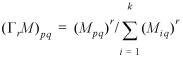Abstract
Detection of protein families in large databases is one of the principal research objectives in structural and functional genomics. Protein family classification can significantly contribute to the delineation of functional diversity of homologous proteins, the prediction of function based on domain architecture or the presence of sequence motifs as well as comparative genomics, providing valuable evolutionary insights. We present a novel approach called TRIBE-MCL for rapid and accurate clustering of protein sequences into families. The method relies on the Markov cluster (MCL) algorithm for the assignment of proteins into families based on precomputed sequence similarity information. This novel approach does not suffer from the problems that normally hinder other protein sequence clustering algorithms, such as the presence of multi-domain proteins, promiscuous domains and fragmented proteins. The method has been rigorously tested and validated on a number of very large databases, including SwissProt, InterPro, SCOP and the draft human genome. Our results indicate that the method is ideally suited to the rapid and accurate detection of protein families on a large scale. The method has been used to detect and categorise protein families within the draft human genome and the resulting families have been used to annotate a large proportion of human proteins.
INTRODUCTION
Genome projects are generating enormous amounts of sequence data (1) that need to be effectively analysed. The goal of functional genomics is to determine the function of proteins predicted from these sequencing projects (2–4). To achieve this goal, computational approaches can assist the classification of functional genomics targets. In particular, it is well known that members of the same protein family may possess similar or identical biochemical functions (5). Protein families can be defined as those groups of molecules which share significant sequence similarity (6). To detect a protein family, algorithms should take into account all similarity relationships in a given arbitrary set of sequences, a process that is defined as ‘sequence clustering’ (7). This approach is usually based on grouping homologous proteins together via a similarity measure obtained from direct sequence comparison. Ideally, the resulting clusters should correspond to protein families, whose members are related by a common evolutionary history (8). Well characterised proteins within a family can hence allow one to reliably assign functions to family members whose functions are not known or not well understood (7). The detection of protein families is also instrumental in the field of comparative genomics (9). Families may be specific to certain taxonomic groups or widespread across all domains of life (10), facts that can provide evolutionary insights into the underlying biology of organisms (11).
Many methods are currently available for the clustering of proteins into families. These methods generally rely on sequence similarity measures such as those obtained by BLAST (12) or other database search methods (13). One problem that these methods face is the detection of the multi-domain structure of many protein families (14). Proteins containing multiple domains can confound these methods and result in the incorrect grouping of proteins into families (15). The presence of a shared domain within a group of proteins does not necessarily imply that these proteins perform the same biochemical function (16). Ideally, these types of proteins should be classified into a single family only if they exhibit highly similar domain architectures. Apart from these relatively large, independently folded, protein domains (17), it has been realised that smaller, quite widespread protein modules exacerbate the problem even further (18). Many proteins sharing these so-called ‘promiscuous domains’ (e.g. SH2, WD40, DnaJ) (19) are known to have very different functions. Proteins assigned to a protein family purely on the basis of a promiscuous domain are unlikely to share a common evolutionary history with other members of that family.
The problem of detecting and classifying multi-domain proteins has been addressed by a number of approaches, which rely on the detection of individual domains using BLAST reports (20), domain database dictionaries (21,22) or iterative sequence comparison (23). Some of these methods rely on manual intervention for family assignment of multi-domain proteins (24). All the above mentioned techniques suffer from a number of drawbacks: they can be either too computationally intensive, somewhat inaccurate or not fully automatic, hence they do not allow the reliable and automatic detection of protein families within very large data sets, such as the human genome (25). Given the ever-increasing amount of genome sequence information (over half a million protein sequences) (26), it is imperative that protein sequence clustering methods be as robust and automatic as possible.
Despite significant progress in sequence clustering, new challenges have emerged, due to the availability of large eukaryotic genomes, in terms of their size and complexity (27). In particular, eukaryotic protein families constitute a bottleneck for most methods. Many eukaryotic proteins contain large numbers of protein domains (28,29), each of which needs to be detected and resolved by an efficient clustering algorithm. The iterative automatic domain detection algorithms (23) suffer from an excessive and unpredictable number of additional sequence comparison steps, which renders them somewhat impractical when using modest computational resources. Another approach would be to detect proteins with very similar domain architectures (30), rather than attempting to detect each domain individually. The assumption is that proteins with near-identical sets of domains may have very similar biochemical roles (5,31).
Previously, we developed the GeneRAGE algorithm for the clustering of proteins within complete genomes (23). This algorithm utilises a comprehensive system of error detection and correction using the Smith–Waterman dynamic programming alignment algorithm (32). The problem of multi-domain proteins is addressed using sequence comparison to detect domains using a domain detection algorithm (also based on Smith–Waterman). This algorithm was developed and tested on protein families within relatively small data sets, such as prokaryotic genomes (33). Given such data sets, the algorithm effectively and accurately identifies protein families and also correctly detects multi-domain proteins (34). When the algorithm is applied to larger data sets, such as those obtained from eukaryotic organisms, some of the above mentioned problems become apparent. The detection of protein domains using GeneRAGE becomes hampered to a large extent by promiscuous domains, peptide fragments (representing incomplete database entries) and proteins of complex domain structure. Domains such as a ‘response regulator’ domain from two-component systems (35) cause proteins with vastly differing functions (such as heat shock factors and phytochromes) (36) to be assigned incorrectly to the same family (37).
Given the difficulty of detecting such domains accurately and the ever-increasing amount of eukaryotic data available, we have re-approached sequence clustering using an elegant mathematical approach based on probability and graph flow theory. Sequence similarity search algorithms have previously benefited from such approaches, for example hidden Markov model-based search algorithms provide very sensitive detection of distant protein sequence similarity (38). An ideal method would require sequence similarity relationships as input and be able to rapidly detect clusters solely using this information, without being led astray by the complex modular domain structure of eukaryotic proteins. Traditionally, most methods deal with similarity relationships in a pairwise manner, while graph theory allows the classification of proteins into families based on a global treatment of all relationships in similarity space simultaneously. To this end, we present the TRIBE-MCL algorithm as an efficient and reliable method for sequence clustering. TRIBE-MCL is based on the Markov cluster (MCL) algorithm, previously developed for graph clustering using flow simulation (39). This approach for protein sequence clustering is astonishingly fast and highly accurate. It avoids most of the problems mentioned above and has already been successfully utilised for the clustering of large data sets and family annotation of the draft human genome (25) (see also www.ensembl.org).
