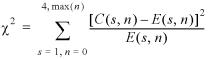Abstract
Understanding of protein–DNA interactions is crucial for prediction of DNA-binding specificity of transcription factors and design of novel DNA-binding proteins. In this paper we develop a novel approach to analysis of protein–DNA interactions. We bring together two sources of information: (i) structures of protein–DNA complexes (PDB/NDB database) and (ii) experimentally obtained sites recognized by DNA-binding proteins. Sites are used to compute conservation (information content) of each base pair, which indicates relative importance of the base pair in specific recognition. The main result of this study is that conservation of base pairs in a site exhibits significant correlation with the number of contacts the base pairs have with the protein. In particular, base pairs that have more contacts with the protein are more conserved in evolution. As natural as it is, this result has never been reported before. We also observe that for most of the studied proteins, hydrogen bonds and hydrophobic interactions alone cannot explain the pattern of evolutionary conservation in the binding site suggesting cumulative contribution of different types of interactions to specific recognition. Implications for prediction of the DNA-binding specificity are discussed.
INTRODUCTION
Protein–DNA interactions are central for the regulation of gene expression in a cell. Up to 10% of predicted genes in the newly sequenced bacterial genomes are believed to be transcription factors (1). The DNA-binding specificity of these factors and, hence, sites they bind are not known. The DNA-binding specificity of transcription factors, if possible to predict, can provide a great deal of information about the network of gene regulation in a cell. Unfortunately, our understanding of the energetics of protein–DNA recognition is sparse.
Much progress has been made since the first DNA-binding protein was isolated (2). The most detailed picture of protein–DNA interactions comes from more than 200 X-ray and NMR solved structures of protein–DNA complexes (3). As this information was accumulated, structures have been thoroughly examined by the authors. Protein–DNA complexes have been studied by chemical modifications (for review see 4) and site-specific mutagenesis (5,6), and binding motifs and interactions have been classified (7–10). Recently, three groups (11–14) extensively studied representative protein–DNA complexes: chemical and physical properties of the interfaces, their polarity, size, shape and packing. Several other groups (15–17) studied protein–DNA complexes by an approach borrowed from the field of protein folding. By ignoring atomic details of the structures, they derived a knowledge-based potential of residue–nucleotide interactions. The research is aimed at ab initio prediction of protein–DNA specificity and was successfully applied to certain zinc-finger proteins (17). Although X-ray and NMR structures give us the most detailed picture of protein–DNA interactions, the structures are missing information about the energetics of the interactions and relative importance of different residues and nucleotides in the recognition.
By mutating the protein and the DNA site one can explore the relative importance of different residues and nucleotides in protein–DNA recognition. These experiments are labor-intensive, making it impossible to study all possible mutations of a few residues and corresponding base pairs. An enormous number of such mutations, however, have already been tested in the ‘natural laboratory’ by molecular evolution. Families of homologous proteins tell us about mutations that were tolerated by the protein, while alignments of footprinted or computationally derived DNA sites tell us about tolerated nucleotide substitutions. Clearly, nucleotides that were conserved in evolution are more important than those that had been frequently altered. Although evolutionary information does not provide us with ‘ΔΔG’ for every base pair substitution, it reveals the relative importance of different residues and nucleotides in the protein–DNA recognition. Naturally, this evolutionary information complements a high-resolution picture of the protein–DNA interface provided by the NMR and the X-ray crystallography.
In this study we combine structural information for the protein–DNA complexes with the evolutionary information of corresponding footprinted DNA sites. We focus on the bacterial transcription factors because they usually bind the DNA independently and, unlike the eukaryotic factors, no large protein complexes are formed. Besides, many footprinted sites for bacterial transcription factors are available in the DPI database (18). The goal is to identify and understand primary determinants of specific DNA recognition by proteins.
We study how conservation of nucleotides in the DNA site is linked to the structural role of base pairs in the protein–DNA complex. In these complexes we compute the number of interactions every base pair has with the protein and compare this number with the degree of conservation of this base pair in footprinted and SELEX-generated sites (5,19,20). Despite differences observed previously between the natural and the SELEX sites (21), we observe that the base pairs having more interactions with the protein are more conserved in the binding sites. As natural as it is, this result has never been reported before. Perhaps the lack of organization of sites in a single database (18,22,23) prevented systematic comparison between the sites and the protein–DNA structures.
It is surprising that evolutionary conservation of base pairs in the sites correlates so strongly with the number of protein contacts, given that different types of interactions contribute differently to the binding energy. An important and unexpected result is that the pattern of hydrogen bonds and the pattern of hydrophobic interactions do not correlate well with the evolutionary conservation in most of studied proteins, suggesting cumulative contribution of different types of interactions in determining specific recognition.
