen H., Lin,R.L., Schiltz,L., Chakravarti,D., Nash,A., Nagy,L., Privalsky,M.L., Nakatani,Y. and Evans,R.M. (1997) Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric complex with P/CAF and CBP/p300. Cell, 90, 569–580. [DOI] [PubMed] [Google Scholar]
- 61.McInerney E.M., Rose,D.W., Flynn,S.E., Westin,S., Mullin,T.M., Krones,A., Inostroza,J., Torchia,J., Nolte,R.T., Assa-Munt,N. et al. (1998) Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev., 12, 3357–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blobel G.A., Nakajima,T., Eckner,R., Montminy,M. and Orkin,S.H. (1998) CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc. Natl Acad. Sci. USA, 95, 2061–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsang A.P., Visvader,J.E., Turner,C.A., Fujuwara,Y., Yu,C., Weiss,M.J., Crossley,M. and Orkin,S.H. (1997) FOG, a multitype zinc finger protein as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell, 90, 109–119. [DOI] [PubMed] [Google Scholar]
- 64.Gu W. and Roeder,R.G. (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell, 90, 595–606. [DOI] [PubMed] [Google Scholar]
- 65.Imhof A., Yang,X.J., Ogryzko,V.V., Nakatani,Y., Wolffe,A.P. and Ge,H. (1997) Acetylation of general transcription factors by histone acetyltransferases. Curr. Biol., 7, 689–692. [DOI] [PubMed] [Google Scholar]
- 66.Chen H., Lin,R.J., Xie,W., Wilpitz,D. and Evans,R.M. (1999) Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell, 98, 675–686. [DOI] [PubMed] [Google Scholar]
- 67.Kouzarides T. (2000) Acetylation: a regulatory modification to rival phosphorylation? EMBO J., 19, 1176–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang W. and Bieker,J.J. (1998) Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc. Natl Acad. Sci. USA, 95, 9855–9860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang W., Kadam,S., Emerson,B.M. and Bieker,J.J. (2001) Site-specific acetylation by p300 or CREB binding protein regulates erythroid Kruppel-like factor transcriptional activity via its interaction with the SWI-SNF complex. Mol. Cell. Biol., 21, 2413–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang S., Qiu,Y., Shi,Y., Xu,Z. and Brandt,S.J. (2000) P/CAF-mediated acetylation regulates the function of the basic helix–loop–helix transcription factor TAL1/SCL. EMBO J., 19, 6792–6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen C.J., Deng,Z., Kim,A.Y., Blobel,G.A. and Lieberman,P.M. (2001) Stimulation of CREB binding protein nucleosomal histone acetyltransferase activity by a class of transcriptional activators. Mol. Cell. Biol., 21, 476–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kraus W.L., Manning,E.T. and Kadonaga,J.T. (1999) Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol. Cell. Biol., 19, 8123–8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Archer T.K., Lefebvre,P., Wolford,R.G. and Hager,G.L. (1992) Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation [Erratum, Science (1992) 256, 161]. Science, 255, 1573–1576. [DOI] [PubMed] [Google Scholar]
- 74.Lee D., Hwang,S.G., Kim,J. and Choe,J. (2002) Functional interaction between p/CAF and human papillomavirus E2 protein. J. Biol. Chem., 277, 6483–6489. [DOI] [PubMed] [Google Scholar]
- 75.Recillas-Targa F., Bell,A.C. and Felsenfeld,G. (1999) Positional enhancer-blocking activity of the chicken beta-globin insulator in transiently transfected cells. Proc. Natl Acad. Sci. USA, 96, 14354–14359. [DOI] [PMC free article] [PubMed] [Google Scholar]