MATERIALS AND METHODS
Data handling
A FASTA file containing all sequences that are to be clustered into families is assembled. This file is filtered using an accurate and sensitive compositional bias detection algorithm, CAST (40), then compared against itself using BLAST (12). The all-against-all sequence similarities generated by this analysis are parsed and stored in a square matrix.
Algorithm
This matrix represents sequence similarities as a connection graph. Nodes of the graph represent proteins, and edges represent sequence similarity that connects such proteins. A weight is assigned to each edge by taking the average pairwise –log10(E-value) (12), resulting in a symmetric matrix. We have found that this simple weighting scheme produces reliable results. Other more complex schemes may be devised in future research, for example length-based weighting. These weights are transformed into probabilities associated with a transition from one protein to another within this graph. This matrix is passed through iterative rounds of matrix multiplication and matrix inflation (see below) until there is little or no net change in the matrix. The final matrix is then interpreted as a protein family clustering. The inflation value parameter of the MCL algorithm is used to control the granularity (or ‘tightness’) of these clusters.
Availability
The original MCL algorithm and additional information is available at http://members.ams.chello.nl/svandong/thesis/index.html. The additional modules for protein sequence analysis can be obtained from the authors on request; more information is also available at http://www.ebi.ac.uk/research/cgg/services/tribe/.
MARKOV CLUSTERING OF SEQUENCE SIMILARITIES
Data representation
Sequence similarity relationships within a given protein data set can be represented as a square matrix, whose elements contain similarity metrics for any pair of proteins in the data set. These elements can be binary numbers (23) or real numbers [e.g. E-values from BLAST (12)]. Alternatively, this matrix can be considered as a weighted graph, whose nodes (vertices) represent proteins and connections (edges) represent similarity relationships. It has been realised that such graphs are an elegant and concise way of representing sequence similarity relationships (41). Furthermore, these graphs are amenable to graph clustering algorithms, developed in the fields of mathematics and computer science. Such algorithms include single-linkage clustering and k-means (42), with which we have extensively experimented, before choosing the MCL algorithm, because of its relevance, elegance and efficiency.
The algorithm
The MCL algorithm is an algorithm designed specifically for the settings of simple graphs and weighted graphs (43). It has previously been used in the field of computational graph clustering (39,43,44). Given that it is possible to represent biological sequence similarity relationships in terms of these graphs (23,41), it is possible to use an algorithm such as MCL for biological sequence clustering.
Natural clusters in a graph are characterised by the presence of many edges between the members of that cluster, and one expects that the number of ‘higher-length’ (longer) paths between two arbitrary nodes in the cluster is high. In particular, this number should be high, relative to node pairs lying in different natural clusters. A different angle on this is that random walks on the graph will infrequently go from one natural cluster to another, based on graph transition probability estimates.
The MCL algorithm finds cluster structure in graphs by a mathematical bootstrapping procedure. The process deterministically computes (the probabilities of) random walks through the sequence similarity graph, and uses two operators transforming one set of probabilities into another. It does so using the language of stochastic matrices (also called Markov matrices) which capture the mathematical concept of random walks on a graph.
The MCL algorithm simulates random walks within a graph by alternation of two operators called expansion and inflation. Expansion coincides with taking the power of a stochastic matrix using the normal matrix product (i.e. matrix squaring). Inflation corresponds with taking the Hadamard power of a matrix (taking powers entrywise), followed by a scaling step, such that the resulting matrix is stochastic again, i.e. the matrix elements (on each column) correspond to probability values.
Definition of the inflation operator
A column stochastic matrix is a non-negative matrix with the property that each of its columns sums to 1. Given such a matrix M Œ Rk×k, M ≥ 0, and a real number, r > 1, the column stochastic matrix resulting from inflating each of the columns of M with power coefficient r is written GrM, and Gr is called the inflation operator with power coefficient r. Formally, the action of Gr: Rk×k → Rk×k is defined by:
Each column j of a stochastic matrix M corresponds with node j of the stochastic graph associated with M. Row entry i in column j (i.e. the matrix entry Mij) corresponds with the probability of going from node j to node i. It is observed that for values of r > 1, inflation changes the probabilities associated with the collection of random walks departing from one particular node (corresponding with a matrix column) by favouring more probable walks over less probable walks.
Expansion corresponds to computing random walks of ‘higher length’, which means random walks with many steps. It associates new probabilities with all pairs of nodes, where one node is the point of departure and the other is the destination. Since higher length paths are more common within clusters than between different clusters, the probabilities associated with node pairs lying in the same cluster will, in general, be relatively large as there are many ways of going from one to the other. Inflation will then have the effect of boosting the probabilities of intra-cluster walks and will demote inter-cluster walks. This is achieved without any a priori knowledge of cluster structure. It is simply the result of cluster structure being present. This property of the algorithm lends itself well to the problem of biological sequence comparison (see below).
Eventually, iterating expansion and inflation results in the separation of the graph into different segments. There are no longer any paths between these segments and the collection of resulting segments is simply interpreted as a clustering. The inflation operator can be altered using the parameter r. Increasing this parameter has the effect of making the inflation operator stronger, and this increases the granularity or ‘tightness’ of clusters.
Cast in the language of stochastic flow, we can state that expansion causes flow to dissipate within clusters whereas inflation eliminates flow between different clusters. Expansion and inflation represent different tidal forces which are alternated until an equilibrium state is reached. An equilibrium state takes the form of a so-called doubly idempotent matrix, i.e. a matrix that does not change with further expansion or inflation steps. The graph associated with such a matrix consists of different connected directed components. Each component is interpreted as a cluster, and has a star-like form, with one attractor in the centre and arcs going from all nodes of that component to the attractor. In theory, attractor systems with more than one attractor may occur (these do not change the cluster interpretation). Also, nodes may exist that are connected to different stars, which is canonically interpreted as cluster overlap, or in other words nodes may belong to multiple clusters (39,43,44).
