Abstract
Difficulty with attention is an important symptom in many conditions in psychiatry, including neurodiverse conditions such as autism. There is a need to better understand the neurobiological correlates of attention and leverage these findings in healthcare settings. Nevertheless, it remains unclear if it is possible to build dimensional predictive models of attentional state in a sample that includes participants with neurodiverse conditions. Here, we use 5 datasets to identify and validate functional connectome-based markers of attention. In dataset 1, we use connectome-based predictive modeling and observe successful prediction of performance on an in-scan sustained attention task in a sample of youth, including participants with a neurodiverse condition. The predictions are not driven by confounds, such as head motion. In dataset 2, we find that the attention network model defined in dataset 1 generalizes to predict in-scan attention in a separate sample of neurotypical participants performing the same attention task. In datasets 3–5, we use connectome-based identification and longitudinal scans to probe the stability of the attention network across months to years in individual participants. Our results help elucidate the brain correlates of attentional state in youth and support the further development of predictive dimensional models of other clinically relevant phenotypes.
Keywords: fingerprinting, functional connectivity, individual differences, machine learning, predictive modeling
Introduction
Autism spectrum disorder (hereafter “autism”) affects approximately 1% of children in the world (Zeidan et al. 2022) and is characterized by impairments in social communication and interaction as well as restricted and repetitive behaviors and atypical responses to sensory information (American Psychiatric Association 2013). An important symptom in autism with widespread individual differences is difficulty with attention. Between ~40% and 80% (Gadow et al. 2006; Lee and Ousley 2006) of individuals with autism have co-occurring attention symptoms, affecting quality of life (Masi et al. 2017). In addition, other neurodiverse individuals, like those with attention-deficit/hyperactivity disorder (ADHD) and/or the broader autism phenotype (Ingersoll 2010), also have difficulties with attention (Gerdts and Bernier 2011; American Psychiatric Association 2013). Given the impact, there has been much recent work investigating the neurobiological correlates of state- and trait-related attention in neurodiverse populations through the use of functional magnetic resonance imaging (fMRI). Of particular interest have been functional connectivity studies, in which measures of synchrony of the blood oxygen level-dependent signal are calculated between different regions of interest (Biswal et al. 1995). Group-based functional connectivity studies—comparing those with a neurodiverse condition like autism (Di Martino et al. 2013; Keehn et al. 2013; Fitzgerald et al. 2015) or ADHD (Qiu et al. 2011; Di Martino et al. 2013; Posner et al. 2013; Hoekzema et al. 2014) to neurotypical participants—have helped advance understanding of the brain correlates of attention. In particular, the default mode network, which plays a role in mediating aspects of attention, has been noted to be consistently affected in those with autism and ADHD (recently reviewed in Harikumar et al. 2021).
While collectively these studies have proven useful, they have largely failed to make a clinical impact. Aside from issues associated with participant head motion in this population (Yerys et al. 2009) and smaller samples (Marek et al. 2022), another potential reason is a lack of prediction-based studies focusing on individuals. Studies using cross-dataset prediction—building and validating models in one sample, then testing it in a separate sample (Scheinost et al. 2019)—are rare in the neuroimaging literature, despite their potential clinical utility (Gabrieli et al. 2015). Besides clinical applications, a prediction-based approach holds promise for avoiding statistical issues hindering generalizability (Yarkoni and Westfall 2017; Yarkoni 2020) and avoids the general lack of reliability of simple association studies (Marek et al. 2022) (though issues of validity can still occur with prediction approaches, especially with confounds related to symptom severity). Finally, a prediction-based framework can offer insights into populations undergoing significant developmental changes, particularly in youth (Rosenberg et al. 2018) and align with the goals of interrogating symptom dimensions among diverse individuals to aid further understanding of mental disorders (Insel et al. 2010).
Based on the importance of attention and the need for prediction studies focusing on individual differences, we set out to test if it is possible to build predictive models of sustained attention phenotypes based on an in-scan attention task in a sample of youth with autism and other neurodiverse conditions, as well as neurotypical controls. There are numerous reasons it might not be possible to generate a predictive model in a youth sample comprising many patients, including difficulties with task completion and issues with scan compliance (Yerys et al. 2009). Difficulty obtaining high-quality data can also be an issue in young participants (e.g. Horien et al. 2020).
In addition, there are potential issues related to building brain–behavior models in neurodiverse populations. For example, some have suggested that brain differences among those with a neurodiverse condition compared to neurotypical participants (Ross and Margolis 2019) might make it difficult to dimensionally model a phenotype using measures of functional organization. (Though it should be noted that neurodiversity exists on a spectrum (Armstrong 2015), and the distinction between what constitutes neurotypical vs. neurodiverse is somewhat artificial.) Furthermore, cognitive processes like attention are supported by complex, brain-wide correlates (Kessler et al. 2016; Rosenberg, Finn, et al. 2016a). Such distributed network markers, despite their complexity, are responsive to methylphenidate (Rosenberg, Zhang, et al. 2016b), as well as other pharmacological agents (Rosenberg et al. 2020; Chamberlain and Rosenberg 2022), suggesting that if identified, a distributed network marker of sustained attentional state might offer clinical utility.
With these factors in mind, we address 3 main issues in this work. We aim to (i) determine if attention-based predictive models can be generated in a sample of youth (some of whom have neurodiverse conditions), (ii) test if such a model generalizes out of sample, and (iii) interrogate neuroanatomy of the network model and assess the stability in individual participants across time. Using connectome-based predictive modeling (CPM) (Finn et al. 2015; Rosenberg, Finn, et al. 2016a; Shen et al. 2017; Beaty et al. 2018; Greene et al. 2018; Hsu et al. 2018; Yoo et al. 2018; Rapuano et al. 2020; Rohr et al. 2020; Boyle et al. 2022), we show that we are indeed able to predict performance on an in-scan sustained attention task in novel subjects based on functional connectivity data. The predictions are robust to factors such as in-scanner head motion, Autism Diagnostic Observation Schedule (ADOS) scores, age, sex, and intelligence quotient (IQ) scores. Crucially, we find the network model generalizes out of sample, increasing confidence in the model. In line with other dimensional work in neurodiverse populations (Lake et al. 2019; Rohr et al. 2020; Xiao et al. 2021), we observe that the brain correlates identified by the model are complex and distributed across broad swaths of cortical, subcortical, and cerebellar regions. Using connectome-based identification (ID) (Finn et al. 2015; Kaufmann et al. 2017; Vanderwal et al. 2017; Waller et al. 2017; Amico and Goni 2018; Graff, Tansey, Ip, et al. 2022a; Graff, Tansey, Rai, et al. 2022b), we perform exploratory analyses testing the longitudinal stability of the network model in individual participants. In sum, our data suggest that robust network markers of attentional state can be generated in youth and add to the growing literature suggesting the power of dimensional approaches in modeling brain–behavior relationships.
Materials and methods
Description of datasets
We used 5 independent datasets (Table 1) in this study. The first dataset consisted of youths with autism and other neurodiverse conditions (e.g. ADHD, anxiety, broader autism phenotype, bipolar disorder) as well as typically developing children and has been described previously (hereafter “neurodiverse sample”) (Horien et al. 2020). Participants were scanned on a 3T Siemens Prisma System. See Supplementary Material for exclusion criteria and imaging parameters for the neurodiverse sample. A second dataset of neurotypical adults was used as a test dataset (hereafter “validation sample”) and is described elsewhere (Rosenberg, Finn, et al. 2016a). Participants were scanned on a 3T Siemens Trio TIM system.
Table 1.
Demographic and imaging characteristics of samples used in this study.
| Measure | Neurodiverse sample | Utah | UM | Pitt | Validation sample |
|---|---|---|---|---|---|
| Number of participants (males) | 70 (39) | 16 (16) | 27 (4) | 44 (21) | 25 (12) |
| Number of participants with a neurodevelopmental or psychiatric condition | 33 total 7 = ADHD 2 = anxiety disorder 20 = autism 3 = BAP 1 = bipolar |
- | - | - | - |
| Age in years, mean (standard deviation) | 11.59 (2.87) | 22.81 (7.59) | 64.7 (7.3) | 16.42 (2.55) | 22.79 (3.54) |
| Time between scans in years, mean (standard deviation) | - | 2.56 (0.28) | 0.30 (0.061) | 1.69 (0.28) | - |
| Scan duration in minutes (volumes) | 10 min (two 5-min runs; 600 total volumes) | 8 min (240) | 5 min (150) | 5 min (200) | 36-min task (three 12-min runs; 824 total volumes); 6-min rest (360 total volumes) |
| TR in seconds | 1 | 2 | 2 | 1.5 | 1 |
| IQ, mean (standard deviation) | 107.23 (15.93) | - | - | - | - |
ADHD, attention-deficit/hyperactivity disorder; BAP, broader autism phenotype; ADOS, Autism Diagnostic Observations Schedule; IQ, intelligence quotient, TR, repetition time.
Three additional datasets from the Consortium for Reliability and Reproducibility (CoRR) (Zuo et al. 2014) were used to assess stability of the network model: the University of Pittsburgh School of Medicine dataset, the University of Utah dataset, and the University of McGill dataset (hereafter, “Pitt,” “Utah,” and “UM,” respectively). Full details of the Pitt and UM datasets can be found elsewhere (Hwang et al. 2013; Orban et al. 2015). All scans were acquired using Siemens 3-T Tim Trio scanners; all participants were neurotypical.
All datasets were collected in accordance with the institutional review board or research ethics committee at each site. Where appropriate, informed consent was obtained from the parents or guardians of participants. Written assent was obtained from children aged 13–17 years; verbal assent was obtained from participants under the age of 13 years.
Gradual onset continuous performance task description
Participants in the neurodiverse sample completed the gradual onset continuous performance task (gradCPT) (Esterman et al. 2013; Rosenberg et al. 2013; Rosenberg, Finn, et al. 2016a). The gradCPT is an assessment of sustained attention and inhibition abilities that has been shown to produce a range of performance scores across neurotypical participants (Esterman et al. 2013; Rosenberg et al. 2013). In the task, participants viewed grayscale pictures of cities and mountains presented at the center of the screen. Images gradually transitioned from one to the next every 1,000 ms. Subjects were told to respond by pressing a button for city scenes and to withhold button presses for mountain scenes. City scenes occurred randomly 90% of the time. As in previous studies (Esterman et al. 2013; Rosenberg, Finn, et al. 2016a), accuracy was emphasized without reference to speed, and performance was quantified using d’ (sensitivity), the participant’s hit rate minus false alarm rate. Participant d’ scores were calculated for scan 1 and scan 2 individually, as well as the average across both scans. Participants in the validation sample also completed gradCPT with the same parameters as above, except scene transitions took 800 ms; resting-state data were also collected in this sample.
