Abstract
The HNH motif was originally identified in the subfamily of HNH homing endonucleases, which initiate the process of the insertion of mobile genetic elements into specific sites. Several bacteria toxins, including colicin E7 (ColE7), also contain the 30 amino acid HNH motif in their nuclease domains. In this work, we found that the nuclease domain of ColE7 (nuclease-ColE7) purified from Escherichia coli contains a one-to-one stoichiometry of zinc ion and that this zinc-containing enzyme hydrolyzes DNA without externally added divalent metal ions. The apo-enzyme, in which the indigenous zinc ion was removed from nuclease-ColE7, had no DNase activity. Several divalent metal ions, including Ni2+, Mg2+, Co2+, Mn2+, Ca2+, Sr2+, Cu2+ and Zn2+, re-activated the DNase activity of the apo-enzyme to various degrees, however higher concentrations of zinc ion inhibited this DNase activity. Two charged residues located at positions close to the zinc-binding site were mutated to alanine. The single-site mutants, R538A and E542A, showed reduced DNase activity, whereas the double-point mutant, R538A + E542A, had no observable DNase activity. A gel retardation assay further demonstrated that the nuclease-ColE7 hydrolyzed DNA in the presence of zinc ions, but only bound to DNA in the absence of zinc ions. These results demonstrate that the zinc ion in the HNH motif of nuclease-ColE7 is not required for DNA binding, but is essential for DNA hydrolysis, suggesting that the zinc ion not only stabilizes the folding of the enzyme, but is also likely to be involved in DNA hydrolysis.
INTRODUCTION
The HNH motif was first identified based on the consensus sequence observed in several group I intron-encoded homing endonucleases (1–3). These homing endonucleases make DNA breaks at specific sites and initiate the homing process, which moves mobile introns from intron-containing alleles to intronless alleles of cognate genes (4–6). Subsequent sequence comparison in gene data banks revealed a family of these type of proteins widespread in all phylogenic kingdoms, including bacteriophage, bacteria, virus, yeast mitochondria and plant chloroplasts. The biggest subfamily among the HNH proteins are site-specific homing endonucleases, such as the group I homing endonucleases I-HmuI (7), I-HmuII, I-HmuIII (8), I-TevIII (9) and IA2 (10); and the group II homing endonucleases Cpc (11), Avi (11), PetD (12), COXI al1 and COXI al2 (13). Other HNH proteins with known functions include the bacteria restriction enzyme McrA (14), repair enzyme MutS (13), and protein toxins that contain DNase activity, such as DNase-type colicins (15,16) and pyocins (17).
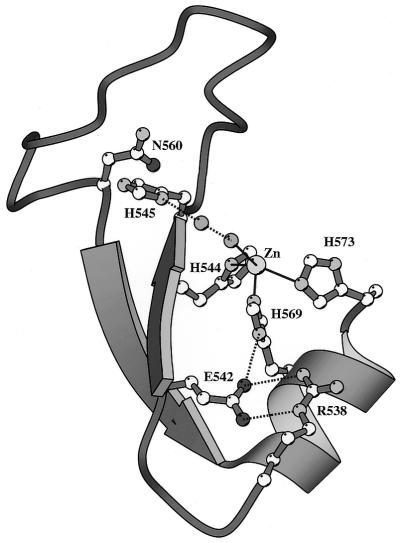
The E-group DNase-type colicins, including ColE2, ColE7, ColE8 and ColE9, are protein toxins secreted by Escherichia coli that kill other E.coli and closely related bacteria (18). They share high sequence identity and all contain an HNH motif in their cytotoxic nuclease domains. After these colicins traverse the membranes of bacteria cells, they digest DNA chromosomes in target cells and lead to cell death (16). An immunity protein (Im) bound specifically to the nuclease domain of colicin (nuclease-ColE) is co-expressed with colicin to inhibit its DNase activity, and thus protects the host cell from the cytotoxic effect. The crystal structures of two DNase-type colicins, nuclease-ColE7 in complex with immunity E7 protein (Im7) (19) and nuclease-ColE9 in complex with Im9 (20), have been reported. These structures revealed the topology of the HNH motif of two antiparallel β-strands linked to a C-terminal α-helix with a metal ion, similar to that of a classic zinc finger motif (Fig. 1). A Zn2+ ion is located at the center of the HNH motif in ColE7 and a Ni2+ ion is located at the center of the HNH motif of ColE9. The zinc ion in the HNH motif of ColE7 is bound to three conserved histidine residues and one water molecule in a distorted tetrahedral geometry.
Figure 1.
The crystal structural model of the HNH motif in the DNase domain of ColE7 (19). This motif has a topology similar to that of the classical zinc finger motif with two antiparallel β-strands linked to an α-helix by a Zn2+ ion. The Zn2+ ion is bound to three histidine residues, His544, His569 and His573, and a water molecule in a distorted tetrahedral geometry.
Since all the HNH family proteins with known function carry endonuclease activity, it is very likely that the HNH motif is involved in DNA hydrolysis. However, how the HNH motif mediates its function in DNA hydrolysis and the role of the zinc ion in the HNH motif are still elusive. Divalent metal ions have been shown to play essential roles in activating DNase activity in many restriction enzymes and nucleases (21–23). A divalent metal ion may function as (i) a general base to activate the attacking water (24), (ii) a Lewis acid to stabilize the pentacovalent phosphate group (24,25), or (iii) a general acid to activate a water molecule, which provides a proton for the leaving group (25). The metal ion in the HNH motif of ColE9 may play a structural role to stabilize the HNH motif, as was suggested by the finding that zinc-bound nuclease-ColE9 is more thermally stable than the apo-nuclease-ColE9 (26). However, a structural comparison between the active sites of the Serratia nuclease, the His–Cys box homing nuclease I-PpoI and the HNH motif of ColE9, revealed a similar ββα folding, with the divalent metal ions in the three proteins superimposed at a similar position (27). The metal ions in the Serratia nuclease and I-PpoI have been proposed to be involved in catalytic pathways (25,28,29), implying that the zinc ion in the HNH motif also participates in DNA hydrolysis. A recent review article compared the structures of several nuclease-type colicins, and the authors suggested that the metal-ion-binding site in the HNH motif is the DNase-active site based solely on the finding that a phosphate ion is directly bound to the Ni2+ ion in the crystal structure of nuclease-ColE9 (30). Therefore, it is necessary to further clarify the role of divalent metal ions in the HNH motif of DNase-type colicins for non-specific DNA hydrolysis.