MATERIALS AND METHODS
For our analysis we selected all bacterial transcription factors for which (i) a sufficient number of footprinted sites in the DPI database (18) and (ii) a high-resolution structure of a protein–DNA complex (24) are both available. Only five proteins, all from Escherichia coli, satisfy these criteria: Crp, PurR, TrpR, Ihf and MetJ. For each structure we computed the number of contacts ni each base pair i has with the protein, i.e. the number of heavy atoms that are at a distance less than or equal to Rcutoff from a protein atom. To focus on the specific interactions of the DNA with the protein, we excluded atoms belonging to the sugar–phosphate DNA backbone because they do not depend on the DNA sequence. We also computed the number of hydrogen bonds ![]() (including water-mediated) and the number of hydrophobic interactions
(including water-mediated) and the number of hydrophobic interactions ![]() each base pair has with the protein. Hydrogen bonds were computed using NUCPLOT/HBPLUS (25,26). Two chemical groups are said to have a hydrophobic interaction if both have a CHARMM (27) group-charge less than 0.3 and they are separated by less than Rcutoff. Hydrogen bonds and hydrophobic interactions with the sugar–phosphate DNA backbone were not taken into account. We varied Rcutoff in a range from 3.5 to 5 Å and studied how the value of Rcutoff influences the results (see Results). Although certain interactions can be classified as hydrogen bonds and hydrophobic interactions, most of the contacts between a base pair and a protein cannot be easily classified. These interactions include contacts between hydrophobic and polar groups, polar and polar, charged and polar groups, etc. We did not consider these groups separately in this study.
each base pair has with the protein. Hydrogen bonds were computed using NUCPLOT/HBPLUS (25,26). Two chemical groups are said to have a hydrophobic interaction if both have a CHARMM (27) group-charge less than 0.3 and they are separated by less than Rcutoff. Hydrogen bonds and hydrophobic interactions with the sugar–phosphate DNA backbone were not taken into account. We varied Rcutoff in a range from 3.5 to 5 Å and studied how the value of Rcutoff influences the results (see Results). Although certain interactions can be classified as hydrogen bonds and hydrophobic interactions, most of the contacts between a base pair and a protein cannot be easily classified. These interactions include contacts between hydrophobic and polar groups, polar and polar, charged and polar groups, etc. We did not consider these groups separately in this study.
Aligned footprinted sites collected from the literature were obtained from DPI database (18). For each site we computed variability Si (sequence entropy) (28) at position i as
where fi(x) is a frequency of nucleotide x in position i of the site. To match Si and ni we manually aligned the DNA sequence from the PDB file with the collection of sites. In most cases the alignment is gapless and unambiguous due to high similarity between the PDB sequence and the consensus sequence.
In the case of a palindromic site, relative orientation of the site and the DNA sequence were chosen as follows. In Crp, the DNA sequence in the structure is palindromic, while the sites are not perfectly palindromic, with one half-site more conserved. We chose the orientation such that a more conserved half-site is aligned with a half-site in the PDB sequence which has more interactions with the protein. In PurR, while the sites are not perfectly palindromic, the DNA sequence in the structure is palindromic and the structure of the complex is perfectly symmetric. So, the choice of orientation is irrelevant. In Ihf, the site is not palindromic and the choice of orientation is unambiguous. In MetJ and TrpR, the DNA sequence in the structure is palindromic. Although the structures are not perfectly symmetric, vectors n are almost symmetric leading to very little change in correlation r upon different orientations. In this case we kept the orientation given in the PDB file.
To compute the correlation between S and n we used three different measures: the linear correlation coefficient r, χ2 association (29) and 2 × 2 association measure γ (30). The correlation coefficient measures the degree of linear correlation between S and n, while χ2 and γ can identify a non-linear association between the variables. For all three measures we computed statistical significance Pr, Pχ2, Pγ as the probability of observed association under the null hypothesis of independence. For example, to computed Pr we randomly shuffled the S vector 1000 times and computed r for each shuffled S and original n. Then, Pr is computed as a fraction of observations with r(Sshuffled, n) ≤ r(S, n). The statistical significance of χ2 and γ are computed the same way (31).
Both χ2 and γ measure the association between categorical variables, hence to use χ2 and γ one needs to group variables into classes. To compute χ2 we grouped S into four bins: [0, log 1.2], [log 1.2, log 2], [log 2, log 3], [log 3, log 4]. There is no need to bin the number of contacts n, as it is a discrete variable. If C(s, n) is 4 × max(n) matrix with the number of base pairs that have S in one of the four classes s, and n interactions, then
Where E(s, n) is the expected number of such base pairs given marginal distributions of s and n.
Similarly, to compute γ we built a 2 × 2 table by classifying positions as being variable (Si > Scut) versus conserved (Si ≤ Scut) and as strongly involved (ni > ncut) versus slightly involved (ni < ncut) in interactions with the protein. To eliminate ambiguity in setting the cutoffs, Scut and ncut, we used medians of S and n accordingly. This way we obtained a 2 × 2 variability-involvement frequency table ρ,
ρ11 = number of positions with Si > Scut and ni > ncut
ρ12 = number of positions with Si ≤ Scut and ni > ncut
ρ21 = number of positions with Si > Scut and ni ≤ ncut
ρ22 = number of positions with Si ≤ Scut and ni ≤ ncut 3
Then the association between S and n is measured as (30)
Missing values of ni were set to 0. Missing values of si were set to log4.
RESULTS
Table 1 summarizes the results for all five proteins. Strikingly, all the proteins except MetJ exhibit a strong negative correlation between the variability S and the number of protein–DNA interactions n. In other words, base pairs that have more interactions with the protein are more conserved. Importantly, interactions of all types were counted together.
