Abstract
This study examined the single-nucleotide polymorphism heritability and genetic correlations of cognitive abilities and brain structural measures (regional subcortical volume and cortical thickness) in middle-aged and elderly East Asians (Korean) from the Gwangju Alzheimer’s and Related Dementias cohort study. Significant heritability was found in memory function, caudate volume, thickness of the entorhinal cortices, pars opercularis, superior frontal gyri, and transverse temporal gyri. There were 3 significant genetic correlations between (i) the caudate volume and the thickness of the entorhinal cortices, (ii) the thickness of the superior frontal gyri and pars opercularis, and (iii) the thickness of the superior frontal and transverse temporal gyri. This is the first study to describe the heritability and genetic correlations of cognitive and neuroanatomical traits in middle-aged to elderly East Asians. Our results support the previous findings showing that genetic factors play a substantial role in the cognitive and neuroanatomical traits in middle to advanced age. Moreover, by demonstrating shared genetic effects on different brain regions, it gives us a genetic insight into understanding cognitive and brain changes with age, such as aging-related cognitive decline, cortical atrophy, and neural compensation.
Keywords: subcortical volume, SNP heritability, middle aged to older adults, cortical thickness, cognitive abilities
Introduction
Due to the increase in life expectancy, a growing number of individuals experience significant challenges related to cognitive dysfunction, and age-related cognitive decline and cognitive disorders, such as Alzheimer’s disease (AD), have arguably become a serious issue in public health. The genetic studies investigating their etiology have revealed that age-related cognitive dysfunction is influenced by genetic factors (Fan et al. 2019; Mollon et al. 2021), but the genetic underpinnings have not yet been fully elucidated (Collins and Williams-Gray 2016; Fan et al. 2019; Mollon et al. 2021). Accordingly, numerous studies have been performed to elucidate the genetic basis of age-related cognitive dysfunction, and among those studies, the exploration of genetic influences on brain structures and cognitive abilities has often been attempted since abnormal brain morphology and impaired cognitive ability are intertwined in the pathophysiology of cognitive dysfunction (Goh et al. 2011; Salthouse 2011; Thompson et al. 2020).
Neuroimaging genetic studies have repeatedly found that neur-oanatomical phenotypes such as intracranial volume, subcortical volume, and cortical thickness are highly heritable in childhood to adulthood, with genetic factors accounting for up to >90% of their variability (Lessov-Schlaggar et al. 2012; Batouli et al. 2014; Brouwer et al. 2014; Hibar et al. 2015; Ge et al. 2016; Lukies et al. 2017; Satizabal et al. 2019; Zhao et al. 2019; Biton et al. 2020; Grasby et al. 2020; Pizzagalli et al. 2020), and the heritability differs depending on the brain area. Behavioral genetic research has shown that general cognitive ability is substantially heritable across the life course, with approximately half of the variance in general cognition attributed to genetic factors (Haworth et al. 2010; Davies et al. 2011, 2015; Greenwood et al. 2011; Kraljević et al. 2021; Mollon et al. 2021). Specific cognitive abilities such as attention, executive function, and memory have also been found to be heritable (Greenwood et al. 2011; Robinson et al. 2015; Lemvigh, Brouwer, Pantelis, et al. 2020; Mollon et al. 2021). These heritability studies have demonstrated that the genetic contribution to cognition and brain structure changes with age by showing that the heritability is moderated by age and have suggested that the heritability variation may be attributed to the different stages in developmental or aging trajectories (Lessov-Schlaggar et al. 2012; Batouli et al. 2014; Lukies et al. 2017; Mollon et al. 2021).
However, no studies have investigated the genetic contributions to brain structure and cognitive function simultaneously in middle-aged and older adults, who are the most vulnerable to age-related neurodegenerative diseases and are in the middle of the aging process, experiencing cortical atrophy and cognitive decline (Grady 2012; Montembeault et al. 2012). Furthermore, although it is known that people from different ethnic groups can have different genetic backgrounds and could be subject to different nongenetic factors, no studies based on the populations of East Asian ancestry have investigated the heritability of cognitive function and brain structure together in middle-to-late adulthood. To fill these research gaps, in this study, we estimated the single-nucleotide polymorphism (SNP) heritability of cognitive abilities and brain structural measures (subcortical volume and cortical thickness) in middle-aged and elderly East Asians. Furthermore, we calculated the genetic correlations among these cognitive and brain structural measures. For heritability and genetic correlation analyses, we performed a genome-wide complex trait analysis (GCTA) (Yang, Lee, et al. 2011; Yang, Manolio, et al. 2011). We hope that our study can offer a better understanding of the genetic influences on the cognitive abilities and brain structures in middle-aged and older adults and provide insights into the genetic underpinnings of age-related cognitive dysfunction.
Materials and methods
Study participants
The study participants included 2,159 middle-aged to elderly (41–105 years) subjects who were enrolled in the Gwangju Alzheimer’s and Related Dementias study in Korea. All subjects underwent comprehensive clinical and neuropsychological assessments and brain magnetic resonance (MR) imaging. Participants were stratified into 3 categories of cognitive status, including normal cognition, mild cognitive impairment (MCI), and AD, based on the National Institute Neurological and Communicative Disorders and Stroke–Alzheimer’s Disease and Research Disorders Association (McKhann et al. 1984) diagnostic criteria. All diagnoses were evaluated by dementia specialists in neurology and psychiatry at the Chosun University Hospital and Chonnam National University Hospital, Gwangju, Republic of Korea. Excluded subjects included individuals with a focal lesion on brain MRI, history of head trauma, or psychiatric disorders which could affect their mental function.
The study protocol was approved by the Institutional Review Board of Chosun University Hospital, Korea (CHOSUN 2013-12-018-070). All volunteers or authorized guardians for cognitively impaired individuals gave written informed consent before participation.
Cognitive measures
Cognitive functions were measured using the Seoul Neuropsychological Screening Battery (SNSB) (Ahn et al. 2010; Kang et al. 2019), which assesses 5 domains of cognition, including attention, language, visuospatial, memory, and frontal/executive function. Each domain comprises the following subtests: (i) attention: Digit Span Test (forward and backward) (Kang et al. 2002); (ii) language: comprehension, repetition, and Korean-Boston Naming Test (Kang et al. 1999); (iii) visuospatial function: Rey Complex Figure Test-copy (Meyers and Meyers 1995) and Clock Drawing Test; (iv) memory: Seoul Verbal Learning Test and Rey Complex Figure Test (Meyers and Meyers 1995); and (v) frontal/executive function: go/no-go, phonemic fluency, Korean-Color Word Stroop Test color reading (Lee et al. 2000), Digit Symbol Coding (Joy et al. 2004), and Korean-Trail Making Test-Elderly’s version Part B (Yi et al. 2007).
A composite score for each cognitive domain was calculated. For memory, language, visuospatial, and frontal function domains, the z-score of each test was calculated based on the age-, sex-, and education-specific normative data presented in the SNSB. The average of the z-scores of tests comprising the domain was used as the composite score for each domain. For the attention domain, the sum of the forward and backward Digit Span Test raw scores was calculated as the composite score. The composite cognitive domain scores were then used as measures of cognitive abilities in all analyses.
MR imaging and preprocessing
Brain MR imaging was performed using a 3.0-T scanner (Skyra, Siemens; 20-channel head coil; MPRAGE sagittal view; time to repetition (TR) = 2,300 ms; time to echo (TE) = 2.143 ms; inversion time (TI) = 900 ms; flip angle (FA) = 9°; field of view (FoV) = 256 mm × 256 mm; matrix = 320 × 320; slice thickness = 0.8 mm) and a 1.5-T scanner (Avanto, Siemens; 12-channel head coil; MPRAGE axial view; TR = 1,800 ms; TE = 3.43 ms; TI = 1,100 ms; FA = 15°; FoV = 224 mm × 224 mm; matrix = 256 × 256; slice thickness = 0.9 mm).
T1-weighted images were processed in an automated pipeline using FreeSurfer software version 5.3.0 (Fischl 2012). In short, the procedure included: motion correction, image normalization, removal of nonbrain tissue, Talairach transformation, white matter (WM) and gray matter (GM) subcortical segmentation, intensity normalization, tessellation of the GM and WM boundaries, automated topology correction, and surface deformation.
Subcortical volumes were calculated with FreeSurfer’s automated procedure for volumetric measures. These procedures resulted in the extraction of volumes for 8 bilateral subcortical regions, including the amygdala, caudate, hippocampus, nucleus accumbens, pallidum, putamen, thalamus, and lateral ventricles.
Cortical thickness was calculated as the shortest distance between the GM/WM boundary and pial surface at each vertex across the cortical mantle and was measured in millimeters (mm). In addition to vertex-based reconstruction, FreeSurfer automatically parcellated the cortex into 31 gyral-based regions of interest per hemisphere according to the Desikan-Killiany-Tourvill atlas (Klein and Tourville 2012). For each of the 68 cortical parcellations, FreeSurfer calculated the average cortical thickness (in mm).