In this work, we characterize the requirement of a divalent metal ion in the HNH motif of ColE7 for DNA hydrolysis and DNA binding. We found that the nuclease-ColE7 is active in buffers with low concentrations of zinc ions but higher concentrations of zinc ions function as inhibitors for the DNase activity. A variety of divalent metal ions are active cofactors for nuclease-ColE7. The apo-nuclease-ColE7 without a metal ion bound at the HNH motif, binds to DNA, but does not hydrolyze DNA. Our results suggest that the zinc ion in the HNH motif of ColE7 is likely to be involved in DNA hydrolysis.
MATERIALS AND METHODS
Purification of the DNase domain of ColE7
The expression vector pQE-30 (Qiagen, Germany) containing a six-histidine affinity tag at the N-terminus of the cloning site was used to overexpress nuclease-ColE7/Im7 complex. The detailed procedure for the construction and protein purification of the complex was previously described (19,31). The nuclease-ColE7 was further dissociated from Im7 by lowering the pH of the buffer to 3 to denature the purified complex and the two proteins were applied to a SP-Sepharose column (Amersham, USA) and eluted with buffers of pH 3 and 7, respectively. Greater than 98% homogeneity of the nuclease-ColE7 was obtained as shown in sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS–PAGE). The mass spectrometry analysis gave a molecular weight of 16 708 Da as compared with the calculated molecular weight of 16 707 Da (molecular weight of nuclease-ColE7 plus zinc). The contents of metal elements in the nuclease-ColE7/Im7 complex and the free nuclease-ColE7 were measured on a Jarrell-Ash model ICP 9000 inductively coupled plasma atomic emission spectrophotometer. The protein concentrations were estimated based on the absorbance at 280 nm (ɛ280 = 22 190 M-1 cm-1 for the nuclease-ColE7/Im7 complex and 12 665 M-1 cm-1 for the free nuclease-ColE7).
Optimization of the enzymatic activity of nuclease-ColE7
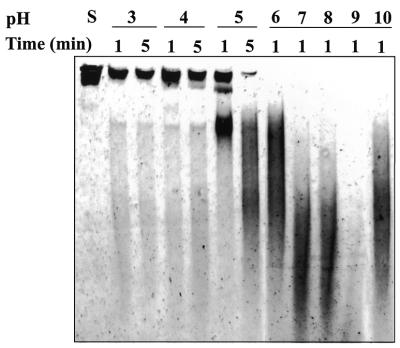
Plasmid DNA, pUC18, was used as a substrate for the DNase activity test. The supercoiled DNA of pUC18 was obtained by the alkaline lysis method (32). For the determination of the optimal temperature for DNase activity of nuclease-ColE7, 100 nM of the purified DNase domain and 0.1 µg pUC18 DNA were incubated for 20 min in a digestion buffer containing 10 mM MgCl2 and 10 mM Tris–HCl (pH 7.25) in a series of designated temperatures (from 0 to 70°C). The digested patterns of the pUC18 DNA at distinct incubation temperatures were examined by 0.8% agarose gel electrophoresis. The degrees of completion of DNA digestion were measured by a Personal densitometer SI and Image QuaNT (Molecular Dynamics, Sunnyvale, USA) after agarose gel electrophoresis. The reactions were repeated three times and were stopped with the gel loading buffer containing 250 mM EDTA, 0.25% bromophenol blue and 30% glycerol. The optimal pH for the enzymatic reaction of nuclease-ColE7 was determined in a similar way in that 6 µM of the purified nuclease-ColE7 and 6.4 nM of pUC18 DNA were incubated for 1 or 5 min in the digestion buffers of pH ranging from 3 to 10 at 37°C. These reactions were stopped with the addition of EDTA to a final concentration of 250 mM and 600 µAU protease K (Qiagen). The sample was then loaded onto a running Novex® 4–20% TBE polyacrylamide gel (Invitrogen). The digestion pattern is shown in Figure 2.
Figure 2.
The pH-dependent DNase activities of nuclease-ColE7. The DNase activity of nuclease-ColE7 in solutions with different pH varying from 3 to 10, was determined based on the DNA digestion patterns observed in a 4–20% TBE polyacrylamide gel using pUC18 plasmid DNA as the substrate. The DNase activity is indicated by the decreased amount of substrate DNA (the lane labeled with S). The nuclease-ColE7 has the optimal activity at ∼pH 9 in which most of the DNA substrates were digested.
EDTA treatment of the purified nuclease-ColE7
To examine the effect of a single divalent metal ion in the activation of nuclease-ColE7, preparation of a divalent metal-free nuclease domain is necessary. A 10 ml solution of the purified nuclease domain (0.17 mg/ml) was incubated with 1 M divalent metal chelating agent EDTA at room temperature for 1 h. The EDTA-treated nuclease-ColE7 was dialyzed against 1 l of Tris–HCl buffer overnight at 4°C with three changes of buffer. A residual EDTA concentration of ∼1 µM remained in the solution which would extract any contaminated divalent metal ions in solution. After dialysis, the volume of EDTA-treated protein was concentrated to 3 ml (0.4 mg/ml) with an Ultrafree-0.5 centrifugal filter (Millipore, Bedford, MA, USA). The enzymatic activity of the EDTA-treated nuclease-ColE7 was determined by the method described in the following section.
Characterization of the effects of divalent metal ions on DNase activity
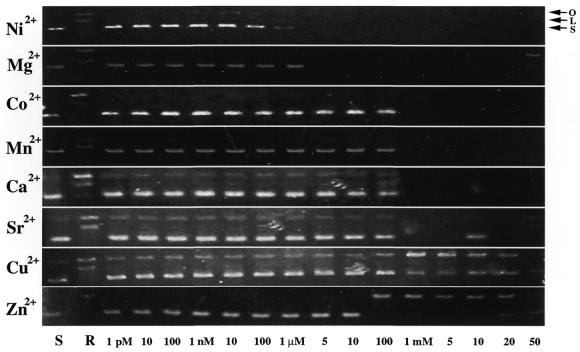
Eight divalent metal ions (Ni2+, Co2+, Mg2+, Mn2+, Ca2+, Sr2+, Cu2+ and Zn2+) were used for these tests. The concentrations of divalent metal ions used for activation of nuclease-ColE7 with or without EDTA treatment ranged from 1 pM to 50 mM. A supercoiled form of pUC18 DNA (0.1 µg) and purified nuclease-ColE7 (100 nM) were used for the enzymatic reaction throughout the experiments. The procedure for the activity assay is described in the above section ‘Optimization of the enzymatic activity of nuclease-ColE7’. The digestion patterns of DNA were stained with ethidium bromide and monitored by agarose gel.