Table 1. The correlation between the evolutionary variability of the site S and the number of protein–DNA interactions n for Rcutoff = 4.5 Å.
| Factor | PDB | Msites | r | Pr | χ2 | Pχ2 | γ | Pγ |
|---|---|---|---|---|---|---|---|---|
| Crp | 1run | 49 | –0.77 | <10–3 | 44.7 | <10–3 | –0.87 | 0.003 |
| PurR | 1bdh | 22 | –0.61 | <0.002 | 49.4 | <10–3 | –0.77 | 0.020 |
| Ihf | 1ihf | 26 | –0.74 | <10–3 | 46.6 | <10–3 | –0.81 | 0.020 |
| TrpR | 1rcs | 13 | –0.47 | 0.004 | 43.5 | <10–3 | –0.41 | 0.182 |
| TrpR SELEX | 1rcs | 58 | –0.61 | <10–3 | 85.5 | <10–3 | –1.00 | <10–3 |
| MetJ | 1mj2 | 15 | –0.34 | 0.068 | 25.1 | <10–3 | –0.50 | 0.234 |
| MetJ SELEX holo | 1mj2 | 74 | –0.67 | 0.001 | 15.2 | 0.239 | –0.56 | 0.202 |
| MetJ SELEX apo | 1mj2 | 55 | –0.62 | 0.003 | 18.2 | 0.074 | –0.78 | 0.046 |
PDB, Protien Data Bank identifier of the structure used to compute n. Msites, number of footprinted sites used to compute S. Other quantities are defined in the text.
As the number of contacts n depends on the value of Rcutoff , we studied how this parameter influences our results. Cutoffs for atomic interactions typically range from 3.5 to 5 Å (16,32,33). Table 2 shows correlation r and association γ computed using different values of Rcutoff. Although the qualitative picture does not change much upon variation of the cutoff, the trend is that a greater cutoff provides a somewhat higher correlation. Using a single cutoff for all types of atoms and groups and all types of interactions is clearly a simplification, as different chemical groups have different effective radii and interactions of a different nature (electrostatic, hydrophobic, etc.) have different ranges (34,35).
Table 2. Correlation between S and n for different values of contact cutoff.
| Factor | Correlations coefficient | γ | ||||||
|---|---|---|---|---|---|---|---|---|
| Rcutoff | 3.5 | 4.0 | 4.5 | 5.0 | 3.5 | 4.0 | 4.5 | 5.0 |
| Crp | –0.71 | –0.71 | –0.77 | –0.81 | –0.92 | –0.94 | –0.87 | –0.87 |
| PurR | –0.32 | –0.50 | –0.61 | –0.72 | –0.59 | –0.77 | –0.77 | –0.84 |
| Ihf | –0.68 | –0.72 | –0.74 | –0.76 | –0.74 | –0.81 | –0.81 | –0.77 |
| TrpR | –0.46 | –0.47 | –0.47 | –0.47 | –0.41 | –0.41 | –0.41 | –0.42 |
| TrpR SELEX | –0.55 | –0.60 | –0.61 | –0.49 | –1.00 | –1.00 | –1.00 | –1.00 |
| MetJ | –0.23 | –0.32 | –0.34 | –0.31 | –0.14 | –0.68 | –0.50 | –0.50 |
| MetJ SELEX holo | –0.63 | –0.61 | –0.65 | –0.67 | –0.86 | –0.69 | –0.56 | –0.95 |
| MetJ SELEX apo | –0.59 | –0.57 | –0.62 | –0.61 | –1.00 | –0.90 | –0.78 | –0.94 |
Statistically significant values (P < 2.5%) are shown in bold.
To examine the contribution of different types of interactions we compute the number of hydrogen bonds (including water-mediated ones) (25) and the number of hydrophobic interactions formed by each base pair with the protein. Two groups are said to form a hydrophobic interaction if they are in contact (r < Rcutoff) and both interacting groups are hydrophobic (see Materials and Methods). As water-mediated hydrogen bonds are included, certain nucleotides can have hydrogen bonds with a protein while having no direct interaction as defined by r < Rcutoff. Table 3 presents correlations between S and the number of hydrogen bonds and hydrophobic interactions. Surprisingly, correlations obtained for any single type of interaction are weaker than correlations obtained for all types taken together. Note that aside from hydrogen bonds and hydrophobic interactions, there are many more contacts between nucleotides and amino acids. These include interactions between polar groups, polar and hydrophobic groups, charged groups, etc. (36–38). A detailed examination of these types of interactions is beyond the scope of this study.
Table 3. The correlation between the evolutionary variability of the site (s) and the number of specific protein–DNA interactions: hydrogen bonds and hydrophobic interactions.
| Factor | N | NHB | NHF | Correlation coefficient | ||
|---|---|---|---|---|---|---|
| n | nHB | nHF | ||||
| Crp | 150 | 6 | 14 | –0.77 | –0.50 | –0.50 |
| PurR | 220 | 14 | 30 | –0.61 | –0.35 | –0.60 |
| Ihf | 206 | 4 | 11 | –0.74 | –0.10 | –0.76 |
| TrpR | 215 | 3 | 17 | –0.47 | –0.07 | –0.35 |
| TrpR SELEX | 215 | 3 | 17 | –0.61 | –0.24 | –0.47 |
| MetJ | 128 | 9 | 11 | –0.34 | –0.28 | 0.03 |
| MetJSELEX holo | 128 | 9 | 11 | –0.67 | –0.67 | –0.43 |
| MetJ SELEX apo | 128 | 9 | 11 | –0.62 | –0.66 | –0.43 |
Below we consider separately each studied protein–DNA complex.