With respect to convergence, it can be proven that the process simulated by the algorithm converges quadratically around the equilibrium states. In practice, the algorithm starts to converge noticeably after 3–10 iterations. Global convergence is very hard to prove; it is conjectured that the process always converges if the input graph is symmetric (39,43,44). This conjecture is supported by results concerning the matrix iterands. For symmetric input graphs, it is true that all iterands have real spectrum (the set of eigenvalues), and that all iterands resulting from expansion have non-negative spectrum and are diagonally symmetric to a positive semi-definite matrix. It can be shown that these matrices have a structural property which associates a directed acyclic graph (DAG) with each of them. It turns out that inflation strengthens (in a quantitative sense) this structural property and will never change the associated DAG, whereas expansion is in fact able to change the associated DAG. This is a more mathematical view on the ‘tidal forces’ analogy mentioned earlier. DAGs generalise the star graphs associated with MCL limits, and the spectral properties of MCL iterands and MCL limits can be related via the inflation operator. These results imply that the equilibrium states can be viewed as a set of extreme points of the set of matrices that are diagonally similar to a positive semi-definite matrix. This establishes a close relationship between the MCL iterands, MCL limits, and cluster (and DAG) structure in graphs (45).
The MCL algorithm also associates return probabilities (or loops) with each node in the initial input graph. The flow paradigm underlying MCL naturally requires this, and it can be motivated in terms of the spectral and structural properties mentioned earlier. As for the weights that are chosen, experience shows that a ‘neutral’ value works well. In the implementation used, ‘neutral’ is chosen as a weight (in principle different for each node) that will not change when the inflation operator is applied to the stochastic column associated with the node. It is possible to choose larger weights (see Fig. 2), and this will increase cluster granularity. The effect is secondary, however, to that of varying the inflation parameter, and the algorithm is not very sensitive to changes in the loop weights.
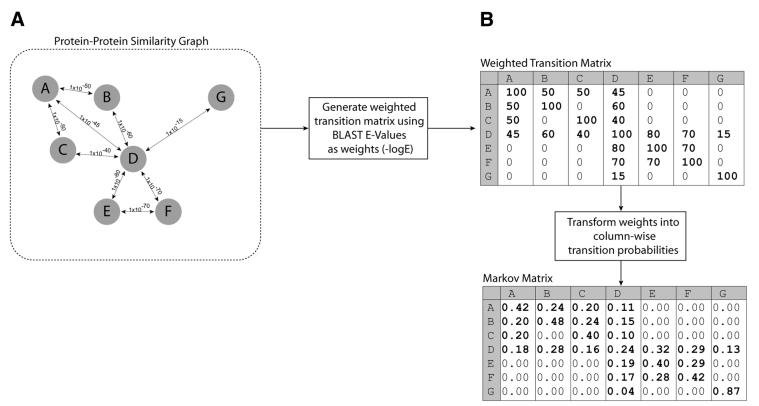
Figure 2.
(A) Example of a protein–protein similarity graph for seven proteins (A–F), circles represent proteins (nodes) and lines (edges) represent detected BLASTp similarities with E-values (also shown). (B) Weighted transition matrix and associated column stochastic Markov matrix for the seven proteins shown in (A). For explanations, please see text.
A very important asset of the algorithm is its ‘bootstrapping’ nature, retrieving cluster structure via the imprint made by this structure on the flow process. Further key benefits of the algorithm are (i) it is not misled by edges linking different clusters; (ii) it is very fast and very scalable; (iii) it has a natural parameter for influencing cluster granularity; (iv) the mathematics associated with the algorithm shows that there is an intrinsic relationship between the process it simulates and cluster structure in the input graph (45); and (v) its formulation is simple and elegant.
From the definition of the MCL algorithm it is seen that it is based on a very different paradigm than any linkage-based algorithm. One possible view of this is that MCL, although based on similarities between pairs, recombines these similarities (via expansion) and is thus affected by similarities on the level of sets (as generalising pairs). Alternating expansion with inflation turns out to be an appropriate way of exploiting this recombination property.
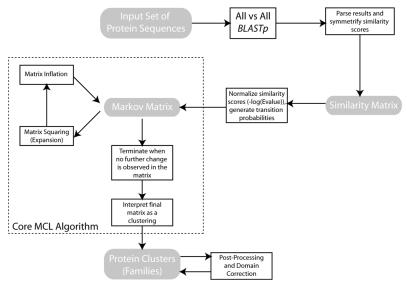
The structure of the MCL algorithm is fully described in Figure 1. The algorithm sets out by computing the graph of random walks of an input graph, yielding a stochastic matrix. It then alternates the expansion operator that squares a matrix using the usual matrix product with the inflation operator. Inflation is done by raising each matrix entry to a given power and rescaling the matrix so that it becomes stochastic again. Alternation continues until an equilibrium state is reached in the form of a so-called doubly idempotent matrix.
Figure 1.
Flowchart of the TRIBE-MCL algorithm.
Application of the MCL algorithm to biological graphs
The section above describes the MCL algorithm in a general fashion. In this section, we describe how the algorithm relates to the clustering of proteins into protein families. Biological graphs may be represented as follows (Fig. 2A): (i) nodes of the graph represent a set of proteins that we would like to assign to families; (ii) edges within the graph represent similarity between these proteins; (iii) edges are weighted according to a sequence similarity score obtained from an algorithm such as BLAST.
A Markov matrix (Fig. 2B) is constructed, representing transition probabilities from any protein in the graph to any other protein for which a similarity has been detected. Each column of the matrix represents a given protein, and each entry in a column represents a similarity between this protein and another protein. Diagonal elements are set arbitrarily to a ‘neutral’ value as described above. The entries in the Markov matrix are probabilities generated from weighted sequence similarity scores (e.g. from BLAST). This Markov matrix is supplied to the MCL algorithm. Initial expansion of the Markov matrix simulates random walks, which allow one to measure ‘flow’ in the graph. Areas of high flow indicate that a large number of random walks go through this area. The MCL algorithm uses iterative rounds of expansion and inflation (explained earlier) to promote flow through the graph where it is strong, and remove flow where it is weak. This process terminates when equilibrium has been reached, i.e. further rounds of expansion and inflation leave the matrix unaltered.