Measurement of the DNA cleavage using the fluorophore-labeled oligonucleotides
The 30 base pair (bp) long oligonucleotide labeled with a fluorophore of hetrachloro-6-carboxyfluorescein and a quencher of BHQ-1™ (DNaseAlert™ QC system DNA substrate; Ambion Inc., USA) was used to measure the DNase activity of nuclease-ColE7. Fluorescence measurements were routinely carried out by mixing 1 µl of 6 µM nuclease-ColE7, 10 µl of 100 mM Tris–HCl buffer (pH 8.0), 10 µl of 2 µM DNaseAlert™ QC system DNA substrate and various concentrations of ZnCl2 or MgCl2 (BioChemika grade; Fluka, Switzerland) in a plate-well incubated at 40°C. The increased fluorescence emission intensity resulting from the DNA cleavage was measured on a NUNC™ black 96-well plate with a Fluoroskan Ascent plate reader (equipped with a 538 nm excitation filter, a 584 nm emission filter and a thermostatted temperature option; ThermoLabsystems, Finland). Fluorescence kinetic data were collected at 60 s intervals over a period of 80 min at 40°C. All the assay component volumes (100 µl) remained the same for all the experiments. The initial velocity was calculated only from the data points from the first 3–8 min after enzyme addition. Data were analyzed using SigmaPlot 6.0 (SPSS Inc., Chicago, IL, USA).
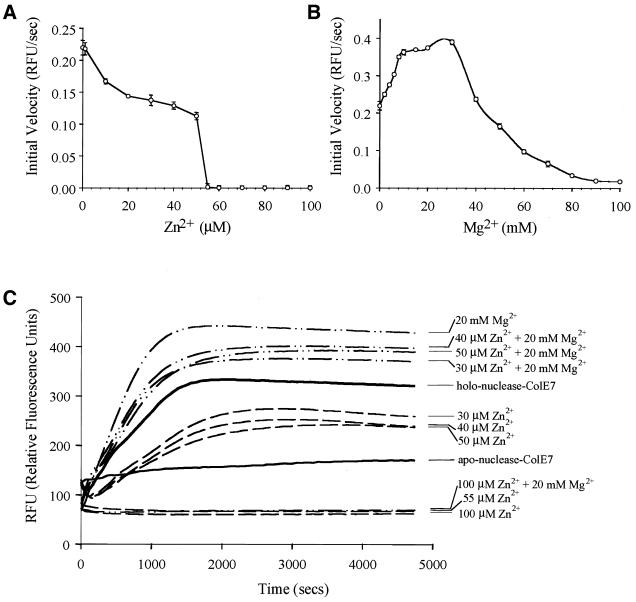
The holo-nuclease-ColE7 used in this assay (see Fig. 5C) was obtained by supplement of zinc ions during purification processes and extra zinc ions were removed by a desalting column (Amersham) followed by dialysis against Milli-Q water. The apo-enzyme of nuclease-ColE7 was obtained using a Chelex® 100 chelating ion exchange resin (Bio-Rad Laboratories, Switzerland) to remove Zn2+, followed by elution with 1 M imidazole (BioChemika grade; Fluka). Apo-nuclease-ColE7 was then dialyzed against metal-free water.
Figure 5.
Dependence of nuclease-ColE7 DNase activity on the concentrations of Zn2+ and Mg2+ measured by a fluorescent method using the fluorophore and quencher-labeled oligonucleotide as a substrate. (A) The initial velocity of DNA cleavage for the holo-nuclease-ColE7 was varied with Zn2+ ion concentration. The zinc ion inhibited the DNA cleavage and nuclease-ColE7 had no detectable DNase activity when Zn2+ concentrations (55 µM) were more than ∼1000-fold of enzyme concentration (60 nM). (B) The initial velocity of DNA cleavage for the zinc-containing holo-nuclease-ColE7 was varied with Mg2+ ion concentration. (C) The time course of fluorescence emission intensity caused by nuclease-ColE7 in buffers containing different divalent metal ions. The zinc-containing holo-nuclease-ColE7 had moderate activity; the apo-enzyme had no DNase activity; Zn2+ inhibited and Mg2+ (<40 mM) activated DNase activity of the holo-enzyme.
Construction of mutant nuclease-ColE7 and DNase-activity assay
PCR-mediated site-directed mutagenesis was performed as previously described (33). Plasmid pColE7-K317 (34) was used as a template for PCR-mediated site-directed mutagenesis to create mutations at the active site of the DNase domain of ColE7. The crystal structure of the DNase domain of ColE7 in complex with Im7 (19) showed that the three residues Arg538, Glu542 and His569 are situated near the metal-binding site of the DNase domain and that His569 is directly bound to the zinc ion. In order to verify the critical residues for the DNase activity of the domain, a strategy was used to generate mutants at the designated residues: R538A, E542A and R538A + E542A. The resultant mutagenic plasmids were designated as pColE538, pColE542 and pColE538–542, respectively. The sequences of the mutagenic oligonucleotide primer pairs are listed as follows:
(i) R538A: T2A-538U, 5′-TCAGGGAAGGCAACTTCATTCG-3′; T2A-538D, 5′-AAGTTGCCTTCCCTGAAACATC-3′
(ii) E542A: T2A-542U, 5′-TTCATTCGCGCTTCATCATGAG-3′; T2A-542D, 5′-ATGAAGCGCGAATGAAGTTCTC-3′
(iii) R538A + E542A: T2A-53842U, 5′-GAAGGCAACTTCATTCGCGCTTCATC-3′; T2A-53842D, 5′-AAGCGCGAATGAAGTTGCCTT-3′
The sites for the change of residues are underlined.
For analysis of the DNase activity of the mutant nuclease-ColE7, 0.1 µg of supercoiled pUC18 plasmid DNA was incubated with 100 nM of each of the purified mutant nuclease-ColE7 in the presence of 10 mM MgCl2 and 10 mM Tris–HCl, pH 7.25 for 20 min at 37°C. The methods used for gel electrophoresis of the digested DNA and determination of the relative nuclease-ColE7 activity by densitometry were the same as described in the above section ‘Optimization of the enzymatic activity of nuclease-ColE7’.