The Pitt, Utah, and UM subjects completed only resting-state scans that were spaced apart at longer time intervals (months to years between scans; Table 1).
Preprocessing
The preprocessing strategy for the neurodiverse sample has been described previously (Greene et al. 2018; Horien et al. 2019). Preprocessing steps were performed using BioImage Suite (Joshi et al. 2011) unless otherwise indicated, and included: skull stripping the 3D magnetization prepared rapid gradient echo images using optiBET (Lutkenhoff et al. 2014) and performing linear and non-linear transformations to warp a 268-node functional atlas from Montreal Neurological Institute space to single subject space (Greene et al. 2018). Functional images were motion-corrected using SPM8 (https://www.fil.ion.ucl.ac.uk/spm/software/spm8/). Covariates of no interest were regressed from the data, including linear, quadratic, and cubic drift, a 24-parameter model of motion (Satterthwaite et al. 2013), mean cerebrospinal fluid signal, mean white matter signal, and the global signal. Data were temporally smoothed with a zero-mean unit-variance low-pass Gaussian filter (approximate cutoff frequency of 0.12 Hz). The results of skull-stripping, non-linear, and linear registrations were inspected visually after each step.
We used previously preprocessed data for the Utah, UM, Pitt, and validation samples; the preprocessing approach has been described elsewhere (Horien et al. 2019; Rosenberg, Finn, et al. 2016a). (See below for more about how motion was controlled in all analyses in all samples.)
Node and network definition
We used a 268-node functional atlas (Finn et al. 2015). For each participant, the mean time-course of each region of interest (“node” in graph theoretic terminology) was calculated, and the Pearson correlation coefficient was calculated between each pair of nodes to achieve a symmetric 268 × 268 matrix of correlation values representing “edges” (connections between nodes) in graph theoretic terminology. We transformed the Pearson correlation coefficients to z-scores via a Fisher transformation and only considered the upper triangle of the matrix, yielding 35,778 unique edges for whole-brain CPM analyses. For certain analyses, we grouped the 268 nodes into the 10 functional networks described by Horien et al. (2019).
Connectome-based predictive modeling
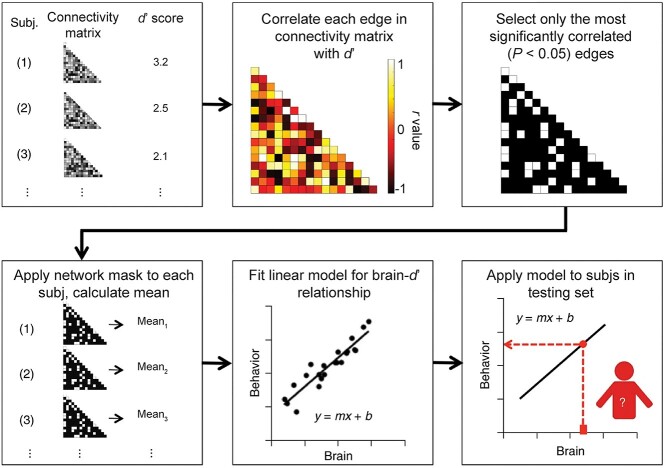
To predict gradCPT performance (d’) from the brain data (connectivity matrices) in the neurodiverse sample, we used CPM (Shen et al. 2017) (Fig. 1). Briefly, using 10-fold cross-validation, connectivity matrices from gradCPT and d’ scores were divided into an independent training set including subjects from 9 folds and a testing set including the left-out fold (Box 1). In the training set, linear regression was used to relate edge strength to d’ (Box 2). The edges most strongly related to d’ were selected (using a feature selection threshold of P = 0.05; Box 3) for both a “positive-association network” (in which increased connectivity was associated with a higher d’ score) and a “negative-association network” (in which decreased connectivity was associated with a higher d’ score). Mean network strength (Box 4) was calculated in both the positive-association and negative-association networks, and the difference between these network strengths was calculated (“combined network strength”), as in previous work (Greene et al. 2018):
Fig. 1.
A schematic of CPM. Figure adapted with permission from Shen et al. (2017).
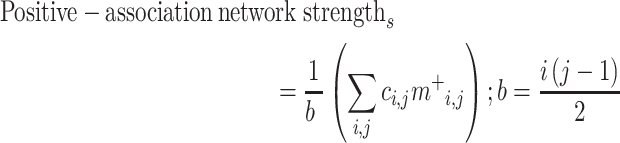 |
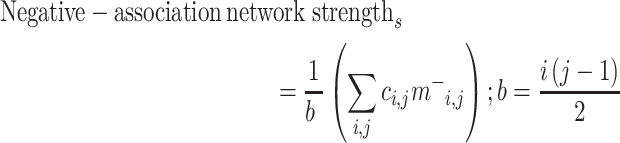 |
 |
where 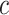 is the connectivity matrix for subject s and
is the connectivity matrix for subject s and 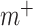 and
and 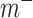 are binary matrices indexing the edges
are binary matrices indexing the edges 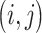 that survived the feature selection threshold for the positive-association and negative-association networks, respectively. (Recall that
that survived the feature selection threshold for the positive-association and negative-association networks, respectively. (Recall that 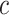 and
and 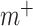 and
and 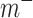 comprise only the upper triangle of the connectivity matrix, as specified above.) Throughout the text, we refer to the edges in the positive-association and negative-association network as comprising the “attention network.” We call attention to the fact that for both the positive- and negative-association networks, mean network strength was calculated, as opposed to summed network strength (Shen et al. 2017).
comprise only the upper triangle of the connectivity matrix, as specified above.) Throughout the text, we refer to the edges in the positive-association and negative-association network as comprising the “attention network.” We call attention to the fact that for both the positive- and negative-association networks, mean network strength was calculated, as opposed to summed network strength (Shen et al. 2017).
A linear model was then generated relating combined network strength to d’ scores in the training data (Box 5). In the final step, combined network strength was calculated for the left-out participants in the testing set, and the model was applied to generate d’ predictions for these left-out subjects (Box 6). We conducted the main CPM analyses by constructing an average connectivity matrix per participant across the 2 gradCPT runs; behavioral data were averaged as well, as in previous work using gradCPT (Rosenberg, Finn, et al. 2016a). (See “Multiverse analysis and CPM” section below for how the effects of arbitrary choices were assessed.)
As in Scheinost et al. (2021), model performance was assessed by comparing the similarity between predicted and observed gradCPT d’ scores using both Spearman’s correlation (to avoid distribution assumptions) and root mean square error (defined as: 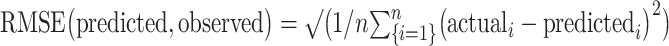 ). We performed 1,000 iterations of a given CPM analysis and selected the median-performing model; we report this in the main text when discussing model performance. To calculate significance, we randomly shuffled participant labels and attempted to predict gradCPT d’ scores. We repeated this 1,000 times and calculated the number of times a permuted predictive accuracy was greater than the median of the unpermuted predictions to achieve a nonparametric P-value:
). We performed 1,000 iterations of a given CPM analysis and selected the median-performing model; we report this in the main text when discussing model performance. To calculate significance, we randomly shuffled participant labels and attempted to predict gradCPT d’ scores. We repeated this 1,000 times and calculated the number of times a permuted predictive accuracy was greater than the median of the unpermuted predictions to achieve a nonparametric P-value:
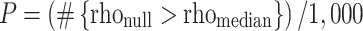 |
where #{rhonull > rhomedian} indicates the number of permuted predictions numerically greater than or equal to the median of the unpermuted predictions (Scheinost et al. 2021). Note that all P-values reported for CPM performance are from this permutation testing procedure. We also report the variance explained (R-squared) between predicted and observed d’ scores.
In addition, we performed CPM using only the edges in conventionally defined resting-state networks, as well as after performing a “lesion” analysis, in which we removed all edges within and between a given resting-state network. Prediction performance was quantified as above. Results of each model were also compared to the “main” whole brain-based CPM model reported in Fig. 2A (obtained with the P < 0.05 feature selection threshold) using Steiger’s z test (Steiger 1980).
Fig. 2.
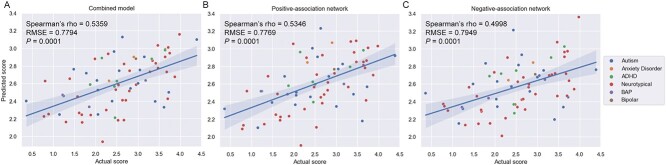
In-scan sustained attention task performance (gradCPT d’) can be predicted in a sample of neurodiverse youth using CPM. A) Results from the combined network model. B) Results from the positive-association network. C) Results from the negative-association network. For all plots, results are shown for a feature selection threshold of 0.05. Actual gradCPT d’ scores are indicated on the x-axis; predicted scores, on the y-axis. Higher d’ scores indicate better performance on the task and imply better sustained attention. A regression line and 95% confidence interval are shown. ADHD, attention-deficit/hyperactivity disorder; BAP, broader autism phenotype; P = P-value; RMSE, root mean square error.
Multiverse analysis and CPM
To determine if CPM findings were robust, we used a multiverse approach to explore how results were affected by different analytical choices (Steegen et al. 2016). The goal of this approach is not to determine what CPM pipeline gives the “best” prediction; rather, it is to gather converging evidence across a range of analytical scenarios to determine how modeling choices affect results. Specifically, we altered the feature selection threshold used to select significant edges (P = 0.1, P = 0.05, P = 0.01, P = 0.005, P = 0.001); we tested CPM using a combined network model versus testing the positive-association and negative-association networks separately; we tested the effect of controlling for participant age, sex, IQ, ADOS (Lord et al. 2012) calibrated severity score, and head motion as described previously, by using partial correlation at the feature selection step (Scheinost et al. 2021; Dufford et al. 2022); and we also built models using data from gradCPT run 1, gradCPT run 2, and average gradCPT data. To ensure this approach did not result in false positives, the Benjamini–Hochberg procedure (Benjamini and Hochberg 1995) was applied to control for multiple comparisons (correcting for 13 CPM tests). In addition, we tested if it was possible to successfully predict d’ using only the neurodiverse individuals; we used leave-one-out cross-validation instead of 10-fold (due to the smaller sample size) to generate CPM models and quantified prediction performance as above.