Crp
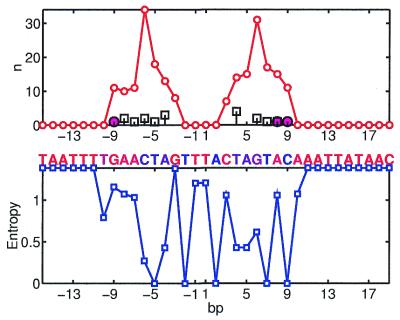
Figure 1 presents Si and ni for the complex of Catabolite gene activator protein (CAP) with its site. CAP is a homodimer. The binding site of each domain can be seen as the region of high ni and low Si on the figure. Interestingly, the ‘right’ site is slightly less conserved and indeed it has fewer interactions with the protein. Most of the interactions are formed by Arg-180, Arg-185 and Glu-181 in both chains. They form both hydrogen bonds and hydrophobic interactions (by Cβ, Cγ atoms interacting with the CH3 group of T).
Figure 1.
Crp site. (Top) Thin line shows the number of interactions n base pairs have with the protein. The number of hydrogen bonds formed by a base pair (including water-mediated bonds) is shown by large circles. The number of hydrophobic interactions between a base pair and the protein is shown by squares. (Bottom) Variability (entropy) in the footprinted DNA sites. The ‘consensus’ (most frequent) nucleotides are shown by letters above the plot. The color of a letter indicates its conservation from blue (conserved) to red (variable).
The hydrogen bonding pattern nHB and the hydrophobic pattern nHF of interactions exhibit significant, but much weaker correlations with S (see Table 3).
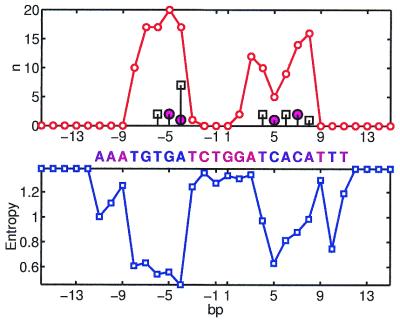
PurR
For purine repressor, both S and n are very symmetric (Fig. 2). However, the perfect symmetry of n is the result of the X-ray structure that was built assuming the 2-fold symmetry of the molecule (39). The correlation between S and n is statistically significant, but not very high (–0.61).
Figure 2.
PurR site; notation as in Figure 1.
A few outliers can be seen on Figure 2, e.g. base pairs AT in positions –3 and 3 are very conserved, but have very few interactions with the protein. Most other positions show a regular trend: S decreases as n increases. On the protein side, residues that have most of the contacts with the bases are Thr-14, Arg-24, Leu-52, Ala-49 and Ala-53. Both hydrogen bonding and hydrophobic interactions are involved in recognition. The hydrogen pattern has a low correlation with conservation, while the hydrophobic one exhibits high and significant correlation with S, suggesting the importance of the hydrophobic interactions in specific recognition by PurR.
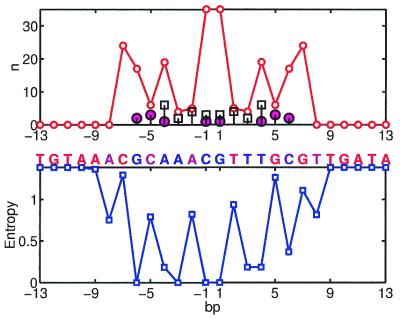
Ihf
Integration host factor (IHF) is known to bend DNA 160° at the binding site. The site consists of two regions: a 5′ region with no clear consensus and a 3′ region with a significant but very small consensus. Accordingly, the X-ray structure of the IHF complex shows very few, if any, protein–DNA contacts in the 5′ region and tight protein–DNA interactions in the 3′ region (40). Our analysis brings quantitative support to these observations. Figure 3 shows the number of protein–DNA interactions and variability of the base pairs in the IHF site. Our results indicate that conservation in the 3′ region can be very well explained by direct protein interactions with the DNA. Two peaks in n correspond to the regions where two proline residues (one from each protein chain) intercalate the DNA. Four arginines, Arg-59 and Arg-62 from both chains A and B, form almost as many interactions with the bases as intercalating prolines. Most of the other interactions in these regions are formed by Lys-65 (chains A and B), Ile-72 (chain A), Asn-63 (chains A and B) and Gly-61 (chains A and B). While arginines are involved in direct and water-mediated hydrogen bonding, prolines and isoleucines form hydrophobic interactions with the bases. Two out of three hydrogen bonds with the bases, however, are formed by a non-conserved G at position –4 and a non-conserved C at position 3. Position –4 is occupied by G in only 15% of the sites (T is the most frequent) and position 3 is occupied by C in 19% (G is the most frequent; Fig. 3) indicating that hydrogen bonding of these base pairs does not lead to strong specificity. Another hydrogen bond and several non-bonded interactions are formed by Arg-46 (chain B) with base pairs at positions –10. . . –13. These interactions are also apparently non-specific as base pairs at these positions are not conserved. In summary, a –0.74 correlation is observed in the IHF site, while the hydrogen binding pattern alone cannot explain observed conservation. In contrast, hydrophobic interactions dominate in the specific recognition exhibiting the correlation of –0.76.