Genotyping
Before the quality control and imputation procedures, 2,162 participants were genotyped using an Affymetrix customized KoreanChip (Moon et al. 2019; Seo et al. 2019). Genotype data were processed using PLINK (Purcell et al. 2007) and ONETOOL (Song et al. 2018). SNPs were excluded if the genotype call rate was <0.95 or not in the Hardy–Weinberg equilibrium (HWE) (P < 1 × 10−5). Subjects were also excluded if they were duplicated (identity-by-state >0.9), the genotype call rate was <0.95, or there was an inconsistency between recorded and genetic sex. After applying these filters, 2,159 subjects and 685,742 SNPs remained.
Genotypes were prephased using SHAPEIT (Delaneau et al. 2014) and were then imputed using the 1000 Genomes Phase 3 reference panel and IMPUTE2 (Howie et al. 2009). Imputed SNPs were excluded in cases with the INFO score <0.5, a genotype call rate <0.98, minor allele frequencies <0.01, or a P-value for the HWE <1 × 10−6. We finally pruned SNPs that were in linkage disequilibrium using a cutoff of r2 > 0.5, windows of 50 SNPs, and a step size of 5 SNPs. The filtering after imputation was made using PLINK as well. As a result, 2,159 subjects with genotype data of 604,613 SNPs were further analyzed.
SNP heritability and genetic correlation analysis
GCTA was employed to estimate the heritability of cognitive function, regional brain subcortical volume, and cortical thickness from the individual genotypes. Only genotypes on the autosomal chromosomes were used to calculate the genetic relationship matrix with GCTA. Heritability was calculated using a linear mixed model after adjusting for the effects of age, sex, and the first 10 principal components (PCs) from the genomic relationship matrix as fixed covariates. PCs were obtained using EIGENSOFT (Price et al. 2006). Additionally, the heritability estimate for each autosomal chromosome was calculated. Since we used a sample from the AD study cohort, there was an ascertainment bias (AB) that the proportion of individuals with MCI or AD was relatively higher than the real-world population of middle-aged to older adults. Therefore, the heritability estimates that were found to be significant were adjusted for the AB by assuming the prevalence of MCI and AD in the middle-aged to elderly (>age of 60 years) population in Korea, which was 30.52% (Lee et al. 2011; Korea 2020). The genetic correlation was estimated with the bivariate genomic-restricted maximum likelihood method (Yang, Manolio, et al. 2011), as implemented in the GCTA software, for pairs of phenotypes with significant heritability.
Results
Subject characteristics
Table 1 shows the demographic and clinical characteristics of 2,159 participants. The mean age is 72.8, and 60.2% of them are female. There were 805 (37.3%) cognitively normal individuals, 1,247 (57.7%) MCI patients, and 107 (5.0%) AD-type dementia patients. The proportions of apolipoprotein E (APOE) ε2 and ε4 carriers were, respectively, 11.6% and 19.5%. The mean scores of each domain of neuropsychological assessment were 8.83 (standard deviation [SD] = 2.29) for attention, −0.15 (SD = 0.63) for language, 0.05 (SD = 0.96) for visuospatial function, −0.29 (SD = 0.76) for memory, and − 0.29 (SD = 0.77) for frontal function.
Table 1.
Descriptive characteristics of the study sample.
| Characteristics (n = 2159) | Mean (SD) or n (%) |
|---|---|
| Demographic data | |
| Age | 72.77 (5.42) |
| Sex | |
| Female | 1,300 (60.2) |
| Male | 859 (39.8) |
| AD diagnosis | |
| NC | 805 (37.3) |
| MCI | 1,247 (57.7) |
| AD | 107 (5.0) |
| Genetic traits | |
| APOE ε2 carrier | 250 (11.6) |
| APOE ε4 carrier | 422 (19.5) |
| Neuropsychological scores | |
| Attention | 8.83 (2.29) |
| Language | −0.15 (0.63) |
| Visuospatial | 0.05 (0.96) |
| Memory | −0.29 (0.76) |
| Frontal | −0.29 (0.77) |
Note: SD, standard deviation; AD, Alzheimer’s disease; NC, normal cognition; MCI, mild cognitive impairment; APOE, apolipoprotein E.
SNP heritability of cognitive function
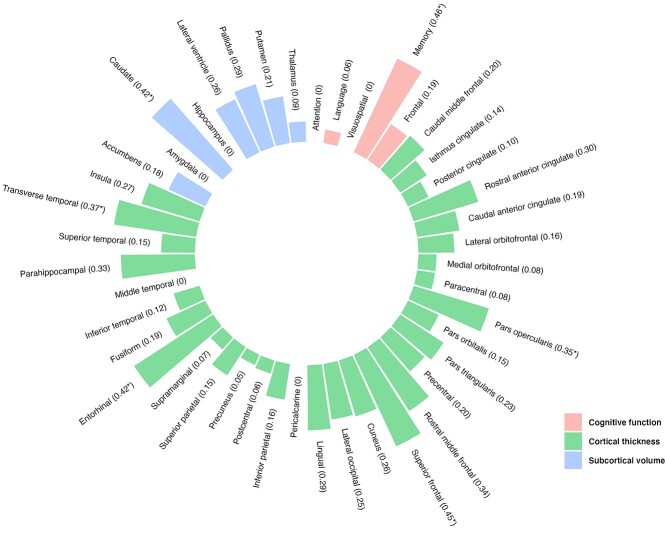
Estimates of the SNP heritability of cognitive function are presented in Table 2 and Fig. 1. Significant heritability was observed only for memory function (h2SNP = 0.46, P = 0.01). Additionally, for memory function, the heritability after adjusting for AB is presented in Supplementary Table S1 (h2SNP adjusted for AB = 0.51) and the heritability estimates for each chromosome are described in Supplementary Table S2 and Supplementary Fig. S1. There was a significant heritability on chromosome 13 (h2SNP = 0.10) and 19 (h2SNP = 0.06). However, no significant heritability was found for other cognitive functions (attention: h2SNP = 0.00, P = 0.50; language: h2SNP = 0.06, P = 0.39; visuospatial function: h2SNP = 0.00, P = 0.50; frontal function: h2SNP = 0.19, P = 0.19).
Table 2.
SNP heritability of cognitive function in the whole sample.
| Cognitive function | Vp | Vg | Heritability | ||
|---|---|---|---|---|---|
| h 2 SNP | SE | P-value | |||
| Attention | 4.65 | 0.00 | 0.00 | 0.21 | 0.50 |
| Language | 0.36 | 0.02 | 0.06 | 0.22 | 0.39 |
| Visuospatial | 0.86 | 0.00 | 0.00 | 0.22 | 0.50 |
| Memory | 0.52 | 0.24 | 0.46 | 0.21 | 0.01a |
| Frontal | 0.52 | 0.10 | 0.19 | 0.22 | 0.19 |
Note: All analyses are adjusted for age, sex, and 10PCs. Vp, total phenotypic variance; Vg, genetic variance; h2SNP, single-nucleotide polymorphism (SNP)-based heritability estimates; SE, standard error.
aIndicates significant heritability at P < 0.05.
Fig. 1.
Heritability estimates of cognitive function and brain structural measures (regional subcortical volume and cortical thickness).
SNP heritability of regional subcortical volume
Estimates of the SNP-based heritability of regional subcortical volume are presented in Table 3 and Fig. 1. We found a significant heritability estimate for caudate volume (h2SNP = 0.42, P = 0.02). For caudate volume, the heritability after adjusting for AB is described in Supplementary Table S1 (h2SNP adjusted for AB = 0.46) and the chromosome-specific heritability is illustrated in Supplementary Table S3 and Supplementary Fig. S2. Significant heritability was found on chromosome 18 (h2SNP = 0.08). However, no significant heritability was observed for other subcortical volumes (nucleus accumbens: h2SNP = 0.18, P = 0.20; amygdala: h2SNP = 0.00, P = 0.50; hippocampus: h2SNP = 0.00, P = 0.50; lateral ventricle: h2SNP = 0.26, P = 0.10; pallidus: h2SNP = 0.29, P = 0.07; putamen: h2SNP = 0.21, P = 0.17: thalamus: h2SNP = 0.09, P = 0.34).
Table 3.
SNP heritability of regional subcortical volumes in the whole sample.
| Subcortical volume | Vp | Vg | Heritability | ||
|---|---|---|---|---|---|
| h 2SNP | SE | P-value | |||
| Nucleus accumbens | 8,661.59 | 1,586.84 | 0.18 | 0.22 | 0.20 |
| Amygdala | 58,779.65 | 0.07 | 0.00 | 0.22 | 0.50 |
| Caudate | 282,936.24 | 119,219.77 | 0.42 | 0.21 | 0.02a |
| Hippocampus | 243,318.62 | 0.28 | 0.00 | 0.21 | 0.50 |
| Lateral ventricle | 56,537,981.92 | 14,822,716.05 | 0.26 | 0.21 | 0.10 |
| Pallidus | 58,011.30 | 16,930.84 | 0.29 | 0.21 | 0.07 |
| Putamen | 523,003.68 | 108,740.51 | 0.21 | 0.21 | 0.17 |
| Thalamus | 959,118.40 | 82,143.45 | 0.09 | 0.21 | 0.34 |
Note: All analyses are adjusted for age, sex, and 10PCs. Vp, total phenotypic variance; Vg, genetic variance; h2SNP, single-nucleotide polymorphism (SNP)-based heritability estimates; SE, standard error.
aIndicates significant heritability at P < 0.05.