In a biological sense, we expect that members of a protein family will be more similar to each other than to proteins in another family. Experiments using the Bio-Layout graph visualisation algorithm (41) have shown this to be true for most protein similarity graphs. Because of this property of biological graphs, flow within protein families is strong, i.e. a random walk starting at any given protein in a family is more likely to linger within this family than to cross to another family. Flow between protein families will be weaker than flow within a family as there are relatively few (if any) paths that cross two distinct protein families. Inter-family paths represent either sequence similarity relationships due to multi-domain proteins or mere false positive similarity detections. These properties of biological similarity graphs make them ideally suited to the MCL algorithm. The iterative rounds of inflation and expansion remove this weak flow across protein families, and promote the stronger flow within protein families. This boot-strapping procedure allows protein families hidden in the graph to become visible by gradually stripping the graph down to its basic components as detected by stochastic flow.
Many of the problems that normally hinder protein sequence clustering are eliminated by the MCL approach. Proteins possessing a promiscuous domain, which is present in many functionally unrelated proteins, are normally very difficult to cluster correctly. Promiscuous domains will connect a member of a given protein family to all members of that family and possibly to other protein families. Because these inter-family connections are still far fewer than intra-family connections, the algorithm gradually eliminates these inter-family similarities and detects protein families accurately. The algorithm requires no a priori knowledge of protein domains, and clusters proteins into families purely based on observed relationships through the entire similarity graph. However, proteins containing different domains or sets of domains will have very different sequence similarity patterns, and hence we expect the MCL algorithm to cluster proteins with different domain structures into distinct families. We have extensively validated the performance of the algorithm in terms of speed and accuracy. We have also assessed the performance of the algorithm in terms of the quality of protein family descriptions, based on database annotations.
VALIDATION OF THE ALGORITHM
In order to test the effectiveness of protein family detection using TRIBE-MCL, we have performed extensive validation using the InterPro protein domain database (46) and the Structural Classification of Proteins (SCOP) database (47). These databases contain extensive information relating to protein domains and structures. Ideally, clusters detected by the TRIBE-MCL algorithm should have similar domain architectures, including sequence patterns and protein folds, based on InterPro and SCOP, respectively.
InterPro validation
The InterPro database (46) is a collection of protein domains and functional signatures from multiple databases such as PRINTS (48), PFAM (21) and PROSITE (49). This well curated database contains a vast amount of information relating to protein domains and sequence motifs. It is possible to obtain InterPro information for many entries in the SwissProt database (50). In order to validate our clustering algorithm, we took the SwissProt protein database (80 000 proteins) and clustered it into 8332 families using the TRIBE-MCL algorithm. This analysis took ∼5 min to complete on a Sun Ultra 10 workstation. Protein family and domain information for each SwissProt protein was extracted (if available) from the InterPro database. Of the 8332 families, 1821 families contain four or more members with corresponding InterPro annotations. Families that do not contain four or more annotated members are discarded. For each of the 1821 families, we determine the domain structure of annotated members of that family, according to InterPro domain classifications, and retain the most frequently occurring domain combination.
This analysis is performed in order to determine which families exhibit robust domain combinations in contrast to less well defined protein families which may display disparate (or even conflicting) domain architecture. Interestingly, 1583 families (out of 1821 or 87%) display full correspondence of domain structure across all annotated members. When individual proteins are considered, we count the proportion of proteins with identified InterPro domain combinations identical to the most frequently occurring domain combination of the cluster they belong to. The number of proteins with this property is 14 188 out of a total of 14 409 proteins considered (98%); this value can be considered as an estimate of the classification precision according to InterPro. This result illustrates that although the algorithm has no fixed concept of protein domains, the resulting families have a very consistent domain structure, indicating accurate and meaningful clustering. Although the second set was also clustered using TRIBE-MCL, no validation was possible due to limited available annotation of the detected families from InterPro.
SCOP validation
The SCOP database is a collection of well characterised proteins for which three-dimensional structures are available (47). These proteins have been expertly classified into families based on their folding patterns and a variety of other information. Given that family information for these proteins is well understood and accurately represented in SCOP, it was decided to cluster all proteins in the PDB (18 248 entries) into protein families using the TRIBE-MCL algorithm at multiple inflation values (corresponding to different cluster granularity). This analysis detected 1167 families (inflation value 1.1)—and with increasing inflation values of 2, 3, 4 and 5 the number of families is 1395, 1606, 1672 and 1761, respectively. For each set of clusters (i.e. families), we determine the most frequently occurring SCOP annotation, as above. We also count the number of distinct clusters containing identical SCOP annotations in the same way. Similar to the InterPro validation, we calculate the total number of proteins in clusters with SCOP classifications consistent with the cluster SCOP assignment. For higher inflation values (i.e. tighter clustering), this precision estimate is highest: 87% for inflation value 5 decreasing to 79% for the lowest inflation value of 1.1. These results further indicate that the clustering obtained by TRIBE-MCL is accurately and consistently assigning proteins into families, despite the fact that this classification relies on structural similarities, which are not all readily detectable at the sequence level.