Figure 3.
Ihf site; notation as in Figure 1.
TrpR
Only four natural footprinted sites are available for TrpR in the DPI database. However, 13 TrpR sites were found by McGuire et al. (41) in the genomes of E.coli and Haemophilus influenzae. We used these 13 sites for our analysis. Although significant as judged by r and χ2 (but not by γ), the correlation between S and n is weak (Fig. 4). Both n and S are symmetric and exhibit the distinct pattern of highly conserved A–7C– 6T–5A– 4 and T4A5G6T7. Base pairs C– 6 and G6 have the largest number of interactions with the protein. Both half-sites form multiple hydrophobic interactions with the protein and very few hydrogen bonds. Another conserved base pair is G·C9. It has 11 interactions with the protein and a single hydrogen bond. However, mutations that eliminate this hydrogen bond have a minor effect on the stability of the complex (42). Both hydrogen bonds and hydrophobic interactions alone show no significant correlation with evolutionary conservation. Perhaps other types of interactions (including non-direct readout) determine the specific recognition by TrpR (43,44).
Figure 4.
Trp site; notation as in Figure 1.
When the sites obtained by SELEX are used to compute S, the correlation between S and n becomes much stronger with γ = –1 and r = –0.61. Conservation in the SELEX sequences is localized around the GNACTAG consensus that corresponds to the binding half-site of one of the two protein domains. The rest of the sequences exhibit no conservation. This pronounced pattern gives rise to a higher correlation. Half-site bound by the second protein domain does not show any conservation in SELEX, while exhibiting this conservation in the natural sites. Perhaps only one domain was effectively binding randomized sequences in the SELEX experiment.
Another reason for this inconsistency between the number of interactions and the natural sites could result from different modes of binding observed in Trp repressor, which exhibits both dimer and tandem binding (42,45). To avoid interference between overlapping sites we used the structure of a Trp dimer for our analysis, while the pattern of conservation may arise from the combination of tandem and dimer binding modes.
MetJ
MetJ binds to arrays of two to five adjacent copies of an 8 bp ‘metbox’ sequence. Naturally occurring operators differ from the consensus sequence to a greater extent as the number of metboxes increases. This makes the motif obtained from the individual 8 bp sites very weak, exhibiting no significant correlation with the number of direct protein–DNA complexes. However, the conservation pattern of SELEX-derived sites does correlate with the number of interactions between the base pairs and the protein. Protein–DNA hydrophobic interactions are not present in this complex. The pattern of hydrogen bonding, however, exhibits a very strong and significant correlation with the conservation, suggesting an important role of hydrogen bonds in the specific recognition of the MetJ site.
Predictions
Based on the observed correlation one can make certain predictions. If a protein–DNA complex is available but recognition motif is unknown, one can compute the number of contacts per base pair and predict the most conserved ones. On the contrary, when many footprinted sites are known and the structure of the complex has not been solved, one can predict which base pairs form most of the interactions with the protein. Such predictions can be verified by future structural work.
For example, a high-resolution structure of Rob transcription factor bound to its site has been solved recently. However, very few sites have been footprinted for this factor, making it difficult to derive a motif and assess the relative importance of base pairs in the site. The DNA fragments in the PDB (1d5y) file is
TGACAGCACTGAATGTCAAAG-
-CTGTCGTGACTTACAGTTTCA
Judging by the number of contacts with the protein, we predict GC5CG6AT7 to be the most conserved base pairs (underlined above) as they have 15, 33 and 15 contacts, correspondingly (Rcutoff = 4.5 Å).
Another example is LexA, a transcription factor regulating a number of genes involved in the response to DNA damage. Although many footprinted sites are available for LexA, no structure of LexA protein–DNA complex has been solved. LexA has a consensus sequence TACTGTATATATATACAGTA with most conserved C–8T–7G–6 and C6A7T8 (underlined above). We predict that these base pairs have more contacts with the protein then others.
DISCUSSION
Here we introduced a novel approach to study protein–DNA interactions. This approach is based on two sources of information: structural information contained in the high-resolution protein–DNA complexes, and evolutionary information in the form of DNA sites of the DNA-binding proteins. The use of evolutionary information gives an enormous advantage: it allows us to find conserved base pairs and hence reveal protein–DNA interactions that are more important for specific recognition. The question addressed here is whether patterns of relative conservation in the DNA site can be rationalized using structural information. The main result of the study is that a statistically significant correlation was observed between the number of protein–base pair interactions and the conservation of this base pair. In other words, direct interactions between protein and DNA can explain very well the pattern of conservation of the DNA sites.
The origin of this correlation is clear: some of the direct interactions between the nucleotides and the protein are stabilizing the complex; then mutations of a base pair which has more interactions are more destabilizing for the complex and, hence, are eliminated in evolution. For the same reason amino acids that have more interactions within a protein (buried residues) are more conserved. Although this result for amino acids in proteins has been known for decades (46) it was quantified only recently (47). A similar result for base pairs in protein–DNA complexes is reported here for the first time.