SNP heritability of regional cortical thickness
Table 4 and Fig. 1 show the SNP-based heritability of regional cortical thickness. Significant heritability was found for the pars opercularis (h2SNP = 0.35, P = 0.04), superior frontal gyri (h2SNP = 0.45, P = 0.02), entorhinal cortices (h2SNP = 0.42, P = 0.03), and transverse temporal gyri (h2SNP = 0.37, P = 0.04). For these cortical regions showing significant heritability, the heritability estimates after correcting AB are presented in Supplementary Table S1 (pars opercularis, h2SNP adjusted for AB = 0.39; superior frontal gyri, h2SNP adjusted for AB = 0.50; entorhinal cortices, h2SNP adjusted for AB = 0.46; transverse temporal gyri, h2SNP adjusted for AB = 0.41). Supplementary Tables S4–S7 and Supplementary Figs. S3–S6 show the heritability estimates by chromosome. Significant chromosome-specific heritability was found on chromosomes 5 (h2SNP = 0.13) and 16 (h2SNP = 0.11) for the pars opercularis (Supplementary Table S4 and Supplementary Fig. S3); on chromosomes 5 (h2SNP = 0.19), 17 (h2SNP = 0.09), and 19 (h2SNP = 0.11) for the superior frontal gyri (Supplementary Table S5 and Supplementary Fig. S4); on chromosomes 13 (h2SNP = 0.08), 18 (h2SNP = 0.10), and 20 (h2SNP = 0.10) for the entorhinal cortices (Supplementary Table S6 and Supplementary Fig. S5); and on chromosomes 2 (h2SNP = 0.13) and 6 (h2SNP = 0.08) for the transverse temporal gyri (Supplementary Table S7 and Supplementary Fig. S6). On the other hand, significant heritability was not found for cortical thickness of other brain regions (caudal middle frontal: h2SNP = 0.20, P = 0.17; isthmus cingulate: h2SNP = 0.14, P = 0.26; posterior cingulate: h2SNP = 0.10, P = 0.31; rostral anterior cingulate: h2SNP = 0.30, P = 0.08; caudal anterior cingulate: h2SNP = 0.19, P = 0.19; lateral orbitofrontal: h2SNP = 0.16, P = 0.21; medial orbitofrontal: h2SNP = 0.08, P = 0.35; paracentral: h2SNP = 0.08, P = 0.35; pars orbitalis: h2SNP = 0.15, P = 0.23; pars triangularis: h2SNP = 0.23, P = 0.14; precentral: h2SNP = 0.20, P = 0.17; rostral middle frontal: h2SNP = 0.34, P = 0.06; cuneus: h2SNP = 0.26, P = 0.10; lateral occipital: h2SNP = 0.25, P = 0.13; lingual: h2SNP = 0.29, P = 0.09; pericalcarine: h2SNP = 0.00, P = 0.50; inferior parietal: h2SNP = 0.16, P = 0.23; postcentral: h2SNP = 0.06, P = 0.39; precuneus: h2SNP = 0.05, P = 0.40; superior parietal: h2SNP = 0.15, P = 0.25; supramarginal: h2SNP = 0.07, P = 0.37; fusiform: h2SNP = 0.19, P = 0.19; inferior temporal: h2SNP = 0.12, P = 0.29; middle temporal: h2SNP = 0.00, P = 0.50; parahippocampal: h2SNP = 0.33, P = 0.07; superior temporal: h2SNP = 0.15, P = 0.24; insula: h2SNP = 0.27, P = 0.10).
Table 4.
SNP heritability of regional cortical thickness in the whole sample.
| Cortical thickness | Vp | Vg | Heritability | |||
|---|---|---|---|---|---|---|
| h 2 SNP | SE | P-value | ||||
| Cingulate | ||||||
| Caudal middle frontal | 0.09 | 0.02 | 0.20 | 0.21 | 0.17 | |
| Isthmus cingulate | 0.13 | 0.02 | 0.14 | 0.21 | 0.26 | |
| Posterior cingulate | 0.10 | 0.01 | 0.10 | 0.21 | 0.31 | |
| Rostral anterior cingulate | 0.14 | 0.04 | 0.30 | 0.21 | 0.08 | |
| Caudal anterior cingulate | 0.07 | 0.01 | 0.19 | 0.21 | 0.19 | |
| Frontal | ||||||
| Lateral orbitofrontal | 0.10 | 0.02 | 0.16 | 0.21 | 0.21 | |
| Medial orbitofrontal | 0.09 | 0.01 | 0.08 | 0.21 | 0.35 | |
| Paracentral | 0.22 | 0.02 | 0.08 | 0.21 | 0.35 | |
| Pars opercularis | 0.08 | 0.03 | 0.35 | 0.21 | 0.04a | |
| Pars orbitalis | 0.11 | 0.02 | 0.15 | 0.21 | 0.23 | |
| Pars triangularis | 0.08 | 0.02 | 0.23 | 0.21 | 0.14 | |
| Precentral | 0.16 | 0.03 | 0.20 | 0.21 | 0.17 | |
| Rostral middle frontal | 0.09 | 0.03 | 0.34 | 0.22 | 0.06 | |
| Superior frontal | 0.08 | 0.04 | 0.45 | 0.21 | 0.02a | |
| Occipital | ||||||
| Cuneus | 0.09 | 0.02 | 0.26 | 0.21 | 0.10 | |
| Lateral occipital | 0.13 | 0.03 | 0.25 | 0.22 | 0.13 | |
| Lingual | 0.06 | 0.02 | 0.29 | 0.22 | 0.09 | |
| Pericalcarine | 0.07 | 0.00 | 0.00 | 0.21 | 0.50 | |
| Parietal | ||||||
| Inferior parietal | 0.13 | 0.02 | 0.16 | 0.21 | 0.23 | |
| Postcentral | 0.09 | 0.01 | 0.06 | 0.21 | 0.39 | |
| Precuneus | 0.10 | 0.01 | 0.05 | 0.21 | 0.40 | |
| Superior parietal | 0.11 | 0.02 | 0.15 | 0.21 | 0.25 | |
| Supramarginal | 0.10 | 0.01 | 0.07 | 0.21 | 0.37 | |
| Temporal | ||||||
| Entorhinal | 0.52 | 0.22 | 0.42 | 0.22 | 0.03a | |
| Fusiform | 0.16 | 0.03 | 0.19 | 0.21 | 0.19 | |
| Inferior temporal | 0.16 | 0.02 | 0.12 | 0.21 | 0.29 | |
| Middle temporal | 0.16 | 0.00 | 0.00 | 0.21 | 0.50 | |
| Parahippocampal | 0.34 | 0.11 | 0.33 | 0.22 | 0.07 | |
| Superior temporal | 0.13 | 0.02 | 0.15 | 0.22 | 0.24 | |
| Transverse temporal | 0.22 | 0.08 | 0.37 | 0.21 | 0.04a | |
| Insula | ||||||
| Insula | 0.10 | 0.03 | 0.27 | 0.21 | 0.10 | |
Note: All analyses are adjusted for age, sex, and 10PCs. Vp, total phenotypic variance; Vg, genetic variance; h2SNP, single-nucleotide polymorphism (SNP)-based heritability estimates; SE, standard error.
aIndicates significant heritability at P < 0.05.
Genetic correlations
Table 5 presents the genetic and environmental correlations between the traits showing significant heritability in the univariate SNP heritability analysis. Significantly positive genetic correlations were found between caudate volume and entorhinal cortices thickness (rG = 0.571, P = 0.03), between pars opercularis thickness and superior frontal gyri thickness (rG = 0.823, P = 0.03), and between superior frontal gyri thickness and transverse temporal gyri thickness (rG = 0.887, P = 0.01).
Table 5.
Genetic correlations between variables showing significant SNP heritability in univariate GCTA heritability analysis.
| Traits | Cognitive function | Subcortical volume | Cortical thickness | |||
|---|---|---|---|---|---|---|
| Memory | Caudate | Pars opercularis | Superior frontal | Entorhinal | Transverse temporal | |
| Memory | ||||||
| Caudate | 0.092 | |||||
| Pars opercularis | 0.167 | - | ||||
| Superior frontal | 0.223 | - | 0.823a | |||
| Entorhinal | 0.208 | 0.571a | −0.105 | 0.298 | ||
| Transverse temporal | −0.236 | - | 0.721 | 0.887a | 0.286 | |
Note: All analyses are adjusted for age, sex, and 10PCs. SNP, single-nucleotide polymorphism; GCTA, genome-wide complex trait analysis.
aIndicates significant heritability at P < 0.05.