Testing generalizability of the attention network
To assess if the network model of attention generalized out of sample, we defined a consensus positive-association network and consensus negative-association network as edges that appear in at least 6/10 folds in 600/1,000 iterations of CPM. This resulted in 922 edges in the positive-association network and 896 edges in the negative-association network (we note the size of these networks is consistent with other CPM networks that have generalized (e.g. Rosenberg, Finn, et al. 2016a; Rosenberg, Zhang, et al. 2016b; Yip et al. 2019). Using the combined network strength in the consensus networks (as above for CPM), we determined model coefficients across the neurodiverse sample, as in Rosenberg, Finn, et al. (2016a), Ju et al. (2020), and Dufford et al. (2022). We then applied the network masks and model coefficients to the validation sample to generate d’ predictions.
As above for CPM analyses, model performance was assessed by comparing the similarity between predicted and observed gradCPT d’ scores using both Spearman’s correlation and by calculating RMSE. Nonparametric P-values were computed as for CPM. As above, we used a similar multiverse approach to ensure results were not driven by subject age, sex, or head motion; the Benjamini–Hochberg procedure (Benjamini and Hochberg 1995) was again used to control for multiple comparisons. To further ensure results were robust, we tested a range of summary networks of varying sizes (i.e. from stringent cases where an edge must appear in 10/10 folds and 1,000/1,000 iterations, to more liberal thresholds where an edge must appear in 3/10 folds and 300/1,000 iterations, moving in intervals of 1 fold and 100 iterations for each summary network). Testing various summary networks is crucial, given the arbitrary nature of summarizing a network model and the researcher degrees of freedom (Wicherts et al. 2016) involved in such a task.
Connectome-based ID
To test the stability of the predictive model over time in a given individual, we used connectome-based ID (Finn et al. 2015) and the Pitt, Utah, and UM samples. (See Supplemental Fig. 1 for a schematic of connectome-based ID.) Briefly, after selecting only the edges in the positive-association and negative-association networks (i.e. the same consensus edges used in the cross-dataset test—the 922 and 896 edges in the positive-association and negative-association networks, respectively), a database was created consisting of all subjects’ matrices from scan 1. In an iterative process, a connectivity matrix from a given subject was then selected from scan 2 and denoted as the target. Pearson correlation coefficients were calculated between the target connectivity matrix and all the matrices in the database. If the highest Pearson correlation coefficient was between the target subject in one session and the same subject in the second session (i.e. within-subject correlation > all other between-subject correlations), this was recorded as a correct identification. The process was repeated until identifications had been performed for all subjects and database–target combinations. We averaged both database–target pairs (because these can be reversed) for a dataset to achieve an average ID rate. To calculate P-values, we randomly shuffled subject identities and reperformed ID for 1,000 iterations and compared the actual ID rates to this null distribution (Finn et al. 2015; Horien et al. 2018, 2019):
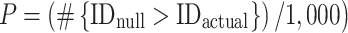 |
where #{IDnull > IDactual} indicates the number of permuted ID rates numerically greater than or equal to the actual ID rate obtained using the original data.
We also assessed if connections inside the attention network were more or less stable than connections in the rest of the brain. We generated 1,000 summary networks comprising edges outside of the consensus attention network (and the same size as the positive-association and negative-association networks). Connectome-based ID was performed using the random networks, and we compared ID results to those obtained using the original consensus attention network. P-values were obtained as follows:
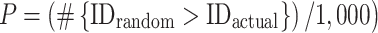 |
where #{IDrandom > IDactual} indicates the number of random ID rates numerically greater than or equal to the actual ID rate obtained using the original data.
In the Pitt dataset, incomplete scan coverage during the functional runs resulted in 158/922 and 145/896 edges missing in the positive-association and negative-association networks, respectively; we performed ID with the remaining edges.
Motion control considerations
In-scanner head motion has been shown to affect estimates of functional connectivity (Satterthwaite et al. 2013; Power et al. 2015) and brain–behavior relationships (Siegel et al. 2017). We therefore adopted a rigorous motion control strategy during scanning acquisition and in all analyses.
Specifically, all participants underwent an intensive mock scan protocol 5 days prior to scanning (along with a refresher training period on the day of the scan). We have previously shown in this same sample that the mock scan protocol significantly lowers motion artifact (Horien et al. 2020). In the present paper, such an approach led to 100% of the sample having a mean frame-to-frame displacement (FFD) < 0.24 mm (Table 2), well below other samples of youth (e.g. Casey et al. 2018). Further, 95.7% of the sample (67/70 of the final participants used in analyses) contained data with a mean FFD < 0.2 mm, a typical threshold used for determining high- versus low-motion data in youth and/or those with a mental health condition (Yip et al. 2019; Ju et al. 2020; Lichenstein et al. 2021). (See Supplemental Fig. 2 for a histogram of mean FFD values in the neurodiverse sample.)
Table 2.
Quantifying the number of participants in the neurodiverse sample with mean FFD values below motion thresholds.
| Motion threshold (mean FFD, mm) | Number of participants below threshold | Percentage of participants below threshold |
|---|---|---|
| 0.24 | 70 | 100% |
| 0.20 | 67 | 95.7% |
| 0.15 | 59 | 84.3% |
| 0.10 | 43 | 61.4% |
As in other work using the gradCPT (Rosenberg, Finn, et al. 2016a; Rosenberg et al. 2020), no censoring of the functional data was performed to avoid removing a different number of timepoints across participants (hence leaving different numbers of behaviorally relevant button presses across participants—a potential confound when building models with task fMRI data given that we are interested in predicting d’ scores from button presses). In addition, similar to other recent CPM papers (i.e. Lake et al. 2019; Scheinost et al. 2021; Dufford et al. 2022), steps were taken during CPM and connectome-based ID analyses to limit the effects of motion. Specifically, we adjusted the CPM model for each participant’s mean FFD over the course of gradCPT and found that motion was not driving predictions (i.e. the model still successfully predicted gradCPT d’ scores when controlling for motion (Spearman’s rho = 0.54, RMSE = 0.78, P = 0.0001)). Next, when testing if the network model generalized in the test sample, we again controlled for in-scanner head motion and found that models were not confounded by mean FFD (i.e. successful prediction was again achieved, Spearman’s rho = 0.67, P = 0.0008). In addition, the fact that the model from the neurodiverse sample generalizes to predict d’ in the validation sample (with slightly faster trials; neurodiverse inter-trial interval of 1,000 ms, validation sample inter-trial interval of 800 ms) increases confidence that model success is not due to participant head motion yoked to stimuli presentation. Specifically, any overfitting in the neurodiverse sample due to motion artifact would impact model performance in the validation sample. Because the model generalizes in the validation sample with slightly different timing parameters, this suggests that head motion is not driving the results.
In connectome-based ID, we focused our analyses on only the low-motion longitudinal subjects previously described in Horien et al. (2019). These participants have previously been used in whole-brain and canonical network-based ID analyses, and it was shown that connectome-based ID results were not driven by head motion. From this low-motion sample (i.e. all participants had a mean FFD < 0.1 mm for all resting-state scans), we further considered how within-participant self-correlations derived from the connectome-based ID process related to in-scanner head motion in the present analyses. Across all 3 datasets, we found there were no statistically significant relationships between within-participant correlation scores and head motion in 5/6 cases (range of rho values: −0.0964 to 0.1236; P > 0.53 across all samples; Supplemental Table 1). The only statistically significant relationship we observed was in Pitt, and there was a negative association (high attention network: rho = −0.3636, P = 0.0153), indicating higher head motion in this sample was associated with lower within-subject self-correlation scores (in line with previous results) (Horien et al. 2018; Graff, Tansey, Ip, et al. 2022a; Graff, Tansey, Rai, et al. 2022b). These results suggest that head motion is not acting as a confound in the connectome-based ID results.
In sum, while head motion is always a concern in functional connectivity analyses of brain–behavior relationships, the present data suggest it is not driving the findings described here.
Code and data availability
Preprocessing was carried out using software freely available here: (https://medicine.yale.edu/bioimaging/suite/). CPM code is available here: (https://github.com/YaleMRRC/CPM). The parcellation, the attention network models, and the connectome-based ID code are available here: (https://www.nitrc.org/frs/?group_id=51). Data from the longitudinal samples are openly available through CoRR (http://fcon_1000.projects.nitrc.org/indi/CoRR/html/). All other data is available from the authors upon request.
Results
Prediction of in-scan attention scores in the neurodiverse sample
In the neurodiverse sample, there were no differences between neurodiverse and neurotypical participants in in-scanner head motion (t(68) = 0.77, P = 0.4437) or gradCPT d’ (t(68) = −0.60, P = 0.5487). Across the sample, we observed that motion and d’ were negatively correlated (r = − 0.35, P = 0.0034); we hence adopted a rigorous motion control strategy to ensure motion was not driving the brain–behavior models (Methods, “Motion control considerations”).
Next, using CPM, we built a model using within-dataset cross-validation to predict unseen participants’ gradCPT d’ scores from functional connectivity data (in which higher d’ scores indicate better task performance and imply better sustained attentional state). The model successfully predicted gradCPT d’ scores in the neurodiverse dataset (feature selection threshold of 0.05, Spearman’s rho = 0.54, RMSE = 0.78, P = 0.0001, corrected; Fig. 2A; Table 3). There was no statistically significant difference in model error (i.e. between actual d’ scores and those predicted by the model) between neurodiverse and neurotypical participants (t(68) = 0.87, P = 0.3855). Prediction performance was also high when we calculated the R2 between predicted and observed d’ scores (R2 = 29.12%, P = 0.0000014; see Supplemental Table 2 for R2 for all models reported in this section).
Table 3.