Note that the observed correlation, although statistically significant, is not very strong. There are many outliers, i.e. non-conserved base pairs with many interactions as well as conserved base pairs with very few interactions. The correlation reflects a general tendency of base pairs with more interactions to be more conserved, but this rule has many exceptions.
Another result concerns the role of hydrogen bonds that are believed to dominate in determining the specificity and stability of protein–DNA complexes. Our results, on the contrary, indicate that hydrogen bonds alone cannot explain the pattern of conservation in most cases and, hence, are not the primary determinants of specific recognition. Only when hydrogen bonds, hydrophobic and other interactions are taken together does this number correlate with patterns of conservation.
Our analysis is based on an assumption that a DNA-binding protein forms similar structural complexes with different sites. In particular, we assumed that the number of contacts a base pair has with the protein in the crystal structure stays the same in all the sites bound by the protein. In general, the number and the nature of interactions changes when the DNA sequence of the site is altered. Recently, Pabo and co-workers (48) showed that the same protein Zif-268 could shift its contacts when it interacts with different sequences. To assess how such shift can affect our results we computed the number of interactions n per base pair for the structure of Zif-268 binding two different sequences (48) (PDB accession codes 1G2F and 1G2D). We found that, in spite of the shift, the number of interactions per base pair does not change drastically. The correlation between ![]() and
and ![]() is 0.80. . . 0.86 for Rcutoff range from 3.5 to 5 Å (taking into account base pairs with at least one interaction). This example shows that although interactions of the same protein with two different sequences can be different, the profile of the number of interactions does not change much. Unfortunately, it is rare to have a structure of the same protein binding different DNA sequences and, hence, the magnitude of this effect cannot be assessed easily. One possible approach is to build computer models of the same protein binding different sequences and assess structural changes in the complexes subject to minimization of energy and molecular dynamics. We are currently working in this direction.
is 0.80. . . 0.86 for Rcutoff range from 3.5 to 5 Å (taking into account base pairs with at least one interaction). This example shows that although interactions of the same protein with two different sequences can be different, the profile of the number of interactions does not change much. Unfortunately, it is rare to have a structure of the same protein binding different DNA sequences and, hence, the magnitude of this effect cannot be assessed easily. One possible approach is to build computer models of the same protein binding different sequences and assess structural changes in the complexes subject to minimization of energy and molecular dynamics. We are currently working in this direction.
Contribution of different types of interaction is another important issue. The nature of protein–DNA interactions is very complex and involves many types of interactions: hydrogen bonds, hydrophobic interactions (4), electrostatic interactions (38), effects of ‘indirect readout’ related to water extrusion (43,49) and local DNA bending and twisting (50). In this study we focused on the contribution of hydrogen bonds and hydrophobic interactions and did not consider separate contributions of other types of interactions, such as electrostatic interactions, CH. . . O hydrogen bonds (36), cation-π interactions (37), etc. Our results suggest that although a single type of interaction (hydrogen bonds or hydrophobic interactions) can rationalize conservation in one protein, these interactions do not work for another protein. In contrast, a parameter n, which includes all types of interactions, works uniformly better for all proteins. It is surprising that such a simple parameter as the number of all direct interactions (that does not take into account even the different strength of interactions) is able to explain the patterns of conservation in the DNA-binding sites. This result makes us believe that more complex models of protein–DNA energetics would be able to predict binding motifs of the DNA-binding proteins. However, to be successful, such methods need to concentrate on interactions with conserved nucleotides, rather than on all protein–DNA interactions. A similar focus on more conserved interactions in prediction of protein structures was very productive (51). Another analogy is profiles constructed using multiple sequence alignments. By weighing conserved amino acids more than variable ones, such profiles achieved very high sensitivity in detecting remote homologs.
In summary, we have studied five different bacterial transcription factors and have demonstrated that the number of interactions a base pair has with the protein significantly correlates with conservation of this base pair. We have also shown that neither hydrogen bonds nor hydrophobic interactions dominate in determining this correlation. The contribution of these interactions varies for different transcription factors.
Acknowledgments
ACKNOWLEDGEMENTS
L.A.M. was supported the William F. Milton Fund. M.S.G. is supported by grants from the Russian Fund of Basic Research (99-04-48247 and 00-15-99362), INTAS (99-1476) and HHMI (55000309).
REFERENCES
- 1.Stover C., Pham,X., Erwin,A., Mizoguchi,S., Warrener,P., Hickey,M., Brinkman,F., Hufnagle,W., Kowalik,D., Lagrou,M., Garber,R., Goltry,L., Tolentino,E., Westbrock-Wadman,S., Yuan,Y., Brody,L., Coulter,S., Folger,K., Kas,A., Larbig,K., Lim,R., Smith,K., Spencer,D., Wong,G., Wu,Z. and Paulsen,I. (2000) Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature, 406, 959–664. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert W. and Muller-Hill,B. (1967) The lac operator is DNA. Proc. Natl Acad. Sci. USA, 58, 2415–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman H., Zardecki,C. and Westbrook,J. (1998) The nucleic acid database: a resource for nucleic acid science. Acta Crystallogr. D. Biol. Crystallogr., 54, 1095–1104. [DOI] [PubMed] [Google Scholar]
- 4.Larson C. and Verdine,G. (1996) The chemistry of protein-DNA interactions. In Hecht,S.M. (ed.), Bioorganic Chemistry: Nucleic Acids. Oxford University Press, Oxford, UK, pp. 324–346.