Discussion
In the present study, we evaluated the SNP heritability and genetic correlations of cognitive abilities and brain structural measures in middle-aged to older adults from South Korea. We found that memory function, caudate volume, and cortical thickness of the entorhinal cortices, pars opercularis, superior frontal gyri, and transverse temporal gyri are heritable. We observed 3 significant genetic correlations between (i) the caudate volume and the thickness of the entorhinal cortices; (ii) the thickness of the superior frontal gyri and pars opercularis; and (iii) the thickness of the superior frontal and transverse temporal gyri. This result confirmed previous findings regarding the heritability of cognitive abilities and regional brain structures, supporting the notion that genetic factors play a great role in cognitive and neuroanatomical traits in middle to advanced age. Furthermore, by demonstrating shared genetic effects on different brain regions, it gives us a genetic insight into understanding cognitive and brain changes with age such as aging-related cognitive decline, cortical atrophy, and neural compensation.
In our heritability analysis, memory function was found to be moderately heritable (46%) among various cognitive functions. This result is in agreement with prior research findings showing that memory function exhibits moderate-to-high heritability (twin-based heritability: 30%–62%) until old age (Johansson et al. 1999;Wilson et al. 2011; Reynolds and Finkel 2015) and that the heritability of memory function increases with age, whereas other cognitive traits, such as attention, language, and visuospatial function, show a declining pattern of heritability with age (Wilson et al. 2011; Reynolds and Finkel 2015). In previous genome-wide association studies (GWAS), several variants of genes such as APOE, BDNF, WWC1, NECTIN2, TOMM40, and H2ACP1 were found to be associated with memory function and modulation of age-related cognitive decline (Kauppi et al. 2011; Persson et al. 2013; Lamb et al. 2015; Li et al. 2017). Among them, genes, including NECTIN2, TOMM40, and H2ACP1, are located on chromosome 13 or 19 (Arpawong et al. 2017; de la Fuente et al. 2021) where significant chromosome-specific heritability of memory function was found in our study, and the effects of several genes, such as APOE, BDNF, and WWC1, on memory function have been reported to increase with age (Li et al. 2010; Kauppi et al. 2011; Persson et al. 2013; Papenberg et al. 2015). Our result confirms the previous finding that memory function is substantially influenced by multiple genes. This may indicate that the high level of heritability of memory function in middle-aged to older adults may result from the expression of multiple genes affecting memory function or magnifying effects of memory-related genes as age increases.
In the heritability analysis of regional subcortical volumes, we observed moderate heritability (42%) of the caudate volume, which is in line with existing findings that the caudate volume is one of the most highly heritable features among the subcortical structures of healthy adults. According to previous SNP heritability studies, a moderate proportion (34%–57%) of the variance in caudate volume is estimated to be accounted for by the genetic factors in young-to-aged adults (Roshchupkin et al. 2016; Elliott et al. 2018; Satizabal et al. 2019; Biton et al. 2020). Moreover, the caudate volume has shown high twin-based heritability (71%–85%) among subcortical regions in middle-aged to older adults (Kremen et al. 2010; Wen et al. 2016; Satizabal et al. 2019). In previous GWAS, several genes, such as SLC6A3, DRD2, LOC107987256, WDR4, and ZNF521, have been identified as being associated with caudate volume (Durston et al. 2005; Stein et al. 2011; Satizabal et al. 2019; Smith et al. 2021; Brouwer et al. 2022). In particular, the LOC107987256 and ZNF521 genes are located on chromosome 18, where significant chromosome-specific heritability of caudate volume was observed in the present study (Smith et al. 2021). Recently, the adverse genetic effect of DRD2 on caudate volume was found to exist only in older adults but not in young adults (Li et al. 2018), showing an age-specific genetic effect on caudate volume. Based on these previous findings and our results, it can be inferred that the high heritability of caudate volume in middle-aged to older adults may be attributed to the genetic factors associated with caudate volume, such as an increase in the expression of genes involved in caudate volume.
Heritability analysis of the regional cortical thickness revealed significant heritability in the entorhinal cortices (42%), pars opercularis (35%), superior frontal gyri (45%), and transverse temporal gyri (37%), which are mainly located in the frontal and temporal areas. This result is consistent with the previous findings from heritability studies and GWAS. Numerous heritability studies have shown that the brain structures of the frontal and temporal cortices are heritable in adults (twin-based heritability: frontal = 49%–83% and temporal = 40%–70%) and that, compared to other cortical regions, the heritability of these regions is maintained at a relatively high level until old age (twin-based heritability: frontal = 66%–86% and temporal = 55%–92%) (Batouli et al. 2014; Eyler et al. 2014; Wen et al. 2016; Lukies et al. 2017). Also, according to previous GWAS, various genes have been identified to be associated with the neurostructural traits of frontal and temporal cortices, and several genes are located on the chromosomes where significant chromosome-specific heritability of regional cortical thickness was also found in our current study (Zhao et al. 2019; Grasby et al. 2020; Smith et al. 2021). This includes (i) MIR4460 and ADAMTS19-AS1 on chromosome 5 for the pars opercularis; (ii) CCT7P2 and LDHBP3 on chromosome 5 for the superior frontal gyri; (iii) CCBE1 on chromosome 18 for the entorhinal cortices; and (iv) LYPD6B on chromosome 2 as well as MIR4643 and MAP3K7 on chromosome 6 for the transverse temporal gyri. Earlier research has reported that the neurodevelopmental and aging-related changes in cortical structures follow the genetic organization of the cortex, and the genetic influences on structural changes in the frontal and temporal regions are relatively substantial in later life. Moreover, the frontal and temporal areas are known to follow a similar pattern of structural changes with age; these are the regions which are the most affected by aging in elderly people as well as late-maturing brain regions, whose maturation continues even until mid-to-late adulthood (Long et al. 2012; Tamnes et al. 2013; Fjell et al. 2015; Sele et al. 2021). Taken together, our result may indicate that the cortical thickness of the frontal and temporal areas in middle-aged to elderly people is influenced by genetic factors or by an increase in the expression of genes involved in the age-related structural changes of the frontal and temporal areas.
The results of some previous studies are contradictory to the findings of our heritability analysis. With regard to cognitive function, apart from memory function, other cognitive abilities, such as executive function, attention, and language ability, were found to have significant heritability (Davies et al. 2015; Mollon et al. 2021). With regard to the regional subcortical volumes, some subcortical regions that did not have significant heritability according to our results, including the nucleus accumbens, amygdala, hippocampus, pallidum, putamen, and thalamus, were shown to have significant heritability in other studies (Eyler et al. 2014; Wen et al. 2016). With regard to the regional cortical thickness, regions that are not included in the frontal and temporal areas have been shown to have significant heritability (Grasby et al. 2020; Hofer et al. 2020). The inconsistency between the previous reports and our results could be explained by participant characteristics, such as the small sample size, inclusions of only East Asians, and inclusion of elderly people with or without cognitive impairment. First, our sample size was smaller compared to previous studies. This might have led to false negatives where we were not able to detect true effects in our study. A future study with a larger sample may provide more accurate results and help to identify whether the discrepancy between the previous and present results is due to different sample sizes. Second, unlike most previous studies, our study only included participants of East Asian descent. Most previous research has been conducted in European-derived populations, and since the findings have the most relevance for those populations, they might not apply equally to non-European populations. Therefore, the difference in ethnicity could lead to inconsistency with previous findings from other populations, and our findings may provide an understanding of genetic influences in the East Asian populations which are distinguishable from other populations. In future studies, comparative analyses across different populations can help increase the understanding of genetic similarities or differences by ethnicity. Last, in our study, the average age was relatively high, and individuals with cognitive impairment, such as patients with AD, were included. Previous studies and the resource-modulation model indicate that the heritability is likely to diminish once individuals reach very low resource levels; for example, in individuals with dementia or terminal decline, for whom genes may account less for the individual differences in cognitive function and brain traits (Wilson et al. 2011; Papenberg et al. 2015). Consistent with this notion, one study reported that APOE does not affect the progression rate of cognitive decline in clinical AD (MacDonald et al. 2011) or even the rate of decline from preclinical to clinical dementia (Bunce et al. 2004). A replication study without cognitively impaired individuals may help elucidate the reasons for inconsistencies in the findings. However, in contrast to the traits with significant heritability, most of the cognitive and brain structural traits that did not show significant heritability in our study have been commonly reported to have a decreasing heritability pattern with age during late adulthood and a low level of heritability in later life (Giedd et al. 2007; Batouli et al. 2014; Jansen et al. 2015; Reynolds and Finkel 2015; Lemvigh, Brouwer, Sahakian, et al. 2020). As suggested by these previous studies, the traits that did not show significant heritability in our study may be the ones that are likely to be more influenced by the environmental effects with age. This would explain the relative decrease in the genetic contribution to the traits and low heritability in late adulthood.