Testing model performance in various scenarios.
| Condition tested | Spearman’s rho | RMSE | P-value |
|---|---|---|---|
| Mean gradCPT, 0.05 feat. sel. | 0.54 | 0.78 | 0.0001 |
| Mean gradCPT, 0.1 feat. sel. | 0.52 | 0.79 | 0.0001 |
| Mean gradCPT, 0.01 feat. sel. | 0.50 | 0.79 | 0.0001 |
| Mean gradCPT, 0.005 | 0.45 | 0.80 | 0.002 |
| Mean gradCPT, 0.001 | 0.43 | 0.81 | 0.0001 |
| Mean gradCPT, partial age | 0.47 | 0.80 | 0.0001 |
| Mean gradCPT, partial sex | 0.52 | 0.77 | 0.0001 |
| Mean gradCPT, partial IQ | 0.49 | 0.84 | 0.003 |
| Mean gradCPT, partial ADOS | 0.53 | 0.77 | 0.0001 |
| Session 1 | 0.31 | 0.85 | 0.031 |
| Session 2 | 0.27 | 0.87 | 0.073 |
| Mean gradCPT, neurodiverse individuals | 0.37 | 0.82 | 0.001 |
| Mean gradCPT, positive-association network only | 0.53 | 0.78 | 0.0001 |
| Mean gradCPT, negative-association network only | 0.50 | 0.79 | 0.0001 |
The median-performing model after 1,000 iterations of CPM is reported for each condition tested. P-values were obtained from permutation testing. The result from Fig. 2A is shown in the top row. gradCPT, gradual onset continuous performance task; ADOS, Autism Diagnostic Observation Schedule.
To assess the robustness of the d’ prediction, we used a multiverse approach to explore how results were affected by different analytical choices (Steegen et al. 2016). We stress the point of this approach is not to determine what pipeline gives the “best” prediction performance; it is instead to gather converging evidence across a range of analytical scenarios to determine the extent to which arbitrary choices affect CPM results.
As the choice of feature selection threshold is arbitrary, we started by testing a range of thresholds (Table 3), while still controlling for motion. We were able to significantly predict d’ scores in all cases (feature selection threshold of 0.1: Spearman’s rho = 0.52, RMSE = 0.79, P = 0.0001, corrected; feature selection threshold of 0.01: Spearman’s rho = 0.50, RMSE = 0.79, P = 0.0001, corrected; feature selection threshold of 0.005: Spearman’s rho = 0.45, RMSE = 0.80, P = 0.002, corrected; feature selection threshold of 0.001: Spearman’s rho = 0.43, RMSE = 0.81, P = 0.0001, corrected). Interestingly, all models performed quite well, but when more stringent feature selection thresholds were applied (i.e. fewer edges were included in a model), performance decreased.
We next performed analyses adjusting for participant age, sex, IQ, and ADOS score. Each pipeline demonstrated similar prediction performance of d’ (age-adjusted model: Spearman’s rho = 0.47, RMSE = 0.80, P = 0.001, corrected; sex-adjusted model: Spearman’s rho = 0.52, RMSE = 0.77, P = 0.0001, corrected; IQ-adjusted model: Spearman’s rho = 0.49, RMSE = 0.96, P = 0.003, corrected; ADOS-adjusted model: Spearman’s rho = 0.53, RMSE = 0.77, P = 0.0001, corrected). Models were also built for gradCPT scan 1 and gradCPT scan 2 separately (scan 1: Spearman’s rho = 0.31, RMSE = 0.85, P = 0.031, corrected; scan 2: Spearman’s rho = 0.27, RMSE = 0.87, P = 0.073, corrected). Prediction performance dropped in this case, echoing recent results that more data fed into predictive models results in higher accuracies (Taxali et al. 2021). In addition, we restricted our analysis to performing CPM on only neurodiverse individuals (e.g. those with a psychiatric condition, as outlined in Table 1) and again observed successful d’ prediction (Spearman’s rho = 0.37, RMSE = 0.82, P = 0.001, corrected), ensuring the CPM results derived from the entire sample were not being driven by only the neurotypical participants.
Finally, the choice to model attentional state using a “combined network” (used in all analyses above) is also arbitrary. We repeated the CPM prediction of d’ scores and tested the positive-association and negative-association networks. We again observed similar d’ prediction in both cases (positive-association network: Spearman’s rho = 0.53, RMSE = 0.78, P = 0.0001, corrected; negative-association network: Spearman’s rho = 0.50, RMSE = 0.79, P = 0.0001, corrected; Fig. 2B and C).
In all, these results suggest that attentional state prediction is robust in this sample and is not driven by potential confounding factors.
External validation of the attention network
Overfitting—deriving statistical patterns specific to noise in a sample—is a constant concern in machine learning studies. The ultimate test is to assess how well a model works in an independent dataset; we perform such a test here. We determined which edges tended to contribute consistently to successful prediction (922 in the positive-association network and 896 in the negative-association network, 1,818 edges total) and built a consensus model in the neurodiverse sample (Methods, “Testing generalizability of the attention network”). Applying the model to the validation sample comprising subjects completing the same gradCPT, we observed successful prediction of d’ scores (Spearman’s rho = 0.65, RMSE = 1.059 P = 0.0006, corrected; Fig. 3A). The R2 between predicted and observed d’ scores was similarly high (R2 = 42.24%, P = 0.00044).
Fig. 3.
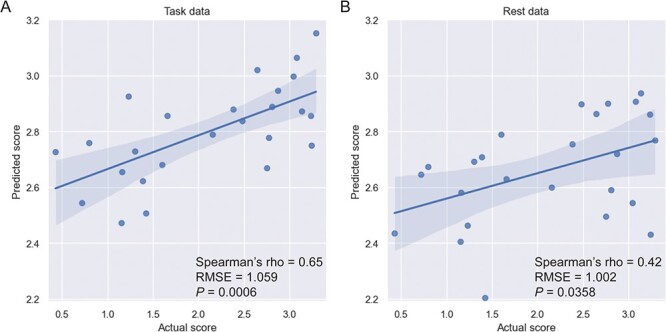
Generalization of the attention network to an independent sample. A) Results using task data. B) Results using rest data. For all plots, actual gradCPT d’ scores are indicated on the x-axis; predicted scores, on the y-axis. Higher d’ scores indicate better performance on the task and imply better sustained attention. A regression line and 95% confidence interval are shown. RMSE, root mean square error; P = P-value.
We repeated analyses controlling for several other variables, adjusting for in-scanner head motion (Spearman’s rho = 0.67, P = 0.0008, corrected), participant sex (Spearman’s rho = 0.61, P = 0.0014, corrected), as well as age (Spearman’s rho = 0.65, P = 0.0009, corrected), and observed similar results. In addition, when we tested both the positive-association and negative-association networks separately, we found each of these networks predicted d’ scores (positive-association network: Spearman’s rho = 0.59, RMSE = 1.094, P = 0.021, corrected; negative-association network: Spearman’s rho = 0.59, RMSE = 1.03, P = 0.0025, corrected). In line with previous work (Rosenberg, Finn, et al. 2016a; Rosenberg et al. 2020), we also tested whether the attention network could be used to predict d’ scores from resting-state data in the adult sample (specifically, using the resting-state data to predict d’ scores from the gradCPT). We again found the model generalized (Spearman’s rho = 0.42, RMSE = 1.002, P = 0.0358, corrected; R2 = 19.80%, P = 0.0258; Fig. 3B).
We further tested the stability of results by altering how consistently an edge had to appear across CPM iterations to be included in the summary attention network (Methods, “Testing generalizability of the attention network”). This resulted in 8 attention summary networks, ranging from ~100 to 3,000 edges. In 7/8 cases, the attention network generalized to predict d’ scores (range of Spearman’s rho = 0.50–0.66; all P < 0.0104 after FDR correction; Supplemental Table 3). The only summary network that did not predict d’ score (Spearman’s rho = 0.23, P = 0.27) was quite small (~100 edges), approximately an order of magnitude smaller than the original attention network tested above (and other networks that have generalized) (Rosenberg, Finn, et al. 2016a; Greene et al. 2018; Yip et al. 2019). These results suggest that generalization in this sample is robust to the arbitrary choices made when defining a summary model.
Neuroanatomy of CPM predictive networks
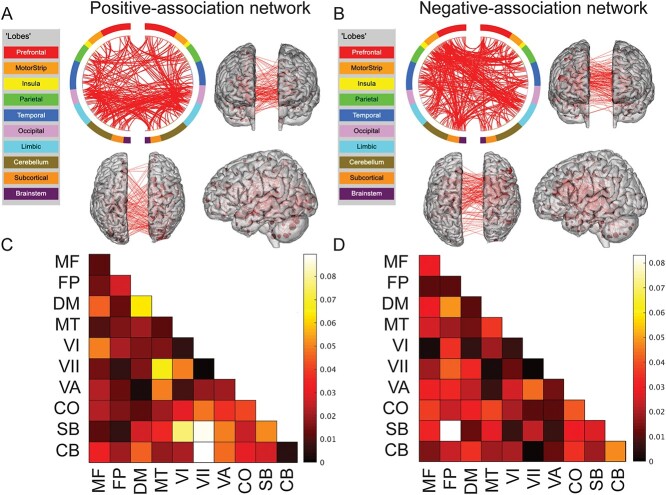
We next performed post hoc visualizations to localize brain connections contributing to the model. Together, the 1,818 total edges comprise 5.08% of the connectome. Similar to other CPM models (Rosenberg, Finn, et al. 2016a; Beaty et al. 2018; Greene et al. 2018; Lake et al. 2019; Ju et al. 2020; Dufford et al. 2022), the predictive edges in the positive-association and negative-association networks comprise complex, distributed networks spanning the entire brain (Fig. 4A and B). In line with task demands, additional visualizations at the network level (Fig. 4C and D) revealed that connections involving subcortical, cerebellar, and visual networks were particularly important. In the positive-association network, for example, the top 3 network pairs containing the greatest proportion of edges involved subcortical, cerebellar, and visual networks. In the negative-association network, a network pair involving the cerebellum (the cerebellar-frontoparietal network) contained the greatest proportion of edges. For completeness, we present the matrices in Fig. 4C and D in Supplemental Fig. 3 without normalizing by network size; subcortical, visual, and cerebellar networks again tended to harbor large numbers of edges. (See also Supplemental Fig. 4 for an additional visualization using circle plots and glass brains.)
Fig. 4.