LARGE-SCALE FAMILY DETECTION
An analysis of the effects of promiscuous domains
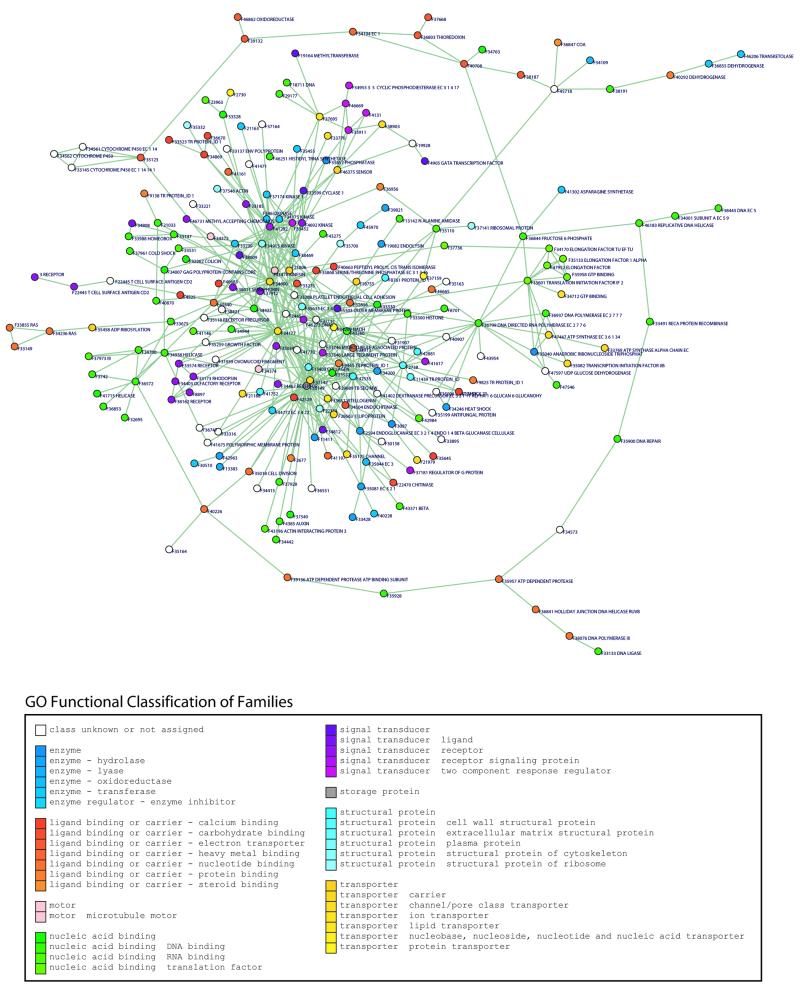
As mentioned above, TRIBE-MCL does not require any explicit knowledge of protein domains to detect protein families. This feature can be used for the analysis of domains that are present in many families, such as promiscuous domains. We have decided to analyse the presence of these domains in the SwissProt database using TRIBE-MCL, and estimate how frequently they occur, based on the presence of InterPro (46) entries in the corresponding SwissProt (50) sequence. In other words, for each protein entry in SwissProt containing a detectable domain with InterPro, we count how many different protein families we have detected that contain this domain. A list of these promiscuous domains in SwissProt is described in Table 1. It is surprising that the largest set of proteins that are interconnected through promiscuous domains comprises of 237 protein families (21 727 sequences in total), corresponding to 22.3% of all SwissProt entries (Fig. 3). Although the spectacular complexity of these interconnected families and the range of their functional properties has been suspected before (14), this is the first time that we obtain a glimpse of this effect at this scale using clustering and visualisation. This effect arises for a number of reasons: first, the family detection is accurate but the corresponding domain is falsely identified by InterPro (e.g. crystallin, shared by 141 families; Table 1); secondly, the presence of these domains in unrelated families represents a meaningful biological phenomenon (e.g. RNA-binding region RNP-1, shared by 110 families; Table 1); thirdly, the granularity of family definition is sometimes too high, resulting in many distantly related families to contain similar motifs (e.g. immunoglobulin and major histocompatibility complex domain, shared by 107 families; Table 1).
Table 1. The top 50 promiscuous domains from InterPro occurring in distinct SwissProt protein families identified by TRIBE-MCL.
| InterPro ID | No. of families | Domain description |
|---|---|---|
| IPR001064 | 141 | Crystallin |
| IPR000504 | 110 | RNA-binding region RNP-1 (RNA recognition motif) |
| IPR003006 | 107 | Immunoglobulin and major histocompatibility complex domain |
| IPR000531 | 97 | TonB-dependent receptor protein |
| IPR003015 | 96 | Myc-type, helix–loop–helix dimerisation domain |
| IPR001680 | 76 | G-protein β WD-40 repeats |
| IPR000561 | 73 | EGF-like domain |
| IPR000169 | 72 | Eukaryotic thiol (cysteine) proteases active sites |
| IPR000255 | 67 | Phosphopantetheine attachment site |
| IPR001899 | 65 | Gram-positive cocci surface protein ‘anchoring’ hexapeptide |
| IPR001450 | 60 | 4Fe–4S ferredoxin, iron–sulfur binding domain |
| IPR000130 | 54 | Neutral zinc metallopeptidases, zinc-binding region |
| IPR000205 | 54 | NAD-binding site |
| IPR001005 | 54 | Myb DNA-binding domain |
| IPR001440 | 52 | TPR repeat |
| IPR001356 | 49 | Homeobox domain |
| IPR000822 | 45 | Zinc finger, C2H2 type |
| IPR001841 | 43 | RING finger |
| IPR000005 | 42 | AraC type helix–turn–helix domain |
| IPR001777 | 42 | Fibronectin type III domain |
| IPR001452 | 38 | Src homology 3 (SH3) domain |
| IPR002290 | 37 | Serine/Threonine protein kinase family active site |
| IPR000886 | 34 | Endoplasmic reticulum targeting sequence |
| IPR001304 | 32 | C-type lectin domain |
| IPR000194 | 31 | ATP synthase α and β subunit, N-terminal |
| IPR002203 | 29 | Protein splicing (intein) |
| IPR000923 | 28 | Type-1 copper (blue) domain |
| IPR001092 | 28 | Helix–loop–helix dimerisation domain |
| IPR001789 | 28 | Response regulator receiver domain |
| IPR001611 | 27 | Leucine-rich repeat |
| IPR001917 | 27 | Aminotransferases class II |
| IPR000063 | 24 | Thioredoxin family |
| IPR002110 | 24 | Ankyrin-repeat |
| IPR001220 | 23 | Legume lectins β |
| IPR003009 | 23 | Proteins binding FMN and related compounds core region |
| IPR000524 | 22 | Bacterial regulatory proteins, GntR family |
| IPR002114 | 22 | Serine phosphorylation site in HPr protein |
| IPR000014 | 21 | PAS domain |
| IPR001478 | 20 | PDZ domain (also known as DHR or GLGF) |
| IPR000792 | 19 | Bacterial regulatory protein, LuxR family |
| IPR001650 | 19 | Helicase C-terminal domain |
| IPR002088 | 19 | Protein prenyltransferases α subunit repeat |
| IPR000644 | 18 | CBS domain |
| IPR002035 | 18 | von Willebrand factor type A domain |
| IPR000047 | 17 | λ and other repressor helix–turn–helix |
| IPR000086 | 17 | NUDIX hydrolase domain |
| IPR001623 | 17 | DnaJ N-terminal domain |
| IPR002223 | 17 | Pancreatic trypsin inhibitor (Kunitz) family |
| IPR000437 | 16 | Prokaryotic membrane lipoprotein lipid attachment site |
| IPR000583 | 16 | Glutamine amidotransferase class II |
Column names: InterPro ID, the InterPro accession number, No. of families, the number of families in which the corresponding domain is present; Domain description, the InterPro description line.