We found a significant genetic correlation between the caudate volume and entorhinal cortical thickness. Both the caudate and entorhinal cortex have been involved in memory function, and they play a role in different memory systems. The caudate is a central structure in the basal ganglia memory system that contributes to habit learning, which refers to the gradual learning of stimulus–response associations over many trials (Knowlton et al. 1996; Jog et al. 1999; Yin and Knowlton 2006). On the other hand, the entorhinal cortex is a pivotal region in the medial temporal lobe (MTL) memory system that supports explicit memories of events or episodes (O'Keefe and Nadel 1978; Eichenbaum 2004). These two memory systems have been known to interact in dynamic ways and compensate for one another when one system starts failing because of neurological dysfunction or brain aging. According to functional neuroimaging studies, during a memory task, an increased activity of the MTL has been observed in individuals who have functional degradation of the caudate (Voermans et al. 2004; Rieckmann and Bäckman 2009; Rieckmann et al. 2010). Furthermore, in recent structural neuroimaging studies, an increase in caudate volume was observed in patients with AD and elderly adults, who had medial temporal cortical atrophy due to AD neuropathology or aging (Persson et al. 2018; Sodums and Bohbot 2020). These previous studies have suggested that increased activity and neuroanatomical structure of the MTL or entorhinal cortex reflect compensation to maintain memory function despite structural or functional impairment of other regions. Taken together, these findings, along with our results, may imply that the memory function maintained by the compensatory interaction between the caudate and entorhinal cortex in individuals who experience aging, or possibly have age-related neurodegeneration, are affected by the genetic factors involved in the neuroanatomical structures of the caudate and entorhinal cortex. In the future, GWAS that investigate the genetic variants simultaneously associated with the regions that cooperatively facilitate memory function, such as the caudate and entorhinal cortex, may provide a better understanding of the mechanism underlying the memory impairment caused by aging or neurodegeneration.
According to our genetic correlation analysis, the cortical thickness of the superior frontal gyri is genetically correlated with the cortical thickness of the pars opercularis and transverse temporal gyri. In agreement with our findings, in various functional and structural neuroimaging studies, it has been reported that the superior frontal area works in conjunction with the pars opercularis and transverse temporal gyri in the cognitive processing that those 2 areas involved in, such as language and speech ability (Nachev et al. 2008; Tremblay and Gracco 2009; Hertrich et al. 2016; Tremblay and Dick 2016). The pars opercularis has been identified as a crucial region for language production (Tremblay and Dick 2016), and the medial superior frontal regions, including the supplementary motor area (SMA) and pre-SMA, support the language production by contributing to the initiation/execution of speech output and higher language processes, such as context-tracking and monitoring of language representations (Nachev et al. 2008; Tremblay and Gracco 2009; Hertrich et al. 2016). The transverse temporal gyrus plays an important role in speech perception (Qian et al. 2017; Eckert et al. 2019; Armstrong et al. 2020), and the prefrontal area, including the superior frontal gyri, has been associated with the process of speech perception such as blocking noise and inhibiting lexical competitors (Wong et al. 2010; Husain et al. 2011). Furthermore, prior neuroimaging research has revealed that people with impairment in the pars opercularis or transverse temporal area caused by aging or neurological lesions show a higher activity or increased structural volume/thickness in the frontal area, including the superior frontal gyri (Wong et al. 2010; Du et al. 2016; Rosemann and Thiel 2020). This suggests that an increase in the activity and structure of the superior frontal region works as a compensatory strategy to maintain the language and speech abilities under challenging circumstances. Accordingly, our findings may indicate that in middle-aged to older adults, age-related decline in language ability, such as word-finding difficulty and problems with speech comprehension or retention of speech ability accompanied by compensation of the superior frontal region, is influenced by shared genetic factors influencing the superior frontal gyri with the pars opercularis and transverse temporal gyri. Additionally, further studies that identify which specific genetic factor or variant is associated with the cortical thickness of those areas may deepen our understanding of the underlying etiology of age-related language problems and may help to develop a preventive treatment for language impairment.
The overall findings of our genetic correlation analyses are in line with the notion that the effects of genetic variance on regional differences in the neuroanatomical structures may partly correspond to functional specialization. Several studies have argued for the genetic covariance of importance for functional specialization by demonstrating the genetic correlations between functionally related regions (Chen et al. 2013; Fjell et al. 2015). Moreover, an interesting observation in our study was that the brain regions showing significant genetic correlations are the areas where the compensatory and cooperative interactions appear to maintain the cognitive ability against the infirmity of the other regions; furthermore, the associated cognitive abilities with the brain areas showing the genetic correlations are known to be cognitive traits that are vulnerable to aging. Together, our results may imply that, in individuals experiencing aging processes, the compensatory interaction or changes within the brain may be driven by the genetic factors involved in neuroanatomical structures. In line with this idea, a recent study suggested that the APOE gene is a moderator of neuronal compensation and that the APOE ε4 carriers recruited the prefrontal area to maintain the memory function when age-related brain changes substantially progressed in older adults (Scheller et al. 2018).
We would like to acknowledge some limitations of our findings. The major problem for our study is a relatively small sample size for a GCTA heritability analysis. Because of this, it was not easy to get sufficient statistical power with a correction for multiple testing. However, the cognitive traits and brain structural traits used in our study were highly correlated and were not independent, thus, a correction for the number of analyses performed could result in an overly conservative significance criterion and multiple testing problems are less serious for our results. Another limitation of our study is that the proportion of individuals with MCI or AD in our sample was relatively higher than that of the general middle-aged to elderly population. We considered samples ascertained from the AD status and it generated AB with a high proportion of MCI or AD. According to previous studies, genetic effects tend to diminish once individuals reach very low brain-resource levels, as observed in individuals with severe cognitive impairment such as AD (Wilson et al. 2011; Papenberg et al. 2015). Therefore, including a relatively high number of individuals with severe cognitive impairment because of the AB might dilute the genetic effects and underestimate the actual heritability. Although the SNP heritability analyses were not performed without individuals with cognitive impairment because the SNP heritability estimates based on GCTA are affected by the sample size, when correcting AB with AD and MCI prevalence, the heritability estimates did not largely change, and they were in the previously reported heritability range. However, in order to yield more stable and accurate results, heritability analyses need to be performed with larger samples that are similar to the real-world proportion of cognitive status in future studies. One final limitation of our findings is that GCTA can detect only the additive effects of common genetic variants and does not capture the effect of rare variants or nonadditive effects, such as gene–gene interaction and gene–environment interaction, whereas the twin-based method captures both additive and nonadditive effects. Therefore, our SNP heritability estimates might be lower than the estimates from twin-based studies, and our estimates may only provide a lower limit of the actual heritability. Thus, we should be cautious about assuming true heritability estimates from our findings. Furthermore, this missing heritability gap between GCTA and twin studies needs to be filled by future studies that compare GCTA and twin study estimates of heritability or explore the effects of rare genetic variants on cognitive function and brain regional structures.
Conclusion
In conclusion, this is the first study to describe the heritability and genetic correlations of cognitive and neuroanatomical traits in middle-aged to elderly East Asians. Overall, the results of our heritability analyses are in accordance with previous findings on the heritability of cognitive function, regional subcortical volume, and cortical thickness in different ethnic groups. The genetic correlation results indicate that in middle-aged to older adults, the brain area showing increased genetic effects on neuroanatomical structures corresponds to the region where brain compensatory work is more likely to appear with increasing age, and the shared genetic effects on brain structures exist within the areas where compensatory interaction takes place. Accordingly, the results may imply that the mechanism underlying age-related brain compensation and cognitive ability maintained by this compensation may be accounted for, partly, by the genetic factors that influence variations in the structure of the brain areas associated with cognitive abilities vulnerable to aging. Our study provides an understanding of the genetic variation in cognitive ability and neuroanatomical structures in middle-aged to older adults and provides an insight into the genetic basis of age-related cognitive decline and neurodegenerative diseases.
Supplementary Material
Acknowledgements
This study was supported by the Korean National Supercomputing Center with supercomputing resources including technical support (KSC-2022-CRE-0319).
Contributor Information
Younghwa Lee, Department of Public Health Sciences, Graduate School of Public Health, Seoul National University, Seoul, Korea.
Jun Young Park, Department of Public Health Sciences, Graduate School of Public Health, Seoul National University, Seoul, Korea.
Jang Jae Lee, Gwangju Alzheimer’s Disease & Related Dementia Cohort Research Center, Chosun University, Gwangju, Korea.
Jungsoo Gim, Gwangju Alzheimer’s Disease & Related Dementia Cohort Research Center, Chosun University, Gwangju, Korea; Department of Biomedical Science, Chosun University, Gwangju, Korea.
Ah Ra Do, Interdisciplinary Program in Bioinformatics, Seoul National University, Seoul, Korea.