Neuroanatomy of CPM predictive networks. A) The consensus positive-association network. B) The consensus negative-association network. For both (A) and (B): A circle plot is shown in the upper left. The top of the circle represents anterior; the bottom, posterior. The left half of the circle plot corresponds to the left hemisphere of the brain. A legend indicating the approximate anatomic “lobe” is shown to the left. The same edges are plotted in the glass brains as lines connecting different nodes; in these visualizations, nodes are sized according to degree, the number of edges connected to that node. Note that to aid in visualization, we have thresholded the matrices to only show nodes with a degree threshold >15 (unthresholded circle plots and glass brains are shown in Supplemental Fig. 4). C) Matrix of the consensus positive-association network. D) Matrix of the consensus negative-association network. For both (C) and (D): The proportion of edges in a given network pair; data have been corrected for differing network size. MF, medial frontal; FP, frontoparietal; DM, default mode; MT, motor; VI, visual I; VII, visual II; VA, visual association; CO, cingulo-opercular; SB, subcortical; CB, cerebellum.
Further investigations into the neurobiology of d’ prediction
To add more biological context to the prediction of d’, we next assessed how CPM performance is impacted by lesioning resting-state networks. Specifically, we eliminated edges within and between specific networks and reperformed CPM using the gradCPT data in the neurodiverse sample. Consistent with the distributed nature of the attention network, we observed little impact when eliminating edges. That is, prediction performance was still quite high—predictions were statistically significant in all cases—and did not drop compared to the original whole-brain model reported in Fig. 2A (Table 4). We also attempted CPM using only the edges within resting-state networks; in all cases, prediction performance was poor (Table 5). In 8/10 cases, a negative correlation between predicted and observed d’ scores was observed—that is, the model tended to predict a high d’ score, when, in fact, the individual had a low d’ score (and vice versa). As a final test, we performed CPM on edges outside the consensus attention network, observing poor prediction performance (Spearman’s rho = −0.79 between predicted and observed d’ scores, RMSE = 1.61, P = 0.0001 compared to the original whole-brain CPM model). Together, these findings highlight the distributed nature of the consensus attention network and reinforce that while some brain areas seem to be important in successful prediction (subcortical, visual, and cerebellar network), the attention network, as a whole, is greater than the sum of its parts for predicting d’.
Table 4.
Results of CPM models with lesions to edges within and between resting-state networks.
| Lesioned model | Performance compared to whole-brain model | ||||
|---|---|---|---|---|---|
| Lesioned network | Spearman’s rho | RMSE | P-value | Steiger’s z | P-value |
| Medial frontal | 0.54 | 0.77 | 0.0001 | 0.11 | 0.91 |
| Frontoparietal | 0.52 | 0.78 | 0.0001 | 0.39 | 0.70 |
| Default mode | 0.52 | 0.79 | 0.0001 | 0.51 | 0.61 |
| Motor | 0.52 | 0.77 | 0.0001 | 0.25 | 0.80 |
| Visual I | 0.54 | 0.77 | 0.0001 | 0.21 | 0.83 |
| Visual II | 0.54 | 0.78 | 0.0001 | 0.004 | 0.99 |
| Visual association | 0.54 | 0.78 | 0.0001 | 0.14 | 0.89 |
| Cingulo-opercular | 0.49 | 0.80 | 0.0001 | 0.84 | 0.40 |
| Subcortical | 0.49 | 0.79 | 0.0001 | 0.84 | 0.40 |
| Cerebellum | 0.51 | 0.79 | 0.0001 | 0.57 | 0.57 |
The median-performing model after 1,000 iterations is shown. P-values for the lesioned models were obtained from permutation testing. Steiger’s z is reported for each lesioned model, compared to the original whole-brain model reported in Fig. 2A.
Table 5.
Using conventionally defined resting-state networks to predict d’ scores.
| Network model | Performance compared to whole-brain model | ||||
|---|---|---|---|---|---|
| Network | Spearman’s rho | RMSE | P-value | Steiger’s z | P-value |
| Medial frontal | −0.31 | 0.92 | 0.99 | 4.70 | 0.0001 |
| Frontoparietal | 0.14 | 0.88 | 0.31 | 2.96 | 0.003 |
| Default mode | −0.05 | 0.90 | 0.63 | 3.78 | 0.0001 |
| Motor | −0.11 | 0.92 | 0.77 | 3.90 | 0.0001 |
| Visual I | −0.22 | 0.92 | 0.99 | 4.56 | 0.0001 |
| Visual II | −0.09 | 0.91 | 0.72 | 3.99 | 0.0001 |
| Visual association | −0.36 | 0.92 | 0.99 | 4.98 | 0.0001 |
| Cingulo-opercular | −0.35 | 0.92 | 0.99 | 5.40 | 0.0001 |
| Subcortical | −0.29 | 0.92 | 0.99 | 4.61 | 0.0001 |
| Cerebellum | 0.35 | 0.84 | 0.23 | 1.92 | 0.055 |
The median-performing model after 1,000 iterations is shown. P-values for the network model were obtained from permutation testing. Steiger’s z is reported for each lesioned model, compared to the original whole-brain model reported in Fig. 2A.
Individual-level stability of predictive network model
An ultimate goal of using individual-level approaches in the clinic is to infer future outcomes based on current data. Because the attention network was defined in relation to a state-based cognitive process that itself fluctuates (Cohen and Maunsell 2011; Esterman et al. 2013; Esterman et al. 2014; Rosenberg et al. 2015; Terashima et al. 2021), it is possible individual connectivity patterns in the attention network might change over time. Hence, we conducted an exploratory analysis using connectome-based ID and longitudinal datasets with months to years between scans, asking: Are connections in the attention network stable enough within individuals to identify a participant from a group?
Across the 3 longitudinal samples, we observed that the attention network results in ID rates well-above chance (ID rate range: 53.4–93.5%; P < 0.0001 across all samples; Supplemental Fig. 5). Specifically, ID rates were high when there were months between scans (UM dataset; 92.6% and 81.5% in the positive-association and negative-association networks, respectively) and when there were years between scans (Utah: positive-association network = 84.4%, negative-association network = 81.3%). ID rates were lower, but still above chance levels, in Pitt (positive-association network = 53.4%, negative-association network = 62.5%). These results suggest that participants retain their unique connectivity signatures in the attention network.
We conducted additional analyses and determined that the high ID rates were not being driven by head motion (Supplemental Table 1), though motion might be contributing to the lower ID rates observed in Pitt, as head motion was significantly correlated with lower participant self-correlation scores. This is consistent with other work demonstrating that higher head motion is associated with lower ID rates (Horien et al. 2018; Graff, Tansey, Ip, et al. 2022a; Graff, Tansey, Rai, et al. 2022b). We also performed ID using other connections in the rest of the brain and observed no differences in ID rates compared to the attention network (Supplemental Fig. 5 and Supplemental Table 4). In all, these results suggest that the attention network tends to retain participant-specific connectivity signatures across months to years because the brain as a whole retains participant-specific connectivity signatures across months to years (see the Supplemental Materials for more about the connectome-based ID results).
Discussion
In this work, we set out to test if it was possible to generate connectome-based predictive models of attentional state in a sample of youth, some of whom were neurodiverse. Using CPM, we were able to build a predictive network model of in-scan sustained attention scores. Crucially, we found the network generalized out of sample, further suggesting that the brain–behavior model we originally identified is a robust marker of attentional state. The network model was spatially complex, comprising connections across the brain. Lastly, we conducted exploratory analyses in 3 open-source samples using the network and connectome-based ID.
The power and potential of dimensional models
Our work adds to the growing literature suggesting it is feasible to use a dimensional approach to model individual differences in brain–behavior relationships in neurodiverse youth. In addition to autism symptoms (Lake et al. 2019), groups have developed dimensional models predictive of behavioral inhibition (Rohr et al. 2020), social affect (Xiao et al. 2021), and adaptive functioning (Plitt et al. 2015) in neurodiverse samples. In all cases, the predictive models comprise complex networks, with connections spanning the entire brain (reviewed in Horien et al. 2022). Nevertheless, subcortical and cerebellar networks tend to emerge as major contributors in these models, regions we also observed as important in our attention network. Furthermore, similar brain areas have also been noted to play a role in attention (Rosenberg, Finn, et al. 2016a; Green et al. 2017; Yoo et al. 2022), consistent with the growing recognition that subcortical and cerebellar circuits are important in mediating cognitive processes (Buckner 2013; Clark et al. 2021).
Beyond helping to hone in on brain areas involved in sustaining attention, the network identified is intriguing from a clinical standpoint. For example, it has been shown that a network connectivity model of attentional state (Rosenberg, Finn, et al. 2016a) is sensitive to methylphenidate (Rosenberg, Zhang, et al. 2016b). It is therefore possible the network identified here may help in tracking changes after administration of a therapeutic. Though more work is needed, it is generally encouraging that markers identified through dimensional analyses appear to be sensitive to clinically useful drugs. There is a need for objective, biological markers in psychiatry, and dimensional approaches could offer a framework to identify quantitative markers to help individuals clinically (McPartland 2021).
Generalizability of the attention network and open science
Much has been written about the reproducibility crisis in biomedical science (Pashler and Wagenmakers 2012; Open Science Collaboration 2015; Baker 2016), as well as the fear results might suffer from a lack of generalizability (Yarkoni 2020), particularly in psychiatry and psychology. We emphasize the present study uses predictive modeling, which is an additional step beyond association studies, that helps reproducibility (Rosenberg, Finn, et al. 2016a; Yarkoni and Westfall 2017; Rosenberg et al. 2018; Scheinost et al. 2019; Poldrack et al. 2020). Testing to ensure results are robust across samples and contexts is also imperative. This effort is especially important in neuroimaging, where hopes have been high for clinical impact, yet there has been little progress translating papers into practice (Chekroud 2017; Chekroud and Koutsouleris 2018). Further, even when findings might be clinically useful, many roadblocks stand in the way of successful implementation (Chekroud and Koutsouleris 2018). It is incumbent on researchers to test findings in multiple samples to avoid having other investigators waste time and resources.
Hence, the fact the attention network generalizes out of sample increases confidence in the original model and opens new opportunities for analysis. That is, the attention network generalized from a young, neurodiverse sample to an older, neurotypical sample. This finding is in line with the dimensional view of brain–behavior relationships (Insel et al. 2010) and also supports the notion that despite developmental changes in brain function, a “core” brain network architecture associated with sustaining attention is likely present (Yoo et al. 2022). An important next step to further assess generalizability will be testing if the network can generalize to predict different aspects of state- and trait-related attention phenotypes. The fact that the network predicted gradCPT d’ scores using gradCPT data indicates that the model is capturing behavioral variance attributable to attentional state; that we could also use rest data to predict the same gradCPT d’ scores suggests the model is capturing variance related to trait-based individual differences in attention. (We note both task and resting-state data were used to predict d’ as a measure of assessing internal reliability—previous work has indicated both data types should yield successful predictions, with task-based predictions tending to outperform rest-based predictions. As such, our data are consistent with earlier reports (Greene et al. 2018; Jiang et al. 2020).