Gel retardation assays
A 27 bp DNA was synthesized and purified by a chromatographic method using Nensorb™ resin (DuPont, USA). A solution containing 50, 5.0 or 0.5 µM of nuclease-ColE7 and 6.7 µM dsDNA in 50 mM HEPES (pH 7.5), 150 mM NaCl, with or without 1.25 mM EDTA was incubated for 2 min at 25°C. 180 µAU protease K (Qiagen) was added in a reaction mixture and incubated at 54°C for 1 h to digest nuclease-ColE7. These samples were then loaded onto a running Novex® 20% TBE polyacrylamide gel (Invitrogen) with a length of 12 cm, equilibrated in 1× TBE and subjected to 200 V for 45 min at room temperature. Gels were stained with SYBR Green I (Molecular Probes, the Netherlands) and monitored by FUJI Science Imaging System LAS-1000 Plus (Fuji Photo Film, Japan).
RESULTS
Zinc ion binds with a one-to-one stoichiometry to ColE7
The zinc contents in the nuclease-ColE7/Im7 complex and in the free nuclease-ColE7 were measured by atomic emission spectroscopy. The zinc ion was bound with a one-to-one stoichiometry (complex:zinc = 1:0.97) to the nuclease-ColE7/Im7 complex. The amount of other metal ions was negligible. Only residual amounts of zinc ions were associated with the free nuclease-ColE7 with a molar ratio of nuclease-ColE7:zinc = 1:0.05. It is likely that the zinc ions were dissociated from nuclease-ColE7 during the protein purification steps, which separated the nuclease-ColE7 from Im7 by partial denaturation of the complex in low pH buffers. It has been shown that Zn2+ has the highest affinity for ColE9 as compared with two other transition metal ions Ni2+ and Co2+ (26). Therefore, it is well accepted that the DNase-type colicins are zinc-dependent enzymes and one zinc ion is bound to one protein molecule.
Optimization of DNase activity
The endonuclease activity of nuclease-ColE7 was assayed using pUC18 plasmid DNA as a substrate at different temperatures and over a range of pHs. The optimal temperature for the DNase activity of nuclease-ColE7 was ∼50°C. The nuclease-ColE7 started to display weak DNase activity at 4°C, which gradually increased to reach its peak at 50°C (data not shown). The DNase activity of nuclease-ColE7 was tested from pH 3 to 10 and the optimal pH for enzymatic reaction was ∼9 (see Fig. 2). The DNase activity appeared ∼pH 5.0 and gradually increased to a maximum at pH 9, but was inhibited at higher pH (pH >10). Preheating the nuclease-ColE7 at temperatures between 4 and 37°C for up to 12 h did not affect the enzyme activity. However, when nuclease-ColE7 was pretreated at 50°C for 1 h, its DNase activity decreased substantially.
The zinc-containing nuclease-ColE7 is active at low but inactive at higher Zn2+ concentrations
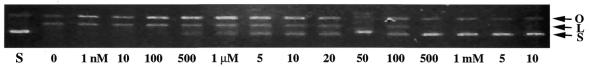
Activation of a Zn2+-containing nuclease-ColE7 was characterized by the topological changes of the substrate DNA (Fig. 3). Without externally added divalent ion, the Zn2+-containing nuclease-ColE7 converted the supercoiled form of DNA into open circular and linear forms of DNA (Fig. 3). In this experiment, the concentration of nuclease-ColE7 was ∼1 nM, and the concentration of zinc ion was increased from 1 nM to 10 mM. Nuclease-ColE7 was most active when the concentration of zinc ion was <1 µM, and it became inactive at higher zinc concentrations, i.e. when the molar ratio of zinc ion to nuclease-ColE7 was >1000-fold, the zinc ion started to inhibit DNase activity. When the Zn2+ concentration reached 500 µM, that is ∼105-fold to the nuclease-ColE7, the zinc ion completely inhibited the DNase activity.
Figure 3.
The effect of various concentrations of zinc ion on the DNase activity of nuclease-ColE7. A plasmid DNA pUC18 (0.1 µg) was used as the substrate to react with 100 nM nuclease-ColE7. The topological changes of the substrate DNA were monitored by 0.8% agarose gel electrophoresis. The supercoiled (S) form of pUC18 plasmid DNA alone was used as negative control (lane S). The effects of enzymatic reactions with various concentrations of zinc ions were monitored by the conversion of the supercoiled form DNA to its corresponding open circular (O) or linear (L) form. The concentration of zinc ion was increased from 1 nM to 10 mM as indicated on the bottom of each lane.
A variety of divalent metal ions activate the DNase activity of ColE7
To characterize the DNase activity of nuclease-ColE7 in the presence of a single metal ion, purified nuclease-ColE7 was treated first with EDTA to remove the zinc ion associated with the enzyme. However, ∼1 µM of EDTA remained in the protein solution, which would extract any residual metal ions to ensure there was no contamination. We found that the EDTA-treated DNase domain completely lost its enzymatic activity (data not shown). However, the activity of the EDTA-treated nuclease-ColE7 resumed to various degrees when different divalent ions were introduced into the assay system (Fig. 4). Ni2+ and Mg2+ were the most potent divalent ions in activating EDTA-treated nuclease-ColE7: the enzyme completely digested DNA when the metal ion concentration exceeded the residual EDTA concentration at ∼1 µM. On the other hand, Co2+, Mn2+, Ca2+ and Sr2+ required a higher concentration (1 mM) for activation. Activation of the enzyme by Cu2+ and Zn2+ was the least obvious. These two metal ions only activated the conversion of the supercoiled form of the substrate DNA to the respective open circular form.
Figure 4.
The effect of various divalent metal ions in the activation of EDTA-treated nuclease-ColE7. Supercoiled pUC18 DNA was used as a negative control (lane S). Digestion of the substrate DNA by the Zn2+-containing nuclease-ColE7 with no externally added divalent ion was used as a reference (lane R). EDTA-treated nuclease-ColE7 was used for digestion of the substrate DNA in the presence of various concentrations (1 pM to 50 mM) of divalent metal ions as indicated in the last row.