Jinyeon Jo, Department of Public Health Sciences, Graduate School of Public Health, Seoul National University, Seoul, Korea.
Juhong Park, Department of Public Health Sciences, Graduate School of Public Health, Seoul National University, Seoul, Korea.
Kangjin Kim, Department of Public Health Sciences, Graduate School of Public Health, Seoul National University, Seoul, Korea.
Kyungtaek Park, Institute of Health and Environment, Seoul National University, Seoul, South Korea.
Heejin Jin, Institute of Health and Environment, Seoul National University, Seoul, South Korea.
Kyu Yeong Choi, Gwangju Alzheimer’s Disease & Related Dementia Cohort Research Center, Chosun University, Gwangju, Korea.
Sarang Kang, Gwangju Alzheimer’s Disease & Related Dementia Cohort Research Center, Chosun University, Gwangju, Korea.
Hoowon Kim, Gwangju Alzheimer’s Disease & Related Dementia Cohort Research Center, Chosun University, Gwangju, Korea; Department of Neurology, Chosun University Hospital, Gwangju, Korea.
SangYun Kim, Department of Neurology, Seoul National University College of Medicine, Seoul, Korea; Department of Neurology, Seoul National University Bundang Hospital, Seongnam, Korea.
Seung Hwan Moon, Department of Nuclear Medicine, Samsung Medical Center, Seoul, Korea.
Lindsay A Farrer, Department of Medicine, Boston University School of Medicine, Boston, MA, United States.
Kun Ho Lee, Gwangju Alzheimer’s Disease & Related Dementia Cohort Research Center, Chosun University, Gwangju, Korea; Department of Biomedical Science, Chosun University, Gwangju, Korea; Dementia Research Group, Korea Brain Research Institute, Daegu, Korea.
Sungho Won, Department of Public Health Sciences, Graduate School of Public Health, Seoul National University, Seoul, Korea; Interdisciplinary Program in Bioinformatics, Seoul National University, Seoul, Korea; RexSoft Inc., Seoul, Korea.
Funding
This work was supported by the Healthcare AI Convergence Research & Development Program through the National IT Industry Promotion Agency of Korea (NIPA) funded by the Ministry of Science and ICT (No. 1711120216), the National Research Foundation (NRF) grant funded by the Korean government (MSIT) (NRF-2021R1A5A1033157), the Basic Science Research Program through the NRF of Korea funded by the Ministry of Education (NRF-2020R1F1A01072033), the Korea Brain Research Institute (KBRI) basic research program through the KBRI funded by the Ministry of Science and ICT (22-BR-03-05), and the National Institute on Aging grants U01-AG062602, R01-AG048927, and U01-AG032984.
Conflict of interest statement: None declared.
Data availability
Due to confidentiality agreements, we are unable to provide a publicly available database. Researchers who wish to access the GARD cohort dataset used in this study can obtain it by contacting the corresponding author, professor Kunho Lee (leekho@chosun.ac.kr).
References
- Ahn HJ, Chin J, Park A, Lee BH, Suh MK, Seo SW, Na DL. Seoul Neuropsychological Screening Battery-dementia version (SNSB-D): a useful tool for assessing and monitoring cognitive impairments in dementia patients. J Korean Med Sci. 2010:25:1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong NM, An Y, Ferrucci L, Deal JA, Lin FR, Resnick SM. Temporal sequence of hearing impairment and cognition in the Baltimore Longitudinal Study of Aging. J Gerontol A Biol Sci Med Sci. 2020:75(3):574–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpawong TE, Pendleton N, Mekli K, McArdle JJ, Gatz M, Armoskus C, Knowles JA, Prescott CA. Genetic variants specific to aging-related verbal memory: insights from GWASs in a population-based cohort. PLoS One. 2017:12(8):e0182448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batouli SA, Sachdev PS, Wen W, Wright MJ, Ames D, Trollor JN. Heritability of brain volumes in older adults: the Older Australian Twins Study. Neurobiol Aging. 2014:35(4):937.e5–937.e18. [DOI] [PubMed] [Google Scholar]
- Biton A, Traut N, Poline JB, Aribisala BS, Bastin ME, Bülow R, Cox SR, Deary IJ, Fukunaga M, Grabe HJ, et al. Polygenic architecture of human neuroanatomical diversity. Cereb Cortex. 2020:30(4):2307–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer RM, Hedman AM, van Haren NE, Schnack HG, Brans RG, Smit DJ, Kahn RS, Boomsma DI, Hulshoff Pol HE. Heritability of brain volume change and its relation to intelligence. NeuroImage. 2014:100:676–683. [DOI] [PubMed] [Google Scholar]
- Brouwer RM, Klein M, Grasby KL, Schnack HG, Jahanshad N, Teeuw J, Thomopoulos SI, Sprooten E, Franz CE, Gogtay N, et al. Genetic variants associated with longitudinal changes in brain structure across the lifespan. Nat Neurosci. 2022:25(4):421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunce D, Fratiglioni L, Small BJ, Winblad B, Bäckman L. APOE and cognitive decline in preclinical Alzheimer disease and non-demented aging. Neurology. 2004:63(5):816–821. [DOI] [PubMed] [Google Scholar]
- Chen CH, Fiecas M, Gutiérrez ED, Panizzon MS, Eyler LT, Vuoksimaa E, Thompson WK, Fennema-Notestine C, Hagler DJ Jr, Jernigan TL, et al. Genetic topography of brain morphology. Proc Natl Acad Sci U S A. 2013:110(42):17089–17094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins LM, Williams-Gray CH. The genetic basis of cognitive impairment and dementia in Parkinson's disease. Front Psych. 2016:7:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G, Tenesa A, Payton A, Yang J, Harris SE, Liewald D, Ke X, Le Hellard S, Christoforou A, Luciano M, et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol Psychiatry. 2011:16:996–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G, Armstrong N, Bis JC, Bressler J, Chouraki V, Giddaluru S, Hofer E, Ibrahim-Verbaas CA, Kirin M, Lahti J, et al. Genetic contributions to variation in general cognitive function: a meta-analysis of genome-wide association studies in the CHARGE consortium (N = 53949). Mol Psychiatry. 2015:20:183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente J, Davies G, Grotzinger AD, Tucker-Drob EM, Deary IJ. A general dimension of genetic sharing across diverse cognitive traits inferred from molecular data. Nat Hum Behav. 2021:5:49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Marchini J, The 1000 Genomes Project C . Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat Commun. 2014:5:3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Buchsbaum BR, Grady CL, Alain C. Increased activity in frontal motor cortex compensates impaired speech perception in older adults. Nat Commun. 2016:7:12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durston S, Fossella JA, Casey BJ, Hulshoff Pol HE, Galvan A, Schnack HG, Steenhuis MP, Minderaa RB, Buitelaar JK, Kahn RS, et al. Differential effects of DRD4 and DAT1 genotype on fronto-striatal gray matter volumes in a sample of subjects with attention deficit hyperactivity disorder, their unaffected siblings, and controls. Mol Psychiatry. 2005:10:678–685. [DOI] [PubMed] [Google Scholar]
- Eckert MA, Vaden KI Jr, Dubno JR. Age-related hearing loss associations with changes in brain morphology. Trends Hear. 2019:23:2331216519857267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenbaum H. Hippocampus: cognitive processes and neural representations that underlie declarative memory. Neuron. 2004:44:109–120. [DOI] [PubMed] [Google Scholar]
- Elliott LT, Sharp K, Alfaro-Almagro F, Shi S, Miller KL, Douaud G, Marchini J, Smith SM. Genome-wide association studies of brain imaging phenotypes in UK Biobank. Nature. 2018:562:210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyler LT, Vuoksimaa E, Panizzon MS, Fennema-Notestine C, Neale MC, Chen CH, Jak A, Franz CE, Lyons MJ, Thompson WK, et al. Conceptual and data-based investigation of genetic influences and brain asymmetry: a twin study of multiple structural phenotypes. J Cogn Neurosci. 2014:26:1100–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Tao W, Li X, Li H, Zhang J, Wei D, Chen Y, Zhang Z. The contribution of genetic factors to cognitive impairment and dementia: Apolipoprotein E gene, gene interactions, and polygenic risk. Int J Mol Sci. 2019:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B. Free Surfer. NeuroImage. 2012:62:774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell AM, Grydeland H, Krogsrud SK, Amlien I, Rohani DA, Ferschmann L, Storsve AB, Tamnes CK, Sala-Llonch R, Due-Tønnessen P, et al. Development and aging of cortical thickness correspond to genetic organization patterns. Proc Natl Acad Sci U S A. 2015:112:15462–15467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge T, Reuter M, Winkler AM, Holmes AJ, Lee PH, Tirrell LS, Roffman JL, Buckner RL, Smoller JW, Sabuncu MR. Multidimensional heritability analysis of neuroanatomical shape. Nat Commun. 2016:7:13291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedd JN, Schmitt JE, Neale MC. Structural brain magnetic resonance imaging of pediatric twins. Hum Brain Mapp. 2007:28:474–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh S, Bansal R, Xu D, Hao X, Liu J, Peterson BS. Neuroanatomical correlates of intellectual ability across the life span. Dev Cogn Neurosci. 2011:1:305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady C. The cognitive neuroscience of ageing. Nat Rev Neurosci. 2012:13:491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasby KL, Jahanshad N, Painter JN, Colodro-Conde L, Bralten J, Hibar DP, Lind PA, Pizzagalli F, Ching CRK, McMahon MAB, et al. The genetic architecture of the human cerebral cortex. Science. 2020:367:eaay6690.32193296 [Google Scholar]