- 5.Fields D., He,Y., Al-Uzri,A. and Stormo,G. (1997) Quantitative specificity of the Mnt repressor. J. Mol. Biol., 271, 178–194. [DOI] [PubMed] [Google Scholar]
- 6.Brown B. and Sauer,R. (1999) Tolerance of Arc repressor to multiple-alanine substitutions. Proc. Natl Acad. Sci. USA, 96, 1983–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrison S. (1991) A structural taxonomy of DNA-binding domains. Nature, 353, 715–719. [DOI] [PubMed] [Google Scholar]
- 8.Pabo C. and Sauer,R. (1992) Transcription factors: structural families and principles of DNA recognition. Annu. Rev. Biochem., 61, 1053–1095. [DOI] [PubMed] [Google Scholar]
- 9.Wintjens R. and Rooman,M. (1996) Structural classification of HTH DNA-binding domains and protein–DNA interaction modes. J. Mol. Biol., 262, 294–313. [DOI] [PubMed] [Google Scholar]
- 10.Sauer R. and Harrison,S. (1996) Interactions of proteins with RNA and DNA. Curr. Opin. Struct. Biol., 6, 51–52. [DOI] [PubMed] [Google Scholar]
- 11.Luscombe N., Laskowski,R. and Thornton,J. (2001) Amino acid-base interactions: a three-dimensional analysis of protein–DNA interactions at an atomic level. Nucleic Acids Res., 29, 2860–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones S., van Heyningen,R., Berman,H. and Thornton,J. (1999) Protein–DNA interactions: a structural analysis. J. Mol. Biol., 287, 877–896. [DOI] [PubMed] [Google Scholar]
- 13.Nadassy K., Wodak,S. and Janin,J. (1999) Structural features of protein–nucleic acid recognition sites. Biochemistry, 38, 1999–2017. [DOI] [PubMed] [Google Scholar]
- 14.Pabo C. and Nekludova,L. (2000) Geometric analysis and comparison of protein–DNA interfaces. J. Mol. Biol., 301, 597–624. [DOI] [PubMed] [Google Scholar]
- 15.Lustig B. and Jernigan,R. (1995) Consistencies of individual DNA base–amino acid interactions in structures and sequences. Nucleic Acids Res., 23, 4707–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kono H. and Sarai,A. (1999) Structure-based prediction of DNA target sites by regulatory proteins. Protein, 35, 114–131. [PubMed] [Google Scholar]
- 17.Mandel-Gutfreund Y. and Margalit,H. (1998) Quantitative parameters for amino acid-base interaction: implications for prediction of protein–DNA binding sites. Nucleic Acids Res., 26, 2306–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robison K., McGuire,A. and Church,G. (1998) A comprehensive library of DNA-binding site matrices for 55 proteins applied to the complete Escherichia coli K-12 genome. J. Mol. Biol., 284, 241–254. [DOI] [PubMed] [Google Scholar]
- 19.Czernik P., Shin,D. and Hurlburt,B. (1994) Functional selection and characterization of DNA binding sites for trp repressor of Escherichia coli. J. Biol. Chem., 269, 27869–27875. [PubMed] [Google Scholar]
- 20.Tuerk C. and Gold,L. (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science, 249, 505–510. [DOI] [PubMed] [Google Scholar]
- 21.Shultzaberger R. and Schneider,T. (1999) Using sequence logos and information analysis of Lrp DNA binding sites to investigate discrepancies between natural selection and SELEX. Nucleic Acids Res., 27, 882–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salgado H., Santos-Zavaleta,A., Gama-Castro,S., Millan-Zarate,D., Diaz-Peredo,E., Sanchez-Solano,F., Perez-Rueda,E., Bonavides-Martinez,C. and Collado-Vides,J. (2001) RegulonDB (version 3.2): transcriptional regulation and operon organization in Escherichia coli K-12. Nucleic Acids Res., 29, 72–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCue L., Thompson,W., Carmack,C., Ryan,M., Liu,J., Derbyshire,V. and Lawrence,C. (2001) Phylogenetic footprinting of transcription factor binding sites in proteobacterial genomes. Nucleic Acids Res., 29, 774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berman H., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T., Weissig,H., Shindyalov,I. and Bourne,P. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDonald I. and Thornton,J. (1994) Satisfying hydrogen bonding potential in proteins. J. Mol. Biol., 238, 777–793. [DOI] [PubMed] [Google Scholar]
- 26.Luscombe N., Laskowski,R. and Thornton,J. (1997) NUCPLOT: a program to generate schematic diagrams of protein-nucleic acid interactions. Nucleic Acids Res., 25, 4940–4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacKerell A., Brooks,C., Brooks,L., Nilsson,L., Roux,B., Won,Y. and Karplus,M. (1998) CHARMM: the energy function and its parameterization with an overview of the program. In Schleyer,R. et al. (eds), The Encyclopedia of Computational Chemistry. John Wiley & Sons, Chichester, pp. 271–277.