The DNase activity of nuclease-ColE7 was further assayed by a more sensitive fluorescent method using smaller oligonucleotides covalently bound with fluorescent dye and quencher as a substrate (35,36). The degradation of the flurophore-labeled oligonucleotides by nuclease-ColE7 resulted in the increase of fluorescence emission intensity, which was monitored by a spectrofluorometer. We found that the DNase activity of nuclease-ColE7 gradually decreased with the increase of Zn2+ concentration and the enzyme completely lost its DNase activity when the Zn2+ concentration (>55 µM) is over ∼1000-fold larger than that of nuclease-ColE7 (60 nM) (Fig. 5A). This inhibitory effect produced by Zn2+ ions is consistent with the results from the topological analysis using plasmid DNA as substrates described in the previous section. On the other hand, Mg2+ activated the DNase activity of the Zn2+-containing holo-nuclease-ColE7 with an initial increase in activity with increasing [Mg2+], followed by a gradual decrease in activity (Fig. 5B). The decrease of activity at high [Mg2+] has been observed for several Mg2+-dependent nucleases and was explained by a model of metal-mediated substrate inhibition (37). The time course of fluorescence emission intensity caused by the DNase activity of nuclease-ColE7 in the presence of Zn2+, Mg2+ or Zn2+ + Mg2+ is shown in Figure 5C. It clearly demonstrates that the apo-enzyme of nuclease-ColE7 had no DNase activity, Zn2+ inhibited the DNase activity, and Mg2+ activated the DNase activity of the Zn2+-containing holo-enzyme.
Mutations at positions close to the metal-binding site eliminated the DNase activity
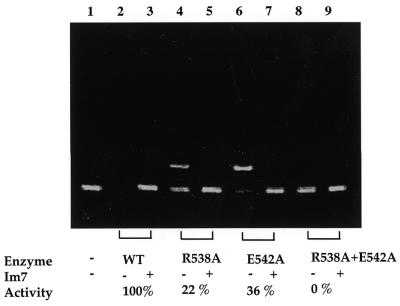
Two residues, Arg538 and Glu542, near the metal-binding site were mutated. In the crystal structure of nuclease-ColE7/Im7, Arg538 forms hydrogen bonds to Glu542 and His569, which binds directly to the zinc ion (Fig. 1). The mutation of the three conserved residues, Arg544 (equivalent to Arg538 in ColE7), Glu548 (equivalent to Glu542) and His575 (equivalent to His569) in ColE9 to alanine, generated inactive mutants in vivo and in vitro (38). However, we found that R538A (22%) and E542A (36%) contained residual DNase activity as compared with the wild-type nuclease-ColE7 using pUC18 plasmid DNA as the substrate (Fig. 6). The double mutant, R538A + E542A, had no observable DNase activity and all the DNA substrate remained intact. The immunity protein Im7 inhibited the DNase activity for these mutants, indicating that the mutants still retained their conformation. Glu542 is ∼50% conserved and Arg538 is only slightly conserved (<10%) in the HNH family proteins, suggesting that they do not play major roles in DNA hydrolysis.
Figure 6.
The DNase activity of nuclease-ColE7 is significantly reduced with mutations at Arg538 and Glu542. Either wild-type or mutated nuclease-ColE7 (100 nM) was incubated with 0.1 µg of the supercoiled form of pUC18 plasmid DNA. An equal molar concentration of Im7 inhibitor to that of nuclease-ColE7 was used to monitor the inhibitory effect of the inhibitor against the mutant-type nuclease-ColE7 protein. The upper and lower bands indicate the open circular and supercoiled form DNA, respectively. The relative DNase activities with respect to the wild-type enzyme measured by densitometry were 22% for R538A, 36% for E542A and 0% for R538A + E542A. The enzyme activity was completely inhibited with the addition of an equal molar concentration of Im7 inhibitor indicating these mutants still folded well. Lanes 1 and 2, pUC18 incubated without and with wild-type nuclease-ColE7, respectively. Lane 3, the DNA incubated together with wild-type nuclease-ColE7 and equal molar concentrations of Im7 inhibitor. Lanes 4, 6 and 8, the DNA incubated with nuclease-ColE7 mutant R538A, E542A and R538A + E542A, respectively. Lanes 5, 7 and 9, have the same conditions as lanes 4, 6 and 8 except for the addition of equal molar concentrations of Im7 inhibitor.
The metal-free enzyme is capable of binding to DNA
EDTA-treated nuclease-ColE7 has no DNase activity as demonstrated in the previous section. An earlier comparison of the circular dichroism spectra and the tryptophan emission fluorescence between the apo- and holoenzyme of the DNase domain of ColE9 showed that the apo-protein retains a fold similar to that of the holoenzyme (26). This finding implies that the zinc ion in the HNH motif of the DNase-type colicin is not required for protein folding. However, it is unknown whether the apo-enzyme can bind to DNA.
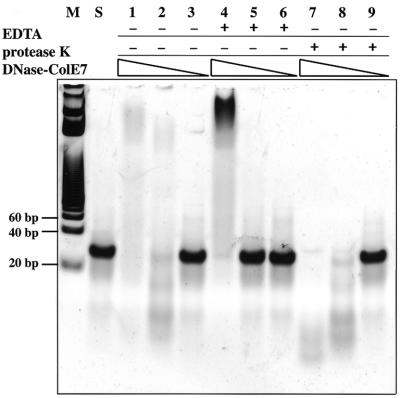
To address this important question, we used a 27 bp dsDNA as the substrate and found that the zinc-containing nuclease-ColE7 digested DNA, but that the metal-free nuclease-ColE7 only bound to DNA as shown in a gel-shift assay (Fig. 7). The concentration of the 27mer DNA was maintained at 6.7 µM while the concentrations of nuclease-ColE7 varied from 50 to 0.5 µM (lanes 1–3, 4–6 and 7–9). The 27mer DNA was digested in lanes 1, 2, 7 and 8 when EDTA was not present, but the DNA was up-shifted by the enzyme when EDTA was added as shown in lane 4. Protease K was added to digest nuclease-ColE7 in lanes 7–9. Compared with lanes 1–3, it appeared that nuclease-ColE7 binds to the smaller fragment DNA products in lane 1 and the DNA products were released from the enzymes in lane 7 after protease K digestion. These results demonstrate that not only is the zinc ion in the HNH motif not required for protein folding, it is also not required for DNA binding.
Figure 7.
A gel retardation assay shows that nuclease-ColE7 hydrolyzed a 27mer DNA in the presence of zinc ions, but up-shifted the DNA in the absence of zinc ions. A solution containing 50 µM(lanes 1, 4, 7), 5 µM (lanes 2, 5, 8) or 0.5 µM (lanes 3, 6, 9) of nuclease-ColE7 was incubated with 6.7 µM of 27 bp of dsDNA for 2 min at 25°C with or without the presence of 1.25 mM EDTA. Protease K was used to digest nuclease-ColE7 after the incubation described above (lanes 7, 8, 9). Lane M, DNA markers; lane S, substrate 27 bp DNA.