Figure 3.
Graph representing the largest interconnected group of protein families from the SwissProt protein database (237 protein families, 21 727 sequences in total). Circles represent protein families, with associated family Ids and annotations (where known). Edges show BLAST similarities between families. Circles are coloured according to the GeneOntology (GO) (52) functional class assignments (where available). This graph was generated using the Bio-Layout graph layout algorithm (41).
Protein family analysis of the draft human genome
The release of the publicly sequenced draft human genome necessitated the need for protein family analysis and functional annotation to be performed and made available for researchers. This type of information can be very useful for locating similar genes to a gene of interest or for assigning function to previously unknown genes. To achieve high-quality annotation automatically, functional descriptions for all genome sequences might be obtained from curated database entries, based on detected sequence similarities and subsequent family assignments. To this end, TRIBE-MCL was applied to a full set of peptides from the EnsEMBL 0.80 release (29 691 proteins) of the draft human genome along with a vertebrate subset of proteins from the SwissProt and SPTrEMBL databases (73 347 entries). All protein similarities within these 103 038 proteins were detected using BLAST (12). This information (over 15 million protein similarities) was used by the TRIBE-MCL algorithm to detect all protein families within these three data sets. Given the pre-computed BLAST-based similarities, the clustering step took ∼15 min on a single CPU of a Compaq ES40 server. Finally, the RLCS algorithm (A.J.Enright, unpublished data) was then used to determine a consensus annotation for each detected protein family.
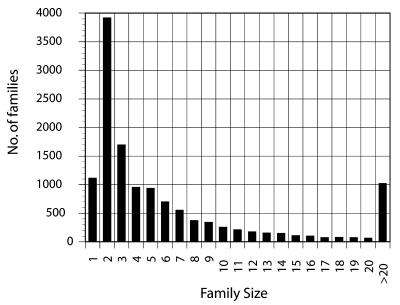
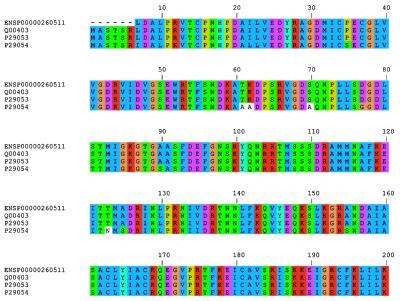
In all, 13 023 protein families were detected, of which 11 481 families (88% of the total) are human specific. On average, each human protein family contains 2.5 members, while there are only 1110 single-member families (3% of the total number of families). The family size distribution has an exponential shape, with hundreds of protein families with more than 20 members (Fig. 4) and 347 families with more than 50 members (data not shown), indicating a high degree of paralogy. Some well known families that are detected are zinc finger-containing proteins, olfactory receptors, members of the ras superfamily of GTPases, myosin, actin, keratin, immunoglobulin, certain ribosomal proteins and multiple kinase types (Table 2). The procedure has detected many well known families and a number of novel families in the human genome whose functions are either unknown or predicted. As an example, we show that the TFIIB family of proteins (51) has been identified correctly, containing the human, rat and Xenopus homologues (Fig. 5). Despite the fact that the quality of clusters is very high, some of the largest families (with more than 1000 members) may contain a number of unrelated members. This usually arises from the presence of multiply repeated sequence patterns and not the presence of individual promiscuous domains. We are working towards the definition of multiple levels of protein family classification, using post-processing of the initial clusters with multiple-threshold clustering.
Figure 4.
Distribution of protein family sizes within the human genome. The x-axis represents family size and the y-axis (bars) indicates the number of paralogous protein families.
Table 2. The 28 largest protein families in the draft human genome recorded in Ensembl 0.80 together with their automatically derived consensus annotations and the total number of sequences (from Ensembl, SwissProt and SPTrembl) that they contain.
| Ensembl 080 family | Automatic annotation | No. of peptides |
|---|---|---|
| ENSF00000002017 | Zinc finger protein | 1743 |
| ENSF00000002558 | Class II histocompatibility antigen, β chain | 1497 |
| ENSF00000004397 | Cytochrome B | 1231 |
| ENSF00000004396 | Cytochrome B | 1122 |
| ENSF00000002016 | Olfactory receptor | 975 |
| ENSF00000002557 | Class I histocompatibility antigen, α chain precursor | 814 |
| ENSF00000004395 | Cytochrome B fragment | 782 |
| ENSF00000004394 | Cytochrome B | 731 |
| ENSF00000004718 | Cytochrome C oxidase polypeptide I EC 1.9.3.1 | 648 |
| ENSF00000002556 | HLA Class I histocompatibility antigen, B α chain precursor | 526 |
| ENSF00000006350 | NADH ubiquinone oxidoreductase chain 4 EC 1.6.5.3 | 456 |
| ENSF00000002015 | Myosin heavy chain | 455 |
| ENSF00000004393 | Cytochrome B | 447 |
| ENSF00000004392 | Cytochrome B fragment | 435 |
| ENSF00000006349 | NADH ubiquinone oxidoreductase chain 2 EC 1.6.5.3 | 419 |
| ENSF00000002555 | Class II histocompatibility antigen, α chain | 398 |
| ENSF00000002013 | Protein tyrosine phosphatase, non-receptor type EC 3.1.3.48 protein tyrosine phosphatase | 381 |
| ENSF00000002645 | Haemoglobin chain | 375 |
| ENSF00000002014 | Receptor precursor EC 2.7.1.112 | 368 |
| ENSF00000002012 | Unknown | 355 |
| ENSF00000002009 | Cadherin-related tumour suppressor homologue precursor fat protein homologue | 349 |
| ENSF00000004391 | Cytochrome B | 341 |
| ENSF00000002554 | HLA Class II histocompatibility antigen, β chain precursor | 341 |
| ENSF00000002010 | Protein EC 2.7.1.- | 341 |
| ENSF00000002644 | Haemoglobin α chain | 338 |
| ENSF00000006348 | NADH ubiquinone oxidoreductase chain 2 EC 1.6.5.3 | 328 |
| ENSF00000002011 | EC 3.4.21.- | 327 |
| ENSF00000002553 | Class I histocompatibility antigen, α chain precursor | 317 |
Figure 5.