- 28.Stormo G., Schneider,T. and Gold,L. (1986) Quantitative analysis of the relationship between nucleotide sequence and functional activity. Nucleic Acids Res., 14, 6661–6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeGroot M. (1996) Probability and Statistics. Addison-Wesley Pub. Co., Reading, MA.
- 30.Goodman L. and Kruskal,W. (1979) Measures of association for cross classifications. Springer-Verlag, New York, NY.
- 31.Good P. (1994) Permutation tests: a practical guide to resampling methods for testing hypotheses. Springer-Verlag, New York, NY.
- 32.Miyazawa S. and Jernigan,R.L. (1996) Residue–residue potentials with a favorable contact pair term and an unfavorable high packing density term, for simulation and threading. J. Mol. Biol., 256, 623–644. [DOI] [PubMed] [Google Scholar]
- 33.Hinds D. and Levitt,M. (1994) Exploring conformational space with a simple lattice model for protein structure. J. Mol. Biol., 243, 668–682. [DOI] [PubMed] [Google Scholar]
- 34.Tsai J., Taylor,R., Chothia,C. and Gerstein,M. (1999) The packing density in proteins: standard radii and volumes. J. Mol. Biol., 290, 253–266. [DOI] [PubMed] [Google Scholar]
- 35.Nadassy K., Tomas-Oliveira,I., Alberts,I., Janin,J. and Wodak,S. (2001) Standard atomic volumes in double-stranded DNA and packing in protein–DNA interfaces. Nucleic Acids Res., 29, 3362–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mandel-Gutfreund Y., Margalit,H., Jernigan,R. and Zhurkin,V. (1998) A role for CH. . . O interactions in protein–DNA recognition. J. Mol. Biol., 277, 1129–1140. [DOI] [PubMed] [Google Scholar]
- 37.Wintjens R., Lievin,J., Rooman,M. and Buisine,E. (2000) Contribution of cation-pi interactions to the stability of protein–DNA complexes. J. Mol. Biol., 302, 395–410. [DOI] [PubMed] [Google Scholar]
- 38.Madan B. and Sharp,K. (2001) Hydration heat capacity of nucleic acid constituents determined from the random network model. Biophys. J., 81, 1881–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schumacher M., Choi,K., Zalkin,H. and Brennan,R. (1994) Crystal structure of LacI member, PurR, bound to DNA: minor groove binding. Science, 266, 763–770. [DOI] [PubMed] [Google Scholar]
- 40.Rice P. (1997) Making DNA do a U-turn: IHF and related proteins. Curr. Opin. Struct. Biol., 7, 86–93. [DOI] [PubMed] [Google Scholar]
- 41.McGuire A., Hughes,J. and Church,G. (2000) Conservation of DNA regulatory motifs and discovery of new motifs in microbial genomes. Genome Res., 10, 744–757. [DOI] [PubMed] [Google Scholar]
- 42.Grillo A., Brown,M. and Royer,C. (1999) Probing the physical basis for trp repressor-operator recognition. J. Mol. Biol., 287, 539–554. [DOI] [PubMed] [Google Scholar]
- 43.Shakked Z., Guzikevich-Guerstein,G., Frolow,F., Rabinovich,D., Joachimiak,A. and Sigler,P. (1994) Determinants of repressor/operator recognition from the structure of the Trp operator binding site. Nature, 368, 469–473. [DOI] [PubMed] [Google Scholar]
- 44.Ladbury J., Wright,J., Sturtevant,J. and Sigler,P. (1994) A thermodynamic study of the Trp repressor-operator interaction. J. Mol. Biol., 238, 669–681. [DOI] [PubMed] [Google Scholar]
- 45.Lawson C. and Carey,J. (1993) Tandem binding in crystals of a Trp repressor/operator half-site complex. Nature, 366, 178–182. [DOI] [PubMed] [Google Scholar]
- 46.Branden C. and Tooze,J. (1998) Introduction to Protein Structure. Garland Publishing, Inc., New York, NY.
- 47.Mirny L. and Shakhnovich,E. (1999) Universally conserved residues in protein folds. Reading evolutionary signals about protein function, stability and folding kinetics. J. Mol. Biol., 291, 177–196. [DOI] [PubMed] [Google Scholar]
- 48.Wolfe S., Grant,R., Elrod-Erickson,M. and Pabo,C. (2001) Beyond the “recognition code”: structures of two Cys(2)His(2) zinc finger/TATA box complexes. Structure, 9, 717–723. [DOI] [PubMed] [Google Scholar]
- 49.Janin J. (1999) Wet and dry interfaces: the role of solvent in protein–protein and protein–DNA recognition. Structure Fold Des., 7, R277–R279. [DOI] [PubMed] [Google Scholar]
- 50.Hizver J., Rozenberg,H., Frolow,F., Rabinovich,D. and Shakked,Z. (2001) DNA bending by an adenine–thymine tract and its role in gene regulation. Proc. Natl Acad. Sci. USA, 98, 8490–8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reva B., Skolnick,J. and Finkelstein,A. (1999) Averaging interaction energies over homologs improves protein fold recognition in gapless threading. Protein, 35, 353–359. [PubMed] [Google Scholar]