DISCUSSION
The zinc ion in the HNH motif of nuclease-ColE7 is not required for DNA binding
It has been shown before that the zinc binding to the apo-nuclease-ColE9 increased the proteolytic resistance and melting point of the enzyme; therefore, it was concluded that the transition metal ion in the HNH motif serves a structural role (26). However, in the present work, our results indicate that the zinc ion in the HNH motif of ColE7 also appears to be involved in the catalytic pathway. First, nuclease-ColE7 is bound to zinc ions in a one-to-one stoichiometry, and this holoenzyme is active for DNA hydrolysis. Removal of the zinc ion produces an apo-enzyme with no DNase activity. Secondly, mutations in the vicinity of the metal-binding site impair DNase activity. Thirdly, the apo-enzyme has no DNase activity, but it retains the ability to bind DNA. Since the zinc ion in the HNH motif is not required for DNA binding, but is required for DNA hydrolysis, it is very likely that the zinc ion is involved in DNA hydrolysis.
However, our DNA-binding results for nuclease-ColE7 are different from that of I-CmoeI (39), which is a group I HNH family homing endonuclease encoded by the introns in the Chlamydomonas moewusii chloroplast psbA gene. I-CmoeI requires a metal ion cofactor for DNA binding that it cannot bind to its DNA substrate in the presence of EDTA. The different results observed for the two HNH proteins may be due to the intrinsic difference in the two proteins: I-CmoeI is a site-specific homing endonuclease, whereas nuclease-ColE7 is a non-specific enzyme. In addition, these two proteins only share sequence similarity in the HNH motif. It is also possible that the different experimental conditions used, including the concentrations of proteins, DNA substrates and EDTA, result in the differing binding activities. It will be instructive to find out whether a zinc ion is required for DNA binding for other homing endonucleases and bacteria toxins in the HNH family.
The Zn2+ dependence of ColE7 DNase activity
The metal-dependence study of colicin E9 showed that the addition of Zn2+ did not result in any DNase activity according to Kunitz assays (26). However, in the present work, supercoiled DNA was nicked to open-circular and linear forms when incubated with nuclease-ColE7 in a low Zn2+ concentration range. The EDTA-treated enzyme was reactivated by Zn2+, but the DNase activity was inhibited when the enzyme was incubated in a buffer containing a higher concentration of Zn2+. This phenomenon was further assayed using a fluorophore and quencher-labeled oligonucleotide as a substrate, and a similar result was observed that extra Zn2+ ion inhibited DNase activity. Therefore, the higher concentration of zinc ion (>1000 in molar ratio to enzyme) has an inhibitory effect on the DNase activity. The different results observed for ColE9 (26) might be due to the high Zn2+ concentration (10 µM as compared with ∼1 nM of protein concentration) used in their experiments, which in turn produced an inhibitory effect on DNase activity.
Inhibition of enzyme activity by a relatively higher concentration of Zn2+ has been observed for several zinc-dependent enzymes, including metallo-β-lactamase (40) and methionine aminopeptidase (41). There are two metal-binding sites in metallo-β-lactamase, and Zn2+ binding to the second site produces the inhibitory effect. A previous study (42) and our present work showed that HNH endonucleases are more active in the presence of Mg2+. Mg2+ prefers binding to oxygen-containing ligands, and thus far no structural example has shown that Mg2+ binds to three histidine residues (43). It seems unlikely that Mg2+ binds at the same zinc-binding site in ColE7. In I-PpoI and Serratia nuclease, the Mg2+ binds to an asparagine residue and the scissile phosphate oxygen atoms, and Mg2+ likely participates in protonation of the 3′ oxygen leaving group (25,29,43–45). We suspect that the DNA hydrolysis by ColE7 is catalyzed by more than one metal ion, even though we observed only one metal-binding site in ColE7. We further noted that Mg2+ activated the DNase activity of nuclease-ColE7, but it cannot rescue the Zn2+-suppressed activity that the enzyme had no activity in a buffer containing 100 µM of Zn2+ and 20 mM of Mg2+ (Fig. 5C). This result is consistent with the earlier prediction that Zn2+ can dislodge Mg2+ from its octahedral binding site (46). It will be interesting to investigate the possibility of a second metal-binding site for Mg2+ in ColE7 upon DNA binding.
Possible roles of zinc ion in the HNH motif
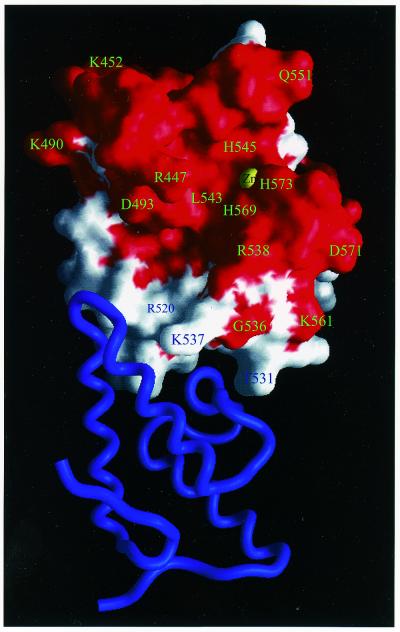
A conserved surface for nuclease-ColE7 is displayed in Figure 8, in which all the conserved residues in bacteria toxins, including ColE2, ColE7, ColE8, ColE9, Pyocin S1 and S2, are colored red, and the non-conserved residues are colored white. The protein–protein interaction surface for immunity proteins is clearly not conserved. This explains the specificity for each pair of colicin and immunity proteins since this surface has to be different for specific recognition by the cognate inhibitor. On the contrary, the DNA-binding site and DNase-active site are likely to be located within a conserved surface because all of these toxins carry a similar DNase activity. We found that the zinc ion in nuclease-ColE7 is indeed located in the middle of a conserved surface exposed to solvent and accessible for DNA hydrolysis.
Figure 8.
The conserved and non-conserved molecular surface of nuclease-ColE7. The molecular surface that contains the conserved residues in the six bacteria toxins in HNH family is colored red, and the non-conserved surface is colored white. Im7 is displayed in a ribbon model in blue. The protein interface is not conserved, therefore the immunity protein specifically recognizes its cognate colicin. On the contrary, the zinc ion is located in the middle of a conserved surface where it is likely that the DNA-binding site and DNase-active site are located.