- Greenwood TA, Beeri MS, Schmeidler J, Valerio D, Raventós H, Mora-Villalobos L, Camacho K, Carrión-Baralt JR, Angelo G, Almasy L, et al. Heritability of cognitive functions in families of successful cognitive aging probands from the Central Valley of Costa Rica. J Alzheimers Dis. 2011:27:897–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haworth CM, Wright MJ, Luciano M, Martin NG, de Geus EJ, van Beijsterveldt CE, Bartels M, Posthuma D, Boomsma DI, Davis OS, et al. The heritability of general cognitive ability increases linearly from childhood to young adulthood. Mol Psychiatry. 2010:15:1112–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertrich I, Dietrich S, Ackermann H. The role of the supplementary motor area for speech and language processing. Neurosci Biobehav Rev. 2016:68:602–610. [DOI] [PubMed] [Google Scholar]
- Hibar DP, Stein JL, Renteria ME, Arias-Vasquez A, Desrivières S, Jahanshad N, Toro R, Wittfeld K, Abramovic L, Andersson M, et al. Common genetic variants influence human subcortical brain structures. Nature. 2015:520:224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer E, Roshchupkin GV, Adams HHH, Knol MJ, Lin H, Li S, Zare H, Ahmad S, Armstrong NJ, Satizabal CL, et al. Genetic correlations and genome-wide associations of cortical structure in general population samples of 22,824 adults. Nat Commun. 2020:11:4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009:5:e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain FT, Pajor NM, Smith JF, Kim HJ, Rudy S, Zalewski C, Brewer C, Horwitz B. Discrimination task reveals differences in neural bases of tinnitus and hearing impairment. PLoS One. 2011:6:e26639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen AG, Mous SE, White T, Posthuma D, Polderman TJ. What twin studies tell us about the heritability of brain development, morphology, and function: a review. Neuropsychol Rev. 2015:25:27–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jog MS, Kubota Y, Connolly CI, Hillegaart V, Graybiel AM. Building neural representations of habits. Science. 1999:286:1745–1749. [DOI] [PubMed] [Google Scholar]
- Johansson B, Whitfield K, Pedersen NL, Hofer SM, Ahern F, McClearn GE. Origins of individual differences in episodic memory in the oldest-old: a population-based study of identical and same-sex fraternal twins aged 80 and older. J Gerontol B Psychol Sci Soc Sci. 1999:54:P173–P179. [DOI] [PubMed] [Google Scholar]
- Joy S, Kaplan E, Fein D. Speed and memory in the WAIS-III Digit Symbol—Coding subtest across the adult lifespan. Arch Clin Neuropsychol. 2004:19:759–767. [DOI] [PubMed] [Google Scholar]
- Kang Y, Kim H, Na D. A short form of the Korean-Boston Naming Test (K-BNT) for using in dementia patients. Korean J Clin Psychol. 1999:18:125–138. [Google Scholar]
- Kang Y, Chin J, Na D. A normative study of the digit span test for the elderly. Korean J Clin Psychol. 2002:21:911–922. [Google Scholar]
- Kang SH, Park YH, Lee D, Kim JP, Chin J, Ahn Y, Park SB, Kim HJ, Jang H, Jung YH. The cortical neuroanatomy related to specific neuropsychological deficits in Alzheimer's continuum. Dement Neurocogn Disord. 2019:18:77–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi K, Nilsson LG, Adolfsson R, Eriksson E, Nyberg L. KIBRA polymorphism is related to enhanced memory and elevated hippocampal processing. J Neurosci. 2011:31:14218–14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A, Tourville J. 101 labeled brain images and a consistent human cortical labeling protocol. Front Neurosci. 2012:6:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton BJ, Mangels JA, Squire LR. A neostriatal habit learning system in humans. Science. 1996:273:1399–1402. [DOI] [PubMed] [Google Scholar]
- Korea MoHaWo . Dementia observatory by City, County and District [Internet]. Sejong-si, Korea: Ministry of Interior and Safety. Available from: https://www.data.go.kr/data/15073342/fileData.do.
- Kraljević N, Schaare HL, Eickhoff SB, Kochunov P, Yeo BTT, Kharabian Masouleh S, Valk SL. Behavioral, anatomical and heritable convergence of affect and cognition in superior frontal cortex. NeuroImage. 2021:243:118561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremen WS, Prom-Wormley E, Panizzon MS, Eyler LT, Fischl B, Neale MC, Franz CE, Lyons MJ, Pacheco J, Perry ME, et al. Genetic and environmental influences on the size of specific brain regions in midlife: the VETSA MRI study. NeuroImage. 2010:49:1213–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb YN, Thompson CS, McKay NS, Waldie KE, Kirk IJ. The brain-derived neurotrophic factor (BDNF) val66met polymorphism differentially affects performance on subscales of the Wechsler Memory Scale–Third Edition (WMS-III). Front Psychol. 2015:6:1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kang Y, Na D. Efficiencies of stroop interference indexes in healthy older adults and dementia patients. Korean J Clin Psychol. 2000:19:807–818. [Google Scholar]
- Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011:88:294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemvigh CK, Brouwer RM, Pantelis C, Jensen MH, Hilker RW, Legind CS, Anhøj SJ, Robbins TW, Sahakian BJ, Glenthøj BY, et al. Heritability of specific cognitive functions and associations with schizophrenia spectrum disorders using CANTAB: a nation-wide twin study. Psychol Med. 2020:52:1101–1114. [DOI] [PubMed] [Google Scholar]
- Lemvigh CK, Brouwer RM, Sahakian BJ, Robbins TW, Johansen LB, Legind CS, Anhøj SJ, Hilker R, Hulshoff Pol HE, Ebdrup BH, et al. Heritability of memory functions and related brain volumes: a schizophrenia spectrum study of 214 twins. Schizophrenia Bulletin Open. 2020:1:sgaa066. [Google Scholar]
- Lessov-Schlaggar CN, Hardin J, DeCarli C, Krasnow RE, Reed T, Wolf PA, Swan GE, Carmelli D. Longitudinal genetic analysis of brain volumes in normal elderly male twins. Neurobiol Aging. 2012:33:636–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SC, Chicherio C, Nyberg L, von Oertzen T, Nagel IE, Papenberg G, Sander T, Heekeren HR, Lindenberger U, Bäckman L. Ebbinghaus revisited: influences of the BDNF Val66Met polymorphism on backward serial recall are modulated by human aging. J Cogn Neurosci. 2010:22:2164–2173. [DOI] [PubMed] [Google Scholar]
- Li H, Lv C, Yang C, Wei D, Chen K, Li S, Zhang Z. SORL1 rs1699102 polymorphism modulates age-related cognitive decline and gray matter volume reduction in non-demented individuals. Eur J Neurol. 2017:24:187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Papenberg G, Kalpouzos G, Bäckman L, Persson J. Influence of the DRD2/ANKK1 Taq1A polymorphism on caudate volume in older adults without dementia. Brain Struct Funct. 2018:223:2653–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X, Liao W, Jiang C, Liang D, Qiu B, Zhang L. Healthy aging: an automatic analysis of global and regional morphological alterations of human brain. Acad Radiol. 2012:19:785–793. [DOI] [PubMed] [Google Scholar]
- Lukies MW, Watanabe Y, Tanaka H, Takahashi H, Ogata S, Omura K, Yorifuji S, Tomiyama N, Osaka University Twin Research G . Heritability of brain volume on MRI in middle to advanced age: a twin study of Japanese adults. PLoS One. 2017:12:e0175800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald SW, Karlsson S, Fratiglioni L, Bäckman L. Trajectories of cognitive decline following dementia onset: what accounts for variation in progression? Dement Geriatr Cogn Disord. 2011:31:202–209. [DOI] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984:34:939–944. [DOI] [PubMed] [Google Scholar]
- Meyers JE, Meyers KR. Rey Complex Figure Test and recognition trial professional manual. Odessa (FL): Psychological Assessment Resources; 1995 [Google Scholar]
- Mollon J, Knowles EEM, Mathias SR, Gur R, Peralta JM, Weiner DJ, Robinson EB, Gur RE, Blangero J, Almasy L, et al. Genetic influence on cognitive development between childhood and adulthood. Mol Psychiatry. 2021:26:656–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montembeault M, Joubert S, Doyon J, Carrier J, Gagnon JF, Monchi O, Lungu O, Belleville S, Brambati SM. The impact of aging on gray matter structural covariance networks. NeuroImage. 2012:63:754–759. [DOI] [PubMed] [Google Scholar]