In addition, determining if the network model generalizes to predict other phenotypes entirely and/or in different study populations could be of interest. To this end, we openly share the attention network model and encourage other investigators to test it widely. By sharing materials, particularly those from neurodiverse participants, we as a community can ensure our findings are relevant for all individuals.
Individual stability of the attention network
Using connectome-based ID, it has been shown that the connectome tends to be individually stable across short time scales (minutes) (Miranda-Dominguez et al. 2014; Finn et al. 2015; Kaufmann et al. 2017; Vanderwal et al. 2017; Waller et al. 2017; Amico and Goni 2018) and longer time scales (months to years) (Miranda-Dominguez et al. 2018; Horien et al. 2019; Demeter et al. 2020; Jalbrzikowski et al. 2020; Ousdal et al. 2020; Graff, Tansey, Ip, et al. 2022a; Graff, Tansey, Rai, et al. 2022b). Further, individual resting-state networks have tended to exhibit high stability (Finn et al. 2015). Here, we tested if a state-based, behaviorally defined network comprising edges across the brain exhibited the same degree of stability.
Encouragingly, we observed the attention network is a stable multivariate marker of connectivity over long time scales, and this did not differ from the rest of the brain in terms of stability. We interpret this finding as a positive for the field, as it suggests that networks for other phenotypes, with different neurobiological correlates, will likely exhibit a high degree of individual stability as well. Indeed, this seems to be the case, as recent work has demonstrated that predictive network models tend to have substantially higher reliability than individual functional connections (Taxali et al. 2021). Future studies could more rigorously assess how stability of the attention network relates to CPM model performance in longitudinal samples, as the datasets used here did not contain assessments of attention. It is important to note that participant identifiability of functional connections/networks is not equivalent to predictive utility (Noble et al. 2017; Finn and Rosenberg 2021; Mantwill et al. 2022). That is, correctly identifying participants at a high rate does not mean that there will be a clear link between the same connectomes and behavior. Nevertheless, focusing on the stability and reliability of measurements is crucial for those conducting functional connectivity studies (Noble et al. 2019), as a lack of reliability can continue to impede the clinical utility of fMRI (Milham et al. 2021).
Limitations and future considerations
Compared to other open-source datasets (e.g. ABCD (Casey et al. 2018), the Human Connectome Project (Van Essen et al. 2013), UK Biobank (Miller et al. 2016)), the samples used here are small. Another limitation is participants in the neurodiverse sample had fairly high IQ scores compared to the population at large (Wingate et al. 2014). While IQ was not shown to be a confounding factor in the predictive model, individuals with lower IQs may have difficulties completing the gradCPT. Future studies could address generalizability of the task and/or network in more varied individuals. In addition, attention is a broad construct, and in this work, we focused on the ability to sustain attentional state. More research could be conducted to determine if it is possible to build dimensional models of other aspects of attention. Relatedly, CPM networks could be derived from other in-scanner tasks (e.g. an emotion task) to investigate the biological specificity of different network markers associated with different (emotional) phenotypes, as well as how these overlap with the attention network described here (while keeping in mind that confounds such as head motion might play a role in both attention and emotion).
The longitudinal samples we used to measure stability of the model did not contain behavioral/clinical data. It is unclear if the predictive network is able to predict attention phenotypes across longer time scales. Studies with the same participants completing gradCPT at multiple time points could help to answer this question. Finally, we have focused on a single phenotype here. An important next step will be to use a multimodal, multidimensional framework in large numbers of individuals—incorporating numerous phenotypes and data types—to generate findings across multiple spatial and temporal scales (Lombardo et al. 2019). Such an approach holds the promise of illuminating the complex biology underlying heterogenous neurodiverse conditions.
Conclusion
In sum, we have shown that it is possible to generate task-based predictive models of in-scan attentional state in a sample of youth and that such a model generalizes. Results support the further development of predictive dimensional models of cognitive phenotypes and suggest that such an approach can yield stable imaging markers.
Supplementary Material
Acknowledgments
The authors thank Hedwig Sarofin and Cheryl McMurray for technical assistance during the MRI scans and Jitendra Bhawnani for technical assistance with task hardware and software.
Contributor Information
Corey Horien, Interdepartmental Neuroscience Program, Yale School of Medicine, New Haven, CT, United States; MD-PhD Program, Yale School of Medicine, New Haven, CT, United States.
Abigail S Greene, Interdepartmental Neuroscience Program, Yale School of Medicine, New Haven, CT, United States; MD-PhD Program, Yale School of Medicine, New Haven, CT, United States.
Xilin Shen, Department of Radiology and Biomedical Imaging, Yale School of Medicine, New Haven, CT, United States.
Diogo Fortes, Yale Child Study Center, New Haven, CT, United States.
Emma Brennan-Wydra, Yale Child Study Center, New Haven, CT, United States.
Chitra Banarjee, Yale Child Study Center, New Haven, CT, United States.
Rachel Foster, Yale Child Study Center, New Haven, CT, United States.
Veda Donthireddy, Yale Child Study Center, New Haven, CT, United States.
Maureen Butler, Yale Child Study Center, New Haven, CT, United States.
Kelly Powell, Yale Child Study Center, New Haven, CT, United States.
Angelina Vernetti, Yale Child Study Center, New Haven, CT, United States.
Francesca Mandino, Department of Radiology and Biomedical Imaging, Yale School of Medicine, New Haven, CT, United States.
David O’Connor, Department of Biomedical Engineering, Yale University, New Haven, CT, United States.
Evelyn M R Lake, Department of Radiology and Biomedical Imaging, Yale School of Medicine, New Haven, CT, United States.
James C McPartland, Yale Child Study Center, New Haven, CT, United States; Department of Psychology, Yale University, New Haven, CT, United States.
Fred R Volkmar, Yale Child Study Center, New Haven, CT, United States; Department of Psychology, Yale University, New Haven, CT, United States.
Marvin Chun, Department of Psychology, Yale University, New Haven, CT, United States.
Katarzyna Chawarska, Yale Child Study Center, New Haven, CT, United States; Department of Statistics and Data Science, Yale University, New Haven, CT, United States; Department of Pediatrics, Yale School of Medicine, New Haven, CT, United States.
Monica D Rosenberg, Department of Psychology, University of Chicago, Chicago, IL, United States; Neuroscience Institute, University of Chicago, Chicago, IL, United States.
Dustin Scheinost, Interdepartmental Neuroscience Program, Yale School of Medicine, New Haven, CT, United States; Department of Radiology and Biomedical Imaging, Yale School of Medicine, New Haven, CT, United States; Yale Child Study Center, New Haven, CT, United States; Department of Statistics and Data Science, Yale University, New Haven, CT, United States.
R Todd Constable, Interdepartmental Neuroscience Program, Yale School of Medicine, New Haven, CT, United States; Department of Radiology and Biomedical Imaging, Yale School of Medicine, New Haven, CT, United States; Department of Neurosurgery, Yale School of Medicine, New Haven, CT, United States.
Funding
This work was supported by the National Institutes of Health (P50MH115716, T32GM007205 to CH and ASG, and TR001864 to ASG).
Conflict of interest statement: JCM consults with Customer Value Partners, Bridgebio, Determined Health, and BlackThorn Therapeutics; has received research funding from Janssen Research and Development; serves on the Scientific Advisory Boards of Pastorus and Modern Clinics; and receives royalties from Guilford Press, Lambert, and Springer.