What is the role of the zinc ion in the HNH motif of ColE7 in DNA hydrolysis? A comparison between the HNH motif and the active site of other nucleases may shed some light on the possible functions of the metal ion. Structural similarity has been observed between the active sites of ColE9, the non-specific nuclease from Serratia and the His–Cys box containing homing endonuclease I-PpoI. An analogous ββα folding was identified in the three enzymes with divalent metal ions (Mg2+ in the Serratia nuclease and I-PpoI, and Zn2+ in ColE7) and several conserved residues in each family superimposed at similar positions (27). The magnesium ion in I-PpoI appears to be involved in stabilizing the phosphoanion transition state and in protonating the 3′ oxygen leaving group as shown in the crystal structures of I-PpoI in complex with DNA substrates (25). The magnesium ion in the Serratia nuclease seems to play a similar role in stabilizing the phosphoanion transition state and helping to protonate the leaving group (28,29). The zinc ion in the HNH motif of ColE7 binds to a water molecule in the crystal structure of nuclease-ColE7/Im7, however, the metal ion in the same position in ColE9 binds to a phosphate ion. Our recent structural data (not shown) also verify that a phosphate ion is directly bound to the zinc ion when the complex crystals are first soaked in phosphate buffer before data collection. This finding indicates that the zinc ion in the HNH motif may be involved in stabilizing the phosphoanion transition state. Nevertheless, the exact role of the zinc ion in the HNH motif is still a matter for discussion and more information is needed to further define the mechanism for DNA hydrolysis catalyzed by the HNH family proteins at the atomic level.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by research grants from the National Science Council of the Republic of China and Academia Sinica to P.-H.L. (NSC89-2311-B001-187), K.-F.C. (NSC89-2320-B010-119) and H.S.Y. (NSC89-2320-B001-055).
REFERENCES
- 1.Shub D.A., Goodrich-Blair,H. and Eddy,S.R. (1994) Amino acid sequence motif of group I intron endonucleases is conserved in open reading frames of group II introns. Trends Biochem. Sci., 19, 402–404. [DOI] [PubMed] [Google Scholar]
- 2.Gorbalenya A.E. (1994) Self-splicing group I and group II introns encode homologous (putative) DNA endonucleases of a new family. Protein Sci., 3, 1117–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalgaard J.Z., Klar,A.J., Moser,M.J., Holley,W.R., Chatterjee,A. and Mian,I.S. (1997) Statistical modeling and analysis of the LAGLIDADG family of site-specific endonucleases and identification of an intein that encodes a site-specific endonuclease of the HNH family. Nucleic Acids Res., 25, 4626–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambowitz A.M. and Belfort,M. (1993) Intorns as mobile genetic elements. Annu. Rev. Biochem., 62, 587–622. [DOI] [PubMed] [Google Scholar]
- 5.Belfort M. and Perlman,P.S. (1995) Mechanisms of intron mobility. J. Biol. Chem., 270, 30237–30240. [DOI] [PubMed] [Google Scholar]
- 6.Chevalier B.S. and Stoddard,B.L. (2001) Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res., 29, 3757–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodrich-Blair H. and Shub,D.A. (1996) Beyond homing: competition between intron endonucleases confers a selective advantage on flanking genetic markers. Cell, 84, 211–221. [DOI] [PubMed] [Google Scholar]
- 8.Goodrich-Blair H. (1994) The DNA polymerase genes of several HMU-bacteriophages have similar group I introns with highly divergent open reading frames. Nucleic Acids Res., 22, 3715–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eddy S.R. and Gold,L. (1991) The phage T4 nrdB intron: a deletion mutant of a version found in the wild. Genes Dev., 5, 1032–1041. [DOI] [PubMed] [Google Scholar]
- 10.Foley S., Bruttin,A. and Brussow,H. (2000) Widespread distribution of a group I intron and its three deletion derivatives in the lysin gene of Streptococcus thermophilius bacteriophages. J. Virol., 74, 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferat J.L. and Michel,F. (1993) Group II self-splicing introns in bacteria. Nature, 364, 358–361. [DOI] [PubMed] [Google Scholar]
- 12.Kuck U. (1989) The intron of a plasmid gene from a green alga contains an open reading frame for a reverse transcriptase-like enzyme. Mol. Gen. Genet., 218, 257–265. [DOI] [PubMed] [Google Scholar]
- 13.Eisen J.A. (1998) A phylogenomic study of the MutS family of proteins. Nucleic Acids Res., 26, 4291–4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hiom K. and Sedgwick,S.G. (1991) Cloning and structural characterization of the mcrA locus of Escherichia coli. J. Bacteriol., 173, 7368–7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pugsley A.P. and Schwartz,M. (1984) Colicin E2 release: lysis, leakage or secretion? Possible role of a phospholipase. EMBO J., 3, 2393–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.James R., Kleanthous,C. and Moore,G.R. (1996) The biology of E colicins: paradigms and paradoxes. Microbiology, 142, 1569–1580. [DOI] [PubMed] [Google Scholar]
- 17.Sano Y. and Kageyama,M. (1993) A novel transposon-like structure carries the genes for pyocin AP41, a Pseudomonas aeruginosa bacteriocin with a DNase domain homology to E2 group colicins. Mol. Gen. Genet., 237, 161–170. [DOI] [PubMed] [Google Scholar]
- 18.Pugsley A.P. and Oudega,B. (1987) Methods for studying colicins and their plasmids. In Hardy,K.G. (ed.), Plasmids—A Practical Approach. IRL Press, Oxford and Washington, pp. 105–161.