Protein sequence alignment of the eukaryotic TFIIB family of proteins detected using TRIBE-MCL, including three members from SwissProt (accession numbers given) and the human TFIIB (51).
The largest family identified (EnsEMBL 0.80 release) is a class of zinc finger-containing transcription factors, while the largest unannotated family contains 355 members (Table 2). All detected protein families were subsequently made available as part of the EnsEMBL 0.80 release. The clustering is fully accessible at www.ensembl.org and is continually being updated with new versions of the EnsEMBL database. It is worth mentioning that the annotations derived from the families detected by TRIBE-MCL are being used as annotations for a large number of human genes at EnsEMBL.
DISCUSSION
We have presented a novel algorithm that generates accurate protein families using the MCL formalism for graph clustering by flow simulation. The actual implementation of the algorithm allows the efficient and rapid clustering of any arbitrary set of protein sequences, given a list of all pairwise similarities obtained by another method, such as BLAST. Because the method does not operate directly on sequences but on a graph that contains similarity information, it avoids the expensive step of sequence alignment. Instead, global patterns of sequence similarity are detected and used to partition the similarity graph into protein families.
The quality of the clustering is impressive, as validated using the available protein domain and structure databases—InterPro and SCOP, respectively. Up to 95% agreement can be obtained in a comparison of the resulting classification using TRIBE-MCL and the manually curated InterPro database. Given the speed and quality of the resulting clusters, TRIBE-MCL has been used to cluster all human genes (from the EnsEMBL project) into annotated protein families. This task would previously have been prohibitively expensive to achieve in such a short period of time. We hope that the method will become widely used by the community and find some other interesting applications.
REFERENCES
- 1.Bernal A., Ear,U. and Kyrpides,N. (2001) Genomes OnLine Database (GOLD): a monitor of genome projects world-wide. Nucleic Acids Res., 29, 126–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsoka S. and Ouzounis,C.A. (2000) Recent developments and future directions in computational genomics. FEBS Lett., 480, 42–48. [DOI] [PubMed] [Google Scholar]
- 3.Eisenberg D., Marcotte,E.M., Xenarios,I. and Yeates,T.O. (2000) Protein function in the post-genomic era. Nature, 405, 823–826. [DOI] [PubMed] [Google Scholar]
- 4.Bork P., Dandekar,T., Diaz-Lazcoz,Y., Eisenhaber,F., Huynen,M. and Yuan,Y. (1998) Predicting function: from genes to genomes and back. J. Mol. Biol., 283, 707–725. [DOI] [PubMed] [Google Scholar]
- 5.Hegyi H. and Gerstein,M. (1999) The relationship between protein structure and function: a comprehensive survey with application to the yeast genome. J. Mol. Biol., 288, 147–164. [DOI] [PubMed] [Google Scholar]
- 6.Dayhoff M.O. (1976) The origin and evolution of protein superfamilies. Fed. Proc., 35, 2132–2138. [PubMed] [Google Scholar]
- 7.Heger A. and Holm,L. (2000) Towards a covering set of protein family profiles. Prog. Biophys. Mol. Biol., 73, 321–337. [DOI] [PubMed] [Google Scholar]
- 8.Fitch W.M. (1973) Aspects of molecular evolution. Annu. Rev. Genet., 7, 343–380. [DOI] [PubMed] [Google Scholar]
- 9.Ouzounis C., Casari,G., Sander,C., Tamames,J. and Valencia,A. (1996) Computational comparisons of model genomes. Trends Biotechnol., 14, 280–285. [DOI] [PubMed] [Google Scholar]
- 10.Ouzounis C. and Kyrpides,N. (1996) The emergence of major cellular processes in evolution. FEBS Lett., 390, 119–123. [DOI] [PubMed] [Google Scholar]
- 11.Rubin G.M., Yandell,M.D., Wortman,J.R., Gabor Miklos,G.L., Nelson,C.R., Hariharan,I.K., Fortini,M.E., Li,P.W., Apweiler,R., Fleischmann,W. et al. (2000) Comparative genomics of the eukaryotes. Science, 287, 2204–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eddy S.R. (1996) Hidden Markov models. Curr. Opin. Struct. Biol., 6, 361–365. [DOI] [PubMed] [Google Scholar]
- 14.Doolittle R.F. (1995) The multiplicity of domains in proteins. Annu. Rev. Biochem., 64, 287–314. [DOI] [PubMed] [Google Scholar]
- 15.Smith T.F. and Zhang,X. (1997) The challenges of genome sequence annotation or ‘the devil is in the details’. Nat. Biotechnol., 15, 1222–1223. [DOI] [PubMed] [Google Scholar]
- 16.Henikoff S., Greene,E.A., Pietrokovski,S., Bork,P., Attwood,T.K. and Hood,L. (1997) Gene families: the taxonomy of protein paralogs and chimeras. Science, 278, 609–614. [DOI] [PubMed] [Google Scholar]
- 17.Enright A.J., Iliopoulos,I., Kyrpides,N.C. and Ouzounis,C.A. (1999) Protein interaction maps for complete genomes based on gene fusion events. Nature, 402, 86–90. [DOI] [PubMed] [Google Scholar]
- 18.Doolittle R.F. and Bork,P. (1993) Evolutionarily mobile modules in proteins. Sci. Am., 269, 50–56. [DOI] [PubMed] [Google Scholar]
- 19.Marcotte E.M., Pellegrini,M., Ng,H.L., Rice,D.W., Yeates,T.O. and Eisenberg,D. (1999) Detecting protein function and protein-protein interactions from genome sequences. Science, 285, 751–753. [DOI] [PubMed] [Google Scholar]
- 20.Guan X. (1997) Domain identification by clustering sequence alignments. Proc. Int. Conf. Intell. Syst. Mol. Biol., 5, 124–130. [PubMed] [Google Scholar]