References
- Amico E, Goni J. The quest for identifiability in human functional connectomes. Sci Rep. 2018:8:8254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association . Diagnostic and statistical manual of mental disorders: DSM-5. Arlington: American Psychiatric Association; 2013 [Google Scholar]
- Armstrong T. The myth of the normal brain: embracing neurodiversity. AMA J Ethics. 2015:17:348–352. [DOI] [PubMed] [Google Scholar]
- Baker M. 1,500 scientists lift the lid on reproducibility. Nature. 2016:533:452–454. [DOI] [PubMed] [Google Scholar]
- Beaty RE, Kenett YN, Christensen AP, Rosenberg MD, Benedek M, Chen Q, Fink A, Qiu J, Kwapil TR, Kane MJ, et al. Robust prediction of individual creative ability from brain functional connectivity. Proc Natl Acad Sci U S A. 2018:115:1087–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995:57:289–300. [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995:34:537–541. [DOI] [PubMed] [Google Scholar]
- Boyle R, Connaughton M, McGlinchey E, Knight SP, De Looze C, Carey D, Stern Y, Robertson IH, Kenny RA, Whelan R. Connectome-based predictive modeling of cognitive reserve using task-based functional connectivity. bioRxiv. 2022: 10.1101/2022.06.01.494342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL. The cerebellum and cognitive function: 25 years of insight from anatomy and neuroimaging. Neuron. 2013:80:807–815. [DOI] [PubMed] [Google Scholar]
- Casey BJ, Cannonier T, Conley MI, Cohen AO, Barch DM, Heitzeg MM, Soules ME, Teslovich T, Dellarco DV, Garavan H, et al. The Adolescent Brain Cognitive Development (ABCD) study: imaging acquisition across 21 sites. Dev Cogn Neurosci. 2018:32:43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain TA, Rosenberg MD. Propofol selectively modulates functional connectivity signatures of sustained attention during rest and narrative listening. Cereb Cortex. 2022:32(23):5362–5375. [DOI] [PubMed] [Google Scholar]
- Chekroud AM. Bigger data, harder questions-opportunities throughout mental health care. JAMA Psychiat. 2017:74:1183–1184. [DOI] [PubMed] [Google Scholar]
- Chekroud AM, Koutsouleris N. The perilous path from publication to practice. Mol Psychiatry. 2018:23:24–25. [DOI] [PubMed] [Google Scholar]
- Clark SV, Semmel ES, Aleksonis HA, Steinberg SN, King TZ. Cerebellar-subcortical-cortical systems as modulators of cognitive functions. Neuropsychol Rev. 2021:31:422–446. [DOI] [PubMed] [Google Scholar]
- Cohen MR, Maunsell JH. When attention wanders: how uncontrolled fluctuations in attention affect performance. J Neurosci. 2011:31:15802–15806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeter DV, Engelhardt LE, Mallett R, Gordon EM, Nugiel T, Harden KP, Tucker-Drob EM, Lewis-Peacock JA, Church JA. Functional connectivity fingerprints at rest are similar across youths and adults and vary with genetic similarity. iScience. 2020;23: 10.1016/j.isci.2019.100801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino A, Zuo XN, Kelly C, Grzadzinski R, Mennes M, Schvarcz A, Rodman J, Lord C, Castellanos FX, Milham MP. Shared and distinct intrinsic functional network centrality in autism and attention-deficit/hyperactivity disorder. Biol Psychiatry. 2013:74:623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufford AJ, Kimble V, Tejavibulya L, Dadashkarimi J, Ibrahim K, Sukhodolsky DG, Scheinost D. Predicting transdiagnostic social impairments in childhood using connectome-based predictive modeling. medRxiv. 2022: 10.1101/2022.04.07.22273518. [DOI] [Google Scholar]
- Esterman M, Noonan SK, Rosenberg M, DeGutis J. In the zone or zoning out? Tracking behavioral and neural fluctuations during sustained attention. Cereb Cortex. 2013:23:2712–2723. [DOI] [PubMed] [Google Scholar]
- Esterman M, Rosenberg MD, Noonan SK. Intrinsic fluctuations in sustained attention and distractor processing. J Neurosci. 2014:34:1724–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn ES, Shen XL, Scheinost D, Rosenberg MD, Huang J, Chun MM, Papademetris X, Constable RT. Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nat Neurosci. 2015:18:1664–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn ES, Rosenberg MD. Beyond fingerprinting: choosing predictive connectomes over reliable connectomes. NeuroImage. 2021:239:118254. [DOI] [PubMed] [Google Scholar]
- Fitzgerald J, Johnson K, Kehoe E, Bokde AL, Garavan H, Gallagher L, McGrath J. Disrupted functional connectivity in dorsal and ventral attention networks during attention orienting in autism spectrum disorders. Autism Res. 2015:8:136–152. [DOI] [PubMed] [Google Scholar]
- Gabrieli JDE, Ghosh SS, Whitfield-Gabrieli S. Prediction as a humanitarian and pragmatic contribution from human cognitive neuroscience. Neuron. 2015:85:11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadow KD, DeVincent CJ, Pomeroy J. ADHD symptom subtypes in children with pervasive developmental disorder. J Autism Dev Disord. 2006:36:271–283. [DOI] [PubMed] [Google Scholar]
- Gerdts J, Bernier R. The broader autism phenotype and its implications on the etiology and treatment of autism spectrum disorders. Autism Res Treat. 2011:2011:545901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff K, Tansey R, Ip A, Rohr C, Dimond D, Dewey D, Bray S. Benchmarking common preprocessing strategies in early childhood functional connectivity and intersubject correlation fMRI. Dev Cogn Neurosci. 2022a:54:101087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff K, Tansey R, Rai S, Ip A, Rohr C, Dimond D, Dewey D, Bray S. Functional connectomes become more longitudinally self-stable, but not more distinct from others, across early childhood. NeuroImage. 2022b:258:119367. [DOI] [PubMed] [Google Scholar]
- Green JJ, Boehler CN, Roberts KC, Chen LC, Krebs RM, Song AW, Woldorff MG. Cortical and subcortical coordination of visual spatial attention revealed by simultaneous EEG-fMRI recording. J Neurosci. 2017:37:7803–7810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene AS, Gao SY, Scheinost D, Constable RT. Task-induced brain state manipulation improves prediction of individual traits. Nat Commun. 2018;9: 10.1038/s41467-018-04920-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harikumar A, Evans DW, Dougherty CC, Carpenter KLH, Michael AM. A review of the default mode network in autism spectrum disorders and attention deficit hyperactivity disorder. Brain Connect. 2021:11:253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekzema E, Carmona S, Ramos-Quiroga JA, Richarte Fernandez V, Bosch R, Soliva JC, Rovira M, Bulbena A, Tobena A, Casas M, et al. An independent components and functional connectivity analysis of resting state fMRI data points to neural network dysregulation in adult ADHD. Hum Brain Mapp. 2014:35:1261–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horien C, Floris DL, Greene AS, Noble S, Rolison M, Tejavibulya L, O'Connor D, McPartland JC, Scheinost D, Chawarska K, et al. Functional connectome-based predictive modeling in autism. Biol Psychiatry. 2022:92(8):626–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horien C, Fontenelle S, Joseph K, Powell N, Nutor C, Fortes D, Butler M, Powell K, Macris D, Lee K, et al. Low-motion fMRI data can be obtained in pediatric participants undergoing a 60-minute scan protocol. Sci Rep-Uk. 2020;10: 10.1038/s41598-020-78885-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horien C, Noble S, Finn ES, Shen X, Scheinost D, Constable RT. 2018. Considering factors affecting the connectome-based identification process: comment on Waller et al. NeuroImage 169:172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horien C, Shen XL, Scheinost D, Constable RT. The individual functional connectome is unique and stable over months to years. NeuroImage. 2019:189:676–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu WT, Rosenberg MD, Scheinost D, Constable RT, Chun MM. Resting-state functional connectivity predicts neuroticism and extraversion in novel individuals. Soc Cogn Affect Neurosci. 2018:13:224–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang K, Hallquist MN, Luna B. The development of hub architecture in the human functional brain network. Cereb Cortex. 2013:23:2380–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingersoll B. Broader autism phenotype and nonverbal sensitivity: evidence for an association in the general population. J Autism Dev Disord. 2010:40:590–598. [DOI] [PubMed] [Google Scholar]
- Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K, Sanislow C, Wang P. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am J Psychiatry. 2010:167:748–751. [DOI] [PubMed] [Google Scholar]
- Jalbrzikowski M, Liu F, Foran W, Klei L, Calabro FJ, Roeder K, Devlin B, Luna B. Functional connectome fingerprinting accuracy in youths and adults is similar when examined on the same day and 1.5-years apart. Hum Brain Mapp. 2020:41:4187–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Zuo N, Ford JM, Qi S, Zhi D, Zhuo C, Xu Y, Fu Z, Bustillo J, Turner JA, et al. Task-induced brain connectivity promotes the detection of individual differences in brain-behavior relationships. NeuroImage. 2020:207:116370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A, Scheinost D, Okuda H, Belhachemi D, Murphy I, Staib LH, Papademetris X. Unified framework for development, deployment and robust testing of neuroimaging algorithms. Neuroinformatics. 2011:9:69–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju Y, Horien C, Chen W, Guo W, Lu X, Sun J, Dong Q, Liu B, Liu J, Yan D, et al. Connectome-based models can predict early symptom improvement in major depressive disorder. J Affect Disord. 2020:273:442–452. [DOI] [PubMed] [Google Scholar]
- Kaufmann T, Alnaes D, Doan NT, Brandt CL, Andreassen OA, Westlye LT. Delayed stabilization and individualization in connectome development are related to psychiatric disorders. Nat Neurosci. 2017:20:513. [DOI] [PubMed] [Google Scholar]
- Keehn B, Shih P, Brenner LA, Townsend J, Muller RA. Functional connectivity for an "island of sparing" in autism spectrum disorder: an fMRI study of visual search. Hum Brain Mapp. 2013:34:2524–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler D, Angstadt M, Sripada C. Growth charting of brain connectivity networks and the identification of attention impairment in youth. JAMA Psychiat. 2016:73:481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake EMR, Finn ES, Noble SM, Vanderwal T, Shen X, Rosenberg MD, Spann MN, Chun MM, Scheinost D, Constable RT. The functional brain organization of an individual allows prediction of measures of social abilities transdiagnostically in autism and attention-deficit/hyperactivity disorder. Biol Psychiatry. 2019:86:315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DO, Ousley OY. Attention-deficit hyperactivity disorder symptoms in a clinic sample of children and adolescents with pervasive developmental disorders. J Child Adolesc Psychopharmacol. 2006:16:737–746. [DOI] [PubMed] [Google Scholar]
- Lichenstein SD, Scheinost D, Potenza MN, Carroll KM, Yip SW. 2021. Dissociable neural substrates of opioid and cocaine use identified via connectome-based modelling. Mol Psychiatry. 26:4383–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo MV, Lai MC, Baron-Cohen S. Big data approaches to decomposing heterogeneity across the autism spectrum. Mol Psychiatry. 2019:24:1435–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord CRM, DiLavore PC, Risi S, Gotham K, Bishop S. Autism diagnostic observation schedule. Second ed. Torrance, CA: Western Psychological Services; 2012 [Google Scholar]