- 19.Ko T.-P., Liao,C.-C., Ku,W.-Y., Chak,K.-F. and Yuan,H.S. (1999) The crystal structure of the DNase domain of colicin E7 in complex with its inhibitor Im7 protein. Structure, 7, 91–102. [DOI] [PubMed] [Google Scholar]
- 20.Kleanthous C., Kuhlmann,U.C., Pommer,A.J., Ferguson,N., Radford,S.E., Moore,G.R., James,R. and Hemmings,A.M. (1999) Structural and mechanistic basis of immunity toward endonuclease colicins. Nature Struct. Biol., 6, 243–252. [DOI] [PubMed] [Google Scholar]
- 21.Pingoud A. and Jeltsch,A. (1997) Recognition and cleavage of DNA by type-II restriction endonucleases. Eur. J. Biochem., 246, 1–22. [DOI] [PubMed] [Google Scholar]
- 22.Pan C.Q., Ulmer,J.S., Herzka,A. and Lazarus,R.A. (1998) Mutational analysis of human DNase I at the DNA binding interface: Implications for DNA recognition, catalysis and metal ion dependence. Protein Sci., 7, 628–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosfield D.J., Guan,Y., Haas,B.J., Cunningham,R.P. and Tainer,J.A. (1999) Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell, 98, 397–408. [DOI] [PubMed] [Google Scholar]
- 24.Chevalier B.S., Monnat,R.J. and Stoddard,B.L. (2001) The homing endonuclease I-CreI uses three metals, one of which is shared between the two active sites. Nature Struct. Biol., 8, 312–316. [DOI] [PubMed] [Google Scholar]
- 25.Galburt E.A., Chevalier,B., Tang,W., Jurica,M.S., Flick,K.E., Monnat,R.J. and Stoddard,B.L. (1999) A novel endonuclease mechanism directly visualized for I-PpoI. Nature Struct. Biol., 6, 1096–1099. [DOI] [PubMed] [Google Scholar]
- 26.Pommer A.J., Kuhlmann,U.C., Cooper,A., Hemmings,A.M., Moore,G.R., James,R. and Kleanthous,C. (1999) Homing in on the role of transition metals in the HNH motif of colicin endonuclease. J. Biol. Chem., 274, 27153–27160. [DOI] [PubMed] [Google Scholar]
- 27.Kuhlmann U.C., Moore,G.R., James,R., Kleanthous,C. and Hemmings,A.M. (1999) Structural parsimony in endonuclease active site: should the number of homing endonuclease families be redefined? FEBS Lett., 463, 1–2. [DOI] [PubMed] [Google Scholar]
- 28.Lunin V.Y., Levdikov,V.M., Shlyapnikov,S.V., blagova,E.V., Lunin,V.V., Wilson,K.S. and Mikhailov,A.M. (1997) Three-dimensional sructure of Serratia marcescens nuclease at 1.7 Å resolution and mechanism of its action. FEBS Lett., 412, 217–222. [DOI] [PubMed] [Google Scholar]
- 29.Miller M.D., Cai,J. and Krause,K.L. (1999) The active site of Serratia endonuclease contains a conserved magnesium-water cluster. J. Mol. Biol., 288, 975–987. [DOI] [PubMed] [Google Scholar]
- 30.Kleanthous C. and Walker,D. (2001) Immunity proteins: enzyme inhibitors that avoid the active site. Trends Biochem. Sci., 26, 624–631. [DOI] [PubMed] [Google Scholar]
- 31.Chak K.-F., Hsieh,S.-Y., Liao,C.-C. and Kan,L.-S. (1998) Change of thermal stability of colicin E7 triggerred by acidic pH suggests the existence of unfolded intermediate during the membrane-translocation phase. Proteins Struct. Func. Genet., 32, 17–25. [DOI] [PubMed] [Google Scholar]
- 32.Birnboim H.C. (1983) A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol., 100, 243–255. [DOI] [PubMed] [Google Scholar]
- 33.Lu F.-M. and Chak,K.-F. (1996) Two overlapping SOS-boxes in ColE operons are responsible for the viability of cells harboring the Col plasmid. Mol. Gen. Genet., 251, 407–411. [DOI] [PubMed] [Google Scholar]
- 34.Chak K.-F., Kuo,W.-S., Lu,F.-M. and James,R. (1991) Cloning and characterization of the ColE7 plasmid. J. Gen. Microbiol., 137, 91–100. [DOI] [PubMed] [Google Scholar]
- 35.Trubetskoy V.S., Hagstrom,J.E. and Budker,V.G. (2002) Self-quenched covalent fluorescent dye-nucleic acid conjugates as polymeric substrates for enzymatic nuclease assays. Anal. Biochem., 300, 22–26. [DOI] [PubMed] [Google Scholar]
- 36.Kelemen B.R., Klink,T.A., Behlke,M.A., Eubanks,S.R., Leland,P.A. and Raines,R.T. (1999) Hypersensitive substrates for ribonucleases. Nucleic Acids Res., 27, 3696–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cowan J.A. (1998) Metal activation of enzymes in nucleic acid biochemistry. Chem. Rev., 98, 1067–1087. [DOI] [PubMed] [Google Scholar]
- 38.Garinot-Schneider C., Pommer,A.J., Moore,G.R., Kleanthous,C. and James,R. (1996) Identification of putative active-site residues in the DNase domain of colicin E9 by ramdom mutagenesis. J. Mol. Biol., 260, 731–742. [DOI] [PubMed] [Google Scholar]
- 39.Drouin M., Lucas,P., Otis,C., Lemieux,C. and Turmel,M. (2000) Biochemical characterization of I-CmoeI reveals that this H–N–H homing endonuclease shares functional similarities with H–N–H colicins. Nucleic Acids Res., 28, 4566–4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valladares M.H., Felici,A., Weber,G., Adolph,H.W., Zeppezauer,M., Rossolini,G.M., Amicosante,G., Frere,J.-M. and Galleni,M. (1997) Zn(II) dependence of the aeromonas hydrophila AE036 metallo-β-lactamase activity and stability. Biochemistry, 36, 11534–11541. [DOI] [PubMed] [Google Scholar]
- 41.Walker K.W. and Bradshaw,R.A. (1998) Yeast methionine aminopeptidase I can utilize either Zn2+ or Co2+ as a cofactor: a case of mistaken identity? Protein Sci., 7, 2684–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pommer A.J., Wallis,R., Moore,G.R., James,R. and Kleanthous,C. (1998) Enzymatic characterization of the nuclease domain from the bacterial toxin colicin E9 from Escherichia coli.Biochem. J., 334, 387–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dudev T., Cowan,J.A. and Lim,C. (1999) Competitive binding in magnesium coordination chemistry: water versus ligands of biological interest. J. Am. Chem. Soc., 121, 7665–7673. [Google Scholar]
- 44.Higgins C.F., Peltz,S.W. and Jacobson,A. (1992) Turnover of mRNA in prokaryotes and lower eukaryotes. Curr. Opin. Genet. Dev., 2, 739–747. [DOI] [PubMed] [Google Scholar]
- 45.Friedhoff P., Meiss,G., Kolmes,B., Pieper,U., Gimadutdinow,O., Urbanke,C. and Pingoud,A. (1996) Kinetic analysis of the cleavage of natural and synthetic substrates by the Serratia nuclease. Eur. J. Biochem., 241, 572–580. [DOI] [PubMed] [Google Scholar]
- 46.Dudev T. and Lim,C. (2001) Metal selectivity in metalloproteins: Zn2+ vs. Mg2+. J. Phys. Chem. B, 105, 4446–4452. [Google Scholar]