- 21.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Howe,K.L. and Sonnhammer,E.L. (2000) The Pfam protein families database. Nucleic Acids Res., 28, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corpet F., Gouzy,J. and Kahn,D. (1998) The ProDom database of protein domain families. Nucleic Acids Res., 26, 323–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enright A.J. and Ouzounis,C.A. (2000) GeneRAGE: a robust algorithm for sequence clustering and domain detection. Bioinformatics, 16, 451–457. [DOI] [PubMed] [Google Scholar]
- 24.Tatusov R.L., Natale,D.A., Garkavtsev,I.V., Tatusova,T.A., Shankavaram,U.T., Rao,B.S., Kiryutin,B., Galperin,M.Y., Fedorova,N.D. and Koonin,E.V. (2001) The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res., 29, 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 26.Iliopoulos I., Tsoka,S., Andrade,M.A., Janssen,P., Audit,B., Tramontano,A., Valencia,A., Leroy,C., Sander,C. and Ouzounis,C.A. (2001) Genome sequences and great expectations. Genome Biol., 2, INTERACTIONS0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Birney E., Bateman,A., Clamp,M.E. and Hubbard,T.J. (2001) Mining the draft human genome. Nature, 409, 827–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hegyi H. and Bork,P. (1997) On the classification and evolution of protein modules. J. Protein Chem., 16, 545–551. [DOI] [PubMed] [Google Scholar]
- 29.Apic G., Gough,J. and Teichmann,S.A. (2001) Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J. Mol. Biol., 310, 311–325. [DOI] [PubMed] [Google Scholar]
- 30.Apic G., Gough,J. and Teichmann,S.A. (2001) An insight into domain combinations. Bioinformatics, 17 (Suppl. 1), S83–S89. [DOI] [PubMed] [Google Scholar]
- 31.Ouzounis C.A. and Karp,P.D. (2000) Global properties of the metabolic map of Escherichia coli. Genome Res., 10, 568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith T.F. and Waterman,M.S. (1981) Identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. [DOI] [PubMed] [Google Scholar]
- 33.Janssen P.J., Audit,B. and Ouzounis,C.A. (2001) Strain-specific genes of Helicobacter pylori: distribution, function and dynamics. Nucleic Acids Res., 29, 4395–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coulson R.M., Enright,A.J. and Ouzounis,C.A. (2001) Transcription-associated protein families are primarily taxon-specific. Bioinformatics, 17, 95–97. [DOI] [PubMed] [Google Scholar]
- 35.Stock A.M., Robinson,V.L. and Goudreau,P.N. (2000) Two-component signal transduction. Annu. Rev. Biochem., 69, 183–215. [DOI] [PubMed] [Google Scholar]
- 36.Chang C. and Meyerowitz,E.M. (1994) Eukaryotes have ‘two-component’ signal transducers. Res. Microbiol., 145, 481–486. [DOI] [PubMed] [Google Scholar]
- 37.Yeh K.C., Wu,S.H., Murphy,J.T. and Lagarias,J.C. (1997) A cyanobacterial phytochrome two-component light sensory system. Science, 277, 1505–1508. [DOI] [PubMed] [Google Scholar]
- 38.Eddy S.R. (1998) Profile hidden Markov models. Bioinformatics, 14, 755–763. [DOI] [PubMed] [Google Scholar]
- 39.Van Dongen S. (2000) Graph clustering by flow simulation. PhD Thesis, University of Utrecht, The Netherlands.
- 40.Promponas V.J., Enright,A.J., Tsoka,S., Kreil,D.P., Leroy,C., Hamodrakas,S., Sander,C. and Ouzounis,C.A. (2000) CAST: an iterative algorithm for the complexity analysis of sequence tracts. Complexity analysis of sequence tracts. Bioinformatics, 16, 915–922. [DOI] [PubMed] [Google Scholar]
- 41.Enright A.J. and Ouzounis,C.A. (2001) BioLayout—an automatic graph layout algorithm for similarity visualization. Bioinformatics, 17, 853–854. [DOI] [PubMed] [Google Scholar]
- 42.Michalski R.S., Bratko,I. and Kubat,M. (1998) Machine Learning and Data Mining. Wiley, New York, NY.
- 43.Van Dongen S. (2000) A new cluster algorithm for graphs. Report No. INS-R0010, Center for Mathematics and Computer Science (CWI), Amsterdam.
- 44.Van Dongen S. (2000) Performance criteria for graph clustering and Markov cluster experiments. Report No. INS-R0012, Center for Mathematics and Computer Science (CWI), Amsterdam.
- 45.Van Dongen S. (2000) A stochastic uncoupling process for graphs. Report No. INS-R0011, Center for Mathematics and Computer Science (CWI), Amsterdam.
- 46.Apweiler R., Attwood,T.K., Bairoch,A., Bateman,A., Birney,E., Biswas,M., Bucher,P., Cerutti,L., Corpet,F., Croning,M.D. et al. (2001) The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res., 29, 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lo Conte L., Ailey,B., Hubbard,T.J., Brenner,S.E., Murzin,A.G. and Chothia,C. (2000) SCOP: a structural classification of proteins database. Nucleic Acids Res., 28, 257–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Attwood T.K., Flower,D.R., Lewis,A.P., Mabey,J.E., Morgan,S.R., Scordis,P., Selley,J.N. and Wright,W. (1999) PRINTS prepares for the new millennium. Nucleic Acids Res., 27, 220–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hofmann K., Bucher,P., Falquet,L. and Bairoch,A. (1999) The PROSITE database, its status in 1999. Nucleic Acids Res., 27, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan S. and Richmond,T.J. (1998) Eukaryotic transcription factors. Curr. Opin. Struct. Biol., 8, 41–48. [DOI] [PubMed] [Google Scholar]
- 52.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T. et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]