- Moon S, Kim YJ, Han S, Hwang MY, Shin DM, Park MY, Lu Y, Yoon K, Jang HM, Kim YK, et al. The Korea Biobank Array: design and identification of coding variants associated with blood biochemical traits. Sci Rep. 2019:9:1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachev P, Kennard C, Husain M. Functional role of the supplementary and pre-supplementary motor areas. Nat Rev Neurosci. 2008:9:856–869. [DOI] [PubMed] [Google Scholar]
- O'Keefe J, Nadel L. The hippocampus as a cognitive map. Oxford (United Kingdom): Clarendon Press; 1978. [Google Scholar]
- Papenberg G, Lindenberger U, Backman L. Aging-related magnification of genetic effects on cognitive and brain integrity. Trends Cogn Sci. 2015:19:506–514. [DOI] [PubMed] [Google Scholar]
- Persson N, Lavebratt C, Wahlin A. Synergy effects of HbA1c and variants of APOE and BDNFVal66Met explains individual differences in memory performance. Neurobiol Learn Mem. 2013:106:274–282. [DOI] [PubMed] [Google Scholar]
- Persson K, Bohbot VD, Bogdanovic N, Selbaek G, Braekhus A, Engedal K. Finding of increased caudate nucleus in patients with Alzheimer's disease. Acta Neurol Scand. 2018:137:224–232. [DOI] [PubMed] [Google Scholar]
- Pizzagalli F, Auzias G, Yang Q, Mathias SR, Faskowitz J, Boyd JD, Amini A, Rivière D, McMahon KL, de Zubicaray GI, et al. The reliability and heritability of cortical folds and their genetic correlations across hemispheres. Commun Biol. 2020:3:510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006:38:904–909. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007:81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian ZJ, Chang PD, Moonis G, Lalwani AK. A novel method of quantifying brain atrophy associated with age-related hearing loss. Neuroimage Clin. 2017:16:205–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds CA, Finkel D. A meta-analysis of heritability of cognitive aging: minding the "missing heritability" gap. Neuropsychol Rev. 2015:25:97–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieckmann A, Bäckman L. Implicit learning in aging: extant patterns and new directions. Neuropsychol Rev. 2009:19:490–503. [DOI] [PubMed] [Google Scholar]
- Rieckmann A, Fischer H, Bäckman L. Activation in striatum and medial temporal lobe during sequence learning in younger and older adults: relations to performance. NeuroImage. 2010:50:1303–1312. [DOI] [PubMed] [Google Scholar]
- Robinson EB, Kirby A, Ruparel K, Yang J, McGrath L, Anttila V, Neale BM, Merikangas K, Lehner T, Sleiman PM, et al. The genetic architecture of pediatric cognitive abilities in the Philadelphia Neurodevelopmental Cohort. Mol Psychiatry. 2015:20:454–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosemann S, Thiel CM. Neuroanatomical changes associated with age-related hearing loss and listening effort. Brain Struct Funct. 2020:225:2689–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roshchupkin GV, Gutman BA, Vernooij MW, Jahanshad N, Martin NG, Hofman A, McMahon KL, van der Lee SJ, van Duijn CM, de Zubicaray GI, et al. Heritability of the shape of subcortical brain structures in the general population. Nat Commun. 2016:7:13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salthouse TA. Neuroanatomical substrates of age-related cognitive decline. Psychol Bull. 2011:137:753–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satizabal CL, Adams HHH, Hibar DP, White CC, Knol MJ, Stein JL, Scholz M, Sargurupremraj M, Jahanshad N, Roshchupkin GV, et al. Genetic architecture of subcortical brain structures in 38,851 individuals. Nat Genet. 2019:51:1624–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller E, Schumacher LV, Peter J, Lahr J, Wehrle J, Kaller CP, Gaser C, Klöppel S. Brain aging and APOE ε4 interact to reveal potential neuronal compensation in healthy older adults. Front Aging Neurosci. 2018:10:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sele S, Liem F, Merillat S, Jäncke L. Age-related decline in the brain: a longitudinal study on inter-individual variability of cortical thickness, area, volume, and cognition. NeuroImage. 2021:240:118370. [DOI] [PubMed] [Google Scholar]
- Seo S, Park K, Lee JJ, Choi KY, Lee KH, Won S. SNP genotype calling and quality control for multi-batch-based studies. Genes Genomics. 2019:41:927–939. [DOI] [PubMed] [Google Scholar]
- Smith SM, Douaud G, Chen W, Hanayik T, Alfaro-Almagro F, Sharp K, Elliott LT. An expanded set of genome-wide association studies of brain imaging phenotypes in UK Biobank. Nat Neurosci. 2021:24:737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodums DJ, Bohbot VD. Negative correlation between grey matter in the hippocampus and caudate nucleus in healthy aging. Hippocampus. 2020:30:892–908. [DOI] [PubMed] [Google Scholar]
- Song YE, Lee S, Park K, Elston RC, Yang HJ, Won S. ONETOOL for the analysis of family-based big data. Bioinformatics. 2018:34:2851–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, Hibar DP, Madsen SK, Khamis M, McMahon KL, de Zubicaray GI, Hansell NK, Montgomery GW, Martin NG, Wright MJ, et al. Discovery and replication of dopamine-related gene effects on caudate volume in young and elderly populations (N=1198) using genome-wide search. Mol Psychiatry. 2011:16:927–937 881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamnes CK, Walhovd KB, Dale AM, Østby Y, Grydeland H, Richardson G, Westlye LT, Roddey JC, Hagler DJ Jr, Due-Tønnessen P, et al. Brain development and aging: overlapping and unique patterns of change. NeuroImage. 2013:68:63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Jahanshad N, Ching CRK, Salminen LE, Thomopoulos SI, Bright J, Baune BT, Bertolín S, Bralten J, Bruin WB, et al. ENIGMA and global neuroscience: A decade of large-scale studies of the brain in health and disease across more than 40 countries. Transl Psychiatry. 2020:10:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay P, Dick AS. Broca and Wernicke are dead, or moving past the classic model of language neurobiology. Brain Lang. 2016:162:60–71. [DOI] [PubMed] [Google Scholar]
- Tremblay P, Gracco VL. Contribution of the pre-SMA to the production of words and non-speech oral motor gestures, as revealed by repetitive transcranial magnetic stimulation (rTMS). Brain Res. 2009:1268:112–124. [DOI] [PubMed] [Google Scholar]
- Voermans NC, Petersson KM, Daudey L, Weber B, Van Spaendonck KP, Kremer HP, Fernández G. Interaction between the human hippocampus and the caudate nucleus during route recognition. Neuron. 2004:43:427–435. [DOI] [PubMed] [Google Scholar]
- Wen W, Thalamuthu A, Mather KA, Zhu W, Jiang J, de Micheaux PL, Wright MJ, Ames D, Sachdev PS. Distinct genetic influences on cortical and subcortical brain structures. Sci Rep. 2016:6:32760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RS, Barral S, Lee JH, Leurgans SE, Foroud TM, Sweet RA, Graff-Radford N, Bird TD, Mayeux R, Bennett DA. Heritability of different forms of memory in the Late Onset Alzheimer's Disease Family Study. J Alzheimers Dis. 2011:23:249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong PC, Ettlinger M, Sheppard JP, Gunasekera GM, Dhar S. Neuroanatomical characteristics and speech perception in noise in older adults. Ear Hear. 2010:31:471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011:88:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Manolio TA, Pasquale LR, Boerwinkle E, Caporaso N, Cunningham JM, de Andrade M, Feenstra B, Feingold E, Hayes MG, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011:43:519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H, Chin J, Lee B, Kang Y, Na D. Development and validation of Korean version of Trail Making Test for elderly persons. Dement Neurocognitive Disord. 2007:6:54–66. [Google Scholar]
- Yin HH, Knowlton BJ. The role of the basal ganglia in habit formation. Nat Rev Neurosci. 2006:7:464–476. [DOI] [PubMed] [Google Scholar]
- Zhao B, Ibrahim JG, Li Y, Li T, Wang Y, Shan Y, Zhu Z, Zhou F, Zhang J, Huang C, et al. Heritability of regional brain volumes in large-scale neuroimaging and genetic studies. Cereb Cortex. 2019:29:2904–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Luo T, Li T, Li Y, Zhang J, Shan Y, Wang X, Yang L, Zhou F, Zhu Z, et al. Genome-wide association analysis of 19,629 individuals identifies variants influencing regional brain volumes and refines their genetic co-architecture with cognitive and mental health traits. Nat Genet. 2019:51:1637–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Due to confidentiality agreements, we are unable to provide a publicly available database. Researchers who wish to access the GARD cohort dataset used in this study can obtain it by contacting the corresponding author, professor Kunho Lee (leekho@chosun.ac.kr).