- Lutkenhoff ES, Rosenberg M, Chiang J, Zhang K, Pickard JD, Owen AM, Monti MM. Optimized brain extraction for pathological brains (optiBET). PLoS One. 2014:9:e115551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantwill M, Gell M, Krohn S, Finke C. Brain connectivity fingerprinting and behavioural prediction rest on distinct functional systems of the human connectome. Commun Biol. 2022:5:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek S, Tervo-Clemmens B, Calabro FJ, Montez DF, Kay BP, Hatoum AS, Donohue MR, Foran W, Miller RL, Hendrickson TJ, et al. Reproducible brain-wide association studies require thousands of individuals. Nature. 2022:603:654–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masi A, DeMayo MM, Glozier N, Guastella AJ. An overview of autism spectrum disorder, heterogeneity and treatment options. Neurosci Bull. 2017:33:183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPartland JC. Refining biomarker evaluation in ASD. Eur Neuropsychopharmacol. 2021:48:34–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milham MP, Vogelstein J, Xu T. Removing the reliability bottleneck in functional magnetic resonance imaging research to achieve clinical utility. JAMA Psychiat. 2021:78(6):587–588. [DOI] [PubMed] [Google Scholar]
- Miller KL, Alfaro-Almagro F, Bangerter NK, Thomas DL, Yacoub E, Xu J, Bartsch AJ, Jbabdi S, Sotiropoulos SN, Andersson JL, et al. Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat Neurosci. 2016:19:1523–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Dominguez O, Mills BD, Carpenter SD, Grant KA, Kroenke CD, Nigg JT, Fair DA. Connectotyping: model based fingerprinting of the functional connectome. PLoS One. 2014:9:e111048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Dominguez O, Feczko E, Grayson DS, Walum H, Nigg JT, Fair DA. Heritability of the human connectome: a connectotyping study. Netw Neurosci. 2018:2:175–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble S, Spann MN, Tokoglu F, Shen X, Constable RT, Scheinost D. Influences on the test-retest reliability of functional connectivity MRI and its relationship with behavioral utility. Cereb Cortex. 2017:27:5415–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble S, Scheinost D, Constable RT. A decade of test-retest reliability of functional connectivity: a systematic review and meta-analysis. NeuroImage. 2019:203:116157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Open Science Collaboration . PSYCHOLOGY. Estimating the reproducibility of psychological science. Science. 2015:349:aac4716. [DOI] [PubMed] [Google Scholar]
- Orban P, Madjar C, Savard M, Dansereau C, Tam A, Das S, Evans AC, Rosa-Neto P, Breitner JCS, Bellec P, et al. Test-retest resting-state fMRI in healthy elderly persons with a family history of Alzheimer's disease. Sci Data. 2015;2: 10.1038/sdata.2015.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ousdal OT, Kaufmann T, Kolskar K, Vik A, Wehling E, Lundervold AJ, Lundervold A, Westlye LT. Longitudinal stability of the brain functional connectome is associated with episodic memory performance in aging. Hum Brain Mapp. 2020:41:697–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashler H, Wagenmakers EJ. Editors' introduction to the special section on replicability in psychological science: a crisis of confidence? Perspect Psychol Sci. 2012:7:528–530. [DOI] [PubMed] [Google Scholar]
- Plitt M, Barnes KA, Wallace GL, Kenworthy L, Martin A. Resting-state functional connectivity predicts longitudinal change in autistic traits and adaptive functioning in autism. Proc Natl Acad Sci USA. 2015:112:E6699–E6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack RA, Huckins G, Varoquaux G. Establishment of best practices for evidence for prediction a review. JAMA Psychiat. 2020:77:534–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posner J, Rauh V, Gruber A, Gat I, Wang Z, Peterson BS. Dissociable attentional and affective circuits in medication-naive children with attention-deficit/hyperactivity disorder. Psychiatry Res. 2013:213:24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Schlaggar BL, Petersen SE. 2015. Recent progress and outstanding issues in motion correction in resting state fMRI. Neuroimage. 105:536–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu MG, Ye Z, Li QY, Liu GJ, Xie B, Wang J. Changes of brain structure and function in ADHD children. Brain Topogr. 2011:24:243–252. [DOI] [PubMed] [Google Scholar]
- Rapuano KM, Rosenberg MD, Maza MT, Dennis NJ, Dorji M, Greene AS, Horien C, Scheinost D, Todd Constable R, Casey BJ. Behavioral and brain signatures of substance use vulnerability in childhood. Dev Cogn Neurosci. 2020:46:100878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr CS, Kamal S, Bray S. Building functional connectivity neuromarkers of behavioral self-regulation across children with and without autism spectrum disorder. Dev Cogn Neuros-Neth. 2020;41: 10.1016/j.dcn.2019.100747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M, Noonan S, DeGutis J, Esterman M. Sustaining visual attention in the face of distraction: a novel gradual-onset continuous performance task. Atten Percept Psycho. 2013:75:426–439. [DOI] [PubMed] [Google Scholar]
- Rosenberg MD, Casey BJ, Holmes AJ. Prediction complements explanation in understanding the developing brain. Nat Commun. 2018;9: 10.1038/s41467-018-02887-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg MD, Finn ES, Constable RT, Chun MM. Predicting moment-to-moment attentional state. NeuroImage. 2015:114:249–256. [DOI] [PubMed] [Google Scholar]
- Rosenberg MD, Finn ES, Scheinost D, Papademetris X, Shen XL, Constable RT, Chun MM. A neuromarker of sustained attention from whole-brain functional connectivity. Nat Neurosci. 2016a:19:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg MD, Zhang S, Hsu WT, Scheinost D, Finn ES, Shen XL, Constable RT, Li CSR, Chun MM. Methylphenidate modulates functional network connectivity to enhance attention. J Neurosci. 2016b:36:9547–9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg MD, Scheinost D, Greene AS, Avery EW, Kwon YH, Finn ES, Ramani R, Qiu M, Constable RT, Chun MM. Functional connectivity predicts changes in attention observed across minutes, days, and months. Proc Natl Acad Sci U S A. 2020:117:3797–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Margolis RL. Research domain criteria: strengths, weaknesses, and potential alternatives for future psychiatric research. Mol Neuropsychiatry. 2019:5:218–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite TD, Elliott MA, Gerraty RT, Ruparel K, Loughead J, Calkins ME, Eickhoff SB, Hakonarson H, Gur RC, Gur RE, et al. An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. NeuroImage. 2013:64:240–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinost D, Dadashkarimi J, Finn ES, Wambach CG, MacGillivray C, Roule AL, Niendam TA, Pine DS, Brotman MA, Leibenluft E, et al. Functional connectivity during frustration: a preliminary study of predictive modeling of irritability in youth. Neuropsychopharmacology. 2021:46:1300–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheinost D, Noble S, Horien C, Greene AS, Lake EMR, Salehi M, Gao SY, Shen XL, O'Connor D, Barron DS, et al. Ten simple rules for predictive modeling of individual differences in neuroimaging. NeuroImage. 2019:193:35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen XL, Finn ES, Scheinost D, Rosenberg MD, Chun MM, Papademetris X, Constable RT. Using connectome-based predictive modeling to predict individual behavior from brain connectivity. Nat Protoc. 2017:12:506–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel JS, Mitra A, Laumann TO, Seitzman BA, Raichle M, Corbetta M, Snyder AZ. 2017. Data Quality Influences Observed Links Between Functional Connectivity and Behavior. Cereb Cortex. 27:4492–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steegen S, Tuerlinckx F, Gelman A, Vanpaemel W. Increasing transparency through a multiverse analysis. Perspect Psychol Sci. 2016:11:702–712. [DOI] [PubMed] [Google Scholar]
- Steiger JH. Tests for comparing elements of a correlation matrix. Psychol Bull. 1980:87:245. [Google Scholar]
- Taxali A, Angstadt M, Rutherford S, Sripada C. Boost in test-retest reliability in resting state fMRI with predictive modeling. Cereb Cortex. 2021:31:2822–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima H, Kihara K, Kawahara JI, Kondo HM. Common principles underlie the fluctuation of auditory and visual sustained attention. Q J Exp Psychol (Hove). 2021:74:705–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, Ugurbil K, Consortium WU-MH . The WU-Minn Human Connectome Project: an overview. NeuroImage. 2013:80:62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderwal T, Eilbott J, Finn ES, Craddock RC, Turnbull A, Castellanos FX. Individual differences in functional connectivity during naturalistic viewing conditions. NeuroImage. 2017:157:521–530. [DOI] [PubMed] [Google Scholar]
- Waller L, Walter H, Kruschwitz JD, Reuter L, Muller S, Erk S, Veer IM. Evaluating the replicability, specificity, and generalizability of connectome fingerprints. NeuroImage. 2017:158:371–377. [DOI] [PubMed] [Google Scholar]
- Wicherts JM, Veldkamp CL, Augusteijn HE, Bakker M, van Aert RC, van Assen MA. Degrees of freedom in planning, running, analyzing, and reporting psychological studies: a checklist to avoid p-hacking. Front Psychol. 2016:7:1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingate M, Kirby RS, Pettygrove S, Cunniff C, Schulz E, Ghosh T, Robinson C, Lee LC, Landa R, Constantino J, et al. Prevalence of autism spectrum disorder among children aged 8 years - autism and developmental disabilities monitoring network, 11 sites, United States, 2010. MMWR Surveill Summ. 2014:63. [PubMed] [Google Scholar]
- Xiao J, Chen H, Shan X, He C, Li Y, Guo X, Chen H, Liao W, Uddin LQ, Duan X. Linked social-communication dimensions and connectivity in functional brain networks in autism spectrum disorder. Cereb Cortex. 2021:31:3899–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarkoni T. The generalizability crisis. Behav Brain Sci. 2020:1–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarkoni T, Westfall J. Choosing prediction over explanation in psychology: lessons from machine learning. Perspect Psychol Sci. 2017:12:1100–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerys BE, Jankowski KF, Shook D, Rosenberger LR, Barnes KA, Berl MM, Ritzl EK, Vanmeter J, Vaidya CJ, Gaillard WD. The fMRI success rate of children and adolescents: typical development, epilepsy, attention deficit/hyperactivity disorder, and autism spectrum disorders. Hum Brain Mapp. 2009:30:3426–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip SW, Scheinost D, Potenza MN, Carroll KM. Connectome-based prediction of cocaine abstinence. Am J Psychiatry. 2019:176:156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo K, Rosenberg MD, Hsu WT, Zhang S, Li CR, Scheinost D, Constable RT, Chun MM. Connectome-based predictive modeling of attention: comparing different functional connectivity features and prediction methods across datasets. NeuroImage. 2018:167:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo K, Rosenberg MD, Kwon YH, Lin Q, Avery EW, Sheinost D, Constable RT, Chun MM. A brain-based general measure of attention. Nat Hum Behav. 2022:6:782–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidan J, Fombonne E, Scorah J, Ibrahim A, Durkin MS, Saxena S, Yusuf A, Shih A, Elsabbagh M. Global prevalence of autism: a systematic review update. Autism Res. 2022:15:778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo XN, Anderson JS, Bellec P, Birn RM, Biswal BB, Blautzik J, Breitner JCS, Buckner RL, Calhoun VD, Castellanos FX, et al. An open science resource for establishing reliability and reproducibility in functional connectomics. Sci Data. 2014;1: 10.1038/sdata.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Preprocessing was carried out using software freely available here: (https://medicine.yale.edu/bioimaging/suite/). CPM code is available here: (https://github.com/YaleMRRC/CPM). The parcellation, the attention network models, and the connectome-based ID code are available here: (https://www.nitrc.org/frs/?group_id=51). Data from the longitudinal samples are openly available through CoRR (http://fcon_1000.projects.nitrc.org/indi/CoRR/html/). All other data is available from the authors upon request.